Significance
Our work provides insights into how cells solve the problem of delivering nickel, a toxic metal, to the active site of a metalloenzyme such as urease. Urease, a nickel-containing enzyme, is a virulence factor for Helicobacter pylori, which infects half of the human population and causes peptic ulcers. Supported by structural and biochemical evidence, we present a paradigm on how a metallochaperone UreG couples GTP hydrolysis/binding to allosterically control the binding/release of nickel ions and to switch protein-binding partners along the metal-delivery pathway so that the nickel ions are passing from one metallochaperone to another, without releasing the “free” toxic metal to the cytoplasm.
Keywords: metallochaperones, urease maturation, G protein, Helicobacter pylori, nickel
Abstract
The ability of metallochaperones to allosterically regulate the binding/release of metal ions and to switch protein-binding partners along the metal delivery pathway is essential to the metallation of the metalloenzymes. Urease, catalyzing the hydrolysis of urea into ammonia and carbon dioxide, contains two nickel ions bound by a carbamylated lysine in its active site. Delivery of nickel ions for urease maturation is dependent on GTP hydrolysis and is assisted by four urease accessory proteins UreE, UreF, UreG, and UreH(UreD). Here, we determined the crystal structure of the UreG dimer from Klebsiella pneumoniae in complex with nickel and GMPPNP, a nonhydrolyzable analog of GTP. Comparison with the structure of the GDP-bound Helicobacter pylori UreG (HpUreG) in the UreG2F2H2 complex reveals large conformational changes in the G2 region and residues near the 66CPH68 metal-binding motif. Upon GTP binding, the side chains of Cys66 and His68 from each of the UreG protomers rotate toward each other to coordinate a nickel ion in a square-planar geometry. Mutagenesis studies on HpUreG support the conformational changes induced by GTP binding as essential to dimerization of UreG, GTPase activity, in vitro urease activation, and the switching of UreG from the UreG2F2H2 complex to form the UreE2G2 complex with the UreE dimer. The nickel-charged UreE dimer, providing the sole source of nickel, and the UreG2F2H2 complex could activate urease in vitro in the presence of GTP. Based on our results, we propose a mechanism of how conformational changes of UreG during the GTP hydrolysis/binding cycle facilitate urease maturation.
Nearly half of all enzymes contain metals in their active sites (1, 2). Cells have evolved mechanisms to maintain metal homeostasis and to ensure metalloenzymes receive the correct metal ions (3, 4). Metals at the top of the Irving–Williams series such as nickel, copper, and zinc, can form more stable protein–metal complexes than weaker metals such as magnesium and can inactivate enzymes that require the less competitive metals to function (5, 6). Therefore, the free cytoplasmic concentrations of these competitive metal ions are tightly controlled and are kept at subnanomolar concentrations to avoid cytotoxicity (5, 7). One strategy of delivering the correct metal to a metalloenzyme is through specific protein–protein interactions with metallochaperones (4). Typically, metal ions are passed from one metallochaperone to another before they are eventually inserted into the catalytic site of metalloenzymes (8–10). Conceptually, this scheme requires the ability of metallochaperones to allosterically regulate the binding/release of metal ions and to change protein-binding partners along the metal delivery pathway.
Urease, catalyzing the hydrolysis of urea into ammonia and carbon dioxide, contains two nickel ions bound by a carbamylated lysine residue in its active site (11). The enzyme is a virulence factor for Helicobactor pylori because the pathogen uses the neutralizing ammonia released for its survival in acidic stomach (12). Delivery of nickel and urease maturation are assisted by four urease accessory proteins, namely, UreE, UreF, UreG, and UreH (or UreD in other species) (13–17). The urease maturation is dependent on GTP hydrolysis and involves the formation of an activation complex containing UreF, UreG, UreH, and apourease (13, 18–23). UreF can form a UreF2H2 complex with UreH in a 2:2 stoichiometry (24). The formation of the UreF2H2 complex is essential to the recruitment of UreG to form the UreG2F2H2 complex (20, 24, 25). UreG belongs to the G3E family of SIMIBI (signal recognition particle, MinD, and BioD) class GTPases (26, 27), and undergoes a GTP-dependent dimerization upon binding of nickel (20). We have previously shown that the Ni/GTP-bound UreG dimer, providing the sole source of nickel, can activate urease in vitro in the presence of the UreF2H2 complex (20), which can induce conformational changes in apourease that are essential for nickel delivery (13, 28). It has been shown recently that nickel can be transferred from another metallochaperone UreE to UreG by forming a UreE2G2 complex (29). In this study, we determined the crystal structure of Klebsiella pneumoniae UreG in complex with nickel and Guanylyl imidodiphosphate (GMPPNP). Together with our biochemical and mutagenesis studies, we have demonstrated how conformational changes induced by the GTP hydrolysis/binding cycle in the metallochaprone UreG facilitate urease maturation by modulating its binding to the nickel ion and protein-binding partners.
Result
Crystal Structure of KpUreG in Complex with Nickel and GMPPNP.
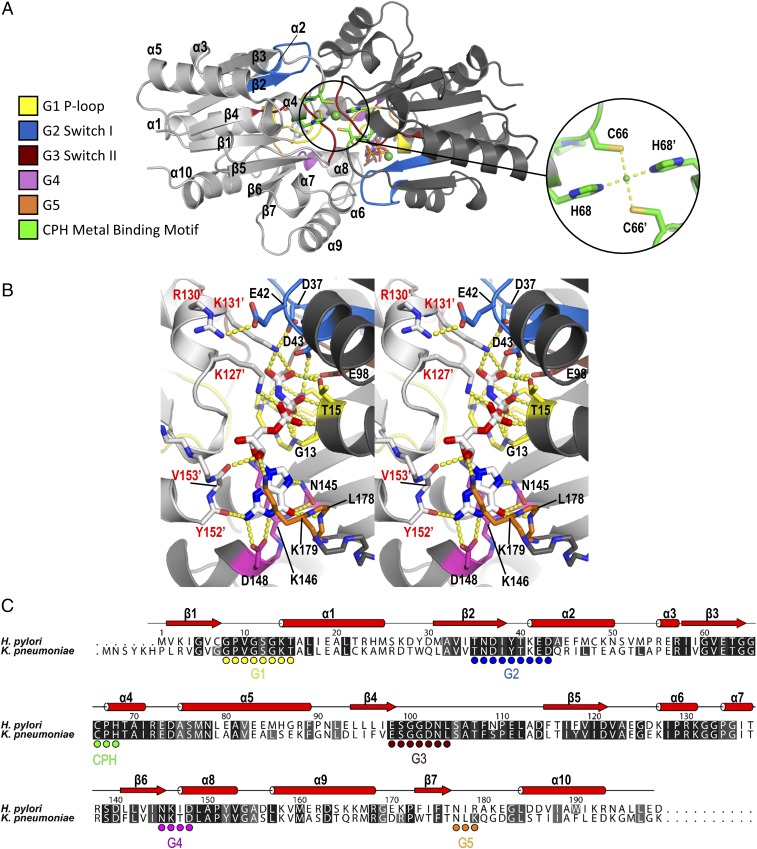
We have previously determined the structure of H. pylori GDP-bound UreG (HpUreG) in the UreG2F2H2 complex (20). To understand the conformational changes of UreG upon binding of nickel and GTP, we determined the crystal structure of UreG from K. pneumoniae (KpUreG) in complex with GMPPNP and nickel to a resolution of 1.8 Å (SI Appendix, Table S1 and Fig. 1A, PDB ID code 5XKT). To improve the quality of crystals, a truncated KpUreG(∆N4∆C1) was used for structure determination. KpUreG, sharing 64% sequence identity with HpUreG, is six residues longer (Fig. 1C). To avoid confusion, we will use the sequence numbering of HpUreG in subsequent structural comparison.
Fig. 1.
Crystal structure of KpUreG in complex with GMPPNP and nickel. (A) The structure of KpUreG (PDB ID code 5XKT) was solved as a dimer in complex with GMPPNP and nickel at a resolution of 1.8 Å. Cys66 and His68 from each of the two UreG protomers (colored in light and dark gray) coordinate a nickel ion in a square-planer geometry. Conserved motif (G1–G5) and CPH metal binding motif are colored as indicated. (B) A stereodiagram showing the interaction between KpUreG and GMPPNP. GMPPNP is sandwiched between the two KpUreG protomers and forms a network of hydrogen bonds (yellow dotted lines) with residues of the G1–G5 motifs. (C) Sequence alignment of KpUreG and HpUreG. The G1–G5 and the CPH metal-binding motifs are indicated as circles. Residues are numbered according to the HpUreG sequence. Apostrophes denote residues from the opposite protomer.
