Abstract
MicroRNAs have been shown to play an important role in stem cell fate determination and self-renewal. However, the role of miRNAs in neural stem cells (NSCs) remains poorly understood. In this study, we showed that miR-346, a less characterized microRNA, promoted NSCs proliferation, differentiation and apoptosis by targeting KLF4, a core transcriptional factor in stem cell fate determination. Our data suggested that miR-346 could directly target the 3’-untranslated region of KLF4. Overexpression of miR-346 decreased KLF4 expression at both mRNA and protein levels in NSCs. More importantly, Overexpression of miR-346 repressed NSC proliferation and induced the expression of lineage markers including GFAP and Tuj1. Additionally, overexpression of miR-346 promoted apoptosis of NSCs. In concert, suppressing its expression by an antisense RNA, anti-miR-346, promoted NSC proliferation, and meanwhile inhibited its differentiation and apoptosis. We also showed that the effects of miR-346 overexpression could be reversed by re-expression of KLF4. Taken together, Those data suggest that miR-346 is a novel miRNA that regulates NSC proliferation and differentiation by targeting KLF4.
Keywords: miR-346, KLF4, NSC
Introduction
Neural stem cells (NSCs) are a group of self-renewing, undifferentiated precursor cells that retain the abilities to differentiate to both glial (astrocytes and oligodendrocytes) and neuronal lineages [1]. NSCs are mainly located in the subventricular zone and the subgranular zone of the brain, both in the adult and developing mammalian [2,3]. Recently, studies indicated that NSCs can serve as cell replacement therapies for neurological disorders, such as Alzheimer’s disease, Parkinson’s disease, Huntington’s disease, and spinal cord injuries [4-7]. Additionally, engineered NSCs are emerging as a promising new therapeutic strategy for cancer therapy attributed to their tumor-homing properties [8]. Despite of the great prospect for clinically intervention, it is still a long distance before clinical applications due to the limited understandings of the underlying mechanisms of NSCs self-renewal and differentiation.
MiRNAs (microRNAs) are a large family of endogenous single-stranded non-coding RNAs with, 19-23 nucleotides in length, that modulate gene expression post-transcriptionally by either targeting the 3’-untranslated regions (UTRs) of aimed mRNA for degradation or directly inhibiting their translation [9]. As far as we know, about 30% of the human genome, that is upto 10,000 genes, could be potentially be regulated by miRNAs [10,11]. Growing studies evidences show that miRNAs play pivotal roles in a wide area of biological processes, including cell development, proliferation, differentiation, invasion, and apoptosis [12]. Recently it was reported that miRNAs have been shown to playedact an important role in stem cell fate determination and self-renewal by controlling the expression of stem cell regulators, such as Nanog [13-16]. However, the precise role of miRNAs in neural stem cells remains poorly understood.
The Krüppel-like transcription factor (KLF) families have been reported to regulate a diverse array of cellular processes, including development, differentiation, proliferation, and apoptosis [17,18]. KLF4, a member of this family, was known to be one of the four transcriptional factors (OCT3/4, SOX2, KLF4, and c-MYC), essential for reprogramming differentiated cells into induced pluripotent stem (iPS) cells [19-25]. As an transcriptional factor, it promotes or repress gene expressions depending on the interaction partners and the context of the binding sites. For example, human telomerase reverse transcriptase (hTERT), the dominate factor maintaining telomere length in human cells, was identified as a target gene of KLF4 in embryonic stem cells (ESCs) [26,27]. KLF4 is required for maintaining hTERT expression in human ESCs and cancer cells by directly activating its transcription [27]. lectin galactoside binding soluble 3 (Lgals3), also known as galectin-3, implicated in a broad range of biological processes, from chemotaxis and inflammation to fibrosis and apoptosis, was another newly identified target gene of KLF4 [28]. In spite, there are many other targets of KLF4, including p21, a cell-cycle related gene [29,30].
MiR-346, located in the second intron of the glutamate receptor ionotropic delta 1 (GRID1) gene, was firstly reported to be redundant in follicular thyroid carcinoma (FTC) [31,32]. Further studies showed that miR-346 also regulates other physiological and pathological processes, including cell differentiation, carcinogenesis and inflammatory response [33-37]. However, the role of miR-346 in NSCs was still unknown. Here, we reported that miR-346 is a novel regulator of cell proliferation and differentiation of NSCs by targeting KLF4, a crucial transcriptional factor of stem cells. Our study give new insight into NSCs regulation by miRNAs, and also suggested a role of KLF4 in NSCs regulation.
Materials and methods
Cell isolation and cultures
NSCs used in this study were isolated from the forebrains of C57/BL6 mice (male, 8-10 week old) using a Percoll gradient-centrifugation manner. The animal experimental procedures were fully complied with the guidelines of Institutional Animal Care and Use Committee of Xi’an Jiaotong University. The cells were maintained in DMEM/F12 medium (Invitrogen, Carlsbad, CA, USA) supplemented with 1 mM L-glutamine (Sigma, St. Louis, MO, USA), 1% N2 supplement (Gibco, Rockville, MD, USA), 20 ng/ml epidermal growth factor (EGF, PeproTech, Rocky Hill, NJ, USA), 20 ng/ml basic fibroblast growth factor (FGF, PeproTech), 50 ng/ml heparin (Sigma) and 1% penicillin and streptomycin (Invitrogen). For astrocyte differentiation, NSCs were grown in DMEM/F12 medium containing 1% N2 supplement, 5 mM forskolin (Sigma) and 0.5% fetal bovine serum (FBS; Gibco) for 3 days. For neural differentiation, NSCs were treated with 1 mM retinoic acid (Sigma) and 0.5% FBS for 3 days in 1% N2 supplemented medium.
