Abstract
Auxin response factors (ARFs) play important roles in regulating plant growth and development and response to environmental stress. An exhaustive analysis of the CaARF family was performed using the latest publicly available genome for pepper (Capsicum annuum L.). In total, 22 non-redundant CaARF gene family members in six classes were analyzed, including chromosome locations, gene structures, conserved motifs of proteins, phylogenetic relationships and Subcellular localization. Phylogenetic analysis of the ARFs from pepper (Capsicum annuum L.), tomato (Solanum lycopersicum L.), Arabidopsis and rice (Oryza sativa L.) revealed both similarity and divergence between the four ARF families, and aided in predicting biological functions of the CaARFs. Furthermore, expression profiling of CaARFs was obtained in various organs and tissues using quantitative real-time RT-PCR (qRT-PCR). Expression analysis of these genes was also conducted with various hormones and abiotic treatments using qRT-PCR. Most CaARF genes were regulated by exogenous hormone treatments at the transcriptional level, and many CaARF genes were altered by abiotic stress. Systematic analysis of CaARF genes is imperative to elucidate the roles of CaARF family members in mediating auxin signaling in the adaptation of pepper to a challenging environment.
Keywords: abiotic stresses, auxin, auxin response factor, Capsicum annuum, expression pattern
1. Introduction
Auxin plays crucial roles in several aspects of plant growth and developmental processes, including embryogenesis, vascular elongation, apical dominance, root architecture, flower and fruit development, organ initiation and patterning, secondary metabolism, and responses to environmental stimuli [1,2,3]. Auxin response factors (ARFs) and auxin/indole-3-acetic acid (Aux/IAA) are important transcription factor families that can modulate/regulate the expression of auxin early-responsive gene families including Small Auxin Up RNA (SAUR) and Gretchen Hagen3 (GH3) gene families [4]. ARFs regulate the expression levels of auxin response genes by binding to the promoters of auxin response elements (AuxREs): TGTCTC or some variant of this element (e.g., TGTCCC or TGTCAC) [5,6].
Most ARFs consists of three conserved domains: a B3 DNA binding domain (DBD) which can recognize AuxREs in the promoter of auxin-responsive genes in N-terminal; a variable middle region (MR) that may function as an activation or repression domain; and a conserved Carboxy-Terminal Domains (CTD), which is participated in protein interaction with Aux/IAA proteins [5,7]. In recent years, several mutants of ARF genes have been screened to study their genetic functions. For example, the leaf senescence of the atarf2 mutant is delayed in Arabidopsis [8]. The genes AtARF2, 7 and 19 were induced by senescence, and mutations in AtARF7 and 19 enhanced atarf2 phenotypes [9]. Atarf3 mutation caused deviant floral meristem and reproductive organs [10]. ETTIN (ETT/AtARF3) and AtARF4 control the development of floral organs and leaves in Arabidopsis [11]. Abnormal formation of vascular bundle and embryonic axis are observed in the atarf5 mutant [12,13]. AtARF6 and AtARF8 affect the timing of flower maturation [14]. Atarf7 mutation blocks the hypocotyl response to blue light and reduces the auxin response [15]. AtARF7/AtARF19 double mutant affect lateral root formation and have abnormal gravitropism in both hypocotyls and roots [16,17]. AtARF10 targeted by AtmicroRNA160 functions in the regulation of seed germination and post-germination [18,19]. In rice (Oryza sativa), OsARF12 is involved in iron homeostasis [20]. Another ARF, OsARF16, plays a role in efficient utilization of iron and is also required for iron deficiency response [21,22]. SlARF3 is involved in the formation of trichomes and epidermal cells in tomato [23]. SlARF7 moderates the auxin response during fruit growth and acts as a negative regulator of fruit set until pollination and fertilization have taken place [24].
Auxin plays a vital role in the spatiotemporal coordination of plant tolerance to stress [25,26]. Auxin signaling is initiated via activation of transcriptional response by the ARF transcription factors. Since AtARF1 was first cloned from Arabidopsis, 17 members in Solanum lycopersicon, 19 in Citrus sinensis, 25 in rice, 24 in Medicago truncatula, 31 in Zea mays, 39 in Populus trichocarpa and 35 in Gossypium raimondii have been identified [6,27,28,29,30,31,32]. However, little is known about the ARF family genes in pepper (Capsicum annuum L.). Pepper is an economically important agricultural Solanaceous vegetable and is susceptible to abiotic stresses, including salinity and extreme temperature. In this study, we provide comprehensive information on the 22 CaARF genes, and investigate their expression patterns after exposure to salt and extreme temperatures, as well as various hormone treatments.
2. Results
2.1. Genome-Wide Identification of CaARF Genes from Pepper
After an extensive BLAST search, 22 non-redundant ARF genes were identified in pepper. These CaARFs were named as CaARF1–CaARF22 according to their locations on chromosomes (Table 1, Figure 1). Information on CaARF gene families such as gene names, gene IDs, open reading frame (ORF) lengths, number of exons, chromosomal localization, domains and parameters for the deduced polypeptides are listed in Table 1. The number of ARF genes in pepper is similar to that in Arabidopsis (23), rice (25) and M. truncatula (24), and more than the number in tomato (17). The ORF sizes for the CaARF genes varied from 621 bp (CaARF14) to 3384 bp (CaARF9), and the sizes of deduced CaARF proteins were varied 206 from 1125 amino acids. The predicted molecular weights of CaARF proteins ranged from 23,695.03 Da (CaARF14) to 124,451.92 Da (CaARF9). The predicted pI ranged from 5.190 (CaARF4) to 10.386 (CaARF6). Pair-wise analysis of CaARF proteins indicated that the overall identity fell in a range from 22.39% (between CaARF5 and CaARF22) to 90.65% (between CaARF6 and CaARF22) (Figure S1).
Table 1.
Characteristics of CaARF genes in pepper (Capsicum annuum).
| Gene | Locus ID | ORF Length(bp) | No. of Exons | Chromosome | Domains | Deduced Polypeptid | ||
|---|---|---|---|---|---|---|---|---|
| No. | Length(aa) | Mw(Da) | pI | |||||
| CaARF1 | Capana01g000109 | 1869 | 13 | 1 | DBD, ARF, PB1 | 622 | 69,361.85 | 6.608 |
| CaARF2 | Capana02g001847 | 2487 | 11 | 2 | DBD, ARF | 828 | 90,404.2 | 7.4 |
| CaARF3 | Capana03g000640 | 2535 | 14 | 3 | DBD, ARF, PB1 | 844 | 94,051.49 | 6.528 |
| CaARF4 | Capana04g000259 | 2808 | 9 | 4 | DBD, ARF, PB1 | 935 | 102,999.67 | 5.19 |
| CaARF5 | Capana04g001778 | 1749 | 2 | 4 | DBD, ARF | 582 | 64,136.74 | 6.707 |
| CaARF6 | Capana04g002497 | 660 | 6 | 4 | DBD, ARF | 219 | 24,895.03 | 10.386 |
| CaARF7 | Capana05g001658 | 3288 | 13 | 5 | DBD, ARF, PB1 | 1095 | 120,930.6 | 6.207 |
| CaARF8 | Capana06g000514 | 2055 | 4 | 6 | DBD, ARF | 684 | 76,040.43 | 5.988 |
| CaARF9 | Capana07g000865 | 3384 | 15 | 7 | DBD, ARF, PB1 | 1127 | 124,451.92 | 6.417 |
| CaARF10 | Capana07g000989 | 2745 | 16 | 7 | DBD, ARF, PB1 | 914 | 102,060.74 | 5.772 |
| CaARF11 | Capana08g001056 | 2049 | 13 | 8 | DBD, ARF, PB1 | 682 | 76,246.12 | 6.411 |
| CaARF12 | Capana08g001774 | 1980 | 14 | 8 | DBD, ARF, PB1 | 659 | 73,752.73 | 6.273 |
| CaARF13 | Capana09g000452 | 2463 | 16 | 9 | DBD, ARF | 820 | 90,274.66 | 5.938 |
| CaARF14 | Capana09g001199 | 621 | 7 | 9 | DBD, ARF | 206 | 23,695.03 | 8.465 |
| CaARF15 | Capana09g002164 | 2082 | 3 | 9 | DBD, ARF | 693 | 76,014.18 | 6.753 |
| CaARF16 | Capana10g001461 | 1134 | 2 | 10 | DBD, ARF | 377 | 42,514.83 | 7.345 |
| CaARF17 | Capana11g000076 | 2067 | 4 | 11 | DBD, ARF | 688 | 76,060.18 | 8.296 |
| CaARF18 | Capana11g000105 | 2259 | 12 | 11 | DBD, ARF | 752 | 83,911.87 | 6.353 |
| CaARF19 | Capana12g001354 | 2499 | 14 | 12 | DBD, ARF, PB1 | 832 | 92,780.98 | 6.568 |
| CaARF20 | Capana00g000236 | 1833 | 7 | 0 | DBD, ARF | 610 | 68,804.93 | 6.586 |
| CaARF21 | Capana00g001906 | 2676 | 14 | 0 | DBD, ARF, PB1 | 891 | 98,644.16 | 6.208 |
| CaARF22 | Capana00g004127 | 1986 | 9 | 0 | DBD, ARF | 661 | 74,277.79 | 6.294 |
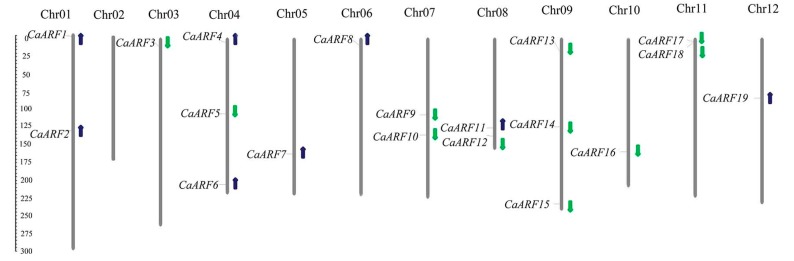
Figure 1.
