Specific changes in phospholipid content are a hallmark of the membranes of maturing endosomes and phagosomes, but is it unclear how this is controlled. Naufer et al. now show that acidification of the lumen of endosomes and phagosomes triggers dissociation of the Vps34 lipid kinase from these organelles, which terminates PtdIns(3)P synthesis and signaling.
Abstract
Phagocytosis of filamentous bacteria occurs through tubular phagocytic cups (tPCs) and takes many minutes to engulf these filaments into phagosomes. Contravening the canonical phagocytic pathway, tPCs mature by fusing with endosomes. Using this model, we observed the sequential recruitment of early and late endolysosomal markers to the elongating tPCs. Surprisingly, the regulatory early endosomal lipid phosphatidylinositol-3-phosphate (PtdIns(3)P) persists on tPCs as long as their luminal pH remains neutral. Interestingly, by manipulating cellular pH, we determined that PtdIns(3)P behaves similarly in canonical phagosomes as well as endosomes. We found that this is the product of a pH-based mechanism that induces the dissociation of the Vps34 class III phosphatidylinositol-3-kinase from these organelles as they acidify. The detachment of Vps34 stops the production of PtdIns(3)P, allowing for the turnover of this lipid by PIKfyve. Given that PtdIns(3)P-dependent signaling is important for multiple cellular pathways, this mechanism for pH-dependent regulation of Vps34 could be at the center of many PtdIns(3)P-dependent cellular processes.
Introduction
Phagocytosis is triggered when specialized receptors on the surface of phagocytes engage ligands on the target particle, activating the remodeling of the underlying actin cytoskeleton to extend membrane pseudopodia around the particle. This process culminates with the enclosure of the particle within a de novo organelle, the phagosome (Swanson, 2008; Flannagan et al., 2012; Gray and Botelho, 2017). Phagosomes then mature by sequentially fusing with and acquiring properties of early endosomes, late endosomes, and lysosomes (Fairn and Grinstein, 2012; Flannagan et al., 2012; Levin et al., 2015; Gray and Botelho, 2017). Hence, phagosomes transiently acquire molecular traits characteristic of early endosomes such as the GTPase Rab5 and phosphatidylinositol-3-phosphate (PtdIns[3]P), followed by the attainment of endolysosomal markers such as the GTPase Rab7 and the Lysosomal-associated membrane proteins (LAMPs; Via et al., 1997; Vieira et al., 2002, 2003; Harrison et al., 2003; Fairn and Grinstein, 2012; Flannagan et al., 2012; Gray and Botelho, 2017). The end product of maturation is the transformation of a nascent inert phagosome into a phagolysosome, where the sequestered particle is digested.
The transient nature of the early phagosome is a poorly characterized process. We do not understand the mechanisms that coordinate the timing of the association and dissociation of early endosomal factors with maturing phagosomes. For example, PtdIns(3)P synthesis, predominantly by class III phosphatidylinositol (PtdIns) 3-kinase Vps34, was recently shown to be coupled to the removal of phosphatidylinositol-4,5-bisphosphate (PtdIns[4,5]P2) from nascent phagosomes in Caenorhabditis elegans (Vieira et al., 2001; Thi and Reiner, 2012; Cheng et al., 2015; Backer, 2016). Yet it is unclear if this mechanism is present in mammalian phagocytes. Once synthesized, PtdIns(3)P typically lasts on phagosomes for only 5 to 10 min, during which it recruits various effectors to the phagosomal (and endosomal) membrane, including the early endosome antigen 1 (EEA1), which mediates phagosome fusion with early endosomes (Ellson et al., 2001; Fratti et al., 2001; Vieira et al., 2001). Subsequently, PtdIns(3)P is removed from maturing phagosomes through a process partly encoded by PtdIns(3)P itself, which recruits PIKfyve, a lipid kinase that converts PtdIns(3)P into phosphatidylinositol-3,5-bisphosphate (PtdIns[3,5]P2), a major regulator of lysosomes (Sbrissa et al., 1999, 2002; Ho et al., 2012). In fact, inhibition of PIKfyve delays phagosome divestment of PtdIns(3)P (Hazeki et al., 2012; Kim et al., 2014). PtdIns(3)P removal from phagosomes may also be catalyzed by myotubularins, and/or by inactivation and dissociation of Vps34 from membranes (Nandurkar and Huysmans, 2002; Robinson and Dixon, 2006). Nonetheless, it is unknown what governs the timing of PtdIns(3)P removal from phagosomes, or, for that matter, from endosomes.
Strikingly, the phagosome formation and maturation processes briefly summarized here are the product of studies using model targets, mainly latex beads and red blood cells (Champion et al., 2008). However, phagocytes encounter targets of disparate morphology and size, including parasites, molds, yeasts, bacteria, and abiotic targets (Doshi and Mitragotri, 2010; Paul et al., 2013). Targets of filamentous morphology can present a hurdle for phagocytosis. Indeed, some bacterial species adopt a filamentous morphology to evade phagocytosis (Justice et al., 2008; Yang et al., 2016), and macrophages fail to efficiently internalize filamentous targets when they are engaged by their long axis (Champion et al., 2008). Nevertheless, phagocytosis of filamentous bacteria proceeds successfully when macrophages capture and engulf filaments by one of their ends (Möller et al., 2012).
We have previously characterized the phagocytosis of filamentous Legionella (Prashar et al., 2013). Because of its length, which can easily surpass the length of the cell, Legionella filaments are pulled into the cell to form tubular phagocytic cups (tPCs) that often coil within the cytoplasm. Hence, full enclosure of the particle occurs over time periods that dramatically exceed those for the uptake of model spheroidal targets (Prashar et al., 2013). Strikingly, the pericytoplasmic portions of tPCs sequentially fuse with endosomes and lysosomes before sealing in a process that resembles the maturation of canonical phagosomes (Prashar et al., 2013). Thus, tPCs present an interesting model to investigate how phagocytosis and phagosome maturation are coordinated.
Here, we report a persistent association of PtdIns(3)P on tPCs despite a divestment of other early endosomal markers and acquisition of endolysosomal proteins, suggesting that signals that normally couple PtdIns(3)P removal with maturation fail on the tPCs. Notably, PtdIns(3)P was eventually eliminated by PIKfyve upon complete enclosure of the filamentous target or from the tPC regions that extended beyond a length of 20 µm, indicating that tPCs were competent for such a signal under specific conditions. Investigating the interplay between PtdIns(3)P loss and acidification in tPCs, canonical phagosomes, and endosomes, we found that acidification signals the cessation of PtdIns(3)P synthesis by triggering the dissociation of the Vps34 complex from membranes. Overall, we present evidence of a novel pH-based mechanism that modulates the association of Vps34 with endomembranes and thereby controls PtdIns(3)P synthesis.
Results
Noncanonical persistence of PtdIns(3)P on tPCs during maturation
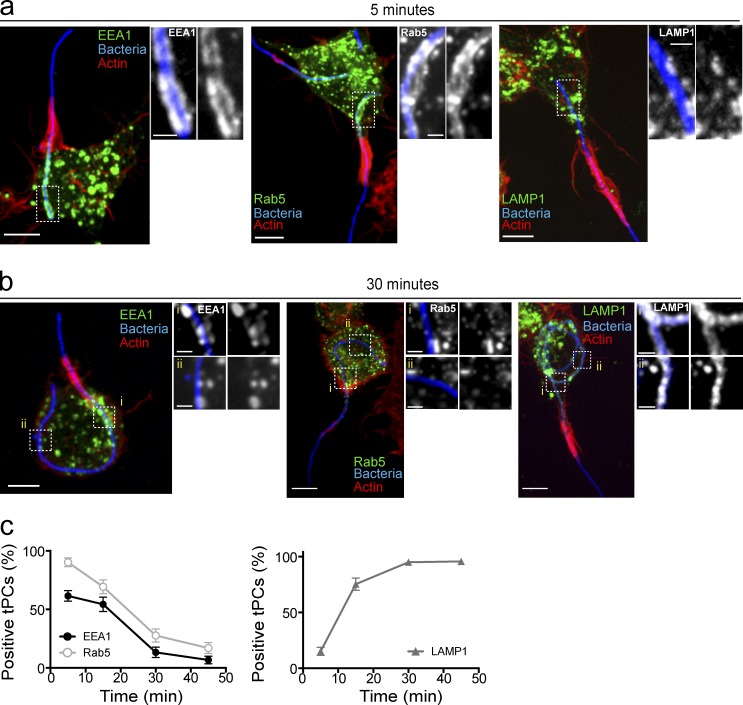
During canonical maturation, phagosomes transiently acquire the small GTPase Rab5 and PtdIns(3)P, enabling the recruitment of EEA1 to tether early endosomes and phagosomes to prime their fusion (Horiuchi et al., 1997; Simonsen et al., 1998; Christoforidis et al., 1999; Fratti et al., 2001; Vieira et al., 2003; Tian et al., 2008; Mishra et al., 2010). This is then followed by nearly simultaneous loss of Rab5, PtdIns(3)P, and EEA1 from phagosomes and attainment of endolysosome factors such as the LAMP proteins (Pitt et al., 1992; Horiuchi et al., 1997; Fratti et al., 2001; Vieira et al., 2001, 2003; Harrison et al., 2003; Huynh et al., 2007; Fairn and Grinstein, 2012). Thus, we examined the dynamics of these maturation markers during phagocytosis of filamentous bacteria by macrophages. To this end, RAW264.7 macrophages (referred to as RAW cells herein) were presented for 5 or 30 min with killed filamentous Legionella pneumophila to avoid the effect of its toxins on phagocytosis. These two time points generate tPCs with spatiotemporally distinct regions (see Fig. S1 a for the anatomy of tPCs). At 5 min, short, nascent tPCs containing only a proximal tPC region were formed. In comparison, 30 min of phagocytosis formed extended tPCs with an older, distal tPC region and a nascent, proximal tPC region.
As early as 5 min after the onset of phagocytosis, both Rab5 and EEA1 were recruited to the nascent tPCs (Fig. 1 a). As expected, this region was devoid of LAMP1 (Fig. 1 a). However, by 30 min of internalization, the entire tPC no longer contained Rab5 and EEA1, whereas LAMP1 became ubiquitously present (Fig. 1 b). Altogether, these results indicate that the tPC behaves like a canonical phagosome by first acquiring the early endosomal markers, EEA1 and Rab5, followed by their dissociation and subsequent acquisition of LAMP1 in a temporally regulated process (Fig. 1 c).
Figure 1.
Early endosomal markers are recruited to tPCs. (a and b) tPCs fuse with endocytic compartments acquiring features of mature phagolysosomes. Representative confocal imaging of RAW macrophages ingesting filamentous bacteria (blue) 5 min (a) and 30 min (b) after onset of phagocytosis. Images illustrate the recruitment of EEA1 (left set), GFP-Rab5 (center set), and LAMP1 (right set) to the tPC. Actin jackets, denoted by F-actin accumulation (red), delineate the top border of the tPCs. Images in main panels show merged z-stacks, and images to the right of main panels show magnified single planes from framed regions showing recruitments of early endosomal markers to the tPCs. RAW cells were either transiently expressing GFP-Rab5 or immunolabeled for EEA1 or LAMP1. (c) Number of tPCs positive for EEA1, Rab5, and LAMP1 recruitment over time from panels a and b. tPCs were scored positive for markers if labeling was observed along the entirety of the cup. Data shown are means ± SEMs of percentages from three independent experiments (n = 50 at each time point). Bars: 5 µm; (enlarged areas) 1 µm.
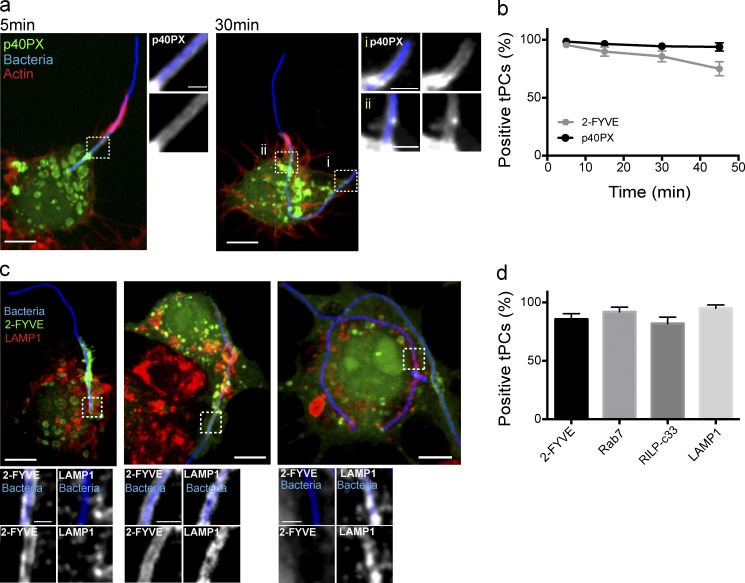
We then followed the dynamics of PtdIns(3)P during the biogenesis of tPCs using the 2FYVE-GFP and p40PX-GFP probes (Gillooly et al., 2000; Ellson et al., 2001; Kanai et al., 2001; Vieira et al., 2001). Strikingly, and unlike Rab5 and EEA1, both GFP probes persisted on tPCs even 45 min after the onset of the phagocytosis (Fig. 2, a and b; and Fig. S1 b). In fact, after 5 min of “early phagocytosis,” the PtdIns(3)P probes colocalized with LAMP1 and other lysosomal marks such as Rab7 and RILP-C33-GFP, a probe for GTP-bound Rab7 (Fig. 2, c and d). Importantly, the PtdIns(3)P probes were stripped from the tPCs after phagosome closure (Fig. 2 c), as is typical of canonical phagosomes. Our results indicate that PtdIns(3)P persists on LAMP1-positive tPCs long after the disappearance of other early endosome markers. Thus, tPCs represent an unique tool that decouples signals that coordinate PtdIns(3)P depletion and maturation of phagosomes and endolysosomal compartments.
Figure 2.
