Abstract
Orofacial clefts are one of the most common birth defects, affecting 1–2 per 1000 births, and have a complex etiology. High-resolution array-based comparative genomic hybridization has increased the ability to detect copy number variants (CNVs) that can be causative for complex diseases such as cleft lip and/or palate. Utilizing this technique on 97 nonsyndromic cleft lip and palate cases and 43 cases with cleft palate only, we identified a heterozygous deletion of Isthmin 1 in one affected case, as well as a deletion in a second case that removes putative 3′ regulatory information. Isthmin 1 is a strong candidate for clefting, as it is expressed in orofacial structures derived from the first branchial arch and is also in the same “synexpression group” as fibroblast growth factor 8 and sprouty RTK signaling antagonist 1a and 2, all of which have been associated with clefting. CNVs affecting Isthmin 1 are exceedingly rare in control populations, and Isthmin 1 scores as a likely haploinsufficiency locus. Confirming its role in craniofacial development, knockdown or clustered randomly interspaced short palindromic repeats/Cas9-generated mutation of isthmin 1 in Xenopus laevis resulted in mild to severe craniofacial dysmorphologies, with several individuals presenting with median clefts. Moreover, knockdown of isthmin 1 produced decreased expression of LIM homeobox 8, itself a gene associated with clefting, in regions of the face that pattern the maxilla. Our study demonstrates a successful pipeline from CNV identification of a candidate gene to functional validation in a vertebrate model system, and reveals Isthmin 1 as both a new human clefting locus as well as a key craniofacial patterning gene.
Keywords: branchial arches, cleft lip and palate, copy number variation, craniofacial development, Xenopus laevis
CLEFT lip and/or palate (CL/P) are common birth defects that cause significant morbidity and can impose a substantial financial burden resulting from surgical, nutritional, dental, speech, medical, and behavioral interventions (Wehby and Cassell 2010). CL/P can occur as part of a more complex chromosomal, Mendelian, or teratogenic syndrome, or can be an isolated finding [nonsyndromic (NS); as reviewed in Leslie and Marazita (2013)]. Recent work indicates that 60% of CL/P cases are NS (Genisca et al. 2009), and while some of the genetic and environmental triggers for syndromic CL/P have been identified, the explanations for these more common NS cases have remained more elusive.
Copy number variants (CNVs) are now considered to be common causes of disease (Glessner et al. 2009; Greenway et al. 2009; Mefford et al. 2009; Mefford and Eichler 2009; Rosenfeld et al. 2010, 2013; Bassuk et al. 2013), playing a prominent role in neurodevelopmental disorders (Girirajan et al. 2010; Marshall et al. 2017; Yuen et al. 2017), birth defects in general (Mefford et al. 2008; Lu et al. 2012; Bassuk et al. 2013), and CL/P in particular (Osoegawa et al. 2008; Shi et al. 2009; Younkin et al. 2014, 2015; Cao et al. 2016; Conte et al. 2016; Klamt et al. 2016; Cai et al. 2017). Previous studies investigating the role of CNVs in NSCL/P have identified two deletions (one overlapping MGAM on chromosome 7q34, and the second overlapping ADAM3A and ADAM5 on chromosome 8p11), which are overtransmitted in cleft vs. control trios (Younkin et al. 2015), and one region on 7p14.1 in which de novo deletions occur more frequently in probands with clefts than controls (Younkin et al. 2014; Klamt et al. 2016). In addition, deletions overlapping several genes previously implicated in CL/P, such as SATB2, MEIS2, SUMO1, TBX1, and TFAP2A, have been reported (Shi et al. 2009; Conte et al. 2016). However, systematic studies identifying rare, higher effect size CNVs followed by functional analysis in vertebrate model organisms have not been explored for NSCL/P.
Craniofacial development in vertebrates results from the coordinated growth and convergence of facial prominences, which respond to a complex, tightly regulated series of molecular signals [reviewed in Twigg and Wilkie (2015)]. In the early stages of development, a subset of neural crest cells arising near the midbrain–hindbrain boundary (MHB; also called the isthmus or isthmic organizer) migrate to populate the branchial arches (BAs), including the first BA, which forms the mandible and maxilla. The Isthmin 1 (ism1) gene was originally identified in an unbiased screen of secreted factors expressed in the Xenopus gastrula (called xIsm) (Pera et al. 2002). It is expressed prominently in the MHB, as well as in the nascent mesoderm, neural tube, and pharyngeal (BA) arches in the mouse, chick, and Xenopus embryos (Pera et al. 2002; Osorio et al. 2014). Fgf8 (human ortholog FGF8), spry1a (human ortholog SPRY1), and spry2 (human ortholog SPRY2) are coexpressed with ism1 in the isthmus, BAs, and ear vesicle in Xenopus (Christen and Slack 1997; Pera et al. 2002; Panagiotaki et al. 2010; Wang and Beck 2014), placing them in the same synexpression group. Notably, synexpression group members have been shown to function in the same biological process (Niehrs and Pollet 1999; Niehrs and Meinhardt 2002). Intriguingly, each of these genes, with the exception of ism1, has been previously implicated in clefting (Vieira et al. 2005; Goodnough et al. 2007; Riley et al. 2007; Welsh et al. 2007; Thomason et al. 2008; Fuchs et al. 2010; Goudy et al. 2010; Mangold et al. 2010; Yang et al. 2010; Green et al. 2015; Simioni et al. 2015; Conte et al. 2016). Conditional expression of Spry1 in the neural crest causes facial clefting and cleft palate in mice, and deletions of SPRY1 have been identified in three patients with cleft palate only (Yang et al. 2010; Conte et al. 2016). SPRY2 is found near the NSCL/P-associated genome-wide association study signal at 13q31.1 (Ludwig et al. 2012), and rare point mutations in this gene have been identified in individuals with NSCL/P (Vieira et al. 2005). Additionally, mice carrying a deletion of Spry2 have cleft palate, which has been shown to be complementary to (but independent of) Fgf8 signaling during craniofacial morphogenesis (Goodnough et al. 2007; Welsh et al. 2007). FGF8 itself (in addition to impaired FGF signaling in general) contributes to NSCL/P in humans (Riley et al. 2007; Simioni et al. 2015), and is altered in a Tp63-deficient mouse model of facial clefting (Thomason et al. 2008). The strong evidence for each of these synexpression group members in craniofacial morphogenesis and clefting implicate ISM1 as a plausible clefting candidate in humans.
Due to the high conservation of orofacial development between humans and Xenopus and the well-established use of Xenopus as a model organism, this vertebrate has become an effective model to study orofacial development and defects (Dickinson and Sive 2006; Dickinson 2016; Chen et al. 2017; Dubey and Saint-Jeannet 2017). Elegant bead implantation and transplantation studies in Xenopus from the Sive laboratory have helped characterize specific cellular mechanisms involved in craniofacial development, including the role of the Kinin–Kallikrein and Wnt/planar cell polarity pathways in mouth formation (Jacox et al. 2014, 2016). Moreover, a study using Xenopus to model the effect of depleted retinoic acid signaling (which leads to clefting in humans) revealed decreased expression of the homeobox genes lhx8 and msx2 (corresponding with a failure of dorsal anterior cartilage formation), resulting in a midline orofacial cleft (Kennedy and Dickinson 2012). Interestingly, FGF8b has been shown to mediate lhx8, msx1, and msx2 expression in the chick embryo through regulation of retinoic acid signaling, suggesting a potential mechanism behind the specification of first BA derivatives via signals derived from the isthmic organizer (Kennedy and Dickinson 2012; Shimomura et al. 2015).
Using high-resolution microarray-based Comparative Genomic Hybridization (aCGH) in a clefting cohort, we report here the identification of a rare heterozygous deletion that removes Isthmin 1 (ISM1), in addition to a deletion in a second unrelated case that potentially removes 3′ regulatory information. Additionally, sequencing of ISM1 in two CL/P cohorts identified a novel mutation in cases that is absent in control populations. ISM1 scores as a haploinsufficiency locus, and morpholino knockdown as well as clustered randomly interspaced short palindromic repeat (CRISPR)/Cas9 deletion in Xenopus laevis resulted in mild to severe craniofacial dysmorphologies including clefting phenotypes, demonstrating that ISM1 is critical in patterning craniofacial structures. Finally, knockdown of ism1 reduced the craniofacial expression of a known clefting locus, lhx8. Collectively, these data provide compelling evidence that ISM1 is a new craniofacial patterning locus.
Materials and Methods
Patient material
For the aCGH experiments, a total of 140 unrelated individuals with clefts [97 cleft lip and palate (CLP) and 43 cleft palate only (CPO)] were processed, with 130 individuals analyzed after passing bioinformatic quality controls. All cases were NS and seen during surgical screening as part of the Operation Smile medical missions in the Philippines (Murray et al. 1997). We used an additional 245 NSCL/P cases from the US (IA) and 275 NSCL/P cases from the Philippines for direct sequencing of the coding regions of the gene, as well as a set of 344 Filipino control samples. All participants included in this study were recruited following signed informed consent obtained in compliance with Institutional Review Board (IRB) No. 199804081 (Philippines) and IRB No. 199804080 (IA).
aCGH
aCGH was performed as recommended by the array manufacturer (Roche NimbleGen cgh_cnv_userguide_v7p0) on 140 unrelated Filipinos with clefts. Briefly, 1 μg of case DNA was labeled with Cy3-coupled nonamers and 1 μg of control DNA (from an unaffected Filipino male) was labeled with Cy5-coupled nonamers. Next, 34 μg of each labeled DNA was cohybridized to a Roche NimbleGen human whole-genome tiling microarray (Human CGH 2.1 M Whole-Genome Tiling v2.0D Array) and the array was washed, scanned, and analyzed.