KpUreG adopts a SIMIBI-like fold with a seven-stranded β-sheet sandwiched by 10 α-helices, and exists as a dimer in the crystal structure. The invariant CPH metal binding motif from each protomer of KpUreG is juxtaposed to bind a nickel ion, which is coordinated by Cys66 and His68 in a square-planar geometry (SI Appendix, Fig. S1 A and C). KpUreG contains the canonical G1–G5 motifs for guanine nucleotide recognition (Fig. 1 B and C). One molecule of GMPPNP was bound to each of the two KpUreG protomers at the dimerization interface, forming extensive hydrogen bonds with residues of the G1–G5 motifs (Fig. 1B and SI Appendix, Fig. S1D). Phosphate groups of GMPPNP are wrapped around by G1 residues that constituted the P loop. The guanine base is sandwiched between the aliphatic side chains of Lys146 and Lys179, forming canonical hydrogen bonds to Asp148 of the G4 motif and to backbone amides of the G5 motif (Fig. 1B). Anomalous difference electron density maps revealed that a nickel ion occupies the magnesium-binding site in each of the nucleotide-binding pockets (SI Appendix, Fig. S1B). Noteworthy, the nickel ion is coordinated in an octahedral geometry, which is commonly observed for magnesium ion in GTPases (30, 31). Presumably, the nickel ions, being a more competitive ion, displaced the active site magnesium ions in the crystallization conditions used. Our result suggests that while nickel ion is required for the formation of the nickel-charged UreG dimer, excess nickel ions should be inhibitory because they could displace the magnesium ions in the active sites.
GTP Binding Induces Large Conformational Changes in UreG.
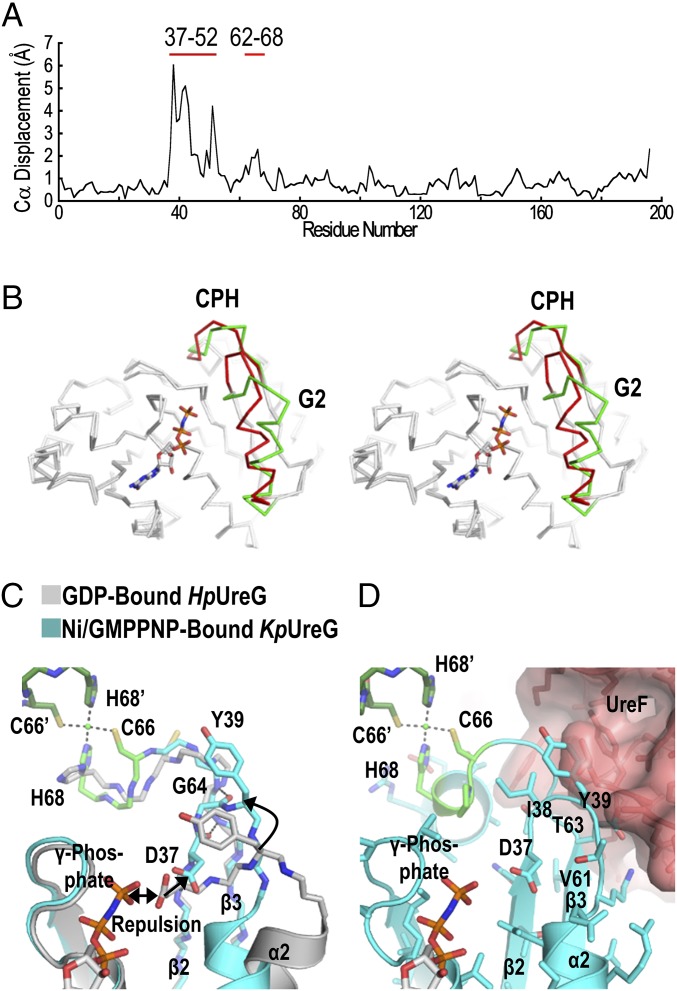
To understand the GTP-induced conformational changes, the structure of Ni/GMPPNP-bound KpUreG was compared with the crystal structure of GDP-bound HpUreG in complex with UreF2H2 (20). Most residues of the Ni/GMPPNP-bound KpUreG were superimposable with the GDP-bound HpUreG (Fig. 2 A and B), suggesting that the two proteins are structurally homologous with each other. Notably, large conformational changes were observed in the G2 region (β2 and α2; residues 37–52) and in residues near the CPH nickel-binding motif (residues 62–68) (Fig. 2B and Movie S1).
Fig. 2.
Conformational changes of UreG upon GTP binding. (A) The structures of the GDP-bound HpUreG (4HI0, chain F) and the Ni/GMPPNP-bound KpUreG (5XKT, chain A) were superimposed and values of Cα displacement were plotted. (B) Stereodiagram highlighting significant conformational changes found in the G2 region (residues 37–52) and near the CPH metal-binding motif (residues 62–68), which are colored in green and red for the GDP-bound HpUreG and the Ni/GMPPNP-bound KpUreG, respectively. (C) Charge–charge repulsion between Asp37 and the γ-phosphate group elicits conformational changes that are propagated to the CPH metal-binding site. The repulsion pushes the Asp37 away from the nucleotide-binding site so that Asp37 and Ile38 (β2 strand) form backbone hydrogen bonds with Val61, Thr63, and Gly64 (β3 strand). This zip-up motion of the β2 and β3 strands propagates conformational changes to the CPH metal-binding motif and causes helix-2 to tilt by ∼35° toward the nucleotide-binding site. (D) Moreover, Tyr39 of the G2 region moves toward the UreF2H2-binding site, introducing steric clashes that promote dissociation of UreG from the UreG2F2H2 complex. Residues from the opposite protomer are indicated by apostrophes.
Taking a closer look at the nucleotide-binding pocket reveals that the invariant residue Asp37 (or Asp43 in KpUreG) plays an instrumental role in initiating the GTP-dependent conformational changes. Upon GTP binding, charge–charge repulsion between the γ-phosphate and Asp37 pushes the invariant residue away from the nucleotide-binding pocket (Fig. 2C), causing Asp37 and Ile38 to form backbone hydrogen bonds with Val61, Thr63, and Gly64. As a result, β2 and β3 of Ni/GMPPNP-bound KpUreG is extended (Fig. 2C). This “zip-up” motion of β2 and β3 strands propagates the conformational changes to the CPH metal-binding motif, which is located at the end of β3. The Cys66 and His68 of each protomer move in an opposite direction toward the metal binding site and coordinate a nickel ion at the dimer interface. Asp102 of the G3 motif moves toward the CPH motif and forms hydrogen bonds to Asn103 and His68, stabilizing the square-planar coordination geometry of the CPH motif (SI Appendix, Fig. S2A). At the same time, GTP binding causes helix-2 to tilt by ∼35° toward the nucleotide-binding site, bringing Glu42 to form a salt bridge with Arg130 of the opposite protomer (SI Appendix, Fig. S2B). In addition, the opposite protomer of UreG undergoes a rigid-body movement toward the bound nucleotide so that Tyr152 and Val153 of the opposite protomer form two extra hydrogen bonds to the guanine ring of GMPPNP (SI Appendix, Fig. S2C).
We have previously shown that addition of nickel ions and GTP can dissociate UreG from the UreG2F2H2 complex (20). The crystal structure of Ni/GMPPNP-bound KpUreG determined here explains how the conformational changes of UreG induce the dissociation. As shown in Fig. 2 C and D, the zip-up motion of β2 and β3 causes Tyr39 of the G2 motif to protrude into the UreF2H2-binding site, introducing steric clashes that break the interaction between UreG and the UreF2H2 complex.
Double Mutation D37A/E42A Abolishes the Formation of the Nickel-Charged HpUreG Dimer, GTPase Activity, and in Vitro Activation of Urease.