PtdIns(3)P coexists with late endosomal markers at tPCs. (a) p40PX-GFP (green) recruitment to the base of tPCs in RAW macrophages engulfing filamentous bacteria (blue) at indicated time points. Actin jackets, denoted by F-actin accumulation (red), delineate the top border of the phagocytic cup. Main panels show representative images of merged z-stacks, and images to the right of each main panel show magnified single planes from framed regions showing recruitments of p40PX-GFP to tPCs. (b) Recruitment of 2-FYVE-GFP and p40PX-GFP, indicative of the presence of PtdIns(3)P around tPCs. Cells transiently expressing either 2-FYVE-GFP or p40PX-GFP were scored for partially internalized filamentous bacteria positive for PtdIns(3)P. Data shown are means ± SEMs from three independent experiments (n = 30 for each time point). (c) RAW macrophages expressing 2-FYVE-GFP were challenged with filamentous bacteria, fixed, permeabilized, and immunostained for LAMP1. Left and middle panels show partially internalized bacteria. Right panel depicts fully internalized bacteria. Main panels are merged z-stacks, and images at the bottom show single planes from framed regions. Bars: 5 µm; (enlarged areas) 1 µm. (d) Number of tPCs positive for 2-FYVE, LAMP1, Rab7, and RILP-C33 recruitment at 30 min after the onset of phagocytosis. Cells expressing 2-FYVE-GFP, GFP-Rab7, or RILP-C33-GFP or immunostained for LAMP1 were scored for partially internalized filamentous bacteria. tPCs were scored positive for markers if labeling was observed along the entirety of the cup. Data shown are means ± SEMs of percentages from three independent experiments (n = 30 for each).
Vps34 activity is responsible for PtdIns(3)P persistence at tPCs
The persistence of PtdIns(3)P observed on tPCs could be the consequence of continuous production of the lipid or its limited turnover (Fig. S1 c). Although it is generally considered that PtdIns(3)P is produced by the class III PtdIns 3-kinase, Vps34 (Backer, 2008, 2016; Jeschke et al., 2015), 5- and/or 4-phosphatases acting on the class I PtdInsP 3-kinase products, phosphatidylinositol-3,4-bisphosphate and phosphatidylinositol-3,4,5-trisphosphate, can also generate PtdIns(3)P (Shin et al., 2005). Conversely, PtdIns(3)P turnover is driven by PIKfyve, which phosphorylates PtdIns(3)P to produce phosphatidylinositol-3,5-bisphosphate (PtdIns(3,5)P2; Ho et al., 2012; McCartney et al., 2014; Jin et al., 2016) and by myotubularins that convert it to PtdIns (Robinson and Dixon, 2006; Cao et al., 2008).
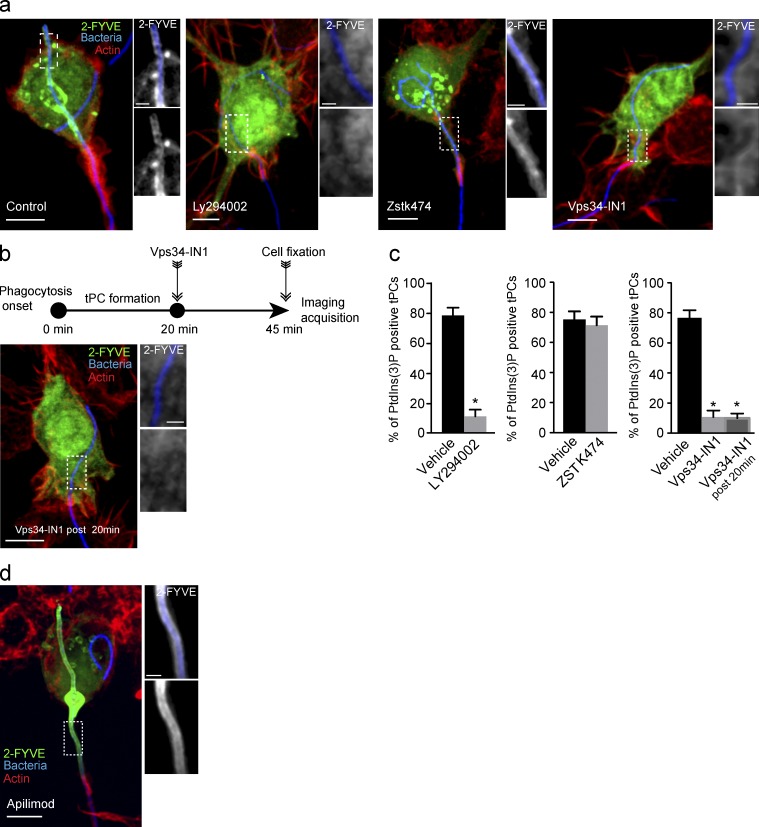
Therefore, we next sought to investigate the source of PtdIns(3)P persistence on tPCs. To this end we used inhibitors against different classes of PtdIns 3-kinases. The treatment of macrophages with the pan–PtdIns 3-kinase inhibitor LY294002 (Gharbi et al., 2007) clearly abrogated PtdIns(3)P production on tPCs (Fig. 3, a and c). To distinguish whether class I or class III PtdIns 3-kinases contributed to PtdIns(3)P synthesis on tPCs, we treated cells with their respective inhibitors, ZSTK474 or Vps34-IN1 (Kong and Yamori, 2007; Bago et al., 2014). Strikingly, Vps34-IN1 abrogated 2FYVE-GFP recruitment to tPCs, whereas ZSTK474 did not (Fig. 3, a and c), thus suggesting that Vps34 is the primary kinase responsible for the presence of PtdIns(3)P on tPCs.
Figure 3.
PtdIns(3)P synthesis at tPCs is driven by the class III PtdIns 3-kinase, Vps34. (a) RAW cells expressing 2-FYVE-GFP (green) were treated with 1 µM DMSO (vehicle), 100 µM LY294002, 1 µM ZSTK474, or 1 µM Vps34-IN1. After these treatments, cells were allowed to engulf filamentous bacteria for 30 min, followed by fixation and staining for F-actin jackets. (b) RAW cells expressing 2-FYVE-GFP underwent phagocytosis for 20 min, followed by treatment with 1 µM Vps34-IN1 and then fixed after 25 min of treatment. Cells were stained as in panel b. (c) Cells from experiments in panels b and c were scored for the presence of PtdIns(3)P, detected via 2-FYVE accumulation at tPCs. Data shown are means ± SEMs from three independent experiments (n = 35 for each). P < 0.05. Bars, 5 µm. (d) RAW cells expressing 2-FYVE-GFP (green) were treated with 10 nM apilimod for 1 h before the phagocytosis. Representative phenotype from 90 cells analyzed in three independent experiments.
Does the PtdIns(3)P present on the tPCs undergo turnover? To assess this, we allowed phagocytosis to proceed and tPCs to accumulate PtdIns(3)P before treating macrophages with VPS34-IN1, 20 min after the onset of phagocytosis. Significantly, 2FYVE-GFP rapidly detached from elongating tPCs after the addition of VPS34-IN1, demonstrating that PtdIns(3)P was undergoing turnover during tPC biogenesis (Fig. 3, b and c). We found that PIKfyve plays a role in this process as the treatment of macrophages with apilimod, a specific inhibitor of PIKfyve, strongly increased the levels of 2FYVE-GFP associated with tPCs (Fig. 3 d). Altogether these results indicate that the persistence of PtdIns(3)P on tPCs is the consequence of its continuous synthesis by Vps34 and not a defect in its turnover. Thus, tPCs are competent for PtdIns(3)P synthesis and turnover, but unlike canonical phagosomes, PtdIns(3)P synthesis seems to persist over time.
PtdIns(3)P is removed from the distal ends of tPCs in a target length–dependent manner
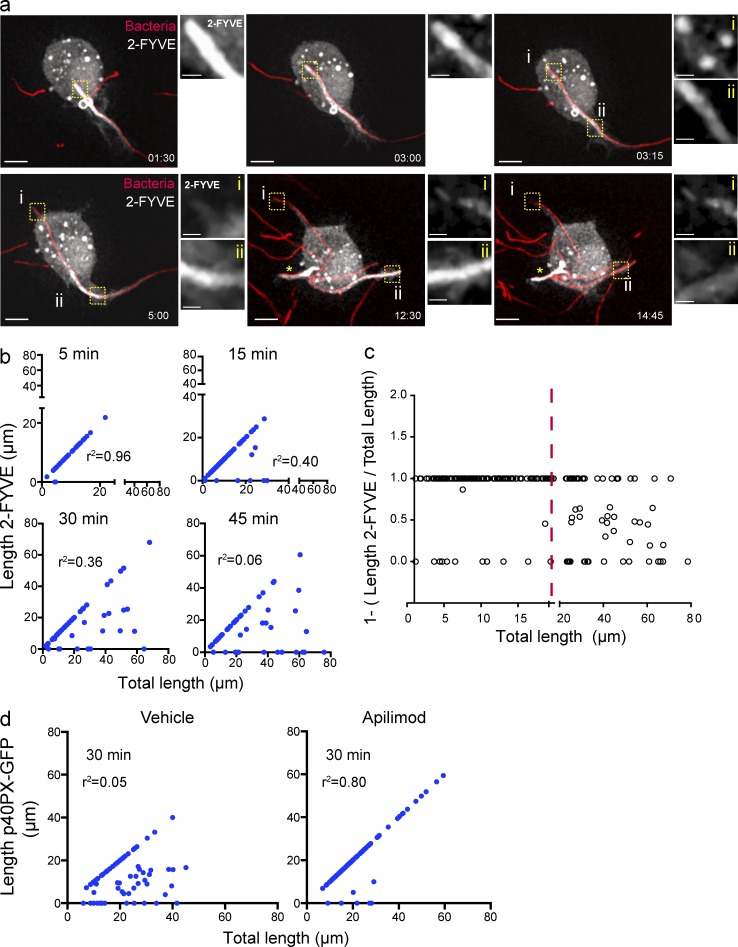
We proceeded to characterize in detail the fate of PtdIns(3)P on tPCs by time-lapse video microscopy. Fig. 4 a and Video 1 depict the phagocytosis of a filamentous bacterium by a macrophage. As the target was internalized, 2FYVE-GFP was present all along the tPC (Fig. 4 a, 1:30 and 3:00 min frames). Strikingly, this distribution was altered once a threshold length was surpassed, at which point 2FYVE-GFP began to disappear from the distal end of the tPCs (Fig. 4 a, inset i in 3:15, 5:00, and 12:30 min frames), while persisting in the nascent proximal tPC region (Fig. 4 a, inset ii in 3:15, 5:00, and 12:30 min frames). Eventually, 2FYVE-GFP dissociated completely from phagosomes upon full enclosure (Fig. 4 a, 14:45 min frame). We confirmed these observations using fixed cell imaging by correlating the total length of the tPC to its length that was positive for 2FYVE-GFP or p40PX-GFP. As shown in Fig. 4 b and Fig. S1 d, early in phagocytosis, the total length of the tPCs tended to be decorated with the PtdIns(3)P probes. However, the strong correlation between these parameters was lost as GFP-fluorescence began to disappear from the most internalized portion of tPCs, usually longer than 20 µm (Fig. 4, b and c). Hence, PtdIns(3)P persistence predominates within the proximal regions of tPCs but is eliminated if the distal regions become >20 µm in length. Importantly, this phenomenon depended on PIKfyve activity, because PtdIns(3)P probes remained associated with distal tPCs in cells incubated with apilimod (Fig. 4 d). Therefore, PtdIns(3)P production may be surpassed by its consumption by PIKfyve activity in the distal tPCs.
Figure 4.
PtdIns(3)P dynamics on the tPCs. (a) RAW macrophage expressing 2-FYVE-GFP (white), engulfing a filamentous bacteria (red), selected frames from Video 1. Images to the right of the main panels are magnifications of the framed regions in the main panels depicting 2-FYVE-GFP in tPCs. For the first two images, the area magnified corresponds to the most distal tip of the tPC containing filamentous bacteria. In the 4 last micrographs (times 3:15 to 14:45), panel i is the most distal tip of the tPC and ii is the proximal tPC. Bars: 5 µm; (enlarged areas) 1 µm. (b) RAW macrophages expressing 2-FYVE were challenged with filamentous bacteria and fixed after 5, 15, 30, and 45 min of phagocytosis. The length of filamentous bacteria positive for 2-FYVE was plotted against the bacterial length internalized at each time point. The corresponding correlation coefficients (r2) were calculated. (c) Regions of tPCs that surpass a length of 20 µm (red line) tend to be devoid of 2FYVE-GFP. The ratio of 2FYVE-GFP–positive length to the total length of the tPC is shown. A ratio of 1 indicates that the entire tPC is decorated with 2FYVE-GFP, a ratio <1 indicates that a portion is devoid of this probe, and a ratio of 0 indicates that the tPC is completely divested of the probe. Data from three independent experiments (n = 30 for each time point). (d) RAW macrophages expressing p40PX-GFP, treated with 10 nM apilimod or 0.1% DMSO (vehicle) for 1 h, were allowed to engage in phagocytosis of pHrodo-conjugated filamentous bacteria for 30 min of phagocytosis. The length of filamentous bacteria positive for p40PX-GFP was measured in three independent experiments, n = 30 for each condition, and plotted as described in panel b.
PtdIns(3)P loss at the tPCs correlates with their acidification
Our observations suggested that longer tPCs acquired a signaling gradient that allows for PtdIns(3)P removal. We previously showed that tPCs acquire vacuolar H+-ATPases (V-ATPases) and that small molecules, including H+, can diffuse out of the tPC to the extracellular space. Because acidification is considered a hallmark of PtdIns(3)P-negative phagolysosomes, we postulated that H+ leakage across the actin jacket could lead to failed acidification of the tPCs (Prashar et al., 2013). However, once a tPC becomes very long, a H+ gradient could be produced from its distal end to its proximal end, and perhaps luminal acidification serves to signal PtdIns(3)P termination from membranes. We therefore investigated if the acidification of tPCs affected PtdIns(3)P persistence.
