Abstract
Twenty-five genome-wide association studies (GWAS) of asthma were published between 2007 and 2016, the largest with a sample size of 157242 individuals. Across these studies, 39 genetic variants in low linkage disequilibrium (LD) with each other were reported to associate with disease risk at a significance threshold of P<5 × 10−8, including 31 in populations of European ancestry. Results from analyses of the UK Biobank data (n=380 503) indicate that at least 28 of the 31 associations reported in Europeans represent true-positive findings, collectively explaining 2.5% of the variation in disease liability (median of 0.06% per variant). We identified 49 transcripts as likely target genes of the published asthma risk variants, mostly based on LD with expression quantitative trait loci (eQTL). Of these genes, 16 were previously implicated in disease pathophysiology by functional studies, including TSLP, TNFSF4, ADORA1, CHIT1 and USF1. In contrast, at present, there is limited or no functional evidence directly implicating the remaining 33 likely target genes in asthma pathophysiology. Some of these genes have a known function that is relevant to allergic disease, including F11R, CD247, PGAP3, AAGAB, CAMK4 and PEX14, and so could be prioritized for functional follow-up. We conclude by highlighting three areas of research that are essential to help translate GWAS findings into clinical research or practice, namely validation of target gene predictions, understanding target gene function and their role in disease pathophysiology and genomics-guided prioritization of targets for drug development.
Introduction
Asthma is a common and chronic inflammatory disease of the airways, specifically affecting the bronchi and bronchioli. Bronchial inflammation, which results in airway narrowing and shortness of breath symptoms, is generally caused by innate and adaptive immune responses to inhaled viruses and/or allergens.1 These and other environmental exposures are strong risk factors for disease onset and exacerbations but, based on twin studies, account for less than half of the overall disease liability.2, 3, 4 The remaining disease risk is largely explained by inherited genetic factors, with gene-by-environment interaction effects also thought to play a role. Given this high heritability, there has been a long-standing interest in identifying specific genetic risk factors for asthma, initially through linkage analysis (19895 to 20106) and subsequently through candidate-gene association studies (since 19957). Linkage studies were largely (if not entirely) unsuccessful because this approach is only adequately powered with realistic sample sizes to identify very large genetic effects,8 which we now know do not exist for asthma. On the other hand, most published candidate-gene studies suffered from a number of methodological limitations (for example, small number of samples and genetic markers tested),9 and so the reported candidate gene associations have been largely discounted.
In 2004, it became feasible to genotype hundreds of thousands of genetic variants in a single experiment.10 The development of genotyping arrays enabled the design of genome-wide association studies (GWAS), the first published soon after in 200511. In the years that followed, not only the cost of genotyping arrays decreased substantially, but also methods for the analysis of GWAS data were developed and refined; notably, these included statistical approaches to account for population structure and to infer individual genotypes for genetic variants not present in the genotyping arrays.12 As a result, GWAS including data from thousands of individuals genotyped for millions of genetic variants became a reality, at last providing a powerful tool to identify genetic associations with disease risk. For asthma, the first GWAS was published in 2007 by Moffatt et al.13 Since then, and until the end of 2016, 24 additional GWAS of asthma were published. In this review, we summarize and interpret the key genetic findings from these studies, specifically addressing the following questions: are any published risk variants likely to be false-positive associations? How much variation in disease liability do they explain? What are the likely target genes of those risk variants? Do those genes point to potential new mechanisms underlying disease pathophysiology? Lastly, we conclude by highlighting areas of research that are essential to help translate genetic findings into clinical research or practice.
Summary of genetic associations reported in asthma GWAS performed between 2007 and 2016
We searched the NHGRI-EBI catalog of published GWAS14 to identify studies that tested the association between genetic variants and asthma risk, between 2007 and 2016. We used the search term ‘Asthma’ and applied no filters. The search was performed on the 2nd of August 2017, and returned 73 unique studies, which were individually reviewed for inclusion in our analysis. Of these, 48 (66%) were excluded (Supplementary Table 1), most (34 studies) because the phenotype tested in the GWAS was not asthma, but instead an asthma-related trait (for example, lung function). Studies were also commonly excluded because they were based on DNA pooling (five studies) or reported genetic interactions (for example, gene-by-environment; four studies) rather than main effects. We extracted data from the GWAS Catalog for the remaining 25 studies,13, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38 which were included for analysis (Supplementary Table 2).
The definition of asthma was not always the same across the 25 published GWAS. For example, six GWAS ascertained asthma cases with disease onset in childhood, whereas in other GWAS cases had more severe symptoms or co-morbid allergies (Supplementary Table 2). The smallest GWAS included 66 cases and 42 controls,31 and the largest 28 399 cases and 128 843 controls.15 For most studies (18 of 25), the primary GWAS included exclusively individuals of European descent; two studies were based on populations of Asian ancestry,27, 29 two of Latino ancestry,18, 36 two of African ancestry16, 35 and one included multiple ancestries.28 Across these 25 studies, 73 unique genetic variants were reported to associate with disease risk at a genome-wide significance threshold of P<5 × 10−8 (listed per study in Supplementary Table 3). Some single nucleotide polymorphisms (SNPs) were located in close proximity, and so we used the clump procedure in PLINK39 to determine which were likely to represent independent associations and which were simply correlated with previously published variants. Specifically, we assigned each SNP into groups of correlated variants based on pairwise linkage disequilibrium (LD). LD was estimated using data from the 1000 Genomes Project (release 20130502_v5a), separately for individuals of European, Asian and African ancestry, as appropriate. Using a conservative LD threshold of r2>0.05, there were 31 groups of correlated risk variants reported in GWAS of European ancestry (Table 1 and Supplementary Table 4), with seven, one and three variants reported in GWAS of Asian, African and Latino ancestry, respectively (Supplementary Table 5). One variant reported in Asians (rs1837253) and two (rs9272346 and rs907092) in Latinos were in strong LD (r2>0.8) with risk variants reported in Europeans, and so do not represent independent associations. Therefore, in total, 39 genetic variants (31 in Europeans and 8 additional in other ancestries) in low LD with each other were reported to associate with asthma risk in GWAS published between 2007 and 2016. In the sections below, we focus on the 31 associations reported in Europeans.
Table 1. Variants in low LD with each other (r 2<0.05) reported to associate with asthma risk in GWAS conducted between 2007 and 2016 in populations of European ancestry.
| Index | Chr | Bp | Contexta |
First association reported with P<5 × 10−8 |
Correlated SNPs reported in other asthma GWAS b | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Top SNP | Effect allele | OR | P-value | PMID | Year | |||||
| 1 | 1 | 10557251 | [PEX14] | rs662064 | T | 0.94 | 3.2E-08 | 27182965 | 2016 | No |
| 2 | 1 | 154426264 | [IL6R] | rs4129267 | T | 1.09 | 2.4E-08 | 21907864 | 2011 | No |
| 3 | 1 | 161159147 | PPOX-[]-ADAMTS4 | rs4233366 | T | 1.09 | 4.8E-15 | 27182965 | 2016 | No |
| 4 | 1 | 167433420 | [CD247] | rs1723018 | G | 0.95 | 1.4E-08 | 27182965 | 2016 | No |
| 5 | 1 | 173152036 | TNFSF18—[]-TNFSF4 | rs6691738 | T | 0.94 | 2.9E-08 | 27182965 | 2016 | No |
| 6 | 1 | 197325908 | [CRB1] | rs2786098 | A | 0.63 | 8.6E-09 | 20032318 | 2009 | No |
| 7 | 1 | 203100504 | [ADORA1] | rs6683383 | T | 1.06 | 1.1E-08 | 27182965 | 2016 | No |
| 8 | 2 | 8458080 | [LINC00299] | rs13412757 | G | 1.06 | 1.3E-08 | 27182965 | 2016 | No |
| 9 | 2 | 102986222 | [IL18R1] | rs3771166 | A | 0.87 | 3.4E-09 | 20860503 | 2010 | Yes |
| 10 | 2 | 242698640 | [D2HGDH] | rs34290285 | G | 1.11 | 1.8E-15 | 27182965 | 2016 | No |
| 11 | 3 | 188402471 | [LPP] | rs73196739 | T | 0.92 | 6.5E-09 | 27182965 | 2016 | No |
| 12 | 4 | 38799710 | [TLR1] | rs4833095 | T | 1.20 | 5.0E-12 | 24388013 | 2013 | Yes |
| 13 | 5 | 59369794 | [PDE4D] | rs1588265 | G | 0.60 | 2.5E-08 | 19426955 | 2009 | No |
| 14 | 5 | 110401872 | SLC25A46—[]-TSLP | rs1837253 | C | 1.19 | 7.3E-10 | 21804549 | 2011 | Yes |
| 15 | 5 | 131969874 | [RAD50] | rs6871536 | C | 1.14 | 2.4E-09 | 21907864 | 2011 | Yes |
| 16 | 5 | 141529762 | [NDFIP1] | rs200634877 | I | 0.94 | 2.5E-08 | 27182965 | 2016 | No |
| 17 | 6 | 31322197 | [HLA-B] | rs2428494 | T | 0.92 | 1.4E-16 | 27182965 | 2016 | No |
| 18 | 6 | 32728261 | HLA-DQA1-[]-HLA-DQB1 | rs17843604 | T | 1.16 | 1.7E-10 | 20860503 | 2010 | Yes |
| 19 | 6 | 90985198 | [BACH2] | rs58521088 | T | 0.93 | 7.1E-11 | 27182965 | 2016 | No |
| 20 | 7 | 105658451 | [CDHR3] | rs6967330 | A | 1.45 | 1.4E-08 | 24241537 | 2013 | Yes |
| 21 | 8 | 81291879 | MIR5708—[]—ZBTB10 | rs7009110 | T | 1.14 | 4.0E-09 | 24388013 | 2013 | Yes |
| 22 | 9 | 6190076 | RANBP6—[]-IL33 | rs1342326 | C | 1.20 | 9.2E-10 | 20860503 | 2010 | Yes |
| 23 | 10 | 9049253 | GATA3---[]---SFTA1P | rs12413578 | T | 0.89 | 8.1E-12 | 27182965 | 2016 | No |
| 24 | 11 | 76270683 | WNT11—[]-LRRC32 | rs7130588 | G | 1.09 | 1.8E-08 | 21907864 | 2011 | Yes |
| 25 | 12 | 57509055 | [STAT6] | rs3001426 | T | 0.94 | 1.4E-10 | 27182965 | 2016 | No |
| 26 | 14 | 68749927 | [RAD51B] | rs3784099 | G | 0.94 | 1.6E-08 | 27182965 | 2016 | No |
| 27 | 15 | 61069988 | [RORA] | rs11071559 | T | 0.85 | 3.8E-09 | 21907864 | 2011 | Yes |
| 28 | 15 | 67446785 | [SMAD3] | rs744910 | A | 0.89 | 3.9E-09 | 20860503 | 2010 | Yes |
| 29 | 16 | 11228712 | [CLEC16A] | rs62026376 | C | 1.17 | 1.0E-08 | 24388013 | 2013 | Yes |
| 30 | 17 | 38069949 | [GSDMB] | rs7216389 | T | 1.45 | 9.0E-11 | 17611496 | 2007 | Yes |
| 31 | 22 | 37534034 | [IL2RB] | rs2284033 | A | 0.89 | 1.2E-08 | 20860503 | 2010 | No |
Abbreviations: OR, odds ratio; PMID, PubMed identifier.
