Significance
Therapies that activate the host immune system have shown tremendous promise for a variety of solid tumors. However, in most cancer types, fewer than half of patients respond to these immunotherapies. We propose epigenetic therapy as a mechanism to sensitize tumors to immune checkpoint therapy. We have shown that inhibiting DNA methylation triggers a viral defense pathway in tumors. Here we show that epigenetic therapy in a mouse model of ovarian cancer increases the numbers of activated immune cells, and that this is dependent on the interferon antiviral response. The combination of epigenetic therapy and immune checkpoint blockade leads to the greatest reduction in tumor burden and increase in survival, and may hold the greatest promise for patients.
Keywords: 5-azacytidine, histone deacetylase inhibitors, type I interferon, ovarian cancer, immunosuppression
Abstract
Ovarian cancer is the most lethal of all gynecological cancers, and there is an urgent unmet need to develop new therapies. Epithelial ovarian cancer (EOC) is characterized by an immune suppressive microenvironment, and response of ovarian cancers to immune therapies has thus far been disappointing. We now find, in a mouse model of EOC, that clinically relevant doses of DNA methyltransferase and histone deacetylase inhibitors (DNMTi and HDACi, respectively) reduce the immune suppressive microenvironment through type I IFN signaling and improve response to immune checkpoint therapy. These data indicate that the type I IFN response is required for effective in vivo antitumorigenic actions of the DNMTi 5-azacytidine (AZA). Through type I IFN signaling, AZA increases the numbers of CD45+ immune cells and the percentage of active CD8+ T and natural killer (NK) cells in the tumor microenvironment, while reducing tumor burden and extending survival. AZA also increases viral defense gene expression in both tumor and immune cells, and reduces the percentage of macrophages and myeloid-derived suppressor cells in the tumor microenvironment. The addition of an HDACi to AZA enhances the modulation of the immune microenvironment, specifically increasing T and NK cell activation and reducing macrophages over AZA treatment alone, while further increasing the survival of the mice. Finally, a triple combination of DNMTi/HDACi plus the immune checkpoint inhibitor α-PD-1 provides the best antitumor effect and longest overall survival, and may be an attractive candidate for future clinical trials in ovarian cancer.
Ovarian carcinoma is the leading cause of death from gynecological malignancies in the United States (1). Epithelial ovarian cancer is characterized by an immunosuppressive microenvironment (2), and patient responses to immunotherapy have been disappointing (3). In particular, the response of ovarian cancer to the immune checkpoint inhibitors α-PD-1 or α-PD-L1 has thus far been modest in comparison with the robust responses observed for melanoma, non-small cell lung cancer, and renal cell cancers (4). New treatment strategies are needed to reverse the immunosuppressive microenvironment of ovarian cancer and sensitize these tumors to immune checkpoint blockade.
One promising approach for reversing the tumor immune evasion phenotype and improving the efficacy of immune checkpoint therapy of cancers in general and ovarian cancers in particular is epigenetic therapy. The first suggestion for this approach came from a clinical observation in trials involving patients with advanced, pretreated non-small cell lung cancer (NSCLC) (5). These individuals initially received the DNA methyltransferase inhibitor (DNMTi) Vidaza (5-azacytidine; AZA) and the histone deacetylase inhibitor (HDACi) entinostat (MS275). This coupling was based on preclinical data that DNMTis and HDACis in combination enhance reactivation of aberrantly silenced genes in tumor cells and cause reductions in tumor burden that are more effective with the combined agents (6–9). After disease progression, these patients entered the first trials of immune checkpoint therapy for NSCLC, and several of these patients had durable responses compared with those who received the immunotherapy alone (10). Clinical trials are being conducted to validate this preliminary observation (10), but preclinical work to optimize this combinatorial approach of HDACis and DNMTis has not been explored.
Compelling data are emerging to show that this combinatorial approach in preclinical studies induces immune signaling in both tumor and immune cells. Studies by our group and others have shown that DNMTis can up-regulate a wide range of genes involved in immune signaling in breast, lung, colon, and ovarian cancer cells (11–15). A particularly important component of this in ovarian and colon cancer cells is that DNMTis can induce an up-regulation of a cytosolic sensing double-stranded RNA (dsRNA) antiviral pathway which results in type I IFN production and downstream signaling for up-regulation of IFN-stimulated genes, including immune cell attracting chemokines and cytokines (11, 14). Furthermore, epigenetic changes, specifically de novo methylation, have been shown to play an important role in the promotion of CD8+ T-cell exhaustion, which is a barrier to immune cell activation during checkpoint inhibition therapy (16). While this study showed that demethylation is important for T-cell reactivation, our data in the present study show that treatment of isolated tumor cells with DNMT inhibitors is sufficient to increase immune cell numbers in the tumor microenvironment. Taken together, both the tumor and the immune microenvironment are responsive to epigenetic therapy.
Others have utilized mouse models of ovarian cancer to explore how epigenetic therapy affects the response to immune checkpoint blockade, finding that the DNMTi decitabine can increase activated immune cells in ovarian ascites and sensitize tumors to α-CTLA4 (cytotoxic T lymphocyte-associated protein 4) (14). Similarly, another study showed that epigenetic therapy induced tumor expression of CXCL10, which led to an increase in T effector cells in the tumor microenvironment and to an improved response to α-PD-L1 therapy (15). However, the precise mechanisms underlying their findings and the link to a DNMTi/HDACi–induced antiviral, cellular response remain to be clarified.
Recently, using human NSCLC cells and a mouse model of this disease, our group has approached this question by trying to elucidate the best HDACis to use based on their pharmacologic half-lives and Ki effects on key members of the HDAC family. Moreover, we have optimized the ability to combine DNMTis and HDACis to develop a chronic, tolerable, low-dose schema to keep constant pressure on reversing not only DNA methylation abnormalities but the repressive chromatin events which accompany these. A potent regimen has been developed for inducing preclinical antitumor responses and reversing tumor evasion (17). We now employ this improved regimen in a mouse model of ovarian cancer, outlining not only a potent potential paradigm for an ovarian cancer clinical trial combining epigenetic with immune checkpoint therapy but, importantly, mechanistically tying induction of viral defense to the above antitumor responses by finding, in a mouse model of ovarian cancer, that DNMTis and HDACis increase both the number and activation of immune cells in the tumor microenvironment, reduce tumor burden, and extend overall survival of the mice. This modulation of the tumor microenvironment stems in part from an AZA-induced, IFN-mediated, up-regulation of an immune gene signature including genes involved in viral defense, chemokines, cytokines, IFN signaling, and cancer testis antigens (11, 13). We also now verify that type I IFN signaling caused by AZA-induced immune signaling is required for this drug to increase the number of CD45+ immune cells in the tumor microenvironment and activation of CD8+ T cells and natural killer (NK) cells. Moreover, we demonstrate that when HDACis are combined with AZA in vivo, these drugs act on the immune compartment to further improve the activation of CD8+ T cells and NK cells and to decrease myeloid cells to create a less immunosuppressed tumor microenvironment. Together, the actions of these drugs on tumor cells and the tumor microenvironment indicate that combination epigenetic therapy may offer an approach to optimize immune checkpoint therapy for patients with ovarian cancer, and that AZA, the HDACi givinostat (ITF2357; ITF), and α-PD-1 may hold the most promise.
Results
Pretreatment of Tumor Epithelial Cells ex Vivo.
To study how DNMTis and HDACis directly affect tumor epithelial cells to regulate response and immune cell interactions, we pretreated cultured, syngeneic mouse ovarian surface epithelial cancer cells, ID8-VEGF-Defensin (18–21), with epigenetic agents, injected the cells into untreated mice, and analyzed ascites volume as a measure of tumor burden (22). The ID8 parental cells were derived from ovarian surface epithelium instead of from the fallopian tube, which is hypothesized to be the origin of most ovarian tumors (23, 24). While genetic models of the development of ovarian cancer in the fallopian tube exist (25, 26), the ID8 model is one of very few routinely available syngeneic murine models of ovarian cancer (27) that can be transplanted into immunocompetent mice and creates an immunosuppressed tumor microenvironment (22), making it well suited to our studies. Parental ID8 cells (21) were modified to overexpress VEGF and Defensin (18, 20) to make them more aggressive and more like some human ovarian cancers (28, 29). The ID8-VEGF-Defensin cells also consistently generate hemorrhagic ascites ∼4 wk after transplantation of only 250,000 cells. The ascites generated by the ID8-VEGF-Defensin cells is easily drained for analysis of tumor and immune cells, and is representative of tumor burden (18, 22, 30–34). The number of tumor cells in the ascites fluid correlates positively with the volume drained, while the number of immune cells is inversely correlated, showing that the increase in tumor cells is not a general increase in cellularity of the fluid (Fig. S1 A and B).
