Abstract
Pharmacologically difficult targets, such as MYC transcription factors, represent a major challenge in cancer therapy. For the childhood cancer neuroblastoma, amplification of the oncogene MYCN is associated with high-risk disease and poor prognosis. Here, we deployed genome-scale CRISPR-Cas9 screening of MYCN-amplified neuroblastoma and found a preferential dependency on genes encoding the polycomb repressive complex 2 (PRC2) components EZH2, EED, and SUZ12. Genetic and pharmacological suppression of EZH2 inhibited neuroblastoma growth in vitro and in vivo. Moreover, compared with neuroblastomas without MYCN amplification, MYCN-amplified neuroblastomas expressed higher levels of EZH2. ChIP analysis showed that MYCN binds at the EZH2 promoter, thereby directly driving expression. Transcriptomic and epigenetic analysis, as well as genetic rescue experiments, revealed that EZH2 represses neuronal differentiation in neuroblastoma in a PRC2-dependent manner. Moreover, MYCN-amplified and high-risk primary tumors from patients with neuroblastoma exhibited strong repression of EZH2-regulated genes. Additionally, overexpression of IGFBP3, a direct EZH2 target, suppressed neuroblastoma growth in vitro and in vivo. We further observed strong synergy between histone deacetylase inhibitors and EZH2 inhibitors. Together, these observations demonstrate that MYCN upregulates EZH2, leading to inactivation of a tumor suppressor program in neuroblastoma, and support testing EZH2 inhibitors in patients with MYCN-amplified neuroblastoma.
Keywords: Oncology
Keywords: Epigenetics
Introduction
The recent characterization of tumor genomes has revealed that pediatric cancers generally have lower mutational rates than adult cancers (1). Moreover, mutations in readily targetable proteins, such as kinases, are exceptions in these pediatric malignancies. Rather, pediatric cancers typically possess a “master” oncogenic event, such as MLL fusion proteins in infant leukemias (2), EWS/FLI1 fusions in Ewing sarcoma (3), or MYCN amplification in neuroblastoma (4). These events often involve transcription factors, a class of proteins typically difficult to “drug.” Functional genomic screens offer an opportunity to address this challenge through the identification of new, and potentially druggable, dependencies in pediatric cancers. In fact, genome-scale shRNA screening has already led to the discovery of new dependencies. For example, shRNA screening identified dependencies on the CDK4/cyclin D1 complex in Ewing sarcoma (5) and the PI3K pathway in osteosarcoma (6), and more recently, shRNA screening identified mutations in the SWI/SNF subunits as candidate biomarkers of response to EZH2 inhibition (7). ShRNA screening, however, has the significant limitation of off-target effects due to unintended silencing of transcripts complementary to the seed sequence of the shRNAs (8). Even though mathematical correction approaches to this problem have improved hit calling, alternative solutions could be transformative. The more recently described CRISPR-Cas9 (clustered regularly interspaced short palindromic repeats–CRISPR-associated protein 9) screening approach provides a new and powerful tool for high-throughput assessment of gene dependencies in mammalian systems with the hope of reduced off-target effects compared with shRNA (9–12).
Over the last 3 years, CRISPR-Cas9 has been applied to mammalian screens, including screens to identify mechanisms of resistance or enhanced sensitivity to drugs (9, 13), to identify mediators of immune response (14), and to identify enhancer elements (15). To date, however, this approach has not been systematically applied to pediatric cancers. In this study, we report a CRISPR-Cas9–based systematic functional genomics screen in a childhood cancer, specifically high-risk, MYCN-amplified neuroblastoma.
Neuroblastoma is the most common extracranial solid tumor diagnosed in children, and the majority of patients present with high-stage disease. MYCN amplification occurs in 25% of neuroblastoma tumors and is the best-characterized single gene alteration reliably linked to high-risk neuroblastoma (4, 16, 17). In mice, targeted expression of MYCN to the neural crest leads to neuroblastoma, strongly supporting MYCN as a master oncogene in this disease (18). Patients with MYCN-amplified neuroblastoma have poor 5-year event-free survival, suffer from toxicities of current intensive therapy, and have high rates of relapse and death. Therefore, novel therapeutic approaches are needed (19). Multiple approaches to targeting MYCN in neuroblastoma are under consideration, including the use of aurora kinase A or PI3K inhibitors to destabilize and degrade the protein and the use of epigenetic modifiers, such as BET bromodomain inhibitors, to impair the expression and function of MYCN (20–23). We sought to identify alternative druggable dependencies in MYCN-amplified neuroblastoma through an unbiased genome-scale CRISPR-Cas9 screen.
Genes that are differentially essential in a specific context rather than in all cancer cell lines represent potential candidates for targeted therapeutics. To identify genes more essential in neuroblastoma compared with the other cancer cell lines, we analyzed the loss-of-function CRISPR-Cas9–based screen of 341 cancer cell lines, including 9 MYCN-amplified and 2 MYCN-nonamplified neuroblastoma cell lines (24). Our analysis of the cancer dependency screen nominated the polycomb repressive complex 2 (PRC2) as a top dependency in a subset of human neuroblastoma cell lines compared with all other cancers in the screen, thus nominating the PRC2 complex as a druggable pathway in neuroblastoma.
Results
Genome-scale CRISPR-Cas9 screening reveals dependency on the PRC2 complex components EZH2, EED, and SUZ12 in a subset of neuroblastoma.
To identify specific dependencies in high-risk, MYCN-amplified neuroblastoma, we analyzed our genome-scale CRISPR-Cas9 loss-of-function screening of a collection of 341 human cancer cell lines from 26 tumor types (24), using the Avana library (25), which contains 73,372 guides with approximately 4 guides per gene. Eleven neuroblastoma cell lines, CHP-212, IMR-32, Kelly, KP-N-YN, MHH-NB-11, NB1, SK-N-BE(2), SK-N-AS, SK-N-DZ, SK-N-FI, and SIMA, were included in the screen, and all but SK-N-FI and SK-N-AS are MYCN amplified. Cell lines stably expressing Cas9 were generated and Cas9 activity confirmed as previously described (26). Stable cell lines were then transduced with the Avana library and passaged in triplicate for 21 days, and the relative enrichment or depletion of guides was assessed using massively parallel sequencing and compared with the initial plasmid pool. Data were normalized across cell lines, and a gene-level dependency score was calculated using the recently described CERES algorithm that takes into account the increased “cutting toxicity” observed in regions of copy number gain (24, 26).
We then identified preferential genetic dependencies in neuroblastoma cell lines by applying independent component analysis (ICA) (27, 28) to the CRISPR-Cas9 data. In contrast to the more common method of principal component analysis (29, 30), which identifies orthogonal components described by linear factors, ICA is an unsupervised method for multivariate data deconvolution into a set of independent components described by nonlinear factors. Therefore, ICA is often used for the analysis of biological data that involve higher-order associations, such as identifying tumor-related pathways in transcriptional data sets, classifying disease lineages, characterizing transcriptional regulators, and identifying disease-specific biomarkers (31–37). Thus, we applied ICA to rank-normalized CRISPR-Cas9 screening data and identified the top 3 independent components (IC1, IC2, and IC3) that were significantly depleted in neuroblastoma compared with other cancer cell lines screened (Figure 1A).
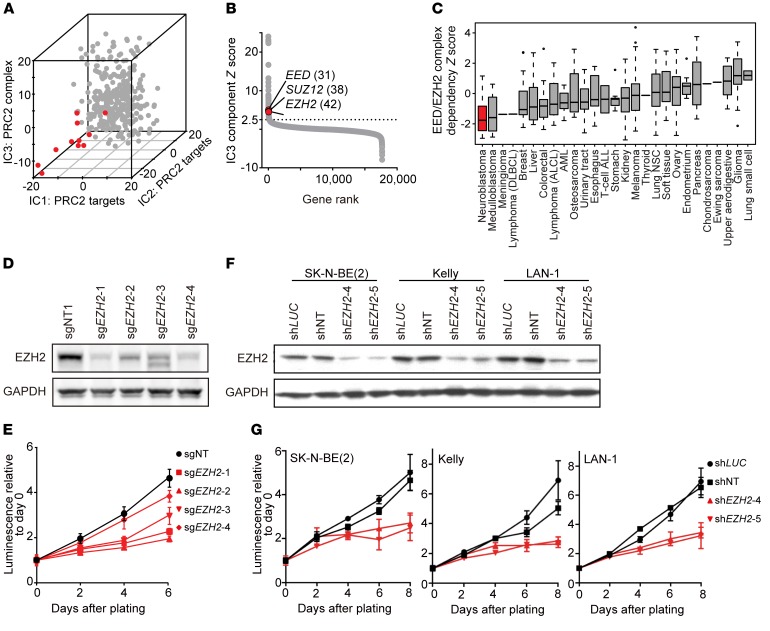
Figure 1. Genome-scale CRISPR-Cas9 screen reveals neuroblastoma dependency on the PRC2 complex components EZH2, EED, and SUZ12.
(A) Projection of the 341 cancer cell lines on the top 3 independent components (IC1, IC2, IC3) that are enriched for depletion in neuroblastoma (red) versus other cell lines (gray). (B) Rank of IC3 component genes by the gene Z score in the component with rank in parentheses. (C) Neuroblastoma is the cancer type with the most depletion in the CRISPR-Cas9 screen for the EED/EZH2 complex based on single-sample enrichment analysis. (D and E) Immunoblot (D) and cell viability assay (E) after CRISPR-Cas9 knockout of EZH2 with 4 EZH2 sgRNAs in SK-N-BE(2). Results are representative of 3 independent experiments; mean ± SD of 8 technical replicates is shown. (F and G) Immunoblot (F) and cell viability assay (G) after shRNA-mediated suppression of EZH2 in SK-N-BE(2), Kelly, and LAN-1. Results are representative of 3 independent experiments; mean ± SD of 8 technical replicates is shown.
Further investigation showed that the leading-edge genetic dependencies for each of these top 3 components (Supplemental Table 1A; supplemental material available online with this article; https://doi.org/10.1172/JCI90793DS1) were enriched in PRC2 gene signatures. The PRC2 complex has histone methyltransferase activity and is responsible for trimethylating histone H3 on lysine 27 (H3K27me3), causing transcriptional repression. The PRC2 complex genes EZH2, EED, and SUZ12 ranked among the top 50 genetic dependencies in the leading edge of the independent component IC3 (Figure 1B). In addition, a single-sample enrichment analysis for rank-normalized CERES gene effects across the mammalian protein complexes in the CORUM database (38) demonstrated that neuroblastoma is the cancer type most dependent on the EED/EZH2 complex compared with all other cancer types screened (Figure 1C and Supplemental Figure 1A). The EZH2, SUZ12, and EED dependency scores demonstrate individual dependency across subsets of neuroblastoma cell lines in the CRISPR-Cas9 screening data and demonstrate significant pairwise correlation (Supplemental Figure 1, B and C). Taken together, the results of these analyses support a PRC2-dependent role for EZH2 in a subset of neuroblastoma.
EZH2 validates as a dependency in neuroblastoma using genetic approaches.
We next confirmed the pooled screen results for EZH2 by analyzing the effects of genetic deletion or suppression of EZH2 in MYCN-amplified neuroblastoma cell lines. We focused on EZH2 because it is imminently druggable, with multiple small-molecule inhibitors already in early-phase studies in the clinic. We transduced SK-N-BE(2) cells with the 4 EZH2 single guide RNAs (sgRNAs) used in the CRISPR-Cas9 screen or a control sgRNA. EZH2 sgRNAs knocked out EZH2 and decreased H3K27me3 levels as shown by immunoblotting (Figure 1D and Supplemental Figure 1D). The CRISPR-Cas9 EZH2 knockout resulted in decreased cell viability over time as compared with the control guide (Figure 1E).
