Abstract
Background:
Ebola viral disease is a severe and mostly fatal disease in humans caused by Ebola virus. This virus belongs to family Filoviridae and is a single-stranded negative-sense virus. There is no single treatment for this disease which puts forth the need to identify new therapy to control and treat this fatal condition. Curcumin, one of the bioactives of turmeric, has proven antiviral property.
Objective:
The current study evaluates the inhibitory activity of curcumin, bisdemethoxycurcumin, demethoxycurcumin, and tetrahydrocurcumin against Zaire Ebola viral proteins (VPs).
Materials and Methods:
Molecular simulation of the Ebola VPs followed by docking studies with ligands comprising curcumin and related compounds was performed.
Results:
The highest binding activity for VP40 is −6.3 kcal/mol, VP35 is −8.3 kcal/mol, VP30 is −8.0 kcal/mol, VP24 is −7.7 kcal/mol, glycoprotein is −7.1 kcal/mol, and nucleoprotein is 6.8 kcal/mol.
Conclusion:
Bisdemethoxycurcumin shows better binding affinity than curcumin for most VPs. Metabolite tetrahydrocurcumin also shows binding affinity comparable to curcumin. These results indicate that curcumin, curcuminoids, and metabolite tetrahydrocurcumin can be potential lead compounds for developing a new therapy for Ebola viral disease.
SUMMARY
Curcumin, bisdemethoxycurcumin, and demethoxycurcumin are active constituents of turmeric. Tetrahydrocurcumin is the major metabolite of curcumin formed in the body after consumption and absorption of curcuminoids
Curcuminoids have proven antiviral activity
Bisdemethoxycurcumin showed maximum inhibition of Ebola viral proteins (VPs) among the curcuminoids in the docking procedure with a docking score as high as −8.3 kcal/mol
Tetrahydrocurcumin showed inhibitory activity against Ebola VPs close to that of curcumin’s inhibitory action.
Abbreviations Used: EBOV: Ebola virus, GP: Glycoprotein, NP: Nucleoprotein, NPT: Isothermal-isobaric Ensemble, amount of substance (N), pressure (P) and temperature (T) conserved, NVE: Canonical ensemble, amount of substance (N), volume (V) and temperature (T) conserved, VP: Viral protein.
Keywords: Bisdemethoxycurcumin, curcumin, demethoxycurcumin, docking, Ebola virus, tetrahydrocurcumin
INTRODUCTION
The epidemic of Ebola virus (EBOV) that occurred in 2014 in West Africa has been the largest outbreak since the first report of the disease in 1976. Thousands to millions of potential cases were reported with a mortality rate up to 74%.[1]
There have been 15 outbreaks since the first report of EBOV in 1976. EBOV is considered a rare virus, but there has been slow progress in prevention and treatment.[2,3]
EBOV is a zoonotic filovirus, consisting of enveloped, nonsegmented negative-stranded RNA. Up till now, five species have been identified: Zaire ebolavirus, Bundibugyo ebolavirus, Tai Forest ebolavirus, Sudan ebolavirus, and Reston ebolavirus.[4,5]
The virus name is derived from the Ebola River in the Democratic Republic of Congo (formerly Zaire), where the first case of hemorrhagic fever was recorded in 1976. The prevalence of the disease was reported simultaneously in Southern Sudan and in Northern Zaire and hence the name of the two different species SEBOV and ZEBOV, respectively.[6]
EBOV belongs to the family Filoviridae and the name Filoviridae comes from the Latin word “filum” or thread. Indeed, the virion shape resembles a twisted thread when viewed under an electron microscope. Among the viruses of the Ebolavirus genus, Z. ebolavirus has the highest fatality rate (up to 90%).[6]
EBOV particle has a uniform diameter of about 80 nm and the length from 970 to 1200 nm. The core consists of one molecule of linear, nonsegmented, single-stranded, negative-sense RNA. This RNA segment is helically wound and bound to nucleoprotein (NP), viral protein 35 (VP35), VP30, and L proteins.[6]
This helical structure is surrounded by an outer envelope of specific glycoprotein (GP) spikes. Viral matrix protein, VP40 and VP24, are located between the nucleocapsid (NC) and the outer envelope.[6]
The viral genome is approximately 19 kb in length. It is the longest among all viruses belonging to the order Mononegavirales.[6]
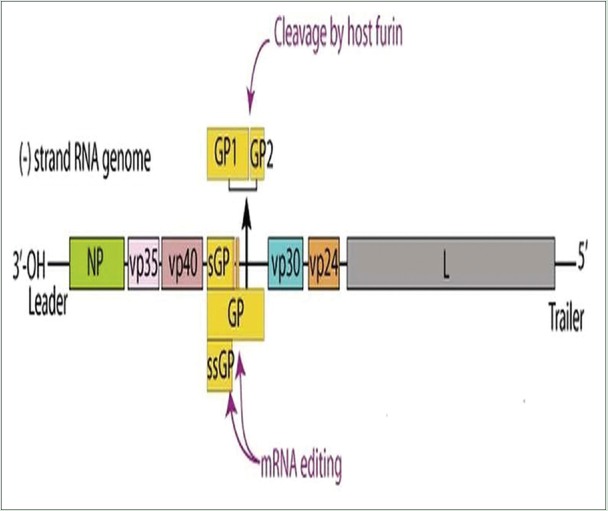
The Ebola VPs are arranged sequentially in the following manner [Figure 1], 3’- NP, polymerase cofactor (VP35), matrix protein (VP40), GP, protein VP30, matrix protein (VP24), RNA-dependent RNA polymerase (L), and one nonstructural small GP[6,7] [Figure 1].
