Abstract
Cryptococcosis is a systemic infection caused by species of the encapsulated yeast Cryptococcus. The disease may occur in immunocompromised and immunocompetent hosts and is acquired by the inhalation of infectious propagules present in the environment. Cryptococcus is distributed in a plethora of ecological niches, such as soil, pigeon droppings, and tree hollows, and each year new reservoirs are discovered, which helps researchers to better understand the epidemiology of the disease. In this review, we describe the ecoepidemiology of the C. gattii species complex focusing on clinical cases and ecological reservoirs in developing countries from different continents. We also discuss some important aspects related to the antifungal susceptibility of different species within the C. gattii species complex and bring new insights on the revised Cryptococcus taxonomy.
Keywords: Cryptococcus, cryptococcosis, reservoirs, developing countries, ecoepidemiology
1. Introduction
Cryptococcosis is a systemic fungal disease caused by yeasts belonging to the Cryptococcus neoformans/C. gattii species complexes [1], affecting both immunocompetent and immunocompromised hosts and causing devastating diseases [2]. Cryptococcal meningitis is the most common mycosis associated with acquired immune deficiency syndrome (AIDS) patients with significant morbidity and mortality especially in sub-Saharan Africa, Asia, and Latin America [3]. It is estimated that approximately 225,000 new cryptococcal meningitis cases occur globally each year, the majority of which (73%) occur in sub-Saharan Africa [3].
C. gattii sensu lato (s.l.) is an emerging pathogen, initially considered an endemic disease, affecting patients living in tropical and subtropical zones [4]. However, over time, the geographic distribution of C. gattii s.l. infections expanded to temperate climate regions including Canada and the USA [4,5,6]. In addition, many ecological niches have been investigated globally in an attempt to elucidate the environmental reservoirs [7,8,9].
In this review, we describe the ecological distribution of the C. gattii species complex and highlight the environmental reservoirs of this pathogen in developing countries. We also discuss some important points about the antifungal susceptibility of this species complex and changes in the Cryptococcus taxonomy that has recently been debated among researchers and clinicians.
2. Cryptococcus gattii Species Complex Distribution in Developing Countries
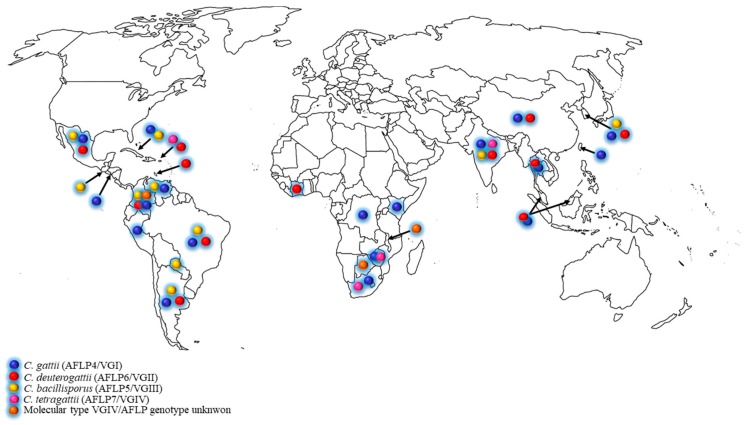
The C. gattii species complex was initially found in tropical and subtropical areas [10], but during the past two decades, the expansion to temperate climate regions was increasingly reported [5,6,11,12,13,14,15]. The ecological niches of the C. gattii species complex has been thoroughly investigated, and many global studies revealed that a plethora of tree species may be colonized by these pathogenic fungi [1,7,8,9,13,14,16,17,18,19,20]. The distribution of C. gattii species complex in developing countries is shown in the Figure 1 and Table 1. Based on these data, it became clear that the C. gattii species complex is not associated to a specific tree genus but that it has a predilection for plant/wood debris in general.
Figure 1.
Distribution of clinical and environmental Cryptococcus gattii species complex in developing countries.
Table 1.
Cryptococcus gattii species complex in developing countries.
| Continent | Species | Source | Country |
|---|---|---|---|
| Latin America | C. gattii s.s. | Clinical, Environmental, Veterinary | Argentina [21,22,23,24], Brazil [25,26,27,28], Colombia [29,30], Cuba [31], Honduras [32], Mexico [33,34], Peru [35] |
| C. deuterogattii | Clinical, Environmental, Veterinary | Brazil [18,25,26,36,37,38,39,40,41,42,43,44], Colombia [29,30], French Guiana [45], Mexico [33,34], Puerto Rico [46] | |
| C. bacillisporus | Clinical, Environmental | Argentina [23], Brazil [25], Colombia [29,30,47], Cuba [48], Guatemala [48], Mexico [33,34], Paraguay [48], Venezuela [48] | |
| C. tetragattii | Environmental | Colombia [35], Mexico [33,34], Puerto Rico [46] | |
| Africa | C. gattii s.l. | Clinical, Environmental | Botswana [49], Rwanda [50], South Africa [51,52,53], Zambia [54] |
| C. gattii s.s. | Clinical | D.R. Congo [32], Kenya [55], South Africa [56], Zimbabwe [57,58] | |
| C. deuterogattii | Clinical | Ivory Coast [59] | |
| C. tetragattii | Clinical, Veterinary | Botswana [60], Malawi [60], South Africa [57]; Zimbabwe [57,58] | |
| Asia | C. gattii s.l. | Clinical, Environmental | China [61,62,63], India [64,65,66,67,68,69], Taiwan [70] |
| C. gattii s.s. | Clinical, Environmental | China [71,72,73,74,75], India [76,77,78], Korea [79], Malaysia [80,81], Taiwan [82], Thailand [8,83] | |
| C. deuterogattii | Clinical | China [71,74,75], India [84], Korea [79,85], Malaysia [80,81], Thailand [83,86] | |
| C. bacillisporus | Clinical, Environmental | India [78], Korea [79,85] | |
| C. tetragattii | Clinical, Veterinary | India [87] |
s.s.: sensu stricto; s.l.: sensu lato.
2.1. Latin America
C. gattii sensu stricto (s.s.) (genotype AFLP4/VGI) is a major aetiologic agent of cryptococcosis among immunocompetent patients from Brazil [25,26,27], Colombia [29], Mexico [33,34], Honduras [32], and Peru [35] and has caused pneumonia in a renal transplant patient from Argentina [21]. This pathogen has also been involved with a fatal infection in Cuba in an imported cheetah from South Africa [31]. In nature, this species was found in Psittaciformes excreta in Brazil [28] and some tree species, such as Tipuana tipu, Grevillea robusta, and Eucalyptus spp. in Argentina [22,23,24] and Ficus spp. in Colombia [30].
C. deuterogattii (genotype AFLP6/VGII) has been isolated from clinical samples and has been involved in meningitis, cutaneous diseases, and lung infection in immunocompetent and HIV-positive patients from Brazil [25,26,36,37,38], Colombia [29], Mexico [33,34], and French Guiana [45]. From Brazil, it was also reported causing disease in dogs [39,40]. C. deuterogattii is found in a variety of ecological niches, being isolated from tree detritus in Puerto Rico [46], from Moquilea tomentosa, Plathymenia reticulata, and Senna sianea in Brazil [18,41,42] and from Eucalyptus spp. in Colombia [30]. In addition, C. deuterogattii was also isolated from indoor dust from typical wooden houses in Amazonas, Brazil, and from Guettarda acreana trees [43,44].
The species C. bacillisporus (genotype AFLP5/VGIII) has been isolated from clinical samples from Colombian immunocompetent patients [29], as well as from patients in Mexico [33,34], Cuba [88], Brazil [25], Guatemala, Paraguay, and Venezuela [48]. In the environment, Corymbia ficifolia and Ficus spp. trees have been reported as reservoirs in Colombia [30,47] and Tipuana tipu in Argentina [23].
C. tetragattii (genotype AFLP7/VGIV) has been found in Puerto Rico from tree detritus [46] and the molecular type VGIV has also been found in México and Colombia, but AFLP or MLST genotyping has not been performed to differentiate C. tetragattii (genotype AFLP7/VGIV) and C. decagattii (genotype AFLP10/VGIV) from each other [33,34,35]. Some of these isolates have been recently investigated and shown to belong to C. decagattii rather than to C. tetragattii [1].
