Abstract
miRandola (http://mirandola.iit.cnr.it/) is a database of extracellular non-coding RNAs (ncRNAs) that was initially published in 2012, foreseeing the relevance of ncRNAs as non-invasive biomarkers. An increasing amount of experimental evidence shows that ncRNAs are frequently dysregulated in diseases. Further, ncRNAs have been discovered in different extracellular forms, such as exosomes, which circulate in human body fluids. Thus, miRandola 2017 is an effort to update and collect the accumulating information on extracellular ncRNAs that is spread across scientific publications and different databases. Data are manually curated from 314 articles that describe miRNAs, long non-coding RNAs and circular RNAs. Fourteen organisms are now included in the database, and associations of ncRNAs with 25 drugs, 47 sample types and 197 diseases. miRandola also classifies extracellular RNAs based on their extracellular form: Argonaute2 protein, exosome, microvesicle, microparticle, membrane vesicle, high density lipoprotein and circulating. We also implemented a new web interface to improve the user experience.
INTRODUCTION
miRNAs are small non-coding RNAs (ncRNAs) (21–23 nt long) that regulate gene expression at the post-transcriptional level by binding to messenger RNAs (mRNAs) and inhibiting their translation into proteins or by binding to other ncRNAs (1).
First discovered in 1993 in Caenorhabditis elegans (2), miRNAs had tremendous impact on the study of gene expression regulation and regulatory networks. Since their discovery as post-transcriptional regulators (3), specific links have been discovered between miRNA and human pathologies (4–7), and further studies indicate the utility of some miRNAs as biomarkers for cancer and other diseases (5). Some miRNA-targeted therapeutics have been tested in clinical trials, including a miRNA mimic of the tumor suppressor miR-34, which reached phase I clinical trials for treating cancer, and antimiRs for miR-122, which reached phase II trials for treating hepatitis (8).
Recently, miRNAs were shown to be present in human body fluids (9), resulting in the potential use of these small RNAs as non-invasive biomarkers (9,10). The great potential of extracellular miRNAs as biomarkers is their high stability in plasma, serum, saliva, urine and many other fluids (9–12). This stability is due to the formation of complexes of extracellular miRNAs in membrane-bound vesicles such as exosomes, which offers them protection from RNAses (13,14). Moreover, miRNAs can be found complexed with the Argonaute2 (Ago2) protein, part of the RNA-induced silencing complex (RISC) responsible of the RNA silencing mediated by miRNAs (15,16). miRNAs also complex with high density lipoprotein (HDL) (17), which provides the mechanism for a new pathway for intercellular communication. In fact, miRNAs transported by HDL can be delivered to recipient cells, where they can alter the expression of their targets (17).
Given the rising interest in miRNAs and more generally other ncRNAs as potential biomarkers, we present an updated version of the miRandola database (11,12), with extensive addition of curated publications, new visualizations and additional functionality in the web interface.
DATA COLLECTION AND CONTENT
The majority of the data were extracted from the literature in PubMed (https://www.ncbi.nlm.nih.gov/pubmed). This included information regarding: the RNA type, sample type, experimental procedures, associated diseases, extracellular RNA forms and other metadata regarding publications, including a summary of the results of the study. Some articles were collected from two publicly available resources, ExoCarta (18) and Vesiclepedia (19), which are two manually curated databases specialized in collecting information on extracellular vesicles.
The first version of the database made use of human biocurators that searched in PubMed using several keywords such as ‘microRNA’, ‘circulating’ and ‘extracellular’ and then manually extracted the relevant information. For this new version, we introduced text-mining-assisted curation to identify and prioritize papers for manual curation. The text mining approach will help our internal curators to increase the update frequency of the database to at least twice a year.
To identify terms of interest, the text-mining software uses dictionaries of human ncRNAs (20), diseases (21) and keywords that indicate extracellular RNA forms. We performed the text mining on more than 26 million entries in PubMed using the tagger software (22). We scored pairs of these terms by summing scores for all co-occurrences of the terms in the same sentence, paragraph and abstract with decreasing weights. We then normalized these scores, and we calculated the geometric mean of RNA-disease and RNA-circulating scores, which we took as the final score for the combined association between circulating RNA and disease. Scientific articles that contain all three types of terms were given the same score as the triple (RNA-extracellular form-disease). Scores were then used to rank articles and facilitate the manual curation.
