ABSTRACT
The migration of chromosomes during mitosis is mediated primarily by kinesins that bind to the chromosomes and move along the microtubules, exerting pulling and pushing forces on the centrosomes. We report that a DNA replication protein, Sld5, localizes to the centrosomes, resisting the microtubular pulling forces experienced during chromosome congression. In the absence of Sld5, centriolar satellites, which normally cluster around the centrosomes, are dissipated throughout the cytoplasm, resulting in the loss of their known function of recruiting the centrosomal protein, pericentrin. We observed that Sld5-deficient centrosomes lacking pericentrin were unable to endure the CENP-E- and Kid-mediated microtubular forces that converge on the centrosomes during chromosome congression, resulting in monocentriolar and acentriolar spindle poles. The minus-end-directed kinesin-14 motor protein, HSET, sustains the traction forces that mediate centrosomal fragmentation in Sld5-depleted cells. Thus, we report that a DNA replication protein has an as yet unknown function of ensuring spindle pole resistance to traction forces exerted during chromosome congression.
KEYWORDS: GINS, kinesin CENP-E, multipolarity, centriolar satellites, chromosome congression, microtubule forces
INTRODUCTION
A multitude of microtubule-dependent motor proteins mediate the alignment of chromosomes to the spindle equator in eukaryotic cells (1, 2). The two classes of motor proteins that promote chromosome congression are kinesins and dyneins, which interact with kinetochores of duplicated chromosomes and move directionally along the dynamic microtubules (3, 4). Congression can occur either before or after biorientation, i.e., when the sister kinetochores are attached to the ends of the microtubules emanating from opposite spindle poles (5, 6). Congression of bioriented chromosomes is mediated primarily by members of the kinesin-8 family that promote depolymerization-coupled pulling at the leading sister kinetochore (7). On the other hand, the congression of mono-oriented chromosomes is dependent on the plus-end-directed kinesin motor, CENP-E, as well as on chromokinesins that propel the unaligned chromosome arms away from the spindle poles (1, 8–10). CENP-E associated with the free sister kinetochore of the mono-oriented chromosomes attaches laterally to the lattice of preexisting microtubule fibers and then slides the chromosomes toward the spindle equator (6). After congression to the equatorial plate, the unattached sister kinetochore makes an end-on attachment to the microtubule fibers to complete chromosome biorientation (11, 12).
Apart from kinesin-7 motor proteins, another family of motor proteins, known as chromokinesins, that are present on the chromosome arms function to generate polar ejection forces to push the chromosome arms away from the spindle poles, thereby aiding in chromosome biorientation (13). At the minus ends of microtubules, the kinesin-14 family motor proteins mediate the anchoring of microtubules to the centrosomes (14). Thus, the combined actions of different kinesin motor proteins mediate the chromosome movements and maintain spindle pole organization in mitosis. During the congression of chromosomes, one end of the microtubule is bound to the kinetochore, while the other end is bound to the centrosomes, resulting in a significant pulling force on the centrosomes. To resist the microtubule-mediated forces, centrosomes possess a robust architecture, perturbation of which results in centrosome fragmentation. It has been reported that perturbations of kinetochore and centrosomal proteins disrupt the balance of motor forces, leading to spindle pole fragmentation (15–17).
It is believed that centrosomal integrity is regulated by bodies called centriolar satellites that cluster around the centrosomes. Centriolar satellites are spherical granules approximately 100 nm in diameter that regulate the centrosomal composition either by sequestering centrosomal proteins or by altering the dynein-dependent transport of centrosomal proteins (18). Many proteins have been identified at the centriolar satellites, but significant among them is PCM-1, which is required for the assembly of centriolar satellites, where it serves as a scaffold for other proteins. It has been reported that PCM-1 binds to the major pericentriolar matrix protein, pericentrin, which forms the lattice in which γ-tubulin complexes are embedded to facilitate microtubule nucleation (19). Inhibition of PCM-1 results in reduced levels of many centrosomal proteins, demonstrating that it is required for maintaining centrosomal integrity. Recent reports indicate that centriolar satellites undergo dynamic modulations during stress, although the consequences of such adaptations are not clearly understood.
Apart from the well-defined role of DNA replication proteins in genome duplication, recent studies have shown that they play diverse cellular roles. A mutant of Dpb2, the noncatalytic subunit of Saccharomyces cerevisiae DNA polymerase ε, demonstrates abnormalities in the transcription of genes, while the roles of TopBP1 and replication protein A in checkpoint signaling are well established (20, 21). Thus, it is being realized that DNA replication proteins are involved in diverse pathways in eukaryotic cells, such as maintenance of heterochromatin, checkpoint signaling, and regulation of gene expression (22, 23). Recent studies have also demonstrated that proteins known to function in DNA replication localize to the centrosomes (24, 25). Apart from their localization to the centrosomes, it has been observed that the depletion of DNA replication proteins results in supernumerary centrosomes, indicating a requirement for them in the maintenance of centrosome numbers (23, 26, 27). However, the physiological function of replication proteins in preventing centrosomal instability has remained elusive. In the present study, we examine the role of a GINS subunit, Sld5, in maintaining spindle pole integrity (28, 29). We report that the DNA replication factor Sld5 has an independent role in maintaining the centrosome structure by resisting the microtubule-mediated forces during mitosis.
RESULTS
Sld5 localizes to centrosomes.
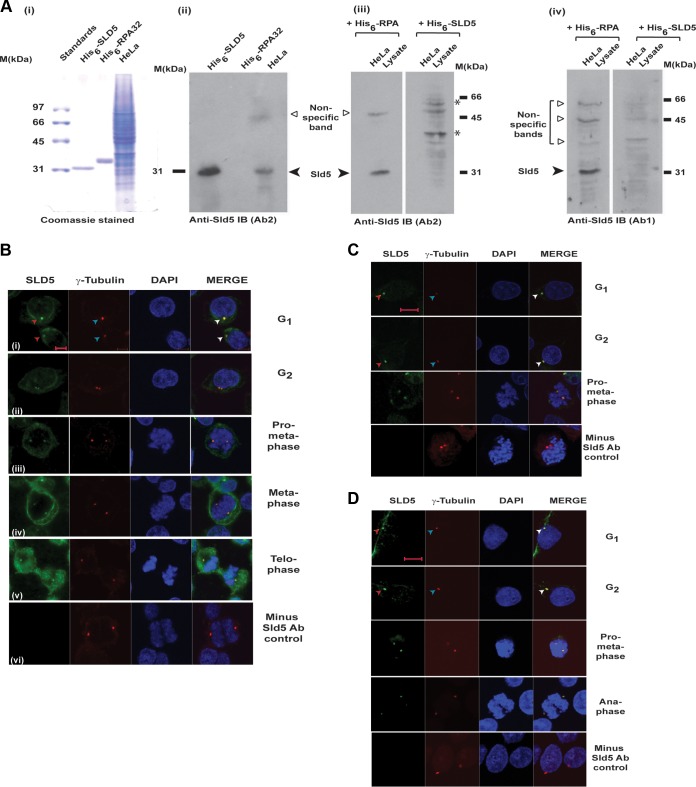
In eukaryotes, the tetrameric GINS complex (comprising Sld5, Psf1, Psf2, and Psf3) is involved in both the initiation and elongation stages of DNA replication. The Sld5 subunit is vital for the stability of the GINS complex, with its inactivation resulting in an M phase delay (30). We raised an antibody (Ab1) against His6-tagged Sld5 expressed in E. coli cells and purified on a nickel-nitrilotriacetic acid (NTA) column, which recognized the endogenous protein from HeLa cell lysates (Fig. 1A). Preincubation with His-Sld5 but not His-RPA protein led to the loss of the Sld5 immunoblotting signal observed at 31 kDa, establishing the specificity of the antibodies used (Fig. 1A, panels iii and iv). Cells were prepermeabilized to remove the nuclear fraction of Sld5, and we assayed its subcellular localization by immunofluorescence. γ-Tubulin served as a marker of centrosomes, and we observed that Sld5 colocalized with it during interphase, as well as mitosis (Fig. 1B, panels i to v). Removal of anti-Sld5 antibody abolished the Alexa Fluor 488 signal, ruling out nonspecificity of the secondary antibody, as well as bleed-through of the Alexa Fluor 555 signal (Fig. 1B, panel vi). The localization of Sld5 to centrosomes was confirmed with two other antibodies raised against different regions of Sld5 (Ab2 and Ab3) (Fig. 1C and D). We observed that these antibodies also marked the centrosomes during interphase, as well as different mitotic phases. Preincubation with bacterially expressed Sld5 protein, but not a control protein, inhibited the centrosomal localization of anti-Sld5 antibody, confirming that the antibody specifically recognized Sld5 protein at centrosomes (Fig. 2A). Coimmunofluorescence with Ab1 anti-Sld5 antibody without prepermeabilization displayed the expected nuclear localization of Sld5 (Fig. 2B). To further authenticate the localization of Sld5, asynchronous HeLa cells were transfected with SLD5 small interfering RNA (siRNA) on three consecutive days, which specifically led to a decrease in the Sld5 protein and RNA (Fig. 2C and D). RNA interference (RNAi)-mediated depletion of Sld5 resulted in loss of the immunofluorescent signal of anti-Sld5 antibody (Ab1) at the centrosomes of both interphase and mitotic cells, confirming Sld5 localization (Fig. 2E).
FIG 1.
Sld5 colocalizes with γ-tubulin at centrosomes. Sld5 localization to centrosomes was confirmed by immunofluorescence assays with multiple antibodies. (A) His6-tagged Sld5 protein expressed in E. coli was injected into rabbits to produce anti-Sld5 antibody. (i) His6-tagged Sld5 (0.5 μg) and His6-tagged RPA32 (0.5 μg) purified on a nickel-NTA column and 15 μg HeLa cell lysate were resolved by SDS-PAGE and stained with Coomassie blue. (ii) Alternatively, they were probed with Ab2 anti-Sld5 antibody. (iii) Ab2 anti-Sld5 antibody was incubated with 5 ng/μl His6-Sld5 or control His6-RPA protein, and the blots were developed with the same exposure time. Preincubation with His6-Sld5 but not His6-RPA protein led to the loss of Sld5 immunoblot (IB) signal observed at 31 kDa. Note that the nonspecific bands did not significantly change due to preincubation with His-Sld5 protein. Due to the presence of bacterial protein, some nonspecific sticking occurred (bands marked by asterisks), which was absent in the initial Ab2 immunoblot. (iv) Specificity of Ab1 antibody was demonstrated as explained for blot iii. (B) HeLa cells were prepermeabilized to remove the nuclear fraction of Sld5, followed by coimmunofluorescence assays with rabbit anti-Sld5 (Ab1) and mouse anti-γ-tubulin antibodies, in combination with anti-rabbit Alexa Fluor 488- and anti-mouse Alexa Fluor 555-conjugated antibodies, respectively. DNA was stained with DAPI. The right column is a merge of Alexa Fluor 488, Alexa Fluor 555, and DAPI images. (i to v) Cells in interphase (i and ii) and different phases of mitosis (iii to v). (vi) Immunofluorescence assays carried out in the absence of anti-Sld5 antibody, with other conditions and antibodies remaining similar, ruled out nonspecificity of secondary Alexa Fluor 488-conjugated antibody, as well as bleed-through of the Alexa Fluor 555 signal. (C and D) HeLa cells in different phases of the cell cycle were prepermeabilized to remove the nuclear fraction of Sld5, followed by coimmunofluorescence assay with either Ab3 anti-Sld5 antibody (C) or Ab2 anti-Sld5 antibody (D). Centrosomes are marked by arrowheads. Scale bars, 10 μm.