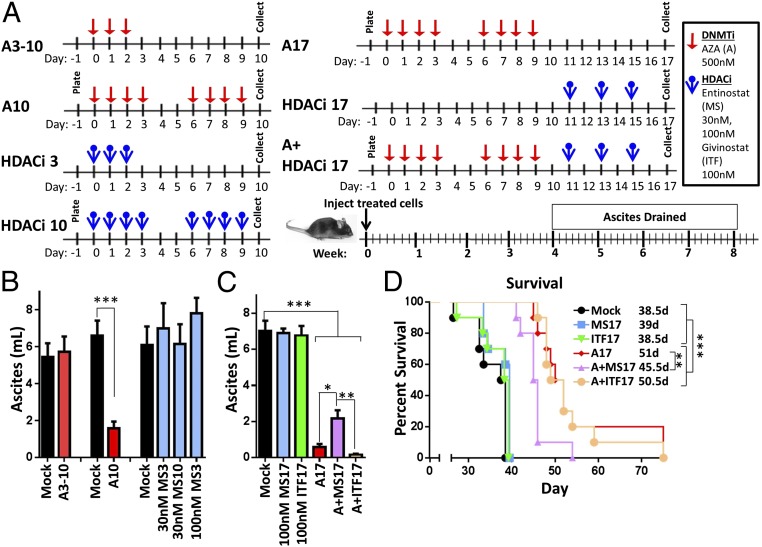
Ten or 17 (A10, A17), but not 3 (A3-10), days of AZA pretreatment of tumor cells (Fig. 1A) led to significantly less ascites (Fig. 1B), reflected as a reduction in weight gain (Fig. S1C) and increased mouse survival compared with vehicle (mock)- or HDACi-pretreated tumor cells (Fig. 1 and Fig. S1 C and D). Combination pretreatment of AZA and entinostat (A+MS17) or AZA and givinostat (A+ITF17) decreased ascites compared with mock, but did not decrease ascites (Fig. 1C) or improve survival (Fig. 1D) compared with AZA treatment alone. The slight antagonistic effect of AZA+MS17 compared with AZA alone is not observed when the whole mouse is treated, and may be due to effects on the tumor cells that are not observed in an in vivo, immunocompetent setting. Overall, the decrease in ascites volume and increase in survival with AZA or AZA+HDACi pretreatment appear to be driven by an AZA-mediated effect on tumor cells.
Fig. 1.
Pretreatment of tumor epithelial cells with AZA and transplantation into untreated C57BL/6 mice lead to decreased tumor-associated ascites and increased overall survival. (A) Treatment schematic for in vitro treatment of cultured ID8-VEGF-Defensin cells (AZA, 500 nM; MS275, 30 or 100 nM; ITF, 100 nM). A, AZA; ITF, givinostat; MS, entinostat. (B and C) Ascites volume drained from mice 4 to 5 wk after pretreated tumor injection. Mean ± SEM is shown. A10, MS3, MS10: n = 7 to 30 mice, three biological replicates; A3-10: n = 9 mice, two biological replicates; MS17, ITF17, A17, A+MS17, and A+ITF17: n = 9 or 10 mice, one biological replicate. Statistical outliers were removed using Peirce’s criterion, and significance was determined by Mann–Whitney t test. (D) Survival of mice in days, with median survival shown. Significance was determined using a log-rank (Mantel–Cox) test. *P < 0.05, **P < 0.01, ***P < 0.001.
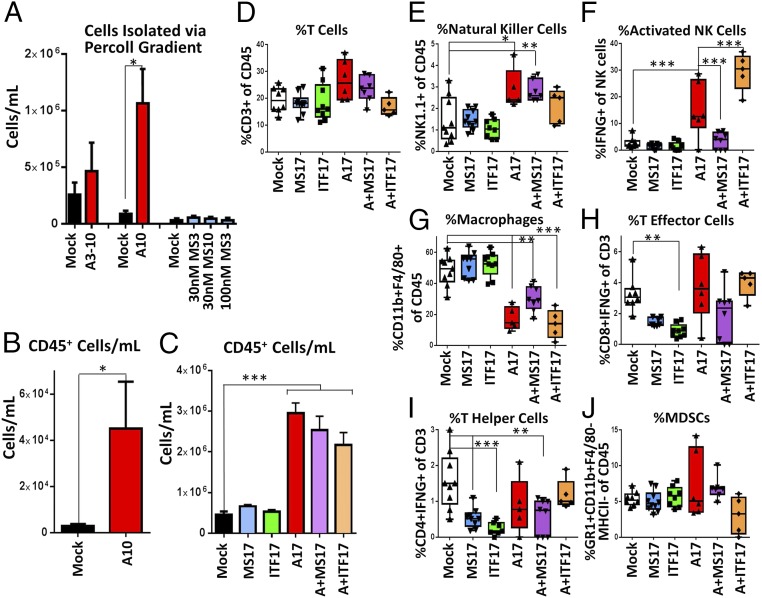
AZA pretreatment of tumor epithelial cells in these ex vivo, pretreatment studies led to changes in the immune microenvironment, with increased numbers of immune cells (CD45+) in the ascites of A10 pretreated tumor cells (Fig. 2 A and B). Entinostat or givinostat treatment of cultured tumor cells did not alter the number of CD45+ cells per mL (Fig. 2C), but did lead to decreases in the percentage of T effector and T helper cells (Fig. 2 I and J) in the ascites. Neither the addition of givinostat (A+ITF17) nor entinostat (A+MS17) to AZA increased the number of CD45+ cells above AZA treatment alone (Fig. 2C); however, there were some significant changes in the percentage of activated NK cells (Fig. 2F). These changes are particularly interesting in light of the fact that the immune cells, associated with the tumor microenvironment, were not treated in the ex vivo, pretreatment model.
Fig. 2.
Pretreatment of tumor epithelial cells with AZA and an HDACi leads to alterations in the numbers and activation of immune cell populations in tumor-associated ascites. ID8-VEGF-Defensin cells were pretreated (AZA, 500 nM; entinostat, 30 or 100 nM; givinostat, 100 nM) and injected into mice. Cells were analyzed from the drained ascites fluid (Fig. 1 A–C). (A) Immune cells per mL separated via Percoll gradient (n = 6 to 12 mice, two or three biological replicates). (B) CD45+ cells per mL identified via Percoll gradient and FACS (n = 6 to 11 mice, two biological replicates). Mean ± SEM is shown, and significances were determined by Mann–Whitney t test. (C–J) All cells from ascites were analyzed via FACS (n = 5 to 9 mice, one biological replicate). Mean ± SEM is shown, and significances were determined by one-way ANOVA. (C) CD45+ cells per mL of ascites. (D–J) Response of immune cell subpopulations to tumor cells pretreated ex vivo with AZA (A10). *P < 0.05, **P < 0.01, ***P < 0.001.
AZA-Induced Immune Signaling in Tumor Cells.
Treatment with AZA at doses that degrade its molecular target, DNA methyltransferase 1, in ID8-VEGF-Defensin cells (Fig. S1 E–G) caused an up-regulation of an immune-related viral defense signature in these murine cells, as was previously described for human ovarian cancer cells (11) (Fig. S2A). This expression, also seen in cancer testis antigens (CTAs), was especially robust with prolonged treatment (Fig. S2 A and B). HDACis have been shown to synergize with DNMT inhibitors to reexpress silenced genes in cancer (7, 9), but entinostat (MS275) or givinostat (ITF) treatment (HDACi17) alone or in combination with AZA caused only small or moderate changes in the expression of the antiviral and CTA genes in these mouse ovarian cancer cells (Fig. S2 C and D).
We and others have previously demonstrated that AZA treatment of human ovarian carcinoma cell lines induced the expression of RNA from endogenous retroviruses (ERVs), which led to increased IFN signaling and viral defense gene expression (11, 14). We now demonstrate in mouse ovarian ID8-VEGF-Defensin cells that AZA significantly increased several mERVs in both cultured tumor cells (Fig. S2E) and tumor cells sorted from ascites from treated C57BL/6 mice (Figs. S2F and S4A and Table S1). While the mERVs are increased early in this treatment (day 3), they sharply decrease at later time points (days 4, 7, and 10). This is reminiscent of the increase and subsequent decrease in ERV transcripts observed in ref. 11. We hypothesize that antiviral proteins up-regulated by the IFN response may destroy the mERV RNA. Interestingly, in vivo (Fig. S2F), we observe a long-term increase in ERV transcripts. We have previously shown that these ERVs, silenced by DNA methylation, lose methylation and increase transcription upon DNMTi treatment (11).
Lastly, AZA treatment in vitro (A10) and in vivo (Fig. S4A) altered the secreted protein levels of chemokines and cytokines. In an array of 40 chemokines and cytokines, CD54, IL-1RA, CXCL10, CCL2, and CCL5 were significantly up-regulated in vitro, while CXCL12 was down-regulated. In the ascites fluid of AZA-treated mice, IL-1RA was significantly down-regulated, while CXCL10 and CXCL1 were significantly up-regulated. Only CXCL10 was significantly increased by AZA in both the media and the ascites fluid (Fig. S2 G and H).
Treatment of Mice with AZA, HDACi, and α-PD-1.