We next used shRNA as a complementary approach to test the consequences of suppressing EZH2 in neuroblastoma cell lines. We used 2 previously validated EZH2 shRNAs and extended our testing to additional neuroblastoma cell lines not in the primary screen. Knockdown of EZH2 effectively depleted EZH2 levels in all 3 cell lines as shown by immunoblotting (Figure 1F), led to a decrease of H3K27me3 levels (Supplemental Figure 1D), and resulted in markedly decreased cell viability over time as compared with 2 control shRNAs (Figure 1G).
To confirm that the effect of CRISPR-Cas9–mediated disruption is on-target, we performed a rescue experiment following knockout of EZH2, with a codon-optimized WT EZH2 or a codon-optimized catalytically inactive mutant EZH2 with 3 amino acid mutations in the methyltransferase domain (EZH2-TM) (7). We first transduced SK-N-BE(2) cells with a doxycycline-inducible WT EZH2 or EZH2-TM expression plasmid, then disrupted the endogenous EZH2 gene using CRISPR-Cas9 and induced expression of codon-optimized EZH2 or EZH2-TM with doxycycline. Upon overexpression of exogenous EZH2, but not EZH2-TM, we observed a rescue of the deleterious effects of EZH2 knockout on cell viability (Supplemental Figure 1, E and F). These data confirmed the dependency of human neuroblastoma SK-N-BE(2) cells on EZH2 methyltransferase activity.
Pharmacological inhibition of EZH2 inhibits neuroblastoma growth in vitro and promotes apoptosis.
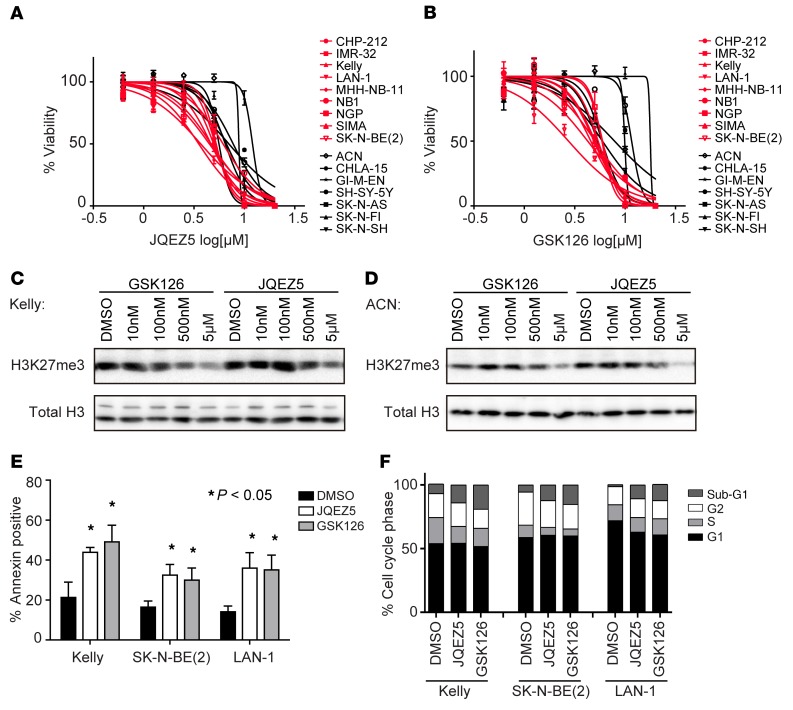
To complement the genetic perturbation studies, and to further support that neuroblastoma cells are dependent on the enzymatic activity of EZH2, we tested 2 selective small-molecule inhibitors of EZH2: JQEZ5 (39) and GSK126 (40). Nine MYCN-amplified and 7 MYCN-nonamplified neuroblastoma cell lines were cultured with serially diluted concentrations of EZH2 inhibitors, and cell viability was measured following 5 days of treatment (Figure 2, A and B). Concentration-dependent inhibition of H3K27me3 was confirmed by immunoblotting of histone extractions (Figure 2, C and D, and Supplemental Figure 2A). The range of growth inhibition IC50 was 2–8 μM for the sensitive neuroblastoma cell lines (Figure 2, A and B), a concentration range at which we saw a decrement of H3K27me3, a marker of target inhibition (Figure 2, C and D, Supplemental Figure 2A). An area-under-concentration-response curve (AUC) was calculated as a measure of drug sensitivity (41). The sensitivity scores for the 2 EZH2 inhibitors, JQEZ5 and GSK126, were positively correlated across the 16 neuroblastoma cell lines tested (Spearman correlation coefficient = 0.72, P = 0.015). There was a modest but statistically significant AUC difference in sensitivity to pharmacological inhibition of EZH2 between MYCN-amplified and MYCN-nonamplified cell lines (Mann-Whitney test P = 0.0079 for GSK126 at day 5 and Mann-Whitney test P = 0.0003 for JQEZ5 at day 5; Supplemental Figure 2B). The MYCN-nonamplified neuroblastoma cell lines SK-N-FI and ACN were the most resistant to small-molecule EZH2 inhibition in vitro (average AUC = 130) compared with the MYCN-amplified neuroblastoma cells (average AUC = 84) or the other MYCN-nonamplified cells (average AUC = 100). Moreover, sensitivity to pharmacological inhibition by GSK126 at day 5 was positively correlated with EZH2 depletion in the CRISPR-Cas9 data (Spearman correlation coefficient = 0.77, P = 0.015); correlation with JQEZ5, however, did not achieve statistical significance at day 5 (Spearman correlation coefficient = 0.38, P = 0.308) (Supplemental Table 1B).
Figure 2. Pharmacological inhibition of EZH2 inhibits neuroblastoma growth in vitro.
(A and B) Neuroblastoma cells treated with the EZH2 inhibitor JQEZ5 (A) or GSK126 (B) for 5 days in vitro. Results are representative of 3 independent experiments; data represent mean ± SD of 4 technical replicates. (C and D) Immunoblot showing target inhibition by JQEZ5 or GSK126 in the neuroblastoma cell lines Kelly (C) and ACN (D). (E) Percent of annexin V–positive cells in the cell lines treated with 3 μM JQEZ5, GSK126, or DMSO control for 8–10 days. Mean ± SD of 3 technical replicates is shown. *P < 0.05 by 2-tailed Student’s t test. (F) Cell cycle analysis in neuroblastoma cell lines treated with 3 μM JQEZ5, GSK126, or DMSO for 7 days. Results are representative of 3 independent experiments.
To further characterize the phenotypic consequences of EZH2 inhibition for neuroblastoma cells, we determined the effects of JQEZ5 and GSK126 on apoptosis and cell cycle. Both JQEZ5 and GSK126 treatment induced apoptosis, as confirmed by a significant increase in the annexin V–positive population (Figure 2E and Supplemental Figure 2C) and an increase in the sub-G1 fraction (Figure 2F) 7 days after treatment.
EZH2 inhibition is efficacious in vivo.
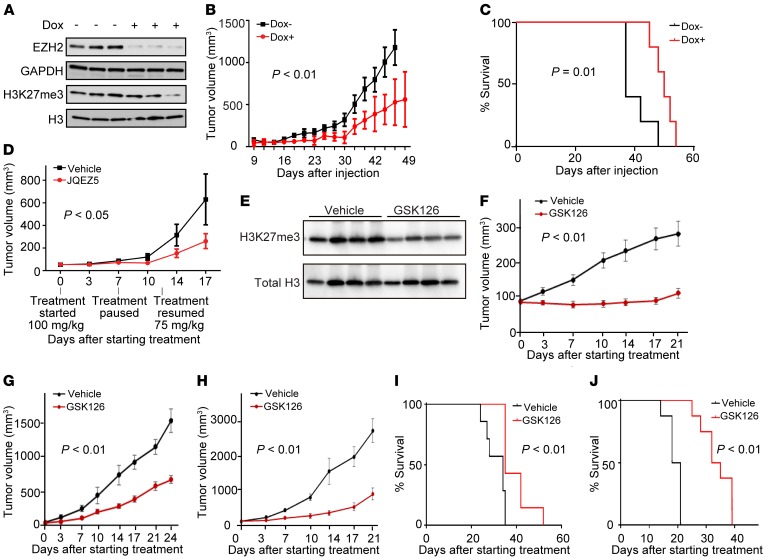
In vitro studies do not always predict in vivo dependencies. Therefore, we next tested the effects of both genetic and chemical inhibition of EZH2 in MYCN-amplified neuroblastoma mouse models. First, we generated immunocompromised mice bearing flank xenografts of a human neuroblastoma cell line, NGP, which were stably transfected with a tetracycline-inducible shRNA targeting EZH2. In this model, inducible EZH2 suppression after the initial establishment of the neuroblastoma xenograft led to decreased tumor growth as well as prolonged survival (Figure 3, A–C), indicating that EZH2 is critical to tumor growth in this neuroblastoma model in vivo.
Figure 3. Genetic and pharmacological inhibition of EZH2 inhibits growth in vivo.
(A) Tumor EZH2 and H3K27me3 levels in mice implanted with the human neuroblastoma cell line NGP expressing a doxycycline-inducible shEZH2 after 20 days of doxycycline or control treatment in vivo. Proteins and histones were extracted from tumors in 3 control mice and in 3 doxycycline-treated mice. GAPDH is a control for EZH2, and H3 is a control for H3K27me3. (B) Tumor volume in mice (n = 5 for each group) implanted with NGP cells expressing a doxycycline-inducible shEZH2. Results are representative of 2 independent experiments; mean ± SEM is shown. P calculated using 2-way ANOVA. (C) Kaplan-Meier curves with log-rank (Mantel-Cox) test showing overall survival of mice (n = 5 for control and n = 5 for doxycycline treated) implanted with the neuroblastoma cell line NGP expressing a doxycycline-inducible shEZH2. Results are representative of 2 independent experiments. (D) Tumor volume in mice (n = 10 for each group) treated with JQEZ5 or vehicle in a mouse xenograft model of the neuroblastoma cell line Kelly. Mean ± SEM is shown. P calculated using 2-way ANOVA. (E) H3K27me3 levels in mice with human neuroblastoma cell line CHP-212 xenograft after 10 days of GSK126 or vehicle treatment. Histones were extracted from circulating white blood cells in 4 GSK126-treated mice and 4 vehicle-treated mice. (F–H) Tumor volume over 21-day treatment with vehicle or 150 mg/kg/d GSK126 treatment in a mouse xenograft model of CHP-212 (F, n = 8 each), SK-N-BE(2) (G, n = 7 each), and SH-SY-5Y (H, n = 8 each). Mean ± SEM is shown. P calculated using 2-way ANOVA. (I and J) Kaplan-Meier curves show overall survival of mice with SK-N-BE(2) (I, n = 7 each) or SH-SY-5Y (J, n = 8 each). P calculated using log-rank (Mantel-Cox) test.