Chromosomal distribution analysis of CaARF family genes in pepper. MapInspect software was used to draw the location images of CaARF genes. Pepper chromosomes were arranged in blocks by its relative length. The chromosome number is indicated at the top of each chromosome. 19 MtARF genes were among eight chromosomes except chromosomes 2. The direction of transcriptions were indicated by arrows. The scale is megabases (Mb).
2.2. Chromosomal Distributions
Based on the start positions of the CaARF genes on chromosomes, we mapped all 22 genes onto the chromosomes (Table 1 and Figure 1). There were 19 CaARF genes unevenly mapped on 11 out of 12 pepper chromosomes, except for chromosome 2. Three unmapped CaARF genes were present on a pseudo-chromosome (chr00). Chromosomes 3, 5, 6, 10 and 12 contained only one CaARF gene; two genes were located on chromosomes 1, 7, 8 and 11; and three CaARF genes were distributed on chromosomes 4 and 9 (Table 1, Figure 1).
2.3. Phylogenetic Relationship Analyses and Gene Structure
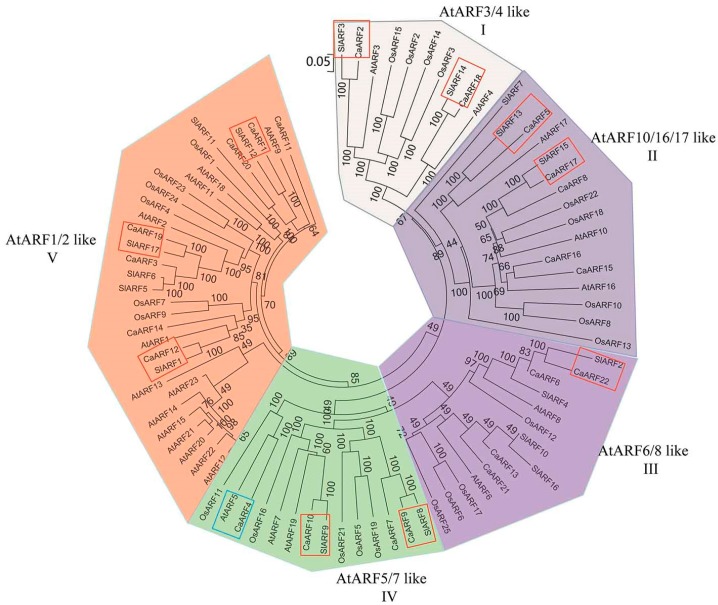
The biological functions of several ARF genes have been reported in Arabidopsis, rice and tomato [8,14,15,20,24]. Investigations into the evolutionary relationships of ARF proteins among pepper, Arabidopsis, tomato and rice would booster our understanding of the possible biological functions of these genes. The amino acid alignments of the ARF family proteins from Arabidopsis, tomato, rice and pepper were performed using the Clustal Omega program. A phylogenetic tree comprising 87 ARF genes from Arabidopsis (23), tomato (17), rice (25) and pepper (22) was constructed using MEGA6.0 software with the neighbor-joining method (Figure 2). The phylogenetic distribution showed that the ARF family could be classified into five major classes: I (AtARF3/4-like), II (AtARF10/16/17-like), III (AtARF6/8/10-like), IV (AtARF5/7-like), and V (AtARF1/2-like). CaARF2 and CaARF18 from pepper were clustered in class I; 5 members (CaARF5, 8, 15, 16 and 17 in class II; CaARF6, 13, 21 and 22 in class III. CaARF4, 7, 9 and 10 in class IV; and CaARF1, 3, 11, 12, 14, 19 and 20 in class V. Based on the phylogenetic tree, 10 sister-gene pairs were identified between tomato and pepper: CaARF1/SlARF12, CaARF2/SlARF3, CaARF5/SlARF13, CaARF9/SlARF8, CaARF10/SlARF9, CaARF18/SlARF14, CaARF12/SlARF1, CaARF19/SlARF17, CaARF17/SlARF15 and CaARF22/SlARF2. A sister gene pair was identified between Arabidopsis and pepper: CaARF4/AtARF5.
Figure 2.
Phylogenetic relationship analysis of ARF family between Pepper, tomato, Arabidopsis and rice. All branches were marked with bootstrap values. The orthologous genes between pepper and tomato were indicated by red boxes. The orthologous genes between pepper and Arabidopsis were indicated by blue boxes.
The intron-exon structures of CaARF genes were determined by comparison of cDNA sequences with genomic DNA sequences. This gene sequence structure revealed introns in all CaARF genes. There were 2 to 16 exons in CaARF genes (Figure S2).
2.4. Analysis of Protein Structure and Prediction of miRNA Targets among CaARFs
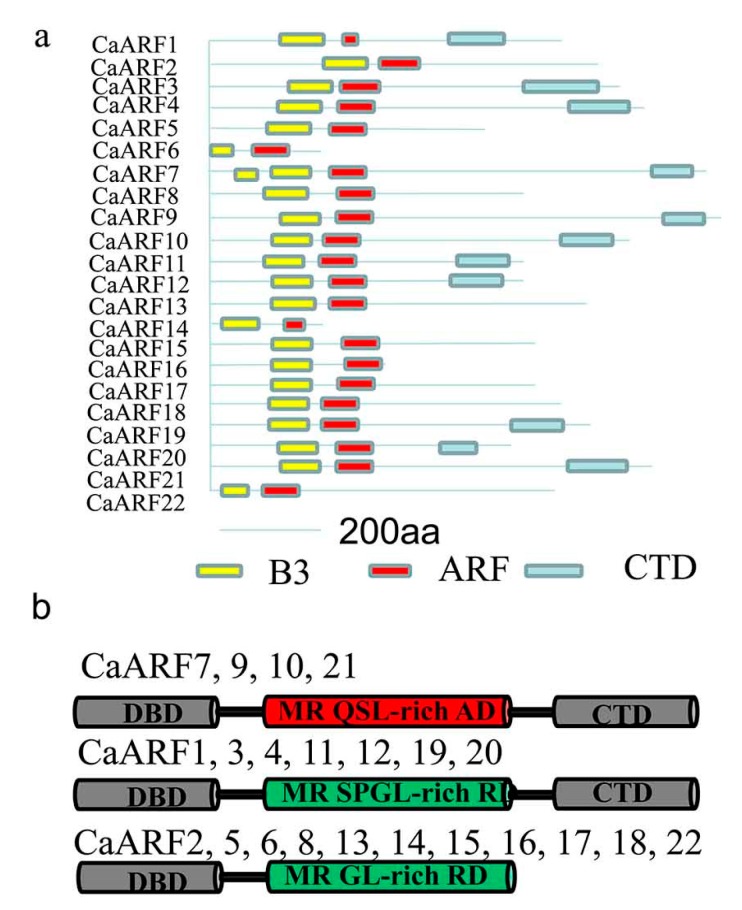
The 22 CaARF proteins were divided into three groups, based on their variable middle region (MR) amino acid composition and the presence or absence of CTD: (1) ARF proteins with a B3 DNA binding domain (DBD), activator MR (MR enriched with glutamine (Q), serine (S), and leucine (L)) and a CTD; (2) ARF proteins with a DBD, repressor MR (MR enriched with S, L, proline (P) and glycine (G)) and a CTD; and (3) ARF proteins with a DBD and repressor MR, but no CTD (Figure 3). The CaARF family encodes four putative transcriptional activators: CaARF7, 9, 10 and 21; and seven putative transcriptional repressors: CaARF1, 3, 4, 11, 12, 19 and 20. Eleven CaARF proteins did not contain a CTD: CaARF2, 5, 6, 8, 13, 14, 15, 16, 17, 18 and 22 (Figure 3).
