Abstract
Generation of haploid gametes by meiosis is a unique property of germ cells and is critical for sexual reproduction. Leaving mitosis and entering meiosis is a key step in germ cell development. Several inducers or intrinsic genes are known to be important for meiotic initiation, but the regulation of meiotic initiation, especially at the protein level, is still not well understood. We constructed a comparative proteome profile of female mouse fetal gonads at specific time points (11.5, 12.5, and 13.5 days post coitum), spanning a critical window for initiation of meiosis in female germ cells. We identified 3666 proteins, of which 473 were differentially expressed. Further bioinformatics analysis showed that these differentially expressed proteins were enriched in the mitochondria, especially in the electron transport chain and, notably, 9 proteins in electron transport chain Complex I were differentially expressed. We disrupted the mitochondrial electron transport chain function by adding the complex I inhibitor, rotenone to 11.5 days post coitum female gonads cultured in vitro. This treatment resulted in a decreased proportion of meiotic germ cells, as assessed by staining for histone γH2AX. Rotenone treatment also caused decreased ATP levels, increased reactive oxygen species levels and failure of the germ cells to undergo premeiotic DNA replication. These effects were partially rescued by adding Coenzyme Q10. Taken together, our results suggested that a functional electron transport chain is important for meiosis initiation. Our characterization of the quantitative proteome of female gonads provides an inventory of proteins, useful for understanding the mechanisms of meiosis initiation and female fertility.
Germ cells are among the most specific class of cells in the mammalian organism and are critical for transfer of genetic material from one generation to the next. Meiosis is the special form of cell division unique to germ cells. The correct regulation of meiosis allows germ cells to give rise to haploid gametes necessary for sexual reproduction. A pivotal step in transformation of germline stem cells into differentiated gametes involves their departure from the mitotic cell cycle and onset of the meiotic cell cycle. In mice, the timing of germ cell entry into meiosis differs between female and male embryos (1). The first meiotic germ cells appear in female gonads at about 13.5 days post coitum (dpc)1 (2, 3). In contrast, male germ cells do not enter meiosis during fetal life but, instead, arrest in the G0/G1 phase of mitosis, at about 13.5 dpc (4–6).
It has been long considered that retinoic acid (RA), an inducer originating from mesonephros, is essential for the initiation of meiosis in the ovary (7). But this theory was questioned by Kumar et al. (1), thereafter the debate on whether RA was the principal driver of meiotic entry in mouse fetal ovaries, and where does the RA come from, have been conducted over the recent years (8–11), but without reaching definite conclusions. Besides RA, several intrinsic factors, such as Dazl, Msx1/2, and Dmrt1, have also been reported to be related to the initiation of meiosis in fetal ovaries (10). However, the complex mechanisms by which germ cells choose to commence meiotic cell division are not well understood, how female oogonia switch from mitosis to meiosis remains a key question in reproductive biology. To deeply understand the molecular mechanisms involved in meiosis initiation, we analyzed 11.5, 12.5, and 13.5 dpc female gonads, a timeframe spanning the critical window for initiation of meiosis in female germ cells. Using 6-plex isobaric mass tagging technology combined with liquid chromatography-tandem mass spectrometry (LC-MS3), we generated the first differential proteome dataset from female fetal gonads, thus providing a molecular description of female gonads at the protein level. We successfully identified 3666 proteins and, among them, 473 proteins were differentially expressed. Further bioinformatics analysis showed that these differentially expressed proteins were enriched in the mitochondria, especially in the electron transport chain, with 9 proteins in electron transport chain Complex I differentially expressed among gonads at ages 11.5–13.5 dpc. These results indicated that the electron transport chain was closely linked to meiosis initiation. To confirm this interpretation, we used the Complex I inhibitor, rotenone (ROT) (12, 13), to disrupt the function of the mitochondrial electron transport chain. Our experiments indicated that suppressing electron transport chain activity inhibited female germ cell entry into meiosis.
EXPERIMENTAL PROCEDURES
Animals and Gonad Collection
All animal work was approved by the ethics committee of Nanjing Medical University (China). Timed mating was conducted with CD1 mice and noon on the day when the mating plug was observed was designated as 0.5 dpc. Embryos were collected at 11.5, 12.5, and 13.5 dpc. To confirm the specific times of meiosis initiation in the female gonadal germ cells, we examined expression of the meiosis markers Stra8, Sycp3 and Dmc1 (14) by quantitive PCR in embryonic gonads at 11.5, 12.5, and 13.5 dpc. The reference gene was 18s rRNA and primer sequences are listed in supplemental Table S1. The sex of the gonad is apparent by macroscopic observation at 12.5 and 13.5 dpc. The 11.5 dpc embryos were genotyped to determine sex using primers to detect Sry (15), with β-actin as an internal control.
Proteomic Sample Preparation and Digestion
Proteomic sample preparation and digestion were performed as described (16). Mouse embryonic female gonads from 11.5, 12.5, and 13.5 dpc were collected and lysed using protein extraction buffer (7 m urea, 2 m Thiourea, 65 mm DTT and 1% (v/v) protease inhibitor mixture) and subsequently subjected to centrifugation at 40,000 × g for 1 h. The protein concentration was measured by Bradford assay. Seventy micrograms protein was used for proteomic digestion and tandem mass tag (TMT) labeling. In brief, cysteine residues were reduced in 200 mm DTT and alkylated in 375 mm IAA. Protein lysates were cleaned up by acetone precipitation and resuspended in 200 mm triethylammonium bicarbonate (TEAB), followed by trypsin digestion at 37 °C overnight.
TMT Labeling
A detailed description of the procedures followed of TMT 6-plex labeling has been published previously (16). Briefly, TMT Label Reagents were equilibrated to room temperature, each aliquot was resuspended in 41 μl anhydrous ACN and 42 μl was added to protein digestion mixture dissolved in 200 mm TEAB. After 60 min reaction time at room temperature, 8 μl 5% hydroxylamine was added to each tube and samples incubated for 15 min. The aliquots were then pooled and solvent was removed under vacuum.
Strong Cation Exchange (SCX) Fractionation
The TMT labeled peptide mixture was resuspended in SCX chromatography Buffer A (10 mm NH4COOH, 5% ACN, pH 2.7) and loaded onto a cation ion exchange column (1 mm ID × 10 cm packed with Poros 10 S, DIONEX, Sunnyvale, CA) and chromatographed on the UltiMate® 3000 HPLC systems at a flow rate of 50 μl/min. The following linear gradient conditions were used, with Buffers A and B: 0 to 56% Buffer B (800 mm NH4COOH, 5% ACN, pH 2.7) for 30 min; 56 to 100% B for 1 min; 100% B for 3 min; 100 to 0% B for 1 min; 0% B for 20 min before the next run. Effluents were monitored at 214 nm based on the UV light trace and fractions collected every 2 min (17–20).
Mass Spectrometry Analysis
The 40 fractions analyzed were sequentially loaded onto a μ-precolumnTM cartridge (0.3 × 5 mm, 5 μm, 100 A; DIONEX) at a flow rate of 0.3 μl/min. The trap column effluent was then transferred to a reverse-phase microcapillary column (0.075 × 150 mm, Acclaim® PepMap100 C18 column, 3 μm, 100 A; DIONEX). The reverse-phase separation of peptides was performed using the following mobile phase components: 2% ACN, 0.5% acetic acid (Buffer A) and 80% ACN, 0.5% acetic acid (Buffer B). The gradient elution conditions (193 min total) were: 4% to 9% Buffer B for 3 min; 9% to 33% Buffer B for 170 min; 33% to 50% Buffer B for 10 min; 50% to 100% Buffer B for 1 min; 100% Buffer B for 8 min; 100% to 4% Buffer B for 1 min.
Peptides were detected with a hybrid dual-cell quadrupole linear ion trap-orbitrap mass spectrometer (LTQ Orbitrap Velos, Thermo Fisher Scientific, Waltham, MA) using a data-dependent Top8-MS3 method as previously described (21). For each cycle, one full MS scan of mass/charge ratio (m/z) = 400 to 1800 was acquired in the Orbitrap at a resolution of 30,000 or 60,000. CID MS/MS spectra were acquired from the survey scan for the 8 most intense ions (as determined by X-calibur mass spectrometer software in real time). After each CID MS/MS, the most intense fragment ion in an m/z range between 110 and 160% of the precursor m/z was selected for HCD-MS3. Lock mass, with siloxane (m/z = 445.120025) as an internal standard, was used in all runs to calibrate orbitrap MS precursor masses. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (22) partner repository with the dataset identifier PXD005670.
