Abstract
Supramolecular peptide nanofibers are attractive for applications in vaccine development due to their ability to induce strong immune responses without added adjuvants or associated inflammation. Here, we report that self-assembling peptide nanofibers bearing CD4+ or CD8+ T cell epitopes are processed through mechanisms of autophagy in antigen-presenting cells (APCs). Using standard in vitro antigen presentation assays, we confirmed loss and gain of the adjuvant function using pharmacological modulators of autophagy and APCs deficient in multiple autophagy proteins. The incorporation of microtubule-associated protein 1A/1B-light chain-3 (LC3-II) into the autophagosomal membrane, a key biological marker for autophagy, was confirmed using microscopy. Our findings indicate that autophagy in APCs plays an essential role in the mechanism of adjuvant action of supramolecular peptide nanofibers.
Introduction
Biomaterials constructed from de novo designed peptides that assemble into supramolecular nanofibers and hydrogels have enormous potential for applications in biology and medicine.1,2 Self-assembling peptide biomaterials have been used as multivalent scaffolds for applications in tissue engineering, regenerative medicine, and drug delivery due to their biocompatibility, ease of synthesis, and the rich chemistry with which the primary sequence can be manipulated to impart structure or function.3−5 In recent years, β-sheet rich peptide nanofibers have attracted considerable attention as immune adjuvants for applications in vaccine development and immunotherapy due to their ability to induce strong antibody and cellular responses to conjugated antigens.6,7 Unlike traditional depot-forming adjuvants (alum or Freund’s), peptide nanofibers are noninflammatory and vaccination does not lead to inflammation at the injection site (recruitment of neutrophils, eosinophils, monocytes, etc.).8 Several studies have confirmed the efficacy of peptide nanofiber-based vaccines in animal models of infectious disease, cancer, and drug addiction; however, the immunological mechanisms that drive the adjuvant potential have not yet been elucidated.9−14 In this study, we investigated intracellular mechanisms in macrophages (MΦs) and dendritic cells (DCs) that are involved in processing of peptide nanofibers and presentation of antigens to CD4+ and CD8+ T cells.
Previous studies have shown that changes in physicochemical properties of peptide nanofibers could lead to loss or gain of adjuvant function. Targeted amino acid substitutions (F → P) in the core of the self-assembling peptide Q11 (QQKFQFQFEQQ) led to loss of self-assembly and Ab responses, confirming that supramolecular assembly is required for adjuvant activity.12 Recently, Wen et al. designed Q11-based peptides with a broad range of physicochemical properties and found that despite presenting a competent epitope, negatively charged nanofibers were not taken up by antigen-presenting cells (APCs) such as dendritic cells (DCs) and macrophages and lacked adjuvant activity.15 In comparison, positively charged nanofibers significantly enhanced uptake by APCs and immune responses.15 Our lab reported that changing the chirality of the self-assembling domain KFE8 (FKFEFKFE) from the natural l-form to protease-resistant d-form did not impact self-assembly but significantly enhanced in vivo persistence and the Ab response, suggesting a role for intracellular proteases in the processing of nanofibers.16 Taken together, these studies suggest that innate cellular mechanisms in APCs involved in the clearance and degradation of cytoplasmic contents, and immune recognition and antigen presentation, possibly play a role in the mechanism of action of peptide nanofibers.
An evolutionarily conserved mechanism that cells use to degrade cytoplasmic proteins and organelles and maintain cellular homeostasis is “autophagy”.17 Autophagy is crucial for the clearance of fibrillar protein aggregates implicated in neurodegenerative diseases such as Alzheimer’s and Huntington’s18,19 and a component of innate immunity that is involved in host defense elimination of pathogens.20 Autophagy has been identified as a route by which cytoplasmic and nuclear antigens are delivered to major histocompatibility complex (MHC) class II molecules for presentation to CD4+ T cells and MHC class I cross-presentation of tumor antigens to CD8+ T cells.21 Macroautophagy, a general form of autophagy, targets the clearance of damaged organelles, cytosolic proteins, and invasive microbes by sequestering them within cytosolic double-membrane vesicles termed autophagosomes.17 Autophagosomes then deliver the cytoplasmic components to lysosomes, which degrade the contents through proteolysis and antigens loaded onto MHC class II molecules for presentation.21 This presumably explains why the antibody response to peptide nanofibers is completely abolished in the absence of CD4+ T cells (which help B cells that have encountered the same antigen to make antibodies).12 In this study, we investigated and confirmed a role for autophagy in the mechanism of adjuvant action of peptide nanofibers.
Results and Discussion
To assess MHC class II antigen presentation, we used a peptide fragment from Ag85B protein of Mycobacterium tuberculosis (Mtb), a well-known immunodominant protein, which induces cell proliferation and production of IL-2 and interferon-γ in lymphocytes from Mtb-infected C57BL6 mice. Kariyone et al. used a series of 15-amino-acid peptides that overlapped each other by five amino acids and identified an immunodominant MHC class II antigenic epitope FQDAY-NAAGGHNAVF (aa 240−254) that requires processing by antigen-presenting cells (referred to as Ag85B peptide here).23 Further transgenic CD4+ T cells that recognize Ag85B peptide are available (BB7 hybridoma) for use in in vitro antigen-presentation assays. Similarly, MHC class I epitope OVA (SIINFEKL) and its cognate OT-I cells (CD8+ T cells with receptors specific for OVA) were used to gain potential insights into differences in antigen presentation between soluble and fibrillized antigens.