Figure 3.
Analysis of ARF protein structures and domains. (a) Depiction of the domain structure of each CaARF protein sequence. The B3 DNA bindingdomain (BDB), auxin response factor domain (ARF) and Carboxy-Terminal Domains (CTD) are colored in yellow, red and light blue, respectively. (b) Three kinds of CaARF protein structures. DBD, DNA-binding domain; CTD, C-terminal dimerization domain; MR, middle region; RD, repression domain, showed in green color ; AD, activation domain, showed in red color; Q, glutamine; S, serine; L, leucine; P, proline; G, glycine.
To further investigate conserved motifs, the Multiple Expectation Maximization for Motif Elicitation (MEME) software was employed to characterize the distribution of motifs in 22 CaARF proteins. There 15 motifs identified and mapped to the CaARF amino acid sequences (Figure S3). CaARF7 and CaARF9 contained the greatest number (14 motifs), and CaARF14 the fewest motifs (five motifs). Motifs 1, 2 and 7 corresponded to the B3-DBDs; motifs5, 8, 12 and 15 corresponded to ARF domain; and motifs 4 and 10 corresponded to the OPCA-like motif((D-x-D/E-x-D-xn-D/E) known as the octicosapeptide repeat, p40phox and budding yeast Cdc24p, a typical PKC interaction domain (OPCA) motif) and conserved lysine motif of the PB1 domain, which function in protein-protein interaction with Aux/IAA [33,34].
Previous studies showed that expression levels of several ARF genes were regulated by miRNA in Arabidopsis. For example, AtARF6 and 8 were targeted by AtmiRNA167 [35,36,37,38]; and AtARF10, 16, and 17 regulated by AtmiRNA160 [18,19]. To explore whether there was potential regulation of CaARF genes by miRNA, putative target sites were searched using the BLASTN algorithm. Target sites of AtmiRNA160 (UGCCUGGCUCCCUGGAUGCCA) were predicted within the mRNA region of CaARF5, 8, 15 and 17 (Figure S4a). Additionally, target sites of AtmiRNA 167 (UCAAGCUGCCUGCAUGAUCUA) were predicted within the mRNA region of CaARF13, 21 and 22 (Figure S4b). The phylogenetic tree placed CaARF13, 21 and 22, AtARF6 and 8 in class III; and CaARF5, 8, 15, and 17, AtARF10, 16 and 17 in class II. The results of miRNA targets analysis suggested that miRNA160-/167-mediated post-transcriptional regulation of ARFs was conserved between pepper and Arabidopsis.
2.5. Analyses of Tissue-Specific Expressions and Subcellular Location
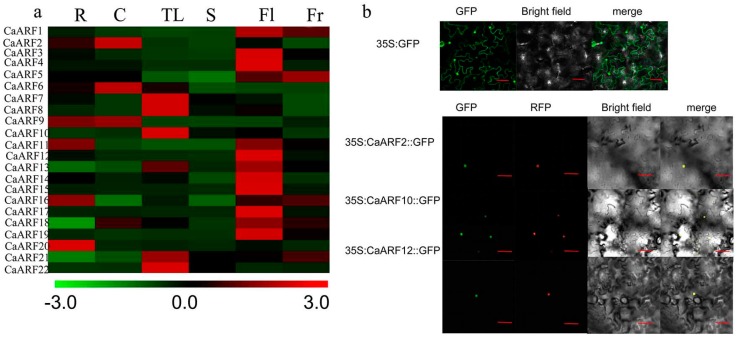
To probe the biological function of CaARF genes, quantitative real-time RT-PCR (qRT-PCR) was used to determine the spatial specificity expression pattern of 22 CaARF genes in six pepper organs: roots, cotyledons, true leaves, stems, flowers and fruits) (Figure 4a). Transcripts of the 22 CaARF genes were detected in different organs and tissues, indicating multiple functions of CaARF genes in pepper growth and development. Some CaARF genes exhibited organ/tissue-specific expression patterns. CaARF20 was especially expressed in fruits. CaARF2 and CaARF6 were highly expressed in cotyledons. CaARF7, 8, 10, 21 and 22 were expressed more strongly in true leaves than in the other tissues/organs. CaARF1, 3, 4, 12, 14, 15, 17, 18 and 19 were highly expressed in flowers compared with other tissues.
Figure 4.
Tissues-specific expressions analysis of CaARF genes and Subcellular localization of three selected CaARF proteins. (a) Expression patterns of CaARF genes in six tissues. R: roots; C: cotyledons; TL: true leaves; S: stems; Fl: flowers; Fr: fruits. Levels of different colours were shown on expression scale of each CaARF genes. The value of CaACTIN defines as 1000 and the relative mRNA level of individual genes was normalized with respect to the CaACTIN gene. Heat map was draw by MeV software was using the average log10 (expression value) (b) CaARFs and GFP fusion proteins were transiently expressed in tobacco epidermis cells under the CaMV35S promoter. From top to bottom: 35S:GFP, 35S:CaARF2::GFP, 35S:CaARF10::GFP, 35S:CaARF12::GFP. Left to right: GFP fluorescence, red fluorescence of nuclear marker (NSL-mCherry), bright-field, merged microscope images. Bar = 50 μm.
According to previous reports, most ARF proteins were nuclear-localized. Based on the amino acid composition and conserved motifs analysis, CaARF2, CaARF10 and CaARF12 from three different groups were fused in-frame to the N-terminus of the green fluorescence protein (GFP). The subcellular location of CaARF2, CaARF10 and CaARF12 in the epidermis cells of tobacco leaves showed that these proteins were nuclear-localized (Figure 4b).
