Abstract
During infection, antigen-specific T cells undergo tightly regulated developmental transitions controlled by transcriptional and post-transcriptional regulation of gene expression. We found that the microRNA miR-31 was strongly induced by activation of the T cell antigen receptor (TCR) in a pathway involving calcium and activation of the transcription factor NFAT. During chronic infection with lymphocytic choriomeningitis virus (LCMV) clone 13, miR-31-deficent mice recovered from clinical disease, while wild-type mice continued to show signs of disease. This disease phenotype was explained by the presence of larger numbers of cytokine-secreting LCMV-specific CD8+ T cells in miR-31-deficent mice than in wild-type mice. Mechanistically, miR-31 increased the sensitivity of T cells to type I interferons, which interfered with effector T cell function and increased the expression of several proteins related to T cell dysfunction during chronic infection. These studies identify miR-31 as an important regulator of T cell exhaustion in chronic infection.
The control of pathogens and cancers requires proper functioning of the adaptive immune system. CD8+ T cells are a crucial component of such immune responses; they are able to undergo massive population expansion to produce a large pool of effector cells that eliminate virus-infected or transformed cells. CD8+ T cells continuously integrate many distinct signals through the T cell antigen receptor (TCR) as well as through co-stimulatory, co-inhibitory, cytokine and chemokine receptors. These environmental signals either promote pathogen clearance and long-term memory or can result in immune-system dysfunction and T cell exhaustion. These disparate outcomes are dependent on changes in multiple networks of genes encoding proteins that affect the survival, proliferation and effector function of cells1.
MicroRNAs (miRNAs) are a class of small non-coding RNAs that regulate gene expression by translational repression and mRNA destabilization. Deletion of the key miRNA-biosynthetic enzyme Dicer during thymic T cell development markedly diminishes the abundance of CD8+ T cells in the periphery2,3, and deletion of Dicer in mature CD8+ T cells impairs the effector response of CD8+ T cells in vivo4. Individual miRNAs have important roles in the activation of CD8+ T cells. The miR-17-92 cluster regulates the proliferation of CD8+ T cells after activation and shapes effector and memory differentiation5. Other miRNAs shape signaling from TCRs and cytokine receptors. miR-146 is part of a negative feedback loop induced by the TCR and pro-inflammatory cytokines that inhibits activation of the transcription factor NF-κB6,7. In contrast, miR-155 enhances pro-inflammatory cytokine signaling by repressing the signaling inhibitor SOCS1 and promotes the population expansion of anti-viral CD8+ T cells by blocking the anti-proliferative effect of type I interferons8,9. Alterations in miRNA expression can also act downstream of cytokine signaling in CD8+ T cells. For example, signaling via interleukin 2 (IL-2) and inflammatory cytokines drives the differentiation of CD8+ T cells by repressing the expression of miR-150 and miR-139 (ref. 10). However, the function of most miRNAs expressed by CD8+ T cells remains unknown.
Here we report that the expression of miR-31 was strongly induced following the activation of T cells via TCR signaling through pathways dependent on calcium and the transcription factor NFAT. The expression of miR-31 promoted T cell dysfunction by enhancing the expression of multiple inhibitory molecules, particularly during exposure to type I interferons. During chronic viral infection, miR-31 promoted the dysfunction of CD8+ T cells and impaired viral control in a T cell–intrinsic manner. These results identify a role for miR-31 in promoting the exhaustion of CD8+ T cells.
RESULTS
TCR triggering induces sustained expression of miR-31
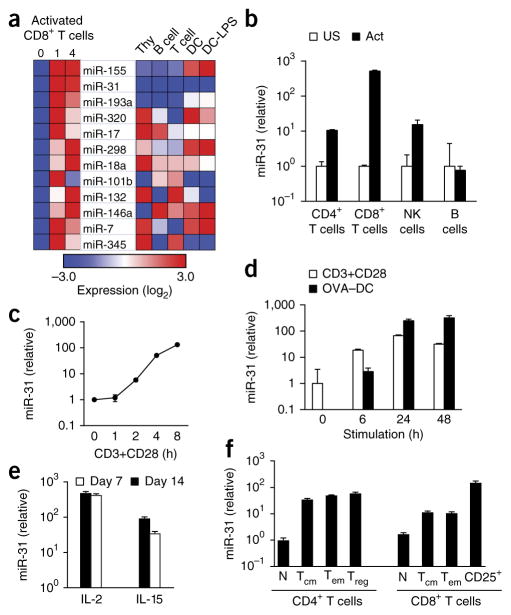
To globally define miRNA expression during T cell activation and differentiation, we used a bead-based assay to profile miRNAs in CD8+ T cells before and after activation via a complex of the TCR and its invariant chain CD3 with an antibody to CD3 (anti-CD3) and an antibody to the co-stimulatory receptor CD28 (anti-CD28). These profiling data revealed dynamic regulation of miRNA expression during T cell activation (Fig. 1a, Supplementary Fig. 1 and Supplementary Table 1). Several of the miRNAs upregulated following activation, including miR-146a, miR-155 and miR-17–miR-18a from the miR-17-92 cluster, have established functions in CD8+ T cells5–9, while many of the other upregulated miRNAs do not have annotated functions in T cells. Of particular interest, miR-31 was not expressed in total thymocytes or naive CD8+ T cells, but its expression was substantially upregulated following the activation of CD8+ T cells (Fig. 1a). Consistent with the profiling data, quantification of miR-31 expression by quantitative PCR (qPCR) indicated upregulation of >100-fold in the expression of miR-31 in CD8+ T cells following stimulation with anti-CD3 plus anti-CD28 relative to its expression in naive CD8+ T cells (Fig. 1b). We also observed upregulation of ~10-fold in the expression of miR-31 in CD4+ T cells following activation with anti-CD3 plus anti-CD28 and in natural killer (NK) cells following stimulation with the phorbol ester PMA and ionomycin, relative to its expression in the corresponding unstimulated cell populations (Fig. 1b). However, miR-31 expression was not induced in CD19+ B cells following stimulation with an antibody specific for immunoglobulin M and a CpG oligo-nucleotide (Fig. 1b). Kinetic studies demonstrated a rapid increase in miR-31 expression in CD8+ T cells stimulated with anti-CD3 plus anti-CD28, with induction of ~100-fold at 8 h after activation relative to the expression before activation (Fig. 1c). Stimulation of naive OT-I CD8+ T cells (which have transgenic expression of an ovalbumin (OVA)-specific TCR) with dendritic cells (DCs) and OVA peptide also rapidly induced an increase of >100-fold in the expression of miR-31 relative to its expression in unstimulated CD8+ T cells (Fig. 1d). These results indicated that miR-31 expression was induced in activated CD4+ and CD8+ T cells, as well as in NK cells, with the highest expression in activated CD8+ T cells.
Figure 1.
miR-31 is induced during T cell activation. (a) miRNAs (middle) with an increase in expression of more than twofold following activation of CD8+ T cells for 1–4 d with anti-CD3 plus anti-CD28, relative to that of unstimulated cells (day 0) (left), and expression of those miRNAs in total thymocytes (Thy), B cells, T cells, bone-marrow-derived DCs (DC) and LPS-treated DCs (DC-LPS) (right), presented as row-normalized relative expression (log2). (b) qPCR analysis of miR-31 in CD4+ or CD8+ T cells activated (Act) for 24 h with anti-CD3 plus anti-CD28, NK cells activated for 24 h with PMA and ionomycin, and B cells activated for 24 h with an antibody to immunoglobulin M and CpG; results are presented relative to those of the unstimulated counterparts (US). (c) qPCR analysis of miR-31 in CD8+ T cells activated for 0–8 h (horizontal axis) with anti-CD3 plus anti-CD28 (CD3+CD28); results are presented relative to those at 0 h. (d) qPCR analysis of mir-31 in OT-I T cells activated for 0–48 h (horizontal axis) with anti-CD3 plus anti-CD28 (CD3+CD28) or by co-culture with OVA-peptide-pulsed DCs (OVA–DC) (T cell/DC ratio, 1:1) (key); results presented as in c. (e) qPCR analysis of mir-31 in OT-I CD8+ T cells activated for 48 h by OVA-peptide-pulsed splenocytes, then treated for 7 or 14 d (key) with IL-2 or IL-15 (horizontal axis); results are presented relative to those of untreated cells. (f) qPCR analysis of mir-31 in naive (CD44loCD62L+) T cells (N), effector memory (CD44hiCD62L−) T cells (TEM), central memory (CD44hiCD62L+) T cells (TCM), regulatory (CD4+CD25+GITR+) T cells (Treg), and activated CD8+ (CD8+CD25+) T cells (CD25+), with all CD4+ or CD8+ subsets (horizontal axis) sorted ex vivo; results are presented relative to those of naive T cells. Data are representative of one experiment (a), three experiments (b; error bars, s.d.) or two experiments (c–f; error bars (d–f), s.d.).
To investigate whether miR-31 expression was maintained in activated T cells, we quantified its expression 1 and 2 weeks following stimulation. Naive CD8+ T cells isolated from OT-I mice were stimulated for 72 h with OVA peptide, followed by culture with IL-2 or IL-15. The expression of miR-31 remained high for 2 weeks in CD8+ T cells cultured with IL-2 (Fig. 1e). In contrast, its expression progressively decreased in CD8+ T cells cultured with IL-15, although it remained higher at 7 d and 14 d in those cells than in naive CD8+ T cells (Fig. 1e). Treatment of naive CD8+ T cells with the cytokines IL-2, IL-7 or IL-15 in the absence of TCR stimulation did not induce the expression of miR-31 (Supplementary Fig. 2a–c), which indicated that those cytokines were not sufficient to induce mir-31 expression.
We also investigated the expression of miR-31 ex vivo. Populations of CD4+ or CD8+ effector and effector-memory T cells, as well as central memory T cells, had higher expression of miR-31 than that of their naive counterparts; the highest expression was observed in activated CD25+ CD8+ T cells (Fig. 1f). T cell activation induces degradation of Argonaute proteins (which are essential for the assembly and function of the miRNA-containing RNA-induced silencing complex) and remodeling of the miRNA repertoire11. We confirmed induction of miR-31 expression in activated T cells through the use of the small nucleolar RNA sno234 as a standard (Supplementary Fig. 2d–f). These results indicated that increased miR-31 expression was a feature of effector CD4+ and CD8+ T cell subsets as well as memory CD4+ and CD8+ T cell subsets.
