Mitotic spindle component BuGZ is known to undergo phase separation. Huang et al. show that BuGZ promotes Aurora A phosphorylation and activation and that this is inhibited when BuGZ phase separation is disrupted.
Abstract
Protein phase separation or coacervation has emerged as a potential mechanism to regulate biological functions. We have shown that coacervation of a mostly unstructured protein, BuGZ, promotes assembly of spindle and its matrix. BuGZ in the spindle matrix binds and concentrates tubulin to promote microtubule (MT) assembly. It remains unclear, however, whether BuGZ could regulate additional proteins to promote spindle assembly. In this study, we report that BuGZ promotes Aurora A (AurA) activation in vitro. Depletion of BuGZ in cells reduces the amount of phosphorylated AurA on spindle MTs. BuGZ also enhances MCAK phosphorylation. The two zinc fingers in BuGZ directly bind to the kinase domain of AurA, which allows AurA to incorporate into the coacervates formed by BuGZ in vitro. Importantly, mutant BuGZ that disrupts the coacervation activity in vitro fails to promote AurA phosphorylation in Xenopus laevis egg extracts. These results suggest that BuGZ coacervation promotes AurA activation in mitosis.
Introduction
Spindle formation is regulated by many spindle assembly factors (SAFs) that localize along the spindle. For example, the SAF Aurora A (AurA), a mitotic kinase, localizes along spindle microtubules (MTs), with the highest concentration found at spindle poles. AurA promotes spindle assembly by phosphorylating other SAFs (Nikonova et al., 2013; Fu et al., 2015; Lim et al., 2016). Studies have shown that AurA is activated by its interacting SAFs such as TPX2 (Bayliss et al., 2003; Eyers et al., 2003; Tsai et al., 2003; Tsai and Zheng, 2005), Ajuba (Hirota et al., 2003; Sabino et al., 2011; Bai et al., 2014), and HEF1 (Pugacheva and Golemis, 2005), and is inactivated by phosphatases (Zeng et al., 2010). Among these proteins, the mechanism by which TPX2 activates AurA is best understood. Allosteric AurA activation is achieved when TPX2 binds to a conserved hydrophobic groove of the protein, resulting in AurA assuming an active conformation (Zorba et al., 2014).
AurA can also be activated by autophosphorylation of its threonine 288 (T288; Walter et al., 2000; Littlepage et al., 2002). Recent studies show that autophosphorylation of AurA involves two interacting kinase molecules that add the phosphate group to each other on T288. However, only a small fraction of AurA forms stable dimers in vitro with an estimated Kd >300 µM (Zorba et al., 2014). Therefore, dimer-mediated AurA autophosphorylation may be inefficient unless some AurA-interacting SAFs can promote AurA dimer formation. Indeed, AurA binds to the centrosome protein Cep192, which can concentrate AurA at centrosomes to promote AurA phosphorylation and activation (Joukov et al., 2010, 2014). Despite these studies, the control of AurA activity and localization on spindles remains incompletely understood (Kufer et al., 2002; Sardon et al., 2008).
Bub3-interacting and GLE-2–binding sequence containing ZNF207 (BuGZ) was identified as a component of the spindle matrix (Ma et al., 2009). Through RNAi-mediated screens, two independent studies show that BuGZ interacts with and stabilizes Bub3 to promote MT–kinetochore interaction and chromosome alignment in mitosis (Jiang et al., 2014; Toledo et al., 2014). BuGZ localization at kinetochores depends on Bub3 (Toledo et al., 2014; Dai et al., 2016). Interestingly, BuGZ contains a nuclear localization signal (NLS) at its N terminus, and it is concentrated in the interphase nucleus, where it promotes proper mRNA splicing, although the mechanism remains unknown (Wan et al., 2015). The NLS of BuGZ interacts with importins, which helps to stabilize BuGZ by preventing its interaction with an E3 ubiquitin ligase, Ubr5. During metaphase, a high concentration of RanGTP dislodges importins from BuGZ, leading to Ubr5-mediated BuGZ degradation. This in turn aids Bub3 reduction and facilitates metaphase-to-anaphase transition (Jiang et al., 2015a).
BuGZ is also enriched on spindles, and it enhances MT assembly as part of the spindle matrix during spindle formation independent of its function at kinetochores (Jiang et al., 2015b). BuGZ undergoes coacervation, which in turn promotes the assembly of both the spindle MTs and spindle matrix (Jiang et al., 2015b). The N-terminal 92 amino acids of BuGZ directly bind to tubulin, and via BuGZ coacervation, tubulin is greatly concentrated in the spindle matrix formed in Xenopus laevis egg extracts or in BuGZ coacervates formed in vitro. This explains in part how BuGZ can promote spindle assembly (Jiang et al., 2015b).
In this study, we report that the two zinc fingers of BuGZ directly bind to AurA and that BuGZ coacervation appears to promote AurA activation during spindle assembly.
