Abstract
Microbial communities are accompanied by a diverse array of viruses. Through infections of abundant microbes, these viruses have the potential to mediate competition within the community, effectively weakening competitive interactions and promoting coexistence. This is of particular relevance for host-associated microbial communities, because the diversity of the microbiota has been linked to host health and functioning. Here, we study the interaction between two key members of the microbiota of the freshwater metazoan Hydra vulgaris. The two commensal bacteria Curvibacter sp. and Duganella sp. protect their host from fungal infections, but only if both of them are present. Coexistence of the two bacteria is thus beneficial for Hydra. Intriguingly, Duganella sp. appears to be the superior competitor in vitro due to its higher growth rate when both bacteria are grown separately, but in co-culture the outcome of competition depends on the relative initial abundances of the two species. The presence of an inducible prophage in the Curvibacter sp. genome, which is able to lytically infect Duganella sp., led us to hypothesize that the phage modulates the interaction between these two key members of the Hydra microbiota. Using a mathematical model, we show that the interplay of the lysogenic life cycle of the Curvibacter phage and the lytic life cycle on Duganella sp. can explain the observed complex competitive interaction between the two bacteria. Our results highlight the importance of taking lysogeny into account for understanding microbe–virus interactions and show the complex role phages can play in promoting coexistence of their bacterial hosts.
Keywords: bacterial competition, bacteria–phage interactions, mathematical modelling, lysogenic and lytic cycles
1. Introduction
Microbial communities are often highly diverse and it is increasingly being recognized that this diversity is key to the ecological functions of these communities [1,2]. It is thus crucially important to understand the mechanisms allowing for the coexistence of microbial species found in natural communities.
Bacteriophages, the viruses of bacteria, are found across virtually all habitats harbouring microbial communities. Their ubiquity and the observation that globally they are responsible for the turnover of vast amounts of microbial biomass every second [3–5] have led to the suggestion that they may play an important role in structuring bacterial communities and maintaining microbial diversity [6]. In particular, theoretical and experimental studies show that they can regulate competitively dominant species or even specific strains via a mechanism termed ‘kill the winner’ [7–9]. This allows for the coexistence of less competitive species that would otherwise be excluded, thus promoting diversity.
This is, in particular, relevant for microbes living in and on animal and plant hosts (the microbiota) as there is growing evidence that the composition and functioning of the microbiota and the well-being of the host are closely intertwined. In particular, high microbial diversity has been found to correlate with healthier hosts [10–12], in line with previous results relating diversity and community functioning [13,14].
As in many other environments, it has been shown that microbiota are accompanied by an abundant community of viruses [15–19]. While the majority of previous studies have focused on lytic viruses, recent studies have pointed to the prominent role of lysogeny in microbiota–phage interactions [20,21] and, in fact, an estimated 5% of the human gut bacterial gene content codes for prophage proteins [22]. This suggests that the relationship between phages and bacteria in the microbiota goes beyond predatory or parasitic interactions, and in fact temperate phages have been implicated in increasing the competitive fitness of their lysogenic hosts [23] and driving microbial evolution [24,25].
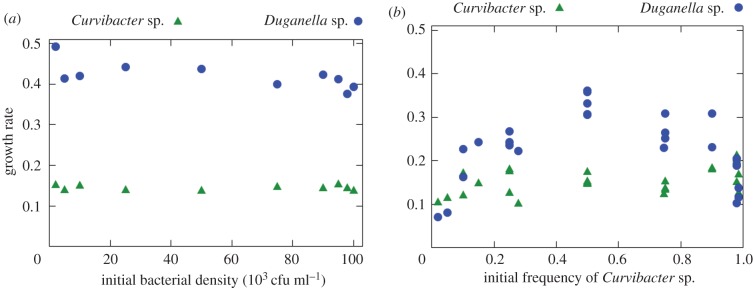
Here, we investigate how phages mediate the competition between two key members of the natural microbiota of the freshwater polyp Hydra vulgaris. The two bacterial species, Curvibacter sp. and Duganella sp., are able to protect the polyp against fungal infections and, interestingly, this antifungal activity is strongly synergistic and greatly diminished when one of the species is absent [26]. Coexistence of the two species is thus beneficial for the host. However, as shown by Li et al. [27] Duganella sp. appears to have a much higher growth rate than Curvibacter sp. when both are grown in monoculture in vitro (figure 1a), suggesting that it is competitively dominant. But when both species are grown in co-culture at varying initial proportions, the growth rate of Duganella shows a strong nonlinear dependence on the initial Curvibacter sp. frequency. Intriguingly, Duganella sp. growth is suppressed at both low and high initial Curvibacter sp. frequencies, but not at intermediate frequencies (figure 1b).
Figure 1.