CNV detection and quality control
BioDiscovery’s Nexus Copy Number FASST2 Segmentation Algorithm, a Hidden Markov Model-based approach, was used to make initial copy number calls. The significance threshold for segmentation was set at 1.0E−6, also requiring a minimum of three probes per segment and a maximum probe spacing of 1000 between adjacent probes before breaking a segment. The log2 ratio thresholds for single-copy gain and single-copy loss were set at 0.3 and −0.3, respectively. A 3:1 sex chromosome gain threshold was set to 1.2 and a 4:1 sex chromosome gain threshold was set to 1.7. NimbleGen’s DEVA segMNT algorithm, which minimizes squared error relative to the segment means, was used as a second algorithm for all copy number calls after data extraction, LOESS spatial correction, and background correction. Default parameters were used, apart from setting the minimum segment difference to 0.3 and requiring a minimum of three probes per segment, to more similarly call CNVs when compared to Nexus. X-shift values in females (hybed against a male control) were adjusted by the median X-shift across all females in the cohort. Finally, CNV calls from Nexus and DEVA were compared using the BEDTOOLS intersect function. Only calls with a 50% reciprocal overlap were retained for further analysis. Microarray data were quality controlled based on several data metrics generated by DEVA and Nexus. DEVA was used to calculate experimental metrics that include interquartile density, ratio range, signal range, mean empty, mean experimental, and mean random [for a detailed discussion of these metrics see Brophy et al. (2013)]. Nexus also calculated a surrogate measure of noise (quality score). Lastly, following the calculation of the high-quality CNV regions (CNVRs), the number of duplications and deletions (and total CNVRs) was used as a quality control metric. The mean and SD were calculated for each metric across all arrays. Any array that had six or more metrics falling outside of two SD of the mean was excluded from further analysis.
Filtering Nexus calls to identify rare CNVs
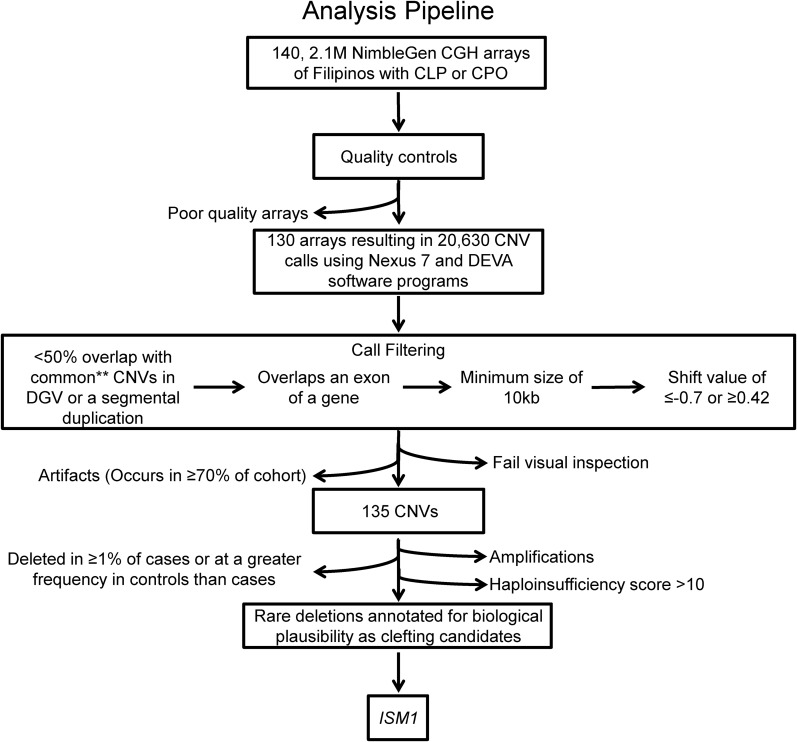
We identified a total of 20,630 CNVs called by both Nexus and DEVA from the 130 arrays passing quality controls (see CNV detection and quality control). We then compared the CNV calls to an in-house curated list of CNVs from the Database of Genomic Variants (DGV). We included all studies containing ≥ 100 individuals within the DGV that used CGH or SNP arrays. Any CNVs occurring at a frequency of < 1% within each study were removed from the list of likely benign variants, as these are defined as rare occurrences and are more likely to contribute to a disease phenotype. We then filtered our CNV calls to include only CNVs that (1) had ≤ 50% overlap with these likely benign variants or a segmental duplication, (2) overlapped an exon of a gene, (3) spanned ≥ 10 kb, and (4) had log2 median shift values of ≤ −0.7 or ≥ 0.42 for deletions and amplifications, respectively. All calls were visually inspected and false positives were removed. We also excluded one gene (IQCA1) that was overlapped by a CNV in 75% of the samples, since these calls were likely due to a control-specific CNV or a Filipino-specific polymorphism. Using these criteria, we identified 51 deletions and 83 amplifications (Supplemental Material, Table S1 in File S1). For our functional analysis, we focused on deletions overlapping protein-coding genes occurring at a frequency of < 1%; these CNVs overlapped 42 genes (Figure 1 and Table S2 in File S1).
Figure 1.
Bioinformatic analysis pipeline leading to the identification of ISM1 as a clefting candidate. CNVs from 130 high-quality CGH arrays were called by Nexus 7 Software and compared to calls made by DEVA software using the BEDTOOLS 50% reciprocal overlap function. Calls were retained if they were called by both programs, did not overlap in a list of curated benign variants (**) from DGV or segmental duplications ≥ 50%, overlapped an exon of a protein-coding gene, spanned 10 kb, had a shift value of −0.7 or 0.42, were overlapped by a deletion in < 1% of cases, were likely haploinsufficiency loci, and had not been previously implicated in clefting. These deletions were assessed in the Mouse Genome Informatics, National Center for Biotechnology Information, and PubMed databases for biological plausibility, and one gene (ISM1) was selected for functional follow-up. All calls were visually inspected and artifacts were removed. CGH, comparative genomic hybridization; CLP, cleft lip and palate; CNV, copy number variant; CPO, cleft palate only; DGV, Database of Genomic Variants.
ISM1 deletion breakpoints
Upon identification of deletions in or near ISM1 by aCGH, we performed a validation by independent methods using long-range PCR (LRPCR) followed by direct sequencing of the PCR product. We used flanking primers [designed using Primer3; (Rozen and Skaletsky 2000)] ranging from 500 bp to 3 kb on each side of the potential breakpoints, as given by the aCGH analysis by Nexus Copy Number. LRPCR was performed using Takara LA Taq following the manufacturer’s protocol. The PCR products were analyzed on a 1.5% agarose gel and sent to Functional Biosciences (http://functionalbio.com/web/index.php) for direct sequencing to identify the exact coordinates of the breakpoints (chromosome 20:12,737,592–13,341,144 for the coding deletion and chromosome 20:13,281,543–13,293,591 for the 3′ deletion; based on the hg19 genome build).
Direct sequencing of ISM1 in cases and controls
Primers were designed using Primer3 (Rozen and Skaletsky 2000) for the six coding exons of ISM1. PCR conditions and primers are available upon request. Sequencing reactions were performed by Functional Biosciences (http://functionalbio.com/web/index.php). Polyphen 2, SIFT, MutationTaster, MutationAssessor, FATHMM Prediction, and FATHMM-MKL Coding Prediction within dbNSFP (Liu et al. 2016) were used to predict whether the variants detected were deleterious, and phastCons and phyloP LRT scores were obtained from the University of California, Santa Cruz Genome Browser.
Sequencing of clefting loci in two cases
All exons of ISM1, FGF8, and SPRY2 were sequenced in the two cases harboring CNVs overlapping or near ISM1, as per above, with the following modification: sequencing reactions were performed by the Carver Center for Genomics in the Department of Biology, University of Iowa. Primers are available upon request.
Xenopus embryos
Adult X. laevis females were induced by injecting human chorionic gonadotropin. Eggs were collected in high-salt 1.2× Marc's Modified Ringer's (MMR) rinsed with 0.3× MMR [10× MMR: 1 M NaCl, 18 mM KCl, 20 mM CaCl2, 10 mM MgCl2, and 150 mM HEPES (pH 7.6)], drained, and then fertilized using a 1.5 ml sperm suspension in 0.3× MMR for 10 min before washing in 0.1× MMR. After 1 hr, the embryos were dejellied in 2% cysteine in 0.1× MMR (pH 7.9) for 4 min before washing the embryos with 0.1× MMR. Embryos were reared in 0.1× MMR to the desired developmental stage.
Whole-mount in situ hybridization
Full-length ism1.L cDNA in pCMV-SPORT6 was obtained commercially (TransOMIC Technologies). Antisense probes were synthesized from the plasmid and diluted to 1 μg/ml in hybridization buffer before use. Whole-mount in situ hybridization was performed as previously described (Hulstrand and Houston 2013). To assess the reduction in lhx8 levels upon ism1 knockdown, we compared lhx8 maxillary expression levels on both sides of the embryos, with one side injected with ism1 morpholino (6 or 12 ng) and the other side subjected to a standard control needle prick. We used ImageJ software to assess the mean intensities of signal for both sides, and then divided the ism1 knockdown value by the control value to arrive at a fold change. A minimum of six embryos were used for each group.
mRNA synthesis
The coding region of the full-length ism1.L cDNA (accession number BC160753; GE/Dharmacon) was amplified by PCR and cloned into pCR8/GW/TOPO (Invitrogen, Carlsbad, CA). Clones were sequenced and inserted into a custom pCS2-HA/GW vector using LR recombination (Invitrogen). Template RNA was linearized using NotI for sense transcription and capped mRNAs were synthesized using the SP6 mMESSAGE mMACHINE kit (Ambion).