Protein Identification, Quantification and Bioinformatics Analysis
The raw files were processed with MaxQuant (23) software (version 1.2.2.5) using the IPI (International Protein Index) (24) mouse proteome database (version 3.83; 60010 entries). A common contaminants database was also included for quality control. The reverse strategy was used to estimate the false discovery rate (FDR). Enzyme specificity was set to be Trypsin/P (no P restriction) and the maximum missed cleavage sites permitted were two. The minimum peptide length required was six. Except for TMT labels, carbamidomethyl (C) was set as a fixed modification. Variable modifications were Oxidation (M) and Acetyl (Protein N-term). Mass tolerance for precursor ions and fragment ions were both set to 20 ppm. The site, peptide and protein FDR values were all set to 0.01. Protein quantification was calculated by combining MaxQuant identification results with a local modified Libra algorithm (25). Principal Components Analysis (PCA) was conducted in MetaboAnalyst 3.0 and the data were filtered using mean intensity values and Pareto scaled (26). To identify proteins that were significantly different among the time points, HemI 1.0 was used to perform hierarchical clustering analysis (27). The Database for Annotation, Visualization and Integrated Discovery (DAVID) (28, 29) (http://david.abcc.ncifcrf.gov/) was used for functional annotation of proteins and gene ontology (GO) was used to classify these proteins, including their involvement in cellular components and biological processes. We used the GO analyses results to build a diagram of differentially expressed proteins according to the GO Biological Process terms. The diagram was generated by connecting DE proteins to represent direct and indirect biological relationships, using open source software (Cytoscape; Cytoscape Consortium, San Diego, CA).
Gonad Culture
Gonads were isolated from 11.5 dpc embryos by dissecting in physiological saline at room temperature and collected into warmed Dulbecco's Minimal Eagle's Medium (DMEM) containing 10% fetal calf serum (FCS; Thermo Fisher Scientific) and 50 μg/ml ampicillin at 37 °C, equilibrated with 95% air, 5% CO2. ROT and Coenzyme Q10 (CoQ10) were purchased from Sigma-Aldrich (St. Louis, MO). After culture for 24 h in ROT (0.1 μm) followed by culture for 48 h in their normal medium, gonads were collected for further study. Reactive oxygen species (ROS) and ATP detection were performed with ROS Advanced Fluoro Assay Kit (Genmed Scientifics, Wilmington, DE) and ATP Assay Kit (Beyotime, Beijing, China), respectively.
Immunofluorescence Staining
For whole mount and cryosection staining, gonads were fixed with 4% paraformaldehyde for 30 min, blocked with 5% bovine serum albumin (BSA) for 2 h and incubated with primary antibodies overnight at 4 °C, as described previously (20). Paraffin-embedded gonad sections were dewaxed and dehydrated. Endogenous peroxidase activity was inhibited by incubating sections with 3% hydrogen peroxide for 15 min. Antigen retrieval was performed by boiling samples in 0.01 m sodium citrate buffer (pH 6.0) for 10 min. The sections were incubated with 5% BSA in PBS for 2 h at room temperature, then with primary antibody at 4 °C overnight. Next, sections were washed 3 times and incubated with secondary antibody for 2 h. The nuclei were stained with DAPI and sections were mounted on slides and examined. Images were captured on a Zeiss laser confocal microscope (LSM 710, Carl Zeiss, Oberkochen, Germany). Antibodies against DDX4 (diluted 1:1000; Abcam, Cambridge, UK), AXAN2 (diluted 1:500; Abcam), GATM (diluted 1:500; Santa Cruz Biotechnology, Dallas, TX), RBPMS2 (diluted 1:500, Santa Cruz Biotechnology), UCHL1 (diluted 1:100; Abcam), SOAT1 (diluted 1:100; Abcam), ALDH1A1 (diluted 1:100; Abcam), AFP (diluted 1:100, ABclonal, Woburn, MA), ALDH1A2 (diluted 1:500; Abcam), COFILIN (diluted 1:100; Abcam), GNPDA1 (diluted 1:500; Abcam), CPT1A (diluted 1:500; Abcam), CAV1 (diluted 1:100; BD, Transduction Laboratories TM, Canada), γH2AX (diluted 1:500, Abcam), OCT4 (diluted 1:200, Santa Cruz Biotechnology) were used, where indicated.
Quantitative PCR
Gene expression was quantified with SYBR Premix Ex TaqTM kit (TaKaRa, Dalian, China) in an ABI 7300 real-time PCR instrument (Applied Biosystems, Foster City, CA, USA), using the standard curve method with 18s rRNA as the reference gene. The primers used are listed in supplemental Table S1. Amplification was performed in 20-μl reaction volumes, according to the manufacturer's instructions.
DNA Content Analysis of Germ Cells from Female Gonads
A 12.0 dpc culture was treated with ROT (0.1 μm) or ROT (0.1 μm) adding CoQ10 (20 μm) for 24 h while other samples were incubated, in parallel, in normal medium or with DMSO as a vehicle control for 24 h. Cultures were then collected at 14.5 dpc and incubated in 50 ml trypsin solution at 37 °C for 5 min and then washed briefly in PBS. Cell suspensions were placed on slides to dry, then fixed in 4% paraformaldehyde for 20 min. Immunostaining was performed with a rabbit antibody to DDX4 (1:1000 dilution) as described above. DAPI and DDX4 staining images were taken with a Zeiss laser confocal microscope (Lsm710, Carl Zeiss). All image sets were processed automatically using the software program CellProfiler (http://www.cellprofiler.org) to ensure consistent, unbiased results (30).
Experimental Design and Statistical Rationale
For the proteomics screen, ∼800 female gonad pairs were collected at the age of 11.5 dpc and more than 500 female gonad pairs were resected from female mouse embryos at the ages of 12.5 and 13.5 dpc. 4 biological replicates were used in each stage. For identification of differentially expressed (DE) proteins among stages, a protein with a fold change greater than 2 and a false discovery rate (Q value) less than 0.01 by the SAM (Significance Analysis of Microarrays) algorithm in J-Express (31) were considered significant (16, 21). For gonad culture, gonads were cultured in medium containing ROT (0.1 μm) or ROT (0.1 μm) adding CoQ10 (20 μm) for 24 h followed by culture in their normal medium, gonads were collected at 14.5 dpc for further study. Other samples were incubated, in parallel, in normal medium or with DMSO as a vehicle control for 24 h. All experiments were conducted at least three times and results within each experiment are described using mean ± S.E. Statistically significant (*p < 0.05, **p < 0.01, ***p < 0.001) were determined by using the two sample Student's t test.
RESULTS
Identification of the Specific Time Points of Meiosis Initiation in Female Gonadal Germ Cells
Consistent with previous literature (2, 3, 7, 30, 32), we confirmed that our selected time points (11.5, 12.5, and 13.5 dpc) were representative of female germ cell meiosis initiation in embryonic gonads. The female germ cells continued mitotic division after colonizing the gonads and entered meiosis from 13.5 dpc (Fig. 1). Stra8, Sycp3, and Dmc1 are markers of meiosis (14) and we examined their expression in 11.5, 12.5, and 13.5 dpc embryonic gonads to verify the specific times of meiosis initiation in female gonadal germ cells. Stra8, as a gatekeeper for germ cell entry into meiosis over the years, is required for pre-meiotic DNA replication and the subsequent events of meiotic prophase in germ cells of embryonic ovaries (30, 33). As expected, gene expression levels of Stra8 in 13.5 dpc gonads were significantly higher than in 11.5 dpc gonads. Similarly, expression of Sycp3 and Dmc1 were gradually increased from 11.5 to 13.5 dpc (Fig. 1A). Immunofluorescence staining also showed no germ cells (stained by DDX4) entering meiosis in female gonads at 12.5 dpc, whereas at 13.5 dpc, about 13% of the germ cells were detected by staining for γH2AX, the accepted meiotic cell marker (33, 34). This percentage increased to 73% at 14.5 dpc (Fig. 1B). These results suggested that the female germ cells began entering meiosis at 13.5 dpc and that most had entered meiosis by 14.5 dpc.