Results and discussion
BuGZ contributes to AurA kinase activation in mitosis
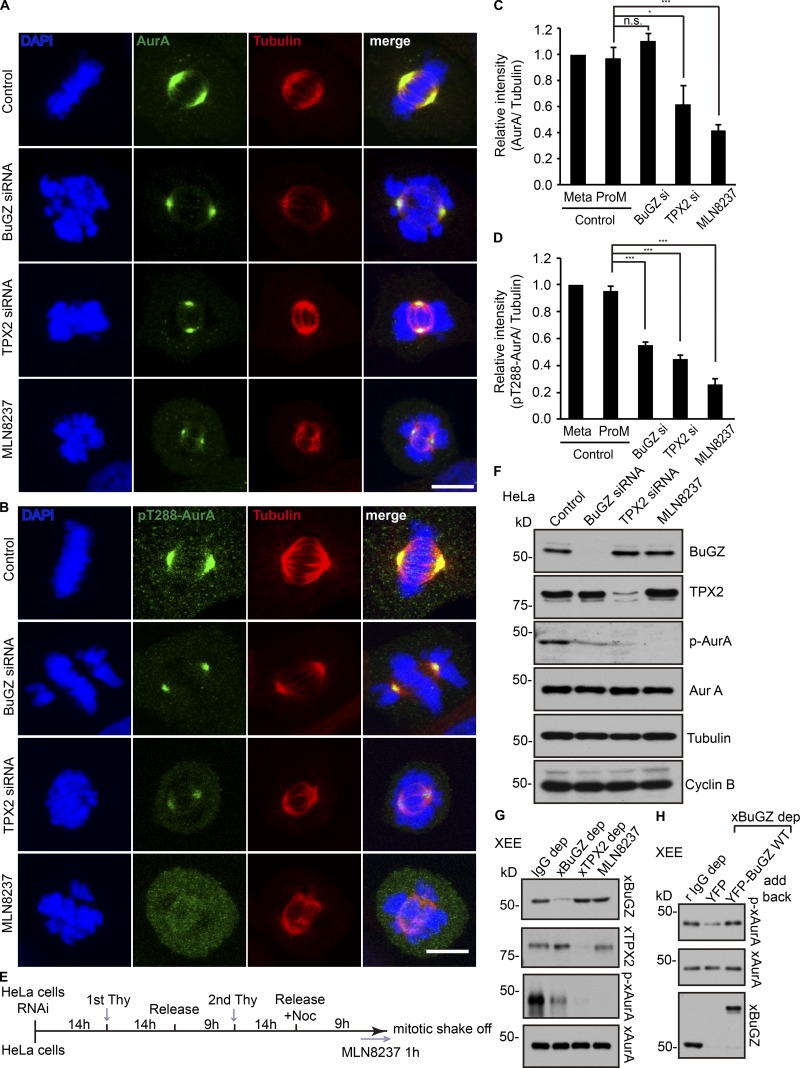
Because BuGZ promotes MT polymerization from AurA beads in cytostatic factor–arrested (CSF) Xenopus egg extracts (XEEs) in the presence of RanGTP (Tsai and Zheng, 2005; Ma et al., 2009; Goodman et al., 2010; Jiang et al., 2015b), we asked whether BuGZ regulates AurA activity in cells during mitosis. We depleted BuGZ by treating HeLa cells with siRNA. As controls, we treated cells using control siRNA, TPX2 siRNA, or an AurA inhibitor MLN8237. Cells were immunostained with tubulin and AurA antibodies. BuGZ depletion resulted in defects in both spindle assembly and chromosome alignment as expected (Jiang et al., 2014, 2015b; Toledo et al., 2014), whereas TPX2 depletion and MLN8237 treatment disrupted spindle assembly (Fig. 1, A and B; Nikonova et al., 2013). We quantified total AurA immunostaining intensity on MTs right next to the centrosome at spindle poles and normalized AurA intensity against MT intensity. We found that depletion of BuGZ did not cause a reduction of AurA on the spindle, whereas cells depleted of TPX2 or treated with MLN8237 showed a significant reduction of AurA compared with control cells with aligned metaphase chromosomes, incompletely aligned chromosomes, or misaligned chromosomes (Fig. 1 C).
Figure 1.
Effects of BuGZ on AurA kinase activity in mitosis. (A–D) Cells were transfected with control, BuGZ, or TPX2 siRNA (si) or were treated with AurA inhibitor MLN8237 (100 nM) followed by staining with antibodies to total AurA (A) or p-AurA (B). Bars, 10 µm. The immunostaining intensity of total AurA (C) and p-AurA (D) in control siRNA–treated cells in metaphase or prometaphase (ProM, containing misaligned chromosomes or thick chromosome bars) and cells in the experimental groups containing misaligned chromosomes were quantified. 75–152 total cells from three independent experiments in each experiment were measured and quantified. Error bars indicate SEM. One-way ANOVA: *, P < 0.05; ***, P < 0.001. (E) Scheme of cell synchronization and mitotic shakeoff. (F) Cells transfected with siRNA were synchronized, collected, and analyzed by Western blotting with indicated antibodies. (G) XEEs were depleted (dep) with unimmunized rabbit IgG (IgG), xBuGZ, or xTPX2 antibodies or were treated with MLN8237. xAurA was immunoprecipitated and analyzed by Western blotting using indicated antibodies. (H) XEEs were depleted of xBuGZ and supplemented by purified YFP or YFP-xBuGZ followed by AurA immunoprecipitation and Western blotting. Noc, nocodazole; Thy, thymidine.
We then immunostained the above-treated cells using an antibody recognizing the T288 phosphorylated active AurA (p-AurA; Fig. 1 B; Walter et al., 2000. By quantifying p-AurA as described in the previous paragraph, we found that BuGZ depletion resulted in a significant reduction of p-AurA on spindle MTs (Fig. 1 D). As expected, MLN8237 treatment or TPX2 depletion also diminished the amount of p-AurA on spindle MTs (Fig. 1 D). DMSO treatment (control for MLN8237; not depicted) exhibited similar levels of p-AurA as the control RNAi-treated cells.
We next synchronized the siRNA-treated HeLa cells by double thymidine block (Fig. 1 E). Upon release, control siRNA–treated cells progressed through mitosis as judged by the rise and fall of the levels of cyclin B along with BuGZ, TPX2, AurA, and p-AurA (Fig. S1). The peak p-AurA level was observed at 9 h after release when the cyclin B activity was high. We collected control-, BuGZ-, or TPX2 siRNA–treated cells at 9 h after the second thymidine block. For the control, we added MLN8237 at 8 h after the second thymidine block and released to inhibit AurA kinase activity (Fig. 1 E). We found that BuGZ depletion caused a significant reduction of p-AurA, similar to cells depleted of TPX2 or treated with MLN8237 (Fig. 1 F). Removal of xBuGZ from XEEs also diminished the level of p-xAurA, which can be rescued by the addition of purified YFP-xBuGZ (Fig. 1, G and H). Thus, BuGZ activates AurA in mitosis.
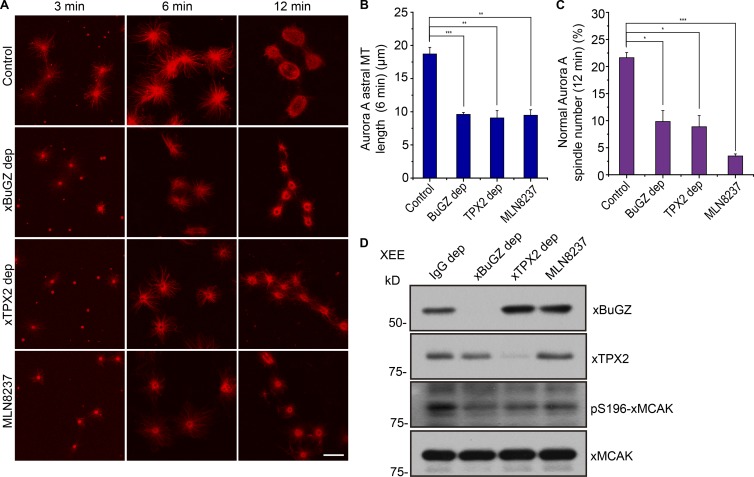
Depletion of xBuGZ or xTPX2 in the XEEs or treating XEEs with MLN8237 all resulted in a reduction of MT aster length and spindle assembly from xAurA beads (Fig. 2, A–C). Previous research has shown that AurA phosphorylation on serine 196 of xMCAK reduces MCAK MT depolymerase activity (Zhang et al., 2008). We found that egg extracts depleted of xBuGZ or xTPX2 or treated by MLN8237 all exhibited reduced p196-xMCAK levels (Fig. 2 D). This suggests that xBuGZ- and xTPX2-mediated AurA activation contributes to increased MT assembly through inhibiting xMCAK. Consistently, we found that addition of an inhibitory antibody to xMCAK into egg extract depleted of xBuGZ or xTPX2 or that was treated with MLN8237 significantly rescued aster MT length and spindle assembly induced by AurA beads and RanGTP (Fig. S2). Thus, BuGZ contributes to AurA kinase activation during mitosis, which may in turn promote spindle assembly.