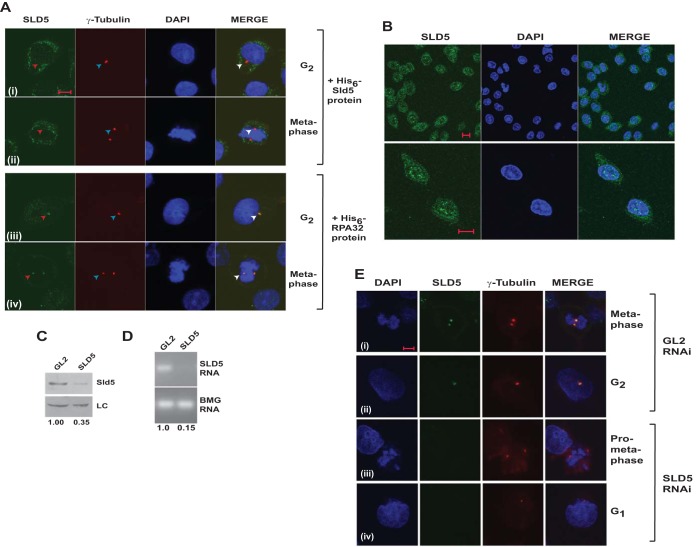
FIG 2.
Localization of Sld5 to centrosomes was confirmed by RNAi-mediated depletion. (A) Coimmunofluorescence assay of HeLa cells, as described in the legend to Fig. 1, was carried out in the presence of 5 ng/μl bacterially purified proteins. Preincubation with His6-Sld5 protein but not His6-RPA32 inhibited the localization of anti-Sld5 antibody to the centrosomes (red arrowheads). The top two rows demonstrate that after His6-Sld5 preincubation, the anti-Sld5 antibody was absent at the location where γ-tubulin stained, confirming that the antibody specifically binds to endogenous Sld5. (B) Coimmunofluorescence assay with Ab1 anti-Sld5 antibody without prepermeabilization showing nuclear localization of Sld5. The images were captured at different magnifications to show Sld5 localization in the entire field (top row) or individual cells (bottom row). (C) HeLa cells were transfected on three consecutive days with control GL2 or SLD5 siRNA, and the lysates were immunoblotted with anti-Sld5 antibody to confirm its specificity. LC, loading control showing equal protein loads in different lanes; the numbers indicate levels of Sld5 relative to control GL2 siRNA-transfected cells. (D) The decrease of SLD5 mRNA was confirmed by reverse transcriptase PCR. The numbers indicate the SLD5 mRNA levels following Sld5 depletion relative to control GL2 siRNA-transfected cells. Beta-2 microglobulin (BMG) served as the internal RNA-loading control. (E) HeLa cells were transfected on three consecutive days with control GL2 or SLD5 siRNA and costained for Sld5 (green), γ-tubulin (red), and DNA (blue) to confirm centrosomal localization of Sld5. (i and iii) Sld5 signal at mitotic centrosomes. (ii and iv) Sld5 signal at interphase centrosomes. Scale bars, 10 μm.
Sld5-depleted cells arrest in mitosis with defects in spindle pole organization and chromosome congression.
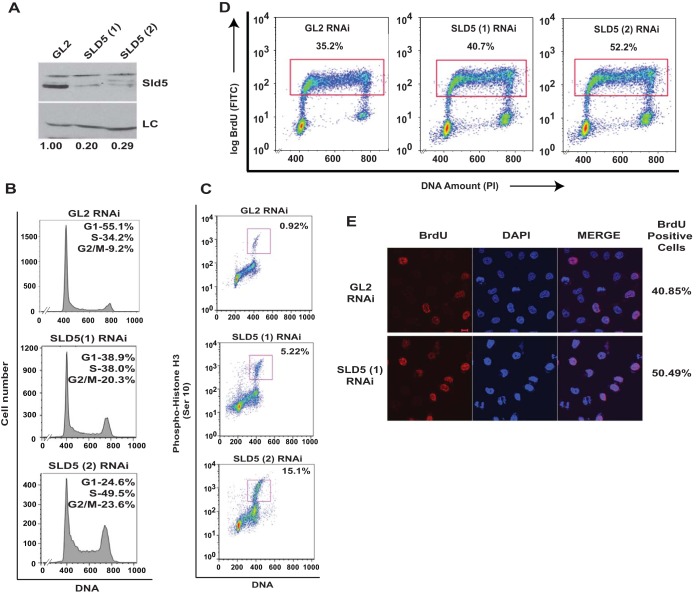
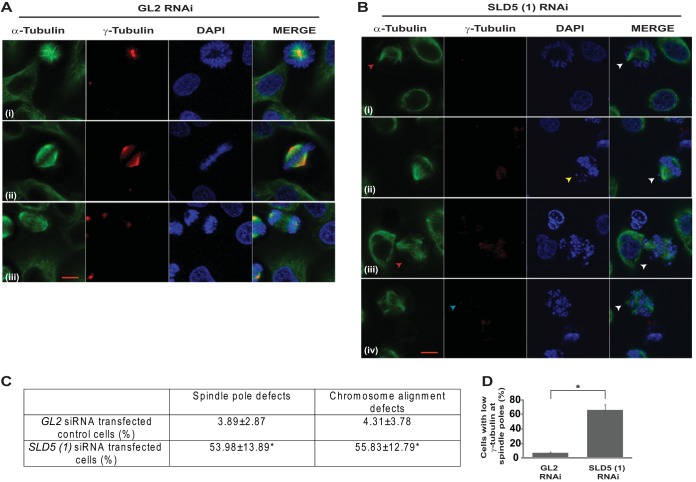
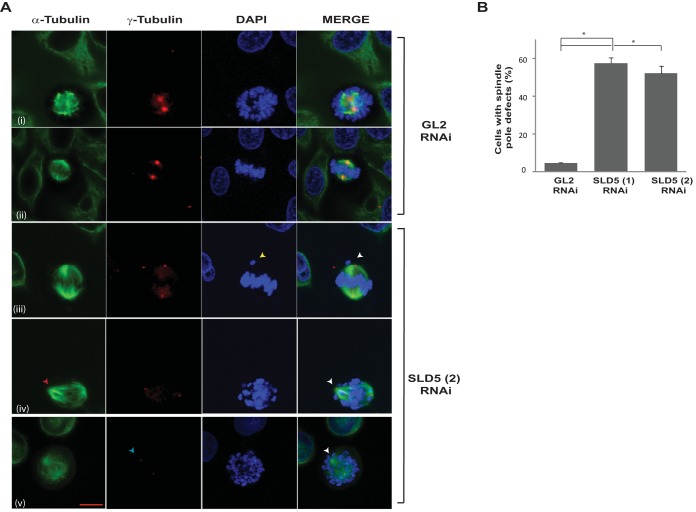
To rule out off-target effects of RNAi, we transfected another siRNA duplex, SLD5 (2), which targets a different region (nucleotides [nt] 339 to 363) of SLD5 mRNA (Fig. 3A). After 24 h of SLD5 siRNA transfection, we analyzed the cell cycle distribution, which displayed an increase in S phase (Fig. 3B). Moreover, the levels of the mitotic marker phospho-histone H3 (Ser 10) were significantly elevated, exhibiting accumulation in M phase (Fig. 3C). The accumulation in S phase indicated a delay in completing replication, as assayed by bromodeoxyuridine (BrdU) incorporation in flow cytometry and immunofluorescence assays (Fig. 3D and E). To assess its possible mitotic role, mitotic spindles and centrosomes of HeLa cells depleted of Sld5 were visualized by staining with anti-α-tubulin and anti-γ-tubulin antibodies, respectively. We observed the following mitotic aberrations: (i) asymmetric mitotic spindles, (ii) multipolar spindle poles, and (iii) chromosome congression failure with multiple unaligned chromosomes (Fig. 4A and B). Quantification revealed that the spindle pole organization and chromosome alignment defects after Sld5 depletion were significantly greater than in the control cells (Fig. 4C). Strikingly, the levels of γ-tubulin at centrosomes were significantly reduced in Sld5-depleted cells, suggesting loss of centrosomal integrity (Fig. 4D). Overexpression of Sld5 to rescue the phenotype resulted in multiple pleiotropic effects, as has been previously reported for other DNA replication proteins (31–35). To ensure specificity, HeLa cells were transfected with SLD5 (2) siRNA, which resulted in similar chromosome congression and spindle pole defects. More than 50% of the cells displayed unaligned or lagging chromosomes and asymmetric mitotic spindles (Fig. 5).
FIG 3.
Depletion of Sld5 leads to accumulation in M phase. (A) HeLa cells were transfected on three consecutive days with control GL2, SLD5 (1), or SLD5 (2) (targeting a different region of Sld5) siRNA, and the lysates were immunoblotted with anti-Sld5 antibody to confirm the specificity of RNAi-mediated depletion. LC, loading control showing equal protein loads in different lanes; the numbers indicate levels of Sld5 relative to control GL2 siRNA-transfected cells. (B and C) Transfected cells were harvested 24 h after the last transfection, followed by staining with propidium iodide alone (B) or in combination with anti-phospho-histone H3 (Ser 10) antibody (C), which marks the mitotic cells. The percentages in panel B show cell cycle distribution, while panel C shows cells in M phase. (D) Sld5 depletion causes S phase delay. Transfected cells were pulsed with BrdU for 30 min, followed by staining with anti-BrdU antibody conjugated to fluorescein isothiocyanate (FITC), along with propidium iodide (PI). The dot plots show BrdU incorporation (y axes) and DNA content (x axes), and the percentages of cells incorporating BrdU. (E) Transfected cells were pulsed with BrdU for 20 min, followed by staining with anti-BrdU (red) antibody. The coimmunofluorescence images display BrdU incorporation in different samples, while DNA was stained with DAPI (blue). Scale bar, 10 μm.
FIG 4.
Depletion of Sld5 at centrosomes leads to spindle pole defects. (A and B) HeLa cells transfected with control GL2 or SLD5 (1) siRNA were costained for α-tubulin (green) and γ-tubulin (red) and with DAPI for DNA (blue). GL2 siRNA-transfected cells in different mitotic phases displayed normal chromosome congression and spindle pole formation; however, Sld5-depleted cells displayed asymmetric mitotic spindles (red arrowhead in panel B, row i), chromosome congression failure (yellow arrowhead in panel B, row ii), multipolar spindles (red arrowhead in panel B, row iii), and decreased γ-tubulin staining at centrosomes (blue arrowhead in panel B, row iv). (C) Quantification of spindle pole and chromosome alignment defects due to Sld5 depletion (expressed as a percentage of total mitotic cells). The data are represented as means and SD; P values were calculated using a two-tailed t test and show that SLD5 (1) siRNA-transfected samples were significantly different from control GL2 siRNA-transfected samples (*, P < 0.05). (D) The γ-tubulin levels in the GL2 or SLD5 (1) siRNA-transfected cells were surveyed to identify the spindle poles with low levels of γ-tubulin staining, which was confirmed with NIS Elements imaging software (version 3.22.00) for multiple examples to be less than 50% of the mean intensity of the signal observed in the control cells. The data are represented as the means and SD of the results of three independent experiments, with more than 40 cells analyzed in each sample. Scale bar, 10 μm. *, P < 0.05.
FIG 5.
Knockdown with an independent siRNA confirmed that Sld5 depletion leads to multiple mitotic aberrations. (A) Control GL2 or SLD5 (2) (targeting a different region of Sld5) siRNA-transfected HeLa cells were costained for α-tubulin (green) and γ-tubulin (red) and for DNA with DAPI (blue). Control GL2 siRNA-transfected cells in different mitotic phases are shown. Examples of Sld5-depleted cells display an asymmetric mitotic spindle (red arrowhead in row iv), chromosome congression failure (yellow arrowhead in row iii), and decreased γ-tubulin staining at centrosomes (blue arrowhead in row v). (B) Quantification of spindle pole defects due to Sld5 depletion by siRNAs targeting different regions of Sld5 (expressed as percentages of total mitotic cells). The data are represented as the means and SD of the results of two independent experiments, with more than 40 cells analyzed in each sample. The P values were calculated using a two-tailed t test and show that the SLD5 siRNA-transfected samples were significantly different from control GL2 siRNA-transfected samples. *, P < 0.05. Scale bar, 10 μm.
Sld5 depletion-induced centrosomal aberrations are not a consequence of DNA damage.
