Significance
From seasonal variation in the color of butterfly wings to trees bending toward the light, organisms often change in response to their environment. These changes, known as phenotypic plasticity, can result from differences in how genes are expressed among environments. Mutations causing environment-specific changes in gene expression provide raw material for phenotypic plasticity, but their frequency, effect size, and direction of effects among environments are not well understood. This study shows that mutations in the promoter of a yeast metabolic gene often display environment-dependent effects on gene expression and that these environment-dependent effects have been shaped by selection in natural populations.
Keywords: gene-by-environment interactions, cis-regulation, mutation, polymorphism, gene expression
Abstract
Phenotypic plasticity is an evolvable property of biological systems that can arise from environment-specific regulation of gene expression. To better understand the evolutionary and molecular mechanisms that give rise to plasticity in gene expression, we quantified the effects of 235 single-nucleotide mutations in the Saccharomyces cerevisiae TDH3 promoter (PTDH3) on the activity of this promoter in media containing glucose, galactose, or glycerol as a carbon source. We found that the distributions of mutational effects differed among environments because many mutations altered the plastic response exhibited by the wild-type allele. Comparing the effects of these mutations with the effects of 30 PTDH3 polymorphisms on expression plasticity in the same environments provided evidence of natural selection acting to prevent the plastic response in PTDH3 activity between glucose and galactose from becoming larger. The largest changes in expression plasticity were observed between fermentable (glucose or galactose) and nonfermentable (glycerol) carbon sources and were caused by mutations located in the RAP1 and GCR1 transcription factor binding sites. Mutations altered expression plasticity most frequently between the two fermentable environments, with mutations causing significant changes in plasticity between glucose and galactose distributed throughout the promoter, suggesting they might affect chromatin structure. Taken together, these results provide insight into the molecular mechanisms underlying gene-by-environment interactions affecting gene expression as well as the evolutionary dynamics affecting natural variation in plasticity of gene expression.
Phenotypic plasticity, which is the ability of an organism to develop different phenotypes in different environments, has been observed for diverse traits in diverse species (1, 2). This plasticity can facilitate adaptation to dynamically changing environments (3) but is not always adaptive (4, 5). Indeed, the role of natural selection in the evolution of phenotypic plasticity has long been the subject of debate (reviewed in ref. 6). For selection to act on plasticity, populations must exhibit genetic variation responsible for differences in plasticity among individuals (7). Such genetic variation, frequently detected as significant gene-by-environment (GxE) interactions in studies of quantitative traits (8), results from the mutational process generating variation in phenotypic plasticity, natural selection changing frequencies of alleles based on their effects on fitness, and genetic drift changing allele frequencies stochastically. Understanding how new mutations generate variation in plasticity and how selection acts on this variation are thus key for understanding the origin, maintenance, and evolution of phenotypic plasticity (9–12).
One common source of phenotypic plasticity is environment-specific regulation of gene expression (reviewed in ref. 13), which has been described for many organisms in response to different environmental cues (e.g., refs. 14–18). Standing genetic variation altering these environment-specific responses also appears to be common (e.g., refs. 18–22). For example, a study comparing gene expression in two strains of the baker’s yeast Saccharomyces cerevisiae grown in media containing glucose or ethanol as a carbon source found that 79% of genes showed expression differences attributed to the environment and 47% of genes had expression affected by quantitative trait loci (QTL) with evidence of significant interactions with the environment (20). QTL that alter plasticity of gene expression (and therefore contribute to GxE interactions) appear to be common in other systems as well (18, 22, 23). In most cases, it remains unclear whether the variation in gene expression plasticity observed has resulted from the neutral accumulation of mutations or has been shaped by the action of natural selection.
To separate the contributions of mutation and selection, mutation accumulation experiments have been used to isolate the effects of spontaneous mutations on a variety of quantitative traits (reviewed in refs. 11, 12, 24), including gene expression (e.g., refs. 25–28). By comparing trait variation among lines in which nonlethal mutations have accumulated in the near absence of selection, these studies estimate the mutational variance (Vm), which is the phenotypic variance added to a population each generation by new mutations. The Vm was found to differ among environments (indicating GxE interactions) for some traits (29–32) but not others (33, 34), demonstrating that the propensity of new mutations to alter plasticity differs depending on the trait and environments considered. Because mutation accumulation experiments allow variation to accumulate throughout the genome, trait variation observed in these studies cannot easily be tied to a particular mutation(s) (28, 35, 36). Recent studies have provided more direct links between specific mutations and variation in gene expression by using random or targeted mutagenesis to mutate the cis-regulatory region (promoter or enhancer) of a focal gene and then using high-throughput RNA sequencing or fluorescence reporters to determine the effects of these mutations on expression (37–41). By empirically describing the effects of new mutations on gene expression, a neutral model is generated that can be compared with the effects of genetic variants segregating in natural populations to infer the effects of natural selection (26, 42–44). To date, all such comparisons have been made between the effects of mutations and polymorphisms in a single environment; however, a similar approach can be used to disentangle the relative contributions of mutation and selection to variation in gene expression plasticity if the effects of mutations and polymorphisms are measured in multiple environments.
Here, we use this approach to examine the effects of mutation and selection on plasticity of a cis-regulatory sequence controlling gene expression by measuring the effects of 235 mutations and 30 polymorphisms in the S. cerevisiae TDH3 promoter (PTDH3) in media containing one of three different carbon sources (glucose, galactose, and glycerol). This promoter comes from the TDH3 gene, which encodes a glycolytic enzyme (glyceraldehyde-3-phosphate dehydrogenase) involved in the metabolism of fermentable sugars such as glucose and galactose as well as nonfermentable carbon sources such as glycerol (Fig. 1A). These three carbon sources are likely to be encountered and used by natural populations of S. cerevisiae (albeit at unknown frequencies), since the genome contains conserved sets of genes for metabolizing glucose (45), galactose (46), and glycerol (47). Genes required specifically for metabolism of galactose or glycerol are repressed in the presence of glucose, which appears to be the preferred carbon source for S. cerevisiae (48). We found that activity of the wild-type PTDH3 displays plasticity among environments containing glucose, galactose, or glycerol as a carbon source. We also found that the distributions of effects of cis-regulatory mutations on promoter activity varied among these three environments because of differences in the frequency, magnitude, and direction of GxE effects observed between pairs of environments. When we compared the effects of these mutations with the effects of PTDH3 polymorphisms segregating in natural populations, we found that mutations tended to cause larger differences in promoter activity between the two fermentable environments than polymorphisms, suggesting that natural selection has preferentially eliminated genetic variants that increase expression plasticity between glucose and galactose. Finally, we found that positions in the promoter of mutations with environment-dependent effects differed among pairs of environments. Mutations with different effects on expression between fermentable and nonfermentable carbon sources clustered in previously characterized transcription factor binding sites, whereas mutations with different effects between the two fermentable carbon sources were distributed more evenly throughout the promoter, suggesting that they affect chromatin state.
Fig. 1.
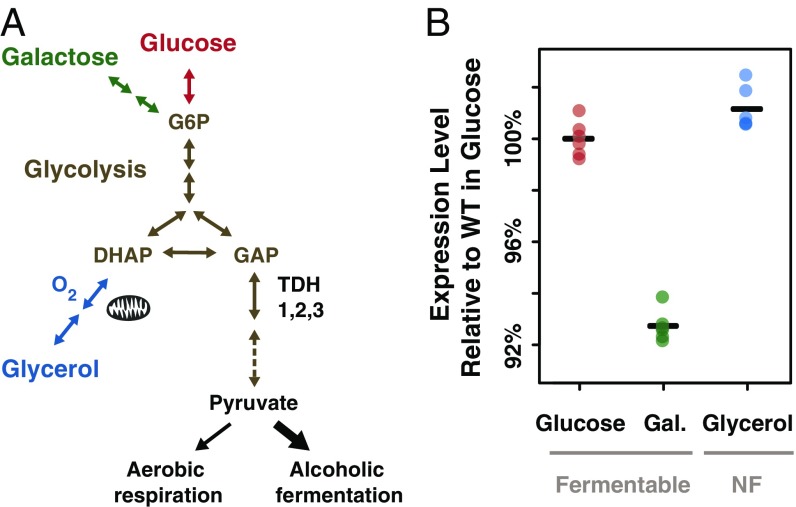
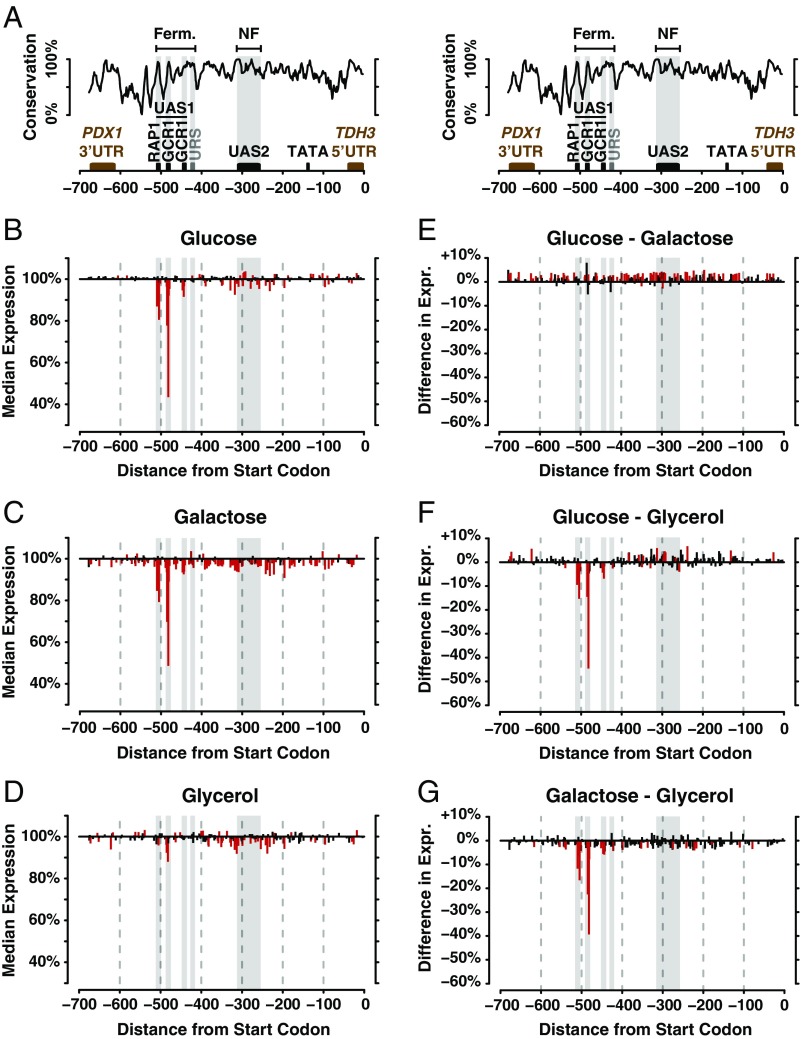
TDH3 functions in central metabolism, and its promoter activity is plastic among environments. (A) TDH3 encodes one of S. cerevisiae’s three glyceraldehyde-3-phosphate dehydrogenase proteins involved in glycolysis and required for metabolism of fermentable carbon sources such as glucose and galactose and of the nonfermentable carbon source glycerol after aerobic conversion in dihydroxyacetone phosphate (DHAP) in mitochondria. Greater detail is provided in the Yeast Pathways Database (https://pathway.yeastgenome.org/) (46, 47, 83). (B) Activity of wild-type PTDH3 in media containing glucose (red), galactose (green), and glycerol (blue) is shown. Colored dots represent the median fluorescence level of each of the six replicate populations, and black bars indicate the mean of the six median fluorescent levels observed for each environment. NF, nonfermentable.