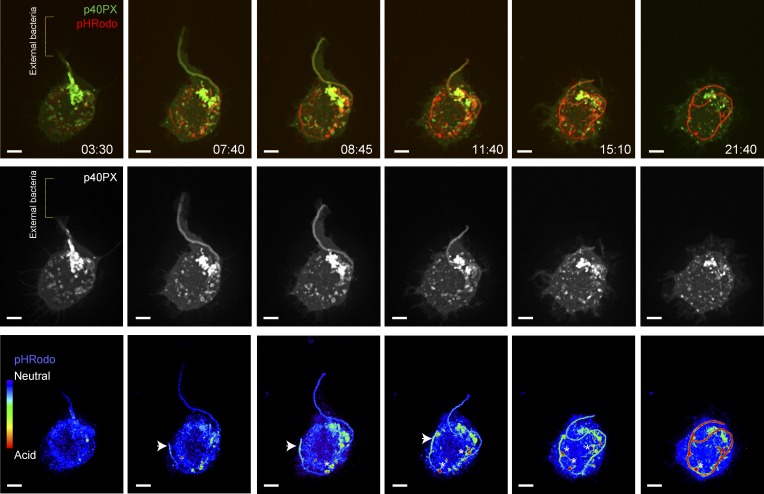
To this end, we conjugated the pH indicator pHrodo to filamentous bacteria and used them as targets for phagocytosis. This approach allowed us to detect the occurrence of a pH gradient along the lumen of the tPCs as they elongated over time (Videos 2 and 3; selected frames in Fig. 5 and Fig. S2 a). Acidification of the tPCs, depicted by the increase of fluorescence emission in the pHrodo channel, occurred only after considerable portions of long filaments were engulfed by the macrophage, in the most distal regions of tPCs (Fig. 5, 8:45 and 11:40 frames, arrowheads), extending outward as the internalization of the bacteria proceeded to ultimately yield an acidic phagosome (Fig. 5, 21:40 frame; and Fig. S2 a). The way tPCs acidify must be the result of a balance between H+ leakage across the diffusion barriers associated with the actin jacket and the activity of the V-ATPase pumps. As tPCs grow in length, H+ leakage may not be fast enough to dissipate the acidification of their distal end. Furthermore, long tPCs could reach the perinuclear zone where they can fuse with more acidic lysosomes (Johnson et al., 2016).
Figure 5.
Loss of PtdIns(3)P is correlated with the acidification of distal tPCs. Evolution of p40PX-GFP and pHrodo fluorescence in growing tPCs. RAW macrophages expressing p40PX-GFP (top, green; middle, white) were challenged with pHrodo-conjugated filamentous bacteria and imaged live by simultaneous acquisition of both fluorescence emissions. Micrographs are selected frames from Video 2. pHrodo fluorescence is shown in red in the top row of micrographs and in false-rainbow palette in bottom one. Blue in the rainbow palette indicates neutral pH and red indicates acidic pH. Brackets in the panels from the left indicate position of external filamentous bacteria, detected in bright light field. Arrowheads in lower panels point to the distal tip of the tPCs. Asterisks indicate bacterial-associated pHrodo fluorescence extracted into cytoplasmic vesicles as the tPCs remodel during internalization. Bars, 5 µm.
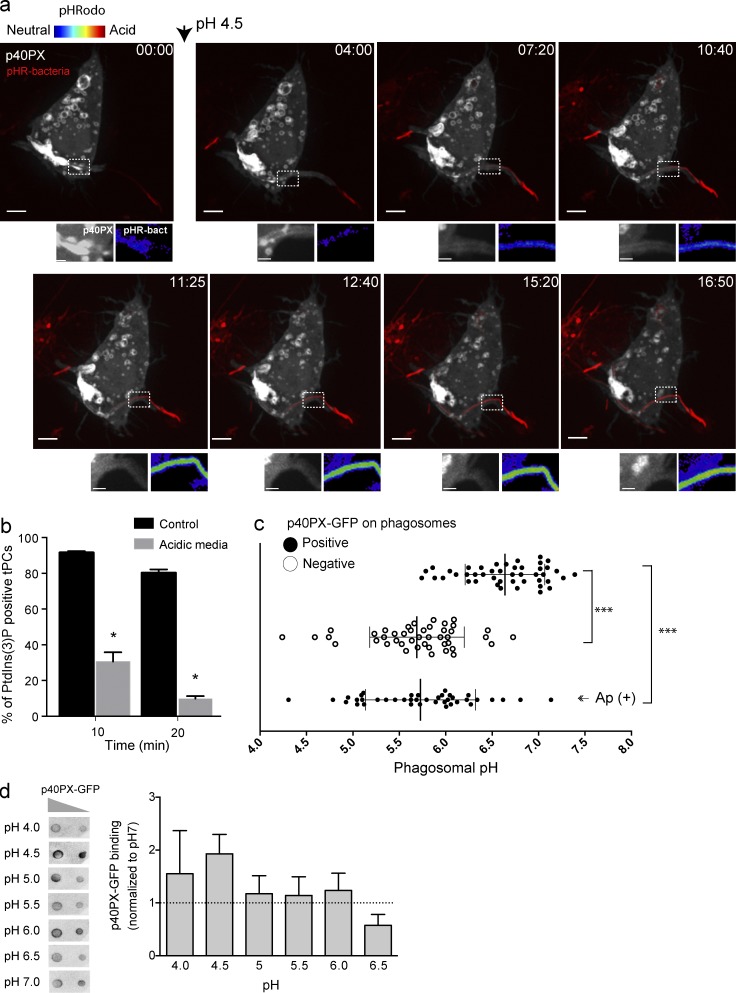
Importantly, our results in Fig. 5 and Fig. S2 a showed that PtdIns(3)P levels and acidification in tPCs were inversely related, where acidic regions were divested of PtdIns(3)P. We confirmed this phenomenon by forcing the acidification as well as neutralizing tPCs and subsequently following the fate of PtdIns(3)P in this compartment. For tPC acidification, we bathed macrophages in culture media adjusted to pH 4.0. We previously showed that this treatment allows for the rapid acidification of the lumen of tPCs (Prashar et al., 2013). Video 4 and Fig. 6 a show that as the acidification of tPC progressed, pHrodo fluorescence increased, and this was accompanied by the disappearance of the PtdIns(3)P probe from the proximal tPC (see magnified areas in Fig. 6 a). In contrast, the proximal tPC in control cells retained PtdIns(3)P (Fig. 6 b). Alternatively, we hindered the acidification of tPCs with the proton pump inhibitor concanamycin A (ConA). As shown in Fig. S2 (b and c), this treatment stabilized PtdIns(3)P in distal regions of tPCs and abated turnover of PtdIns(3)P from fully enclosed filamentous phagosomes.
Figure 6.
Forced acidification divests PtdIns(3)P reporters from tPCs. (a) Selected frames from a time-lapse sequence from Video 4 showing a RAW macrophage expressing p40PX-GFP (white) engulfing a pHrodo-conjugated bacteria (turning red as it acidifies). Arrowhead indicates the time at which cells were exposed to culture media at pH 4.5. Images at the bottom of main panels show magnifications of the framed areas. Right panels show pHrodo signal (false-rainbow palette) and left panels show p40PX-GFP. Bars, 5 µm. (b) RAW cells expressing 2FYVE-GFP were challenged with filamentous bacteria in neutral or acidic conditions. After 10 and 20 min of phagocytosis, cells were imaged for accumulation of 2FYVE-GFP at tPCs. Data shown are means ± SEMs from three independent experiments (n = 20 for each; *, P < 0.05). (c) RAW cells expressing p40PX-GFP were challenged with pHrodo-conjugated filamentous bacteria. The phagosomal pH was determined for p40PX-GFP–positive and –negative phagosomes after 15 and 30 min of phagocytosis, respectively. (Ap+) cells were treated with 10 nM apilimod 1 h before phagocytosis and pH measured 30 min of phagocytosis. Data represent 15 phagosomal pH values ± SD from three independent experiments. ANOVA test was used to compare each group; ***, P < 0.001. (d) Binding of p40PX-GFP to PtdIns(3)P does not depend on pH. (Left) Protein-lipid overlay (PLO) using recombinant p40PX-GFP-Hisx6 and membranes containing 400 and 200 pmols of PtdIns(3)P. This is a representative of three independent experiments. (Right) Densitometry of spots from PLOs expressed relative to that observed at pH7, expressed as means ± SEMs of three independent experiments.
Our observations suggest that the acidification of the phagosomal lumen may provide a trigger to remove PtdIns(3)P from maturing phagosomes. Thus, we next sought to determine the pH at which PtdIns(3)P was eliminated from the phagosomal membranes. We measured the pH of phagosomes holding bacteria in the range of 10 to 15 µm in length, which permits near uniform recruitment and dissociation of 2FYVE-GFP from the tPC/phagosome membrane. Using pHrodo-Legionella, we clustered phagosomes as being p40PX-GFP–positive or –negative and measured its pH by calibrating pHrodo emission as described in the section Materials and methods (Fig. S4 a). Strikingly, p40PX-GFP–positive phagosomes had a mean pH of 6.6, whereas p40PX-GFP–divested phagosomes had a pH mean of 5.7. By comparing the range of pH values, we estimate that phagosomes that hit pH 6.2 tend to lose the probe (Fig. 6 c). PtdIns(3)P loss was not a result of acid-induced disruption of protein-lipid interaction as recombinant expressed p40PX and 2FYVE domains still bound to PtdIns(3)P in a lipid overlay assay under acidic conditions (Fig. 6 d; Fig. S3, b and c; and Fig. S4 a). In fact, the probes bound to PtdIns(3)P blots more effectively in media set to pH < 6.5 (Fig. 6 d), consistent with Lee et al. (2005) and He et al. (2009). In addition, p40PX-GFP remained associated with enclosed phagosomes in apilimod-treated cells despite significant acidification (Fig. 6 c). Altogether, our findings show a causality between pH and the fate of PtdIns(3)P fate in tPCs.
pH controls the lifespan of PtdIns(3)P in phagosomes
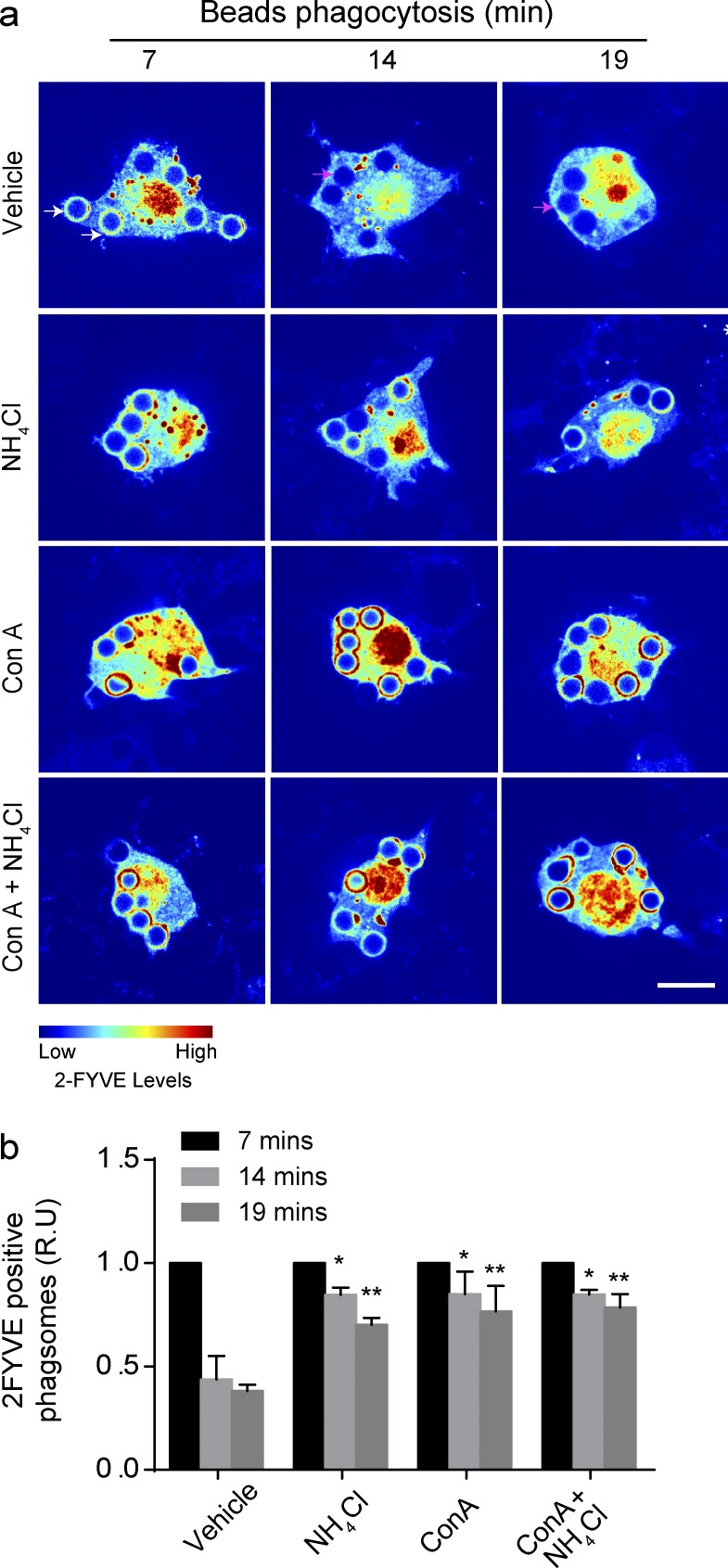
We next investigated the impact of pH on the dynamics of PtdIns(3)P on phagosomes containing latex bead. As expected, the number of PtdIns(3)P-positive phagosomes in macrophages diminished over time (Fig. 7 a). In comparison, PtdIns(3)P persisted on phagosomes neutralized by treating macrophages with NH4Cl, or with ConA, or a combination of both treatments (Fig. 7, a and b).
Figure 7.
pH neutralization causes PtdIns(3)P to persists in phagosomes. (a) RAW cells expressing 2FYVE-GFP (rainbow) were treated with 0.1% DMSO (vehicle), 1 µM ConA, 10 mM NH4Cl, or both ConA and NH4Cl for 15 min before the phagocytosis of IgG-opsonized beads. White and magenta arrows depict examples of 2FYVE-positive and -negative phagosomes, respectively. (b) The phagosome/cytosol ratio of 2FYVE-GFP fluorescence for each time point indicated, normalized to 7 min, represented as relative units (R.U). Data shown are means ± SEMs from three independent experiments (n = 30 for each). ANOVA test was used to compare each treatment condition to vehicle. For each time point; *, P < 0.05; **, P < 0.01. Bars, 5 µm.