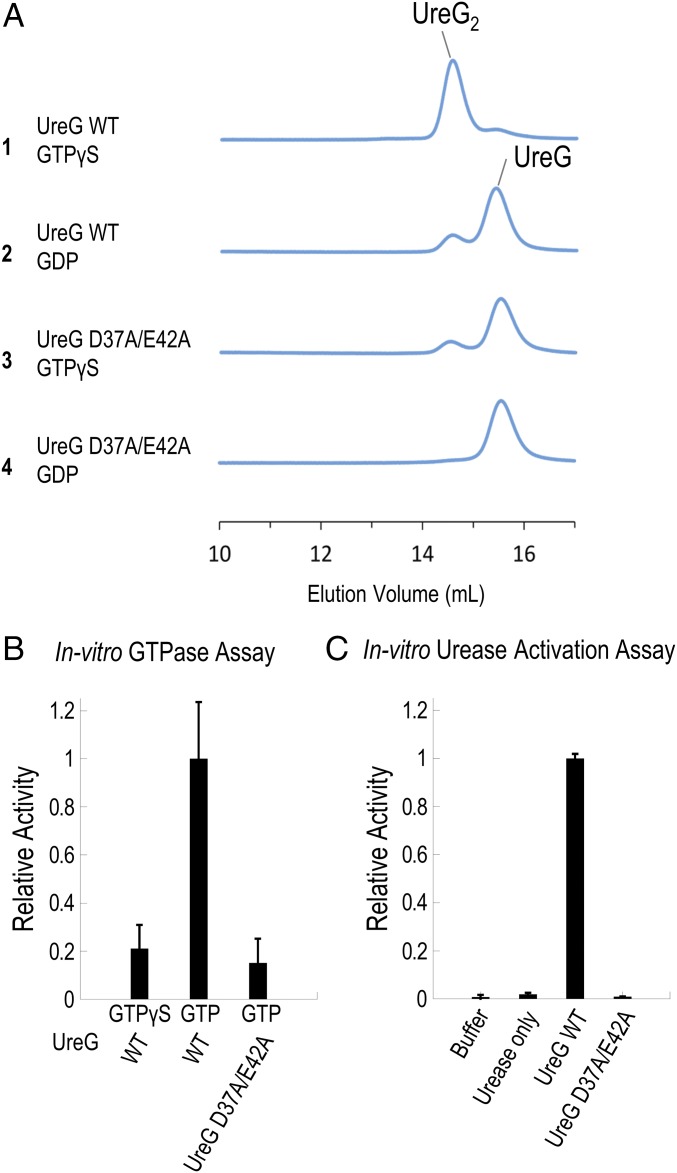
As discussed above, a structural comparison suggests that charge–charge repulsion between Asp37 and the γ-phosphate group of GTP is important in initiating the GTP-dependent conformational changes that bring Glu42 to form a salt bridge with Arg130 with the opposite protomer (SI Appendix, Fig. S2B). We argued that these residues are important in inducing the conformational changes of UreG from a GDP-bound state to a GTP-bound state. To test this hypothesis, we created a double mutant, D37A/E42A, of HpUreG and tested its ability to dimerize upon addition of nickel ions and GTP. It is expected that the double mutant should favor the GDP-bound state conformation even in the presence of GTP or its analog. Circular dichroism spectra of the D37A/E42A mutant, collected at 25–42 °C, were similar to those of the wild-type HpUreG, suggesting that the mutant was folded and stable at these temperatures (SI Appendix, Fig. S9). Based on the size-exclusion chromatography/static light-scattering (SEC/SLS) experiments, we showed that the elution profile of the D37A/E42A mutant in the presence of GTPγS was similar to that of wild-type HpUreG in the presence of GDP, suggesting that the double mutant failed to undergo GTP-dependent dimerization of HpUreG (Fig. 3A). We further showed that D37A/E42A HpUreG abolished its in vitro GTPase activity (Fig. 3B) and its ability to activate urease in vitro (Fig. 3C). Taken together, our results are consistent with the conclusion that Asp37 and Glu42 are essential for the GTP-induced conformational changes that lead to UreG dimerization, which is in turn important for GTPase activity and urease activation.
Fig. 3.
Double mutation D37A/E42A abolishes the formation of the nickel-charged HpUreG dimer, GTPase activity, and urease activation. (A) Protein samples of 30 µM H. pylori UreG (WT or mutant) were mixed with 45 µM nickel ion and 300 µM GTPγS/GDP and were analyzed using SEC/SLS. The wild-type UreG mainly existed as a dimer in the presence of GTPγS (injection 1), but as a monomer in the presence of GDP (injection 2). In contrast, the D37A/E42A mutant mainly existed as a monomer regardless of addition of GTPγS or GDP (injections 3 and 4). (B) GTP hydrolysis was followed by the amount of phosphate released using the malachite green assay as described in Materials and Methods. A total of 5 µM of UreG (WT or D37A/E42A mutant) was incubated with 300 µM of GTP/GTPγS in 2 mM MgSO4, 10 mM potassium bicarbonate, 4 µM NiSO4, 200 mM NaCl, 1 mM TCEP, 20 mM Hepes pH 7.5 buffer at 37 °C for 60 min. The hydrolysis rates were determined and analyzed by linear regression using the PRISM program (GraphPad Software). The hydrolysis rate of the wild-type HpUreG was significantly different from those of the double mutant and the GTPγS control (P < 0.01), while there were no significant differences between the mutant and the GTPγS control. Moreover, the slope of the regression lines for the double mutant and the GTPγS were not significantly deviated from zero. Relative activity was normalized using the hydrolysis rate of wild-type HpUreG (43 ± 7 nM phosphate/µM UreG/min). (C) A total of 10 µM apourease was activated by 40 µM UreG (WT or mutant) and 20 µM UreF2H2 complex in 2 mM MgSO4, 10 mM potassium bicarbonate, 45 µM NiSO4, 300 µM GTP, and 20 mM Hepes pH 7.5, 200 mM NaCl, 1 mM TCEP, at 37 °C for 20 min. Urease activity was determined by measuring the amount of ammonia released. In the “urease only” control, no urease accessory protein was added, while in the “buffer” control, no apourease or urease accessory proteins were added. Relative activity was normalized using the activity of urease activated by wild-type HpUreG (304 ± 5 µmol NH3/mg urease/min).
D37A/E42A Double Mutation Greatly Reduces the GTP-Dependent Dissociation of HpUreG from UreG2F2H2 and the Formation of the UreE2G2 Complex.
Next, we tested whether Asp37 and Glu42 are essential for the dissociation of HpUreG from the UreG2F2H2 complex. We added GDP or GTPγS to the UreG2F2H2 complex in a buffer containing nickel (SI Appendix, Fig. S3). In the presence of GDP, both wild-type and D37A/E42A HpUreG were able to form the UreG2F2H2 complex. Upon addition of GTPγS, the majority of wild-type HpUreG dissociated from the UreG2F2H2 complex to form the UreG dimer (SI Appendix, Fig. S3). In contrast, the dissociation of UreG was greatly reduced in the D37A/E42A mutant.
In a previous study, UreG was shown to interact with UreE to form a UreE2G2 complex in the presence of GTP, but to form a UreE2G complex in the presence of GDP (29). We hypothesized that the conformational changes induced upon GTP binding will cause UreG to prefer a 2:2 stoichiometry in forming a complex with UreE. Therefore, we tested whether the D37A/E42A mutations will affect the formation of the UreE2G2 complex. H. pylori UreG and UreE were added in equal molar ratio in a buffer containing nickel and GDP/GTPγS (SI Appendix, Fig. S4). In the presence of GDP, both wild-type and D37A/E42A HpUreG mainly formed a 2:1 complex with UreE (SI Appendix, Fig. S4, cyan lines). Consistent with previous findings, wild-type HpUreG was able to form complex with UreE in a 2:2 stoichiometry in the presence of GTPγS (SI Appendix, Fig. S4, red lines). In the case of the D37A/E42A mutant, the GTP-dependent formation of the UreE2G2 complex was greatly reduced. Taken together, our results suggest that the double mutation, D37A/E42A, prevents the GTP-dependent conformational changes of UreG that are essential for the dissociation of UreG from the UreG2F2H2 complex and the formation of UreE2G2 complex.
HpUreG Swaps Protein-Binding Partners During the GTP Hydrolysis/Binding Cycle.
