Abstract
In this report we investigated, within a group of closely related single domain camelid antibodies (VHHs), the relationship between binding affinity and neutralizing activity as it pertains to ricin, a fast-acting toxin and biothreat agent. The V1C7-like VHHs (V1C7, V2B9, V2E8, and V5C1) are similar in amino acid sequence, but differ in their binding affinities and toxin-neutralizing activities. Using the X-ray crystal structure of V1C7 in complex with ricin’s enzymatic subunit (RTA) as a template, Rosetta-based homology modeling coupled with energetic decomposition led us to predict that a single pairwise interaction between Arg29 on V5C1 and Glu67 on RTA was responsible for the difference in ricin toxin binding affinity between V1C7, a weak neutralizer, and V5C1, a moderate neutralizer. This prediction was borne out experimentally: substitution of Arg for Gly at position 29 enhanced V1C7’s binding affinity for ricin, whereas the reverse (i.e., Gly for Arg at position 29) diminished V5C1’s binding affinity by >10 fold. As expected, the V5C1R29G mutant was largely devoid of toxin-neutralizing activity. However, the toxin-neutralizing activity of the V1C7G29R mutant was not correspondingly improved, indicating that in the V1C7 family binding affinity alone does not account for differences in antibody function. V1C7 and V5C1, as well as their respective point mutants, recognized indistinguishable epitopes on RTA, at least at the level of sensitivity afforded by hydrogen-deuterium mass spectrometry. The results of this study have implications for engineering therapeutic antibodies because they demonstrate that even subtle differences in epitope specificity can account for important differences in antibody function.
Keywords: antibody engineering, structure-based protein modeling, epitope mapping, vaccine
INTRODUCTION
The eventual success of a given subunit vaccine for most infectious diseases and biothreat agents will depend on its capacity to elicit a robust antibody response against key protective epitopes on the target antigen and minimize off target interactions with non-neutralizing or “decoy” epitopes 1–3. Ricin, a toxic glycoprotein from the castor bean (Ricinus communis), is a biothreat agent for which no licensed therapeutic or prophylactic countermeasures currently exist. Ricin is a heterodimer consisting of an enzymatic subunit (RTA) linked via a single disulfide bond to a binding subunit (RTB), a Gal/GalNAc-specific lectin that promotes attachment, endocytosis, and retrograde transport of ricin in eukaryotic cells 4. In the endoplasmic reticulum (ER), the disulfide bond between RTA and RTB is reduced by protein disulfide isomerase (PDI) 5, after which RTA is retro-translocated across the ER membrane. Ricin’s cytotoxic effects are due to RTA, an RNA N-glycosidase whose substrate is a conserved A residue within the sarcin-ricin loop (SRL) of eukaryotic 28S rRNA 6. Depurination of the SRL results in ribosome-inactivation and programmed cell death by apoptosis 7.
RTA is the focus of current efforts to develop countermeasures against ricin toxin: two enzymatically-inactive forms of RTA are being pursued as possible ricin toxin subunit vaccines 8–10, while a number of RTA-specific monoclonal antibodies (mAbs) are being evaluated as possible therapeutics 11–13. However, the mechanisms by which antibodies neutralize (or fail to neutralize) ricin toxin in vitro and in vivo are not fully understood. For example, how do factors like binding affinity and epitope specificity contribute to overall toxin-neutralizing activity (TNA)? RTA-specific monoclonal (mAbs) and single-domain camelid (sdAbs or VHHs) antibodies are particularly powerful tools to begin to address these questions 13–21. Antibody R70 (UNIVAX 70/138) was one of the first RTA-specific mAbs to be shown to neutralize ricin toxin in vitro and in vivo 22. R70 recognizes a conserved immunodominant epitope defined by a solvent exposed α-helix, referred to as α-helix B, which encompasses residues 97–107 23,24. Other mAbs and VHHs like E5 (Figure 1A) have been identified that compete with R70 for binding to RTA and/or that have been shown by X-ray crystallography to contact α-helix B directly 17,21,25–27. Although these antibodies recognize overlapping epitopes, which we refer to as epitope Cluster I, on the surface of RTA, their TNA (IC50s) range from potent (5 nM) to weak (>330 nM) to undetectable 21,26,27. Because differences in binding affinities (KD) only partially account for these differences in TNA, we have proposed that epitope specificity, notably contact with α-helix B, may be the primary factor in determining whether a Cluster I antibody can inactivate ricin 26,27.
Figure 1. Epitope cluster, alignments, and comparative 3D structures of the V1C7 family of VHHs.
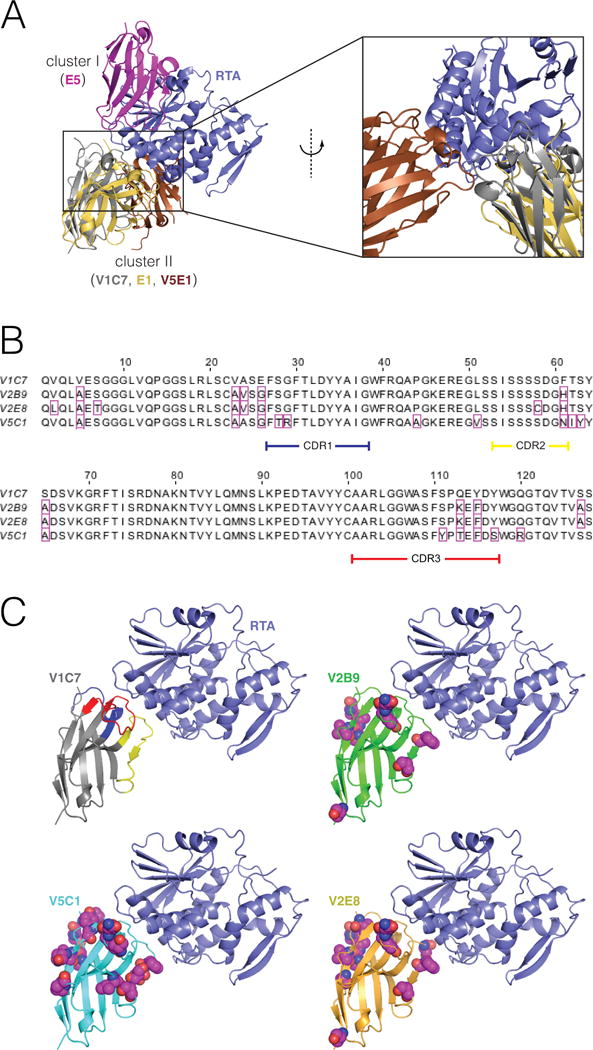
(Panel A) Representative binding modes for epitope clusters I and II on the surface of RTA. VHHs were assigned to cluster I or II based on whether they were competitively inhibited from binding to ricin by the mouse mAbs PB10 (which is similar to UNIVAX R70) or SyH7, respectively. These are the only two clusters for which our group has published X-ray structures of bound VHHs. VHH E5 binds to a cluster-I epitope. V1C7 and E1 bind to a cluster-II epitopes; these two VHHs have a 69% overall sequence identity 25% over the CDRs. (Panel B) Primary amino acid sequence alignment of the V1C7 family of VHHs. Residues that differ from V1C7 are boxed in magenta. V1C7 has a sequence identity of >87% overall and of >81% over the CDRs with any other member of the family. (Panel C) Proposed structures of the four VHHs in complex with RTA. The X-ray structure of V1C7 in complex with RTA was solved at 1.8-Å resolution (PDB: 5J56). The structures of V2B9, V2E8, and V5C1 are computational models having the same backbone structure as V1C7. Residues that differ from their V1C7 counterparts are represented as spheres (carbon: magenta; oxygen: red; nitrogen: blue; sulfur: yellow).
So-called epitope cluster II on RTA was originally defined by the mAb SyH7 and now includes three other mAbs, PA1, PH12, and TB12 19,20. SyH7 is roughly equivalent to R70 in its capacity to neutralize ricin in vitro and in vivo 19. We recently identified three VHHs, V5E1, E1 and V1C7 that we assigned to cluster II, because they were competitively inhibited from binding to ricin by SyH7 28. The three VHHs are interesting because their different toxin-neutralizing activities: V5E1 has strong TNA, E1 has moderate TNA, and V1C7 weak TNA. The X-ray crystal structures of the VHH–RTA complexes demonstrated that V5E1 recognizes an epitope that is distinct from that of E1 and V1C7 (Figure 1A), suggesting that V5E1 contacts critical residues associated with ricin neutralizing activity whereas E1 and V1C7 do not. However, this interpretation is confounded by their differences in binding affinities: V5E1 has picomolar binding affinity for RTA, while E1 and V1C7 have nanomolar affinities. This leaves open the possibility that binding affinity alone is sufficient to explain the observed difference in ricin neutralizing activity. Indeed, there are examples in the literature where increased antibody binding affinities correspond to enhanced neutralizing activities 17,29–31. Without a better understanding of the contributions of binding affinity and epitope specificity to ricin toxin-neutralizing activity information, it is not possible to rationally design more effective therapeutic antibodies.
The recent identification of three additional V1C7-like VHHs from our ricin-specific alpaca phage-display single domain library (D. Vance, J. Tremblay, C. Shoemaker, and N. Mantis, manuscript in preparation) afforded an opportunity to address the contribution of binding affinity as it relates to toxin-neutralizing activity. As will be described in the Results section, the three V1C7-like VHHs, V2B9, V2E8, and V5C1, share considerable DNA and amino acid sequence identity with V1C7 and most certainly arose from a common B cell progenitor. Moreover, the VHH antibodies display a range of binding affinities and toxin-neutralizing activities. Thus, we reasoned that we could use the V1C7–RTA complex to model the structure of the other VHHs of the family, and then predict which specific residues are responsible for the observed differences in binding affinity. These predictions could then be examined experimentally.