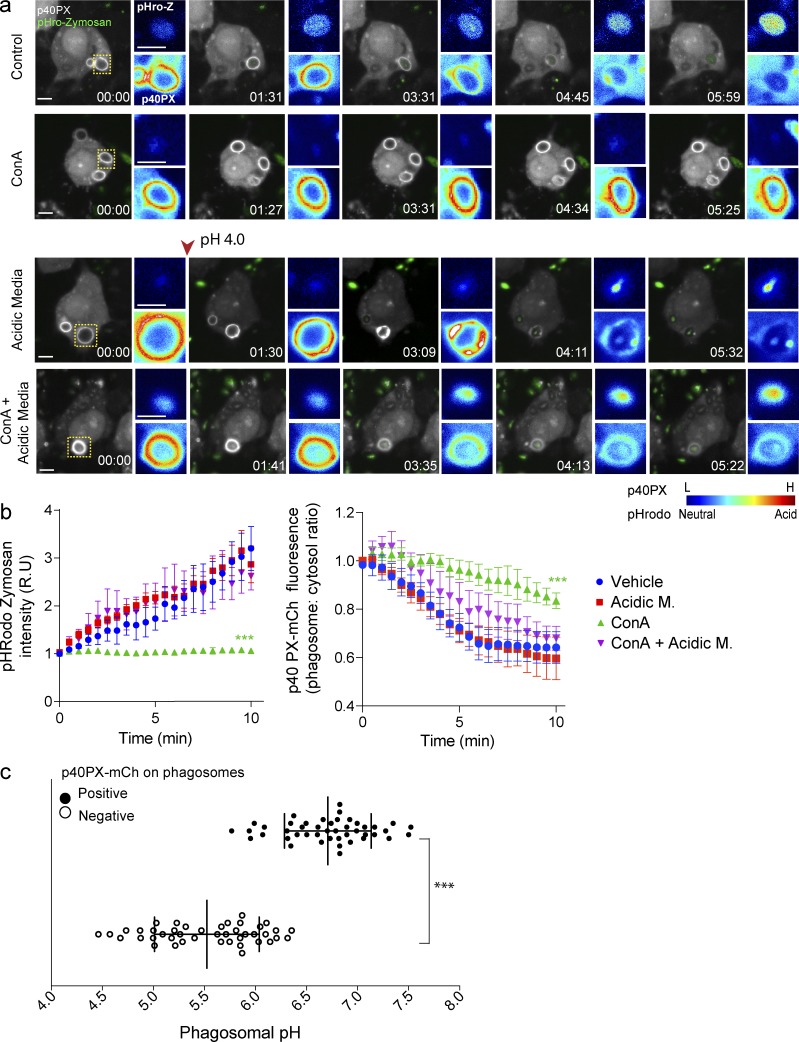
To complement this experiment, we followed phagosome acidification and 2FYVE-GFP dynamics in macrophages engulfing pHrodo-conjugated zymosan particles. In resting cells, pHrodo-associated fluorescence within phagosomes increased over 10 min and correlated with the progressive depletion of phagosome-associated 2FYVE-GFP (Fig. 8, a and b). As for latex beads, ConA caused a dramatic extension of phagosome-associated 2FYVE-GFP, though phagosomes eventually became devoid of 2FYVE-GFP, suggesting that additional mechanisms that signal depletion of PtdIns(3)P exist (Fig. 8, a and b). In comparison, exposure of ConA-treated cells to acidic media acidified the phagosomes and caused the dissociation of 2FYVE-GFP from phagosomes. We recognize that treating cells with media set to pH 4.0 will acidify both the cytosol and the lumen of phagosomes, albeit the cytosol did not reach pH 4.0 as could be inferred by the retention of GFP fluorescence in the cytoplasm, which is quenched below pH 5.0 (Haupts et al., 1998). However, when we take into account that ConA alone prolonged the lifetime of 2FYVE-GFP–positive phagosomes, our data collectively suggest that luminal pH controls PtdIns(3)P depletion from the membranes of canonical phagosomes.
Figure 8.
p40PX-mCh recruitment to zymosan-containing phagosomes is pH dependent. (a) Live-cell imaging of p40PX-mCh–expressing (white) RAW cells, internalizing pHrodo-conjugated zymosan particles (green), either in culture media (vehicle) or in acidic media (pH 4.0), with or without 1 µM ConA. The arrowhead indicates the switch to acidic media, 30 s after the onset of the experiment. Images were acquired every 15–30 s. Images to the right of the main panels show fluorescence intensities (rainbow) for pHrodo-zymosan (top) and p40PX-mCh (bottom) phagosomes framed in the main panels. Blue indicates low fluorescence levels for p40PX-mCh and pHrodo (neutral pH), whereas red indicates high-emission signal for p40PX-mCh and pHrodo (acidic pH). (b) Progression of pHRodo and p40PX-mCh fluorescence intensities in zymosan-containing phagosomes for the experimental conditions described in panel a. Each time point was normalized to 0 mins, represented as relative units (R.U). Data shown are normalized means ± SEMs from 3 independent experiments (n = 10–15 phagosomes for each). ANOVA test was used to compare each treatment condition to control; ***, P < 0.001. Bars, 5 µm. (c) RAW cells expressing p40PX-mCh were challenged with pHrodo-conjugated zymosan and pH determined for p40PX-mCh–positive and –negative phagosomes. Data represent 15 phagosomal pH values ±SD from three independent experiments. Each phagosome was internally calibrated (***, P < 0.001).
We then measured the fluorescence of pHrodo-labeled zymosan particles in p40PX-positive and p40PX-negative phagosomes followed by calibration with buffers of known pH and ionophores (Fig. 8 c and Fig. S4 c). As with bacteria, phagosomes containing or devoid of PtdIns(3)P had a median pH of 6.7 and 5.5, respectively. By examining the boundary of the SD between these conditions, we estimate that phagosomes tend to become negative for the lipid at pH 6.1 to 6.3. Moreover, tracking 3 individual phagosomes over time, we could infer that PtdIns(3)P accumulates in newly formed phagosomes as they acidify to pH 6.5 and begins to decay as the pH further acidifies. Indeed, PtdIns(3)P gradually disappeared below this pH until it could no longer be detected at pH 5.5 (Fig. S4 d). Thus, altogether our findings strongly demonstrated that pH controls the lifespan of PtdIns(3)P in phagosomes.
Organellar pH controls the association of Vps34 with membranes
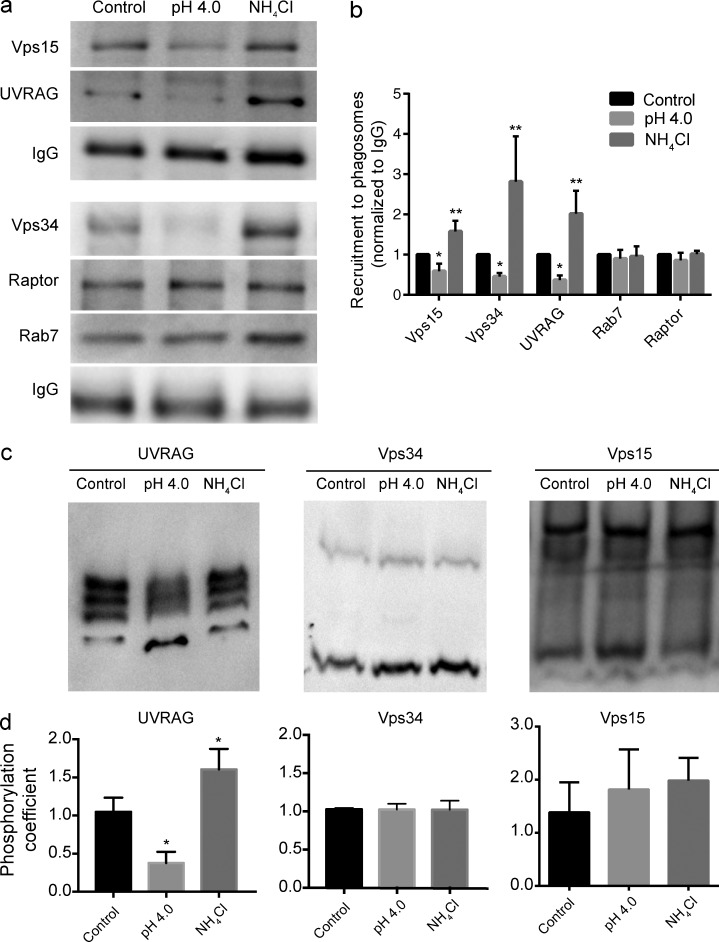
Our observations support a model in which acidification signals depletion of PtdIns(3)P from phagosomal and endosomal membranes. This is not likely a result of turnover machinery because PtdIns(3)P can be depleted from Vps34-inhibited tPCs and further stabilized by PIKfyve inhibition. Thus, we postulated that pH-triggered PtdIns(3)P depletion may occur by inactivation and/or dissociation of Vps34. Vps34 binds to endophagosomal membranes as part of a complex formed by Vps15 (p150/PIK3R4), Beclin-1, and UVRAG (Rostislavleva et al., 2015; Backer, 2016). To investigate if pH affects the association of these subunits with phagosomes, we purified phagosomes containing latex beads and immunoblotted for Vps34, Vps15, and UVRAG. As shown in Fig. 9 (a and b), all three subunits detached from phagosomes after exposure of cells to acidic media, but bound more intensely to phagosomes treated with NH4Cl (ConA pretreatment significantly inhibited phagocytosis, precluding sufficient yield of phagosomes to run Western blot analyses; not depicted). In comparison, the endolysosomal proteins Rab7 and Raptor remained associated to phagosomes regardless of treatment (Fig. 9, a and b). To further demonstrate that pH can impact the biochemical state of the Vps34 complex, we examined the phosphorylation state of its subunits (Kim et al., 2013; Munson et al., 2015). To this end, we used Phos-tag mobility shift gels to better resolve multiple phospho-isoforms. Acidic media and NH4Cl caused a change in the distribution of phosphorylated UVRAG species relative to resting cells, whereas Vps34 and Vps15 phosphorylation levels were unchanged (Fig. 9, c and d). This showed that the Vps34 complex is controlled by the pH in the phagosomal and cytoplasmic milieu.
Figure 9.
The Vps34 complex II association to phagosomes is controlled by pH. (a) Vps34 complex II subunits in isolated latex beads phagosomes. RAW macrophages in culture media (control), acidic culture media (pH 4.0), or cell-culture medium with 10 mM NH4Cl were allowed to internalize IgG-opsonized latex beads for 20 min. After this period, phagosomes were isolated and immunoblotted for the presence of Vps15, UVRAG, Vps34, Raptor, and Rab7. Human IgG was used as a control for number of phagosomes loaded. (b) Protein densitometric quantification for blots shown in panel a. Human IgG was used to normalize phagosomal loading. Data shown are means ± SEMs from four to six independent experiments; *, P < 0.05; **, P < 0.01. (c) Phos-Tag SDS-PAGE determination of the phosphorylation states of UVRAG, Vps34, and Vps15 in phagosomes isolated as described in panel a. (d) The phosphorylation coefficient was used to quantify the preferred states of phosphorylation. This was done by calculating an internal ratio of the most (top) to least (bottom) phosphorylated species. Data shown are means ± SEMs from three or four independent experiments. *, P < 0.05.
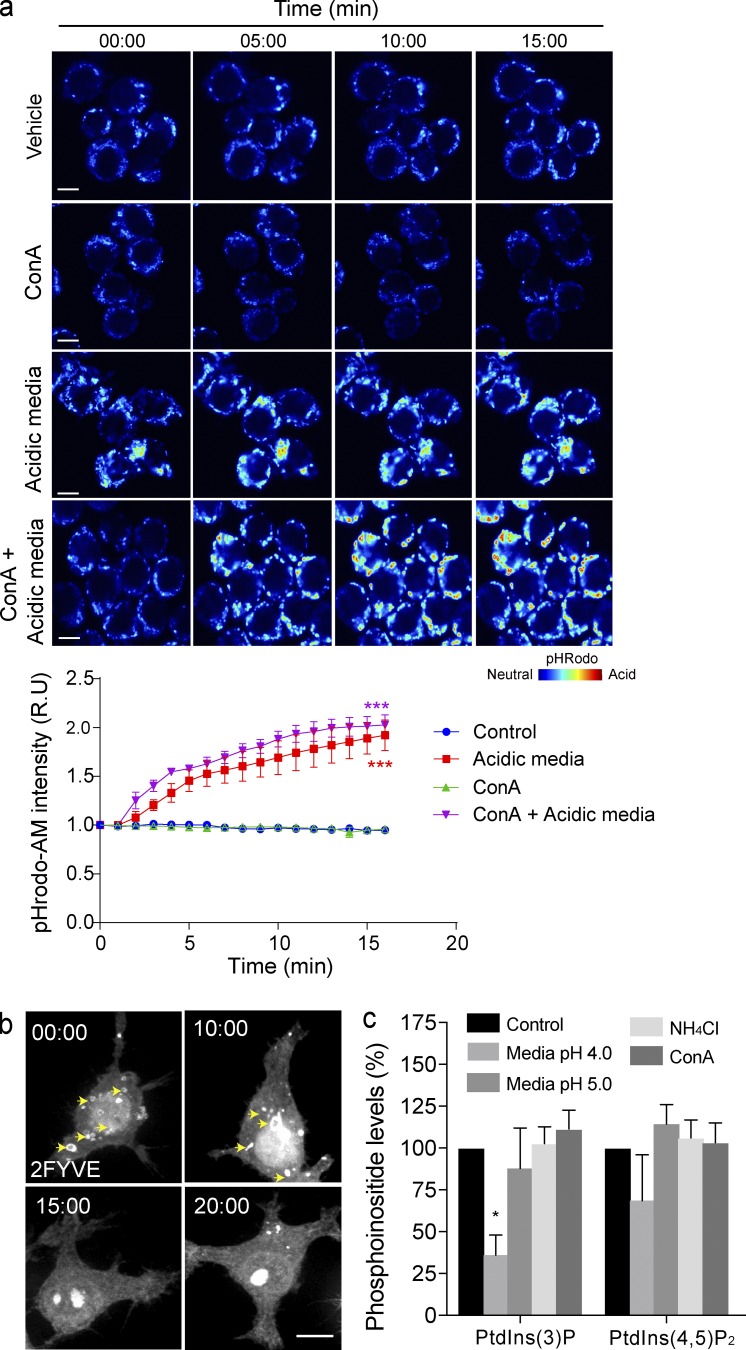
Finally, we speculated that pH-dependent dissociation of Vps34 may also apply to endosomes and modulate cellular levels of PtdIns(3)P. To corroborate this hypothesis, we bathed cells in acidified media, with the caveat that this treatment acidifies both cytosol and organelles (Fig. 10 a). Nevertheless, this treatment caused dissociation of 2FYVE-GFP from endosomes (Fig. 10 b). Importantly, by metabolically labeling cells with 3H-myo-inositol, we observed a significant reduction in PtdIns(3)P levels in cells at pH 4.0, whereas phosphatidylinositol-4,5-bisphosphate suffered a smaller decrease (Fig. 10 c). Conversely, PtdIns(3)P levels were slightly augmented or unchanged in cells treated with NH4Cl or ConA (Fig. 10 c).