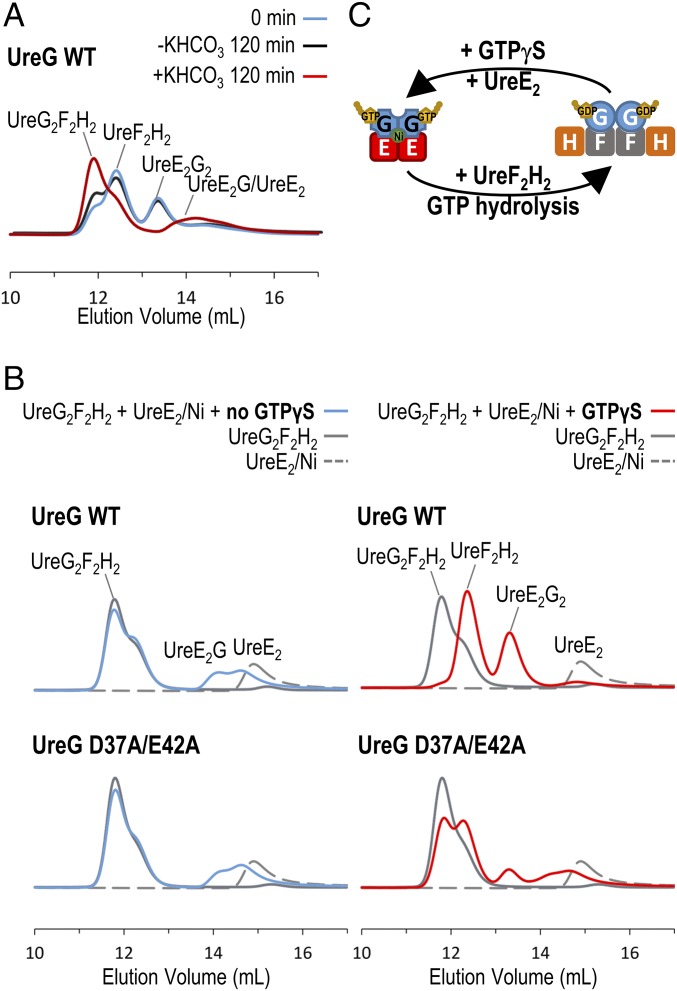
Our structural studies suggest that UreG exists in two distinct conformational states: the GDP-bound and the GTP-bound state. UreG prefers to form the UreG2F2H2 complex with UreF and UreH in the presence of GDP (20) (SI Appendix, Fig. S3), but prefers to form the UreE2G2 complex with UreE in the presence of GTPγS (29) (SI Appendix, Fig. S4). These observations suggest that GTP hydrolysis should change the conformational state of UreG and cause it to change protein-binding partners. To test this hypothesis, we first prepared a UreE2G2 complex by mixing nickel-charged H. pylori UreE dimer (UreE2/Ni) with HpUreG in the presence of GTP (SI Appendix, Fig. S5). Upon activation of GTP hydrolysis by addition of KHCO3, the UreE2G2 complex was dissociated into UreE2G and a monomeric HpUreG (SI Appendix, Fig. S6). When the UreE2G2 complex was mixed with the UreF2H2 complex, HpUreG was displaced from UreE2G2 and formed the UreG2F2H2 complex upon addition of KHCO3 (Fig. 4A).
Fig. 4.
UreG swaps protein-binding partners during the GTP hydrolysis/binding cycle. (A) Equal molar ratio (15 µM) mixture of H. pylori UreE2G2/Ni and UreF2H2 complexes (cyan) was incubated in 2 mM MgSO4, 1 mM GTP, 0.2 mM TCEP, 100 mM NaCl, 20 mM Hepes, pH 7.2 with (red) or without (black) 10 mM KHCO3 at 37 °C for 120 min. The protein samples were then analyzed by SEC/SLS. Upon activation of GTP hydrolysis by KHCO3 (red), the UreE2G2 complex disappeared and the majority of the UreF2H2 complex was converted to the UreG2F2H2 complex. (B) A total of 15 µM UreG2F2H2 complex (gray solid lines) and 15 µM UreE2/Ni dimer (gray dotted lines) was added to 2 mM MgSO4, 0.2 mM TCEP, 100 mM NaCl, 20 mM Hepes, pH 7.2 buffer with (red lines) or without (cyan lines) 300 µM GTPγS. The protein samples were analyzed by SEC/SLS. In the absence of GTPγS (Left), only a small amount of UreG dissociated from the UreG2F2H2 complex to form the UreE2G complex. In the presence of GTPγS (Right), wild-type UreG completely dissociated from the UreG2F2H2 complex to form the UreE2G2 complex with the UreE dimer. For the UreG D37A/E42A mutant, the GTP-dependent swapping of protein-binding partners was greatly abolished. (C) Our results suggest that the preference of protein-binding partners is dictated by conformational changes in UreG induced by GTP binding/hydrolysis.
Next, we tested whether the UreE2G2 complex can be regenerated from the UreG2F2H2 complex by addition of GTPγS. We prepared a H. pylori GDP-bound UreG2F2H2 complex and mixed it with the nickel-charged UreE dimer (Fig. 4B). We showed that the majority of the UreG2F2H2 complex remained intact despite the fact that small amounts of HpUreG were dissociated from the UreG2F2H2 complex to form the UreE2G complex (Fig. 4B, Left). This observation suggests that UreG prefers to form a complex with UreF2H2 over UreE2 in its GDP-bound conformation. In contrast, addition of GTPγS to the UreG2F2H2 complex and the nickel-charged UreE dimer resulted in the formation of the UreF2H2 and UreE2G2 complexes (Fig. 4B, Right). We further showed that HpUreG could also switch from the UreG2F2H2 complex to the UreE2G2 complex in the absence of nickel ion (SI Appendix, Fig. S7), suggesting that the swapping of protein-binding partners is only dependent on GTP but not on nickel. Noteworthy, the ability of HpUreG to swap protein-binding partners was greatly reduced by the D37A/E42A mutations (Fig. 4B, Right), which presumably favors the GDP-bound state of UreG. Taken together, our results are consistent with the conclusion that the conformational changes upon GTP hydrolysis/binding dictate the protein-binding partners of UreG (Fig. 4C).
In Vitro Urease Activation Assay Suggests That UreE Is the Nickel Source for Urease Maturation.
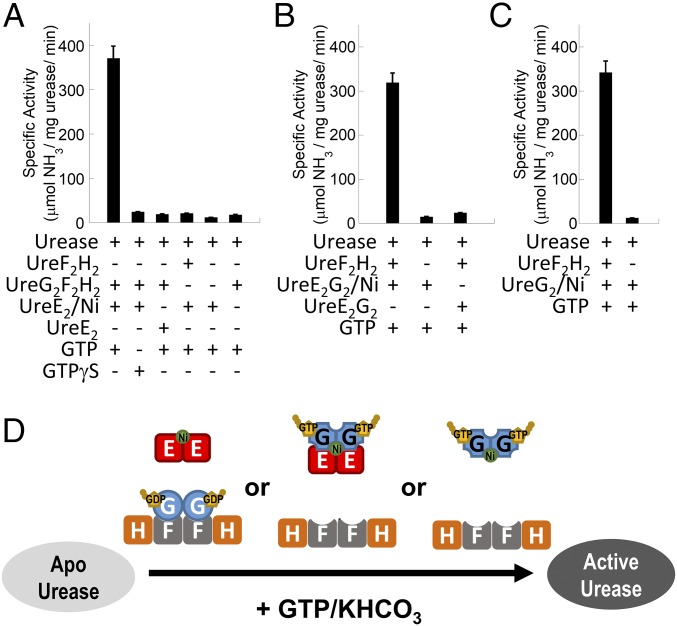
We have established an in vitro urease activation assay, in which purified samples of urease accessory proteins are added to the apourease to test how they affect H. pylori urease activation (20). We showed that purified nickel-charged UreG dimers (UreG2/Ni), which provide the sole source of nickel, can activate urease in vitro in the presence of the UreF2H2 complex (20). It has been shown that nickel ions can be transferred from UreE to UreG via the formation of the UreE2G2 complex (29), suggesting that UreE should be the source of nickel for urease activation. To test this hypothesis, we added a purified sample of nickel-charged H. pylori UreE dimer (UreE2/Ni) to the UreG2F2H2 complex and apourease and showed that the urease was activated in the presence of GTP, but not in the presence of GTPγS (Fig. 5A). The activation was nickel dependent because adding apo-UreE2 without the bound nickel (UreE2) failed to activate urease (Fig. 5A). Moreover, the activation requires HpUreG because the nickel-charged UreE dimer was not able to activate urease in the presence of the UreF2H2 complex (Fig. 5A).
Fig. 5.