2.2. Africa
On the African continent, most of the literature data consists in descriptions of the C. neoformans species complex’s ecological distribution and clinical involvement. But only few studies have found new C. gattii species complex members. In South Africa, HIV-positive and HIV-negative children and adults were reported to have cryptococcosis caused by C. gattii s.l., but no genotyping was performed to determine the species [51,52,53]. The same holds true for Botswana [49] and Rwanda, where cryptococcal meningitis cases with C. gattii s.l. were found [50]. Environmental niches of C. gattii s.l. were investigated in Zambia, where positive samples were found in Colophospermum mopane, Julbernadia globiflora, Eucalyptus spp., Brachystegia spp., fig tree, and feces from Hyrax midden, but no genotyping was performed [54].
C. gattii s.s. (genotype AFLP4/VGI) has been recovered from HIV-positive patients, bird droppings, Acacia xanthophloea, and Eucalyptus saligna from Kenya [55], from HIV-positive patients with meningitis from Zimbabwe [57,58], D.R. Congo [32], and South Africa [56]. The species C. deuterogattii (genotype AFLP6/VGII) was reported causing cryptococcosis in HIV-positive patients from Ivory Coast [59]. In addition, C. tetragattii (genotype AFLP7/VGIV) was found to be a major cause of meningitis in HIV-positive patients in Zimbabwe [57,58] and was reported to cause cryptococcosis in patients from Botswana, Malawi, and South Africa [57,60]. A South African veterinary C. tetragattii (genotype AFLP7/VGIV) isolate was closely related to environmental C. tetragattii isolates from Colombia, Puerto Rico, and Spain [57].
2.3. Asia
There have been many studies performed in developing Asian countries reporting C. gattii s.l. causing human diseases and the ecological niche of this species complex. Unfortunately, most of the isolates were not genotyped. Chen and colleagues (2000) performed a study in Taiwan with clinical cases of cryptococcosis during the 1980s and 1990s [70]. Infections by C. gattii s.l. occurred in 35.6% of patients during the study period. The cryptococcosis cases included both immunocompetent and immunocompromised patients with a predominance of central nervous system (CNS) diseases [70]. In China, C. gattii s.l. was isolated from a surgical wound [61]. In addition, in India, it was isolated from HIV-positive and HIV-negative patients [62,63]. Environmental niches of C. gattii s.l. in India were recognized being tree hollows of Syzygium cumini, Ficus religiosa, Polyalthia longifolia, Azadirachta indica, Cassia fistula, Mimusops elengi, and Cassia marginata [64,65,66,67,68], and flowers, bark, and detritus of Eucalyptus camaldulensis and E. tereticornes [69].
C. gattii s.s (genotype AFLP4/VGI) was isolated from patients with meningitis in Malaysia [80,81] and India [76]; this species was also reported in clinical samples from Korean, Taiwanese, and Thai patients [79,82,83]. China has reported cryptococcosis cases in HIV-positive, HIV-negative, and immunocompetent patients [71,72,73,74,75]. The environmental source of Thai C. gattii s.s. is decaying wood inside a Castanopsis argyrophylla hollow [8], and in India this species was isolated from tree hollows [77,78].
C. deuterogattii (genotype AFLP6/VGII) has been isolated from chronic meningitis in Malaysia [80,81], HIV-negative patients from Korea and India [79,84,85], and immunocompetent patients in China [71,74,75]. In Thailand, this species was reported causing disease in HIV-positive and HIV-negative patients, as well as causing primary cutaneous cryptococcosis [83,86]. However, the environmental niche of this species has not been reported.
C. bacillisporus (genotype AFLP5/VGIII) was found causing diseases in patients from Korea [79,85], which is interesting because in Asia C. gattii s.s. and C. deuterogattii have a predominance among clinical samples. In India, the first environmental C. bacillisporus isolate was recovered from decaying wood of Manilkara hexandra [78].
C. tetragattii (genotype AFLP7/VGIV) was isolated in India from several clinical sources, including an HIV-positive patient with meningitis, cutaneous lesions, and granulomas in HIV-negative patients. All these isolates were genetically similar to C. tetragattii found in Botswana, Africa. However, only one patient had previously travelled to Egypt [87].
A hypothesis to explain the differences in geographic distribution of C. gattii/C. neoformans species complexes was put forward by Casadevall and colleagues (2017). These authors hypothesized that it may be attributed to the breakup of the supercontinent Pangea. The physical separation of Cryptococcus species complexes was an important point for its speciation [89]. In addition, it was suggested that environmental events, such as wind, ocean currents, and animals, would be involved, driving the more recent speciation of Cryptococcus species complexes [89]. Despite all epidemiological studies carried out, there are many countries where the presence of cryptococcal molecular genotypes has not yet been explored [90].
3. Antifungal Susceptibility among the C. gattii Species Complex
Among cryptococcal species, different antifungal susceptibility patterns have been observed. In general, the C. gattii species complex shows higher minimum inhibitory concentrations (MICs) of azoles than isolates from the C. neoformans species complex [68,91,92]. In addition, C. gattii s.l. clinical isolates from Taiwan showed higher amphotericin B and flucytosine MIC values than C. neoformans s.l. clinical isolates [70]. In Brazil, C. deuterogattii (genotype AFLP6/VGII) clinical isolates showed higher MIC values for flucytosine [93,94] and fluconazole than C. neoformans s.s. (genotype AFLP1/VNI) [93].
However, different antifungal susceptibility profiles are also present within species of the C. gattii species complex. C. deuterogattii (genotype AFLP6/VGII) has higher geometric mean MICs for flucytosine, fluconazole, voriconazole, itraconazole, posaconazole, and isavuconazole than C. gattii s.s. (genotype AFLP4/VGI) [95]. Lockhart and colleagues (2012) investigated the correlation of C. gattii species complex and its antifungal susceptibility [96]. C. deuterogattii (genotype AFLP6/VGII) had the highest geometric mean MIC for fluconazole, followed by C. bacillisporus (genotype AFLP5/VGIII), genotype VGIV (AFLP non-genotyped), while C. gattii s.s. (genotype AFLP4/VGI) had the lowest among species [96]. Trilles and colleagues (2012) also observed that C. deuterogattii isolates had higher MICs of azoles than C. gattii s.s. [93]. An Indian study showed that clinical and environmental C. gattii s.l. isolates had high fluconazole MICs [68]. However, despite these differences in antifungal susceptibility among cryptococcal species, the initial cryptococcosis therapy is the same; the clinical management changes are according to presentations and immune status, but does not consider the species involved in the disease [97].
Another important point is the phenomenon of heteroresistance, the ability of adaptation to a high concentrations of drugs, observed in C. gattii s.l. to itraconazole and fluconazole [98,99]. The development of heteroresistance is related to phenotypic changes, such as a decrease in cell and capsule size, low ergosterol content in the cell wall, less susceptibility to oxidative stress, and a great ability to proliferate inside macrophages [98,99]. This intrinsic mechanism present in members of the C. neoformans/C. gattii species complexes may contribute to a relapse of cryptococcosis during maintenance therapy [98,100]. However, the clinical importance of heteroresistance is not yet clear and requires further investigation [100,101].
4. The C. gattii Species Complex: Four Molecular Types, Five Genotypes or Five Species?
The taxonomy of the tremellomycetous yeasts has recently been revised [102,103]. Since the genus Cryptococcus was described, it has grown out as a highly polyphyletic one that contained more than 100 species within the orders Filobasidiales, Tremellales, and Trichosporonales [102,103]. The taxonomic revision of the genus Cryptococcus has been extensively discussed over the past two decades. At the 6th International Conference on Cryptococcus and Cryptococcosis (ICCC) debate, “How many species and varietal states are there?” [104], different hypotheses were discussed about the status of the C. neoformans/C. gattii species complex: Should the situation be kept in a “two-species division”? [105,106]. Should it be divided into six species? [107,108]. Eight? [109]. The hypotheses were supported based on different opinions about the definition of species. The first one was supported by the idea that phenetic, biological, and cladistic species concepts need to be used together to proper classify the agents of cryptococcosis, because genetic variation as shown by the molecular types does not always reflect their biological characteristics [105,106,110]. However, the second hypothesis was based on phylogenetic support that included analysis of mitochondrial, ribosomal, and nuclear genes to investigate the relationship among the various C. neoformans and C. gattii genotypes. The different genotypes clustered in six monophyletic lineages for all loci studied, suggesting that C. neoformans serotype A and D represent two different species and that C. gattii genotypes represent four individual taxa [107,108]. The third hypothesis goes a little further, considering that each genotype within C. neoformans and C. gattii has sufficient genotypic variation to be considered a different species [109].