Figure 10.
PtdIns(3)P on endosomes is controlled by pH. (a) The cytoplasmic pH of pHrodo-AM–loaded RAW macrophages over time. RAW macrophages were incubated in either culture media (vehicle) or acidic media (pH 4.0), with or without 1 µM ConA. pHrodo-AM fluorescence intensity is shown in rainbow palette. Bars, 5 µm. Graph illustrates the changes in fluorescence intensity over time. Each time point was normalized to 0 mins, represented as relative units (R.U). Data shown are means ± SEMs from three independent experiments (n > 50 cells for each condition). ANOVA test was used to compare each treatment condition to control; ***, P < 0.001. (b) RAW macrophages expressing 2FYVE-GFP (white) show PtdIns(3)P containing endosomes (arrows). 2FYVE-GFP–positive endosomes disappeared when macrophages were exposed to acidic cell-culture medium (pH 4.0) for the time points indicated (representative of the independent experiments). Bars, 5 µm. (c) Determination of PtdIns(3)P level in RAW macrophages. Cells were incubated with 3H-myo-inositol overnight and subjected to the conditions indicated for 20 min. After inositol isolation and separation, levels of PtdIns(3)P and PtdIns(4,5) were determined. For each treatment, PtdInsP levels were normalized to the parent PtdIns peak and compared with control. Data shown are means ± SEMs from four independent experiments. *, P < 0.05.
In conclusion, our observations show that organelles use a H+ gradient across their membranes to signal PtdIns(3)P depletion from their surface by triggering dissociation of the Vps34 complex. Such a mechanism couples the progressive acidification of phagosomes and endosomes to disappearance of PtdIns(3)P from these organelles as they mature. However, when tPCs remain open to the extracellular space, H+ leaks into the extracellular space, permitting PtdIns(3)P to persist on the tPC membrane, despite the loss of other early endosomal markers and acquisition of endolysosomal properties. When tPCs become sufficiently long or seal into a phagosome, then a H+ gradient is generated that signals PtdIns(3)P depletion.
Discussion
tPCs decouple PtdIns(3)P depletion from phagosome maturation
There are several aspects in the tPC biogenesis that contravene the canonical maturation pathway of phagosomes, as we reported here and in Prashar et al. (2013). First, endosomal and lysosomal markers are recruited at the level of the phagocytic cup before its scission from the plasma membrane, and this phenomenon is uncoupled from the acidification of the compartment. Second, whereas the early phagosomal proteins Rab5 and EEA1 arrive in a timely manner to incipient tPCs, and disappear as the phagocytic cups elongate, PtdIns(3)P persists on tPCs. Third, PtdIns(3)P overlaps with endolysosomal markers as long as the pH of the tPCs remain near neutral. These observations have several important implications toward our understanding of the mechanisms that impart identity to early endosomal and phagosomal compartments.
For one, the data presented here show that regulation of Rab5 and PtdIns(3)P, two of the most important organizers of early endosome/phagosome identity, can be decoupled such that Rab5 can be inactivated whereas PtdIns(3)P remains present (Behnia and Munro, 2005; Stenmark, 2009; Posor et al., 2015). Rab5 encodes its own inactivation by interacting with Mon/CCZ, which recruits and activates the GTPase Rab7 as endosomes mature (Rink et al., 2005; Nordmann et al., 2010). Indeed, tPCs are competent for this transition as Rab5 is eliminated and Rab7 is recruited. Thus, unlike what we propose for PtdIns(3)P depletion, we speculate that Rab5 to Rab7 conversion is independent of organellar pH and proceeds during tPC biogenesis. Second, our observations suggest that PtdIns(3)P removal is not a prerequisite for phagosomes to become a phagolysosome. Instead, we purport that the opposite likely occurs where progression into a phagolysosome, through the accompanying acidification, signals for PtdIns(3)P elimination. In fact, others have demonstrated that phagosomes can acidify before acquisition of LAMP proteins (Geisow et al., 1981; Bouvier et al., 1994; Blanchette et al., 2009; Lu and Zhou, 2012), and our own observations with zymosan demonstrate that phagosomes acidify at rates faster than those reported for acquisition of LAMP1 (Vieira et al., 2002; Kinchen and Ravichandran, 2008; Flannagan et al., 2012). Thus collectively, we envision a model in which phagosome acidification, and perhaps endosome acidification, likely precede fusion with lysosomes and that acidification arrests PtdIns(3)P synthesis to deplete it from maturing phagosomes and endosomes.
Termination of PtdIns(3)P signaling on phagosomes and endosomes
PtdIns(3)P is a critical regulator that orchestrates many signaling events during the maturation of endosomes and phagosomes. Proteins like EEA1 and Hrs (hepatocyte growth factor–regulated tyrosine kinase substrate) are recruited to PtdIns(3)P-positive endosomes, autophagosomes, and phagosomes to regulate membrane fusion and recycling, as well as protein sorting to the Golgi and targeting into multivesicular bodies (Schink et al., 2013; Levin et al., 2015). Although PtdIns(3)P synthesis is necessary for proper endosome and phagosome maturation, the loss of PtdIns(3)P is also considered a hallmark in the maturation of phagosomes and endosomes (Fairn and Grinstein, 2012). The transient association of PtdIns(3)P with endosomes and canonical phagosomes has been explained in terms of a burst in its synthesis mediated by Vps34, followed by downstream conversion of PtdIns(3)P by the enzymes PIKfyve and myotubularins (Nandurkar and Huysmans, 2002; Cao et al., 2008; Hazeki et al., 2012; Ho et al., 2012; Zolov et al., 2012; Kim et al., 2014; McCartney et al., 2014; Jin et al., 2016).
Here we show that PtdIns(3)P accumulates on tPCs in Vps34-dependent manner, and persists on tPCs for well over 30 min, rather than the typical lifespan of 5 to 10 min on canonical phagosomes. We propose that rapid cessation of PtdIns(3)P from canonical phagosomes and endosomes requires the dissociation and/or inactivation of Vps34 from membranes. Otherwise, a sustained production and persistence of PtdIns(3)P on membranes occurs, as in tPCs. Vps34 and PtdIns(3)P association with phagosomes correlated well during manipulation of phagosomal pH. The neutralization of pH delayed the loss of PtdIns(3)P and enhanced the levels of Vps34, Vps15, and UVRAG associated with phagosomes, whereas forced acidification depleted both PtdIns(3)P and Vps34 complexes from these organelles. Our data with endosomes and cellular levels of PtdIns(3)P argue that this mechanism applies to endosomal PtdIns(3)P as well. Overall, we propose that the dissociation of Vps34 complex from maturing endolysosomes and phagosomes is essential to terminate PtdIns(3)P signaling and that this dissociation depends on organelle acidification. Because maturing endolysosomes and phagosomes progressively acidify, this circuit would couple maturation, timed by acidification, to the dissociation of Vps34 and depletion of PtdIns(3)P. In other words, it is tantalizing to consider that the identity of the cytosolic leaflet of endosomes and phagosomes may be subservient to their luminal identity defined by pH.
How might pH govern Vps34 and PtdIns(3)P dynamics?
We provide evidence that as phagosomes/endosomes acidify during maturation, the pH-gradient across the membrane serves as a trigger to displace Vps34 from membranes. This raises the question of how luminal pH might control cytoplasmic complexes and lipids. We envision at least two mechanisms. First, a transmembrane protein could act as a sensor by undergoing a pH-dependent conformational change. Parallels to this exist, such as pH-dependent conformational changes in bacterial toxins that causes their insertion into endosomal membranes. The V-ATPase itself may also act as a pH sensor (Liger et al., 1998; Qa’Dan et al., 2000). Notably, the V-ATPase is a major regulator of mTOR activity, which senses and transmits the luminal concentration of amino acids within lysosomes (Bar-Peled and Sabatini, 2014; Marshansky et al., 2014; Stransky and Forgac, 2015). Second, the membranes of endosomes and phagosomes allow diffusion of protons to the cytoplasm (Lukacs et al., 1990; Demaurex, 2002; Marshansky, 2007). Thus, proton leakage could generate localized changes in the pH around these organelles that could be potentially sensed by pH-sensitive signaling proteins (Casey et al., 2010; Koivusalo et al., 2010; Johnson and Casey, 2011). Alternatively, the H+ gradient could control the activity of another channel or transporter such as those for Ca2+, which in turn could act as cytoplasmic intermediate for controlling the localization of Vps34 (Ishida et al., 2013; Xu and Ren, 2015). For instance, the lysosomal mucolipin TRP channel 3 responds to pH by releasing Ca2+ (Miao et al., 2015). The two-pore channel 2 releases Ca2+ by integrating luminal pH of lysosomes and the second messenger nicotinic acid adenine dinucleotide phosphate (Pitt et al., 2010; Zhu et al., 2010).
Furthermore, the phosphorylation pattern of UVRAG, but not Vps34 or Vps15, could be toggled such that higher-order phosphorylation was enhanced upon alkalinization relative to acidification. This suggests that pH can regulate Vps34 complex through a pH-dependent kinase that remains to be identified. Recently, mTOR was shown to phosphorylate UVRAG to enhance the activity of Vps34 activity during autophagic lysosome reformation (Munson et al., 2015). Tantalizingly, these authors also suggest that additional kinases exist that act on UVRAG.
Overall, the work presented here has uncovered a mechanism showing that PtdIns(3)P signal termination is controlled by the acidification of phagosomes and endosomes. Ultimately, this mechanism may be of relevance for the many cellular processes governed by PtdIns(3)P and its effector proteins. Indeed, PtdIns(3)P-binding proteins bearing PX and FYVE domains play roles not only in vesicle trafficking and protein sorting along the recycling, endosomal, and phagosomal pathways but also in cytokinesis, metabolic signaling, autophagosomes, and the production of reactive oxygen species by NADPH oxidase (Tian et al., 2008; Berger et al., 2010; Bohdanowicz and Grinstein, 2013).
Materials and methods
Reagents and antibodies
DMEM and FBS were purchased from Wisent Inc. Alexa Fluor–conjugated secondary antibodies, phalloidin, LysoSensor green, and pHrodo red succinimidyl ester were from Life Technologies Inc. Skim milk was purchased from BioShop Canada Inc., and PFA was obtained from Canemco & Marivac. Rat polyclonal (1D4B) anti-LAMP1 antibodies were from Developmental Studies Hybridoma Bank, and goat polyclonal anti-EEA1 (N-19) antibody was from Santa Cruz Biotechnology, Inc. Rabbit polyclonal anti–L. pneumophila antibody was provided by C. Guyard (Ontario Public Health Agency, Toronto, Canada). For phagosome isolation and Phos-Tag Western blotting, we used primary rabbit antibodies to Vps15 (Bethyl Laboratories), EEA1, Rab7, and Vps34 (Cell Signaling), and UVRAG (EMD Millipore), all at 1:1,000 dilution. Secondary HRP-conjugated donkey anti–rabbit IgG antibodies and donkey anti–human antibodies were diluted to 1:10,000 (Bethyl Laboratories).
DNA constructs
The construct 2FYVE-GFP encodes two tandem copies of the FYVE domain of EEA1 fused to GFP (Gillooly et al., 2000), and the plasmid p40PX-GFP or mCherry encodes the PX domain of p40Phox fused to GFP or mCherry (Kanai et al., 2001). The plasmids GFP-Rab7, GFP-Rab5, and RILP-C33-GFP, respectively, encode WT Rab7, Rab5, and the C-terminal half of RILP cloned into pEGFP-C1 vector (Roberts et al., 1999; Bucci et al., 2000; Colucci et al., 2005).
For the lipid overlay assay, the PX domain of p40Phox and the tandem FYVE domain of EEA1 fused to GFP were PCR-amplified from p40PX-EGFP (Kanai et al., 2001) and pEGFP1-2FYVE (Gillooly et al., 2000) constructs using Gateway compatible primers (Table 1). The amplified products were then cloned into pDEST42 and pDEST15 expression vectors (Gateway Technology, Invitrogen), resulting in PX-GFP-V5-His6 and GST-GFP-2FYVE, respectively. The GFP sequence alone was PCR amplified and cloned into pDEST15 to generate GST-GFP to be used as control. Plasmid sequences were confirmed by standard DNA sequencing. Constructs were expressed in Escherichia coli BL-21 DE3 cells and induced with 0.3 mM IPTG for 12 h at 20°C for protein purification (Fig. S3).
Table 1. List of Gateway compatible primers used for PCR amplification.
| Primers | Sequences |
|---|---|
| PX-GFP-Gtwy-Forward | 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTTCGAAGGAGATAGAACCATGGCTGTGGCCCAGCAGCTG-3′ |
| PX-GFP-Gtwy-Reverse | 5′-GGGGACCACTTTGTACAAGAAAGCTGGGTCCTTGTACAGCTCGTCCATGCC-3′ |
| GFP-2FYVE-Gtwy-Forward | 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTTCGAAGGAGATAGAACCATGGTGAGCAAGGGCGAGGAG-3′ |
| GFP-2FYVE-Gtwy-Reverse | 5′-GGGGACCACTTTGTACAAGAAAGCTGGGTCTTATCCTTGCAAGTCATTGAA-3′ |
| GFP-Gtwy-Forward | 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTTCGAAGGAGATAGAACCATGGTGAGCAAGGGCGAGGAG-3′ |
| GFP-Gtwy-Reverse | 5′-GGGGACCACTTTGTACAAGAAAGCTGGGTCCTTGTACAGCTCGTCCATGCC-3′ |
Cell culture and transfection
The mouse macrophage cell line RAW264.7, obtained from the American Type Culture Collection, was cultured in DMEM medium supplemented with 10% heat-inactivated FBS at 37°C and 5% CO2. RAW cells were transfected using FuGENE HD (Promega Corp.) according to the manufacturer’s instructions, and cells were used after overnight expression of constructs.