A previous study seeking to enhance binding affinity for a different mAb–ricin complex relied on docking to first predict the binding mode, then identify mutations to vary the binding affinity 32. While such mutations were indeed identified, it is difficult in the absence of structural confirmation to be certain that the underlying basis for the altered binding affinity was indeed as designed. Rather than docking, we instead predicted the three-dimensional (3D) structure of each VHH variant by homology modeling 33. This approach begins from the expectation that the 3D structure of a given protein will closely resemble that of other highly homologous proteins (e.g., those with >25–30% sequence identity 34). Thus, the 3D structure of a homologous protein, when available, can serve as a template for modeling the protein of interest. Most of today’s successful homology modeling methods combine templates from multiple proteins 35,36, since each template may provide distinct insights into different portions of the input sequence. This is especially true of antibody modeling, where templates for the framework regions can be combined with templates for each of the CDR loops 37–41. These general principles of antibody modeling are also expected to hold for VHHs 38,42, although a dedicated assessment for VHH modeling has yet to occur.
Here we report on the homology modeling of the V1C7 VHH family, and show how it led to identification of a residue pairwise interaction—between Arg29 on V5C1 and Glu67 on RTA—that was demonstrated to enhance binding affinity and be necessary, but not sufficient, for V5C1’s higher TNA. Ultimately, a better understanding of the relationship between binding affinity and toxin-neutralizing activity holds important implications for design of novel therapeutic antibodies.
MATERIALS AND METHODS
VHHs, toxins, and other reagents
The RTA-specific VHHs were identified from the so-called HobJo alpaca single domain antibody phage displayed library by repeated rounds of affinity enrichment on ricin or RTA (D. Vance, C. Shoemaker, and N. Mantis, manuscript in preparation) 21. Ricin toxin (RCA-II) and biotinylated ricin were obtained from Vector Laboratories (Burlingame, CA). All other chemicals and reagents were purchased from Sigma-Aldrich (St. Louis, MO), unless noted otherwise. Hydrogen exchange studies were conducted with an inactivated mutant of RTA (V76M; Y80A), which is known as RiVax43. The two point mutations do not alter the tertiary structure of RTA, as reported by Legler and colleagues 44. RiVax was expressed and purified as described 45.
ELISA and surface plasmon resonance (SPR)
For competition ELISAs, Nunc-Immuno plates (ThermoScientific, Swedesboro, NJ) were coated overnight with murine IgGs (1 μg/mL) and then blocked with 2% goat serum in phosphate buffered saline. Ricin (1 μg/mL) was applied to bind to the mAb-coated well, and probed with E-tagged VHHs. The plates were developed with anti-E-horseradish peroxidase (Bethyl, Montgomery, TX) followed by SureBlue Peroxidase Substrate (KPL). The reaction was quenched with 1M phosphoric acid, and absorbance was read at 450 nm using a VersaMax Microplate Reader (Molecular Devices, Sunnyvale, CA). The dissociation constants (KD) for each VHH were determined by SPR using the ProteOn XPR36 (Bio-Rad Inc. CA, USA) using ProteOn GLC sensor chips coated with ricin at high (4 μg/ml: L1) or low (2 μg/ml: L2) density, as described 27. PBS with 0.005% Tween-20 (pH 7.4) was used as the dilution and running buffer. All analyses were conducted at 25°C.
Computational modeling of VHH–RTA complexes
We modeled the structure of each VHH using the known crystal structure of V1C7 as the only template, under the assumption that the four members of the family share a common structure as well as a common RTA binding mode given their high sequence similarity. Each VHH aligns with sequence identity of >87% (and no gaps) to V1C7, as determined by the EMBOSS Needle implementation of the Needleman–Wunsch algorithm 46,47. This extremely close similarity gave us confidence that each of these family members would share the binding mode of V1C7, as opposed to adopting one of the several other binding modes that have been observed for VHHs bound to RTA 27.
Threading of each VHH sequence onto the template was carried out using the Rosetta partial_thread application. We preserved the bound pose of the VHH relative to RTA from the crystal structure of the V1C7–RTA complex (PDB: 5J56), generating an ensemble of 5,000 energetically minimized conformations using the Rosetta relax application 48; this procedure searches for alternate side-chain rotamers on both RTA and VHH, and also includes gradient-based minimization of their backbones and side-chains, and the relative orientation between them. The disulfide bond between VHH residues Cys22 and Cys100 was enforced throughout, and the pwSHO term was included to describe polar solvation. This modeling approach yielded models for all three VHHs that were structurally very similar to the V1C7 template, as expected.
The average VHH–RTA interaction energy over the ensemble of 5,000 low-energy models was then used as an estimate of the relative RTA binding affinity. The VHH–RTA interaction energy of each model was computed as the difference between the total energy of the complex and the total energy of the two monomers taken in isolation. We selected a representative model of each VHH–RTA ensemble as the “center” of the largest cluster formed by the Rosetta cluster application, where clustering (by RMSD) was limited to the 1,000 models of lowest VHH–RTA interaction energy. In the analysis of representative models, we identified the most favorable residue-pair interactions at the VHH–RTA interface using the Rosetta interface_energy application.
HX-MS Analysis
Hydrogen exchange was performed using an H/DX PAL robot (LEAP Technologies, Carrboro, North Carolina). MS measurements were conducted using a QTOF mass analyzer (Agilent 6530, Santa Clara, California) with Agilent 1260 Infinity LC System. For HX, 4 μL of RiVax prepared at 20 µM was incubated with 36 μL deuterated buffer (20 mM sodium phosphate, 150 mM sodium chloride, pD 7.7) at 25 °C for 13 s – 24 h. For experiments in the presence of VHH, RiVax was prepared at 20 µM with a VHH concentration of 40 µM. To account for differences in measuring pD with a pH meter, a value 0.4 units was added to the pH meter reading 49. Labeling at each time point was performed in triplicate. The exchange was quenched by a 1:1 dilution into quench buffer (4 M Gdn-HCl, 0.2 M phosphate, pH 2.5) at 0 °C for 60 s. Fifty five µL of the quenched sample was then injected into the 100 μL sample loop of the refrigerated compartment of the H/DX PAL, maintained at 0 °C, containing a pepsin column (50 mm × 2.1 mm), a C8 trap (Poroshell 120 SB-C8, 2.1× 5 mm, 2.7 μm particle diameter), and a reversed phase column (ZORBAX 300SB-C18, 2.1 × 50 mm, 1.8 μm particle diameter). The immobilized pepsin column was prepared in-house as described elsewhere 50. The quenched sample was carried through the pepsin column using 0.1% formic acid at 200 μL min–1 over 3 min. The resulting peptic peptides were desalted on the C8 trap for an additional 60 s. The LC method used a 15 min segmented gradient from 1% B to 95% B, with mobile phase A consisting of 0.1% formic acid and mobile phase B consisting of 0.1% formic acid in 90% acetonitrile/10% water flowing at 200 μL min–1. A second 15 min gradient similar to the first was used to minimize peptide carry over from the trap and analytical column. To minimize carryover in the immobilized pepsin column, the pepsin column was washed as described previously 51. MS/MS analysis was used to generate a peptide map of RiVaxs that covered 100% of the RiVax sequence with 138 peptides. The peptides are numbered sequentially from the N-terminus to the C-terminus (Supplemental Table S1).
The HX data were processed using HDExaminer software (Sierra Analytics, Modesto, California). The mass increase of peptides at each time point was exported from HDExaminer. For each peptide, data from all time points was combined into a single value representing the average difference in HX between bound and free states, normalized for peptide length as described 52
| (1) |
where is the number of HX labeling times, is the theoretical maximum deuteration, is the average mass of the peptide in the bound state and is the average mass of the peptide in the free state averaged over technical replicates. The subscript i denotes ith HX exchange time. For each peptide, was determined by counting the number of exchangeable amides after neglecting the first two N-terminal residues that undergo rapid back-exchange. The propagated standard error in was estimated using equation (2) where denotes the sample standard deviations from triplicate technical replicates ( ) of and . To set the confidence interval, we have taken a “3σ” confidence interval for statistically significant changes in HX, which is . Strong protection was defined as and intermediate protection as statistically significant and .
| (2) |
Cloning, expression, and purification of V1C7G29R variant and RTA for crystallization
PCR amplicons corresponding to V1C7 (residues 1–136) with a Gly to Arg mutation at residue 29 and RTA (residues 1–267) were subcloned into the N-terminally deca-histidine pSUMO expression vector using a standard ligase independent cloning protocol 26. Both proteins were expressed in E. coli strain BL21(DE3)-pRARE. The transformed bacteria were grown at 37°C in TB medium and induced at 20°C with 0.1 mM IPTG at an OD600 of 0.6 for ~16 hours. After induction, cells were harvested and resuspended in 20 mM Tris-Cl pH 7.5 and 150 mM NaCl. The cell suspension was sonicated and centrifuged at 30,000 × g for 30 minutes. After centrifugation, the protein-containing supernatant was purified by nickel-affinity and size-exclusion chromatography on an AKTAxpress system (GE Healthcare), which consisted of a 1 mL nickel affinity column followed by a Superdex 200 16/60 gel filtration column. The elution buffer consisted of 0.5M imidazole in HEPES pH 7.6 and 150 mM NaCl, and the gel filtration buffer consisted of 20 mM HEPES pH 7.6, 150mM NaCl, and 20mM imidazole. Fractions containing protein were pooled and subjected to TEV protease cleavage (1:20 weight ratio) for 3 hours at room temperature to remove the decahistidine-maltose binding protein tag. The cleaved protein was passed over a 1mL Ni-NTA agarose (Qiagen) gravity column to remove the added TEV protease, cleaved residues, and uncleaved fusion protein. To generate VHH–RTA protein complexes, RTA was mixed in a 1:1 molar stoichiometry with the purified V1C7G29R variant and incubated on ice for 1 h. Purified VHH–RTA complex was concentrated to a final total concentration of 10 mg/ml for crystallization experiments.
Crystallization and data collection
Both versions of the V1C7G29R–RTA complex crystals were grown by sitting drop vapor diffusion at 20°C using a protein to reservoir volume ratio of 1:1 with total drop volumes of 0.4 μL. Crystals of the V1C7G29R–RTA complex were grown under two independent conditions with the first condition containing a high salt concentration of 1.6 M ammonium dihydrogen phosphate along with 20% glycerol and 100 mM Tris pH 8.5 as the buffer. The second crystallization condition for the V1C7G29R–RTA complex contained 8% PEG 4000 and 100 mM sodium acetate pH 4.6. All crystals were flash frozen in liquid nitrogen after a short soak in the appropriate crystallization buffers supplemented with 20–25% ethylene glycol. Data were collected at the 24-ID-E beamline at the Advanced Photon Source, Argonne National Labs. All data was indexed, merged, and scaled using HKL2000 53, then converted to structure factors using CCP4 54.