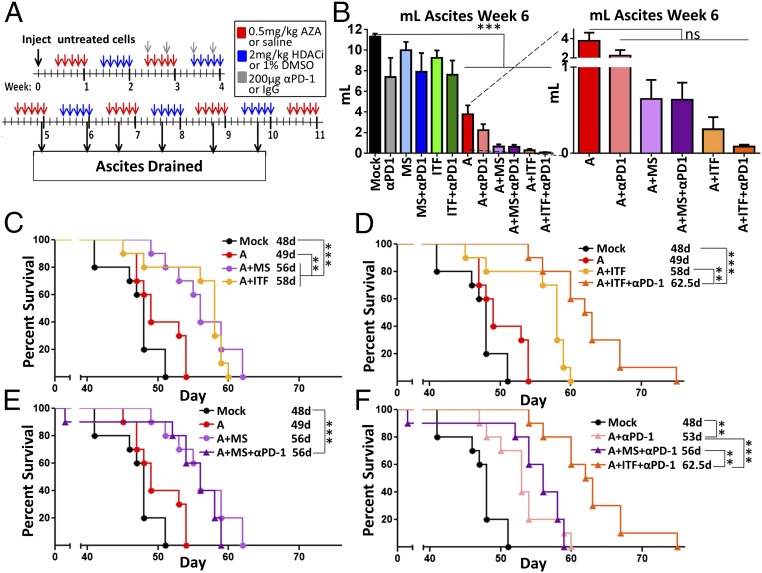
Having shown that pretreatment of ovarian tumor cells led to increased immune cells in the tumor microenvironment and improved survival of the mice, we next asked whether the addition of α-PD-1 would provide added benefit. Mice bearing ovarian tumors treated intraperitoneally with AZA, an HDACi, and α-PD-1 had improved overall survival, decreased tumor burden, and alterations of immune cell populations that would promote immune cell killing of tumor cells (Figs. 3 and 4). AZA as a single agent or in combination with either HDACi or α-PD-1 significantly reduced ascites volume (Fig. 3B, expansion), while HDACis or HDACis+α-PD-1 were ineffective (Fig. 3B). Mirroring these ascites data, α-PD-1, HDACis alone, or HDACis plus α-PD-1 did not affect survival (Fig. S3B), while the combination of AZA with either HDACi significantly improved this key parameter over AZA treatment alone (Fig. 3C). This is in contrast to the ex vivo treatment model, where the addition of HDACi to AZA did not affect the tumor burden or overall survival (Fig. 1 C and D). The improved survival with in vivo treatment may reflect the effects of the epigenetic drugs on the host immune cells. Adding α-PD-1 to AZA+ITF further significantly increased survival over AZA+ITF or AZA+MS+α-PD-1 (Fig. 3 D–F). In summary, the triple combination of AZA+ITF+α-PD-1 was the most effective at decreasing ascites volume and increasing overall survival (Fig. 3F).
Fig. 3.
Addition of immune checkpoint inhibition to epigenetic therapy in an intact mouse model decreases tumor burden and increases survival. (A) In vivo treatment schematic of AZA (0.5 mg/kg), entinostat (2 mg/kg), givinostat (2 mg/kg), and α-PD-1 (200 μg per mouse). (B) Volume of ascites fluid drained at week 6. Mean ± SEM is shown and significances were determined by one-way ANOVA. All significances are compared with mock; **P < 0.01, ***P < 0.001; ns, not significant; n = 8 to 10 mice per group. (C–F) Survival of the mice in days, with median survival shown. Significances were determined by log-rank (Mantel–Cox) test; n = 10 mice per group.
Fig. 4.
Epigenetic therapy and α-PD-1 increase the number and activation of immune cells in the tumor microenvironment. Mice were treated as described in Fig. 3A. Cells from ascites fluid drained at week 6.5 were analyzed via FACS. Median, 25th and 75th percentiles, and range are plotted for each experimental arm, and significances were determined by Mann–Whitney t test. Significances compared with mock are marked with *, and significances compared with AZA are marked with #. */#P < 0.05, **/##P < 0.01, ***/###P < 0.001. (A) % T effector cells (CD8+IFN-γ+) of T cells. (B) % T helper cells (CD4+IFN-γ+) of T cells. (C) % activated NK cells (NK1.1+, IFN-γ+) of NK1.1+ cells. (D) % myeloid-derived suppressor cells (GR-1+, CD11b+, F4/80−, MHCII−) of CD45+ cells. (E) % macrophages (CD11b+, F4/80+) of CD45+ cells. (F) % CD3+ T cells of CD45+ cells. (A–C and F) n = 4 to 9 mice per group. (D and E) n = 2 to 9 mice per group.
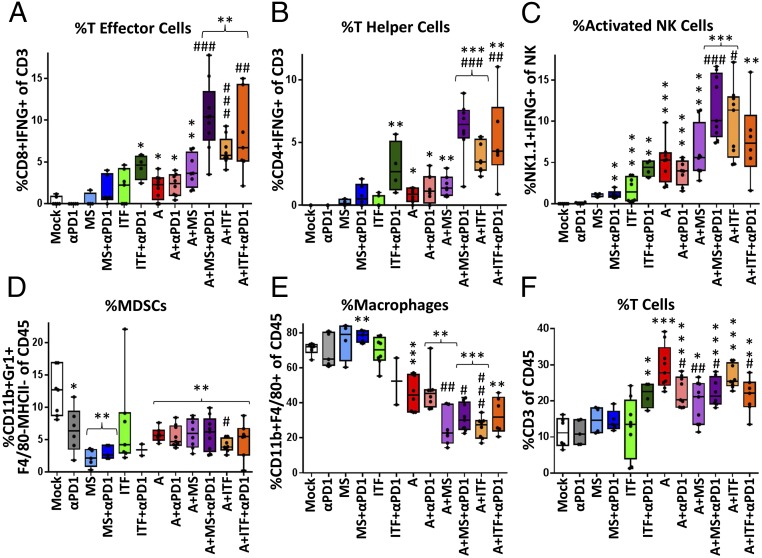
Immune cell subpopulations in the ascites fluid of tumor-bearing mice were changed by epigenetic therapy and α-PD-1, but immune cells in nonmalignant tissues, such as the spleen, were not affected (Fig. S4B). In the tumor microenvironment, AZA treatment moderately but significantly increased the percentage of activated CD4+ and CD8+ T cells and NK cells and, when combined with ITF or either HDACi+α-PD-1, this activation was markedly enhanced (Fig. 4 A–C). All treatments containing AZA led to an increase in the percentage of T cells (Fig. 4F), though this effect was less consistently observed in replicate experiments than the increases in activation of CD8+ T and NK cells. None of the treatment groups altered the percentages of T regulatory, CD4+, CD8+, CD4+PD-1+, or CD8+PD-1+cells or the CD4/CD8 ratio (Fig. S5). The addition of givinostat or either HDACi+α-PD-1 to AZA therapy increased the activation of key tumor-killing subsets of immune cells.
Myeloid-derived suppressor cells (MDSCs), which aid in tumor immune evasion (35), were significantly decreased by almost all therapies, with the exception of ITF and ITF+α-PD-1; however, this exception may be due to limitations in sample number and high variability (Fig. 4D). All treatments containing AZA decreased the percentage of macrophages, which can influence tumor growth (36) (Fig. 4E). Compared with AZA alone, the addition of an HDACi significantly decreased the percentage of macrophages further. Overall, increased numbers of CD45+ immune cells, increased activation of CD8+ T cells and NK cells, and decreases in macrophages and MDSCs were the most consistent, significant changes resulting from epigenetic therapy. Differences were observed between the changes of the immune cell subpopulations in the ex vivo (Fig. 2 D–J) and in vivo (Fig. 4) models. This was not unexpected, as the immune cells were exposed to drug in the in vivo model but not in the ex vivo, pretreatment model. Further investigation is needed regarding the response of immune cell populations to epigenetic treatment.
In support of the fact that in vivo epigenetic treatment alters host immune cell populations in the tumor microenvironment, we find that AZA treatment increases expression of viral defense genes in CD8+ and CD4+ T cells and in CD11b+ myeloid cells in the ascites fluid of treated, tumor-bearing mice (Fig. S6). Ingenuity analysis also identified IFN-associated genes as top upstream regulators of the transcriptional program in these in vivo AZA-treated cells (Table S2). These results suggest that the AZA-induced increase in gene expression of IFN-associated genes in both tumor and host immune cells may be an integral component of the improved outcome of mice treated with epigenetic therapy and immune checkpoint inhibitors.
In summary, the combinations of epigenetic agents, as well as the addition of α-PD-1 to AZA+HDACi, increase the numbers and activation of key tumor-killing immune cells in the tumor microenvironment and decrease numbers of MDSCs and macrophages (Fig. 4). We hypothesize that these effects contribute to the reduction in tumor burden and survival when treating with AZA, its combination with HDACis, and importantly these agents combined with α-PD-1.
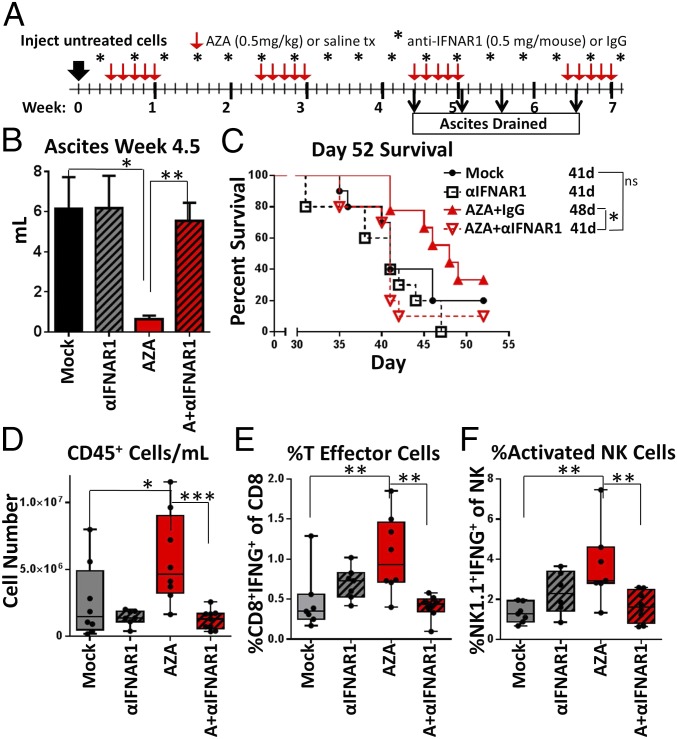
Blockade of IFNAR1 Inhibits the Actions of AZA.