2.6. Expression Patterns of CaARF Genes in Response to Various Abiotic Stress Treatments
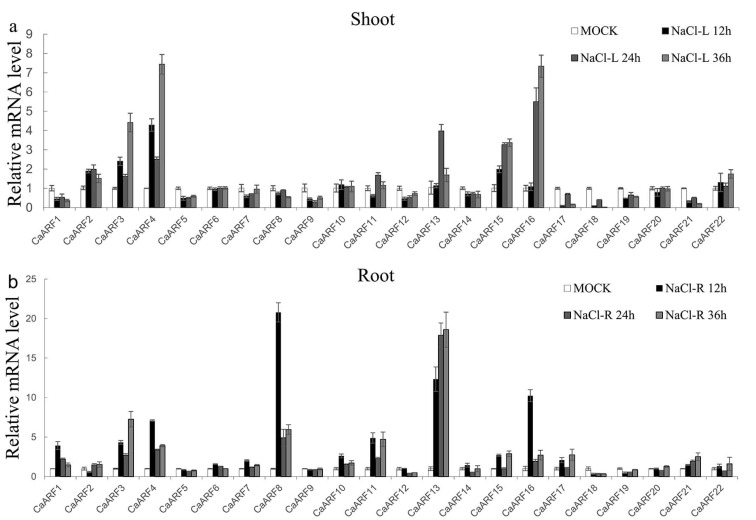
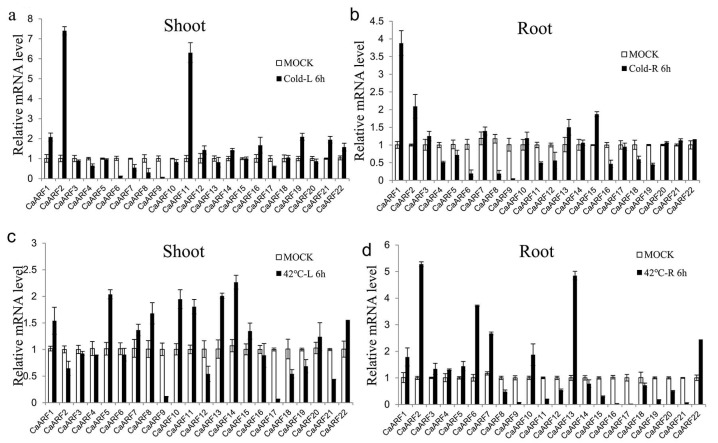
Pepper is sensitive to stress factors, such as extreme temperature, salinity and osmotic stresses [39,40]. Many studies have shown that auxin and its signaling are involved in stress responses, including to drought, cold and salt [36]. In this study, expression levels of CaARF genes under NaCl, cold (4 °C) and heat stress (42 °C) treatments were determined by qRT-PCR to investigate their potential roles in pepper responses to various abiotic stresses. Under these stresses, the majority of CaARF genes exhibited different expression patterns. Under the NaCl treatment, 10 (45.45%) CaARF genes were down-regulated in shoots and nine (40.9%) were up-regulated in roots (Figure 5). Following the 4 °C treatment, the expression levels of CaARF4, 6, 8 and 17 were declined in both shoots and roots; CaARF1 and 2 were up-regulated in both shoots and roots; and CaARF11 was up-regulated in shoots and down-regulated in roots (Figure 6a). CaARF2, 9, 12, 17, 18, 19 and 21 were down-regulated in shoots after high temperature (42 °C) treatment. CaARF2, 6, 7, 13 and 22 were induced (value > 2) in roots after 42 °C treatment (Figure 6b).
Figure 5.
Expression profiles analysis of CaARF family genes in response to salt stress in both shoots (a) and roots (b). Expression levels of CaARF family genes were analysed by qRT-PCR using 4-week-old pepper seedlings, which were treated with 200 mM NaCl for 36 h. The relative expression levels were normalized to a value of 1 in mock seedlings. Standard deviations were shown with error bars.
Figure 6.
Expression profiles analysis of CaARF family genes in response to cold and heat stress. The expression levels of CaARF genes in cold (a) and heat (c) treated shoots were compared to mock shoots as relative mRNA levels. The expression levels of CaARF genes in cold (b) and heat (d) treated shoots were compared to mock shoots as relative mRNA levels. Expression levels of CaARF family genes were analysed by qRT-PCR using 4-week-old pepper seedlings, which were grown under 4 or 42 °C for 6 h, respectively. The relative expression levels were normalized to a value of 1 in mock seedlings. Standard deviations were shown with error bars.
2.7. Expression of CaARF Genes in Response to Various Hormone Treatments
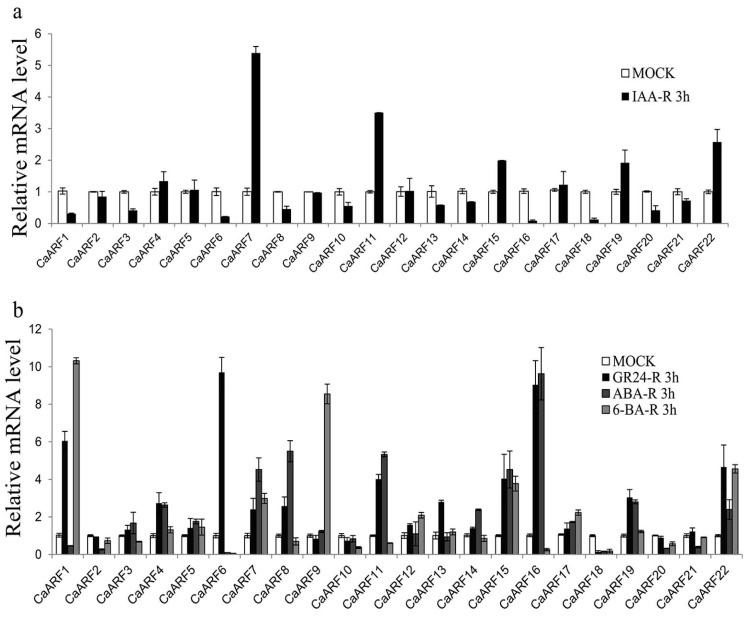
The expression patterns of CaARF genes in response to exogenous IAA stimulation were analyzed using qRT-PCR. The expression levels of CaARF 11, 15, 19 and CaARF22 were induced by IAA treatment (Figure 7a). To investigate whether the CaARF gene family was involved in other hormone signaling pathways, we also analyzed expression profiles of these genes under other various hormone treatments. The transcriptional expression of the CaARF family genes in pepper seedlings were respectively tested using qRT-PCR following treatments with a synthetic analogue of strigolactone (GR24), abscisic acid (ABA) and 6-benzylaminopurine (6-BA) treatments. Following the GR24 treatment, expression levels of CaARF4, 6, 7, 8, 11, 13, 15, 16, 19 and 22 were up-regulated (>2 fold) (Figure 7b). The ABA treatment resulted in down-regulation of CaARF1, 2, 6, 10, 18, 20 and 21. The 6-BA treatment induced up-regulation of CaARF1, 7, 9, 12, 15, 17 and 22 (>2 fold) (Figure 7b). Our results suggested that expression levels of some CaARF genes were responsive to these hormones.
Figure 7.
qRT-PCR analysis of CaARF family genes in roots under various hormone treatments. (a) Expression patterns of CaARF family genes family genes under 10 μM IAA treatment for 3 h. (b) Expression profiles analysis of CaARF family genes under 10 μM GR24, 1 μM ABA, 1 μM 6-BA for 3 h, respectively. Error bars represent standard deviations from 3 biological replicates.
3. Discussion
The phytohormone auxin is involved in regulating organogenesis and patterning processes during plant growth and development [37]. The auxin signaling pathway is mainly composed of two types of transcription factors: ARF and Aux/IAAs [4,38]. The ARFs directly bind to AuxREs in the promoters of down-stream target genes and regulate their expression during development [41]. Genome-wide characterization and analysis of CaARFs would help determine their mechanisms in pepper growth and development. We identified 22 ARF genes in pepper, a similar number to that of Arabidopsis (23). Domain analysis of CaARF proteins provided useful information for predicting their biological functions in pepper. ARFs binding to AuxRE (TGTCTC) in the promoters of auxin-responsive genes rely on the DBD domain. The ARF transcription factors function as activators or repressors, determined by the amino acid composition of MR [42]. The ratio of activator/repressor among ARF proteins in pepper was only 0.22 less than half of that in Arabidopsis (0.59) or rice (0.56) [43]. The CTD/PB1 domain of ARF protein is involved in resembling domains III and IV of Aux/IAA proteins [44]. The ratio of CTD-truncated CaARFs (50%) was much higher than that in soybean (15.68%), Arabidopsis (17.39%), Brassica rapa (22.58%) and rice (24%) [45,46]. These results suggested that some ARF genes in pepper might be regulated in an auxin independent way. Recently, similar work has been published by Zhang et al. [47]. The reason of the differences about the number (19 CaARF genes) and domain structure of the CaARF proteins obtained by Zhang et al. (2017) might be that different databases were used in this study [47]. Zhang et al. (2017) used the pepper genome database (https://solgenomics.net/organism/Capsicum_annuum/genome), SMART, Motif Scan (http://myhits.isb-sib.ch/cgi-bin/PFSCAN) and the MEME web server, while the Pepper Genomics Database 2.0, SMART and MEME were used in our study.