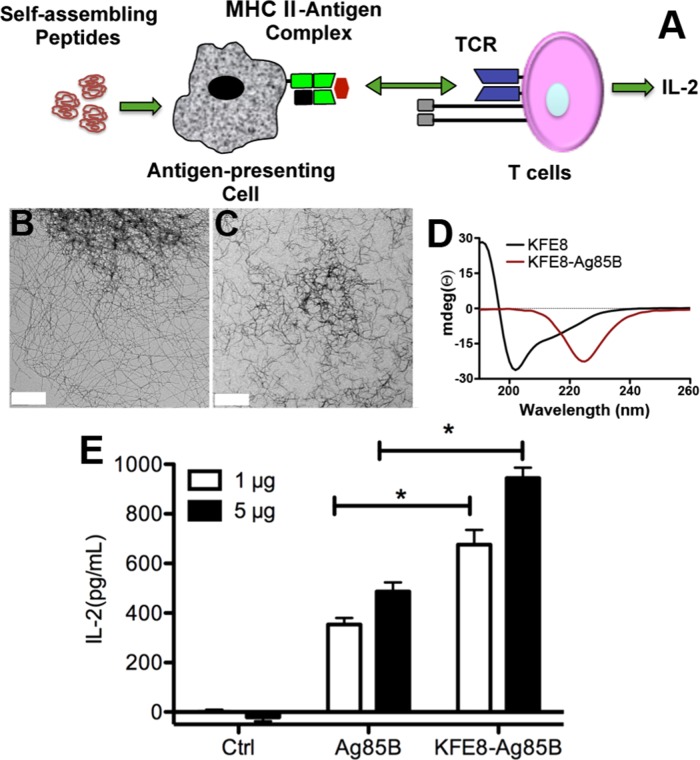
The amphipathic peptide KFE8 (FKFEFKFE) was used as the self-assembling domain because it is the most studied and well characterized with abundant data pertaining to sequence length and pattern variation of amino acids on self-assembly.22 Also, we have previously shown that KFE8 is a powerful immune adjuvant in mice, raising robust antibody and cellular immune responses.9,10,12,16 The fusion peptide KFE8-Ag85B was synthesized by linking the epitopes using a proteolytic amino acid spacer (GGAAY). Transmission electron microscopy (TEM) data indicated that KFE8-Ag85B assembled into robust fibrillar architectures with shorter morphology compared to that of KFE8 (Figure 1B,C). Interestingly, a structural shift was observed from cross-β secondary structure with signals at 218 (β-sheet) and 205 nm (from π−π effects) for KFE8 to a pure β-sheet structure (220−230 nm) for KFE8-Ag85B (Figure 1D) and KFE8-OVA (Figure S6). This is not surprising because conjugation of an epitope to the N- or C-terminus of a self-assembling domain could potentially impact its assembly and spatial organization, and such transitions have been reported before for other epitope-bearing peptides.16
Figure 1.
Conjugation of peptide antigens to self-assembling domains enhances antigen presentation by macrophages. Schematic depicting in vitro antigen-presentation assay (A). Negative stain transmission electron micrographs of KFE8 nanofibers (B) and KFE8-Ag85B nanofibers (C). Scale bar is 400 nm. Circular dichroism spectroscopy showing predominantly cross-β structure of KFE8 and a pure β sheet structure for KFE8-Ag85B (D). Interleukin (IL)-2 production by BB7 cells is significantly higher in MΦs treated with KFE8-Ag85B nanofibers compared to that in MΦs treated with soluble Ag85B at two different doses tested (E). *p < 0.05 using Student’s t-test (n = 4 per group).
In an in vitro antigen presentation assay, BB7 or OT-I cells release IL-2 in response to Ag85B–MHC II recognition or OVA-MHC I recognition on MΦs, respectively, which can be quantified using ELISA (Figure 1A).24 To investigate the effect of self-assembly on antigen processing and presentation of CD4+ T cell epitopes, bone marrow-derived MΦs from wild-type (WT) mice were first treated with BCG, which leads to the rapid expression of intracellular antigen processing and presentation machinery. The cells were washed and overlaid with medium containing soluble Ag85B or KFE8-Ag85B, and the amount of antigen (Ag85B) was kept constant in both groups. Following overlay with BB7 cells, IL-2 production was found to be significantly higher in MΦs treated with KFE8-Ag85B compared to that in MΦs treated with Ag85B at two different concentrations tested (Figure 1E). A similar effect was also observed in bone marrow-derived DCs (Figure S1). To confirm that BCG-derived Ag85B was not a confounding factor in the observed IL-2 response, we conducted the assay without first infecting MΦs and observed similar results (Figure S2). However, an overall decrease in the amount of IL-2 was observed compared to that in BCG-treated groups, presumably due to the lack of pattern recognition receptors that activate additional innate immune mechanisms. In MΦs treated with bare KFE8 nanofibers, IL-2 production was negligible, suggesting that the nanofibers did not induce any bystander activation and that IL-2 production by BB7 cells was antigen-specific (Figure S2). These data indicate that self-assembly into supramolecular nanofibers imparts physical properties to peptides that enhance antigen processing and presentation by MΦs and DCs.