Reagents and transfections
The miR-346 mimics and miR-346 inhibitor (anti-miR-346) and their controls miR-NC and anti-miR-NC were synthesized by Shanghai GenePharma Co. Ltd (Shanghai, China). Before transfection, NSCs were seeded into a 6-well plates at a density of 1×105 per well and cultured overnight. Then, miRNAs mimics or pcDNA3.1 vectors were transfected using Lipofectamine 2000 (Invitrogen, CA, USA) according to the manufacturer’s instructions.
Luciferase reporter assay
The cDNA fragment of KLF4 3’-UTR containing the miR-346 binding site was amplified and then, subcloned into the pmirGLO vector (Promega, Madison, WI, USA). For the detection of luciferase activity, human embryonic kidney (HEK) 293 cells were seeded into a 48-well plate 24 h before transfection. 10 ng pmirGLO-KLF4 recombinant vectors and 50 nM miR-346 mimics were cotransfected into the cells using Lipofectamine 2000. The luciferase intensity was measured with a Dual Luciferase Assay System (Promega) in accordance with the manufacturer’s protocol 24 h later.
RNA extraction and qPCR analysis
Total RNA of cells were isolated using miRNAeasy mini kit (QIAGEN, Dusseldorf, Germany). First-strand cDNA was synthesized by M-MLV reverse transcriptase (Clontech, Palo Alto, CA, USA). Quantitative PCR was performed on an ABI 7500 thermocycler (Applied Biosystems) using SYBRGreen MasterMix (Bio-Rad, Hercules, CA, USA) according to the manufacturer’s instructions. GAPDH was served as an internal control.
Western blot analysis
Total protein from cells were lyzed by RIPA buffer. The β-actin was regarded as the endogenous normalizer. Primary antibodies including anti-KLF4, anti-hTERT, anti-LGALS3, anti-p21 and anti-β-actin were purchased from Santa Cruz Biotechnology Inc. (Santa Cruz, CA, USA).
Cell proliferation assay
Cell proliferation was detected by a BrdU incorporation assay using a commercial kit (Millipore, Billerica, MA, USA) according to the manufacturer’s instructions.
Apoptosis detection
Cells were seeded into a 6-well plate with complete medium. 24 h later, cells were transfected with miRNA mimics, anti-miRNAs, their controls or left untreated (mock) as indicated. After 40 h, cells were serum-starved overnight in DMEM/F12 medium containing 0.1% FBS. Then all cells were collected by centrifugation, and stained with 0.5 μg annexin V for 20 min in the dark, followed by incubation with 10 μg of Propidium Iodide (PI) for 5 min. The cells were then analyzed on a FACS Calibur flow cytometer (Beckman Coulter).
Data analysis
Data were shown as means ± standard deviation (SD). Statistical analyses were performed by SPSS version 19.0 (SPSS Inc., Chicago, IL, USA) using one-way ANOVA analysis. Correlations were assessed by Spearman’s rank correlation coefficients. Results were considered statistically significant at p < 0.05.
Results
KLF4 was a potential target gene of miR-346
To investigate whether miR-346 is involved in regulating NSC differentiation, its expression was determined during NSC differentiation in vitro. qPCR analysis showed that the level of miR-346 was gradually increased along with NSC differentiation (Figure 1A), whereas the mRNA level of KLF4 decreased in this process (Figure 1B). Therefore, we attempted to find potential target sequences of miR-346 in KLF4 gene. By bioinformatic algorithms, we found there was a predicted target site located in the 3’-UTR of KLF4 mRNA (Figure 1C). Further analysis suggested that this sequence was conserved between species (Figure 1D). To confirm this hypothesis, we constructed reporter vectors harbor either the wild-type miR-346 target sequences in KLF4 3’UTR or mutant ones (Figure 1C). Luciferase reporter assay showed that miR-346 mimics transfection significantly decreased the expression of wild-type reporter vector, but not that of the mutant one (Figure 1E). Correlation analysis showed that miR346 expression was inversely correlated with KLF4 expression (Figure 1F). These results suggested that KLF4 was a potential target of miR-346.
Figure 1.
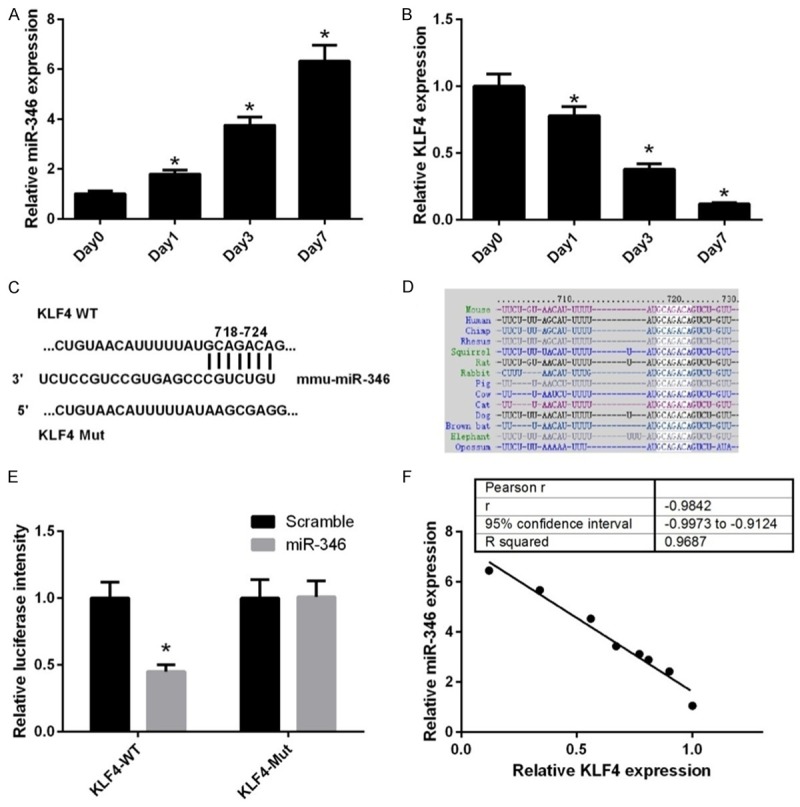
miR-346 expression inversely correlates with KLF4 expression during NSC differentiation. qPCR analysis of miR-346 (A) and KLF4 mRNA (B) expression in NSCs during differentiation over a period of 7 days. Day 0 represents undifferentiated cells and was normalized to a value of 1. *p < 0.05. (C) Diagram of the predicted miR-346 targeting site within the 3’UTR of KLF4. (D) Bioinformatics analysis of the conservation of the target sequences of miR-346 in the 3’UTR of KLF4 acrossing species. (E) The recombinant pmirGLO vectors containing WT or MT 3’UTR of KLF4 were co-transfected with miR-NC or the miR-346 mimics. *p < 0.05 vs. miR-NC. (F) Correlation of miR-346 expression to KLF4 mRNA expression in NSCs during differentiation. R-squared = 0.9687, p < 0.001. Data were shown as means ± standard deviation (SD) and were processed by one-way ANOVA analysis. Correlations were assessed by Spearman’s rank correlation coefficients. All experiments were repeated at least three times independently.