The induction of miR-31 is dependent on calcium signaling
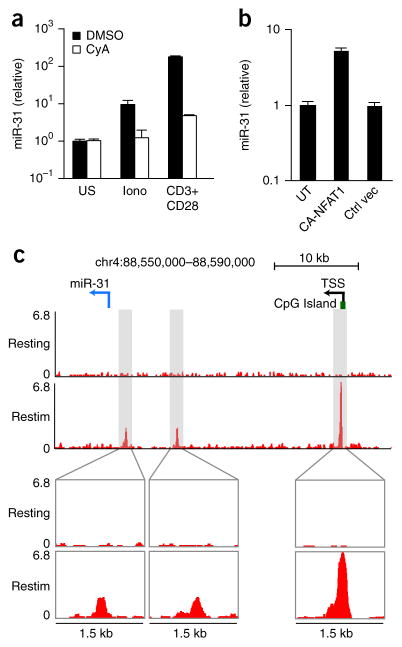
The rapid induction of miR-31 after T cell activation places the gene encoding miR-31 (Mir31) among the early-response genes, many of which are transcriptionally activated through the calcium-NFAT pathway. Treatment of CD8+ T cells with ionomycin, a calcium ionophore, was sufficient to induce miR-31 expression, albeit at a lower level than that induced by stimulation with anti-CD3 plus anti-CD28, while inhibition of calcium signaling with cyclosporine A blocked the induction of miR-31 following treatment with ionomycin or stimulation with anti-CD3 plus anti-CD28 (Fig. 2a). Expression (via a lentiviral vector) of constitutively active NFAT1 containing serine-to-alanine substitutions of key residues within the regulatory domain12 in CD8+ T cells induced expression of miR-31 to levels similar to those achieved with ionomycin treatment (Fig. 2b). Genome-wide analysis of the binding of NFAT1 by chromatin immunoprecipitation in CD8+ T cells stimulated with PMA and ionomycin revealed three binding peaks upstream of the miR-31 hairpin on mouse chromosome 4 (Fig. 2c), one of which was located near a CpG island homologous to the promoter of the human gene encoding miR-31 (MIR31HG)13,14. The binding of NFAT1 to these genomic elements was not observed in resting CD8+ T cells or in CD8+ T cells stimulated with PMA and ionomycin and also treated with cyclosporin A (Fig. 2c). These data indicated that calcium signaling and subsequent NFAT transcriptional activity were linked to the induction of miR-31 via TCR activation in CD8+ T cells.
Figure 2.
mir-31 is induced by signaling via calcium and NFAT. (a) qPCR analysis of miR-31 in CD8+ T cells stimulated for 24 h with ionomycin (Iono) or with anti-CD3 plus anti-CD28 (horizontal axis) and treated with vehicle (DMSO) or cyclosporin A (CyA) (key); results are presented relative to those of unstimulated cells. (b) qPCR analysis of mR-31 in T cells infected with a lentiviral vector driving expression of constitutively active NFAT1 (CA-NFAT1) or a control lentiviral vector (Ctrl vec); results are presented relative to those of untransduced CD8+ T cells (UT). (c) Chromatin-immunoprecipitation analysis of NFAT1 in CD8+ T cells activated with anti-CD3 plus anti-CD28, followed by population expansion for 6 d in medium containing IL-2, then left untreated (Resting) or re-stimulated for 1 h with PMA and ionomycin (Restim) (left margin). Below, enlargement of three areas of enrichment for NFAT-1 binding detected in re-stimulated cells by the HOMER suite of tools for motif discovery. Top, positions of the pre-miR-31 hairpin (blue arrow, location and direction of transcription), the CpG island, and the predicted transcription start site (TSS) of Mir31. Data are representative of two experiments (error bars (a,b), s.d.).
Identification of miR-31 target genes in CD8+ T cells
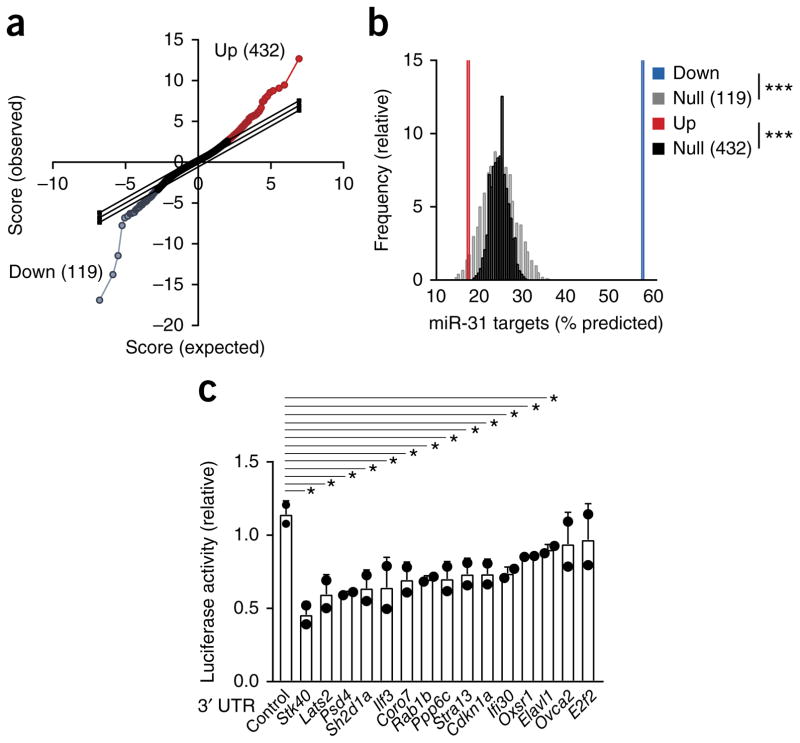
miRNAs repress many target mRNAs, but accurate prediction of their targets is challenging due to the partial complementarity of mammalian miRNA–mRNA pairs and limited understanding of the factors that regulate the binding of miRNAs15. Because regulation by a miRNA reduces the abundance of the majority of target mRNAs, we used expression profiling16,17 of OT-I CD8+ T cells transduced with a lentiviral vector expressing miR-31 and enhanced green fluorescent protein (eGFP) (lenti-miR31), or a control vector expressing eGFP alone (lenti-eGFP) to identify targets of miR-31. Transduction of cells with lenti-miR31 resulted in significant changes in mRNA expression relative to that of cells transduced with lenti-eGFP (Fig. 3a). The interaction between miRNAs and repressed target mRNAs occurs mainly in the 3′ untranslated region (UTR) of the target mRNAs18. The mRNAs downregulated in CD8+ T cells transduced with lenti-miR31 relative to their expression in CD8+ T cells transduced with lenti-eGFP showed considerable enrichment for predicted miR-31-binding sites (relative to the abundance of mRNAs that were expressed but not regulated differentially): 57% of mRNAs with reduced abundance (68 of 199) contained a predicted binding site in the 3′ UTR (Fig. 3b). We further validated 15 targets of miR-31 by cloning their 3′ UTRs downstream of a luciferase reporter vector; 13 of these 3′ UTRs resulted in lower luciferase activity in the presence of exogenous miR-31 than that of cells transduced with a luciferase vector without an inserted 3′ UTR (Fig. 3c). Several of these (Ppp6c, Lats2, Oxsr1, Elavl1 (ref. 19) and Stk40)20–23 have been reported as targets of miR-31, while eight mRNAs (Psd4, Sh2d1a, Ilf3, Coro7, Rab1b, Stra13, Cdkn1a and Ifi30) represented newly identified targets of miR-31 (Fig. 3c). These results identified previously unknown targets of miR-31 in activated mouse CD8+ T cells.
Figure 3.
Microarray profiling reveals targets of miR-31 in primary CD8+ T cells. (a) Microarray analysis of gene expression in CD8+ T cells transduced with lenti-miR31 relative to that of cells transduced with lenti-eGFP (T scores): diagonal lines indicate expected null distribution (middle line) and significance cutoffs (top and bottom lines); colors indicate probes significantly downregulated (blue) or upregulated (red), as determined by the SAM (significance analysis of microarrays) technique; in parentheses, number of upregulated probes (Up; top right) or downregulated probes (Down; bottom left). (b) Frequency of predicted miR-31 target genes among the 119 downregulated probes and 432 upregulated probes in a (key), and matching null distributions generated from probes not expressed differentially (Null). ***P < 0.001, up- or downregulated probe versus the corresponding null distribution (one-sample t-test). (c) Luciferin bioluminescence assay of cells transduced with a luciferase reporter gene containing no 3′ UTR (Control) or the 3′ UTR of predicted miR-31 target genes (horizontal axis), in the presence of exogenous miR-31; results are presented relative to those obtained in the absence of exogenous miR-31. Each symbol represents an individual experiment. *P = 0.008, 0.02, 0.007, 0.02, 0.04, 0.03, 0.01, 0.03. 0.03, 0.03, 0.01, 0.02 or 0.04 (left to right), versus control reporter (unpaired one-tailed Student’s t-test). Data are representative of one experiment (a,b) or three experiments (c; mean + s.d.).
miR-31 promotes CD8+ T cell dysfunction
To investigate the function of miR-31 in vivo, we deleted Mir31 in the germline of C57BL/6 mice by crossing mice with loxP-flanked Mir31 alleles (Mir31fl/fl) to mice expressing Cre recombinase under the control of the promoter of the gene encoding the glyco-protein Zp3 (Zp3-Cre) to generate Mir31fl/flZp3-Cre mice (called ‘Mir31−/−’ mice here; Supplementary Fig. 3a). We also deleted Mir31 conditionally in T cells by crossing Mir31fl/fl mice with mice in which Cre is expressed under the control of the T cell–specific Cd4 promoter (Cd4Cre; Mir31fl/flCd4Cre). Activation of CD8+ cells from Mir31−/− and Mir31fl/flCd4Cre mice with anti-CD3 plus anti-CD28 demonstrated complete loss of miR-31 expression relative to that of activated wild-type or Mir31fl/fl CD8+ T cells (Supplementary Fig. 3b,c). Mir31−/− mice were born at the expected Mendelian ratios, displayed no anatomical alterations and had cellularity and mature T cell subset composition in the thymus, spleen and peripheral lymph nodes similar to that of wild-type mice (data not shown).