(a) In monoculture, the growth rates of Curvibacter sp. and Duganella sp. do not depend on the initial densities. (b) In co-culture, the growth rate of Duganella sp. changes nonlinearly with the initial frequency of Curvibacter sp., showing a maximum at intermediate frequencies and inhibition especially at low and high frequencies of Curvibacter sp. The growth rate of Curvibacter sp., on the other hand, does not substantially depend on the initial frequencies. (Adapted from Li et al. [27].) (Online version in colour.)
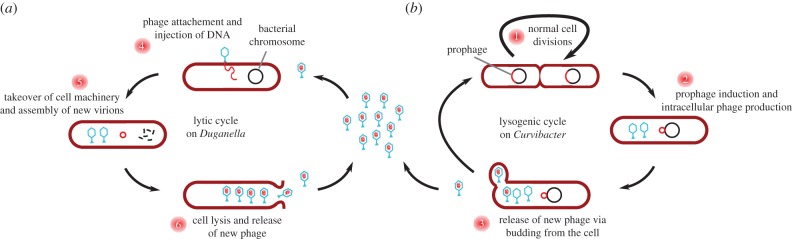
A recent study has shown that different Hydra species harbour a diverse virome and it has been suggested that phages play a role in regulating the Hydra-associated microbiota [28,29]. Indeed we found an intact prophage sequence in the genome of Curvibacter sp., which could be reactivated and isolated from bacterial cultures (figure 2). Interestingly, the phage can infect and lytically replicate on Duganella sp. This leads us to hypothesize that the prophage acts as a self-replicating weapon against Duganella sp., thereby playing a key role in modulating the competition between the two bacterial species in the observed non-intuitive way.
Figure 2.
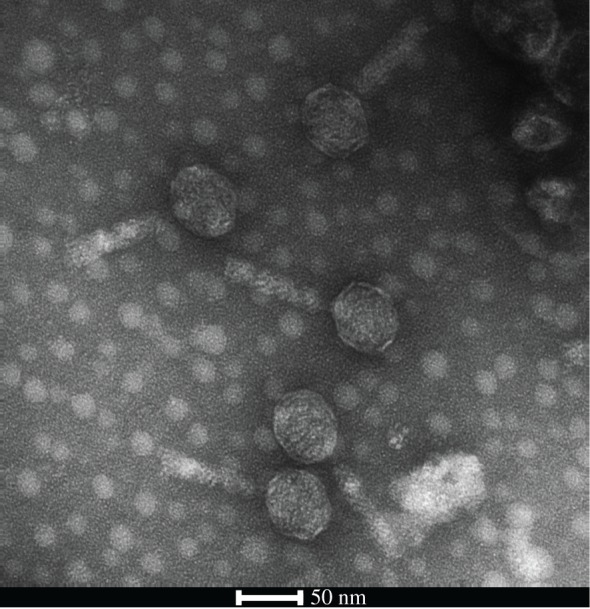
Transmission electron micrograph of mature phages isolated from Curvibacter sp. showing the icosahedral head containing the DNA and the tail fibre.
To test this hypothesis, we build a mechanistic bacteria–phage model where, crucially, both the lysogenic cycle on Curvibacter sp. and the lytic cycle on Duganella sp. contribute to phage population growth. Our model shows that the interplay between the two phage life cycles explains the observed frequency-dependent interactions between the two bacteria and that neither life cycle alone can give rise to the observed competitive interactions.
2. Material and methods
2.1. Isolation of Curvibacter phage
Curvibacter sp. was grown in R2A broth medium and upon reaching the exponential growth phase mitomycin C was added at a final concentration of 0.05 μg ml−1 to induce the prophage. After an inoculation time of 16 h, bacterial cells were removed by filtration (pore size 0.2 μm). Bacteriophages were further purified by CsCl density gradient ultracentrifugation [28]. Purified phages were negatively stained in 2% (w/v) aqueous uranyl acetate and visualized by transmission electron microscopy using a Tecnai BioTWIN at 80 kV and magnifications of 40 000–100 000×. Infectivity of Duganella sp. by Curvibacter phage was tested by spot assays according to the protocol described by Adams [30].
3. The model
We start by deriving a model of the interactions between the two bacteria and the phage in a well-mixed batch growth culture with a fixed amount of initial nutrients. This allows us to make predictions about the impact of phages on both bacterial species in order to understand the nonlinear competition between them in vitro.
We denote the densities of the two bacteria (cells ml−1) with C (Curvibacter sp.) and D (Duganella sp.), and the density (particles ml−1) of free phages with P. In the absence of phages both bacteria take up nutrients N (μg ml−1) and grow according to Monod kinetics,
| 3.1 |
with maximum growth rates rC,D (h−1), respectively. The half-saturation constant H (μg ml−1) and nutrient conversion efficiency c are assumed to be the same for both bacteria.
