Abstract
Circadian rhythms govern various kinds of physiological and behavioral functions of the living organisms, and disruptions of the rhythms are highly detrimental to health. Although several databases have been built for circadian genes, a resource for comprehensive post-transcriptional regulatory information of circadian RNAs and expression patterns of disease-related circadian RNAs is still lacking. Here, we developed CirGRDB (http://cirgrdb.biols.ac.cn) by integrating more than 4936 genome-wide assays, with the aim of fulfilling the growing need to understand the rhythms of life. CirGRDB presents a friendly web interface that allows users to search and browse temporal expression patterns of interested genes in 37 human/mouse tissues or cell lines, and three clinical disorders including sleep disorder, aging and tumor. More importantly, eight kinds of potential transcriptional and post-transcriptional regulators involved in the rhythmic expression of the specific genes, including transcription factors, histone modifications, chromatin accessibility, enhancer RNAs, miRNAs, RNA-binding proteins, RNA editing and RNA methylation, can also be retrieved. Furthermore, a regulatory network could be generated based on the regulatory information. In summary, CirGRDB offers a useful repository for exploring disease-related circadian RNAs, and deciphering the transcriptional and post-transcriptional regulation of circadian rhythms.
INTRODUCTION
Circadian rhythms represent evolutionary adaptations of living organisms to the daily cyclical changes in their environments (1,2). In mammals, the rhythms are generated by cell-autonomous transcriptional feedback loops composed of positive transcriptional activators and negative feedback elements, resulting in rhythmic expression of several core clock genes and numerous clock-controlled genes (CCGs) (3–5). Genome-wide transcriptional profiling have revealed thousands of circadian-expressed transcripts, the majority of which are essential for mammalian physiology and behavior (6–10). Thus, through the CCGs, circadian rhythms govern a wide variety of behavioral, physiological and metabolic functions (11–14). Persistent abnormalities in circadian rhythms are highly detrimental to health, possibly leading to aberrant metabolism and cellular proliferation, and then increasing the risk of cancers, obesity, diabetes, accelerated aging and mental illnesses (15–21). Therefore, identifying the genes that are expressed with a circadian cycle and uncovering their regulators will not only improve the understanding of the rhythms of life, but will also provide valuable insights into potential treatments and therapies for clinical disorders, which are associated with disturbances of circadian rhythms (22–24).
Genome-wide oscillations of CCGs require the coordinated control of numerous regulators at multiple levels (7). Besides the core circadian feedback elements, numerous transcriptional and post-transcriptional regulators have been shown to be involved in the oscillations of core clock genes and CCGs (25,26). For example, CLOCK, a central component of the circadian pacemaker, can work as a histone acetyltransferase and directly alters histone acetylation of specific lysine residues around their DNA binding sites (27,28). Recently, histone deacetylase 3 (HDAC3) is suggested to be another critical component for the circadian system through regulating the activation and repression processes in a deacetylase activity-independent manner (29). Furthermore, data obtained by chromatin immunoprecipitation sequencing (ChIP-Seq) demonstrated the circadian fluctuation of various histone modifications, such as H3K4me3, H3K9ac, H3K27ac, H3K36me3 and H3K4me1, correlated with the rhythmic expression of their co-localized CCGs (9). Rhythmic enhancer RNAs (eRNAs) have also been speculated to harbor enhancers to form chromatin loops, which facilitates the rhythmic binding of transcription factors (TFs) to regulate the expression of CCGs at the transcriptional level (30–32). However, only ∼20% of cycling mRNAs have corresponding rhythmic nascent RNA (6,7,33), indicating the existence of another type of regulation for the cycling of the remaining rhythmic mRNAs. Following the development of diverse sequencing technologies, the results of several studies have strongly suggested that post-transcriptional regulation may be implicated in 70∼80% of cycling of RNAs. For example, two rhythmic RNA-binding proteins (RBPs), CIRBP and RBM3, can regulate several clock genes in response to temperature fluctuations by binding to 3′UTRs of these genes (34,35). Additionally, miRNA and RNA editing are both viewed as being the key mechanisms in the circadian clockwork (9,36). To acquire an overall view of the regulation of genes involved in circadian expression, it is imperative to elucidate the regulators of RNA cycling at both the transcriptional and post-transcriptional level (37).