miR-346 regulates the expression of KLF4 and NSC proliferation and differentiation
Previously, we have shown that the KLF4 may be a direct target of miR-346. To further verify this assumption, we performed gain and loss-of-function assays using the miR-346 mimics and anti-miR-346. Overexpression of miR-346 in NSCs significantly decreased the mRNA levels of KLF4, whereas suppression of miR-346 expression by its inhibitor anti-miR-346, apparently increased KLF4 expression (Figure 2A). These results were further confirmed by western blot assay (Figure 2B). In summary, these results further confirmed that KLF4 was a direct target of miR-346. Given that KLF4 was a core regulator of stem cells, we hypothesized that miR-346 may have a role in NSCs. Overexpression of miR-346 repressed proliferation of NSCs, whereas suppression of miR-346 markedly enhanced their proliferation (Figure 2C). Additionally, miR-346 overexpression significantly promoted expression of the astrocyte marker, glial fibrillary acidic protein (GFAP) in NSCs treated with forskolin and FBS (Figure 2D). Similarly, The neuronal marker neuronal class III b-tubulin (Tuj1) was also upregulated by miR-346 overexresion when treated with retinoic acid and FBS (Figure 2E). In contrast, the marker of neuroepithelial stem cell, neuroepithelial stem cell protein (Nestin) was depressed by miR-346 overexpression (Figure 2F). In concert, suppression of miR-346 exhibited opposite effects to miR-346 overexpression on NSCs differentiation (Figure 2D-F). Collectively, these results indicated that miR-346 promotes NSCs differentiation and suppresses their proliferation.
Figure 2.
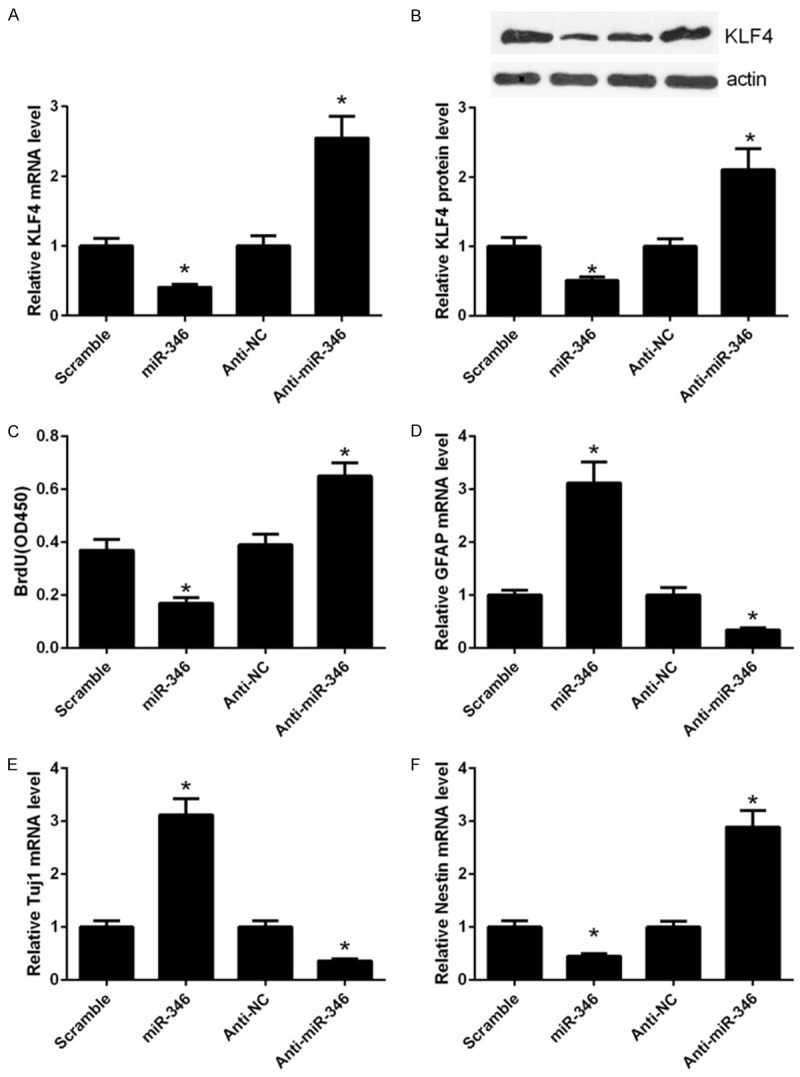
miR-346 regulates KLF4 expression and NSC proliferation and differentiation. (A) qPCR analysis of KLF4 mRNA expression in NSCs transfected with the miR-346 mimics or antimiR-346. (B) Western blot analysis of KLF4 protein expression in NSCs transfected with the miR-346 mimics or anti-miR-346. The relative protein expression of KLF4 was quantified using Image-Pro Plus 6.0. After transfection for 48 h, cells were harvested for analysis. *p < 0.05 vs. miR-NC, &p < 0.05 vs. anti-miR-NC. (C) Proliferation of NSCs was detected by the BrdU assay. NSCs were transfected with the miR-346 mimics or anti-miR-346 for 48 h followed by detection with BrdU assay. (D) qPCR analysis of GFAP expression in NSCs. NSCs were transfected with the miR-346 mimics or anti-miR-346 in the presence with 5 mM forskolin and 0.5% FBS and cultured for 3 days. (E) qPCR analysis of Tuj1 expression in NSCs transfected with the miR-346 mimics or anti-miR-346 in the presence with 1 mM retinoic acid and 0.5% FBS for 3 days. (F) qPCR analysis of Nestin expression in NSCs transfected with the miR-346 mimics or anti-miR-346. Cells were cultured in the maintaining medium for 3 days before analysis. *p < 0.05 vs. miR-NC, &p < 0.05 vs. anti-343 miR-NC. Data were shown as means ± standard deviation (SD) and were processed by one-way ANOVA analysis. All experiments were repeated at least three times independently.