Fig. 1.
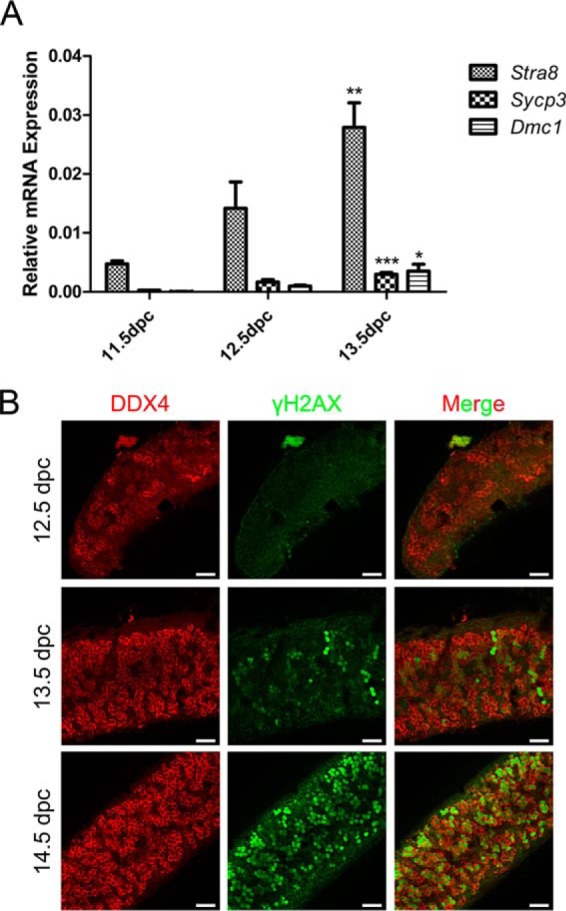
Verification of the specific time points of initiation of female gonad germ cell meiosis. A, Expression levels of meiosis markers (Stra8, Sycp3, Dmc1) were significantly increased from 11.5 to 13.5 dpc. B, Immunofluorescence staining of DDX4 (germ cell marker, red) and γH2AX (meiosis cell marker, green) showed that germ cells began entering meiosis at 13.5 dpc and most had entered meiosis by 14.5 dpc. Error bars indicate S.E. and similar experiments were each repeated at least three times (*p < 0.05, **p < 0.01, ***p < 0.001, Student's t test). Bar, 50 μm.
Identification of Differentially Expressed Proteins During Meiosis Initiation in Female Mouse Gonads
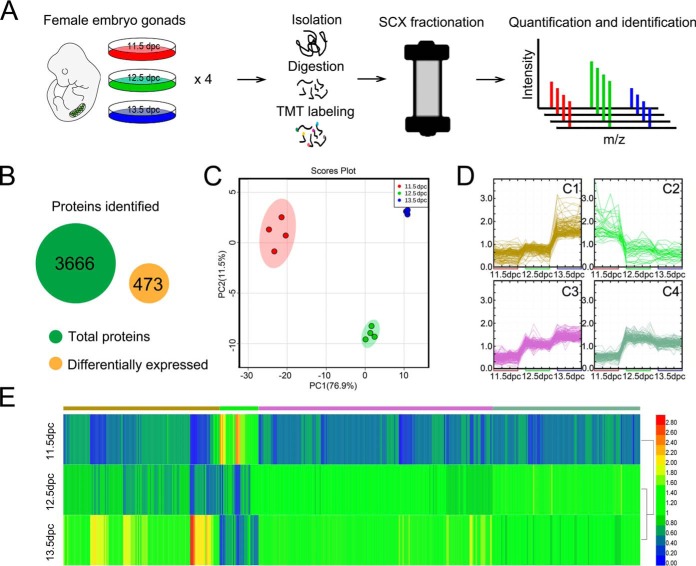
We selected three time points (11.5, 12.5, and 13.5 dpc) to construct a specific proteome profile of early gonadal development (Fig. 2A). A total of 3,666 proteins were identified and were quantified with at least one unique peptide. About 68.4% of the proteins were quantified with at least two unique peptides. Thus, we obtained the first quantitative profiles of protein expression during meiosis initiation. We found that 473 proteins were differentially expressed (DE) with at least a 2-fold significant change (Q value <0.01) among samples (Fig. 2B, supplemental Table S2). The results of the PCA indicated clear differences among the three groups, with samples from the same group clustering together. This demonstrated replicate reproducibility and distinctive patterns, findings subsequently verified by unbiased hierarchical clustering analysis (Fig. 2C). The 473 DE proteins were grouped into four classes (C1, C2, C3 and C4), according to four distinct expression patterns, and relatively discrete chronological boundaries emerged, using k-means clustering algorithm (35, 36) by J-Express software (31). The four classes indicated four specific and unique expression patterns (Fig. 2D, supplemental Table S2): evidently increase at 13.5 dpc (128 proteins, C1), evidently decrease from 12.5 dpc (32 proteins, C2), steady increase across all time points (192 proteins, C3), and increase at 12.5 dpc but slightly decrease at 13.5 dpc (121 proteins, C4). Unbiased hierarchical clustering confirmed that global protein expression could distinguish these three time points and four classes (Fig. 2E). We also found that, among these 473 DE proteins, a large percentage belonged to C1 and C3, indicating that most DE proteins related to meiosis initiation were upregulated with time.
Fig. 2.
Proteomics of gonads from 11.5 to 13.5 dpc and global proteomics analyses. A, Schematic of the experimental design for proteomics, with assay results for gonads from 11.5 to 13.5 dpc. B, Peptides corresponding to 3666 proteins. C, Principal components analysis (PCA) of 11.5, 12.5, and 13.5 dpc. For each stage, there were 4 replicates. D, Cluster analysis of the 473 DE proteins, grouped into four clusters (C1, C2, C3, and C4). For each stage, there were 4 replicates. E, Unbiased hierarchical clustering of DE protein expression.
Because each gonad was very small and yielded little protein, only limited quantities of material were available to verify the LC-MS3 results for identified proteins by Western blotting. Therefore, we randomly selected 12 proteins (AXAN2, GATM, RBPMS2, UCHL1, ALDH1A1, AFP, ALDH1A2, COFILIN, GNPDA1, CAV1, CPT1A, and SOAT1) on the basis of their expression patterns to verify the LC-MS3 findings by immunofluorescence staining, a “semi-quantitative” method to gain insight into relative expression levels. Through the comparison of fluorescence intensity, we can find that these results (shown in supplemental Fig. S1) were essentially in agreement with the TMT quantification. Notably, ALDH1A1, a RA-synthesizing enzyme, recently reported to provide a source of endogenous meiosis-inducing RA in the fetal ovary (11, 37), was found in our study. And the result of ALDH1A1 immunofluorescence confirmed an upregulation expression from 11.5 to 13.5dpc (supplemental Fig. S1), it is in accordance with previous studies (37, 38) and indicated the important role of ALDH1A1 and endogenous RA during meiosis initiation.
Gene Annotation of Differentially Expressed Proteins
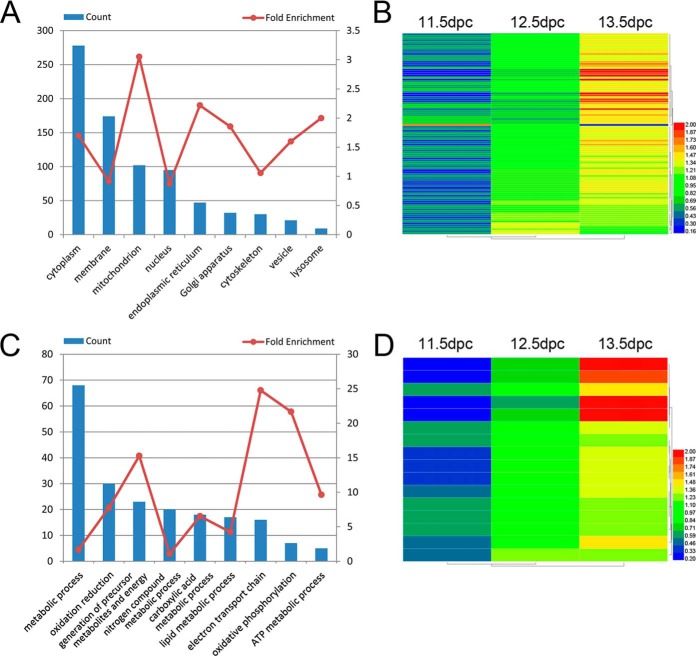
Bioinformatics analyses were performed to determine subcellular localizations of the DE proteins based on gene ontology annotation, using DAVID. Those DE proteins with highest fold enrichment were annotated as mitochondrion, followed by endoplasmic reticulum, lysosome and Golgi apparatus (Fig. 3A) (detailed information is provided in supplemental Table S3). Among the 102 mitochondrion-enriched DE proteins, most were associated with the electron transport chain, oxidative phosphorylation and generation of precursor metabolites and energy (Fig. 3C, supplemental Table S4). Unbiased hierarchical clustering of 102 mitochondrion-enriched and 16 electron transport chain-enriched DE proteins showed that these proteins primarily mapped to patterns C1 and C3, indicating that the mitochondria and electron transport chain were closely linked to meiosis initiation (Figs. 3B and 3D).