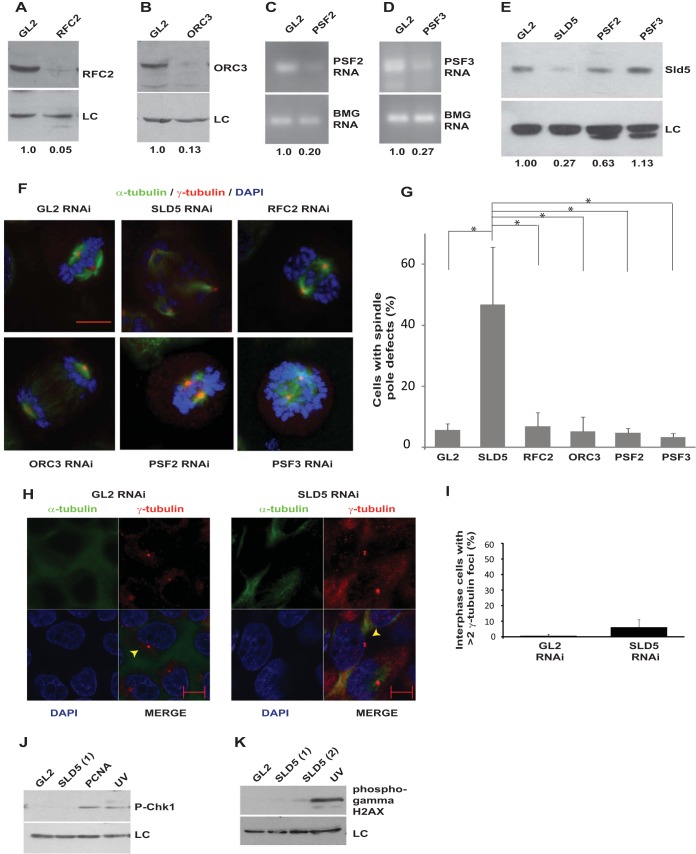
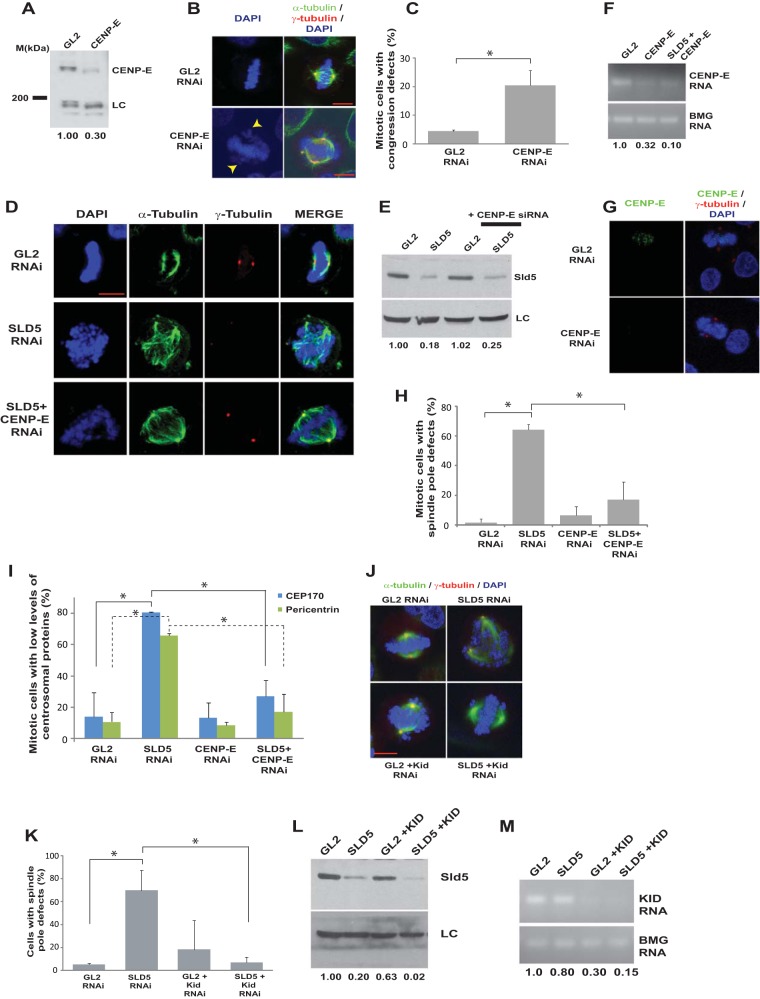
Depletion of DNA replication proteins, such as Orc3 and Rfc2, did not cause spindle pole aberrations, ruling out the possibility that the observed phenotype was a general effect of DNA replication stalling (Fig. 6A, B, F, and G). To evaluate the roles of other subunits of the GINS complex in centrosomal stability, we depleted Psf2 and Psf3. Since antibodies against Psf2 and Psf3 did not work in immunoblotting assays, we evaluated the decrease in PSF2 and PSF3 mRNAs, which were decreased in comparison to control cells (Fig. 6C and D). We did not observe spindle pole defects after PSF2 and PSF3 RNAi. There was a moderate decrease in Sld5 protein after PSF2 RNAi, and thus, it appears that a critical threshold in the Sld5 level is required for the mitotic functions, which is maintained in the case of PSF2 RNAi but not SLD5 RNAi (Fig. 6E to G) (36). We wanted to ascertain the cause of the multipolarity induced after Sld5 depletion. It has been reported that multiple spindle poles can arise through various mechanisms. Cytokinesis failure can lead to centrosome amplification, but we did not observe tetraploid cells or an increase in DNA content due to Sld5 depletion, ruling out this possibility (Fig. 3B and D) (37). It has been reported that ionizing-radiation-induced DNA damage causes centrosome amplification in cells arrested in G2 phase, and therefore, it is possible that Sld5 depletion results in replication defects that lead to centrosomal aberrations (38, 39). However, Sld5-depleted cells did not display supernumerary centrosomes during interphase, establishing that the phenotype was not due to centrosome overduplication (Fig. 6H and I). It has been independently shown that if cells are forced into mitosis with damaged DNA, the centrosomes break, forming multipolar spindles (40). In the case of Sld5 depletion, the DNA damage pathway was not activated, excluding DNA damage as the cause of the observed mitotic aberrations (Fig. 6J and K).
FIG 6.
Sld5 depletion-induced centrosomal aberrations are not caused by DNA damage. (A and B) HeLa cells were transfected with siRNA duplexes targeting replication factor RFC2 or ORC3, and the lysates were immunoblotted with the indicated antibodies. The numbers indicate levels of RFC2 or ORC3 relative to control GL2 siRNA-transfected cells. (C and D) HeLa cells were transfected with siRNA duplexes targeting GINS subunit PSF2 or PSF3, and the decrease in mRNAs was confirmed by reverse transcriptase PCR. The numbers indicate the mRNA levels following specific siRNA depletion relative to control GL2-transfected cells. BMG served as the internal RNA-loading control. (E) Immunoblotting of control GL2, PSF2, or PSF3 siRNA-transfected samples with anti-Sld5 antibody confirmed that depletion of PSF2 or PSF3 did not significantly decrease Sld5 protein levels, contrary to what had been reported previously (36). The numbers indicate levels of Sld5 relative to control GL2 siRNA-transfected cells. (F) HeLa cells transfected with siRNA duplexes targeting replication factor RFC2, ORC3, PSF2, or PSF3 were costained for α-tubulin (green) and γ-tubulin (red) and for DNA with DAPI (blue). Merged images are shown. (G) Quantification of spindle pole defects due to RNAi depletion (expressed as a percentage of total mitotic cells). The data are represented as the means and SD of the results of three independent experiments, with more than 20 cells analyzed in each sample. The P values were calculated using a two-tailed t test and show that the SLD5 samples were significantly different from control GL2, RFC2, ORC3, PSF2, and PSF3 samples (*, P < 0.05). The single-factor ANOVA results showed that there was a significant difference in spindle pole defects between the different transfected samples [F(5, 12) = 13.0; ANOVA, P < 0.001]. (H) Sld5 depletion does not cause centrosome overduplication in interphase. HeLa cells were transfected with control GL2 or SLD5 siRNA and costained for α-tubulin (green), γ-tubulin (red), and DNA (blue); representative images of cells in interphase are shown. Centrosomes are marked by arrowheads. (I) Quantification of the experiment shown in panel H demonstrating that depletion of Sld5 did not cause an increase in the centrosome number in interphase cells (expressed as a percentage of total interphase cells). The data are represented as the means and SD of the results of two independent experiments, with more than 20 cells analyzed in each sample. The t test showed that the SLD5 samples were not significantly different from the GL2 samples (P = 0.25). (J and K) Immunoblotting of control GL2, SLD5 (1), SLD5 (2), or PCNA siRNA-transfected samples to detect phospho-Chk1 (Ser 345) or phospho-γ-H2AX (Ser 139) confirmed that the DNA damage checkpoint was not activated after Sld5 depletion. LC, loading control showing equal protein loads in different lanes; the numbers indicate levels of Sld5 relative to control GL2 siRNA-transfected cells. Scale bars, 10 μm.
Sld5-deficient centrosomes fragment during chromosome congression.
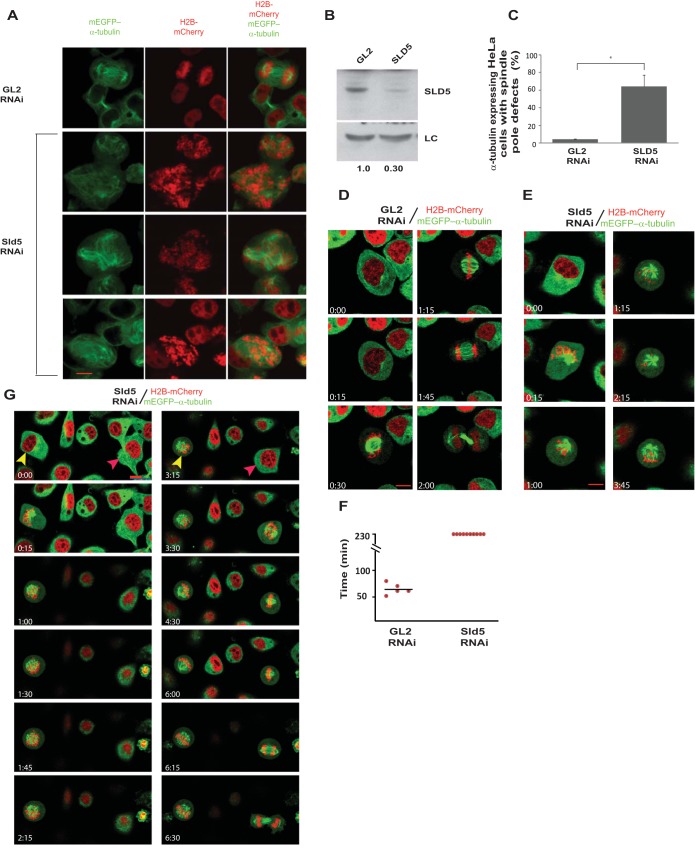
To establish the nature and timing of mitotic aberrations, HeLa cells stably coexpressing a red chromatin marker (core histone H2B fused to monomeric cherry [H2B-mCherry]) and a green marker (enhanced green fluorescent protein [EGFP]) for microtubules (mEGFP–α-tubulin) were transfected for three consecutive days with SLD5 siRNA, which resulted in chromosomal congression, as well as spindle pole defects (Fig. 7A to C). To understand the mitotic defects, the transfections were followed by live-cell imaging for almost 4 h. Control GL2 siRNA-transfected cells displayed normal spindle pole organization and progressed from prometaphase to cytokinesis in an average time of 64 ± 5.09 min (Fig. 7D and F; see Movie S1 in the supplemental material). On the other hand, Sld5-depleted cells appeared normal until chromosome condensation in prophase, followed by an abnormal prometaphase, during which multiple spindle poles were formed, and up to the end of the imaging period, the cells did not progress to anaphase (Fig. 7E and F; see Movie S2 in the supplemental material). Live-cell imaging capture of cells transfected with SLD5 siRNA revealed that a minor population of cells did complete mitosis, indicating poorly transfected cells (Fig. 7G, red arrowhead). On the other hand, after control GL2 siRNA transfection, almost all cells completed mitosis without developing any chromosomal or spindle pole defects. This demonstrates that the observed mitotic defects are due to Sld5 depletion, and we selected those cells to study Sld5-depletion-induced mitotic defects.
FIG 7.