To assess whether pathways of autophagy were involved in the processing and presentation of KFE8-Ag85B nanofibers, MΦs were pretreated with pharmacological modulators before incubation with nanofibers and loss or gain of IL-2 was measured. Data indicated significant loss of IL-2 in MΦs pretreated with the autophagy inhibitor 3-methyl adenine (3MA),25 which blocks the formation of autophagosomes by inhibiting phosphatidylinositol 3-kinases (PI3K) (Figure 2A). IL-2 levels from 3MA-treated MΦs were comparable to those of soluble Ag85B (Figure 2A), and addition of 3MA did not impact IL-2 production by MΦs treated with soluble Ag85B (Figure S3). Also, addition of bafilomycin to MΦs, an inhibitor of vacuolar H+ ATPase (V-ATPase)26 (prevents fusion between autophagosomes and lysosomes) also resulted in reduced IL-2 production (Figure S3). Interestingly, bafilomycin also caused loss of IL-2 production in MΦs treated with soluble Ag85B (Figure S3). On the contrary, higher levels of IL-2 were detected in MΦs treated with rapamycin, a direct inhibitor of mTOR, which is a major negative regulatory axis of autophagy (Figure 2A).27 The addition of rapamycin has been shown to enhance the presentation of soluble antigens and is approved by the FDA for clinical use. These findings demonstrate a role for macroautophagy in processing of peptide nanofibers in APCs and presentation to CD4+ T cells.
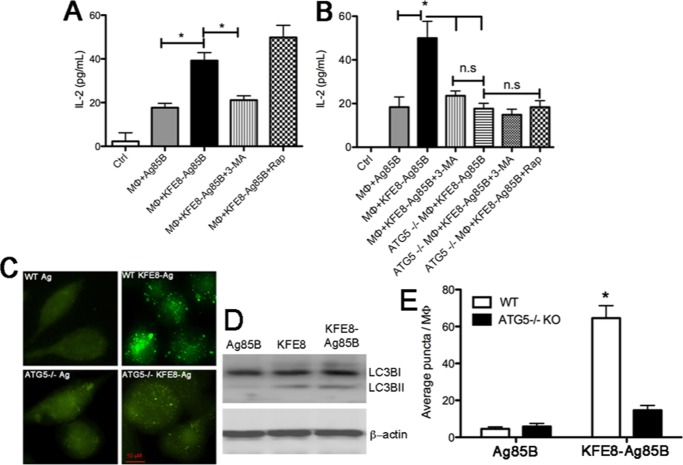
Figure 2.
Autophagy is essential for antigenic-processing and presentation of self-assembling peptide nanofibers. IL-2 production by MΦs treated with KFE8-Ag85B nanofibers was significantly reduced in the presence of the autophagy inhibitor 3MA, whereas treatment with rapamycin (inducer of autophagy) enhanced IL-2 production (A). Data demonstrating that IL-2 production is significantly impaired in WT MΦs treated with the autophagy inhibitor 3MA and Atg5–/– MΦs treated with KFE8-Ag85B (B). Addition of 3MA or rapamycin to Atg5–/– MΦs treated with KFE8-Ag85B did not impact IL-2 production (B). *p < 0.05 by analysis of variance (ANOVA) using Tukey’s post-hoc test (n = 3 per group, data representative of two experiments with similar results). Representative fluorescent microscopy images (C). Macrophage lysates tested for microtubule associated with light chain (LC3B) using western blot (day 1 post treatment) also demonstrated increased expression of the lapidated form of LC3 (LC3B-II) by bare as well as Ag85B conjugated nanofiber, hence further confirming the induction of autophagy by nanofibers (D). Quantification of autophagy in WT or Atg5–/– MΦs (E), indicating significantly more LC3B puncta in WT MΦs treated with KFE8-Ag85B compared to that in WT MΦs treated with soluble Ag85B. Data shown is the average of five fields from each group.
The execution of autophagy involves a set of evolutionarily conserved gene products, known as the autophagy-related (Atg) proteins.17 To further confirm our findings with pharmacological modulators of autophagy, the involvement of Atg proteins that play a role in the formation of the isolation membrane and the autophagosomes was investigated. One of the key autophagy proteins that controls autophagosome formation is Atg5.28 Using a fully reconstituted giant unilamellar vesicle system and recombinant proteins, it has been shown that the conserved Atg5–Atg12/Atg16 complex is essential for autophagosome formation and that membrane binding is mediated by Atg5.28 We cultured bone marrow-derived MΦs from Atg5–/– mice and conducted in vitro antigen presentation assays using soluble Ag85B or KFE8-Ag85B nanofibers, as described above. Data indicated significantly lower levels of IL-2 in Atg5–/– MΦs treated with KFE8-Ag85B nanofibers and was comparable to WT MΦs pretreated with 3MA or soluble Ag85B (Figure 2B). Addition of 3MA or rapamycin to Atg5–/– MΦs cultured with KFE8-Ag85B nanofibers did not result in loss or gain of function, as expected (Figure 2B). Further studies were conducted in Atg7-deficient DCs to exclude the possibility that impaired antigen presentation of peptide nanofibers was not limited to MΦs or Atg5. Data indicated significantly reduced IL-2 production in Atg7–/– DCs treated with Ag85B nanofibers; however, a similar loss of IL-2 was also observed in DCs treated with soluble Ag85B (Figure S4). Although the exact mechanism of Atg7–/– in processing and presentation of soluble or fibrillized antigens is beyond the scope of this study, the data clearly indicate that MΦs and DCs deficient in autophagy-related proteins exhibit impaired antigen processing and presentation of peptide nanofibers. To confirm whether autophagy is triggered only by peptide nanofibers that are phagocytosed/endocytosed from the environment or also by nanofibers that form within the cell, we used a second self-assembling domain NapFFKYp (Nap = napthylalanine, Yp = phosphotyrosine). Unlike KFE8, which assembles in the presence of salt-containing buffers, assembly of NapFFKYp (referred hereafter as NapFFK) is triggered by the cleavage of the phosphate group on the tryptophan side chain by intracellular phosphatases.29 Therefore, NapFFK nanofibers form inside the macrophages and allow investigation of autophagy in response to intracellular aggregates. Interestingly, IL-2 production in Atg5–/– MΦs was also impaired when treated with Ag85B-NapFFK peptide, which suggests that autophagy is engaged by peptide nanofibers acquired from the extracellular environment and also intracellular peptide aggregates (Figure S5).