Structure determination and refinement
The structure of each VHH–RTA complex was solved by molecular replacement using the program Phaser 55. Molecular replacement calculations were performed using the coordinates of the ricin A chain as a search model for RTA (PDB: 1RTC) for both of the V1C7G29R–RTA complexes. The VHH coordinates used as a search model for each V1C7G29R–RTA complex were those of the native V1C7 (PDB: 5J56) with all three of the CDRs removed from the search model. The resulting phase information was used to manually insert the Arg 29 mutation into the native V1C7 molecule followed by iterative manual building of each model using the program COOT 56. All structural refinement was done employing the PHENIX package 57. During refinement a cross-validation test set was created from a random 5% of the reflections. Data collection and refinement statistics are listed in Supplemental Table S2. Molecular graphics were prepared using PyMOL (Schrodinger) (DeLano Scientific LLC, Palo Alto, CA).
Accession numbers
The structures generated in this study were deposited in the Protein Data Bank (PDB; http://www.rcsb.org/pdb/) under accession numbers 5U4L for the V1C7G29R variant crystallized under high salt conditions and 5U4M for the V1C7G29R variant crystallized without salt as described in Supplemental Table S2.
RESULTS
Alignment and functional characterization of the V1C7 family of VHHs
In an effort to isolate ricin-specific VHHs beyond those described in previous manuscripts, we subjected the so-called HobJo phage-displayed single domain VHH library to a series of pannings using different ricin targets (D. Vance, C. Shoemaker, and N. Mantis, manuscript in preparation). Through these screens we isolated three VHHs, V2B9, V2E8, and V5C1, that shared considerable amino acid sequence identity with V1C7 (Figure 1B). VHHs V2B9, V2E8, and V5C1 differ from V1C7 by 9, 12, and 16 amino acid residues, respectively, with those differences occurring in both framework regions and CDRs 1, 2, and 3. Phylogenetic analysis using MegAlign confirmed that V1C7 was most distantly related to V5C1, while V2E8 and V2B9 were most similar to each other and more closely related to V1C7 than V5C1 (data not shown). By comparison, we have recently solved the structures of several groups of VHHs bound to RTA with nearly identical structural epitopes, but very different primary sequences, particularly in the CDR3. VHHs E5 and F5, for example, have nearly identical structural epitopes but differ in primary amino acid sequence by more than 31 residues with nine insertions/deletions26.
Competitive ELISAs with SyH7 and other cluster-II mAbs indicated that all V1C7-like VHHs bind overlapping epitopes on RTA (Figure 2), which was expected considering the sequence similarity of the V1C7-like VHHs to each other. Yet, despite their sequence similarities, the V1C7 VHH family members differed in their binding affinities and toxin-neutralizing activities (TNAs). V5C1 had the highest binding affinity for ricin (0.4 nM) (Table 1; Figure 3A; Supplemental Figure S1–2), as well as the most potent TNA of the four V1C7 family members (Figure 3B). As noted above, V5C1 differs from V1C7 by 16 residues, 7 of which are localized to CDR1–3. V2E8 had the weakest binding affinity for ricin (~5.7 nM) and the least potent TNA, while antibody V2B9 was functionally the most similar to V1C7, possibly because the CDRs of V2B9 and V1C7 differ by just 3 residues.
Figure 2. Epitope cluster competition profiles with the V1C7 family of VHHs.
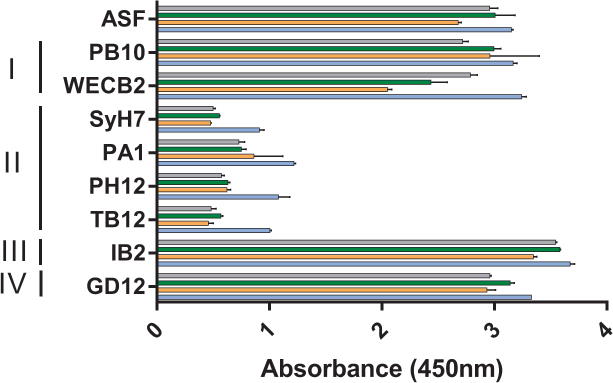
Ricin was captured on microtiter plates by the surrogate receptor, asialofetuin (ASF; top row), or individual mAbs representing epitope clusters I–IV, as indicated by the vertical bars (left). Cluster III is defined by IB2 and cluster IV by GD12, as described 20. The plates were then probed with V1C7 (gray), V2B9 (green), V2E8 (orange), or V5C1 (blue), as described in the Materials and Methods. The values shown are the average (with standard deviation) of a single representative experiment done in triplicate.
Table 1.
Binding affinities of V1C7 VHHs
| VHH | KDa |
|---|---|
| V1C7 | 2.24 |
| V1C7G29R | 0.39 |
| V2B9 | 1.03 |
| V2E8 | 5.73 |
| V5C1 | 0.37 |
| V5C1R29G | 4.08 |
Dissociation constants (nM) were determined by SPR.
Figure 3. Relative toxin-neutralizing activities of the V1C7 family of VHHs.
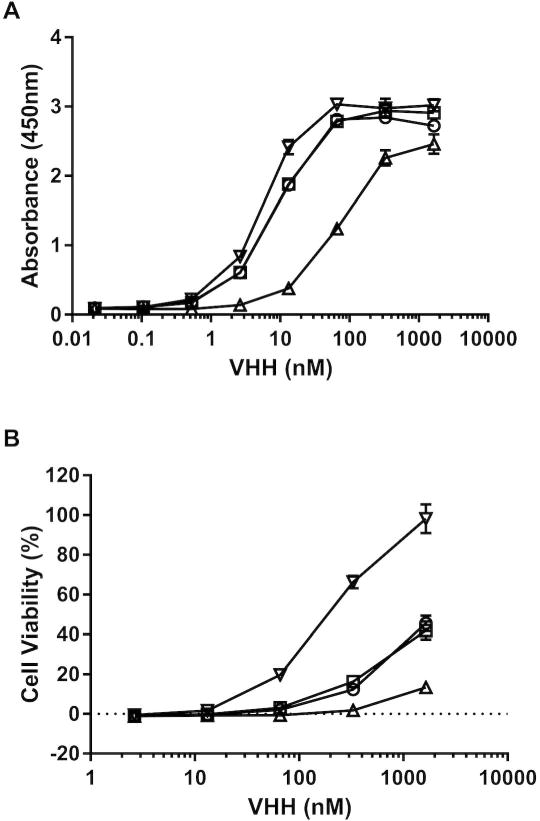
(Panel A) Relative affinities of V1C7 (circles), V2B9 (squares), V2E8 (triangles), and V5C1 (inverted triangles) for ricin toxin, as determined by ELISA in which ricin was captured onto plastic surface by ASF. (Panel B) Ricin toxin-neutralizing activities associated with the V1C7-like VHHs (symbols as in Panel A), determined in a Vero cell cytotoxicity assay (see Materials and Methods). The values shown are the average (with standard deviation) of two separate experiments each done in triplicate.
Using homology modeling in Rosetta to identify the structural basis of differential RTA binding affinities
At first approximation, these results suggest that within the V1C7 family of VHHs there is a direct relationship between ricin binding affinity and TNA. Accordingly, we predicted that increasing V1C7’s binding affinity for ricin toxin to be more like that of V5C1 would result in a concomitant increase in TNA. In the absence of X-ray crystal structures of V5C1, V2B9, and V2E8 in complex with RTA, we sought to investigate the structural basis of differential RTA binding affinities of the V1C7 family via homology modeling. Specifically, with the Rosetta software suite 58 we used the X-ray crystal structure of the V1C7–RTA complex (PDB: 5J56) as a template to build ensembles of 5,000 homology models of RTA bound to V2B9, V2E8, and V5C1. As a reference, we also built a corresponding ensemble of models for V1C7–RTA. For all four VHHs, models remained similar to the V1C7–RTA template after the energy minimization stage of the modeling procedure (Figure S3), meeting our expectation that the sequence similarity of V1C7 family members should translate into a common RTA binding mode.
The relative RTA binding affinity for each of the four VHHs was estimated based on the energy of the interaction between VHH and RTA as averaged over the ensemble of models. As shown in Figure 4A, the average interaction energy for V5C1 was at least 1.9 Rosetta Energy Units (REUs) lower (i.e., more favorable) than that of the other three VHHs, whose average interaction energies fall within a comparatively narrow range. While the ranking of V1C7, V2B9, and V2E8 did not precisely match the experimentally defined affinity constants (Table 1), Rosetta did correctly predict that V5C1 has the highest binding affinity among the four related VHHs.
Figure 4. Computational modeling predicts V5C1 to have the highest RTA binding affinity due to Gly29Arg substitution.
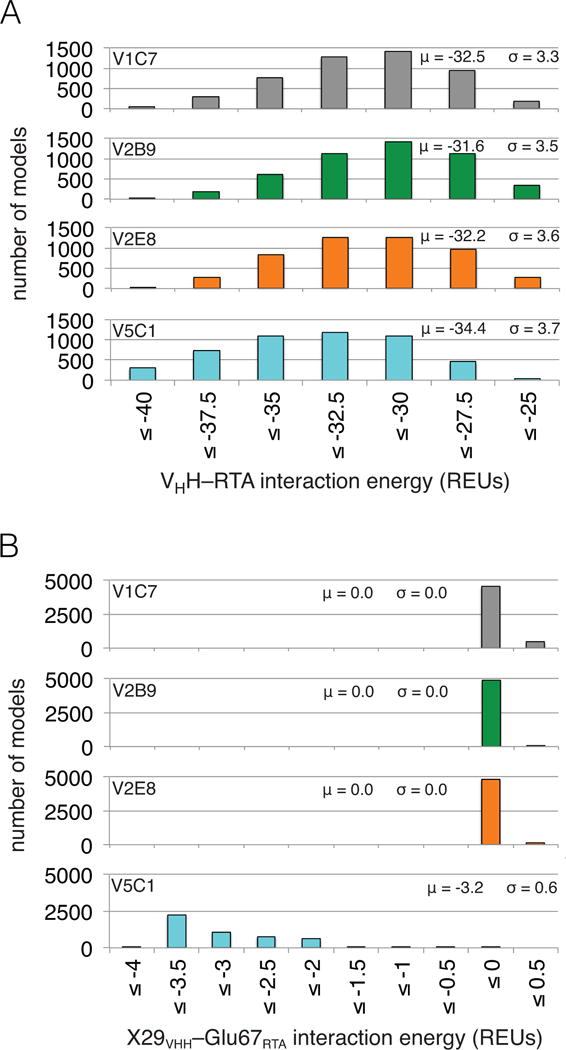
(Panel A) Distributions of VHH–RTA interaction energy over ensembles of 5,000 computational models (μ: average; σ: standard deviation). The V5C1 models have lower interaction energies than the models of any other VHH, by an average margin of 1.9 REUs or more. (Panel B) Distributions of X29VHH–Glu67RTA interaction energy (X = Gly for V1C7, V2B9, and V2E8; X = Arg for V5C1) over the ensembles of models. While the Arg29V5C1–Glu67RTA interaction is favorable (−3.2 REUs on average), the Gly29VHH–Glu67RTA interaction is essentially absent (0.0 REUs on average).