Results and Discussion
Plasticity of PTDH3 Activity.
To measure PTDH3 activity in living cells, we quantified the expression of a PTDH3-YFP reporter gene inserted in the genome of the laboratory strain YPW1 that contains a wild-type allele of PTDH3 fused to the coding sequence of Venus YFP (49). The fluorescence level of YPW1 was measured after growth in rich media containing glucose, galactose, or glycerol as a carbon source to quantify differences in promoter activity among environments and measure plasticity. For each environment, the level of fluorescence of at least 10,000 cells was quantified by flow cytometry in each of six replicate populations, and the median fluorescence of each sample was divided by the average fluorescence measured among the six replicates in glucose (“Plasticity.WT” worksheet in Dataset S1). We observed statistically significant differences in expression of the reporter gene (i.e., expression plasticity) between each pair of environments (Fig. 1B; t tests: Pglu-gal = 3.2 × 10−9, Pglu-gly = 0.025, Pgal-gly = 6.1 × 10−9). Surprisingly, a larger difference in expression level was observed between the two fermentable carbon sources, glucose and galactose (7.3% lower expression in galactose), than between one of the fermentable carbon sources (glucose) and the nonfermentable (glycerol) carbon source (1.2% higher expression in glycerol), despite the fact that growth in the two fermentable carbon sources involves more similar metabolic processes than growth in fermentable and nonfermentable environments (Fig. 1A).
Environment-Dependent Distributions of Mutational Effects.
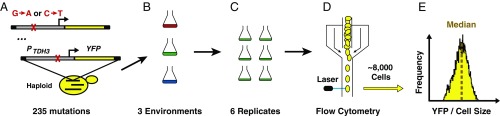
To determine how new mutations altered the plastic response observed for the wild-type allele of PTDH3, we examined 235 mutant versions of the reporter gene created by using site-directed mutagenesis to change one of the 241 G’s and C’s in PTDH3 into an A or T, respectively, in each strain (42) (Fig. 2A). Each of these 235 mutant genotypes, as well as the unmutated “wild-type” genotype, was then grown in six replicate populations in media containing glucose, galactose, or glycerol as a carbon source (Fig. 2 B and C). The activity of the PTDH3-YFP reporter gene was assayed in ∼8,000 cells, on average, in each population using flow cytometry (Fig. 2D). The fluorescence level of individual cells was normalized by cell size using a procedure that took into account the small differences observed in the relationships between fluorescence and cell size among carbon sources (Materials and Methods). After controlling for technical variation, the median fluorescence level normalized by cell size was calculated for each population and used as a proxy for promoter activity (Fig. 2E and “All.Mutations.Data” worksheet in Dataset S1).
Fig. 2.
Experimental approach used to measure the activity of 235 alleles of PTDH3 in three carbon source environments. (A) Site-directed mutagenesis was used to change G’s to A’s and C’s to T’s in the PTDH3 driving expression of a YFP in each of 235 strains (42). (B) Each of these strains, plus a strain carrying the wild-type PTDH3-YFP allele, was then grown in three environments containing different carbon sources. (C) Six replicate populations for each genotype were analyzed in each type of media. (D) Fluorescence of individual cells in each population was assessed using flow cytometry. (E) After correcting for differences in cell size (as described in Materials and Methods), the median fluorescence was determined for each population.
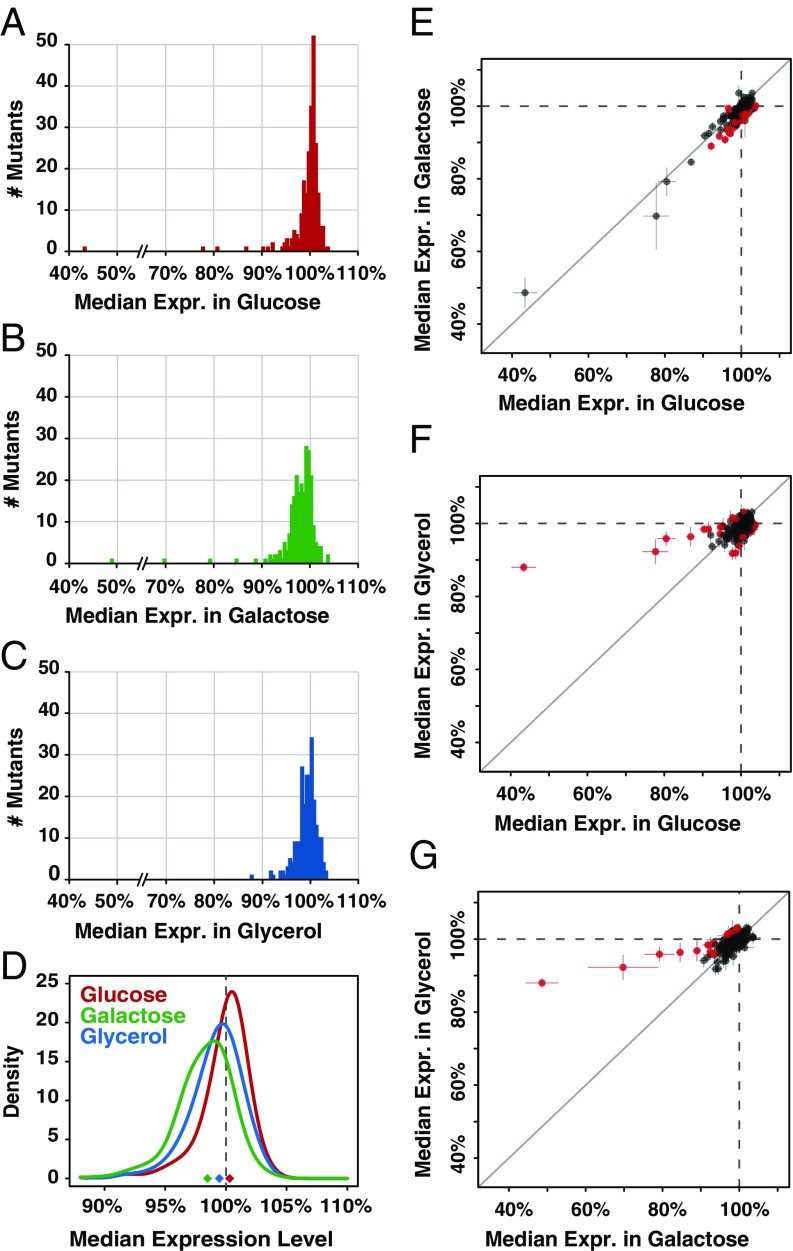
To quantify changes in plasticity caused by mutations, we controlled for the plasticity of the wild-type PTDH3 allele (Fig. 1B) by dividing the median fluorescence level of each replicate population of the 235 mutant genotypes by the average median fluorescence measured for the wild-type allele in the corresponding environment. The mean relative fluorescence from six replicates was then used to estimate the effect of each genotype on promoter activity in each environment, with variability among replicates used to calculate the 95% confidence interval around this mean. Using this relative measure of expression, we found that the distributions of effects of the 235 mutant genotypes on PTDH3-YFP expression were different between glucose and galactose [Fig. 3 A and B; Kolmogorov–Smirnov (KS) test with permutations: D = 0.43, P < 5 × 10−6], between glucose and glycerol (Fig. 3 A and C; KS test with permutations: D = 0.23, P < 5 × 10−6), and between galactose and glycerol (Fig. 3 B and C; KS test with permutations: D = 0.22, P < 5 × 10−6). In galactose, the distribution of mutational effects was more skewed toward decreases in expression (median effect = −1.5% relative to wild type) than in glucose (median effect = +0.3% relative to wild type, permutation test: Pglu-gal < 5 × 10−6) or glycerol (median effect = −0.5% relative to wild type; Fig. 3D; permutation tests: Pglu-gal < 5 × 10−6, Pglu-gly = 5 × 10−6, Pgly-gal < 5 × 10−6). This mutational bias toward lower expression suggests that new mutations in PTDH3 will tend to further decrease TDH3 expression in galactose relative to its expression in glucose or glycerol, amplifying the plasticity observed for the wild-type allele.
Fig. 3.
Effects of 235 cis-regulatory mutations on activity of PTDH3 in glucose, galactose, and glycerol. Histograms show the median activity (averaged across six replicate populations) for the 235 mutant PTDH3 alleles relative to the average activity of the wild-type promoter in the same environment for cells grown in glucose (A), galactose (B), or glycerol (C). (D) Curves showing the kernel density estimates computed from the distributions of mutational effects measured in glucose (red), galactose (green), and glycerol (blue). Diamonds indicate the median effect size of the 235 mutations, with colors corresponding to the carbon sources. The vertical dotted line shows the expression level conferred by the wild-type promoter in glucose. Comparisons of effects of TDH3 cis-regulatory mutations between glucose and galactose (E), glucose and glycerol (F), and galactose and glycerol (G) are shown. The median activity of mutant promoters is expressed relative to the average activity of the wild-type allele in each environment, with dots showing the average activity across six replicates and error bars showing 95% confidence intervals. Dots colored red showed evidence of a statistically significant GxE interaction based on t tests corrected for multiple testing using the Benjamini–Hochberg FDR adjustment (Padj < 0.01). Expr., expression.