Our expression data show that AZA treatment leads to increased IFN signaling and viral defense gene expression in ID8-VEGF-Defensin cells (Fig. S2A). We therefore questioned the role and importance that IFN signaling plays in the AZA-induced decrease in tumor burden and alterations in immune cells observed in Figs. 3 and 4. To test this hypothesis, we used an antibody targeting IFN alpha and beta receptor subunit 1 (α-IFNAR1). Mice harboring ID8-VEGF-Defensin tumor cells were injected with α-IFNAR1 (i.p., 0.5 mg per mouse) every 3 d and simultaneously treated with AZA or vehicle control (mock) (Fig. 5A). The AZA-mediated reduction in ascites volume routinely observed in our experiments was inhibited by treatment with α-IFNAR1 (Fig. 5B), and total numbers of CD45+ cells in the ascites were not increased as with the AZA and IgG control treatment but remained near mock values (Fig. 5D). Likewise, activation of CD8+ T effector cells and NK cells in response to AZA treatment was also completely blocked by α-IFNAR1 (Fig. 5 E and F), and there was no survival benefit for mice treated with AZA and α-IFNAR1 (Fig. 5C). Previous in vitro studies have demonstrated that up-regulation of a dsRNA sensing pathway by AZA triggers the activation of an intact type I IFNAR1 (11, 14), and here the use of α-IFNAR1 prevented the AZA-induced increase in expression normally observed for antiviral genes, such as ISG15, IFIT1, and ICAM1, in vitro (Fig. S7 A–C). Our α-IFNAR1 data indicate that the type I IFN response is, indeed, required for effective in vivo antitumorigenic actions of 5-azacytidine, including reduced tumor burden, extended survival, and increased numbers and activation of immune cells.
Fig. 5.
Blockade of IFNAR1 inhibits the actions of AZA. (A) Treatment schematic for the mice. Mice were treated with AZA (0.5 mg/kg) or saline as described in Fig. 3. Anti-IFNAR1 was injected i.p. (0.5 mg per mouse) every 3 d, beginning 1 d before the AZA regimen. (B) Volume of ascites drained from the mice at week 4.5. Mean ± SEM is shown, and significances were determined by Mann–Whitney t test; n = 8 to 10 mice per group. (C) Survival of the mice in days, with median survival shown. Significances were determined by log-rank (Mantel–Cox) test; n = 10 mice per group. (D–F) Median, 25th and 75th percentiles, and range are plotted, and significances were determined by Mann–Whitney t test; n = 6 to 9 mice per group. (D) CD45+ cells per mL of ascites. (E) % T effector cells (CD8+IFNγ+) of CD3+ T cells. (F) % activated NK cells (NK1.1+, IFNγ+) of NK1.1+ cells. *P < 0.05, **P < 0.01, ***P < 0.001.
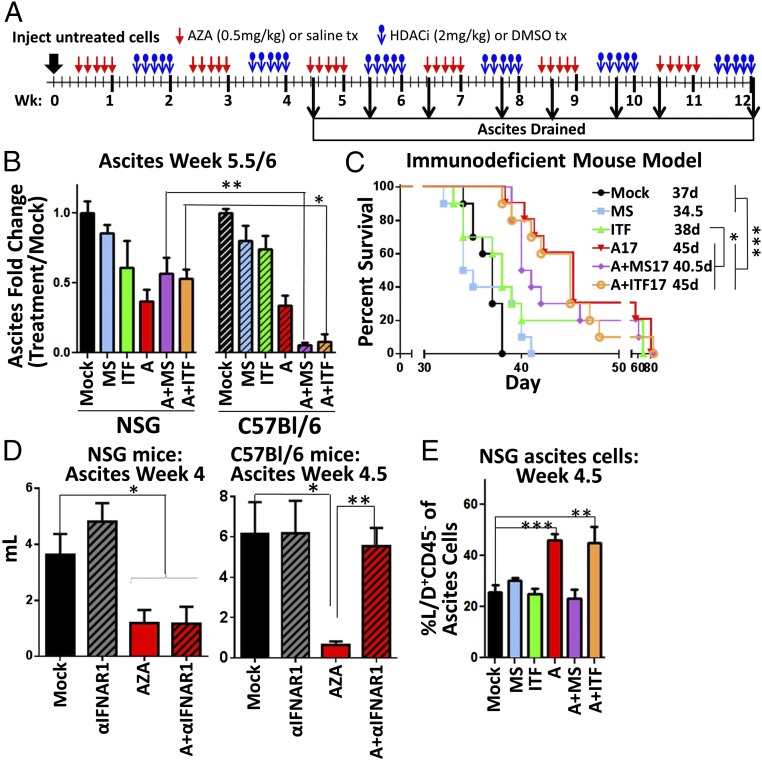
AZA+HDACi Efficacy Requires a Treated Immune System.
To further assess the role of the immune cells in the antitumorigenic response, we compared the response to epigenetic agents in treated immunodeficient NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ (NSG) mice that lack functional B, T, and NK cells (37) (Fig. 6A) with the response in the treated immunocompetent mice (Fig. 3). In the NSG immunodeficient mice, AZA treatment reduced ascites volume and increased survival as in the C57BL/6 mice, and HDACi treatment alone did not significantly affect ascites volume or survival in either mouse model. Combination treatment was more effective than AZA as a single agent in the treated immunocompetent C57BL/6 mice (Fig. 6 B and C) and not in the immunodeficient mice or in the pretreatment model (Fig. 1 C and D), suggesting that when combined with AZA, HDACis may act on the immune microenvironment to reduce tumor burden. Taken together, these data imply that AZA can act on both tumor and immune cells, while the added benefit of the combination with the HDACi may rely on the treatment and presence of an intact host immune microenvironment. Specifically, the activation of T and NK cells and decreases in macrophages were all significantly enhanced by the HDACi addition to AZA, and may be responsible for the reduction in tumor burden in the treated mice in an immune-intact setting (Fig. 4 A–C and E). Interestingly, α-IFNAR1 treatment (Fig. 5A) did not inhibit the reduction in ascites burden in AZA-treated NSG mice (Fig. 6D), as it did in the C57BL/6 mice, shown again here for comparison, suggesting that the role IFN signaling is playing in the response to AZA is dependent on functional immune cells in the tumor microenvironment. These data also suggest that there are additional mechanisms responsible for the antitumorigenic effect of AZA that are not dependent on IFN signaling.
Fig. 6.
AZA+HDACi combination therapy is less effective at reducing tumor burden and increasing survival in an immunodeficient mouse model. (A) Treatment schematic for in vivo treatment of NSG mice with AZA and HDACis entinostat or givinostat. (B) Fold change in ascites volume drained at week 5.5 (NSG) or 6 (C57BL/6). The C57BL/6 data from Fig. 3B are shown here for direct comparison; n = 3 to 10 mice per group. (C) NSG mouse survival in days, with median survival shown. Significances were determined by a log-rank (Mantel–Cox) test; n = 10 mice per group. (D) Ascites volume drained at week 4.5 or 4 from C57BL/6 or NSG mice, respectively, treated with AZA and anti-IFNAR1 as shown in Fig. 5 A and B; n = 8 to 10 mice per group. (E) % dead, CD45−, nonimmune ascites cells (Live/Dead stain+, CD45−) from NSG ascites fluid; n = 5 to 10 mice per group. (B, D, and E) Mean ± SEM is shown, and significances were determined by Mann–Whitney t test. *P < 0.05, **P < 0.01, ***P < 0.001.
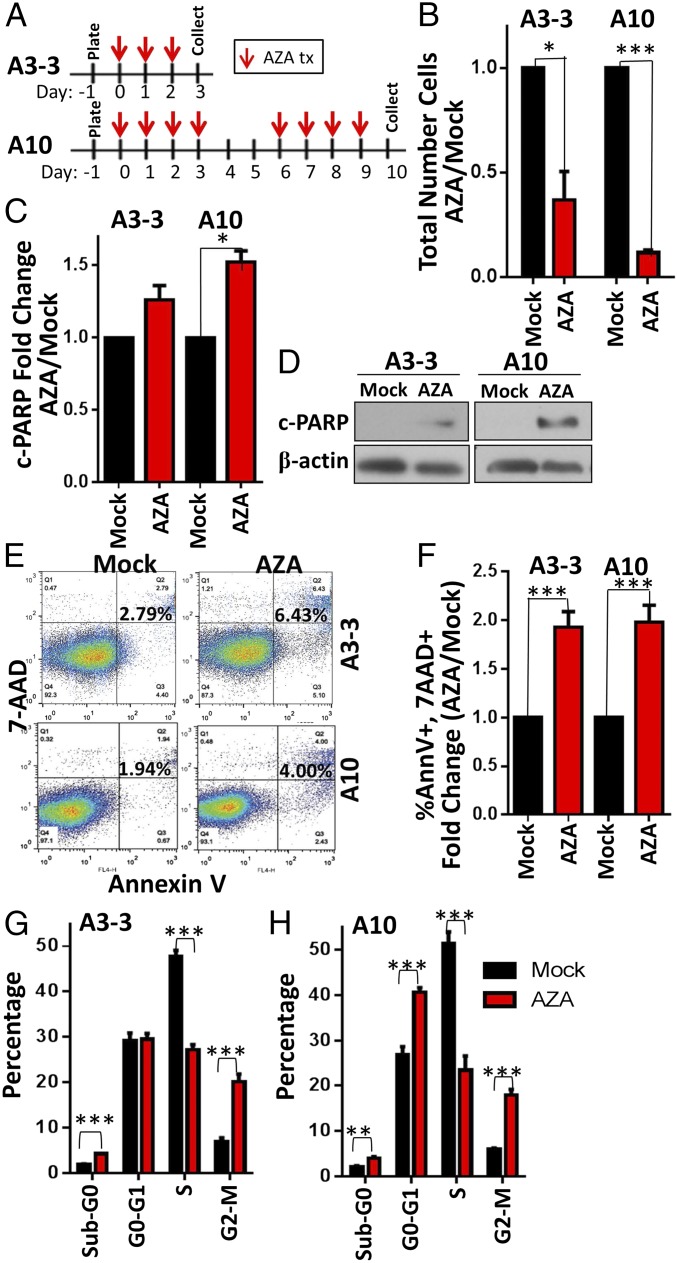
AZA Has Direct Antitumorigenic Effects.