Figure 1.
Arrangement of Ebola viral genome
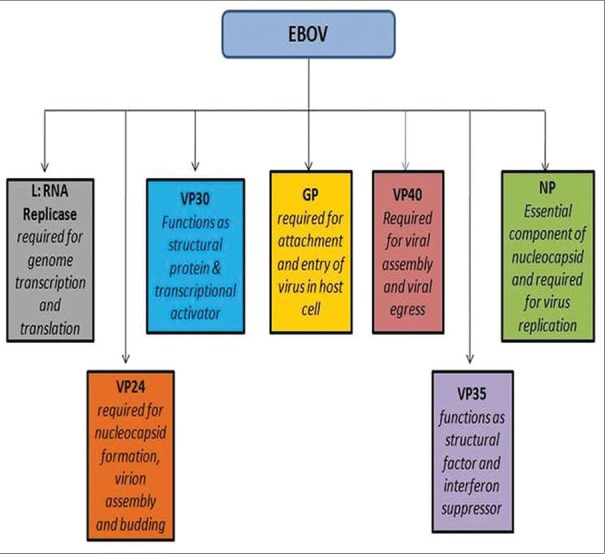
Each of these VPs plays different roles in structure, assembly, infection, and replication of the virus [Figure 2].
Figure 2.
Target Ebola viral proteins and the prime roles in Ebola virus infection
Unlike most viral infections, which remain asymptomatic and subclinical for quite some time, EBOV contrastingly shows high pathogenicity. Most viruses have high affinity to specific tissues, but EBOV can replicate in a wide range of human cells.[8]
EBOV first disables immune system and then attacks the vascular system as well as other organs. EBOV evokes severe illness associated with fever, myalgia, gastrointestinal symptoms (vomiting and diarrhea), hypotension, and abdominal ache. Hemorrhagic manifestations include rash on the skin, hemorrhages from mucosal linings, and bleeding from the gastrointestinal and genitourinary tract.[4,9,10] In severe conditions, patients develop hypovolemic shock and multiorgan failure.
There is no specific treatment for EBOV. Antiviral drugs such as brincidofovir and favipiravir have been identified. ZMapp is another experimental treatment for Ebola viral disease which consists of monoclonal antibodies that serve as antiviral therapy.[11]
Another drug BCX4430 (developed by BioCryst), a new synthetic adenosine analog, inhibits infection of different filoviruses in human cells.[12] TKM-Ebola and AVI-6002 are identified for the treatment of EBOV disease and exert their action via gene silencing. Vector-based vaccines are also being developed.[13]
Turmeric (Curcuma longa) and its polyphenolic compound curcumin are identified with antimicrobial properties traditionally.[14] Curcumin or diferuloylmethane and other curcuminoids – bisdemethoxycurcumin and demethoxycurcumin – constitute the main phytochemicals of C. longa. Tetrahydrocurcumin is the major metabolite of curcumin [Figure 3a-d].
Figure 3.