Phenotypic diversity within the C. gattii species complex is also supporting the division of five species. Capsule and cell size showed to be variable within the complex, C. gattii s.s. (genotype AFLP4/VGI) had the largest capsules but smaller cells compared to the other species, while C. deuterogattii (genotype AFLP6/VGII) has the largest cells but smaller capsules [111]. All species in the C. gattii species complex have the ability to grow at 25, 30, and 35 °C, but with variable tolerance to 37 °C [1,111]. C. deuterogattii (genotype AFLP6/VGII) has the highest thermotolerance to 37 °C, while C. gattii s.s. (genotype AFLP4/VGI), C. bacillisporus (genotype AFLP/VGIII), and C. tetragattii (genotype AFLP7/VGIV) have less growth at 37 °C than 30 °C [1,111,112]. There is no significant difference in tolerance to oxidative or osmotic stresses among species [111,112].
The understanding of genetic diversity is an important step for the discovery of previously unrecognized phenetic differences [111]. The exact moment that individuals in an ancestral species are split into progeny species is not recognized for any method of species delimitation, because this process needs time until the changes in morphology, mating behavior, or gene sequences may be recognized in the progeny species [113]. Phylogenomic analyses calculated the time since divergence of the C. neoformans species complex and the C. gattii species complex to be ~34 million years ago (mya) [114]; the divergence between C. deuterogattii (AFLP6/VGII) and the other species of the C. gattii species complex occurred ~12 mya [114,115]; and the divergence of C. neoformans (AFLP1/VNI/AFLP1; VNII/AFLP1A/1B) and C. deneoformans (VNIV/AFLP2), ~24 mya [115]. The divergence among the species within the C. neoformans/C. gattii species complexes occurred recently, and will most likely continue as an ongoing process. The occurrence of interspecies hybrids may also be attributed to the recent divergence event, because species currently hybridizing are most likely the youngest [116].
Although a revision of the cryptococcal taxonomy has been published, part of the cryptococcal research community is not fully in favor of using the ‘seven species recognition’. Some investigators believe that it will lead to taxonomic instability due to the fact that there are most likely more species present. Many points have been discussed, including the number of isolates used, the use of phylogenetic approaches for species delineation, the accommodation of hybrids in the new taxonomy, and the fact that the new names may cause confusion between the published literature and clinical practice [110]. With these points of view, Kwon-Chung and colleagues (2017) suggested the use of the “C. neoformans species complex” and the “C. gattii species complex” as an intermediate step, instead of using the seven species nomenclature, until biological and clinical relevant differences become clear [110]. Although, according to Hagen and colleagues (2017), it is important to consider the presence of different species inside the complexes to avoid delay in the clinical progress [117].
5. Final Remarks
Clinical and environmental occurrence of the C. gattii species complex is related to geographic location, which may be attributed to the (micro)climate, or even a lack of diagnosis/environmental isolation. Cryptococcosis in most developing countries is underreported and the precise burden of cryptococcosis caused by the C. gattii species complex is uncertain. In addition, not all clinical laboratories differentiate the pathogenic Cryptococcus species. In the environment, many tree species have been described as a reservoir, proving that the C. gattii species complex has no tree species-specific relation, and is widely spread in the environment.
C. gattii species complex members differ in phenotypic traits, as capsule and cell size, thermotolerance and antifungal susceptibility. Many studies have demonstrated higher MICs of azoles for members of the C. gattii species complex compared to the C. neoformans species complex. Difference in antifungal susceptibility has also been observed within the C. gattii species complex, with C. deuterogattii (genotype AFLP6/VGII) being less susceptible to azoles than C. gattii s.s (genotype AFLP4/VGI). However, in vitro antifungal susceptibility does not correlate to in vivo susceptibility. Clinical manifestations in patients with C. gattii s.l. infections tend to be more severe than C. neoformans. In the former, cerebral involvement causes more hydrocephalus, focal CNS signs, as well as papilledema, ataxia, hearing loss, altered mentation, and neurological sequelae. Usually, meningo-encephalitis caused by C. gattii s.l. is followed by higher intracranial pressures, sometimes irresponsible to multiple LPs and/or CNF shunts. Simultaneous pulmonary involvement in >50% of patients is also observed, and mass lesions (cryptococcomas) are associated to a prolonged clinical course and respond slowly to therapy.
A new taxonomy of the polyphyletic genus Cryptococcus has been published, including the medical important species complexes C. neoformans and C. gattii. In addition, there are different opinions about the new classification. The presence of genetic differences within the C. gattii species complex needs to be considered in future studies to correlate genotypic and phenotypic traits of each species to diseases clinical presentation.
Acknowledgments
Patricia F. Herkert is financially supported by “Coordenação de Aperfeiçoamento de Pessoal de Nível Superior” and “Conselho Nacional de Desenvolvimento Científico e Tecnológico”, Brazil. Jacques F. Meis received research grants from Astellas, Basilea, Gilead Sciences, and Merck. Flávio Queiroz-Telles received grants for advisory boards and clinical research from Astellas, Gilead, MSD, Pfizer, and United Medical.
Author contributions
Patricia F. Herkert and Ferry Hagen designed the study and wrote the first draft; Rosangela L. Pinheiro and Marisol D. Muro analyzed the data; Jacques F. Meis and Flávio Queiroz-Telles revised the paper. All authors contributed to the writing and approved the final manuscript.
Conflicts of Interest
The other authors declare no conflict of interest.
References
- 1.Hagen F., Khayhan K., Theelen B., Kolecka A., Polacheck I., Sionov E., Falk R., Parnmen S., Lumbsch H.T., Boekhout T. Recognition of seven species in the Cryptococcus gattii/Cryptococcus neoformans species complex. Fungal Genet. Biol. 2015;78:16–48. doi: 10.1016/j.fgb.2015.02.009. [DOI] [PubMed] [Google Scholar]