Even in the absence of tumor-killing immune cells in the NSG model, we noted increased numbers of dead cells in the CD45− (nonimmune cell) population with AZA and AZA+ITF treatment, the two groups with the longest median survival (Fig. 6E). This could be explained by the fact that the antitumor effects of AZA can be mediated through mechanisms that are not immune- or IFN-dependent, such as apoptosis and disruption of the cell cycle (11, 38–40). Indeed, 3 or 10 d of in vitro treatment of the tumor cells (A3-3, A10) with 500 nM AZA caused a significant decrease in cultured cell numbers (Fig. 7 A and B) associated with signs of apoptosis reflected by increased cleaved-PARP protein levels and percentage of cells positive for annexin V and 7-AAD (Fig. 7 C–F). These data confirm that nanomolar doses of AZA induce a low level of apoptosis in cancer cells, as we have previously described (11, 38), which appears to be too small to account for the large decrease in tumor cells observed in culture (Fig. 7B). A more important factor in the decrease in tumor cell number could be that A3 and A10 treatment in vitro decreased the percentage of tumor cells in S phase and increased those in G2-M arrest (Fig. 7 G and H), as has been observed in other models (39, 40). In summary, AZA directly affects intrinsic, antitumorigenic mechanisms in the tumor cells, leading to increased apoptosis and cell-cycle arrest. However, the changes in the cell cycle and small increases in apoptosis were not inhibited in vitro by treatment with the α-IFNAR1 blocking antibody described previously (Fig. S7 D and E), suggesting that while IFN signaling is important for the recruitment and activation of immune cells, AZA also has non–immune-related and IFN-independent effects.
Fig. 7.
Ex vivo treatment of ID8-VEGF-Defensin cells with low-dose AZA decreases viable cell number, increases apoptosis, and disrupts the cell cycle. (A) Three or 10 d in vitro treatment with 500 nM AZA. (B) Total number of cells relative to mock; n = 3. (C) Quantification of c-PARP levels in AZA-treated cells relative to mock; n = 3. (D) A representative Western blot of c-PARP levels. (E and F) Percentage of annexin V+ and 7-AAD+ apoptotic cells. Representative flow cytometry data are shown (E) along with quantification (F); n = 3. (G and H) Cell-cycle analysis, determined by BrdU incorporation and 7-AAD staining of DNA content; n = 3. Mean ± SEM is shown, and significances were determined by Mann–Whitney t test. *P < 0.05, **P < 0.01, ***P < 0.001.
Overall, our data demonstrate that AZA reduces tumor burden and increases the number of immune cells in the tumor microenvironment, in part through effects on the tumor cells themselves. AZA treatment up-regulates immune gene expression in tumor cells and in immune cells, and type I IFN signaling is required for some antitumorigenic effects of in vivo AZA, such as decreased ascites burden, extended survival, and activation of immune cells. When tumor-bearing mice are treated in vivo, the addition of an HDACi to AZA further reduces tumor burden and increases survival, perhaps due to an increase in activated T and NK cells and a decrease in macrophages. Finally, the combination of AZA, givinostat, and α-PD-1 was the most effective in improving overall survival.
Discussion
The use of different treatment models in this study has enabled us to understand how 5-azacytidine and HDACis act individually and in combination on ovarian tumor epithelial cells and immune cells in the microenvironment to establish antitumor responses and to enhance immune checkpoint therapy. Low doses of AZA, but not HDACis, directly induce multiple antitumorigenic mechanisms in tumor cells, most notably increased immune signaling, increased apoptosis, and disruptions of the cell cycle, as well as increasing immune cell activation in the tumor microenvironment via type I IFN signaling. When an HDACi, especially givinostat, is combined with AZA in vivo, these agents can enhance the activation of specific immune subsets such as T and NK cells. Our data now show that the addition of an HDACi to a DNMTi may be optimal to achieve a maximal sensitization to checkpoint inhibitors. We hypothesize that other DNMTis such as decitabine may behave similarly to AZA used in this study, as decitabine has also been shown to trigger an antiviral response in tumor cells (11, 41). The triple combination of AZA, givinostat, and α-PD-1 extended median survival by 14.5 d compared with mock-treated mice. This change is significantly greater than the benefit derived from paclitaxel treatment, a common chemotherapy for ovarian cancer, in this fast-growing model (42). The addition of givinostat, but not entinostat, to AZA was able to sensitize the tumors to α-PD-1 therapy. The reasons why givinostat outperforms entinostat in overall survival when combined with AZA and α-PD-1 are currently under investigation. Other studies have shown that HDAC inhibition can affect tumor-associated immune cells, with one finding that B cells as well as IFN-γ receptor signaling in the tumor cells were important for the antitumorigenic effect of the HDACis vorinostat and panobinostat in syngeneic models of colon cancer and lymphoma (43).
In our study, blockade of type I IFN signaling with an antibody against IFNAR1 impairs the antitumorigenic effects of AZA. Although it had been observed that AZA treatment could stimulate IFN pathway induction by viral defense pathway signaling, it was not known which in vivo consequences would be linked to this pathway up-regulation. Importantly, our data now demonstrate that many of the antitumorigenic actions of AZA in ovarian cancer are mediated via IFN α, β receptor subunit 1, including decreased tumor burden, increased CD45+ immune cells in the tumor microenvironment, and increased activation of CD8+ T and NK cells. We show that type I IFN signaling is responsible for the antitumor effects of AZA in immunocompetent mice (Fig. 6D). However, AZA has an effect that leads to decreased tumor burden in NSG mice that is not mediated by the IFN response (Fig. 6D). Thus, we conclude that the antiviral response caused by AZA primarily reduces tumor burden by recruitment and activation of immune cells to kill tumor cells in this model. Separately (Fig. S7), we show that AZA also has IFN-independent effects such as cell cycle disruptions in ID8-VEGF-Defensin cells (Fig. S7 F and G), which may account for the antitumorigenic effect of AZA in the NSG mice. AZA induced low levels of apoptosis that were not significantly affected by blocking IFNAR1 (Fig. S7 D and E), in contrast to previous work from our group and others that indicated an intrinsic apoptotic effect of AZA in human ovarian tumor cells (11, 14). Thus, in this mouse model, the primary effect of the AZA-induced IFN response is through recruitment and activation of host immune cells, while AZA also causes IFN-independent signaling to inhibit tumor cell growth.
Interestingly, P53 was identified as an upstream regulator of signaling induced by AZA in immune cells. P53 and DNMTs may cooperate to regulate repetitive elements that when transcribed trigger the IFN response (44). P53 is known to transcriptionally regulate specific IFN-stimulated genes, including IRF9 and TLR3. In addition, STAT1 and PKR up-regulate P53, leading to transcription of proapoptotic genes (45). Previous work has shown up-regulation of P53 through the DNA damage repair pathway by the DNMTi decitabine (46). Thus, P53 signaling may be activated by either the IFN response or DNA damage in the immune cells in question.
Others have tested whether the IFN response can play a role in sensitization to immune checkpoint blockade, as stabilization of IFNAR1 improves the efficacy of α-PD-1 therapy (47) and the loss of IFN-γ pathway genes is a mechanism of resistance to anti–CTLA-4 (48). IFN-γ has also been shown to suppress genes related to M2-like function in macrophages (49). On the other hand, prolonged tumor IFN signaling has been shown to induce resistance to immune checkpoint blockade over time (50), implying that perhaps the timing and duration of the response are important. Our study provides a greater understanding of how IFN signaling may sensitize tumors to immune checkpoint blockade.
The AZA-induced increases in immune signaling observed in ID8-VEGF-Defensin mouse ovarian cancer cells are consistent with our results for AZA-treated human ovarian cancer cells (11, 13). Thus, in both the human and murine tumor cells, AZA treatment leads to an increase in the expression of endogenous retroviral transcripts, viral defense genes, cancer testis antigens, and chemokines/cytokines (11, 13, 14). The chemokine CXCL10 was significantly increased in both the cell-culture media of AZA-treated murine cells and the ascites fluid of AZA-treated mice. CXCL10 has been identified in humanized models of ovarian cancer (15), validating the relevance of the ID8-VEGF-Defensin mouse model of ovarian cancer to human disease and suggesting that these chemokines/cytokines may play a role in the AZA-induced recruitment of immune cells to the tumor-associated ascites (51–53). In fact, CXCL10 is linked to decreased tumor burden in the ID8 model (54).
Our work now provides mechanistic insight for previous studies showing that epigenetic agents may alter the tumor-associated microenvironment to potentially sensitize tumors to immunotherapy. Our study defines the cellular targets of AZA and HDACis using a more comprehensive panel of immune cells and proves a requirement for type I IFN signaling in the AZA-induced immune response. We have also shown that the addition of an HDACi to AZA can increase the activation of immune subsets, using doses of epigenetic therapy that are clinically relevant and can be immediately applied in clinical trials. The preclinical data in this manuscript helped to initiate a Celgene-sponsored, phase II randomized study of pembrolizumab with or without epigenetic modulation with CC-486 (oral AZA) in patients with platinum-resistant or -refractory epithelial ovarian, fallopian tube, or primary peritoneal cancer (55). This trial started enrollment in December 2016 and will hopefully reveal that addition of a DNMTi to checkpoint inhibitor therapy provides benefit beyond that of immunotherapy alone. The addition of an HDACi to the DNMTi in future trials may provide more benefit, and our data suggest that this combination can most effectively decrease the immunosuppression in the ovarian tumor microenvironment to sensitize to immune checkpoint blockade therapy.