As an increasing number of cycling genes and transcriptional and post-transcriptional regulators have been identified due to multiple ‘omic’ approaches, it is imperative to build a platform to collect and integrate the multi-layer information. Several databases have been built for circadian genes, such as CGDB (http://cgdb.biocuckoo.org/index.php) (38), CircaDB (http://circadb.hogeneschlab.org/) (39), and CircadiOmics (http://circadiomics.igb.uci.edu/) (40). CGDB hosts circadian genes curated from published small-scale or high-throughput experimental data of multiple organisms; CircaDB focuses on gene expression profiles of different mouse tissues; CircadiOmics integrates genomic, transcriptomic, proteomic and metabolomic data sets from the livers of wild-type and Clock mutant mice. However, disease-related circadian RNAs and their expression patterns, and comprehensive post-transcriptional regulatory information of circadian RNAs such as histone modifications, eRNAs, RBPs and miRNA, are still lacking from these databases. In the present study, we developed CirGRDB by integrating >4936 genome-wide assays from 96 circadian transcriptome/regulatome profiling datasets, with the aim of fulfilling the growing need to understand the rhythms of life. The database offers a user-friendly web interface to search, visualize and interpretate circadian genes and multiple transcriptional and post-transcriptional regulatory events in both humans and mice. In general, CirGRDB is designed to provide bench researchers substantial convenience to explore the underlying regulatory mechanism for oscillating genes and disease-related circadian RNAs.
DATA COLLECTION AND PROCESSING
Data collection
CirGRDB focuses on the identification and annotation of circadian RNAs and their regulators from the available human and mouse transcriptome/regulatome datasets. Firstly, 2249 assays from 10 circadian datasets of human sleep disorder, aging, epidermal stem cell and cancer cell line were obtained from the ArrayExpress/SRA/GEO database (Supplementary Table S1). We also collected 2259 assays from 50 genome-wide circadian transcriptional profiling datasets in 25 mouse tissues/cell lines of wild type, loss of function of core clock genes, tumor-bearing, sleep restriction, aging or fasting strains under normal light-dark cycle or constant darkness. Moreover, we incorporated 60 regulatome assays from six human cell lines. In addition, we included 368 assays from 30 genome-wide circadian datasets of eight kinds of potential regulators, including TFs, histone modifications, chromatin accessibility, eRNAs, miRNAs, RNA editing, RBPs and RNA methylation, from mouse livers, muscles, and two cell lines, respectively. Detailed information is listed in Supplementary Table S1. Due to the lack of direct regulatory information, we adopted the latest published methods to predict the potential role of rhythmic miRNA in RNA cycling.
Identification of the expression pattern of coding genes
For microarray data sets, we performed standard analysis process including quality control, pre-processing, normalization by using simpleaffy, genefilter, oligo (41) and limma (42) packages across different platforms. To avoid distortion of the results by noise, we filtered out uninformative probes (low variance, expressed uniformly close to background detection levels, without Entrez Gene identifiers, or have duplicated Entrez Gene identifiers). Finally, normalized log2-transformed expression values were obtained. For RNA-Seq data sets, after removing adapters and low quality bases of RNA-Seq datasets, we implemented STAR (43) to map reads to mouse genome mm10 or human genome hg38. Data sets with high mapping rate (more than 70%) were selected for further analysis. Then, featureCounts (44) was used to count the reads according to the coordinates of genes in GENCODE (human V24 and mouse V11) (45). Gene expression levels were normalized using the TMM method by edgeR (46). Genes with RPKM (reads per kilobase of exon model per million mapped reads) value >0.5 in at least one third of the samples (33) were used for further analysis. Rhythmically expressed genes were identified using LSPR (47). Genes expressed with period between 22 and 26 h and a cutoff of P <0.05 and amplitude >0.10 were defined as rhythmic-expressed genes.
Identification and prediction of rhythmic binding regions, histone modifications and DNase I hypersensitive sites
ChIP-Seq datasets of 23 TFs related to circadian rhythm and 8 histone modifications, and datasets of DNase I hypersensitive sites were mapped to human genome hg38 or mouse genome mm10. MACS_1.4.0rc2 (48) was employed to identify peaks, and subdivided by Peaksplitter (49). The highest peak was selected as the master peak, and signals of master peaks were normalized into counts per million (CPM) (7). LSPR (47) was then implemented to identify regions of rhythmic binding or histone modifications. For ChIP-Seq datasets only detected two-time points within 24-hour, differential regions were identified using diffReps (50). The regions obtained above were then annotated to human or mouse genome using HOMER (51).