- 2.May R.C., Stone N.R.H., Wiesner D.L., Bicanic T., Nielsen K. Cryptococcus: From environmental saprophyte to global pathogen. Nat. Rev. Microbiol. 2015;14:106–117. doi: 10.1038/nrmicro.2015.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rajasingham R., Smith R.M., Park B.J., Jarvis J.N., Govender N.P., Chiller T.M., Denning D.W., Loyse A., Boulware D.R. Global burden of disease of HIV-associated cryptococcal meningitis: An updated analysis. Lancet Infect. Dis. 2017;17:873–881. doi: 10.1016/S1473-3099(17)30243-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chaturvedi V., Chaturvedi S. Cryptococcus gattii: A resurgent fungal pathogen. Trends Microbiol. 2011;19:564–571. doi: 10.1016/j.tim.2011.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kidd S.E., Hagen F., Tscharke R.L., Huynh M., Bartlett K.H., Fyfe M., MacDougall L., Boekhout T., Kwon-Chung K.J., Meyer W. A rare genotype of Cryptococcus gattii caused the cryptococcosis outbreak on Vancouver Island (British Columbia, Canada) Proc. Natl. Acad. Sci. USA. 2004;101:17258–17263. doi: 10.1073/pnas.0402981101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Datta K., Bartlett K.H., Baer R., Byrnes E., Galanis E., Heitman J., Hoang L., Leslie M.J., MacDougall L., Magill S.S., et al. Spread of Cryptococcus gattii into Pacific Northwest Region of the United States. Emerg. Infect. Dis. 2009;15:1185–1191. doi: 10.3201/eid1508.081384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cogliati M., D’Amicis R., Zani A., Montagna M.T., Caggiano G., De Giglio O., Balbino S., De Donno A., Serio F., Susever S., et al. Environmental distribution of Cryptococcus neoformans and C. gattii around the Mediterranean basin. FEMS Yeast Res. 2016;16:fow045. doi: 10.1093/femsyr/fow045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Khayhan K., Hagen F., Norkaew T., Puengchan T., Boekhout T., Sriburee P. Isolation of Cryptococcus gattii from a Castanopsis argyrophylla tree hollow (Mai-Kaw), Chiang Mai, Thailand. Mycopathologia. 2017;182:365–370. doi: 10.1007/s11046-016-0067-7. [DOI] [PubMed] [Google Scholar]
- 9.Vélez N., Escandón P. Report on novel environmental niches for Cryptococcus neoformans and Cryptococcus gattii in Colombia: Tabebuia guayacan and Roystonea regia. Med. Mycol. 2017;55 doi: 10.1093/mmy/myw138. [DOI] [PubMed] [Google Scholar]
- 10.Kwon-Chung K.J., Bennett J.E. High prevalence of Cryptococcus neoformans var. gattii in tropical and subtropical regions. Zentralbl. Bakteriol. Mikrobiol. Hyg. A. 1984;257:213–218. [PubMed] [Google Scholar]
- 11.Galanis E., MacDougall L., Kidd S., Morshed M., The British Columbia Cryptococcus gattii working group Epidemiology of Cryptococcus gattii, British Columbia, Canada, 1999–2007. Emerg. Infect. Dis. 2010;16:251–257. doi: 10.3201/eid1602.090900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hagen F., Boekhout T. The search for the natural habitat of Cryptococcus gattii. Mycopathologia. 2010;170:209–211. doi: 10.1007/s11046-010-9313-6. [DOI] [PubMed] [Google Scholar]
- 13.Chowdhary A., Randhawa H.S., Boekhout T., Hagen F., Klaassen C.H., Meis J.F. Temperate climate niche for Cryptococcus gattii in Northern Europe. Emerg. Infect. Dis. 2012;18:172–174. doi: 10.3201/eid1801.111190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Colom M.F., Hagen F., Gonzalez A., Mellado A., Morera N., Linares C., García D.F., Peñataro J.S., Boekhout T., Sánchez M. Ceratonia siliqua (carob) trees as natural habitat and source of infection by Cryptococcus gattii in the Mediterranean environment. Med. Mycol. 2012;50:67–73. doi: 10.3109/13693786.2011.574239. [DOI] [PubMed] [Google Scholar]
- 15.Rosenberg J.F., Haulena M., Hoang L.M.N., Morshed M., Zabek E., Raverty S.A. Cryptococcus gattii type VGIIa infection in harbor seals (Phoca vitulina) in British Columbia, Canada. J. Wildl. Dis. 2016;52:677–681. doi: 10.7589/2015-11-299. [DOI] [PubMed] [Google Scholar]
- 16.Kidd S.E., Chow Y., Mak S., Bach P.J., Chen H., Hingston A.O., Kronstad J.W., Bartlett K.H. Characterization of environmental sources of the human and animal pathogen Cryptococcus gattii in British Columbia, Canada, and the Pacific Northwest of the United States. Appl. Environ. Microbiol. 2007;73:1433–1443. doi: 10.1128/AEM.01330-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hagen F., Chowdhary A., Prakash A., Yntema J.-B., Meis J.F. Molecular characterization of Cryptococcus gattii genotype AFLP6/VGII isolated from woody debris of divi-divi (Caesalpinia coriaria), Bonaire, Dutch Caribbean. Rev. Iberoam. Micol. 2014;31:193–196. doi: 10.1016/j.riam.2013.10.007. [DOI] [PubMed] [Google Scholar]
- 18.Anzai M.C., dos Santos Lazéra M., Wanke B., Trilles L., Dutra V., de Paula D.A.J., Nakazato L., Takahara D.T., Simi W.B., Hahn R.C. Cryptococcus gattii VGII in a Plathymenia reticulata hollow in Cuiabá, Mato Grosso, Brazil. Mycoses. 2014;57:414–418. doi: 10.1111/myc.12177. [DOI] [PubMed] [Google Scholar]
- 19.Linares C., Colom M.F., Torreblanca M., Esteban V., Romera Á., Hagen F. Environmental sampling of Ceratonia siliqua (carob) trees in Spain reveals the presence of the rare Cryptococcus gattii genotype AFLP7/VGIV. Rev. Iberoam. Micol. 2015;32:269–272. doi: 10.1016/j.riam.2014.11.002. [DOI] [PubMed] [Google Scholar]
- 20.Chowdhary A., Randhawa H.S., Prakash A., Meis J.F. Environmental prevalence of Cryptococcus neoformans and Cryptococcus gattii in India: An update. Crit. Rev. Microbiol. 2012;38:1–16. doi: 10.3109/1040841X.2011.606426. [DOI] [PubMed] [Google Scholar]
- 21.Cicora F., Petroni J., Formosa P., Roberti J. A rare case of Cryptococcus gattii pneumonia in a renal transplant patient. Transpl. Infect. Dis. 2015;17:463–466. doi: 10.1111/tid.12371. [DOI] [PubMed] [Google Scholar]
- 22.Refojo N., Perrotta D., Brudny M., Abrantes R., Hevia A.I., Davel G. Isolation of Cryptococcus neoformans and Cryptococcus gattii from trunk hollows of living trees in Buenos Aires City, Argentina. Med. Mycol. 2009;47:177–184. doi: 10.1080/13693780802227290. [DOI] [PubMed] [Google Scholar]
- 23.Mazza M., Refojo N., Bosco-Borgeat M.E., Taverna C.G., Trovero A.C., Rogé A., Davel G. Cryptococcus gattii in urban trees from cities in North-eastern Argentina. Mycoses. 2013;56:646–650. doi: 10.1111/myc.12084. [DOI] [PubMed] [Google Scholar]
- 24.Cattana M.E., de los Ángeles Sosa M., Fernández M., Rojas F., Mangiaterra M., Giusiano G. Native trees of the Northeast Argentine: Natural hosts of the Cryptococcus neoformans-Cryptococcus gattii species complex. Rev. Iberoam. Micol. 2014;31:188–192. doi: 10.1016/j.riam.2013.06.005. [DOI] [PubMed] [Google Scholar]
- 25.Trilles L., dos Santos Lazéra M., Wanke B., Oliveira R.V., Barbosa G.G., Nishikawa M.M., Morales B.P., Meyer W. Regional pattern of the molecular types of Cryptococcus neoformans and Cryptococcus gattii in Brazil. Mem. Inst. Oswaldo Cruz. 2008;103:455–462. doi: 10.1590/S0074-02762008000500008. [DOI] [PubMed] [Google Scholar]
- 26.Martins L.M.S., Wanke B., dos Santos Lazéra M., Trilles L., Barbosa G.G., de Macedo R.C.L., do Amparo Salmito Cavalcanti M., Eulálio K.D., Castro J.A., Silva A.S., et al. Genotypes of Cryptococcus neoformans and Cryptococcus gattii as agents of endemic cryptococcosis in Teresina, Piauí (northeastern Brazil) Mem. Inst. Oswaldo Cruz. 2011;106:725–730. doi: 10.1590/S0074-02762011000600012. [DOI] [PubMed] [Google Scholar]
- 27.Nascimento E., Bonifácio da Silva M.E.N., Martinez R., von Zeska Kress M.R. Primary cutaneous cryptococcosis in an immunocompetent patient due to Cryptococcus gattii molecular type VGI in Brazil: A case report and review of literature. Mycoses. 2014;57:442–447. doi: 10.1111/myc.12176. [DOI] [PubMed] [Google Scholar]