Materials and Methods
Cell-Culture Treatments.
MOSE ID8-Defb29/Vegf-a (ID8-VEGF-Defensin) cells, which tested negative for mycoplasm in December 2016, were grown in RPMI medium and treated with AZA (500 nM) at the same “low dose” established by Tsai et al. (38). The A3 treatment paradigm had AZA in the media on days 0, 1, and 2, while the A10 paradigm was also treated with AZA on days 3, 6, 7, 8, and 9. Cells were split on days 3 and 6 and harvested on day 10 (A3 and A10).
Entinostat (MS275) treatment followed the AZA paradigms above, but with 100 nM MS275 for 3 d or 30 nM MS275 for 3 or 10 d (HDACi3 or 10).
For the sequential combination of AZA and HDACi treatment, cells were treated with the A10 paradigm described above, followed by three doses of 100 nM MS275 or ITF. The cells were collected on day 17 (AZA17, HDACi17, and AZA+HDACi17; Fig. 1A). AZA (Sigma) was suspended in 0.9% saline. Entinostat (Syndax Pharmaceuticals) and givinostat (Selleckchem) were both suspended in DMSO and diluted 1:1,000 in media, so that the percentage of DMSO did not exceed 0.1%.
Gene Expression Analysis.
RNA extraction, RNA quality analysis, hybridization to Agilent 4 × 44K Human Gene Expression v2 arrays, and analysis of the arrays were done as previously described (13). In some cases, tumor cells were isolated from ascites fluid by FACS, and RNA was isolated using the Qiagen RNeasy Micro Kit (74004). After total cellular RNA was extracted using the TRIzol method (Life Technologies), RNA concentration was determined using the NanoDrop machine and software (Thermo Fisher Scientific). One microgram total RNA was used to generate cDNA with the QuantiTect Reverse Transcription Kit (Qiagen). Quantitative reverse-transcription PCR (q-RT-PCR) of ISG15, IFIT1, and ICAM1 mRNA was performed using TaqMan assays or Custom TaqMan Gene Expression Array Cards (Life Technologies) and the Applied Biosystems 7500 Fast Real-Time PCR System and software. TBP was used as a reference gene. The ΔΔCT method was used to calculate relative expression levels. Reverse transcriptase negative cDNA synthesis reactions were performed for at least one sample per plate.
Mouse Endogenous Retrovirus and SINE q-PCR.
Cells with A10 treatment were collected on days 3, 4, 7, and 10. RNAs were DNaseI-treated, and cDNA synthesis was performed (High-Capacity cDNA Reverse Transcription Kit; Thermo Fisher Scientific). Expression of nine mERV genes (two IAP gag genes, an IAP-LTR, Mtv 7/8/9 sequences specific for C57BL/6 mouse strains, and placental mERVs including syncytin-A, mErv-3, Peg11, and Mart8) and the B1 SINE gene was quantified by q-PCR (ABI 7300) (see Table S1 for primer sequences and q-PCR methodology). Mouse housekeeping genes 18S rRNA, β-actin, and GAPDH were used for normalization (Table S1). Ascites tumor cells were sorted using FACS at week 4.5 following in vivo AZA treatment (Fig. S6A). We examined gene expression for all nine mERVs and B1 SINE gene expression in sorted ascites tumor cells. The full q-PCR protocol was previously described (56, 57).
All Mouse Experiments.
Tumor burden was assessed via measurement of body weight and the amount of ascites drained from the mice at the point where they had gained 20 to 30% of their body weight. Statistical outliers were removed using Peirce’s criterion. Mice were cared for in accordance with the policies of The Johns Hopkins University Animal Care and Use Committee. n = 10 mice for all treatment groups. While each experimental group started with 10 mice, at the time ascites was drained some mice had died, and other treated mice did not have ascites yet. Furthermore, some mice only yielded enough cells to run one panel for flow cytometry, which led to decreases in mouse numbers and differences between groups in some figures.
Mouse Experiments with ex Vivo Epigenetic Treatment of Cancer Cells.
Single-agent therapy.
Cells [2.5 × 105 (A10) or 5 × 105 (HDACi3, HDACi10, and A3)] were injected i.p. into 8- to 10- (A10) or 6- to 8- (HDACi3, HDACi10, and A3) wk-old female C57BL/6 mice. Immune cells were isolated from the ascites fluid via a Percoll gradient and stained for FACS.
Combination therapy.
Cells (2.5 × 105) treated with A17, HDACi17, or A+HDACi17 schedules were injected i.p. into 8- to 10-wk-old female C57BL/6 mice. All cells from the ascites fluid were filtered and stained for FACS.
Mouse Experiments with in Vivo Treatment.
ID8-VEGF-Defensin cells (2.5 × 105) were injected i.p. into 8- to 10-wk-old female C57BL/6NHsd (C57BL/6) mice or NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ (NSG) mice 3 d after injection. AZA (0.5 mg/kg) or saline was given i.p. for 5 d a week. The following week, 2 mg/kg givinostat or entinostat or 1% DMSO in saline was injected i.p. for 5 d. For the rest of the experiment, the treatment alternated AZA/HDACi every other week. α-PD-1 (200 μg per mouse) was given on days 17, 20, 24, and 27 after injection in the C57BL/6 mouse experiment. α-PD-1 (1 mg/mL in saline) has been described previously (58, 59). Blocking of IFNAR1 was achieved with the anti-mouse IFNAR1 antibody (clone MAR1-5A3), injected every 3 d (0.5 mg per mouse) (60). Anti-IFNAR1 and the mouse IgG isotype control were purchased from Leinco Technologies and diluted in PBS.
Flow Cytometry.
Ascites was drained or spleens were collected from 5 to 10 mice per group and incubated in ACK buffer (Thermo Fisher Scientific) to lyse red blood cells for 10 min and then washed. Ascites from each mouse was individually lysed and prepared for flow cytometry. The mononuclear cells collected were cultured for 4 h in RPMI with 5% FBS and in the presence of Cell Stimulation Mixture (plus protein transport inhibitors; eBioscience). Cells were then washed and stained for cell-surface markers including Live/Dead (eBioscience), CD45 (BD Biosciences), CD3 (BD Biosciences), CD4 (BD Biosciences), CD8 (BD Biosciences), PD-1 (eBioscience), NK1.1 (BD Biosciences), F4/80 (BioLegend), MHC II (isotype control 400627; BioLegend), GR1 (isotype control 400635; BioLegend), and CD11b (BioLegend). After incubation, the cells were permeabilized (FoxP3 staining buffers; eBioscience). Intracellular staining was performed for FoxP3 (isotype control 12-4321; eBioscience) and IFN-γ (isotype control 554686; BD Biosciences). Flow cytometry acquisition was performed on an LSR II cytometer (BD Biosciences), and data were analyzed using FlowJo software version 10.2.
Chemokine and Cytokine Array.
Cultured cells were treated with an A10 treatment schedule, and media were collected at day 10. Ascites from mice treated with AZA as described above was collected at week 4 after injection of cells. Cells were removed from the ascites and the supernatant was collected. Media and ascites samples were analyzed with the Proteome Profile Array, Mouse Cytokine Panel A (R&D Systems) according to the manufacturer’s instructions.
Cell-Cycle and Apoptotic Analysis.
Cells were treated with an A10 treatment schedule and collected on days 3 and 10. For cell-cycle analysis, BrdU (10 μM; Sigma) was incubated with cells for 2 h. Cells were fixed, treated with DNase (300 μg/mL), and stained with anti-BrdU (BioLegend) and 7-AAD (Life Technologies). For apoptosis analysis, cells were stained for FACS and measured as apoptotic based on positive annexin V (eBioscience) and 7-AAD (Life Technologies) staining. Flow cytometry was performed on a FACSCalibur cytometer (BD Biosciences), and data were analyzed with FlowJo v10 software.
Western Blots.
Protein extracts were quantified and immunoblotted using the 4 to 20% Mini-PROTEAN TGX gel system (Bio-Rad) and PVDF membranes (Millipore). β-Actin or GAPDH was used as a loading control. Antibodies used were as follows: polyclonal rabbit anti-mouse cleaved PARP (1:1,000; Cell Signaling), polyclonal rabbit anti-DNMT1 (1:1,000; Sigma), mouse anti–β-actin (1:10,000; Sigma), and polyclonal rabbit anti-GAPDH (1:10,000; Trevigen). Band intensities were quantified using Adobe Photoshop Elements 6.0.
RNA Extraction and Sequencing Library Generation for Immune Cells.
CD8+ and CD4+ T cells and CD11b+ myeloid cells were sorted using a FACSAria II from ascites derived from mock- and AZA-treated mice. Approximately 10,000 cells were collected for each sorted population, based on viability and size and lineage exclusion. Cells were pelleted at 300 × g for 10 min. The supernatant was carefully removed and 100 µL of Arcturus PicoPure extraction buffer (Thermo Fisher Scientific) was added. Total RNA was extracted using the Arcturus PicoPure RNA Isolation Kit according to the manufacturer’s protocol. Low-input RNA-sequencing libraries were generated from 200 pg of total RNA using the SMART-Seq v4 Ultra Low Input RNA Kit (Clontech). All samples were subjected to 13 PCR amplification cycles to minimize PCR biases. Amplified cDNA libraries were later fragmented through sonication to obtain 200- to 500-bp fragments. Standard Illumina sequencing libraries were prepared using the ThruPLEX DNA-Seq Kit (Rubicon Genomics) according to the manufacturer’s protocol. Sample barcoded libraries were sequenced on Illumina’s NextSeq 500 instrumentation using the NextSeq 300 Cycle Kit, High Output, V1 reagents (Illumina), and data analysis workflow bcl2fastq v2.17.1.14 to obtain 150-bp paired-end reads.