If the top SNP is located within the boundaries of a gene, then the gene name is shown inside square brackets. Otherwise, the two nearest genes (upstream and downstream) are listed, with the distance to each represented by the number of '−' between the square bracket and the gene name.
SNPs reported in other asthma GWAS and with (1) r2>0.05 with top SNP and (2) P<5 × 10−8 are listed in Supplementary Table 4.
Are any of the published risk variants for asthma likely to represent false-positive associations?
GWAS, even when conducted using strict quality control procedures and an appropriate genome-wide significance threshold,40 are not completely protected from false-positive associations. That is, there is always a small chance that a new genome-wide significant association with asthma risk might not be a true-positive association and instead arise by chance or because of unaccounted methodological biases. As such, whenever possible, it is important to perform an independent and adequately powered replication study to confirm novel associations. This is not always feasible, particularly as GWAS become larger, because there might not be sufficiently large studies available for replication that were not included in the discovery stage.
In the last few years, ~500 000 individuals from the UK have been deeply phenotyped and genotyped as part of the UK Biobank study.41 The genotype data for the full dataset has just been made publicly available,42 providing a unique and timely opportunity to test if the 31 associations with asthma risk reported in Europeans between 2007 and 2016 are reproducible. Briefly, we analyzed data for 380503 unrelated individuals (kinship coefficient indicating <3rd degree relatedness) with (a) European ancestry, confirmed based on analysis of allele sharing with individuals from the 1000 Genomes Project; and (b) non-missing information for field 6152 of the touchscreen questionnaire: ‘Has a doctor ever told you that you have had any of the following conditions?’. A total of 44 003 individuals selected ‘Asthma’ when answering that question, and so were considered as cases. On the other hand, 336 500 individuals did not select ‘Asthma’ and so were considered as controls. Mean age was 56.7 (range 38–72), with 54% of participants being female. We tested the association between individual SNPs and case-control status using SNPTEST, including age, sex and SNP chip as covariates in the model. We adjusted the association results for an LD Score intercept43 of 1.073, estimated using 1.2 million HapMap3 SNPs. In this analysis, we were able to test all 31 reported SNPs (all with imputation information >0.98), either directly (30 SNPs) or through a proxy SNP (rs166079 instead of rs200634877, r2=0.75). Of these, 28 (90%) had a statistically significant (P<0.05/31 SNPs=0.0016) and directionally consistent (same predisposing allele as originally reported) association with disease risk (Table 2 and Figure 1), thereby confirming the original findings as true-positive associations.
Table 2. Association between 31 variants reported in GWAS of European ancestry and self-reported doctor-diagnosed asthma in the UK Biobank study (44 003 cases and 336500 controls).
| Index | SNP | Chr | Bp | Context | Effect allele | MAF | OR | P-value | Asthma h2 explained (%) |
|---|---|---|---|---|---|---|---|---|---|
| 1 | rs662064 | 1 | 10557251 | [PEX14] | C | 0.31 | 1.04 | 9.7E-006 | 0.02 |
| 2 | rs4129267 | 1 | 154426264 | [IL6R] | T | 0.41 | 1.03 | 5.8E-006 | 0.02 |
| 3 | rs4233366 | 1 | 161159147 | PPOX-[]-ADAMTS4 | T | 0.27 | 1.04 | 4.7E-007 | 0.02 |
| 4 | rs1723018 | 1 | 167433420 | [CD247] | G | 0.41 | 0.95 | 5.7E-012 | 0.04 |
| 5 | rs6691738 | 1 | 173152036 | TNFSF18—[]-TNFSF4 | G | 0.29 | 1.04 | 5.8E-007 | 0.02 |
| 6 | rs2786098 | 1 | 197325908 | [CRB1] | G | 0.22 | 1.00 | 0.5827 | 0.00 |
| 7 | rs6683383 | 1 | 203100504 | [ADORA1] | A | 0.33 | 0.95 | 5.9E-011 | 0.04 |
| 8 | rs13412757 | 2 | 8458080 | [LINC00299] | A | 0.34 | 0.94 | 1.8E-013 | 0.05 |
| 9 | rs3771166 | 2 | 102986222 | [IL18R1] | A | 0.38 | 0.90 | 7.4E-040 | 0.15 |
| 10 | rs34290285 | 2 | 242698640 | [D2HGDH] | A | 0.26 | 0.89 | 1.5E-039 | 0.15 |
| 11 | rs73196739 | 3 | 188402471 | [LPP] | T | 0.17 | 0.94 | 4.1E-010 | 0.03 |
| 12 | rs4833095 | 4 | 38799710 | [TLR1] | C | 0.21 | 0.92 | 1.3E-017 | 0.06 |
| 13 | rs1588265 | 5 | 59369794 | [PDE4D] | G | 0.31 | 1.01 | 0.2859 | 0.00 |
| 14 | rs1837253 | 5 | 110401872 | SLC25A46—[]-TSLP | C | 0.26 | 1.12 | 3.4E-041 | 0.16 |
| 15 | rs6871536 | 5 | 131969874 | [RAD50] | C | 0.19 | 1.10 | 1.0E-024 | 0.09 |
| 16 | rs166079 | 5 | 141528959 | [NDFIP1] | T | 0.38 | 1.04 | 1.8E-008 | 0.03 |
| 17 | rs2428494 | 6 | 31322197 | [HLA-B] | A | 0.47 | 1.12 | 6.1E-055 | 0.20 |
| 18 | rs17843604 | 6 | 32620283 | HLA-DQA1-[]-HLA-DQB1 | T | 0.42 | 1.18 | 1.9E-105 | 0.41 |
| 19 | rs58521088 | 6 | 90985198 | [BACH2] | T | 0.35 | 0.93 | 2.5E-021 | 0.08 |
| 20 | rs6967330 | 7 | 105658451 | [CDHR3] | A | 0.17 | 1.01 | 0.2233 | 0.00 |
| 21 | rs7009110 | 8 | 81291879 | MIR5708—[]—ZBTB10 | C | 0.38 | 0.94 | 9.6E-018 | 0.06 |
| 22 | rs1342326 | 9 | 6190076 | RANBP6—[]-IL33 | C | 0.16 | 1.15 | 6.4E-048 | 0.17 |
| 23 | rs12413578 | 10 | 9049253 | GATA3---[]---SFTA1P | T | 0.11 | 0.86 | 1.0E-033 | 0.13 |
| 24 | rs7130588 | 11 | 76270683 | WNT11—[]-LRRC32 | G | 0.36 | 1.10 | 8.9E-033 | 0.12 |
| 25 | rs3001426 | 12 | 57509055 | [STAT6] | C | 0.45 | 1.07 | 1.4E-021 | 0.08 |
| 26 | rs3784099 | 14 | 68749927 | [RAD51B] | A | 0.28 | 1.06 | 1.4E-012 | 0.04 |
| 27 | rs11071559 | 15 | 61069988 | [RORA] | T | 0.13 | 0.91 | 7.3E-017 | 0.06 |
| 28 | rs744910 | 15 | 67446785 | [SMAD3] | A | 0.48 | 0.95 | 3.5E-014 | 0.05 |
| 29 | rs62026376 | 16 | 11228712 | [CLEC16A] | T | 0.25 | 0.92 | 7.7E-023 | 0.08 |
| 30 | rs7216389 | 17 | 38069949 | [GSDMB] | T | 0.48 | 1.11 | 1.7E-044 | 0.16 |
| 31 | rs2284033 | 22 | 37534034 | [IL2RB] | A | 0.43 | 0.97 | 4.2E-004 | 0.01 |
| Total | 2.52 | ||||||||
| Median | 0.06 |
Abbreviations: MAF, minor allele frequency; OR, odds ratio; h2: heritability.
Three SNPs with a P>0.05 are highlighted in gray.
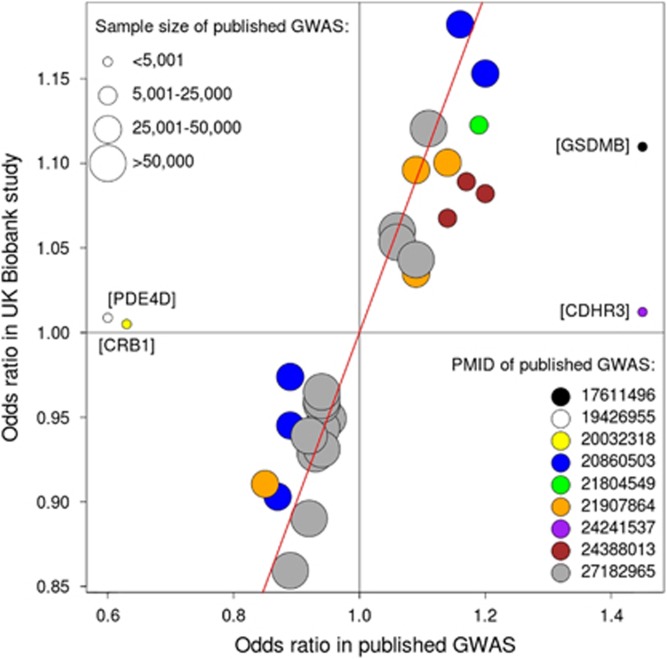
Figure 1.
Effect size (odds ratio) for 31 previously reported asthma risk SNPs, comparing results reported in the original GWAS describing each association with those obtained in the analysis of the UK Biobank study. Each SNP is represented by a circle, with its size and color indicating total GWAS sample size (as per Supplementary Table 2) and PubMed identifier (PMID), respectively. The red diagonal line represents equality of odds ratio between published GWAS and UK Biobank (that is, x=y). The genomic location of the four SNPs that deviate markedly from the diagonal are shown next to the respective circle. For three of these (SNPs in PDE4D, CRB1 and CDHR3), the association in the UK Biobank study had a P-value>0.05 (see Table 2).
For the remaining three SNPs, results from this UK Biobank analysis of self-reported doctor-diagnosed asthma did not support an association with disease risk, and so it is possible that they represent false-positive associations. These associations are located in/near DENND1B/CRB1,34 PDE4D37 and CDHR320. Another explanation for the lack of association with these three SNPs is that the case-control definition we used in the UK Biobank analysis is not a good proxy for that used in the original studies. For example, the original association with rs6967330 in the CDHR3 gene,20 which was subsequently supported by results from Pickrell et al.15 (rs6959584, P=2 × 10−8, r2=0.72 with rs6967330), was found when studying asthma cases with childhood onset and severe exacerbations. If such an association is specific to that subgroup of asthmatics, and if these only represent a small fraction of the UK Biobank asthma cases, then the power to replicate the original association might have been low. In this respect, it is noteworthy that the direction of effect in the UK Biobank for rs6967330 was the same as originally reported. In conclusion, at least 28 of the 31 SNPs previously reported in GWAS of European ancestry have a significant and consistent association with self-reported doctor-diagnosed asthma in the UK Biobank study and so represent bona fide asthma risk variants. It was beyond the scope of this review to report associations found in the UK Biobank study that were not located in previously reported asthma risk loci. Studies that report the full results from the UK Biobank study will be reported elsewhere in the near future.
How much variation in disease liability is explained by the published asthma risk variants and by others yet to be discovered?