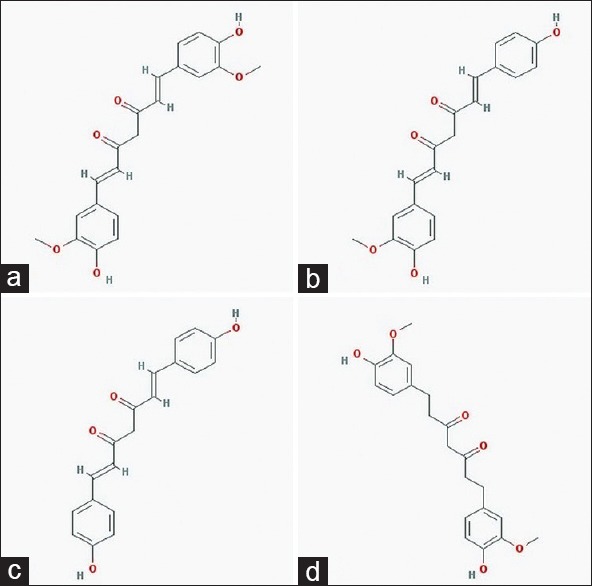
Structures of lead compounds, (a) curcumin, (b) demethoxycurcumin, (c) bisdemethoxycurcumin, (d) tetrahydrocurcumin
Curcumin has broad-spectrum antimicrobial properties.[15] Chen et al. have reported that curcumin specifically inhibits infectivity of enveloped virus by inhibiting hemagglutination activity.[16]
Curcumin also interferes in virus application by acting on specific VPs such as integrase and protease.[17,18] Kutluay et al. also reported that curcumin might inhibit herpes simplex virus infection, and this was perhaps mediated by preventing recruitment of RNA polymerase II by immediate-early gene promoters.[19]
In addition to antiviral activities, Sordillo and Helson have hypothesized that curcumin may help counteract cytokine storm in Ebola fever.[20]
Curcumin analogs have also been identified with antiviral activity; sometimes showing better activity and sometimes showing lesser but comparable affinity.[21,22]
Tetrahydrocurcumin is the major metabolite of curcumin, and the researchers are trying to assess whether tetrahydrocurcumin is superior to curcumin at a molecular level.[23]
This study focuses on an in silico approach towards assessing the inhibitory effect of curcumin and related compounds on EBOV proteins with the aim of identifying potential leads for the prevention and treatment of Ebola fever.
This is the first study to investigate the antiviral activity of bisdemethoxycurcumin, demethoxycurcumin, and tetrahydrocurcumin against Ebola VPs.
MATERIALS AND METHODS
Protein structure
X-ray crystal structures of VP40 (Protein Data Bank [PDB] ID: 1H2D), VP30 (PDB ID: 2I8B), VP35 (PDB ID: 4IBB), VP24 (PDB ID: 4M0Q) NP (PDB ID: 4QAZ), and GP (PDB ID: 2EBO) were obtained from PDB.
Protein preparation was performed using Swiss PDB Viewer. Swiss PDB Viewer is a free package written by a group at Swiss Institute of Bioinformatics and developed by Nicolas Guex.[24]
It is a comprehensive molecular modeling package providing a variety of visualization options as well as allowing homology modeling and energy minimization package. All water molecules, hetero atoms, ligands, and RNA segments were removed.
Visual molecular dynamics (VMD) is a molecular modeling and visualization computer program developed primarily as a tool for viewing and analyzing the results of molecular dynamics simulations.[25]
Using VMD, hydrogen atoms were added to each of the structures, and protein structure files and PDB files were prepared for simulation. Proteins were solvated in a water box of 10 Å and ions were added to neutralize the system.
Molecular dynamics simulation
Minimization and equilibration of all proteins were performed using NAMD 2.11. Molecular dynamics simulations were computed by utilizing the CHARMM force field. NAMD incorporates the particle mesh Ewald algorithm which simplifies the computational complexity.[26]
The primary stages involved minimizing the systems with and without position restraints but using periodic boundary conditions. The latter stages involved equilibration under position restrained dynamic simulation (NVE and NPT) at 310 K for 300 ps.
Active-site prediction
Potential active sites were then identified using AutoLigand and AutoDock Tools. Water molecules and ions were removed from the simulated structures and stored as.pdb files.
AutoDock Tools offers a complete molecular viewer and graphical support for all steps required for setup and analysis of docking runs.[27]
AutoDock Tools were used to prepare.pdbqt files. Hydrogen atoms and Gasteiger charges were added to all structures. AutoLigand is a tool used to identify and characterize ligand-binding sites.
1 Å grid and map files were generated by AutoGrid. AutoLigand GUI was used, and a single file was generated starting at the user-specified point; in this case, it involved coordinates entered in x, y, z boxes. Ten potential active sites for each protein were identified.
Ligand preparation
The three-dimensional structures of lead compounds, namely, curcumin, bisdemethoxycurcumin, demethoxycurcumin, and tetrahydrocurcumin, were obtained from PubChem.
SDF files were converted to.pdb and.pdbqt files. Hydrogen bonds and Gasteiger charges were added using AutoDock Tools.
Molecular docking
AutoDock Vina was used to dock curcumin and its related compounds on each of the receptor EBOV proteins.
AutoDock Vina is an open-source program for doing molecular docking, and it makes extensive use of Python script. It was designed and implemented by Dr. Oleg Trott in the Molecular Graphics Laboratory at The Scripps Research Institute.[28]
The ligands were docked on various potential active sites on each simulated VP to identify the best fit with best dock score and minimum root mean square deviation values.
Discovery Studio Visualizer was used to visualize the resultant docked structures and study interactions of various pose of ligand with target. Discovery Studio Visualizer is a free viewer that is designed to offer interactive environment for viewing and editing various files as well as for displaying plots and other graphical presentations of data.[29]
RESULTS AND DISCUSSION
Each of the VPs (VP40, VP30, VP35, VP24, GP, and NP) plays an important role in EBOV invasion and replication.