Fig. 3.
GO term enrichment analysis and unbiased hierarchical clustering. A, GO term enrichment analysis for cellular components of 437 DE proteins. B, Unbiased hierarchical clustering of expression of 102 mitochondrion-enriched DE proteins. C, GO term enrichment analysis for biological processes of 102 mitochondrion-enriched DE proteins. D, Unbiased hierarchical clustering of expression of 16 electron transport chain-enriched DE proteins.
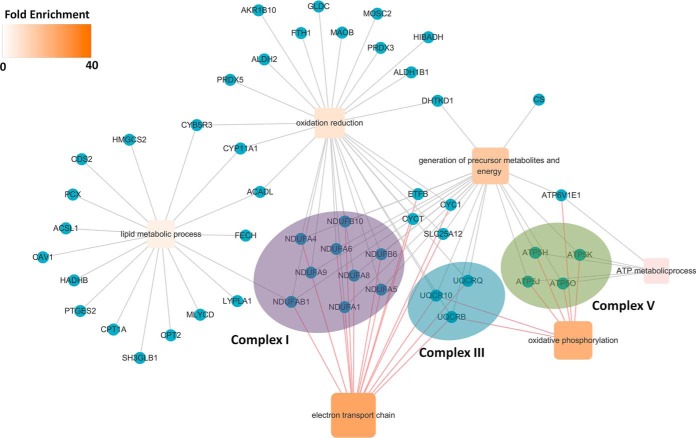
In particular, some of these significant GO terms were closely linked to energy metabolism, so these associated DE proteins were categorized and integrated into these GO biological process terms (Fig. 4, supplemental Table S4). Electron transport chain consists of four major multisubunit complexes designated as NADH ubiquinone reductase (complex I), succinate ubiquinone reductase (complex II), ubiquinol cytochrome c reductase (complex III), and cytochrome-c oxidase (complex IV). In addition, there are 0.5 unit ATP synthase (also called complex V) (39), but GO analyses could not refine these DE proteins to each complex. WikiPathways (http://www.wikipathways.org) is an open, collaborative platform for capturing and disseminating models of biological pathways for data visualization and analysis (40, 41). This database has a detailed gene list of five complexes of electron transport chain (WP295: Electron Transport Chain (Mus musculus), http://www.wikipathways.org/index.php/Pathway:WP295) (supplemental Table S5), thus we further mapped these DE proteins to the mouse electron transport chain, and found that 9 DE proteins were assigned to Complex I, 3 to Complex III, 4 to Complex V, and none to Complexes II and IV. The results suggested that the electron transport chain, especially Complex I, was closely related to meiosis initiation. Therefore, we used ROT, a specific Complex I inhibitor, in further experiments.
Fig. 4.
Differentially expressed proteins enriched for the GO biological process term mapping to the electron transport chain. Circles represent DE proteins (blue), whereas squares represent the GO biological process terms. The square color intensity indicated the fold enrichment of the GO biological process terms. Purple, blue and green ellipses correspond to Complex I, III, and V of electron transport chain (refer to WikiPathways: WP295) respectively. Cytoscape was used to present and visualize the diagram.
Disruption of the Electron Transport Chain Inhibited Female Germ Cell Entry Into Meiosis and CoQ10 Partially Rescued This Phenotype
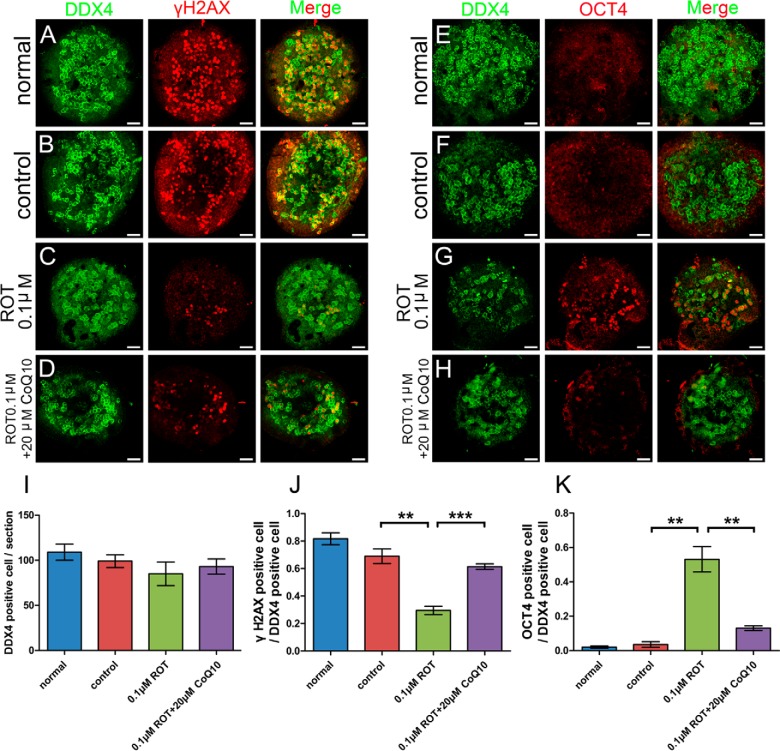
To explore the relationship between the electron respiratory chain and meiosis initiation, we cultured 11.5 dpc female gonads in vitro, treated them with ROT for 24 h and then transferred them to normal culture medium for an additional 2 d. ROT is a type B inhibitor of Complex I, blocking the electron supply to quinol (QH2)-cytochrome c oxidoreductase, resulting in inhibition of oxidative phosphorylation (12). In the ROT treated group, the number of total germ cells (DDX4 positive) was no different than in the control and normal groups, whereas the proportion of cells positive for γH2AX, an accepted meiotic cell marker (33, 34, 42), was significantly lower (Fig. 5A–5C, 5I, 5J). In addition, with ROT treatment, the proportion of cells positive for OCT4, an oogonia marker (43), was clearly increased (Fig. 5E–5G, 5K). These results indicated that the proportion of germ cells entering meiosis decreased with ROT treatment, with more germ cells becoming arrested at the oogonia stage.
Fig. 5.
Disruption of the electron transport chain inhibits entry of female germ cells into meiosis, and CoQ10 rescues the phenotype. Rotenone (0.1 μm) treated (C, G) organ cultures showed decreases in the number of meiotic cells, based on staining for γH2AX (meiotic cell marker), compared with in the normal (A) and control (DMSO) (B) groups; oogonia were clearly increased, based on staining for OCT4 (oogonia marker), compared with staining in the normal (E) and control (DMSO) (F) group; whereas, total germ cell numbers were not different, based on counts of DDX4 (germ cell marker) positive cells (I). Compared with the normal (A, E) and control (B, F) groups, adding 20 μm CoQ10 (D, H) could rescue the phenotype. Bar graphs (I–K) showed the results of stained cell counting. Error bars indicate S.E. and similar experiments were each repeated at least three times (**p < 0.01, ***p < 0.001, Student's t test). Bar, 50μm.