Next, we studied the effects of enzymatic inhibition of EZH2 in MYCN-amplified neuroblastoma in vivo. JQEZ5 was first evaluated in a Kelly cell line xenograft model established by subcutaneous injection into NOD-SCID–IL-2Rγnull (NSG) mice. Tumor-bearing mice were treated with 150 mg/kg of JQEZ5 or vehicle delivered by daily intraperitoneal injection (n = 10 per group) for 7 days, followed by a 5-day no-drug holiday secondary to weight loss, and then treatment with 75 mg/kg of JQEZ5 or vehicle until sacrifice. JQEZ5 treatment significantly diminished tumor volume compared with control treatment (Figure 3D).
Because of the observed JQEZ5-associated weight loss, we then tested the effect of GSK126 in vivo. After 10 days of GSK126 treatment in mice, the levels of H3K27me3 were significantly reduced (Figure 3E) in mononuclear cells. In the mouse xenograft models, GSK126 treatment inhibited the tumor growth of all 3 human neuroblastoma cell line models tested, 2 MYCN-amplified, CHP-212 and SK-N-BE(2), and 1 MYCN-nonamplified, SH-SY-5Y (Figure 3, F–H). The daily treatment of GSK126 also significantly prolonged the survival of mice in the SK-N-BE(2) and SH-SY-5Y xenograft models (Figure 3, I and J). Taken together, these studies suggest that neuroblastoma tumors in vivo are dependent on the enzymatic activity of EZH2.
MYCN directly activates EZH2 expression.
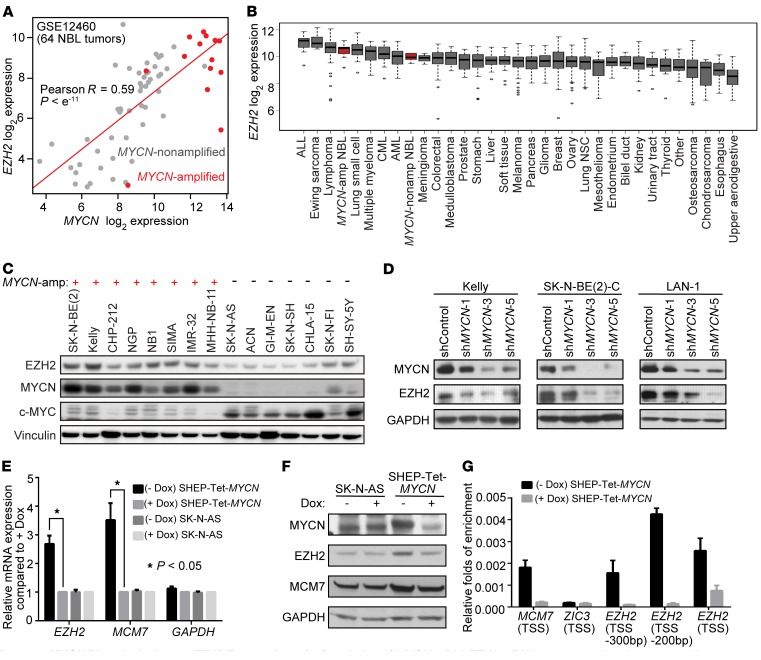
We then focused on the mechanism regulating EZH2 expression in human neuroblastoma cells. EZH2 emerged as one of the top genes in our in silico neuroblastoma MYCN-coexpression network analysis. The transcription level of EZH2 was highly correlated with that of MYCN in primary neuroblastoma patient samples and was higher in MYCN-amplified neuroblastoma tumors compared with MYCN-nonamplified neuroblastoma tumors (Figure 4A). Moreover, data generated from the Cancer Cell Line Encyclopedia (42) showed that MYCN-amplified neuroblastoma cell lines express EZH2 at higher levels than MYCN-nonamplified neuroblastoma cell lines and show higher expression of EZH2 than the majority of cells from other lineages (Figure 4B). Immunoblotting also indicated that EZH2 protein levels were significantly higher in MYCN-amplified than in MYCN-nonamplified neuroblastoma cell lines (Figure 4C).
Figure 4. MYCN directly activates EZH2 expression.
(A) Correlation of MYCN mRNA and EZH2 mRNA expression in primary neuroblastoma (NBL) tumor samples based on Affymetrix data GSE12460. P computed with the rcorr function (R-CRAN, Hmisc library). (B) EZH2 mRNA expression across Cancer Cell Line Encyclopedia cancer cell lines with neuroblastoma cell lines in red. (C) Immunoblot of protein levels of EZH2, MYCN, and MYC in MYCN-amplified compared with MYCN-nonamplified neuroblastoma cells. (D) Effect of MYCN suppression on EZH2 protein levels. Results are representative of 2 independent experiments. (E and F) Effect of conditional overexpression of MYCN on EZH2 expression at the transcription level (E) and the protein level (F) in the MYCN-nonamplified neuroblastoma cell line SHEP. The SHEP-Tet-MYCN cell line was generated by stable transduction of SHEP with a Tet-off MYCN construct. MCM7 is a known MYCN target gene. SK-N-AS is an unmodified MYCN-nonamplified neuroblastoma cell line. Mean ± SEM of 3 independent experiments shown. *P < 0.05 by 2-tailed Student’s t test. (G) ChIP–quantitative PCR with MYCN antibody showing the enrichment of MYCN in the promoter region of EZH2 in MYCN-overexpressing SHEP-Tet-MYCN cells. MCM7 is a positive control, and ZIC3 is a negative control. TSS, transcription start site. Results are representative of 2 independent experiments; mean ± SD of 3 technical replicates is shown.
MYCN stimulates tumor cell growth through activation of an oncogenic transcriptional program (20, 43). Here, we hypothesized that MYCN also positively regulates the expression of EZH2 in neuroblastoma. To test this, we first suppressed endogenous MYCN with 3 different MYCN-targeting shRNAs and observed a substantial decrease in EZH2 at the protein level in all 3 human neuroblastoma cell lines tested (Figure 4D). To further address the effect of MYCN on EZH2 expression, we next took advantage of a previously established conditional overexpression system in which a doxycycline-repressible MYCN construct was integrated into the genome of the human MYCN-nonamplified neuroblastoma cell line SHEP so that the engineered SHEP-Tet-MYCN cell line overexpresses MYCN in the absence of doxycycline. We confirmed that the overexpression of MYCN led to an increase of both EZH2 mRNA and EZH2 protein levels (Figure 4, E and F). Doxycycline did not affect the level of EZH2 in the SK-N-AS control (Figure 4, E and F).
There are 2 putative MYCN binding sites, so-called E-boxes, on the EZH2 promoter region. Therefore, we hypothesized that EZH2 is a direct target gene of MYCN. To test the hypothesis, we performed ChIP–quantitative PCR experiments with MYCN antibodies in the SHEP-Tet-MYCN cell line. Our ChIP–quantitative PCR demonstrated enrichment of MYCN on the EZH2 gene promoter under conditions of MYCN overexpression (Figure 4G), supporting a role for MYCN in activating the transcription of EZH2 directly.
EZH2 target loci are associated with H3K27me3 and gene repression in neuroblastoma.
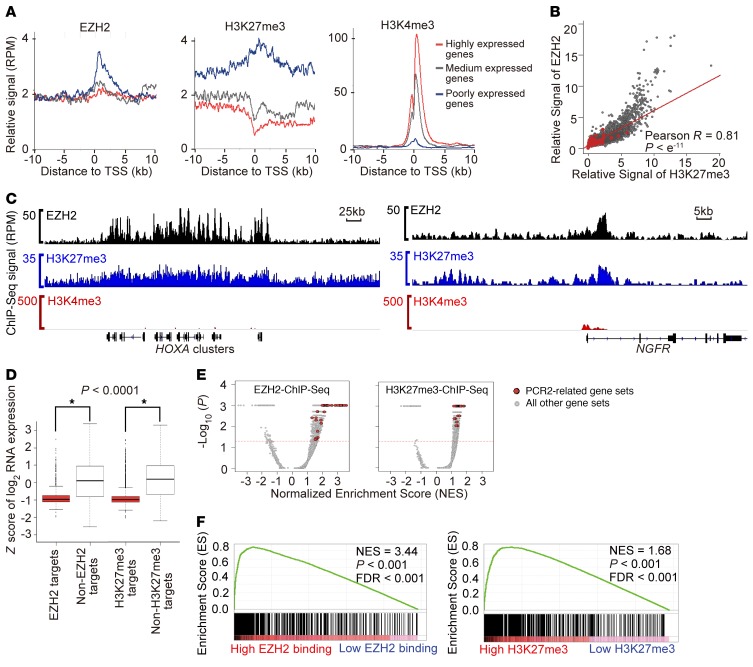
EZH2 exhibits a PRC2-dependent function in transcriptional repression in B cell lymphoma and lung cancer (40, 44), as well as a PRC2-independent function in transcriptional activation in a subtype of prostate and breast cancers (45–47). To evaluate PRC2 dependence, we identified direct EZH2 target genes in human neuroblastoma cell lines by ChIP followed by massively parallel sequencing (ChIP-Seq). In addition, we identified the genome-wide localization of the repressive transcriptional histone mark H3K27me3 and the active histone mark H3K4me3 in neuroblastoma by ChIP-Seq. Globally, EZH2 was enriched at transcriptional start sites. The height of the EZH2 peak was anticorrelated with the amount of mRNA transcribed from the respective locus (Figure 5A and Supplemental Figure 3A). Globally, H3K27me3 also peaked around transcriptional start sites and was anticorrelated with gene transcription, although with broader peaks than EZH2. In contrast, H3K4me3 peaked sharply at transcription start sites and positively correlated with gene expression (Figure 5A and Supplemental Figure 3A). The binding of EZH2 with H3K27me3 marks on gene promoters in neuroblastoma was highly correlated (Pearson R = 0.82). Unlike in castration-resistant prostate cancer cells (45), however, there were no EZH2 “solo” sites defined by high EZH2 binding and low H3K27me3 (Figure 5B, Supplemental Figure 3B, and Supplemental Figure 4).
Figure 5. Genome-wide EZH2 binding pattern in MYCN-amplified neuroblastoma Kelly cells.
(A) Metagene analysis showing the average ChIP-Seq binding signals of EZH2, H3K27me3, or H3K4me3 for 500 randomly selected genes in each of 3 categories based on the gene expression level. The x axis shows the distance in kilobases to the transcription start site (TSS). The y axis shows signal in reads per million (RPM). (B) Scatter plot of the genome-wide correlation between EZH2 relative binding signal and H3K27me3 relative binding signal in the promoter region. P computed with the rcorr function (R-CRAN, Hmisc library). (C) Gene track showing high binding signal for EZH2 and H3K27me3 with low binding signal for H3K4me3 in 2 published validated EZH2 targets. (D) Box plots of expression values for EZH2 target genes versus non-EZH2 target genes and H3K27me3 target genes versus non-H3K27me3 target genes in Kelly. *P < o.ooo1 using 2-tailed Student’s t test. (E) GSEA volcano plot enrichment of published PRC2 target signatures from MSigDB version 5.1 among genes with high EZH2 binding signal (left) or high H3K27me3 binding signal (right) in Kelly cells. (F) Enrichment of Benporath PRC2 target signature among genes with high EZH2 promoter binding signal (left) or high H3K27me3 promoter binding signal (right) in Kelly cells.
We then took a closer look at the gene tracks of expected EZH2 target genes in neuroblastoma cell lines. We saw strong binding peaks of EZH2 and H3K27me3 but minimal peaks for H3K4me3 at the well-known PRC2 target HOXA cluster genes in neuroblastoma. We also observed a high enrichment of EZH2 and H3K27me3, but not H3K4me3, at the promoter region of NGFR, a known EZH2 target and a tumor suppressor in neuroblastoma (Figure 5C, Supplemental Figure 3C, and ref. 48).