We incorporate the two distinct phage life cycles as depicted in figure 3 in the following way. The lytic life cycle generally involves utilization and killing of the host cell and thus conforms to the classical analogy of phages as predators of their hosts. Consequently, for phage reproduction via lytic infections of Duganella sp. we assume that phage adsorption and infection follow mass action kinetics with adsorption rate ϕ (h−1), which leads to lysis and loss of the infected host cell resulting in the release of β (cell−1) new phages. We include phage production via the lysogenic cycle with the function d(C, D), which describes the induction rate of the prophage in Curvibacter sp. and subsequent release of new virions via budding through the cell wall, which does not kill the host cell. Note that this process potentially depends on the densities of both bacteria. The lysogenic cycle is characterized by integration of the phage genetic information into the genome of the host cell, which commonly renders the prophage-carrying host cell immune to infections by related phages [31]. Owing to this superinfection inhibition, we assume Curvibacter sp. to be completely resistant to lytic infections by the phage. Free phages are assumed to decay with a rate m (h−1).
Figure 3.
Population growth of free phages is the sum of phage replication via the lytic life cycle on Duganella sp. (a) and the lysogenic life cycle on Curvibacter sp. (b). The lysogenic cycle is characterized by the integration of the phage DNA into the Curvibacter sp. chromosome as a prophage. Cells divide normally (1) and induction of the prophage leads to the production of virions (2). Mature phages are released by non-lethal extrusion or budding (3). The lytic cycle is initiated by infection of a Duganella sp. cell by a free phage (4), followed by phage replication (5) and release of the new phages (6). The last step results in the lysis and death of the Duganella host cell. (Online version in colour.)
With this the model describing the dynamics of nutrients, bacteria and phages reads
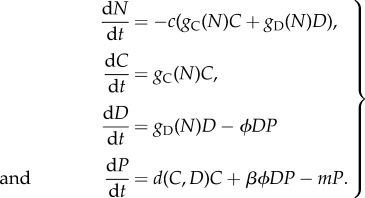 |
3.2 |
The key feature of this model is that phage population growth is the sum of lytic reproduction on Duganella sp. and lysogenic phage production from Curvibacter sp.
So far, we have not explicitly defined the prophage induction rate d(C, D). Although the induction of lysogenic phages can have a substantial impact on bacterial population dynamics, the factors that induce prophages and modulate the rate of induction are still largely unknown [32]. Some of the known factors that can activate prophages include the host's SOS response following cell damage, external triggers and spontaneous induction by stochastic gene expression [33]. As our results do not depend on the particular mechanism modulating the induction rate, in the following section we aim to deduce the mathematically simplest form of d(C, D) that is consistent with the experimental data.
3.1. Approximating the prophage induction rate
First, we need to clarify how the induction rate d(C, D) influences the Duganella sp. growth rate. If nutrients are not limiting, i.e. during exponential growth, we have gD(N) ≈ rD and thus the Duganella sp. growth rate simplifies to
| 3.3 |
The growth rate is the difference between the Duganella sp. growth rate in monoculture and the losses imposed by the phage. We approximate those losses by assuming that phage dynamics are much faster than bacterial dynamics, implying that the phage density quickly reaches its asymptotic value P* for which dP/dt = 0, where the bacterial densities C and D are essentially free parameters. Solving for this equilibrium value and inserting it into the growth rate (3.3) of Duganella sp. yields
| 3.4 |
The term in the brackets corresponds to the exponential growth rate of Duganella sp. in the presence of Curvibacter sp., where the indirect effect of Curvibacter sp. is mediated by the phage. Denoting this term with G and rewriting it using the frequency f = C/B of Curvibacter sp. in the total bacterial population B = C + D gives
| 3.5 |
Note that G(0) = rD recovers the growth rate of Duganella sp. in monoculture.
Now we can ask: what is the simplest form of the prophage induction rate d(f) that would lead to a hump-shaped growth rate G(f) of Duganella sp. as observed in the experiments (figure 1b)? The condition that G(f) attains its maximum at some intermediate Curvibacter sp. frequency implies that
| 3.6 |
for some 0 < f < 1.
Now, if the prophage induction rate is independent of Curvibacter sp. frequency, we have d(f) = p with some constant p > 0. However, in this case d′(f) = 0 and thus
| 3.7 |
for all f. This shows that a constant induction rate does not allow for a hump-shaped growth rate of Duganella sp.
A slightly more complex case is given by a linearly increasing induction rate, namely d(f) = pf, with rate constant p (particles h−1 cell−1). In this case, G′(f) = 0 has two solutions, namely f = 0 and
| 3.8 |
Thus, under the assumption of fast phage and slow bacteria dynamics, a hump-shaped growth rate of Duganella sp. is possible if the prophage induction rate increases linearly with Curvibacter sp. frequency. Accordingly, in the following analysis of the model, we set d(f) = pf.