Sld5-deficient centrosomes fragment during chromosome congression. (A) HeLa cells stably coexpressing a red chromatin marker (H2B-mCherry) and a marker for microtubules (mEGFP–α-tubulin) were transfected on three consecutive days with control GL2 or SLD5 siRNA, followed by fixation for visualization of spindle pole defects. Multiple fields of SLD5 siRNA-transfected cells are shown. (B) Immunoblotting confirmed depletion of Sld5 in cells coexpressing H2B-mCherry and mEGFP–α-tubulin. LC, loading control showing equal protein loads in different lanes; the numbers indicate levels of Sld5 relative to control GL2 siRNA-transfected cells. (C) Quantification of spindle pole defects observed in GL2 or SLD5 siRNA-transfected cells coexpressing H2B-mCherry and mEGFP–α-tubulin. The data are represented as the means and SD of the results of two independent experiments, with more than 20 cells analyzed in each sample (*, P ≤ 0.05). (D and E) HeLa cells stably coexpressing H2B-mCherry and mEGFP–α-tubulin were transfected on three consecutive days with control GL2 or SLD5 siRNA, followed by live-cell imaging for almost 4 h. Selected frames at the indicated time points are shown (live-cell image capture is shown in Movies S1 and S2 in the supplemental material). Note that control cells progressed from interphase to cytokinesis, whereas SLD5 siRNA-transfected cells were arrested in an abnormal prometaphase until the end of the imaging period. (F) Quantification of the time taken by GL2 or SLD5 siRNA-transfected cells to progress from prometaphase to cytokinesis. Each point represents a single cell, while the mean is shown as a horizontal bar. Live-cell imaging is presented up to 230 min, at which time the Sld5-depleted cells were alive but had not progressed to anaphase. (G) HeLa cells stably coexpressing H2B-mCherry and mEGFP–α-tubulin were transfected on three consecutive days with SLD5 siRNA, followed by live-cell imaging. The captured images show a cell (yellow arrowheads) that did not complete mitosis and developed chromosomal and spindle pole defects. A small fraction of the cells (red arrowheads) completed mitosis, albeit slowly (mitosis started at time point 3:15 and was complete by 6:30). Note that almost all GL2 siRNA-transfected cells completed mitosis without developing chromosomal and spindle pole defects, establishing that the mitotic defects were due to Sld5 depletion. Scale bars, 10 μm.
Sld5 depletion leads to the dissipation of centriolar satellite protein PCM-1 in interphase cells.
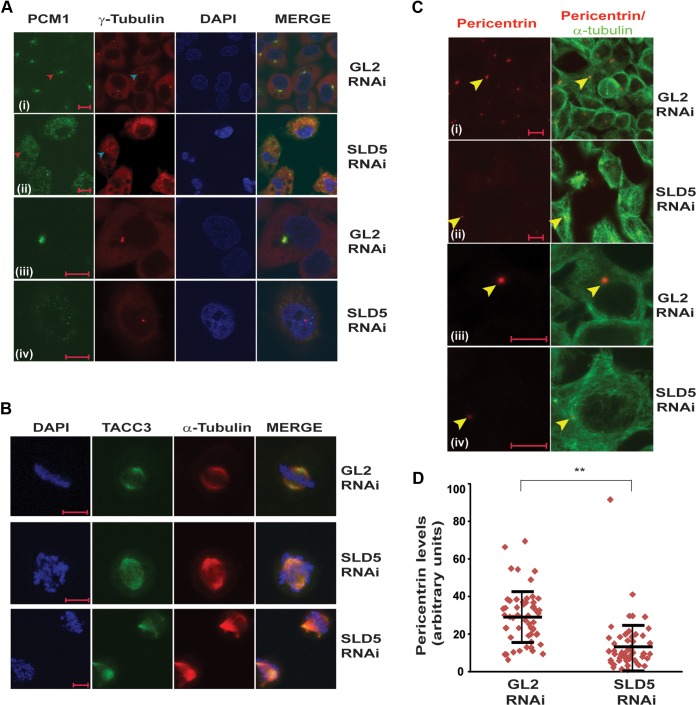
We observed that in control GL2 siRNA-transfected cells, PCM-1 displayed a typical pericentrosomal pattern of localization in interphase cells (Fig. 8A, rows i and iii). On the other hand, depletion of Sld5 resulted in dispersed localization of PCM-1 with loss of pericentrosomal accumulation (Fig. 8A, rows ii and iv). It was previously reported that PCM-1 binds to the major pericentriolar matrix protein, pericentrin, and its inhibition results in reduced levels of pericentrin at the interphase centrosomes (41). We analyzed the effect of PCM-1 dispersion due to Sld5 depletion on the levels of pericentrin in interphase centrosomes. We observed that pericentrin levels were significantly reduced in interphase centrosomes in Sld5-depleted cells (Fig. 8C and D). However, localization and levels of another centrosomal and microtubule-stabilizing protein, TACC3, were not significantly altered after Sld5 depletion, emphasizing that specific centrosomal proteins were affected (Fig. 8B) (42).
FIG 8.
Sld5 depletion leads to dispersion of centriolar satellite protein PCM-1, causing a decrease in pericentrin at centrosomes. (A) HeLa cells were transfected with control GL2 or SLD5 siRNA and costained for pericentriolar protein PCM-1 (green), γ-tubulin (red), and DNA (blue). Note that control cells displayed a dense, granular localization of PCM-1 around centrosomes, whereas Sld5-depleted cells displayed a diffuse cytosolic signal of PCM-1 in interphase. For GL2 or SLD5 siRNA-transfected samples, representative images of the entire field (i and ii) or individual cells (iii and iv) are shown. (B) Sld5 depletion does not alter the levels of TACC3, a protein involved in microtubule nucleation and stabilization of γ-tubulin ring complex. HeLa cells were transfected with control GL2 or SLD5 siRNA and costained for TACC3 (green), α-tubulin (red), and DNA (blue). (C) HeLa cells were transfected with control GL2 or SLD5 siRNA, and a coimmunofluorescence assay was performed to display α-tubulin (green) and pericentrin (red). For GL2 or SLD5 siRNA-transfected samples, representative images of the entire field (i and ii) or individual cells (iii and iv) are shown. Note that interphase cells in SLD5 siRNA-transfected samples displayed significantly reduced levels of pericentrin at the centrosomes (arrowheads). (D) Quantification of the pericentrin levels observed in panel C. Each point represents the pericentrin signal of an individual cell, whereas the horizontal bars represent the means of pericentrin signal in GL2 or SLD5 siRNA-transfected samples. Variability within each sample is shown by the SD (error bars). More than 50 cells were analyzed in each sample (**, P < 0.001). Scale bars, 10 μm.
Thus, in the absence of Sld5, there is a loss of PCM1-mediated recruitment of pericentrin to centrosomes during interphase. This results in a weakened centrosome that is susceptible to the microtubule-mediated traction forces prevalent during mitosis.
Sld5 depletion leads to centriole splitting and PCM fragmentation.
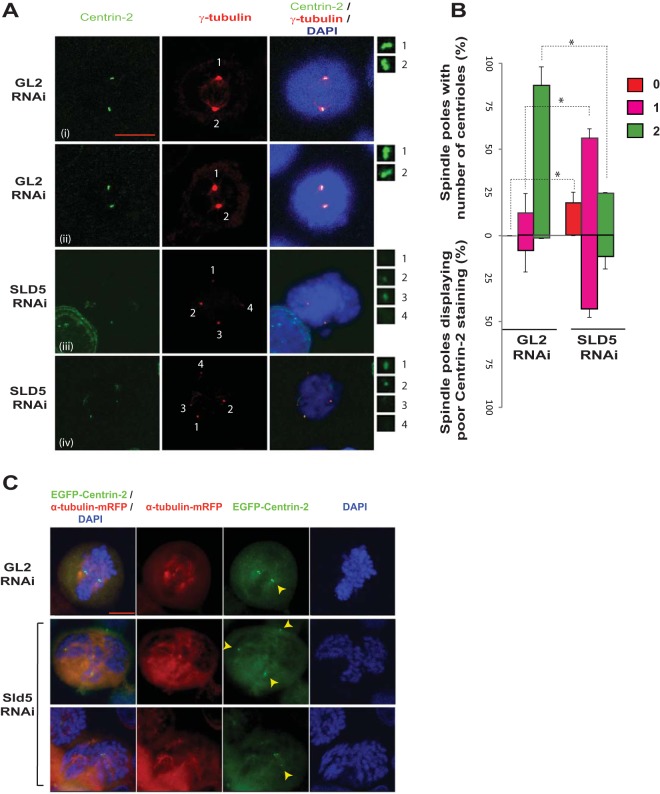
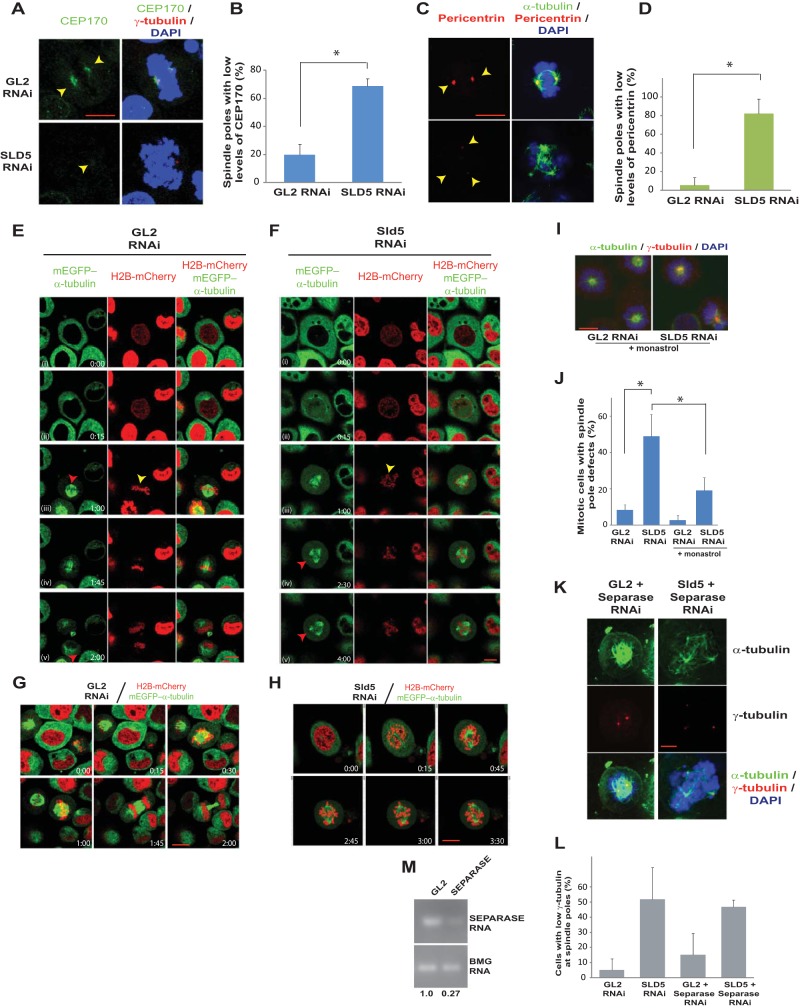
We ruled out the possibility that the supernumerary spindle poles formed after Sld5 depletion arise due to either cytokinesis failure or centrosome overduplication, and we next examined if Sld5 depletion-induced spindle pole defects were an outcome of centriole splitting. We carried out immunofluorescence assays of mitotic cells with an anti-centrin-2 antibody that stains individual centrioles. As expected, most of the spindle poles (87%) in control GL2 siRNA-transfected samples displayed two centrin-2 foci at centrosomes (Fig. 9A and B). On the other hand, only 24% of the spindle poles in Sld5-depleted samples had the complete pair of centrioles: 57% of the spindle poles were monocentriolar, and 19% were acentriolar, implying that they were a product of centriole splitting or PCM fragmentation. Apart from a decrease in the number of centrioles per centrosome, we observed that the signal intensity of centrin-2 was reduced in mitotic cells after Sld5 depletion, indicating its loss at the centrosomes (Fig. 9B). To rule out the possibility that the observed phenotype was an artifact of anti-centrin-2 antibody, we utilized a HeLa cell line that stably coexpresses a centriole marker (centrin-2 fused to monomeric green fluorescent protein [GFP], EGFP–centrin-2) and a marker for microtubules (α-tubulin fused to monomeric red fluorescent protein[RFP], α-tubulin–m-RFP). The cells were transfected on three consecutive days with SLD5 or GL2 siRNA, and 24 h later, the cells were fixed and EGFP–centrin-2 signal was evaluated. We observed that while the control cells displayed a doublet of EGFP–centrin-2 foci at centrosomes, the Sld5-depleted cells displayed spindle poles either negative for EGFP–centrin-2 signal or with one EGFP–centrin-2 focus (Fig. 9C). Therefore, using independent assays, we established that Sld5 depletion leads to centrosome fragmentation, resulting in monocentriolar and acentriolar spindle poles. Next, we analyzed the levels of two centrosomal proteins, namely, Cep170 and pericentrin, that localize to the subdistal appendages and pericentriolar region, respectively, to determine the effect of Sld5 depletion on these proteins (43, 44). We observed that the levels of Cep170 and pericentrin were significantly reduced at the centrosomes after Sld5 depletion (Fig. 10A to D). We observed that the overall levels of pericentrin or CEP170 were not significantly decreased in Sld5-depleted cells, indicating that Sld5 depletion affects the localization of pericentrin and CEP170 to the centrosomes (data not shown). Similar observations have been previously reported for Nedd1 depletion, which results in centrosomes with decreased levels of pericentrin (45). Thus, our data indicate that Sld5 is required in order to retain the structural proteins at the centrosomes. Moreover, we observed a physical association of Sld5 with other centrosomal proteins, which supports its role in maintaining centrosomal integrity (data not shown).