The key biological marker to identify autophagy in mammalian systems is the microtubule-associated protein 1A/1B-light chain-3 (LC3).30 During autophagy, the C-terminal glycine of cytosolic LC3-1 is lipidated to form LC3-II, which is then recruited and tightly bound to the autophagosomal membrane.30 To visually confirm autophagic processes in MΦs treated with Ag85B nanofibers, WT or Atg5–/– MΦs were first treated with soluble or fibrillar Ag85B, stained with LC3-II antibody, and analyzed using fluorescence microscopy to detect autophagosomes. Data indicated numerous green fluorescent punctate structures in WT MΦs treated with KFE8-Ag85B nanofibers but none in WT or Atg5–/– MΦs treated with soluble Ag85B (Figure 2C). To further confirm the effect of nanofibers on autophagy, we measured the levels of microtubule-associated light chain-3 (LC3B) protein in mouse macrophages after treatment with soluble Ag85B, bare KFE8 nanofiber, and KFE8-Ag85B nanofibers. During the induction of autophagy, a cytosolic form of LC3B (LC3B-I) is conjugated to phosphatidylethanolamine to form LC3-phosphatidylethanolamine conjugate (LC3B-II), which is recruited to autophagosomal membranes. When analyzed through western blot using antibodies against LC3B, both bare KFE8 nanofiber and KFE8-Ag85B nanofibers showed increased lipidation of LC3B in mouse macrophages as compared to that of soluble Ag85B (Figure 2D). Detection and the number of LC3-II-positive punctate structures in the cell are considered to be an accurate autophagosome number and marker for autophagy. Quantification of puncta showed significantly higher numbers of LC3IIB puncta in WT MΦs treated with KFE8-Ag85B nanofibers compared to controls (Figure 2E). Some puncta were also detected in Atg5–/– MΦs treated with Ag85B nanofibers, presumably due to other ongoing cellular processes that involve Ag85B nanofibers (Figure 2E). This is evident from the fact that IL-2 production was not fully abrogated either in WT MΦs treated with autophagy inhibitors, Atg5–/– MΦs, or Atg7–/– DCs, suggesting alternate mechanisms could be engaged in the intracellular processing of peptide nanofibers.
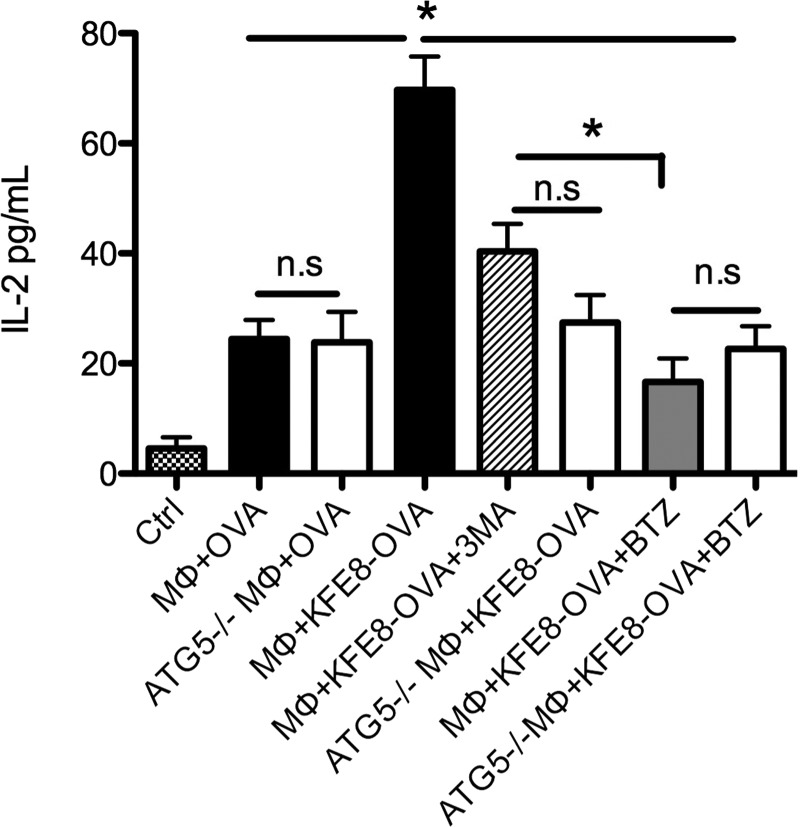
Emerging evidence suggests that macroautophagy could contribute to antigen processing for MHC class I presentation. Using virus-infected MΦs, it was demonstrated that viral proteins are not processed only by the “classical” MHC class I presentation pathway involving the proteosome.30 In late stages of infection, autophagosomes form from the membrane of the outer nuclear leaflet around virus particles and inhibition of Atg5 by siRNA leads to loss of virus-specific CD8+ T cell stimulation.31 This suggests that macroautophagic cargo can gain access to MHC class I presentation. To assess a role for macroautophagy in MHC class I presentation of peptide nanofibers, we used the OVA peptide (SIINFEKL, chicken egg ovalbumin 257–264) linked to the self-assembling domain KFE8 and T cells from OT-I transgenic mice (receptors specific for OVA). Data indicated significantly higher IL-2 production in MΦs treated with KFE8-OVA compared with soluble control and was significantly reduced in the presence of 3MA or Atg5–/– MΦs (Figure 3). This suggests a role for macroautophagy in the processing of peptide nanofibers and MHC class I presentation, and presumably peptide fragments resulting from macroautophagic processes are further degraded in the proteasomes prior to MHC-loading and presentation. This was confirmed using the proteosomal inhibitor bortezomib (BTZ), which led to a significant reduction in IL-2 in MΦs treated with nanofibers compared to that in MΦs treated with the autophagy inhibitor 3MA (Figure 3). No differences in IL-2 levels were detected in WT or Atg5–/– MΦs treated with soluble OVA or in Atg5–/– MΦs treated with 3MA, BTZ, or rapamycin (Figure 3).