Embryo microinjections
Fertilized embryos were injected essentially as previously described (Hulstrand and Houston 2013). Embryos were transferred into 0.5× MMR containing 2% Ficoll400 (GE Bioscience) and injected with morpholino oligonucleotides (MOs) or mRNAs into the animal hemisphere at the two- to four-cell stages. Antisense oligos complimentary to the ism1 5′-UTR and translation start site to block translation (5′-GCCAGTCGCAACATCCTCTTGATGC-3′), or complimentary to the exon 1/intron 1 spice junction to disrupt the splicing of ism1 (5′-TGTATGTGGAATGGACTAACCTGTA-3′), were synthesized (Gene Tools). Capped mRNAs were injected at the two-cell stage followed by either MO at the four-cell stage for rescue experiments. The standard control oligo (Gene Tools) was used as a negative control for all injection experiments (5′-CCTCCTTACCTCAGTTACAATTTATA-3′).
CRISPR/Cas9 mutagenesis
Mutations in ism1 were generated in F0 embryos using a CRISPR/Cas9 injection strategy. Guide RNAs were designed against exon 1 of X. laevis ism1.L [5′-GCTGGAGTTGGAGGAGCTAT(CGG)-3′; the protospacer adjacent motif (PAM) site is in parentheses]. Ism1.L is the only homeolog remaining following speciation through allopolyploidization (Session et al. 2016) and, thus, Xenopus is functionally diploid for this gene. Genome editing was performed using Alt-R reagents from IDT (Coralville, IA). Custom CRISPR RNAs (crRNAs) were synthesized and annealed with a common trans-acting crRNA (tracrRNA), incubated with an equimolar amount of Cas9 protein (IDT, 1.5–3 µM each final concentration) at 37° for 10 min. Fertilized eggs were injected with ∼6 nl of this mix at the one-cell stage (300 pg RNA/1.5 ng Cas9). The presence of mutations was verified by PCR amplification of exon 1 DNA, followed by T7 endonuclease assays or cloning and sequencing of PCR products (Figure S1 in File S1).
Immunostaining
Embryos for immunostaining were fixed in MOPS, EGTA, MgSO4 and formaldehyde (MEMFA) and washed into 1× PBS as previously described (Hulstrand and Houston 2013). Embryos were washed in PBS-Tween (PBT) (1× PBS, 0.2% BSA, and 0.5% Triton X-100) and blocked for 4 hr at room temperature in PBT + 2.5% BSA. Samples were washed in PBT again and incubated in anti-E-cadherin mAb 8C2 (1:5 dilution; Developmental Studies Hybridoma Bank) overnight at 4°. Embryos were washed for 1 hr in PBT five times. Alexa 488-conjugated goat anti-mouse IgG secondary antibodies and Alexa 568-Phalloidin were diluted 1:500 in 1× PBT and incubated overnight at 4°. Embryos were washed five times for 1 hr in PBT followed by 1 hr in 1× PBS before imaging on an SP2 confocal microscope.
Data availability
Plasmids are available upon request. Raw CNV array .tiff, Nexus data summary .txt files, and DEVA segMNT .txt files are publicly available at the Gene Expression Omnibus (GSE100845).
Results
aCGH identifies deletions in the ISM1 genomic interval
We analyzed aCGH data from 130 NS CLP and CPO Filipino cases that passed quality controls (see Materials and Methods) to identify potentially disease-associated rare CNVs. Data analysis was performed using Nexus Copy Number (version 7.5; BioDiscovery, Hawthorne, CA), DEVA (version 1.2; Roche NimbleGen, Madison, WI), and several in-house custom Python scripts, as described previously [see Materials and Methods and Brophy et al. (2013)]. Since we wished to identify key genes involved in craniofacial patterning that could be validated in vertebrate model organisms, we focused on variants that were represented in < 1% of the cohort and that were exceedingly rare or absent in the control population, since disease-causing genomic variants of high effect size are known to be rare in the population (Kaiser 2012). Overall, 51 coding deletions and 83 coding amplifications (Table S1 in File S1), overlapping at least one exon of 200 genes (Table S2 in File S1), were identified when compared to an in-house curated list of benign variants occurring at a frequency of ≥ 1% in the DGV (see Materials and Methods). One individual harbored a 2.4 Mb duplication of 22q11.21, which is a commonly reported pathogenic variant in 22q11.2 duplication syndrome (Ensenauer et al. 2003) and presents as a spectrum of phenotypic abnormalities with or without CL/P. We revisited this individual’s clinical file and were unable to rule out the possibility of a 22q11.2 duplication syndrome vs. a NS cleft due to insufficient phenotyping. Aside from the 22q11.2 duplication locus, four deletions (IMMP2L, PTPRD, CDH1, and NOSIP) and five amplifications (IFIT2, SLC46A1, RFC1, TULP4, and DMD) overlapped genes previously associated with clefting (Tables S1 and S2 in File S1), providing evidence that our pipeline was effective in identifying clefting loci.
Next, we elected to follow up novel clefting candidates that were overlapped by a deletion in < 1% of the case cohort (since deletions are likely more deleterious than amplifications), had a haploinsufficiency score ≤ 10 (predicted as likely haploinsufficent in DECIPHER; https://decipher.sanger.ac.uk/), and were overlapped by CNVs at a higher frequency in cases vs. controls (Figure 1). Nine genes passed all filters and fit these criteria, so we assessed the biological plausibility of each gene’s involvement in clefting by searching the Mouse Genome Informatics (http://www.informatics.jax.org/) database for expression in the mouth, palate, and face, and the National Center for Biotechnology Information and PubMed databases for known biological function. Four of the genes, Cadherin 1 (CDH1), Protein inhibitor of activated STAT 2 (PIAS2), UDP-glucose pyrophosphorylase 2 (UGP2), and Isthmin 1 (ISM1) fulfilled these criteria (Table S2 in File S1). Variants in CDH1 have been previously implicated in CL/P in individuals with hereditary diffuse gastric cancer (Frebourg et al. 2006), and therefore CDH1 was not considered for further functional validation. Although PIAS2 was sequenced for damaging variants in 192 Europeans with NSCLP along with other genes encoding Small ubiquitin-like modifier proteins, no such variants were identified (Carta et al. 2012), and thus additional support for its involvement for clefting is needed. UGP2 has been identified as a biomarker for gallbladder cancer and metastatic hepatocellular carcinoma (Tan et al. 2014; Wang et al. 2016), and thus was not a compelling clefting candidate. The remaining locus, ISM1, was particularly intriguing since this gene is in the same synexpression group as FGF8, SPRY1, and SPRY2, which themselves are associated with orofacial clefting (see the Introduction and Discussion), is expressed in the oral mucosa in humans (Valle-Rios et al. 2014), and interacts with αVβ5 integrins, a family of integrins that causes cleft palate in mice when mutated (Aluwihare et al. 2009). Thus, we selected ISM1 for functional follow-up.
ISM1 is a haploinsufficiency locus
Large sequencing and microarray studies have identified numerous highly constrained or haploinsufficient regions of the genome (Petrovski et al. 2013; Zarrei et al. 2015; Ruderfer et al. 2016), and only four deletions overlapping the coding region of ISM1 have been reported in control populations within the DGV (http://dgv.tcag.ca). Notably, of these four, only one deletion CNV event is from a study containing > 40 individuals, and in that study the deletion encompassed the first exon of ISM1 in one patient out of 873 (Uddin et al. 2015). This is significant given that, as of mid-2017, the DGV had identified over six million sample-level CNVs from control populations of over 70 studies. Data from the DECIPHER database and the ClinGen resource (https://decipher.sanger.ac.uk/; https://www.clinicalgenome.org/) further support the likelihood that ISM1 is intolerant to copy number reductions. Specifically, there are only three entries within the ClinGen resource of deletion CNVs spanning ISM1, and two of those are in excess of 6 Mb and encompass numerous other genes. The one deletion CNV event in ClinGen that is under this threshold (∼570 kb) was labeled a variant of uncertain significance and did not contain detailed phenotypic information (nssv584541). In DECIPHER, there are five deletion CNV events spanning ISM1, three of which are > 3 Mb in size and two that are in the range of 500–600 kb. The latter two were both found to be inherited, and in patients with either no phenotypic data provided or minimal data without a mention of clefting. Finally, there are only two deletions involving ISM1 noted in the CNV calls from the ExAC database (data from > 60,000 individuals; http://exac.broadinstitute.org/), and the gene itself is predicted to be haploinsufficient (percentage haploinsufficiency of 8.20 as reported by DECIPHER) (Huang et al. 2010). Taken together, these data suggest ISM1 is likely a haploinsufficiency locus.