FIG 9.
Sld5 depletion leads to centriole splitting and PCM fragmentation. (A) HeLa cells were transfected with control GL2 or SLD5 siRNA and costained for centrin-2 (green) and γ-tubulin (red) with specific antibodies, as well as for DNA with DAPI (blue). The centrosomes are numbered, and the insets show magnifications of the centrin-2 signal at the indicated poles. Note that in the control GL2 sample, one γ-tubulin spot coincides with a doublet of centrin-2 foci. Each centrin-2 focus marks one centriole and has been identified by staining with anti-centrin-2 antibody (i and ii). Examples of Sld5-depleted cells are shown in rows iii and iv. In rows iii and iv, centrosomes of mitotic cells (marked 1 to 4) show centrosome splitting and PCM fragmentation. (B) Quantification of the experiment shown in panel A. Spindle poles in GL2 or SLD5 siRNA-transfected samples were surveyed for the number of centrioles, as shown in the upper half of the bar graph. The lower half of the graph shows the percentages of monocentriolar and bicentriolar spindle poles that displayed low levels of centrin-2 staining in GL2 or SLD5 siRNA-transfected samples. The level of centrin-2 staining was surveyed at each monocentriolar and bicentriolar spindle pole to identify the poles with low levels of centrin-2 staining, which was confirmed by NIS Elements software for multiple examples to be less than 50% of the mean intensity of the signal observed in control cells. The data are represented as the means and SD of the results of two independent experiments, with more than 40 spindle poles analyzed in each sample (*, P < 0.05). (C) Centriole disengagement in HeLa cells stably coexpressing EGFP–centrin-2 and α-tubulin–m-RFP. Cells were transfected on three consecutive days with control GL2 or SLD5 siRNA, fixed, and imaged to capture EGFP, RFP, and DAPI signals. Centrioles, as identified by centrin-2 staining, are marked by arrowheads. Scale bar, 10 μm.
FIG 10.
Spindle pole fragmentation due to Sld5 depletion is preceded by the presence of unaligned chromosomes. (A to D) HeLa cells were transfected with control GL2 or SLD5 siRNA and costained for two structural centrosomal proteins, Cep170 (A, green) or pericentrin (C, red), in combination with γ-tubulin (red) or α-tubulin (green), respectively, as indicated. GL2 or SLD5 siRNA-transfected cells were surveyed to identify spindle poles with low levels of Cep170 or pericentrin staining, which was confirmed by NIS Elements software for multiple examples to be less than 50% of the mean intensity of the signal observed in the control cells. Quantification of panels A and C is shown in panels B and D, respectively. The data are represented as the means and SD of the results of two independent experiments, with more than 20 cells analyzed in each sample for the levels of Cep170 and pericentrin signals (*, P < 0.05). (E and F) HeLa cells stably coexpressing H2B-mCherry and mEGFP–α-tubulin were transfected on three consecutive days with control GL2 or SLD5 siRNA, followed by live-cell imaging. Selected frames at the indicated time points are shown (live-cell capture is shown in Movies S3 and S4 in the supplemental material). In row iii of the control GL2 samples, symmetric spindle poles (red arrowhead) and chromosomes aligned at the metaphase plate (yellow arrowhead) that result in equivalent cytokinesis are visible. In row iii of the Sld5-depleted samples, lagging or unaligned chromosomes are visible (yellow arrowhead), though the spindle poles appear to be normal. In rows iv and v, the appearance of supernumerary spindle poles is marked by red arrowheads. (G and H) Live-cell imaging of GL2 or SLD5 siRNA-transfected cells stably coexpressing H2B-mCherry and mEGFP–α-tubulin obtained from an independent experiment (see Movies S5 and S6 in the supplemental material) different from that shown in panels E and F. In panel H, lagging chromosomes are visible, followed by the appearance of supernumerary spindle poles. (I) Spindle pole defects observed after monastrol treatment, demonstrating that centriole splitting occurs after centrosome separation at prophase. Shown are merged images of control GL2 or SLD5 siRNA-transfected cells costained for α-tubulin (green) and γ-tubulin (red) and for DNA with DAPI (blue) after incubation with monastrol for 4 h. (J) Quantification of the spindle pole defects observed in panel I. The data are represented as the means and SD of the results of three independent experiments, with more than 20 cells analyzed in each sample (*, P < 0.05). (K and L) Codepletion of separase does not prevent spindle pole fragmentation in Sld5-depleted cells. HeLa cells were transfected with control GL2, SLD5, or SEPARASE siRNA as indicated, with the combined concentration brought to 80 nM with GL2 siRNA. The cells were fixed and costained for α-tubulin (green), γ-tubulin (red), and DNA (blue). The quantification is shown in panel L. The data are represented as the means and SD of the results of two independent experiments, with more than 20 cells analyzed in each sample. The t test showed that SLD5-plus-SEPARASE samples were not significantly different from the SLD5 samples (P = 0.77). (M) Decrease of SEPARASE mRNA confirmed by reverse transcriptase PCR. The numbers indicate the mRNA levels following specific siRNA-mediated depletion relative to control GL2 siRNA-transfected cells. BMG served as the internal RNA-loading control. Scale bars, 10 μm.
Spindle pole fragmentation due to Sld5 depletion is preceded by the presence of unaligned chromosomes.
To understand the formation of supernumerary spindle poles, we monitored chromosome and spindle pole dynamics during mitosis. As expected, in control GL2 siRNA-transfected cells, chromosomes congressed to form the metaphase plate, which was followed by cell division (Fig. 10E; see Movie S3 in the supplemental material). Though a bipolar spindle was formed in Sld5-depleted cells, unaligned chromosomes that did not congress to the equatorial plate were observed (Fig. 10F; see Movie S4 in the supplemental material). This was followed by formation of asymmetric spindle poles that subsequently progressed to multipolarity (also illustrated by other examples shown in Fig. 10G and H). The cells were arrested at an abnormal prometaphase stage where chromosome congression and segregation were not completed. Congression of unaligned chromosomes requires the activity of motor proteins that associate with sister kinetochores or chromosomal arms and move along the microtubules, thereby sliding the chromosomes toward the spindle equator (3, 6, 46). Since unaligned chromosomes were the first aberrations to appear after Sld5 depletion, we wanted to examine the role of these forces in spindle pole fragmentation. When Eg5 kinesin was inhibited after Sld5 depletion by incubation with monastrol, we observed that cells formed monopolar short spindles with closely spaced centrosomes, indicating that spindle pole fragmentation occurs after centrosome separation in Sld5-depleted cells (Fig. 10I and J). It has been reported that separase activity during a mitotic delay augments premature centriole disengagement, eventually progressing to supernumerary spindle poles (47, 48). Codepletion of separase did not prevent Sld5 depletion-induced spindle pole defects, ruling out its role in mediating the observed mitotic aberrations (Fig. 10K to M). Moreover, we observed that in Sld5-depleted cells, defects in chromosome congression occurred before spindle pole fragmentation, which is unlike cohesion fatigue, where centriole splitting precedes other aberrations. Thus, spindle pole fragmentation due to Sld5 depletion is preceded by the presence of unaligned chromosomes.
Sld5-depleted centrosomes fragment due to the traction forces generated by kinesin-7 family motor proteins, as well as chromokinesins.
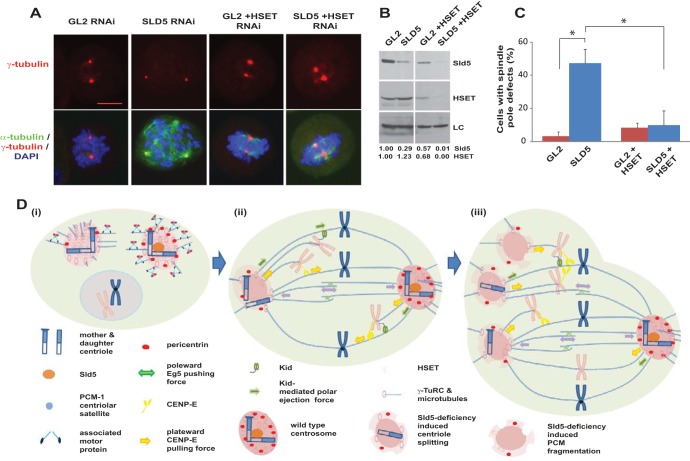
Since misaligned chromosomes appeared before spindle pole fragmentation in Sld5-depleted cells, we hypothesized that the congression forces might have a role in the observed aberrations. CENP-E is a plus-end-directed kinesin-7 family motor protein, and its depletion led to congression defects but not spindle pole fragmentation (Fig. 11A to C) (49). Codepletion of CENP-E suppressed the spindle pole aberrations caused by Sld5 depletion (Fig. 11D to H). There was also a restoration in the levels of γ-tubulin, Cep170, and pericentrin at the spindle poles, indicating that the loss of centrosomal proteins in the absence of Sld5 is mediated by CENP-E (Fig. 11D and I). Since energy-driven chromosome movement is the only known activity of CENP-E, we concluded that the loss of spindle pole integrity upon Sld5 depletion was due to CENP-E-mediated forces. Next, we wanted to investigate the role of chromokinesins in the observed mitotic defects. RNAi-mediated depletion of Kid resulted in congression defects with multiple unaligned chromosomes due to loss of the natural congression process; however, the spindle pole fragmentation observed after Sld5 depletion was significantly suppressed (Fig. 11J to M). This indicates that depletion of Sld5 per se did not lead to loss of spindle pole integrity but that it occurred because Sld5-depleted centrosomes were unable to withstand the CENP-E- and Kid-mediated forces. A minus-end-directed kinesin-14 motor protein, HSET, binds to the centrosomal protein, Cep215, thereby mediating the association between centrosomes and spindle poles. We wanted to check if HSET had a role in mediating the microtubular traction forces that fragment centrosomes in the absence of Sld5. We observed that codepletion of HSET suppressed the spindle pole fragmentation due to Sld5 depletion (Fig. 12A to C). HSET depletion resulted in short spindles, a phenotype expected from the known functions of HSET (50). Thus, HSET anchors the microtubules at the centrosomes, sustaining the traction forces that mediate centrosomal fragmentation in Sld5-depleted cells.
FIG 11.