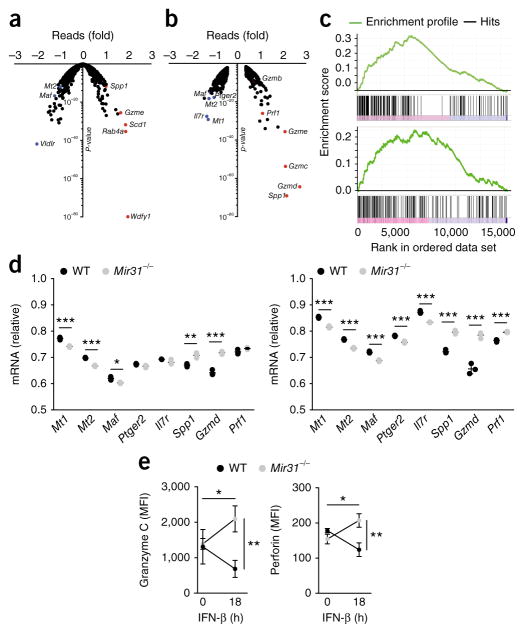
Ingenuity-pathway analysis of all mRNAs expressed differentially in CD8+ T cells transduced with lenti-miR31 relative to their expression in CD8+ T cells transduced with lenti-eGFP revealed correlations with pathways linked to type I interferons, including alterations in the expression of Ifna2 mRNA (which encodes the interferon IFN-α2), and Irf3 mRNA and Irf7 mRNA (which encode the transcription factors IRF3 and IRF7, respectively) (Supplementary Table 2). We therefore analyzed CD8+ T cells from wild-type and Mir31−/− mice by RNA-based next-generation sequencing (RNA-seq) on day 7 following in vitro stimulation with anti-CD3 plus anti-CD28 and culture in IL-2, with or without stimulation with IFN-β for 18 h before analysis. Mir31−/− CD8+ T cells had significantly higher expression of mRNAs encoding T cell effector molecules, including perforin (Prf1) and several granzymes (Gzmd, Gzmc, Gzme, Gzmg and Gzmb), as well as osteopontin (Spp1), than that of wild-type cells, with these changes in gene expression being more prominent following stimulation with IFN-β (Fig. 4a,b). In addition, several genes encoding products known to promote T cell dysfunction, including intracellular metallothioneins (Mt1 and Mt2), which contribute to T cell dysfunction in mouse tumors, as well as the transcription factor c-Maf (Maf) and the receptor for prostaglandin E2 (Ptger2), which contribute to CD8+ T cell dysfunction in a model of chronic infection with lymphocytic choriomeningitis virus (LCMV) clone 13 (refs. 1,24,25), had higher expression in wild-type CD8+ T cells than in Mir31−/− CD8+ T cells (Fig. 4a,b). Consistent with those findings, gene-set–enrichment analysis identified ‘LCMV Armstrong versus clone 13’ as well as ‘Effector versus exhausted CD8+ T cell’ as major gene-expression signatures at 0 h and 18 h, respectively, after stimulation with IFN-β (Fig. 4c). Validation of those RNA-seq data by qPCR indicated that Maf mRNA and Ptger2 mRNA had lower expression in Mir31−/− CD8+ T cells than in wild-type CD8+ T cells following treatment with IFN-β (Fig. 4d). The expression of mRNA encoding perforin (Prf1 mRNA) was higher in Mir31−/− CD8+ T cells than in wild-type CD8+ T cells only following stimulation with IFN-β (Fig. 4d). Stimulation with IFN-β also increased the differences in the expression of granzyme-encoding mRNAs as well osteopontin-encoding mRNA (Spp1) in Mir31−/− CD8+ T cells relative to their expression in wild-type CD8+ T cells (Fig. 4d). Flow cytometry showed that treatment with IFN-β also resulted in a greater abundance of per-forin and granzyme C in Mir31−/− CD8+ T cells than in wild-type CD8+ T cells (Fig. 4e). Immunoblot analysis indicated only small differences between Mir31−/− CD8+ T cells and wild-type CD8+ T cells in phosphorylation of the transcription factor STAT1 following treatment with IFN-β (Supplementary Fig. 4a–d). Also, there were only small differences between those cells in their expression of the transcription factors STAT1, STAT2 and IRF9 and of SOCS1 (Supplementary Fig. 4e). Type 1 interferons are important for the initial activation of CD8+ T cells26. However, miR-31 is expressed only following TCR activation and might therefore regulate the effects of secondary or chronic exposure to type I interferons. In models of chronic viral infection, continued exposure to type I interferons impairs T cell responses and drives disease progression27–29.
Figure 4.
miR-31 limits CD8+ T cell effector programs and enhances exhaustion following stimulation with IFN-β. (a,b) RNA-seq analysis of wild-type and Mir31−/− CD8+ T cells activated with beads coated with anti-CD3 plus anti-CD28 and then cultured for 7 d in IL-2, then left unstimulated (a) or stimulated for 18 h with IFN-β (b); results for Mir31−/− cells are presented relative to those of wild-type cells (red and blue dots indicate relevant genes or genes with the most differential expression (labels adjacent)). (c) Gene-set–enrichment analysis of cells at 0 h (top) and 18 h (bottom) after stimulation with IFN-β, highlighting the ‘LCMV Armstrong versus clone 13’ program (top; normalized enrichment score = 5.728; FDR q value, <0.001) and ‘Effector versus Exhausted T cell’ program (bottom; normalized enrichment score = 3.509; FDR q value, <0.001). (d) qPCR analysis of genes encoding products associated with T cell dysfunction and effector-memory programs (horizontal axes), assessed in wild-type (WT) and Mir31−/− T cells (key) at 0 h (left) and 18 h (right) after stimulation with IFN-β; results are presented relative to those of the control gene Gapdh. (e) Flow-cytometry analysis of granzyme C (left) and perforin (right) in wild-type and Mir31−/− cells T cells (key) at 0 h and 18 h after stimulation with IFN-β (horizontal axes); results are presented as mean fluorescence intensity (MFI). *P < 0.05, **P < 0.01 and ***P < 0.001 (unpaired two-tailed Student’s t-test). Data are from one experiment (a–c) or two independent experiments (d,e) with n = 3 cultures from three mice (mean ± s.d. in d,e).
To address which targets of miR-31 might regulate sensitivity to type I interferons, we transduced the 7678 mouse T cell hybridoma with lentiviral vectors containing short hairpin RNA (shRNA) targeting each of four miR-31-regulated genes (Lats2, Stk40, Ppp6c and Sh2d1a) or a control lentiviral vector and assessed expression of the activation marker CD69, which is induced by type I interferons. Transduction of cells with lenti-miR31 or with shRNA for silencing the miR-31 target Ppp6c enhanced their sensitivity to IFN-β relative to that of cells transduced with the respective control lentiviral vector (Supplementary Fig. 5a). Ppp6c is a phosphatase that regulates cytokine signaling through the kinase Map3k7 and has been linked to inhibition of type I interferon signaling in a high-throughput screen30–32. In contrast, transduction of the 7678 T cell hybridoma with shRNA targeting Lats2, Stk40 or Sh2d1a had no effect on responsiveness to IFN-β (Supplementary Fig. 5a). To confirm those findings in primary T cells, we transduced Mir31−/− OT-I T cells with lenti-miR31, shRNA targeting Ppp6c or the respective control lentiviral vector. Transduction with lenti-miR31 or shRNA targeting Ppp6c increased the sensitivity of CD8+ T cells to IFN-β relative to that of CD8+ T cells transduced with the respective control lentiviral vector (Supplementary Fig. 5b). We also measured Ppp6c by immunoblot analysis of OT-I CD8+ T cells activated with OVA peptide and observed approximately four-fold lower expression of Ppp6c in wild-type CD8+ T cells than in Mir31−/− T cells (Supplementary Fig. 5c,d). These results suggested a role for miR-31 in ‘tuning’ the sensitivity of CD8+ T cells to type I interferon signaling.
miR-31 inhibits T cell responses to chronic infection
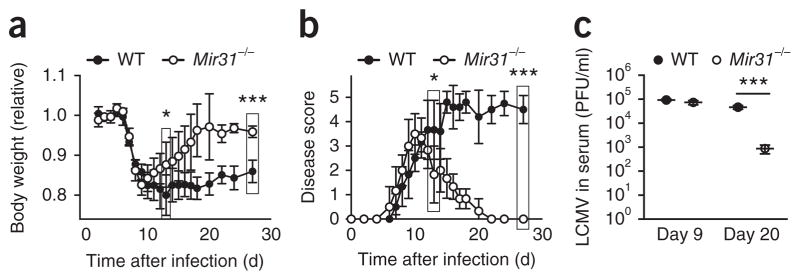
In adult mice, LCMV Armstrong strain causes an acute infection that is cleared by day 8 after infection, while LCMV clone 13 causes a chronic infection that persists >90 d after infection. Both expression of the inhibitory receptor PD-1 and signaling via type I interferons are critical for the chronic infection of mice with LCMV clone 13 (refs. 33–35), while blockade of chronic signaling via type I interferons results in control of infection with LCMV clone 13 (refs. 34,35). We next investigated if miR-31 has a role in controlling the chronicity of infection with LCMV clone 13. The initial disease course was similar in wild-type mice and Mir31−/− mice infected with LCMV clone 13, with all mice displaying disease symptoms at days 9–11 after infection (Fig. 5a,b). Wild-type mice exhibited signs of chronic disease and wasting, while Mir31−/− mice showed no signs of disease and had recovered ~95% of their initial body weight by day 20 after infection (Fig. 5a,b). We next sought to determine whether disease resolution was associated with control of the virus. During the acute phase of the infection (day 9 after infection), wild-type mice and Mir31−/− mice had similar titers of circulating virus (Fig. 5c). However, by day 20 after infection, viral titers were significantly (>1.5-log) lower in the serum of Mir31−/− mice than in that of wild-type mice (Fig. 5c). We also detected lower levels of viral RNA in tissue samples from Mir31−/− mice than in those from wild-type mice at day 30 after infection (Supplementary Fig. 6a). These results suggested that miR-31 impaired the control of infection with LCMV clone 13.
Figure 5.
Mir31−/− mice undergo a faster recovery and show better viral control in a chronic-LMCV-infection model. (a) Body weight of wild-type mice (n = 4) and Mir31−/− mice (n = 6) (key) infected intravenously with LCMV clone 13 (1 × 106 plaque-forming units), assessed daily before (day 0) and after infection (time, horizontal axis); results are presented relative to original body weight (day 0). *P = 0.02 (day 13) and ***P = 0.003 (day 27) (unpaired two-tailed Student’s t-test). (b) Disease scores of wild-type mice (n = 4) and Mir31−/− mice (n = 6) as in a. * P = 0.024 (day 13) and ***P = 4.5 × 10−8 (day 27) (unpaired two-tailed Student’s t-test). (c) Viral load in the serum of mice as in a (n = 3 per group), assessed by plaque assay at days 9 and 20 after infection (horizontal axis) and presented as plaque-forming units (PFU) per ml serum. Each symbol represents an individual mouse; small horizontal lines indicate the mean (± s.d.). ***P = 0.0008 (unpaired two-tailed Student’s t-test). Data are representative of two experiments (mean ± s.d. in a,b).