As previously reported, ROT is a type B inhibitor of Complex I and can block oxidative phosphorylation which, in turn, can lead to intracellular ATP depletion and generation of ROS (12, 13, 44). CoQ10 is a well-known antioxidant and can decrease ROS levels (45, 46). After adding CoQ10, we found that the proportion of γH2AX positive cells was significantly higher than in the group treated with only ROT, whereas the proportion of OCT4 positive cells was decreased (Fig. 5D, 5H, 5I–5K). These results demonstrated that CoQ10 partially rescued the severe phenotype caused by disruption of electron transport by ROT.
Because of the severe effects of ROT on female germ cell meiosis initiation, we investigated whether it also affected female somatic cells. We performed immunofluorescence staining of GATA4, which is a transcriptional factor specially expressed in somatic cells in genital ridges (47, 48), the results indicated that the number of somatic cells in female gonads was not noticeably affected by ROT (supplemental Fig. S2A). We also tested whether ROT played a role in male gonads. In 12.0 dpc male gonads cultured in vitro for 24 h, immunofluorescence showed that the morphology of the testis cords was not affected by ROT (supplemental Fig. S2B).
As previously reported (12, 13, 44), ROT disrupts the electron transport chain, causing intracellular ATP depletion and ROS generation. To confirm this, we measured ATP and ROS, finding that ATP was decreased and ROS increased in the ROT treated group. In the presence of CoQ10, this effect was partially rescued, with ATP significantly increased and ROS significantly decreased (supplemental Fig. S3).
Disruption of the Electron Transport Chain Prevented Female Germ Cells from Undergoing Premeiotic DNA Replication
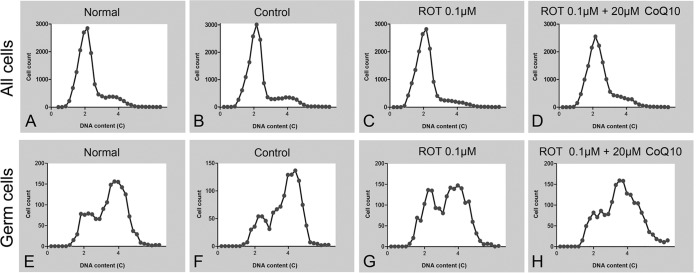
To explore how ROT affected meiotic initiation, we first analyzed DNA contents of entire female gonads, in which germ cells are greatly outnumbered by somatic cells. In the normal and control groups, the proportion of 4C cells increased in 14.5 dpc gonads (Fig. 6A–6B), reflecting an increased number of germ cells having completed premeiotic DNA replication. In contrast, the ROT treated group contained few, if any, 4C cells and their numbers did not increase (Fig. 6C), implying that ROT prevented germ cells from undergoing premeiotic DNA replication. To corroborate and refine this model, we used a germ cell specific antibody (against DDX4) to label the germ cell population in our experiments, while also analyzing DNA content. We observed that a substantial fraction of germ cells in the normal and control groups were 4C in 14.5 dpc gonads (Fig. 6E–6F), again indicating that an increased number of germ cells had completed premeiotic DNA replication by 14.5 dpc. In ROT treated female gonads, in contrast, only a very small fraction of germ cells were 4C at 14.5 dpc (Fig. 6G). When ROT treated gonads were also treated with CoQ10, the number of 4C germ cells was increased (Fig. 6H), again showing that CoQ10 could rescue the ROT-induced phenotype. Together, our findings indicated that, in the female gonads, once the electron transport chain was disrupted, germ cells could not undergo premeiotic DNA replication or subsequent events associated with meiotic prophase.
Fig. 6.
Disruption of the electron transport chain inhibits female germ cells from undergoing premeiotic DNA replication. Histograms of cellular DNA content in female gonads, 12.0 to 14.5 dpc cultures. (A–D) All cells from (A) normal, (B) control, (C) 0.1 μm rotenone, (D) 0.1 μm rotenone plus 20 μm CoQ10 groups. (E–H) Germ cells from (E) normal, (F) control, (G) 0.1 μm rotenone, (H) 0.1 μm rotenone plus 20 μm CoQ10 groups. Total numbers of cells plotted: A, 15,701; B, 15,929; C, 14,423; D, 16,741; E, 1,575; F, 1,960; G, 1,315; H, 1,699.
CoQ10 Inhibited ROT-induced Changes in Expression of Cell Cycle Related Factors, such as p53 and p21, in Germ Cells
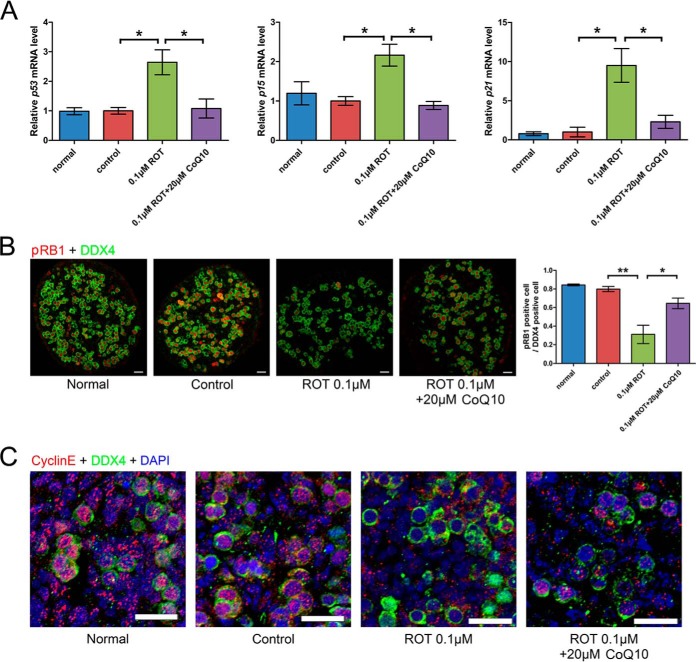
Next, we investigated the potential mechanisms leading to defect of meiosis initiation. As the results in Fig. 5 indicated, germ cells entering meiosis decreased and more germ cell cycle arrest occurred in the ROT treated group. To further validate this result, we examined expression of p53, p15, and p21. With ROT treatment, expression of p53, p15, and p21 was significantly higher than in controls. Addition of CoQ10 led to decreased expression of p53, p15, and p21 compared with in the group receiving only ROT (Fig. 7A). By immunofluorescence, pRB1-positive cells were clearly fewer in number with ROT than in control and normal groups and their numbers increased when CoQ10 was present (Fig. 7B). Immunofluorescence also showed that there were fewer cyclin E-positive germ cells in the ROT treated than in the normal and control groups and that CoQ10 partially rescued this effect. There were, however, no changes in cyclin-E positive somatic cells with ROT (Fig. 7C).
Fig. 7.
Rotenone can lead to germ cell cycle arrest, but CoQ10 can rescue this phenotype. A, Relative expression of p53, p15 and p21 in normal, control, 0.1 μm rotenone and 0.1 μm rotenone plus 20 μm CoQ10. B, Immunofluorescence results showing pRB1-positive germ cells clearly decreased after treatment with 0.1 μm rotenone, compared with in normal and control groups; addition of CoQ10 rescued the phenotype. C, Immunofluorescence also showed that cyclin E-positive germ cells were decreased in the rotenone treated group, compared with in normal and control groups, and CoQ10 could partially rescue this phenotype. Cyclin E-positive somatic cells were not detectably changed in the rotenone-treated group. Error bars indicate S.E. and similar experiments were each repeated at least three times (*p < 0.05, **p < 0.01, Student's t test). Bar, 50 μm.
DISCUSSION
Eukaryotic cells typically contain hundreds of maternally inherited mitochondria, and the mitochondrial and nuclear genomes interact to fuel and perform metabolism (49). Mitochondria provide energy for eukaryotic cells via oxidative phosphorylation (OXPHOS), producing much of the cellular ATP in nonproliferating cells. OXPHOS is accomplished by the five enzymes of the mitochondrial respiratory chain, Complex I (CI, NADH: ubiquinone reductase) is the largest of the ETC (electron transport chain) complexes and is recognized as the major respiratory chain site of ROS production. In addition, one-third of cellular ATP is synthetized during oxidation of intramitochondrial NADH by Complex I (50–53). Compounds that inhibit Complex I activity are divided into three fundamental types, including type B, which includes ROT. ROT is a classical, high affinity and irreversible Complex I inhibitor that blocks the electron supply to quinol (QH2)-cytochrome c oxidoreductase, inhibiting oxidative phosphorylation. This blockage, in turn, leads to intracellular ATP depletion and ROS generation (12, 13, 44).