Sld5-depleted centrosomes fragment due to CENP-E- and Kid-mediated forces. (A) HeLa cells transfected with control GL2 or CENP-E siRNA were immunoblotted with anti-CENP-E antibody to confirm RNAi depletion. The numbers indicate levels of CENP-E protein relative to control GL2 siRNA-transfected cells. (B and C) GL2 or CENP-E siRNA-transfected samples were costained for α-tubulin (green) and γ-tubulin (red), whereas TOTO-3 was used to stain the nucleus (pseudocolor blue). The arrowheads point to unaligned chromosomes observed in CENP-E-depleted samples. Quantification of the congression defects is shown in panel C. (D) HeLa cells were transfected with control GL2, SLD5, or CENP-E siRNA as indicated, with the combined concentration brought to 80 nM with GL2 siRNA. The cells were fixed and costained for α-tubulin (green) and γ-tubulin (red), whereas TOTO-3 was used to stain the nucleus (pseudocolor blue). (E) The transfected cells were immunoblotted with anti-Sld5 antibody to confirm RNAi depletion. The numbers indicate levels of Sld5 relative to control GL2 siRNA-transfected cells. (F) Decrease of CENP-E mRNA confirmed by reverse transcriptase PCR. The numbers indicate the CENP-E mRNA levels following siRNA-mediated depletion relative to control GL2 siRNA-transfected cells. BMG served as the internal RNA-loading control. (G and H) HeLa cells transfected with control GL2, SLD5, or CENP-E siRNA, as described for panel D, were stained with anti-CENP-E antibody to confirm RNAi depletion. Quantification of the spindle pole defects is shown in panel H. (I) HeLa cells transfected with different siRNAs, as indicated, were surveyed to identify mitotic cells with low levels of Cep170 or pericentrin staining, which was confirmed by NIS Elements software for multiple examples to be less than 50% of the mean intensity of the signal observed in control cells. (J and K) HeLa cells transfected with control GL2, SLD5, or KID siRNA, as indicated, with the combined concentration brought to 80 nM with GL2 siRNA. The cells were fixed and costained for α-tubulin (green) and γ-tubulin (red) and for DNA with DAPI (blue). Note that inhibition of KID activity resulted in multiple chromosome congression defects. Quantification of the spindle pole defects is shown in panel K. (L) The transfected samples were immunoblotted with anti-Sld5 antibody to confirm RNAi depletion. The numbers indicate levels of Sld5 protein in different samples relative to control GL2 siRNA-transfected cells. (M) Decrease of KID mRNA confirmed by reverse transcriptase PCR. The numbers indicate the KID mRNA levels following siRNA-mediated depletion relative to control GL2-transfected cells. BMG served as the internal RNA-loading control. Quantification of data is represented as the means and SD of the results of two independent experiments, with more than 20 cells analyzed in each sample. *, P < 0.05. LC, loading control showing equal protein loads in different lanes. Scale bars, 10 μm.
FIG 12.
Inhibition of HSET prevents spindle pole fragmentation in Sld5-depleted cells. (A) HeLa cells transfected with control GL2, SLD5, or HSET siRNA, as indicated, with the combined concentration brought to 80 nM with GL2 siRNA. The cells were fixed and costained for α-tubulin (green) and γ-tubulin (red) and for DNA with DAPI (blue). Note that HSET depletion resulted in shortened or asymmetric spindle poles. Scale bar, 10 μm. (B) The transfected samples were immunoblotted with anti-Sld5 and anti-HSET antibodies to confirm RNAi depletion. LC, loading control showing equal protein loads in different lanes; the numbers indicate levels of Sld5 and HSET proteins in different samples relative to control GL2 siRNA-transfected cells. (C) Quantification of the spindle pole defects shown in panel A, represented as the means and SD of the results of three independent experiments, with more than 20 cells analyzed in each sample (*, P < 0.05). (D) Schematic representation of spindle pole fragmentation in the absence of Sld5. (i) Depletion of Sld5 leads to the dissipation of centriolar satellite protein PCM-1 in interphase cells, resulting in the loss of its known function of recruiting the centrosomal protein pericentrin. (ii and iii) During alignment of chromosomes in prometaphase, kinesin, CENP-E, and the chromokinesin Kid cause the unaligned chromosomes to migrate away from the spindle poles, exerting a significant pulling force on the centrosomes. While a wild-type centrosome (shown as the right pole) is able to resist this force, a Sld5-deficient centrosome (shown as the left pole) splits, forming monocentriolar or acentriolar spindle poles, as depicted in diagram iii. The minus-end-directed kinesin-14 motor protein, HSET, anchors the microtubules at the centrosomes.
DISCUSSION
Previous studies have demonstrated that DNA replication proteins localize to the centrosomes and their depletion results in spindle pole aberrations. However, their physiological role in maintaining centrosomal homeostasis has remained ambiguous (23, 24, 27). We report that Sld5 ensures centrosomal resistance to microtubule-mediated forces during chromosome alignment. Depletion of Sld5 results in the dissipation of centriolar satellites, leading to the loss of their known function of recruiting the centrosomal protein pericentrin. This results in a “weakened” centrosome that is stable during interphase but is unable to resist the pulling forces generated by CENP-E and Kid during mitosis, culminating in centriole splitting and PCM fragmentation (a model is shown in Fig. 12D). Previous reports have shown that centrosomal proteins withstand the microtubule-mediated forces, thereby preserving spindle pole integrity, but such a role for a DNA replication protein has not been reported (15–17).
Live-cell imaging of Sld5-depleted cells demonstrated that most of the mitotic cells were arrested at the prometaphase stage with the formation of disorganized spindles. Cells with multipolar spindles formed after Sld5 depletion are unlikely to exit mitosis and produce viable progeny, exemplifying the interlinking of chromosome duplication and segregation to prevent propagation of aberrant cells (51). Progression to cancer is accompanied by counteracting protective mechanisms, such as clustering of supernumerary centrosomes to assemble pseudobipolar spindles (52). Though our study emphasizes the essential requirement for Sld5 in preventing spindle pole defects, its role in preserving centrosomal integrity is yet to be established. We have observed that in the absence of Sld5, the levels of pericentrin are reduced at the interphase centrosomes, and therefore, one of the possibilities is that Sld5 is required for the recruitment of PCM proteins, in the absence of which the microtubule-mediated forces fragment the centrosomes in mitosis. Analyzing the changes in the pericentriolar matrix after depletion of Sld5 would aid in understanding its role in the maintenance of spindle pole integrity.
Centriolar satellites are present only in interphase cells and diminish when cells enter mitosis (53). PCM-1 resides in the centriolar satellite, which clusters around the centrosomes and plays a key role in maintaining the composition of centrosomal proteins. It has been reported that inhibition of PCM-1 by different approaches, such as microinjection of antibodies, expression of a PCM-1 deletion mutant, and depletion by siRNA, results in reduced levels of pericentrin at the interphase centrosomes (41). In contrast to unperturbed cells, where PCM-1 displays typical dense pericentrosomal localization, we observed that Sld5 depletion leads to dispersion of PCM-1 throughout the cytoplasm in interphase cells. We have also shown that Sld5 depletion results in decreased levels of pericentrin at the interphase centrosomes. We propose that Sld5 depletion-triggered changes in PCM-1 cause a reduction of pericentrin at the centrosomes. Previous studies have shown that pericentrin loss is observed after inhibition of both dynactin and PCM-1, and therefore, it has been proposed that PCM-1 has a role in the microtubule-dependent recruitment of pericentrin (41). We believe that loss of PCM-1-mediated pericentrin recruitment to the centrosomes observed after Sld5 depletion results in a centrosomal structure that is susceptible to the microtubule-mediated traction forces prevalent during mitosis.
Emerging evidence suggests that centriolar satellites serve as a platform for dynamic regulation of centrosomes in response to a variety of cues. Centriolar satellites undergo remodeling during mitosis that results in their dissolution and redistribution to either the centrosome or cytoplasm (19). Recent studies have demonstrated that cellular stresses, such as UV radiation, induce rapid removal of a subset of centriolar satellite proteins, including PCM-1 (54). It seems that Sld5 depletion at the centrosomes results in loss of centriolar satellites, thereby triggering a sequence of events in mitosis that culminates in centrosomal fragmentation. Though Sld5 depletion led to loss of pericentrin at the interphase centrosomes, we did not observe a similar decrease of γ-tubulin during interphase. Previous reports also indicated that upon PCM-1 depletion, γ-tubulin levels remain largely unaltered during interphase (41). Though it has been reported that depletion of PCM-1 inhibits microtubule anchoring to the centrosomes, pericentrin depletion per se had no significant effect on microtubule organization. In the case of Sld5 depletion, it appears that dispersion of PCM-1 leads primarily to the loss of pericentrin without altering microtubular forces significantly.
The GINS complex has a well-described function in DNA replication. It associates with Cdc45 and the MCMs to form the CMG complex, which functions as the replicative helicase (55). Targeted disruption of Sld5 in mice results in lethality at the peri-implantation stage, indicating that Sld5 is essential for embryogenesis. Since Sld5 is a part of the four-subunit complex, it raises the prospect of the entire GINS complex being required for maintaining centrosome integrity. There have been conflicting reports on the effect of GINS depletion on mitotic progression. It has been previously observed that ectopically expressed Psf2, a partner protein of Sld5 in the GINS complex, localizes to the mitotic spindles and that its depletion causes chromosome misalignment, indicating an unknown function in mitosis (55, 56). Similarly, mutations in Drosophila Sld5 led to an M phase delay with the appearance of polycentric chromosomes and chromosome breakage (30). However, Psf2 depletion in untransformed human dermal fibroblasts did not alter mitotic exit, suggesting a differential requirement in untransformed cells (57). We did not observe mitotic aberrations after Psf2 depletion, implying that the centrosomal stability is a function unique to Sld5 rather than the entire GINS complex, though it is possible that the differences in the observations were due to varying efficiencies of RNAi depletion.
It has been reported that depletion of Sld5 leads to accumulation at the G1/S transition, resulting in decreased cell proliferation (36). However, in our study, the Sld5-depleted cells showed accumulation in the S phase, indicating that the cells move into the S phase with low levels of Sld5 and take longer to complete replication, reflecting its function in DNA replication elongation. The differences from the previous study with respect to the cell cycle phase in which the cells primarily accumulate are possibly due to RNAi efficiencies. While in our study it appears that a minimum threshold of Sld5 was retained to allow cells to enter the S phase, on the other hand, the depletion of Sld5 to less than 10% in the previous study led to G1/S accumulation. The decrease of DNA replication proteins below a critical threshold causes the replication fork to collapse, resulting in DNA damage; however, unlike the previous study, we did not observe activation of the DNA damage checkpoint after Sld5 depletion, indicating that fork collapse was largely averted. It seems that the hypomorphic effect of SLD5 depletion aided in the discovery of an unknown mitotic function. The discovery of a mitotic function of Sld5 raises the question of what the possible advantage could be for mammalian cells to assign dual functions to a DNA replication protein. The consequences of a decrease in the levels of replication proteins for mammalian cell proliferation have been paradoxical. A decrease of up to 95% in the levels of MCMs did not stall replication but resulted in chromosomal instability, such as breaks, gaps, and aberrant rearrangements (58). It is possible that in the absence of a mitotic block, the Sld5-depleted cells would have continued proliferation and incorporated DNA lesions in successive replication cycles. Therefore, by inherently requiring a minimal level of Sld5 for mitotic progression, the cellular protective machinery may have devised a mechanism to prevent aberrant cells from progressing into the next cell cycle. One would have to override the Sld5 depletion-induced M phase block to test such a hypothesis. In summary, we conclude that the DNA replication factor, Sld5, ensures resistance to CENP-E- and Kid-mediated traction forces exerted during chromosome congression, thereby preserving spindle pole integrity.
MATERIALS AND METHODS
Fixed-cell and time-lapse imaging.
HeLa cells were obtained from the cell repository of the National Centre for Cell Science, Pune, India, and authenticated by short-tandem-repeat (STR) analysis, which showed a 94% match with the ATCC STR database. HeLa cells stably coexpressing mCherry-tagged histone H2B and mEGFP-tagged α-tubulin were obtained from Daniel Gerlich (Institute of Molecular Biotechnology, Vienna, Austria). HeLa cells stably coexpressing EGFP–centrin-2 and α-tubulin–m-RFP were obtained from Patrick Meraldi (University of Geneva, Geneva, Switzerland). For live-cell imaging, HeLa cells stably coexpressing mCherry-tagged histone H2B and mEGFP-tagged α-tubulin were transfected with specific siRNAs, followed by live-cell imaging. The cells were placed on a live-cell imaging stage (37°C with 5% CO2), and images were captured at 10-min intervals with a 63×/1.4-numerical-aperture objective of a Zeiss Observer Z1 inverted fluorescence microscope using a Zeiss LSM 510 confocal laser scanning microscope. Fluorophores, mEGFP and mCherry, were excited with a 488-nm laser line (from an argon-krypton laser) and a 543-nm laser line (with a helium-neon laser), and emissions were detected using a 505- to 550-nm band-pass filter and a 560-nm long-pass filter, respectively. Similarly, for fixed HeLa cells, the fluorophores Alexa Fluor 488, Alexa Fluor 555, and 4′,6-diamidino-2-phenylindole (DAPI) were excited with 488-nm, 543-nm, and tunable multiphoton laser lines, respectively.