Overexpression of miR-346 induced apoptosis of NSCs
Nest we tested whether miR-346 influence cell apoptosis of NSCs. FACS analysis suggested that overexpression of miR-346 apparently induced apoptosis of NSCs, and its inhibitor, anti-miR-346 decreased their apoptosis (Figure 3A). Preventative pictures were revealed in Figure 3B. These results indicated that miR-346 could promote cell apoptosis of NSCs.
Figure 3.
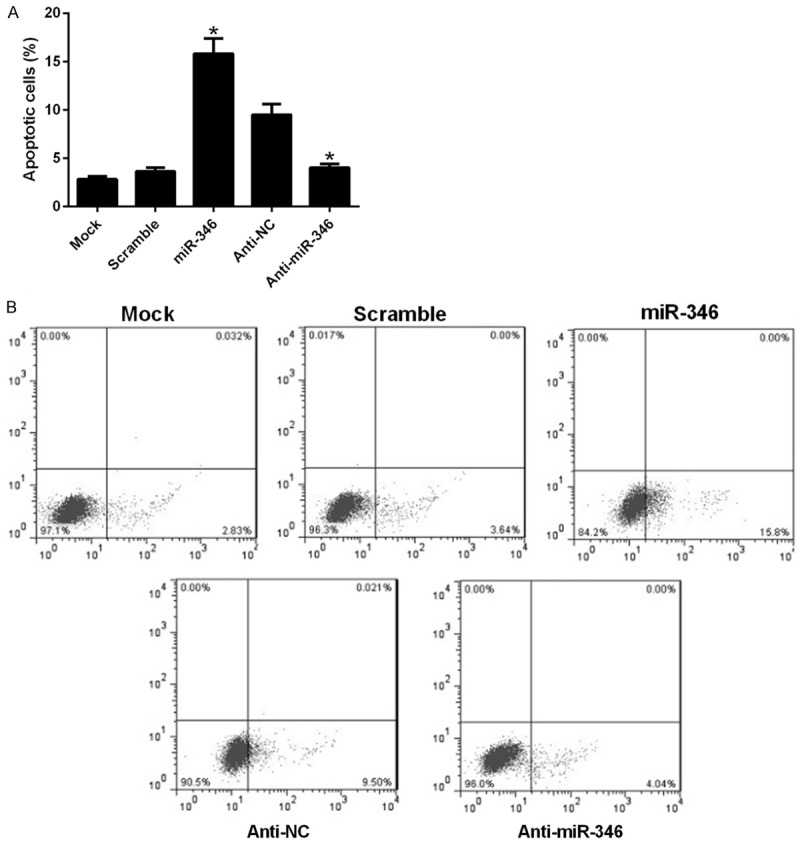
miR-346 regulates cell apoptosis of NSCs. NSCs were transfected with a scramble miRNA, miR-346 mimics, anti-NC, anti-miR-346 or left untreated (mock). 48 h later, cells were harvested after serum-starvation over night for FACS analysis. Stastics results (A) and representative pictures (B). Data were shown as means ± standard deviation (SD) and were processed by one-way ANOVA analysis. All experiments were repeated at least three times independently. *p < 0.05.
MiR-346 regulated the target genes of KLF4
Given that miR-346 could directly target KLF4 and suppressed its expression, we inferred that miR-346 may aslo regulate the downstream targets of KLF4. qPCR analysis revealed that hTERT, Lgals3 and p21 expressions were decreased after overexpression of miR-346 (Figure 4A). These results were further verified by western blot analysis (Figure 4B). These results supported the conclusion that KLF4 was a target of miR-346.
Figure 4.