Figure 2.
BuGZ contributes to MT assembly and reduces S196 phosphorylated xMCAK. (A) MT asters and spindles assemble around AurA beads at the indicated time points in mock-depleted XEEs or XEEs depleted (dep) of xBuGZ or xTPX2 or treated with MLN8237. Bar, 10 µm. (B and C) MT aster lengths (B) and percentages of bipolar spindles (C) were quantified. Approximately 50 (B) and ∼200 (C) AurA bead structures were quantified in three independent experiments. (D) XEE mock-depleted by unimmunized rabbit IgG, xBuGZ, or xTPX2 antibodies or treated with MLN8237 were analyzed by Western blotting probing with the indicated antibodies. The means and SEM are graphed. Student’s t test: *, P < 0.05; **, P < 0.01; ***, P < 0.001.
The two zinc fingers of BuGZ recognize the kinase domain of AurA
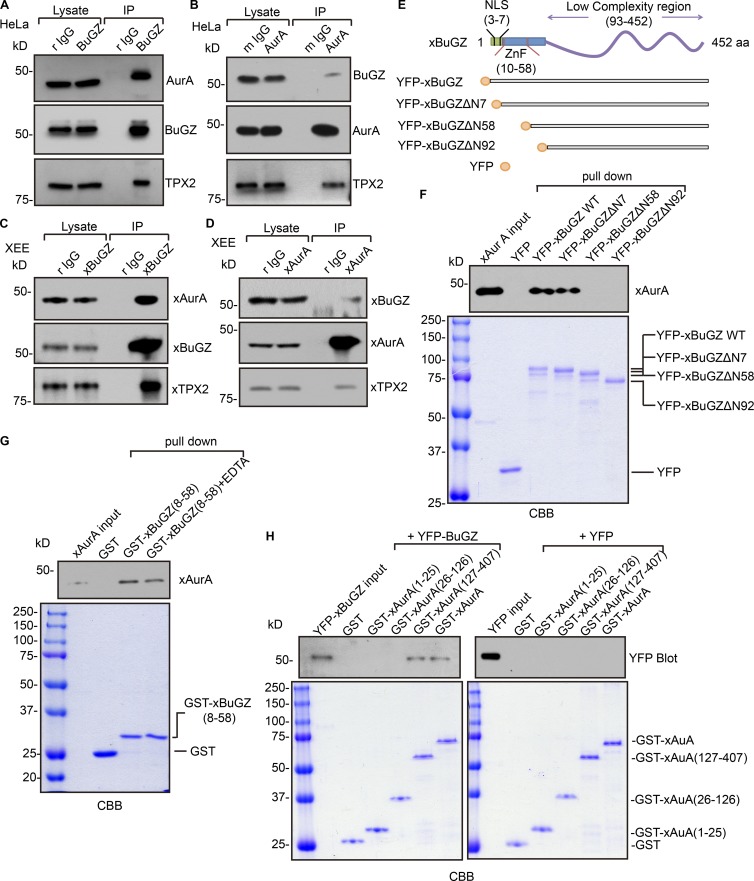
To study whether BuGZ interacts with AurA, HeLa cells were synchronized using a double thymidine block and released into media containing nocodazole. Mitotic cells were collected for immunoprecipitation by BuGZ or AurA antibodies. BuGZ and AurA reciprocally immunoprecipitated one another and pulled down TPX2 (Fig. 3, A and B). xBuGZ and xAurA also interacted with one another in immunoprecipitations from XEEs (Fig. 3, C and D).
Figure 3.
The zinc fingers of BuGZ directly bind to the kinase domain of AurA. (A and B) Immunoprecipitation (IP) using BuGZ (A) or AurA (B) antibodies in mitotic HeLa cell lysates. Equivalent of 1% of the input lysate and 20% of total immunoprecipitates were loaded. (C and D) Immunoprecipitation using xBuGZ (rabbit polyclonal IgG; r IgG; C) or xAurA (mouse polyclonal IgG; m IgG; D) antibodies in XEEs. Equivalent of 0.2% of the input lysate and 30% of total immunoprecipitates were loaded. (E) Schematic of xBuGZ with NLS, zinc fingers (ZnFs), and the C-terminal region as well as YFP-tagged xBuGZ used in the study. (F) Purified YFP, YFP-xBuGZ, YFP-xBuGZΔ7, YFP-xBuGZΔN58, or YFP-xBuGZΔN92 were incubated with purified xAurA, followed by pulldown with antibody to YFP and then Western blotting and Coomassie Brilliant blue (CBB) staining. (G) Purified GST or GST-xBuGZ(8–58) was incubated with purified xAurA in the presence or absence of EDTA followed by GST pulldown, Western blotting, and Coomassie blue staining. (H) Purified GST, GST-xAurA, GST-xAurA(1–25), GST-xAurA(26–126), or GST-xAurA(127–407) were incubated with YFP or YFP-xBuGZ followed by GST pulldown, Western blotting, and Coomassie blue staining. For G and H, 0.1% of total reaction and 10% of total xBuGZ pulldowns were analyzed by Western blotting. 1% of total reaction and 10% of total pulldowns were analyzed by Coomassie blue staining.
We next performed pulldown experiments with various deletion constructs of YFP-xBuGZ and xAurA (Fig. 3 E). The N terminus of BuGZ contains an NLS followed by two C2H2 zinc fingers from amino acids 10–58, whereas the rest of BuGZ sequence is predicted to have low sequence complexity and to be unstructured (Fig. 3 E). The N-terminal 92 amino acids bind to tubulin (Jiang et al., 2015b). Full-length YFP-xBuGZ or xBuGZ in which the first seven, 58, or 92 amino acids were deleted as well as YFP-xBuGZΔN7, YFP-xBuGZΔN58, or YFP-xBuGZΔN92, respectively, were incubated with purified xAurA. Using anti–GFP antibody–coated beads that recognized YFP, we found that YFP-xBuGZ and xBuGZΔN7 pulled down similar amounts of xAurA, whereas YFP and the other xBuGZ deletions did not (Fig. 3 F). As expected, YFP-xBuGZ, but not YFP-xBuGZΔN92, pulled down purified tubulin (Fig. S3 A). YFP-xBuGZΔN58 also did not pull down tubulin (Fig. S3 A). Interestingly, YFP-xBuGZ pulled down ∼40% more tubulin than that of YFP-xBuGZΔN7 (Fig. S3 A). Thus, the N-terminal seven amino acids of BuGZ contribute to tubulin but not AurA binding.