In addition to differences in their median effects, the distributions of mutational effects displayed different degrees of dispersion depending on the environment (Fig. 3D). To quantify the dispersion of each distribution, we calculated the median absolute deviation (MAD) of the median expression levels observed across the 235 mutant strains in each environment. The MAD is a measure of dispersion less sensitive to rare outliers than the SD. We found that the variability of mutational effects was similar in galactose and glycerol (MADgal = 1.97%, MADgly = 1.75%; permutation test: Pgal-gly = 0.12), but lower in glucose (MADglu = 1.12%) than in galactose (permutation test: Pglu-gal < 5 × 10−6) or glycerol (permutation test: Pglu-gly = 3.3 × 10−4). In addition, the median effect size of mutations on promoter activity was lowest in glucose (median absolute effect of 235 mutations: glucose = 0.92%, galactose = 1.80%, glycerol = 1.16%; permutation tests: Pglu-gal < 5 × 10−6, Pglu-gly = 0.036, Pgal-gly = 3 × 10−5), indicating that mutant promoters conferred expression levels closest to the wild-type promoter in glucose. Taken together, these observations suggest that the activity of PTDH3 is more robust to the effects of new cis-regulatory mutations in glucose, which is the preferred carbon source of S. cerevisiae (48), than in glycerol or galactose.
Frequency, Magnitude, and Direction of GxE Interactions.
Differences in the distributions of mutational effects among the three environments indicated that at least some of these mutations in PTDH3 have environment-dependent effects. To identify these mutations (i.e., to test for GxE interactions), we compared the effects of each mutation relative to the wild-type allele between environments using a series of pairwise t tests with a Benjamini–Hochberg false discovery rate (FDR) correction for multiple testing. We found that the effects of individual mutations were more strongly correlated between the two fermentable carbon sources (Fig. 3E; Pearson correlation coefficient: rglu-gal = 0.94) than between either fermentable carbon source and the nonfermentable glycerol (Fig. 3F, rglu-gly = 0.55; Fig. 3G, rgal-gly = 0.65). However, nearly threefold as many mutations (N) showed statistically significant evidence of GxE interactions for the comparison between the two fermentable carbon sources (glucose and galactose) than for other pairs of environments (Penv1-env2 < 0.01: Nglu-gal = 75, Nglu-gly = 29, Ngal-gly = 26). This disconnect results from differences in the magnitude of significant GxE effects between pairs of environments, with the largest differences observed when comparing fermentable and nonfermentable environments (Δglu-gly = 6.3%, Δgal-gly = 6.0%, Δglu-gal = 3.1%; Mann–Whitney–Wilcoxon tests: PΔglu-gly vs. Δgal-gly = 0.77, PΔglu-gly vs. Δglu-gal = 0.0032, PΔgal-gly vs. Δglu-gal = 0.029). These data indicate that mutations with large GxE effects can arise when plasticity of the wild-type allele is small (glucose vs. glycerol) and mutational effects can be well correlated between environments even when the plasticity of the wild-type allele is large (glucose vs. galactose). Biases in the direction of GxE effects also differed among pairwise comparisons: 73 of 75 mutations with significant GxE effects between glucose and galactose (Fig. 3E) and all 24 mutations with significant GxE effects between glycerol and galactose (Fig. 3G) caused lower expression in galactose (binomial test: P < 10−6 in both cases), whereas no bias in the direction of GxE effects was observed between glucose and glycerol (Fig. 3F; Nglu > gly = 15, Ngly > glu = 14; binomial test: P = 1). The strong directional bias of the GxE effects observed between galactose and other carbon sources suggests that plasticity of expression could increase in the absence of natural selection simply through the random occurrence of cis-regulatory mutations.
Effects of Selection on Expression Plasticity.
Genetic variation for plasticity segregating in natural populations can be influenced both by the mutational process creating new genetic variation and by natural selection filtering genetic variants based on their phenotypic effects. To test for evidence of selection acting on plasticity of PTDH3 activity, we compared the effects of mutations in media containing glucose, galactose, and glycerol with the effects in the same environments of 30 polymorphisms in this promoter that were identified in 85 strains of S. cerevisiae. To quantify the effects of these polymorphisms on promoter activity, 39 haplotypes of PTDH3 that differed from each other by one to 10 polymorphisms (Dataset S2) were cloned upstream of the YFP coding sequence (42), and the fluorescence levels of the resulting strains were measured by flow cytometry after growth of six replicate populations in each of the three carbon source environments. The effect of each of 30 unique polymorphisms (Fig. S1 and “Polymorphisms.Effects” worksheet in Dataset S1) was inferred by dividing the fluorescence measured for a haplotype containing that polymorphism by the average fluorescence measured for an ancestral haplotype that differed by only that polymorphism, as described more fully by Metzger et al. (42) and Duveau et al. (50) (also Materials and Methods). As a group, these 30 polymorphisms caused expression to vary from 94.6–102.5% of the ancestral allele in glucose (Fig. 4A), 96.1–105.5% in galactose (Fig. 4B), and 95.4–103.7% in glycerol (Fig. 4C). The 10 polymorphic G:C → A:T transitions, 13 other types of single-nucleotide polymorphisms (SNPs), and seven indel polymorphisms showed similar median effects on expression level in all three environments (Fig. S1 B–D). Despite the narrow range of effects covered by these PTDH3 polymorphisms, their effects were well correlated between environments (Fig. S2, Pearson correlation coefficients: rglu-gal = 0.58, Pglu-gal = 8.4 × 10−4; rglu-gly = 0.42, Pglu-gly = 0.022; rgal-gly = 0.72, Pgly = 6.9 × 10−6). Evidence of significant GxE effects was observed for four of the 30 polymorphisms between glucose and galactose, two between glucose and glycerol, and two between galactose and glycerol (Fig. S2).
Fig. 4.
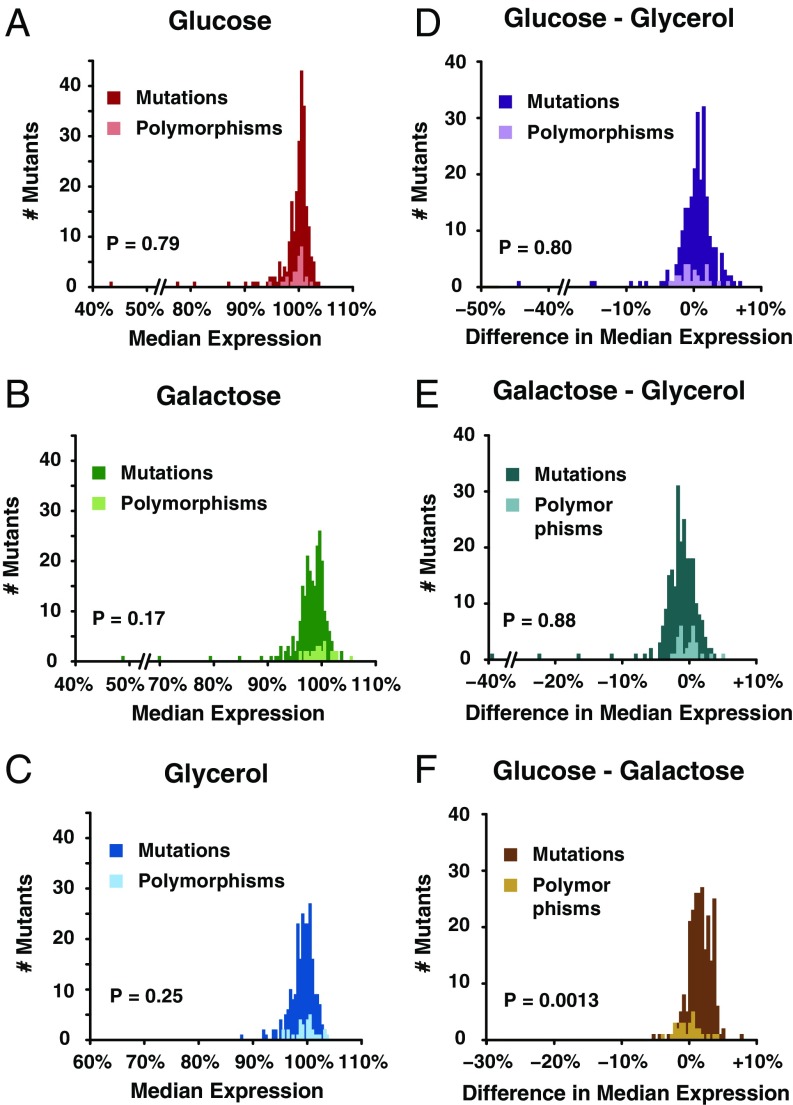
Selection affects natural variation in plasticity of PTDH3 activity. Histograms show the distributions of effects for 235 point mutations and for 30 polymorphisms in PTDH3 upon growth in glucose (A), galactose (B), and glycerol (C). P values represent the significance of the difference between the effects of polymorphisms and mutations obtained using nonparametric resampling tests as described in Materials and Methods. (D–F) Histograms showing the distributions of differences in effects between environments for 235 point mutations and for 30 polymorphisms in PTDH3. The differences of promoter activity were calculated between glucose and glycerol (D), galactose and glycerol (E), and glucose and galactose (F). P values were determined using nonparametric resampling tests to compare the effects of mutations and polymorphisms on plasticity between pairs of environments.
To determine whether selection has maintained genetic variants with a particular subset of effects on expression in natural populations, we compared the distributions of effects of mutations and polymorphisms in each environment with the same nonparametric approach used by Metzger et al. (42). For cells grown in glucose, we found that the distribution of effects of polymorphisms was not significantly different from random sampling of the mutational distribution (Fig. 4A and Fig. S3A; P = 0.43), consistent with the findings of Metzger et al. (42). Likewise, we found that the effects of these polymorphisms in galactose (Fig. 4B and Fig. S3B; P = 0.35) and glycerol (Fig. 4C and Fig. S3C; P = 0.51) were consistent with random sampling of mutations. Therefore, no significant signature of selection acting on the level of activity of PTDH3 was detected in any of the three environments tested.