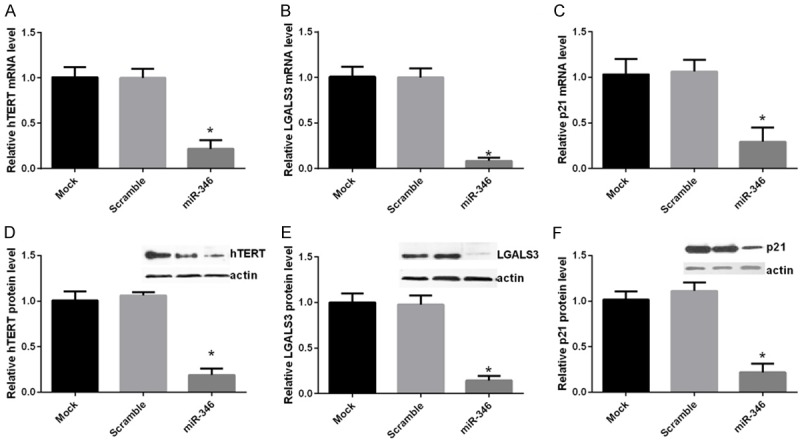
miR-346 regulates target genes of KLF4. NSCs were transfected with a scramble miRNA, miR-346 mimics or left untreated (mock). hTERT, LGALS3 and p21 mRNA and protein levels were determined by qPCR (A-C) or western bolt (D-F) respectively 48 h after transfections. For qPCR analysis, GAPDH was served as a internal control. For western bolt analysis, actin was taken as a loading control. Mock cells and was normalized to a value of 1. Data were shown as means ± standard deviation (SD) and were processed by one-way ANOVA analysis. All experiments were repeated at least three times independently. *p < 0.05.
The functions of miR-346 in NSCs was dependent on KLF4
We have shown that miR-346 play a critical role in proliferation, differentiation and apoptosis of NSCs and KLF4 was a direct target of miR-346. As KLF4 was one of the core regulators of stem cells, we reasoned that miR-346 may exert these influences through KLF4. To verified this assumption, a series of loss and gain function assays were carried out. In NSCs, overexpression of miR-346 remarkably decreased KLF4 mRNA level and this effect could be reversed by transfection of a KLF4 expressing vector (Figure 5A). This result was further confirmed by western blot analysis (Figure 5B). BrdU assay showed that KLF4 overexpression recovered the proliferate capabilities of NSCs, which were dampened by miR-346 overexpression previously (Figure 5C). Similar effects on differentiation were observed. The linage markers, GFAP and Tuj1 were repressed, whereas the stem cells marker, Nestin were upregulated by KLF4 re-expression (Figure 5D-F). Apoptosis of NSCs induced by miR-346 overexpression was reduced by KLF4 overexpression (Figure 6A and 6B). These results suggested that the functions of miR-346 in NSCs was dependent on KLF4.
Figure 5.
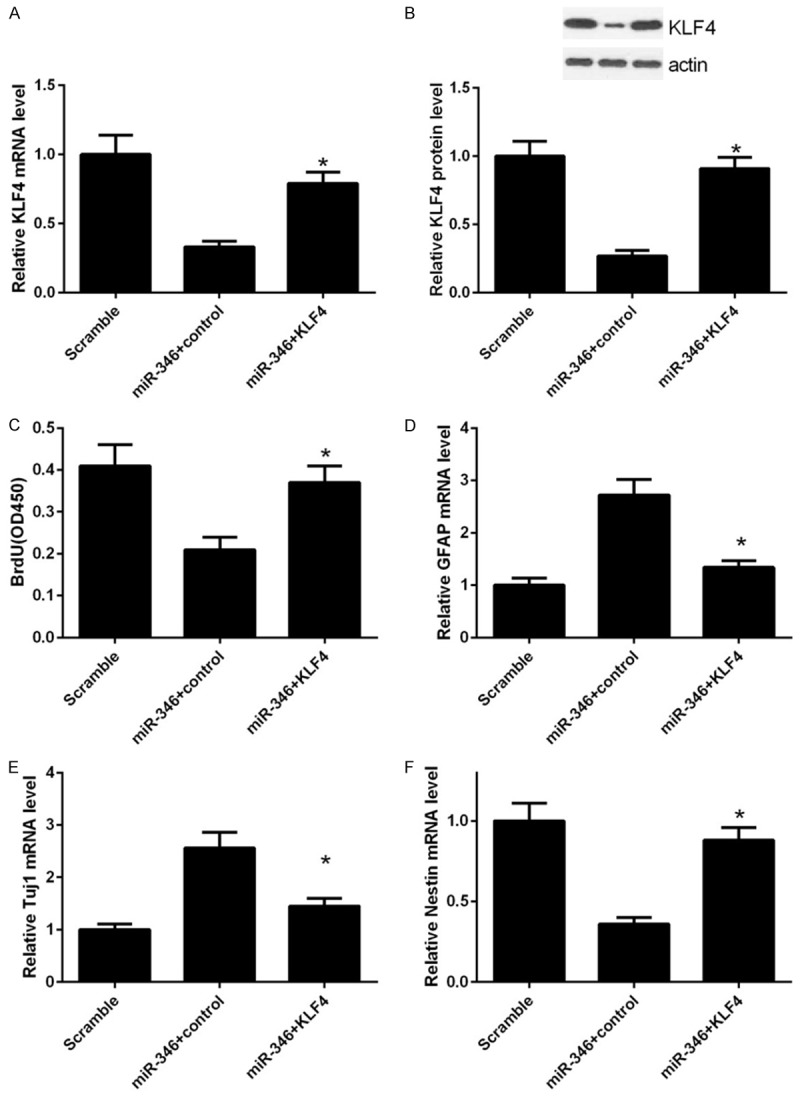
Overexpression of KLF4 conversed effects of miR-346. NSCs were transfected with a scramble miRNA, miR-346 mimics or miR-346 combined with KLF4 respectively. mRNA and protein levels of KLF4 were determined by qPCR (A) or western bolt (B) respectively 48 h after transfections. (C) NSCs were treated as in (A). 24 h later, proliferation was detected by the BrdU incorporation assay. mRNA levels of GFAP (D), Tuj1 (E) and Nestin (F) were determined by qPCR analysis. Data were normalized to GAPDH and shown as means ± standard deviation (SD). Data were processed by one-way ANOVA analysis. All experiments were repeated at least three times independently. *p < 0.05.