A second ISM1 deletion removes putative 3′ regulatory sequences
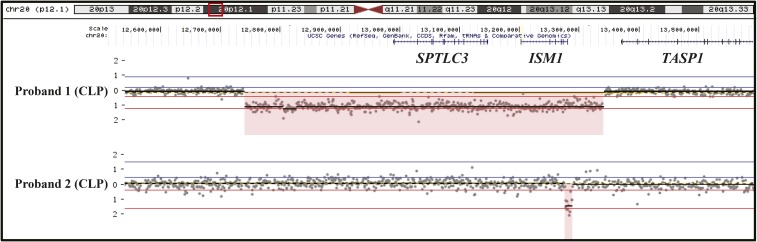
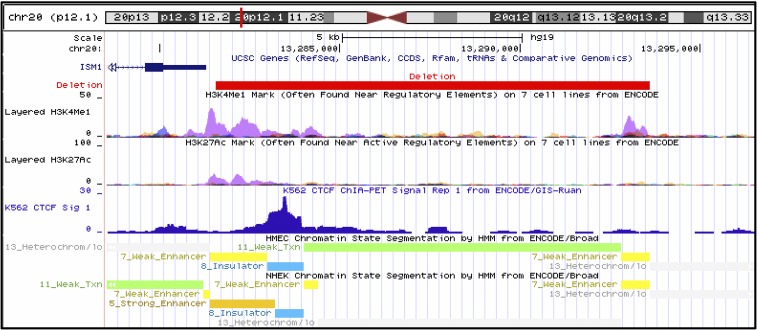
Using either aCGH or LRPCR, we assessed whether any family members also harbored the ISM1 coding deletion at chromosome 20:12,737,592–13,341,144 (hg19), which removed ISM1 and SPTLC3. We found that the deletion was paternally inherited in two brothers (with both the father and brother of the proband being unaffected) (Figure S2 in File S1). Interestingly, while assessing all coding deletions and amplifications passing our pipeline for nearby noncoding variants, we detected one deletion immediately 3′ of ISM1 [chromosome 20: 13,281,543–13,293,591 (hg19)] overlapping a chromatin immunoprecipitation sequencing-validated CTCF-binding site and other putative ISM1 regulatory sequences (Figure 2 and Figure 3), although we are uncertain of the effect that this deletion would have on ISM1 expression. The 3′ deletion of ISM1 was inherited maternally from a mother with cleft lip only (Figure S2 in File S1). This deletion was confirmed in both the unaffected maternal grandfather and two unaffected maternal uncles. We also inspected the ISM1 genomic region for the presence of CNVs for samples that did not pass quality controls (all of which were of sufficient quality for CNV calling within this interval) and confirmed that only two out of 140 probands harbored deletions in or near ISM1. These data are consistent with incomplete penetrance of the phenotype.
Figure 2.
Identification of heterozygous deletions in two unrelated probands with nonsyndromic cleft lip and palate. The deletion detected in proband 1 removes the entirety of ISM1 and SPTLC3, while the deletion in proband 2 occurs just 3′ of ISM1. Plots of aCGH data were generated by Nexus 7 software. aCGH, comparative genomic hybridization; CLP, cleft lip and palate.
Figure 3.
Deletion 3′ to ISM1 deletes regulatory information. CTCF binding sites (K562 blood cell line, dark blue) indicative of an insulator element (HMEC and NHEK epithelial cell lines, teal) are deleted by the 3′ ISM1 deletion (red). In addition, the deletion removes chromatin marks H3K4Me1 and H3K27Ac (indicative of enhancers, both shown in purple), which were also detected within the epithelial cell lines (orange, strong enhancer; yellow, weak enhancer; gray, heterochromatin; green, weak transcription; K562, human erythroleukemia cell line; HMEC, human mammary epithelial cells; NHEK, normal human epidermal keratinocytes).
ISM1 direct sequencing in NSCL/P
The rare coding deletion identified in the ISM1 genomic interval prompted us to look for single-nucleotide variants in patients with NSCL/P. We sequenced each of its six exons in a total of 520 cases [245 individuals of European descent from the US (IA) and 275 Filipinos with NSCL/P] in addition to 344 control subjects from the Philippines. The NHLBI Exome Sequencing Project Variant Server (Exome Variant Server 2017) and the 1000 Genomes Project (1000 Genomes Project Consortium et al. 2012) were used as control references for the cases of European descent. We identified nine missense variants [several of which had never been reported (Table S3 in File S1)]. The most promising variant (N188S) did not appear in any of the 8400 control alleles (while occurring in 1 out of 456 case alleles), is absent from dbSNP, EVS, ExAC and 1000 Genomes, and is predicted to be probably damaging by Polyphen, damaging by MutationTaster and FATHMM MKL, but tolerated by SIFT, FATHMM, and MutationAssessor. However, we note that greater power is needed to convincingly assess the pathogenicity of the variants detected in NSCL/P cases.
Sequencing of clefting loci in ISM1 cases
To assess whether damaging genomic variants in the remaining copies of ISM1 (from the two cases harboring deletions overlapping or near ISM1) might be contributing to the clefting phenotype, we sequenced all exons of ISM1 in both cases and were unable to find any missense variants. We also decided to sequence all exons of FGF8 and SPRY2 (the synexpression group members having the strongest connections to human clefting) to determine whether missense variants in these genes might be contributing to the phenotype. Only one variant was identified (rs504122), a common variant in SPRY2 in the case harboring the deletion overlapping ISM1.
Characterization of ism1 expression during X. laevis craniofacial development
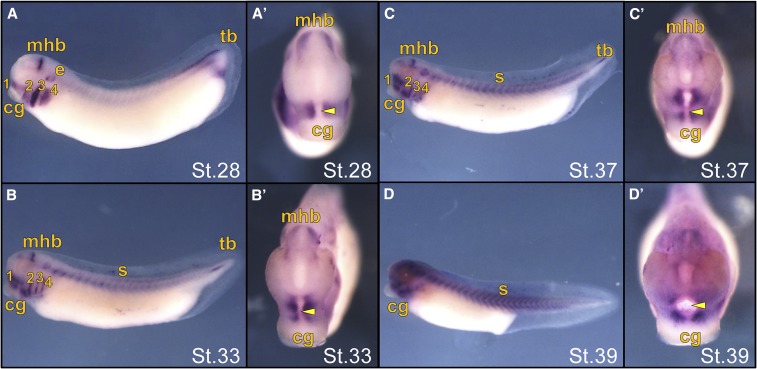
Previous work in X. laevis identified concentrated ism1 expression in the BAs and MHB at stage 30 (Pera et al. 2002). Since craniofacial precursors derive from the first BA, we performed in situ analysis of X. laevis embryos at tailbud and tadpole stages to determine if ism1 is expressed in the developing face (Figure 4). We found that ism1 was strongly expressed in the MHB, BAs, and ear placode (stages 28 and 33; Figure 4, A and B), with decreasing expression in the MHB and BAs (stages 37–39; Figure 4, C and D), but increasing and concentrated expression around the primary mouth in the region of the developing mandible and maxilla, which correlates with growth of the maxillary processes toward the midline (stages 28–39; Figure 4, A’–D’). These results demonstrate that ism1 is expressed in areas critical for craniofacial development in X. laevis embryos.
Figure 4.
In situ hybridization of ism1 expression in late stage (St.) X. laevis embryos. Embryos are shown in a lateral (A–D) or anterior (A’–D’) view with the cement gland (cg) labeled for ventral orientation. (A) St.28 embryo showing strong expression in the branchial arches (ba), midbrain–hindbrain boundary (mhb), ear placode (e), and tailbud (tb), with the yellow arrowheads marking the primitive mouth. (B) St.33 embryos showing decreased expression in the ba, mhb, and tb, with concentrated expression surrounding the primitive mouth (arrowhead) and expression in the somites (s). (C and D) St.37 and 39 embryos showing continued concentrated ism1 expression surrounding the primitive mouth (arrowhead).
Decreased expression of ism1 in X. laevis results in craniofacial dysmorphologies
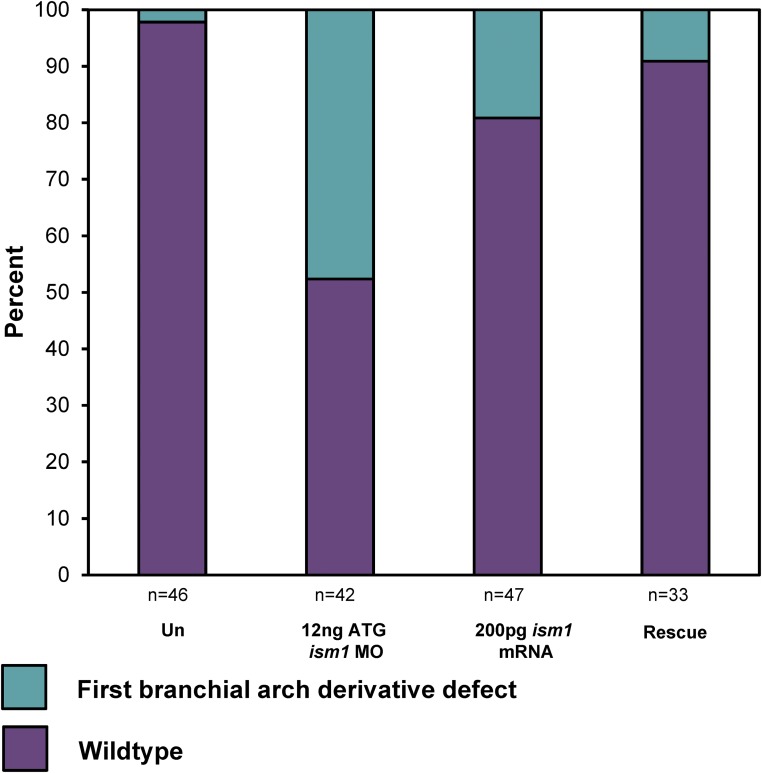
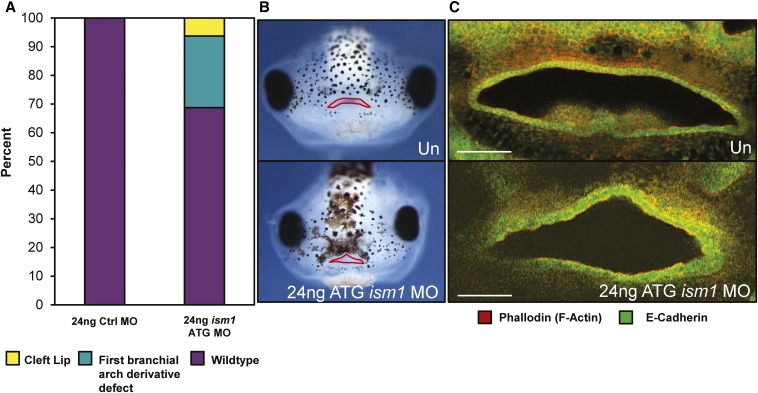
To determine if knockdown of ism1 expression resulted in craniofacial defects in X. laevis embryos, we injected wild-type embryos with MOs targeting the translation start site (ism1 ATG MO) or the exon 1–intron 1 splice junction (ism1 e1i1 MO). Injecting the embryos at a four-cell stage with 12 ng of the ism1 ATG MO (n = 42) resulted in a spectrum of phenotypic abnormalities including a shortened axis (n = 10; 23.8%), an upturned tail (n = 3; 4.2%), a curved tail (n = 5; 6.4%), an abnormal gut (n = 20; 47.6%), loss of one or both eyes (n = 1; 2.4%), and mouth defects (n = 20; 47.6%). These phenotypes were recapitulated with the injection of the ism1 e1i1 MO. Injection of embryos with a control MO failed to produce similar dysmorphologies (n = 21), providing evidence that the phenotypes were due to decreased Ism1 activity and not morpholino toxicity. In addition, injection of HA-tagged ism1 mRNA lacking the MO binding site, and thus impervious to the morpholino, rescued the spectrum of phenotypes in the majority of embryos, including the craniofacial abnormalities (Figure 5). An increased dose of 24 ng ism1 ATG MO (n = 49) resulted in similar yet more penetrant whole-embryo (n = 36; 73.5%), melanocyte localization (n = 39; 79.6%), and craniofacial (n = 38; 77.5%) abnormalities, while an even higher dose of 48 ng (n = 26) resulted in 100% penetrance of the whole-embryo abnormalities in addition to a low percentage with no heads altogether (n = 9; 34.6%). Intriguingly, we observed a cleft-like mouth at a low penetrance in the embryos injected with 24 ng ism1 ATG MO (n = 5; 10.2%), which was not present in any embryos injected with the control MO at the same dose (Figure 6, A and B). Whole-mount antibody staining against F-actin (phalloidin) and E-cadherin were used to further visualize these midline mouth defects (Figure 6C), which have previously been reported as cleft-like phenotypes in Xenopus embryos disrupted for retinoic acid signaling (Kennedy and Dickinson 2012).