Identification of rhythmic-expressed eRNAs and miRNAs
eRNA can activate gene expression by recruiting transcription factors and forming transcription interactions between enhancer and its target promoter (30). Previous studies suggest that eRNA can act as a regulator of rhythmic gene expression (31). For CirGRDB construction, 5724 expressed eRNAs were collected, and 2369 rhythmic eRNAs were identified by LSPR (47). miRNAs could typically bind to 3′UTR of their target mRNAs and recruit protein to inhibit translation or accelerate degradation of mRNAs. About 2% rhythmic mRNA are driven by miRNA (33). We collected 53 and 85 rhythmically expressed miRNAs from two miRNA data sets. multiMiR (52) was implemented to predict target of the obtained rhythmic miRNA. To explore the role of miRNA in rhythmic modulation in mRNA expression, we compared 24-h profile of RNA-Seq datasets between Dicer1 knockout mice and wild-type mice (33).
Identification and collection of RNA editing and RBPs binding sites
RNA editing means the deamination of adenosine into inosine (A-to-I) catalyzed by the ADAR family of proteins. ADARB1, a key player in cycling RNA editing and mRNA rhythms, could control the speed of circadian oscillations (36). We collected 132 rhythmic editing sites, mediated by the rhythmic expression of ADARB1 in mouse liver (36). In addition, we analyzed RNA editing sites in seven circadian transcriptome datasets from 14 mouse tissues and two human tissues using RED-ML (53). Rhythmic editing sites were identified using LSPR (47) and annotated using HOMER (51). RBPs can form ribonucleoprotein (RNP) complexes with RNAs, and play a critical role in RNA biogenesis, stability, transport and cellular localization at the post-transcriptional level (54). Genome-wide binding regions of two rhythmic expressed RNA-binding proteins (RBPs), CIRBP and RBM3 (34,35), were also integrated into our database.
Analysis of regulatory network and functional effect
For each rhythmic genes, we constructed a comprehensive regulatory network using Cytoscape Web (55) based on the collected hierarchical gene regulatory networks from PTHGRN (56), which contained the information of post-translational modifications (PTMs), TFs, epigenetic modifications and gene expression. The functional effect of specific gene was annotated to human disease database, DISEASES (http://diseases.jensenlab.org).
DATABASE CONTENT AND THE WEB INTERFACE
Database content and statistics
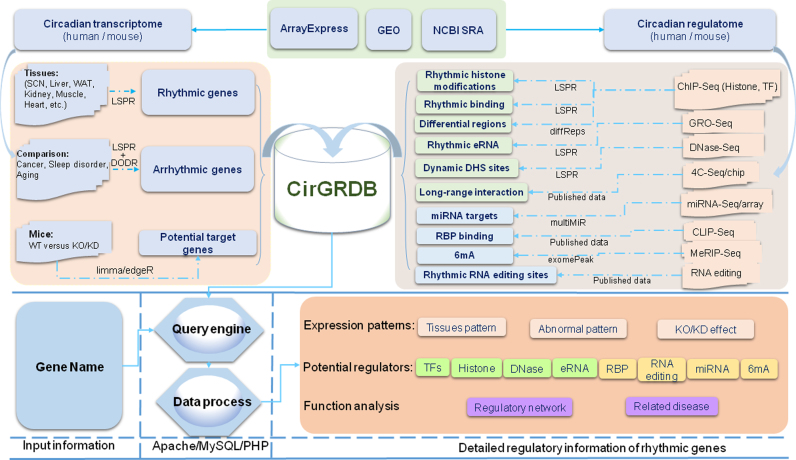
To fulfill the unprecedented need for decoding circadian oscillations from multiple levels, we developed CirGRDB by integrating more than 4936 genome-wide assays. All the metadata of CirGRDB were stored in a MySQL database, and the gene symbols and aliases of rhythmic genes were used as the primary keys to index by MySQL (Figure 1). Open source JavaScript frameworks, jQuery.js, highcharts.js and Cytoscape Web, were employed to display tables and to visualize the oscillations of regulatory elements or gene expression. By employing this database, users can search, browse and annotate circadian genes and their potential regulators, including: (i) temporal expression patterns of circadian genes in 12 human tissues/cell lines and 25 mouse tissues/cell lines under normal light-dark conditions or in constant darkness; (ii) cycling genes related to 3 clinical disorders and corresponding abnormal expression patterns; (iii) eight kinds of potential transcriptional and post-transcriptional regulators involved in the rhythmic expression of the specific genes, such as RNA editing site, binding regions of miRNA and histone modification; (iv) abnormal expression patterns of cycling genes under the loss function of 17 regulators; (v) functional effect and network of target genes. Data retrieved using CirGRDB could be utilized, based on the genes of interest, and users may access the data freely through the web interface (http://cirgrdb.biols.ac.cn).
Figure 1.