We next confirmed that both the EZH2 targets and H3K27me3-enriched genes identified by ChIP-Seq were associated with lower levels of mRNA compared with other genes in the neuroblastoma genome (Figure 5D and Supplemental Figure 3D). Additionally, gene set enrichment analysis (GSEA) showed that EZH2 and H3K27me3 binding in neuroblastoma cells was significantly enriched for PRC2-related signatures in the Molecular Signatures Database (MSigDB), including gene sets identified by ChIP-chip or ChIP-Seq to be bound by H3K27me3, SUZ12, or EED in embryonic stem and neural progenitor cells (Figure 5, E and F; Supplemental Figure 3, E and F; and Supplemental Table 2). Taken together, these analyses also support the PRC2-dependent function of EZH2 in neuroblastoma.
Transcriptional effects of EZH2 inhibition in neuroblastoma.
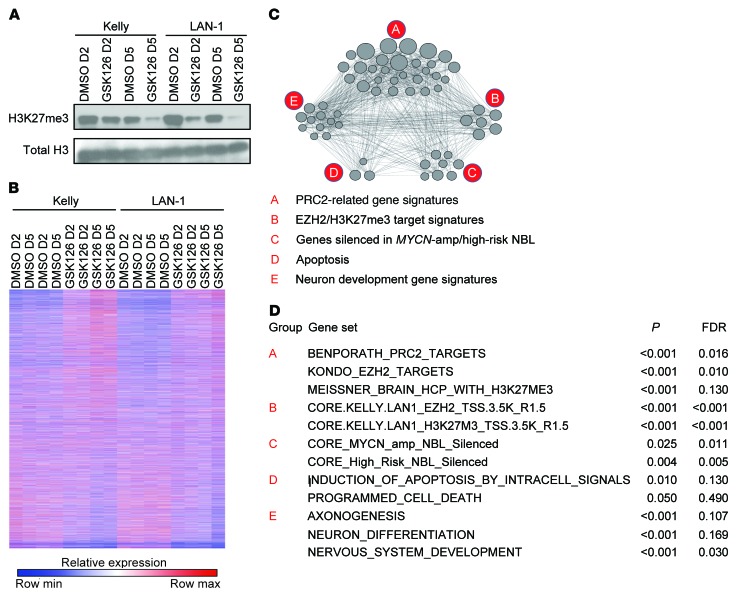
The transcriptional program controlled by EZH2 is highly cell context dependent. While EZH2-regulated transcriptional programs have been established in some cancer types, they have not yet been thoroughly established in the context of neuroblastoma. To study neuroblastoma-specific, EZH2-regulated transcriptional programs, we profiled 2 MYCN-amplified neuroblastoma cell lines, Kelly and LAN-1, treated with 2 μM of the EZH2 inhibitor GSK126 for 2 or 5 days. GSK126 treatment led to decreased trimethylation of histone H3K27 as assessed by immunoblotting (Figure 6A) and more dynamic global gene expression changes at 5 days compared with 2 days (Figure 6B). Our gene expression profiles after treatment with GSK126 were highly enriched for the previously published gene signatures reported in neuroblastoma cell lines after treatment with EPZ-6438, a distinct selective EZH2 inhibitor (Supplemental Figure 5A and ref. 49). Among the genes derepressed with GSK126, PRC2 target gene signatures were strongly enriched (Figure 6, C and D, and Figure 7A). Because these published PRC2 target gene signatures were derived in other cellular contexts, we developed custom neuroblastoma PRC2 target signatures from the top 300 genes with promoter H3K27me3 binding signal and the top 300 genes with promoter EZH2 binding signal in our ChIP-Seq analysis (Supplemental Table 3–5). As expected, the neuroblastoma PRC2 target signatures were strongly enriched among genes derepressed with GSK126 (Figure 6, C and D, and Figure 7B), supporting the effectiveness and selectivity of in vitro treatment of neuroblastoma cells with this type of EZH2 inhibitor.
Figure 6. Transcriptome changes in response to GSK126 treatment.
(A) Immunoblots showing H3K27me3 changes after 2 and 5 days of treatment with 2 μM GSK126 or DMSO. (B) Heatmap of RNA-Seq gene expression following 2 and 5 days of 2 μM GSK126 treatment of neuroblastoma cell lines Kelly and LAN-1. (C) Functional enrichment map depicts the functional groups of the GSEA hits for the GSK126 treatment effect on neuroblastoma cell lines Kelly and LAN-1. The nodes correspond to enriched gene sets. An edge connects 2 nodes if the corresponding gene sets are overlapping. (D) The top enriched gene sets from each functional group depicted in the enrichment map.
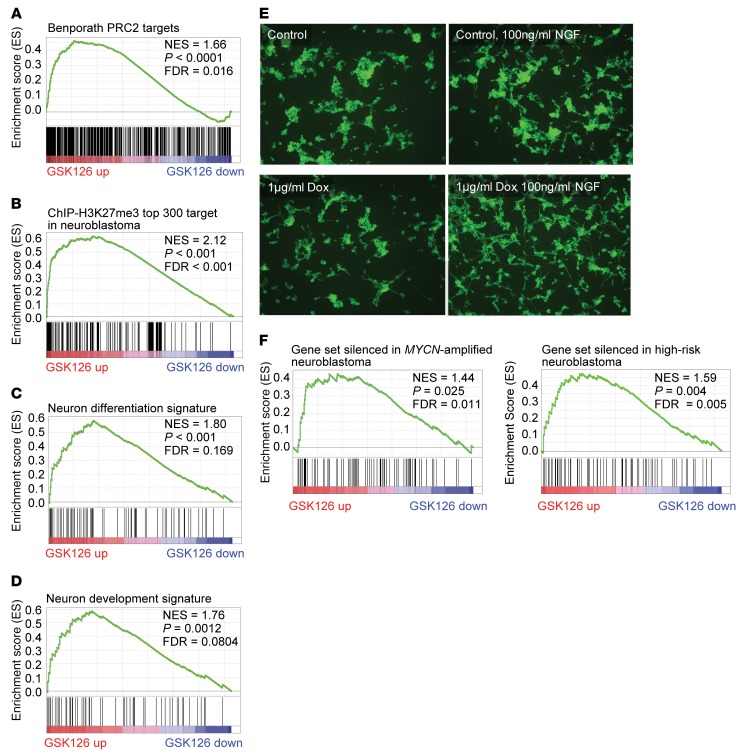
Figure 7. Effect of pharmacological and genetic suppression of EZH2.
(A) GSEA showing enrichment of Benporath PRC2 target signature in genes upregulated by EZH2 inhibitor GSK126. (B) GSEA showing that genes upregulated with GSK126 treatment are enriched for the top 300 genes with highest H3K27me3 ChIP-Seq signal in Kelly and LAN-1 cells. (C) GSEA showing enrichment of neuron differentiation signature in genes upregulated by GSK126. (D) GSEA showing enrichment of neuron development signature in genes upregulated by GSK126. (E) The effect of conditional knockdown of EZH2 on NGF-induced neurite outgrowth in neuroblastoma cell line NGP. Shown is immunocytochemistry with βIII-tubulin antibodies, with photographs representative of 5 fields taken at ×100 original magnification. (F) GSEA showing that genes upregulated with GSK126 treatment are enriched for genes silenced in MYCN-amplified and high-risk neuroblastoma based on primary tumor expression data sets.
In order to further interrogate the functional implications of selective EZH2 inhibition, we queried our gene expression data with the gene signatures in the MSigDB c2 collection. The top-ranked GSK126-upregulated signatures were neuronal differentiation, axon genesis, neuron development, neurite development, and all-trans retinoic acid treatment (Figure 7, C and D, and Supplemental Figure 5B). These results suggest that inhibition of EZH2 in MYCN-amplified neuroblastoma alters the epigenetic landscape to induce a more differentiated transcriptional program. Apoptotic signaling also scored as a top-ranked signature with EZH2 inhibitor treatment in MYCN-amplified neuroblastoma cells, consistent with the induction of apoptosis observed in Figure 2 and Supplemental Figure 2.
We further verified the role of EZH2 in neuroblastoma differentiation by studying the morphological changes induced by shRNA-mediated EZH2 conditional knockdown. One important feature of neural differentiation is the neurite outgrowth induced by nerve growth factor (NGF). Here we treated NGP-dox-shEZH2 cells for 6 days with 100 ng/ml NGF, with or without doxycycline for EZH2 knockdown, and then visualized neurite outgrowth via anti–βIII-tubulin immunocytochemistry (Figure 7E). We demonstrated that EZH2 suppression strongly potentiated NGF-induced neurite outgrowth in neuroblastoma cells in vitro.
To determine the relevance of data generated in cell line models to primary patient tumors, we integrated the gene expression changes upon GSK126 treatment with 2 primary neuroblastoma tumor gene expression data sets, GSE12460 (50) and GSE49711 (51). We derived custom gene sets specifically silenced in primary neuroblastoma tumors associated with high risk or MYCN amplification compared with neuroblastoma with low risk or nonamplified MYCN. GSEA revealed that the gene sets silenced in high-risk or MYCN-amplified neuroblastoma were enriched for genes upregulated with GSK126 treatment of MYCN-amplified neuroblastoma cells (Figure 7F). In addition, single-sample GSEA revealed that gene sets upregulated with GSK126 treatment and gene sets for EZH2 or H3K27me3 targets were enriched for genes silenced in high-risk or MYCN-amplified neuroblastoma (Supplemental Figure 5C). Our findings suggest that EZH2 inhibits a tumor suppressor program in MYCN-amplified, high-risk neuroblastoma.
Integrative analysis reveals new EZH2-repressed tumor suppressors in MYCN-amplified neuroblastoma.
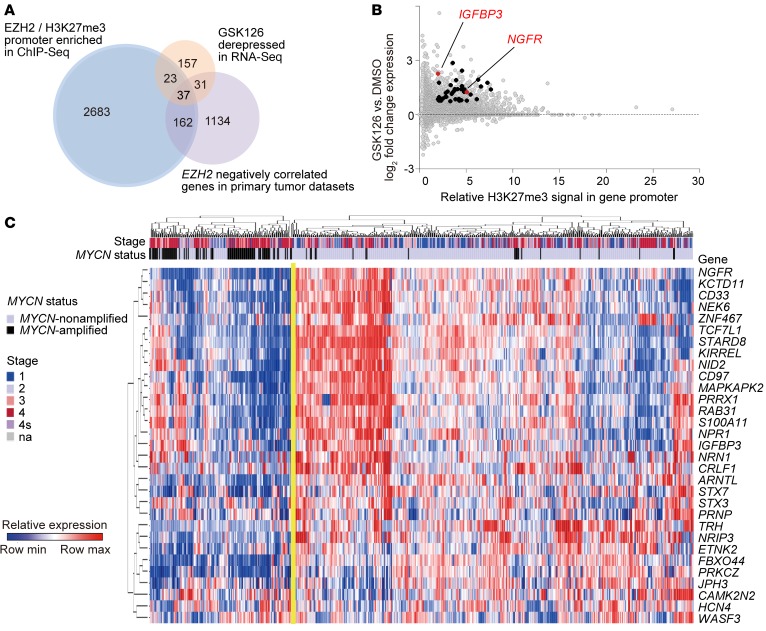
We hypothesized that neuroblastoma tumor suppressors are preferentially silenced in MYCN-amplified, high-risk neuroblastoma via MYCN-driven overexpression of EZH2. To test the hypothesis, we first aimed to identify the EZH2-regulated neuroblastoma tumor suppressor programs by integrating 3 distinct approaches: epigenetic profiling, chemical genomics, and transcriptional network analysis. Correlating histone modification profiles with the gene expression changes for cells treated with GSK126, we identified 60 genes whose promoters were enriched for EZH2 and H3K27me3 and whose expression increased significantly in both the neuroblastoma cell lines Kelly and LAN-1 in response to GSK126 treatment. We further integrated the transcriptional network analysis of primary neuroblastoma tumor expression data and demonstrated that 37 genes among the 60 genes were also negatively correlated with EZH2 gene expression in primary tumor data sets (Figure 8, A–C). Among this list of 37 genes were the previously validated neuroblastoma tumor suppressor NGFR (48, 52) and neuron-specific genes and some tumor suppressor genes validated in other cancer types, such as insulin-like growth factor–binding protein 3 (IGFBP3), a known tumor suppressor gene in non–small cell lung cancer and glioblastoma (refs. 53, 54, and Figure 8B). An unsupervised clustering analysis of primary tumor gene expression data revealed the repression of these 37 EZH2-regulated genes in MYCN-amplified and high-risk neuroblastoma, suggesting that EZH2 inhibits this neuroblastoma tumor suppressor program in poor-outcome neuroblastoma (Figure 8C).