- 28.Abegg M.A., Cella F.L., Faganello J., Valente P., Schrank A., Vainstein M.H. Cryptococcus neoformans and Cryptococcus gattii isolated from the excreta of Psittaciformes in a Southern Brazilian Zoological Garden. Mycopathologia. 2006;161:83–91. doi: 10.1007/s11046-005-0186-z. [DOI] [PubMed] [Google Scholar]
- 29.Lizarazo J., Escandón P., Agudelo C.I., Firacative C., Meyer W., Castañeda E. Retrospective study of the epidemiology and clinical manifestations of Cryptococcus gattii infections in Colombia from 1997–2011. PLoS Neglect. Trop. Dis. 2014;8:e3272. doi: 10.1371/journal.pntd.0003272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Escandón P., Castañeda E. Long-term survival of Cryptococcus neoformans and Cryptococcus gattii in stored environmental samples from Colombia. Rev. Iberoam. Micol. 2015;32:197–199. doi: 10.1016/j.riam.2014.06.007. [DOI] [PubMed] [Google Scholar]
- 31.Illnait-Zaragozí M.T., Hagen F., Fernández-Andreu C.M., Martínez-Machín G.F., Polo-Leal J.L., Boekhout T., Klaassen C.H., Meis J.F. Reactivation of a Cryptococcus gattii infection in a cheetah (Acinonyx jubatus) held in the National Zoo, Havana, Cuba. Mycoses. 2011;54:e889–e892. doi: 10.1111/j.1439-0507.2011.02046.x. [DOI] [PubMed] [Google Scholar]
- 32.Boekhout T., Theelen B., Diaz M., Fell J.W., Hop W.C., Abeln E.C., Dromer F., Meyer W. Hybrid genotypes in the pathogenic yeast Cryptococcus neoformans. Microbiology. 2001;147:891–907. doi: 10.1099/00221287-147-4-891. [DOI] [PubMed] [Google Scholar]
- 33.Olivares L.R.C., Martínez K.M., Cruz R.M.B., Rivera M.A.M., Meyer W., Espinosa R.A.A., Martínez R.L., Santos G.M.R.P.Y. Genotyping of Mexican Cryptococcus neoformans and C. gattii isolates by PCR-fingerprinting. Med. Mycol. 2009;47:713–721. doi: 10.3109/13693780802559031. [DOI] [PubMed] [Google Scholar]
- 34.González G.M., Casillas-Vega N., Garza-González E., Hernández-Bello R., Rivera G., Rodríguez J.A., Bocanegra-Garcia V. Molecular typing of clinical isolates of Cryptococcus neoformans/Cryptococcus gattii species complex from Northeast Mexico. Folia Microbiol. 2016;61:51–56. doi: 10.1007/s12223-015-0409-8. [DOI] [PubMed] [Google Scholar]
- 35.Meyer W., Castañeda A., Jackson S., Huynh M., Castañeda E., the IberoAmerican cryptococcal study group Molecular typing of IberoAmerican Cryptococcus neoformans isolates. Emerg. Infect. Dis. 2003;9:189–195. doi: 10.3201/eid0902.020246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Freire A.K.L., dos Santos Bentes A., de Lima Sampaio I., Matsuura A.B.J., Ogusku M.M., Salem J.I., Wanke B., de Souza J.V.B. Molecular characterisation of the causative agents of cryptococcosis in patients of a tertiary healthcare facility in the state of Amazonas-Brazil: Cryptococcosis in the state of Amazonas-Brazil. Mycoses. 2012;55:e145–e150. doi: 10.1111/j.1439-0507.2012.02173.x. [DOI] [PubMed] [Google Scholar]
- 37.Matos C.S., de Souza Andrade A., Oliveira N.S., Barros T.F. Microbiological characteristics of clinical isolates of Cryptococcus spp. in Bahia, Brazil: Molecular types and antifungal susceptibilities. Eur. J. Clin. Microbiol. Infect. Dis. 2012;31:1647–1652. doi: 10.1007/s10096-011-1488-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Herkert P.F., Hagen F., de Oliveira Salvador G.L., Gomes R.R., Ferreira M.S., Vicente V.A., Muro M.D., Pinheiro R.L., Meis J.F., Queiroz-Telles F. Molecular characterisation and antifungal susceptibility of clinical Cryptococcus deuterogattii (AFLP6/VGII) isolates from Southern Brazil. Eur. J. Clin. Microbiol. Infect. Dis. 2016;35:1803–1810. doi: 10.1007/s10096-016-2731-8. [DOI] [PubMed] [Google Scholar]
- 39.Headley S.A., Di Santis G.W., de Alcântara B.K., Costa T.C., da Silva E.O., Pretto-Giordano L.G., Gomes L.A., Alfieri A.A., Bracarense A.P.F.R.L. Cryptococcus gattii-induced infections in dogs from Southern Brazil. Mycopathologia. 2015;180:265–275. doi: 10.1007/s11046-015-9901-6. [DOI] [PubMed] [Google Scholar]
- 40.De Abreu D.P.B., Machado C.H., Makita M.T., Botelho C.F.M., Oliveira F.G., da Veiga C.C.P., dos Anjos Martins M., de Assis Baroni F. Intestinal lesion in a dog due to Cryptococcus gattii type VGII and review of published cases of canine gastrointestinal cryptococcosis. Mycopathologia. 2016;182:597–602. doi: 10.1007/s11046-016-0100-x. [DOI] [PubMed] [Google Scholar]
- 41.Costa S.P., Lazéra M., Santos W.R., Morales B.P., Bezerra C.C., Nishikawa M.M., Barbosa G.G., Trilles L., Nascimento J.L., Wanke B. First isolation of Cryptococcus gattii molecular type VGII and Cryptococcus neoformans molecular type VNI from environmental sources in the city of Belém, Pará, Brazil. Mem. Inst. Oswaldo Cruz. 2009;104:662–664. doi: 10.1590/S0074-02762009000400023. [DOI] [PubMed] [Google Scholar]
- 42.Alves G.S.B., Freire A.K.L., dos Santos Bentes A., de Souza Pinheiro J.F., de Souza J.V.B., Wanke B., Matsuura T., Jackisch-Matsuura A.B. Molecular typing of environmental Cryptococcus neoformans/C. gattii species complex isolates from Manaus, Amazonas, Brazil. Mycoses. 2016;59:509–515. doi: 10.1111/myc.12499. [DOI] [PubMed] [Google Scholar]
- 43.Fortes S.T., Lazera M.S., Nishikawa M.M., Macedo R.C., Wanke B. First isolation of Cryptococcus neoformans var. gattii from a native jungle tree in the Brazilian Amazon rainforest. Mycoses. 2001;44:137–140. doi: 10.1046/j.1439-0507.2001.00651.x. [DOI] [PubMed] [Google Scholar]
- 44.Brito-Santos F., Barbosa G.G., Trilles L., Nishikawa M.M., Wanke B., Meyer W., Carvalho-Costa F.A., dos Santos Lazéra M. Environmental isolation of Cryptococcus gattii VGII from indoor dust from typical wooden houses in the deep Amazonas of the Rio Negro basin. PLoS ONE. 2015;10:e0115866. doi: 10.1371/journal.pone.0115866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Debourgogne A., Hagen F., Elenga N., Long L., Blanchet D., Veron V., Lortholary O., Carme B., Aznar C. Successful treatment of Cryptococcus gattii neurocryptococcosis in a 5-year-old immunocompetent child from the French Guiana Amazon region. Rev. Iberoam. Micol. 2012;29:210–213. doi: 10.1016/j.riam.2012.01.008. [DOI] [PubMed] [Google Scholar]
- 46.Loperena-Alvarez Y., Ren P., Li X., Bopp D.J., Ruiz A., Chaturvedi V., Rios-Velazquez C. Genotypic characterization of environmental isolates of Cryptococcus gattii from Puerto Rico. Mycopathologia. 2010;170:279–285. doi: 10.1007/s11046-010-9296-3. [DOI] [PubMed] [Google Scholar]
- 47.Escandón P., Sánchez A., Firacative C., Castañeda E. Isolation of Cryptococcus gattii molecular type VGIII, from Corymbia ficifolia detritus in Colombia. Med. Mycol. 2010;48:675–678. doi: 10.3109/13693780903420633. [DOI] [PubMed] [Google Scholar]
- 48.Firacative C., Roe C.C., Malik R., Ferreira-Paim K., Escandón P., Sykes J.E., Castañón-Olivares L.R., Contreras-Peres C., Samayoa B., Sorrell T.C., et al. MLST and Whole-Genome-Based population analysis of Cryptococcus gattii VGIII links clinical, veterinary and environmental strains, and reveals divergent serotype specific sub-populations and distant ancestors. PLoS Neglect. Trop. Dis. 2016;10:e0004861. doi: 10.1371/journal.pntd.0004861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Steele K.T., Thakur R., Nthobatsang R., Steenhoff A.P., Bisson G.P. In-hospital mortality of HIV-infected cryptococcal meningitis patients with C. gattii and C. neoformans infection in Gaborone, Botswana. Med. Mycol. 2010;48:1112–1115. doi: 10.3109/13693781003774689. [DOI] [PubMed] [Google Scholar]