Next, we tested for evidence of selection acting on expression plasticity between environments by comparing the difference of effects between each pair of environments for mutations and polymorphisms using the approach described above. We detected no significant difference in the plasticity of expression conferred by the set of polymorphisms and random sets of mutations between fermentable and nonfermentable environments (glucose and glycerol, P = 0.80, Fig. 4D and Fig. S3D; galactose and glycerol, P = 0.88, Fig. 4E and Fig. S3E); however, we observed a significant difference in plasticity between the two fermentable environments, glucose and galactose (P = 0.0013; Fig. 4F and Fig. S3F), with the 30 polymorphisms showing smaller median differences in expression between glucose and galactose than random sets of 30 mutations [Δpol(glu-gal) = +0.08% vs. Δmut(glu-gal) = +1.64%; permutation test: P = 5.2 × 10−3]. This observation suggests that selection has preferentially eliminated new mutations that cause the largest expression plasticity between glucose and galactose. Selection might have also disfavored mutations with a large impact on plasticity observed between fermentable and nonfermentable environments (Fig. 3 F and G), but the low frequency of these mutations suggests that a larger number of polymorphisms would be needed to detect selection acting on plasticity between fermentable and nonfermentable environments.
Observing evidence of selection acting on plasticity between glucose and galactose without also observing evidence of selection acting on expression levels in either environment individually was surprising. To better understand this result, we examined the effects of mutations and polymorphisms in glucose and galactose more closely. We found that mutations tended to confer slightly higher expression levels than polymorphisms in glucose [difference of median effects: Δglu(mut-pol) = +0.37%] and lower expression levels than polymorphisms in galactose [difference of median effects: Δgal(mut-pol) = −1.0%]. Because these (weak) directional biases were in opposite directions and because polymorphisms had similar effects in glucose and galactose [difference of median effects: Δpol(glu-gal) = +0.08%; permutation test: P = 0.85], the difference in effects between mutations and polymorphisms was larger and easier to detect between the two environments than in either environment alone. These observations suggest that selection might disfavor mutations that increase expression in glucose and/or decrease expression in galactose. The larger difference in median effects between mutations and polymorphisms in galactose than glucose as well as the minimal fitness effects of small increases in TDH3 expression in glucose reported recently (50) suggest that selection against mutations that decrease expression in galactose is more likely to be driving this pattern.
Potential Molecular Mechanisms Underlying Gene-by-Environment Interactions.
One of the reasons that we selected PTDH3 for this work was that prior studies had characterized functional elements within its sequence (Fig. 5A), providing an opportunity to develop hypotheses about the molecular mechanisms underlying plasticity and GxE interactions. Specifically, Kuroda et al. (51) described an upstream activating sequence (UAS1; 426–528 bp upstream of the start codon), as required for expression in a fermentable carbon source (glucose) but not a nonfermentable carbon source (glycerol + lactate). This sequence contains binding sites for the RAP1 and GCR1 proteins, which work together to regulate transcription of glycolytic genes and have different effects on transcription in fermentable and nonfermentable environments (52–54). An upstream repressing sequence (URS; 419–431 bp upstream of the start codon) adjacent to UAS1 was also identified by Kuroda et al. (51) that appeared to repress activity of a second UAS (UAS2) in the fermentable environment assayed (Fig. 5A). This UAS2 (255–309 bp upstream of the start codon) was described as being primarily responsible for activation in the nonfermentable environment tested (51).
Fig. 5.
Locations of mutations affecting activity and plasticity of PTDH3 within the promoter sequence. (A) Summary of previously identified functional elements in PTDH3 is shown. These elements include binding sites for the RAP1 and GCR1 transcription factors as well as sequences shown previously by deletion analysis to be required for expression during growth on a fermentable carbon source, glucose (UAS1 and URS), or a nonfermentable carbon source, glycerol plus lactate (UAS2) (51). For orientation, the TATA box is also indicated, although this sequence was not mutated in any of the strains analyzed. The black curve shows sequence conservation across species of the S. senso stricto genus. The effect of each mutation on the median expression level of PTDH3-YFP relative to expression of the unmutated wild-type allele is shown for cells grown on glucose (B), galactose (C), and glycerol (D). Mutation effects represented in red led to a significant change in expression relative to the wild-type allele (t test: P < 0.01). The difference in effect of each mutation on PTDH3-YFP in each pair of environments relative to the wild-type allele is shown for glucose and galactose (E), glucose and glycerol (F), and galactose and glycerol (G). The mutation represented in red showed a significant difference of effects between the two environments based on t tests corrected for multiple testing using the Benjamini–Hochberg FDR adjustment (Padj < 0.01). Areas shaded in gray correspond to functional elements located directly above these regions in A. Expr., expression; Ferm., fermentable; NF, nonfermentable.
To better understand the environment-specific regulation of PTDH3, we compared the locations of mutations with significant effects on PTDH3 activity with these previously described functional elements. In the two fermentable environments we examined (glucose and galactose), the mutations with the largest effect altered sequences in the RAP1 and GCR1 transcription factor binding sites (Fig. 5 B and C). The effects of mutations in these binding sites were much smaller in the nonfermentable environment (glycerol), although they remained among the sites with the largest effects on expression even in this environment (Fig. 5D). This result is consistent with the UAS1 region playing a larger role in regulating TDH3 expression in fermentable than nonfermentable carbon sources, but it also suggests that activity of UAS1 is not strictly limited to fermentable environments. In glycerol, many mutations that affected the activity of PTDH3 were concentrated near the UAS2 region previously described as required for expression in media containing glycerol and lactate (51), but mutations with significant effects also extended beyond this region (Fig. 5D), indicating that the functional region used for expression in glycerol is larger than the UAS2 region described by Kuroda et al. (51). Mutations in the UAS2 region also impacted expression in glucose and galactose, indicating their function is not restricted to nonfermentable carbon sources. In the repressive URS sequence, one mutation (423 bp upstream of the start codon) increased expression in glucose and another mutation (426 bp upstream of the start codon) increased expression in galactose, but none had significant effects in glycerol (Fig. 5 B–D), consistent with its previous description as a repressor in fermentable carbon sources. Additional mutations in this sequence had significant effects only in galactose, where they decreased expression (Fig. 5 B–D).
To identify molecular mechanisms that might give rise to GxE effects of single-nucleotide changes in PTDH3, we mapped the differences of mutational effects between each pair of environments on the promoter architecture. In the comparison between the two fermentable environments (glucose and galactose), the 75 mutations with significantly different effects between glucose and galactose (73 of which showed higher expression in glucose than galactose) appeared to be distributed randomly throughout the promoter (Fig. 5E). The result of a nearest neighbor statistical test for clustering of mutations was consistent with this observation (SD of distance to nearest neighbor: observed = 6.63 vs. expected = 8.12; P = 0.51). Between either fermentable carbon source (glucose or galactose) and the nonfermentable glycerol, mutations with the largest GxE effects (expression differences ≥2% between environments) appeared to be concentrated in previously identified functional elements (Fig. 5 F and G). In the comparison between glucose and glycerol (Fig. 5F), permutation testing confirmed that the previously identified UAS1 sequence required for expression on fermentable carbon was significantly enriched for mutations showing evidence of GxE interactions (Fig. 5F; number of mutations with significant GxE effects in UAS1: observed = 9 vs. expected = 4.4; P = 0.009), as was the UAS2 element previously shown to be required for expression during growth on glycerol plus lactate (number of mutations with significant GxE effects in UAS2: observed = 6 vs. expected = 3.0; P = 0.027). In the comparison between galactose and glycerol (Fig. 5G), significant enrichment of mutations showing evidence of GxE interactions was observed for the UAS1 region (number of mutations with significant GxE effects in UAS1: observed = 9 vs. expected = 4.1; P = 0.001) but not for the UAS2 region (number of significant GxE effects in UAS2: observed = 1 vs. expected = 3.0; P = 0.94).
Taken together, these data suggest that different molecular mechanisms underlie GxE interactions in different pairs of environments. Transcription factor binding sites for RAP1 and GCR1 in UAS1, and potentially binding sites for other transcription factors in UAS2, appear to regulate TDH3 expression differently in fermentable (glucose or galactose) and nonfermentable (glycerol) environments. The fact that mutations with the largest GxE effects between either pair of fermentable and nonfermentable carbon sources affected the RAP1 and GCR1 binding sites (Fig. 5 F and G) is consistent with this hypothesis. By contrast, mutations with significant GxE interactions between the two fermentable environments (glucose and galactose) were distributed throughout PTDH3 (Fig. 5E), suggesting that they might affect more widespread environment-specific chromatin structure rather than specific binding sites. Pavlović and Hörz (55) identified a nucleosome-free region in PTDH3 extending from the 5′ end of UAS1 to the 3′ end of UAS2 in both glucose and glycerol but did not examine chromatin structure in galactose. We hypothesize that this region exhibits altered nucleosome positioning when cells are grown in galactose. Differences in nucleosome occupancy between these environments may reduce expression of the wild-type allele in galactose relative to glucose (Fig. 1B), while maintaining the strong correlation of mutational effects observed between these two environments (Fig. 3E). Changes in chromatin structure across PTDH3 in galactose could also explain why so many mutations distributed throughout the promoter reduced expression 2–5% only in galactose (Fig. 5C). Consistent with this model of environment-specific nucleosome occupancy, prior work has shown that activity of promoters occupied by nucleosomes tends to be affected by mutations distributed throughout the whole promoter sequence, whereas activity of promoters with a nucleosome-free region upstream of the transcription start site tends to be affected by a smaller number of mutations clustered in transcription factor binding sites (56). In addition, differences in nucleosome positioning have been observed for several promoters between yeast cells grown in glucose and galactose (57–59).
Conclusions
By characterizing the effects of 235 single-nucleotide changes in PTDH3 on its activity in three different environments, we have shown that changes in the environment can modify the distribution of expression phenotypes generated by new cis-regulatory mutations. We observed mutational biases toward decreased PTDH3 activity in galactose and (to a lesser extent) in glycerol, but not in glucose, suggesting that the random accumulation of mutations would tend to increase the plastic response of PTDH3 activity in these environments relative to the wild-type allele of the promoter. In addition, the variability of effects observed among the 235 PTDH3 mutations was lower in glucose than in galactose or glycerol, indicating that mutational robustness was greatest in the environment containing the preferred carbon source of S. cerevisiae (48). Such mutational robustness of promoter activity could be the result of adaptation to a commonly experienced environment (60–63) or fluctuating selection (64, 65), although nonadaptive processes could also account for this result (66–69). Because mutations create the phenotypic variation necessary for the action of other evolutionary forces and because organisms are constantly faced with environmental changes, differences in the distributions of mutational effects among environments can play an important role in trait evolution (12, 70–72).