3.2. Results
For a wide range of parameter values and initial conditions (N0, C0, D0 > 0 and P0 ≥ 0) the population dynamics described by equations (3.2) result in the following general pattern (see the electronic supplementary material, figure S1, for an example): after inoculation of the co-culture when nutrients are not limiting, both bacteria grow exponentially. Curvibacter sp. keeps growing until nutrients near depletion ( ), at which point its growth rate approaches zero and its density reaches a final value C*, which depends on the initial density D0 of Duganella sp. and the initial amount N0 of nutrients. Duganella sp., on the other hand, will initially also grow exponentially, but in addition to the nutrient levels its population growth is affected by the losses imposed by phage infections.
), at which point its growth rate approaches zero and its density reaches a final value C*, which depends on the initial density D0 of Duganella sp. and the initial amount N0 of nutrients. Duganella sp., on the other hand, will initially also grow exponentially, but in addition to the nutrient levels its population growth is affected by the losses imposed by phage infections.
To avoid the confounding effects of nutrient depletion and to keep the model close to the experimental set-up, we restrict our analysis to the exponential growth phase when nutrients are not yet limiting. We define the end of this phase as the time when Curvibacter sp. growth rate falls below a certain threshold ɛ > 0, because Curvibacter sp. growth rate is only influenced by the current nutrient level. In particular, we numerically calculate the Duganella sp. growth rate during the exponential phase as
| 3.9 |
over the time interval 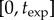 for which
for which
| 3.10 |
holds. In exactly the same manner we obtain the growth rate rC of Curvibacter sp.
During this time interval the phage is the only factor influencing Duganella sp. growth rate and the question is whether the growth dynamics of the phage alone can give rise to the observed nonlinear interaction between the two bacteria. To address this question, it is important to note that the growth of the phage population is determined by both Curvibacter sp. and Duganella sp. densities via prophage induction and lytic reproduction, respectively. As we will now show, the contributions of these two pathways to phage growth behave very differently as the relative initial abundances of the two bacteria change.
3.2.1. Contribution of the lytic cycle
The lytic life cycle is known to be especially effective at high host densities, when each phage particle quickly finds a new host. In this case, even a small amount of phage can initiate rapid replication resembling a chain reaction, leading to a quick and notable decline or even collapse of the bacterial host population. If the host density is too low, however, the losses through phage decay are not outweighed by reproduction; the phage can thus not invade the host population and is eventually lost from the system. In our system, the so-called ‘replication threshold’ [34–36]—i.e. the minimum host density that allows positive phage growth—can be calculated explicitly by observing that, if there is only lytic reproduction (d(f) = 0 for all f), we have
| 3.11 |
This implies that, with lytic reproduction alone, the phage can grow and have a substantial impact on Duganella sp. growth only if the initial Curvibacter sp. frequency f is relatively low. Conversely, as long as the relative abundance of Curvibacter sp. in the total population is too high, the phage does not find enough suitable Duganella sp. hosts and cannot persist. Note that the replication threshold increases with total bacterial population density B and approaches 1 for very high densities, implying that it is most relevant in the early stages of exponential growth when bacterial densities are still low, which is exactly the case we consider here. In particular, because Duganella sp. is growing faster than Curvibacter sp. when it is rare, it will eventually reach the replication threshold of the phage, but it does so too late for the phage to grow to substantial densities before nutrients are depleted.
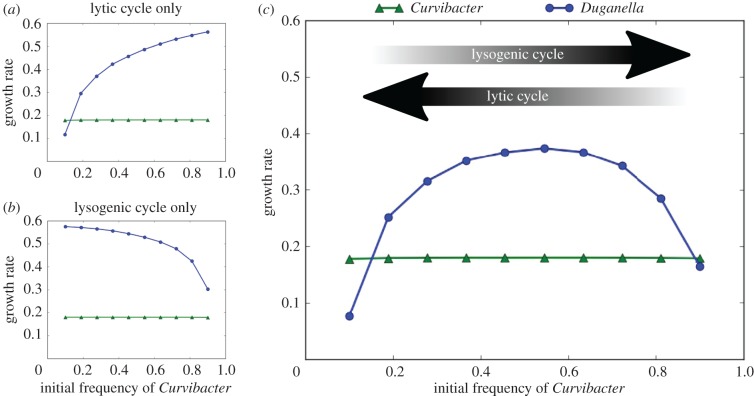
See figure 4a for a numerical example of this frequency-dependent impact of lytic phage growth on the two bacteria, which shows the bacterial growth rates as defined above for different initial frequencies of Curvibacter sp. and Duganella sp. in the case of no lysogenic production. Note that rC is constant for all initial Curvibacter sp. frequencies, reflecting that during the exponential growth phase Curvibacter sp. growth rate is independent of both the phage and the presence of Duganella sp.
Figure 4.