Construction and content of CirGRDB. Circadian datasets were downloaded from ArrayExpress (http://www.ebi.ac.uk/arrayexpress/), Gene Expression Omnibus (GEO, https://www.ncbi.nlm.nih.gov/gds/) or Sequence Read Archive (SRA, https://www.ncbi.nlm.nih.gov/sra/). Potential regulators involved in the rhythmic expression of the specific genes were identified using an integrated periodicity detection algorithm, LSPR. WT: wild-type; KO: knockout; KD: knockdown; ChIP-Seq: chromatin immunoprecipitation sequencing; SCN: suprachiasmatic nucleus; WAT: white adipose tissue; eRNA: enhancer RNAs; DHS: DNase I hypersensitive sites; TFs: transcription factors; RBP: RNA-binding proteins.
Web interface
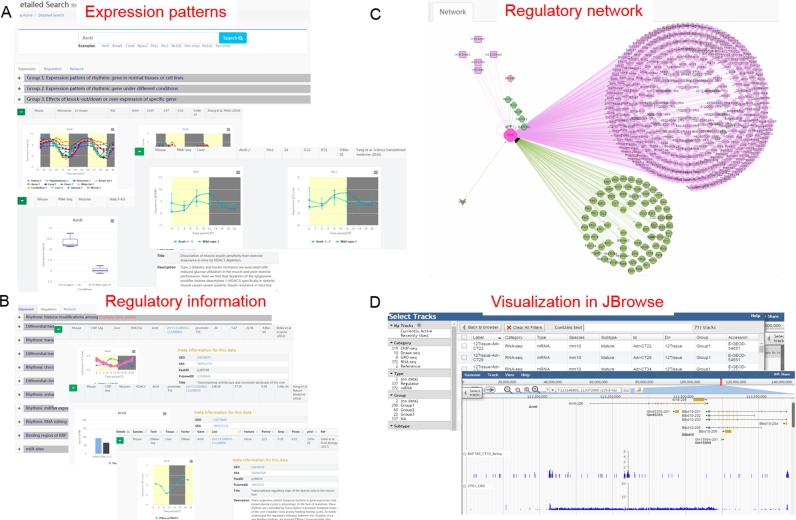
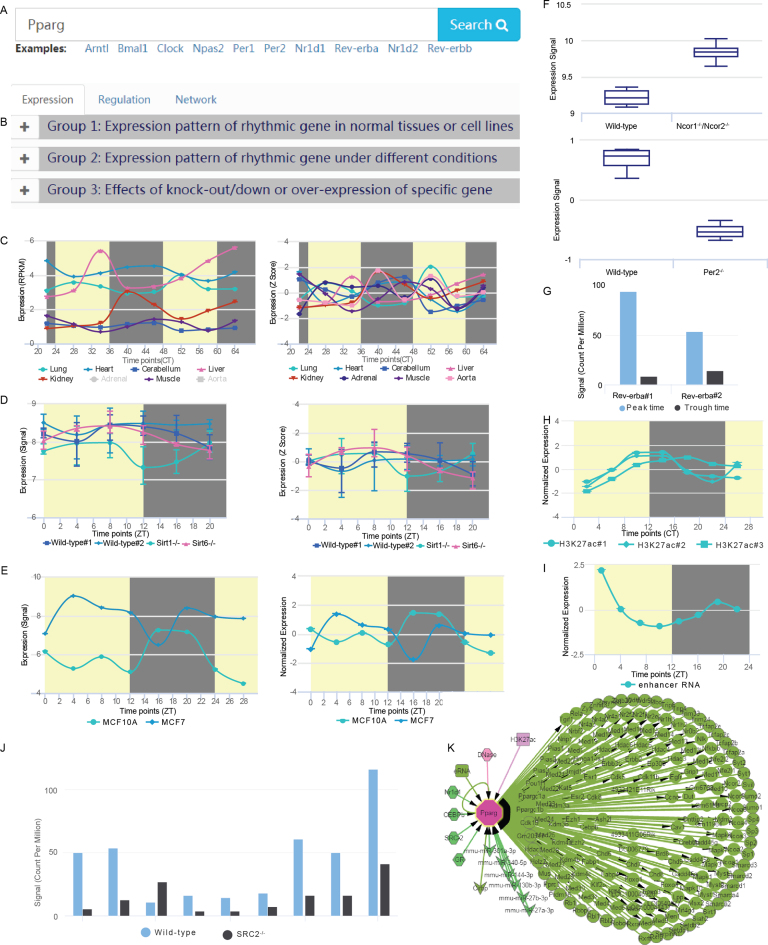
A user-friendly web interface for CirGRDB was constructed, and users can input the gene symbol or alias of interest to retrieve its rhythmic expression patterns and eight kinds of regulatory information involved in its expression (Figure 2). Briefly, user can retrieve three groups of transcriptome profiles including Group1: Expression pattern of rhythmic gene in normal tissues or cell lines; Group2: Expression pattern of rhythmic gene under different conditions; Group3: Effects of knock-out/down or over-expression of specific gene (Figure 2A). Then, user can also retrieve 8 kinds of regulatory information including transcriptional factors, histone modifications, enhancer RNAs, chromatin accessibility, miRNAs, RNA binding proteins, RNA editing and 6 mA (Figure 2B). Furthermore, a regulatory network and disease information relating to rhythmic genes are shown in the figures and tables below (Figure 2C). In addition, user can visualize these 3 groups of transcriptome profiles and regulatory signals in CirGRDB with JBrowse (Figure 2D). For example, taking the gene Pparg as the input (Figure 3A, B) to the search web interface will instantly get the ‘Expression’ (Figure 3C–F), ‘Regulation‘ (Figure 3G–J), and ‘Network’ information (Figure 3K) related to Pparg. In more detail, Pparg was expressed rhythmically in several tissues (Figure 3C), and different expression patterns of PPARG