Figure 6.
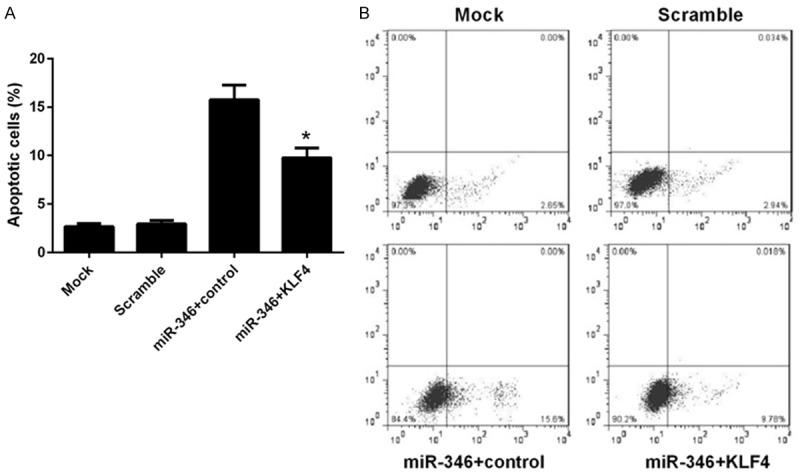
Overexpression of KLF4 represses miR-346 induced apoptosis of NSCs. NSCs were transfected with a scramble miRNA, miR-346 mimics combined with KLF4 expressing vector or a control vector, or left untreated (mock). 48 h later, cells were harvested after serum-starvation over night for FACS analysis. Stastics results (A) and representative pictures (B). Data were shown as means ± standard deviation (SD) and were processed by one-way ANOVA analysis. All experiments were repeated at least three times independently. *p < 0.05.
Discussion
MiR-346, a less characterized miRNA, can target different genes such as SRCIN1 and GSK-β to exert distinct biological effects, including cell differentiation, carcinogenesis and inflammatory response [33-37]. However, the role of miR-346 in stem cells remains largely unknown. In this study, we showed that miR-346 positively regulate NSCs differentiation in a KLF4-dependent manner. Besides, we aslo showed that it regulated proliferation and apoptosis of NSCs. The underlying mechanism also involved KLF4.
KLF4 was known as one of the four famous transcriptional factors essential for reprogramming different kinds of differentiated cells into iPS cells [19-25]. However, its role in NSCs remains poorly understood. In this study, our primary data suggested that KLF4 may also have an important role in maintaining the stem cell properties of NSCs, as suppression of its expression by miRNA (miR-346) led to differentiation of NSCs and re-expression of it would converse this effects (Figure 5). Additional work were still needed to verify this conclusion. As a critical transcriptional factor, KLF4 has been shown to be intensively regulated. To date, it has been reported to be regulated by several miRNAs, including miR-10b, miR-32, miR-7a, 7b, miR-375, miR-367, miR-200b [39-44]. However, most of them were characterized in tumor cells. Here, we reported another miRNA, miR-346, as a novel regulator of KLF4 in NSCs. In conclusion, our study identified a new miRNA, miR-346, involved in NSCs regulation, and unveiled the underlying mechanism, that was by directly target a stem cell transcriptional factor KLF4. This study also added new insights into the regulation of KLF4 and its function in NSCs.
Acknowledgements
The Project was Supported by Natural Science Basic Research Plan in Shaanxi Province of China (Program No. 2017JM8103).
Disclosure of conflict of interest
None.
References
- 1.Weiss S, van der Kooy D. CNS stem cells: where’s the biology (a.k.a. beef)? J Neurobiol. 1998;36:307–314. doi: 10.1002/(sici)1097-4695(199808)36:2<307::aid-neu14>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- 2.Shi Y, Sun G, Zhao C, Stewart R. Neural stem cell self-renewal. Crit Rev Oncol Hematol. 2008;65:43–53. doi: 10.1016/j.critrevonc.2007.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brand AH, Livesey FJ. Neural stem cell biology in vertebrates and invertebrates: more alike than different? Neuron. 2011;70:719–729. doi: 10.1016/j.neuron.2011.05.016. [DOI] [PubMed] [Google Scholar]
- 4.Zhu N, Lin J, Wang K, Wei M, Chen Q, Wang Y. Huperzine A protects neural stem cells against Abeta-induced apoptosis in a neural stem cells and microglia co-culture system. Int J Clin Exp Pathol. 2015;8:6425–6433. [PMC free article] [PubMed] [Google Scholar]
- 5.Rossignol J, Fink K, Davis K, Clerc S, Crane A, Matchynski J, Lowrance S, Bombard M, Dekorver N, Lescaudron L, Dunbar GL. Transplants of adult mesenchymal and neural stem cells provide neuroprotection and behavioral sparing in a transgenic rat model of Huntington’s disease. Stem Cells. 2014;32:500–509. doi: 10.1002/stem.1508. [DOI] [PubMed] [Google Scholar]
- 6.Gonzalez C, Bonilla S, Flores AI, Cano E, Liste I. An update on human stem cell-based therapy in Parkinson’s disease. Curr Stem Cell Res Ther. 2016;11:561–568. doi: 10.2174/1574888x10666150531172612. [DOI] [PubMed] [Google Scholar]
- 7.You Y, Che L, Lee HY, Lee HL, Yun Y, Lee M, Oh J, Ha Y. Antiapoptotic effect of highly secreted GMCSF from neuronal cell-specific GMCSF overexpressing neural stem cells in spinal cord injury model. Spine (Phila Pa 1976) 2015;40:E1284–1291. doi: 10.1097/BRS.0000000000001080. [DOI] [PubMed] [Google Scholar]