We then fused the two zinc fingers of BuGZ, amino acids 8–58, to GST (GST-xBuGZ(8–58)) and incubated it with purified xAurA in the presence or absence of 2 mM EDTA to chelate zinc. We found that GST-xBuGZ(8–58) pulled down more xAurA in the absence of EDTA than in its presence (Fig. 3 G). However, addition of EDTA greatly enhanced the binding of GST-xBuGZ(8–58) to tubulin (Fig. S3 B). Thus, AurA favors binding to the zinc finger fold of BuGZ, whereas tubulin binding involves the charged residues in the zinc finger sequence that could become less accessible upon the formation of zinc finger fold. It will be interesting to further understand whether free zinc in cells could regulate BuGZ interaction with tubulin and AurA.
We also created GST-AurA fusion constructs of full-length (amino acids 1–407), the N-terminal 1–25 or 26–126 amino acids, and the C-terminal kinase domain (amino acids 127–407; GST-xAurA, GST-xAurA(1–25), GST-xAurA(26–126), and GST-xAurA(127–407), respectively). We found that only GST-xAurA and GST-xAurA(127–407) pulled down YFP-xBuGZ (Fig. 3 H). Thus, the kinase domain of AurA directly interacts with the zinc fingers of BuGZ.
AurA incorporation into the BuGZ liquid droplets in vitro
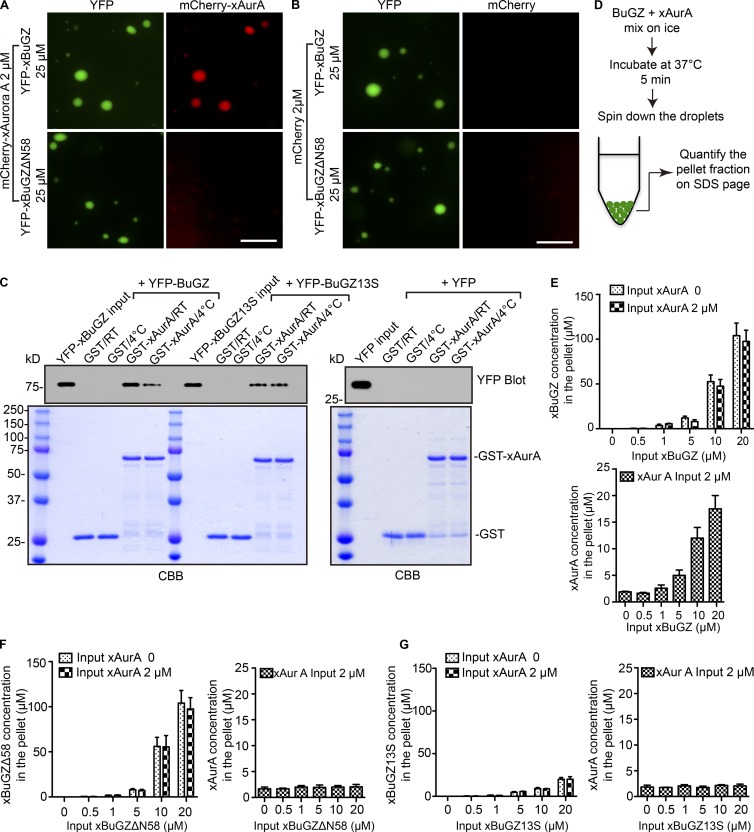
We asked whether the binding of AurA to the zinc fingers of BuGZ allows AurA to incorporate into the liquid droplets formed by BuGZ coacervation. To visualize BuGZ liquid droplets and AurA, purified YFP-xBuGZ or YFP-xBuGZΔN58 was mixed with purified mCherry-xAurA or mCherry, incubated at 37°C, and imaged. mCherry-xAurA but not mCherry incorporated into and became concentrated in the droplets formed by YFP-xBuGZ but not by YFP-xBuGZΔN58 (Fig. 4, A and B). To test whether the temperature-dependent coacervation of BuGZ enhanced its binding to AurA, we incubated GST-xAurA–coated beads with YFP, YFP-xBuGZ, or YFP-xBuGZ13S (a mutant xBuGZ with reduced phase transition activity at all temperatures in vitro; Jiang et al., 2015b) at RT or 4°C followed by washing the GST beads at the respective temperatures. We found that more YFP-xBuGZ bound to GST-xAurA at RT than at 4°C, whereas YFP did not bind at either temperature (Fig. 4 C). The amount of YFP-xBuGZ bound to GST-xAurA at 4°C was similar to those of YFP-xBuGZ13S bound to beads at both temperatures (Fig. 4 C). Thus, temperature-dependent coacervation of BuGZ facilitates its binding to AurA.
Figure 4.
Effects of BuGZ phase separation on AurA binding. (A and B) Purified YFP-xBuGZ or YFP-xBuGZΔN58 was incubated to allow coacervation in the presence of purified mCherry-xAurA (A) or mCherry (B). Bars, 10 µm. (C) Purified YFP, YFP-xBuGZ, or YFP-xBuGZ13S were incubated with purified GST or GST-xAurA at RT or 4°C followed by GST pulldown, Western blotting (0.1% of total reaction and 10% of total YFP-xBuGZ and YFP-xBuGZ13S pulldowns were analyzed), and Coomassie Brilliant blue (CBB) staining (1% of the total reaction and 10% of total pulldowns were analyzed). (D) Schematic of droplet spindown assay. (E–G) Quantification of YFP-xBuGZ and xAurA (E), YFP-xBuGZΔN58 and xAurA (F), and YFP-xBuGZ13S and xAurA (G) in the pellets (X axis) against the initial input xBuGZ proteins (Y axis). Error bars indicate SEM.
To estimate the amount of AurA in the BuGZ droplets, we used a droplet spin down assay (Fig. 4 D; Jiang et al., 2015b). Increasing concentrations of purified YFP-xBuGZ, YFP-xBuGZΔN58, or YFP-xBuGZ13S were incubated with or without 2 µM xAurA. The droplets were pelleted by centrifugation, and the amount of xBuGZ and xAurA in the pelleted droplets was quantified (Fig. 4, E–G) and plotted against the initial xBuGZ concentration in the reaction. As expected, YFP-xBuGZ and YFP-xBuGZΔN58 were greatly concentrated in the pellets, whereas the phase transition–deficient YFP-xBuGZ13S failed to do so (Fig. 4, E–G). xAurA was only enriched in the droplets formed by YFP-xBuGZ (Fig. 4, E–G). Thus, by binding to the zinc fingers of BuGZ, AurA can be incorporated and enriched into the BuGZ liquid droplets formed in vitro.