Immunoblotting and immunofluorescence.
For Western blotting, the whole-cell lysates from cells of almost equal confluence were prepared in a proportionate volume of Laemmli buffer and denatured at 95°C, followed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). The gel was transferred onto a nitrocellulose membrane blocked with 3% bovine serum albumin (BSA) prepared in 1× Tris-buffered saline with Tween 20 (TBST). The membrane was then incubated with the appropriate antibody, washed, and probed with horseradish peroxidase (HRP)-conjugated secondary antibody. Enhanced chemiluminescence was used to visualize the protein bands. For indirect immunofluorescence of α-tubulin, γ-tubulin, pericentrin, PCM-1, TACC3, and Cep170, HeLa cells grown on coverslips were fixed with 4% formaldehyde for 10 min, followed by fixation with ice-cold methanol for 5 min. For visualization of centrioles with an anti-centrin-2 antibody, the cells were prepermeabilized in extraction buffer (80 mM PIPES, 1 mM MgCl2, 1 mM EGTA, and 0.5% Triton X-100) for 2 min, followed by fixation with ice-cold methanol for 2 min. To visualize kinetochores with anti-CENP-E antibody, the cells were fixed with ice-cold methanol for 10 min. Samples were blocked with 3% bovine serum albumin or 10% fetal bovine serum (FBS) for 30 min and then stained with specific primary antibody for 1 h, followed by incubation with secondary antibody for 1 h. Finally, the cells were visualized under the microscope after mounting with Vectashield mounting reagent containing DAPI, which stains the nucleus. The secondary antibodies used were conjugated to Alexa Fluor 488 or Alexa Fluor 555 and were purchased from Invitrogen. We assayed the intensity of the Alexa Fluor 488 or Alexa Fluor 555 signal with NIS Elements imaging software (version 3.22.00) for quantification of the levels of γ-tubulin, centrin-2, Cep170, and pericentrin signals at the spindle poles. We took multiple examples from each sample and confirmed that the mean intensity of Alexa Fluor signal for spindle poles marked as low intensity was less than 50% of the average mean intensity of the signal observed in control transfected cells.
Antibodies.
Anti-Sld5 antibody (Ab1) was produced using recombinant His-tagged Sld5 [cloned in the pET28a (+) vector] and purified on a nickel-nitrilotriacetic acid (NTA) column (Qiagen). Rabbits were injected intramuscularly with recombinant protein, along with complete Freund's adjuvant, followed by a second injection of Sld5 protein mixed with incomplete Freund's adjuvant after 2 weeks, and the blood was collected after 30 days. Antibody Ab2, raised against amino acids 1 to 198 of human Sld5, was procured from Abcam (catalog number ab101346), while antibody Ab3, raised against amino acids 1 to 63 of human Sld5, was procured from Sigma (catalog number HPA024663). The following antibodies were used for Western blotting. Antibodies against Sld5, Orc3, Rfc2, CENP-E, HSET, and pericentrin were purchased from Abcam. Anti-β-actin and anti-phospho-γ-H2AX (Ser 139) were obtained from Santa Cruz Biotechnology. Anti-phospho-histone H3 (Ser 10), anti-phospho-Chk1 (Ser 317), anti-phospho-Chk2 (Thr 68), anti-PCNA, and anti-Myc were procured from Cell Signaling Technology. Antibody against hemagglutinin (HA) tag was procured from Sigma. For indirect immunofluorescence assays, the following antibodies were used. Antibodies against γ-tubulin, α-tubulin, pericentrin, Cep170, CENP-E, and TACC3 were obtained from Abcam. Anti-centrin-2 antibody was procured from Santa Cruz Biotechnology. Anti-PCM-1 antibody was procured from Cell Signaling Technology. The secondary antibodies used were conjugated to Alexa Fluor 488, Alexa Fluor 555, or Alexa Fluor 594 and were purchased from Invitrogen.
Cell culture, cell cycle analysis, drug treatment, and cloning.
Cell lines were maintained at 37°C in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% FBS and 1% antibiotic and antimycotic solution. The HeLa cell lines stably expressing H2B-mCherry and EGFP–α-tubulin were grown in DMEM with selective antibiotics (puromycin and G418; Sigma). For cell cycle analysis, the cells were fixed with 70% ethanol after washing with 1× phosphate-buffered saline (PBS). After fixation, the cell pellet was resuspended in 1× PBS with 0.1% Triton X-100, 20 μg/ml RNase A, and 70 μg/ml propidium iodide, and the stained cells were analyzed by flow cytometry. The flow cytometry data were acquired on a Becton Dickinson FACSCalibur machine using CellQuest Pro software. The cell cycle analysis was done by the Dean-Jett-Fox method. The fitting models of G0/G1, G2/M, and S phase populations have been described in the FlowJo software. Quantity One software (version 4.6.3; Bio-Rad) was utilized to evaluate the levels of specific proteins, which were expressed after normalization with the protein-loading control. The results are presented as means and standard deviations (SD), and Student's t test was used for statistical analysis. A P value of less than 0.05 was considered significant unless otherwise indicated. A significant difference in spindle pole defects was calculated by one-way analysis of variance (ANOVA), using a threshold of an F distribution cumulative probability of 0.001. For UV irradiation, the medium was removed and the cells were washed with 1× PBS, followed by exposure to 100 J/m2 UV radiation in uncovered dishes with UV-C using a UV cross-linker (CL-1000; UVP). For inhibition of Eg5, HeLa cells after RNAi were treated with 100 μM monastrol (Sigma) for 4 h, followed by fixation for indirect immunofluorescence assay. For cloning of human Sld5 in the pET28a (+) vector, Sld5 was cloned using NheI and NotI restriction sites. Full-length Sld5 cDNA was amplified by PCR and subcloned into a cytomegalovirus (CMV) promoter-driven plasmid, pCDNA3, which carries a sequence encoding the hemagglutinin tag at the N terminus of the insert. Similarly, Sld5 cDNA was cloned into a murine leukemia virus long terminal repeat (LTR)-driven plasmid, pMX-puro-3NLS-GST-HA. For expression of Sld5, HeLa cells were transfected with pCDNA3-SLD5 and harvested 48 h later. 293T cells were transfected with pMX-puro-SLD5, along with helper plasmids (which express the viral vesicular stomatitis virus G [VSV-G] envelope protein, as well as the Gag and Pol proteins) to generate viral particles. To obtain a stable HeLa cell line expressing Sld5, HeLa cells were infected with the viral particles and selected with 1 μg/ml puromycin after 48 h.
RNAi silencing, cell synchronization, and reverse transcriptase PCR.
Transfection of small interfering RNA targeting the endogenous genes was carried out using Lipofectamine 2000 (Invitrogen). Specific siRNA duplexes (40 to 80 nM) were transfected on three consecutive days, and the cells were harvested 24 h after the last transfection for immunoblotting or reverse transcriptase PCR. For reverse transcriptase PCR, RNA was isolated using the standard TRIzol method, and 1 μg of RNA was used for cDNA synthesis. Details of the protocols and DNA primers used are available on request. The siRNA sequences used for targeting the human genes were as follows: GL2, CGUACGCGGAAUACUUCGA; SLD 5(1), AAAUGGAGAUGGAGAGGAU; SLD5(2), GCUGGAGAGCAAGCCUGAGAUUGUA; PSF2, GCGAUUAACCUGAAACAAA; PSF3, AGAAAGGACUGAAUGACUU; ORC3, CCCCAAACAGGCAUGUAUC; PCNA, GGAUCUUAGGCAUUCUUAA; RFC2, CUAGAGGUCUUUGCAAGGG; CENP-E(1), AAACACUUACUGCUCUCCAGU; CENP-E(2), ACUCUUACUGCUCUCCAGU; NS2, UAAGGCUAUGAAGAGAUAC; SEPARASE, GCGGAAUGCUGCUGCACAA; KID, AAGAUUGGAGCUACUCGUCGU; HSET, UCAGAAGCAGCCCUGUCAA.
Supplementary Material
ACKNOWLEDGMENTS
We thank Ritu Shekhar, Sheetal Uppal, Nasir Imam, and Sunder Bisht for helping with various parts of the manuscript. We thank Daniel Gerlich and Patrick Meraldi for the stable cell lines.
This study was supported by sanctioned projects from DBT and DST, Government of India, and a NET fellowship to M.K.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/MCB.00371-17.