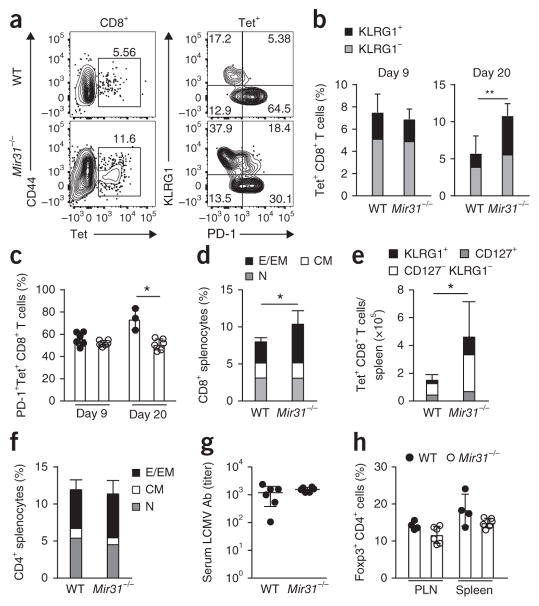
We next investigated whether early disease resolution and improved viral control were associated with enhanced anti-viral CD8+ T cell responses. The frequency of CD8+ T cells positive for a tetramer (Tet) of LCMV glycoprotein (amino acids 33–41) in blood was similar in wild-type mice and Mir31−/− mice on day 8 after infection with LCMV clone 13 (Fig. 6a). However, on day 20 after infection, Mir31−/− mice had a higher frequency of Tet+ CD8+ T cells in blood than that of wild-type mice (Fig. 6a,b). At day 20 after infection, more Tet+CD8+ T cells in the blood of Mir31−/− mice than in that of wild-type mice expressed the effector T cell marker KLRG1, and fewer Tet+CD8+ cells in the blood of Mir31−/− mice than in that of wild-type mice expressed PD-1 (Fig. 6a–c). At day 30 after infection, there was a greater frequency of CD8+ T cells in the spleen of Mir31−/− mice than in that of wild-type mice, an increase driven by the population expansion of CD44+CD62L− CD8+ effector and effector-memory T cells (Fig. 6d). Tetramer labeling indicated population expansion of Tet+KLRG1+ CD8+ T cells and Tet+KLRG1−CD127− CD8+ T cells in Mir31−/− mice relative to the size of those populations in wild-type mice at 30 d after infection, while the frequency of Tet+CD127+ CD8+ T cells in Mir31−/− mice was similar to that of wild-type mice (Fig. 6e).
Figure 6.
Enhanced LCMV-specific CD8+ T cell responses in miR-31 deficient mice. (a) Flow cytometry of CD8+ T cells from the blood of wild-type and Mir31−/− mice (left margin) on day 20 after infection with LCMV clone 13 (left), and surface expression of KLRG1 and PD-1 on Tet+ CD8+ T cells gated as at left (right). Numbers adjacent to outlined areas (left) indicate percent CD44+Tet+ cells; numbers in quadrants (right) indicate percent cells in each. (b) Frequency of KLRG1+ and KLRG1− cells (key) among Tet+ CD8+ T cells in the blood of wild-type mice (n = 4) and Mir31−/− mice (n = 6) (horizontal axis) on days 9 and 20 (above plots) after infection as in a. **P = 0.002 (unpaired two-tailed Student’s t-test). (c) Frequency of PD-1+Tet+ CD8+ T cells in wild-type mice (n = 3) and Mir31−/− mice (n = 6) (key) on days 9 and 20 (horizontal axis) after infection as in a. *P = 0.02 (unpaired two-tailed Student’s t-test). (d) Flow cytometry of effector and effector-memory (CD44+CD62L−) CD8+ T cells (E-EM), central memory (CD44+CD62L+) CD8+ T cells (CM) and naive (CD44− CD62L+) CD8+ T cells (N) (key) in the spleen of wild-type mice (n = 4) and Mir31−/− mice (n = 6) (horizontal axis) on day 30 after infection as in a. *P = 0.03 (unpaired two-tailed Student’s t-test). (e) Quantification of total KLRG1+, CD127−KLRG1− and CD127+ cells (key) among Tet+ CD8+ T cells in the spleen of mice as in d. *P = 0.04 (unpaired two-tailed Student’s t-test). (f) Flow cytometry of CD4+ T cell populations, gated as in d (key), in the spleen mice as in d. (g) ELISA of LCMV-specific antibodies (LCMV Ab) in the serum of wild-type and Mir31−/− mice (horizontal axis) on day 20 after infection with LCMV clone 13. (h) Frequency of Foxp3+ CD4+ T cells in peripheral lymph nodes (PLN) and spleen (horizontal axis) of wild-type and Mir31−/− mice (key) on day 30 after infection with LCMV clone 13. Each symbol (c,g,h) represents an individual mouse; small horizontal lines (g) indicate the mean (± s.d.). Data are representative of two experiments (error bars (b–f,h), s.d.).
We did not observe a difference between wild-type mice and Mir31−/− mice in their CD44+CD62L− CD4+ effector and effector-memory T cells (Fig. 6f). Also, we did not detect differences between wild-type mice and Mir31−/− mice in their titers of LCMV-specific antibodies (Fig. 6g). CD4+ Foxp3+ regulatory T cells undergo population expansion during infection with LCMV clone 13 and inhibit the response of CD8+ T cells to LCMV36,37. miR-31 is known to inhibit the differentiation of CD4+ regulatory T cells in humans through direct regulation of expression of the transcription factor Foxp3 and inhibits the differentiation of induced regulatory T cells in mice through repression of the G-protein-coupled receptor Gprc5a38,39. We therefore investigated the role of miR-31 in regulating the expression of Foxp3 in mice. We found that a luciferase reporter linked to the mouse Foxp3 3′ UTR was not repressed by miR-31 (Supplementary Fig. 6b). We also did not observe differences in the frequency of Foxp3+ CD4+ T cells isolated from the spleen of Mir31−/− mice versus those isolated from that of wild-type mice at baseline or at day 30 following infection with LCMV clone 13 (Fig. 6h and Supplementary Fig. 6c). In mice challenged with influenza virus (strain X31-gp33), weight loss, viral burden in the lungs and the frequency of influenza-virus-specific CD8+ T cells were similar in Mir31+/− mice and Mir31−/− mice on day 8 following infection (Supplementary Fig. 7); this confirmed that miR-31 inhibited CD8+ T cell responses during chronic infection but not during acute infection.
T cell–intrinsic effect of miR-31 in chronic LCMV infection
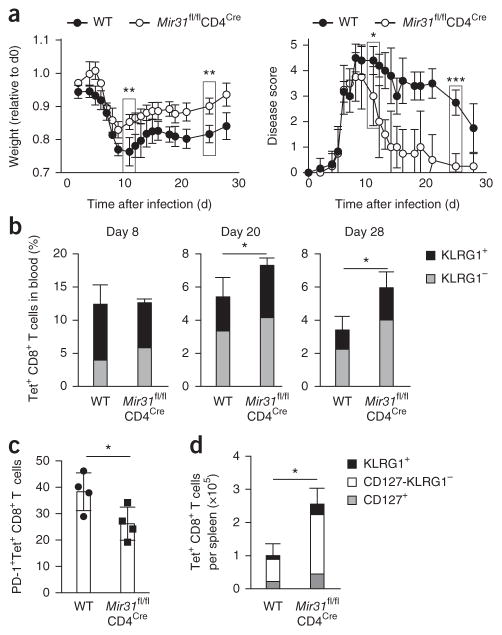
To isolate T cell–intrinsic effects of miR-31, we infected Mir31fl/flCd4Cre mice with LCMV clone 13. The disease course in Mir31fl/flCd4Cre mice was similar to that of Mir31−/− mice following infection with LCMV clone 13, with rapid resolution of disease, in contrast to the chronic disease observed in wild-type mice (Fig. 7a). Mir31fl/flCd4Cre mice had a greater frequency of circulating Tet+ CD8+ T cells at day 20 after infection, and lower expression of PD-1, compared with that of wild-type mice (Fig. 7b,c). We also observed a greater abundance of total Tet+KLRG1+ CD8+ T cells and Tet+KLRG1−CD127− CD8+ T cells in the spleen of Mir31fl/flCd4Cre mice on 28 after infection relative to the abundance of such cells in the spleen of wild-type mice (Fig. 7d). These results demonstrated that miR-31 promoted CD8+ T cell dysfunction and the chronicity of infection with LCMV clone 13 in a T cell–intrinsic manner.
Figure 7.
T cell–intrinsic effect of miR-31 on anti-viral CD8+ T cell responses during chronic infection with LCMV. (a) Body weight (left) and disease scores (right) of wild-type and Mir31fl/flCd4Cre mice (n = 4 per group) infected intravenously with LCMV clone 13 (1 × 106 plaque-forming units), assessed daily before (day 0) and following infection; weight is presented relative to starting body weight (day 0). **P = 0.003 (day 12) and **P = 0.001 (day 25), body weight; *P = 0.01 (day 12) and ***P = 0.0004 (day 25), disease score (unpaired two-tailed Student’s t-test). (b) Frequency of KLRG1+ and KLRG1− cells (key) among Tet+ CD8+ T cells in the blood of mice as in a (n = 4 per group), assessed by flow cytometry at days 8, 20 and 28 after infection. *P = 0.02 (day 20) and *P = 0.03 (day 28) (unpaired two-tailed Student’s t-test). (c) Frequency of PD-1+Tet+ CD8+ T cells in the blood of mice as in a (n = 4 per group), assessed by flow cytometry at day 20 after infection treated. *P = 0.04 (unpaired two-tailed Student’s t-test). (d) Quantification of KLRG1+, CD127−KLRG1− and CD127+ cells (key) among Tet+ CD8+ T cells in the spleen of mice treated as in a (n = 4 per group), assessed by flow cytometry. Each symbol (c) represents an individual mouse. *P = 0.002 (unpaired two-tailed Student’s t-test). Data are representative of one experiment (error bars, s.d.).