In vitro urease activation assays suggest that nickel-charged UreE dimer provides the nickel source for urease maturation. The in vitro urease activation assay was performed by incubating 10 µM H. pylori apourease with 20 µM of H. pylori urease accessory proteins/complexes as indicated at 37 °C for 20 min in 20 mM Hepes pH 7.5 buffer containing 1 mM GTP or GTPγS, 2 mM MgSO4, 10 mM potassium bicarbonate, 200 mM NaCl, and 1 mM TCEP. Urease activity was measured by the amount of ammonia released. Protein samples of urease accessory proteins/complexes were prepared and analyzed by SEC/SLS (SI Appendix, Fig. S8). Nickel-charged UreE dimer (UreE2/Ni) and nickel-charged UreE2G2 (UreE2G2/Ni) complex were prepared and analyzed by atomic absorption spectroscopy (SI Appendix, Fig. S5). (A) Apourease was activated only when 20 µM nickel-charged UreE dimer, providing the sole source of nickel, was added with the presence of 20 µM UreG2F2H2 complex. (B) Apourease was activated when 20 µM nickel-charged UreE2G2 complex (UreE2G2/Ni), providing the sole source of nickel, was added with the presence of 20 µM UreF2H2 complex. (C) Apourease (10 µM) was activated when 20 µM nickel-charged UreG dimer (UreG2/Ni), providing the sole source of nickel, was added with the presence of 20 µM UreF2H2 complex. (D) Schematic diagram summarizing the combination of urease accessory proteins/complexes that can activate urease in the in vitro assay. Either UreE2/Ni, UreE2G2/Ni, or UreG2/Ni can provide the nickel source for urease activation.
Our results also suggest that UreG switches from the UreG2F2H2 complex to the UreE2G2 complex upon GTP binding (Fig. 4B and SI Appendix, Fig. S7). So, we hypothesized that the resulting UreE2G2 and UreF2H2 complexes are essential to urease activation. To test this hypothesis, the nickel-charged H. pylori UreE2G2 and UreF2H2 complexes were added to the apourease (Fig. 5B). Our results showed the nickel-charged UreE2G2 complex (UreE2G2/Ni) was able to activate urease in the presence of UreF2H2 (Fig. 5B). The activation was nickel dependent because the UreE2G2 complex without the bound nickel failed to activate urease (Fig. 5B). The activity of the urease activated by UreE2G2/Ni was similar to that activated by UreG2/Ni (Fig. 5C). Taken together, our results suggest that the nickel-charged UreE dimer, providing the sole source of nickel, can activate urease via the formation of the nickel-charged UreE2G2 complex.
To show that protein–protein interactions are essential to urease activation, we set up the protein components of the in vitro urease activation assay in either side of a dialysis membrane in a two-chamber dialyzer as indicated in SI Appendix, Fig. S10. The dialysis membrane allows diffusion of nickel ions but prevents direct protein–protein interactions across the membrane. Consistent with what was observed in Fig. 5A, urease was activated when UreE2/Ni, providing the sole source of nickel ions, was added to the chamber on the right where UreG2F2H2 and apourease were present (SI Appendix, Fig. S10, A1). On the other hand, urease activation was greatly abolished when UreE2/Ni was separated from UreG2F2H2 and apourease by the dialysis membrane (SI Appendix, Fig. S10, A2), suggesting that the interactions of UreE2/Ni with other proteins in the system are essential to urease activation. Moreover, our results do not support the alternative hypothesis that nickel ions are released from UreE2/Ni into the solution, and then, through diffusion, picked up by UreG for urease activation. As a control, we showed that free nickel ions, if present, in the left chamber could diffuse across the dialysis membrane and activate urease in the right chamber with UreG2F2H2 (SI Appendix, Fig. S10, C1). While addition of Ni/GTP induces the dissociation of UreG2F2H2 into UreG2/Ni and UreF2H2 that could activate urease in vitro (20), it is unlikely to be physiologically relevant because cytoplasmic free nickel ions are kept at subnanomolar concentrations to avoid cytotoxicity (5, 7). Interestingly, urease activation was inhibited when apo-UreE2 was added to the left chamber (SI Appendix, Fig. S10, C2), presumably due to the removal of free nickel ions from the solution. Taken together, our results reinforce the suggestion that the delivery of nickel ions for urease activation requires interactions of UreE2/Ni with other urease accessory proteins.
Similarly, urease was activated when UreG2/Ni, providing the sole source of nickel ions, was added to the right chamber where UreF2H2 and apourease were present (SI Appendix, Fig. S10, B1), but not when UreG2/Ni was added to the left chamber (SI Appendix, Fig. S10, B2). These observations suggest that interactions between UreG2/Ni and UreF2H2/apourease are essential to urease activation. Moreover, addition of free nickel ions failed to activate the urease in the absence of UreG (SI Appendix, Fig. S10, C3), suggesting that UreG is required for the activation.
Discussion
In this study, we determined the crystal structure of the KpUreG dimer in complex with nickel ions and a nonhydrolyzable analog of GTP, GMPPNP. We showed that UreG exists in two distinct conformational states: the GTP-bound state and the GDP-bound state. Structural comparison reveals that GTP binding induces conformational changes in the G2 region, which are propagated to the CPH nickel-binding motif. The main theme of this study was to understand how conformational changes of UreG play essential roles in urease maturation.
First, our work provides structural insights into why GTP-dependent conformational changes would induce nickel binding and why GTP hydrolysis would promote nickel release that is essential for urease maturation. In the crystal structure of the H. pylori GDP-bound UreG in the UreG2F2H2 complex (20), Cys66 and His68 are pointing away from each other and are not in a position to chelate a nickel ion (SI Appendix, Fig. S2A). Upon GTP binding, Cys66 and His68 from both protomers of UreG form a square-planar coordination that chelates a nickel ion at the dimeric interface (Fig. 1A and SI Appendix, Fig. S2). Since the square-planar coordination is preferred for Ni2+ (for having a d8 electron configuration) but not for other ions such as Zn2+ (31), it justifies the observation that UreG has a stronger affinity toward Ni2+ than Zn2+ in the presence of GTP (20, 29). GTP hydrolysis reverts UreG to the GDP-bound conformational state and promotes nickel release.
Second, we showed that the conformational state of UreG dictates the formation of different protein complexes that are involved in urease maturation. It has been shown that HpUreG dissociates from the UreG2F2H2 complex in the presence of nickel ions and GTPγS (20). Here we showed that binding of GTP induces conformational changes in the G2 region so that the invariant residue Tyr39 of UreG makes steric clashes with UreF (Fig. 2D), facilitating the dissociation of UreG from the UreG2F2H2 complex. Interestingly, we showed that the UreE dimer can take the UreG from the UreG2F2H2 complex in the presence of GTPγS (Fig. 4B and SI Appendix, Fig. S7) to form the UreE2G2 complex.
That conformational changes are essential for UreG to change protein partners is further supported by mutagenesis studies. We identified that the invariant residues Asp37 and Glu42 play important roles in the conformational changes upon GTP binding. In the GDP-bound state of UreG, Asp37 partially occupies the γ-phosphate–binding pocket (Fig. 2C). Binding of GTP creates charge–charge repulsion between the γ-phosphate group of GTP and Asp37, which induces large conformational changes in the G2 region (Fig. 2). Notably, helix-2 turns ∼35° toward the nucleotide-binding site, bringing Glu42 to form an intermolecular salt bridge with another invariant residue, Arg130, that stabilizes the formation of the UreG dimer. The D37A/E42A mutations greatly abolished GTP-dependent dimerization of UreG and prevented dissociation of UreG from the UreG2F2H2 complex to form the UreE2G2 complex (Figs. 3 and 4). Apparently, the double mutation of D37A/E42A locks the conformation of UreG in the GDP-bound state even in the presence of GTP.
We have previously shown that the nickel-charged UreG dimer can activate urease in vitro, likely via the formation of an activation complex with UreF2H2 and urease (13, 19, 20, 24). It has been suggested that the nickel-charged UreG dimer gets its nickel from UreE. Yang and coworkers have demonstrated that UreE can receive its nickel from HypA and can form a UreE2G2 complex with UreG in the presence of GTP (29, 32, 33). Mutagenesis (29) and modeling (34) studies suggest that the metal binding sites of UreE and UreG should face toward each other and the nickel ion can be transferred from UreE to UreG within the UreE2G2 complex (29). Here, we provided direct evidence that the nickel-charged UreE dimer is the source of nickel, for it can activate urease in the presence of UreG2F2H2 and GTP (Fig. 5A). Our results also show that the nickel-charged UreE2G2 complex, which provides the sole source of nickel in the in vitro assay, can activate urease in the presence of UreF2H2 (Fig. 5B).