Growth rates obtained from the model during the exponential growth phase of Curvibacter sp. and Duganella sp. for different initial frequencies of Curvibacter sp. (a,b) The growth rates if only the lytic (a) or lysogenic (b) phage life cycle is taken into account. (c) Bacterial growth rates if both life cycles are taken into account. The arrows indicate at which ends of the initial frequency spectrum the two different phage life cycles are most effective. Parameter values for this example: N0 = 100, B0 = 105, P0 = 3.5 × 104, c = 10−5, rC = 0.125, rD = 0.4, H = 0.05, ϕ = 10−8, β = 50, m = 0.25, p = 0.05. (Online version in colour.)
3.2.2. Contribution of the lysogenic cycle
Let us now turn to phage reproduction via the lysogenic pathway (denoted by d(C, D) in equation (3.2)). It is clear that the contribution of the lysogenic pathway to phage growth increases as the frequency of Curvibacter sp. in the total population increases. However, lysogenic production alone is generally not enough to sustain the phage at high densities and lytic reproduction is still necessary for the phage to reach densities at which it has a visible impact on Duganella sp. growth. We can illustrate how the contribution of the lysogenic pathway increases with Curvibacter sp. frequency by setting the density of initially present phages P0 to zero. In this case, the founder population of phages is introduced solely via induction of the prophage from Curvibacter sp., emphasizing the contribution of the lysogenic pathway.
In this scenario, if the initial relative abundance of Curvibacter sp. is too low, the production of phages via prophage induction is not sufficient to provide a large enough phage population for it to have a substantial impact on Duganella sp. before nutrients are depleted. Only when the initial frequency of Curvibacter sp. is sufficiently high does the lysogenic cycle produce enough phages to suppress Duganella sp. growth (figure 4b).
3.2.3. Combined effects of the lytic and lysogenic cycles
Now, if both pathways are active and phage population growth is the sum of the lysogenic and lytic life cycles, the two patterns described above combine to give the observed nonlinear dependence of the Duganella sp. growth rate on the initial composition of the bacterial population (figure 4c). Essentially, the two effects appear superimposed on each other, with the Duganella sp. growth rate being suppressed at both low and high initial Curvibacter sp. frequencies.
4. Discussion
In this study, we considered how temperate phages affect the interactions between bacteria in a frequency-dependent manner. We developed a mathematical model that takes into account both the lysogenic and the lytic life cycles of temperate phages. Crucially, our model shows that the two pathways show very different efficiencies as the host population composition changes.
At low frequencies of the prophage-carrying Curvibacter sp., the main route of phage production is via the lytic pathway, resulting in a significant decrease of the Duganella sp. growth rate (figure 4). In this case, the phage has a high chance of encountering a susceptible Duganella sp. cell, which allows the phage to spread rapidly and lyse a great portion of the Duganella sp. population in the process. The mechanism provides an efficient way for Curvibacter sp. to bounce back from very low frequencies, thus preventing extinction and preserving diversity. A similar use of temperate phages has been shown to confer a competitive advantage on prophage carriers in bacterial competition [37], in particular allowing rare invaders to spread and persist in susceptible resident populations [38,39]. The effectiveness of temperate phages as either a persistence or invasion strategy relies on the fact that the lytic pathway is particularly efficient when the resistant prophage carriers are rare and susceptible hosts are abundant, allowing the rapid spread of the phage as a self-replicating weapon. This is in contrast with antimicrobial toxins, which are most effective when the toxin producers are at high densities [38].
At high relative abundances of Curvibacter sp., on the other hand, the lytic pathway is very ineffective as the majority of the bacterial population are resistant lysogens. In this case, lytic replication alone is not able to sustain the phage population, and thus without the lysogenic production from Curvibacter sp. the phage would be lost from the system. But through increased lysogenic reproduction at high Curvibacter sp. densities the phage is not only able to persist, but in fact it can lyse a significant portion of the Duganella sp. population and thus diminish its growth rate (figure 4).
At intermediate Curvibacter sp. frequencies, however, neither life cycle is particularily effective, which allows Duganella sp. to grow relatively unaffected by the phage. Taken together, this gives rise to the observed hump-shaped dependence of Duganella sp. population growth on the initial relative abundance of Curvibacter sp. Crucially, our results also show that only by taking both life cycles into account can the observed growth rates be recovered (figure 4).
We deduced that a prophage induction rate that is linearly increasing with Curvibacter sp. frequency is consistent with the experimental data. Such a concerted prophage induction is, for example, possible via quorum-sensing mechanisms and this has been hypothesized to allow phages to sense favourable conditions [40]. However, we want to emphasize that our results do not depend on the precise mechanism of prophage induction. In particular, the induction rate does not necessarily have to be linearly increasing and we do not expect prophage induction rates to follow a linear function in nature. Rather, our aim was to show in a ‘proof-of-principle’ manner an example of minimal complexity that gives rise to the observed bacterial growth dynamics.
Our result provides a potential mechanism behind the finding that weak competitive interactions are the dominant type of interactions within host-associated microbial communities [41]. In line with the ‘killing the winner’ mechanism [42], the phage imposes a top-down control of the otherwise stronger competitor Duganella sp., potentially alleviating the competitive interactions between the two bacterial species and allowing them to coexist within the microbiota of Hydra vulgaris.