Twin studies have estimated the heritability of asthma to be between 55 and 74% in adults,2, 3 with even larger estimates reported in young children.4, 44 The aim of GWAS is to identify variants that contribute to this heritability. So it is important to understand to what extent the heritability of asthma is explained by the asthma risk variants discovered to date, and how much heritability remains to be discovered. To answer the first question, we estimated the total variance in disease liability explained in the UK Biobank study by each of the 31 published risk variants, using the formula var(g)/(var(g)* (π2)/3) described by Pawitan et al.45 This is often referred to as the SNP heritability. In this formula, var(g) for each SNP is given by 2p*(1-p)*(log(OR))2, where p and OR are respectively the frequency and odds ratio for the effect allele, while π is the mathematical constant pi. Using this formula, we found that the median SNP heritability was 0.06% (range 0 to 0.41% Table 2), while the sum of the 31 SNP heritabilities was 2.5%. That is, in the UK Biobank study, 2.5% of the variation in asthma liability is explained by the 31 asthma risk variants discovered to date.
The second question of interest is how much heritability is likely to be explained by risk variants that remain to be discovered. One approach to address this question might be to simply subtract the SNP heritability explained by the 31 published associations (2.5%) from the overall asthma heritability estimates reported in twin studies (e.g. 55%). Therefore, potentially, asthma risk variants yet to be discovered could account for at least ~52% (55%–2.5%) of disease liability. However, heritability estimates from twin studies can be inflated, for example, because of violations of study design assumptions.46 Thus, such estimates should be taken as the upper boundary of the total variation in disease liability that is explained by genetic variants.
A more conservative approach to address the same question involves first estimating the disease heritability that is explained collectively by all genetic variants studied in a GWAS, not just those with a strong association with disease risk. This is referred to as the SNP-based disease heritability, which can be estimated using for example GCTA,47 BOLT-REML48 or LD Score regression.43 In theory, the SNP-based heritability can be lower than the twin-based heritability if genetic variants not tested (or not well tagged) in GWAS contribute to disease risk, which could be the case for uncommon variants (e.g. with minor allele frequency [MAF] <1%). Differences in heritability estimates could also arise if the twin-based heritability estimate is inflated, as discussed above. When we applied the LD Score regression approach to genome-wide results from the UK Biobank GWAS analysis described above (n=380 503), we found that the overall SNP-based heritability for asthma was 14%, with a standard error (SE) of 1%. This estimate was obtained based on results for 1.2 million common, well imputed HapMap3 SNPs; therefore, it can be considered as the lower boundary of the total variation in disease liability that is explained by common SNPs. If we consider this estimate, then genetic risk variants yet to be discovered, and that could be identified in larger asthma GWAS of common variants, are likely to account for about 11.5% (14%–2.5%) of the variation in disease liability. This conclusion was supported by the observation that the overall asthma SNP-based heritability obtained after removing from the UK Biobank GWAS the 31 published SNPs (and all variants in LD with them, r2>0.05) was 12% (SE=1%).
Have we found fewer asthma risk variants than expected based on GWAS sample size?
The largest asthma GWAS published between 2007 and 2016 included 157 242 individuals and identified 27 independent associations with disease risk. This figure is smaller than reported by GWAS of some complex diseases using similar or smaller sample sizes (Table 3). For example, with a similar sample size (n=150 064), Ripke et al.49 found 128 independent associations with schizophrenia (SCZ), which is 4.7-fold greater than those found for asthma. What underlies this difference in GWAS yield? First, the power to detect an association with a SNP depends on the proportion of variance in disease liability it explains. As discussed above, this can be calculated from the risk allele frequency and the odds ratio. For example, power is about the same to detect an association with two SNPs, one with a risk allele of frequency 0.5 and odds ratio of 1.2, and the other with a risk allele of frequency 0.01 and odds ratio of 2.5—both SNPs explain about 0.5% of variation in disease liability (see formula in section above). When comparing two diseases, one needs to consider that the power to detect an association with a SNP that explains the same proportion of variance in disease liability depends on the disease prevalence.50, 51 Using the formula derived by Yang et al.,51 if we consider a SNP that explains 0.05% of the variation in disease liability, then the expected non-centrality parameter (NCP; which reflects power, but is linearly related to sample size) for the SCZ GWAS listed in Table 3 is 102 (assuming a disease prevalence of 1%). This is 2.6-fold greater than obtained for the similar-sized asthma GWAS (NCP=40, assuming a prevalence of 15%). In other words, although the overall sample size is the same, the SCZ GWAS has substantially greater power to detect an association with such a SNP when compared to the asthma GWAS. Another consideration is the genetic architecture (number of risk loci, their frequency and effect size), which is unknown and may differ across different diseases.52 For example, asthma might have a smaller number of common risk variants than SCZ, or SNP effects might be smaller. So to adequately compare the number of associations reported for different diseases (and quantitative traits), one needs to consider not just the sample size but also disease lifetime risk and the likely genetic architecture, as highlighted previously.50, 53
Table 3. Number of associations reported in published GWAS of other polygenic diseases, which were based on a similar or smaller sample size than the largest asthma GWAS published.
| Disease | N cases | N controls | N Total | N of independent associations in discovery GWAS (P<5x10−8) | PMID | Disease prevalence | NCP for a SNP with h2=0.05% |
|---|---|---|---|---|---|---|---|
| Atopic dermatitis | 18900 | 84166 | 103066 | 21 | 26482879 | 20% | 24 |
| Asthma | 28399 | 128843 | 157242 | 27 | 27182965 | 15% | 40 |
| Type 2 diabetes | 26676 | 132532 | 159208 | 42 | 28566273 | 5% | 52 |
| Schizophrenia | 36989 | 113075 | 150064 | 128 | 25056061 | 1% | 102 |
| Rheumatoid arthritis | 29880 | 73758 | 103638 | 101 | 24390342 | 1% | 77 |
Abbreviations: PMID, PubMed identifier; NCP: non-centrality parameter (which reflects power); h2: heritability.
All studies are based on the analysis of 1000 Genome Project SNPs imputed from GWAS arrays (not including fine-mapping arrays).
Another factor that influences power, and that could have a more severe effect for some diseases than others, is disease heterogeneity and misclassification. If the genetic architecture of asthma is not homogeneous, and instead has components that are specific to clinically distinct subtypes, then lumping all individuals who reported ever having asthma in the same case group could lead to underestimation of SNP heritabilities, which decreases power. As examples of this, the risk allele for susceptibility variants near ORMDL3 is significantly more common in cases with childhood onset asthma,6, 54 while the association near CDHR3 might be specific to children with early onset and severe exacerbations,20 as discussed above. Similarly, including in the control group individuals who do not have asthma but suffer from other common allergic diseases (for example, hay fever) can significantly decrease power to detect associations with SNPs that affect allergies in general, not just asthma.55 Shared genetic effects are expected to be widespread amongst common allergic diseases, with pairwise genetic correlations estimated to be >50% in twin studies.2, 4, 56 One approach to capitalize on this high genetic correlation is to define cases as those suffering from two or more allergic diseases, and controls as those who do not suffer from any allergic disease.55 We have shown empirically that the estimated SNP effects are indeed larger with such approach, and so power is increased.19 Another approach that also increases power to detect shared genetic effects is to define cases as those who suffer from any allergic disease.55 We have recently performed a large GWAS using this approach (n=360 838) and identified 136 independent associations for allergic disease.57 Of note, the expected NCP in this study design for a SNP with a 0.05% heritability and assuming a 30% disease prevalence was 123, which is comparable to the SCZ GWAS that identified 128 independent associations.
Lastly, if gene-by-environment interactions have a substantial contribution to asthma risk, then ignoring the relevant environmental exposures in GWAS could also result in underestimated SNP effects. For example, gene-by-environment interactions have been reported for variants in the ORMDL3 locus, with larger SNP effects found in children with rhinovirus wheezing illness in early life58 or exposed to tobacco smoke in early life.59 The latter interaction, however, was not replicated in a large independent study.60 A small number of genome-wide interaction analyses between environmental exposures and asthma risk have been published,61, 62, 63, 64 but are not discussed in this review. For reviews of this topic, see for example.65, 66 Thus, in summary, fewer genetic associations have been reported for asthma as compared to some complex diseases with GWAS of similar or smaller size. This likely reflects lower statistical power arising from the relatively high disease prevalence, but also study design limitations, such as not accounting for disease subtypes, information from genetically-correlated diseases and gene-by-environment interaction effects.
What are the likely target genes of the published asthma risk variants?
The aim of asthma GWAS per se is to identify genetic variants associated with disease risk. A genetic variant is associated with asthma risk most likely because that same variant, or another in strong LD with it (for example, r2>0.8), affects the protein sequence or the transcription patterns of a gene (that is, the ‘target gene’) that plays a role in disease pathophysiology. Therefore, knowing which variants are associated with disease risk might highlight specific genes and molecular pathways dysregulated in asthma, and ultimately help better understand why asthma develops in the first place. How do we identify the likely target genes of risk variants? First, we can determine if the associated variant, or another in strong LD with it, is a non-synonymous coding variant, using for example ANNOVAR.67 If we focus on the 28 published risk variants that had a reproducible association with asthma in the UK Biobank study (Table 2), and also include the additional correlated risk variants reported in other asthma GWAS (Supplementary Table 4), then this approach identifies eight likely target genes: GSDMA, GSDMB, HLA-DQA1, HLA-DQB1, IL1RL1, IL6R, TLR1 and ZPBP2 (Supplementary Table 6).
A second approach that can be used to identify likely target genes is to determine if the associated SNPs are in strong LD with variants that have been reported to associate with variation in gene expression levels, known as expression quantitative trait loci (eQTL). A plethora of eQTL studies have been reported in recent years, including 39 conducted using tissues relevant to asthma pathophysiology (Supplementary Table 7), for example, whole-blood, lung, skin and individual immune cell types, such as CD4+ T cells. For each of these studies, we extracted eQTL results (that is, SNP, gene and P-value) from the original publication, keeping only associations in cis (that is, SNP within 1 Mb of gene) that were significant at a conservative threshold of P<2.3 × 10−9, which corresponds to a Bonferroni correction for testing each of 21472 genes68 for association with 1000 independent SNPs.69 In each study and for each gene, we then used the clump procedure in PLINK to reduce the published list of eQTLs (which typically includes many correlated variants) to the subset of strongest eQTLs that were in low LD with each other (r2<0.05), which we refer to as sentinel eQTLs. Finally, we asked if any of the 28 variants with a reproducible association with asthma risk in the UK Biobank study (Table 2), or the additional correlated risk variants reported in other asthma GWAS (Supplementary Table 4), were in strong LD (r2>0.8) with a sentinel eQTL. Using this approach, we found 48 likely target genes (Table 4 and Supplementary Table 8). Of note, these include all eight genes with non-synonymous variants listed above, indicating that for these variation in both protein sequence and transcript levels might be important determinants of cellular function and disease risk.