Focusing on the effects of individual mutations, we found that ∼10–30% of the 235 mutations examined showed significant GxE interactions affecting PTDH3 activity, depending on the pair of environments. This observation indicates that mutations in PTDH3 readily generate genetic variation affecting expression plasticity, providing the raw material needed for natural selection to alter plasticity (7, 73, 74). The mutations tested in PTDH3 increased the difference in expression between glucose and galactose more than the polymorphisms examined, suggesting that natural selection has eliminated alleles that increase the plasticity beyond a certain degree. Because only two mutations significantly reduced plasticity between this pair of environments, we were unable to determine whether reduced plasticity between glucose and galactose is more likely to be neutral, deleterious, or beneficial. Therefore, we cannot distinguish between a hypothesis of directional selection favoring minimal plasticity and a hypothesis of stabilizing selection maintaining a particular degree of plasticity. The wild-type allele and all but one of the 30 polymorphisms examined were recently shown to confer maximal fitness in the glucose-based medium tested (50), but the fitness effects of PTDH3 alleles will also need to be determined in galactose to discriminate between these two different evolutionary scenarios.
In addition to advancing our understanding of how mutation and selection interact to maintain expression plasticity in natural populations, the single-nucleotide resolution of our data allowed us to identify potential molecular mechanisms underlying plasticity and GxE interactions of PTDH3. These data suggest that the condition-dependent use of transcription factor binding sites is primarily responsible for differences in regulation of TDH3 expression between fermentable and nonfermentable environments, whereas differences in chromatin structure might play a larger role in generating plasticity between the two fermentable environments tested. Prior studies hypothesizing mechanisms for GxE interactions have focused primarily on variation in context-dependent cis-regulatory sequences (19, 21, 75). Characterizing the distributions of mutational effects in multiple environments for other promoters and including mutations in the trans-acting factors that interact with these promoters will be necessary to draw general conclusions about the mechanisms affecting the evolution of gene expression plasticity.
Materials and Methods
Yeast Strains.
Construction of the unmutagenized reference strain YPW1 containing the PTDH3-YFP reporter gene is described fully by Gruber et al. (49). This reporter gene contains a 678-bp PTDH3 sequence fused to the coding sequence for YFP Venus optimized for expression in S. cerevisiae (76) and the CYC1 (cytochrome c isoform 1) terminator, and is inserted near the SWH1 pseudogene on chromosome 1 of strain BY4724 (77) at position 199270. All strains included in this study were derived from BY4724, and therefore are auxotrophs for uracil and lysine and haploids with MATa mating type. Construction of 236 mutant strains, each containing a single G:C → A:T transition in the PTDH3 region of this reporter gene, was accomplished by PCR-mediated site-directed mutagenesis, as described by Metzger et al. (42). One of these strains, YPW519, was excluded from this study because it appeared to have acquired a secondary mutation that increased expression of PTDH3-YFP. To identify natural haplotypes of PTDH3, the 678-bp promoter region was sequenced in 85 isolates of S. cerevisiae (78), identifying 28 distinct haplotypes (42). These haplotypes differed from each other by one to 13 polymorphisms. Each of these haplotypes was PCR-amplified and inserted upstream of the YFP coding sequence in the YPW1 genetic background, as described by Metzger et al. (42). Fifteen additional haplotypes of PTDH3 that differed from one of the 28 natural haplotypes by only a single polymorphism were then constructed by PCR-mediated site-directed mutagenesis of a natural haplotype and cloned upstream of the YFP coding sequence. Together, these 15 haplotypes plus the 28 haplotypes observed among the isolates sampled resulted in a set of 39 PTDH3 haplotypes in which each haplotype differed from at least one other haplotype by as little as one polymorphism. The remaining four haplotypes, all of which were observed in one or more natural isolates, differed from the most similar haplotype by more than one polymorphism and were excluded from this study. Relationships among these haplotypes were represented in a haplotype network as described below and by Metzger et al. (42). In all, these 39 haplotypes contained 30 unique polymorphisms in PTDH3 (Dataset S2).
Growth Conditions.
The fluorescence levels of strains included in this study were quantified in four consecutive experiments. The goal of the first experiment was to quantify the plasticity of expression of the wild-type PTDH3 allele by measuring (in parallel) the fluorescence of strain YPW1 after growth in media containing glucose, galactose, or glycerol as a carbon source. The other three experiments measured the fluorescence of the 236 cis-regulatory alleles with point mutations in PTDH3 and the 39 PTDH3 haplotypes with different polymorphisms in each of the three carbon source environments. Fluorescence data for samples grown in rich media containing glucose originally collected for and described in a study by Metzger et al. (42) were reanalyzed for this study using scripts modified to allow identical analyses in multiple environments.
We started the first experiment by reviving strain YPW1 that contained the wild-type PTDH3-YFP reporter gene and strain BY4724 used to correct for autofluorescence from frozen glycerol stocks onto YPG agar plates (10 g/L yeast extract, 20 g/L peptone, 3% vol/vol glycerol, and 20 g/L agar). After 48 h of growth at 30 °C, we filled 12 wells of a deep 96-well plate with 0.5 mL of liquid YPG, 12 wells with 0.5 mL of YPD (10 g/L yeast extract, 20 g/L peptone, and 20 g/L d-glucose) and 12 wells with 0.5 mL of YPGal (10 g/L yeast extract, 20 g/L peptone, and 20 g/L galactose). Other treatments were applied to the remaining wells, but these data were not used in this study (a full description is provided in the “Layout.Plasticity.txt” worksheet in Dataset S3). Six wells containing YPG were inoculated with strain YPW1, and the six other wells were inoculated with strain BY4724 to a density of ∼1.5 × 10−5 cells per milliliter. The plate was then incubated at 30 °C with constant orbital shaking at (9 × g). Each well contained a 3-mm glass bead that maintained cells in suspension during growth. After 22 h of growth, YPW1 and BY4724 were each inoculated in six wells containing YPD and six wells containing YPGal to a density of ∼1.5 × 105 cells per milliliter. The plate was then incubated for another 20 h at 30 °C with constant shaking. We delayed the inoculation of samples in YPD and YPGal so that all samples reached a density above 6 × 107 cells per milliliter simultaneously a few hours before the end of the experiment, despite the slower growth rate in the nonfermentable YPG (∼170 min per division) compared with the fermentable YPD (∼85 min per division) and YPGal (∼95 min per division). Samples were grown in parallel in the three carbon source environments to avoid previously observed day-to-day variation in flow cytometer sensitivity that complicates quantitative comparisons of absolute fluorescence levels between experiments. After growth, samples were diluted to ∼2.5 × 106 cells per milliliter in a clean plate containing 0.5 mL per well of synthetic complete (SC) medium lacking arginine (SC-arginine) with the same carbon source used for growth, and fluorescence levels of single cells were quantified by flow cytometry as described below.
For the three experiments performed separately in media containing glucose, glycerol, or galactose, yeast strains were arrayed and kept frozen at −80 °C in eight 96-well plates before use. These experiments included the 236 mutant strains with point mutations in the copy of PTDH3 driving expression of YFP and the 39 strains with natural and reconstructed haplotypes of PTDH3 driving expression of YFP (used to quantify the effects of 30 unique polymorphisms on fluorescence levels), as well as appropriate controls such as YPW1, with the wild-type allele of PTDH3 driving expression of YFP and BY4724 used to correct for autofluorescence. Other strains not used for this study were also included in these experiments. Importantly, each plate contained 20 replicates of the control strain YPW1 at fixed positions used to correct for technical variation in fluorescence levels. Apart from these controls, the position of other strains was fully randomized to avoid systematic positional bias in fluorescence levels for different types of mutants (a full description is provided in the “Layout.Mutations.Polymorphisms” worksheet in Dataset S3). Before each experiment, samples from each of the eight frozen plates were transferred to an OmniTray containing YPG agar medium. After 48 h of growth, samples from each OmniTray were transferred with a V&P Scientific pintool to three 96-well plates containing 0.5 mL of YPD (glucose), 0.5 mL of YPG (glycerol), or 0.5 mL of YPGal (galactose) per well (with one 3-mm glass bead per well), for a total of 24 plates in each experiment (three replicates of the eight initial plates). Plates were incubated at 30 °C with constant shaking at (9 × g) during 20 h for samples grown in YPD or YPGal and during 42 h for samples grown in YPG. After growth, 20 μL of cell culture was diluted into 0.5 mL of synthetic complete medium lacking arginine (with the same carbon source used for population growth) in a clean 96-well plate that was immediately run through the flow cytometer for quantification of fluorescence levels as described below. Twenty-four additional 96-well plates were inoculated the next day from the same OmniTrays stored at 4 °C. These samples were grown and their fluorescence was scored following the same procedure as described above so that, in total, fluorescence was measured for six replicates of the eight initial plates (i.e., six replicate populations of each mutant strain). For the experiment performed in YPD, we quantified fluorescence for nine replicate populations of each sample but only analyzed data from six replicates to keep the number of replicates consistent between environments. For the experiment performed in YPGal, two plates were accidently flipped during manipulations required for the experiment and data associated with these plates were excluded from our analysis.
Quantification of cis-Regulatory Activity.
Activity of the PTDH3-YFP reporter gene was measured by flow cytometry using similar experimental procedures as in the study by Metzger et al. (42) and a similar analysis pipeline as in the study by Duveau et al. (50). After dilution in SC-arginine, cells were directly sampled from 96-well plates using an IntelliCyt HyperCyt Autosampler and passed through a Becton Dickinson Accuri C6 flow cytometer using a flow rate of 14 μL⋅min−1 and core size of 10 μm. A blue laser (λ = 488 nm) was used for excitation of YFP, and fluorescence was acquired from the FL1 channel using a 533/30-nm optical filter. Liquid cultures of each strain were sampled for 2–3 s each, with ∼20,000 events recorded on average. Data were then processed with custom scripts using flowClust (3.0.0) and flowCore (1.26.3) packages in R (3.0.2) to remove artifacts such as debris and other noncell events (79, 80) as well as cell doublets. Samples with fewer than 800 events following processing were excluded from further analysis. Next, we defined a measure of fluorescence level that was independent of cell size, which was complicated by the fact that the positive relationship between fluorescence intensity (FL1.A) and cell size (forward scatter; FSC.A) was not the same for samples collected in glucose, glycerol, and galactose. To correct for cell size homogeneously in the three environments, we transformed the log10(FSC.A) and log10(FL1.A) data for each sample using a rotation around the centroid and with an angle determined iteratively so that the intercept of the linear regression of the transformed values of log10(FSC.A) and log10(FL1.A) would be close to zero (below 0.01). The fluorescence level for each event was then calculated as the ratio of the transformed value of log10(FL1.A) over the transformed value of log10(FSC.A), and the fluorescence level for each sample was calculated as the median fluorescence across all events (or cells) in the sample.