Phagocytosis assays
For the phagocytosis of filamentous bacteria, L. pneumophila expressing red fluorescent protein were prepared and killed with PFA as described in Prashar et al. (2013). Filamentous bacteria were opsonized with 0.1 mg/ml anti-Legionella antibody for 1 h at room temperature (RT) and incubated with RAW cells, precooled at 15°C for 5 min at a ratio of 1 cell/40 filamentous bacteria. Attachment was synchronized by spinning bacteria onto cells at 300 g for 5 min at 15°C. Unattached filaments were removed by washing with PBS, and the cells were then moved to a tissue culture incubator to allow phagocytosis to proceed to indicated time periods (Prashar et al., 2013).
For the phagocytosis of latex beads, 3 µm poly(styrene/divinylbenzene) beads (Bangs Laboratories) were opsonized at a ratio of 1:7 with 2.86 mg/ml IgG from human serum (Sigma-Aldrich) in PBS, gyrating for 1 h at RT. Beads were then centrifuged and washed 3 times with PBS. The final opsonized bead mixture in PBS was added at 1:10 per well. Phagocytosis synchronization was accomplished through centrifugation at 500 g for 2 min at 4°C. Unbound beads were washed off with PBS, and cells were then incubated in cell culture conditions.
For phagocytosis of pHrodo-conjugated zymosan bioparticles (Life Technologies), particles were suspended in culture medium as per the manufacturer’s instructions. Zymosan particles were added to cells at 0.125 mg/ml in culture medium. Phagocytosis was then synchronized through centrifugation at 500 g for 5 min at 4°C. After synchronization, cells were incubated for 5 min at 15°C to allow for zymosan particle coordination to receptors, and unattached particles were washed off with PBS. Cells were then incubated at cell culture conditions.
Pharmacological inhibitors
Before phagocytosis, cells were washed and incubated in culture media containing one of the following PtdIns 3-kinase inhibitors: 100 µM LY294002 (Sigma-Aldrich) for 30 min, 1 µM ZSTK474 (Selleckchem) for 10 min, and 1 µM Vps34-IN1 (Bago et al., 2014) for 10 min. Five minutes after synchronized attachment of targets, cells were washed to remove unbound filamentous bacteria, and media-containing inhibitor was re-added. Internalization was allowed to proceed for an additional 25 min at 37°C, after which the cells were fixed using 4% PFA for 20 min. For inhibition of the V-ATPase, cells were pretreated with 1 µM ConA (BioShop) in DMSO for 15 min and/or 10 mM NH4Cl. After pretreatments, cells were incubated with these compounds during phagocytosis assays for the indicated time periods. Similarly, for PIKFyve inhibition, cells were treated with 10 nM apilimod (Toronto Research Chemicals) for 1 h before phagocytosis and during phagocytosis for the time points indicated.
Preparation of pHrodo-conjugated filamentous bacteria
PFA-killed filamentous bacteria were washed three times with 250 mM glycine in PBS, pH 7.2 (quench buffer), and then conjugated to pHrodoT Red succinimidyl ester (Life Technologies) as per the manufacturer’s directions. In brief, 109 filamentous bacteria were resuspended in 100 mM sodium bicarbonate buffer, pH 8.0, and incubated with pHrodo at a final concentration of 0.5 mM. After 45 min at RT, the unbound pHrodo was removed by three washes in PBS. pHrodo-conjugated filamentous bacteria were finally resuspended in PBS before use in phagocytic assays.
Immunofluorescence and fluorescence labeling
Filamentous actin was stained with Alexa Fluor–conjugated phalloidin (Invitrogen) diluted to 1:500 in PBS. Immunofluorescence of RAW cells and bacteria was performed as in Prashar et al. (2013). Early endosomes were labeled with anti-EEA1 (1:30), whereas late endosomes and lysosomes were labeled with anti-LAMP1 antibody (1:25) in solutions containing 5% skim milk/PBS. Fluorescence secondary antibodies (Invitrogen) were used at 1:1,000 in 5% skim milk for 1 h, RT. The coverslips were mounted using fluorescence mounting medium (Dako).
pH calibration of phagosomes
After synchronized phagocytosis, each internalized bioparticle, pHrodo Red–conjugated filamentous bacteria, or pHrodo Green–conjugated zymosan underwent pH calibrations (Fig. S4, a and d). For internal pH calibrations, the same cell samples were bathed sequentially in isotonic pH calibration buffers (ThermoFisher), ranging from pH 7.5, 6.5, 5.5, and 4.5, containing 20 µM of valinomycin and 20 µM of nigericin. To ensure complete luminal homogenization at each pH, buffers were incubated for 10 min before image acquisition using a spinning disc confocal microscope (Quorum Technologies) fitted with an Andor iXON 897 EMCCD camera. A calibration curve was generated that relates the background-corrected mean fluorescence to calibration pH. This callibration curve was then fit onto a linear function or onto a one-phase exponential decay function. The resulting function was used to transform initial phagosomal fluorescence measurements to a luminal pH value.
Microscopy
Images were acquired using a spinning disc confocal microscope consisting of a DMI6000B inverted fluorescence microscope (Leica) equipped with Hamamatsu EM-CCD and ORCA-R2 cameras and spinning disc confocal scan head, an ASI motorized XY stage, and an Improvision Piezo Focus Drive (Quorum Technologies Canada). The equipment was controlled by MetaMorph acquisition software (Molecular Devices, LLC). Unless otherwise indicated, images were acquired using a 63× oil immersion objective, NA 1.4. For live-cell imaging, phagocytosis assays were performed using a stage incubator (Live Cell Instrument) set at 37°C/5% CO2. Unless otherwise stated, macrophages were presented with opsonized filamentous bacteria for 15 min. After this period, cells were washed with PBS and incubated in FluoroBrite DMEM for imaging.
Image processing, deconvolution (90% confidence interval), and analysis were performed with Volocity (PerkinElmer Inc.) and/or ImageJ (v. 1.47 bundled with 64-bit Java). Images were processed with Adobe Photoshop and Illustrator (Adobe Systems Inc.).
Phagosome isolation
Phagosomes containing latex beads were isolated from macrophages in 6-well tissue culture plates after 20 min of phagocytosis. Cells were incubated on ice and washed with ice-cold PBS and scraped in 1 ml of ice-cold homogenization buffer (20 mM Tris, pH 7.4, 1 mM MgCl2, 1 mM CaCl2, 1 µg/ml RNase, 1 µg/ml DNase, 1 mM AEBSF, and 1:250 protease inhibitor cocktail [Sigma-Aldrich]) per well. Cells were pelleted at 500 g for 5 min at 4°C, resuspended with homogenization buffer, and lysed through a 22-gauge needle by syringing 10 to 15 times. Samples were centrifuged at 1200 g for 5 min at 4°C. The resulting pellets were suspended in 200 µl of ice-cold PBS and carefully transferred onto the sucrose gradients. Sucrose gradients were prepared freshly, before phagosome isolation, by centrifugation of 1 ml 60% sucrose (in PBS) at 22,000 g for 1 h at 4°C and stored at 4°C until use. Cell lysates were applied onto sucrose gradients and centrifuged at 22,000 g for 15 min at 4°C. Phagosomes were withdrawn from the sucrose layer and washed with ice-cold PBS.
Determination of phagosome-associated proteins and Phos-tag gel assays
Proteins associated with the isolated latex beads-containing phagosomes were extracted and resolved by Western blot. To this end, phagosomes were resuspended in Laemmli buffer supplemented with 1:100 protease inhibitor cocktail (Sigma-Aldrich). Proteins were then separated in a 10% SDS-PAGE, followed by protein transfer to a polyvinylidene difluoride membrane. Membranes were then immunoblotted using primary and secondary antibodies prepared in 5% skim milk in Tris-buffered saline buffer with 0.1% Tween 20 at the dilutions indicated. Proteins were detected using enhanced chemiluminescence, where protein loading was normalized to levels of human IgG (opsonin) in isolated phagosomes. Alternatively, we used Phos-tag gels to detect differential phosphorylation states of phagosomal markers. To this end, isolated phagosomes were resuspended in Laemmli buffer supplemented with 1:100 protease inhibitor cocktail (Sigma-Aldrich) and PhosSTOP phosphatase inhibitor cocktail (Sigma-Aldrich). SDS-PAGE gels were prepared with one fifth the strength of Phos-tag, as recommended by the manufacturer (Wako Chemicals USA). A 5.5% SDS-PAGE was used for Vps15 and Vps34, whereas UVRAG was assessed in samples run in a 7.8% SDS-PAGE. After the recommended electrophoresis settings, gels were agitated in 10 mM EDTA, pH 8.0, prepared in dH2O three times for 20 min, and then rinsed three times in transfer buffer (Bio-Rad) for 10 min, after which routine protein transfer to polyvinylidene difluoride membrane, immunoblotting, and chemiluminescence detection conditions were used.
Phosphoinositide labeling and HPLC-coupled flow scintillation
Cells were incubated in tritium labeling medium (inositol-free DMEM [MP Biomedical]) supplemented with 10 µCi/ml myo-(2-3H[N]) inositol (Perkin Elmer), 4 mM l-glutamine (Sigma-Aldrich), insulin-transferrin-selenium-ethanolamine (Gibco), 10% dialyzed FBS (Gibco), 20 mM Hepes (Gibco), and penicillin-streptomycin mix (Sigma-Aldrich) for 24 h. After incubation, tritium labeling medium was removed and replaced with cell culture medium to perform experimental conditions for 20 min. Cells were lysed on ice with 4.5% perchloric acid (vol/vol) for 15 min and pelleted at 12,000 g for 10 min. Pellets were washed with 0.1 M EDTA, pH 8.0, and sonicated in 50 µl of water. Samples were incubated in phospholipid deacylation reagent (45.7% methanol, 10.7% methylamine, and 11.4% 1-butanol [vol/vol]) for 50 min at 53°C. Samples were subsequently dried in speed-vacuum and resuspended in water by sonication twice before a final drying step for storing them. To resume analysis, dried samples were resuspended in a 1.5:1 ratio of water to extraction reagent (1-butanol/ethyl ether/ethyl formate [20:4:1]), vortexed for 5 min, and centrifuged for 2 min. The aqueous layer was carefully isolated and extracted twice more, then vacuum-dried and resuspended in 50 µl of water. For each sample, equal counts of tritium were separated by HPLC (Agilent Technologies) through an anion exchange 4.6 × 250-mm column (Phenomenex) and subjected to a gradient of water and 1 M (NH4)2HPO4, pH 3.8. For the HPLC separation protocol, see Ho et al. (2016). The radiolabeled eluate was detected with a 1:2 ratio of eluate to scintillation fluid by β-RAM 4 (LabLogic) and analyzed by Laura 4 software. PtdIns(3)P and phosphatidylinositol-4,5-bisphosphate levels were normalized against the parent PtdIns peak.
Recombinant protein purification for lipid overlay assay
For purification of PX-GFP-V5-His6 and GFP-V5-His6, cells were pelleted and resuspended in lysis buffer (50 mM NaH2PO4, pH 8.0, 300 mM NaCl, 10 mM imidazole, 0.05% Tween-20, 1 mM phenylmethane sulfonyl fluoride, 1× protease inhibitor cocktail [Roche], and 3 µg/ml pepstatin) and sonicated. Lysates were cleared for 30 min at 15,000 g, and the supernatant was collected and incubated with Ni-NTA beads (QIAGEN) preequilibrated with lysis buffer for 1 h at 4°C. The mixture was then loaded into a chromatography column (BioRad) and washed four times with wash buffer (50 mM NaH2PO4, pH 8.0, 300 mM NaCl, 10 mM imidazole, and 0.05% Tween 20). The fusion protein was eluted with elution buffer (50 mM NaH2PO4 pH 8.0, 300 mM NaCl, 250 mM imidazole, and 0.05% Tween 20) and concentrated using an Amicon Ultra-15 centrifugal filter.
For purification of GST-GFP-2FYVE and GST-GFP, cells were pelleted and resuspended in GST lysis buffer A (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 5 mM DTT, 0.05% NP-40, 1 mM phenylmethane sulfonyl fluoride, and 1 mM benzamidine). Cells were sonicated and lysates were cleared for 40 min at 20,000 g. The supernatant was collected and incubated with Glutathione Sepharose 4B (GE Healthcare) preequilibrated with lysis buffer B (50 mM Tris-HCl, 150 mM NaCl, 3 mM DTT, 0.05% NP-40) for 1.5 h at 4°C. The mixture was then loaded into a chromatography column (BioRad) and washed four times with wash buffer A (25 mM Tris-HCl, pH 7.5, 750 mM NaCl, 1 mM DTT, and 0.1% NP-40) and once with wash buffer B (25 mM Tris-HCl, pH 7.5, 150 mM NaCl, and 1 mM DTT). Fusion proteins were eluted with elution buffer (25 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1 mM DTT, and 30 mM reduced glutathione) and concentrated using an Amicon Ultra-15 centrifugal filter and then analyzed by 8% SDS-PAGE preloaded with 2,2,2-tricholoethanol (Fig. S3 a).
Protein-lipid overlay assay
Lipids (Avanti Polar) stocks were dissolved in chroloform/methanol/water (20:9:1 vol/vol). The indicated amount of lipids were spotted on a nitrocellulose membrane (GE Healthcare) and allowed to dry at RT for 1 h. The membranes were then blocked for 1 h in Tris-HCl 50 mM, pH 7.5, 150 mM NaCl, and 0.1% Tween 20 buffer containing 3% fat-free BSA and then incubated at 4°C overnight with 5 µg/ml of GST-GFP-2FYVE, GST-GFP, or PX-GFP-V5-His6 in pH calibration buffers (140 mM KCl, 1 mM MgCl2,1 mM CaCl2, 5 mM glucose, 3% fat free BSA, and 10 mM of the appropriate buffer covering a pH ranging from 7.5 to 4.0). The following buffers were used: pH 4.0, acetate-acetic acid; pH 4.5, acetate-acetic acid; pH 5.0, acetate-acetic acid; pH 5.5, 2-(N-morpholino) ethanesulfonic acid (MES); pH 6.0, MES; pH 6.5, MES; pH 7.0, Hepes; pH 7.5, TBS. Buffers were adjusted to proper pH using either 1 M KOH or 1 M HCl. The membranes were then washed, and GFP-tagged proteins were detected with anti-GFP antibody (Roche) by chemiluminescence using an Imager 600 (GE Healthcare). Densitometry of the blots was performed using ImageJ. Experiments were repeated at least three times.
