Abstract
MethBank (http://bigd.big.ac.cn/methbank) is a database that integrates high-quality DNA methylomes across a variety of species and provides an interactive browser for visualization of methylation data. Here, we present an updated implementation of MethBank (version 3.0) by incorporating more DNA methylomes from multiple species and equipping with more enhanced functionalities for data annotation and more friendly web interfaces for data presentation, search and visualization. MethBank 3.0 features large-scale integration of high-quality methylomes, involving 34 consensus reference methylomes derived from a large number of human samples, 336 single-base resolution methylomes from different developmental stages and/or tissues of five plants, and 18 single-base resolution methylomes from gametes and early embryos at multiple stages of two animals. Additionally, it is enhanced by improving the functionalities for data annotation, which accordingly enables systematic identification of methylation sites closely associated with age, sites with constant methylation levels across different ages, differentially methylated promoters, age-specific differentially methylated cytosines/regions, and methylated CpG islands. Moreover, MethBank provides tools to estimate human methylation age online and to identify differentially methylated promoters, respectively. Taken together, MethBank is upgraded with significant improvements and advances over the previous version, which is of great help for deciphering DNA methylation regulatory mechanisms for epigenetic studies.
INTRODUCTION
DNA methylation, as a major epigenetic modification, plays important roles in human diseases and aging (1–4), embryonic development of animals (5–8) as well as growth and development of plants (9–12). Recently, studies in human have found that DNA methylation biomarkers of aging are associated closely with mortality rates and incidence of cardio-metabolic disease (13,14). Accordingly, identification of DNA methylation states for healthy people at different ages is able to provide references and controls for studying DNA methylation regulatory mechanisms and investigating epigenetic biomarkers for diseases (15–17). Additionally, DNA methylation exerts substantial impacts on animal embryonic development (5,7) as well as responds to stress (9) and development processes in plants (11). To make it short, recent studies on DNA methylation conducted in a wide variety of species have generated vast amounts of data, presenting the necessity for comprehensive integration of methylation data that would be of great help for systematic exploration of DNA methylation signatures in epigenetic studies.
MethBank (http://bigd.big.ac.cn/methbank), with the first version released in 2014, is a database dedicated to integrating high-quality DNA methylomes across a variety of species and providing an interactive browser for visualization of high-resolution DNA methylation data (18). With the rapid advancements in high-throughput sequencing technologies and the ever-growing amount of methylation data as mentioned above, here we present the upgraded version of MethBank (v3.0) and describe its major updates in the past three years. In contrast to its first release that was primarily focused on DNA methylation reprogramming in early embryonic development in two model organisms (zebrafish and mouse), MethBank 3.0 integrates 34 consensus reference methylomes (CRMs) compiled from 4577 healthy human samples at different ages, 336 single-base resolution methylomes (SRMs) of five plants and 18 SRMs of two animals. Moreover, it features systematic identification of not only gene methylation profiles but also differentially methylated promoters (DMPs), age-specific differentially methylated regions/cytosines (DMRs/DMCs) and methylation sites in close association with age, etc. and equips with more enhanced functionalities for data annotation and more friendly web interfaces for data presentation, search and visualization.
MATERIALS AND METHODS
Data sources
Whole-genome bisulfite sequencing data with genome coverage > 30X on Illumina sequencing platform of animals and plants are downloaded from Sequence Read Archive (SRA) (19) and Genome Sequence Archive (GSA) (20). Considering the higher resolution of HumanMethylation450 (450K; 482 421 CpG sites) BeadChip than HumanMethylation27 (27K; 27 578 CpG sites) BeadChip (21,22), MethBank only collects 450K array datasets from peripheral blood of healthy people with known age in Gene Expression Omnibus (GEO) (13,23–28). All datasets used in MethBank 3.0 are summarized in Supplementary Table S1.
Data analysis
For whole-genome bisulfite sequencing (WGBS) data of animals and plants (Supplementary Figure S1), sequencing read quality is evaluated by FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc), low quality read ends are filtered and adapter sequences are removed via Trim Galore (http://www.bioinformatics.babraham.ac.uk/projects/trim_galore). WBSA (Web service for Bisulfite Sequencing data Analysis) (29) is used to align reads to genome, identify cytosine context, and annotate the methylation levels of cytosine sites. A CpG island is defined as a sequence ≥200 bp with C + G content ≥ 50% and CpG observed/CpG expected ≥0.6. A CpG island with average methylation level >0.6 is defined as a methylated CpG island (mCpGI). The promoter region is defined as the region 2000 bp upstream of transcription start site (TSS) for animals and 1500 bp upstream of TSS for plants. DMPs are identified via Fisher's exact test with FDR corrected P-values <0.01 on the condition that the delta methylation levels of the promoters between two samples are greater than 0.1 for C/CH (H = A, C or T) and 0.2 for CG. In animals, only CG sequence context is considered.