BuGZ coacervation property contributes to AurA activation
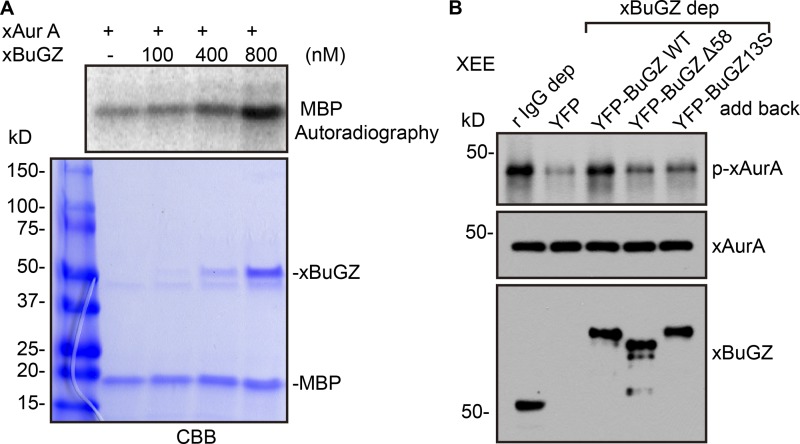
To assess whether BuGZ directly promotes AurA phosphorylation, xAurA purified from SF9 cells was incubated with myelin basic protein (MBP) and an increasing concentration of xBuGZ. The purified xAurA had a low kinase activity, whereas xBuGZ addition activated AurA in a dosage-dependent manner (Fig. 5 A). Because we had estimated ∼700 nM of BuGZ on spindles (Jiang et al., 2015b), the amount of BuGZ needed to activate AurA in vitro suggests that the amount of spindle-localized BuGZ is sufficient to activate AurA in vivo. Next, we determined whether the coacervation activity of BuGZ and the binding of BuGZ to AurA indeed contributed to AurA activation in mitosis. We depleted endogenous xBuGZ from XEEs, added back endogenous levels of YFP, YFP-xBuGZ, YFP-xBuGZΔN58, or YFP-xBuGZ13S, and immunoprecipitated xAurA. Western blotting analyses using an antibody that recognized both p-T288–AurA and p-T295–xAurA (p-xAurA; Table S1; Tsai et al., 2003) showed that xBuGZ depletion resulted in a significant reduction of active AurA as judged by the amount of p-xAurA with YFP addback, which was rescued by YFP-xBuGZ but not by YFP-xBuGZΔN58 or YFP-xBuGZ13S (Fig. 5 B). This suggests that BuGZ coacervation contributes to AurA activation in mitosis.
Figure 5.
Effects of BuGZ phase transition and binding to AurA on kinase activity. (A) Purified xAurA (cleaved from mCherry-xAurA) was incubated from purified xBuGZ (cleaved from YFP-xBuGZ) and MBP to assay AurA kinase activity in vitro. CBB, Coomassie Brilliant blue. (B) XEE was mock depleted or depleted (dep) with BuGZ antibody, and the BuGZ-depleted XEE was supplemented with the indicated purified proteins followed by xAurA immunoprecipitation and Western blotting analyses. r IgG, rabbit polyclonal IgG.
Using XEEs, we show that both the zinc fingers and the coacervation property of BuGZ are required to promote AurA activation. Curiously, although the interaction between BuGZ and AurA allows AurA to be enriched in the BuGZ coacervates in vitro, depletion of BuGZ in cells did not reduce the amount of total AurA along spindle MTs. Because BuGZ depletion reduced the amount of p-AurA in cells and egg extracts, BuGZ must promote AurA activation by means other than concentrating the kinase on spindle MTs. AurA dimer formation can greatly stimulate intermolecular AurA phosphorylation and activation (Zorba et al., 2014). Therefore, BuGZ coacervation could limit AurA diffusion in the spindle, which would aid dimer formation and AurA activation. BuGZ coacervation may also drive two AurA molecules together through a depletion attraction effect (Marenduzzo et al., 2006). Alternatively, BuGZ coacervation may exclude phosphatases, thereby maintaining AurA activation (Su et al., 2016). Although our data suggest that BuGZ coacervation activates AurA, it remains possible that the binding of BuGZ to the kinase domain by itself could prevent the N terminus of AurA to block the access of the phosphorylation sites, thereby enhancing the interacting kinase dimer to phosphorylate one another or the binding of TPX2 to AurA. More studies are needed to uncover the detailed mechanism of AurA activation by BuGZ in mitosis.
Materials and methods
Constructs
The constructs for YFP-xBuGZ and its mutant expression using baculovirus in SF9 cells were described previously (Jiang et al., 2015b). For the details of other plasmid construction and antibody information, see Table S1.
Cell culture, siRNA transfection, and inhibitor treatments
HeLa cells were cultured in DMEM (Thermo Fisher Scientific) supplemented with 10% (vol/vol) FBS (Biochrom). The cells were grown at 37°C in a 5% CO2 humidified chamber. To synchronize HeLa cells, cells were first treated with 2 mM thymidine for 14 h and then were washed with PBS. After releasing into fresh medium for 9 h, the cells were washed and incubated in 2 mM thymidine for another 14 h, followed by releasing into medium containing 330 nM nocodazole for 9 h. To inhibit AurA activity, MLN8237 (100 nM; Selleckchem) was added for 1 h after 8 h of release from the second thymidine block. Cell lysates were made by collecting mitotic cells using mitotic shakeoff. To check the synchrony of the double thymidine block, the cells were released from the second thymidine block into fresh medium. Cell lysates were made at the indicated time points and used for Western blot analysis.
For BuGZ knockdown, we used our previously published BuGZ siRNA oligonucleotide (5′-GCCUGCUACACUUACAACAACUAGU-3′; Jiang et al., 2014, 2015b), control oligonucleotide (12935-300; Thermo Fisher Scientific), and a TPX2 oligonucleotide (SI02665082; QIAGEN). In brief, 40 pM of each siRNA oligonucleotide was transfected into cells using Lipofectamine RNAiMAX (Invitrogen). After 60 h transfection, cells were treated with 10 µM MG132 for 1 h before fixation for 12 min in PTEMF (20 mM Pipes-KOH, pH 6.8, 0.2% Triton X-100, 10 mM EGTA, 2 mM MgCl2, and 4% formaldehyde) and blocked with 1% (wt/vol) BSA in PBS, followed by primary antibody incubation overnight at 4°C and secondary antibody incubation.
MT structures assembled in XEEs were fixed by ice-cold methanol for 10 min followed by PBS wash and mounting. An ECLIPSE E800 microscope (Nikon) or a TCS SP5 laser confocal microscope (Leica Microsystems) was used for imaging. For confocal microscopy, 63 Å, 1.4 NA oil objective lenses (Leica Microsystems) was used, and the cells were imaged by scanning optical sections at ∼0.5-µm intervals. To quantify spindle MT intensity or the ratio of AurA or p-AurA versus the MT intensities, images at the same exposure time and setting were captured using a 60×, 1.4 NA oil objective on an ECLIPSE E800 microscope. For controls, metaphase or prometaphase spindles (with misaligned or incompletely congressed chromosomes) were quantified, whereas for BuGZ-, TPX2 RNAi–, or MLN8237-treated cells, bipolar spindles with misaligned or incompletely congressed chromosomes were quantified. One 15 × 15 pixel region right next to the spindle pole was chosen to measure the mean immunostaining intensity of AurA, p-AurA, and tubulin. The intensities for AurA or p-AurA were normalized against the tubulin intensity. Images were cropped in Photoshop (CS5.1; Adobe), sized, and placed using Illustrator (CS6; Adobe) as shown in Fig. 1.