Paired-end RNA-sequencing reads were trimmed to remove Illumina’s adapter sequences. Sequencing reads were further processed to remove poor-quality reads and/or reads mapping to mouse rRNA and tRNA sequences using the Array Studio package (www.omicsoft.com/array-studio). The following criteria were used to remove poor-quality reads: trim reads with base quality score (Sanger quality score) <10; filter out reads if trim length is <25 bp; filter out reads if maximal base quality score is <15; filter out reads if average quality score is <10; filter out reads if polyAGCT rate is ≥80%; and filter out the pair if either read fails the filtering criteria. Sequencing reads were aligned to the mouse reference genome (Build38) using OSA version 4 (61). To obtain transcript count data, RSEM package (62) and National Center for Biotechnology Information mouse RefSeq gene model (release July 2015) annotations were used. Transcripts with zero counts in more than two-thirds of the samples were discarded from downstream analysis to reduce noise in the expression data. Filtered count data were later normalized using quantile normalization, and differentially expressed transcripts were identified using Limma Voom (63). A P-value cutoff of 0.05 was used to classify transcripts as differentially expressed in the treatment condition. Upstream regulator and pathway analyses of differentially expressed transcripts were performed using Qiagen’s Ingenuity Pathway Analysis (www.qiagen.com/ingenuity).
Statistical Analysis.
Data were graphed in GraphPad Prism 5.0, and significance was determined by a Mann–Whitney t test or by multiple pairwise comparisons using the one-way ANOVA test with Bonferroni correction. Significances in survival data were determined by Mantel–Cox (log-rank) test. Differences were deemed significant with a P value of less than 0.05. Outliers were removed from ascites volume datasets and ascites immune cell datasets using Peirce’s criterion (64). Significances are shown as *P < 0.05, **P < 0.01, and ***P < 0.001.
Supplementary Material
Acknowledgments
We acknowledge Ada Tam for her help with flow cytometry, Peter Ordentlich from Syndax Pharmaceuticals for generously providing entinostat, and Robert Schreiber of the Washington University in St. Louis School of Medicine for assistance in obtaining the IFNAR1 blocking antibody. We also greatly acknowledge Mrs. Elizabeth Stiegler for her expert technical assistance. This work was supported by the Defense Health Program, through the Department of Defense Ovarian Cancer Research Program, under Teal Innovator Award OC130454/W81XWH-14-1-0385. Research reported in this publication was supported by the National Cancer Institute under Awards F32CA183214 and K99CA204592 (to K.B.C.) and Award P30CA006973. This work was also supported in part by Janssen, the SWCRF Collaboration for a Cure Grant, Irving Hansen Fund, and Dr. Miriam and Sheldon G. Adelson Medical Research Foundation. Opinions, interpretations, conclusions, and recommendations are those of the authors and are not necessarily endorsed by the Department of Defense. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Conflict of interest statement: S.B.B. and C.A.Z. have a collaborative research agreement with Janssen. V.B., K.R.W., G.S.C., and K.E.B. are employed by Janssen.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1712514114/-/DCSupplemental.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 2.Preston CC, Goode EL, Hartmann LC, Kalli KR, Knutson KL. Immunity and immune suppression in human ovarian cancer. Immunotherapy. 2011;3:539–556. doi: 10.2217/imt.11.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chester C, Dorigo O, Berek JS, Kohrt H. Immunotherapeutic approaches to ovarian cancer treatment. J Immunother Cancer. 2015;3:7. doi: 10.1186/s40425-015-0051-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brahmer JR, et al. Safety and activity of anti-PD-L1 antibody in patients with advanced cancer. N Engl J Med. 2012;366:2455–2465. doi: 10.1056/NEJMoa1200694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Juergens RA, et al. Combination epigenetic therapy has efficacy in patients with refractory advanced non-small cell lung cancer. Cancer Discov. 2011;1:598–607. doi: 10.1158/2159-8290.CD-11-0214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arrowsmith CH, Bountra C, Fish PV, Lee K, Schapira M. Epigenetic protein families: A new frontier for drug discovery. Nat Rev Drug Discov. 2012;11:384–400. doi: 10.1038/nrd3674. [DOI] [PubMed] [Google Scholar]
- 7.Zahnow CA, et al. Inhibitors of DNA methylation, histone deacetylation, and histone demethylation: A perfect combination for cancer therapy. Adv Cancer Res. 2016;130:55–111. doi: 10.1016/bs.acr.2016.01.007. [DOI] [PubMed] [Google Scholar]
- 8.Allis CD, Caparros M-L, Jenuwein T, Reinberg D, Lachlan M. Epigenetics. 2nd Ed Cold Spring Harbor Lab Press; Cold Spring Harbor, NY: 2015. [Google Scholar]
- 9.Cameron EE, Bachman KE, Myöhänen S, Herman JG, Baylin SB. Synergy of demethylation and histone deacetylase inhibition in the re-expression of genes silenced in cancer. Nat Genet. 1999;21:103–107. doi: 10.1038/5047. [DOI] [PubMed] [Google Scholar]
- 10.Wrangle J, et al. Alterations of immune response of non-small cell lung cancer with azacytidine. Oncotarget. 2013;4:2067–2079. doi: 10.18632/oncotarget.1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chiappinelli KB, et al. Inhibiting DNA methylation causes an interferon response in cancer via dsRNA including endogenous retroviruses. Cell. 2015;162:974–986, and errata (2016) 164:1073 and (2017) 169:361. doi: 10.1016/j.cell.2015.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Matei D, et al. Epigenetic resensitization to platinum in ovarian cancer. Cancer Res. 2012;72:2197–2205. doi: 10.1158/0008-5472.CAN-11-3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li H, et al. Immune regulation by low doses of the DNA methyltransferase inhibitor 5-azacitidine in common human epithelial cancers. Oncotarget. 2014;5:587–598. doi: 10.18632/oncotarget.1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roulois D, et al. DNA-demethylating agents target colorectal cancer cells by inducing viral mimicry by endogenous transcripts. Cell. 2015;162:961–973. doi: 10.1016/j.cell.2015.07.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peng D, et al. Epigenetic silencing of TH1-type chemokines shapes tumour immunity and immunotherapy. Nature. 2015;527:249–253. doi: 10.1038/nature15520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ghoneim HE, et al. De novo epigenetic programs inhibit PD-1 blockade-mediated T cell rejuvenation. Cell. 2017;170:142–157.e19. doi: 10.1016/j.cell.2017.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Topper et al., Epigenetic therapy ties MYC depletion to reversing immune evasion and treating lung cancer. Cell, in press. [DOI] [PMC free article] [PubMed]
- 18.Conejo-Garcia JR, et al. Tumor-infiltrating dendritic cell precursors recruited by a beta-defensin contribute to vasculogenesis under the influence of Vegf-A. Nat Med. 2004;10:950–958. doi: 10.1038/nm1097. [DOI] [PubMed] [Google Scholar]
- 19.Hung CF, et al. Vaccinia virus preferentially infects and controls human and murine ovarian tumors in mice. Gene Ther. 2007;14:20–29. doi: 10.1038/sj.gt.3302840. [DOI] [PubMed] [Google Scholar]
- 20.Zhang L, et al. Generation of a syngeneic mouse model to study the effects of vascular endothelial growth factor in ovarian carcinoma. Am J Pathol. 2002;161:2295–2309. doi: 10.1016/s0002-9440(10)64505-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roby KF, et al. Development of a syngeneic mouse model for events related to ovarian cancer. Carcinogenesis. 2000;21:585–591. doi: 10.1093/carcin/21.4.585. [DOI] [PubMed] [Google Scholar]
- 22.Duraiswamy J, Freeman GJ, Coukos G. Therapeutic PD-1 pathway blockade augments with other modalities of immunotherapy T-cell function to prevent immune decline in ovarian cancer. Cancer Res. 2013;73:6900–6912. doi: 10.1158/0008-5472.CAN-13-1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee Y, et al. A candidate precursor to serous carcinoma that originates in the distal fallopian tube. J Pathol. 2007;211:26–35. doi: 10.1002/path.2091. [DOI] [PubMed] [Google Scholar]
- 24.Singh N, et al. Primary site assignment in tubo-ovarian high-grade serous carcinoma: Consensus statement on unifying practice worldwide. Gynecol Oncol. 2016;141:195–198. doi: 10.1016/j.ygyno.2015.10.022. [DOI] [PubMed] [Google Scholar]