For Illumina Infinium HumanMethylation450 BeadChip data (Supplementary Figure S2), Minfi (source (‘https://bioconductor.org/biocLite.R’)) is implemented to upload the raw intensity data into R (version 3.3.2). Cytosine methylation beta values are calculated as
 |
PCA (Principal Component Analysis) is used to remove outlier samples. BMIQ (Beta MIxture Quantile dilation) (30) is used to correct the bias of probes coming from two different designs. In addition, DNA methylation levels of age-related CpG sites at the same age in different batches are assumed to be similar. Consequently, samples at the same age in different batches are grouped together, and batch effects are removed in each group using L/S batch adjustment separately. All 4577 samples are divided into 12 age groups by clustering analysis. In CRMs, the methylation level for a specific CpG site is the median of methylation levels of all the samples in any specific age group. The reference ranges of methylation level in healthy population are defined from the first quantile to the third quantile. The Spearman correlation between DNA methylation level and age is measured for each CpG site. The CpGs with correlation coefficients ≥0.6 are considered to be sites that are closely associated with age. Following a published procedure (31), the methylation cytosine is defined as an age-specific DMC if the linear model considering a fixed effect for age and a random effect for gender fits the data better than that without considering a fixed effect for age, satisfying with the Bonferroni corrected P ≤ 1.03E–7 (0.05/485K CpGs, F-test) as well as the effect size ≥ 20%. Multiple comparisons are then performed to determine which age group each DMC belongs to. A CpG site is identified as a site with constant methylation levels across different ages, if it has no statistically significant difference in the mean methylation (Scheffe test) between any two age groups and the absolute difference of methylation levels between any two age groups is ≤0.1. An age-specific DMR is defined as a region covering at least three age-specific DMCs with an inter-CpG distance ≤1000 bp.
NEW FEATURES AND UPDATES
MethBank 3.0 features large-scale integration of CRMs compiled from 450K data of humans (Table 1), SRMs from WGBS data of plants and animals (Table 2). Compared to its first release in 2014 that only included 18 SRMs from gametes and early embryos at multiple stages of two animal species (zebrafish and mouse), MethBank 3.0 additionally integrates 34 CRMs at different age groups of humans and 336 SRMs from different developmental stages and/or tissues of five plants. Furthermore, it is enhanced not only by equipping with more friendly web interfaces for data presentation, search and visualization but also improving the functionalities for data annotation, which leads to systematic identification of genome-wide methylation profiles, DMCs, DMRs, DMPs and methylation sites in close association with age, etc. Detailed statistics of database contents in MethBank 3.0 are summarized in Tables 1 and 2.
Table 1. Data statistics of human 450K data in MethBank 3.0.
| Data item | Number |
|---|---|
| CRMsa | 34 |
| Sites closely associated with age | 692 |
| Sites with relatively constant methylation levels across different ages | 2371 |
| Age-specific DMCsb | 53 680 |
| Age-specific DMRsc | 1716 |
| Annotated genes | 984 842 |
aConsensus Reference Methylome.
bDifferentially Methylated Cytosine.
cDifferentially Methylated Region.
Table 2. Data statistics of WGBS data in MethBank 3.0.
| Species | SRMsa | DMPsb | mCpGIsc | Genes related to mCpGIs | Annotated genes |
|---|---|---|---|---|---|
| Mouse | 9 | 108 215 | 4645 | 4526 | 526 338 |
| Zebrafish | 9 | 6572 | 56 204 | 33 679 | 331 708 |
| Rice | 172 | 46 674 | 528 463 | 90 893 | 6 049 625 |
| Soybean | 112 | 89 036 | 40 955 | 5110 | 4 794 282 |
| Tomato | 40 | 54 387 | 53 353 | 2254 | 979 927 |
| Cassava | 8 | NAd | 6126 | 378 | 198 522 |
| Common bean | 4 | NAd | 4079 | 178 | 81 766 |
aSingle-base Resolution Methylome.
bDifferentially Methylated Promoter.
cMethylated CpG Island.
dNot available due to limited sample.