in human breast cancer cell line and normal breast cell line were observed (Figure 3E). Knockout of Ncor1/Ncor2 results in increased expression of Pparg in liver (Figure 3E), whereas knockout of Per2 results in decreased expression of Pparg in liver (Figure 3F). H3K27ac peaks nearby Pparg show similar expression pattern with Pparg (Figure 3H), and eRNA nearby Pparg shows rhythmic expression pattern (Figure 3I). The regulatory network of Pparg was also constructed and visualized based on regulatory information collected from PTHGRN (Figure 3K).
Figure 2.
Web interface of CirGRDB. (A) Detailed search of target gene and three groups of expression patterns. (B) Eight kinds of regulatory factors related to gene of interest. Network obtained (C), visualization of 3 groups of transcriptome, and regulatory signals with Jbrowse (D).
Figure 3.
An example of data access in CirGRDB. ‘CirGRDB’ search (A, B) enables integrative viewing of the expression patterns (C–F), multiple potential regulatory information (G–J) and network (K) of Pparg. User can retrieve ‘Expression’, ‘Regulation’ and ‘Network’ information related to Pparg (B). Expression patterns of Pparg with raw expression level or normalized (Z score) in eight mouse tissues (C), and in livers of wild type, Sirt1−/−, and Sirt6−/− mice (D) at different circadian time (CT) or zeitgeber time (ZT). Different expression patterns of PPARG in human breast cancer cell line (MCF7) and normal breast cell line (MCF10A) were observed (E). Knockout of Ncor1/Ncor2 results in increased expression of Pparg in liver, whereas knockout of Per2 results in decreased expression of Pparg in liver (F). Differential binding of Rev-erba between rhythmic expression time points (G). Peak time represents time point with the highest binding of Rev-erba, while trough time represents time point with the lowest binding of Rev-erba. H3K27ac peaks nearby Pparg show similar expression pattern with Pparg (H). Enhancer RNA (eRNA) nearby Pparg shows rhythmic expression pattern (I). Knockout of SRC2 results in deactivation of DNase I hypersensitive sites (J). Network of input gene was constructed based on PTHGRN database (K).