To elucidate the structural basis for the observed differences in binding affinity among the V1C7 family of VHHs, we considered four representative models selected by clustering each ensemble on the basis of structural similarity. Using the four representative models we decomposed the VHH–RTA interaction energy to pairwise residue interactions, and identified the interactions that were ranked most favorable. As expected, a number of favorable interactions that were common among all four VHHs were identified. For example, the interaction between Asp59VHH and Arg197RTA was the most favorable pairwise interaction for all four VHHs, with values ranging from −4.6 to −4.4 REUs. Two other interactions, Glu114VHH–Gln160RTA and Glu114VHH–Thr163RTA, always ranked within the five most favorable interactions, while Ser/Cys58VHH–Glu145RTA and Ser55VHH–Glu146RTA always ranked among the ten most favorable interactions. For each VHH, at least eight of the ten most favorable interactions with RTA were mediated by antibody elements CDR2 or CDR3. The most conspicuous difference between V5C1 and the other VHHs was the interaction between RTA residue 67 and VHH residue 29, located within CDR1. Residue 29 is an Arg in the case of V5C1, but a Gly in V1C7, V2B9, and V2E8. While Arg29V5C1 has a favorable interaction of −3.6 REUs with Glu67RTA, no interaction with Glu67RTA occurs when VHH position 29 is occupied by a Gly (Figure 5A).
Figure 5. Computational modeling and X-ray assessment of the Arg29VHH–Glu67RTA interaction.
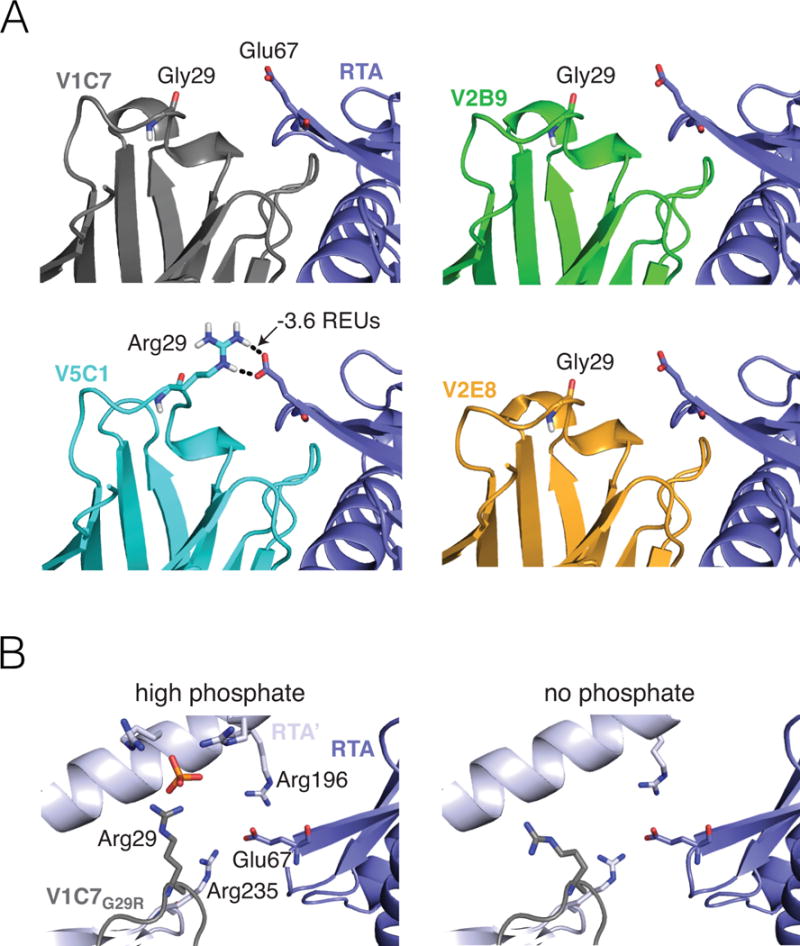
(Panel A) A representative computational model chosen by clustering the V5C1 ensemble by structural similarity has a favorable interaction of −3.6 REUs between Arg29V5C1 and Glu67RTA. Corresponding representative models for V1C7, V2B9, and V2E8 have no interaction between Gly29VHH and Glu67RTA. Broken lines indicate hydrogen bonds. (Panel B) Methodological artefacts prevent X-ray crystallography from confirming the importance of the Arg29VHH–Glu67RTA interaction in the V1C7G29R mutant. Crystallization in the presence of 1.6 M ammonium dihydrogen phosphate (left) causes Arg29VHH to interact with a phosphate molecule rather than Glu67RTA. Meanwhile, Glu67RTA engages two arginine residues (Arg196 and Arg235) from a symmetry-related RTA molecule (RTA’); this interaction is also observed under crystallization conditions devoid of phosphate (right).
We therefore examined whether the energetic gap in favor of the Arg29V5C1–Glu67RTA interaction over the Gly29VHH–Glu67RTA interaction was conserved over the entire ensembles of models (Figure 4B). We found that the presence of Gly at position 29 in VHHs V1C7, V2B9, and V2E8 resulted in no significant interaction energy with Glu67RTA (i.e., 0.0 REUs on average), as compared to a favorable energy (i.e., −3.2 REUs on average) for the Arg29V5C1–Glu67RTA interaction. Further, the magnitude of the Arg29V5C1–Glu67RTA interaction fully explains the observed difference in VHH–RTA interaction energy between V5C1 and the other V1C7 family members (Figure 4A, B). This favorable interaction between Arg29V5C1 and Glu67RTA is mostly promoted by electrostatic effects, which contribute −2.9 REUs on average. In Rosetta, electrostatic effects are further split into those from hydrogen bonding versus those from generic longer-ranged effects. Our models show that hydrogen bonding contributes 24% of the electrostatic energy between Arg29V5C1 and Glu67RTA (−0.7 REUs on average), with 97% of models containing at least one Arg29V5C1–Glu67RTA hydrogen bond. Thus, the long side chain of Arg at position 29 may allow V5C1 to effectively salt-bridge Glu67RTA while keeping the same CDR1 backbone conformation as V1C7’s.
Based on this analysis, we reasoned that if Arg at position 29 is indeed crucial to V5C1’s observed affinity for RTA, then reverting this position to Gly would be expected to worsen V5C1’s dissociation constant (KD). Conversely, assuming that V5C1 and V1C7 interact with RTA through a conserved binding mode (as in our models), we expected that introduction of a Gly29Arg mutation into V1C7 would enhance binding affinity for RTA. To examine this computationally, we applied homology modeling to V5C1R29G and V1C7G29R (Figure 6A). As expected, the ensemble of homology models built for V5C1R29G had an average VHH–RTA interaction energy 2.4 REUs worse than V5C1. Conversely, homology modeling indicated that V1C7G29R had an improvement of 2.5 REUs in average interaction energy compared to V1C7. By decomposing into pairwise energetic contributions, we found that the changes in the overall interaction energies for both V5C1R29G and V1C7G29R can be attributed to the interaction between VHH residue 29 and Glu67RTA. While this specific interaction was completely lost by V5C1R29G, costing an average of 3.2 REUs relative to V5C1 for this residue pair, it was gained by V1C7G29R, for an average improvement of 3.1 REUs relative to V1C7 (Figure 6B). These opposing energetic consequences support our prediction that the Gly29Arg mutation is both necessary and sufficient to explain the difference in RTA binding affinity between V1C7 and V5C1.
Figure 6. The Arg29VHH–Glu67RTA interaction is predicted to be both necessary and sufficient to enhance RTA binding affinity.