Figure 5.
Knockdown of ism1 results in whole-embryo and craniofacial defects, which are rescued by ism1 mRNA. Whole-embryo knockdown of ism1 with 12 ng translation blocking (ATG) morpholino (MO) results in first branchial arch-derivative defects and is rescued with the injection of 200 pg ism1 mRNA lacking the MO binding site (P ≤ 0.001, Fisher’s Exact Test).
Figure 6.
Morpholino knockdown of ism1 in X. laevis embryos results in clefting phenotypes. (A) Quantification of embryos injected with 24 ng control morpholino (Ctrl MO) vs. 24 ng ism1 translation-blocking (ATG) MO shows craniofacial anomalies and clefting phenotypes in the ism1-altered group. (B) Faces of stage 43 embryos which are uninjected (Un, top) or injected with 24 ng ism1 ATG MO and exhibiting a cleft (bottom). Mouths have been outlined in red. (C) Phalloidin (red) and E-cadherin (green) staining to detect cell boundaries and epithelial cells, respectively, of the mouth of Un (top) or 24 ng ism1 ATG MO-injected (bottom) embryos.
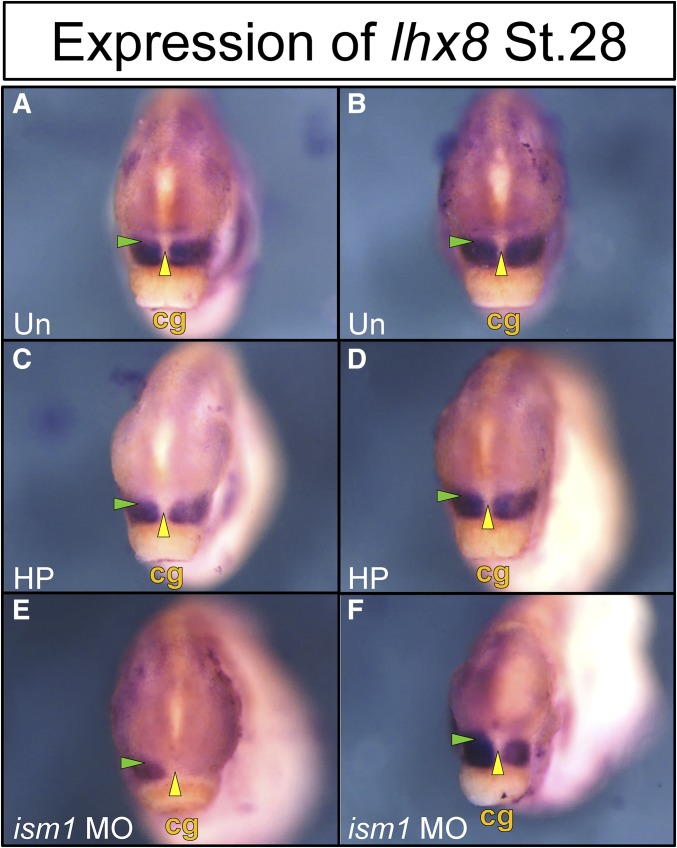
Decreased expression of lhx8 and msx2 in the maxillary and nasal prominences has been reported in retinoic acid-deficient embryos (Kennedy and Dickinson 2012). Since human LHX8 was previously shown to be associated with human clefting (Yildirim et al. 2014), we assessed lhx8 expression in ism1 knockdown stage 30 embryos (Figure 7). Due to the fact that lhx8 is expressed at too low an overall level to be detected using our current techniques [and that we would need to significantly increase our number of injected embryos (Kennedy and Dickinson 2012)], we instead elected to quantify the relative differences in the lhx8 in situ signals in half-embryo knockdowns. Comparison of the ism1 MO knockdown side injected with 6 ng ATG MO to the control needle-prick side resulted in significantly reduced lhx8 expression in the developing maxilla (mean of 1.4-fold reduction; Figure 7, E and F), whereby 12 ng ATG MO resulted in an even more significant reduction (average of 2.1-fold reduction, with some embryos showing almost complete absence of lhx8 in the knockdown half). Collectively, these data suggest that Ism1 may either regulate specific subsets of BA signaling networks or affect cellular processes such as cell migration.
Figure 7.
Expression of lhx8 decreases with knockdown of ism1. Embryos are shown in an anterior view with the cement gland (cg) labeled for ventral orientation. Control uninjected (Un) embryos or embryos pricked with the injection needle on half (HP) show strong lhx8 expression in the first branchial arch at stage 28 (A–D) surrounding the primitive mouth (yellow arrowhead), as well as in the maxillary prominences (green arrowhead). Knockdown of ism1 with 12 ng ATG morpholino (MO) in half of the animal results in undetectable (E) or decreased (F) branchial arch expression of lhx8, especially in the maxillary prominence.
To further confirm a role for ism1 in craniofacial morphogenesis, we generated intragenic deletions of ism1 in Xenopus using CRISPR/Cas9 in F0 embryos, which were confirmed by PCR of dysmorphic embryos followed by Sanger sequencing (Figure S1 in File S1). Notably, all dysmorphic animals harboring ism1 deletions recapitulated the phenotypic spectrum of the ism1 MO-depleted embryos including a short axis, curved tail, abnormal gut, aberrant eye development, and abnormal melanocyte localization (data not shown), as well as the craniofacial phenotypes, including small or absent mouths (Figure S3 in File S1). The observation of the same range of phenotypes with injection of ATG MO, e1i1 MO, or CRISPR/Cas9 mutation of ism1, in addition to the successful rescue of the MO knockdown phenotypes with ism1 mRNA, confirms that our results are specifically due to the knockdown of ism1 and not off-target effects.
Discussion
Our study describes a successful pipeline from CNV-based disease gene discovery to functional characterization in a vertebrate model system, resulting in the identification of ISM1 as a new clefting and craniofacial patterning locus. Importantly, we specifically sought to identify copy number losses that would more likely be of higher effect size to identify craniofacial genes playing key roles in facial patterning. Thus, we focused on deletions that were present in < 1% of the cohort, were extremely rare or absent in control populations, and altered the coding sequence of genes having high haploinsufficiency scores. This analysis strategy revealed a deletion of ISM1 in a NSCLP case. Intriguingly, we also identified a deletion 3′ of ISM1 that removes a conserved CTCF site just downstream of its polyadenylation site as well as additional sequences of high regulatory potential, although we cannot assess the effect of the 3′ regulatory deletion on ISM1 expression in the developing face and, thus, are uncertain of its pathogenicity. However, it is important to note that no other noncoding CNVs were identified near our other final clefting candidates. Additionally, Sanger sequencing of ISM1 in a collection of cases with NSCLP from two cohorts relative to controls detected several missense variants, one of which appears to be promising due to its absence in control populations (N188S).