DISCUSSION AND PERSPECTIVES
Several databases including CGDB (38) and CircaDB (39) have been built for circadian genes curated from published small-scale or high-throughput experimental data, but disease-related circadian RNAs, and their post-transcriptional regulators are not included into these databases. CircadiOmics (40) is another database that integrates genomic, transcriptomic, proteomic and metabolomic data sets from the livers of wild-type and Clock mutant mice, and several public data sets of ChIP-Seq/ChIP-ChIP. However, information on circadian time point, which is crucial for circadian researches, are still absent from these ChIP-Seq/ChIP-ChIP data sets. Thus, disease-related circadian RNAs and their expression patterns, and comprehensive post-transcriptional regulatory information of circadian RNAs such as eRNAs, RBPs and miRNA, are still lacking from the published databases. This study builds the first platform for genome-wide deciphering both transcriptional and post-transcriptional regulation of circadian RNAs using 96 circadian transcriptome/regulatome profiling datasets. In this study, eight kinds of regulatory elements were collected and predicted, including TFs, histone modifications, chromatin accessibility, eRNAs, miRNAs, RBPs, RNA editing and RNA methylation, to construct a regulatory network of rhythmic genes at multiple regulatory layers. An integrated analysis revealed that circadian clock was endogenously oscillated by interconnected hierarchies consisting of various transcriptional and post-transcriptional regulators (32). The present study provides a highly useful resource for the further analysis of temporal patterns of gene expression and functionality. Currently, there is a lack of sets of regulatory data relating to circadian rhythms in mice and human tissues other than liver, such as the SCN, kidney, colon and heart, hence CirGRDB is limited to regulatory information in this well-studied mouse tissue. In the nucleus, the chromatin remodelers, dynamic transcriptional factors, non-coding RNAs and epigenetic modifications might control 3–30% of oscillating transcripts at the transcriptional level through the formation of a three-dimensional circadian interactome (32,57). With increasing numbers of studies into the regulatory mechanisms of circadian clocks and the prevalence of high-throughput sequencing, more regulatory sequencing datasets, such as three-dimensional interactions, rhythmic TFs and RBP bindings, and histone modifications in different tissues, will become available. Emerging mass spectrometry methodologies will also provide unprecedented opportunities for the identification of the rhythmic expression of proteins and subsequent post-translational modifications. We will keep CirGRDB updated as above information emerges. It is anticipated that CirGRDB will be a useful repository for deciphering the regulation of circadian rhythms, and that it will provide valuable insights into novel therapies for circadian-related disorders of humans.
Supplementary Material
SUPPLEMENTARY DATA
Supplementary Data are available at NAR online.
FUNDING
National Natural Science Foundation of China [31371187, 31200888, 31271266]. Funding for open access charge: National Natural Science Foundation of China [31371187].
Conflict of interest statement. None declared.
REFERENCES
- 1. Bass J., Takahashi J.S.. Circadian integration of metabolism and energetics. Science. 2010; 330:1349–1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Green C.B., Takahashi J.S., Bass J.. The meter of metabolism. Cell. 2008; 134:728–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Albrecht U., Sun Z.S., Eichele G., Lee C.C.. A differential response of two putative mammalian circadian regulators, mper1 and mper2, to light. Cell. 1997; 91:1055–1064. [DOI] [PubMed] [Google Scholar]
- 4. Hogenesch J.B., Ueda H.R.. Understanding systems-level properties: timely stories from the study of clocks. Nat. Rev. Genet. 2011; 12:407–416. [DOI] [PubMed] [Google Scholar]
- 5. Sun Z.S., Albrecht U., Zhuchenko O., Bailey J., Eichele G., Lee C.C.. RIGUI, a putative mammalian ortholog of the Drosophila period gene. Cell. 1997; 90:1003–1011. [DOI] [PubMed] [Google Scholar]
- 6. Menet J.S., Rodriguez J., Abruzzi K.C., Rosbash M.. Nascent-Seq reveals novel features of mouse circadian transcriptional regulation. eLife. 2012; 2012:1–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Koike N., Yoo S.H., Huang H.C., Kumar V., Lee C., Kim T.K., Takahashi J.S.. Transcriptional architecture and chromatin landscape of the core circadian clock in mammals. Science. 2012; 338:349–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Rey G., Cesbron F., Rougemont J., Reinke H., Brunner M., Naef F.. Genome-wide and phase-specific DNA-binding rhythms of BMAL1 control circadian output functions in mouse liver. PLoS Biol. 2011; 9:e1000595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Vollmers C., Schmitz R.J., Nathanson J., Yeo G., Ecker J.R., Panda S.. Circadian oscillations of protein-coding and regulatory RNAs in a highly dynamic mammalian liver epigenome. Cell Metab. 2012; 16:833–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhang E.E., Kay S.A.. Clocks not winding down: unravelling circadian networks. Nat. Rev. Mol. Cell. Biol. 2010; 11:764–776. [DOI] [PubMed] [Google Scholar]