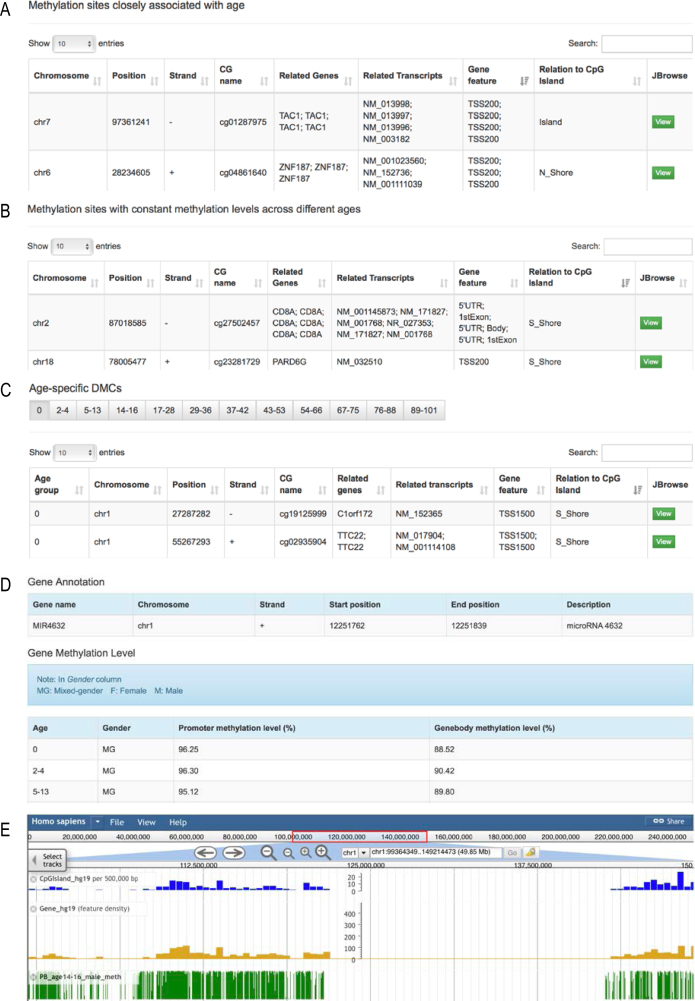
For human, MethBank integrates a comprehensive collection of HumanMethylation450 BeadChip datasets from 4577 peripheral blood samples of healthy people at different ages (from newborn to 101 years old). Based on clustering analysis, all collected samples are divided into 12 groups in terms of age, namely, 0 (newborn; n = 258), 2–4 (n = 13), 5–13 (n = 56), 14–16 (n = 180), 17–28 (n = 424), 29–36 (n = 297), 37–42 (n = 201), 43∼53 (n = 760), 54–66 (n = 1299), 67–75 (n = 602), 76–88 (n = 453) and 89–101 (n = 34), accordingly leading to 34 CRMs for all age groups with each considering three cases by gender (that is, male, female and mixed; except for the 2–4 age group due to limited samples). As each site has a reference range of methylation level in healthy population, MethBank offers CRMs for healthy people at different age groups, which are of great helpfulness to investigate the difference of methylation profiles by comparison with specific samples under investigation, for example, from cancer patients. Considering that aberrant DNA methylation is a potential cause of age-related diseases (32,33), MethBank detects 692 methylation sites that are closely associated with age (Figure 1), helpful for investigating aging states and studying regulatory mechanisms of DNA methylation during aging. Meanwhile, MethBank identifies 2371 methylation sites that have relatively constant methylation levels across different ages in healthy people, which potentially can be used as putatively candidate biomarkers to predict specific diseases (34). In addition, MethBank provides a list of 53 680 age-specific DMCs and 1716 age-specific DMRs (Table 1), presumably bearing great utility for fully capturing epigenetic signatures during human aging. To support information search and exploration, MethBank also allows users to search gene methylation profiles in all age groups. It is also able to visualize all methylation profiles in the methylome browser and download all annotated data and analysis results.
Figure 1.
Screenshots of human methylation data. (A) Sites closely associated with age. (B) Sites with constant methylation levels across different ages. (C) Age-specific differentially methylated cytosines. (D) Methylation states at promoter and gene body as well as basic information, by searching the human gene MIR4632. (E) Methylation levels in a genomic region from 99 364 349 to 149 214 473 in human chromosome 1.