- 8.Bago JR, Sheets KT, Hingtgen SD. Neural stem cell therapy for cancer. Methods. 2016;99:37–43. doi: 10.1016/j.ymeth.2015.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rajewsky N. microRNA target predictions in animals. Nat Genet. 2006;38(Suppl):S8–13. doi: 10.1038/ng1798. [DOI] [PubMed] [Google Scholar]
- 11.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 12.He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 13.Garg N, Po A, Miele E, Campese AF, Begalli F, Silvano M, Infante P, Capalbo C, De Smaele E, Canettieri G, Di Marcotullio L, Screpanti I, Ferretti E, Gulino A. microRNA-17-92 cluster is a direct Nanog target and controls neural stem cell through Trp53inp1. EMBO J. 2013;32:2819–2832. doi: 10.1038/emboj.2013.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou Y, Jiang H, Gu J, Tang Y, Shen N, Jin Y. MicroRNA-195 targets ADP-ribosylation factorlike protein 2 to induce apoptosis in human embryonic stem cell-derived neural progenitor cells. Cell Death Dis. 2013;4:e695. doi: 10.1038/cddis.2013.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li X, Feng R, Huang C, Wang H, Wang J, Zhang Z, Yan H, Wen T. MicroRNA-351 regulates TMEM 59 (DCF1) expression and mediates neural stem cell morphogenesis. RNA Biol. 2012;9:292–301. doi: 10.4161/rna.19100. [DOI] [PubMed] [Google Scholar]
- 16.Morgado AL, Xavier JM, Dionisio PA, Ribeiro MF, Dias RB, Sebastiao AM, Sola S, Rodrigues CM. MicroRNA-34a modulates neural stem cell differentiation by regulating expression of synaptic and autophagic proteins. Mol Neurobiol. 2015;51:1168–1183. doi: 10.1007/s12035-014-8794-6. [DOI] [PubMed] [Google Scholar]
- 17.Bhattacharya R, Senbanerjee S, Lin Z, Mir S, Hamik A, Wang P, Mukherjee P, Mukhopadhyay D, Jain MK. Inhibition of vascular permeability factor/vascular endothelial growth factormediated angiogenesis by the Kruppel-like factor KLF2. J Biol Chem. 2005;280:28848–28851. doi: 10.1074/jbc.C500200200. [DOI] [PubMed] [Google Scholar]
- 18.Fisch S, Gray S, Heymans S, Haldar SM, Wang B, Pfister O, Cui L, Kumar A, Lin Z, Sen-Banerjee S, Das H, Petersen CA, Mende U, Burleigh BA, Zhu Y, Pinto YM, Liao R, Jain MK. Kruppel-like factor 15 is a regulator of cardiomyocyte hypertrophy. Proc Natl Acad Sci U S A. 2007;104:7074–7079. doi: 10.1073/pnas.0701981104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aoi T, Yae K, Nakagawa M, Ichisaka T, Okita K, Takahashi K, Chiba T, Yamanaka S. Generation of pluripotent stem cells from adult mouse liver and stomach cells. Science. 2008;321:699–702. doi: 10.1126/science.1154884. [DOI] [PubMed] [Google Scholar]
- 20.Maherali N, Sridharan R, Xie W, Utikal J, Eminli S, Arnold K, Stadtfeld M, Yachechko R, Tchieu J, Jaenisch R, Plath K, Hochedlinger K. Directly reprogrammed fibroblasts show global epigenetic remodeling and widespread tissue contribution. Cell Stem Cell. 2007;1:55–70. doi: 10.1016/j.stem.2007.05.014. [DOI] [PubMed] [Google Scholar]
- 21.Nakagawa M, Koyanagi M, Tanabe K, Takahashi K, Ichisaka T, Aoi T, Okita K, Mochiduki Y, Takizawa N, Yamanaka S. Generation of induced pluripotent stem cells without Myc from mouse and human fibroblasts. Nat Biotechnol. 2008;26:101–106. doi: 10.1038/nbt1374. [DOI] [PubMed] [Google Scholar]
- 22.Okita K, Ichisaka T, Yamanaka S. Generation of germline-competent induced pluripotent stem cells. Nature. 2007;448:313–317. doi: 10.1038/nature05934. [DOI] [PubMed] [Google Scholar]
- 23.Stadtfeld M, Maherali N, Breault DT, Hochedlinger K. Defining molecular cornerstones during fibroblast to iPS cell reprogramming in mouse. Cell Stem Cell. 2008;2:230–240. doi: 10.1016/j.stem.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 25.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 26.Greider CW, Blackburn EH. Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell. 1985;43:405–413. doi: 10.1016/0092-8674(85)90170-9. [DOI] [PubMed] [Google Scholar]
- 27.Wong CW, Hou PS, Tseng SF, Chien CL, Wu KJ, Chen HF, Ho HN, Kyo S, Teng SC. Kruppellike transcription factor 4 contributes to maintenance of telomerase activity in stem cells. Stem Cells. 2010;28:1510–1517. doi: 10.1002/stem.477. [DOI] [PubMed] [Google Scholar]
- 28.Ma J, Yao Y, Wang P, Liu Y, Zhao L, Li Z, Li Z, Xue Y. MiR-152 functions as a tumor suppressor in glioblastoma stem cells by targeting Kruppel-like factor 4. Cancer Lett. 2014;355:85–95. doi: 10.1016/j.canlet.2014.09.012. [DOI] [PubMed] [Google Scholar]