Statistical analysis
Data shown are mean ± SEM from at least three independent experiments unless otherwise stated. Statistical analysis was performed using a two-tailed Student’s t test or ANOVA using GraphPad Prism software (GraphPad Software, Inc.) and SigmaPlot software (Systat Software Inc.). A 95% confidence interval was used to determine statistical significance, and P ≤ 0.05 was considered to be statistically significant.
Online supplemental material
Fig. S1 a shows a schematic of the tPC. Fig. S1 b shows 2FYVE-GFP distribution in tPCs. Fig. S1 c shows possible pathways for PtdIns(3)P production at tPCs. Fig. S1 d shows the length of filamentous bacteria positive for p40PX-GFP versus the bacterial length internalized at each time point. Fig. S2 a shows PtdIns(3)P and acidification of tPCs measured in selected frames from Video 2 (related to Fig. 5). Fig. S2, b and c, Effect of ConA treatment in PtdIns(3)P retention in tPCs. Fig. S3 (a–c) shows that recombinant GST-GFP-2FYVE and PX-GFP-V5-His6 bind specifically to PtdIns(3)P. Fig. S3 c shows protein lipid overlays at the indicated range of pH. Fig. S4 a, pH calibration curves for Legionella-containing phagosomes. Fig. S4 b shows protein-lipid overlay of GST-GFP-2FYVE onto PtdIns(3)P. Fig. S4 c shows pH calibration curves for zymosan-containing phagosomes. Fig. S4 d shows the evolution of phagosomal pH and p40PX-mCh in zymosan-containing phagosomes. Video 1 shows PtdIns(3)P dynamics in tPCs. Videos 2 and 3 show the correlation between acidification and loss of PtdIns(3)P in tPCs. Video 4 shows that the exposure of macrophages to acidic media induces the loss of PtdIns(3)P in tPCs.
Supplementary Material
Acknowledgments
This work was supported by a grant from the Natural Sciences and Engineering Research Council to M.R. Terebiznik. R.J. Botelho is funded by the Natural Sciences and Engineering Research Council and the Canadian Institutes of Health Research. V. Zaremberg is funded by the Natural Science and Engineering Research Council. This work was also supported by an Early Researcher Award from the Government of Ontario and contributions from Ryerson University to R.J. Botelho. R.J. Botelho holds a Canada Research Chair in Biomedical Sciences. A. Naufer was supported by a Natural Sciences and Engineering Research Council A.G. Bell Award. V.E.B. Hipolito was supported by an Ontario Graduate Scholarship, Canada Graduate Scholarship, and Ryerson Graduate Fellowship. We would also like to thank the Centre for Neurobiology of Stress at University of Toronto Scarborough for the use of their imaging facility.
The authors declare no competing financial interests.
Author contributions: A. Naufer, V.E.B. Hipolito, S. Ganesan, and A. Prashar carried out and analyzed experiments and contributed to the writing of the manuscript. V. Zaremberg, R.J. Botelho, and M.R. Terebiznik designed experiments, wrote the manuscript, and supervised the project.
Footnotes
Abbreviations used:
- ConA
- concanamycin A
- LAMP
- Lysosomal-associated membrane protein
- MES
- 2-(N-morpholino) ethanesulfonic acid
- PtdIns
- phosphatidylinositol
- PtdIns(3)P
- phosphatidylinositol-3-phosphate
- tPC
- tubular phagocytic cup
References
- Backer J.M. 2008. The regulation and function of Class III PI3Ks: novel roles for Vps34. Biochem. J. 410:1–17. 10.1042/BJ20071427 [DOI] [PubMed] [Google Scholar]
- Backer J.M. 2016. The intricate regulation and complex functions of the Class III phosphoinositide 3-kinase Vps34. Biochem. J. 473:2251–2271. 10.1042/BCJ20160170 [DOI] [PubMed] [Google Scholar]
- Bago R., Malik N., Munson M.J., Prescott A.R., Davies P., Sommer E., Shpiro N., Ward R., Cross D., Ganley I.G., and Alessi D.R.. 2014. Characterization of VPS34-IN1, a selective inhibitor of Vps34, reveals that the phosphatidylinositol 3-phosphate-binding SGK3 protein kinase is a downstream target of class III phosphoinositide 3-kinase. Biochem. J. 463:413–427. 10.1042/BJ20140889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-Peled L., and Sabatini D.M.. 2014. Regulation of mTORC1 by amino acids. Trends Cell Biol. 24:400–406. 10.1016/j.tcb.2014.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behnia R., and Munro S.. 2005. Organelle identity and the signposts for membrane traffic. Nature. 438:597–604. 10.1038/nature04397 [DOI] [PubMed] [Google Scholar]
- Berger S.B., Romero X., Ma C., Wang G., Faubion W.A., Liao G., Compeer E., Keszei M., Rameh L., Wang N., et al. . 2010. SLAM is a microbial sensor that regulates bacterial phagosome functions in macrophages. Nat. Immunol. 11:920–927. 10.1038/ni.1931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchette C.D., Woo Y.H., Thomas C., Shen N., Sulchek T.A., and Hiddessen A.L.. 2009. Decoupling internalization, acidification and phagosomal-endosomal/lysosomal fusion during phagocytosis of InlA coated beads in epithelial cells. PLoS One. 4:e6056 10.1371/journal.pone.0006056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohdanowicz M., and Grinstein S.. 2013. Role of phospholipids in endocytosis, phagocytosis, and macropinocytosis. Physiol. Rev. 93:69–106. 10.1152/physrev.00002.2012 [DOI] [PubMed] [Google Scholar]
- Bouvier G., Benoliel A.M., Foa C., and Bongrand P.. 1994. Relationship between phagosome acidification, phagosome-lysosome fusion, and mechanism of particle ingestion. J. Leukoc. Biol. 55:729–734. [DOI] [PubMed] [Google Scholar]
- Bucci C., Thomsen P., Nicoziani P., McCarthy J., and van Deurs B.. 2000. Rab7: a key to lysosome biogenesis. Mol. Biol. Cell. 11:467–480. 10.1091/mbc.11.2.467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao C., Backer J.M., Laporte J., Bedrick E.J., and Wandinger-Ness A.. 2008. Sequential actions of myotubularin lipid phosphatases regulate endosomal PI(3)P and growth factor receptor trafficking. Mol. Biol. Cell. 19:3334–3346. 10.1091/mbc.E08-04-0367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casey J.R., Grinstein S., and Orlowski J.. 2010. Sensors and regulators of intracellular pH. Nat. Rev. Mol. Cell Biol. 11:50–61. 10.1038/nrm2820 [DOI] [PubMed] [Google Scholar]
- Champion J.A., Walker A., and Mitragotri S.. 2008. Role of particle size in phagocytosis of polymeric microspheres. Pharm. Res. 25:1815–1821. 10.1007/s11095-008-9562-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng S., Wang K., Zou W., Miao R., Huang Y., Wang H., and Wang X.. 2015. PtdIns(4,5)P2 and PtdIns3P coordinate to regulate phagosomal sealing for apoptotic cell clearance. J. Cell Biol. 210:485–502. 10.1083/jcb.201501038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christoforidis S., McBride H.M., Burgoyne R.D., and Zerial M.. 1999. The Rab5 effector EEA1 is a core component of endosome docking. Nature. 397:621–625. 10.1038/17618 [DOI] [PubMed] [Google Scholar]
- Colucci A.M., Campana M.C., Bellopede M., and Bucci C.. 2005. The Rab-interacting lysosomal protein, a Rab7 and Rab34 effector, is capable of self-interaction. Biochem. Biophys. Res. Commun. 334:128–133. 10.1016/j.bbrc.2005.06.067 [DOI] [PubMed] [Google Scholar]
- Demaurex N. 2002. pH Homeostasis of cellular organelles. News Physiol. Sci. 17:1–5. [DOI] [PubMed] [Google Scholar]
- Doshi N., and Mitragotri S.. 2010. Macrophages recognize size and shape of their targets. PLoS One. 5:e10051 10.1371/journal.pone.0010051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellson C.D., Anderson K.E., Morgan G., Chilvers E.R., Lipp P., Stephens L.R., and Hawkins P.T.. 2001. Phosphatidylinositol 3-phosphate is generated in phagosomal membranes. Curr. Biol. 11:1631–1635. 10.1016/S0960-9822(01)00447-X [DOI] [PubMed] [Google Scholar]
- Fairn G.D., and Grinstein S.. 2012. How nascent phagosomes mature to become phagolysosomes. Trends Immunol. 33:397–405. 10.1016/j.it.2012.03.003 [DOI] [PubMed] [Google Scholar]
- Flannagan R.S., Jaumouillé V., and Grinstein S.. 2012. The cell biology of phagocytosis. Annu. Rev. Pathol. 7:61–98. 10.1146/annurev-pathol-011811-132445 [DOI] [PubMed] [Google Scholar]
- Fratti R.A., Backer J.M., Gruenberg J., Corvera S., and Deretic V.. 2001. Role of phosphatidylinositol 3-kinase and Rab5 effectors in phagosomal biogenesis and mycobacterial phagosome maturation arrest. J. Cell Biol. 154:631–644. 10.1083/jcb.200106049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisow M.J., D’Arcy Hart P., and Young M.R.. 1981. Temporal changes of lysosome and phagosome pH during phagolysosome formation in macrophages: studies by fluorescence spectroscopy. J. Cell Biol. 89:645–652. 10.1083/jcb.89.3.645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gharbi S.I., Zvelebil M.J., Shuttleworth S.J., Hancox T., Saghir N., Timms J.F., and Waterfield M.D.. 2007. Exploring the specificity of the PI3K family inhibitor LY294002. Biochem. J. 404:15–21. 10.1042/BJ20061489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillooly D.J., Morrow I.C., Lindsay M., Gould R., Bryant N.J., Gaullier J.M., Parton R.G., and Stenmark H.. 2000. Localization of phosphatidylinositol 3-phosphate in yeast and mammalian cells. EMBO J. 19:4577–4588. 10.1093/emboj/19.17.4577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray M., and Botelho R.J.. 2017. Phagocytosis: Hungry, Hungry Cells. Methods Mol. Biol. 1519:1–16. 10.1007/978-1-4939-6581-6_1 [DOI] [PubMed] [Google Scholar]
- Harrison R.E., Bucci C., Vieira O.V., Schroer T.A., and Grinstein S.. 2003. Phagosomes fuse with late endosomes and/or lysosomes by extension of membrane protrusions along microtubules: role of Rab7 and RILP. Mol. Cell. Biol. 23:6494–6506. 10.1128/MCB.23.18.6494-6506.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haupts U., Maiti S., Schwille P., and Webb W.W.. 1998. Dynamics of fluorescence fluctuations in green fluorescent protein observed by fluorescence correlation spectroscopy. Proc. Natl. Acad. Sci. USA. 95:13573–13578. 10.1073/pnas.95.23.13573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazeki K., Nigorikawa K., Takaba Y., Segawa T., Nukuda A., Masuda A., Ishikawa Y., Kubota K., Takasuga S., and Hazeki O.. 2012. Essential roles of PIKfyve and PTEN on phagosomal phosphatidylinositol 3-phosphate dynamics. FEBS Lett. 586:4010–4015. 10.1016/j.febslet.2012.09.043 [DOI] [PubMed] [Google Scholar]
- He J., Vora M., Haney R.M., Filonov G.S., Musselman C.A., Burd C.G., Kutateladze A.G., Verkhusha V.V., Stahelin R.V., and Kutateladze T.G.. 2009. Membrane insertion of the FYVE domain is modulated by pH. Proteins. 76:852–860. 10.1002/prot.22392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho C.Y., Alghamdi T.A., and Botelho R.J.. 2012. Phosphatidylinositol-3,5-bisphosphate: no longer the poor PIP2. Traffic. 13:1–8. 10.1111/j.1600-0854.2011.01246.x [DOI] [PubMed] [Google Scholar]
- Ho C.Y., Choy C.H., and Botelho R.J.. 2016. Radiolabeling and Quantification of Cellular Levels of Phosphoinositides by High Performance Liquid Chromatography-coupled Flow Scintillation. J. Vis. Exp. 107:. 10.3791/53529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horiuchi H., Lippé R., McBride H.M., Rubino M., Woodman P., Stenmark H., Rybin V., Wilm M., Ashman K., Mann M., and Zerial M.. 1997. A novel Rab5 GDP/GTP exchange factor complexed to Rabaptin-5 links nucleotide exchange to effector recruitment and function. Cell. 90:1149–1159. 10.1016/S0092-8674(00)80380-3 [DOI] [PubMed] [Google Scholar]
- Huynh K.K., Eskelinen E.L., Scott C.C., Malevanets A., Saftig P., and Grinstein S.. 2007. LAMP proteins are required for fusion of lysosomes with phagosomes. EMBO J. 26:313–324. 10.1038/sj.emboj.7601511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida Y., Nayak S., Mindell J.A., and Grabe M.. 2013. A model of lysosomal pH regulation. J. Gen. Physiol. 141:705–720. 10.1085/jgp.201210930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeschke A., Zehethofer N., Lindner B., Krupp J., Schwudke D., Haneburger I., Jovic M., Backer J.M., Balla T., Hilbi H., and Haas A.. 2015. Phosphatidylinositol 4-phosphate and phosphatidylinositol 3-phosphate regulate phagolysosome biogenesis. Proc. Natl. Acad. Sci. USA. 112:4636–4641. 10.1073/pnas.1423456112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin N., Lang M.J., and Weisman L.S.. 2016. Phosphatidylinositol 3,5-bisphosphate: regulation of cellular events in space and time. Biochem. Soc. Trans. 44:177–184. 10.1042/BST20150174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson D.E., and Casey J.R.. 2011. Cytosolic H+ microdomain developed around AE1 during AE1-mediated Cl-/HCO3- exchange. J. Physiol. 589:1551–1569. 10.1113/jphysiol.2010.201483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson D.E., Ostrowski P., Jaumouillé V., and Grinstein S.. 2016. The position of lysosomes within the cell determines their luminal pH. J. Cell Biol. 212:677–692. 10.1083/jcb.201507112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Justice S.S., Hunstad D.A., Cegelski L., and Hultgren S.J.. 2008. Morphological plasticity as a bacterial survival strategy. Nat. Rev. Microbiol. 6:162–168. 10.1038/nrmicro1820 [DOI] [PubMed] [Google Scholar]