- 50.Bogaerts J., Rouvroy D., Taelman H., Kagame A., Aziz M.A., Swinne D., Verhaegen J. AIDS-associated cryptococcal meningitis in Rwanda (1983–1992): Epidemiologic and diagnostic features. J. Infect. 1999;39:32–37. doi: 10.1016/S0163-4453(99)90099-3. [DOI] [PubMed] [Google Scholar]
- 51.Karstaedt A.S., Crewe-Brown H.H., Dromer F. Cryptococcal meningitis caused by Cryptococcus neoformans var. gattii, serotype C, in AIDS patients in Soweto, South Africa. Med. Mycol. 2002;40:7–11. doi: 10.1080/mmy.40.1.7.11. [DOI] [PubMed] [Google Scholar]
- 52.Morgan J., McCarthy K.M., Gould S., Fan K., Arthington-Skaggs B., Iqbal N., Stamey K., Hajjeh R.A., Brandt M.E. Cryptococcus gattii infection: Characteristics and epidemiology of cases identified in a South African province with high HIV seroprevalence, 2002–2004. Clin. Infect. Dis. 2006;43:1077–1080. doi: 10.1086/507897. [DOI] [PubMed] [Google Scholar]
- 53.Meiring S.T., Quan V.C., Cohen C., Dawood H., Karstaedt A.S., McCarthy K.M., Whitelaw A.C., Govender N.P., Group for Enteric. Respiratory and Meningeal disease Surveillance in South Africa (GERMS-SA) A comparison of cases of paediatric-onset and adult-onset cryptococcosis detected through population-based surveillance, 2005–2007. AIDS. 2012;26:2307–2314. doi: 10.1097/QAD.0b013e3283570567. [DOI] [PubMed] [Google Scholar]
- 54.Vanhove M., Beale M.A., Rhodes J., Chanda D., Lakhi S., Kwenda G., Molloy S., Karunaharan N., Stone N., Harrison T.S., et al. Genomic epidemiology of Cryptococcus yeasts identifies adaptation to environmental niches underpinning infection across an African HIV/AIDS cohort. Mol. Ecol. 2016;26:1991–2005. doi: 10.1111/mec.13891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kangogo M., Bader O., Boga H., Wanyoike W., Folba C., Worasilchai N., Weig M., Groß U., Bii C.C. Molecular types of Cryptococcus gattii/Cryptococcus neoformans species complex from clinical and environmental sources in Nairobi, Kenya. Mycoses. 2015;58:665–670. doi: 10.1111/myc.12411. [DOI] [PubMed] [Google Scholar]
- 56.Van Wyk M., Govender N.P., Mitchell T.G., Litvintseva A.P. Multilocus sequence typing of serially collected isolates of Cryptococcus from HIV-infected patients in South Africa. Land GA, ed. J. Clin. Microbiol. 2014;52:1921–1931. doi: 10.1128/JCM.03177-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nyazika T.K., Hagen F., Meis J.F., Robertson V.J. Cryptococcus tetragattii as a major cause of cryptococcal meningitis among HIV-infected individuals in Harare, Zimbabwe. J. Infect. 2016;72:745–752. doi: 10.1016/j.jinf.2016.02.018. [DOI] [PubMed] [Google Scholar]
- 58.Nyazika T.K., Herkert P.F., Hagen F., Mateveke K., Robertson V.J., Meis J.F. In vitro antifungal susceptibility profiles of Cryptococcus species isolated from HIV-associated cryptococcal meningitis patients in Zimbabwe. Diagn. Microbiol. Infect. Dis. 2016;86:289–292. doi: 10.1016/j.diagmicrobio.2016.08.004. [DOI] [PubMed] [Google Scholar]
- 59.Kassi F.K., Drakulovski P., Bellet V., Krasteva D., Gatchitch F., Doumbia A., Kouakou G.A., Delaporte E., Reynes J., Mallié M., et al. Molecular epidemiology reveals genetic diversity among 363 isolates of the Cryptococcus neoformans and Cryptococcus gattii species complex in 61 Ivorian HIV-positive patients. Mycoses. 2016;59:811–817. doi: 10.1111/myc.12539. [DOI] [PubMed] [Google Scholar]
- 60.Litvintseva A.P., Thakur R., Reller L.B., Mitchell T.G. Prevalence of clinical isolates of Cryptococcus gattii serotype C among patients with AIDS in Sub-Saharan Africa. J. Infect. Dis. 2005;192:888–892. doi: 10.1086/432486. [DOI] [PubMed] [Google Scholar]
- 61.Xiujiao X., Ai’e X. Two cases of cutaneous cryptococcosis. Mycoses. 2005;48:238–241. doi: 10.1111/j.1439-0507.2005.01079.x. [DOI] [PubMed] [Google Scholar]
- 62.Capoor M.R., Mandal P., Deb M., Aggarwal P., Banerjee U. Current scenario of cryptococcosis and antifungal susceptibility pattern in India: A cause for reappraisal. Mycoses. 2008;51:258–265. doi: 10.1111/j.1439-0507.2007.01478.x. [DOI] [PubMed] [Google Scholar]
- 63.Patil R.T. Meningitis due to Cryptococcus gattii in an immunocompetent patient. J. Clin. Diagn. Res. 2013;7:2274–2275. doi: 10.7860/JCDR/2013/6770.3492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Randhawa H.S., Kowshik T., Preeti Sinha K., Chowdhary A., Khan Z.U., Yan Z., Xu J., Kumar A. Distribution of Cryptococcus gattii and Cryptococcus neoformans in decayed trunk wood of Syzygium cumini trees in north-western India. Med. Mycol. 2006;44:623–630. doi: 10.1080/13693780600860946. [DOI] [PubMed] [Google Scholar]
- 65.Randhawa H.S., Kowshik T., Chowdhary A., Preeti Sinha K., Khan Z.U., Sun S., Xu J. The expanding host tree species spectrum of Cryptococcus gattii and Cryptococcus neoformans and their isolations from surrounding soil in India. Med. Mycol. 2008;46:823–833. doi: 10.1080/13693780802124026. [DOI] [PubMed] [Google Scholar]
- 66.Khan Z.U., Randhawa H.S., Kowshik T., Chowdhary A., Chandy R. Antifungal susceptibility of Cryptococcus neoformans and Cryptococcus gattii isolates from decayed wood of trunk hollows of Ficus religiosa and Syzygium cumini trees in north-western India. J. Antimicrob. Chemother. 2007;60:312–316. doi: 10.1093/jac/dkm192. [DOI] [PubMed] [Google Scholar]
- 67.Girish Kumar C.P., Prabu D., Mitani H., Mikami Y., Menon T. Environmental isolation of Cryptococcus neoformans and Cryptococcus gattii from living trees in Guindy National Park, Chennai, South India. Mycoses. 2010;53:262–264. doi: 10.1111/j.1439-0507.2009.01699.x. [DOI] [PubMed] [Google Scholar]
- 68.Gutch R.S., Nawange S.R., Singh S.M., Yadu R., Tiwari A., Gumasta R., Kavishwar A. Antifungal susceptibility of clinical and environmental Cryptococcus neoformans and Cryptococcus gattii isolates in Jabalpur, a city of Madhya Pradesh in Central India. Braz. J. Microbiol. 2015;46:1125–1133. doi: 10.1590/S1517-838246420140564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bedi N.G., Nawange S.R., Singh S.M., Naidu J., Kavishwar A. Seasonal prevalence of Cryptococcus neoformans var. grubii and Cryptococcus gattii inhabiting Eucalyptus terreticornis and Eucalyptus camaldulensis trees in Jabalpur City of Madhya Pradesh, Central India. J. Med. Mycol. 2012;22:341–347. doi: 10.1016/j.mycmed.2012.09.001. [DOI] [PubMed] [Google Scholar]
- 70.Chen Y.C., Chang S.C., Shih C.C., Hung C.C., Luhbd K.T., Pan Y.S., Hsieh W.C. Clinical features and in vitro susceptibilities of two varieties of Cryptococcus neoformans in Taiwan. Diagn. Microbiol. Infect. Dis. 2000;36:175–183. doi: 10.1016/S0732-8893(99)00137-6. [DOI] [PubMed] [Google Scholar]
- 71.Lui G. Cryptococcosis in apparently immunocompetent patients. QJM. 2006;99:143–151. doi: 10.1093/qjmed/hcl014. [DOI] [PubMed] [Google Scholar]