Sexual reproduction requires formation of specialized germ cells by meiosis, critical for transferring genetic material from one generation to the next. During female embryonic development, once germ cells have reached the gonads, they cease mitosis and enter prophase of the first meiosis at about 13.5 dpc (3, 54). How oogonia are switched from mitosis to meiosis in mice remains a key question in reproductive biology. To further study the mechanisms of meiosis initiation in female germ cells, we used TMT6-MS3 technology to perform comparative proteomic studies. In this manner, we investigated changes in individual protein levels during meiosis initiation in the female mouse, generating a novel data set of 3666 proteins from the female gonads. A total of 473 proteins were differentially expressed with at least a 2-fold significant change (Q value <0.01) among the time points 11.5, 12.5, and 13.5 dpc. Further bioinformatics analysis showed that these DE proteins were enriched in mitochondria, especially in the electron transport chain, with 9 DE proteins in Complex I. The DE proteins enriched in the mitochondria primarily mapped to the patterns C1 and C3, suggesting that the electron transport chain was closely related to meiosis initiation. To confirm this interpretation, we used the Complex I inhibitor ROT to disrupt mitochondrial electron transport chain function. In 11.5 dpc female gonads cultured in vitro, ROT treatment did not affect germ cell number, but numbers of meiotic germ cells and oogonia were significantly decreased and increased, respectively, compared with in the control groups. Interestingly, male gonads were unaffected by ROT. These results indicated that disruption of the electron transport chain inhibited entrance of female germ cells into meiosis. Disruption of the respiratory chain by ROT was reported to lead to high levels of ROS (12, 13, 44), but the well-known antioxidant CoQ10 can decrease ROS levels (45). Therefore, we added CoQ10 along with ROT and found that it could partially rescue its effects. To explore why ROT affected meiotic initiation, we analyzed DNA content in all cells, both germ and somatic, of the female gonads. In the ROT treated gonads, 4C germ cells were decreased and 2C germ cells increased. This indicated that, in the female gonads, once the electron transport chain was disrupted, the germ cells could not undergo premeiotic DNA replication.
Female germ cells leaving mitosis and entering meiosis is a key step in oogenesis. Before meiosis initiation, germ cells leave the G1 phase of mitosis and enter the premeiotic S phase, undergo premeiotic DNA replication and then enter meiosis (55). The premeiotic G1-S transition is not well understood, but extensive literature about the mitotic cell cycle enabled us to conduct a preliminary investigation of this process. The cell cycle is usually divided into four phases, G1, S, G2, and M, regulated by cyclin/cyclin-dependent kinase (CDK) complexes. Cyclin d-CDK4/6 and cyclin E-CDK2 complexes were reported to be involved in G1, especially the cyclin E-CDK2 complex, important for entry into S phase (56, 57). p53 is a transcription factor responding to a variety of stresses to cause cell cycle arrest (58, 59). p15 is a CDK4/6 inhibitor and p21 is a CDK2 inhibitor. RB1 (retinoblastoma protein) is a member of the “Pocket Protein” family and plays an important role in suppressing the G1-S transition through regulation of E2F-responsive genes. RB1 activity is regulated by phosphorylation and, when active, RB1 binds to and inhibits E2F in G0 and early G1. In proliferating cells, RB1 phosphorylation releases E2F, which then induces genes that mediate S phase entry (60, 61). Thus, to further validate the cell cycle arrest in germ cells, we examined p53, p15 and p21 and found that their expression was significantly higher in ROT treated than in control groups, and was significantly decreased by adding CoQ10. Because male gonads were cell cycle arrested at 13.5–14.5 dpc, their levels of p53, p15 and p21 were higher at those times than at 11.5–12.5 dpc (59). Therefore, our results proved that cell cycle arrest of germ cells occurred before entering meiosis with ROT treatment, as compared with in control groups. By immunofluorescence, pRB1-positive germ cell numbers were decreased in the ROT treated group, further confirming germ cell arrest before entering meiosis. In addition, cyclin E-positive germ cells were decreased with ROT treatment, demonstrating that germ cells were arrested at the G1-S transition and, thereby, prevented from undergoing premeiotic DNA replication.
In line with published literature (57, 62, 63), environmental conditions such as nutrient availability and oxidative stress status can also control the cell cycle. Impaired ATP production or significantly higher ROS levels can cause cell cycle arrest. Different levels and durations of ROS exposure influence cell cycle progression differently. Prolonged exposure to ROS could lead to growth arrest and ROS can also influence the cell cycle by regulating MAPK or PI3K signaling pathways. In some reports, decreased ATP levels activated AMPK, leading to p53 activation and cell cycle arrest (64–66). In our study, we found that ATP levels decreased and ROS levels increased in gonads cultured with ROT, compared with in control groups. These effects of ROT were also attenuated by CoQ10. In previous reports, yeast cells showed distinct metabolic requirements at different phases of meiosis. In acetate sporulation medium, the diploid yeast Saccharomyces cerevisiae entered meiosis. The sporulation process required maximal ATP production at the early phase and minimal ATP production at the middle and late stages (67). In our experiments, when the electron transport chain was blocked by ROT, ATP levels decreased and meiosis initiation was inhibited. This disruption of electron transport by moderate ROT treatment caused intracellular ATP to decrease to levels still sufficient to maintain cell survival, but not to enable progression through the cell cycle. This indicated that more energy is required for meiosis initiation than for routine survival. It should be noted that, because we used whole gonadal tissue rather than isolated germ cells, effects of interfering factors in our findings cannot be excluded.
In conclusion, we performed a comparative proteome analysis of female mouse fetal gonads during the critical time window for meiotic initiation in female germ cells. Bioinformatics analysis and further functional exploration suggested a close relationship between the electron transport chain and meiosis initiation. That is, disruption of the electron transport chain prevented female germ cells from entering meiosis by inducing germ cell arrest, suppressing their ability to undergo premeiotic DNA replication before meiosis.
DATA AVAILABILITY
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD005670.
Supplementary Material
Footnotes
* This work was supported by National Nature Science Foundation of China (31471107), National Key Research and Development Program of China (2017YFC1001301, 2016YFA0503300) and China National Basic Research Program (2013CB911400).
 This article contains supplemental Figures and Tables.
This article contains supplemental Figures and Tables.
1 The abbreviations used are:
- dpc
- days post coitum
- ROT
- rotenone
- RA
- retinoic acid
- TMT
- tandem mass tag
- TEAB
- triethylammonium bicarbonate
- SCX
- strong cation exchange
- HCD
- higher-energy collisional dissociation
- FDR
- false discovery rate
- DE
- differentially expressed
- PCA
- principal components analysis
- CoQ10
- Coenzyme Q10
- ROS
- reactive oxygen species
- ETC
- electron transport chain.