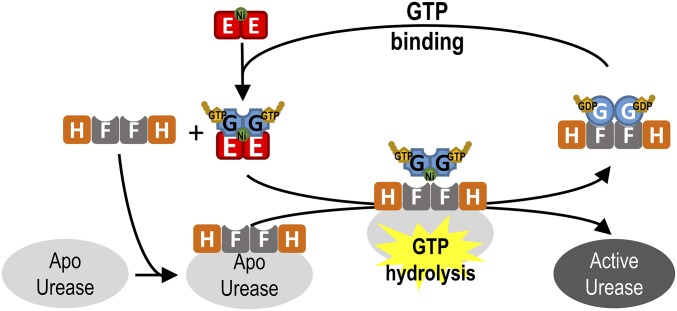
Taken together, the ability of UreG to form different protein complexes during the GTP hydrolysis/binding cycle provides a mechanism of how GTP-dependent conformational changes in UreG facilitate urease maturation (Fig. 6 and Movie S1). GTP binding induces conformational changes in UreG, causing it to dissociate from the UreG2F2H2 complex and to form the UreE2G2 complex with the nickel-charged UreE dimer (Fig. 6). Moreover, the dissociation of the UreG2F2H2 complex also yields the UreF2H2 complex, which can form a complex with the apourease (18, 35) making it ready for activation by either UreG2/Ni or UreE2G2/Ni (Fig. 5 B and C). GTP hydrolysis reverts UreG to its GDP-bound state, disrupting the square-planar coordination by Cys66/His68 and promoting release of nickel ion. It is unclear how the nickel ion released from UreG eventually reaches the catalytic site of urease. Recently, it has been suggested that the nickel ion may pass to UreF, and then go through a tunnel of UreH to the urease (36–38). After GTP hydrolysis-dependent activation of urease, the GDP-bound UreG2F2H2 is regenerated (Fig. 4A), which is ready for the next round of urease activation (Fig. 6).
Fig. 6.
How conformational changes in UreG during the GTP hydrolysis/binding cycle facilitate urease maturation. GTP binding induces conformational changes in UreG that destabilize the UreG2F2H2 complex, causing UreG to dissociate from the complex and form the UreE2G2 complex with the nickel-charged UreE dimer. After receiving its nickel within the UreE2G2 complex, the nickel-charged UreG dimer is recruited to form the activation complex with apourease and UreF2H2. GTP hydrolysis induces conformational changes in the CPH motif of UreG, disrupting the square-planar coordination by Cys66/His68, and, hence, promotes release of the nickel ion for urease maturation. UreG, now in its GDP-bound state, prefers to form the UreG2F2H2 complex, which is now ready to receive its nickel from UreE2/Ni for another round of urease maturation.
It is unclear whether the UreE2G2 complex can directly activate urease by forming a bigger activation complex with UreF2H2 and apourease or whether the activation requires the dissociation of the nickel-charged UreG dimer from the UreE2G2 complex. It has been suggested that the UreG–UreE interaction involves the protein surfaces near the nickel binding site of UreG, which is buried in the UreG2F2H2 complex (29, 34, 39). Moreover, we have previously demonstrated that the nickel-charged UreG dimer can form a complex with UreF2H2 and apourease (20) and the interaction between UreG and UreF2H2 is essential to the urease activation (20, 24, 25). It is, therefore, likely that after receiving its nickel ion, the nickel-charged UreG dimer will dissociate from the UreE2G2 complex and activate urease by the formation of a complex with apourease and UreF2H2 (Fig. 6). It is currently not known how the nickel-charged UreG dimer interacts with apourease and UreF2H2 in the activation complex and what triggers the GTP hydrolysis during urease maturation. Presumably, premature GTP hydrolysis in the absence of UreF2H2/apourease would result in losing the nickel ion to the solution. As a result, GTP hydrolysis of UreG is likely triggered by the formation of the activation complex with UreF2H2 and apourease. It has been suggested that binding of UreF2H2 to urease can induce large conformational changes in urease (18, 19, 28), which may promote the recruitment of the nickel-charged UreG dimer from the UreE2G2 complex to the activation complex, where GTP hydrolysis is triggered for urease maturation (Fig. 6). Future structural studies on the activation complexes with apourease and urease accessory proteins such as UreG and UreF2H2 may help to fill in the knowledge gap here.
Materials and Methods
Protein Expression and Purification.
H. pylori apourease, UreF2H2 complex, and UreG and its mutant were expressed and purified as described previously (20, 24). KpUreG was cloned into an in-house designed pRSETA-HisSUMO vector and expressed as an N-terminal HisSUMO-tagged fusion protein in Escherichia coli. The procedures for purification of HpUreG were used to purify KpUreG (20). Both HpUreG and KpUreG formed a stable dimer in the presence of Ni and GTP. The Ni/GTP-bound UreG dimers were prepared as described previously (20).
H. pylori UreE was cloned into pGEX-6p1 vector and expressed as an N-terminal GST-tagged fusion protein in E. coli. The transformed bacteria were grown to OD600 0.5 and induced with 1 mM isopropyl beta-d-1-thiogalactopyranoside at 18 °C overnight. Cells were resuspended in 20 mM Hepes pH 7.5, 200 mM NaCl and 1 mM Tris(2-carboxyethyl)phosphine hydrochloride (TCEP) (buffer A) and lysed by sonication. After removal of cell debris by centrifugation at 20,000 × g, 60 min, the cell lysate was loaded onto a 5-mL GSTrap column (GE Healthcare) preequilibrated with buffer A. After extensive washing with buffer A, the GST-tagged UreE was eluted using 10 mM glutathione in buffer A. The GST-tag was cleaved using PreScission Protease (GE Healthcare) and the protein sample was dialyzed in 20 mM Tris pH 7.5, 50 mM NaCl and 1 mM TCEP (buffer B). The protein sample was loaded onto a 5-mL HiTrap-SP column (GE Healthcare) preequilibrated with buffer B, and UreE was eluted using 500 mM NaCl, 20 mM Tris pH 7.5 and 1 mM TCEP. To remove any bound metal in the UreE sample, 1 mM EDTA was added followed by gel filtration chromatography using a HiLoad Superdex 75 PG column (GE Healthcare) preequilibrated with buffer A.
Protein samples of nickel-charged UreE dimer were prepared by adding 1 mM NiSO4 to 200 µM sample of UreE. Excess nickel in the protein sample was removed by a HiTrap Desalting column (GE Healthcare) preequilibrated with buffer A. To prepare the nickel-charged UreE2G2 complex, equal molar ratio (∼100 µM) of nickel-charged UreE dimer and UreG was mixed in the presence of 2 mM MgSO4 and 1 mM GTP, followed by gel filtration chromatography using a Superdex 200 Increase 10/300 gel filtration column (GE Healthcare). The amount of bound nickel in UreE2 and UreE2G2 was estimated by atomic absorption spectroscopy (SI Appendix, Fig. S5). To prepare UreE2G2 complex without the bound nickel, apo-UreE2 and UreG were mixed instead. The UreG2F2H2 complex was prepared as described previously (20). The molecular weight of all protein samples prepared were analyzed by size-exclusion chromatography/static light-scattering (SI Appendix, Fig. S8).
Protein Crystallization and Structure Determination.
Purified KpUreG was dialyzed into 20 mM Tris buffer pH 7.5 containing 0.5 mM TCEP and concentrated to 14 mg/mL for crystallization. A total of 2 mM GMPPNP, 4 mM MgSO4, and 2 mM NiSO4 was added to the protein sample before crystallization. Full-length KpUreG was crystallized but crystals were of poor diffraction quality. A truncated construct KpUreG(∆N4∆C1) was used for crystallization to improve crystal quality. The protein was crystallized in 100 mM Hepes pH 7.5, 1.8 M (NH4)2SO4, and 3% dioxane at 16 °C using the hanging-drop-vapor-diffusion setup. Crystals were cryoprotected by soaking in a 1:1 mix of mother liquor with 3.4 M sodium malonate pH 7.0 solution, and flash frozen in liquid nitrogen. Diffraction data were collected using an in-house rotating anode X-ray generator (Rigaku FRE+) and a RAXIS IV imaging plate detector. Diffraction data were indexed and integrated using XDS (40) and scaled with AIMLESS (41) as programmed in Xia2 (42). Initial phases were determined by the molecular replacement method using the structure of HpUreG found in the UreG2F2H2 complex (PDB ID Code 4HI0) using the program PHENIX.AUTOMR (43). Initial models were build using PHENIX.AUTOBUILD and ARP/wARP (44) followed by iterative rounds of manual building using COOT (45) and refinement using PHENIX.REFINE (43). Correctness of the final models was checked using MOLPROBITY (46). To confirm the position of bound nickel in the crystals, diffraction data were also collected at the nickel peak wavelength using beamline I02 of the Diamond Light Source (SI Appendix, Fig. S1). For this dataset, the protein was crystallized in 100 mM Hepes pH 7.5, 1.8 M (NH4)2SO4 and 4% ethylene glycol at 16 °C. The anomalous difference electron density was generated by PHENIX.REFINE. Figures of protein structures were created using PyMOL (www.pymol.org).