XEE assays
Spindle assembly assays were based on previous studies (Tsai and Zheng, 2005; Ma et al., 2009; Goodman et al., 2010; Jiang et al., 2015b). In brief, 20 µl of protein A DynaBeads slurry (Invitrogen) was incubated with 30 µg of Xenopus AurA (also called Eg2) antibody for at least 2 h at RT. After washing with XB buffer (10 mM Hepes, 50 mM sucrose, 100 mM KCl, 1 mM MgCl2, 0.1 mM CaCl2, and 5 mM EGTA, pH 7.7) three times, the beads were resuspended in 50 µl of XB buffer. To induce spindle assembly, 1 µl of the resuspended AurA beads was added into 100 µl of egg extracts that were mock depleted, xBuGZ depleted, xTPX2 depleted, or AurA inhibited by MLN8237. For AurA bead assays, egg extracts were incubated at 4°C with rotation for 30 min followed by addition of purified RanL43E (1 mg/ml final), energy mix (10 mM creatine phosphate, 0.5 mM Na-ATP, 0.5 mM MaCl2, and 50 mM EGTA, pH 7.7), and rhodamine tubulin (1 µM final) to the egg extracts with beads and then incubated at RT for 3–12 min. After incubation, 10 µl egg extracts were then diluted with 1 ml BRB80 buffer (80 mM Pipes, 1 mM MgCl2, and 1 mM EGTA, pH 6.8) containing 30% glycerol. The mixture was spun through a 2-ml cushion of BRB80 with 40% glycerol onto coverslips. The MT structures on coverslips were fixed with ice-cold methanol for 10 min and then washed with PBS three times and mounted, followed by visualization and quantification. Aster MT length was measured from the center of AurA beads to the tip of the longest MT bundle. The percentages of bipolar AurA bead spindles were also quantified by counting more than six random fields using an ECLIPSE E800 microscope and a 60× 1.4 NA oil objective lens. The means were compared with a Student’s t test and considered significant if P < 0.05.
Protein expression, purification, and interaction studies
The xBuGZ WT and mutant proteins were expressed in SF9 cells according to the Bac-to-Bac Baculovirus Expression System (Version E; Invitrogen) and the protocol described by Jiang et al. (2015b). For His-mCherry-xAurA protein, it was expressed and purified similarly to YFP-xBuGZ but using 150 mM NaCl in the His-binding buffer (20 mM phosphate buffer, pH 7.4, 1 mM MgCl2, 1 mM β-mercaptoethanol, 0.01% Triton X-100, 150 mM NaCl, and 25 mM imidazole). To cleave the YFP or mCherry tag from proteins, samples were digested at 4°C for 16 h by adding Ac–tobacco etch virus protease (Invitrogen). Proteins were eluted from the column in a low-salt elution buffer (20 mM phosphate buffer, pH 7.4, 1 mM MgCl2, 1 mM β-mercaptoethanol, 0.01% Triton X-100, 50 mM NaCl, and 500 mM imidazole). The proteins were exchanged into XB buffer using a PD-10 column (GE Healthcare) and further concentrated with an Amicon Ultra 30K device (EMD Millipore) at 4°C. The protein was divided into 10-µl aliquots, snap-frozen in liquid nitrogen, and stored at −80°C.
GST and GST-tagged proteins were expressed and purified from BL21 (DE3) using glutathione agarose (Sigma-Aldrich) according to the manufacturer’s instructions. For each protein expression, we picked a single Escherichia coli colony to inoculate a 3-ml Luria–Bertani culture containing the appropriate antibiotic. After shaking at 37°C overnight, the culture was added to 10 ml of prewarmed Luria–Bertani medium with antibiotic for further incubation at 37°C with shaking. Once the OD600 reached 0.8, IPTG (final concentration 1 mM) was added to the medium to induce protein expression and incubated with shaking at 16°C for 18 h. The E. coli were harvested by centrifugation at 5,000 rpm for 10 min at 4°C. The bacterial pellet was resuspended in 1 ml of prechilled (on ice) PBST (PBS plus 1% Triton X-100) containing 1 mM PMSF and a 1:100 dilution of the protease cocktail (4-(2-Aminoethyl)benzenesulfonyl fluoride hydrochloride [AEBSF], bestatin, E-64, pepstatin A, and phosphoramidon; Sigma-Aldrich). The mixture was then sonicated on ice until the lysate became nonturbid (∼5 min), followed by centrifugation at 12,000 rpm for 15 min at 4°C. The supernatant was incubated with 20 µl glutathione agarose beads with rotation at 4°C for 1 h, and then the beads were washed with 3 ml PBST and 3 ml PBS sequentially. The washed agarose beads were then incubated with various proteins to study the interaction.
To define the binding site of xAurA on xBuGZ, 5 µg purified YFP or YFP-tagged WT xBuGZ, xBuGZΔΝ7, xBuGZΔΝ58, or xBuGZΔΝ92 and 5 µg xAurA (cleaved from mCherry-xAurA) were incubated with GFP-agarose beads (ChromoTek) in 200 µl PBS buffer containing 0.1% Triton X-100 with rotation at 4°C for 4 h. The agarose beads were washed with 1 ml prechilled (on ice) PBS buffer with 0.1% Triton X-100 three times and then with 1 ml of prechilled PBS buffer for another three times. To define the binding site of xBuGZ on xAurA, glutathione agarose beads coated with ∼4 µg GST, GST-tagged full-length xAurA, xAurA(1–25) amino acids, xAurA(26–126) amino acids, or xAurA(127–407) amino acids were incubated with 5 µg purified YFP or YFP-xBuGZ and washed as above. Sample volumes loaded for Coomassie Brilliant blue staining or for Western blotting analyses are described in Figs. 3, 4, and S3.