A phylogenetic tree was constructed to analyse the relationship of ARF members between pepper, tomato, rice and Arabidopsis (Figure 2). The phylogenetic tree not only helped elucidate the phylogenetic relationships of ARF proteins, but also allowed speculation on putative functions of the CaARF proteins based on the functional clades previously described in tomato, rice and Arabidopsis. All of the CaARF genes were distributed in five classes, I–V, which were homologous to AtARF3 and 4; AtARF10, 16 and 17; AtARF6 and 8; AtARF5 and 7; and AtARF1 and 2, respectively (Figure 2). The phylogenetic tree also showed 10 sister-gene pairs with high bootstrap values (≥99%) between pepper and tomato. The phylogenetic relationships of these pepper and tomato genes suggested the putative biological functions of the identified CaARFs [29,30]. Analysis of the protein motifs showed that different classes of CaARFs had a conserved structure. Motif 4 and 10, located in the PB1 domain, included a conserved lysine motif and an OPCA-like motif found only in the ARF family, indicating the evolutionary conservation of ARF function [33,48]. All 22 CaARF proteins have B3 and ARF domains that were predicted by SMART (Figure 3a). Many studies have shown that miRNA plays an important role in post-transcriptional gene regulation by combining with complementary targets. The phylogenetic tree showed that CaARF5, 8, 15 and 17, AtARF10, 16, and 17 were in class II; and these four CaARF genes contained a target site for miRNA160. Additionally, CaARF13, 21 and 22, AtARF6 and 8 were clustered in class III, were a target for miRNA167.
Comprehensive expression patterns analysis in different organs/tissues using qRT-PCR helped us screen for CaARF genes with potentially distinct functions (Figure 4). Our data showed that most CaARF genes were highly variably expressed in all six organs/tissues. Their expression patterns suggest that the encoded proteins may perform diverse functions. ARF genes have previously been shown to be involved in regulating plant growth and development [8,15]. AtARF4 was reported to be involved in flower patterning [49]. In pepper, CaARF18 expression was significantly higher in flowers than in other studied organs, and was closely related to AtARF4 in class I; these findings indicate that CaARF18 might play a crucial role in flower development. In Arabidopsis, auxin induces lateral roots and leaf expansion by activating ARF7 and 19 in Arabidopsis [50]. In pepper, CaARF10 belonged to class IV with AtARF7 and 19, and CaARF10 expression was much higher in pepper leaves than in other organs. These observations suggest that CaARF10 likely regulates auxin-induced leaf expansion. A clear increase in transcriptional level of most CaARF genes was observed in adventitious root growth [47].
Phytohormones are involved in the responses of various plants to environmental stimuli and stress by altering the expression levels of many ARF genes [6,51]. However, evaluation of pepper’s adaptability to its environment is very limited. In this study, an expression profile of the CaARF family genes in response to various abiotic stresses and several hormones was created. Pepper is considered to be extremely sensitive to salt, cold and high temperature. However, few studies have examined the responses of pepper to abiotic stresses and related signal transduction. Genome-wide expression analysis showed that the transcriptional level of many ARF genes changes when plants response to abiotic stresses [52,53,54]. Auxin plays a key role in plant responses to abiotic stresses through complex metabolic and signaling networks [55]. In banana, many MaARF genes at the transcriptional level can respond to abiotic stresses [56]. More than half of the GmARF genes are dehydration responsive have been reported in soybean [54]. In this study, we found that many CaARF genes can respond to abiotic stresses including salt, cold and heat at the transcriptional level. Our data indicated essential roles of these genes in response to abiotic stresses in pepper, providing many excellent candidate genes for further studies.
Since ARFs regulate the expression of auxin response genes, it would be interesting to determine the response of CaARF genes to exogenous IAA treatment. It has been reported that AtARF4, 5, 16, 19 and OsARF1 and 23 slightly increased under auxin treatment, whereas OsARF5, 14 and 21 decreased slightly [27,28,57]. In our study, expression of CaARF genes changed rapidly under exogenous auxin treatment compared with the control. In pepper, 12 of the 22 CaARF genes were down-regulated by IAA treatment in the roots. In addition to auxin, crosstalk between hormone and abiotic stress signaling has been reported in some plant species [52,58]. Strigolactones, a group of plant hormones, were recently described to be involved in the repression of shoot branching. Their signaling is required for auxin-dependent stimulation of secondary growth in plants [59]. In Arabidopsis, auxin acts upstream of ABSCISICACID INSENSITIVE 3 (ABI3) by recruiting AtARF10 and 16 to control expression of ABI3 during seed germination [60]. Auxin and cytokinin play an antagonistic role in plant development; however, their interaction is much more complicated, depending on dose and cell type. Recent studies have begun to elucidate the molecular mechanisms involved in auxin–cytokinin interaction in biosynthesis, inactivation/degradation, transport and signal transduction [61,62]. In soybean, miRNA160 promotes auxin activity by suppressing the levels of the ARF10/16/17 family of repressor ARF transcription factors during nodule development. High miR160 levels promote auxin activity and suppress cytokinin activity, but low miR160 levels did the opposite [63].
4. Materials and Methods
4.1. Plant Material and Treatments
Pepper (Capsicum annuum L.) seeds, sown in perlite beds after sterilized by 1% sodium hypochlorite for 30 min. The seedlings were grown in a greenhouse under the following conditions: 16 h light (600 μE m2 s−1) at 26 °C, 8 h dark at 18 °C, and the relative humidity was 60%. Seedlings were irrigated by half-strength of Hoagland solution (pH 5.6). Seedlings at the four-true leaf stages were used for treatments. For salt treatment, plants were soaked in half-strong Hoagland nutrient solution containing 200 mM NaCl. For cold and hot treatments, seedlings were subjected to 4 °C or 42 °C Light incubator, respectively. For hormone treatments, plants were soaked in nutrient solution containing indole-3-acetic acid (IAA, 10 μM), rac-GR24 (10 μM), abscisic acid (ABA, 1 μM), or 6-Benzylaminopurine (6-BA, 1 μM), respectively. Plants grown in normal half-strong Hoagland nutrient solution were used as control. The root and shoot samples of pepper plants were harvested for RNA extraction. For tissue-specific expression analysis, roots, cotyledons, true leaves and stems samples were collected from two true leaf stages; for flower samples, flowers were collected after opening; for fruits samples, fruits were harvested at 15 days after anthesis. All experiments were repeated for 3 times.
4.2. Identification of ARF Family Genes in Pepper
The sequences of CaARF were collected by homology screening against the Pepper Genomics Database 2.0 (http://peppersequence.genomics.cn/page/species/index.jsp). The known sequences of Arabidopsis ARFs (AtARFs) and tomato ARFs (SlARF) were used as queries. Information on AtARFs and SlARF was downloaded from the TAIR database (The Arabidopsis Information Resource, http://www.arabidopsis.org/) and the SGN database (The tomato Information Resource, http://solgenomics.net), respectively. Then, candidate genes were identified based on the hidden Markov model (HMM) profiles of the ARF gene family (Pfam 02362:B3 DNA binding domain (B3-DBD); Pfam 02309: AUX/IAA family (PB1); Pfam 06507:ARF (AUX_RESP)). To exclude redundant genes, all candidate CaARF genes were checked manually. All the obtained sequences are identified as unique sequences for further studies. The DNAstar tool (http://www.dnastar.com/) was employed to predict protein molecular weight (Mw) and isoelectric point (pI) of each CaARF proteins.
4.3. Sequence Analysis, Genome Distribution, Phylogenetic Tree Building and Gene Structures
B3-DBD, ARF and BP1 domains were analyzed using SMART (http://smart.embl-heidelberg.de/) and the Multiple Expectation Maximization for Motif Elicitation (MEME) web server (http://meme.nbcr.net/meme/cgi-bin/meme.cgi). Optimum motif width was set from 6 to 200, and the maximum number of motifs was set to 15. The chromosomal location data of CaARF family genes were obtained from Pepper Genomics Database. A map of the distribution of CaARF family genes was drawn by MapInspect software (http://www.softsea.com/review/MapInspect.html) based on the chromosomal position of each CaARF gene.