Size-Exclusion Chromatography/Static Light Scattering.
SEC/SLS was used to obtain the elution profile and to estimate the molecular weight of protein complexes of urease accessory proteins. H. pylori urease accessory proteins were used in all SEC/SLS experiments. Protein samples were injected to a Superdex 200 Increase 10/300 gel filtration column (GE Healthcare) attached to a downstream miniDawn light scattering detector and an Optilab DSP refractometer (Wyatt Technologies), and preequilibrated with 20 mM Hepes pH 7.2, 100 mM NaCl, 0.2 mM TCEP (buffer C). Data were analyzed using the ASTRA software provided by the manufacturer.
For the studies of UreG dimerization (Fig. 3A) and UreE/UreG interaction (SI Appendix, Fig. S4), 100 µl of 30 µM protein samples (UreE and/or UreG) in 2 mM MgSO4, 45 µM NiSO4, and 300 µM GTPγS or GDP were mixed and incubated at room temperature for 10 min, before they were injected into the Superdex 200 Increase 10/300 column and analyzed by SEC/SLS. For the study of Ni/GTP-dependent dissociation of UreG from the UreG2F2H2 complex (SI Appendix, Fig. S3), 100 µL of 15 µM of purified UreG2F2H2 complex in 2 mM MgSO4 and 300 µM GTPγS or GDP with 45 µM NiSO4 were analyzed. For the study of effect of GTP hydrolysis on the urease accessory protein complexes (Fig. 4A), nickel-charged UreE2G2 complex and the UreF2H2 complex were mixed in equal molar ratio (∼15 µM) and incubated at room temperature for 10 min before the protein samples were injected into the Superdex 200 Increase 10/300 gel filtration column with or without prior incubation of 10 mM potassium bicarbonate for 120 min at 37 °C. To test whether UreG or its mutant can swap protein-binding partners from UreF2H2 to UreE2 upon GTP binding, (Fig. 4B), the UreG2F2H2 complex and the nickel-charged UreE dimer were mixed in equal molar ratio (∼15 µM) with or without 300 µM GTPγS and incubated at room temperature for 10 min before the protein samples were analyzed by SEC/SLS.
GTPase Assay.
To investigate the effect of the HpUreG mutant (D37A/E42A) on GTPase activity (Fig. 3B), 200 µL of 5 µM of HpUreG (WT/mutant) was incubated in 2 mM MgSO4, 300 µM GTP (or GTPγS), 10 mM potassium bicarbonate, 4 µM NiSO4, 200 mM NaCl, 1 mM TCEP, 20 mM Hepes pH 7.5 buffer for 20, 40, and 60 min at 37 °C. Phosphate released was measured using a colorimetric assay based on malachite green as described (47).
In Vitro Urease Activation Assay.
H. pylori urease accessory proteins and apourease were used for all in vitro urease activation assays. To investigate the effect of the UreG mutant (D37A/E42A) on urease activation (Fig. 3C), an in vitro urease activity assay using purified proteins was used. A total of 10 µM H. pylori apourease, 20 µM UreF2H2 complex, and 40 µM UreG (WT/mutant) were incubated in 20 mM Hepes pH 7.5, 200 mM NaCl, 1 mM TCEP, 2 mM MgSO4, 10 mM potassium bicarbonate, 45 µM NiSO4, and 300 µM GTP at 37 °C for 20 min. Urease activity was then determined by incubating the activated enzyme with 50 mM urea for 30 min at 37 °C and the ammonia released was measured using a phenol/hypochlorite reaction (48).
To investigate whether the nickel-charged UreE dimer, providing the sole source of nickel, can activate urease in vitro (Fig. 5A), 20 µM of nickel-charged UreE dimer was added to 10 µM of apourease with/without 20 µM of UreG2F2H2 complex in the assay buffer (20 mM Hepes pH 7.5, 200 mM NaCl, 1 mM TCEP, 2 mM MgSO4, 10 mM potassium bicarbonate, and 1 mM GTP). To investigate whether the nickel-charged UreE2G2 complex or nickel-charged UreG dimer can activate urease in vitro (Fig. 5 B and C), 20 µM of nickel-charged UreE2G2 complex or nickel-charged UreG dimer was added to 10 µM of apourease with/without 20 µM of UreF2H2 complex in the assay buffer. In all cases, activity of the activated urease was measured by the amount of ammonia released in 30 min at 37 °C using 50 mM urea as substrate (20).
To investigate whether protein–protein interactions are essential to the urease activation, urease accessory proteins (UreE2/Ni, UreG2/Ni, apo-UreE2, apo-UreG, UreG2F2H2, and UreF2H2) and apourease were added, as indicated in SI Appendix, Fig. S10, to either side of a dialysis membrane with a molecular weight cutoff of 6–8 kDa (Spectrum Labs) in a two-chamber dialyzer (Bioprobes, Ltd.). In the experiments reported in SI Appendix, Fig. S10C, 20 μM NiSO4 was also added to the left chamber. The buffer in both chambers contained 20 mM Hepes pH 7.5, 200 mM NaCl, 1 mM TCEP, 2 mM MgSO4, and 1 mM GTP. After equilibration at 4 °C for 16 h, 10 mM KHCO3 was added to both chambers to activate the GTP hydrolysis required for urease activation. After incubation at 37 °C for 1 h, urease activity was determined as described above.
Circular Dichroism.
Circular dichroism spectra of wild-type and D37A/E42A HpUreG were measured with protein samples in 5 mM sodium phosphate buffer at pH 7.5 using a 0.5-mm path length cuvette with a JASCO J810 spectropolarimeter equipped with a Peltier-type temperature control unit.
Supplementary Material
Acknowledgments
We thank Ms. Shu Nga Lui for her help in protein expression and purification and Dr. Yu-Wai Chen of King's College London and Dr. Pierre Aller of the Diamond Light Source for their help in diffraction data collection. This work was supported by grants from the Research Grants Council of Hong Kong (14117314 and AoE/M-05/12) and direct grants from The Chinese University of Hong Kong (3132814 and 3132815).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.wwpdb.org (PDB ID code 5XKT).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1712658114/-/DCSupplemental.