Figure 8. Integrative analysis reveals EZH2 function.
(A) Schematic representation of the integrative analysis to identify EZH2-regulated neuroblastoma tumor suppressors. Venn diagrams present the number of hits identified by the integrative analysis studies. (B) Scatter plot showing whole-genome promoter binding H3K27me3 relative signal (x axis) versus whole-genome transcriptome changes in response to GSK126 (y axis). Highlighted are the 37 genes with high H3K27me3 binding, upregulated by GSK126 and anticorrelated with EZH2 expression in primary neuroblastoma transcriptome data sets. (C) Heatmap of the EZH2 neuroblastoma signature developed by the integrative analysis depicted in A across 562 annotated neuroblastoma tumors.
We further assessed whether EZH2-regulated genes in neuroblastoma, such as IGFBP3, function as tumor suppressors in this disease context. IGFBP3 has not been reported before as a tumor suppressor in neuroblastoma. Protein levels of IGFBP3 are generally higher in MYCN-nonamplified neuroblastoma cell lines than in MYCN-amplified neuroblastoma cell lines (Figure 9A). Our ChIP-Seq and RNA-Seq studies revealed that IGFBP3 is enriched for binding of both EZH2 and H3K27me3 (Figure 9B), and the transcript level of IGFBP3 was increased significantly after treatment with the EZH2 inhibitor GSK126 (Figure 8B). We next overexpressed IGFBP3 in the MYCN-amplified neuroblastoma cell line SK-N-BE(2) and performed a luminescence-based in vitro viability assay 6 days after transduction. NGFR served as a positive control and EGFP as a negative control. We determined that the overexpression of IGFBP3 resulted in markedly decreased cell viability over time as compared with the negative control in SK-N-BE(2) (Figure 9, C and D). Moreover, we showed that the overexpression of IGFBP3 inhibited the in vivo growth of neuroblastoma and prolonged the survival of mice in a mouse xenograft study using engineered human neuroblastoma SK-N-BE(2) cells (Figure 9, E and F). We expanded our in vitro viability assay to 2 additional MYCN-amplified cell lines, CHP-212 and LAN-1, and 2 MYCN-nonamplified cell lines, ACN and SH-SY-5Y. IGFBP3 overexpression inhibited the cell viability in all 4 cell lines. Interestingly, ACN, which is less sensitive to EZH2 inhibitors, is also less sensitive to the overexpression of IGFBP3 compared with other tested neuroblastoma cell lines (Figure 9, G and H). We further demonstrated that IGFBP3 overexpression sensitized SK-N-BE(2) to small-molecule EZH2 inhibitors in vitro (Figure 9I). Overall, our in vitro and in vivo studies confirm the tumor suppressor potential of IGFBP3 in some neuroblastomas and further support the notion that EZH2 inhibits a tumor suppressor program in MYCN-amplified, poor-outcome neuroblastoma.
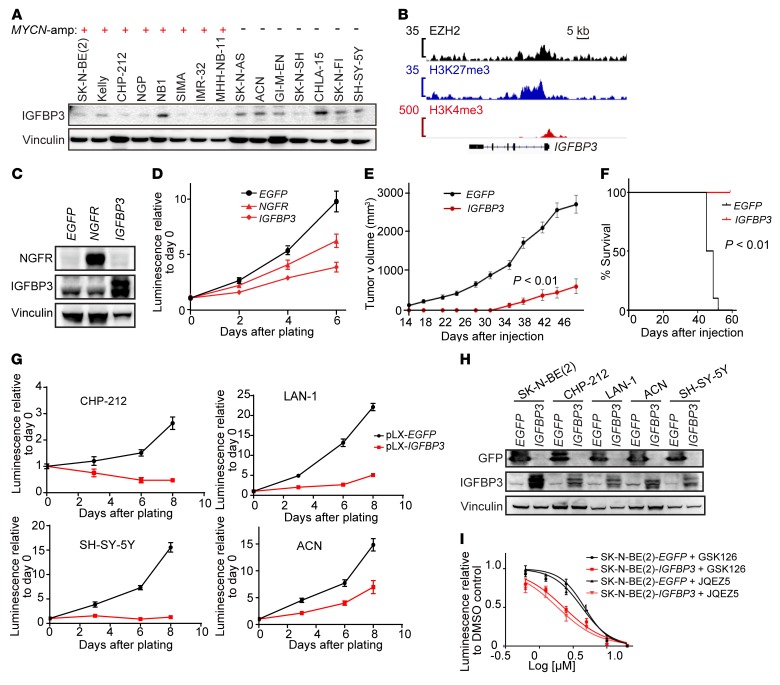
Figure 9. EZH2-regulated gene IGFBP3 functions as a neuroblastoma tumor suppressor.
(A) The protein expression level of IGFBP3, a representative gene in the neuroblastoma EZH2 signature, in human neuroblastoma cell lines with or without MYCN amplification. (B) EZH2, H3K27me3, and H3K4me3 binding signal at the promoter of IGFBP3 in Kelly cells. (C) Immunoblot showing the overexpression of EGFP (negative control), NGFR (positive control), and IGFBP3 in SK-N-BE(2) cell line. (D) Cell viability assay after overexpression of EGFP, NGFR, or IGFBP3 in SK-N-BE(2). Results are representative of 3 independent experiments; mean ± SD of 8 technical replicates is shown. (E) Tumor volume in mouse xenograft model of SK-N-BE(2) with or without IGFBP3 overexpression (n = 10). P calculated with 2-way ANOVA. (F) Kaplan-Meier curves show survival of mice with xenografts of SK-N-BE(2) with or without IGFBP3 overexpression, up to 56 days after injection. P calculated using log-rank (Mantel-Cox) test. (G and H) Cell viability assay (G) and immunoblotting (H) after overexpression of IGFBP3 in CHP-212, LAN-1, ACN, and SH-SY-5Y. Results are representative of 3 independent experiments; data in G represent mean ± SD of 8 technical replicates. (I) Effect of overexpression of IGFBP3 on SK-N-BE(2)’s response to EZH2 inhibitors. Shown is a representative of 2 independent experiments; mean ± SD of 8 technical replicates is shown.
The combination of EZH2 and histone deacetylase inhibition is synergistic in neuroblastoma.
Curative cancer therapy usually requires the combination of multiple drugs. Indeed, while we demonstrate efficacy of EZH2 inhibitors in 3 mouse models of neuroblastoma, they were not curative, suggesting that drug combinations with EZH2 inhibitors will be necessary. To find clinically relevant drugs that are synergistic with EZH2 inhibitors in neuroblastoma, we tested in vitro the combination of an EZH2 inhibitor, GSK126, with a panel of small molecules, including a differentiation agent (cis-retinoic acid), a hypomethylating agent (decitabine), a PI3K inhibitor (GDC0941), a histone deacetylase (HDAC) inhibitor (panobinostat), various ALK inhibitors (lorlatinib, crizotinib, and PD0325901), and various cytotoxic agents (etoposide, irinotecan, and temozolomide). In this study, the HDAC inhibitor panobinostat scored as the most synergistic with GSK126 (Figure 10A). The synergy of GSK126 and panobinostat is strong across a broad concentration range in all 4 of the MYCN-amplified neuroblastoma cell lines tested (Figure 10B and Supplemental Figure 6).
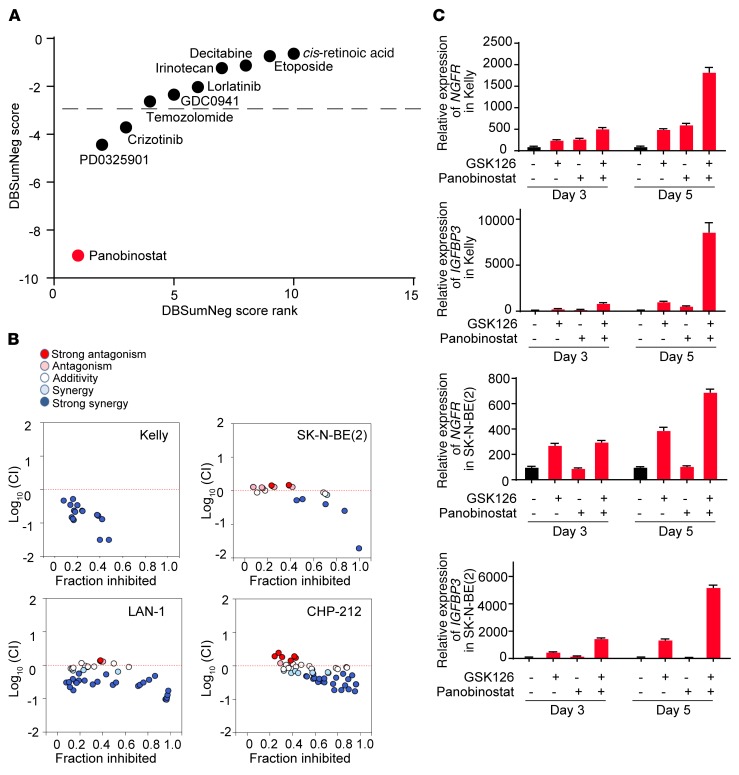
Figure 10. Drug synergy of EZH2 inhibitor GSK126 and small-molecule compounds.
(A) Delta Bliss Sum Negative (DBSumNeg) score of drug synergy analysis with GSK126 treatment in Kelly cells on day 6 in vitro. The synergistic combinations were estimated based on DBSumNeg score cutoff < –3. Results are representative of 3 independent experiments. (B) Combination index analysis of GSK126 and panobinostat in 4 neuroblastoma cell lines. Dotted red line is the line of additivity between antagonism (>0) and synergy (<0). Light blue is significant synergy (log10 [CI] < –0.10), and dark blue is strong synergy (log10 [CI] < –0.22). Results are representative of 2–3 independent experiments. (C) Relative mRNA expression changes upon combination treatment of GSK126 and panobinostat, compared with single-agent treatment. All expression is normalized to DMSO. Results are representative of 3 independent experiments; mean ± SD of 4 technical replicates is shown.