For the experiment designed to determine the plasticity of activity of the wild-type promoter, we subtracted the average autofluorescence measured for the six replicates of nonfluorescent strain BY4724 in each environment from the fluorescence levels measured for strain YPW1 (wild-type PTDH3-YFP) in the corresponding environments. After autofluorescence correction, we divided the median fluorescence measured for each YPW1 replicate in each environment by the average fluorescence measured for the six replicates of YPW1 in YPD (glucose). We finally calculated the mean relative fluorescence across the six replicates of YPW1 in each environment to determine the plasticity of activity of the wild-type PTDH3. The custom script describing the plasticity analysis is provided as Dataset S4.
For the other experiments that included a larger number of plates, control samples were used to test and correct for technical variation creating differences among days and plates, as well as rows and columns within a plate. To do this, we fit YFP fluorescence from the control samples [median log10(FL1.A)/log10(FSC.A) across all cells] to a linear model (fluorescence ∼ day + run + replicate + plate + row + column + block + stack + depth + order) using the lm function in R. Effects in this model correspond to the day on which a plate was run; the replicate number for that plate on that day; the run that uniquely identified each physical plate run on the flow cytometer; the plate number corresponding to one of the eight arrays, row, column, and block in which a sample was run (due to software limitations, data were acquired from each plate in multiple blocks); the stack in which a plate was cultured; the depth of the plate in this stack; and the order in which a plate was run within the replicate. The function step in R was used to choose the model with the best explanatory power based on the Akaike information criterion (AIC) and to drop the factors that did not significantly impact fluorescence. The factors “run” and “block” were the main factors affecting fluorescence; therefore, we used the linear model “fluorescence ∼ day + run” to estimate the effects of these two factors and to correct the fluorescence levels of all samples accordingly. We then subtracted the average autofluorescence measured across the six replicates of nonfluorescent strain BY4724 from the fluorescence of all samples. Finally, the fluorescence of each sample was divided by the mean fluorescence measured across replicates of the reference strain YPW1 in the same environment. The mean relative fluorescence measured across the six replicates of each strain was used as a measure of the effects of the 235 mutations on PTDH3 activity in glucose, galactose, and glycerol. Importantly, our measure of mutational effects was independent of the plasticity of expression observed between environments, allowing us to test for GxE interactions simply by comparing the effects of a mutation between two environments. The custom script used to perform these analyses is provided as Dataset S5.
Effects of Individual Polymorphisms.
The effects of polymorphisms were measured in each environment as described by Metzger et al. (42) and Duveau et al. (50). Using parsimony, a haplotype network (“Haplotype.Network.txt” worksheet in Dataset S3) was generated for the 39 PTDH3 haplotypes described above that each differed from another haplotype by exactly one polymorphism. The most likely ancestral state of PTDH3 was inferred using PTDH3 sequences of other species in the Saccharomyces senso strictus genus and more distantly related strains of S. cerevisiae, and then used to polarize the network (42). Parsimony and maximum likelihood methods were used to construct this haplotype network. Conservation across the 678-bp PTDH3 shown in Fig. 5 and Fig. S1 was determined by comparing sequences of the seven species of the S. stricto sensus genus in 20-bp sliding windows using ConSurf (42, 81, 82). The activity of these promoter haplotypes was measured in parallel with the activity of the 235 mutant promoters after growth in YPD, YPG, and YPGal as described above. After correcting for autofluorescence, we divided the fluorescence measured for each replicate of each haplotype by the average fluorescence across the six replicates of the parental haplotype that differed by a single polymorphism in the haplotype network. The mean relative fluorescence across the six replicates of each haplotype represented the effects of individual polymorphisms on promoter activity. In seven instances, the effect of the same polymorphism was tested in two different pairs of haplotypes. In such cases, after verifying the absence of epistatic interaction (50), we averaged the effects of the polymorphism measured with the two pairs of haplotypes. Overall, this approach allowed us to quantify the effects of 30 unique polymorphisms in PTDH3. Ten of these polymorphisms were G:C-to-A:T transitions (the same type of change tested in the collection of 235 mutations), 13 were other types of SNPs, and 7 were small indels ranging in size from 1 to 13 bp. We observed no significant difference of effects among these three classes of polymorphism in any environment (Fig. S1 B–D).
Test for Selection.
We tested for evidence of selection acting on the average activity of PTDH3 in each environment or on the plasticity of activity between environments using the method developed by Metzger et al. (42). First, the distributions of effects of mutations (or the distributions of differences of effects of mutations between environments) were converted into probability density functions. Then, these functions were used to compute the log-likelihood of 200,000 sets of 30 mutational effects randomly drawn with replacement from the empirical distributions of 235 mutational effects. The log-likelihood value for the effects (differences in effects between environments) of the 30 polymorphisms was then calculated, and a two-sided P value was calculated as twice the proportion of random sets of mutational effects showing a more extreme log-likelihood value than the one observed for polymorphisms. P values below 0.05 indicated that the effects of the 30 polymorphisms were statistically different from the effects of a random set of 30 mutations, suggesting a role of natural selection in shaping the phenotypic effects of polymorphisms. In addition to this test for selection, we compared the median effects of polymorphisms with the median effects of random sets of 30 mutations using permutation tests. For these tests, we randomly picked a set of 30 mutational effects (or 30 differences of mutational effects between environments) and calculated the difference A between the median effect for this random set and the median effect measured across the 30 polymorphisms. The effects of the 30 mutations and 30 polymorphisms were then shuffled, and a randomized difference B of median effects between the two permuted sets was calculated. Lastly, we calculated the difference D equal to A − B. After 200,000 repetitions of this procedure, we calculated the frequency of D values that were above zero and the frequency of D values that were below zero. The two-sided P value of the test was calculated as twice the minimal frequency.
Statistical Analyses.
All statistical tests were performed using R 3.2.3. To compare the activity of wild-type PTDH3 in different environments, we performed t tests on the fluorescence levels measured for the six replicate populations of YPW1 in each pair of environments. To compare the distribution of mutational effects for different pairs of environments, we performed KS tests using the R function ks.test followed by 200,000 permutations to determine the distribution of D-statistics (supremum difference between the two empirical distribution functions) expected under the null hypothesis of no plasticity. For each permutation, we shuffled the two environment labels in which the effects of each of the 235 mutations were measured (the number of possible permutations was 2235), and the D-statistic was calculated for the two randomized distributions. The P value of the KS test was determined as the proportion of the 100,000 permuted distributions that showed a greater D-statistic than the one obtained from the observed distributions of mutational effects.
We used nonparametric permutation tests to compare the median effects of mutations for different pairs of environments. For each permutation, we generated two random sets of 235 mutational effects by shuffling the effects measured in the two environments of interest for each mutation (the number of possible permutations was 2235). We then calculated the difference of median effects between the two random sets and repeated the procedure 200,000 times to generate a null distribution of differences of medians. The two-sided P value of the test was calculated as twice the proportion of random sets that showed a more extreme difference of median effects than the observed difference between the two environments. We used similar permutation tests to compare the MAD (a measure of dispersion) of mutation effects between environments. To do so, for each pair of environments, we first centered the effects of the 235 mutations on the same median. This transformation did not affect the variability of mutational effects (MAD) and ensured that the permutation test was unaffected by differences in median effects of mutations between environments. We then used the same permutation procedure as described above to generate 200,000 differences of MAD between randomized sets of mutation effects and calculated P values as twice the proportion of random MAD differences that were more extreme than the observed difference of MAD across all 235 mutations between the two environments.
We tested for significant GxE interactions using t tests (t.test function in R) to compare the effect of each mutation in each pair of environments. A Benjamini–Hochberg FDR correction was then applied to control for multiple testing, and mutations with adjusted P values below 0.01 were considered to show statistically significant GxE effects (i.e., these mutations showed different effects in the two environments tested). We compared the magnitude of GxE effects across the 235 mutations between pairs of environments using Mann–Whitney–Wilcoxon tests (wilcox.test function in R).
A nearest-neighbor approach was used to test for nonrandom positioning of GxE effects in PTDH3. First, we determined the positions in the promoter of mutations with significant differences of effects between two environments. For each of these mutations, we calculated the distance to the closest mutation with significant GxE effects in the promoter (nearest neighbor). Then, we computed the SD of the minimal distance across all mutations with significant GxE effects between the two environments. Lastly, similar SDs of minimal distance were calculated for 100,000 random sets of mutations (picked among the 235 mutations created in the promoter). The number of random mutations drawn for each set was equal to the number of mutations with significant GxE effects for which nonrandom positioning was being tested. A two-sided P value of the test was calculated as twice the proportion of randomized SDs of distance to nearest neighbor that were more extreme than the observed SD. A P value below 0.05 can be obtained either if the mutations with GxE effects tend to cluster in the promoter or if they tend to be more homogeneously spaced than expected by chance.
We used a resampling method to test for enrichment or depletion of GxE effects in functional elements of PTDH3. A total of 100,000 sets of mutations of the same size as the number of significant GxE effects were randomly drawn without replacement from the mutations we created in the promoter, excluding mutations in functional elements that were not being tested. We then calculated the proportion (P) of random sets of mutations that contained more mutations in the tested functional element than the observed number of GxE effects falling in this functional element. The P value of the test was calculated as twice the lowest value between P and 1 − P.
Access to Data and Analysis Scripts.
Flow cytometry data used in this work are available through the Flow Repository (https://flowrepository.org). Repository ID FR-FCM-ZZBN contains data from the study by Metzger et al. (42) used to quantify the effects of the mutations and polymorphisms in PTDH3 on expression after growth in YPD (glucose); repository ID FR-FCM-ZY8D contains data collected to analyze expression of these genotypes in YPGal (galactose); repository ID FR-FCM-ZY8B contains data collected to analyze expression of these genotypes in YPG (glycerol); and repository ID FR-FCM-ZY9T contains data used to quantify the plasticity of activity of the wild-type PTDH3 in glucose, galactose, and glycerol. The R scripts used to perform the analyses described above are provided as Datasets S4 and S5. Data used by these scripts are provided in Dataset S3, and data produced by these scripts are included in Dataset S1.