Altogether, the collected data consisted of 314 articles (see Supplementary Table and website), a notably higher number of papers compared with the first version of the database (n = 89) (12) and the previous short update (n = 119) (11). For details on the database content see Table 1.
Table 1. Comparison between the latest 2017 version and the previous version of miRandola.
| Previous version | miRandola 2017 | |
|---|---|---|
| *Papers | 119 | 314 |
| Entries | 2276 | 3283 |
| microRNAs | 590 | 1002 |
| lncRNAs | 0 | 12 |
| CircRNAs | 0 | 8 |
| Extracellular RNA forms | 4 | 7 |
| Drugs | 6 | 25 |
| Organisms | 1 | 14 |
| Sample types | 23 | 47 |
| Visualization tool | No | Yes |
| External data | ExoCarta | ExoCarta and Vesiclepedia |
| **Text-mining-assisted curation | No | Yes |
*See the supplementary table.
**See the manuscript for more details.
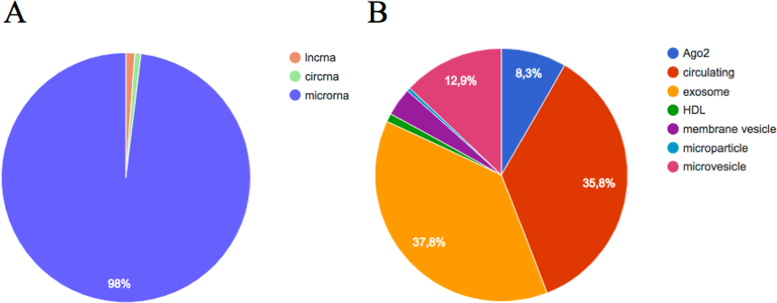
The database aims to present a comprehensive list of all known extracellular ncRNAs. Still, the majority of studies included in this version of miRandola focus on extracellular miRNAs, since they are the most investigated type of ncRNA. Although, in this new version, we started to collect information on two new RNA classes, namely long ncRNAs (lncRNAs) of more than 200 nt, and circular RNAs (circRNAs), which are transcripts that form a continuous loop. These RNA classes represent a small portion of the entries in the database (counts are reported in Table 1 and Figure 1A) and future updates will introduce additional information. Figure 1B shows the variety of extracellular forms that the ncRNAs are found complexed with, including Ago2, exosomes and HDL. More than 35% of RNAs have been annotated only as ‘circulating’, indicating that authors did not specify whether the RNA was complexed with known extracellular forms (Figure 1B).
Figure 1.
Descriptive statistics of the database. (A) Number of RNAs across RNA classes; (B) Number of RNAs across extracellular RNA forms.
Recently, some well-known extracellular lncRNAs have been used as potential non-invasive biomarkers. Probably the most famous example is PCA3 (also known as DD3), which is highly overexpressed in most types of prostate cancer cells and detectable in urine (23). This new non-invasive biomarker shows great potential to improve patient care by reducing the number of biopsies (23).
The function of circRNAs is largely unknown, but some studies have shown that they are able to act as a natural ‘sponge’, by binding and down-regulating miRNAs (24,25). Since circRNAs are stable molecules (26), they have been proposed as novel non-invasive biomarker candidates (26).
DATABASE DEVELOPMENT AND WEB INTERFACE
Data were collected and are maintained in a MySQL database running on an Apache server. The redesigned web interface was implemented using PHP and JavaScript (via the libraries AngularJS and D3.js). Furthermore, Bootstrap is used as front-end framework for faster and easier web development, allowing compatibility with web browsers. In this new version of the database we implemented new functionalities to explore and visualize data making the website more dynamic.
Starting from the home page of the database (see Supplementary Figure S1A), users can quickly search for the name of the RNA of interest by typing it into a search bar. We have implemented an autocomplete function in order to facilitate the search (see Supplementary Figure S1B). After clicking on the RNA of interest, users will have an overview of the extracellular forms in which the RNA has been found, and are able to click on the specific term to browse the results.
The user can browse by the following data types (see Supplementary Figure S2A): ‘miRNAs’, ‘lncRNAs’, ‘circRNAs’, ‘Diseases’, ‘exRNA forms’, ‘Samples’, ‘Drugs’ and ‘Organisms’. For instance, after clicking on ‘miRNAs’, a summary table will be shown (see Supplementary Figure S2B) with miRNA identifiers as reported in literature, but also with the official miRNA identifiers annotated in the last version of the miRNA registry miRBase (27).