Molecular dynamics simulation of each of the VPs was performed to optimize the structure and reach a global minimum [Figure 4]. Active sites were identified. Each of the curcuminoids and tetrahydrocurcumin was docked against simulated structures of Ebola VPs.
Figure 4.
Simulation results of target protein: (a) Target protein prior to simulation. (b) Target protein after simulation on reaching global minimum
The docking interaction of the ligands with target proteins as well as their docking scores was studied. Table 1 lists the docking score of each ligand with each Ebola VP. Table 2 lists the interactions of ligands with each Ebola VP receptor.
Table 1.
Comparison of binding efficacy based on AutoDock Vina docking score (kcal/mol) of curcumin and other derivatives with all six Ebola viral proteins
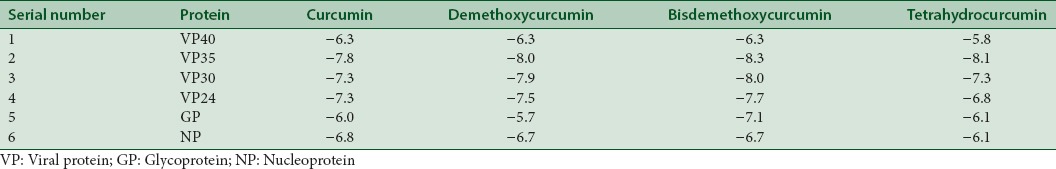
Table 2.
List of interactions of curcumin and natural derivatives with active site residues of each of the six Ebola viral proteins
Curcumin, curcuminoids as well as tetrahydrocurcumin show potential inhibitory action against all VPs of EBOV as evidenced by the docking scores.
Comparing the docking scores of all target proteins, it is observed that bisdemethoxycurcumin shows better binding affinity than curcumin. Interestingly, metabolite tetrahydrocurcumin shows comparable binding affinity as curcumin, thus throwing some light on the possibility to explore this compound as an antiviral agent against EBOV.
Interactions of curcumin and derivatives with viral protein 40
Curcumin and other curcuminoids bind to the VP40 with a binding affinity of −6.3 kcal/mol. Curcumin forms hydrogen bonds with residues ALA128 and ASN130. It forms other noncovalent interactions with PRO131, GLU160, PRO165 GLN159 [Figure 5a].
Figure 5.
Ligand interactions of (a) curcumin with viral protein 40, (b) bisdemethoxycurcumin with viral protein 35, (c) bisdemethoxycurcumin with viral protein 30, (d) bisdemethoxycurcumin with viral protein 24, (e) bisdemethoxycurcumin with glycoprotein, (f) curcumin with nucleoprotein
Tetrahydrocurcumin showed the lowest binding energy of −5.8kcal/mol for VP40. It formed hydrogen bonds with GLN159, VAL166, and PRO169. Other noncovalent interactions were seen with residues PRO131 VAL166, ALA128 PRO165.
VP40 is the most abundantly expressed filoviral protein. It facilitates assembly of the virus at the plasma membrane.[30] Exogenous expression of VP40 alone is sufficient to induce the formation of filamentous virus-like particles (VLPs) in mammalian cells.[31]
VP40 is involved in various aspects of filovirus virion assembly and budding.
VP40 is a 326 amino acid protein and has two significant domains. The N-terminal domain and the C-terminal domain (CTD) are required for formation of VP40 dimer as well as subsequent trafficking to and interaction with the plasma membrane.[30]
At the plasma membrane, VP40 forms butterfly-shaped dimers, which then reorganize into linear hexamers, and these are essential building blocks of the viral matrix. These are vital for virion assembly and budding.[32]
Curcumin inhibits VP40 by forming hydrogen bonds with residues ALA128 and ASN130. Residues ALA128 and ASN130 lie in the N-terminal domain; mutation of these residues or disruption of function can affect plasma membrane localization, VP40 oligomerization, and VLP formation.[33,34]
Interactions of curcumin and derivatives with viral protein 35
Bisdemethoxycurcumin shows highest binding affinity of −8.3 kcal/mol with VP35. It forms hydrogen bond with THR335, VAL294, and other noncovalent interactions with PRO316 PRO293 VAL294 LEU249 [Figure 5b].
Tetrahydrocurcumin shows a binding affinity of −8.1 kcal/mol and forms hydrogen bonds with residues THR335 VAL294 LEU249 SER253. Demethoxycurcumin shows binding affinity of 8.0 kcal/mol. Curcumin shows lowest binding affinity of the lot with −7.8 kcal/mol.