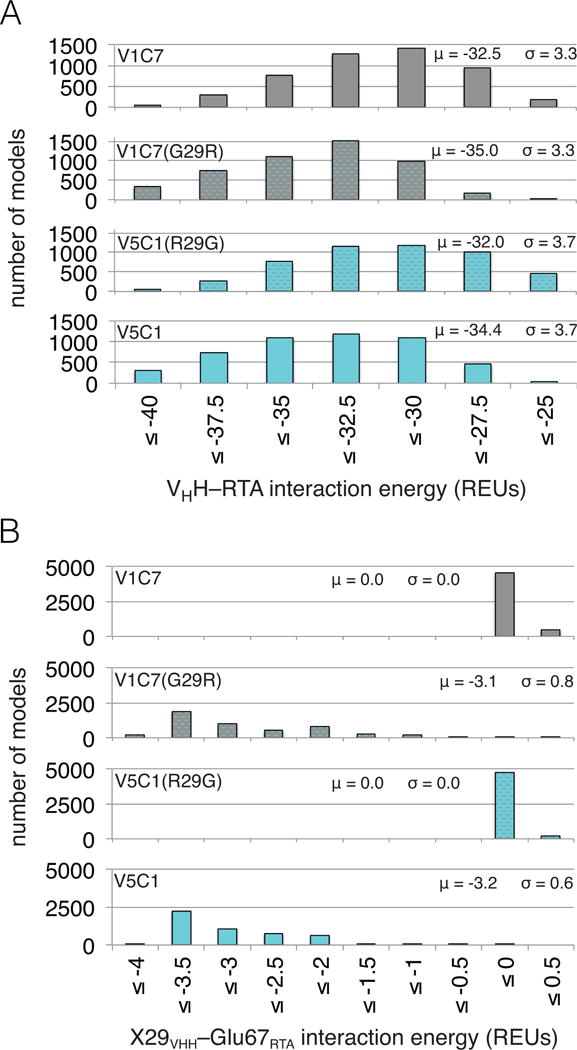
(Panel A) Distributions of VHH–RTA interaction energy over ensembles of 5,000 computational models for V1C7, V5C1, and respective single-point mutants at VHH position 29, named V1C7G29R and V5C1R29G. The Gly29Arg mutation in V1C7G29R causes an average 2.5-REU shift to more favorable interaction energies, which become more similar to those of V5C1. Conversely, because of the Arg29Gly mutation, the interaction energies of V5C1R29G shift by 2.4 REU toward less favorable values, approximating more closely those of V1C7. (Panel B) Distributions of X29VHH–Glu67RTA interaction energy over the ensembles of models; X = Gly for V1C7 and V5C1R29G, X = Arg for V5C1 and V1C7G29R. The Arg29VHH–Glu67RTA interaction of V1C7G29R is as favorable as that of V5C1 (−3.1 and −3.2 REUs on average, respectively). Such an interaction is favorable enough to explain V1C7G29R’s enhancement in VHH–RTA interaction energy (−2.5 REUs on average) and, therefore, the prediction that V1C7G29R and V5C1 have similar RTA binding affinities. On the other hand, absence of the Gly29VHH–Glu67RTA interaction (μ = 0.0) can explain, for the most part, why V1C7 and V5C1R29G are predicted to bind RTA similarly weakly.
Contribution of V5C1 residue Arg29 to RTA binding affinity and toxin-neutralizing activity
To examine the contribution of residue Arg29 to the binding affinity of V5C1 to RTA experimentally, we generated both of the V5C1R29G and V1C7G29R mutants. The engineered VHHs were expressed in E. coli and purified to homogeneity. Their binding affinity to RTA was then characterized by SPR (Figures S1–2). As shown in Table 1, the dissociation constant (KD) of the V5C1R29G mutant was reduced by >10 fold, as compared to V5C1 itself, an observation consistent with Arg29 being important in binding to RTA. Conversely, introduction of arginine at position 29 (Gly29Arg) within the context of V1C7 resulted in a >5-fold increase in binding affinity for ricin. In the Vero cell cytotoxicty assay, the toxin-neutralizing capacity of the V5C1R29G mutant was essentially abolished compared to V5C1 itself, which supports the notion that binding affinity is a critical determinant of neutralizing activity (Figure 7). Surprisingly however, the TNA of the V1C7G29R mutant was unchanged compared to V1C7, despite its having a binding affinity that is significantly improved over V1C7. Thus, differences in binding affinity cannot completely account for the observed differences in TNA within the V1C7 family of antibodies.
Figure 7. Neutralization capacity of V1C7 and V5C1 point mutants.
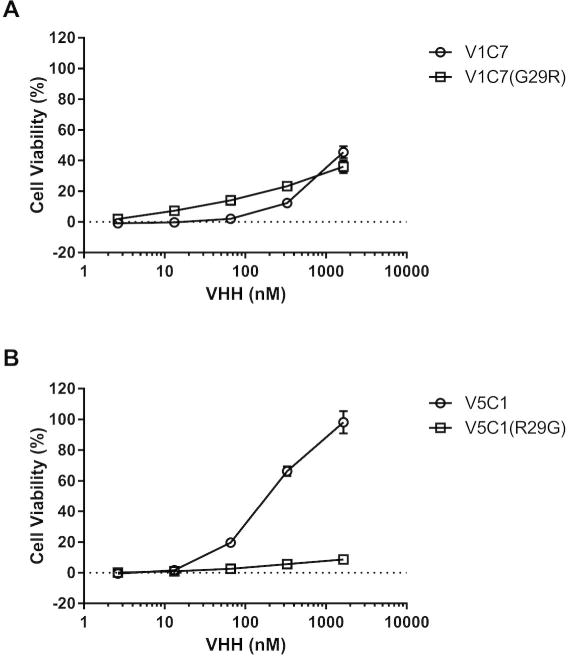
The two VHH point mutants at position 29 were compared against their parental VHHs in a Vero cell cytotoxicity assay (see Materials and Methods). (Panel A) Ricin toxin-neutralizing activities associated with V1C7 versus V1C7G29R. (Panel B) Ricin toxin-neutralizing activities associated with V5C1 versus V5C1R29G. The values shown are the average (with standard deviation) of two separate experiments each done in triplicate.
Since binding affinities cannot account for the differences in TNA between V1C7 and V5C1, we reasoned that there must be subtle differences in epitope specificity between the antibodies. To test this hypothesis, we applied HX-MS for epitope mapping studies of V1C7 and V5C1, as well as the corresponding V1C7G29R and V5C1R29G mutants. By HX-MS, we found that V1C7 strongly protected RiVax residues 60–68 (β-strand D and its adjacent β-turn) and residues 152–168 (α-helix D, α-helix E, and a β-turn between them), which is consistent with V1C7’s epitope as defined by X-ray crystallography (Figure 8; Figure S4). V5C1 had virtually identical protection profiles, except that V1C7 showed slightly more protection across two additional peptides encompassing residues 58 and 69. V5C1R29G and V1C7G29R were also indistinguishable from their parental VHHs and similar, if not identical, to each other. Thus, any differences in epitope specificity are not apparent at the resolution provided by HX-MS.
Figure 8. VHH protection analysis of RTA, as defined by HX-MS.
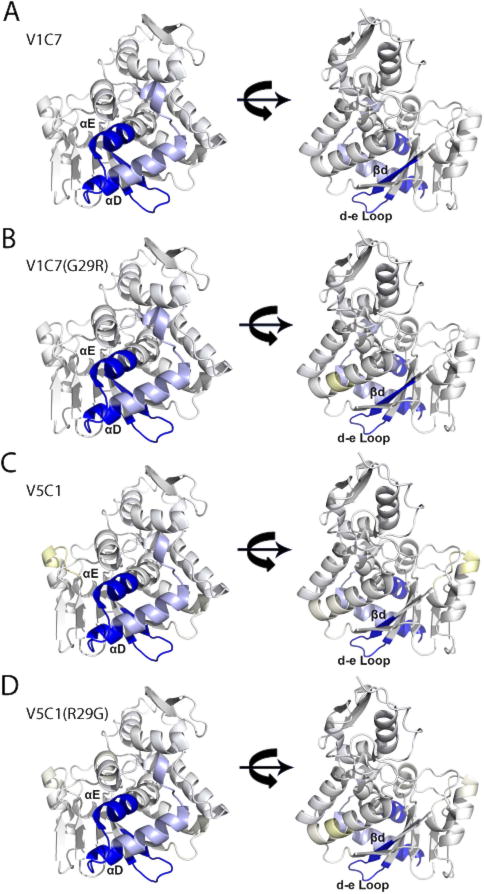
Shown are the results of HX-MS for (Panel A) V1C7, (Panel B) V1C7G29R, (Panel C) V5C1 (Panel D) V5C1R29G mapped onto the solvent accessible surface of RTA (PDB: 3SRP). Strong protection is colored dark blue, intermediate protection light blue, and intermediate deprotection light yellow, as defined in the Materials and Methods section and Supplemental Figure S4.
X-ray crystallography of V1C7G29R–RTA complexes
The results from our computational modeling studies suggested that the enhanced binding affinity of V1C7G29R for ricin is due to the interaction between the acquired Arg29 side chain and RTA’s Glu67. In an effort to corroborate this result, we solved the X-ray crystal structure of the V1C7G29R mutant in complex with RTA. Initially we solved the structure using crystallization conditions that contained 1.6 M ammonium dihydrogen phosphate (pH 8.5); however, we found that the high phosphate concentration resulted in a phosphate molecule binding on the surface of V1C7G29R and forming a salt-bridge with Arg29 (Figure 5B). The interaction between the bound phosphate and Arg29 positioned this residue ~9 Å away from Glu67RTA, thereby abrogating any potential interaction between Arg29 and Glu67RTA (Figure S5A). In this crystal structure, Glu67RTA also interacted with two arginine residues (Arg196 and Arg235) from a symmetry-related RTA molecule, further usurping the possible interaction between Arg29 and Glu67RTA (Figure S5B). We therefore solved a second crystal structure of the V1C7G29R–RTA complex using conditions devoid of phosphate (pH 4.6). The overall structures of the V1C7G29R–RTA complex with phosphate and without phosphate were very similar, as evidenced by an RMSD of 0.3 Å between the two structures. However, even in the absence of phosphate, Arg29 was still too distant (~10.5 Å) from Glu67RTA to allow salt bridge formation (Figure 5B; Figure S5C). We suspect that Arg29 was distant from Glu67RTA because this crystal form was isomorphous with the high-phosphate crystal form, thus possessed similar crystal contacts. As a result, we found that Glu67RTA interacted with the same symmetry-related RTA residues Arg196 and Arg235, indicating why we did not observe interaction between Arg29 and Glu67RTA in this crystal structure either (Figure S5D). For these reasons, the crystal structures of the V1C7G29R–RTA complex did not provide insight into the role of the Arg29VHH–Glu67RTA interaction in solution.
DISCUSSION
The V1C7 family of VHHs has proved to be particularly amenable for investigation into the relationship between binding affinity and TNA, for three reasons. First, the four family members were isolated from the same phage-displayed VHH library and, based on amino acid sequence and phylogenetic alignment, likely derived from the same progenitor B cell. For example, V1C7 and V5C1 differ by only 16 residues, and seven of these are situated within the CDRs. Despite their sequence similarities, however, the four V1C7-like antibodies display a range of binding affinities (0.4–5.7 nM) and TNAs. Second, we had recently solved the X-ray crystal structure of V1C7 in complex with RTA at 1.8 Å resolution 28, thereby providing an excellent template to be used as a starting point for homology modeling. Finally, because the V1C7 family consists of single domain antibodies that lack light-chain elements, our modeling studies were confined to a single protein–protein interface (the VHH–RTA interface). Analogous efforts directed towards a family based on a conventional mAb, such as PB10 or SyH7, would have to contend with modeling interactions of RTA with both VH and VL 59.