- 72.Chen J., Varma A., Diaz M.R., Litvintseva A.P., Wollenberg K.K., Kwon-Chung K.J. Cryptococcus neoformans strains and infection in apparently immunocompetent patients, China. Emerg. Infect. Dis. 2008;14:755–762. doi: 10.3201/eid1405.071312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Feng X., Yao Z., Ren D., Liao W., Wu J. Genotype and mating type analysis of Cryptococcus neoformans and Cryptococcus gattii isolates from China that mainly originated from non-HIV-infected patients. FEMS Yeast Res. 2008;8:930–938. doi: 10.1111/j.1567-1364.2008.00422.x. [DOI] [PubMed] [Google Scholar]
- 74.Dou H.-T., Xu Y.-C., Wang H.-Z., Li T.-S. Molecular epidemiology of Cryptococcus neoformans and Cryptococcus gattii in China between 2007 and 2013 using multilocus sequence typing and the DiversiLab system. Eur. J. Clin. Microbiol. Infect. Dis. 2015;34:753–762. doi: 10.1007/s10096-014-2289-2. [DOI] [PubMed] [Google Scholar]
- 75.Wu S.-Y., Lei Y., Kang M., Xiao Y.-L., Chen Z.-X. Molecular characterisation of clinical Cryptococcus neoformans and Cryptococcus gattii isolates from Sichuan province, China. Mycoses. 2015;58:280–287. doi: 10.1111/myc.12312. [DOI] [PubMed] [Google Scholar]
- 76.Lahiri Mukhopadhyay S., Bahubali V.H., Manjunath N., Swaminathan A., Maji S., Palaniappan M., Parthasarathy S., Chandrashekar N. Central nervous system infection due to Cryptococcus gattii sensu lato in India: Analysis of clinical features, molecular profile and antifungal susceptibility. Mycoses. 2017;60:749–757. doi: 10.1111/myc.12656. [DOI] [PubMed] [Google Scholar]
- 77.Randhawa H.S., Kowshik T., Chowdhary A., Prakash A., Khan Z.U., Xu J. Seasonal variations in the prevalence of Cryptococcus neoformans var. grubii and Cryptococcus gattii in decayed wood inside trunk hollows of diverse tree species in north-western India: A retrospective study. Med. Mycol. 2011;49:320–323. doi: 10.3109/13693786.2010.516457. [DOI] [PubMed] [Google Scholar]
- 78.Chowdhary A., Prakash A., Randhawa H.S., Kathuria S., Hagen F., Klaassen C.H., Meis J.F. First environmental isolation of Cryptococcus gattii, genotype AFLP5, from India and a global review: Cryptococcus gattii, genotype AFLP5, from India. Mycoses. 2013;56:222–228. doi: 10.1111/myc.12039. [DOI] [PubMed] [Google Scholar]
- 79.Park S.H., Choi S.C., Lee K.W., Kim M.-N., Hwang S.M. Genotypes of clinical and environmental isolates of Cryptococcus neoformans and Cryptococcus gattii in Korea. Mycobiology. 2015;43:360. doi: 10.5941/MYCO.2015.43.3.360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tay S.T., Lim H.C., Tajuddin T.H., Rohani M.Y., Hamimah H., Thong K.L. Determination of molecular types and genetic heterogeneity of Cryptococcus neoformans and C. gattii in Malaysia. Med. Mycol. 2006;44:617–622. doi: 10.1080/13693780600857330. [DOI] [PubMed] [Google Scholar]
- 81.Tay S.T., Rohani M.Y., Soo Hoo T.S., Hamimah H. Epidemiology of cryptococcosis in Malaysia: Epidemiology of Cryptococcus neoformans. Mycoses. 2010;53:509–514. doi: 10.1111/j.1439-0507.2009.01750.x. [DOI] [PubMed] [Google Scholar]
- 82.Liaw S.-J., Wu H.-C., Hsueh P.-R. Microbiological characteristics of clinical isolates of Cryptococcus neoformans in Taiwan: Serotypes, mating types, molecular types, virulence factors, and antifungal susceptibility. Clin. Microbiol. Infect. 2010;16:696–703. doi: 10.1111/j.1469-0691.2009.02930.x. [DOI] [PubMed] [Google Scholar]
- 83.Kaocharoen S., Ngamskulrungroj P., Firacative C., Trilles L., Piyabongkarn D., Banlunara W., Poonwan N., Chaiprasert A., Meyer W., Chindamporn A. Molecular epidemiology reveals genetic diversity amongst isolates of the Cryptococcus neoformans/C. gattii species complex in Thailand. PLoS Neglect. Trop. Dis. 2013;7:e2297. doi: 10.1371/journal.pntd.0002297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Jain N., Wickes B.L., Keller S.M., Fu J., Casadevall A., Jain P., Ragan M.A., Banerjee U., Fries B.C. Molecular epidemiology of clinical Cryptococcus neoformans strains from India. J. Clin. Microbiol. 2005;43:5733–5742. doi: 10.1128/JCM.43.11.5733-5742.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Choi Y.H., Ngamskulrungroj P., Varma A., Sionov E., Hwang S.M., Carriconde F., Meyer W., Litvintseva A.P., Lee W.G., Shin J.H., et al. Prevalence of the VNIc genotype of Cryptococcus neoformans in non-HIV-associated cryptococcosis in the Republic of Korea: Molecular epidemiology of cryptococcosis in Korea. FEMS Yeast Res. 2010;10:769–778. doi: 10.1111/j.1567-1364.2010.00648.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Leechawengwongs M., Milindankura S., Sathirapongsasuti K., Tangkoskul T., Punyagupta S. Primary cutaneous cryptococcosis caused by Cryptococcus gattii VGII in a tsunami survivor from Thailand. Med. Mycol. Case Rep. 2014;6:31–33. doi: 10.1016/j.mmcr.2014.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Cogliati M., Chandrashekar N., Esposto M.C., Chandramuki A., Petrini B., Viviani M.A. Cryptococcus gattii serotype-C strains isolated in Bangalore, Karnataka, India: Cryptococcus gattii in Bangalore. Mycoses. 2012;55:262–268. doi: 10.1111/j.1439-0507.2011.02082.x. [DOI] [PubMed] [Google Scholar]
- 88.Illnait-Zaragozí M.T., Ortega-Gonzalez L.M., Hagen F., Martínez-Machin G.F., Meis J.F. Fatal Cryptococcus gattii genotype AFLP5 infection in an immunocompetent Cuban patient. Med. Mycol. Case Rep. 2013;2:48–51. doi: 10.1016/j.mmcr.2013.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Casadevall A., Freij J.B., Hann-Soden C., Taylor J. Continental drift and speciation of the Cryptococcus neoformans and Cryptococcus gattii species complexes. mSphere. 2017;2:e00103-17. doi: 10.1128/mSphere.00103-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Engelthaler D.M., Meyer W. Furthering the continental drift speciation hypothesis in the pathogenic Cryptococcus species complexes. mSphere. 2017;2:e00241-17. doi: 10.1128/mSphere.00241-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Espinel-Ingroff A., Aller A.I., Canton E., Castanon-Olivares L.R., Chowdhary A., Cordoba S., Cuenca-Estrella M., Fothergill A., Fuller J., Govender N., et al. Cryptococcus neoformans-Cryptococcus gattii species complex: An international study of wild-type susceptibility endpoint distributions and epidemiological cutoff values for fluconazole, itraconazole, posaconazole, and voriconazole. Antimicrob. Agents Chemother. 2012;56:5898–5906. doi: 10.1128/AAC.01115-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Espinel-Ingroff A., Chowdhary A., Gonzalez G.M., Guinea J., Hagen F., Meis J.F., Thompson G.R., Turnidge J. Multicenter study of isavuconazole MIC distributions and epidemiological cutoff values for the Cryptococcus neoformans-Cryptococcus gattii species complex using the CLSI M27-A3 broth microdilution method. Antimicrob. Agents Chemother. 2015;59:666–668. doi: 10.1128/AAC.04055-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Trilles L., Meyer W., Wanke B., Guarro J., Lazéra M. Correlation of antifungal susceptibility and molecular type within the Cryptococcus neoformans/C. gattii species complex. Med. Mycol. 2012;50:328–332. doi: 10.3109/13693786.2011.602126. [DOI] [PubMed] [Google Scholar]
- 94.Favalessa O.C., De Paula D.A.J., Dutra V., Nakazato L., Tadano T., dos Santos Lazera M., Wanke B., Trilles L., Walderez Szeszs M., Silva D., et al. Molecular typing and in vitro antifungal susceptibility of Cryptococcus spp from patients in Midwest Brazil. J. Infect. Dev. Ctries. 2014;8 doi: 10.3855/jidc.4446. [DOI] [PubMed] [Google Scholar]