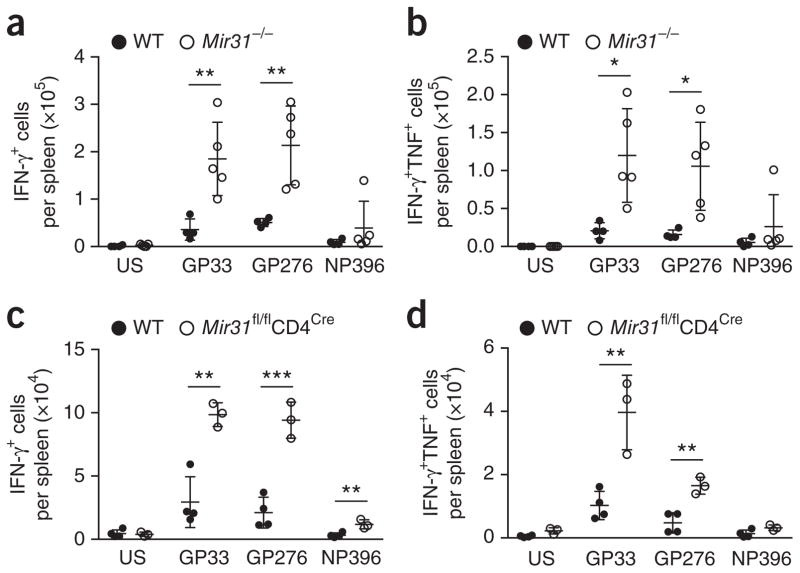
Because CD8+ T cells progressively lose the ability to secrete the cytokines IL-2 and TNF and, finally, IFN-γ40 during chronic viral infection, we assessed the functionality of antigen-specific CD8+ T cells in wild-type, Mir31−/− and Mir31fl/flCd4Cremice on day 28 or 30 after infection with LCMV clone 13. Splenocytes from infected mice were pulsed ex vivo with the peptides GP33 and GP276 (from LCMV glycoprotein) and NP396 (from LCMV nucleoprotein), followed by intracellular cytokine staining for IFN-γ, TNF and IL-2 in CD8+ T cells. We found significantly more IFN-γ+ CD8+ T cells responsive to stimulation with the epitopes GP33 and GP276 in the spleen of Mir31−/− mice and Mir31fl/flCd4Cre mice than in that of wild-type mice (Fig. 8a,c). We also observed more polyfunctional IFN-γ+TNF+ CD8+ T cells in the spleen of Mir31−/− mice and Mir31fl/flCd4Cre mice than in that of wild-type mice (Fig. 8b,d). It has been suggested that miR-31 promotes the secretion of IL-2 by T cells40,41. However, we did not observe differences in the secretion of IL-2 from wild-type, Mir31−/− or Mir31fl/flCd4Cre CD8+ T cells in response to stimulation with LCMV peptide (data not shown), and stimulation with PMA plus ionomycin induced a similar frequency of IL-2+ CD4+ T cells and IL-2+ CD8+ T cells in all mouse strains (Supplementary Fig. 8a). Comparison of cytokine secretion in response to GP33 peptide versus total Tet+ T cells indicated that Mir31−/− mice had a more functional effector pool than that of wild-type mice: Mir31−/− mice had 1.9-fold more IFN-γ+ CD8+ T cells than wild-type mice had, even after correction for a larger population of Tet+ CD8+ T cells (Supplementary Fig. 8b). These results indicated that miR-31 impaired the function of CD8+ T cells in a T cell–intrinsic manner during chronic infection with LCMV clone 13.
Figure 8.
Enhanced production of cytokines by LCMV-specific CD8+ T cells in the absence of miR-31. Quantification of IFN-γ+ CD8+ T cells (a,c) or IFN-γ+TNF+ CD8+ T cells (b,d) among splenocytes obtained from wild-type mice (n = 4) or Mir31−/− mice (n = 6) at day 30 after infection with LCMV clone 13 (a,b) or obtained from wild-type and Mir31fl/flCd4Cre mice (n = 4 per group) at day 28 after infection with LCMV clone 13 (c,d), then left unstimulated (US) or pulsed for 5 h with GP33, GP276 or NP396 peptide (10 μg/ml) (horizontal axes) in the presence of brefeldin A and assessed by intracellular staining; results are presented as total positive cells per spleen. Each symbol represents an individual mouse; small horizontal lines the mean (± s.d.). **P = 0.008 (GP33) and **P = 0.006 (GP276) (a); *P = 0.005 (GP33) or *P = 0.009 (GP276) (b); **P =0.002(GP33), ***P = 0.0007 (GP276) or **P = 0.008 (NP396) (c); and **P = 0.005 (GP33) or **P = 0.003 (GP276) (d) (unpaired two-tailed Student’s t-test). Data are representative of two experiments (a,b) or one experiment (c,d).
DISCUSSION
Here we have shown substantial induction of miR-31 following T cell activation via a process involving calcium signaling and binding of NFAT to the locus encoding miR-31. During chronic infection with LCMV clone 13, Mir31−/− mice had a larger and more functional antigen-specific CD8+ T cell response than that of wild-type mice, which was associated with recovery from infection and control of viral replication. Gene-expression studies revealed that the absence of miR-31 diminished the expression of genes encoding products associated with T cell dysfunction, including c-Maf, the prostaglandin E2 receptor and metallothioneins, and also enhanced T cell effector programs. Those differences appeared in particular after treatment with type I interferons. Our data demonstrated that miR-31 promoted CD8+ T cell exhaustion during the chronic stage of infection.
Early during viral infection, type I interferons promote the priming of anti-viral CD8+ T cells. Treatment of mice with exogenous type I interferons at very early time points following infection with LCMV enhances anti-viral CD8+ T cell responses and improves viral control42. However, sustained production of type I interferons during infection with LCMV clone 13 inhibits T cell–mediated clearance of the virus by suppressing proliferation and increasing the expression of PD-1 by T cells43–45. Antibody blockade of the receptor for IFN-α and IFN-β during chronic infection with LCMV has been shown to promote viral clearance34,35. Mechanistically, blockade of that receptor is associated with reduced expression of the PD-1 ligand PD-L1. We showed that shRNA-mediated knockdown of the miR-31 target Ppp6c increased the sensitivity of CD8+ T cells to type I interferons in vitro and thus was a phenocopy of the effects of miR-31 expression in these cells. The phosphatase Ppp6c has been identified as a negative regulator of interferon signaling in mouse embryonic fibroblasts29. Given that miRNAs typically target many different mRNAs, it is possible that miR-31 targets other mRNAs (in addition to Ppp6c) that contribute to the regulation of interferon signaling in T cells. Also, the contribution of Ppp6c to the in vivo phenotype in Mir31−/− mice remains to be determined.
We did not observe a diminished CD8+ T cell response to acute infection with influenza virus or LCMV in miR-31-deficient mice. While the induction of mir-31 occurred rapidly after T cell activation, the in vivo effects of miR-31 deficiency were evident only after a substantial delay. That pattern is consistent with what has been observed for other activation-induced miRNAs, such as miR-155 (ref. 8). In part, this delay is probably due to the kinetics of miRNA expression and the relative stability of the proteins encoded by the targets of miRNA. Argonaute proteins, which are the effectors of the miRNA-induced silencing complex, are post-transcriptionally downregulated following T cell activation through ubiquitination and proteasome-mediated degradation. This mechanism might substantially delay the miRNA-mediated regulation of mRNA targets11.
The expression of miR-31 increases the abundance of proteins that contribute to T cell dysfunction. The abundance of mRNA encoding prostaglandin E2 receptor (Ptger2) increased in CD8+ T cells during the chronic stage of infection with LCMV clone 13, particularly for T cells with high expression of PD-1. In vitro studies have shown that prostaglandin E2 inhibits the function of cytotoxic T lymphocytes and strongly suppresses the production of cytokines. Inhibition of the production of prostaglandin E2 with an inhibitor of the cyclooxygenase COX-2 has demonstrated synergy with blockade of PD-1 and improved T cell function24. Our data showed that expression of miR-31 was able to increase expression of the receptor for prostaglandin E2 in CD8+ T cells following stimulation with type I interferons. Delineation of the dysfunction signature of tumor-infiltrating CD8+ T cells at the single-cell level has demonstrated that metallothioneins substantially contribute to T cell dysfunction. Metallothionein-deficient mice have a significantly lower tumor burden and stronger CD8+ T cell function than that of metallothionein-sufficient mice25. Both Mt1 and Mt2 (which encode metallothioneins) had higher expression in Mir31+/+ T cells than in Mir31−/− T cells. Maf (which encodes the proto-oncoprotein c-Maf) has also been shown to be overexpressed by T cells during the chronic stage of infection with LCMV clone 13 (ref. 1). However, the expression of several T cell effector molecules was higher in Mir31−/− T cells than in Mir31+/+ T cells, particularly following exposure to type I interferons. Such data explain the enhanced functionality of CD8+ T cells in Mir31−/− mice and the improved clinical course following infection with LCMV clone 13.
Consistent with our findings for mice, stimulation via the TCR induces miR-31 expression in human T cells46. Blocking miR-31 expression in human T cells might prove therapeutically useful for boosting immune responses during chronic viral infection. Moreover, miR-31 might also contribute to human autoimmune conditions characterized by strong signaling via type I interferons, such as systemic lupus erythematosus47. Low expression of miR-31 by circulating T cells has been associated with systemic lupus erythematosus48, which suggests that T cells might escape exhaustion and promote autoimmunity through low expression of miR-31. Finally, the function of miR-31 might also be relevant to cancer, in which T cell dysfunction represents a substantial block to anti-tumor immunity.
ONLINE METHODS
Animals
C57BL/6J mice, C57BL/6-Tg(Zp3-cre)93Knw/J mice, Tg(Cd4-cre)1Cwi/BfluJ (CD4-cre) mice and C57BL/6-Tg(TcraTcrb)1100Mjb/J (OT-I) mice were obtained from Jackson Laboratories. Mir31fl/fl conditional knockout mice were produced on the C57BL/6J background at the Brigham and Women’s Hospital Transgenic Core Facility. Complete or conditional Mir31-knockout mice were produced by crossing Mir31fl/fl mice to Zp3-cre (complete) or CD4-cre (conditional) knockout strains. All mice were maintained in specific-pathogen-free barrier facilities at the Dana-Farber Cancer Institute or at Charles River Laboratories. All experiments were performed in compliance with federal laws and institutional guidelines and were approved by the Animal Care and Use Committee of the Dana-Farber Cancer Institute (Protocol #04-103).
Infection
Age- and sex-matched mice were infected intravenously with LCMV clone 13 (1 × 106 plaque-forming units). LCMV clone 13 was propagated on BHK cells as previously described49. After infection, mice were weighed daily, and body weight was calculated as a percentage of initial weight on day 0. Illness scores were measured by assignment of score of 1 point for each of the following conditions: ruffled fur, hunched back, visually lethargic, lethargic to touch and weight loss exceeding 15% (ref. 50). Viral load was assayed in serum by plaque assay on Vero cells and in tissues by qRT-PCR for viral GP RNA as described49. For infection with influenza virus, age- and sex-matched mice were anesthetized by intraperitoneal injection of 2.5% Avertin (Sigma-Aldrich) and were infected intranasally with recombinant influenza virus expressing the LCMV glycoprotein peptide of amino acids 33–41 (GP33-41; X31-GP33; 1.6 × 105 TCID50). Recombinant influenza virus containing the LCMV GP33-41 epitope inserted into the neuraminidase protein stalk region was provided by R. Webby (St. Jude Children’s Research Hospital, Memphis, TN)51–53. These recombinant influenza virus strains replicate and exhibit pathogenicity similar to that of their non-recombinant counterparts52. Viral titers in lungs were determined by qPCR as described51.