While there is no indication that the phage lysogenizes Duganella sp., there is a possibility that over a longer time span Duganella sp. becomes resistant to the phage via lysogenization. This would imply a shift from its short-term use as a weapon to a more classical parasitic lifestyle, a shift that has been reported to occur after a few days of competition between phage-free strains of Escherichia coli and strains lysogenized by the temperate phage λ [43]. Over longer time spans, antagonistic coevolution between bacteria and phages will also probably contribute to bacterial resistance in the microbiota [44], further adding to the complexity of bacterial competition mediated by phages.
The observation that in vivo the protection of the Hydra host against fungal infections is mediated by the coexistence of both bacterial species [26] suggests that an active regulation of the induction of the Curvibacter sp. prophage could be beneficial for Hydra. And, intriguingly, it has recently been shown that Hydra vulgaris is able to modify the quorum-sensing signals of Curvibacter, thereby changing the phenotype of a bacterial colonizer [45]. This is an example of a mechanism by which a host could potentially manipulate bacterial gene expression, including prophage induction. More generally, the growing recognition of phages as regulators of the microbiota [46,47] leads to the interesting hypothesis that the ability to manipulate the induction of prophages to promote the coexistence of synergistic bacteria is itself an evolvable host trait.
In conclusion, we found that, by taking the lysogenic and lytic life cycles of temperate phages into account, a minimal model is able to explain the observed frequency-dependent outcome of competition between two key members of the Hydra vuglaris microbiota. Our study elucidates the inherently complex effects of temperate phages on bacterial competition and population dynamics, which are shown to be highly dependent on the composition of the host population. Future studies are aimed at identifying the precise mechanisms of Curvibacter prophage regulation and the quantification of its induction rate, which will shed further light on the role of temperate phages in host-associated microbial communities.
Supplementary Material
Data accessibility
The growth rate data shown in figure 1 are available in the data supplement of Li et al. [27].
Authors' contributions
X.-Y.L. and M.S. developed and analysed the mathematical model. T.L. carried out laboratory work and obtained microscopic images. X.-Y.L., T.L. and M.S. conceived the study. All the authors designed the study, contributed to writing the manuscript and gave their final approval for publication.
Competing interests
We declare we have no competing interests.
Funding
This work was supported by the Collaborative Research Centre 1182 ‘Origin and Function of Metaorganisms’ granted by the Deutsche Forschungsgemeinschaft DFG. T.L. acknowledges funding from the Volkswagen Foundation (funding programme ‘Experiment!—In search of bold research ideas’). T.C.G.B. gratefully appreciates support from the Canadian Institute for Advanced Research (CIFAR).
References
- 1.Bell T, Newman JA, Silverman BW, Turner SL, Lilley AK. 2005. The contribution of species richness and composition to bacterial services. Nature 436, 1157–1160. ( 10.1038/nature03891) [DOI] [PubMed] [Google Scholar]
- 2.Hu J, et al. 2016. Probiotic diversity enhances rhizosphere microbiome function and plant disease suppression. mBio 7, e01790–16 ( 10.1128/mBio.01790-16) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ashelford KE, Day MJ, Fry JC. 2003. Elevated abundance of bacteriophage infecting bacteria in soil. Appl. Environ. Microbiol. 69, 285–289. ( 10.1128/AEM.69.1.285-289.2003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Suttle CA. 2007. Marine viruses—major players in the global ecosystem. Nat. Rev. Microbiol. 5, 801–812. ( 10.1038/nrmicro1750) [DOI] [PubMed] [Google Scholar]
- 5.Srinivasiah S, Bhavsar J, Thapar K, Liles M, Schoenfeld T, Wommack KE. 2008. Phages across the biosphere: contrasts of viruses in soil and aquatic environments. Res. Microbiol. 159, 349–357. ( 10.1016/j.resmic.2008.04.010) [DOI] [PubMed] [Google Scholar]
- 6.Weinbauer MG, Rassoulzadegan F. 2004. Are viruses driving microbial diversification and diversity? Environ. Microbiol. 6, 1–11. ( 10.1046/j.1462-2920.2003.00539.x) [DOI] [PubMed] [Google Scholar]
- 7.Thingstad TF. 2000. Elements of a theory for the mechanisms controlling abundance, diversity, and biogeochemical role of lytic bacterial viruses in aquatic systems. Limnol. Oceanogr. 45, 1320–1328. ( 10.4319/lo.2000.45.6.1320) [DOI] [Google Scholar]