Each table can be filtered on a term of interest (see Supplementary Figure S2B), and can also be sorted. After clicking on a specific RNA of interest (see Supplementary Figure S2C and B), a results table is displayed (Figure 2).
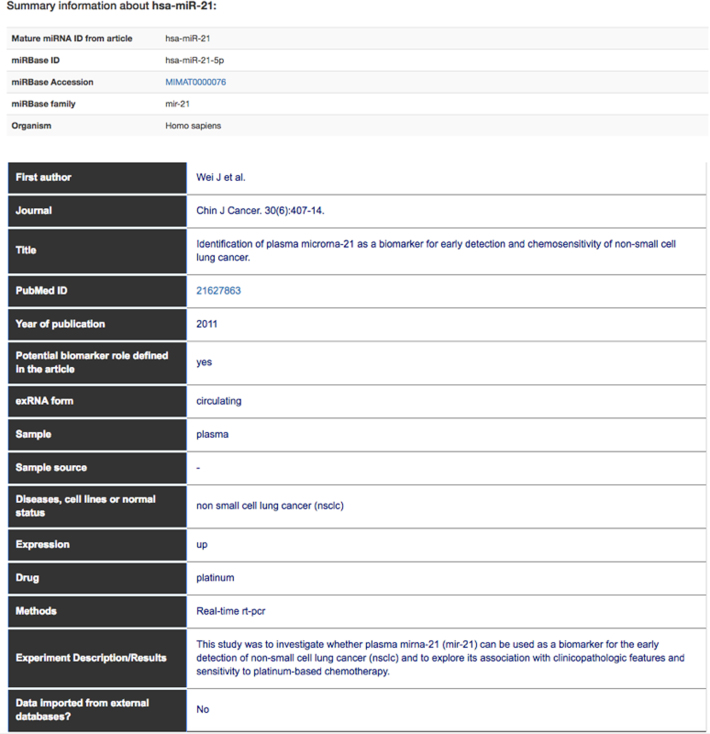
Figure 2.
Results table containing the core information of the database. In this example, we report one result for hsa-miR-21.
The results table contains annotations specific to the selected RNA and other details such as publication identifier, reporting title, publication year, first author and journal. We show associations to diseases, sample types, the extracellular RNA type, the RNA expression level and the drug used in the experiment. We also report the methods used to verify the expression or other relevant techniques, and a short description of the results. The results table contains a field called ‘Potential biomarker role defined in the article’, indicating whether the selected RNA has a potential role as a biomarker, as stated in the published article.
Users can also use the ‘Search’ section (see Supplementary Figure S3) to search for pairs of terms such as ‘hsa-miR-21’ and ‘non-small cell lung cancer’.
All the data in miRandola are available in the ‘Download’ section of the database.
VISUALIZATION
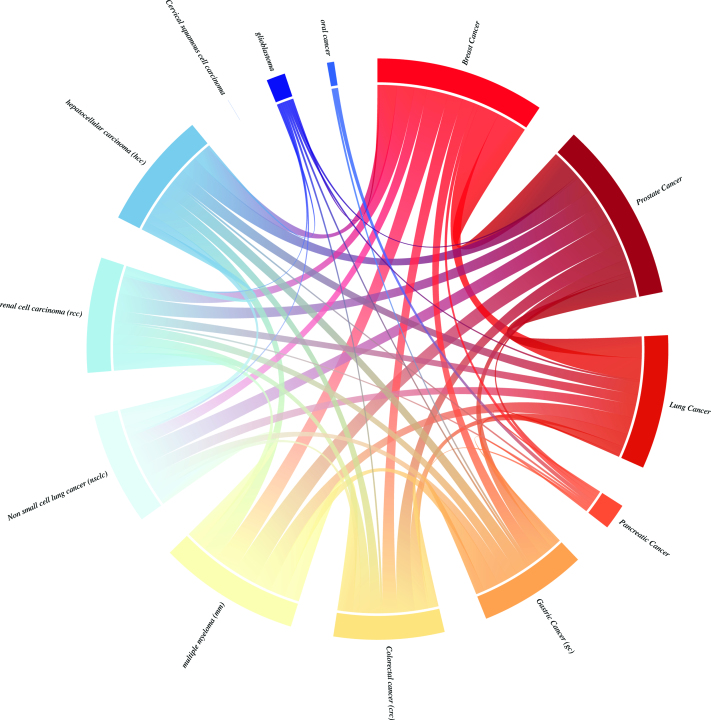
In this new version of miRandola, we introduce a visualization to show RNA-disease co-occurrences extracted from literature, and a circos plot (Figure 3) that shows how many RNAs are shared between the most representative tumor types in our database. This plot reveals that most tumors share several extracellular RNAs, with the exception of ‘Cervical squamous cell carcinoma’ for which we have no evidence of RNAs that are shared with any of the other tumor types. This common signature can be used to help identify common non-invasive biomarkers for many types of cancer.