Ebola VP35 is multifunctional, serving as a component of the viral RNA polymerase complex, as a structural/assembly factor, and as a suppressor of host interferon (IFN) responses.[35] It also functions as an RNAi silencing suppressor.[36]
VP35 contains an N-terminal coiled-coil domain and a C-terminal dsRNA-binding region.[37,38] Oligomerization through the N-terminal oligomerization domain is required for virus replication, whereas the C-terminal region of VP35 interacts with host factors to block signaling that triggers IFN responses.[39]
Structural analyses revealed that a stretch of about 120 residues was required to form an independently folded domain which facilitates IFN inhibitory function.[39] This domain was termed the IFN inhibitory domain (IID). VP35 contributes to viral escape from host innate immunity and is required for EBOV virulence.[39]
Bisdemethoxycurcumin interacts with the IID domain of VP35, and this activity may prevent shutting down of immune responses early in the infection.[39]
Interactions of curcumin and derivatives with viral protein 30
Bisdemethoxycurcumin shows highest binding affinity of −8.0 kcal/mol with VP30. It forms hydrogen bonds with GLN 185 and has other noncovalent interactions with PHE181 CYS251 LEU144 ALA246 LEU147 [Figure 5c].
Demethoxycurcumin shows the next best binding affinity for VP30 of −7.9 kcal/mol followed by curcumin and tetrahydrocurcumin at −7.3 kcal/mol.
VP30 serves as a transcriptional activator. The N-terminal portion of VP30 contains phosphorylation sites, a zinc-binding site, and an RNA-binding site.[40]
The C-terminal domain of VP30 forms a conserved dimer of two globular, α-helical domains.[41]
Hartlieb et al. reported that C-terminal domain of VP30 contains the binding site for the viral NP and together they are involved in the synthesis of EBOV RNA.[41] In addition to transcription initiation activity, VP30 is a structural protein. Bisdemethoxycurcumin interacts with the residues present in the cavity of the dimers of C-terminal domain.
Interactions of curcumin and derivatives with viral protein 24
Bisdemethoxycurcumin shows the highest binding affinity of −7.7 kcal/mol with VP24. It forms hydrogen bonds with ARG154 GLU46 and has other noncovalent interactions with LEU158 PHE230 ILE45 [Figure 5d].
Demethoxycurcumin shows a binding affinity of −7.5 kcal/mol, followed by curcumin with −7.3 kcal/mol. Tetrahydrocurcumin shows minimum but considerable binding affinity of −6.8 kcal/mol.
The VP24 protein of EBOV is thought to be a secondary matrix protein and a minor component of virions. VP24 has many structural features commonly associated with viral matrix proteins. Therefore, it may have a role in virus assembly and budding.[42]
VP24 inhibits IFN signaling.[43] VP24, in conjunction with NP and VP35, is required for the formation of nucleocapsid.[44]
Hoenen et al. demonstrated that VP24-deficient infectious VLPs have less VP30 than VLPs containing VP24, and these VP24-deficient infectious VLPs cannot support the initial transcription of the EBOV genome in VLP-infected cells.[45]
This suggests that presence of VP24 is extremely necessary for development of a fully functional nucleocapsid. Inhibition of VP24 by curcuminoids can subsequently hamper the formation of a functional nucleocapsid and deter viral infection.
Interactions of curcumin and derivatives with glycoprotein
Bisdemethoxycurcumin shows a binding affinity of −7.1 kcal/mol with GP. It forms hydrogen bonds with LEU593 ARG596 and has other noncovalent interactions with TRP597 ILE603 ARG596 [Figure 5e].
Curcumin shows a binding affinity of 6.0 kcal/mol and tetrahydrocurcumin shows a binding affinity of 6.1 kcal/mol. Demethoxycurcumin shows least binding affinity of −5.7 kcal/mol.
EBOV GP is the only VP found on the surface of the Ebola virion and is, therefore, responsible for mediating attachment and entry of the virus into host cells. It is the primary target for host neutralizing antibodies and used for the design of vaccines and entry inhibitors.[46,47]
Binding of curcuminoids to GP can destabilize GP and prevent fusion of viral membrane with host cells.
Interactions of curcumin and derivatives with nucleoprotein
Curcumin shows the highest binding energy of −6.8 kcal/mol with NP. It forms hydrogen bonds with ARG559 and has other noncovalent interactions with LEU561 PHE630 ALA562, LEU558 [Figure 5f].
Demethoxycurcumin and bisdemethoxycurcumin show a binding energy of −6.7 kcal/mol while tetrahydrocurcumin has a binding affinity of −6.1 kcal/mol.
NP of EBOV is the largest NP of the nonsegmented negative-stranded RNA viruses with 739 amino acids. It plays a central role in virus replication.