- 8.Brockhurst MA, Fenton A, Roulston B, Rainey PB. 2006. The impact of phages on interspecific competition in experimental populations of bacteria. BMC Ecol. 6, 19 ( 10.1186/1472-6785-6-19) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thingstad TF, Pree B, Giske J, Våge S. 2015. What difference does it make if viruses are strain-, rather than species-specific? Front Microbiol. 6, 320 ( 10.3389/fmicb.2015.00320) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clemente JC, Ursell LK, Parfrey LW, Knight R. 2012. The impact of the gut microbiota on human health: an integrative view. Cell 148, 1258–1270. ( 10.1016/j.cell.2012.01.035) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lozupone CA, Stombaugh JI, Gordon JI, Jansson JK, Knight R. 2012. Diversity, stability and resilience of the human gut microbiota. Nature 489, 220–230. ( 10.1038/nature11550) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Le Chatelier E, et al. 2013. Richness of human gut microbiome correlates with metabolic markers. Nature 500, 541–546. ( 10.1038/nature12506) [DOI] [PubMed] [Google Scholar]
- 13.Naeem S. 2002. Ecosystem consequences of biodiversity loss: the evolution of a paradigm. Ecology 83, 1537–1552. ( 10.1890/0012-9658(2002)083%5B1537:ECOBLT%5D2.0.CO;2) [DOI] [Google Scholar]
- 14.Tilman D, Isbell F, Cowles JM. 2014. Biodiversity and ecosystem functioning. Annu. Rev. Ecol. Evol. Syst. 45, 471–493. ( 10.1146/annurev-ecolsys-120213-091917) [DOI] [Google Scholar]
- 15.Minot S, Sinha R, Chen J, Li H, Keilbaugh SA, Wu GD, Lewis JD, Bushman FD. 2011. The human gut virome: inter-individual variation and dynamic response to diet. Genome Res. 21, 1616–1625. ( 10.1101/gr.122705.111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stern A, Mick E, Tirosh I, Sagy O, Sorek R. 2012. CRISPR targeting reveals a reservoir of common phages associated with the human gut microbiome. Genome Res. 22, 1985–1994. ( 10.1101/gr.138297.112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Norman JM, et al. 2015. Disease-specific alterations in the enteric virome in inflammatory bowel disease. Cell 160, 447–460. ( 10.1016/j.cell.2015.01.002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang J, Gao Y, Zhao F. 2015. Phage-bacteria interaction network in human oral microbiome. Environ. Microbiol. 18, 2143–2158. ( 10.1111/1462-2920.12923) [DOI] [PubMed] [Google Scholar]
- 19.Manrique P, Bolduc B, Walk ST, van der Oost J, de Vos WM, Young MJ. 2016. Healthy human gut phageome. Proc. Natl Acad. Sci. USA 113, 10 400–10 405. ( 10.1073/pnas.1601060113) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reyes A, Haynes M, Hanson N, Angly FE, Heath AC, Rohwer F, Gordon JI. 2010. Viruses in the faecal microbiota of monozygotic twins and their mothers. Nature 466, 334–338. ( 10.1038/nature09199) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pride DT, Salzman J, Haynes M, Rohwer F, Davis-Long C, White RA 3rd, Loomer P, Armitage GC, Relman DA. 2012. Evidence of a robust resident bacteriophage population revealed through analysis of the human salivary virome. ISME J. 6, 915–926. ( 10.1038/ismej.2011.169) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qin J, et al. 2010. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464, 59–65. ( 10.1038/nature08821) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bossi L, Fuentes JA, Mora G, Figueroa-Bossi N. 2003. Prophage contribution to bacterial population dynamics. J. Bacteriol. 185, 6467–6471. ( 10.1128/JB.185.21.6467-6471.2003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.De Paepe M, Tournier L, Moncaut E, Son O, Langella P, Petit MA. 2016. Carriage of λ latent virus is costly for its bacterial host due to frequent reactivation in monoxenic mouse intestine. PLoS Genet. 12, e1005861 ( 10.1371/journal.pgen.1005861) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Obeng N, Pratama AA, Elsas JD. 2016. The significance of mutualistic phages for bacterial ecology and evolution. Trends Microbiol. 24, 440–449. ( 10.1016/j.tim.2015.12.009) [DOI] [PubMed] [Google Scholar]
- 26.Fraune S, Anton-Erxleben F, Augustin R, Franzenburg S, Knop M, Schröder K, Willoweit-Ohl D, Bosch TCG. 2015. Bacteria–bacteria interactions within the microbiota of the ancestral metazoan Hydra contribute to fungal resistance. ISME J. 9, 1543–1556. ( 10.1038/ismej.2014.239) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li X-Y, Pietschke C, Fraune S, Altrock PM, Bosch TCG, Traulsen A. 2015. Which games are growing bacterial populations playing? J. R. Soc. Interface 12, 20150121 ( 10.1098/rsif.2015.0121) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grasis JA, et al. 2014. Species-specific viromes in the ancestral holobiont Hydra. PLoS ONE 9, e109952 ( 10.1371/journal.pone.0109952) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bosch TCG, Grasis JA, Lachnit T. 2015. Microbial ecology in Hydra: why viruses matter. J. Microbiol. 53, 193–200. ( 10.1007/s12275-015-4695-2) [DOI] [PubMed] [Google Scholar]