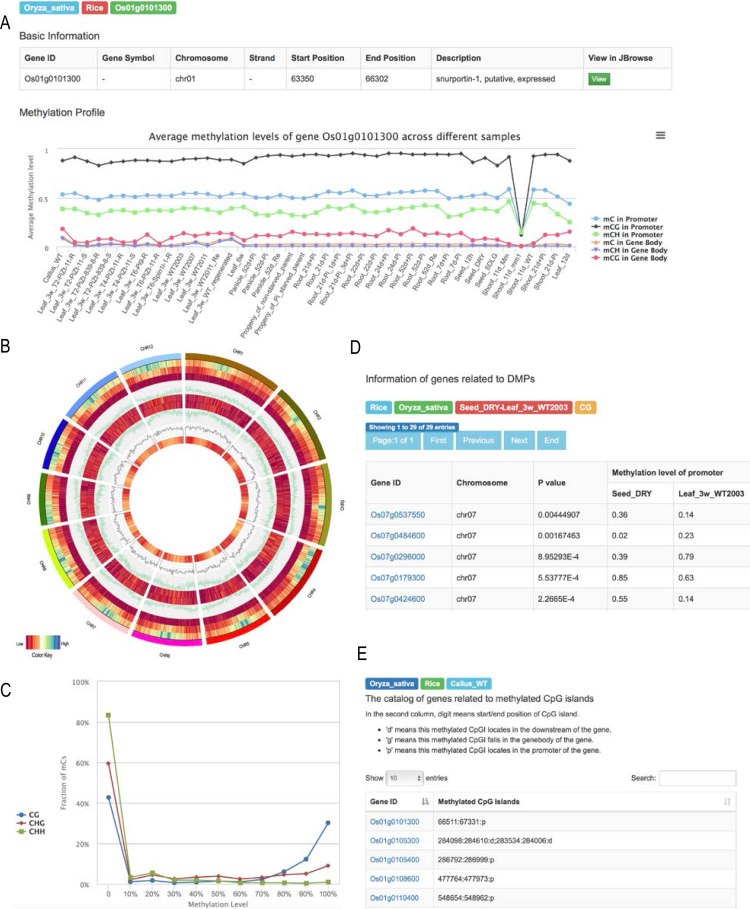
It has been reported that non-CG methylation accounts for a considerable proportion in plants (35–38) and thus is considered to play vital roles during plant growth and development (12). For plants, therefore, both CG and non-CG methylations are taken into account in the upgraded version of MethBank. Specifically, MethBank 3.0 incorporates SRMs differentiated C, CG, CHG and CHH contexts from five economically important crops, including 172 SRMs for Oryza sativa (rice), 112 for Glycine max (soybean), 8 for Manihot esculenta (cassava), 4 for Phaseolus vulgaris (common bean), and 40 for Solanum lycopersicum (tomato). Moreover, to enable in-depth investigation of DNA methylation data, it identifies a large number of DMPs (46 674 for rice, 89 036 for soybean, 54 387 for tomato) between a range of conditions and detects genes related to mCpGIs (90 893 for rice, 5110 for soybean, 2254 for tomato, 378 for cassava, and 178 for common bean) for all collected samples (Table 2). For each gene, MethBank visualizes its methylation levels in promoter and gene body for different sequence contexts under all available stages and tissues (Figure 2). Additionally, detailed metadata of all raw data, such as sequencing platform, submitters, year of submission, number of experiments, mapping rate, genome coverage and bisulfite conversion rate, is summarized for each dataset in each species as well. To better display genome-wide methylation status for each species, MethBank depicts whole-genome methylation alteration in three sequence contexts for all chromosomes and provides the distribution of methylation levels and percentage of methylcytosines identified in different sequence contexts.
Figure 2.
Screenshots of rice methylation data. (A) Methylation profiles at promoter and gene body considering different sequence contexts, by taking rice gene Os01g0101300 as an example. (B) Whole-genome methylation alterations in different sequence contexts, with 10 layers from outer to inner, namely, chromosomes, distributions of CG methylation levels, CHG methylation levels, CHH methylation levels, Watson strand genes, methylation levels in Watson strand genes, methylation levels in Crick strand genes, Crick strand genes, CpG islands, and methylation level in CpG islands. (C) Distribution of methylation levels under different sequence contexts. (D) Gene list related to differentially methylated promoters between seed and leaf in the CG context. (E) Catalog of genes related to methylated CpG islands of callus tissue.