Integral to the success of our study was the development of a modeling approach tailored for VHHs, and to the V1C7 family in particular. Current state-of-the-art antibody structure prediction tools are typically very accurate for framework (FR) and CDR H1 and H2 elements, because there are straightforward sequence-to-structure rules to identify reliable templates 40,60,61. In contrast, accurately modeling CDR H3 elements, especially those >10 residues in length, remains a formidable challenge because their conformational variability is too large to be covered by the templates available in the PDB. These elements are generally better modeled by physics-based, or “de novo”, strategies 41,62,63. While the general principles of conventional immunoglobulin structure prediction should also apply to VHHs 42, neither established methods for VHH modeling nor rules for applying state-of-the-art methods to single domain antibodies currently exist. For these reasons, and given the high sequence similarity among V1C7 family members, we modeled the newly discovered members of this family (i.e., V2B9, V2E8, and V5C1) with the assumption that they should have the same structure as V1C7; then by using this template to generate thousands of energy-minimized VHH–RTA models, we sought to capture any small structural differences between family members.
Having successfully recognized V5C1 as the family member with highest binding affinity, this computational approach may also prove to be a generally useful tool for predicting relative binding affinity, at least among closely related antibodies. In the context of searching for potent neutralizers, computational modeling could be used to prioritize VHH sequences of high predicted affinity for subsequent experimental characterization. The successful deconstruction that allowed us to identify the Gly29Arg mutation as responsible for V5C1’s improved affinity may also inspire additional future applications. In particular, the ability to provide structural insights via computational modeling may facilitate rational design of improved VHHs, as shown here for the V1C7G29R mutant. Whether used prospectively for prediction, or retrospectively to rationalize experimental observations, this approach may accelerate epitope mapping and development of VHHs with enhanced binding affinity.
It is evident, however, from our study that the predictive power of the computational method we employed is subject to two important limitations. The first relates to sensitivity. While the method correctly separated the tighter binders (i.e., V5C1 and V1C7G29R) from the weaker binders (V1C7, V2B9, V2E8, and V5C1R29G), it was unable to capture more subtle differences in relative binding affinities. For example, we were unable to discriminate V1C7, V2B9, and V2E8 from one another, even though their experimentally-defined binding affinities differ by more than 5-fold. The accuracy needed to recapitulate these subtle differences in binding affinity may be possible through much more expensive (and extensive) computational approaches, such as free energy perturbation methods 64.
The second limitation, which is inherent in our homology modeling scheme, is the assumption that the four V1C7-like antibodies, by virtue of their similar amino acid sequences, adopt essentially the same binding mode with respect to RTA. This assumption, in retrospect, is largely validated by a number of the findings in our study. These are the close similarity between the models after energy minimization and the V1C7 template; the identification from the resulting models of the critical Arg29VHH–Glu67RTA interaction; the virtually indistinguishable HX-MS protection profiles of V1C7, V5C1, and their mutants; and finally, an X-ray structure of the V1C7G29R–RTA complex that is essentially the same as that of the V1C7–RTA complex, showing that a stronger, V5C1-like binding affinity is possible even by V1C7’s binding mode, and therefore suggesting that the same binding mode is shared by V5C1 as well.
That said, if the assumption of a common binding mode were strictly true, then the observed differences in TNA would simply be explained by differences in side-chain interactions at the interface with RTA. But the Gly29Arg mutation, although it made V1C7G29R a strong binder like V5C1, was insufficient for V1C7G29R to also become a stronger neutralizer like V5C1. This suggests that V5C1 achieves higher toxin neutralization not just through the increase of binding affinity, but also from having a slightly different epitope specificity compared to V1C7. We presently cannot quantify precisely the degree of overlap between the V1C7 and V5C1 epitopes. Nonetheless, in light of our HX-MS analysis, we expect them to overlap even more extensively than the epitopes of V1C7 and the previously identified cluster-II VHH E1, since V1C7 is far more similar in sequence to V5C1 than to E1 (Figure 1).
Unfortunately, attempts to define V5C1’s exact epitope using X-ray crystallography proved unsuccessful. Therefore, the specific interaction points between V5C1 and RTA responsible for toxin neutralization remain unknown. While we successfully employed HX-MS to localize the epitopes on RTA recognized by V5C1 and V1C7, as well as the V5C1R29G and V1C7 G29R mutants, HX-MS did not afford sufficient sensitivity to decipher different degrees of contact among the four antibodies; this is an inherent limitation of HX-MS as compared to X-ray crystallography. We postulate, however, that V5C1’s CDR3 element is responsible for the enhanced toxin-neutralizing capacity observed in the Vero cell assays. V5C1 and V1C7 differ by four amino acids over CDR3 (residues 101–117; Figure 1). Thus, we speculate that V5C1’s CDR3 assumes a slightly different conformation compared to V1C7. Indeed, it is recognized that CDR3 conformations predicted by structure-based modeling are often incorrect, even for state-of-the-art methods, because of the prevalence of non-canonical structures in this region 40. In this case, the CDR3 of V5C1 might assume an alternative conformation relative to V1C7 that allows it to engage with RTA’s α-helices D and E in a different manner. Such a conformation may be preferred over that of V1C7 because of the lack of interaction of Gln160RTA with Tyr111V5C1 and Thr113V5C1, as opposed to Ser111V1C7 and Gln113V1C7 (Figure S6). Even in the absence of a clear CDR3 conformational change, the Tyr111V5C1 side chain by virtue of its larger size, unlike Ser at that same position in V1C7, might be involved in toxin neutralization.
Members of the V1C7 family of VHHs are defined based on competitive ELISAs with SyH7, as targeting the epitope cluster II on the surface of RTA 20,28. However, all cluster II antibodies are not created equal in terms of toxin-neutralizing potential. As noted in the current study, V1C7 constitutes a weak neutralizer, while V5C1 is considered a moderate neutralizer. V5E1, a recently described cluster II VHH, on the other hand, has an IC50 of 0.5 nM and is therefore one of the most potent toxin-neutralizing antibodies described to date 28. It is not known exactly how any of the cluster II antibodies neutralize ricin toxin, although evidence suggests they may interfere with toxin uptake 28, intracellular trafficking 65, and/or liberation of RTA from RTB by protein disulfide isomerase (PDI) in the endoplasmic reticulum 66. When SyH7 was complexed with ricin and then applied to Vero cells in culture, we observed robust uptake of antibody–toxin complexes into cells but a marked reduction (>50%) in the amount of toxin that gained access to the trans-Golgi network (TGN) 65. Similarly, when tested in an in vitro PDI-mediated reduction assay, SyH7 prevented by ~75% reduction of the single disulfide bond that links RTA and RTB 66. We therefore propose that cluster II antibodies neutralize ricin by occupying a face or “hotspot” on RTA involved in interacting with host proteins that are required to mediate retrograde transport and entry of RTA into the cytoplasm. Furthermore, we speculate that antibodies like SyH7 and V5E1 target the core of the cluster II neutralizing hotspot, whereas antibodies like V1C7 and V5C1 likely bind at the fringe. Ultimately, discerning which exact residues on RTA are critical for neutralizing ricin will require the solution of additional antibody–ricin X-ray crystal structures.
As a final point of discussion, it is interesting to consider how future ricin vaccine designs might be influenced by the results presented in our study. B cell ontogeny and antibody maturation in response to protein subunit vaccines like RiVax are driven by somatic hypermutation (SHM) and affinity selection that takes place in germinal centers (GC) 67. Following a second vaccination, B cells are positively selected in the GC based on their affinity for antigen, a process that is dependent on follicular dendritic cells (FDCs) and T follicular helper cells. B cells that exit the GC following affinity maturation will become plasma cells that produce the specific antibodies found in circulation. In the case of the candidate RTA-based ricin vaccine, RiVax, the onset of high affinity ricin-specific antibodies titers with low or often undetectable neutralizing activities. Toxin-neutralizing antibodies do eventually appear, but they lag by several weeks and their appearance is not accompanied by a measurable increase in overall serum IgG9,68,69. We propose that structure-based computational modeling, similar to that employed here and in our previous studies 45, might be used to identify immunodominant epitopes on RTA that trigger the onset of high-affinity, non-neutralizing antibodies. Ablation of immunodominant non-neutralizing epitopes through a process known as “resurfacing” would be expected to result in a dampening of non-neutralizing antibody levels and a proportional increase in neutralizing titers following vaccination. Indeed, structure-based vaccine design approaches have already shown promise for subunit vaccines against respiratory syncytial virus 70, HIV 71, and a number of other medically important pathogens 72.
Supplementary Material
Acknowledgments
We gratefully acknowledge the 24-ID-C and 24-ID-E beamline staff at the Advanced Photon Source for their assistance in data collection. We thank the Wadsworth Center’s Immunology Core for assistance with SPR and Agilent Technologies for an equipment loan. This work was supported by Contract No. HHSN272201400021C from the National Institutes of Allergy and Infectious Diseases.
Footnotes
Conflicts of Interest: The authors declare that they have no conflicts of interest with the contents of this article.
Author Contributions: AB, DJV, YR, MJR, SKA, and RTT conducted experiments described in paper; AB, DJV, MJR, CRM, DBV, DDW, JK, and NJM analyzed the results and wrote the paper. All authors reviewed the results and approved the final version of the manuscript.