Our laboratory has previously demonstrated that HDAC1/2 inhibition induces neuroblastoma differentiation (55). Moreover, HDAC2 scored as one of the top neuroblastoma-specific dependency genes in the genome-scale CRISPR-Cas9 screen. To explore the mechanism of synergy between HDAC inhibitors and EZH2 inhibitors, quantitative gene expression analysis was performed on the neuroblastoma cell lines Kelly and SK-N-BE(2) to assess the expression level of the neuroblastoma tumor suppressor genes NGFR and IGFBP3 (Figure 10C). In both cell lines, expression levels of NGFR and IGFBP3 were increased in GSK126 single treatments compared with the untreated cells, and they were further increased with the combined treatment of GSK126 and panobinostat compared with either single treatment alone, consistent with the synergy observed between HDAC inhibitors and EZH2 inhibitors.
Discussion
Cancer is driven, in part, by the interplay of regulatory transcription factors and dynamic alterations in chromatin structure. Previous studies of MYCN-amplified neuroblastoma have focused on the role of MYCN in transcriptional activation and the super-enhancer machinery to drive a neuroblastoma oncogenic program (20, 43, 56, 57). The role of MYCN in repressing tumor suppressor programs in neuroblastoma has not been as well explored. Our genome-scale CRISPR-Cas9 screen revealed a preferential dependency of neuroblastoma cells on the transcriptional silencing machinery of the PRC2 complex. Our mechanistic studies determined that MYCN directly drives the transcriptional repressor EZH2 in neuroblastoma, providing a direct link between MYCN overexpression and the repression of tumor suppressor programs in this disease. Specifically, EZH2 represses a neuronal differentiation program in MYCN-amplified neuroblastoma, enhancing the undifferentiated phenotype characteristic of neuroblastoma. Indeed, high-risk neuroblastoma is one of the first malignancies shown to be responsive to differentiation agents, such as 13-cis-retinoic acid (58), suggesting that enhancement of neuronal differentiation programs through alternative approaches, such as EZH2 inhibition, may promote therapeutic benefit.
EZH2 is a SET domain–containing histone methyltransferase that catalyzes methylation of H3K27 (59). It is a key component of PRC2, which represses gene expression in development (60). H3K27 di- and trimethylation is associated with transcriptional repression and heterochromatin formation. Gain-of-function and loss-of-function mutations of EZH2, overexpression of EZH2, and loss-of-function mutations of the EZH2-antagonizing enzyme UTX have been implicated in many adult cancers, such as lymphoma (40), breast (61, 62), myeloid (63), and prostate cancers (64). More recently, Dardenne et al. described a MYCN-induced, EZH2-mediated transcriptional program in neuroendocrine prostate cancer (65). Despite the fact that massively parallel sequencing of neuroblastoma tumors has not revealed frequent mutations or focal amplifications of EZH2, recently published work has suggested a role for EZH2 in neuroblastoma (49, 66, 67).
We have taken a complementary, unbiased genome-scale functional approach. A remarkable advantage of genome-scale loss-of-function CRISPR-Cas9 screens is the ability to comprehensively survey gene dependencies and identify those that are context-specific based on lineage or genotype. In our unbiased screen, the PRC2 complex and PRC2 targets were enriched in the independent components that significantly differentiate the genetic dependencies in neuroblastoma cell lines from other cancer cell lines. Henrich et al. recently used an integrative genome-scale approach analyzing methylomes, transcriptomes, and copy number variations to identify a link between MYCN and EZH2 (49). By applying single-sample GSEA on each cell line, our study demonstrated that, on average, neuroblastoma cell lines are more sensitive to knockout of PRC2 complex members than other cell lines screened. Such a screening approach provides orthogonal and compelling evidence that targeting PRC2, and EZH2 in particular, could be a treatment strategy in neuroblastoma, and overall enables the prioritization of targets for therapeutic intervention both within a disease and across diseases.
Our study suggests that the neural crest lineage disease neuroblastoma is sensitive to EZH2 inhibition in tumors with MYCN amplification. Consistent with our results in human neuroblastoma, Tsubota et al. demonstrated the importance of EZH2 in early-stage spheroid cultures of transgenic mouse neuroblastoma cells by demonstrating that MYCN and PRC2 transcriptional signatures are present, that PRC2 inhibition suppressed in vitro sphere formation and in situ tumor growth, and that MYCN drives expression of EZH2 in this model (66). Additional biomarkers of response to EZH2 inhibitors, however, are likely to be relevant, and this question will require further investigation. Indeed, in our own study, the MYCN-nonamplified neuroblastoma cell line SK-N-AS was also sensitive to EZH2 knockout, and several of the lines were sensitive to EZH2 chemical inhibition, including the SH-SY-5Y line in vivo. Recently, several studies have revealed specific sensitivity to EZH2 inhibition in cancers with ARID1A or other mutations of the BAF complex (7, 68). Resistance to EZH2 inhibition in adult cancers with co-occurrence of BAF complex and RAS pathway mutations was also described (7). While our study was not powered to specifically assess these potential biomarkers of response, we do not see any obvious correlations. For example, the neuroblastoma cell line MHH-NB-11 has an ARID1A SNP predicted to alter splicing and lacks a RAS pathway mutation, but is less sensitive than the other neuroblastoma lines tested to EZH2 inhibition both by small molecules and by CRISPR-Cas9 knockout. Moreover, 2 of the more EZH2 inhibitor–sensitive cell lines, SK-N-AS and CHP-212, have NRAS mutations. The role of PRC2 is highly cell context dependent, and in this case, the neural crest lineage and/or MYCN amplification may outweigh the potential contributions of other collaborating gene mutations, such as RAS or BAF complex–related gene mutations.
Moreover, in some disease contexts, the role of EZH2 is thought to be largely PRC2 independent, such as in castration-resistant prostate cancer (45), and Bate-Eya et al. suggested a histone methyltransferase–independent role for EZH2 in neuroblastoma (67). In our study, however, multiple lines of evidence suggest that the role of EZH2 in some MYCN-amplified neuroblastomas is primarily PRC2 dependent. First, the catalytically dead EZH2 does not rescue cell survival in EZH2-knockout SK-N-BE(2) neuroblastoma cells (Supplemental Figure 1, E and F). Second, dependencies of all 3 members of the PRC2 complex, EZH2, EED, and SUZ12, were correlated across neuroblastoma lines in the CRISPR-Cas9 screen (Supplemental Figure 1, B and C). Third, the concentrations of GSK126 and JQEZ5 at which H3K27me3 levels were repressed were consistent with their growth-inhibitory concentrations (Figure 2). Additionally, the transcriptional and chromatin studies are highly enriched for PRC2 signatures, and PRC2 targets include neuroblastoma tumor suppressors.
We have also revealed new potential tumor suppressor programs regulated by EZH2 in neuroblastoma. Prior studies linked EZH2 to the aberrant repression of tumor suppressor genes in neuroblastoma, such as CASZ1, CLU, RUNX3, and NGFR (48, 69), while Henrich et al. reported the hypermethylation of tumor suppressor DCL1 in neuroblastoma (49). The new potential tumor suppressor genes identified in our study are marked by H3K27me3, upregulated by an EZH2 inhibitor, and poorly expressed in primary MYCN-amplified neuroblastoma tumors. For example, the EZH2-regulated gene IGFBP3 has been shown to induce apoptosis and to impair growth in vitro and in vivo and was a validated tumor repressor of glioblastoma and non–small cell lung cancer (53, 54, 70). Moreover, the level of IGFBP3 expression negatively correlated with patient survival in glioblastoma (54). In this study, we identified a tumor suppressor function of IGFBP3 in the context of neuroblastoma. Other genes that we have identified as candidates, but have not yet validated, such as KCTD11 and PRKCZ, have also been shown to possess tumor suppressor function in other cancer types (71, 72). The current study reveals an underlying mechanism of the function of EZH2 in neuroblastoma and lays a foundation for the future study of additional neuroblastoma tumor suppressors.
We also explored whether clinical and preclinical drugs may be synergistic with EZH2 inhibitors, as our in vivo studies suggest that EZH2 inhibition alone may not be sufficient to cure neuroblastoma. Henrich et al. looked at transcriptional consequences of EZH2 inhibitors and DNA methyltransferase inhibitors (49). We found, however, that synergy with a combination of EZH2 inhibitors and HDAC inhibitors was the most striking in terms of effects on cell viability, supporting the further development of combination therapy to target both PRC2 and histone acetylation in neuroblastoma.
In summary, we have established that the inhibition of the polycomb protein histone methyltransferase EZH2 has an antitumor effect in MYCN-amplified neuroblastoma through derepressing the EZH2-mediated epigenetic silencing of a neuroblastoma tumor suppressor program. We assessed the clinical relevance of EZH2 inhibition, both in vitro and in vivo, using multiple perturbation approaches, such as selective small-molecule EZH2 inhibitors, shRNA-mediated knockdown, and CRISPR-Cas9 knockout. We also studied the epigenetic networks and their response to EZH2 inhibition in MYCN-amplified neuroblastoma and explored the underlying molecular mechanisms of the sensitivity to EZH2 inhibitors using biochemical, genomic, and epigenomic approaches. Highly specific EZH2 inhibitors are currently in clinical trials for patients with various malignancies, and our studies provide strong support for the testing of EZH2 inhibitors in children with high-risk neuroblastoma, including MYCN amplification. Additionally, they support a broader, large-scale effort to apply CRISPR-Cas9 screening systematically to generate a definitive cancer dependency map, particularly for those cancers, such as the pediatric malignancies, defined by simple genomes.
Methods
Cell lines and compounds.
Human neuroblastoma cell lines SK-N-BE(2), SK-N-AS, CHP-212, SIMA, Kelly, MHH-NB-11, IMR-32, NB1, SK-N-FI, and SK-N-SH were provided by the Broad Institute. SK-N-BE(2)-C and SH-SY-5Y were obtained from the American Type Culture Collection, and LAN-1, SHEP-Tet-MYCN, and NGP were provided by Rani George (Dana-Farber Cancer Institute). The identities of all lines were verified by small tandem repeat profiling at the Dana-Farber Cancer Institute and the Broad Institute. Neuroblastoma cell lines were treated with the EZH2 inhibitors JQEZ5 (39) and GSK126 (Excess Biosciences M60071-2). Compounds tested in the synergy study include decitabine (Selleck S1200), cis-retinoic acid (Sigma-Aldrich R3255), etoposide (Selleck S1225), irinotecan (Selleck S2217), lorlatinib (Sigma-Aldrich PZ0271), GDC0941 (Selleck S1065), temozolomide (Selleck S1237), crizotinib (Selleck S1068), PD0325901 (Selleck S1036), and panobinostat (Selleck S1030). Details are provided in the Supplemental Methods.
Protein extraction and immunoblotting.
Whole cell lysates were extracted from cells by lysis with Cell Signaling Lysis Buffer (Cell Signaling Technology) containing cOmplete, EDTA-free Protease Inhibitor Cocktail (Roche Diagnostics) and PhosSTOP Phosphatase Inhibitor (Roche Diagnostics). Histones were extracted by overnight acid extraction using an Abcam protocol (http://www.abcam.com/protocols/histone-extraction-protocol-for-western-blot). Immunoblots were run as previously described (20). Primary antibodies included anti-MYCN (Santa Cruz Biotechnology sc-53993), anti-EZH2 (Leica Biosystems NCL-L-EZH2), anti-MYC (Cell Signaling Technology 9402S), anti-IGFBP3 (Cell Signaling Technology 13216S), anti-vinculin (Abcam 18058), anti-MCM7 (Cell Signaling Technology 4018S), anti-H3K27me3 (Millipore 07-449), anti–total H3 (Millipore 06-755), and anti-GAPDH (Santa Cruz Biotechnology sc-365062). See complete unedited blots in the supplemental material.