Table 4. Likely target genes of published asthma risk variants identified based on LD with sentinel eQTLs.
| Index | GWAS SNP |
Results for sentinel eQTL in strongest LD with GWAS SNP |
eQTL reported in other studies? a | |||||
|---|---|---|---|---|---|---|---|---|
| eQTL | LD (r2) | Target gene | P-value | Study | Tissue | |||
| 1 | rs662064 | rs668805 | 1.00 | PEX14 | 1.2E-019 | Zeller | Monocytes | Yes |
| 1 | rs662064 | rs12028449 | 0.81 | DFFA | 1.2E-012 | Westra | Whole blood | No |
| 2 | rs4129267 | rs4537545 | 0.93 | IL6R | 2.0E-029 | Westra | Whole blood | Yes |
| 3 | rs4233366 | rs4233366 | 1.00 | FCER1G | 5.0E-215 | Jansen | Whole blood | Yes |
| 3 | rs4233366 | rs4233366 | 1.00 | B4GALT3 | 1.5E-026 | GTEx | Fibroblasts | No |
| 3 | rs4233366 | rs4233366 | 1.00 | ADAMTS4 | 1.8E-024 | GTEx | Fibroblasts | No |
| 3 | rs4233366 | rs4233366 | 1.00 | PPOX | 6.2E-012 | GTEx | Fibroblasts | No |
| 3 | rs4233366 | rs4233366 | 1.00 | F11R | 3.4E-011 | Fehrmann | Whole blood | No |
| 3 | rs4233366 | rs4233366 | 1.00 | USF1 | 3.4E-011 | Fehrmann | Whole blood | Yes |
| 3 | rs4233366 | rs2070901 | 0.96 | TOMM40L | 7.3E-010 | Naranbhai | Neutrophils | No |
| 4 | rs1723018 | rs2988279 | 0.93 | CD247 | 3.1E-062 | Zhernakova | Whole blood | Yes |
| 5 | rs6691738 | rs7553711 | 0.99 | TNFSF4 | 1.9E-035 | Yao | Whole blood | No |
| 7 | rs6683383 | rs6683383 | 1.00 | ADORA1 | 6.0E-096 | Zeller | Monocytes | Yes |
| 7 | rs6683383 | rs3766568 | 1.00 | CHIT1 | 6.6E-030 | Zhernakova | Whole blood | Yes |
| 7 | rs6683383 | rs10920570 | 1.00 | MYBPH | 5.2E-018 | Zeller | Monocytes | Yes |
| 7 | rs6683383 | rs17464408 | 0.99 | RP11-335O13.7 | 8.1E-021 | Zhernakova | Whole blood | Yes |
| 7 | rs6683383 | rs7555556 | 0.98 | PPFIA4 | 8.1E-162 | Zhernakova | Whole blood | Yes |
| 9 | rs10173081 | rs3771180 | 1.00 | MFSD9 | 1.7E-015 | Yao | Whole blood | No |
| 9 | rs10173081 | rs11674302 | 0.80 | IL18RAP | 7.9E-091 | Yao | Whole blood | No |
| 9 | rs10173081 | rs10189629 | 0.80 | IL1RL1 | 3.0E-012 | Zhernakova | Whole blood | No |
| 9 | rs3771166 | rs11688559 | 1.00 | AC007278.3 | 4.3E-249 | Zhernakova | Whole blood | No |
| 12 | rs4833095 | rs12233670 | 0.98 | TLR1 | 2.8E-057 | Battle | Whole blood | Yes |
| 14 | rs1438673 | rs7723819 | 0.86 | WDR36 | 2.4E-031 | Yao | Whole blood | No |
| 14 | rs1438673 | rs10073816 | 0.85 | TSLP | 3.3E-012 | Zhernakova | Whole blood | Yes |
| 14 | rs1438673 | rs2289277 | 0.84 | CTC-551A13.2 | 7.0E-029 | Zhernakova | Whole blood | Yes |
| 14 | rs1438673 | rs10051830 | 0.82 | CAMK4 | 1.8E-016 | Yao | Whole blood | No |
| 15 | rs2244012 | rs2246176 | 0.99 | SLC22A5 | 8.7E-014 | Westra | Whole blood | No |
| 16 | rs166079 | rs12655465 | 1.00 | NDFIP1 | 6.8E-115 | Zhernakova | Whole blood | Yes |
| 17 | rs2428494 | rs2428494 | 1.00 | MICB | 2.3E-011 | Walsh | Whole blood | No |
| 18 | rs9268516 | rs9268400 | 0.99 | HLA-DRB6 | 6.2E-011 | Dinarzo | Whole blood | No |
| 18 | rs9272346 | rs9272346 | 1.00 | HLA-DQB1 | <4.9E-324 | Zeller | Monocytes | Yes |
| 18 | rs9272346 | rs9272346 | 1.00 | HLA-DRB5 | 2.1E-121 | Westra | Whole blood | Yes |
| 18 | rs9272346 | rs9272346 | 1.00 | TAP2 | 4.1E-011 | Westra | Whole blood | No |
| 18 | rs9273373 | rs1063355 | 0.99 | HLA-DQA1 | 3.6E-154 | Raj | Monocytes | Yes |
| 18 | rs9273373 | rs3134993 | 0.95 | HLA-DQB1-AS1 | 5.6E-058 | GTEx | Lung | No |
| 18 | rs9273373 | rs1063349 | 0.92 | HLA-DQB2 | 1.1E-089 | Geuvadis | LCLs | No |
| 18 | rs9273373 | rs9272545 | 0.87 | HLA-DQA2 | 1.1E-022 | Quach | Monocytes | Yes |
| 27 | rs10519068 | rs11633029 | 0.86 | RP11-554D20.1 | 3.7E-017 | Zhernakova | Whole blood | Yes |
| 28 | rs56375023 | rs17293632 | 0.98 | AAGAB | 1.7E-013 | Zhernakova | Whole blood | Yes |
| 30 | rs11078927 | rs12946510 | 0.80 | GSDMA | <2.2E-016 | Hao | Lung | No |
| 30 | rs11655198 | rs9903250 | 1.00 | RP11-94L15.2 | 5.2E-064 | Zhernakova | Whole blood | Yes |
| 30 | rs11655198 | rs11655198 | 1.00 | ORMDL3 | 7.4E-041 | Kasela | CD8 T-cells | Yes |
| 30 | rs11655198 | rs2305479 | 0.97 | IKZF3 | 5.6E-021 | Zhernakova | Whole blood | Yes |
| 30 | rs11655198 | rs8067378 | 0.96 | ZPBP2 | 2.4E-017 | Grundberg | LCLs | Yes |
| 30 | rs2271308 | rs1053651 | 0.99 | STARD3 | 1.4E-016 | Fairfax | Monocytes | Yes |
| 30 | rs2271308 | rs1053651 | 0.99 | PGAP3 | 9.9E-014 | Yao | Whole blood | No |
| 30 | rs2271308 | rs4795388 | 0.83 | PPP1R1B | 4.1E-010 | Andiappan | Neutrophils | No |
| 30 | rs4794820 | rs4794820 | 1.00 | GSDMB | 1.1E-296 | Zhernakova | Whole blood | Yes |
GWAS SNPs located in the same locus are highlighted in alternating white/gray shading.
Results from other eQTL studies that provide support for the same target gene are listed in Supplementary Table 8.
For many asthma risk variants (12 of 28, or 43%), the two approaches described above failed to identify any likely target gene. This could arise, for example, if the effect of those variants on gene expression is (a) specific to tissues, cell types and/or cellular conditions (e.g., hypoxia) for which eQTL information is not available at present; or (b) too weak to be detected with the sample sizes included in published eQTL studies and at the conservative significance level that we used. In this case, the likely target genes could potentially be identified using functional studies; these can include, for example, experiments to determine the effect of risk variants on promoter-enhancer chromatin interactions, promoter activity or transcription factor binding. We recently used such approaches to identify PAG1 as a likely target gene of the asthma risk variants located on chromosome 8q2170, for which no eQTL support was available at the strict significance threshold used above. In summary, at least 49 genes (including PAG1) are likely targets of published asthma risk variants, most (90%) identified based on the LD between risk variants and eQTLs.
Do any of the likely target genes represent potential new players in the pathophysiology of asthma and allergic disease more generally?
To address this question, we performed a PubMed query using the HGNC-approved gene symbols listed in Table 4, as well as all known aliases (Supplementary Table 9), and the allergy-related terms ‘asthma OR rhinitis OR eczema OR atopic OR dermatitis OR allergy OR allergi* OR hayfever OR 'hay fever'’. We downloaded results from the PubMed query in XML format and then counted the number of unique articles in that file citing both the allergy-related terms and the gene name or aliases in the title, abstract or keyword fields. On the basis of results from this query, we classified the 49 genes into three groups (Table 5). Group one consisted of nine genes co-mentioned frequently (5 or more studies) with those allergy-related terms prior to 2007, the year the first GWAS of any allergy-related trait was published. The genes were TSLP, IL1RL1, TNFSF4, TLR1, HLA-DQB1, HLA-DQB2, HLA-DQA1, ADORA1 and TAP2. For these genes, GWAS findings did not provide the first clue for a key role in disease pathophysiology.
Table 5. Number of publications co-mentioning gene/alias terms and allergy-related terms.
| Gene | Prior to 2007 | Since 2007 |
|---|---|---|
| Group 1: commonly co-cited before 2007 | ||
| TSLP | 21 | 814 |
| IL1RL1 | 16 | 260 |
| TNFSF4 | 9 | 50 |
| TLR1 | 8 | 43 |
| HLA-DQB1 | 30 | 31 |
| HLA-DQB2 | 28 | 30 |
| HLA-DQA1 | 13 | 13 |
| ADORA1 | 12 | 5 |
| TAP2 | 9 | 2 |
| Group 2: commonly co-cited after 2007 | ||
| ORMDL3 | 0 | 115 |
| GSDMB | 0 | 57 |
| ZPBP2 | 0 | 18 |
| IL6R | 2 | 15 |
| GSDMA | 0 | 15 |
| CHIT1 | 2 | 14 |
| IKZF3 | 0 | 11 |
| FCER1G | 1 | 9 |
| SLC22A5 | 0 | 7 |
| WDR36 | 0 | 6 |
| IL18RAP | 1 | 5 |
| HLA-DQA2 | 0 | 5 |
| NDFIP1 | 0 | 5 |
| Group 3: uncommonly/not co-cited | ||
| F11R | 2 | 3 |
| HLA-DRB5 | 1 | 2 |
| MICB | 1 | 2 |
| STARD3 | 1 | 2 |
| PGAP3 | 0 | 2 |
| PPP1R1B | 0 | 2 |
| AAGAB | 2 | 1 |
| USF1 | 2 | 1 |
| ADAMTS4 | 0 | 1 |
| B4GALT3 | 0 | 1 |
| DFFA | 0 | 1 |
| PAG1 | 0 | 1 |
| AC007278.3 | 0 | 0 |
| CAMK4 | 0 | 0 |
| CD247 | 0 | 0 |
| CTC-551A13.2 | 0 | 0 |
| HLA-DQB1-AS1 | 0 | 0 |
| HLA-DRB6 | 0 | 0 |
| MFSD9 | 0 | 0 |
| MYBPH | 0 | 0 |
| PEX14 | 0 | 0 |
| PPFIA4 | 0 | 0 |
| PPOX | 0 | 0 |
| RP11-335O13.7 | 0 | 0 |
| RP11-554D20.1 | 0 | 0 |
| RP11-94L15.2 | 0 | 0 |
| TOMM40L | 0 | 0 |
Group two consisted of 13 genes co-mentioned frequently with allergy-related terms since, but not before, 2007: ORMDL3, GSDMB, ZPBP2, IKZF3, GSDMA, IL6R, CHIT1, FCER1G, SLC22A5, WDR36, IL18RAP, HLA-DQA2 and NDFIP1. The first five are located in the same asthma risk locus, and were first suspected to contribute to asthma pathophysiology because of GWAS findings. Of the remaining eight genes, for five it can be argued that functional studies provided the first suggestion of a key role in asthma/allergies, namely IL6R,71 CHIT1,72 FCER1G,73 IL18RAP74 and NDFIP175. But that is unlikely to be the case for the other three genes, SLC22A5 (organic cation transporter involved in pulmonary absorption of asthma-related drugs76, 77), WDR36 (nucleolar protein involved in processing of 18S rRNA78) and HLA-DQA2 (HLA class II molecule expressed in epidermal Langerhans cells79).