References
- 1.Malpede BM, Tolia NH. Malaria adhesins: structure and function. Cell Microbiol. 2014;16(5):621–631. doi: 10.1111/cmi.12276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Morales JF, Yu B, Perez G, Mesa KA, Alexander DL, Berman PW. Fragments of the V1/V2 domain of HIV-1 glycoprotein 120 engineered for improved binding to the broadly neutralizing PG9 antibody. Mol Immunol. 2016;77:14–25. doi: 10.1016/j.molimm.2016.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rappuoli R, Bottomley MJ, D’Oro U, Finco O, De Gregorio E. Reverse vaccinology 2.0: Human immunology instructs vaccine antigen design. J Exp Med. 2016;213(4):469–481. doi: 10.1084/jem.20151960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sandvig K, Skotland T, van Deurs B, Klokk TI. Retrograde transport of protein toxins through the Golgi apparatus. Histochem Cell Biol. 2013;140(3):317–326. doi: 10.1007/s00418-013-1111-z. [DOI] [PubMed] [Google Scholar]
- 5.Spooner RA, Watson PD, Marsden CJ, Smith DC, Moore KA, Cook JP, Lord JM, Roberts LM. Protein disulphide-isomerase reduces ricin to its A and B chains in the endoplasmic reticulum. Biochem J. 2004;383(Pt 2):285–293. doi: 10.1042/BJ20040742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Endo Y, Mitsui K, Motizuki M, Tsurugi K. The mechanism of action of ricin and related toxic lectins on eukaryotic ribosomes. The site and the characteristics of the modification in 28 S ribosomal RNA caused by the toxins. J Biol Chem. 1987;262(12):5908–5912. [PubMed] [Google Scholar]
- 7.Tesh VL. The induction of apoptosis by Shiga toxins and ricin. Curr Top Microbiol Immunol. 2012;357:137–178. doi: 10.1007/82_2011_155. [DOI] [PubMed] [Google Scholar]
- 8.Pittman PR, Reisler RB, Lindsey CY, Guerena F, Rivard R, Clizbe DP, Chambers M, Norris S, Smith LA. Safety and immunogenicity of ricin vaccine, RVEc, in a Phase 1 clinical trial. Vaccine. 2015;33(51):7299–7306. doi: 10.1016/j.vaccine.2015.10.094. [DOI] [PubMed] [Google Scholar]
- 9.Roy CJ, Brey RN, Mantis NJ, Mapes K, Pop IV, Pop LM, Ruback S, Killeen SZ, Doyle-Meyers L, Vinet-Oliphant HS, Didier PJ, Vitetta ES. Thermostable ricin vaccine protects rhesus macaques against aerosolized ricin: Epitope-specific neutralizing antibodies correlate with protection. Proc Natl Acad Sci U S A. 2015;112(12):3782–3787. doi: 10.1073/pnas.1502585112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vance DJ, Mantis NJ. Progress and challenges associated with the development of ricin toxin subunit vaccines. Expert review of vaccines. 2016;15(9):1213–1222. doi: 10.1586/14760584.2016.1168701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Noy-Porat T, Rosenfeld R, Ariel N, Epstein E, Alcalay R, Zvi A, Kronman C, Ordentlich A, Mazor O. Isolation of Anti-Ricin Protective Antibodies Exhibiting High Affinity from Immunized Non-Human Primates. Toxins (Basel) 2016;8(3) doi: 10.3390/toxins8030064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pincus SH, Smallshaw JE, Song K, Berry J, Vitetta ES. Passive and active vaccination strategies to prevent ricin poisoning. Toxins (Basel) 2011;3(9):1163–1184. doi: 10.3390/toxins3091163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Van Slyke G, Sully EK, Bohorova N, Bohorov O, Kim D, Pauly MH, Whaley KJ, Zeitlin L, Mantis NJ. Humanized Monoclonal Antibody That Passively Protects Mice against Systemic and Intranasal Ricin Toxin Challenge. Clin Vaccine Immunol. 2016;23(9):795–799. doi: 10.1128/CVI.00088-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Anderson GP, Liu JL, Hale ML, Bernstein RD, Moore M, Swain MD, Goldman ER. Development of antiricin single domain antibodies toward detection and therapeutic reagents. Anal Chem. 2008;80(24):9604–9611. doi: 10.1021/ac8019398. [DOI] [PubMed] [Google Scholar]
- 15.Castelletti D, Fracasso G, Righetti S, Tridente G, Schnell R, Engert A, Colombatti M. A dominant linear B-cell epitope of ricin A-chain is the target of a neutralizing antibody response in Hodgkin’s lymphoma patients treated with an anti-CD25 immunotoxin. Clin Exp Immunol. 2004;136(2):365–372. doi: 10.1111/j.1365-2249.2004.02442.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Colombatti M, Pezzini A, Colombatti A. Monoclonal antibodies against ricin: effects on toxin function. Hybridoma. 1986;5(1):9–19. doi: 10.1089/hyb.1986.5.9. [DOI] [PubMed] [Google Scholar]
- 17.Legler PM, Compton JR, Hale ML, Anderson GP, Olson MA, Millard CB, Goldman ER. Stability of isolated antibody-antigen complexes as a predictive tool for selecting toxin neutralizing antibodies. MAbs. 2017;9(1):43–57. doi: 10.1080/19420862.2016.1236882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maddaloni M, Cooke C, Wilkinson R, Stout AV, Eng L, Pincus SH. Immunological characteristics associated with the protective efficacy of antibodies to ricin. J Immunol. 2004;172(10):6221–6228. doi: 10.4049/jimmunol.172.10.6221. [DOI] [PubMed] [Google Scholar]
- 19.O’Hara JM, Neal LM, McCarthy EA, Kasten-Jolly JA, Brey RN, 3rd, Mantis NJ. Folding domains within the ricin toxin A subunit as targets of protective antibodies. Vaccine. 2010;28:7035–7046. doi: 10.1016/j.vaccine.2010.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.O’Hara JM, Kasten-Jolly JC, Reynolds CE, Mantis NJ. Localization of non-linear neutralizing B cell epitopes on ricin toxin’s enzymatic subunit (RTA) Immunol Lett. 2014;158(1–2):7–13. doi: 10.1016/j.imlet.2013.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vance DJ, Tremblay JM, Mantis NJ, Shoemaker CB. Stepwise engineering of heterodimeric single domain camelid VHH antibodies that passively protect mice from ricin toxin. J Biol Chem. 2013;288(51):36538–36547. doi: 10.1074/jbc.M113.519207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lemley PV, Amanatides P, Wright DC. Identification and characterization of a monoclonal antibody that neutralizes ricin toxicity in vitro and in vivo. Hybridoma. 1994;13(5):417–421. doi: 10.1089/hyb.1994.13.417. [DOI] [PubMed] [Google Scholar]
- 23.Lebeda FJ, Olson MA. Prediction of a conserved, neutralizing epitope in ribosome-inactivating proteins. Int J Biol Macromol. 1999;24(1):19–26. doi: 10.1016/s0141-8130(98)00059-2. [DOI] [PubMed] [Google Scholar]
- 24.Vance DJ, Mantis NJ. Resolution of two overlapping neutralizing B cell epitopes within a solvent exposed, immunodominant alpha-helix in ricin toxin’s enzymatic subunit. Toxicon. 2012;60(5):874–877. doi: 10.1016/j.toxicon.2012.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dai J, Zhao L, Yang H, Guo H, Fan K, Wang H, Qian W, Zhang D, Li B, Wang H, Guo Y. Identification of a novel functional domain of ricin responsible for its potent toxicity. J Biol Chem. 2011;286(14):12166–12171. doi: 10.1074/jbc.M110.196584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rudolph MJ, Vance DJ, Cassidy MS, Rong Y, Shoemaker CB, Mantis NJ. Structural analysis of nested neutralizing and non-neutralizing B cell epitopes on ricin toxin’s enzymatic subunit. Proteins. 2016;84(8):1162–1172. doi: 10.1002/prot.25062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rudolph MJ, Vance DJ, Cheung J, Franklin MC, Burshteyn F, Cassidy MS, Gary EN, Herrera C, Shoemaker CB, Mantis NJ. Crystal Structures of Ricin Toxin’s Enzymatic Subunit (RTA) in Complex with Neutralizing and Non-Neutralizing Single-Chain Antibodies. J Mol Biol. 2014 doi: 10.1016/j.jmb.2014.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rudolph MJ, Vance DJ, Cassidy MS, Rong Y, Mantis NJ. Structural Analysis of Single Domain Antibodies Bound to a Second Neutralizing Hot Spot on Ricin Toxin’s Enzymatic Subunit. J Biol Chem. 2017;292(3):872–883. doi: 10.1074/jbc.M116.758102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dai K, Khan SN, Wang Y, He L, Guenaga J, Ingale J, Sundling C, O’Dell S, McKee K, Phad G, Corcoran M, Wilson R, Mascola JR, Zhu J, Li Y, Karlsson Hedestam GB, Wyatt RT. HIV-1 Vaccine-elicited Antibodies Reverted to Their Inferred Naive Germline Reveal Associations between Binding Affinity and in vivo Activation. Sci Rep. 2016;6:20987. doi: 10.1038/srep20987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nowakowski A, Wang C, Powers DB, Amersdorfer P, Smith TJ, Montgomery VA, Sheridan R, Blake R, Smith LA, Marks JD. Potent neutralization of botulinum neurotoxin by recombinant oligoclonal antibody. Proc Natl Acad Sci U S A. 2002;99(17):11346–11350. doi: 10.1073/pnas.172229899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu H, Pfarr DS, Tang Y, An LL, Patel NK, Watkins JD, Huse WD, Kiener PA, Young JF. Ultra-potent antibodies against respiratory syncytial virus: effects of binding kinetics and binding valence on viral neutralization. J Mol Biol. 2005;350(1):126–144. doi: 10.1016/j.jmb.2005.04.049. [DOI] [PubMed] [Google Scholar]
- 32.Luo L, Luo Q, Guo L, Lv M, Lin Z, Geng J, Li X, Li Y, Shen B, Qiao C, Feng J. Structure-based affinity maturation of a chimeric anti-ricin antibody C4C13. J Biomol Struct Dyn. 2014;32(3):416–423. doi: 10.1080/07391102.2013.771380. [DOI] [PubMed] [Google Scholar]
- 33.Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol. 1993;234(3):779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 34.Rost B. Twilight zone of protein sequence alignments. Protein Eng. 1999;12(2):85–94. doi: 10.1093/protein/12.2.85. [DOI] [PubMed] [Google Scholar]