Figure 3.
Autophagy is involved in the processing and presentation of MHC class I antigens linked to self-assembling peptides. Significantly higher levels of IL-2 are detected in cultures treated with KFE8-OVA compared to those in cultures treated with soluble OVA peptide. No loss of IL-2 is observed in Atg5–/– macrophages compared to that observed in WT when treated with soluble OVA, but in the presence of 3MA or in Atg5–/– macrophages treated with OVA-KFE8 a significant drop in IL-2 is observed. Also, treatment with proteasome inhibitor BTZ also leads to loss of IL-2 in OVA-KFE8 treated macrophages. Addition of rapamycin to Atg5–/– macrophages treated with OVA-KFE8 does not enhance IL-2 production as expected. *p < 0.05 by ANOVA using Tukey post-hoc test (n = 3 wells/group, data representative of two experiments with similar results).
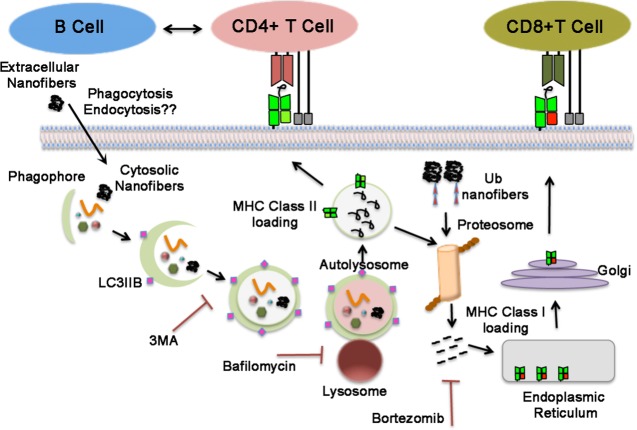
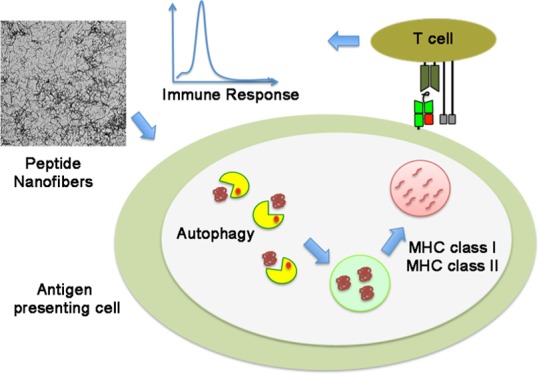
Normally, autophagy always occurs at low basal levels to maintain metabolic homeostasis in the cells.17 However, under stressful situations, this process may be upregulated whereby the occurrences of diseases like diabetes, neurodegeneration, myopathies etc.19 is prevented. It has been demonstrated that vaccines that engage autophagy are essential for the generation and survival of influenza virus-specific memory B cells in mice and triggering autophagy in APCs to process and present pathogen-derived MHC II-restricted antigens that can stimulate cell-mediated immunity in the host.24,32 Enhancing basal autophagy using FDA-approved pharmacological interventions (rapamycin) or inducing cytosolic aggregation of antigens (Ag85B in BCG) has been shown to enhance T helper type 1-mediated host protection against M. tuberculosis (Mtb).24 On the basis of our data, we believe that following uptake by antigen-presenting cells, nanofibers are readily sequestered into the phaghophores, which expands to form a double-membrane vesicle called the autophagosome. The autophagosome eventually fuses with the lysosome, resulting in the formation of an autolysosome, where the sequestered nanofibers are degraded by lysosomal proteases, leading to MHC class II presentation and activation of CD4+T cells (Figure 4). We are tempted to hypothesize that similar processes are involved in MHC class I presentation and that following macroautophagy and lysosomal degradation, the vacuolar contents are further processed by the proteasome to generate peptides for loading onto MHC class I molecules (Figure 4). However, further studies are required to verify the exact mechanism.
Figure 4.
Schematic shows autophagy mechanisms involved in the processing of peptide nanofibers in antigen-presenting cells and MHC class I and MHC class II presentation.
Future studies aim to investigate how self-assembly can be controlled to vary physicochemical properties (size, shape, and stability) of peptide nanofibers and how changes in physical properties of peptide nanofibers influence autophagy-mediated adjuvant potential and immunogenicity. Other pathways of autophagy in antigen-presenting cells remain to be investigated and an extended understanding of how peptide nanofiber vaccines are processed and presented in vivo will be crucial for their clinical translation in the near future. Further, triggering other immunological pathways in conjunction with autophagy through incorporation of TLR agonists could boost the immune response and vaccine efficacy.