Lastly, group three was composed of 27 genes co-mentioned infrequently with allergy-related terms, both before and after 2007. To our knowledge, only two of these genes have been suggested to play a role in allergic disease through functional studies: USF180 and STARD381. However, many have a known function (Supplementary Table 10) that is directly relevant to allergic disease pathophysiology, such as F11R82, 83, MICB84, CD24785, 86, PGAP387, AAGAB,88 CAMK489 and PEX1490. In summary, of the 49 likely target genes of published asthma risk variants, only 16 were previously implicated by functional studies in disease pathophysiology.
Are there examples of genetic findings subsequently translated into clinical research or practice?
Based on the Thomson Reuters CortellisTM Drug database, drugs against five of the 49 likely target genes of asthma risk variants are being considered for clinical development (Table 6). Six of these drugs are being (or have been) tested in clinical trials of asthma. To our knowledge, only one of these clinical studies was motivated directly by results from a GWAS, our clinical trial of tocilizumab (TCZ) in participants with mild to moderate asthma. In 2011, we reported the association between a variant in the IL6R gene (rs4129267) and asthma risk.26 A consistent association with this variant was later reported also for eczema91, 92 and asthma severity.93 We and others noted that the disease protective allele (rs4129267:C) was strongly associated with decreased protein levels of the soluble form of the receptor (sIL-6R),94 but increased mRNA levels of the full length IL6R transcript,95, 96, 97 which encodes for the membrane bound form of the receptor (mIL-6R). That is, decreased asthma risk is associated with decreased sIL-6R but increased mIL-6R. By extension, decreased disease risk is likely associated with decreased IL-6 trans-signaling (which requires sIL-6R and is mainly pro-inflammatory) but increased IL-6 classic signaling (which requires mIL-6R and is thought to be mainly regenerative and protective).98 On the basis of these genetic findings, it was not immediately obvious what to expect from a drug such as TCZ (approved therapeutic for rheumatoid arthritis and other auto-immune diseases), which blocks both sIL-6R and mIL-6R. A small case study published in 2011 reported decreased clinical activity of atopic dermatitis in three patients treated with TCZ for up to 12 months.99 This was the first suggestion of a protective effect of TCZ in allergic conditions. To characterize the effect of TCZ in asthma, in 2013 we performed pre-clinical studies using mouse models of acute allergic asthma.100 In these studies, we found that TCZ had a protective effect on allergen-induced airway inflammation only when the experimental model used resulted in increased levels of sIL-6R in the airways, and so that was likely to involve activation of the IL-6 trans-signaling pathway. When that was not the case, dual receptor blockade resulted in worse airway inflammation when compared to control mice. On the basis of the genetic findings and the mouse studies, in 2014 we initiated a clinical trial of tocilizumab in participants with mild asthma, specifically those with CT or TT genotype for rs4129267, as these have markedly increased levels of sIL-6R levels.94 Results from this trial are expected to be published in 2018. On the other hand, if our prediction from the genetic studies and findings from the mouse studies are correct, then a drug that blocks sIL-6R but not mIL-6R might be more desirable. There is one such drug (sgp130Fc, also known as FE301 or olamkicept), which was shown to be safe in phase 1 clinical trials101 and is now in phase 2 trials for Crohn's disease (Stefan Rose-John, personal communication). Future studies that test the safety and efficacy of this drug in asthma patients are warranted.
Table 6. Drugs against likely target genes of asthma risk variants considered for clinical development.
| Gene | Drug name | Originator company | Indications (approved or in trials) | Target-based actions | Highest status | Clinical trials in asthma |
|---|---|---|---|---|---|---|
| ADORA1 | Adenoscan | King Pharmaceuticals R&D Inc | Coronary artery disease | Adenosine A1 (and A2) receptor agonist | Launched | NA |
| ADORA1 | Trabodenoson | Inotek Pharmaceuticals Corp | Ocular hypertension; Open angle glaucoma; Optic nerve disorder | Adenosine A1 receptor agonist | Phase 3 Clinical | NA |
| ADORA1 | Neladenoson bialanate | Bayer AG | Cardiac failure | Adenosine A1 receptor partial agonist | Phase 2 Clinical | NA |
| ADORA1 | PBF-680 | Palobiofarma SL | Asthma; COPD | Adenosine A1 receptor antagonist | Phase 2 Clinical | NCT02635945 |
| IKZF3 | Iberdomide | Celgene Corp | Multiple myeloma; Systemic lupus erythematosus | Aiolos inhibitor | Phase 2 Clinical | NA |
| IL1RL1 | RG-6149 | Amgen Inc | Asthma | IL-33 receptor antagonist | Phase 2 Clinical | NCT01928368 |
| IL1RL1 | Nerofe | Immune System Key Ltd | Autoimmune disease; Cancer; Diabetes mellitus; Myocardial infarction | IL-33 receptor agonist | Phase 1 Clinical | NA |
| IL1RL1 | GSK-3772847 | Janssen Research & Development LLC | Asthma | IL-33 receptor antagonist | Phase 2 Clinical | NCT03207243 |
| IL6R | Tocilizumab | Chugai Pharmaceutical Co Ltd | Asthma; Chronic lymphocytic leukemia; Motor neurone disease; Rheumatoid arthritis; Scleroderma; and others. | IL-6 receptor antagonist | Launched | ACTRN12614000123640 |
| IL6R | Sarilumab | Regeneron Pharmaceuticals Inc | Arthritis; Juvenile rheumatoid arthritis; Rheumatoid arthritis; Uveitis | IL-6 receptor antagonist | Launched | NA |
| IL6R | Siltuximab | Janssen Biotech Inc | Castlemans disease; Multiple myeloma | IL-6 antagonist | Launched | NA |
| IL6R | Sirukumab | Janssen Biotech Inc | Asthma; Major depressive disorder; Polymyalgia rheumatica; Rheumatoid arthritis; Temporal arteritis | IL-6 antagonist | Pre-registration | NCT02794519 |
| IL6R | SA-237 | Chugai Pharmaceutical Co Ltd | Neuromyelitis optica | IL-6 antagonist | Phase 3 Clinical | NA |
| IL6R | Olamkicept | Conaris Research Institute AG | Inflammatory bowel disease | IL-6/sIL-6R inhibitor | Phase 2 Clinical | NA |
| IL6R | Vobarilizumab | Ablynx NV | Rheumatoid arthritis; Systemic lupus erythematosus | IL-6 receptor antagonist | Phase 2 Clinical | NA |
| TSLP | Tezepelumab | Amgen Inc | Asthma; Atopic dermatitis | TSLP antagonist | Phase 2 Clinical | NCT01405963, NCT02054130 |
Opportunities and challenges
In this section, we highlight three broad research areas that should be addressed in the short-term to help translate findings from asthma GWAS into clinical research or practice.
Improving and confirming target gene predictions
As discussed above, eQTL information provides a valuable tool to identify a set of genes for which variation in SNP genotype for an asthma risk SNP is likely to directly cause variation in transcription levels. However, this approach has some caveats, of which we highlight two. First, SNP effects on gene expression can be tissue-specific102, context-dependent103 and/or relatively small. For these reasons, some genes might not be predicted to be target genes because appropriate (right tissue, context and sample size for that gene) eQTL studies have not been performed. For example, to our knowledge, the largest eQTL study conducted using airway epithelial cells was based on just 105 individuals104 (compared to 5311 for whole-blood105), and so our understanding of the genetic control of gene expression in this relevant cell type is still very poor. We thus consider essential to continue expanding eQTL studies to include more relevant tissues, diverse cellular stimuli and larger sample sizes. The second caveat of this approach is that a genetic overlap between risk variants and eQTLs can often arise by chance, because eQTLs are widespread.106 We therefore argue that eQTL information should be used to generate predictions that can be subsequently tested by functional studies. However, such studies are laborious and time consuming, often deployed one locus at a time (for example, refs. 70, 107, 108). As the number of known asthma risk variants increases in the near future, efficient validation of target gene predictions will require high-throughput approaches, such as capture Hi–C,109 multiplexed reporter assays110 and genome editing.111 Ideally, these studies should be performed in relevant primary cell types, which at present is still technically challenging.
Understanding target gene function and their role in disease pathophysiology
As highlighted above, for many genes it is unclear how variation in gene expression or protein sequence ultimately leads to variation in asthma risk. Therefore, for these, there is an opportunity to make new discoveries regarding the molecular mechanisms underlying the regulation of gene expression, and the impact that this might have on cellular function and disease pathophysiology. The best examples of this to date are ORMDL3112, 113, 114, 115, 116, 117, 118 and GSDMB,119 for which recent functional and animal studies have provided the first insights into their contribution to disease pathophysiology. Similarly, we have recently been interested in understanding how variation in PAG1 expression might contribute to asthma pathophysiology. Our functional studies suggested that decreased PAG1 expression is associated with decreased disease risk, contrary to what was expected based on the widely accepted contribution of this transmembrane adaptor protein to the development of immune responses.70 We are currently testing this possibility using mouse models of experimental asthma. Various models have been described,120 including those that mimic mechanisms underlying acute allergic asthma;121, 122 chronic asthma;123 and viral-induced asthma124 in humans. It is not always straightforward to decide which model is most appropriate to use, and different models can result in different findings,100 presumably because of the different underlying pathophysiology. In this respect, understanding the contribution of gene expression to cellular function might provide the best clue as to which models to begin with. An additional caveat of these models might be encountered when comparing germline knock-out mice against wild-type mice, because of possible genetic compensation mechanisms in the former, that is, the up-regulation of functionally related genes.125 Potential solutions include temporally controlled gene deletion126 or using therapeutic agents that block gene activity. Mouse models are also laborious and so are invariably applied on a gene-by-gene basis, which constitutes a significant bottleneck when dealing with the rapidly increasing number of asthma risk genes. Nonetheless, despite the potential limitations of these models, they will continue to represent essential tools to understand the contribution of target genes of risk variants to asthma pathophysiology.
Genomics-guided prioritization of targets for drug development
GWAS findings, in combination with eQTL information, can provide important clues into the predicted directional effect of gene expression on disease risk. This, in turn, can be used to inform drug repositioning or the development of novel drugs: specifically, if the disease protective allele of an asthma risk variant is associated with decreased (increased) gene expression, then the prediction is that a drug that also decreases (increases) gene expression should similarly have a protective effect on asthma symptoms. For example, the disease-protective allele rs1438673:T19 on chromosome 5q22.1 is strongly associated with decreased expression of TSLP in skin127 (beta=−0.48, P=2 × 10−15) and whole-blood103 (beta=−8.8, P=10−18). The same direction of effect on gene expression is observed in at least eight different tissues studied by the GTEx project,127 including adipose, colon and esophagus. Thus, based on these genetic findings, we can postulate that TSLP antagonists (but not agonists) might improve asthma symptoms. This is consistent with the known contribution of TSLP to disease pathophysiology128 and with results from a recent clinical trial of an anti-TSLP antibody.129
This is not always so straightforward, as the IL6R example discussed above illustrates. Other examples that are harder to interpret occur when the directional effect of a risk variant on gene expression is not the same, or is poorly characterized, in different relevant tissues. For example, the asthma protective allele rs6683383:A is strongly associated with increased expression of ADORA1 in whole-blood (for example, beta=0.40, P=10−44 130), suggesting that agonists and not antagonists (such as PBF-680, which is currently being tested in a phase 2 trial in asthma: NCT02635945) might be beneficial in asthma. But it is possible that rs6683383:A has the opposite effect on ADORA1 expression in other tissues relevant to asthma, such as airway epithelial cells. This example illustrates the importance of characterizing the genetic control of gene expression in multiple relevant tissues and cell types. In this regard, projects such as GTEx127 but with a focus on asthma-related tissues and cell types could prove very useful. Tissues and cell types could be selected based on results from tissue-specific expression131 or SNP heritability132 enrichment analyses. For example, the latter suggests that SNPs associated with allergic disease are enriched amongst enhancers in Th17 cells;57, 133 yet, to our knowledge, no eQTL study has been performed on this cell type.