- 11. Hood S., Amir S.. The aging clock: circadian rhythms and later life. J. Clin. Invest. 2017; 127:437–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sahar S., Sassone-Corsi P.. Metabolism and cancer: the circadian clock connection. Nat. Rev. Cancer. 2009; 9:886–896. [DOI] [PubMed] [Google Scholar]
- 13. Takahashi J.S. Transcriptional architecture of the mammalian circadian clock. Nat. Rev. Genet. 2016; 18:164–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhou J., Yu W., Hardin P.E.. CLOCKWORK ORANGE Enhances PERIOD Mediated rhythms in transcriptional repression by antagonizing E-box binding by CLOCK-CYCLE. PLoS Genet. 2016; 12:e1006430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lewy A.J., Lefler B.J., Emens J.S., Bauer V.K.. The circadian basis of winter depression. Proc. Natl. Acad. Sci. U.S.A. 2006; 103:7414–7419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. LeGates T.A., Altimus C.M., Wang H., Lee H.K., Yang S., Zhao H., Kirkwood A., Weber E.T., Hattar S.. Aberrant light directly impairs mood and learning through melanopsin-expressing neurons. Nature. 2012; 491:594–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Shi S.Q., Ansari T.S., McGuinness O.P., Wasserman D.H., Johnson C.H.. Circadian disruption leads to insulin resistance and obesity. Curr. Biol. 2013; 23:372–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yang G., Chen L., Grant G.R., Paschos G., Song W.L., Musiek E.S., Lee V., McLoughlin S.C., Grosser T., Cotsarelis G. et al. Timing of expression of the core clock gene Bmal1 influences its effects on aging and survival. Sci. Transl. Med. 2016; 8:324ra316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fu L., Pelicano H., Liu J., Huang P., Lee C.. The circadian gene Period2 plays an important role in tumor suppression and DNA damage response in vivo. Cell. 2002; 111:41–50. [DOI] [PubMed] [Google Scholar]
- 20. Takahashi J.S., Hong H.K., Ko C.H., McDearmon E.L.. The genetics of mammalian circadian order and disorder: implications for physiology and disease. Nat. Rev. Genet. 2008; 9:764–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Liu C., Chung M.. Genetics and epigenetics of circadian rhythms and their potential roles in neuropsychiatric disorders. Neurosci. Bull. 2015; 31:141–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Anafi R.C., Francey L.J., Hogenesch J.B., Kim J.. CYCLOPS reveals human transcriptional rhythms in health and disease. Proc. Natl. Acad. Sci. U.S.A. 2017; 114:5312–5317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhang R., Lahens N.F., Ballance H.I., Hughes M.E., Hogenesch J.B.. A circadian gene expression atlas in mammals: implications for biology and medicine. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:16219–16224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Abbondante S., Eckel-Mahan K.L., Ceglia N.J., Baldi P., Sassone-Corsi P.. Comparative circadian metabolomics reveal differential effects of nutritional challenge in the serum and liver. J. Biol. Chem. 2016; 291:2812–2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Beckwith E.J., Yanovsky M.J.. Circadian regulation of gene expression: at the crossroads of transcriptional and post-transcriptional regulatory networks. Curr. Opin. Genet. Dev. 2014; 27:35–42. [DOI] [PubMed] [Google Scholar]
- 26. Miki T., Matsumoto T., Zhao Z., Lee C.C.. p53 regulates Period2 expression and the circadian clock. Nat. Commun. 2013; 4:2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Doi M., Hirayama J., Sassone-Corsi P.. Circadian regulator CLOCK is a histone acetyltransferase. Cell. 2006; 125:497–508. [DOI] [PubMed] [Google Scholar]
- 28. Hirayama J., Sahar S., Grimaldi B., Tamaru T., Takamatsu K., Nakahata Y., Sassone-Corsi P.. CLOCK-mediated acetylation of BMAL1 controls circadian function. Nature. 2007; 450:1086–1090. [DOI] [PubMed] [Google Scholar]
- 29. Shi G., Xie P., Qu Z., Zhang Z., Dong Z., An Y., Xing L., Liu Z., Dong Y., Xu G. et al. Distinct roles of HDAC3 in the core circadian negative feedback loop are critical for clock function. Cell Rep. 2016; 14:823–834. [DOI] [PubMed] [Google Scholar]
- 30. Lam M.T., Li W., Rosenfeld M.G., Glass C.K.. Enhancer RNAs and regulated transcriptional programs. Trends Biochem. Sci. 2014; 39:170–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Fang B., Everett L.J., Jager J., Briggs E., Armour S.M., Feng D., Roy A., Gerhart-Hines Z., Sun Z., Lazar M.A.. Circadian enhancers coordinate multiple phases of rhythmic gene transcription in vivo. Cell. 2014; 159:1140–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Fan Z., Zhao M., Joshi P.D., Li P., Zhang Y., Guo W., Xu Y., Wang H., Zhao Z., Yan J.. A class of circadian long non-coding RNAs mark enhancers modulating long-range circadian gene regulation. Nucleic Acids Res. 2017; 45:5720–5738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Du N.H., Arpat A.B., De Matos M., Gatfield D.. MicroRNAs shape circadian hepatic gene expression on a transcriptome-wide scale. eLife. 2014; 2014:1–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Liu Y., Hu W., Murakawa Y., Yin J., Wang G., Landthaler M., Yan J.. Cold-induced RNA-binding proteins regulate circadian gene expression by controlling alternative polyadenylation. Scientific Rep. 2013; 3:2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Morf J., Rey G., Schneider K., Stratmann M., Fujita J., Naef F., Schibler U.. Cold-inducible RNA-binding protein modulates circadian gene expression posttranscriptionally. Science. 2012; 338:379–383. [DOI] [PubMed] [Google Scholar]