Flow cytometry
Cells were stained with the following monoclonal antibodies: (all from BioLegend; dilutions in parentheses): anti-CD69 (H1.2F3) (1:100), anti-CD25 (PC61) (1:200), anti-KLRG1 (2F1/KLRG1) (1:200), anti-PD-1 (29F.1A12) (1:200), anti-CD8 (53-6.7) (1:400), anti-CD62L (MEL-14) (1:200), anti-CD44 (IM7) (1:400), anti-GITR (YGITR 765) (1:100), anti-CD3 (clone 17A2) (1:400) anti-CD11b (clone M1/70) (1:200), anti-CD11c (clone N418) (1:200) anti-granzyme-B (GB11) (1:100), anti-IFNγ (XMG1.2) (1:100), anti-TNF (MP6-XT22) (1:100), anti-IL-2 (JES6-5H4) (1:100) and anti-FOXP3 (MF-14) (1:100). For LCMV experiments, cells were stained with dextramers of GP33-41 peptide (KAVYNFATC) bound to H-2Db (Immudex). For experiments with influenza virus, cells were stained with H-2Db tetramers loaded with GP33 peptide (KAVYNFATC), NP peptide (ASNENMDAM) or PA peptide (SSLENFRAYV) as described54. For intracellular cytokine staining, splenocytes were incubated in complete RPMI medium for 5 h with no peptide, 10 μg/ml LCMV GP33-41 (KAVYNFATC), GP276-286 (SGVENPGGYCL), NP396-404 (FQPQNGQFI) or 50 ng/ml PMA (phorbol-12-myristate-13-acetate) and 500 nM ionomycin in the presence of 5 μg/ml brefeldin A. Intracellular staining was performed using a BD Cytofix/Cytoperm kit according to the manufacturer’s instructions. For staining of transcription factors, cells were stained and permeabilized with Foxp3 / Transcription Factor Staining Buffer Set (Biolegend) according to manufacturer’s instructions. Data was acquired on a BD Fortessa, and cells were sorted on FACSAria II and III instruments (BD Biosciences). All data were analyzed with FlowJo software (Treestar).
Isolation of immune cell subsets
For in vitro activation, total or naive CD8+ T cells were isolated by negative selection using EasySep beads (Stemcell technologies, Vancouver, BC, Canada). B cells and NK cells were isolated from mouse spleen and peripheral lymph nodes by flow cytometry. B cells were sorted as CD19+B220+ and NK cells were sorted as DX5+CD3−. For miRNA profiling, T cell populations were isolated from spleen and peripheral lymph nodes by flow cytometry. Naive and memory cells were sorted on the basis of the following markers: naive (CD4+ or CD8+, CD62L+CD44loCD25+), central memory (CD4+ or CD8+, CD62L+CD44+CD25−) and effector memory (CD4+ or CD8+, CD62L−CD44+CD25−). Regulatory T cells were sorted as CD4+CD25+GITR+. In vivo–activated CD8+ cells were sorted as CD8+CD25+. T cells activated by co-culture with DCs were purified by flow cytometry as CD3+CD8+CD11b−CD11c−. 7678 T hybridoma cells were isolated as described55.
LCMV antibody ELISA
LCMV antibody titers were determined for serum samples using Smart-M15 enzyme linked immunosorbent assay (Biotech Trading Partners) according to the manufacturer’s instructions.
Cell culture and activation
Cells were cultured in RPMI-1640 medium supplemented with 10% FCS, 50 units/ml penicillin-streptomycin, 10 mM HEPES, 2 mM -glutamine and 55 μM β-mercaptoethanol (’RPMI-10 medium’; all from GIBCO, Carlsbad, CA, USA) at 37 °C with 5% CO2. T cells were activated with 10 μg/ml plate-bound CD3 (clone 17A2) with or without 2 μg/ml soluble CD28 antibody (clone 37.51) (both from eBioscience, San Diego, CA, USA), or 500 nM ionomycin. Cyclosporin A was added at 250 ng/ml. NK cells were activated with PMA (5 ng/ml) and ionomycin (500 nM). B cells were activated with 20 ng/ml IL-2, 1 μg/ml polyclonal goat F(ab′)2 antibody to mouse IgM (Southern Biotech, Birmingham, AL, USA, product number 1022-01) and 3 μg/ml CpG 2006. OT-I CD8+ T cells were activated with splenocytes pulsed with ovalbumin peptide (amino acids 257–264) in the presence of 20 ng/ml IL-2. For type I interferon treatment, cells were incubated with 20 ng/ml (or as indicated in the text) of recombinant mouse IFNβ1 (BioLegend).
To generate DCs, bone marrow cells were isolated from the femurs of C57BL/6 mice, red blood cells were lysed (ACK Lysis Buffer, Gibco) at RT for 5 min and washed, and cells were resuspended in RPMI-1640 (10% FBS and 1% penicillin-streptomycin) supplemented with 20 ng/ml murine GM-CSF (Peprotech,) and plated in 10 ml at a final density of 2 × 105 cells/ml in 10-cm Petri dishes and incubated at 37 °C 5% CO2. After 2 d, 10 ml of additional supplemented RMPI-1640 was added. 2 d later, half of the medium was removed and replenished with supplemented RMPI-1640. After 4 d of further incubation, DCs were stimulated with 1 μg/ml lipopolysaccharide (Sigma-Aldrich) and pulsed with 100 pM ovalbumin peptide (SIINFEKL) (Sigma) overnight. For T cell–activation assays, DCs were then washed, irradiated (2,500 rads), washed, and added to T cells at the indicated ratio.
NFAT1 chromatin immunoprecipitation
For NFAT1 chromatin-immunoprecipitation experiments, P14+ TCRα−/− cells were plated at 1 × 106 cells/ml in six-well plates coated with anti-CD3 (clone 2C11; purified by the A.R. laboratory) and anti-CD28 (clone 37.51; BD-Pharmingen) (1 μg/ml each) by pre-coating with 300 μg/ml goat anti-hamster IgG (catalog 56984; Cappel). After 48 h, cells were removed from the TCR signal and were re-cultured at a density of 5 × 105 cells/ml in media supplemented with 10 U/ml recombinant human IL-2 (rhIL-2). Every 24 h, cells were counted and readjusted to 5 × 105 cells/ml with fresh media containing rhIL-2. On day 6, cells were either left untreated (resting), or re-stimulated with phorbol 12-myristate 13-acetate (PMA; 15 nM; Sigma) and ionomycin (1 μM; Sigma). The protocol for chromatin immunoprecipitation of NFAT1 and computational analysis have been described in detail56 and genome-scale data are available14 The transcription start site for the mouse host gene encoding miR-31 was identified as mm9: chr4:88584380 based on homology to the transcription start site of the human host gene encoding miR-31 (MIR31HG)57, histone H3K4m3 methylation in liver and kidney by ENCODE/LICR58, and spliced expressed sequence tags observed in Mir31−/− CD8+ T-cells (data not shown).
miRNA-specific RT-PCR assay
The synthesis of miRNA specific cDNA from total RNA was carried out using MicroRNA Reverse Transcription Kit (Applied Biosystems, Carlsbad, CA, USA). MiRNA-specific RT-PCRs were performed using Taqman miRNA assays according to the manufacturer’s protocol (Applied Biosystems, Carlsbad, CA, USA). MiRNA expression was calculated using the ΔΔCt method using miR-16 or the ΔCt method using sno234 as expression standards.
Lentiviral vectors
Lentiviral vectors driving expression of shRNAs directed against the luciferase-encoding gene and mouse Lats2, Ppp6c, Stk40 and Sh2d1a were obtained from the RNAi Consortium and subcloned into the pLKO.3G vector. miR-31 precursor sequence was generated by primer extension and was cloned into the pLKO.3G vector. shRNA directed against the luciferase-encoding gene or empty pLKO.3G constructs were used as controls for infection. The cloned mir-31 hairpin sequence is TGCTCCTGTAACTCGGAACTGGAGAG GAGGCAAGATGCTGGCATAGCTGTTGAACTGAGAACCTGCTATGCCA ACATATTGCCATCTTTCCTGTCTGACAGCAGCTTGGCTACCTC.
Constitutively active NFAT1 has been described59 and was subcloned from Addgene plasmid 11792 into the pHAGE lentiviral vector for mammalian expression. Empty pHAGE lentiviral vector was used as an infection control.
miRNA expression profiling
We profiled 279 miRNAs by the Luminex method as described60. Profiling data were normalized by generating a mean array for the expression of each miRNA in all samples. Next, a linear correction for each sample to the mock array was performed. Hierarchical clustering was performed using GenePattern software from the Broad Institute61.
Microarray profiling and miR-31 target gene prediction
CD8+ T cells were purified from OT-I mice by negative selection and were cultured for 2 d in RPMI-10 with 100 ng/ml IL-15 and 1 ng/ml IL-7 (R&D Systems, Minneapolis, MN, USA). Cells were infected with empty or miR-31-expressing pLKO.3G lentiviral vector at a multiplicity of infection of 10:1 by spin-infection in Retronectin-coated plates (Takara Bio, Otsu, Shiga, Japan). After infection, cells were cultured for 3 additional days in RPMI-10 with 10 ng/ml IL-2 (R&D Systems). GFP+ cells from four independent cultures per condition were then sorted by flow cytometry and RNA was isolated using Trizol followed by cleanup with RNeasy columns (Qiagen Valencia, CA, USA). 10 μg RNA per condition was profiled on Affymetrix Mouse Genome 430 2.0 Arrays. Array data analysis was performed using DCHIP including absent/present calls. For probes with 100% present calls in at least one group (22,864), we identified differentially expressed genes using Significance Analysis of Microarrays using standard test statistics and a delta value of 0.582 (refs. 62,63). A master list of miR-31 target genes was generated using the union of mouse miR-31 target predictions from Targetscan, Starbase, Rna22, Pictar and Miranda64–68. To generate matching null distributions for down- or upregulated gene sets, 1 × 104 sets of 119 probes or 432 probes were randomly selected from expressed but not differentially regulated probes, and the frequency of predicted miR-31 targets in each set was calculated. Analysis of statistically associated upstream regulators of differentially expressed genes was performed with Ingenuity Pathway Analysis using standard settings. Microarray data were deposited in the GEO database.