It has a hydrophobic N-terminal half of approximately 350 amino acids and a hydrophilic C-terminal half of approximately 400 amino acids. The N-terminal region is found to be important for RNA binding, and with VP35 and VP24, it helps in the formation of nucleocapsids.[48]
Curcumin binds to NP at the residues are required for the formation of nucleocapsid like structures and viral genome replication, thus disrupting its activity.[49]
CONCLUSION
The results of the study clearly indicate that curcumin, curcuminoids, and tetrahydrocurcumin are effective against multiple targets of EBOV. This is essential in terms of blocking several biochemical pathways simultaneously to deliver faster treatment.
The results indicate that further research should be focused on the antiviral property of other curcuminoids apart from curcumin in Ebola treatment.
This is the first study that investigates the antiviral and inhibitory action of tetrahydrocurcumin, a metabolite of curcumin against Ebola VPs. In some targets, such as VP35, GP, and VP30, tetrahydrocurcumin shows better binding energies than curcumin.
These findings are important in relation to the fact that curcumin is poorly absorbed and has low stability in the body which gives rise to degradation products such as tetrahydrocurcumin. However, tetrahydrocurcumin has antiviral activity suggestive of its therapeutic action.
However, in most targets, bisdemethoxycurcumin and demethoxycurcumin, the curcuminoids that do not receive as much as research focus as curcumin, show better binding energies than curcumin. This highlights the need to focus more research on the therapeutic action as well as the antiviral action of these curcuminoids.
The pleiotropic nature of curcumin and the antiviral action of curcumin, curcuminoids, and its metabolites against Ebola VPs make them potential lead candidates for controlling EBOV infection. Further research in the form of preclinical analysis is warranted not only on stand-alone antiviral activity of these entities but also investigating their synergistic action.
These versatile inhibitors may serve as a new medication for restraining and treating Ebola infection opening avenues for consideration of phytochemicals as therapeutic antiviral agents.
Financial support and sponsorship
Nil.
Conflicts of interest
There are no conflicts of interest
REFERENCES
- 1.Schieffelin JS, Shaffer JG, Goba A, Gbakie M, Gire SK, Colubri A, et al. Clinical illness and outcomes in patients with Ebola in Sierra Leone. N Engl J Med. 2014;371:2092–100. doi: 10.1056/NEJMoa1411680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Leroy EM, Gonzalez JP, Baize S. Ebola and Marburg haemorrhagic fever viruses: Major scientific advances, but a relatively minor public health threat for Africa. Clin Microbiol Infect. 2011;17:964–76. doi: 10.1111/j.1469-0691.2011.03535.x. [DOI] [PubMed] [Google Scholar]
- 3.Zhang L, Wang H. Forty years of the war against Ebola. J Zhejiang Univ Sci B. 2014;15:761–5. doi: 10.1631/jzus.B1400222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goeijenbier M, van Kampen JJ, Reusken CB, Koopmans MP, van Gorp EC. Ebola virus disease: A review on epidemiology, symptoms, treatment and pathogenesis. Neth J Med. 2014;72:442–8. [PubMed] [Google Scholar]
- 5.Martines RB, Ng DL, Greer PW, Rollin PE, Zaki SR. Tissue and cellular tropism, pathology and pathogenesis of ebola and marburg viruses. J Pathol. 2015;235:153–74. doi: 10.1002/path.4456. [DOI] [PubMed] [Google Scholar]
- 6.Zawilińska B, Kosz-Vnenchak M. General introduction into the Ebola virus biology and disease. Folia Med Cracov. 2014;54:57–65. [PubMed] [Google Scholar]
- 7.Hulo C, de Castro E, Masson P, Bougueleret L, Bairoch A, Xenarios I, et al. ViralZone: A knowledge resource to understand virus diversity. Nucleic Acids Res. 2011;39:D576–82. doi: 10.1093/nar/gkq901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Falasca L, Agrati C, Petrosillo N, Di Caro A, Capobianchi MR, Ippolito G, et al. Molecular mechanisms of Ebola virus pathogenesis: Focus on cell death. Cell Death Differ. 2015;22:1250–9. doi: 10.1038/cdd.2015.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bah EI, Lamah MC, Fletcher T, Jacob ST, Brett-Major DM, Sall AA, et al. Clinical presentation of patients with Ebola virus disease in Conakry, Guinea. N Engl J Med. 2015;372:40–7. doi: 10.1056/NEJMoa1411249. [DOI] [PubMed] [Google Scholar]
- 10.Chertow DS, Kleine C, Edwards JK, Scaini R, Giuliani R, Sprecher A, et al. Ebola virus disease in West Africa –Clinical manifestations and management. N Engl J Med. 2014;371:2054–7. doi: 10.1056/NEJMp1413084. [DOI] [PubMed] [Google Scholar]