- Kanai F., Liu H., Field S.J., Akbary H., Matsuo T., Brown G.E., Cantley L.C., and Yaffe M.B.. 2001. The PX domains of p47phox and p40phox bind to lipid products of PI(3)K. Nat. Cell Biol. 3:675–678. 10.1038/35083070 [DOI] [PubMed] [Google Scholar]
- Kim G.H., Dayam R.M., Prashar A., Terebiznik M., and Botelho R.J.. 2014. PIKfyve inhibition interferes with phagosome and endosome maturation in macrophages. Traffic. 15:1143–1163. 10.1111/tra.12199 [DOI] [PubMed] [Google Scholar]
- Kim J., Kim Y.C., Fang C., Russell R.C., Kim J.H., Fan W., Liu R., Zhong Q., and Guan K.L.. 2013. Differential regulation of distinct Vps34 complexes by AMPK in nutrient stress and autophagy. Cell. 152:290–303. 10.1016/j.cell.2012.12.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinchen J.M., and Ravichandran K.S.. 2008. Phagosome maturation: going through the acid test. Nat. Rev. Mol. Cell Biol. 9:781–795. 10.1038/nrm2515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koivusalo M., Welch C., Hayashi H., Scott C.C., Kim M., Alexander T., Touret N., Hahn K.M., and Grinstein S.. 2010. Amiloride inhibits macropinocytosis by lowering submembranous pH and preventing Rac1 and Cdc42 signaling. J. Cell Biol. 188:547–563. (published erratum appears in J. Cell Biol. 189: 385) 10.1083/jcb.200908086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong D., and Yamori T.. 2007. ZSTK474 is an ATP-competitive inhibitor of class I phosphatidylinositol 3 kinase isoforms. Cancer Sci. 98:1638–1642. 10.1111/j.1349-7006.2007.00580.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.A., Eyeson R., Cheever M.L., Geng J., Verkhusha V.V., Burd C., Overduin M., and Kutateladze T.G.. 2005. Targeting of the FYVE domain to endosomal membranes is regulated by a histidine switch. Proc. Natl. Acad. Sci. USA. 102:13052–13057. 10.1073/pnas.0503900102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin R., Grinstein S., and Schlam D.. 2015. Phosphoinositides in phagocytosis and macropinocytosis. Biochim. Biophys. Acta. 1851:805–823. 10.1016/j.bbalip.2014.09.005 [DOI] [PubMed] [Google Scholar]
- Liger D., Nizard P., Gaillard C., vanderSpek J.C., Murphy J.R., Pitard B., and Gillet D.. 1998. The diphtheria toxin transmembrane domain as a pH sensitive membrane anchor for human interleukin-2 and murine interleukin-3. Protein Eng. 11:1111–1120. 10.1093/protein/11.11.1111 [DOI] [PubMed] [Google Scholar]
- Lu N., and Zhou Z.. 2012. Membrane trafficking and phagosome maturation during the clearance of apoptotic cells. Int. Rev. Cell Mol. Biol. 293:269–309. 10.1016/B978-0-12-394304-0.00013-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukacs G.L., Rotstein O.D., and Grinstein S.. 1990. Phagosomal acidification is mediated by a vacuolar-type H(+)-ATPase in murine macrophages. J. Biol. Chem. 265:21099–21107. [PubMed] [Google Scholar]
- Marshansky V. 2007. The V-ATPase a2-subunit as a putative endosomal pH-sensor. Biochem. Soc. Trans. 35:1092–1099. 10.1042/BST0351092 [DOI] [PubMed] [Google Scholar]
- Marshansky V., Rubinstein J.L., and Grüber G.. 2014. Eukaryotic V-ATPase: novel structural findings and functional insights. Biochim. Biophys. Acta. 1837:857–879. 10.1016/j.bbabio.2014.01.018 [DOI] [PubMed] [Google Scholar]
- McCartney A.J., Zhang Y., and Weisman L.S.. 2014. Phosphatidylinositol 3,5-bisphosphate: low abundance, high significance. BioEssays. 36:52–64. 10.1002/bies.201300012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao Y., Li G., Zhang X., Xu H., and Abraham S.N.. 2015. A TRP Channel Senses Lysosome Neutralization by Pathogens to Trigger Their Expulsion. Cell. 161:1306–1319. 10.1016/j.cell.2015.05.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra A., Eathiraj S., Corvera S., and Lambright D.G.. 2010. Structural basis for Rab GTPase recognition and endosome tethering by the C2H2 zinc finger of Early Endosomal Autoantigen 1 (EEA1). Proc. Natl. Acad. Sci. USA. 107:10866–10871. 10.1073/pnas.1000843107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Möller J., Luehmann T., Hall H., and Vogel V.. 2012. The race to the pole: how high-aspect ratio shape and heterogeneous environments limit phagocytosis of filamentous Escherichia coli bacteria by macrophages. Nano Lett. 12:2901–2905. 10.1021/nl3004896 [DOI] [PubMed] [Google Scholar]
- Munson M.J., Allen G.F., Toth R., Campbell D.G., Lucocq J.M., and Ganley I.G.. 2015. mTOR activates the VPS34-UVRAG complex to regulate autolysosomal tubulation and cell survival. EMBO J. 34:2272–2290. 10.15252/embj.201590992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandurkar H.H., and Huysmans R.. 2002. The myotubularin family: novel phosphoinositide regulators. IUBMB Life. 53:37–43. 10.1080/15216540210812 [DOI] [PubMed] [Google Scholar]
- Nordmann M., Cabrera M., Perz A., Bröcker C., Ostrowicz C., Engelbrecht-Vandré S., and Ungermann C.. 2010. The Mon1-Ccz1 complex is the GEF of the late endosomal Rab7 homolog Ypt7. Curr. Biol. 20:1654–1659. 10.1016/j.cub.2010.08.002 [DOI] [PubMed] [Google Scholar]
- Paul D., Achouri S., Yoon Y.Z., Herre J., Bryant C.E., and Cicuta P.. 2013. Phagocytosis dynamics depends on target shape. Biophys. J. 105:1143–1150. 10.1016/j.bpj.2013.07.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitt A., Mayorga L.S., Stahl P.D., and Schwartz A.L.. 1992. Alterations in the protein composition of maturing phagosomes. J. Clin. Invest. 90:1978–1983. 10.1172/JCI116077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitt S.J., Funnell T.M., Sitsapesan M., Venturi E., Rietdorf K., Ruas M., Ganesan A., Gosain R., Churchill G.C., Zhu M.X., et al. . 2010. TPC2 is a novel NAADP-sensitive Ca2+ release channel, operating as a dual sensor of luminal pH and Ca2+. J. Biol. Chem. 285:35039–35046. 10.1074/jbc.M110.156927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posor Y., Eichhorn-Grünig M., and Haucke V.. 2015. Phosphoinositides in endocytosis. Biochim. Biophys. Acta. 1851:794–804. 10.1016/j.bbalip.2014.09.014 [DOI] [PubMed] [Google Scholar]
- Prashar A., Bhatia S., Gigliozzi D., Martin T., Duncan C., Guyard C., and Terebiznik M.R.. 2013. Filamentous morphology of bacteria delays the timing of phagosome morphogenesis in macrophages. J. Cell Biol. 203:1081–1097. 10.1083/jcb.201304095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qa’Dan M., Spyres L.M., and Ballard J.D.. 2000. pH-induced conformational changes in Clostridium difficile toxin B. Infect. Immun. 68:2470–2474. 10.1128/IAI.68.5.2470-2474.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rink J., Ghigo E., Kalaidzidis Y., and Zerial M.. 2005. Rab conversion as a mechanism of progression from early to late endosomes. Cell. 122:735–749. 10.1016/j.cell.2005.06.043 [DOI] [PubMed] [Google Scholar]
- Roberts R.L., Barbieri M.A., Pryse K.M., Chua M., Morisaki J.H., and Stahl P.D.. 1999. Endosome fusion in living cells overexpressing GFP-rab5. J. Cell Sci. 112:3667–3675. [DOI] [PubMed] [Google Scholar]
- Robinson F.L., and Dixon J.E.. 2006. Myotubularin phosphatases: policing 3-phosphoinositides. Trends Cell Biol. 16:403–412. 10.1016/j.tcb.2006.06.001 [DOI] [PubMed] [Google Scholar]
- Rostislavleva K., Soler N., Ohashi Y., Zhang L., Pardon E., Burke J.E., Masson G.R., Johnson C., Steyaert J., Ktistakis N.T., and Williams R.L.. 2015. Structure and flexibility of the endosomal Vps34 complex reveals the basis of its function on membranes. Science. 350:aac7365 10.1126/science.aac7365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sbrissa D., Ikonomov O.C., and Shisheva A.. 1999. PIKfyve, a mammalian ortholog of yeast Fab1p lipid kinase, synthesizes 5-phosphoinositides. Effect of insulin. J. Biol. Chem. 274:21589–21597. 10.1074/jbc.274.31.21589 [DOI] [PubMed] [Google Scholar]
- Sbrissa D., Ikonomov O.C., and Shisheva A.. 2002. Phosphatidylinositol 3-phosphate-interacting domains in PIKfyve. Binding specificity and role in PIKfyve. Endomenbrane localization. J. Biol. Chem. 277:6073–6079. 10.1074/jbc.M110194200 [DOI] [PubMed] [Google Scholar]
- Schink K.O., Raiborg C., and Stenmark H.. 2013. Phosphatidylinositol 3-phosphate, a lipid that regulates membrane dynamics, protein sorting and cell signalling. BioEssays. 35:900–912. 10.1002/bies.201300064 [DOI] [PubMed] [Google Scholar]
- Shin H.W., Hayashi M., Christoforidis S., Lacas-Gervais S., Hoepfner S., Wenk M.R., Modregger J., Uttenweiler-Joseph S., Wilm M., Nystuen A., et al. . 2005. An enzymatic cascade of Rab5 effectors regulates phosphoinositide turnover in the endocytic pathway. J. Cell Biol. 170:607–618. 10.1083/jcb.200505128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonsen A., Lippé R., Christoforidis S., Gaullier J.M., Brech A., Callaghan J., Toh B.H., Murphy C., Zerial M., and Stenmark H.. 1998. EEA1 links PI(3)K function to Rab5 regulation of endosome fusion. Nature. 394:494–498. 10.1038/28879 [DOI] [PubMed] [Google Scholar]
- Stenmark H. 2009. Rab GTPases as coordinators of vesicle traffic. Nat. Rev. Mol. Cell Biol. 10:513–525. 10.1038/nrm2728 [DOI] [PubMed] [Google Scholar]
- Stransky L.A., and Forgac M.. 2015. Amino Acid Availability Modulates Vacuolar H+-ATPase Assembly. J. Biol. Chem. 290:27360–27369. 10.1074/jbc.M115.659128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson J.A. 2008. Shaping cups into phagosomes and macropinosomes. Nat. Rev. Mol. Cell Biol. 9:639–649. 10.1038/nrm2447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thi E.P., and Reiner N.E.. 2012. Phosphatidylinositol 3-kinases and their roles in phagosome maturation. J. Leukoc. Biol. 92:553–566. 10.1189/jlb.0212053 [DOI] [PubMed] [Google Scholar]
- Tian W., Li X.J., Stull N.D., Ming W., Suh C.I., Bissonnette S.A., Yaffe M.B., Grinstein S., Atkinson S.J., and Dinauer M.C.. 2008. Fc gamma R-stimulated activation of the NADPH oxidase: phosphoinositide-binding protein p40phox regulates NADPH oxidase activity after enzyme assembly on the phagosome. Blood. 112:3867–3877. 10.1182/blood-2007-11-126029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Via L.E., Deretic D., Ulmer R.J., Hibler N.S., Huber L.A., and Deretic V.. 1997. Arrest of mycobacterial phagosome maturation is caused by a block in vesicle fusion between stages controlled by rab5 and rab7. J. Biol. Chem. 272:13326–13331. 10.1074/jbc.272.20.13326 [DOI] [PubMed] [Google Scholar]
- Vieira O.V., Botelho R.J., Rameh L., Brachmann S.M., Matsuo T., Davidson H.W., Schreiber A., Backer J.M., Cantley L.C., and Grinstein S.. 2001. Distinct roles of class I and class III phosphatidylinositol 3-kinases in phagosome formation and maturation. J. Cell Biol. 155:19–25. 10.1083/jcb.200107069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira O.V., Botelho R.J., and Grinstein S.. 2002. Phagosome maturation: aging gracefully. Biochem. J. 366:689–704. 10.1042/bj20020691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira O.V., Bucci C., Harrison R.E., Trimble W.S., Lanzetti L., Gruenberg J., Schreiber A.D., Stahl P.D., and Grinstein S.. 2003. Modulation of Rab5 and Rab7 recruitment to phagosomes by phosphatidylinositol 3-kinase. Mol. Cell. Biol. 23:2501–2514. 10.1128/MCB.23.7.2501-2514.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H., and Ren D.. 2015. Lysosomal physiology. Annu. Rev. Physiol. 77:57–80. 10.1146/annurev-physiol-021014-071649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang D.C., Blair K.M., and Salama N.R.. 2016. Staying in Shape: the Impact of Cell Shape on Bacterial Survival in Diverse Environments. Microbiol. Mol. Biol. Rev. 80:187–203. 10.1128/MMBR.00031-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu M.X., Evans A.M., Ma J., Parrington J., and Galione A.. 2010. Two-pore channels for integrative Ca signaling. Commun. Integr. Biol. 3:12–17. 10.4161/cib.3.1.9793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolov S.N., Bridges D., Zhang Y., Lee W.W., Riehle E., Verma R., Lenk G.M., Converso-Baran K., Weide T., Albin R.L., et al. . 2012. In vivo, Pikfyve generates PI(3,5)P2, which serves as both a signaling lipid and the major precursor for PI5P. Proc. Natl. Acad. Sci. USA. 109:17472–17477. 10.1073/pnas.1203106109 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.