RNA extraction and real-time reverse transcriptase PCR.
RNA was extracted from cells with the RNeasy Kit and on-column DNA digestion (Qiagen). Primers and probes for MYCN (Hs00232074_m1), EZH2 (Hs01016789_m1), MCM7 (Hs00428518_m1), IGFBP3 (Hs00181211_m1), NGFR (Hs00609976_m1), GAPDH (Hs02758991_g1), and the control gene RPL13A (Hs01926559_g1) were obtained from Applied Biosystems. Data were collected in technical triplicate, analyzed using the ΔΔCT method, and plotted as a percentage of transcript compared with the negative control condition.
shRNA and CRISPR-Cas9 studies.
Virus was created by transfection of 293T cells with packaging plasmid (pCMV-d8.9), envelope plasmid (VSV-G), and a pLK0.1 hairpin plasmid for shRNA studies or a pLentiCRISPR_v2 CAS9-sgRNA plasmid for CRISPR-Cas9 studies (see Supplemental Table 5 for shRNA and sgRNA target sequences) using Fugene 6 per the manufacturer’s instructions (Promega). Virus was filtered with a 0.4-μM filter before use. Neuroblastoma cell lines were transduced in 10-cm plates with 3 ml virus and 8 μg/ml Polybrene (Sigma-Aldrich) for 2 hours at 37°C, and then 8 ml medium was added to each plate. Three days after transduction, 2 μg/ml [BE(2)-C] or 1 μg/ml (all other cell lines) puromycin (Sigma-Aldrich) was added to select for infected cells. Four to six days after transduction, experiments to evaluate cell growth and protein levels were initiated.
In vivo studies.
Please refer to the Supplemental Methods for details of inducible shRNA experiments and drug treatment experiments.
Statistics.
Statistical differences were determined using 2-tailed Student’s t test, Mann-Whitney test, and 2-way ANOVA where appropriate. Survival data are presented using Kaplan-Meier survival analysis with the log-rank test. A P value less than 0.05 was considered significant. We also used extensive bioinformatics approaches, such as independent component analysis, gene set enrichment analysis, and Chou-Talalay combination index, to analyze CRISPR-Cas9 screening data, ChIP-Seq data, primary tumor gene expression network analysis, RNA-Seq data, and drug synergy screening data.
For all box and whisker plots, the horizontal line in each box indicates the median of the data, whereas the top and bottom of the box represent the upper and lower quartiles, respectively. The whiskers extend to the most extreme point within 1.5 times the interquartile range (IQR) of the box. Data beyond 1.5 times the IQR are depicted as dots. Please refer to the Supplemental Methods for details.
Study approval.
All animal studies were conducted under the auspices of protocols approved by the Animal Care and Use Committees of the National Cancer Institute or Dana-Farber Cancer Institute, and all treatment of mice, including their housing, was in accordance with the institutional guidelines.
Author contributions
LC, LR, ABI, ASC, EJW, VV, NL, FV, DER, GSC, LDA, SP, GJ, WFH, YL, AG, and RL generated data. LC, GA, NVD, VV, LR, FV, BAW, JMKB, AT, RMM, CJT, and KS analyzed data. LC, GA, VV, WCG, NN, FV, DER, AT, TRG, CWMR, WCH, WAW, and KS designed experiments. LC, GA, NVD, VV, CJT, and KS wrote the manuscript. JQ and JEB provided critical reagents. All authors reviewed the manuscript.
Supplementary Material
Acknowledgments
This research was supported with grants from the US National Institute of Neurological Disorders and Stroke (R01-NS088355; to KS and WAW), the National Cancer Institute (U01-CA176058; to WCH), Cookies for Kids’ Cancer (to KS), Cubans Curing Children’s Cancers (4C’s Fund) (to KS and CWMR), and Friends for Life (to KS). LC was supported by a National Cancer Center Post-doctoral Fellowship. NVD is supported by NIH grant T32-CA136432. ABI is a Damon Runyon-Sohn Pediatric Fellow supported by the Damon Runyon Cancer Research Foundation (DRSG-12-15). VV, NL, and CJT are supported by the Intramural Research Program of the NCI, NIH.
Version 1. 12/04/2017
Electronic publication
Version 2. 12/06/2017
Fixed figure resolution in PDF.
Version 3. 01/02/2018
Print issue publication
Footnotes
Conflict of interest: J.E. Bradner is the president of the Novartis Institutes for BioMedical Research. W.C. Hahn reports receiving a commercial research grant from Novartis and is a consultant/advisory board member for the same. K. Stegmaier reports receiving a commercial research grant from Novartis and is a consultant for the same. This study, however, was not supported by Novartis grant funding.
Reference information: J Clin Invest. 2018;128(1):446–462.https://doi.org/10.1172/JCI90793.
Contributor Information
Liying Chen, Email: Liying_chen@dfci.harvard.edu.
Gabriela Alexe, Email: Gabriela_Alexe@DFCI.HARVARD.EDU.
Linda Ross, Email: Linda_Ross@dfci.harvard.edu.
Emily Jue Wang, Email: jue_wang@dfci.harvard.edu.
Veronica Veschi, Email: veronica.veschi@nih.gov.
Norris Lam, Email: norris.lam@nih.gov.
Jun Qi, Email: Jun_Qi@dfci.harvard.edu.
Nicole Nasholm, Email: nicole.nasholm@ucsf.edu.
Francisca Vazquez, Email: vazquez@broadinstitute.org.
Glenn S. Cowley, Email: gscowley@comcast.net.
Sasha Pantel, Email: spantel@broadinstitute.org.
Guozhi Jiang, Email: gjiang@broadinstitute.org.
Yenarae Lee, Email: ylee@broadinstitute.org.
Amy Goodale, Email: agoodale@broadinstitute.org.
Rakela Lubonja, Email: Rakela.Lubonja@gmail.com.
Aviad Tsherniak, Email: aviad@broadinstitute.org.
James E. Bradner, Email: james.bradner@novartis.com.
Carol J. Thiele, Email: ct47a@nih.gov.
Kimberly Stegmaier, Email: kimberly_stegmaier@dfci.harvard.edu.
References
- 1.Lawrence MS, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013;499(7457):214–218. doi: 10.1038/nature12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Neff T, Armstrong SA. Recent progress toward epigenetic therapies: the example of mixed lineage leukemia. Blood. 2013;121(24):4847–4853. doi: 10.1182/blood-2013-02-474833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Delattre O, et al. Gene fusion with an ETS DNA-binding domain caused by chromosome translocation in human tumours. Nature. 1992;359(6391):162–165. doi: 10.1038/359162a0. [DOI] [PubMed] [Google Scholar]
- 4.Schwab M, et al. Amplified DNA with limited homology to myc cellular oncogene is shared by human neuroblastoma cell lines and a neuroblastoma tumour. Nature. 1983;305(5931):245–248. doi: 10.1038/305245a0. [DOI] [PubMed] [Google Scholar]
- 5.Kennedy AL, et al. Functional, chemical genomic, and super-enhancer screening identify sensitivity to cyclin D1/CDK4 pathway inhibition in Ewing sarcoma. Oncotarget. 2015;6(30):30178–30193. doi: 10.18632/oncotarget.4903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Perry JA, et al. Complementary genomic approaches highlight the PI3K/mTOR pathway as a common vulnerability in osteosarcoma. Proc Natl Acad Sci U S A. 2014;111(51):E5564–E5573. doi: 10.1073/pnas.1419260111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim KH, et al. SWI/SNF-mutant cancers depend on catalytic and non-catalytic activity of EZH2. Nat Med. 2015;21(12):1491–1496. doi: 10.1038/nm.3968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gu S, et al. The loop position of shRNAs and pre-miRNAs is critical for the accuracy of dicer processing in vivo. Cell. 2012;151(4):900–911. doi: 10.1016/j.cell.2012.09.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang T, Wei JJ, Sabatini DM, Lander ES. Genetic screens in human cells using the CRISPR-Cas9 system. Science. 2014;343(6166):80–84. doi: 10.1126/science.1246981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shalem O, et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 2014;343(6166):84–87. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Evers B, Jastrzebski K, Heijmans JP, Grernrum W, Beijersbergen RL, Bernards R. CRISPR knockout screening outperforms shRNA and CRISPRi in identifying essential genes. Nat Biotechnol. 2016;34(6):631–633. doi: 10.1038/nbt.3536. [DOI] [PubMed] [Google Scholar]
- 12.Wang T, et al. Identification and characterization of essential genes in the human genome. Science. 2015;350(6264):1096–1101. doi: 10.1126/science.aac7041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ruiz S, et al. A genome-wide CRISPR screen identifies CDC25A as a determinant of sensitivity to ATR inhibitors. Mol Cell. 2016;62(2):307–313. doi: 10.1016/j.molcel.2016.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Parnas O, et al. A genome-wide CRISPR screen in primary immune cells to dissect regulatory networks. Cell. 2015;162(3):675–686. doi: 10.1016/j.cell.2015.06.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Korkmaz G, et al. Functional genetic screens for enhancer elements in the human genome using CRISPR-Cas9. Nat Biotechnol. 2016;34(2):192–198. doi: 10.1038/nbt.3450. [DOI] [PubMed] [Google Scholar]
- 16.Brodeur GM, Seeger RC, Schwab M, Varmus HE, Bishop JM. Amplification of N-myc in untreated human neuroblastomas correlates with advanced disease stage. Science. 1984;224(4653):1121–1124. doi: 10.1126/science.6719137. [DOI] [PubMed] [Google Scholar]
- 17.Seeger RC, et al. Association of multiple copies of the N-myc oncogene with rapid progression of neuroblastomas. N Engl J Med. 1985;313(18):1111–1116. doi: 10.1056/NEJM198510313131802. [DOI] [PubMed] [Google Scholar]
- 18.Weiss WA, Aldape K, Mohapatra G, Feuerstein BG, Bishop JM. Targeted expression of MYCN causes neuroblastoma in transgenic mice. EMBO J. 1997;16(11):2985–2995. doi: 10.1093/emboj/16.11.2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Modak S, Cheung NK. Neuroblastoma: therapeutic strategies for a clinical enigma. Cancer Treat Rev. 2010;36(4):307–317. doi: 10.1016/j.ctrv.2010.02.006. [DOI] [PubMed] [Google Scholar]
- 20.Puissant A, et al. Targeting MYCN in neuroblastoma by BET bromodomain inhibition. Cancer Discov. 2013;3(3):308–323. doi: 10.1158/2159-8290.CD-12-0418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Otto T, et al. Stabilization of N-Myc is a critical function of Aurora A in human neuroblastoma. Cancer Cell. 2009;15(1):67–78. doi: 10.1016/j.ccr.2008.12.005. [DOI] [PubMed] [Google Scholar]
- 22.Moore NF, et al. Molecular rationale for the use of PI3K/AKT/mTOR pathway inhibitors in combination with crizotinib in ALK-mutated neuroblastoma. Oncotarget. 2014;5(18):8737–8749. doi: 10.18632/oncotarget.2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chesler L, et al. Inhibition of phosphatidylinositol 3-kinase destabilizes Mycn protein and blocks malignant progression in neuroblastoma. Cancer Res. 2006;66(16):8139–8146. doi: 10.1158/0008-5472.CAN-05-2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meyers RM, et al. Computational correction of copy-number effect improves specificity of CRISPR-Cas9 essentiality screens in cancer cells. Nat Genet. doi: 10.1038/ng.3984. [published online ahead of print July 10, 2017]. https://doi.org/10.1101/160861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Doench JG, et al. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat Biotechnol. 2016;34(2):184–191. doi: 10.1038/nbt.3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aguirre AJ, et al. Genomic copy number dictates a gene-independent cell response to CRISPR/Cas9 targeting. Cancer Discov. 2016;6(8):914–929. doi: 10.1158/2159-8290.CD-16-0154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Comon P. Independent component analysis, a new concept? Signal Processing. 1994;36(3):287–314. doi: 10.1016/0165-1684(94)90029-9. [DOI] [Google Scholar]