Apart from the 18 SRMs of gametes and early embryos at multiple developmental stages of two model animals (nine for zebrafish and nine for mouse), MethBank 3.0 identifies DMPs between specific development stages and mCpGIs for all collected SRMs. Specifically, it identifies 6572 DMPs, 56 204 mCpGIs and 33 679 genes related to mCpGIs for zebrafish, and 108 215 DMPs, 4645 mCpGIs and 4526 genes related to mCpGIs for mouse, respectively (Table 2), which are helpful for dissecting regulatory mechanisms of DNA methylation reprograming during embryonic development. Similarly, MethBank depicts methylation levels in promoter and gene body for each annotated gene considering all 18 collected gametes and early embryos at multiple developmental stages and illustrates whole-genome CpG methylation levels. All these information can be interactively displayed in the methylome browser. Certainly, MethBank provides detailed metadata for all collected raw data and displays the distribution of CpGs with different methylation levels.
In addition to large-scale data integrated, MethBank is enhanced by equipping with more user-friendly and intuitive web interfaces to facilitate data presentation, search and visualization. For human, users can retrieve methylation profiles in promoter and gene body for any given gene across all age groups with different gender. MethBank also enables users to obtain methylation levels for any specific genomic region across different age groups. For animals and plants, users can search for gene methylation profiles as well as DMPs, mCpGIs and methylation levels for any specific genomic region. In addition, MethBank provides a list of genes related to DMPs between samples in designated environmental conditions or at specific development stages and offers a catalog of mCpGIs and related genes as well as gene methylation profiles for all samples. Regarding data visualization, it is capable of displaying high-resolution methylation profiles in the methylome browser, including 34 CRMs across 12 age groups, 336 SRMs at different developmental stages and/or in tissues of five plants, and 18 SRMs for gametes and different development stages of early embryos of two animals. Compared with the previous release, the updated version of MethBank makes all relevant data publicly downloadable, including DNA methylomes, gene methylation profiles, DMPs, mCpGIs and related genes, methylation sites closely associated with age, sites with constant methylation levels across different ages, age-specific DMCs and DMRs. All annotated data and analysis results are available for download at http://bigd.big.ac.cn/methbank/downloads.
Evidence has accumulated that DNA methylation can be used to predict human methylation age which can reflect the clinical and physiological status of organisms (15,39). Thus, prediction of DNA methylation age is helpful to study age-associated physiological decline and diseases (15,40–43) and also very useful for forensic applications, for example, to estimate the age of a suspect based on blood left on a crime scene (44). Based on large-scale human methylation datasets integrated in MethBank 3.0, we develop an online tool, named Age Predictor, to estimate the DNA methylation age by three machine learning methods (SVM, Random Forest and Elastic Net) and provide the corresponding web services for online analysis. Specifically, methylation sites that are identified to be closely associated with age are used for age prediction (see details in Supplementary Text S1). In addition, a bootstrap process is adopted, with the purpose to provide the confidence interval of the predicted age. To enable users to run the Age Predictor in a convenient way, MethBank allows to upload raw data or processed data or specify a GEO sample ID. The comparison between Age Predictor and two popular approaches (24,41) shows that Age Predictor achieves higher accuracy (see Supplementary Table S2; unpublished results which will be summarized into another paper). Additionally, we develop another tool named IDMP (Identification of Differentially Methylated Promoter) to identify DMPs via Fisher's exact test and FDR correction (see details in Supplementary Text S2), which is publicly accessible and downloadable through the home page of MethBank.
DISCUSSION AND FUTURE PLANS
Since its inception in 2014, MethBank is dedicated to integrating genome-wide DNA methylation data for a wide variety of species. In contrast to the previous version that contains 18 SRMs of embryonic development of two animals, MethBank 3.0 is upgraded significantly by incorporating 34 CRMs from healthy people at different age groups and 336 SRMs under different developments and stress conditions of five economically important plants. In addition, MethBank is significantly enhanced by equipping with more user-friendly web interfaces for data presentation, search and visualization and improving the functionalities for data annotation, which accordingly enables identification of not only gene methylation profiles but also DMCs, DMRs, DMPs, methylation sites in close association with age and sites with constant methylation levels across different ages. It consequently allows users to retrieve genome-wide methylation profiles as well as specific gene/regional methylation levels across all collected samples and also provides two tools for methylation age prediction and DMP identification. As an important resource in the BIG Data Center (45), MethBank will be frequently upgraded and improved. We will integrate more high-quality methylomes from a wider range of species and develop new functionalities that allow users to submit analyzed data to MethBank. Taken together, the ultimate goal of MethBank is to serve as a public methylation databank by the community, for the community and of the community.