The multiple sequence alignment file of ARF proteins from pepper, tomato, Arabidopsis and rice was generated by ClustalW program with the default parameters. Information on SlARF, AtARF and OsARF proteins was according to previous reports [6,27,32]. A neighbour-joining tree was constructed by MEGA6.0 (http://www.megasoftware.net/) with the p-distance and complete deletion parameters. Bootstrap analysis was calculated from 1000 iterations. Exon-intron structures of CaARF family genes were employed by Gene Structure Display Server (GSDS) tool (http://gsds.cbi.pku.edu.cn/) according to the full-length coding sequence and genome from Pepper Genomics Database.
4.4. Subcellular Localization
The ORFs of CaARF2, 10 and 12 were cloned from pepper cDNA and integrated into the pEASY T1 vector (code: CT101-01, Transgen, Beijing, China). Then they were ultimately digested with Kpn I and Sal I, and ligated into the pCAMBIA1301 vector which contains a GEP gene. 35S: All fusion vectors were transiently expressed in epidermal cells of N. benthamiana leaves by agrobacterium-mediated transformation, respectively. NSL-mCherry was used as a nuclear marker. Micrographs were taken using two-photon microscopy LSM710 scanning system (Carl Zeiss, Oberkochen, Germany).
4.5. RNA Isolation and Quantitative Real Time-Polymerase Chain Reaction (qRT-PCR)
Total RNA was extracted from 0.05 g of samples using RNAprep pure Plant Kit (code: DP432, Tiangen, Beijing, China) according to the manufacturer’s instructions. DNase I was used to remove DNA contamination. The same amount of total RNA (1 μg) was used in each assay. Reverse transcription was performed using PrimeScript™ RT reagent Kit (code: RR047, TaKaRa, Kyoto, Japan) according to the manufacturer’s protocol. QRT-PCR was performed on LightCycler480 instrument (Roche, Basel, Switzerland) using SYBR® Premix Ex Taq™ (Tli RNaseH Plus) (code: RR420, TaKaRa, Japan) with the primers listed in Table S1. The CaACTIN (Capana12g001934) was used as an internal standard. The relative expression levels were calculated using the 2−ΔΔCt method [64]. MeV software was employed to draw a heat map using the average log10 (expression value) to visualize the tissues-specific expression data. All the expression analyses were carried out with three biological repeats.
5. Conclusions
In conclusion, 22 ARF gene members in pepper were identified, and comprehensive information was collected. The conserved domains, phylogenetic relationship, the amino acid compositions, the gene structures, subcellular localizations and miRNA targets of CaARFs were analyzed in detail. The analysis of the expression patterns of CaARF genes in different tissues and organs will enable us to study the expression of those ARF genes in specific regions or in a limited time regulated manner. The responsiveness of the CaARF genes to various stresses and hormones suggests that CaARFs are involved in the pepper seedlings’ tolerance to abiotic stresses.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (Grant No. 31701967), the Natural Science Foundation of Zhejiang Province, China (Grant No. LQ17C150003), and the young talent training project of Zhejiang Academy of Agricultural Sciences (2016R23R08E05).
Abbreviations
| ARF | Auxin response factor |
| Aux/IAA | Auxin/indole-3-acetic acid |
| GH3 | Gretchen Hagen3 |
| SAUR | Small Auxin Up RNA |
| AuxREs | auxin response elements |
| DBD | DNA-binding domain |
| AD | Activation domain |
| RD | Repression domain |
| CTD | C-terminal dimerization domain |
| ABA | Abscisic acid |
| 6-BA | 6-benzylaminopurine |
| HMM | Hidden Markov model |
| MEME | Multiple Expectation Maximization for Motif Elicitation |
| GFP | Green fluorescent protein |
| ORF | Open reading frame |
Supplementary Materials
Supplementary materials can be found at www.mdpi.com/1422-0067/18/12/2719/s1.
Author Contributions
Chenliang Yu conceived and designed the research. Yihua Zhan and Xuping Feng performed the experiments; Chendong Sun analysed the data; Chenliang Yu and Zong-An Huang contributed to writing the manuscript. All authors read and approved the final manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Ljung K. Auxin metabolism and homeostasis during plant development. Development. 2013;140:943–950. doi: 10.1242/dev.086363. [DOI] [PubMed] [Google Scholar]
- 2.Farzinebrahimi R., Taha R.M., Rashid K., Yaacob J.S. The effect of various media and hormones via suspension culture on secondary metabolic activities of (Cape jasmine) Gardenia jasminoides ellis. Sci. World J. 2014;2014:407284. doi: 10.1155/2014/407284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Santner A., Estelle M. Recent advances and emerging trends in plant hormone signalling. Nature. 2009;459:1071–1078. doi: 10.1038/nature08122. [DOI] [PubMed] [Google Scholar]
- 4.Guilfoyle T.J., Hagen G. Auxin response factors. Curr. Opin. Plant Biol. 2007;10:453–460. doi: 10.1016/j.pbi.2007.08.014. [DOI] [PubMed] [Google Scholar]
- 5.Hagen G., Guilfoyle T. Auxin-responsive gene expression: Genes, promoters and regulatory factors. Plant Mol. Biol. 2002;49:373–385. doi: 10.1023/A:1015207114117. [DOI] [PubMed] [Google Scholar]
- 6.Shen C., Yue R., Sun T., Zhang L., Xu L., Tie S., Wang H., Yang Y. Genome-wide identification and expression analysis of auxin response factor gene family in Medicago truncatula. Front. Plant Sci. 2015;6:73. doi: 10.3389/fpls.2015.00073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ulmasov T., Hagen G., Guilfoyle T.J. Activation and repression of transcription by auxin-response factors. Proc. Natl. Acad. Sci. USA. 1999;96:5844–5849. doi: 10.1073/pnas.96.10.5844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lim P.O., Lee I.C., Kim J., Kim H.J., Ryu J.S., Woo H.R., Nam H.G. Auxin response factor 2 (ARF2) plays a major role in regulating auxin-mediated leaf longevity. J. Exp. Bot. 2010;61:1419–1430. doi: 10.1093/jxb/erq010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ellis C.M., Nagpal P., Young J.C., Hagen G., Guilfoyle T.J., Reed J.W. AUXIN RESPONSE FACTOR1 and AUXIN RESPONSE FACTOR2 regulate senescence and floral organ abscission in Arabidopsis thaliana. Development. 2005;132:4563–4574. doi: 10.1242/dev.02012. [DOI] [PubMed] [Google Scholar]
- 10.Sessions A., Nemhauser J.L., McColl A., Roe J.L., Feldmann K.A., Zambryski P.C. ETTIN patterns the Arabidopsis floral meristem and reproductive organs. Development. 1997;124:4481–4491. doi: 10.1242/dev.124.22.4481. [DOI] [PubMed] [Google Scholar]
- 11.Finet C., Fourquin C., Vinauger M., Berne-Dedieu A., Chambrier P., Paindavoine S., Scutt C.P. Parallel structural evolution of auxin response factors in the angiosperms. Plant J. 2010;63:952–959. doi: 10.1111/j.1365-313X.2010.04292.x. [DOI] [PubMed] [Google Scholar]
- 12.Hamann T., Benkova E., Baurle I., Kientz M., Jurgens G. The Arabidopsis BODENLOS gene encodes an auxin response protein inhibiting MONOPTEROS-mediated embryo patterning. Genes Dev. 2002;16:1610–1615. doi: 10.1101/gad.229402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Krogan N.T., Ckurshumova W., Marcos D., Caragea A.E., Berleth T. Deletion of MP/ARF5 domains III and IV reveals a requirement for Aux/IAA regulation in Arabidopsis leaf vascular patterning. New Phytol. 2012;194:391–401. doi: 10.1111/j.1469-8137.2012.04064.x. [DOI] [PubMed] [Google Scholar]