Immunoprecipitation and Western blotting analyses
Mitotic HeLa cells or CSF egg extracts were used to perform reciprocal immunoprecipitations to determine the interaction between AurA and BuGZ. HeLa cells were synchronized by double thymidine block release followed by adding nocodazole for 16 h as described in the Cell culture, siRNA transfection, and inhibitor treatments section. The mitotic cell lysates were made by collecting cells using mitotic shakeoff and resuspended in 200 µl prechilled PBST and protease inhibitors (at one tablet/10 ml; cOmplete mini EDTA-free protease inhibitor cocktail tablets; 11836170001; Roche). The mixture was then sonicated on ice, followed by centrifugation at 13,000 rpm for 15 min at 4°C. 10 µg of antibody to BuGZ (rabbit polyclonal; HPA017013; Sigma-Aldrich) or AurA (mouse monoclonal; 610938; BD), or 10 µg of unimmunized rabbit IgG (12-370; EMD Millipore) or mouse IgG (12-371; EMD Millipore) were used for immunoprecipitation using 200 µl mitotic HeLa cell lysates for each experiment. Each antibody was preincubated with a 20 µl slurry of protein A DynaBeads (10007D; Thermo Fisher Scientific) for 2 h followed by washes with PBST. The beads were added to the cell lysate and incubated at 4°C for 4 h. After washing three times with lysis buffer, the beads were resuspended in SDS sample buffer and boiled for 5 min. For Western blotting analyses, 1% of input lysate and 20% of immunoprecipitates were loaded. To study the interaction between xBuGZ and xAurA, 100 µl of the CSF egg extracts were incubated with protein A DynaBeads coated with 10 µg xBuGZ, xAurA, or rabbit unimmunized at 4°C for 2 h followed by washing three times with 800 µl XB buffer each. The DynaBeads were resuspended in SDS sample buffer and boiled for 5 min. For Western blotting analyses, 0.2% of input lysate and 30% of resuspended immunoprecipitates were used. For detailed antibodies and dilutions used, see Table S1.
Assays for how BuGZ phase transition enhances its concentration of AurA and interaction with AurA
To estimate protein concentrations in the BuGZ droplets, different concentrations of YFP-xBuGZ or YFP-xBuGZΔN58 with or without xAurA (cleaved of its mCherry tag) were prepared in the XB buffer and placed on ice (each reaction was 200 µl total). The protein samples were then incubated at 37°C for 5 min. The droplets were pelleted by centrifugation at 2,000 rpm for 5 min in a benchtop microfuge. As the YFP-xBuGZ droplets formed a visible yellow pellet, the supernatant could be removed. 180 µl of the supernatant was removed, followed by carefully removing the remaining supernatant as much as possible without perturbing the pellet. 50 µl XB buffer was added to resuspend each pellet, followed by addition of 50 µl of 2× SDS sample buffer. 5 µl was analyzed by SDS-PAGE followed by Coomassie blue staining. The amount of xAurA and xBuGZ in the pellet was quantified using BSA as a standard. To study whether BuGZ phase transition enhanced its binding to AurA, we incubated glutathione agarose beads with 10 µg purified GST-xAurA and 1 µM YFP, YFP-xBuGZ, or YFP-xBuGZ13S in 200 µl PBST at 4°C or RT for 30 min, followed by washing the beads with 1 ml PBST for two times and then PBS buffer for one time. The proteins retained on the beads were analyzed.
AurA kinase assay in vitro
To verify whether BuGZ can activate AurA, we used 100 ng xAurA (cleaved from mCherry-xAurA purified from SF9 cells) and 1 µg MBP as substrate. The protein mixture was incubated with or without xBuGZ (purified from SF9 with the YFP tag cleaved) in phosphorylation buffer containing 20 mM Hepes, pH 7.5, 10 mM MgCl2, 5 mM EGTA, 1 mM DTT, 10 nM ATP, and 5 µCi of [γ32]P-ATP (PerkinElmer) in a total volume of 30 µl. The kinase reaction was performed at 30°C for 10 min and was terminated by the addition of 10 µl of 4× SDS sample buffer. The samples were analyzed on a 4–20% gradient SDS-PAGE. The gel was stained with Coomassie blue, dried, and imaged by PhosphorImager (GE Healthcare).
Statistical analyses
Statistical analyses are described in each figure legend.
Online supplemental material
Fig. S1 shows validation of cell synchronization. Fig. S2 shows how xMCAK inhibition counteracted the effect of xBuGZ depletion in egg extracts. Fig. S3 shows how tubulin binding to BuGZ involved the first 58 amino acids including the zinc finger sequence and was reduced in the presence of EDTA. Table S1 shows lists of protein expression constructs (including primers used in their construction) as well as antibodies used in this study.
Supplementary Material
Acknowledgments
We thank Dr. Shusheng Wang for constructing the mCherry-AurA plasmid for baculovirus-mediated protein expression in SF9 cells, Ona Martin and Lynne Hugendubler for technical support, and the members of the Zheng laboratory for critical reading of the manuscript.
This study was supported by National Institutes of Health grants R01 GM056312 and R01 GM106023 to Y. Zheng, National Natural Science Foundation of China grant 31420103916 to X. Zhu and Y. Zheng, National Institutes of Health R01 GM 059618 to C.E. Walczak, and by Ligue Nationale Contre le Cancer to C. Prigent.
The authors declare no competing financial interests.
Author contributions: conceptualization: Y. Zheng and Y. Huang; method: Y. Huang and T. Li; data analyses: Y. Huang and T. Li; writing, original draft: Y. Huang; review, editing: Y. Huang, T. Li, S.C. Ems-McClung, C.E. Walczak, C. Prigent, X. Zhu, X. Zhang, and Y. Zheng.