- 35.Modi V, Xu Q, Adhikari S, Dunbrack RL. Assessment of template-based modeling of protein structure in CASP11. Proteins. 2016 doi: 10.1002/prot.25049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roy A, Kucukural A, Zhang Y. I-TASSER: a unified platform for automated protein structure and function prediction. Nat Protoc. 2010;5(4):725–738. doi: 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dunbar J, Krawczyk K, Leem J, Marks C, Nowak J, Regep C, Georges G, Kelm S, Popovic B, Deane CM. SAbPred: a structure-based antibody prediction server. Nucleic Acids Res. 2016;44(W1):W474–478. doi: 10.1093/nar/gkw361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Leem J, Dunbar J, Georges G, Shi J, Deane CM. ABodyBuilder: Automated antibody structure prediction with data-driven accuracy estimation. MAbs. 2016;8(7):1259–1268. doi: 10.1080/19420862.2016.1205773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marcatili P, Rosi A, Tramontano A. PIGS: automatic prediction of antibody structures. Bioinformatics. 2008;24(17):1953–1954. doi: 10.1093/bioinformatics/btn341. [DOI] [PubMed] [Google Scholar]
- 40.Teplyakov A, Luo J, Obmolova G, Malia TJ, Sweet R, Stanfield RL, Kodangattil S, Almagro JC, Gilliland GL. Antibody modeling assessment II. Structures and models Proteins. 2014;82(8):1563–1582. doi: 10.1002/prot.24554. [DOI] [PubMed] [Google Scholar]
- 41.Weitzner BD, Kuroda D, Marze N, Xu J, Gray JJ. Blind prediction performance of RosettaAntibody 3.0: grafting, relaxation, kinematic loop modeling, and full CDR optimization. Proteins. 2014;82(8):1611–1623. doi: 10.1002/prot.24534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Steeland S, Puimege L, Vandenbroucke RE, Van Hauwermeiren F, Haustraete J, Devoogdt N, Hulpiau P, Leroux-Roels G, Laukens D, Meuleman P, De Vos M, Libert C. Generation and characterization of small single domain antibodies inhibiting human tumor necrosis factor receptor 1. J Biol Chem. 2015;290(7):4022–4037. doi: 10.1074/jbc.M114.617787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smallshaw JE, Richardson JA, Pincus S, Schindler J, Vitetta ES. Preclinical toxicity and efficacy testing of RiVax, a recombinant protein vaccine against ricin. Vaccine. 2005;23(39):4775–4784. doi: 10.1016/j.vaccine.2005.04.037. [DOI] [PubMed] [Google Scholar]
- 44.Legler PM, Brey RN, Smallshaw JE, Vitetta ES, Millard CB. Structure of RiVax: a recombinant ricin vaccine. Acta Crystallogr D Biol Crystallogr. 2011;67(Pt 9):826–830. doi: 10.1107/S0907444911026771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wahome N, Sully E, Singer C, Thomas JC, Hu L, Joshi SB, Volkin DB, Fang J, Karanicolas J, Jacobs DJ, Mantis NJ, Middaugh CR. Novel Ricin Subunit Antigens With Enhanced Capacity to Elicit Toxin-Neutralizing Antibody Responses in Mice. J Pharm Sci. 2016;105(5):1603–1613. doi: 10.1016/j.xphs.2016.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Needleman SB, Wunsch CD. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J Mol Biol. 1970;48(3):443–453. doi: 10.1016/0022-2836(70)90057-4. [DOI] [PubMed] [Google Scholar]
- 47.Rice P, Longden I, Bleasby A. EMBOSS: the European Molecular Biology Open Software Suite. Trends in genetics : TIG. 2000;16(6):276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- 48.Tyka MD, Keedy DA, Andre I, DiMaio F, Song Y, Richardson DC, Richardson JS, Baker D. Alternate States of Proteins Revealed by Detailed Energy Landscape Mapping. J Mol Biol. 2011;405(2):607–618. doi: 10.1016/j.jmb.2010.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Glasoe PK, Long FA. Use of Glass Electrodes to Measure Acidities in Deuterium Oxide 1,2. The Journal of Physical Chemistry. 1960;64(1):188–190. [Google Scholar]
- 50.Wang L, Pan H, Smith DL. Hydrogen exchange-mass spectrometry: optimization of digestion conditions. Mol Cell Proteomics. 2002;1(2):132–138. doi: 10.1074/mcp.m100009-mcp200. [DOI] [PubMed] [Google Scholar]
- 51.Majumdar R, Manikwar P, Hickey JM, Arora J, Middaugh CR, Volkin DB, Weis DD. Minimizing carry-over in an online pepsin digestion system used for the H/D exchange mass spectrometric analysis of an IgG1 monoclonal antibody. J Am Soc Mass Spectrom. 2012;23(12):2140–2148. doi: 10.1007/s13361-012-0485-9. [DOI] [PubMed] [Google Scholar]
- 52.Rusinga FI, Weis DD. Automated strong cation exchange cleanup to remove macromolecular crowding agents for protein hydrogen exchange mass spectrometry. Anal Chem. 2016 doi: 10.1021/acs.analchem.6b04057. [DOI] [PubMed] [Google Scholar]
- 53.Otwinowski Z, Minor W. Processing of x-ray diffraction data collected in oscillation mode. Methods in Enzymology. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 54.Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AG, McCoy A, McNicholas SJ, Murshudov GN, Pannu NS, Potterton EA, Powell HR, Read RJ, Vagin A, Wilson KS. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr. 2011;67(Pt 4):235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40(Pt 4):658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 4):486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Leaver-Fay A, Tyka M, Lewis SM, Lange OF, Thompson J, Jacak R, Kaufman K, Renfrew PD, Smith CA, Sheffler W, Davis IW, Cooper S, Treuille A, Mandell DJ, Richter F, Ban YE, Fleishman SJ, Corn JE, Kim DE, Lyskov S, Berrondo M, Mentzer S, Popovic Z, Havranek JJ, Karanicolas J, Das R, Meiler J, Kortemme T, Gray JJ, Kuhlman B, Baker D, Bradley P. ROSETTA3: an object-oriented software suite for the simulation and design of macromolecules. Methods Enzymol. 2011;487:545–574. doi: 10.1016/B978-0-12-381270-4.00019-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sully EK, Whaley KJ, Bohorova N, Bohorov O, Goodman C, Kim do H, Pauly MH, Velasco J, Hiatt E, Morton J, Swope K, Roy CJ, Zeitlin L, Mantis NJ. Chimeric plantibody passively protects mice against aerosolized ricin challenge. Clin Vaccine Immunol. 2014;21(5):777–782. doi: 10.1128/CVI.00003-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Morea V, Lesk AM, Tramontano A. Antibody modeling: implications for engineering and design. Methods. 2000;20(3):267–279. doi: 10.1006/meth.1999.0921. [DOI] [PubMed] [Google Scholar]
- 61.North B, Lehmann A, Dunbrack RL., Jr A new clustering of antibody CDR loop conformations. J Mol Biol. 2011;406(2):228–256. doi: 10.1016/j.jmb.2010.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mandell DJ, Coutsias EA, Kortemme T. Sub-angstrom accuracy in protein loop reconstruction by robotics-inspired conformational sampling. Nat Methods. 2009;6(8):551–552. doi: 10.1038/nmeth0809-551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Weitzner BD, Gray JJ. Accurate Structure Prediction of CDR H3 Loops Enabled by a Novel Structure-Based C-Terminal Constraint. J Immunol. 2017;198(1):505–515. doi: 10.4049/jimmunol.1601137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Clark AJ, Gindin T, Zhang B, Wang L, Abel R, Murret CS, Xu F, Bao A, Lu NJ, Zhou T, Kwong PD, Shapiro L, Honig B, Friesner RA. Free Energy Perturbation Calculation of Relative Binding Free Energy between Broadly Neutralizing Antibodies and the gp120 Glycoprotein of HIV-1. J Mol Biol. 2017;429(7):930–947. doi: 10.1016/j.jmb.2016.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yermakova A, Klokk TI, O’Hara JM, Cole R, Sandvig K, Mantis NJ. Neutralizing Monoclonal Antibodies against Disparate Epitopes on Ricin Toxin’s Enzymatic Subunit Interfere with Intracellular Toxin Transport. Sci Rep. 2016;6:22721. doi: 10.1038/srep22721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.O’Hara JM, Mantis NJ. Neutralizing monoclonal antibodies against ricin’s enzymatic subunit interfere with protein disulfide isomerase-mediated reduction of ricin holotoxin in vitro. J Immunol Methods. 2013;395(1–2):71–78. doi: 10.1016/j.jim.2013.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mesin L, Ersching J, Victora GD. Germinal Center B Cell Dynamics. Immunity. 2016;45(3):471–482. doi: 10.1016/j.immuni.2016.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.O’Hara JM, Brey RN, 3rd, Mantis NJ. Comparative efficacy of two leading candidate ricin toxin a subunit vaccines in mice. Clin Vaccine Immunol. 2013;20(6):789–794. doi: 10.1128/CVI.00098-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Vance DJ, Greene CJ, Rong Y, Mandell LM, Connell TD, Mantis NJ. Comparative Adjuvant Effects of Type II Heat-Labile Enterotoxins in Combination with Two Different Candidate Ricin Toxin Vaccine Antigens. Clin Vaccine Immunol. 2015;22(12):1285–1293. doi: 10.1128/CVI.00402-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Joyce MG, Zhang B, Ou L, Chen M, Chuang GY, Druz A, Kong WP, Lai YT, Rundlet EJ, Tsybovsky Y, Yang Y, Georgiev IS, Guttman M, Lees CR, Pancera M, Sastry M, Soto C, Stewart-Jones GB, Thomas PV, Van Galen JG, Baxa U, Lee KK, Mascola JR, Graham BS, Kwong PD. Iterative structure-based improvement of a fusion-glycoprotein vaccine against RSV. Nat Struct Mol Biol. 2016;23(9):811–820. doi: 10.1038/nsmb.3267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Burton DR, Mascola JR. Antibody responses to envelope glycoproteins in HIV-1 infection. Nat Immunol. 2015;16(6):571–576. doi: 10.1038/ni.3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Du L, Tai W, Yang Y, Zhao G, Zhu Q, Sun S, Liu C, Tao X, Tseng CK, Perlman S, Jiang S, Zhou Y, Li F. Introduction of neutralizing immunogenicity index to the rational design of MERS coronavirus subunit vaccines. Nat Commun. 2016;7:13473. doi: 10.1038/ncomms13473. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