Materials and Methods
Peptide Synthesis
Peptides KFE8 (FKFEFKFE), NapFFK (NapFFKYp, Nap = napthylalanine, Yp = phosphotyrosine), Ag85B (FQDAYNAAGGHNAVF), OVA (SIINFEKL), KFE8-Ag85B (KFE8-GGAAY-Ag85B), KFE8-OVA (KFE8-GGAAY-OVA), and NapFFK-Ag85B (NapFFK-GG-Ag85B) were synthesized on a CEM Blue (Matthews, NC) microwave synthesizer using standard solid phase Fmoc chemistry on Rink amide resin. Fomc-protected amino acids with standard side chain protecting groups, fluorescent dyes, and resins were purchased from Novabiochem (Billerica, MA). Peptides were cleaved using cleavage cocktail containing 95% TFA, 2.5% water, and 2.5% triisopropyl silane for 90 min at room temperature (RT) and precipitated in ice-cold diethyl ether. Crude peptides were purified by reverse phase high-performance liquid chromatography (HPLC) using a water/acetonitrile gradient on a C18 column (∼80% purity) and confirmed by matrix-assisted laser desorption ionization time-of-flight mass spectrometry (Figure S7). All peptides were stored at −20 °C until further use.
Transmission Electron Microscopy (TEM)
Stock solutions of 1 mM peptides were allowed to fibrillize in water overnight at room temperature, diluted in phosphate-buffered saline (PBS) to 0.2 mM, and applied to 200 mesh copper grids with carbon support film (Quantifoil). The grids were negatively stained with 2% uranyl acetate, and imaged on a JEM1400 TEM (JEOL) equipped with LaB6 electron gun and Ultrascan 1000 camera (Gatan).
Circular Dichrosim Spectroscopy
A Jasco 815 circular dichroism spectropolarimeter was used with 0.1 cm path length quartz cells. Stock solutions were prepared by dissolving the peptides in ultrapure water (2 mM) and further diluting to a working concentration (0.25–1 mM) in PBS buffer. The wavelength range was 190–260 nm, the scanning speed was 0.5 nm/s, and the bandwidth was 0.5 nm. Each spectrum is the average of three scans. Under the solution conditions described, adequate signal strength was observed at wavelengths up to 200 nm. The solvent background was subtracted and resultant CD signals plotted.
Mouse Primary Bone Marrow-Derived MΦs and DCs
C57BL6 mice (wild type) were purchased from Jackson labs. Autophagy-deficient mice (Atg5flox/flox Lyz-Cre) and their corresponding wild-type controls (Atg5flox/flox) (generated by Dr. Noboru Mizushima, Department of Biochemistry and Molecular Biology, University of Tokyo) were a kind gift from Dr. Rong Fang at UTMB. Dr. Jin Wang at Baylor College of Medicine (Houston) provided the murine Atg7–/– dendritic cells. To generate BMDCs, femurs and tibias from mice were flushed using Iscove’s modification of Dulbecco medium (IMDM) supplemented with 10% fetal bovine serum (FBS), penicillin, gentamycin, and 20 ng/mL mouse recombinant granulocyte-macrophage colony-stimulating factor (GM-CSF). Following RBC lysis with ACK buffer, the cells were cultured for a week in IMDM, with medium changed every 2 days. On day 8, cells were harvested and fractionated using anti-CD11c magnetic beads (Miltenyi Biotech) to separate DCs from MΦs according to manufacturer protocols. MΦs were then plated onto 24-well plates or eight-chambered slides (for microscopy) in IMDM (with GM-CSF) for an additional 4 days, after which the medium was replaced (without GM-CSF). Cells were rested and used as well-differentiated monolayers.
Antigen-Presentation Assays
This protocol has been extensively described in our previous publication in detail.21 Briefly, M. tuberculosis Ag85B epitope-specific T cell hybridoma (BB7) (a kind gift from Drs. Harding and Boom, Case Western Reserve University) was maintained in Dulbecco’s modified Eagle’s medium supplemented with 10% heat-inactivated FBS (HyClone Labs), 50 μM 2-mecaptoethanol, 1 mM sodium pyruvate, 2 mM l-glutamine, 10 mM HEPES buffer, 10 μg/mL penicillin, and 5 μg/mL gentamicin. MΦs were plated in 24-well plates and used as naive cells or were incubated for 16 h with peptides Ag85B or KFE8-Ag85B. The peptide concentration in the medium was adjusted so that all groups received 25 nmol of Ag85B. MΦs were then washed thrice with PBS and overlaid with BB7 T cells at a ratio of 1:10 (MΦs:BB7 cells). The supernatant was collected 24 h later. IL-2 levels were quantified using a sandwich ELISA kit (R&D Systems). For loss and gain of function studies 3-methyl adenine, bafilomycin, or rapamycin were added 30 min prior to incubation with peptides. Inhibition of Lamp2A was carried out by incubating the MΦs in siRNA duplex solution (30 min), followed by siRNA (5 h) and knock down was allowed to progress for 24 h before addition of peptides.
Fluorescence Microscopy
Immunostaining was performed using chamber slides, as described in detail elsewhere.24 Briefly, MΦs were incubated with peptide solutions for 16 h, washed 3 times with PBS, and fixed in 4% fresh paraformaldehyde. Cells were then permeabilized with 0.1% saponin and 0.05% Triton X-100 in PBS (supplemented with 2% normal mouse serum) and stained with primary LC3B antibody for 18 h at 4 °C (1:500, Cat # 3868; Cell Signaling). MΦs were then counterstained with FITC-conjugated secondary antibody (#711-095-152; Jackson ImmunoResearch) for 2 h, washed, fixed with methanol, and examined for LC3B puncta using a fluorescent microscope.