REFERENCES
- 1. Kumar S., Chatzi C., Brade T., Cunningham T. J., Zhao X., and Duester G. (2011) Sex-specific timing of meiotic initiation is regulated by Cyp26b1 independent of retinoic acid signalling. Nat. Commun. 2, 151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Menke D. B., Koubova J., and Page D. C. (2003) Sexual differentiation of germ cells in XX mouse gonads occurs in an anterior-to-posterior wave. Developmental Biol. 262, 303–312 [DOI] [PubMed] [Google Scholar]
- 3. Monk M., and McLaren A. (1981) X-chromosome activity in foetal germ cells of the mouse. J. Embryol. Exp. Morphol. 63, 75–84 [PubMed] [Google Scholar]
- 4. Hilscher B., Hilscher W., Bulthoff-Ohnolz B., Kramer U., Birke A., Pelzer H., and Gauss G. (1974) Kinetics of gametogenesis. I. Comparative histological and autoradiographic studies of oocytes and transitional prospermatogonia during oogenesis and prespermatogenesis. Cell Tissue Res. 154, 443–470 [DOI] [PubMed] [Google Scholar]
- 5. McLaren A. (1984) Meiosis and differentiation of mouse germ cells. Symposia Soc. Exp. Biol. 38, 7–23 [PubMed] [Google Scholar]
- 6. Zhou Q., Li Y., Nie R., Friel P., Mitchell D., Evanoff R. M., Pouchnik D., Banasik B., McCarrey J. R., Small C., and Griswold M. D. (2008) Expression of stimulated by retinoic acid gene 8 (Stra8) and maturation of murine gonocytes and spermatogonia induced by retinoic acid in vitro. Biol. Reproduction 78, 537–545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bowles J., Knight D., Smith C., Wilhelm D., Richman J., Mamiya S., Yashiro K., Chawengsaksophak K., Wilson M. J., Rossant J., Hamada H., and Koopman P. (2006) Retinoid signaling determines germ cell fate in mice. Science 312, 596–600 [DOI] [PubMed] [Google Scholar]
- 8. Griswold M. D., Hogarth C. A., Bowles J., and Koopman P. (2012) Initiating meiosis: the case for retinoic acid. Biol. Reproduction 86, 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Spiller C. M., Bowles J., and Koopman P. (2012) Regulation of germ cell meiosis in the fetal ovary. Int. J. Developmental Biol. 56, 779–787 [DOI] [PubMed] [Google Scholar]
- 10. Feng C. W., Bowles J., and Koopman P. (2014) Control of mammalian germ cell entry into meiosis. Mol. Cellular Endocrinol. 382, 488–497 [DOI] [PubMed] [Google Scholar]
- 11. Bowles J., Feng C. W., Miles K., Ineson J., Spiller C., and Koopman P. (2016) ALDH1A1 provides a source of meiosis-inducing retinoic acid in mouse fetal ovaries. Nat. Commun. 7, 10845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Degli Esposti M. (1998) Inhibitors of NADH-ubiquinone reductase: an overview. Biochim. Biophys. Acta 1364, 222–235 [DOI] [PubMed] [Google Scholar]
- 13. Sherer T. B., Betarbet R., Testa C. M., Seo B. B., Richardson J. R., Kim J. H., Miller G. W., Yagi T., Matsuno-Yagi A., and Greenamyre J. T. (2003) Mechanism of toxicity in rotenone models of Parkinson's disease. J. Neurosci. 23, 10756–10764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bowles J., and Koopman P. (2007) Retinoic acid, meiosis and germ cell fate in mammals. Development 134, 3401–3411 [DOI] [PubMed] [Google Scholar]
- 15. Koopman P., Munsterberg A., Capel B., Vivian N., and Lovell-Badge R. (1990) Expression of a candidate sex-determining gene during mouse testis differentiation. Nature 348, 450–452 [DOI] [PubMed] [Google Scholar]
- 16. Zheng B., Zhao D., Zhang P., Shen C., Guo Y., Zhou T., Guo X., Zhou Z., and Sha J. (2015) Quantitative proteomics reveals the essential roles of stromal interaction molecule 1 (STIM1) in the testicular cord formation in mouse testis. Mol. Cell. Proteomics 14, 2682–2691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Guo X., Zhang P., Qi Y., Chen W., Chen X., Zhou Z., and Sha J. (2011) Proteomic analysis of male 4C germ cell proteins involved in mouse meiosis. Proteomics 11, 298–308 [DOI] [PubMed] [Google Scholar]
- 18. Dayon L., Hainard A., Licker V., Turck N., Kuhn K., Hochstrasser D. F., Burkhard P. R., and Sanchez J. C. (2008) Relative quantification of proteins in human cerebrospinal fluids by MS/MS using 6-plex isobaric tags. Anal. Chem. 80, 2921–2931 [DOI] [PubMed] [Google Scholar]
- 19. Lee M. V., Topper S. E., Hubler S. L., Hose J., Wenger C. D., Coon J. J., and Gasch A. P. (2011) A dynamic model of proteome changes reveals new roles for transcript alteration in yeast. Mol. Syst. Biol. 7, 514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zheng B., Zhou Q., Guo Y., Shao B., Zhou T., Wang L., Zhou Z., Sha J., Guo X., and Huang X. (2014) Establishment of a proteomic profile associated with gonocyte and spermatogonial stem cell maturation and differentiation in neonatal mice. Proteomics 14, 274–285 [DOI] [PubMed] [Google Scholar]
- 21. Wen F. P., Guo Y. S., Hu Y., Liu W. X., Wang Q., Wang Y. T., Yu H. Y., Tang C. M., Yang J., Zhou T., Xie Z. P., Sha J. H., Guo X., and Li W. (2016) Distinct temporal requirements for autophagy and the proteasome in yeast meiosis. Autophagy 12, 671–688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Vizcaino J. A., Deutsch E. W., Wang R., Csordas A., Reisinger F., Rios D., Dianes J. A., Sun Z., Farrah T., Bandeira N., Binz P. A., Xenarios I., Eisenacher M., Mayer G., Gatto L., Campos A., Chalkley R. J., Kraus H. J., Albar J. P., Martinez-Bartolome S., Apweiler R., Omenn G. S., Martens L., Jones A. R., and Hermjakob H. (2014) ProteomeXchange provides globally coordinated proteomics data submission and dissemination. Nat. Biotechnol. 32, 223–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Cox J., and Mann M. (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 [DOI] [PubMed] [Google Scholar]
- 24. Kersey P. J., Duarte J., Williams A., Karavidopoulou Y., Birney E., and Apweiler R. (2004) The International Protein Index: an integrated database for proteomics experiments. Proteomics 4, 1985–1988 [DOI] [PubMed] [Google Scholar]
- 25. Deutsch E. W., Mendoza L., Shteynberg D., Farrah T., Lam H., Tasman N., Sun Z., Nilsson E., Pratt B., Prazen B., Eng J. K., Martin D. B., Nesvizhskii A. I., and Aebersold R. (2010) A guided tour of the Trans-Proteomic Pipeline. Proteomics 10, 1150–1159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Xia J., Psychogios N., Young N., and Wishart D. S. (2009) MetaboAnalyst: a web server for metabolomic data analysis and interpretation. Nucleic Acids Res. 37, W652–W660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Deng W., Wang Y., Liu Z., Cheng H., and Xue Y. (2014) HemI: a toolkit for illustrating heatmaps. PloS one 9, e111988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Huang da W, Sherman B. T., and Lempicki R. A. (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protocols 4, 44–57 [DOI] [PubMed] [Google Scholar]
- 29. Dennis G. Jr, Sherman B. T., Hosack D. A., Yang J., Gao W., Lane H. C., and Lempicki R. A. (2003) DAVID: Database for annotation, visualization, and integrated discovery. Genome Biol. 4, P3. [PubMed] [Google Scholar]
- 30. Baltus A. E., Menke D. B., Hu Y. C., Goodheart M. L., Carpenter A. E., de Rooij D. G., and Page D. C. (2006) In germ cells of mouse embryonic ovaries, the decision to enter meiosis precedes premeiotic DNA replication. Nat. Genetics 38, 1430–1434 [DOI] [PubMed] [Google Scholar]
- 31. Dysvik B., and Jonassen I. (2001) J-Express: exploring gene expression data using Java. Bioinformatics 17, 369–370 [DOI] [PubMed] [Google Scholar]
- 32. McLaren A. (2003) Primordial germ cells in the mouse. Developmental Biol. 262, 1–15 [DOI] [PubMed] [Google Scholar]
- 33. Anderson E. L., Baltus A. E., Roepers-Gajadien H. L., Hassold T. J., de Rooij D. G., van Pelt A. M., and Page D. C. (2008) Stra8 and its inducer, retinoic acid, regulate meiotic initiation in both spermatogenesis and oogenesis in mice. Proc. Natl. Acad. Sci. U.S.A. 105, 14976–14980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mahadevaiah S. K., Turner J. M., Baudat F., Rogakou E. P., de Boer P., Blanco-Rodriguez J., Jasin M., Keeney S., Bonner W. M., and Burgoyne P. S. (2001) Recombinational DNA double-strand breaks in mice precede synapsis. Nat. Genetics 27, 271–276 [DOI] [PubMed] [Google Scholar]