Concluding remarks
In the last 10 years, GWAS have identified the first reproducible associations between common SNPs and asthma risk. In Europeans, these SNPs account for ~2.5% of the variation in disease liability. The risk SNPs identified provide the starting point for functional studies that can help understand how genetic variation at these loci affects gene expression, cellular function and disease pathophysiology. The target genes of published risk SNPs are likely to include many previously unsuspected players in disease pathophysiology, and so might point to new therapeutic opportunities. Larger GWAS of broadly defined asthma have the potential to identify additional common risk variants that explain at least 12% of disease liability. This could translate into many hundreds or thousands of risk variants, if we consider that the mean SNP heritability is likely to be low (for example, 0.01–0.05%). Larger GWAS will also have increased power to detect individual associations with uncommon variants (for example, MAF<1%), and to quantify their overall contribution to disease liability, which has not been addressed to date. Other areas of research that remain largely unexplored in the field of asthma genetics include understanding if different inflammatory subtypes (for example, neutrophilic and eosinophilic asthma) have distinct genetic components and whether disease remission is a heritable trait. The next decade of research might provide new insights into these important questions.
Acknowledgments
This research has been conducted using the UK Biobank Resource. CTV was supported by a PhD scholarship (QPhD2015025) from Equity Trustees (Australia); JAR was supported by a PhD scholarship (SFRH/BD/92907/2013) from Fundacao para a Ciencia e Tecnologia (Portugal); MARF was supported by a Senior Research Fellowship (APP1124501) from the National Health and Medical Research Council (Australia). We also thank Naomi Wray for providing comments on the original manuscript.
Footnotes
The Supplementary Information that accompanies this paper is available on the Clinical and Translational Immunology website (http://www.nature.com/cti)
The authors declare no conflict of interest.
Supplementary Material
References
- Murray CS, Simpson A, Custovic A. Allergens, viruses, and asthma exacerbations. Proc Am Thorac Soc 2004; 1: 99–104. [DOI] [PubMed] [Google Scholar]
- Duffy DL, Martin NG, Battistutta D, Hopper JL, Mathews JD. Genetics of asthma and hay fever in Australian twins. Am Rev Resp Dis 1990; 142 (6 Pt 1): 1351–1358. [DOI] [PubMed] [Google Scholar]
- Thomsen SF, Ulrik CS, Kyvik KO, Ferreira MA, Backer V. Multivariate genetic analysis of atopy phenotypes in a selected sample of twins. Clin Exp Allergy 2006; 36: 1382–1390. [DOI] [PubMed] [Google Scholar]
- van Beijsterveldt CE, Boomsma DI. Genetics of parentally reported asthma, eczema and rhinitis in 5-yr-old twins. Eur Respir J 2007; 29: 516–521. [DOI] [PubMed] [Google Scholar]
- Cookson WO, Sharp PA, Faux JA, Hopkin JM. Linkage between immunoglobulin E responses underlying asthma and rhinitis and chromosome 11q. Lancet 1989; 1: 1292–1295. [DOI] [PubMed] [Google Scholar]
- Bouzigon E, Forabosco P, Koppelman GH, Cookson WO, Dizier MH, Duffy DL et al. Meta-analysis of 20 genome-wide linkage studies evidenced new regions linked to asthma and atopy. Eur J Hum Genet 2010; 18: 700–6. [DOI] [PMC free article] [PubMed]
- Ohe M, Munakata M, Hizawa N, Itoh A, Doi I, Yamaguchi E et al. Beta 2 adrenergic receptor gene restriction fragment length polymorphism and bronchial asthma. Thorax 1995; 50: 353–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira MA. Linkage analysis: principles and methods for the analysis of human quantitative traits. Twin Res 2004; 7: 513–530. [DOI] [PubMed] [Google Scholar]
- Hoffjan S, Nicolae D, Ober C. Association studies for asthma and atopic diseases: a comprehensive review of the literature. Respir Res 2003; 4: 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuzaki H, Dong S, Loi H, Di X, Liu G, Hubbell E et al. Genotyping over 100 000 SNPs on a pair of oligonucleotide arrays. Nat Methods 2004; 1: 109–111. [DOI] [PubMed] [Google Scholar]
- Klein RJ, Zeiss C, Chew EY, Tsai JY, Sackler RS, Haynes C et al. Complement factor H polymorphism in age-related macular degeneration. Science 2005; 308: 385–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet 2007; 39: 906–913. [DOI] [PubMed] [Google Scholar]
- Moffatt MF, Kabesch M, Liang L, Dixon AL, Strachan D, Heath S et al. Genetic variants regulating ORMDL3 expression contribute to the risk of childhood asthma. Nature 2007; 448: 470–473. [DOI] [PubMed] [Google Scholar]
- MacArthur J, Bowler E, Cerezo M, Gil L, Hall P, Hastings E et al. The new NHGRI-EBI Catalog of published genome-wide association studies (GWAS Catalog). Nucleic Acids Res 2017; 45: D896–D901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickrell JK, Berisa T, Liu JZ, Segurel L, Tung JY, Hinds DA. Detection and interpretation of shared genetic influences on 42 human traits. Nat Genet 2016; 48: 709–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White MJ, Risse-Adams O, Goddard P, Contreras MG, Adams J, Hu D et al. Novel genetic risk factors for asthma in African American children: Precision Medicine and the SAGE II Study. Immunogenetics 2016; 68: 391–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barreto-Luis A, Pino-Yanes M, Corrales A, Campo P, Callero A, Acosta-Herrera M et al. Genome-wide association study in Spanish identifies ADAM metallopeptidase with thrombospondin type 1 motif, 9 (ADAMTS9), as a novel asthma susceptibility gene. J Allergy Clin Immunol 2016; 137: 964–966. [DOI] [PubMed] [Google Scholar]
- Galanter JM, Gignoux CR, Torgerson DG, Roth LA, Eng C, Oh SS et al. Genome-wide association study and admixture mapping identify different asthma-associated loci in Latinos: the Genes-environments & Admixture in Latino Americans study. J Allergy Clin Immunol 2014; 134: 295–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira MA, Matheson MC, Tang CS, Granell R, Ang W, Hui J et al. Genome-wide association analysis identifies 11 risk variants associated with the asthma with hay fever phenotype. J Allergy Clin Immunol 2014; 133: 1564–1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnelykke K, Sleiman P, Nielsen K, Kreiner-Moller E, Mercader JM, Belgrave D et al. A genome-wide association study identifies CDHR3 as a susceptibility locus for early childhood asthma with severe exacerbations. Nat Genet 2014; 46: 51–55. [DOI] [PubMed] [Google Scholar]
- Lasky-Su J, Himes BE, Raby BA, Klanderman BJ, Sylvia JS, Lange C et al. HLA-DQ strikes again: genome-wide association study further confirms HLA-DQ in the diagnosis of asthma among adults. Clin Exp Allergy 2012; 42: 1724–1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramasamy A, Kuokkanen M, Vedantam S, Gajdos ZK, Couto Alves A, Lyon HN et al. Genome-wide association studies of asthma in population-based cohorts confirm known and suggested loci and identify an additional association near HLA. PLoS ONE 2012; 7: e44008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Ampleford EJ, Howard TD, Moore WC, Torgerson DG, Li H et al. Genome-wide association studies of asthma indicate opposite immunopathogenesis direction from autoimmune diseases. J Allergy Clin Immunol 2012; 130: 861–8 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan YI, Shrine NR, Soler Artigas M, Wain LV, Blakey JD, Moffatt MF et al. Genome-wide association study to identify genetic determinants of severe asthma. Thorax 2012; 67: 762–768. [DOI] [PubMed] [Google Scholar]
- Karunas AS, Iunusbaev BB, Fedorova I, Gimalova GF, Ramazanova NN, Gur'eva LL et al. Genome-wide association study of bronchial asthma in the Volga-Ural region of Russia. Mol Biol 2011; 45: 992–1003. [PubMed] [Google Scholar]
- Ferreira MA, Matheson MC, Duffy DL, Marks GB, Hui J, Le Souef P et al. Identification of IL6R and chromosome 11q13.5 as risk loci for asthma. Lancet 2011; 378: 1006–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirota T, Takahashi A, Kubo M, Tsunoda T, Tomita K, Doi S et al. Genome-wide association study identifies three new susceptibility loci for adult asthma in the Japanese population. Nat Genet 2011; 43: 893–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torgerson DG, Ampleford EJ, Chiu GY, Gauderman WJ, Gignoux CR, Graves PE et al. Meta-analysis of genome-wide association studies of asthma in ethnically diverse North American populations. Nat Genet 2011; 43: 887–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguchi E, Sakamoto H, Hirota T, Ochiai K, Imoto Y, Sakashita M et al. Genome-wide association study identifies HLA-DP as a susceptibility gene for pediatric asthma in Asian populations. PLoS Genet 2011; 7: e1002170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira MA, McRae AF, Medland SE, Nyholt DR, Gordon SD, Wright MJ et al. Association between ORMDL3, IL1RL1 and a deletion on chromosome 17q21 with asthma risk in Australia. Eur J Hum Genet 2011; 19: 458–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeWan AT, Triche EW, Xu X, Hsu LI, Zhao C, Belanger K et al. PDE11A associations with asthma: results of a genome-wide association scan. J Allergy Clin Immunol 2010; 126: 871–3 e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himes BE, Lasky-Su J, Wu AC, Wilk JB, Hunninghake GM, Klanderman B et al. Asthma-susceptibility variants identified using probands in case-control and family-based analyses. BMC Med Genet 2010; 11: 122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Howard TD, Zheng SL, Haselkorn T, Peters SP, Meyers DA et al. Genome-wide association study of asthma identifies RAD50-IL13 and HLA-DR/DQ regions. J Allergy Clin Immunol 125: 328–335 e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sleiman PM, Flory J, Imielinski M, Bradfield JP, Annaiah K, Willis-Owen SA et al. Variants of DENND1B associated with asthma in children. N Engl J Med 2010; 362: 36–44. [DOI] [PubMed] [Google Scholar]
- Mathias RA, Grant AV, Rafaels N, Hand T, Gao L, Vergara C et al. A genome-wide association study on African-ancestry populations for asthma. J Allergy Clin Immunol 2010; 125: 336–46 e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock DB, Romieu I, Shi M, Sienra-Monge JJ, Wu H, Chiu GY et al. Genome-wide association study implicates chromosome 9q21.31 as a susceptibility locus for asthma in mexican children. PLoS Genet 2009; 5: e1000623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himes BE, Hunninghake GM, Baurley JW, Rafaels NM, Sleiman P, Strachan DP et al. Genome-wide association analysis identifies PDE4D as an asthma-susceptibility gene. Am J Hum Genet 2009; 84: 581–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffatt MF, Gut IG, Demenais F, Strachan DP, Bouzigon E, Heath S et al. A large-scale, consortium-based genomewide association study of asthma. N Engl J Med 2010; 363: 1211–1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 2007; 81: 559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fadista J, Manning AK, Florez JC, Groop L. The (in)famous GWAS P-value threshold revisited and updated for low-frequency variants. Eur J Hum Genet 2016; 24: 1202–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen NE, Sudlow C, Peakman T, Collins R, Biobank UK. UK biobank data: come and get it. Sci Transl Med 2014; 6: 224ed4. [DOI] [PubMed] [Google Scholar]
- Bycroft C, Freeman C, Petkova D, Band G, Elliott LT, Sharp K et al. Genome-wide genetic data on ~500 000 UK Biobank participants. bioRxiv 2017.