Supplementary Material
Acknowledgments
We thank Jonathan Gruber for technical assistance, as well as Petra Vande Zande, Jennifer Lachowiec, Abigail Lamb, and Rebecca Tarnopol for comments on the manuscript. Funding for this work was provided by March of Dimes Grant 5-FY07-181 (to P.J.W.), the Alfred P. Sloan Research Foundation, the National Science Foundation (Grant MCB-1021398), the NIH (Grants 1R01GM108826 and 1R35GM118073), and the University of Michigan. Additional support was provided by NIH Training Grant T32 GM007544 (to D.C.Y.); the University of Michigan Rackham Graduate School, Ecology and Evolutionary Biology Department, and NIH Genome Sciences Training Grant T32 HG000040 (to B.P.H.M.); European Molecular Biology Organization Postdoctoral Fellowship ALTF 1114-2012 (to F.D.); and NIH National Research Service Award 1F32GM115198 (to A.H.-D.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Flow cytometry data used in this work are available through the Flow Repository (https://flowrepository.org). Repository ID FR-FCM-ZZBN contains data from a study by Metzger et al. (42) used to quantify the effects of the mutations and polymorphisms in PTDH3 on expression after growth in YPD (glucose); repository ID FR-FCM-ZY8D contains data collected to analyze expression of these genotypes in YPGal (galactose); repository ID FR-FCM-ZY8B contains data collected to analyze expression of these genotypes in YPG (glycerol); and repository ID FR-FCM-ZY9T contains data used to quantify the plasticity of activity of the wild-type PTDH3 promoter in glucose, galactose, and glycerol.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1713960115/-/DCSupplemental.
References
- 1.Schlichting C, Pigliucci M. Phenotypic Evolution: A Reaction Norm Perspective. Sinauer; Sunderland, MA: 1998. [Google Scholar]
- 2.Whitman DW, Agrawal AA. Phenotypic Plasticity of Insects: Mechanisms and Consequences. CRC Press; Boca Raton, FL: 2009. What is phenotypic plasticity and why is it important? pp. 1–63. [Google Scholar]
- 3.West-Eberhard MJ. Developmental Plasticity and Evolution. Oxford Univ Press; Oxford: 2003. [Google Scholar]
- 4.Ghalambor CK, et al. Non-adaptive plasticity potentiates rapid adaptive evolution of gene expression in nature. Nature. 2015;525:372–375. doi: 10.1038/nature15256. [DOI] [PubMed] [Google Scholar]
- 5.Bradshaw AD. Evolutionary significance of phenotypic plasticity in plants. Adv Genet. 1965;13:115–155. [Google Scholar]
- 6.Hughes KA, Burleson MH, Rodd FH. Is phenotypic plasticity adaptive? In: Rogers JL, Kohler HP, editors. The Biodemography of Human Reproduction and Fertility. Springer; Boston: 2003. pp. 23–42. [Google Scholar]
- 7.Via S, Lande R. Genotype-environment interaction and the evolution of phenotypic plasticity. Evolution. 1985;39:505–522. doi: 10.1111/j.1558-5646.1985.tb00391.x. [DOI] [PubMed] [Google Scholar]
- 8.Falconer DS. The problem of environment and selection. Am Nat. 1952;86:293–298. [Google Scholar]
- 9.Remold SK, Lenski RE. Contribution of individual random mutations to genotype-by-environment interactions in Escherichia coli. Proc Natl Acad Sci USA. 2001;98:11388–11393. doi: 10.1073/pnas.201140198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Latta LC, 4th, et al. The phenotypic effects of spontaneous mutations in different environments. Am Nat. 2015;185:243–252. doi: 10.1086/679501. [DOI] [PubMed] [Google Scholar]
- 11.Martin G, Lenormand T. A general multivariate extension of Fisher’s geometrical model and the distribution of mutation fitness effects across species. Evolution. 2006;60:893–907. [PubMed] [Google Scholar]
- 12.Martin G, Lenormand T. The fitness effect of mutations across environments: Fisher’s geometrical model with multiple optima. Evolution. 2015;69:1433–1447. doi: 10.1111/evo.12671. [DOI] [PubMed] [Google Scholar]
- 13.López-Maury L, Marguerat S, Bähler J. Tuning gene expression to changing environments: From rapid responses to evolutionary adaptation. Nat Rev Genet. 2008;9:583–593. doi: 10.1038/nrg2398. [DOI] [PubMed] [Google Scholar]
- 14.Li Y, et al. Mapping determinants of gene expression plasticity by genetical genomics in C. elegans. PLoS Genet. 2006;2:e222. doi: 10.1371/journal.pgen.0020222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gibson G. The environmental contribution to gene expression profiles. Nat Rev Genet. 2008;9:575–581. doi: 10.1038/nrg2383. [DOI] [PubMed] [Google Scholar]
- 16.Zhou S, Campbell TG, Stone EA, Mackay TF, Anholt RR. Phenotypic plasticity of the Drosophila transcriptome. PLoS Genet. 2012;8:e1002593. doi: 10.1371/journal.pgen.1002593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dayan DI, Crawford DL, Oleksiak MF. Phenotypic plasticity in gene expression contributes to divergence of locally adapted populations of Fundulus heteroclitus. Mol Ecol. 2015;24:3345–3359. doi: 10.1111/mec.13188. [DOI] [PubMed] [Google Scholar]
- 18.Marais Des DL, Hernandez KM, Juenger TE. Genotype-by-environment interaction and plasticity: Exploring genomic responses of plants to the abiotic environment. Annu Rev Ecol Evol Syst. 2013;44:5–29. [Google Scholar]
- 19.Landry CR, Oh J, Hartl DL, Cavalieri D. Genome-wide scan reveals that genetic variation for transcriptional plasticity in yeast is biased towards multi-copy and dispensable genes. Gene. 2006;366:343–351. doi: 10.1016/j.gene.2005.10.042. [DOI] [PubMed] [Google Scholar]
- 20.Smith EN, Kruglyak L. Gene-environment interaction in yeast gene expression. PLoS Biol. 2008;6:e83. doi: 10.1371/journal.pbio.0060083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grishkevich V, Yanai I. The genomic determinants of genotype × environment interactions in gene expression. Trends Genet. 2013;29:479–487. doi: 10.1016/j.tig.2013.05.006. [DOI] [PubMed] [Google Scholar]
- 22.Hodgins-Davis A, Townsend JP. Evolving gene expression: From G to E to GxE. Trends Ecol Evol. 2009;24:649–658. doi: 10.1016/j.tree.2009.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pigliucci M. Genotype-phenotype mapping and the end of the ‘genes as blueprint’ metaphor. Philos Trans R Soc Lond B Biol Sci. 2010;365:557–566. doi: 10.1098/rstb.2009.0241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Halligan DL, Keightley PD. Spontaneous mutation accumulation studies in evolutionary genetics. Annu Rev Ecol Evol Syst. 2009;40:151–172. [Google Scholar]
- 25.Rifkin SA, Houle D, Kim J, White KP. A mutation accumulation assay reveals a broad capacity for rapid evolution of gene expression. Nature. 2005;438:220–223. doi: 10.1038/nature04114. [DOI] [PubMed] [Google Scholar]
- 26.Denver DR, et al. The transcriptional consequences of mutation and natural selection in Caenorhabditis elegans. Nat Genet. 2005;37:544–548. doi: 10.1038/ng1554. [DOI] [PubMed] [Google Scholar]
- 27.Landry CR, Lemos B, Rifkin SA, Dickinson WJ, Hartl DL. Genetic properties influencing the evolvability of gene expression. Science. 2007;317:118–121. doi: 10.1126/science.1140247. [DOI] [PubMed] [Google Scholar]
- 28.Huang W, et al. Spontaneous mutations and the origin and maintenance of quantitative genetic variation. eLife. 2016;5:e14625. doi: 10.7554/eLife.14625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kishony R, Leibler S. Environmental stresses can alleviate the average deleterious effect of mutations. J Biol. 2003;2:14. doi: 10.1186/1475-4924-2-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rutter MT, et al. Fitness of Arabidopsis thaliana mutation accumulation lines whose spontaneous mutations are known. Evolution. 2012;66:2335–2339. doi: 10.1111/j.1558-5646.2012.01583.x. [DOI] [PubMed] [Google Scholar]
- 31.Roles AJ, Rutter MT, Dworkin I, Fenster CB, Conner JK. Field measurements of genotype by environment interaction for fitness caused by spontaneous mutations in Arabidopsis thaliana. Evolution. 2016;70:1039–1050. doi: 10.1111/evo.12913. [DOI] [PubMed] [Google Scholar]
- 32.Fry JD, Heinsohn SL. Environment dependence of mutational parameters for viability in Drosophila melanogaster. Genetics. 2002;161:1155–1167. doi: 10.1093/genetics/161.3.1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chang S-M, Shaw RG. The contribution of spontaneous mutation to variation in environmental response in Arabidopsis thaliana: Responses to nutrients. Evolution. 2003;57:984–994. doi: 10.1111/j.0014-3820.2003.tb00310.x. [DOI] [PubMed] [Google Scholar]