Figure 3.
Circos plot of the most representative tumor types in the database. The plot shows how many RNAs are shared by different tumors.
FUTURE PERSPECTIVE
When the field of extracellular RNAs was still new, miRandola was started as a small project and it soon became a successful database due to the work of a few biocurators and developers. In this new version, the involvement of additional biocurators and the introduction of assisted curation using text mining both contributed to the collection of many more curated articles, and on an ongoing basis, improves our ability to update the database regularly. We emphasize manual curation as an indispensable step, and define it as the fingerprint of miRandola. For this reason, we will evaluate the participation of the scientific community in the curation process in future updates. We aim to develop a new online tool to achieve this goal, giving users the possibility to curate articles that have been identified and prioritized by text mining. After this step, our internal curators will verify the information introduced by the scientific community.
The final goal of miRandola is to be a reference database for all non-invasive biomarkers, and future updates will consider other data such as extracellular DNA, giving a comprehensive panel of disease-specific biomarkers.
DATA AVAILABILITY
miRandola is available at http://mirandola.iit.cnr.it/.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank the editor and the anonymous reviewers for their constructive suggestions and comments. Following the suggestions, we improved the manuscript and the database.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Novo Nordisk Foundation [NNF14CC0001 to F.R., H.V.C., A.S., K.B., L.J.J., S.B.]. Funding for open access charge: Novo Nordisk Foundation [NNF14CC0001].
Conflict of interest statement. None declared.
REFERENCES
- 1. Bartel D.P. MicroRNAs: target recognition and regulatory functions. Cell. 2009; 136:215–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lee R.C., Feinbaum R.L., Ambros V.. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993; 75:843–854. [DOI] [PubMed] [Google Scholar]
- 3. Bartel D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004; 116:281–297. [DOI] [PubMed] [Google Scholar]
- 4. Calin G.A., Dumitru C.D., Shimizu M., Bichi R., Zupo S., Noch E., Aldler H., Rattan S., Keating M., Rai K. et al. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc. Natl. Acad. Sci. U.S.A. 2002; 99:15524–15529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Calin G.A., Ferracin M., Cimmino A., Di Leva G., Shimizu M., Wojcik S.E., Iorio M.V., Visone R., Sever N.I., Fabbri M. et al. A MicroRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N. Engl. J. Med. 2005; 353:1793–1801. [DOI] [PubMed] [Google Scholar]
- 6. Calin G.A., Sevignani C., Dumitru C.D., Hyslop T., Noch E., Yendamuri S., Shimizu M., Rattan S., Bullrich F., Negrini M. et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc. Natl. Acad. Sci. U.S.A. 2004; 101:2999–3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lagana A., Russo F., Sismeiro C., Giugno R., Pulvirenti A., Ferro A.. Variability in the incidence of miRNAs and genes in fragile sites and the role of repeats and CpG islands in the distribution of genetic material. PLoS One. 2010; 5:e11166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Rupaimoole R., Slack F.J.. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat. Rev. Drug Discov. 2017; 16:203–222. [DOI] [PubMed] [Google Scholar]