Footnotes
Abbreviations used:
- BuGZ
- Bub3-interacting and GLE-2–binding sequence containing ZNF207
- CSF
- cytostatic factor–arrested
- MBP
- myelin basic protein
- MT
- microtubule
- NLS
- nuclear localization signal
- SAF
- spindle assembly factor
- XEE
- Xenopus egg extract
References
- Bai M., Ni J., Wu J., Wang B., Shen S., and Yu L.. 2014. A novel mechanism for activation of Aurora-A kinase by Ajuba. Gene. 543:133–139. 10.1016/j.gene.2014.03.048 [DOI] [PubMed] [Google Scholar]
- Bayliss R., Sardon T., Vernos I., and Conti E.. 2003. Structural basis of Aurora-A activation by TPX2 at the mitotic spindle. Mol. Cell. 12:851–862. 10.1016/S1097-2765(03)00392-7 [DOI] [PubMed] [Google Scholar]
- Dai X.X., Xiong H., Zhang M., Sun S., and Xiong B.. 2016. Zfp207 is a Bub3 binding protein regulating meiotic chromosome alignment in mouse oocytes. Oncotarget. 7:30155–30165. 10.18632/oncotarget.9310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyers P.A., Erikson E., Chen L.G., and Maller J.L.. 2003. A novel mechanism for activation of the protein kinase Aurora A. Curr. Biol. 13:691–697. 10.1016/S0960-9822(03)00166-0 [DOI] [PubMed] [Google Scholar]
- Fu J., Bian M., Xin G., Deng Z., Luo J., Guo X., Chen H., Wang Y., Jiang Q., and Zhang C.. 2015. TPX2 phosphorylation maintains metaphase spindle length by regulating microtubule flux. J. Cell Biol. 210:373–383. 10.1083/jcb.201412109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman B., Channels W., Qiu M., Iglesias P., Yang G., and Zheng Y.. 2010. Lamin B counteracts the kinesin Eg5 to restrain spindle pole separation during spindle assembly. J. Biol. Chem. 285:35238–35244. 10.1074/jbc.M110.140749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirota T., Kunitoku N., Sasayama T., Marumoto T., Zhang D., Nitta M., Hatakeyama K., and Saya H.. 2003. Aurora-A and an interacting activator, the LIM protein Ajuba, are required for mitotic commitment in human cells. Cell. 114:585–598. 10.1016/S0092-8674(03)00642-1 [DOI] [PubMed] [Google Scholar]
- Jiang H., He X., Wang S., Jia J., Wan Y., Wang Y., Zeng R., Yates J. III, Zhu X., and Zheng Y.. 2014. A microtubule-associated zinc finger protein, BuGZ, regulates mitotic chromosome alignment by ensuring Bub3 stability and kinetochore targeting. Dev. Cell. 28:268–281. 10.1016/j.devcel.2013.12.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H., He X., Feng D., Zhu X., and Zheng Y.. 2015a RanGTP aids anaphase entry through Ubr5-mediated protein turnover. J. Cell Biol. 211:7–18. 10.1083/jcb.201503122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H., Wang S., Huang Y., He X., Cui H., Zhu X., and Zheng Y.. 2015b Phase transition of spindle-associated protein regulate spindle apparatus assembly. Cell. 163:108–122. 10.1016/j.cell.2015.08.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joukov V., De Nicolo A., Rodriguez A., Walter J.C., and Livingston D.M.. 2010. Centrosomal protein of 192 kDa (Cep192) promotes centrosome-driven spindle assembly by engaging in organelle-specific Aurora A activation. Proc. Natl. Acad. Sci. USA. 107:21022–21027. 10.1073/pnas.1014664107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joukov V., Walter J.C., and De Nicolo A.. 2014. The Cep192-organized aurora A-Plk1 cascade is essential for centrosome cycle and bipolar spindle assembly. Mol. Cell. 55:578–591. 10.1016/j.molcel.2014.06.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kufer T.A., Silljé H.H., Körner R., Gruss O.J., Meraldi P., and Nigg E.A.. 2002. Human TPX2 is required for targeting Aurora-A kinase to the spindle. J. Cell Biol. 158:617–623. 10.1083/jcb.200204155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim N.R., Yeap Y.Y., Ang C.S., Williamson N.A., Bogoyevitch M.A., Quinn L.M., and Ng D.C.. 2016. Aurora A phosphorylation of WD40-repeat protein 62 in mitotic spindle regulation. Cell Cycle. 15:413–424. 10.1080/15384101.2015.1127472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Littlepage L.E., Wu H., Andresson T., Deanehan J.K., Amundadottir L.T., and Ruderman J.V.. 2002. Identification of phosphorylated residues that affect the activity of the mitotic kinase Aurora-A. Proc. Natl. Acad. Sci. USA. 99:15440–15445. 10.1073/pnas.202606599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L., Tsai M.Y., Wang S., Lu B., Chen R., Yates J.R. III, Zhu X., and Zheng Y.. 2009. Requirement for Nudel and dynein for assembly of the lamin B spindle matrix. Nat. Cell Biol. 11:247–256. 10.1038/ncb1832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marenduzzo D., Finan K., and Cook P.R.. 2006. The depletion attraction: an underappreciated force driving cellular organization. J. Cell Biol. 175:681–686. 10.1083/jcb.200609066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikonova A.S., Astsaturov I., Serebriiskii I.G., Dunbrack R.L. Jr., and Golemis E.A.. 2013. Aurora A kinase (AURKA) in normal and pathological cell division. Cell. Mol. Life Sci. 70:661–687. 10.1007/s00018-012-1073-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pugacheva E.N., and Golemis E.A.. 2005. The focal adhesion scaffolding protein HEF1 regulates activation of the Aurora-A and Nek2 kinases at the centrosome. Nat. Cell Biol. 7:937–946. 10.1038/ncb1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabino D., Brown N.H., and Basto R.. 2011. Drosophila Ajuba is not an Aurora-A activator but is required to maintain Aurora-A at the centrosome. J. Cell Sci. 124:1156–1166. 10.1242/jcs.076711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sardon T., Peset I., Petrova B., and Vernos I.. 2008. Dissecting the role of Aurora A during spindle assembly. EMBO J. 27:2567–2579. 10.1038/emboj.2008.173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su X., Ditlev J.A., Hui E., Xing W., Banjade S., Okrut J., King D.S., Taunton J., Rosen M.K., and Vale R.D.. 2016. Phase separation of signaling molecules promotes T cell receptor signal transduction. Science. 352:595–599. 10.1126/science.aad9964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo C.M., Herman J.A., Olsen J.B., Ding Y., Corrin P., Girard E.J., Olson J.M., Emili A., DeLuca J.G., and Paddison P.J.. 2014. BuGZ is required for Bub3 stability, Bub1 kinetochore function, and chromosome alignment. Dev. Cell. 28:282–294. 10.1016/j.devcel.2013.12.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai M.Y., and Zheng Y.. 2005. Aurora A kinase-coated beads function as microtubule-organizing centers and enhance RanGTP-induced spindle assembly. Curr. Biol. 15:2156–2163. 10.1016/j.cub.2005.10.054 [DOI] [PubMed] [Google Scholar]
- Tsai M.Y., Wiese C., Cao K., Martin O., Donovan P., Ruderman J., Prigent C., and Zheng Y.. 2003. A Ran signalling pathway mediated by the mitotic kinase Aurora A in spindle assembly. Nat. Cell Biol. 5:242–248. 10.1038/ncb936 [DOI] [PubMed] [Google Scholar]
- Walter A.O., Seghezzi W., Korver W., Sheung J., and Lees E.. 2000. The mitotic serine/threonine kinase Aurora2/AIK is regulated by phosphorylation and degradation. Oncogene. 19:4906–4916. 10.1038/sj.onc.1203847 [DOI] [PubMed] [Google Scholar]
- Wan Y., Zheng X., Chen H., Guo Y., Jiang H., He X., Zhu X., and Zheng Y.. 2015. Splicing function of mitotic regulators links R-loop-mediated DNA damage to tumor cell killing. J. Cell Biol. 209:235–246. 10.1083/jcb.201409073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng K., Bastos R.N., Barr F.A., and Gruneberg U.. 2010. Protein phosphatase 6 regulates mitotic spindle formation by controlling the T-loop phosphorylation state of Aurora A bound to its activator TPX2. J. Cell Biol. 191:1315–1332. 10.1083/jcb.201008106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Ems-McClung S.C., and Walczak C.E.. 2008. Aurora A phosphorylates MCAK to control ran-dependent spindle bipolarity. Mol. Biol. Cell. 19:2752–2765. 10.1091/mbc.E08-02-0198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zorba A., Buosi V., Kutter S., Kern N., Pontiggia F., Cho Y.J., and Kern D.. 2014. Molecular mechanism of Aurora A kinase autophosphorylation and its allosteric activation by TPX2. eLife. 3:e02667 10.7554/eLife.02667 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.