Our functional studies in X. laevis revealed that depletion of ism1 results in severe perturbation of craniofacial morphogenesis in animals presumed to have an increased percentage of mutant cells in the developing face, in addition to causing reduced expression of a known clefting locus, lhx8 (Zhao et al. 1999; Yildirim et al. 2014). Additional lines of evidence further strengthen the connection of ISM1 orthologs to craniofacial, and specifically lip/palate, development. First, the MHB (also known as the isthmic organizer) is a signaling hub that regulates the expression of other signaling molecules in the region, including secreted proteins such as Wnts (Wnt1 and Wnt8b) (Joyner et al. 2000; Rhinn and Brand 2001; Wurst and Bally-Cuif 2001; Raible and Brand 2004), fibroblast growth factor family members (FGF8, FGF17, and FGF18) (McMahon et al. 1992; Meyers et al. 1998; Reifers et al. 1998; Picker et al. 1999; Belting et al. 2001; Rhinn and Brand 2001; Burgess et al. 2002; Reim and Brand 2002; Chi et al. 2003; Jaszai et al. 2003), Spry family members (SPRY1, SPRY2, and SPRY4) (Panagiotaki et al. 2010; Wang and Beck 2014), and Isthmin 1 (Pera et al. 2002). Signaling from the MHB regulates the expression of genes encoding transcription factors including Hox paralogs, known to play a key role in neural crest and BA patterning, and disruptions in their expression patterns lead to craniofacial dysmorphologies including clefting in X. laevis (Irving and Mason 2000; Trainor et al. 2002; Kennedy and Dickinson 2012). Thus, signals from the isthmus could help control neural crest cell migration into the BAs and drive the specification (or even migration) of primary BA derivatives, which are required for craniofacial development and palate closure (Trainor et al. 2002).
ISM1 is a secreted 60-kDa protein, composed of both a thrombospondin type 1 repeat domain in the central region and an “adhesion-associated domain in MUC4 and other proteins” (AMOP) domain at the C-terminus (Pera et al. 2002). The AMOP domain contains an “RKD” motif that is involved in integrin-dependent cell adhesions (Schwartz et al. 1995; Maubant et al. 2006; Zhang et al. 2011), and thrombospondins can mediate cellular attachment (Kosfeld and Frazier 1993). Unlike the traditional RGD motif in the AMOP domain, the RKD motif of ISM1 has been demonstrated to selectively bind the extracellular surface of αVβ5 integrins, which are involved in vascular permeability and cell migration. Intriguingly, mice that lack two αVβ integrins display cleft palate due to lack of fusion of the palatal shelf (Aluwihare et al. 2009; Zhang et al. 2011; Venugopal et al. 2015), suggesting that ISM1 could play a role in promoting or facilitating cell migration through integrin regulation, either from the MHB to the BAs or from the BAs into the developing face. It is important to point out that ISM1 is a secreted protein, and that CCN1/Cyr61 has been identified as a secreted factor that can induce cell migration through interaction with the αVβ3 class of integrins (Maity et al. 2014), a class which is closely related to αVβ5 (Xiang et al. 2011). Alternatively, ISM1 may be playing a role in the proliferation and/or survival of cranial neural crest cells and/or BA-derived structures. Live imaging of migrating neural crest cells into the arches upon Ism1 knockdown will help resolve this issue.
ism1 is tightly expressed in a nearly identical pattern to known clefting genes such as fgf8 (Riley et al. 2007; Simioni et al. 2015), as well as spry1 (Yang et al. 2010; Conte et al. 2016) and spry2 (Vieira et al. 2005; Ludwig et al. 2012) [both of which cause clefting when altered in mouse models (Goodnough et al. 2007; Yang et al. 2010)]. These genes are thus part of a synexpression group, and genes expressed in highly similar patterns have been shown to work in the same biological process (Niehrs and Pollet 1999; Niehrs and Meinhardt 2002). Importantly, deletion of Fgf8 within the first BA results in incomplete facial development in mice due to partial failure of neural crest cell survival (Trumpp et al. 1999; Tucker et al. 1999), whereas hypomorphic alleles lead to abnormal craniofacial development, including hypoplastic pharyngeal arches (Abu-Issa et al. 2002). These observations, along with the likelihood that ism1 is a haploinsufficiency locus, strongly implicate ism1 as a key molecule involved in craniofacial development, and this was confirmed with the functional validation studies in frogs, which showed that loss of ism1 can produce dysmorphic faces including a clefting phenotype. Future functional studies, including analysis of fibroblast growth factor and retinoic acid signaling markers, as well as identification of ISM1 action, will help reveal the role of ISM1 in this complex process.
In our study, the case harboring a heterozygous deletion which removes ISM1 presented with CL/P craniofacial defects (with other family members carrying the deletion exhibiting incomplete penetrance), possibly due to the strong haploinsufficiency score of ISM1. However, we have also presented compelling evidence that ISM1 plays a broad and prominent role in craniofacial development. These data are likely reconciled by the fact that the remaining ISM1 allele in unaffected family members carrying the deletion resulted in sufficient ISM1 expression to prevent a clefting phenotype, whereas manipulations using morpholinos and CRISPR/Cas9 in frogs led to a more drastic reduction in Ism1, with little or no wild-type Ism1 function to compensate, thus resulting in additional phenotypic abnormalities. It is also possible that hypomorphic variants in additional clefting loci might be contributing to the clefting phenotypes in the affected cases, although we were unable to identify any damaging variants in either of two synexpression group members strongly associated with human clefting (FGF8 and SPRY2).
Supplementary Material
Supplemental material is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.117.300535/-/DC1.
Acknowledgments
We thank Amanda Dickinson for thoughtful discussions, as well as for generously providing in situ probes, and Jason Dierdorff for technical assistance with the aCGH processing. We are ever grateful to the families who participated in this research and the many nurses, doctors, dentists, speech pathologists, and others who provided care both in the US and through Operation Smile in the Philippines. This work was supported by National Institutes of Health grants to J.R.M. (R01 DE-021071), D.W.H. (R01 GM-083999), J.C.M. (R37 DE-08559), and L.A.L. (T32 GM-008629).
Footnotes
Communicating editor: L. Jorde
Literature Cited
- 1000 Genomes Project Consortium. Abecasis G. R., Auton A., Brooks L. D., DePristo M. A., et al. , 2012. An integrated map of genetic variation from 1,092 human genomes. Nature 491: 56–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abu-Issa R., Smyth G., Smoak I., Yamamura K., Meyers E. N., 2002. Fgf8 is required for pharyngeal arch and cardiovascular development in the mouse. Development 129: 4613–4625. [DOI] [PubMed] [Google Scholar]
- Aluwihare P., Mu Z., Zhao Z., Yu D., Weinreb P. H., et al. , 2009. Mice that lack activity of alphavbeta6- and alphavbeta8-integrins reproduce the abnormalities of Tgfb1- and Tgfb3-null mice. J. Cell Sci. 122: 227–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassuk A. G., Muthuswamy L. B., Boland R., Smith T. L., Hulstrand A. M., et al. , 2013. Copy number variation analysis implicates the cell polarity gene glypican 5 as a human spina bifida candidate gene. Hum. Mol. Genet. 22: 1097–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belting H. G., Hauptmann G., Meyer D., Abdelilah-Seyfried S., Chitnis A., et al. , 2001. Spiel ohne grenzen/pou2 is required during establishment of the zebrafish midbrain-hindbrain boundary organizer. Development 128: 4165–4176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brophy P. D., Alasti F., Darbro B. W., Clarke J., Nishimura C., et al. , 2013. Genome-wide copy number variation analysis of a Branchio-oto-renal syndrome cohort identifies a recombination hotspot and implicates new candidate genes. Hum. Genet. 132: 1339–1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess S., Reim G., Chen W., Hopkins N., Brand M., 2002. The zebrafish spiel-ohne-grenzen (spg) gene encodes the POU domain protein Pou2 related to mammalian Oct4 and is essential for formation of the midbrain and hindbrain, and for pre-gastrula morphogenesis. Development 129: 905–916. [DOI] [PubMed] [Google Scholar]
- Cai Y., Patterson K. E., Reinier F., Keesecker S. E., Blue E., et al. , 2017. Copy number changes identified using whole exome sequencing in nonsyndromic cleft lip and palate in a Honduran population. Birth Defects Res. 109: 1257–1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y., Li Z., Rosenfeld J. A., Pursley A. N., Patel A., et al. , 2016. Contribution of genomic copy-number variations in prenatal oral clefts: a multicenter cohort study. Genet. Med. 18: 1052–1055. [DOI] [PubMed] [Google Scholar]
- Carta E., Pauws E., Thomas A. C., Mengrelis K., Moore G. E., et al. , 2012. Investigation of SUMO pathway genes in the etiology of nonsyndromic cleft lip with or without cleft palate. Birth Defects Res. A Clin. Mol. Teratol. 94: 459–463. [DOI] [PubMed] [Google Scholar]
- Chen J., Jacox L. A., Saldanha F., Sive H., 2017. Mouth development. Wiley Interdiscip. Rev. Dev. Biol. DOI: 10.1002/wdev.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi C. L., Martinez S., Wurst W., Martin G. R., 2003. The isthmic organizer signal FGF8 is required for cell survival in the prospective midbrain and cerebellum. Development 130: 2633–2644. [DOI] [PubMed] [Google Scholar]
- Christen B., Slack J. M., 1997. FGF-8 is associated with anteroposterior patterning and limb regeneration in Xenopus. Dev. Biol. 192: 455–466. [DOI] [PubMed] [Google Scholar]
- Conte F., Oti M., Dixon J., Carels C. E., Rubini M., et al. , 2016. Systematic analysis of copy number variants of a large cohort of orofacial cleft patients identifies candidate genes for orofacial clefts. Hum. Genet. 135: 41–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson A. J., 2016. Using frogs faces to dissect the mechanisms underlying human orofacial defects. Semin. Cell Dev. Biol. 51: 54–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson A. J., Sive H., 2006. Development of the primary mouth in Xenopus laevis. Dev. Biol. 295: 700–713. [DOI] [PubMed] [Google Scholar]
- Dubey A., Saint-Jeannet J. P., 2017. Modeling human craniofacial disorders in Xenopus. Curr. Pathobiol. Rep. 5: 79–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ensenauer R. E., Adeyinka A., Flynn H. C., Michels V. V., Lindor N. M., et al. , 2003. Microduplication 22q11.2, an emerging syndrome: clinical, cytogenetic, and molecular analysis of thirteen patients. Am. J. Hum. Genet. 73: 1027–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Exome Variant Server, NHLBI GO Exome Sequencing Project (ESP), Seattle, WA. Available at: http://evs.gs.washington.edu/EVS/). Accessed: March 2017.