Immunoblot analysis
Cells were lysed in RIPA buffer (Cell Signaling Technology, Danvers MA, USA) in the presence of protease inhibitor cocktail (Roche, Basel, Switzerland). Lysed material was clarified by centrifugation at 12,000g for 10 min at 4 °C. Clarified extracts representing 30 × 105 cells per lane were fractionated on a 10% Bis-Tris gel and transferred to a polyvinylidene difluoride membrane using a transfer apparatus according to the manufacturer’s protocol (Bio-Rad, Hercules, CA, USA). Membranes were blocked for 1 h with 5% nonfat milk in TBST (10 mM Tris, pH 8.0, 150 mM NaCl abd 0.5% Tween 20). Membranes were washed 1× in TBST, and incubated with anti-β-actin (clone 13E5, 1:1000 dilution, Cell Signaling Technology, Danvers MA, USA) and anti-PPP6C (clone ab131335, 1:1000 dilution, Abcam, Cambridge, United Kingdom) for 18 h at 4 °C. Membranes were washed three times with TBST and were incubated with a 1:2,500 dilution of horseradish peroxidase–conjugated polyclonal goat antibody to rabbit (catalog number 7074; Cell Signaling Technology, Danvers MA, USA) for 1 h at RT. Membranes were washed three times with TBST, developed with the ECL system (Amersham Biosciences) and visualized on a ChemiDoc imaging system (Bio-Rad, Hercules, CA, USA) per the manufacturer’s protocols. Full images and size marker standards are presented in Supplementary Figure 6.
Luciferase reporter assay
3′ UTR sequences containing predicted target sites and at least 500 base pairs of flanking sequence were cloned downstream of a firefly luciferase reporter in the pcDNA3.1 mammalian expression vector. 293T cells were transfected in a 96-well format with 20 μg Firefly luciferase reporter containing no 3′ UTR (Control) or test 3′ UTRs, 20 μg control pcDNA3.1 Renilla luciferase reporter, and 200 μg of control or miR-31 pLKO.3G vector. Luciferase activity was quantified at 24 h with DualGlo reagents (Promega, Madison, WI, USA).
Type I interferon stimulation of activated CD8+ T cells
CD8+ T cells were isolated by negative selection (EasySep CD8 Negative Isolation Kit, Stem Cell) from the spleens and lymph nodes of wild-type and Mir31−/− mice and were resuspended in RPMI-10 at a density of 1.5 × 106 cells/ml. T cells were then incubated with beads coated with anti-CD3 and anti-CD28 (Dynabeads, Thermo Scientific) at a ratio of 2:1 in RPMI-10 at 37 °C and 5% CO2 for 48 h. After 48 h, beads were removed using a magnetic column (EasySep Magnet; Stem Cell) and the cells were cultured in RPMI-10 supplemented with 10 ng/ml of IL-2 for 5 d. Supplemented medium was replenished as required to maintain a cell density of 1.5 × 106 cells/ml. After 7 d of culture, T cells were washed and stimulated in RPMI-10 supplemented with 20 ng/ml IFN-β (BioLegend) for either 0, 10, and 30 min (immunoblot analysis) or 0, 4 and 18 h (qPCR or flow cytometry).
For immunoblot analysis sample preparation, cells were counted, and equal numbers from each condition were lysed at each time point in RIPA buffer (Sigma-Aldrich) supplemented with 50 mM NaF, protease inhibitor cocktail (P8340, Sigma) and 0.2 mM sodium orthovanadate. Lysates were incubated at 4 °C for 10 min, then running buffer (NuPAGE LDS Sample Buffer, Thermo) was added at a final concentration of 1×, then the sample was heated at 70 °C for 5 min and then stored at 4 °C. Lysate equivalent to 1 × 105 cells was loaded per lane onto TGX gels (Bio-Rad) for fluorescence-based detection of total proteins based on the manufacturer’s recommendations (45 s UV light treatment followed by UV trans-illumination adjusted for optimal signal intensity). A central section of the gel was used for quantification. This total protein signal was used to normalize STAT1 immunoblot signals.
For RNA analysis, RNA was extracted at each time point using a Directzol RNA Mini-Prep Kit (Zymo Research) per manufacturer’s instructions. For RNA-seq analysis, libraries were generated using Illumina stranded mRNA library prep and sequencing using Illumina NS500 Single-End 75bp sequencing. Statistical significance was determined using VIPER analysis. For qPCR analysis, first strand synthesis was then performed using a SuperScript VILO cDNA synthesis kit (Thermo Fisher Scientific), and qPCR was performed using TaqMan Gene Expression Master Mix (Thermo Fisher Scientific) and primer/probes targeted for Gapdh (Mm99999915_g1), STAT1 (Mm0257286_m1), STAT2 (Mm00490880_m1), IRF9 (Mm00492679_m1), SOCS1 (Mm00782550_s1), Mt1 (Mm00496660_g1), Mt2 (Mm00809556_s1), Maf (Mm02581355_s1), Il7r (Mm00434295_m1), Ptger2 (Mm00436051_m1), Spp1 (Mm00436767_m1), Gzmb (Mm00442837_m1), Gzmd (Mm01722569_g1) and Prf1 (Mm00812512_m1) on a C1000 Touch Thermal Cycler (Bio-Rad).
For flow cytometry, cells were stained for surface markers (CD8-BV786, clone 53-6.7, BioLegend) in flow cytometry buffer (5% FBS in 1X PBS) for 30 min at 4 °C. Cells were fixed in 4% paraformaldehyde for 20 min, washed, permeabilized for 20 min (Cytofix/Cytoperm; BD Biosciences) and stained for 30 min at 4 °C for intracellular markers (granzyme C-PE, clone SFC1D8, BioLegend; perforin, clone eBioMAK-D, eBioscience). All antibodies were used at a dilution of 1:1000. Cells were then washed in flow cytometry buffer and analyzed by flow cytometry (BD LSR Fortessa X20).
Statistical analysis
Statistical analysis was performed using Prism software (Graph Pad) unless otherwise stated in figure legends. Unpaired, two-tailed Student’s t-tests were used for comparison of two groups. Two-way ANOVA with Sidak corrections for multiple comparisons was used for comparisons of more than two groups. For gene-set–enrichment analysis, the false-discovery rate (FDR) q value was calculated using the Gene Set Enrichment Analysis software (Broad Institute, MA, USA). Significance was set at P < 0.05.
Data availability
The data that support the findings of this study are available from the corresponding author upon request. Data have been deposited in the GEO database with the following accession codes: microarray, GSE63549; ChIP-seq (chromatin immunoprecipitation followed by deep sequencing), GSE90707; and RNA-seq, GSE98615.
Supplementary Material
Acknowledgments
We thank R. Webby (St. Jude Children’s Research Hospital) for recombinant influenza virus containing the LCMV GP33–41 epitope inserted into the neuraminidase protein stalk region; J. Maiarana for technical assistance; and the Dana Farber Cancer Institute Molecular Biology Core Facility for RNA sequencing. Supported by the US National Institutes of Health (R01CA173750 to K.W.W.; R01DK102165 and R01CA140986 to C.N.; P01AI056299 to A.S.; and R01AI007386 to H.C.) and the Cancer Research Institute (A.N.R.C.).
Footnotes
AUTHOR CONTRIBUTIONS
H.F.M. and K.W.W. designed the study and discussed and analyzed data; H.F.M., A.N.R.C. and K.W.W. wrote the paper; H.F.M. and A.N.R.C. performed experiments and analyzed data; H.-J.K. and J.G. helped with infection experiments; J.P. performed immunoblot analysis; T.A., G.J.M. and A.R. provided NFAT chromatin-immunoprecipitation data; J.L. and T.R.G. performed miRNA profiling; H.C., A.H.S. and C.D.N. provided advice on experimental design and data interpretation; and K.W.W. supervised the study.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
Publisher’s note: springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Wherry EJ, et al. Molecular signature of CD8+ T cell exhaustion during chronic viral infection. Immunity. 2007;27:670–684. doi: 10.1016/j.immuni.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 2.Muljo SA, et al. Aberrant T cell differentiation in the absence of Dicer. J Exp Med. 2005;202:261–269. doi: 10.1084/jem.20050678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cobb BS, et al. T cell lineage choice and differentiation in the absence of the RNase III enzyme Dicer. J Exp Med. 2005;201:1367–1373. doi: 10.1084/jem.20050572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang N, Bevan MJ. Dicer controls CD8+ T-cell activation, migration, and survival. Proc Natl Acad Sci USA. 2010;107:21629–21634. doi: 10.1073/pnas.1016299107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu T, et al. Temporal expression of microRNA cluster miR-17-92 regulates effector and memory CD8+ T-cell differentiation. Proc Natl Acad Sci USA. 2012;109:9965–9970. doi: 10.1073/pnas.1207327109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang L, et al. miR-146a controls the resolution of T cell responses in mice. J Exp Med. 2012;209:1655–1670. doi: 10.1084/jem.20112218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Taganov KD, Boldin MP, Chang KJ, Baltimore D. NF-κB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci USA. 2006;103:12481–12486. doi: 10.1073/pnas.0605298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gracias DT, et al. The microRNA miR-155 controls CD8+ T cell responses by regulating interferon signaling. Nat Immunol. 2013;14:593–602. doi: 10.1038/ni.2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lu LF, et al. Foxp3-dependent microRNA155 confers competitive fitness to regulatory T cells by targeting SOCS1 protein. Immunity. 2009;30:80–91. doi: 10.1016/j.immuni.2008.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Trifari S, et al. MicroRNA-directed program of cytotoxic CD8+ T-cell differentiation. Proc Natl Acad Sci USA. 2013;110:18608–18613. doi: 10.1073/pnas.1317191110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bronevetsky Y, et al. T cell activation induces proteasomal degradation of Argonaute and rapid remodeling of the microRNA repertoire. J Exp Med. 2013;210:417–432. doi: 10.1084/jem.20111717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Monticelli S, Rao A. NFAT1 and NFAT2 are positive regulators of IL-4 gene transcription. Eur J Immunol. 2002;32:2971–2978. doi: 10.1002/1521-4141(2002010)32:10<2971::AID-IMMU2971>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 13.Martinez GJ, et al. The transcription factor NFAT promotes exhaustion of activated CD8+ T cells. Immunity. 2015;42:265–278. doi: 10.1016/j.immuni.2015.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Scott-Browne JP, et al. Dynamic changes in chromatin accessibility occur in CD8+ T cells responding to viral infection. Immunity. 2016;45:1327–1340. doi: 10.1016/j.immuni.2016.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alexiou P, Maragkakis M, Papadopoulos GL, Reczko M, Hatzigeorgiou AG. Lost in translation: an assessment and perspective for computational microRNA target identification. Bioinformatics. 2009;25:3049–3055. doi: 10.1093/bioinformatics/btp565. [DOI] [PubMed] [Google Scholar]
- 16.Baek D, et al. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Selbach M, et al. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 18.Lim LP, et al. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 19.Wu D, Wang B, Shang J, Song J, Zhang H. miR-31 reduces cell growth of papillary thyroid carcinoma by RNA-binding protein HuR. Clin Lab. 2015;61:1625–1634. doi: 10.7754/clin.lab.2015.150404. [DOI] [PubMed] [Google Scholar]