- 11.Qiu X, Wong G, Audet J, Bello A, Fernando L, Alimonti JB, et al. Reversion of advanced Ebola virus disease in nonhuman primates with ZMapp. Nature. 2014;514:47–53. doi: 10.1038/nature13777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Warren TK, Wells J, Panchal RG, Stuthman KS, Garza NL, Van Tongeren SA, et al. Protection against filovirus diseases by a novel broad-spectrum nucleoside analogue BCX4430. Nature. 2014;508:402–5. doi: 10.1038/nature13027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Choi JH, Croyle MA. Emerging targets and novel approaches to Ebola virus prophylaxis and treatment. BioDrugs. 2013;27:565–83. doi: 10.1007/s40259-013-0046-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Araújo CC, Leon LL. Biological activities of Curcuma longa L. Mem Inst Oswaldo Cruz. 2001;96:723–8. doi: 10.1590/s0074-02762001000500026. [DOI] [PubMed] [Google Scholar]
- 15.Moghadamtousi SZ, Kadir HA, Hassandarvish P, Tajik H, Abubakar S, Zandi K, et al. A review on antibacterial, antiviral, and antifungal activity of curcumin. Biomed Res Int. 2014;2014:186864. doi: 10.1155/2014/186864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen TY, Chen DY, Wen HW, Ou JL, Chiou SS, Chen JM, et al. Inhibition of enveloped viruses infectivity by curcumin. PLoS One. 2013;8:e62482. doi: 10.1371/journal.pone.0062482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vajragupta O, Boonchoong P, Morris GM, Olson AJ. Active site binding modes of curcumin in HIV-1 protease and integrase. Bioorg Med Chem Lett. 2005;15:3364–8. doi: 10.1016/j.bmcl.2005.05.032. [DOI] [PubMed] [Google Scholar]
- 18.Mazumder A, Raghavan K, Weinstein J, Kohn KW, Pommier Y. Inhibition of human immunodeficiency virus type-1 integrase by curcumin. Biochem Pharmacol. 1995;49:1165–70. doi: 10.1016/0006-2952(95)98514-a. [DOI] [PubMed] [Google Scholar]
- 19.Kutluay SB, Doroghazi J, Roemer ME, Triezenberg SJ. Curcumin inhibits herpes simplex virus immediate-early gene expression by a mechanism independent of p300/CBP histone acetyltransferase activity. Virology. 2008;373:239–47. doi: 10.1016/j.virol.2007.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sordillo PP, Helson L. Curcumin suppression of cytokine release and cytokine storm. A potential therapy for patients with Ebola and other severe viral infections. In Vivo. 2015;29:1–4. [PubMed] [Google Scholar]
- 21.Singh AK, Misra K. Human papilloma virus 16 E6 protein as a target for curcuminoids, curcumin conjugates and congeners for chemoprevention of oral and cervical cancers. Interdiscip Sci. 2013;5:112–8. doi: 10.1007/s12539-013-0159-8. [DOI] [PubMed] [Google Scholar]
- 22.Ou JL, Mizushina Y, Wang SY, Chuang DY, Nadar M, Hsu WL, et al. Structure-activity relationship analysis of curcumin analogues on anti-influenza virus activity. FEBS J. 2013;280:5829–40. doi: 10.1111/febs.12503. [DOI] [PubMed] [Google Scholar]
- 23.Aggarwal BB, Deb L, Prasad S. Curcumin differs from tetrahydrocurcumin for molecular targets, signaling pathways and cellular responses. Molecules. 2014;20:185–205. doi: 10.3390/molecules20010185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guex N, Peitsch MC. SWISS-MODEL and the Swiss-PdbViewer: An environment for comparative protein modeling. Electrophoresis. 1997;18:2714–23. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 25.Humphrey W, Dalke A, Schulten K. VMD: Visual molecular dynamics. J Mol Graph. 1996;14:33–8. doi: 10.1016/0263-7855(96)00018-5. 27-8. [DOI] [PubMed] [Google Scholar]
- 26.Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, et al. Scalable molecular dynamics with NAMD. J Comput Chem. 2005;26:1781–802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, et al. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J Comput Chem. 2009;30:2785–91. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Trott O, Olson AJ. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31:455–61. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dassault Systèmes. BIOVIA, Discovery Studio Modeling Environment, Release 2017. San Diego: Dassault Systèmes; 2016. [Google Scholar]