- 9. Mitchell P.S., Parkin R.K., Kroh E.M., Fritz B.R., Wyman S.K., Pogosova-Agadjanyan E.L., Peterson A., Noteboom J., O’Briant K.C., Allen A. et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc. Natl. Acad. Sci. U.S.A. 2008; 105:10513–10518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Weber J.A., Baxter D.H., Zhang S., Huang D.Y., Huang K.H., Lee M.J., Galas D.J., Wang K.. The microRNA spectrum in 12 body fluids. Clin. Chem. 2010; 56:1733–1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Russo F., Di Bella S., Bonnici V., Lagana A., Rainaldi G., Pellegrini M., Pulvirenti A., Giugno R., Ferro A.. A knowledge base for the discovery of function, diagnostic potential and drug effects on cellular and extracellular miRNAs. BMC Genomics. 2014; 15(Suppl. 3):S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Russo F., Di Bella S., Nigita G., Macca V., Lagana A., Giugno R., Pulvirenti A., Ferro A.. miRandola: extracellular circulating microRNAs database. PLoS One. 2012; 7:e47786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhang J., Li S., Li L., Li M., Guo C., Yao J., Mi S.. Exosome and exosomal microRNA: trafficking, sorting, and function. Genomics Proteomics Bioinformatics. 2015; 13:17–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Huang X., Yuan T., Tschannen M., Sun Z., Jacob H., Du M., Liang M., Dittmar R.L., Liu Y., Liang M. et al. Characterization of human plasma-derived exosomal RNAs by deep sequencing. BMC Genomics. 2013; 14:319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Arroyo J.D., Chevillet J.R., Kroh E.M., Ruf I.K., Pritchard C.C., Gibson D.F., Mitchell P.S., Bennett C.F., Pogosova-Agadjanyan E.L., Stirewalt D.L. et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:5003–5008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Turchinovich A., Weiz L., Langheinz A., Burwinkel B.. Characterization of extracellular circulating microRNA. Nucleic Acids Res. 2011; 39:7223–7233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Vickers K.C., Palmisano B.T., Shoucri B.M., Shamburek R.D., Remaley A.T.. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat. Cell Biol. 2011; 13:423–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Keerthikumar S., Chisanga D., Ariyaratne D., Al Saffar H., Anand S., Zhao K., Samuel M., Pathan M., Jois M., Chilamkurti N. et al. ExoCarta: a web-based compendium of exosomal cargo. J. Mol. Biol. 2016; 428:688–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kalra H., Simpson R.J., Ji H., Aikawa E., Altevogt P., Askenase P., Bond V.C., Borras F.E., Breakefield X., Budnik V. et al. Vesiclepedia: a compendium for extracellular vesicles with continuous community annotation. PLoS Biol. 2012; 10:e1001450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Junge A., Refsgaard J.C., Garde C., Pan X., Santos A., Alkan F., Anthon C., von Mering C., Workman C.T., Jensen L.J. et al. RAIN: RNA-protein Association and Interaction Networks. Database (Oxford). 2017; 2017, doi:10.1093/database/baw167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pletscher-Frankild S., Palleja A., Tsafou K., Binder J.X., Jensen L.J.. DISEASES: text mining and data integration of disease-gene associations. Methods. 2015; 74:83–89. [DOI] [PubMed] [Google Scholar]
- 22. Pafilis E., Frankild S.P., Fanini L., Faulwetter S., Pavloudi C., Vasileiadou A., Arvanitidis C., Jensen L.J.. The SPECIES and ORGANISMS resources for fast and accurate identification of taxonomic names in text. PLoS One. 2013; 8:e65390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hessels D., Klein Gunnewiek J.M., van Oort I., Karthaus H.F., van Leenders G.J., van Balken B., Kiemeney L.A., Witjes J.A., Schalken J.A.. DD3(PCA3)-based molecular urine analysis for the diagnosis of prostate cancer. Eur Urol. 2003; 44:8–15. [DOI] [PubMed] [Google Scholar]
- 24. Memczak S., Jens M., Elefsinioti A., Torti F., Krueger J., Rybak A., Maier L., Mackowiak S.D., Gregersen L.H., Munschauer M. et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013; 495:333–338. [DOI] [PubMed] [Google Scholar]
- 25. Hansen T.B., Jensen T.I., Clausen B.H., Bramsen J.B., Finsen B., Damgaard C.K., Kjems J.. Natural RNA circles function as efficient microRNA sponges. Nature. 2013; 495:384–388. [DOI] [PubMed] [Google Scholar]
- 26. Memczak S., Papavasileiou P., Peters O., Rajewsky N.. Identification and Characterization of Circular RNAs As a New Class of Putative Biomarkers in Human Blood. PLoS One. 2015; 10:e0141214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kozomara A., Griffiths-Jones S.. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014; 42:D68–D73. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
miRandola is available at http://mirandola.iit.cnr.it/.