- 34.Andrew JR, et al. Abiotic stress does not magnify the deleterious effects of spontaneous mutations. Heredity (Edinb) 2015;115:503–508. doi: 10.1038/hdy.2015.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhu YO, Siegal ML, Hall DW, Petrov DA. Precise estimates of mutation rate and spectrum in yeast. Proc Natl Acad Sci USA. 2014;111:E2310–E2318. doi: 10.1073/pnas.1323011111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Denver DR, et al. Variation in base-substitution mutation in experimental and natural lineages of Caenorhabditis nematodes. Genome Biol Evol. 2012;4:513–522. doi: 10.1093/gbe/evs028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Patwardhan RP, et al. High-resolution analysis of DNA regulatory elements by synthetic saturation mutagenesis. Nat Biotechnol. 2009;27:1173–1175. doi: 10.1038/nbt.1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Patwardhan RP, et al. Massively parallel functional dissection of mammalian enhancers in vivo. Nat Biotechnol. 2012;30:265–270. doi: 10.1038/nbt.2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Melnikov A, et al. Systematic dissection and optimization of inducible enhancers in human cells using a massively parallel reporter assay. Nat Biotechnol. 2012;30:271–277. doi: 10.1038/nbt.2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hornung G, et al. Noise-mean relationship in mutated promoters. Genome Res. 2012;22:2409–2417. doi: 10.1101/gr.139378.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kwasnieski JC, Mogno I, Myers CA, Corbo JC, Cohen BA. Complex effects of nucleotide variants in a mammalian cis-regulatory element. Proc Natl Acad Sci USA. 2012;109:19498–19503. doi: 10.1073/pnas.1210678109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Metzger BPH, Yuan DC, Gruber JD, Duveau F, Wittkopp PJ. Selection on noise constrains variation in a eukaryotic promoter. Nature. 2015;521:344–347. doi: 10.1038/nature14244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smith JD, McManus KF, Fraser HB. A novel test for selection on cis-regulatory elements reveals positive and negative selection acting on mammalian transcriptional enhancers. Mol Biol Evol. 2013;30:2509–2518. doi: 10.1093/molbev/mst134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hodgins-Davis A, Rice DP, Townsend JP. Gene expression evolves under a house-of-cards model of stabilizing selection. Mol Biol Evol. 2015;32:2130–2140. doi: 10.1093/molbev/msv094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lagunas R. Sugar transport in Saccharomyces cerevisiae. FEMS Microbiol Rev. 1993;10:229–242. doi: 10.1016/0378-1097(93)90598-v. [DOI] [PubMed] [Google Scholar]
- 46.Sellick CA, Campbell RN, Reece RJ. Galactose metabolism in yeast-structure and regulation of the leloir pathway enzymes and the genes encoding them. Int Rev Cell Mol Biol. 2008;269:111–150. doi: 10.1016/S1937-6448(08)01003-4. [DOI] [PubMed] [Google Scholar]
- 47.Klein M, Swinnen S, Thevelein JM, Nevoigt E. Glycerol metabolism and transport in yeast and fungi: Established knowledge and ambiguities. Environ Microbiol. 2017;19:878–893. doi: 10.1111/1462-2920.13617. [DOI] [PubMed] [Google Scholar]
- 48.Gancedo JM. Yeast carbon catabolite repression. Microbiol Mol Biol Rev. 1998;62:334–361. doi: 10.1128/mmbr.62.2.334-361.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gruber JD, Vogel K, Kalay G, Wittkopp PJ. Contrasting properties of gene-specific regulatory, coding, and copy number mutations in Saccharomyces cerevisiae: Frequency, effects, and dominance. PLoS Genet. 2012;8:e1002497. doi: 10.1371/journal.pgen.1002497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Duveau F, Toubiana W, Wittkopp PJ. Fitness effects of cis-regulatory variants in the Saccharomyces cerevisiae TDH3 promoter. Mol Biol Evol. 2017;34:2908–2912. doi: 10.1093/molbev/msx224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kuroda S, Otaka S, Fujisawa Y. Fermentable and nonfermentable carbon sources sustain constitutive levels of expression of yeast triosephosphate dehydrogenase 3 gene from distinct promoter elements. J Biol Chem. 1994;269:6153–6162. [PubMed] [Google Scholar]
- 52.Yagi S, Yagi K, Fukuoka J, Suzuki M. The UAS of the yeast GAPDH promoter consists of multiple general functional elements including RAP1 and GRF2 binding sites. J Vet Med Sci. 1994;56:235–244. doi: 10.1292/jvms.56.235. [DOI] [PubMed] [Google Scholar]
- 53.Huie MA, et al. Characterization of the DNA-binding activity of GCR1: In vivo evidence for two GCR1-binding sites in the upstream activating sequence of TPI of Saccharomyces cerevisiae. Mol Cell Biol. 1992;12:2690–2700. doi: 10.1128/mcb.12.6.2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chambers A, Packham EA, Graham IR. Control of glycolytic gene expression in the budding yeast (Saccharomyces cerevisiae) Curr Genet. 1995;29:1–9. doi: 10.1007/BF00313187. [DOI] [PubMed] [Google Scholar]
- 55.Pavlović B, Hörz W. The chromatin structure at the promoter of a glyceraldehyde phosphate dehydrogenase gene from Saccharomyces cerevisiae reflects its functional state. Mol Cell Biol. 1988;8:5513–5520. doi: 10.1128/mcb.8.12.5513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hornung G, Oren M, Barkai N. Nucleosome organization affects the sensitivity of gene expression to promoter mutations. Mol Cell. 2012;46:362–368. doi: 10.1016/j.molcel.2012.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cavalli G, Thoma F. Chromatin transitions during activation and repression of galactose-regulated genes in yeast. EMBO J. 1993;12:4603–4613. doi: 10.1002/j.1460-2075.1993.tb06149.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lee C-K, Shibata Y, Rao B, Strahl BD, Lieb JD. Evidence for nucleosome depletion at active regulatory regions genome-wide. Nat Genet. 2004;36:900–905. doi: 10.1038/ng1400. [DOI] [PubMed] [Google Scholar]
- 59.Weinhandl K, Winkler M, Glieder A, Camattari A. Carbon source dependent promoters in yeasts. Microb Cell Fact. 2014;13:5. doi: 10.1186/1475-2859-13-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wagner GP, Booth G, Bagheri-Chaichian H. A population genetic theory of canalization. Evolution. 1997;51:329–347. doi: 10.1111/j.1558-5646.1997.tb02420.x. [DOI] [PubMed] [Google Scholar]
- 61.Wilke CO, Wang JL, Ofria C, Lenski RE, Adami C. Evolution of digital organisms at high mutation rates leads to survival of the flattest. Nature. 2001;412:331–333. doi: 10.1038/35085569. [DOI] [PubMed] [Google Scholar]
- 62.Montville R, Froissart R, Remold SK, Tenaillon O, Turner PE. Evolution of mutational robustness in an RNA virus. PLoS Biol. 2005;3:e381. doi: 10.1371/journal.pbio.0030381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sanjuán R, Cuevas JM, Furió V, Holmes EC, Moya A. Selection for robustness in mutagenized RNA viruses. PLoS Genet. 2007;3:e93. doi: 10.1371/journal.pgen.0030093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Proulx SR, Nuzhdin S, Promislow DEL. Direct selection on genetic robustness revealed in the yeast transcriptome. PLoS One. 2007;2:e911. doi: 10.1371/journal.pone.0000911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kawecki TJ. The evolution of genetic canalization under fluctuating selection. Evolution. 2000;54:1–12. doi: 10.1111/j.0014-3820.2000.tb00001.x. [DOI] [PubMed] [Google Scholar]
- 66.Siegal ML, Leu J-Y. On the nature and evolutionary impact of phenotypic robustness mechanisms. Annu Rev Ecol Evol Syst. 2014;45:496–517. doi: 10.1146/annurev-ecolsys-120213-091705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.de Visser JAGM, et al. Perspective: Evolution and detection of genetic robustness. Evolution. 2003;57:1959–1972. doi: 10.1111/j.0014-3820.2003.tb00377.x. [DOI] [PubMed] [Google Scholar]
- 68.van Nimwegen E, Crutchfield JP, Huynen M. Neutral evolution of mutational robustness. Proc Natl Acad Sci USA. 1999;96:9716–9720. doi: 10.1073/pnas.96.17.9716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lynch M. The evolution of genetic networks by non-adaptive processes. Nat Rev Genet. 2007;8:803–813. doi: 10.1038/nrg2192. [DOI] [PubMed] [Google Scholar]
- 70.Martin G, Lenormand T. The fitness effect of mutations across environments: A survey in light of fitness landscape models. Evolution. 2006;60:2413–2427. [PubMed] [Google Scholar]
- 71.Stoltzfus A, Yampolsky LY. Climbing mount probable: Mutation as a cause of nonrandomness in evolution. J Hered. 2009;100:637–647. doi: 10.1093/jhered/esp048. [DOI] [PubMed] [Google Scholar]
- 72.Wagner A. The role of robustness in phenotypic adaptation and innovation. Proc Biol Sci. 2012;279:1249–1258. doi: 10.1098/rspb.2011.2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Masel J, King OD, Maughan H. The loss of adaptive plasticity during long periods of environmental stasis. Am Nat. 2007;169:38–46. doi: 10.1086/510212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Murren CJ, et al. Constraints on the evolution of phenotypic plasticity: Limits and costs of phenotype and plasticity. Heredity (Edinb) 2015;115:293–301. doi: 10.1038/hdy.2015.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Harbison CT, et al. Transcriptional regulatory code of a eukaryotic genome. Nature. 2004;431:99–104. doi: 10.1038/nature02800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sheff MA, Thorn KS. Optimized cassettes for fluorescent protein tagging in Saccharomyces cerevisiae. Yeast. 2004;21:661–670. doi: 10.1002/yea.1130. [DOI] [PubMed] [Google Scholar]
- 77.Brachmann CB, et al. Designer deletion strains derived from Saccharomyces cerevisiae S288C: A useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 1998;14:115–132. doi: 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 78.Maclean CJ, et al. Deciphering the genic basis of yeast fitness variation by simultaneous forward and reverse genetics. Mol Biol Evol. 2017;34:2486–2502. doi: 10.1093/molbev/msx151. [DOI] [PubMed] [Google Scholar]
- 79.Lo K, Hahne F, Brinkman RR, Gottardo R. flowClust: A bioconductor package for automated gating of flow cytometry data. BMC Bioinformatics. 2009;10:145. doi: 10.1186/1471-2105-10-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hahne F, et al. flowCore: A bioconductor package for high throughput flow cytometry. BMC Bioinformatics. 2009;10:106. doi: 10.1186/1471-2105-10-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hittinger CT. Saccharomyces diversity and evolution: A budding model genus. Trends Genet. 2013;29:309–317. doi: 10.1016/j.tig.2013.01.002. [DOI] [PubMed] [Google Scholar]
- 82.Ashkenazy H, Erez E, Martz E, Pupko T, Ben-Tal N. ConSurf 2010: Calculating evolutionary conservation in sequence and structure of proteins and nucleic acids. Nucleic Acids Res. 2010;38:W529–W533. doi: 10.1093/nar/gkq399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.McAlister L, Holland MJ. Differential expression of the three yeast glyceraldehyde-3-phosphate dehydrogenase genes. J Biol Chem. 1985;260:15019–15027. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.