- 14.Nagpal P., Ellis C.M., Weber H., Ploense S.E., Barkawi L.S., Guilfoyle T.J., Hagen G., Alonso J.M., Cohen J.D., Farmer E.E., et al. Auxin response factors ARF6 and ARF8 promote jasmonic acid production and flower maturation. Development. 2005;132:4107–4118. doi: 10.1242/dev.01955. [DOI] [PubMed] [Google Scholar]
- 15.Harper R.M., Stowe-Evans E.L., Luesse D.R., Muto H., Tatematsu K., Watahiki M.K., Yamamoto K., Liscum E. The NPH4 locus encodes the auxin response factor ARF7, a conditional regulator of differential growth in aerial Arabidopsis tissue. Plant Cell. 2000;12:757–770. doi: 10.1105/tpc.12.5.757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Narise T., Kobayashi K., Baba S., Shimojima M., Masuda S., Fukaki H., Ohta H. Involvement of auxin signaling mediated by IAA14 and ARF7/19 in membrane lipid remodeling during phosphate starvation. Plant Mol. Biol. 2010;72:533–544. doi: 10.1007/s11103-009-9589-4. [DOI] [PubMed] [Google Scholar]
- 17.Okushima Y., Fukaki H., Onoda M., Theologis A., Tasaka M. ARF7 and ARF19 regulate lateral root formation via direct activation of LBD/ASL genes in Arabidopsis. Plant Cell. 2007;19:118–130. doi: 10.1105/tpc.106.047761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu P.P., Montgomery T.A., Fahlgren N., Kasschau K.D., Nonogaki H., Carrington J.C. Repression of AUXIN RESPONSE FACTOR10 by microRNA160 is critical for seed germination and post-germination stages. Plant J. 2007;52:133–146. doi: 10.1111/j.1365-313X.2007.03218.x. [DOI] [PubMed] [Google Scholar]
- 19.Liu X., Huang J., Wang Y., Khanna K., Xie Z., Owen H.A., Zhao D. The role of floral organs in carpels, an Arabidopsis loss-of-function mutation in MicroRNA160a, in organogenesis and the mechanism regulating its expression. Plant J. 2010;62:416–428. doi: 10.1111/j.1365-313X.2010.04164.x. [DOI] [PubMed] [Google Scholar]
- 20.Qi Y., Wang S., Shen C., Zhang S., Chen Y., Xu Y., Liu Y., Wu Y., Jiang D. OsARF12, a transcription activator on auxin response gene, regulates root elongation and affects iron accumulation in rice (Oryza sativa) New Phytol. 2012;193:109–120. doi: 10.1111/j.1469-8137.2011.03910.x. [DOI] [PubMed] [Google Scholar]
- 21.Shen C.J., Yue R.Q., Sun T., Zhang L., Yang Y.J., Wang H.Z. OsARF16, a transcription factor regulating auxin redistribution, is required for iron deficiency response in rice (Oryza sativa L.) Plant Sci. 2015;231:148–158. doi: 10.1016/j.plantsci.2014.12.003. [DOI] [PubMed] [Google Scholar]
- 22.Shen C.J., Yue R.Q., Yang Y.J., Zhang L., Sun T., Tie S.G., Wang H.Z. OsARF16 is involved in cytokinin-mediated inhibition of phosphate transport and phosphate signaling in rice (Oryza sativa L.) PLoS ONE. 2014;9:e112906. doi: 10.1371/journal.pone.0112906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang X., Yan F., Tang Y., Yuan Y., Deng W., Li Z. Auxin response gene SlARF3 plays multiple roles in tomato development and is involved in the formation of epidermal cells and trichomes. Plant Cell Physiol. 2015;56:2110–2124. doi: 10.1093/pcp/pcv136. [DOI] [PubMed] [Google Scholar]
- 24.De Jong M., Wolters-Arts M., Feron R., Mariani C., Vriezen W.H. The Solanum lycopersicum auxin response factor 7 (SlARF7) regulates auxin signaling during tomato fruit set and development. Plant J. 2009;57:160–170. doi: 10.1111/j.1365-313X.2008.03671.x. [DOI] [PubMed] [Google Scholar]
- 25.Park J.E., Park J.Y., Kim Y.S., Staswick P.E., Jeon J., Yun J., Kim S.Y., Kim J., Lee Y.H., Park C.M. GH3-mediated auxin homeostasis links growth regulation with stress adaptation response in Arabidopsis. J. Biol. Chem. 2007;282:10036–10046. doi: 10.1074/jbc.M610524200. [DOI] [PubMed] [Google Scholar]
- 26.Zahir Z.A., Shah M.K., Naveed M., Akhter M.J. Substrate-dependent auxin production by Rhizobium phaseoli improves the growth and yield of Vigna radiata L. under salt stress conditions. J. Microbiol. Biotechnol. 2010;20:1288–1294. doi: 10.4014/jmb.1002.02010. [DOI] [PubMed] [Google Scholar]
- 27.Wang D., Pei K., Fu Y., Sun Z., Li S., Liu H., Tang K., Han B., Tao Y. Genome-wide analysis of the auxin response factors (ARF) gene family in rice (Oryza sativa) Gene. 2007;394:13–24. doi: 10.1016/j.gene.2007.01.006. [DOI] [PubMed] [Google Scholar]
- 28.Xing H., Pudake R.N., Guo G., Xing G., Hu Z., Zhang Y., Sun Q., Ni Z. Genome-wide identification and expression profiling of auxin response factor (ARF) gene family in maize. BMC Genom. 2011;12:178. doi: 10.1186/1471-2164-12-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li S.B., OuYang W.Z., Hou X.J., Xie L.L., Hu C.G., Zhang J.Z. Genome-wide identification, isolation and expression analysis of auxin response factor (ARF) gene family in sweet orange (Citrus sinensis) Front. Plant Sci. 2015;6:119. doi: 10.3389/fpls.2015.00119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kalluri U.C., Difazio S.P., Brunner A.M., Tuskan G.A. Genome-wide analysis of Aux/IAA and ARF gene families in Populus trichocarpa. BMC Plant Biol. 2007;7:59. doi: 10.1186/1471-2229-7-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sun R., Wang K., Guo T., Jones D.C., Cobb J., Zhang B., Wang Q. Genome-wide identification of auxin response factor (ARF) genes and its tissue-specific prominent expression in Gossypium raimondii. Funct. Integr. Genom. 2015;15:481–493. doi: 10.1007/s10142-015-0437-0. [DOI] [PubMed] [Google Scholar]
- 32.Kumar R., Tyagi A.K., Sharma A.K. Genome-wide analysis of auxin response factor (ARF) gene family from tomato and analysis of their role in flower and fruit development. Mol. Genet. Genom. 2011;285:245–260. doi: 10.1007/s00438-011-0602-7. [DOI] [PubMed] [Google Scholar]
- 33.Korasick D.A., Westfall C.S., Lee S.G., Nanao M.H., Dumas R., Hagen G., Guilfoyle T.J., Jez J.M., Strader L.C. Molecular basis for auxin response factor protein interaction and the control of auxin response repression. Proc. Natl. Acad. Sci. USA. 2014;111:5427–5432. doi: 10.1073/pnas.1400074111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu Z., Ji A., Song J., Chen S. Genome-wide analysis of auxin response factor gene family members in medicinal model plant Salvia miltiorrhiza. Biol. Open. 2016;5:848–857. doi: 10.1242/bio.017178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu M.-F., Tian Q., Reed J.W. Arabidopsis microRNA167 controls patterns of ARF6 and ARF8 expression, and regulates both female and male reproduction. Development. 2006;133:4211–4218. doi: 10.1242/dev.02602. [DOI] [PubMed] [Google Scholar]
- 36.Lee M., Jung J.H., Han D.Y., Seo P.J., Park W.J., Park C.M. Activation of a flavin monooxygenase gene YUCCA7 enhances drought resistance in Arabidopsis. Planta. 2012;235:923–938. doi: 10.1007/s00425-011-1552-3. [DOI] [PubMed] [Google Scholar]
- 37.Farcot E., Lavedrine C., Vernoux T. A modular analysis of the auxin signalling network. PLoS ONE. 2015;10:e0122231. doi: 10.1371/journal.pone.0122231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ulmasov T., Murfett J., Hagen G., Guilfoyle T.J. Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell. 1997;9:1963–1971. doi: 10.1105/tpc.9.11.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schwarz D., Rouphael Y., Colla G., Venema J.H. Grafting as a tool to improve tolerance of vegetables to abiotic stresses: Thermal stress, water stress and organic pollutants. Sci. Hortic. 2010;127:162–171. doi: 10.1016/j.scienta.2010.09.016. [DOI] [Google Scholar]