- 30.Adu-Gyamfi E, Soni SP, Xue Y, Digman MA, Gratton E, Stahelin RV. The Ebola virus matrix protein penetrates into the plasma membrane: A key step in viral protein 40 (VP40) oligomerization and viral egress. J Biol Chem. 2013;288:5779–89. doi: 10.1074/jbc.M112.443960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Harty RN, Brown ME, Wang G, Huibregtse J, Hayes FP. A PPxY motif within the VP40 protein of Ebola virus interacts physically and functionally with a ubiquitin ligase: Implications for filovirus budding. Proc Natl Acad Sci U S A. 2000;97:13871–6. doi: 10.1073/pnas.250277297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bornholdt ZA, Noda T, Abelson DM, Halfmann P, Wood MR, Kawaoka Y, et al. Structural rearrangement of Ebola virus VP40 begets multiple functions in the virus life cycle. Cell. 2013;154:763–74. doi: 10.1016/j.cell.2013.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Madara JJ, Han Z, Ruthel G, Freedman BD, Harty RN. The multifunctional Ebola virus VP40 matrix protein is a promising therapeutic target. Future Virol. 2015;10:537–46. doi: 10.2217/fvl.15.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Adu-Gyamfi E, Soni SP, Jee CS, Digman MA, Gratton E, Stahelin RV. A loop region in the N-terminal domain of Ebola virus VP40 is important in viral assembly, budding, and egress. Viruses. 2014;6:3837–54. doi: 10.3390/v6103837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zampieri CA, Sullivan NJ, Nabel GJ. Immunopathology of highly virulent pathogens: Insights from Ebola virus. Nat Immunol. 2007;8:1159–64. doi: 10.1038/ni1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Haasnoot J, de Vries W, Geutjes EJ, Prins M, de Haan P, Berkhout B. The Ebola virus VP35 protein is a suppressor of RNA silencing. PLoS Pathog. 2007;3:e86. doi: 10.1371/journal.ppat.0030086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reid SP, Cárdenas WB, Basler CF. Homo-oligomerization facilitates the interferon-antagonist activity of the Ebolavirus VP35 protein. Virology. 2005;341:179–89. doi: 10.1016/j.virol.2005.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hartman AL, Towner JS, Nichol ST. A C-terminal basic amino acid motif of Zaire Ebolavirus VP35 is essential for type I interferon antagonism and displays high identity with the RNA-binding domain of another interferon antagonist, the NS1 protein of influenza A virus. Virology. 2004;328:177–84. doi: 10.1016/j.virol.2004.07.006. [DOI] [PubMed] [Google Scholar]
- 39.Leung DW, Ginder ND, Fulton DB, Nix J, Basler CF, Honzatko RB, et al. Structure of the Ebola VP35 interferon inhibitory domain. Proc Natl Acad Sci U S A. 2009;106:411–6. [Google Scholar]
- 40.John SP, Wang T, Steffen S, Longhi S, Schmaljohn CS, Jonsson CB, et al. Ebola virus VP30 is an RNA binding protein. J Virol. 2007;81:8967–76. doi: 10.1128/JVI.02523-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hartlieb B, Muziol T, Weissenhorn W, Becker S. Crystal structure of the C-terminal domain of Ebola virus VP30 reveals a role in transcription and nucleocapsid association. Proc Natl Acad Sci U S A. 2007;104:624–9. doi: 10.1073/pnas.0606730104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Han Z, Boshra H, Sunyer JO, Zwiers SH, Paragas J, Harty RN, et al. Biochemical and functional characterization of the Ebola virus VP24 protein: Implications for a role in virus assembly and budding. J Virol. 2003;77:1793–800. doi: 10.1128/JVI.77.3.1793-1800.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reid SP, Leung LW, Hartman AL, Martinez O, Shaw ML, Carbonnelle C, et al. Ebola virus VP24 binds karyopherin alpha1 and blocks STAT1 nuclear accumulation. J Virol. 2006;80:5156–67. doi: 10.1128/JVI.02349-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huang Y, Xu L, Sun Y, Nabel GJ. The assembly of Ebola virus nucleocapsid requires virion-associated proteins 35 and 24 and posttranslational modification of nucleoprotein. Mol Cell. 2002;10:307–16. doi: 10.1016/s1097-2765(02)00588-9. [DOI] [PubMed] [Google Scholar]
- 45.Hoenen T, Groseth A, Kolesnikova L, Theriault S, Ebihara H, Hartlieb B, et al. Infection of naive target cells with virus-like particles: Implications for the function of Ebola virus VP24. J Virol. 2006;80:7260–4. doi: 10.1128/JVI.00051-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee JE, Fusco ML, Hessell AJ, Oswald WB, Burton DR, Saphire EO. Structure of the Ebola virus glycoprotein bound to an antibody from a human survivor. Nature. 2008;454:177–82. doi: 10.1038/nature07082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mohan GS, Ye L, Li W, Monteiro A, Lin X, Sapkota B, et al. Less is more: Ebola virus surface glycoprotein expression levels regulate virus production and infectivity. J Virol. 2015;89:1205–17. doi: 10.1128/JVI.01810-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Noda T, Hagiwara K, Sagara H, Kawaoka Y. Characterization of the Ebola virus nucleoprotein-RNA complex. J Gen Virol. 2010;91(Pt 6):478–83. doi: 10.1099/vir.0.019794-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Watanabe S, Noda T, Kawaoka Y. Functional mapping of the nucleoprotein of Ebola virus. J Virol. 2006;80:3743–51. doi: 10.1128/JVI.80.8.3743-3751.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]