- Frebourg T., Oliveira C., Hochain P., Karam R., Manouvrier S., et al. , 2006. Cleft lip/palate and CDH1/E-cadherin mutations in families with hereditary diffuse gastric cancer. J. Med. Genet. 43: 138–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs A., Inthal A., Herrmann D., Cheng S., Nakatomi M., et al. , 2010. Regulation of Tbx22 during facial and palatal development. Dev. Dyn. 239: 2860–2874. [DOI] [PubMed] [Google Scholar]
- Genisca A. E., Frías J. L., Broussard C. S., Honein M. A., Lammer E. J., et al. , 2009. Orofacial clefts in the National Birth Defects Prevention Study, 1997-2004. Am. J. Med. Genet. A. 149A: 1149–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girirajan S., Rosenfeld J. A., Cooper G. M., Antonacci F., Siswara P., et al. , 2010. A recurrent 16p12.1 microdeletion supports a two-hit model for severe developmental delay. Nat. Genet. 42: 203–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glessner J. T., Wang K., Cai G., Korvatska O., Kim C. E., et al. , 2009. Autism genome-wide copy number variation reveals ubiquitin and neuronal genes. Nature 459: 569–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodnough L. H., Brugmann S. A., Hu D., Helms J. A., 2007. Stage-dependent craniofacial defects resulting from Sprouty2 overexpression. Dev. Dyn. 236: 1918–1928. [DOI] [PubMed] [Google Scholar]
- Goudy S., Law A., Sanchez G., Baldwin H. S., Brown C., 2010. Tbx1 is necessary for palatal elongation and elevation. Mech. Dev. 127: 292–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green R. M., Feng W., Phang T., Fish J. L., Li H., et al. , 2015. Tfap2a-dependent changes in mouse facial morphology result in clefting that can be ameliorated by a reduction in Fgf8 gene dosage. Dis. Model. Mech. 8: 31–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenway S. C., Pereira A. C., Lin J. C., DePalma S. R., Israel S. J., et al. , 2009. De novo copy number variants identify new genes and loci in isolated sporadic tetralogy of Fallot. Nat. Genet. 41: 931–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang N., Lee I., Marcotte E. M., Hurles M. E., 2010. Characterising and predicting haploinsufficiency in the human genome. PLoS Genet. 6: e1001154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulstrand A. M., Houston D. W., 2013. Regulation of neurogenesis by Fgf8a requires Cdc42 signaling and a novel Cdc42 effector protein. Dev. Biol. 382: 385–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irving C., Mason I., 2000. Signalling by FGF8 from the isthmus patterns anterior hindbrain and establishes the anterior limit of Hox gene expression. Development 127: 177–186. [DOI] [PubMed] [Google Scholar]
- Jacox L., Sindelka R., Chen J., Rothman A., Dickinson A., et al. , 2014. The extreme anterior domain is an essential craniofacial organizer acting through Kinin-Kallikrein signaling. Cell Rep. 8: 596–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacox L., Chen J., Rothman A., Lathrop-Marshall H., Sive H., 2016. Formation of a “pre-mouth array” from the extreme anterior domain is directed by neural crest and Wnt/PCP signaling. Cell Rep. 16: 1445–1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaszai J., Reifers F., Picker A., Langenberg T., Brand M., 2003. Isthmus-to-midbrain transformation in the absence of midbrain-hindbrain organizer activity. Development 130: 6611–6623. [DOI] [PubMed] [Google Scholar]
- Joyner A. L., Liu A., Millet S., 2000. Otx2, Gbx2 and Fgf8 interact to position and maintain a mid-hindbrain organizer. Curr. Opin. Cell Biol. 12: 736–741. [DOI] [PubMed] [Google Scholar]
- Kaiser J., 2012. Human genetics. Genetic influences on disease remain hidden. Science 338: 1016–1017. [DOI] [PubMed] [Google Scholar]
- Kennedy A. E., Dickinson A. J., 2012. Median facial clefts in Xenopus laevis: roles of retinoic acid signaling and homeobox genes. Dev. Biol. 365: 229–240. [DOI] [PubMed] [Google Scholar]
- Klamt J., Hofmann A., Bohmer A. C., Hoebel A. K., Golz L., et al. , 2016. Further evidence for deletions in 7p14.1 contributing to nonsyndromic cleft lip with or without cleft palate. Birth Defects Res. A Clin. Mol. Teratol. 106: 767–772. [DOI] [PubMed] [Google Scholar]
- Kosfeld M. D., Frazier W. A., 1993. Identification of a new cell adhesion motif in two homologous peptides from the COOH-terminal cell binding domain of human thrombospondin. J. Biol. Chem. 268: 8808–8814. [PubMed] [Google Scholar]
- Leslie E. J., Marazita M. L., 2013. Genetics of cleft lip and cleft palate. Am. J. Med. Genet. C. Semin. Med. Genet. 163C: 246–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Wu C., Li C., Boerwinkle E., 2016. dbNSFP v3.0: a one-stop database of functional predictions and annotations for human nonsynonymous and splice-site SNVs. Hum. Mutat. 37: 235–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu W., Bacino C. A., Richards B. S., Alvarez C., VanderMeer J. E., et al. , 2012. Studies of TBX4 and chromosome 17q23.1q23.2: an uncommon cause of nonsyndromic clubfoot. Am. J. Med. Genet. A. 158A: 1620–1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig K. U., Mangold E., Herms S., Nowak S., Reutter H., et al. , 2012. Genome-wide meta-analyses of nonsyndromic cleft lip with or without cleft palate identify six new risk loci. Nat. Genet. 44: 968–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maity G., Mehta S., Haque I., Dhar K., Sarkar S., et al. , 2014. Pancreatic tumor cell secreted CCN1/Cyr61 promotes endothelial cell migration and aberrant neovascularization. Sci. Rep. 4: 4995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangold E., Ludwig K. U., Birnbaum S., Baluardo C., Ferrian M., et al. , 2010. Genome-wide association study identifies two susceptibility loci for nonsyndromic cleft lip with or without cleft palate. Nat. Genet. 42: 24–26. [DOI] [PubMed] [Google Scholar]
- Marshall C. R., Howrigan D. P., Merico D., Thiruvahindrapuram B., Wu W., et al. , 2017. Contribution of copy number variants to schizophrenia from a genome-wide study of 41,321 subjects. Nat. Genet. 49: 27–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maubant S., Saint-Dizier D., Boutillon M., Perron-Sierra F., Casara P. J., et al. , 2006. Blockade of alpha v beta3 and alpha v beta5 integrins by RGD mimetics induces anoikis and not integrin-mediated death in human endothelial cells. Blood 108: 3035–3044. [DOI] [PubMed] [Google Scholar]
- McMahon A. P., Joyner A. L., Bradley A., McMahon J. A., 1992. The midbrain-hindbrain phenotype of Wnt-1-/Wnt-1- mice results from stepwise deletion of engrailed-expressing cells by 9.5 days postcoitum. Cell 69: 581–595. [DOI] [PubMed] [Google Scholar]
- Mefford H. C., Eichler E. E., 2009. Duplication hotspots, rare genomic disorders, and common disease. Curr. Opin. Genet. Dev. 19: 196–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mefford H. C., Sharp A. J., Baker C., Itsara A., Jiang Z., et al. , 2008. Recurrent rearrangements of chromosome 1q21.1 and variable pediatric phenotypes. N. Engl. J. Med. 359: 1685–1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mefford H. C., Cooper G. M., Zerr T., Smith J. D., Baker C., et al. , 2009. A method for rapid, targeted CNV genotyping identifies rare variants associated with neurocognitive disease. Genome Res. 19: 1579–1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers E. N., Lewandoski M., Martin G. R., 1998. An Fgf8 mutant allelic series generated by Cre- and Flp-mediated recombination. Nat. Genet. 18: 136–141. [DOI] [PubMed] [Google Scholar]
- Murray J. C., Daack-Hirsch S., Buetow K. H., Munger R., Espina L., et al. , 1997. Clinical and epidemiologic studies of cleft lip and palate in the Philippines. Cleft Palate Craniofac. J. 34: 7–10. [DOI] [PubMed] [Google Scholar]
- Niehrs C., Meinhardt H., 2002. Modular feedback. Nature 417: 35–36. [DOI] [PubMed] [Google Scholar]
- Niehrs C., Pollet N., 1999. Synexpression groups in eukaryotes. Nature 402: 483–487. [DOI] [PubMed] [Google Scholar]
- Osoegawa K., Vessere G. M., Utami K. H., Mansilla M. A., Johnson M. K., et al. , 2008. Identification of novel candidate genes associated with cleft lip and palate using array comparative genomic hybridisation. J. Med. Genet. 45: 81–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osorio L., Wu X., Zhou Z., 2014. Distinct spatiotemporal expression of ISM1 during mouse and chick development. Cell Cycle 13: 1571–1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panagiotaki N., Dajas-Bailador F., Amaya E., Papalopulu N., Dorey K., 2010. Characterisation of a new regulator of BDNF signalling, Sprouty3, involved in axonal morphogenesis in vivo. Development 137: 4005–4015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pera E. M., Kim J. I., Martinez S. L., Brechner M., Li S. Y., et al. , 2002. Isthmin is a novel secreted protein expressed as part of the Fgf-8 synexpression group in the Xenopus midbrain-hindbrain organizer. Mech. Dev. 116: 169–172. [DOI] [PubMed] [Google Scholar]