- 28. Hyvärinen A, Karhunen J, Oja E. Independent Component Analysis. New York, New York, USA: Wiley; 2001. [Google Scholar]
- 29.Pearson K. LIII. On lines and planes of closest fit to systems of points in space. Philos Mag. 1901;2(11):559–572. doi: 10.1080/14786440109462720. [DOI] [Google Scholar]
- 30. Jolliffe IT. Principal component analysis and factor analysis. In: Principal Component Analysis. New York, New York, USA: Springer-Verlag New York; 2002:150–166. [Google Scholar]
- 31.Frigyesi A, Veerla S, Lindgren D, Höglund M. Independent component analysis reveals new and biologically significant structures in micro array data. BMC Bioinformatics. 2006;7:290. doi: 10.1186/1471-2105-7-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Teschendorff AE, Journée M, Absil PA, Sepulchre R, Caldas C. Elucidating the altered transcriptional programs in breast cancer using independent component analysis. PLoS Comput Biol. 2007;3(8):e161. doi: 10.1371/journal.pcbi.0030161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Biton A, et al. Independent component analysis uncovers the landscape of the bladder tumor transcriptome and reveals insights into luminal and basal subtypes. Cell Rep. 2014;9(4):1235–1245. doi: 10.1016/j.celrep.2014.10.035. [DOI] [PubMed] [Google Scholar]
- 34.Saidi SA, et al. Independent component analysis of microarray data in the study of endometrial cancer. Oncogene. 2004;23(39):6677–6683. doi: 10.1038/sj.onc.1207562. [DOI] [PubMed] [Google Scholar]
- 35.Liao JC, Boscolo R, Yang YL, Tran LM, Sabatti C, Roychowdhury VP. Network component analysis: reconstruction of regulatory signals in biological systems. Proc Natl Acad Sci U S A. 2003;100(26):15522–15527. doi: 10.1073/pnas.2136632100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li H, Zhan M. Unraveling transcriptional regulatory programs by integrative analysis of microarray and transcription factor binding data. Bioinformatics. 2008;24(17):1874–1880. doi: 10.1093/bioinformatics/btn332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen L, et al. Knowledge-guided multi-scale independent component analysis for biomarker identification. BMC Bioinformatics. 2008;9:416. doi: 10.1186/1471-2105-9-416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Herwig R, Hardt C, Lienhard M, Kamburov A. Analyzing and interpreting genome data at the network level with ConsensusPathDB. Nat Protoc. 2016;11(10):1889–1907. doi: 10.1038/nprot.2016.117. [DOI] [PubMed] [Google Scholar]
- 39.Zhang H, et al. Oncogenic deregulation of EZH2 as an opportunity for targeted therapy in lung cancer. Cancer Discov. 2016;6(9):1006–1021. doi: 10.1158/2159-8290.CD-16-0164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McCabe MT, et al. EZH2 inhibition as a therapeutic strategy for lymphoma with EZH2-activating mutations. Nature. 2012;492(7427):108–112. doi: 10.1038/nature11606. [DOI] [PubMed] [Google Scholar]
- 41.Seashore-Ludlow B, et al. Harnessing connectivity in a large-scale small-molecule sensitivity dataset. Cancer Discov. 2015;5(11):1210–1223. doi: 10.1158/2159-8290.CD-15-0235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Barretina J, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483(7391):603–607. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chipumuro E, et al. CDK7 inhibition suppresses super-enhancer-linked oncogenic transcription in MYCN-driven cancer. Cell. 2014;159(5):1126–1139. doi: 10.1016/j.cell.2014.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Geng J, Li X, Zhou Z, Wu CL, Dai M, Bai X. EZH2 promotes tumor progression via regulating VEGF-A/AKT signaling in non-small cell lung cancer. Cancer Lett. 2015;359(2):275–287. doi: 10.1016/j.canlet.2015.01.031. [DOI] [PubMed] [Google Scholar]
- 45.Xu K, et al. EZH2 oncogenic activity in castration-resistant prostate cancer cells is Polycomb-independent. Science. 2012;338(6113):1465–1469. doi: 10.1126/science.1227604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shi B, et al. Integration of estrogen and Wnt signaling circuits by the polycomb group protein EZH2 in breast cancer cells. Mol Cell Biol. 2007;27(14):5105–5119. doi: 10.1128/MCB.00162-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee ST, et al. Context-specific regulation of NF-κB target gene expression by EZH2 in breast cancers. Mol Cell. 2011;43(5):798–810. doi: 10.1016/j.molcel.2011.08.011. [DOI] [PubMed] [Google Scholar]
- 48.Wang C, et al. EZH2 mediates epigenetic silencing of neuroblastoma suppressor genes CASZ1, CLU, RUNX3, and NGFR. Cancer Res. 2012;72(1):315–324. doi: 10.1158/0008-5472.CAN-11-0961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Henrich KO, et al. Integrative genome-scale analysis identifies epigenetic mechanisms of transcriptional deregulation in unfavorable neuroblastomas. Cancer Res. 2016;76(18):5523–5537. doi: 10.1158/0008-5472.CAN-15-2507. [DOI] [PubMed] [Google Scholar]
- 50.Janoueix-Lerosey I, et al. Somatic and germline activating mutations of the ALK kinase receptor in neuroblastoma. Nature. 2008;455(7215):967–970. doi: 10.1038/nature07398. [DOI] [PubMed] [Google Scholar]
- 51.Wang C, et al. The concordance between RNA-seq and microarray data depends on chemical treatment and transcript abundance. Nat Biotechnol. 2014;32(9):926–932. doi: 10.1038/nbt.3001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brodeur GM. Neuroblastoma: biological insights into a clinical enigma. Nat Rev Cancer. 2003;3(3):203–216. doi: 10.1038/nrc1014. [DOI] [PubMed] [Google Scholar]
- 53.Lee HY, et al. Insulin-like growth factor binding protein-3 inhibits the growth of non-small cell lung cancer. Cancer Res. 2002;62(12):3530–3537. [PubMed] [Google Scholar]
- 54.Yang CH, et al. MicroRNA-21 promotes glioblastoma tumorigenesis by down-regulating insulin-like growth factor-binding protein-3 (IGFBP3) J Biol Chem. 2014;289(36):25079–25087. doi: 10.1074/jbc.M114.593863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Frumm SM, et al. Selective HDAC1/HDAC2 inhibitors induce neuroblastoma differentiation. Chem Biol. 2013;20(5):713–725. doi: 10.1016/j.chembiol.2013.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Brockmann M, et al. Small molecule inhibitors of aurora-a induce proteasomal degradation of N-myc in childhood neuroblastoma. Cancer Cell. 2013;24(1):75–89. doi: 10.1016/j.ccr.2013.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wyce A, et al. BET inhibition silences expression of MYCN and BCL2 and induces cytotoxicity in neuroblastoma tumor models. PLoS One. 2013;8(8):e72967. doi: 10.1371/journal.pone.0072967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Thiele CJ, Reynolds CP, Israel MA. Decreased expression of N-myc precedes retinoic acid-induced morphological differentiation of human neuroblastoma. Nature. 1985;313(6001):404–406. doi: 10.1038/313404a0. [DOI] [PubMed] [Google Scholar]
- 59.Cao R, et al. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298(5595):1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- 60.Aloia L, Di Stefano B, Di Croce L. Polycomb complexes in stem cells and embryonic development. Development. 2013;140(12):2525–2534. doi: 10.1242/dev.091553. [DOI] [PubMed] [Google Scholar]
- 61.Bachmann IM, et al. EZH2 expression is associated with high proliferation rate and aggressive tumor subgroups in cutaneous melanoma and cancers of the endometrium, prostate, and breast. J Clin Oncol. 2006;24(2):268–273. doi: 10.1200/JCO.2005.01.5180. [DOI] [PubMed] [Google Scholar]
- 62.Kleer CG, et al. EZH2 is a marker of aggressive breast cancer and promotes neoplastic transformation of breast epithelial cells. Proc Natl Acad Sci U S A. 2003;100(20):11606–11611. doi: 10.1073/pnas.1933744100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Score J, et al. Inactivation of polycomb repressive complex 2 components in myeloproliferative and myelodysplastic/myeloproliferative neoplasms. Blood. 2012;119(5):1208–1213. doi: 10.1182/blood-2011-07-367243. [DOI] [PubMed] [Google Scholar]
- 64.Varambally S, et al. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature. 2002;419(6907):624–629. doi: 10.1038/nature01075. [DOI] [PubMed] [Google Scholar]
- 65.Dardenne E, et al. N-Myc induces an EZH2-mediated transcriptional program driving neuroendocrine prostate cancer. Cancer Cell. 2016;30(4):563–577. doi: 10.1016/j.ccell.2016.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tsubota S, et al. PRC2-mediated transcriptomic alterations at the embryonic stage govern tumorigenesis and clinical outcome in MYCN-driven neuroblastoma. Cancer Res. 2017;77(19):5259–5271. doi: 10.1158/0008-5472.CAN-16-3144. [DOI] [PubMed] [Google Scholar]
- 67.Bate-Eya LT, et al. Enhancer of zeste homologue 2 plays an important role in neuroblastoma cell survival independent of its histone methyltransferase activity. Eur J Cancer. 2017;75:63–72. doi: 10.1016/j.ejca.2016.12.019. [DOI] [PubMed] [Google Scholar]
- 68.Bitler BG, et al. Synthetic lethality by targeting EZH2 methyltransferase activity in ARID1A-mutated cancers. Nat Med. 2015;21(3):231–238. doi: 10.1038/nm.3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Corvetta D, et al. Physical interaction between MYCN oncogene and polycomb repressive complex 2 (PRC2) in neuroblastoma: functional and therapeutic implications. J Biol Chem. 2013;288(12):8332–8341. doi: 10.1074/jbc.M113.454280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ikonen M, et al. Interaction between the Alzheimer’s survival peptide humanin and insulin-like growth factor-binding protein 3 regulates cell survival and apoptosis. Proc Natl Acad Sci U S A. 2003;100(22):13042–13047. doi: 10.1073/pnas.2135111100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Di Marcotullio L, et al. REN(KCTD11) is a suppressor of Hedgehog signaling and is deleted in human medulloblastoma. Proc Natl Acad Sci U S A. 2004;101(29):10833–10838. doi: 10.1073/pnas.0400690101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Galvez AS, et al. Protein kinase Czeta represses the interleukin-6 promoter and impairs tumorigenesis in vivo. Mol Cell Biol. 2009;29(1):104–115. doi: 10.1128/MCB.01294-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.