Western Blot
Soluble Ag85B, bare KFE8 nanofiber, and KFE8-Ag85B treated (24 h) primary mouse macrophage lysates were subjected to western blotting using antibodies specific for mouse LC3-I/LC3-II (Cell Signaling # 3738). Blots were then probed with horseradish peroxidase-conjugated anti-rabbit IgG as a secondary antibody (Cell Signaling # 7074) and assessed via chemiluminescence. A protein loading control of β-actin (Cell Signaling # 4967) was also included.
Statistical Analysis
All experimental data were plotted using GraphPad Prism software and represented as mean ± SEM, and statistical analysis was performed by ANOVA with Tukey or Bonferroni post-hoc test. Statistical significance was assigned at p values <0.05.
Acknowledgments
This work was supported by the National Institutes of Health (R21 AI115302, J.S.R. and J.J.E.) and the Sealy Center for Vaccine Development at UTMB. We would like to thank Dr. Rong Fang at UTMB for Atg5–/– mice. We also thank Michael Woodson and Misha Sherman and the Sealy Center for Structural Biology for assistance with electron microscopy.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsomega.7b00525.
TEM images, CD spectra, HPLC, mass spectra, autophagy inhibitor data, and Atg7–/– knock out controls (PDF)
Author Contributions
∇ J.S.R. and A.K. contributed equally.
The authors declare no competing financial interest.
Supplementary Material
References
- Webber M. J.; Appel E. A.; Meijer E. W.; Langer R. Supramolecular biomaterials. Nat. Mater. 2016, 15, 13. 10.1038/nmat4474. [DOI] [PubMed] [Google Scholar]
- Busseron E.; Ruff Y.; Moulin E.; Giuseppone N. Supramolecular self-assemblies as functional nanomaterials. Nanoscale 2013, 5, 7098. 10.1039/c3nr02176a. [DOI] [PubMed] [Google Scholar]
- Chan K. H.; Lee W. H.; Zhuo S. M.; Ni M. Harnessing supramolecular peptide nanotechnology in biomedical applications. Int. J. Nanomed. 2017, 12, 1171. 10.2147/IJN.S126154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webber M. J.; Kessler J. A.; Stupp S. I. Emerging peptide nanomedicine to regenerate tissues and organs. J. Intern. Med. 2010, 267, 71. 10.1111/j.1365-2796.2009.02184.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arslan E.; Garip I. C.; Gulseren G.; Tekinay A. B.; Guler M. O. Bioactive supramolecular peptide nanofibers for regenerative medicine. Adv. Healthcare Mater. 2014, 3, 1357. 10.1002/adhm.201300491. [DOI] [PubMed] [Google Scholar]
- Wen Y.; Collier J. H. Supramolecular peptide vaccines: tuning adaptive immunity. Curr. Opin. Immunol. 2015, 35, 73. 10.1016/j.coi.2015.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudra J. S.; Tian Y. F.; Jung J. P.; Collier J. H. A self-assembling peptide acting as an immune adjuvant. Proc. Natl. Acad. Sci. U.S.A. 2010, 107, 622. 10.1073/pnas.0912124107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J. J.; Pompano R. R.; Santiago F. W.; Maillat L.; Sciammas R.; Sun T.; Han H. F.; Topham D. J.; Chong A. S.; Collier J. H. The use of self-adjuvanting nanofiber vaccines to elicit high-affinity B cell responses to peptide antigens without inflammation. Biomaterials 2013, 34, 8776. 10.1016/j.biomaterials.2013.07.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrich B. M.; Beasley D. W.; Rudra J. S. Supramolecular peptide hydrogel adjuvanted subunit vaccine elicits protective antibody responses against West Nile virus. Vaccine 2016, 34, 5479. 10.1016/j.vaccine.2016.09.044. [DOI] [PubMed] [Google Scholar]
- Rudra J. S.; Ding Y.; Neelakantan H.; Ding C.; Appavu R.; Stutz S.; Snook J. D.; Chen H.; Cunningham K. A.; Zhou J. Suppression of Cocaine-Evoked Hyperactivity by Self-Adjuvanting and Multivalent Peptide Nanofiber Vaccines. ACS Chem. Neurosci. 2016, 7, 546. 10.1021/acschemneuro.5b00345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Z. H.; Shi L.; Ma J. W.; Sun Z. Y.; Cai H.; Chen Y. X.; Zhao Y. F.; Li Y. M. A totally synthetic, self-assembling, adjuvant-free MUC1 glycopeptide vaccine for cancer therapy. J. Am. Chem. Soc. 2012, 134, 8730. 10.1021/ja211725s. [DOI] [PubMed] [Google Scholar]