- Bulik-Sullivan BK, Loh PR, Finucane HK, Ripke S, Yang J et al, Schizophrenia Working Group of the Psychiatric Genomics C. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet 2015; 47: 291–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullemar V, Magnusson PK, Lundholm C, Zettergren A, Melen E, Lichtenstein P et al. Heritability and confirmation of genetic association studies for childhood asthma in twins. Allergy 2016; 71: 230–238. [DOI] [PubMed] [Google Scholar]
- Pawitan Y, Seng KC, Magnusson PK. How many genetic variants remain to be discovered? PLoS ONE 2009; 4: e7969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinkhuyzen AA, Wray NR, Yang J, Goddard ME, Visscher PM. Estimation and partition of heritability in human populations using whole-genome analysis methods. Annu Rev Genet 2013; 47: 75–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet 2011; 88: 76–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh PR, Bhatia G, Gusev A, Finucane HK, Bulik-Sullivan BK, Pollack SJ et al. Contrasting genetic architectures of schizophrenia and other complex diseases using fast variance-components analysis. Nat Genet 2015; 47: 1385–1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schizophrenia Working Group of the Psychiatric Genomics C. Biological insights from 108 schizophrenia-associated genetic loci. Nature 2014; 511: 421–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wray NR, Pergadia ML, Blackwood DH, Penninx BW, Gordon SD, Nyholt DR et al. Genome-wide association study of major depressive disorder: new results, meta-analysis, and lessons learned. Mol Psychiatry 2012; 17: 36–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Wray NR, Visscher PM. Comparing apples and oranges: equating the power of case-control and quantitative trait association studies. Genet Epidemiol 2010; 34: 254–257. [DOI] [PubMed] [Google Scholar]
- Shi H, Kichaev G, Pasaniuc B. Contrasting the genetic architecture of 30 complex traits from summary association data. Am J Hum Genet 2016; 99: 139–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levinson DF, Mostafavi S, Milaneschi Y, Rivera M, Ripke S, Wray NR et al. Genetic studies of major depressive disorder: why are there no genome-wide association study findings and what can we do about it? Biol Psychiatry 2014; 76: 510–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreiner-Moller E, Strachan DP, Linneberg A, Husemoen LL, Bisgaard H, Bonnelykke K. 17q21 gene variation is not associated with asthma in adulthood. Allergy 2015; 70: 107–114. [DOI] [PubMed] [Google Scholar]
- Ferreira MA. Improving the power to detect risk variants for allergic disease by defining case-control status based on both asthma and hay fever. Twin Res Hum Genet 2014; 17: 505–511. [DOI] [PubMed] [Google Scholar]
- Thomsen SF, Ulrik CS, Kyvik KO, Skadhauge LR, Steffensen I, Backer V. Findings on the atopic triad from a Danish twin registry. Int J Tuberculosis Lung Dis: Off J Int Union Tuberculosis Lung Dis 2006; 10: 1268–1272. [PubMed] [Google Scholar]
- Ferreira MA, Vonk JM, Baurecht H, Marenholz I, Tian C, Hoffman JD, et al. Shared genetic origin of asthma, hay fever and eczema elucidates allergic disease biology. Nat Genet 2017; 49: 1752–7. [DOI] [PMC free article] [PubMed]
- Caliskan M, Bochkov YA, Kreiner-Moller E, Bonnelykke K, Stein MM, Du G et al. Rhinovirus wheezing illness and genetic risk of childhood-onset asthma. N Engl J Med 2013; 368: 1398–1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouzigon E, Corda E, Aschard H, Dizier MH, Boland A, Bousquet J et al. Effect of 17q21 variants and smoking exposure in early-onset asthma. N Engl J Med 2008; 359: 1985–1994. [DOI] [PubMed] [Google Scholar]
- Brauner EV, Loft S, Raaschou-Nielsen O, Vogel U, Andersen PS, Sorensen M. Effects of a 17q21 chromosome gene variant, tobacco smoke and furred pets on infant wheeze. Genes Immun 2012; 13: 94–97. [DOI] [PubMed] [Google Scholar]
- Gref A, Merid SK, Gruzieva O, Ballereau S, Becker A, Bellander T et al. Genome-wide interaction analysis of air pollution exposure and childhood asthma with functional follow-up. Am J Respir Crit Care Med 2017; 195: 1373–1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du R, Litonjua AA, Tantisira KG, Lasky-Su J, Sunyaev SR, Klanderman BJ et al. Genome-wide association study reveals class I MHC-restricted T cell-associated molecule gene (CRTAM) variants interact with vitamin D levels to affect asthma exacerbations. J Allergy Clin Immunol 2012; 129: 368–373, 73 e1-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ege MJ, Strachan DP, Cookson WO, Moffatt MF, Gut I, Lathrop M et al. Gene-environment interaction for childhood asthma and exposure to farming in Central Europe. J Allergy Clin Immunol 2011; 127: 138–144, 44 e1-4. [DOI] [PubMed] [Google Scholar]
- Vonk JM, Scholtens S, Postma DS, Moffatt MF, Jarvis D, Ramasamy A et al. Adult onset asthma and interaction between genes and active tobacco smoking: the GABRIEL consortium. PLoS ONE 2017; 12: e0172716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnelykke K, Ober C. Leveraging gene-environment interactions and endotypes for asthma gene discovery. J Allergy Clin Immunol 2016; 137: 667–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauffmann F, Demenais F. Gene-environment interactions in asthma and allergic diseases: challenges and perspectives. J Allergy Clin Immunol 2012; 130: 1229–1240; quiz 41-2. [DOI] [PubMed] [Google Scholar]
- Yang H, Wang K. Genomic variant annotation and prioritization with ANNOVAR and wANNOVAR. Nat Protoc 2015; 10: 1556–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebhan M, Chalifa-Caspi V, Prilusky J, Lancet D. GeneCards: integrating information about genes, proteins and diseases. Trends Genet 1997; 13: 163. [DOI] [PubMed] [Google Scholar]
- Davis JR, Fresard L, Knowles DA, Pala M, Bustamante CD, Battle A et al. An efficient multiple-testing adjustment for eQTL studies that accounts for linkage disequilibrium between variants. Am J Hum Genet 2016; 98: 216–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicente CT, Edwards SL, Hillman KM, Kaufmann S, Mitchell H, Bain L et al. Long-Range Modulation of PAG1 Expression by 8q21 Allergy Risk Variants. Am J Hum Genet 2015; 97: 329–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doganci A, Eigenbrod T, Krug N, De Sanctis GT, Hausding M, Erpenbeck VJ et al. The IL-6R alpha chain controls lung CD4+CD25+ Treg development and function during allergic airway inflammation in vivo. J Clin Invest 2005; 115: 313–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seibold MA, Donnelly S, Solon M, Innes A, Woodruff PG, Boot RG et al. Chitotriosidase is the primary active chitinase in the human lung and is modulated by genotype and smoking habit. J Allergy Clin Immunol 2008; 122: 944–50 e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M, Tsai M, Tam SY, Jones C, Zehnder J, Galli SJ. Mast cells can promote the development of multiple features of chronic asthma in mice. J Clin Invest 2006; 116: 1633–1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewkowich IP, Rempel JD, HayGlass KT. Prevention of allergen-specific, Th2-biased immune responses in vivo: role of increased IL-12 and IL-18 responsiveness. J Immunol 2005; 175: 4956–4962. [DOI] [PubMed] [Google Scholar]
- Oliver PM, Cao X, Worthen GS, Shi P, Briones N, MacLeod M et al. Ndfip1 protein promotes the function of itch ubiquitin ligase to prevent T cell activation and T helper 2 cell-mediated inflammation. Immunity 2006; 25: 929–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T, Nakanishi T, Haruta T, Shirasaka Y, Keogh JP, Tamai I. Transport of ipratropium, an anti-chronic obstructive pulmonary disease drug, is mediated by organic cation/carnitine transporters in human bronchial epithelial cells: implications for carrier-mediated pulmonary absorption. Mol Pharm 2010; 7: 187–195. [DOI] [PubMed] [Google Scholar]
- Mukherjee M, Cingolani E, Pritchard DI, Bosquillon C. Enhanced expression of Organic Cation Transporters in bronchial epithelial cell layers following insults associated with asthma-Impact on salbutamol transport. Eur J Pharm Sci 2017; 106: 62–70. [DOI] [PubMed] [Google Scholar]
- Skarie JM, Link BA. The primary open-angle glaucoma gene WDR36 functions in ribosomal RNA processing and interacts with the p53 stress-response pathway. Hum Mol Genet 2008; 17: 2474–2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenormand C, Bausinger H, Gross F, Signorino-Gelo F, Koch S, Peressin M et al. HLA-DQA2 and HLA-DQB2 genes are specifically expressed in human Langerhans cells and encode a new HLA class II molecule. J Immunol 2012; 188: 3903–3911. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Nishiyama C, Nishiyama M, Okumura K, Ra C, Ohtake Y et al. A complex composed of USF1 and USF2 activates the human FcepsilonRI alpha chain expression via a CAGCTG element in the first intron. Eur J Immunol 2001; 31: 590–599. [PubMed] [Google Scholar]
- Hannen RF, Michael AE, Jaulim A, Bhogal R, Burrin JM, Philpott MP. Steroid synthesis by primary human keratinocytes; implications for skin disease. Biochem Biophys Res Commun 2011; 404: 62–67. [DOI] [PubMed] [Google Scholar]
- Cera MR, Del Prete A, Vecchi A, Corada M, Martin-Padura I, Motoike T et al. Increased DC trafficking to lymph nodes and contact hypersensitivity in junctional adhesion molecule-A-deficient mice. J Clin Invest 2004; 114: 729–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell LA, Ward C, Kwon M, Mitchell PO, Quintero DA, Nusrat A et al. Junctional adhesion molecule A promotes epithelial tight junction assembly to augment lung barrier function. Am J Pathol 2015; 185: 372–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maasho K, Opoku-Anane J, Marusina AI, Coligan JE, Borrego F. NKG2D is a costimulatory receptor for human naive CD8+ T cells. J Immunol 2005; 174: 4480–4484. [DOI] [PubMed] [Google Scholar]
- Lundholm M, Mayans S, Motta V, Lofgren-Burstrom A, Danska J, Holmberg D. Variation in the Cd3 zeta (Cd247) gene correlates with altered T cell activation and is associated with autoimmune diabetes. J Immunol 2010; 184: 5537–5544. [DOI] [PubMed] [Google Scholar]
- Vales-Gomez M, Esteso G, Aydogmus C, Blazquez-Moreno A, Marin AV, Briones AC et al. Natural killer cell hyporesponsiveness and impaired development in a CD247-deficient patient. J Allergy Clin Immunol 2016; 137: 942–5 e4. [DOI] [PubMed] [Google Scholar]
- Murakami H, Wang Y, Hasuwa H, Maeda Y, Kinoshita T, Murakami Y. Enhanced response of T lymphocytes from Pgap3 knockout mouse: Insight into roles of fatty acid remodeling of GPI anchored proteins. Biochem Biophys Res Commun 2012; 417: 1235–1241. [DOI] [PubMed] [Google Scholar]