Supplementary Material
ACKNOWLEDGEMENTS
We thank anonymous reviewers for their constructive comments, Dr. Ke Chen for valuable discussions on our work and a number of users for reporting bugs and providing suggestions.
Footnotes
Present address: Shixiang Sun, Department of Genetics, Albert Einstein College of Medicine, Bronx, NY 10461, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Strategic Priority Research Program of the Chinese Academy of Sciences [XDB13040500 to W.Z. and Z.Z., XDA08020102 to Z.Z. and S.H.]; National Key Research & Development Program of China [2017YFC0907502 to Z.Z., 2016YFE0206600 to Y.B.]; National Key Research Program of China [2016YFC0901603 to W.Z., 2016YFB0201702 and 2016YFC0901903 to J.X.]; National Programs for High Technology Research and Development [2015AA020108 and 2012AA020409 to Z.Z.]; International Partnership Program of the Chinese Academy of Sciences [153F11KYSB20160008]; Key Program of the Chinese Academy of Sciences [KJZD-EW-L14 to J.X.]; Key Technology Talent Program of the Chinese Academy of Sciences [to W.Z.]; The 100-Talent Program of the Chinese Academy of Sciences [to Y.B. and Z.Z.]. Funding for open access charge: Strategic Priority Research Program of the Chinese Academy of Sciences [XDA08020102].
Conflict of interest statement. None declared.
REFERENCES
- 1. Jones M.J., Goodman S.J., Kobor M.S.. DNA methylation and healthy human aging. Aging Cell. 2015; 14:924–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Yang X., Han H., De Carvalho D.D., Lay F.D., Jones P.A., Liang G.. Gene body methylation can alter gene expression and is a therapeutic target in cancer. Cancer Cell. 2014; 26:577–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Luo Y., Lu X., Xie H.. Dynamic Alu methylation during normal development, aging, and tumorigenesis. Biomed. Res. Int. 2014; 2014:784706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Li J., Xing X., Li D., Zhang B., Mutch D.G., Hagemann I.S., Wang T.. Whole-genome DNA methylation profiling identifies epigenetic signatures of uterine carcinosarcoma. Neoplasia. 2017; 19:100–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Jiang L., Zhang J., Wang J.J., Wang L., Zhang L., Li G., Yang X., Ma X., Sun X., Cai J. et al. Sperm, but not oocyte, DNA methylome is inherited by zebrafish early embryos. Cell. 2013; 153:773–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Branco M.R., King M., Perez-Garcia V., Bogutz A.B., Caley M., Fineberg E., Lefebvre L., Cook S.J., Dean W., Hemberger M. et al. Maternal DNA methylation regulates early trophoblast development. Dev. Cell. 2016; 36:152–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wang L., Zhang J., Duan J., Gao X., Zhu W., Lu X., Yang L., Li G., Ci W., Li W. et al. Programming and inheritance of parental DNA methylomes in mammals. Cell. 2014; 157:979–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Guo H., Hu B., Yan L., Yong J., Wu Y., Gao Y., Guo F., Hou Y., Fan X., Dong J. et al. DNA methylation and chromatin accessibility profiling of mouse and human fetal germ cells. Cell Res. 2017; 27:165–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Secco D., Wang C., Shou H., Schultz M.D., Chiarenza S., Nussaume L., Ecker J.R., Whelan J., Lister R.. Stress induced gene expression drives transient DNA methylation changes at adjacent repetitive elements. Elife. 2015; 4:e09343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hu L., Li N., Xu C., Zhong S., Lin X., Yang J., Zhou T., Yuliang A., Wu Y., Chen Y.R. et al. Mutation of a major CG methylase in rice causes genome-wide hypomethylation, dysregulated genome expression, and seedling lethality. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:10642–10647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhong S., Fei Z., Chen Y.R., Zheng Y., Huang M., Vrebalov J., McQuinn R., Gapper N., Liu B., Xiang J. et al. Single-base resolution methylomes of tomato fruit development reveal epigenome modifications associated with ripening. Nat. Biotechnol. 2013; 31:154–159. [DOI] [PubMed] [Google Scholar]