- 25.Perets R, et al. Transformation of the fallopian tube secretory epithelium leads to high-grade serous ovarian cancer in Brca;Tp53;Pten models. Cancer Cell. 2013;24:751–765. doi: 10.1016/j.ccr.2013.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sherman-Baust CA, et al. A genetically engineered ovarian cancer mouse model based on fallopian tube transformation mimics human high-grade serous carcinoma development. J Pathol. 2014;233:228–237. doi: 10.1002/path.4353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Walton J, et al. CRISPR/Cas9-mediated Trp53 and Brca2 knockout to generate improved murine models of ovarian high-grade serous carcinoma. Cancer Res. 2016;76:6118–6129. doi: 10.1158/0008-5472.CAN-16-1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hata K, Watanabe Y, Nakai H, Hata T, Hoshiai H. Expression of the vascular endothelial growth factor (VEGF) gene in epithelial ovarian cancer: An approach to anti-VEGF therapy. Anticancer Res. 2011;31:731–737. [PubMed] [Google Scholar]
- 29.Li Z, et al. The inflammatory microenvironment in epithelial ovarian cancer: A role for TLR4 and MyD88 and related proteins. Tumour Biol. 2016;37:13279–13286. doi: 10.1007/s13277-016-5163-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Janát-Amsbury MM, Yockman JW, Anderson ML, Kieback DG, Kim SW. Comparison of ID8 MOSE and VEGF-modified ID8 cell lines in an immunocompetent animal model for human ovarian cancer. Anticancer Res. 2006;26:2785–2789. [PubMed] [Google Scholar]
- 31.Baert T, et al. The dark side of ID8-Luc2: Pitfalls for luciferase tagged murine models for ovarian cancer. J Immunother Cancer. 2015;3:57. doi: 10.1186/s40425-015-0102-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Peter S, Bak G, Hart K, Berwin B. Ovarian tumor-induced T cell suppression is alleviated by vascular leukocyte depletion. Transl Oncol. 2009;2:291–299. doi: 10.1593/tlo.09190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Abiko K, et al. PD-L1 on tumor cells is induced in ascites and promotes peritoneal dissemination of ovarian cancer through CTL dysfunction. Clin Cancer Res. 2013;19:1363–1374. doi: 10.1158/1078-0432.CCR-12-2199. [DOI] [PubMed] [Google Scholar]
- 34.Wei H, et al. Combinatorial PD-1 blockade and CD137 activation has therapeutic efficacy in murine cancer models and synergizes with cisplatin. PLoS One. 2013;8:e84927. doi: 10.1371/journal.pone.0084927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ostrand-Rosenberg S, Sinha P. Myeloid-derived suppressor cells: Linking inflammation and cancer. J Immunol. 2009;182:4499–4506. doi: 10.4049/jimmunol.0802740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bingle L, Brown NJ, Lewis CE. The role of tumour-associated macrophages in tumour progression: Implications for new anticancer therapies. J Pathol. 2002;196:254–265. doi: 10.1002/path.1027. [DOI] [PubMed] [Google Scholar]
- 37.Shultz LD, Ishikawa F, Greiner DL. Humanized mice in translational biomedical research. Nat Rev Immunol. 2007;7:118–130. doi: 10.1038/nri2017. [DOI] [PubMed] [Google Scholar]
- 38.Tsai HC, et al. Transient low doses of DNA-demethylating agents exert durable antitumor effects on hematological and epithelial tumor cells. Cancer Cell. 2012;21:430–446. doi: 10.1016/j.ccr.2011.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weller EM, Poot M, Hoehn H. Induction of replicative senescence by 5-azacytidine: Fundamental cell kinetic differences between human diploid fibroblasts and NIH-3T3 cells. Cell Prolif. 1993;26:45–54. doi: 10.1111/j.1365-2184.1993.tb00005.x. [DOI] [PubMed] [Google Scholar]
- 40.Alexander VM, Roy M, Steffens KA, Kunnimalaiyaan M, Chen H. Azacytidine induces cell cycle arrest and suppression of neuroendocrine markers in carcinoids. Int J Clin Exp Med. 2010;3:95–102. [PMC free article] [PubMed] [Google Scholar]
- 41.Liu M, et al. Vitamin C increases viral mimicry induced by 5-aza-2′-deoxycytidine. Proc Natl Acad Sci USA. 2016;113:10238–10244. doi: 10.1073/pnas.1612262113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Janát-Amsbury MM, Yockman JW, Anderson ML, Kieback DG, Kim SW. Combination of local, non-viral IL12 gene therapy and systemic paclitaxel chemotherapy in a syngeneic ID8 mouse model for human ovarian cancer. Anticancer Res. 2006;26:3223–3228. [PubMed] [Google Scholar]
- 43.West AC, et al. An intact immune system is required for the anticancer activities of histone deacetylase inhibitors. Cancer Res. 2013;73:7265–7276. doi: 10.1158/0008-5472.CAN-13-0890. [DOI] [PubMed] [Google Scholar]
- 44.Leonova KI, et al. p53 cooperates with DNA methylation and a suicidal interferon response to maintain epigenetic silencing of repeats and noncoding RNAs. Proc Natl Acad Sci USA. 2013;110:E89–E98. doi: 10.1073/pnas.1216922110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Muller PA, Vousden KH. p53 mutations in cancer. Nat Cell Biol. 2013;15:2–8. doi: 10.1038/ncb2641. [DOI] [PubMed] [Google Scholar]
- 46.Karpf AR, Moore BC, Ririe TO, Jones DA. Activation of the p53 DNA damage response pathway after inhibition of DNA methyltransferase by 5-aza-2′-deoxycytidine. Mol Pharmacol. 2001;59:751–757. [PubMed] [Google Scholar]
- 47.Katlinski KV, et al. Inactivation of interferon receptor promotes the establishment of immune privileged tumor microenvironment. Cancer Cell. 2017;31:194–207. doi: 10.1016/j.ccell.2017.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gao J, et al. Loss of IFN-γ pathway genes in tumor cells as a mechanism of resistance to anti-CTLA-4 therapy. Cell. 2016;167:397–404.e9. doi: 10.1016/j.cell.2016.08.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kang K, et al. Interferon-γ represses M2 gene expression in human macrophages by disassembling enhancers bound by the transcription factor MAF. Immunity. 2017;47:235–250.e4. doi: 10.1016/j.immuni.2017.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Benci JL, et al. Tumor interferon signaling regulates a multigenic resistance program to immune checkpoint blockade. Cell. 2016;167:1540–1554.e12. doi: 10.1016/j.cell.2016.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Carr MW, Roth SJ, Luther E, Rose SS, Springer TA. Monocyte chemoattractant protein 1 acts as a T-lymphocyte chemoattractant. Proc Natl Acad Sci USA. 1994;91:3652–3656. doi: 10.1073/pnas.91.9.3652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xu LL, Warren MK, Rose WL, Gong W, Wang JM. Human recombinant monocyte chemotactic protein and other C-C chemokines bind and induce directional migration of dendritic cells in vitro. J Leukoc Biol. 1996;60:365–371. doi: 10.1002/jlb.60.3.365. [DOI] [PubMed] [Google Scholar]
- 53.Dufour JH, et al. IFN-gamma-inducible protein 10 (IP-10; CXCL10)-deficient mice reveal a role for IP-10 in effector T cell generation and trafficking. J Immunol. 2002;168:3195–3204. doi: 10.4049/jimmunol.168.7.3195. [DOI] [PubMed] [Google Scholar]
- 54.Au KK, et al. CXCL10 alters the tumour immune microenvironment and disease progression in a syngeneic murine model of high-grade serous ovarian cancer. Gynecol Oncol. 2017;145:436–445. doi: 10.1016/j.ygyno.2017.03.007. [DOI] [PubMed] [Google Scholar]
- 55.Fresco R, Glaspy J, Palodichuk C. 2016 Phase II Randomized Study of Pembrolizumab With or Without Epigenetic Modulation with CC-486 in Patients with Platinum-Resistant Epithelial Ovarian, Fallopian Tube or Primary Peritoneal Cancer, NCT02900560 (Natl Libr Med, Bethesda, MD). Clinicaltrials.gov.
- 56.Henke C, et al. Regulation of murine placentogenesis by the retroviral genes syncytin-A, syncytin-B and Peg10. Differentiation. 2013;85:150–160. doi: 10.1016/j.diff.2013.02.002. [DOI] [PubMed] [Google Scholar]
- 57.Henke C, et al. Selective expression of sense and antisense transcripts of the sushi-ichi-related retrotransposon–derived family during mouse placentogenesis. Retrovirology. 2015;12:9. doi: 10.1186/s12977-015-0138-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mathios D, et al. Anti-PD-1 antitumor immunity is enhanced by local and abrogated by systemic chemotherapy in GBM. Sci Transl Med. 2016;8:370ra180. doi: 10.1126/scitranslmed.aag2942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hirano F, et al. Blockade of B7-H1 and PD-1 by monoclonal antibodies potentiates cancer therapeutic immunity. Cancer Res. 2005;65:1089–1096. [PubMed] [Google Scholar]
- 60.Swann JB, et al. Type I IFN contributes to NK cell homeostasis, activation, and antitumor function. J Immunol. 2007;178:7540–7549. doi: 10.4049/jimmunol.178.12.7540. [DOI] [PubMed] [Google Scholar]
- 61.Hu J, Ge H, Newman M, Liu K. OSA: A fast and accurate alignment tool for RNA-seq. Bioinformatics. 2012;28:1933–1934. doi: 10.1093/bioinformatics/bts294. [DOI] [PubMed] [Google Scholar]
- 62.Li B, Dewey CN. RSEM: Accurate transcript quantification from RNA-seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Law CW, Chen Y, Shi W, Smyth GK. voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 2014;15:R29. doi: 10.1186/gb-2014-15-2-r29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ross SM. Peirce’s criterion for the elimination of suspect experimental data. J Eng Technol. 2003;20:38–41. [Google Scholar]
- 65.Munclinger P, Boursot P, Dod B. B1 insertions as easy markers for mouse population studies. Mamm Genome. 2003;14:359–366. doi: 10.1007/s00335-002-3065-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.