- 30.Adams MH. 1959. Bacteriophages, pp. 450–454. New York, NY: Interscience Publishers, Inc. [Google Scholar]
- 31.Berngruber TW, Weissing FJ, Gandon S. 2010. Inhibition of superinfection and the evolution of viral latency. J. Virol. 84, 10 200–10 208. ( 10.1128/JVI.00865-10) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weinbauer MG, Suttle CA. 1999. Lysogeny and prophage induction in coastal and offshore bacterial communities. Aquat. Microbiol. Ecol. 18, 217–225. ( 10.3354/ame018217) [DOI] [Google Scholar]
- 33.Nanda AM, Thormann K, Frunzke J. 2015. Impact of spontaneous prophage induction on the fitness of bacterial populations and host-microbe interactions. J. Bacteriol. 197, 410–419. ( 10.1128/JB.02230-14) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wiggins BA, Alexander M. 1985. Minimum bacterial density for bacteriophage replication: implications for significance of bacteriophages in natural ecosystems. Appl. Environ. Microbiol. 49, 19–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Payne RJH, Jansen VAA. 2000. Phage therapy: the peculiar kinetics of self-replicating pharmaceuticals. Clin. Pharmacol. Ther. 68, 225–230 ( 10.1067/mcp.2000.109520) [DOI] [PubMed] [Google Scholar]
- 36.Payne RJH, Jansen VAA. 2001. Understanding bacteriophage therapy as a density-dependent kinetic process. J. Theor. Biol. 208, 37–48. ( 10.1006/jtbi.2000.2198) [DOI] [PubMed] [Google Scholar]
- 37.Joo J, Gunny M, Cases M, Hudson P, Albert R, Harvill E. 2006. Bacteriophage-mediated competition in Bordetella bacteria. Proc. R. Soc. B 273, 1843–1848. ( 10.1098/rspb.2006.3512) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brown SP, Le Chat L, De Paepe M, Taddei F. 2006. Ecology of microbial invasions: amplification allows virus carriers to invade more rapidly when rare. Curr. Biol. 16, 2048–2052. ( 10.1016/j.cub.2006.08.089) [DOI] [PubMed] [Google Scholar]
- 39.Davies EV, James CE, Kukavica-Ibrulj I, Levesque RC, Brockhurst MA, Winstanley C. 2016. Temperate phages enhance pathogen fitness in chronic lung infection. ISME J. 10, 2553–2555. ( 10.1038/ismej.2016.51) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ghosh D, Roy K, Williamson KE, Srinivasiah S, Wommack KE, Radosevich M. 2009. Acyl-homoserine lactones can induce virus production in lysogenic bacteria: an alternative paradigm for prophage induction. Appl. Environ. Microbiol. 75, 7142–7152. ( 10.1128/AEM.00950-09) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Coyte KZ, Schluter J, Foster KR. 2017. The ecology of the microbiome: networks, competition, and stability. Science 350, 663–666. ( 10.1126/science.aad2602) [DOI] [PubMed] [Google Scholar]
- 42.Winter C, Bouvier T, Weinbauer MG, Thingstad TF. 2010. Trade-offs between competition and defense specialists among unicellular planktonic organisms: the ‘killing the winner’ hypothesis revisited. Microbiol. Mol. Biol. Rev. 74, 42–57. ( 10.1128/MMBR.00034-09) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gama JA, Reis AM, Domingues I, Mendes-Soares H, Matos AM, Dionisio F. 2013. Temperate bacterial viruses as double-edged swords in bacterial warfare. PLoS ONE 8, e59043 ( 10.1371/journal.pone.0059043) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Koskella B. 2013. Phage-mediated selection on microbiota of a long-lived host. Curr. Biol. 23, 1256–1260. ( 10.1016/j.cub.2013.05.038) [DOI] [PubMed] [Google Scholar]
- 45.Pietschke C, Treitz C, Forêt S, Schultze A, Künzel S, Tholey A, Bosch TCG, Fraune S. 2017. Host modification of a bacterial quorum-sensing signal induces a phenotypic switch in bacterial symbionts. Proc. Natl Acad. Sci. USA 114, E8488–E8497. ( 10.1073/pnas.1706879114) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sun CL, Relman DA. 2013. Microbiota's ‘little helpers’: bacteriophages and antibiotic-associated responses in the gut microbiome. Genome Biol. 14, 127 ( 10.1186/gb-2013-14-7-127) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mirzaei MK, Maurice CF. 2017. Ménage à trois in the human gut: interactions between host, bacteria and phages. Nat. Rev. Microbiol. 15, 397–408. ( 10.1038/nrmicro.2017.30) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The growth rate data shown in figure 1 are available in the data supplement of Li et al. [27].