- 36. Terajima H., Yoshitane H., Ozaki H., Suzuki Y., Shimba S., Kuroda S., Iwasaki W., Fukada Y.. ADARB1 catalyzes circadian A-to-I editing and regulates RNA rhythm. Nat. Genet. 2017; 49:146–151. [DOI] [PubMed] [Google Scholar]
- 37. Hardin P.E., Panda S.. Circadian timekeeping and output mechanisms in animals. Curr. Opin. Neurobiol. 2013; 23:724–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Li S., Shui K., Zhang Y., Lv Y., Deng W., Ullah S., Zhang L., Xue Y.. CGDB: a database of circadian genes in eukaryotes. Nucleic Acids Res. 2017; 45:D397–D403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Pizarro A., Hayer K., Lahens N.F., Hogenesch J.B.. CircaDB: a database of mammalian circadian gene expression profiles. Nucleic Acids Res. 2013; 41:D1009–D1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Patel V.R., Eckel-Mahan K., Sassone-Corsi P., Baldi P.. CircadiOmics: integrating circadian genomics, transcriptomics, proteomics and metabolomics. Nat. Methods. 2012; 9:772–773. [DOI] [PubMed] [Google Scholar]
- 41. Carvalho B.S., Irizarry R.A.. A framework for oligonucleotide microarray preprocessing. Bioinformatics. 2010; 26:2363–2367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., Smyth G.K.. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015; 43:e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R.. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics. 2013; 29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Liao Y., Smyth G.K., Shi W.. FeatureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014; 30:923–930. [DOI] [PubMed] [Google Scholar]
- 45. Harrow J., Frankish A., Gonzalez J.M., Tapanari E., Diekhans M., Kokocinski F., Aken B.L., Barrell D., Zadissa A., Searle S. et al. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res. 2012; 22:1760–1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Robinson M.D., McCarthy D.J., Smyth G.K.. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010; 26:139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Yang R., Zhang C., Su Z.. LSPR: an integrated periodicity detection algorithm for unevenly sampled temporal microarray data. Bioinformatics. 2011; 27:1023–1025. [DOI] [PubMed] [Google Scholar]
- 48. Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W. et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 2008; 9:R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Salmon-Divon M., Dvinge H., Tammoja K., Bertone P.. PeakAnalyzer: genome-wide annotation of chromatin binding and modification loci. BMC Bioinformatics. 2010; 11:415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Shen L., Shao N.Y., Liu X., Maze I., Feng J., Nestler E.J.. diffReps: detecting differential chromatin modification sites from ChIP-seq data with biological replicates. PLoS One. 2013; 8:e65598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Heinz S., Benner C., Spann N., Bertolino E., Lin Y.C., Laslo P., Cheng J.X., Murre C., Singh H., Glass C.K.. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell. 2010; 38:576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ru Y., Kechris K.J., Tabakoff B., Hoffman P., Radcliffe R.A., Bowler R., Mahaffey S., Rossi S., Calin G.A., Bemis L. et al. The multiMiR R package and database: integration of microRNA-target interactions along with their disease and drug associations. Nucleic Acids Res. 2014; 42:e133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Xiong H., Liu D., Li Q., Lei M., Xu L., Wu L., Wang Z., Ren S., Li W., Xia M. et al. RED-ML: a novel, effective RNA editing detection method based on machine learning. Gigascience. 2017; 6:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Bentley D.L. Coupling mRNA processing with transcription in time and space. Nat. Rev. Genet. 2014; 15:163–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Lopes C.T., Franz M., Kazi F., Donaldson S.L., Morris Q., Bader G.D.. Cytoscape Web: an interactive web-based network browser. Bioinformatics. 2011; 26:2347–2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Guan D., Shao J., Zhao Z., Wang P., Qin J., Deng Y., Boheler K.R., Wang J., Yan B.. PTHGRN: unraveling post-translational hierarchical gene regulatory networks using PPI, ChIP-seq and gene expression data. Nucleic Acids Res. 2014; 42:W130–W136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Aguilar-Arnal L., Sassone-Corsi P.. Chromatin landscape and circadian dynamics: Spatial and temporal organization of clock transcription. Proc. Natl. Acad. Sci. U.S.A. 2014; 112:6863–6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.