- 20.Creighton CJ, et al. Molecular profiling uncovers a p53-associated role for microRNA-31 in inhibiting the proliferation of serous ovarian carcinomas and other cancers. Cancer Res. 2010;70:1906–1915. doi: 10.1158/0008-5472.CAN-09-3875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ivanov SV, et al. Pro-tumorigenic effects of miR-31 loss in mesothelioma. J Biol Chem. 2010;285:22809–22817. doi: 10.1074/jbc.M110.100354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu X, et al. MicroRNA-31 functions as an oncogenic microRNA in mouse and human lung cancer cells by repressing specific tumor suppressors. J Clin Invest. 2010;120:1298–1309. doi: 10.1172/JCI39566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rutledge H, et al. Identification of microRNAs associated with allergic airway disease using a genetically diverse mouse population. BMC Genomics. 2015;16:633. doi: 10.1186/s12864-015-1732-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen JH, et al. Prostaglandin E2 and programmed cell death 1 signaling coordinately impair CTL function and survival during chronic viral infection. Nat Med. 2015;21:327–334. doi: 10.1038/nm.3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Singer M, Wang C, Cong L, Marjanovic ND, Kowalczyk MS, Zhang H, et al. A distinct gene module for dysfunction uncoupled from activation in tumor-infiltrating T cells. Cell. 2016;166:1500–1511. doi: 10.1016/j.cell.2016.08.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ivashkiv LB, Donlin LT. Regulation of type I interferon responses. Nat Rev Immunol. 2014;14:36–49. doi: 10.1038/nri3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McNally JM, et al. Attrition of bystander CD8 T cells during virus-induced T-cell and interferon responses. J Virol. 2001;75:5965–5976. doi: 10.1128/JVI.75.13.5965-5976.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marshall HD, Urban SL, Welsh RM. Virus-induced transient immune suppression and the inhibition of T cell proliferation by type I interferon. J Virol. 2011;85:5929–5939. doi: 10.1128/JVI.02516-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Terawaki S, et al. IFN-α directly promotes programmed cell death-1 transcription and limits the duration of T cell-mediated immunity. J Immunol. 2011;186:2772–2779. doi: 10.4049/jimmunol.1003208. [DOI] [PubMed] [Google Scholar]
- 30.Stefansson B, Brautigan DL. Protein phosphatase PP6 N terminal domain restricts G1 to S phase progression in human cancer cells. Cell Cycle. 2007;6:1386–1392. doi: 10.4161/cc.6.11.4276. [DOI] [PubMed] [Google Scholar]
- 31.Kajino T, et al. Protein phosphatase 6 down-regulates TAK1 kinase activation in the IL-1 signaling pathway. J Biol Chem. 2006;281:39891–39896. doi: 10.1074/jbc.M608155200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee MN, et al. Identification of regulators of the innate immune response to cytosolic DNA and retroviral infection by an integrative approach. Nat Immunol. 2013;14:179–185. doi: 10.1038/ni.2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Barber DL, et al. Restoring function in exhausted CD8 T cells during chronic viral infection. Nature. 2006;439:682–687. doi: 10.1038/nature04444. [DOI] [PubMed] [Google Scholar]
- 34.Teijaro JR, et al. Persistent LCMV infection is controlled by blockade of type I interferon signaling. Science. 2013;340:207–211. doi: 10.1126/science.1235214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wilson EB, et al. Blockade of chronic type I interferon signaling to control persistent LCMV infection. Science. 2013;340:202–207. doi: 10.1126/science.1235208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Punkosdy GA, et al. Regulatory T-cell expansion during chronic viral infection is dependent on endogenous retroviral superantigens. Proc Natl Acad Sci USA. 2011;108:3677–3682. doi: 10.1073/pnas.1100213108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Penaloza-MacMaster P, et al. Interplay between regulatory T cells and PD-1 in modulating T cell exhaustion and viral control during chronic LCMV infection. J Exp Med. 2014;211:1905–1918. doi: 10.1084/jem.20132577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rouas R, et al. Human natural Treg microRNA signature: role of microRNA-31 and microRNA-21 in FOXP3 expression. Eur J Immunol. 2009;39:1608–1618. doi: 10.1002/eji.200838509. [DOI] [PubMed] [Google Scholar]
- 39.Zhang L, et al. MicroRNA-31 negatively regulates peripherally derived regulatory T-cell generation by repressing retinoic acid-inducible protein 3. Nat Commun. 2015;6:7639. doi: 10.1038/ncomms8639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xue F, et al. miR-31 regulates interleukin 2 and kinase suppressor of ras 2 during T cell activation. Genes Immun. 2013;14:127–131. doi: 10.1038/gene.2012.58. [DOI] [PubMed] [Google Scholar]
- 41.Fan W, et al. Identification of microRNA-31 as a novel regulator contributing to impaired interleukin-2 production in T cells from patients with systemic lupus erythematosus. Arthritis Rheum. 2012;64:3715–3725. doi: 10.1002/art.34596. [DOI] [PubMed] [Google Scholar]
- 42.Wang Y, et al. Timing and magnitude of type I interferon responses by distinct sensors impact CD8 T cell exhaustion and chronic viral infection. Cell Host Microbe. 2012;11:631–642. doi: 10.1016/j.chom.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bahl K, et al. IFN-induced attrition of CD8 T cells in the presence or absence of cognate antigen during the early stages of viral infections. J Immunol. 2006;176:4284–4295. doi: 10.4049/jimmunol.176.7.4284. [DOI] [PubMed] [Google Scholar]
- 44.Marshall HD, Urban SL, Welsh RM. Virus-induced transient immune suppression and the inhibition of T cell proliferation by type I interferon. J Virol. 2011;85:5929–5939. doi: 10.1128/JVI.02516-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Terawaki S, et al. IFN-α directly promotes programmed cell death-1 transcription and limits the duration of T cell-mediated immunity. J Immunol. 2011;186:2772–2779. doi: 10.4049/jimmunol.1003208. [DOI] [PubMed] [Google Scholar]
- 46.Xue F, et al. miR-31 regulates interleukin 2 and kinase suppressor of ras 2 during T cell activation. Genes Immun. 2013;14:127–131. doi: 10.1038/gene.2012.58. [DOI] [PubMed] [Google Scholar]
- 47.Crow MK. Type I interferon in the pathogenesis of lupus. J Immunol. 2014;192:5459–5468. doi: 10.4049/jimmunol.1002795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fan W, et al. Identification of microRNA-31 as a novel regulator contributing to impaired interleukin-2 production in T cells from patients with systemic lupus erythematosus. Arthritis Rheum. 2012;64:3715–3725. doi: 10.1002/art.34596. [DOI] [PubMed] [Google Scholar]
- 49.Welsh RM, Seedhom MO. Lymphocytic choriomeningitis virus (LCMV): propagation, quantitation, and storage. Current Protocols in Microbiology. 2008;Ch 15(Unit 15A):11. doi: 10.1002/9780471729259.mc15a01s8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stamm A, Valentine L, Potts R, Premenko-Lanier M. An intermediate dose of LCMV clone 13 causes prolonged morbidity that is maintained by CD4+ T cells. Virology. 2012;425:122–132. doi: 10.1016/j.virol.2012.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Laidlaw BJ, et al. Cooperativity between CD8+ T cells, non-neutralizing antibodies, and alveolar macrophages is important for heterosubtypic influenza virus immunity. PLoS Pathog. 2013;9:e1003207. doi: 10.1371/journal.ppat.1003207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Decman V, et al. Cell-intrinsic defects in the proliferative response of antiviral memory CD8 T cells in aged mice upon secondary infection. J Immunol. 2010;184:5151–5159. doi: 10.4049/jimmunol.0902063. [DOI] [PubMed] [Google Scholar]
- 53.Mueller SN, et al. Qualitatively different memory CD8+ T cells are generated after lymphocytic choriomeningitis virus and influenza virus infections. J Immunol. 2010;185:2182–2190. doi: 10.4049/jimmunol.1001142. [DOI] [PubMed] [Google Scholar]
- 54.Wherry EJ, Blattman JN, Murali-Krishna K, van der Most R, Ahmed R. Viral persistence alters CD8 T-cell immunodominance and tissue distribution and results in distinct stages of functional impairment. J Virol. 2003;77:4911–4927. doi: 10.1128/JVI.77.8.4911-4927.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Call MJ, et al. In vivo enhancement of peptide display by MHC class II molecules with small molecule catalysts of peptide exchange. J Immunol. 2009;182:6342–6352. doi: 10.4049/jimmunol.0803464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Martinez GJ, et al. The transcription factor NFAT promotes exhaustion of activated CD8+ T cells. Immunity. 2015;42:265–278. doi: 10.1016/j.immuni.2015.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kent WJ, et al. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rosenbloom KR, et al. ENCODE data in the UCSC Genome Browser: year 5 update. Nucleic Acids Res. 2013;41:D56–D63. doi: 10.1093/nar/gks1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Okamura H, et al. Concerted dephosphorylation of the transcription factor NFAT1 induces a conformational switch that regulates transcriptional activity. Mol Cell. 2000;6:539–550. doi: 10.1016/s1097-2765(00)00053-8. [DOI] [PubMed] [Google Scholar]
- 60.Lu J, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 61.Reich M, et al. GenePattern 2.0. Nat Genet. 2006;38:500–501. doi: 10.1038/ng0506-500. [DOI] [PubMed] [Google Scholar]
- 62.Schadt EE, Li C, Ellis B, Wong WH. Feature extraction and normalization algorithms for high-density oligonucleotide gene expression array data. J Cell Biochem Suppl. 2001;37:120–125. doi: 10.1002/jcb.10073. [DOI] [PubMed] [Google Scholar]
- 63.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Miranda KC, et al. A pattern-based method for the identification of MicroRNA binding sites and their corresponding heteroduplexes. Cell. 2006;126:1203–1217. doi: 10.1016/j.cell.2006.07.031. [DOI] [PubMed] [Google Scholar]
- 65.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Betel D, Koppal A, Agius P, Sander C, Leslie C. Comprehensive modeling of microRNA targets predicts functional non-conserved and non-canonical sites. Genome Biol. 2010;11:R90. doi: 10.1186/gb-2010-11-8-r90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yang JH, et al. starBase: a database for exploring microRNA-mRNA interaction maps from Argonaute CLIP-Seq and Degradome-Seq data. Nucleic Acids Res. 2011;39:D202–D209. doi: 10.1093/nar/gkq1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Krek A, et al. Combinatorial microRNA target predictions. Nat Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon request. Data have been deposited in the GEO database with the following accession codes: microarray, GSE63549; ChIP-seq (chromatin immunoprecipitation followed by deep sequencing), GSE90707; and RNA-seq, GSE98615.