- 29.Mori K, Hamanaka H, Oshima Y, Araki Y, Ishikawa F, Nose K, Shibanuma M. A HIC-5- and KLF4-dependent mechanism transactivates p21(Cip1) in response to anchorage loss. J Biol Chem. 2012;287:38854–38865. doi: 10.1074/jbc.M112.377721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chew YC, Adhikary G, Wilson GM, Reece EA, Eckert RL. Protein kinase C (PKC) delta suppresses keratinocyte proliferation by increasing p21(Cip1) level by a KLF4 transcription factor-dependent mechanism. J Biol Chem. 2011;286:28772–28782. doi: 10.1074/jbc.M110.205245. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 31.Zhu Y, Kalbfleisch T, Brennan MD, Li Y. A MicroRNA gene is hosted in an intron of a schizophrenia-susceptibility gene. Schizophr Res. 2009;109:86–89. doi: 10.1016/j.schres.2009.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weber F, Teresi RE, Broelsch CE, Frilling A, Eng C. A limited set of human MicroRNA is deregulated in follicular thyroid carcinoma. J Clin Endocrinol Metab. 2006;91:3584–3591. doi: 10.1210/jc.2006-0693. [DOI] [PubMed] [Google Scholar]
- 33.Oskowitz AZ, Lu J, Penfornis P, Ylostalo J, McBride J, Flemington EK, Prockop DJ, Pochampally R. Human multipotent stromal cells from bone marrow and microRNA: regulation of differentiation and leukemia inhibitory factor expression. Proc Natl Acad Sci U S A. 2008;105:18372–18377. doi: 10.1073/pnas.0809807105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Alsaleh G, Suffert G, Semaan N, Juncker T, Frenzel L, Gottenberg JE, Sibilia J, Pfeffer S, Wachsmann D. Bruton’s tyrosine kinase is involved in miR-346-related regulation of IL-18 release by lipopolysaccharide-activated rheumatoid fibroblast-like synoviocytes. J Immunol. 2009;182:5088–5097. doi: 10.4049/jimmunol.0801613. [DOI] [PubMed] [Google Scholar]
- 35.Semaan N, Frenzel L, Alsaleh G, Suffert G, Gottenberg JE, Sibilia J, Pfeffer S, Wachsmann D. miR-346 controls release of TNF-alpha protein and stability of its mRNA in rheumatoid arthritis via tristetraprolin stabilization. PLoS One. 2011;6:e19827. doi: 10.1371/journal.pone.0019827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen B, Pan W, Lin X, Hu Z, Jin Y, Chen H, Ma G, Qiu Y, Chang L, Hua C, Zou Y, Gao Y, Ying H, Lv D. MicroRNA-346 functions as an oncogene in cutaneous squamous cell carcinoma. Tumour Biol. 2016;37:2765–2771. doi: 10.1007/s13277-015-4046-2. [DOI] [PubMed] [Google Scholar]
- 37.Yan HL, Li L, Li SJ, Zhang HS, Xu W. miR-346 promotes migration and invasion of nasopharyngeal carcinoma cells via targeting BRMS1. J Biochem Mol Toxicol. 2016;30:602–607. doi: 10.1002/jbt.21827. [DOI] [PubMed] [Google Scholar]
- 38.Wang Q, Cai J, Cai XH, Chen L. miR-346 regulates osteogenic differentiation of human bone marrow-derived mesenchymal stem cells by targeting the Wnt/beta-catenin pathway. PLoS One. 2013;8:e72266. doi: 10.1371/journal.pone.0072266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang J, Wang B, Chen LQ, Yang J, Gong ZQ, Zhao XL, Zhang CQ, Du KL. miR-10b promotes invasion by targeting KLF4 in osteosarcoma cells. Biomed Pharmacother. 2016;84:947–953. doi: 10.1016/j.biopha.2016.09.108. [DOI] [PubMed] [Google Scholar]
- 40.Zhao L, Han T, Li Y, Sun J, Zhang S, Liu Y, Shan B, Zheng D, Shi J. The lncRNA SNHG5/miR-32 axis regulates gastric cancer cell proliferation and migration by targeting KLF4. FASEB J. 2017;31:893–903. doi: 10.1096/fj.201600994R. [DOI] [PubMed] [Google Scholar]
- 41.Wang Q, Li G, Li B, Chen Q, Lv D, Liu J, Ma J, Sun N, Yang L, Fei X, Song Q. Sevoflurane represses the self-renewal ability by regulating miR-7a,7b/Klf4 signalling pathway in mouse embryonic stem cells. Cell Prolif. 2016;49:609–617. doi: 10.1111/cpr.12283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang GC, He QY, Tong DK, Wang CF, Liu K, Ding C, Ji F, Zhang H. MiR-367 negatively regulates apoptosis induced by adriamycin in osteosarcoma cells by targeting KLF4. J Bone Oncol. 2016;5:51–56. doi: 10.1016/j.jbo.2016.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mao Q, Quan T, Luo B, Guo X, Liu L, Zheng Q. MiR-375 targets KLF4 and impacts the proliferation of colorectal carcinoma. Tumour Biol. 2016;37:463–471. doi: 10.1007/s13277-015-3809-0. [DOI] [PubMed] [Google Scholar]
- 44.Eggers JC, Martino V, Reinbold R, Schafer SD, Kiesel L, Starzinski-Powitz A, Schuring AN, Kemper B, Greve B, Gotte M. microRNA miR-200b affects proliferation, invasiveness and stemness of endometriotic cells by targeting ZEB1, ZEB2 and KLF4. Reprod Biomed Online. 2016;32:434–445. doi: 10.1016/j.rbmo.2015.12.013. [DOI] [PubMed] [Google Scholar]