References
- 1.Andreini C, Bertini I, Cavallaro G, Holliday GL, Thornton JM. Metal ions in biological catalysis: From enzyme databases to general principles. J Biol Inorg Chem. 2008;13:1205–1218. doi: 10.1007/s00775-008-0404-5. [DOI] [PubMed] [Google Scholar]
- 2.Waldron KJ, Rutherford JC, Ford D, Robinson NJ. Metalloproteins and metal sensing. Nature. 2009;460:823–830. doi: 10.1038/nature08300. [DOI] [PubMed] [Google Scholar]
- 3.Chandrangsu P, Rensing C, Helmann JD. Metal homeostasis and resistance in bacteria. Nat Rev Microbiol. 2017;15:338–350. doi: 10.1038/nrmicro.2017.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Waldron KJ, Robinson NJ. How do bacterial cells ensure that metalloproteins get the correct metal? Nat Rev Microbiol. 2009;7:25–35. doi: 10.1038/nrmicro2057. [DOI] [PubMed] [Google Scholar]
- 5.Foster AW, Osman D, Robinson NJ. Metal preferences and metallation. J Biol Chem. 2014;289:28095–28103. doi: 10.1074/jbc.R114.588145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Macomber L, Hausinger RP. Mechanisms of nickel toxicity in microorganisms. Metallomics. 2011;3:1153–1162. doi: 10.1039/c1mt00063b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Capdevila DA, Edmonds KA, Giedroc DP. Metallochaperones and metalloregulation in bacteria. Essays Biochem. 2017;61:177–200. doi: 10.1042/EBC20160076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zeer-Wanklyn CJ, Zamble DB. Microbial nickel: Cellular uptake and delivery to enzyme centers. Curr Opin Chem Biol. 2017;37:80–88. doi: 10.1016/j.cbpa.2017.01.014. [DOI] [PubMed] [Google Scholar]
- 9.Robinson NJ, Winge DR. Copper metallochaperones. Annu Rev Biochem. 2010;79:537–562. doi: 10.1146/annurev-biochem-030409-143539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Higgins KA, Carr CE, Maroney MJ. Specific metal recognition in nickel trafficking. Biochemistry. 2012;51:7816–7832. doi: 10.1021/bi300981m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pearson MA, Schaller RA, Michel LO, Karplus PA, Hausinger RP. Chemical rescue of Klebsiella aerogenes urease variants lacking the carbamylated-lysine nickel ligand. Biochemistry. 1998;37:6214–6220. doi: 10.1021/bi980021u. [DOI] [PubMed] [Google Scholar]
- 12.Scott DR, Marcus EA, Weeks DL, Sachs G. Mechanisms of acid resistance due to the urease system of Helicobacter pylori. Gastroenterology. 2002;123:187–195. doi: 10.1053/gast.2002.34218. [DOI] [PubMed] [Google Scholar]
- 13.Farrugia MA, Macomber L, Hausinger RP. Biosynthesis of the urease metallocenter. J Biol Chem. 2013;288:13178–13185. doi: 10.1074/jbc.R112.446526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carter EL, Flugga N, Boer JL, Mulrooney SB, Hausinger RP. Interplay of metal ions and urease. Metallomics. 2009;1:207–221. doi: 10.1039/b903311d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ge RG, Wang DX, Hao MC, Sun XS. Nickel trafficking system responsible for urease maturation in Helicobacter pylori. World J Gastroenterol. 2013;19:8211–8218. doi: 10.3748/wjg.v19.i45.8211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim JK, Mulrooney SB, Hausinger RP. The UreEF fusion protein provides a soluble and functional form of the UreF urease accessory protein. J Bacteriol. 2006;188:8413–8420. doi: 10.1128/JB.01265-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee MH, Mulrooney SB, Renner MJ, Markowicz Y, Hausinger RP. Klebsiella aerogenes urease gene cluster: Sequence of ureD and demonstration that four accessory genes (ureD, ureE, ureF, and ureG) are involved in nickel metallocenter biosynthesis. J Bacteriol. 1992;174:4324–4330. doi: 10.1128/jb.174.13.4324-4330.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chang Z, Kuchar J, Hausinger RP. Chemical cross-linking and mass spectrometric identification of sites of interaction for UreD, UreF, and urease. J Biol Chem. 2004;279:15305–15313. doi: 10.1074/jbc.M312979200. [DOI] [PubMed] [Google Scholar]
- 19.Farrugia MA, et al. Analysis of a soluble (UreD:UreF:UreG)2 accessory protein complex and its interactions with Klebsiella aerogenes urease by mass spectrometry. J Am Soc Mass Spectrom. 2013;24:1328–1337. doi: 10.1007/s13361-013-0677-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fong YH, et al. Structure of UreG/UreF/UreH complex reveals how urease accessory proteins facilitate maturation of Helicobacter pylori urease. PLoS Biol. 2013;11:e1001678. doi: 10.1371/journal.pbio.1001678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boer JL, Quiroz-Valenzuela S, Anderson KL, Hausinger RP. Mutagenesis of Klebsiella aerogenes UreG to probe nickel binding and interactions with other urease-related proteins. Biochemistry. 2010;49:5859–5869. doi: 10.1021/bi1004987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carter EL, Hausinger RP. Characterization of the Klebsiella aerogenes urease accessory protein UreD in fusion with the maltose binding protein. J Bacteriol. 2010;192:2294–2304. doi: 10.1128/JB.01426-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Soriano A, Hausinger RP. GTP-dependent activation of urease apoprotein in complex with the UreD, UreF, and UreG accessory proteins. Proc Natl Acad Sci USA. 1999;96:11140–11144. doi: 10.1073/pnas.96.20.11140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fong YH, et al. Assembly of preactivation complex for urease maturation in Helicobacter pylori: Crystal structure of UreF-UreH protein complex. J Biol Chem. 2011;286:43241–43249. doi: 10.1074/jbc.M111.296830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boer JL, Hausinger RP. Klebsiella aerogenes UreF: Identification of the UreG binding site and role in enhancing the fidelity of urease activation. Biochemistry. 2012;51:2298–2308. doi: 10.1021/bi3000897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bange G, Sinning I. SIMIBI twins in protein targeting and localization. Nat Struct Mol Biol. 2013;20:776–780. doi: 10.1038/nsmb.2605. [DOI] [PubMed] [Google Scholar]
- 27.Gasper R, Meyer S, Gotthardt K, Sirajuddin M, Wittinghofer A. It takes two to tango: Regulation of G proteins by dimerization. Nat Rev Mol Cell Biol. 2009;10:423–429. doi: 10.1038/nrm2689. [DOI] [PubMed] [Google Scholar]
- 28.Quiroz-Valenzuela S, Sukuru SCK, Hausinger RP, Kuhn LA, Heller WT. The structure of urease activation complexes examined by flexibility analysis, mutagenesis, and small-angle X-ray scattering. Arch Biochem Biophys. 2008;480:51–57. doi: 10.1016/j.abb.2008.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang X, Li H, Lai TP, Sun H. UreE-UreG complex facilitates nickel transfer and preactivates GTPase of UreG in Helicobacter pylori. J Biol Chem. 2015;290:12474–12485. doi: 10.1074/jbc.M114.632364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wittinghofer A, Vetter IR. Structure-function relationships of the G domain, a canonical switch motif. Annu Rev Biochem. 2011;80:943–971. doi: 10.1146/annurev-biochem-062708-134043. [DOI] [PubMed] [Google Scholar]
- 31.Kuppuraj G, Dudev M, Lim C. Factors governing metal-ligand distances and coordination geometries of metal complexes. J Phys Chem B. 2009;113:2952–2960. doi: 10.1021/jp807972e. [DOI] [PubMed] [Google Scholar]
- 32.Yang X, et al. Nickel translocation between metallochaperones HypA and UreE in Helicobacter pylori. Metallomics. 2014;6:1731–1736. doi: 10.1039/c4mt00134f. [DOI] [PubMed] [Google Scholar]
- 33.Benoit SL, McMurry JL, Hill SA, Maier RJ. Helicobacter pylori hydrogenase accessory protein HypA and urease accessory protein UreG compete with each other for UreE recognition. Biochim Biophys Acta. 2012;1820:1519–1525. doi: 10.1016/j.bbagen.2012.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bellucci M, Zambelli B, Musiani F, Turano P, Ciurli S. Helicobacter pylori UreE, a urease accessory protein: Specific Ni(2+)- and Zn(2+)-binding properties and interaction with its cognate UreG. Biochem J. 2009;422:91–100. doi: 10.1042/BJ20090434. [DOI] [PubMed] [Google Scholar]
- 35.Moncrief MBC, Hausinger RP. Purification and activation properties of UreD-UreF-urease apoprotein complexes. J Bacteriol. 1996;178:5417–5421. doi: 10.1128/jb.178.18.5417-5421.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Farrugia MA, Wang B, Feig M, Hausinger RP. Mutational and computational evidence that a nickel-transfer tunnel in UreD is used for activation of Klebsiella aerogenes urease. Biochemistry. 2015;54:6392–6401. doi: 10.1021/acs.biochem.5b00942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zambelli B, et al. Nickel binding properties of Helicobacter pylori UreF, an accessory protein in the nickel-based activation of urease. J Biol Inorg Chem. 2014;19:319–334. doi: 10.1007/s00775-013-1068-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Musiani F, et al. Protein tunnels: The case of urease accessory proteins. J Chem Theory Comput. 2017;13:2322–2331. doi: 10.1021/acs.jctc.7b00042. [DOI] [PubMed] [Google Scholar]
- 39.Merloni A, et al. Molecular landscape of the interaction between the urease accessory proteins UreE and UreG. Biochim Biophys Acta. 2014;1844:1662–1674. doi: 10.1016/j.bbapap.2014.06.016. [DOI] [PubMed] [Google Scholar]
- 40.Kabsch W. Evaluation of single-crystal X-ray diffraction data from a position-sensitive detector. J Appl Cryst. 1988;21:916–924. [Google Scholar]
- 41.Winter G, Lobley CMC, Prince SM. Decision making in xia2. Acta Crystallogr D Biol Crystallogr. 2013;69:1260–1273. doi: 10.1107/S0907444913015308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Winter G. Xia2: An expert system for macromolecular crystallography data reduction. J Appl Cryst. 2010;43:186–190. [Google Scholar]
- 43.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Langer G, Cohen SX, Lamzin VS, Perrakis A. Automated macromolecular model building for X-ray crystallography using ARP/wARP version 7. Nat Protoc. 2008;3:1171–1179. doi: 10.1038/nprot.2008.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen VB, et al. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Baykov AA, Evtushenko OA, Avaeva SM. A malachite green procedure for orthophosphate determination and its use in alkaline phosphatase-based enzyme immunoassay. Anal Biochem. 1988;171:266–270. doi: 10.1016/0003-2697(88)90484-8. [DOI] [PubMed] [Google Scholar]
- 48.Weatherburn MW. Phenol-hypochlorite reaction for determination of ammonia. Anal Chem. 1967;39:971–974. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.