- Petrovski S., Wang Q., Heinzen E. L., Allen A. S., Goldstein D. B., 2013. Genic intolerance to functional variation and the interpretation of personal genomes. PLoS Genet. 9: e1003709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picker A., Brennan C., Reifers F., Clarke J. D., Holder N., et al. , 1999. Requirement for the zebrafish mid-hindbrain boundary in midbrain polarisation, mapping and confinement of the retinotectal projection. Development 126: 2967–2978. [DOI] [PubMed] [Google Scholar]
- Raible F., Brand M., 2004. Divide et Impera–the midbrain-hindbrain boundary and its organizer. Trends Neurosci. 27: 727–734. [DOI] [PubMed] [Google Scholar]
- Reifers F., Bohli H., Walsh E. C., Crossley P. H., Stainier D. Y., et al. , 1998. Fgf8 is mutated in zebrafish acerebellar (ace) mutants and is required for maintenance of midbrain-hindbrain boundary development and somitogenesis. Development 125: 2381–2395. [DOI] [PubMed] [Google Scholar]
- Reim G., Brand M., 2002. Spiel-ohne-grenzen/pou2 mediates regional competence to respond to Fgf8 during zebrafish early neural development. Development 129: 917–933. [DOI] [PubMed] [Google Scholar]
- Rhinn M., Brand M., 2001. The midbrain–hindbrain boundary organizer. Curr. Opin. Neurobiol. 11: 34–42. [DOI] [PubMed] [Google Scholar]
- Riley B. M., Mansilla M. A., Ma J., Daack-Hirsch S., Maher B. S., et al. , 2007. Impaired FGF signaling contributes to cleft lip and palate. Proc. Natl. Acad. Sci. USA 104: 4512–4517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenfeld J. A., Ballif B. C., Torchia B. S., Sahoo T., Ravnan J. B., et al. , 2010. Copy number variations associated with autism spectrum disorders contribute to a spectrum of neurodevelopmental disorders. Genet. Med. 12: 694–702. [DOI] [PubMed] [Google Scholar]
- Rosenfeld J. A., Coe B. P., Eichler E. E., Cuckle H., Shaffer L. G., 2013. Estimates of penetrance for recurrent pathogenic copy-number variations. Genet. Med. 15: 478–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen S., Skaletsky H., 2000. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 132: 365–386. [DOI] [PubMed] [Google Scholar]
- Ruderfer D. M., Hamamsy T., Lek M., Karczewski K. J., Kavanagh D., et al. , 2016. Patterns of genic intolerance of rare copy number variation in 59,898 human exomes. Nat. Genet. 48: 1107–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz M. A., Schaller M. D., Ginsberg M. H., 1995. Integrins: emerging paradigms of signal transduction. Annu. Rev. Cell Dev. Biol. 11: 549–599. [DOI] [PubMed] [Google Scholar]
- Session A. M., Uno Y., Kwon T., Chapman J. A., Toyoda A., et al. , 2016. Genome evolution in the allotetraploid frog Xenopus laevis. Nature 538: 336–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi M., Mostowska A., Jugessur A., Johnson M. K., Mansilla M. A., et al. , 2009. Identification of microdeletions in candidate genes for cleft lip and/or palate. Birth Defects Res. A Clin. Mol. Teratol. 85: 42–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimomura T., Kawakami M., Okuda H., Tatsumi K., Morita S., et al. , 2015. Retinoic acid regulates Lhx8 expression via FGF-8b to the upper jaw development of chick embryo. J. Biosci. Bioeng. 119: 260–266. [DOI] [PubMed] [Google Scholar]
- Simioni M., Araujo T. K., Monlleo I. L., Maurer-Morelli C. V., Gil-da-Silva-Lopes V. L., 2015. Investigation of genetic factors underlying typical orofacial clefts: mutational screening and copy number variation. J. Hum. Genet. 60: 17–25. [DOI] [PubMed] [Google Scholar]
- Tan G. S., Lim K. H., Tan H. T., Khoo M. L., Tan S. H., et al. , 2014. Novel proteomic biomarker panel for prediction of aggressive metastatic hepatocellular carcinoma relapse in surgically resectable patients. J. Proteome Res. 13: 4833–4846. [DOI] [PubMed] [Google Scholar]
- Thomason H. A., Dixon M. J., Dixon J., 2008. Facial clefting in Tp63 deficient mice results from altered Bmp4, Fgf8 and Shh signaling. Dev. Biol. 321: 273–282. [DOI] [PubMed] [Google Scholar]
- Trainor P. A., Ariza-McNaughton L., Krumlauf R., 2002. Role of the isthmus and FGFs in resolving the paradox of neural crest plasticity and prepatterning. Science 295: 1288–1291. [DOI] [PubMed] [Google Scholar]
- Trumpp A., Depew M. J., Rubenstein J. L., Bishop J. M., Martin G. R., 1999. Cre-mediated gene inactivation demonstrates that FGF8 is required for cell survival and patterning of the first branchial arch. Genes Dev. 13: 3136–3148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker A. S., Al Khamis A., Ferguson C. A., Bach I., Rosenfeld M. G., et al. , 1999. Conserved regulation of mesenchymal gene expression by Fgf-8 in face and limb development. Development 126: 221–228. [DOI] [PubMed] [Google Scholar]
- Twigg S. R., Wilkie A. O., 2015. New insights into craniofacial malformations. Hum. Mol. Genet. 24: R50–R59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uddin M., Thiruvahindrapuram B., Walker S., Wang Z., Hu P., et al. , 2015. A high-resolution copy-number variation resource for clinical and population genetics. Genet. Med. 17: 747–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valle-Rios R., Maravillas-Montero J. L., Burkhardt A. M., Martinez C., Buhren B. A., et al. , 2014. Isthmin 1 is a secreted protein expressed in skin, mucosal tissues, and NK, NKT, and th17 cells. J. Interferon Cytokine Res. 34: 795–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venugopal S., Chen M., Liao W., Er S. Y., Wong W. S., et al. , 2015. Isthmin is a novel vascular permeability inducer that functions through cell-surface GRP78-mediated Src activation. Cardiovasc. Res. 107: 131–142. [DOI] [PubMed] [Google Scholar]
- Vieira A. R., Avila J. R., Daack-Hirsch S., Dragan E., Felix T. M., et al. , 2005. Medical sequencing of candidate genes for nonsyndromic cleft lip and palate. PLoS Genet. 1: e64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Yang Z. L., Zou Q., Yuan Y., Li J., et al. , 2016. SHP2 and UGP2 are biomarkers for progression and poor prognosis of gallbladder cancer. Cancer Invest. 34: 255–264. [DOI] [PubMed] [Google Scholar]
- Wang Y. H., Beck C. W., 2014. Distal expression of sprouty (spry) genes during Xenopus laevis limb development and regeneration. Gene Expr. Patterns 15: 61–66. [DOI] [PubMed] [Google Scholar]
- Wehby G. L., Cassell C. H., 2010. The impact of orofacial clefts on quality of life and healthcare use and costs. Oral Dis. 16: 3–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsh I. C., Hagge-Greenberg A., O’Brien T. P., 2007. A dosage-dependent role for Spry2 in growth and patterning during palate development. Mech. Dev. 124: 746–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurst W., Bally-Cuif L., 2001. Neural plate patterning: upstream and downstream of the isthmic organizer. Nat. Rev. Neurosci. 2: 99–108. [DOI] [PubMed] [Google Scholar]
- Xiang W., Ke Z., Zhang Y., Cheng G. H., Irwan I. D., et al. , 2011. Isthmin is a novel secreted angiogenesis inhibitor that inhibits tumour growth in mice. J. Cell. Mol. Med. 15: 359–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Kilgallen S., Andreeva V., Spicer D. B., Pinz I., et al. , 2010. Conditional expression of Spry1 in neural crest causes craniofacial and cardiac defects. BMC Dev. Biol. 10: 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yildirim Y., Kerem M., Koroglu C., Tolun A., 2014. A homozygous 237-kb deletion at 1p31 identified as the locus for midline cleft of the upper and lower lip in a consanguineous family. Eur. J. Hum. Genet. 22: 333–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Younkin S. G., Scharpf R. B., Schwender H., Parker M. M., Scott A. F., et al. , 2014. A genome-wide study of de novo deletions identifies a candidate locus for non-syndromic isolated cleft lip/palate risk. BMC Genet. 15: 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Younkin S. G., Scharpf R. B., Schwender H., Parker M. M., Scott A. F., et al. , 2015. A genome-wide study of inherited deletions identified two regions associated with nonsyndromic isolated oral clefts. Birth Defects Res. A Clin. Mol. Teratol. 103: 276–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuen R. K. C., Merico D., Bookman M., Howe J. L., Thiruvahindrapuram B., et al. , 2017. Whole genome sequencing resource identifies 18 new candidate genes for autism spectrum disorder. Nat. Neurosci. 20: 602–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarrei M., MacDonald J. R., Merico D., Scherer S. W., 2015. A copy number variation map of the human genome. Nat. Rev. Genet. 16: 172–183. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Chen M., Venugopal S., Zhou Y., Xiang W., et al. , 2011. Isthmin exerts pro-survival and death-promoting effect on endothelial cells through alphavbeta5 integrin depending on its physical state. Cell Death Dis. 2: e153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Guo Y. J., Tomac A. C., Taylor N. R., Grinberg A., et al. , 1999. Isolated cleft palate in mice with a targeted mutation of the LIM homeobox gene lhx8. Proc. Natl. Acad. Sci. USA 96: 15002–15006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Plasmids are available upon request. Raw CNV array .tiff, Nexus data summary .txt files, and DEVA segMNT .txt files are publicly available at the Gene Expression Omnibus (GSE100845).