- 12. Zhou S., Liu X., Zhou C., Zhou Q., Zhao Y., Li G., Zhou D.X.. Cooperation between the H3K27me3 chromatin mark and non-CG methylation in epigenetic regulation. Plant Physiol. 2016; 172:1131–1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Horvath S., Gurven M., Levine M.E., Trumble B.C., Kaplan H., Allayee H., Ritz B.R., Chen B., Lu A.T., Rickabaugh T.M. et al. An epigenetic clock analysis of race/ethnicity, sex, and coronary heart disease. Genome Biol. 2016; 17:171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Teschendorff A.E., Menon U., Gentry-Maharaj A., Ramus S.J., Weisenberger D.J., Shen H., Campan M., Noushmehr H., Bell C.G., Maxwell A.P. et al. Age-dependent DNA methylation of genes that are suppressed in stem cells is a hallmark of cancer. Genome Res. 2010; 20:440–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Marioni R.E., Shah S., McRae A.F., Chen B.H., Colicino E., Harris S.E., Gibson J., Henders A.K., Redmond P., Cox S.R. et al. DNA methylation age of blood predicts all-cause mortality in later life. Genome Biol. 2015; 16:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hao X., Luo H., Krawczyk M., Wei W., Wang W., Wang J., Flagg K., Hou J., Zhang H., Yi S. et al. DNA methylation markers for diagnosis and prognosis of common cancers. Proc. Natl. Acad. Sci. U.S.A. 2017; 114:7414–7419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Xiong Y., Wei Y., Gu Y., Zhang S., Lyu J., Zhang B., Chen C., Zhu J., Wang Y., Liu H. et al. DiseaseMeth version 2.0: a major expansion and update of the human disease methylation database. Nucleic Acids Res. 2017; 45:D888–D895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zou D., Sun S., Li R., Liu J., Zhang J., Zhang Z.. MethBank: a database integrating next-generation sequencing single-base-resolution DNA methylation programming data. Nucleic Acids Res. 2015; 43:D54–D58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kodama Y., Shumway M., Leinonen R. International Nucleotide Sequence Database, C. . The Sequence Read Archive: explosive growth of sequencing data. Nucleic Acids Res. 2012; 40:D54–D56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wang Y., Song F., Zhu J., Zhang S., Yang Y., Chen T., Tang B., Dong L., Ding N., Zhang Q. et al. GSA: genome sequence archive. Genomics Proteomics Bioinformatics. 2017; 15:14–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bibikova M., Barnes B., Tsan C., Ho V., Klotzle B., Le J.M., Delano D., Zhang L., Schroth G.P., Gunderson K.L. et al. High density DNA methylation array with single CpG site resolution. Genomics. 2011; 98:288–295. [DOI] [PubMed] [Google Scholar]
- 22. Sandoval J., Heyn H., Moran S., Serra-Musach J., Pujana M.A., Bibikova M., Esteller M.. Validation of a DNA methylation microarray for 450,000 CpG sites in the human genome. Epigenetics. 2011; 6:692–702. [DOI] [PubMed] [Google Scholar]
- 23. Barrett T., Wilhite S.E., Ledoux P., Evangelista C., Kim I.F., Tomashevsky M., Marshall K.A., Phillippy K.H., Sherman P.M., Holko M. et al. NCBI GEO: archive for functional genomics data sets–update. Nucleic Acids Res. 2013; 41:D991–D995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hannum G., Guinney J., Zhao L., Zhang L., Hughes G., Sadda S., Klotzle B., Bibikova M., Fan J.B., Gao Y. et al. Genome-wide methylation profiles reveal quantitative views of human aging rates. Mol. Cell. 2013; 49:359–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Voisin S., Almen M.S., Zheleznyakova G.Y., Lundberg L., Zarei S., Castillo S., Eriksson F.E., Nilsson E.K., Bluher M., Bottcher Y. et al. Many obesity-associated SNPs strongly associate with DNA methylation changes at proximal promoters and enhancers. Genome Med. 2015; 7:103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tsaprouni L.G., Yang T.P., Bell J., Dick K.J., Kanoni S., Nisbet J., Vinuela A., Grundberg E., Nelson C.P., Meduri E. et al. Cigarette smoking reduces DNA methylation levels at multiple genomic loci but the effect is partially reversible upon cessation. Epigenetics. 2014; 9:1382–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Levine M.E., Lu A.T., Chen B.H., Hernandez D.G., Singleton A.B., Ferrucci L., Bandinelli S., Salfati E., Manson J.E., Quach A. et al. Menopause accelerates biological aging. Proc. Natl. Acad. Sci. U.S.A. 2016; 113:9327–9332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tan Q., Frost M., Heijmans B.T., von Bornemann Hjelmborg J., Tobi E.W., Christensen K., Christiansen L.. Epigenetic signature of birth weight discordance in adult twins. BMC Genomics. 