- Pohler E, Mamai O, Hirst J, Zamiri M, Horn H, Nomura T et al. Haploinsufficiency for AAGAB causes clinically heterogeneous forms of punctate palmoplantar keratoderma. Nat Genet 2012; 44: 1272–1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koga T, Ichinose K, Mizui M, Crispin JC, Tsokos GC. Calcium/calmodulin-dependent protein kinase IV suppresses IL-2 production and regulatory T cell activity in lupus. J Immunol 2012; 189: 3490–3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijayan V, Srinu T, Karnati S, Garikapati V, Linke M, Kamalyan L et al. A new immunomodulatory role for peroxisomes in macrophages activated by the Tlr4 ligand lipopolysaccharide. J Immunol 2017; 198: 2414–2425. [DOI] [PubMed] [Google Scholar]
- Esparza-Gordillo J, Schaarschmidt H, Liang L, Cookson W, Bauerfeind A, Lee-Kirsch MA et al. A functional IL-6 receptor (IL6R) variant is a risk factor for persistent atopic dermatitis. J Allergy Clin Immunol 2013; 132: 371–377. [DOI] [PubMed] [Google Scholar]
- Paternoster L, Standl M, Waage J, Baurecht H, Hotze M, Strachan DP et al. Multi-ancestry genome-wide association study of 21 000 cases and 95 000 controls identifies new risk loci for atopic dermatitis. Nat Genet 2015; 47: 1449–1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins GA, Robinson MB, Hastie AT, Li X, Li H, Moore WC et al. The IL6R variation Asp(358)Ala is a potential modifier of lung function in subjects with asthma. J Allergy Clin Immunol 2012; 130: 510–5 e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melzer D, Perry JR, Hernandez D, Corsi AM, Stevens K, Rafferty I et al. A genome-wide association study identifies protein quantitative trait loci (pQTLs). PLoS Genet 2008; 4: e1000072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dongen J, Jansen R, Smit D, Hottenga JJ, Mbarek H, Willemsen G et al. The contribution of the functional IL6R polymorphism rs2228145, eQTLs and other genome-wide SNPs to the heritability of plasma sIL-6R levels. Behav Genet 2014; 44: 368–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revez JA, Bain L, Chapman B, Powell JE, Jansen R, Duffy DL et al. A new regulatory variant in the interleukin-6 receptor gene associates with asthma risk. Genes Immun 2013; 14: 441–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira RC, Freitag DF, Cutler AJ, Howson JM, Rainbow DB, Smyth DJ et al. Functional IL6R 358Ala allele impairs classical IL-6 receptor signaling and influences risk of diverse inflammatory diseases. PLoS Genet 2013; 9: e1003444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaper F, Rose-John S. Interleukin-6: biology, signaling and strategies of blockade. Cytokine Growth Factor Rev 2015; 26: 475–487. [DOI] [PubMed] [Google Scholar]
- Navarini AA, French LE, Hofbauer GF. Interrupting IL-6-receptor signaling improves atopic dermatitis but associates with bacterial superinfection. J Allergy Clin Immunol 2011; 128: 1128–1130. [DOI] [PubMed] [Google Scholar]
- Ullah MA, Revez JA, Loh Z, Simpson J, Zhang V, Bain L et al. Allergen-induced IL-6 trans-signaling activates gammadelta T cells to promote type 2 and type 17 airway inflammation. J Allergy Clin Immunol 2015; 136: 1065–1073. [DOI] [PubMed] [Google Scholar]
- Cottingham I, PETRI NA. Administration of a selective il-6-trans-signalling inhibitor. 2016. Patent WO2016087941 A1. PCT/IB2015/002459.
- Kasela S, Kisand K, Tserel L, Kaleviste E, Remm A, Fischer K et al. Pathogenic implications for autoimmune mechanisms derived by comparative eQTL analysis of CD4+ versus CD8+ T cells. PLoS Genet 2017; 13: e1006643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhernakova DV, Deelen P, Vermaat M, van Iterson M, van Galen M, Arindrarto W et al. Identification of context-dependent expression quantitative trait loci in whole blood. Nat Genet 2017; 49: 139–145. [DOI] [PubMed] [Google Scholar]
- Luo W, Obeidat M, Di Narzo AF, Chen R, Sin DD, Pare PD et al. Airway epithelial expression quantitative trait loci reveal genes underlying asthma and other airway diseases. Am J Respir Cell Mol Biol 2016; 54: 177–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westra HJ, Peters MJ, Esko T, Yaghootkar H, Schurmann C, Kettunen J et al. Systematic identification of trans eQTLs as putative drivers of known disease associations. Nat Genet 2013; 45: 1238–1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun S, Casparino A, Patsopoulos NA, Croteau-Chonka DC, Raby BA, De Jager PL et al. Limited statistical evidence for shared genetic effects of eQTLs and autoimmune-disease-associated loci in three major immune-cell types. Nat Genet 2017; 49: 600–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berlivet S, Moussette S, Ouimet M, Verlaan DJ, Koka V, Al Tuwaijri A et al. Interaction between genetic and epigenetic variation defines gene expression patterns at the asthma-associated locus 17q12-q21 in lymphoblastoid cell lines. Hum Genet 2012; 131: 1161–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verlaan DJ, Berlivet S, Hunninghake GM, Madore AM, Lariviere M, Moussette S et al. Allele-specific chromatin remodeling in the ZPBP2/GSDMB/ORMDL3 locus associated with the risk of asthma and autoimmune disease. Am J Hum Genet 2009; 85: 377–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javierre BM, Burren OS, Wilder SP, Kreuzhuber R, Hill SM, Sewitz S et al. Lineage-specific genome architecture links enhancers and non-coding disease variants to target gene promoters. Cell 2016; 167: 1369–84 e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tewhey R, Kotliar D, Park DS, Liu B, Winnicki S, Reilly SK et al. Direct identification of hundreds of expression-modulating variants using a multiplexed reporter assay. Cell 2016; 165: 1519–1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pashos EE, Park Y, Wang X, Raghavan A, Yang W, Abbey D et al. Large, diverse population cohorts of hiPSCs and derived hepatocyte-like cells reveal functional genetic variation at blood lipid-associated loci. Cell Stem Cell 2017; 20: 558–70 e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller M, Tam AB, Mueller JL, Rosenthal P, Beppu A, Gordillo R et al. Cutting edge: targeting epithelial ORMDL3 increases, rather than reduces, airway responsiveness and is associated with increased Sphingosine-1-Phosphate. J Immunol 2017; 198: 3017–3022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller M, Rosenthal P, Beppu A, Gordillo R, Broide DH. Oroscomucoid like protein 3 (ORMDL3) transgenic mice have reduced levels of sphingolipids including sphingosine-1-phosphate and ceramide. J Allergy Clin Immunol 2017; 139: 1373–6 e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loser S, Gregory LG, Zhang Y, Schaefer K, Walker SA, Buckley J et al. Pulmonary ORMDL3 is critical for induction of Alternaria-induced allergic airways disease. J Allergy Clin Immunol 2017; 139: 1496–507 e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oyeniran C, Sturgill JL, Hait NC, Huang WC, Avni D, Maceyka M et al. Aberrant ORM (yeast)-like protein isoform 3 (ORMDL3) expression dysregulates ceramide homeostasis in cells and ceramide exacerbates allergic asthma in mice. J Allergy Clin Immunol 2015; 136: 1035–46 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller M, Rosenthal P, Beppu A, Mueller JL, Hoffman HM, Tam AB et al. ORMDL3 transgenic mice have increased airway remodeling and airway responsiveness characteristic of asthma. J Immunol 2014; 192: 3475–3487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha SG, Ge XN, Bahaie NS, Kang BN, Rao A, Rao SP et al. ORMDL3 promotes eosinophil trafficking and activation via regulation of integrins and CD48. Nat Commun 2013; 4: 2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller M, Tam AB, Cho JY, Doherty TA, Pham A, Khorram N et al. ORMDL3 is an inducible lung epithelial gene regulating metalloproteases, chemokines, OAS, and ATF6. Proc Natl Acad Sci USA 2012; 109: 16648–16653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das S, Miller M, Beppu AK, Mueller J, McGeough MD, Vuong C et al. GSDMB induces an asthma phenotype characterized by increased airway responsiveness and remodeling without lung inflammation. Proc Natl Acad Sci USA 2016; 113: 13132–13137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd CM. Building better mouse models of asthma. Curr Allergy Asthma Rep 2007; 7: 231–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry J, Loh Z, Collison A, Mazzone S, Lalwani A, Zhang V et al. Absence of Toll-IL-1 receptor 8/single immunoglobulin IL-1 receptor-related molecule reduces house dust mite-induced allergic airway inflammation in mice. Am J Respir Cell Mol Biol 2013; 49: 481–490. [DOI] [PubMed] [Google Scholar]
- Dullaers M, Schuijs MJ, Willart M, Fierens K, Van Moorleghem J, Hammad H et al. House dust mite-driven asthma and allergen-specific T cells depend on B cells when the amount of inhaled allergen is limiting. J Allergy Clin Immunol 2017; 140: 76–88 e7. [DOI] [PubMed] [Google Scholar]
- McMillan SJ, Lloyd CM. Prolonged allergen challenge in mice leads to persistent airway remodelling. Clin Exp Allergy 2004; 34: 497–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arikkatt J, Ullah MA, Short KR, Zhang V, Gan WJ, Loh Z et al. RAGE deficiency predisposes mice to virus-induced paucigranulocytic asthma. Elife 2017; 6: e21199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Brolosy MA, Stainier DYR. Genetic compensation: a phenomenon in search of mechanisms. PLoS Genet 2017; 13: e1006780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohlmann A, Gotthardt M, Willnow TE, Hammer RE, Herz J. Sustained somatic gene inactivation by viral transfer of Cre recombinase. Nat Biotechnol 1996; 14: 1562–1565. [DOI] [PubMed] [Google Scholar]
- Consortium GT. Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science 2015; 348: 648–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson B, Gauvreau GM. Thymic stromal lymphopoietin: a central regulator of allergic asthma. Expert Opin Ther Targets 2014; 18: 771–785. [DOI] [PubMed] [Google Scholar]
- Gauvreau GM, O'Byrne PM, Boulet LP, Wang Y, Cockcroft D, Bigler J et al. Effects of an anti-TSLP antibody on allergen-induced asthmatic responses. N Engl J Med 2014; 370: 2102–2110. [DOI] [PubMed] [Google Scholar]
- Lloyd-Jones LR, Holloway A, McRae A, Yang J, Small K, Zhao J et al. The genetic architecture of gene expression in peripheral blood. Am J Hum Genet 2017; 100: 228–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells A, Kopp N, Xu X, O'Brien DR, Yang W, Nehorai A et al. The anatomical distribution of genetic associations. Nucleic Acids Res 2015; 43: 10804–10820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finucane HK, Bulik-Sullivan B, Gusev A, Trynka G, Reshef Y, Loh PR et al. Partitioning heritability by functional annotation using genome-wide association summary statistics. Nat Genet 2015; 47: 1228–1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farh KK, Marson A, Zhu J, Kleinewietfeld M, Housley WJ, Beik S et al. Genetic and epigenetic fine mapping of causal autoimmune disease variants. Nature 2015; 518: 337–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.