- 35. Do J. H., and Choi D. K. (2008) Clustering approaches to identifying gene expression patterns from DNA microarray data. Mol. Cells 25, 279–288 [PubMed] [Google Scholar]
- 36. Akopov A. S., Moskovtsev A. A., Dolenko S. A., and Savina G. D. (2013) [Cluster analysis in biomedical researches]. Patologicheskaia fiziologiia i eksperimental'naia terapiia, 84–96 [PubMed] [Google Scholar]
- 37. Mu X., Wen J., Guo M., Wang J., Li G., Wang Z., Wang Y., Teng Z., Cui Y., and Xia G. (2013) Retinoic acid derived from the fetal ovary initiates meiosis in mouse germ cells. J. Cell. Physiol. 228, 627–639 [DOI] [PubMed] [Google Scholar]
- 38. Bowles J., Feng C. W., Knight D., Smith C. A., Roeszler K. N., Bagheri-Fam S., Harley V. R., Sinclair A. H., and Koopman P. (2009) Male-specific expression of Aldh1a1 in mouse and chicken fetal testes: implications for retinoid balance in gonad development. Develop. Dynamics 238, 2073–2080 [DOI] [PubMed] [Google Scholar]
- 39. Lenaz G., and Genova M. L. (2007) Kinetics of integrated electron transfer in the mitochondrial respiratory chain: random collisions vs. solid state electron channeling. Am. J. Physiol. Cell Physiol. 292, C1221–C1239 [DOI] [PubMed] [Google Scholar]
- 40. Kelder T., van Iersel M. P., Hanspers K., Kutmon M., Conklin B. R., Evelo C. T., and Pico A. R. (2012) WikiPathways: building research communities on biological pathways. Nucleic Acids Res. 40, D1301–D1307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kutmon M., Riutta A., Nunes N., Hanspers K., Willighagen E. L., Bohler A., Melius J., Waagmeester A., Sinha S. R., Miller R., Coort S. L., Cirillo E., Smeets B., Evelo C. T., and Pico A. R. (2016) WikiPathways: capturing the full diversity of pathway knowledge. Nucleic Acids Research 44, D488–D494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Guerquin M. J., Duquenne C., Lahaye J. B., Tourpin S., Habert R., and Livera G. (2010) New testicular mechanisms involved in the prevention of fetal meiotic initiation in mice. Developmental Biol. 346, 320–330 [DOI] [PubMed] [Google Scholar]
- 43. Byskov A. G., Hoyer P. E., Yding Andersen C., Kristensen S. G., Jespersen A., and Mollgard K. (2011) No evidence for the presence of oogonia in the human ovary after their final clearance during the first two years of life. Human Reproduction 26, 2129–2139 [DOI] [PubMed] [Google Scholar]
- 44. Greenamyre J. T., Betarbet R., and Sherer T. B. (2003) The rotenone model of Parkinson's disease: genes, environment and mitochondria. Parkinsonism Related Disorders 9, S59–S64 [DOI] [PubMed] [Google Scholar]
- 45. Beyer R. E., Segura-Aguilar J., Di Bernardo S., Cavazzoni M., Fato R., Fiorentini D., Galli M. C., Setti M., Landi L., and Lenaz G. (1996) The role of DT-diaphorase in the maintenance of the reduced antioxidant form of coenzyme Q in membrane systems. Proc. Natl. Acad. Sci. U.S.A. 93, 2528–2532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lenaz G., Bovina C., D'Aurelio M., Fato R., Formiggini G., Genova M. L., Giuliano G., Merlo Pich M., Paolucci U., Parenti Castelli G., and Ventura B. (2002) Role of mitochondria in oxidative stress and aging. Ann. N.Y. Acad. Sci. 959, 199–213 [DOI] [PubMed] [Google Scholar]
- 47. Hu Y. C., Okumura L. M., and Page D. C. (2013) Gata4 is required for formation of the genital ridge in mice. PLoS Genetics 9, e1003629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Efimenko E., Padua M. B., Manuylov N. L., Fox S. C., Morse D. A., and Tevosian S. G. (2013) The transcription factor GATA4 is required for follicular development and normal ovarian function. Developmental Biol. 381, 144–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Shoffner J. M. (1996) Maternal inheritance and the evaluation of oxidative phosphorylation diseases. Lancet 348, 1283–1288 [DOI] [PubMed] [Google Scholar]
- 50. DegliEsposti M., Ballester F., Timoneda J., Crimi M., and Lenaz G. (1990) The oxidation of ubiquinol by the isolated Rieske iron-sulfur protein in solution. Arch. Biochem. Biophys. 283, 258–265 [DOI] [PubMed] [Google Scholar]
- 51. Trumpower B. L. (1981) New concepts on the role of ubiquinone in the mitochondrial respiratory chain. J. Bioenergetics Biomembranes 13, 1–24 [DOI] [PubMed] [Google Scholar]
- 52. Weiss H., Friedrich T., Hofhaus G., and Preis D. (1991) The respiratory-chain NADH dehydrogenase (complex I) of mitochondria. Eur. J. Biochem. 197, 563–576 [DOI] [PubMed] [Google Scholar]
- 53. Walker J. E. (1992) The NADH:ubiquinone oxidoreductase (complex I) of respiratory chains. Quarterly Rev. Biophys. 25, 253–324 [DOI] [PubMed] [Google Scholar]
- 54. Speed R. M. (1982) Meiosis in the foetal mouse ovary. I. An analysis at the light microscope level using surface-spreading. Chromosoma 85, 427–437 [DOI] [PubMed] [Google Scholar]
- 55. Spiller C. M., and Koopman P. (2011) Cell cycle control of germ cell differentiation. Results Problems Cell Differentiation 53, 269–308 [DOI] [PubMed] [Google Scholar]
- 56. Ohtsubo M., Theodoras A. M., Schumacher J., Roberts J. M., and Pagano M. (1995) Human cyclin E, a nuclear protein essential for the G1-to-S phase transition. Mol. Cell. Biol. 15, 2612–2624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Boonstra J., and Post J. A. (2004) Molecular events associated with reactive oxygen species and cell cycle progression in mammalian cells. Gene 337, 1–13 [DOI] [PubMed] [Google Scholar]
- 58. Hasty P., and Christy B. A. (2013) p53 as an intervention target for cancer and aging. Pathobiol. Aging Age Related Dis. 3, 22702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. De Felici M., and Farini D. (2012) The control of cell cycle in mouse primordial germ cells: old and new players. Current Pharmaceutical Design 18, 233–244 [DOI] [PubMed] [Google Scholar]
- 60. Spiller C. M., Wilhelm D., and Koopman P. (2010) Retinoblastoma 1 protein modulates XY germ cell entry into G1/G0 arrest during fetal development in mice. Biol. Reproduction 82, 433–443 [DOI] [PubMed] [Google Scholar]
- 61. Cobrinik D. (2005) Pocket proteins and cell cycle control. Oncogene 24, 2796–2809 [DOI] [PubMed] [Google Scholar]
- 62. Carmeliet P., Dor Y., Herbert J. M., Fukumura D., Brusselmans K., Dewerchin M., Neeman M., Bono F., Abramovitch R., Maxwell P., Koch C. J., Ratcliffe P., Moons L., Jain R. K., Collen D., and Keshert E. (1998) Role of HIF-1alpha in hypoxia-mediated apoptosis, cell proliferation and tumour angiogenesis. Nature 394, 485–490 [DOI] [PubMed] [Google Scholar]
- 63. Owusu-Ansah E., Yavari A., Mandal S., and Banerjee U. (2008) Distinct mitochondrial retrograde signals control the G1-S cell cycle checkpoint. Nat. Genetics 40, 356–361 [DOI] [PubMed] [Google Scholar]
- 64. Chae H. D., Lee M. R., and Broxmeyer H. E. (2012) 5-Aminoimidazole-4-carboxyamide ribonucleoside induces G(1)/S arrest and Nanog downregulation via p53 and enhances erythroid differentiation. Stem Cells 30, 140–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Moiseeva O., Bourdeau V., Roux A., Deschenes-Simard X., and Ferbeyre G. (2009) Mitochondrial dysfunction contributes to oncogene-induced senescence. Mol. Cell. Biol. 29, 4495–4507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Mandal S., Guptan P., Owusu-Ansah E., and Banerjee U. (2005) Mitochondrial regulation of cell cycle progression during development as revealed by the tenured mutation in Drosophila. Developmental Cell 9, 843–854 [DOI] [PubMed] [Google Scholar]
- 67. Ray D., and Ye P. (2013) Characterization of the metabolic requirements in yeast meiosis. PloS One 8, e63707. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD005670.