- Rudra J. S.; Sun T.; Bird K. C.; Daniels M. D.; Gasiorowski J. Z.; Chong A. S.; Collier J. H. Modulating Adaptive Immune Responses to Peptide Self-Assemblies. ACS Nano 2012, 6, 1557. 10.1021/nn204530r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudalla G. A.; Modica J. A.; Tian Y. F.; Rudra J. S.; Chong A. S.; Sun T.; Mrksich M.; Collier J. H. A self-adjuvanting supramolecular vaccine carrying a folded protein antigen. Adv. Healthcare Mater. 2013, 2, 1114. 10.1002/adhm.201200435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudra J. S.; Mishra S.; Chong A. S.; Mitchell R. A.; Nardin E. H.; Nussenzweig V.; Collier J. H. Self-assembled peptide nanofibers raising durable antibody responses against a malaria epitope. Biomaterials 2012, 33, 6476. 10.1016/j.biomaterials.2012.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen Y.; Waltman A.; Han H. F.; Collier J. H. Switching the Immunogenicity of Peptide Assemblies Using Surface Properties. ACS Nano 2016, 10, 9274. 10.1021/acsnano.6b03409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appavu R.; Chesson C. B.; Koyfman A. Y.; Snook J. D.; Kohlhapp F. J.; Zloza A.; Rudra J. S. Enhancing the Magnitude of Antibody Responses through Biomaterial Stereochemistry. ACS Biomater. Sci. Eng. 2015, 1, 601. 10.1021/acsbiomaterials.5b00139. [DOI] [PubMed] [Google Scholar]
- Ryter S. W.; Cloonan S. M.; Choi A. M. Autophagy: a critical regulator of cellular metabolism and homeostasis. Mol. Cells 2013, 36, 7. 10.1007/s10059-013-0140-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng W.; Wei M.; Li S.; Le W. Nanomaterial-modulated autophagy: underlying mechanisms and functional consequences. Nanomedicine 2016, 11, 1417. 10.2217/nnm-2016-0040. [DOI] [PubMed] [Google Scholar]
- Lamark T.; Johansen T. Aggrephagy: selective disposal of protein aggregates by macroautophagy. Int. J. Cell Biol. 2012, 2012, 736905 10.1155/2012/736905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crotzer V. L.; Blum J. S. Autophagy and its role in MHC-mediated antigen presentation. J. Immunol. 2009, 182, 3335. 10.4049/jimmunol.0803458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deretic V. Autophagy as an innate immunity paradigm: expanding the scope and repertoire of pathogen recognition receptors. Curr. Opin. Immunol. 2012, 24, 21. 10.1016/j.coi.2011.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kariyone A.; Higuchi K.; Yamamoto S.; Nagasaka-Kametaka A.; Harada M.; Takahashi A.; Harada N.; Ogasawara K.; Takatsu K. Identification of amino acid residues of the T-Cell epitope of Mycobacterium tuberculosis α antigen critical for Vβ11+Th1 cells. Infect. Immun. 1999, 67, 4312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowerman C. J.; Nilsson B. L. Self-assembly of amphipathic beta-sheet peptides: insights and applications. Biopolymers 2012, 98, 169. 10.1002/bip.22058. [DOI] [PubMed] [Google Scholar]
- Jagannath C.; Lindsey D. R.; Dhandayuthapani S.; Xu Y.; Hunter R. L. Jr.; Eissa N. T. Autophagy enhances the efficacy of BCG vaccine by increasing peptide presentation in mouse dendritic cells. Nat. Med. 2009, 15, 267. 10.1038/nm.1928. [DOI] [PubMed] [Google Scholar]
- Wu Y. T.; Tan H. L.; Shui G.; Bauvy C.; Huang Q.; Wenk M. R.; Ong C. N.; Codogno P.; Shen H. M. Dual role of 3-methyladenine in modulation of autophagy via different temporal patterns of inhibition on class I and III phosphoinositide 3-kinase. J. Biol. Chem. 2010, 285, 10850. 10.1074/jbc.M109.080796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto A.; Tagawa Y.; Yoshimori T.; Moriyama Y.; Masaki R.; Tashiro Y. Bafilomycin A1 prevents maturation of autophagic vacuoles by inhibiting fusion between autophagosomes and lysosomes in rat hepatoma cell line, H-4-II-E cells. Cell Struct. Funct. 1998, 23, 33. 10.1247/csf.23.33. [DOI] [PubMed] [Google Scholar]
- Jung C. H.; Ro S. H.; Cao J.; Otto N. M.; Kim D. H. mTOR regulation of autophagy. FEBS Lett. 2010, 584, 1287. 10.1016/j.febslet.2010.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanov J.; Walczak M.; Ibiricu I.; Schuchner S.; Ogris E.; Kraft C.; Martens S. Mechanism and functions of membrane binding by the Atg5-Atg12/Atg16 complex during autophagosome formation. EMBO J. 2012, 31, 4304. 10.1038/emboj.2012.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y.; Shi J.; Yuan D.; Xu B. Imaging enzyme-triggered self-assembly of small molecules inside live cells. Nat. Commun. 2012, 3, 1033 10.1038/ncomms2040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanida I.; Ueno T.; Kominami E. LC3 and Autophagy. Methods Mol. Biol. 2008, 445, 77. 10.1007/978-1-59745-157-4_4. [DOI] [PubMed] [Google Scholar]
- English L.; Chemali M.; Duron J.; Rondeau C.; Laplante A.; Gingras D.; Alexander D.; Leib D.; Norbury C.; Lippe R.; Desjardins M. Autophagy enhances the presentation of endogenous viral antigens on MHC class I molecules during HSV-1 infection. Nat. Immunol. 2009, 10, 480. 10.1038/ni.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M.; Hong M. J.; Sun H.; Wang L.; Shi X.; Gilbert B. E.; Corry D. B.; Kheradmand F.; Wang J. Essential role for autophagy in the maintenance of immunological memory against influenza infection. Nat. Med. 2014, 20, 503. 10.1038/nm.3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.