- 40.Guo M., Lu J.P., Zhai Y.F., Chai W.G., Gong Z.H., Lu M.H. Genome-wide analysis, expression profile of heat shock factor gene family (CaHsfs) and characterisation of CaHsfA2 in pepper (Capsicum annuum L.) BMC Plant Biol. 2015;15:151. doi: 10.1186/s12870-015-0512-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cho H., Ryu H., Rho S., Hill K., Smith S., Audenaert D., Park J., Han S., Beeckman T., Bennett M.J., et al. A secreted peptide acts on BIN2-mediated phosphorylation of ARFs to potentiate auxin response during lateral root development. Nat. Cell Biol. 2014;16:66–76. doi: 10.1038/ncb2893. [DOI] [PubMed] [Google Scholar]
- 42.Tiwari S.B., Hagen G., Guilfoyle T. The roles of auxin response factor domains in auxin-responsive transcription. Plant Cell. 2003;15:533–543. doi: 10.1105/tpc.008417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mohamed Z., Fu Y., Anne-Laure C.B., Isabelle M., Pierre F., Hua W., Corinne A., Jean-Paul R., Mondher B. Characterization of the tomato ARF gene family uncovers a multi-levels post-transcriptional regulation including alternative splicing. PLoS ONE. 2014;9:e84203. doi: 10.1371/journal.pone.0084203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ulmasov T., Hagen G., Guilfoyle T.J. ARF1, a transcription factor that binds to auxin response elements. Science. 1997;276:1865–1868. doi: 10.1126/science.276.5320.1865. [DOI] [PubMed] [Google Scholar]
- 45.Shen C., Wang S., Bai Y., Wu Y., Zhang S., Chen M., Guilfoyle T.J., Wu P., Qi Y. Functional analysis of the structural domain of arf proteins in rice (Oryza sativa L.) J. Exp. Bot. 2010;61:3971–3981. doi: 10.1093/jxb/erq208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mun J.H., Yu H.J., Shin J.Y., Oh M., Hwang H.J., Chung H. Auxin response factor gene family in Brassica rapa: Genomic organization, divergence, expression, and evolution. Mol. Genet. Genom. 2012;287:765–784. doi: 10.1007/s00438-012-0718-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang H., Cao N., Dong C., Shang Q. Genome-wide identification and expression of ARF gene family during adventitious root development in hot pepper (Capsicum annuum) Hortic. Plant J. 2017;3:151–164. doi: 10.1016/j.hpj.2017.07.001. [DOI] [Google Scholar]
- 48.Nanao M.H., Vinos-Poyo T., Brunoud G., Thevenon E., Mazzoleni M., Mast D., Laine S., Wang S.C., Hagen G., Li H.B., et al. Structural basis for oligomerization of auxin transcriptional regulators. Nat. Commun. 2014;5:3617. doi: 10.1038/ncomms4617. [DOI] [PubMed] [Google Scholar]
- 49.Hunter C., Willmann M.R., Wu G., Yoshikawa M., de la Luz Gutiérrez-Nava M., Poethig S.R. Trans-acting siRNA-mediated repression of ETTIN and ARF4 regulates heteroblasty in Arabidopsis. Development. 2006;133:2973–2981. doi: 10.1242/dev.02491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wilmoth J.C., Wang S., Tiwari S.B., Joshi A.D., Hagen G., Guilfoyle T.J., Alonso J.M., Ecker J.R., Reed J.W. NPH4/ARF7 and ARF19 promote leaf expansion and auxin-induced lateral root formation. Plant J. 2005;43:118–130. doi: 10.1111/j.1365-313X.2005.02432.x. [DOI] [PubMed] [Google Scholar]
- 51.Chen Z., Yuan Y., Fu D., Shen C., Yang Y. Identification and expression profiling of the auxin response factors in Dendrobium officinale under abiotic stresses. Int. J. Mol. Sci. 2017;18:927. doi: 10.3390/ijms18050927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jain M., Khurana J.P. Transcript profiling reveals diverse roles of auxin-responsive genes during reproductive development and abiotic stress in rice. FEBS J. 2009;276:3148–3162. doi: 10.1111/j.1742-4658.2009.07033.x. [DOI] [PubMed] [Google Scholar]
- 53.Wang S.K., Bai Y.H., Shen C.J., Wu Y.R., Zhang S.N., Jiang D.A., Guilfoyle T.J., Chen M., Qi Y.H. Auxin-related gene families in abiotic stress response in Sorghum bicolor. Funct. Integr. Genom. 2010;10:533–546. doi: 10.1007/s10142-010-0174-3. [DOI] [PubMed] [Google Scholar]
- 54.Ha C.V., Le D.T., Nishiyama R., Watanabe Y., Sulieman S., Tran U.T., Mochida K., Dong N.V., Yamaguchi-Shinozaki K., Shinozaki K., et al. The auxin response factor transcription factor family in soybean: Genome-wide identification and expression analyses during development and water stress. DNA Res. 2013;20:511–524. doi: 10.1093/dnares/dst027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Matsui A., Mizunashi K., Tanaka M., Kaminuma E., Nguyen A.H., Nakajima M., Kim J.M., Nguyen D.V., Toyoda T., Seki M. tasiRNA-ARF pathway moderates floral architecture in Arabidopsis plants subjected to drought stress. BioMed Res. Int. 2014;2014:303451. doi: 10.1155/2014/303451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hu W., Zuo J., Hou X., Yan Y., Wei Y., Liu J., Li M., Xu B., Jin Z. The auxin response factor gene family in banana: Genome-wide identification and expression analyses during development, ripening, and abiotic stress. Front. Plant. Sci. 2015;6:742. doi: 10.3389/fpls.2015.00742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang J.W., Wang L.J., Mao Y.B., Cai W.J., Xue H.W., Chen X.Y. Control of root cap formation by microRNA-targeted auxin response factors in Arabidopsis. Plant Cell. 2005;17:2204–2216. doi: 10.1105/tpc.105.033076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Popko J., Hansch R., Mendel R.R., Polle A., Teichmann T. The role of abscisic acid and auxin in the response of poplar to abiotic stress. Plant Biol. 2010;12:242–258. doi: 10.1111/j.1438-8677.2009.00305.x. [DOI] [PubMed] [Google Scholar]
- 59.Agusti J., Herold S., Schwarz M., Sanchez P., Ljung K., Dun E.A., Brewer P.B., Beveridge C.A., Sieberer T., Sehr E.M., et al. Strigolactone signaling is required for auxin-dependent stimulation of secondary growth in plants. Proc. Natl. Acad. Sci. USA. 2011;108:20242–20247. doi: 10.1073/pnas.1111902108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liu X., Zhang H., Zhao Y., Feng Z., Li Q., Yang H.Q., Luan S., Li J., He Z.H. Auxin controls seed dormancy through stimulation of abscisic acid signaling by inducing ARF-mediated ABI3 activation in Arabidopsis. Proc. Natl. Acad. Sci. USA. 2013;110:15485–15490. doi: 10.1073/pnas.1304651110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vanstraelen M., Benkova E. Hormonal interactions in the regulation of plant development. Annu. Rev. Cell. Dev. Biol. 2012;28:463–487. doi: 10.1146/annurev-cellbio-101011-155741. [DOI] [PubMed] [Google Scholar]
- 62.Schaller G.E., Bishopp A., Kieber J.J. The yin-yang of hormones: Cytokinin and auxin interactions in plant development. Plant Cell. 2015;27:44–63. doi: 10.1105/tpc.114.133595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nizampatnam N.R., Schreier S.J., Damodaran S., Adhikari S., Subramanian S. MicroRNA160 dictates stage-specific auxin and cytokinin sensitivities and directs soybean nodule development. Plant J. 2015;84:140–153. doi: 10.1111/tpj.12965. [DOI] [PubMed] [Google Scholar]
- 64.Yu C., Dong W., Zhan Y., Huang Z.A., Li Z., Kim I.S., Zhang C. Genome-wide identification and expression analysis of CLLAX, CLPIN and CLABCB genes families in Citrullus lanatus under various abiotic stresses and grafting. BMC Genet. 2017;18:33. doi: 10.1186/s12863-017-0500-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.