2014; 15:1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Liang F., Tang B., Wang Y., Wang J., Yu C., Chen X., Zhu J., Yan J., Zhao W., Li R.. WBSA: web service for bisulfite sequencing data analysis. PLoS One. 2014; 9:e86707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Teschendorff A.E., Marabita F., Lechner M., Bartlett T., Tegner J., Gomez-Cabrero D., Beck S.. A beta-mixture quantile normalization method for correcting probe design bias in Illumina Infinium 450 k DNA methylation data. Bioinformatics. 2013; 29:189–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Slieker R.C., Bos S.D., Goeman J.J., Bovee J.V., Talens R.P., van der Breggen R., Suchiman H.E., Lameijer E.W., Putter H., van den Akker E.B. et al. Identification and systematic annotation of tissue-specific differentially methylated regions using the Illumina 450k array. Epigenet. Chromatin. 2013; 6:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hinoue T., Weisenberger D.J., Lange C.P., Shen H., Byun H.M., Van Den Berg D., Malik S., Pan F., Noushmehr H., van Dijk C.M. et al. Genome-scale analysis of aberrant DNA methylation in colorectal cancer. Genome Res. 2012; 22:271–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wang Y., Zhang J., Xiao X., Liu H., Wang F., Li S., Wen Y., Wei Y., Su J., Zhang Y.. The identification of age-associated cancer markers by an integrative analysis of dynamic DNA methylation changes. Sci. Rep. 2016; 6:22722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Dong Y., Zhao H., Li H., Li X., Yang S.. DNA methylation as an early diagnostic marker of cancer (review). Biomed. Rep. 2014; 2:326–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Takuno S., Ran J.H., Gaut B.S.. Evolutionary patterns of genic DNA methylation vary across land plants. Nat. Plants. 2016; 2:15222. [DOI] [PubMed] [Google Scholar]
- 36. Wang H., Beyene G., Zhai J., Feng S., Fahlgren N., Taylor N.J., Bart R., Carrington J.C., Jacobsen S.E., Ausin I.. CG gene body DNA methylation changes and evolution of duplicated genes in cassava. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:13729–13734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kim K.D., El Baidouri M., Abernathy B., Iwata-Otsubo A., Chavarro C., Gonzales M., Libault M., Grimwood J., Jackson S.A.. A comparative epigenomic analysis of polyploidy-derived genes in soybean and common bean. Plant Physiol. 2015; 168:1433–1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ingouff M., Selles B., Michaud C., Vu T.M., Berger F., Schorn A.J., Autran D., Van Durme M., Nowack M.K., Martienssen R.A. et al. Live-cell analysis of DNA methylation during sexual reproduction in Arabidopsis reveals context and sex-specific dynamics controlled by noncanonical RdDM. Genes Dev. 2017; 31:72–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Horvath S., Zhang Y., Langfelder P., Kahn R.S., Boks M.P., van Eijk K., van den Berg L.H., Ophoff R.A.. Aging effects on DNA methylation modules in human brain and blood tissue. Genome Biol. 2012; 13:R97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Weidner C.I., Lin Q., Koch C.M., Eisele L., Beier F., Ziegler P., Bauerschlag D.O., Jockel K.H., Erbel R., Muhleisen T.W. et al. Aging of blood can be tracked by DNA methylation changes at just three CpG sites. Genome Biol. 2014; 15:R24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Horvath S. DNA methylation age of human tissues and cell types. Genome Biol. 2013; 14:R115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Petkovich D.A., Podolskiy D.I., Lobanov A.V., Lee S.G., Miller R.A., Gladyshev V.N.. Using DNA methylation profiling to evaluate biological age and longevity interventions. Cell Metab. 2017; 25:954–960.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hannum G., Guinney J., Zhao L., Zhang L., Hughes G., Sadda S., Klotzle B., Bibikova M., Fan J.B., Gao Y. et al. Genome-wide methylation profiles reveal quantitative views of human aging rates. Mol Cell. 2013; 49:359–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Freire-Aradas A., Phillips C., Lareu M.V.. Forensic individual age estimation with DNA: From initial approaches to methylation tests. Forensic Sci. Rev. 2017; 29:121–144. [PubMed] [Google Scholar]
- 45. Zhang Z., Zhao W.M., Xiao J.F., Song S.H., Hao L.L., Li R.J., Ma L.N., Sheng X., Sang J., Wang Y.Q. et al. The BIG Data Center: from deposition to integration to translation. Nucleic Acids Res. 2017; 45:D18–D24. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.