REFERENCES
- 1.Wood KW, Sakowicz R, Goldstein LS, Cleveland DW. 1997. CENP-E is a plus end-directed kinetochore motor required for metaphase chromosome alignment. Cell 91:357–366. doi: 10.1016/S0092-8674(00)80419-5. [DOI] [PubMed] [Google Scholar]
- 2.Goshima G, Vale RD. 2003. The roles of microtubule-based motor proteins in mitosis: comprehensive RNAi analysis in the Drosophila S2 cell line. J Cell Biol 162:1003–1016. doi: 10.1083/jcb.200303022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kops GJ, Saurin AT, Meraldi P. 2010. Finding the middle ground: how kinetochores power chromosome congression. Cell Mol Life Sci 67:2145–2161. doi: 10.1007/s00018-010-0321-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Walczak CE, Cai S, Khodjakov A. 2010. Mechanisms of chromosome behaviour during mitosis. Nat Rev Mol Cell Biol 11:91–102. doi: 10.1038/nrm2832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Skibbens RV, Skeen VP, Salmon ED. 1993. Directional instability of kinetochore motility during chromosome congression and segregation in mitotic newt lung cells: a push-pull mechanism. J Cell Biol 122:859–875. doi: 10.1083/jcb.122.4.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kapoor TM, Lampson MA, Hergert P, Cameron L, Cimini D, Salmon ED, McEwen BF, Khodjakov A. 2006. Chromosomes can congress to the metaphase plate before biorientation. Science 311:388–391. doi: 10.1126/science.1122142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Su X, Arellano-Santoyo H, Portran D, Gaillard J, Vantard M, Thery M, Pellman D. 2013. Microtubule-sliding activity of a kinesin-8 promotes spindle assembly and spindle-length control. Nat Cell Biol 15:948–957. doi: 10.1038/ncb2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rieder CL, Salmon ED. 1994. Motile kinetochores and polar ejection forces dictate chromosome position on the vertebrate mitotic spindle. J Cell Biol 124:223–233. doi: 10.1083/jcb.124.3.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wandke C, Barisic M, Sigl R, Rauch V, Wolf F, Amaro AC, Tan CH, Pereira AJ, Kutay U, Maiato H, Meraldi P, Geley S. 2012. Human chromokinesins promote chromosome congression and spindle microtubule dynamics during mitosis. J Cell Biol 198:847–863. doi: 10.1083/jcb.201110060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Iemura K, Tanaka K. 2015. Chromokinesin Kid and kinetochore kinesin CENP-E differentially support chromosome congression without end-on attachment to microtubules. Nat Commun 6:6447. doi: 10.1038/ncomms7447. [DOI] [PubMed] [Google Scholar]
- 11.Shrestha RL, Draviam VM. 2013. Lateral to end-on conversion of chromosome-microtubule attachment requires kinesins CENP-E and MCAK. Curr Biol 23:1514–1526. doi: 10.1016/j.cub.2013.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cai S, O'Connell CB, Khodjakov A, Walczak CE. 2009. Chromosome congression in the absence of kinetochore fibres. Nat Cell Biol 11:832–838. doi: 10.1038/ncb1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Levesque AA, Compton DA. 2001. The chromokinesin Kid is necessary for chromosome arm orientation and oscillation, but not congression, on mitotic spindles. J Cell Biol 154:1135–1146. doi: 10.1083/jcb.200106093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McDonald HB, Stewart RJ, Goldstein LS. 1990. The kinesin-like ncd protein of Drosophila is a minus end-directed microtubule motor. Cell 63:1159–1165. doi: 10.1016/0092-8674(90)90412-8. [DOI] [PubMed] [Google Scholar]
- 15.Oshimori N, Ohsugi M, Yamamoto T. 2006. The Plk1 target Kizuna stabilizes mitotic centrosomes to ensure spindle bipolarity. Nat Cell Biol 8:1095–1101. doi: 10.1038/ncb1474. [DOI] [PubMed] [Google Scholar]
- 16.Logarinho E, Maffini S, Barisic M, Marques A, Toso A, Meraldi P, Maiato H. 2012. CLASPs prevent irreversible multipolarity by ensuring spindle-pole resistance to traction forces during chromosome alignment. Nat Cell Biol 14:295–303. doi: 10.1038/ncb2423. [DOI] [PubMed] [Google Scholar]
- 17.Kim K, Rhee K. 2011. The pericentriolar satellite protein CEP90 is crucial for integrity of the mitotic spindle pole. J Cell Sci 124:338–347. doi: 10.1242/jcs.078329. [DOI] [PubMed] [Google Scholar]
- 18.Lopes CA, Prosser SL, Romio L, Hirst RA, O'Callaghan C, Woolf AS, Fry AM. 2011. Centriolar satellites are assembly points for proteins implicated in human ciliopathies, including oral-facial-digital syndrome 1. J Cell Sci 124:600–612. doi: 10.1242/jcs.077156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kubo A, Sasaki H, Yuba-Kubo A, Tsukita S, Shiina N. 1999. Centriolar satellites: molecular characterization, ATP-dependent movement toward centrioles and possible involvement in ciliogenesis. J Cell Biol 147:969–980. doi: 10.1083/jcb.147.5.969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dmowski M, Fijalkowska IJ. 2017. Diverse roles of Dpb2, the non-catalytic subunit of DNA polymerase epsilon. Curr Genet 63:983–987. doi: 10.1007/s00294-017-0706-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zou L, Elledge SJ. 2003. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science 300:1542–1548. doi: 10.1126/science.1083430. [DOI] [PubMed] [Google Scholar]
- 22.Knockleby J, Lee H. 2010. Same partners, different dance: involvement of DNA replication proteins in centrosome regulation. Cell Cycle 9:4487–4491. doi: 10.4161/cc.9.22.14047. [DOI] [PubMed] [Google Scholar]
- 23.Prasanth SG, Prasanth KV, Siddiqui K, Spector DL, Stillman B. 2004. Human Orc2 localizes to centrosomes, centromeres and heterochromatin during chromosome inheritance. EMBO J 23:2651–2663. doi: 10.1038/sj.emboj.7600255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ferguson RL, Maller JL. 2008. Cyclin E-dependent localization of MCM5 regulates centrosome duplication. J Cell Sci 121:3224–3232. doi: 10.1242/jcs.034702. [DOI] [PubMed] [Google Scholar]
- 25.Ferguson RL, Pascreau G, Maller JL. 2010. The cyclin A centrosomal localization sequence recruits MCM5 and Orc1 to regulate centrosome reduplication. J Cell Sci 123:2743–2749. doi: 10.1242/jcs.073098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Prasanth SG, Prasanth KV, Stillman B. 2002. Orc6 involved in DNA replication, chromosome segregation, and cytokinesis. Science 297:1026–1031. doi: 10.1126/science.1072802. [DOI] [PubMed] [Google Scholar]
- 27.Hemerly AS, Prasanth SG, Siddiqui K, Stillman B. 2009. Orc1 controls centriole and centrosome copy number in human cells. Science 323:789–793. doi: 10.1126/science.1166745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chang YP, Wang G, Bermudez V, Hurwitz J, Chen XS. 2007. Crystal structure of the GINS complex and functional insights into its role in DNA replication. Proc Natl Acad Sci U S A 104:12685–12690. doi: 10.1073/pnas.0705558104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kamada K, Kubota Y, Arata T, Shindo Y, Hanaoka F. 2007. Structure of the human GINS complex and its assembly and functional interface in replication initiation. Nat Struct Mol Biol 14:388–396. doi: 10.1038/nsmb1231. [DOI] [PubMed] [Google Scholar]
- 30.Gouge CA, Christensen TW. 2010. Drosophila Sld5 is essential for normal cell cycle progression and maintenance of genomic integrity. Biochem Biophys Res Commun 400:145–150. doi: 10.1016/j.bbrc.2010.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vaziri C, Saxena S, Jeon Y, Lee C, Murata K, Machida Y, Wagle N, Hwang DS, Dutta A. 2003. A p53-dependent checkpoint pathway prevents rereplication. Mol Cell 11:997–1008. doi: 10.1016/S1097-2765(03)00099-6. [DOI] [PubMed] [Google Scholar]
- 32.Liu E, Lee AY, Chiba T, Olson E, Sun P, Wu X. 2007. The ATR-mediated S phase checkpoint prevents rereplication in mammalian cells when licensing control is disrupted. J Cell Biol 179:643–657. doi: 10.1083/jcb.200704138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tatsumi Y, Sugimoto N, Yugawa T, Narisawa-Saito M, Kiyono T, Fujita M. 2006. Deregulation of Cdt1 induces chromosomal damage without rereplication and leads to chromosomal instability. J Cell Sci 119:3128–3140. doi: 10.1242/jcs.03031. [DOI] [PubMed] [Google Scholar]
- 34.Canitrot Y, Frechet M, Servant L, Cazaux C, Hoffmann JS. 1999. Overexpression of DNA polymerase beta: a genomic instability enhancer process. FASEB J 13:1107–1111. [DOI] [PubMed] [Google Scholar]
- 35.Chan K, Houlbrook S, Zhang QM, Harrison M, Hickson ID, Dianov GL. 2007. Overexpression of DNA polymerase beta results in an increased rate of frameshift mutations during base excision repair. Mutagenesis 22:183–188. doi: 10.1093/mutage/gel070. [DOI] [PubMed] [Google Scholar]
- 36.Aparicio T, Guillou E, Coloma J, Montoya G, Mendez J. 2009. The human GINS complex associates with Cdc45 and MCM and is essential for DNA replication. Nucleic Acids Res 37:2087–2095. doi: 10.1093/nar/gkp065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Borel F, Lohez OD, Lacroix FB, Margolis RL. 2002. Multiple centrosomes arise from tetraploidy checkpoint failure and mitotic centrosome clusters in p53 and RB pocket protein-compromised cells. Proc Natl Acad Sci U S A 99:9819–9824. doi: 10.1073/pnas.152205299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dodson H, Bourke E, Jeffers LJ, Vagnarelli P, Sonoda E, Takeda S, Earnshaw WC, Merdes A, Morrison C. 2004. Centrosome amplification induced by DNA damage occurs during a prolonged G2 phase and involves ATM. EMBO J 23:3864–3873. doi: 10.1038/sj.emboj.7600393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Inanc B, Dodson H, Morrison CG. 2010. A centrosome-autonomous signal that involves centriole disengagement permits centrosome duplication in G2 phase after DNA damage. Mol Biol Cell 21:3866–3877. doi: 10.1091/mbc.E10-02-0124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hut HM, Lemstra W, Blaauw EH, Van Cappellen GW, Kampinga HH, Sibon OC. 2003. Centrosomes split in the presence of impaired DNA integrity during mitosis. Mol Biol Cell 14:1993–2004. doi: 10.1091/mbc.E02-08-0510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dammermann A, Merdes A. 2002. Assembly of centrosomal proteins and microtubule organization depends on PCM-1. J Cell Biol 159:255–266. doi: 10.1083/jcb.200204023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee MJ, Gergely F, Jeffers K, Peak-Chew SY, Raff JW. 2001. Msps/XMAP215 interacts with the centrosomal protein D-TACC to regulate microtubule behaviour. Nat Cell Biol 3:643–649. doi: 10.1038/35083033. [DOI] [PubMed] [Google Scholar]
- 43.Guarguaglini G, Duncan PI, Stierhof YD, Holmstrom T, Duensing S, Nigg EA. 2005. The forkhead-associated domain protein Cep170 interacts with Polo-like kinase 1 and serves as a marker for mature centrioles. Mol Biol Cell 16:1095–1107. doi: 10.1091/mbc.E04-10-0939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ishikawa H, Kubo A, Tsukita S, Tsukita S. 2005. Odf2-deficient mother centrioles lack distal/subdistal appendages and the ability to generate primary cilia. Nat Cell Biol 7:517–524. doi: 10.1038/ncb1251. [DOI] [PubMed] [Google Scholar]
- 45.Lawo S, Hasegan M, Gupta GD, Pelletier L. 2012. Subdiffraction imaging of centrosomes reveals higher-order organizational features of pericentriolar material. Nat Cell Biol 14:1148–1158. doi: 10.1038/ncb2591. [DOI] [PubMed] [Google Scholar]
- 46.Sharp DJ, Rogers GC, Scholey JM. 2000. Microtubule motors in mitosis. Nature 407:41–47. doi: 10.1038/35024000. [DOI] [PubMed] [Google Scholar]
- 47.Gorbsky GJ. 2013. Cohesion fatigue. Curr Biol 23:R986–R988. doi: 10.1016/j.cub.2013.08.017. [DOI] [PubMed] [Google Scholar]
- 48.Agircan FG, Schiebel E. 2014. Sensors at centrosomes reveal determinants of local separase activity. PLoS Genet 10:e1004672. doi: 10.1371/journal.pgen.1004672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yao X, Abrieu A, Zheng Y, Sullivan KF, Cleveland DW. 2000. CENP-E forms a link between attachment of spindle microtubules to kinetochores and the mitotic checkpoint. Nat Cell Biol 2:484–491. doi: 10.1038/35019518. [DOI] [PubMed] [Google Scholar]
- 50.Cai S, Weaver LN, Ems-McClung SC, Walczak CE. 2009. Kinesin-14 family proteins HSET/XCTK2 control spindle length by cross-linking and sliding microtubules. Mol Biol Cell 20:1348–1359. doi: 10.1091/mbc.E08-09-0971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ganem NJ, Godinho SA, Pellman D. 2009. A mechanism linking extra centrosomes to chromosomal instability. Nature 460:278–282. doi: 10.1038/nature08136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brinkley BR, Cox SM, Pepper DA, Wible L, Brenner SL, Pardue RL. 1981. Tubulin assembly sites and the organization of cytoplasmic microtubules in cultured mammalian cells. J Cell Biol 90:554–562. doi: 10.1083/jcb.90.3.554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kubo A, Tsukita S. 2003. Non-membranous granular organelle consisting of PCM-1: subcellular distribution and cell-cycle-dependent assembly/disassembly. J Cell Sci 116:919–928. doi: 10.1242/jcs.00282. [DOI] [PubMed] [Google Scholar]
- 54.Villumsen BH, Danielsen JR, Povlsen L, Sylvestersen KB, Merdes A, Beli P, Yang YG, Choudhary C, Nielsen ML, Mailand N, Bekker-Jensen S. 2013. A new cellular stress response that triggers centriolar satellite reorganization and ciliogenesis. EMBO J 32:3029–3040. doi: 10.1038/emboj.2013.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.MacNeill SA. 2010. Structure and function of the GINS complex, a key component of the eukaryotic replisome. Biochem J 425:489–500. doi: 10.1042/BJ20091531. [DOI] [PubMed] [Google Scholar]
- 56.Huang HK, Bailis JM, Leverson JD, Gomez EB, Forsburg SL, Hunter T. 2005. Suppressors of Bir1p (Survivin) identify roles for the chromosomal passenger protein Pic1p (INCENP) and the replication initiation factor Psf2p in chromosome segregation. Mol Cell Biol 25:9000–9015. doi: 10.1128/MCB.25.20.9000-9015.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Barkley LR, Song IY, Zou Y, Vaziri C. 2009. Reduced expression of GINS complex members induces hallmarks of pre-malignancy in primary untransformed human cells. Cell Cycle 8:1577–1588. doi: 10.4161/cc.8.10.8535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ibarra A, Schwob E, Mendez J. 2008. Excess MCM proteins protect human cells from replicative stress by licensing backup origins of replication. Proc Natl Acad Sci U S A 105:8956–8961. doi: 10.1073/pnas.0803978105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.