- 95.Hagen F., Illnait-Zaragozi M.-T., Bartlett K.H., Swinne D., Geertsen E., Klaassen C.H.W., Boekhout T., Meis J.F. In vitro antifungal susceptibilities and amplified fragment length polymorphism genotyping of a worldwide collection of 350 clinical, veterinary, and environmental Cryptococcus gattii isolates. Antimicrob. Agents Chemother. 2010;54:5139–5145. doi: 10.1128/AAC.00746-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lockhart S.R., Iqbal N., Bolden C.B., DeBess E.E., Marsden-Haug N., Worhle R., Thakur R., Harris J.R. Epidemiologic cutoff values for triazole drugs in Cryptococcus gattii: Correlation of molecular type and in vitro susceptibility. Diagn. Microbiol. Infect. Dis. 2012;73:144–148. doi: 10.1016/j.diagmicrobio.2012.02.018. [DOI] [PubMed] [Google Scholar]
- 97.Perfect J.R., Dismukes W.E., Dromer F., Goldman D.L., Graybill J.R., Hamill R.J., Harrison T.S., Larsen R.A., Lortholary O., Nguyen M., et al. Clinical practice guidelines for the management of cryptococcal disease: 2010 update by the infectious diseases society of America. Clin. Infect. Dis. 2010;50:291–322. doi: 10.1086/649858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Varma A., Kwon-Chung K.J. Heteroresistance of Cryptococcus gattii to Fluconazole. Antimicrob. Agents Chemother. 2010;54:2303–2311. doi: 10.1128/AAC.00153-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ferreira G.F., Santos J.R.A., da Costa M.C., de Holanda R.A., Denadai Â.M.L., de Freitas G.J.C., Santos Á.R.C., Tavares P.B., Paixão T.A., Santos D.A. Heteroresistance to itraconazole alters the morphology and increases the virulence of Cryptococcus gattii. Antimicrob. Agents Chemother. 2015;59:4600–4609. doi: 10.1128/AAC.00466-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Sionov E., Chang Y.C., Garraffo H.M., Kwon-Chung K.J. Heteroresistance to fluconazole in Cryptococcus neoformans is intrinsic and associated with virulence. Antimicrob. Agents Chemother. 2009;53:2804–2815. doi: 10.1128/AAC.00295-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Falagas M.E., Makris G.C., Dimopoulos G., Matthaiou D.K. Heteroresistance: A concern of increasing clinical significance? Clin. Microbiol. Infect. 2008;14:101–104. doi: 10.1111/j.1469-0691.2007.01912.x. [DOI] [PubMed] [Google Scholar]
- 102.Liu X.-Z., Wang Q.-M., Göker M., Groenewald M., Kachalkin A.V., Lumbsch H.T., Millanes A.M., Wedin M., Yurkov A.M., Boekhout T., et al. Towards an integrated phylogenetic classification of the Tremellomycetes. Stud. Mycol. 2015;81:85–147. doi: 10.1016/j.simyco.2015.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Liu X.-Z., Wang Q.-M., Theelen B., Groenewald M., Bai F.-Y., Boekhout T. Phylogeny of tremellomycetous yeasts and related dimorphic and filamentous basidiomycetes reconstructed from multiple gene sequence analyses. Stud. Mycol. 2015;81:1–26. doi: 10.1016/j.simyco.2015.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Coenjaerts F.E.J. The sixth international conference on Cryptococcus and cryptococcosis: The sixth international conference on Cryptococcus and cryptococcosis (ICCC) FEMS Yeast Res. 2006;6:312–317. doi: 10.1111/j.1567-1364.2006.00068.x. [DOI] [PubMed] [Google Scholar]
- 105.Kwon-Chung J. Is it a one, two, or more species system supported by the major species concept?; Proceedings of the 6th International Conference on Cryptococcus and Cryptococcosis; Boston, MA, USA. 24–28 June 2005; p. 43. [Google Scholar]
- 106.Kwon-Chung K.J., Varma A. Do major species concepts support one, two or more species within Cryptococcus neoformans? FEMS Yeast Res. 2006;6:574–587. doi: 10.1111/j.1567-1364.2006.00088.x. [DOI] [PubMed] [Google Scholar]
- 107.Boekhout T., Bovers M., Fell J.W., Diaz M., Hagen F., Theelen B., Kuramae E. How many species?; Proceedings of the 6th International Conference on Cryptococcus and Cryptococcosis; Boston, MA, USA. 24–28 June 2005; p. 44. [Google Scholar]
- 108.Bovers M., Hagen F., Kuramae E.E., Boekhout T. Six monophyletic lineages identified within Cryptococcus neoformans and Cryptococcus gattii by multi-locus sequence typing. Fungal Genet. Biol. 2008;45:400–421. doi: 10.1016/j.fgb.2007.12.004. [DOI] [PubMed] [Google Scholar]
- 109.Meyer W., Boekhout T., Castañeda E., Karaoglu H., Ngamskulrungroj P., Kidd S., Escandón P., Hagen F., Narszewska K., Velegraki A. Molecular characterization of the Cryptococcus neoformans species complex; Proceedings of the 6th International Conference on Cryptococcus and Cryptococcosis; Boston, MA, USA. 24–28 June 2005; pp. 41–42. [Google Scholar]
- 110.Kwon-Chung K.J., Bennett J.E., Wickes B.L., Meyer W., Cuomo C.A., Wollenburg K.R., Bicanic T.A., Castañeda E., Chang Y.C., Chen J., et al. The case for adopting the “species complex” nomenclature for the etiologic agents of cryptococcosis. mSphere. 2017;2:e00357-16. doi: 10.1128/mSphere.00357-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Fernandes K.E., Dwyer C., Campbell L.T., Carter D.A. Species in the Cryptococcus gattii complex differ in capsule and cell size following growth under capsule-inducing conditions. mSphere. 2016;1:e00350-16. doi: 10.1128/mSphere.00350-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Thompson G.R., Albert N., Hodge G., Wilson M.D., Sykes J.E., Bays D.J., Firacative C., Meyer W., Kontoyiannis D.P. Phenotypic differences of Cryptococcus molecular types and their implications for virulence in a Drosophila model of infection. Infect. Immun. 2014;82:3058–3065. doi: 10.1128/IAI.01805-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Taylor J.W., Jacobson D.J., Kroken S., Kasuga T., Geiser D.M., Hibbett D.S., Fisher M.C. Phylogenetic species recognition and species concepts in fungi. Fungal Genet. Biol. 2000;31:21–32. doi: 10.1006/fgbi.2000.1228. [DOI] [PubMed] [Google Scholar]
- 114.D’Souza C.A., Kronstad J.W., Taylor G., Warren R., Yuen M., Hu G., Jung W.H., Sham A., Kidd S.E., Tangen K., et al. Genome variation in Cryptococcus gattii, an emerging pathogen of immunocompetent hosts. MBio. 2011;2:e00342-10. doi: 10.1128/mBio.00342-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Ngamskulrungroj P., Gilgado F., Faganello J., Litvintseva A.P., Leal A.L., Tsui K.M., Mitchell T.G., Vainstein M.H., Meyer W. Genetic diversity of the Cryptococcus species complex suggests that Cryptococcus gattii deserves to have varieties. PLoS ONE. 2009;4:e5862. doi: 10.1371/annotation/3037bb69-1b8e-4d99-b169-afdf4b74ace2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Mallet J. Hybridization as an invasion of the genome. Trends Ecol. Evol. 2005;20:229–237. doi: 10.1016/j.tree.2005.02.010. [DOI] [PubMed] [Google Scholar]
- 117.Hagen F., Lumbsch H.T., Arsic Arsenijevic V., Badali H., Bertout S., Billmyre R.B., Bragulat M.R., Cabañes F.J., Carbia M., Chakrabarti A., et al. Importance of resolving fungal nomenclature: The case of multiple pathogenic species in the Cryptococcus Genus. mSphere. 2017;2:e00238-17. doi: 10.1128/mSphere.00238-17. [DOI] [PMC free article] [PubMed] [Google Scholar]