Abstract
Ciita was discovered for its role in regulating transcription of major histocompatibility complex class II (MHCII) genes. Subsequently, CIITA was predicted to control many other genes based on reporter and ChIP-seq analysis but few such predictions have been verified in vivo using Ciita−/− mice. Testing these predictions for classical dendritic cells (cDCs) has been particularly difficult, since Ciita−/− mice lack MHCII expression required to identify cDCs. However, recent identification of the cDC-specific transcription factor Zbtb46 allows identification of cDCs independently of MHCII expression. We crossed Zbtb46gfp mice onto the Ciita−/− background and found that all cDC lineages developed in vivo in the absence of Ciita. We then compared the complete transcriptional profile of wild-type and Ciita−/− cDCs to define the physiological footprint of CIITA for both immature and activated cDCs. We find that CIITA exerts a highly restricted control over only the MHCII, H2-DO and H2-DM genes, in DC1 and DC2 cDC subsets, but not over other proposed targets, including Ii. These findings emphasize the caveats needed in interpreting transcription factor binding sites identified by in vitro reporter analysis, or by ChIP-seq, which may not necessarily indicate their functional activity in vivo.
Keywords: ciita, dendritic cell, transcription factor, zbtb46
Introduction
Ciita was identified as a major regulator of transcription of MHCII genes based on restoration of MHCII expression in bare lymphocyte syndrome [1]. CIITA interacts with several proteins that bind DNA directly in SXY modules, including the RFX complex, RFX5 and RFXAP [2], CREB [3, 4], NFY [5], and TAFII32 [6–10]. The SXY module was identified at the promoter of numerous genes in the MHC locus and was thus thought to restrict the activity of CIITA to this region of the genome [11]. Subsequently, whole-genome sequencing and ChIP-seq in CIITA-deficient B cell lines have revealed SXY modules and CIITA binding sites across the genome, expanding the putative genomic footprint of CIITA [10, 12, 13]. Bioinformatic analyses and in vitro promoter assays have identified genes with proximal CIITA-binding sites that are putatively regulated by CIITA and that are predicted to have functions related to antigen processing and presentation [14–16]. These studies imply CIITA might function as a master regulator of this process beyond its role in MHCII expression. For example, Plxna1, RAB4B, and Btn2a2 have been proposed to be involved in T cell-DC interactions, cDC antigen processing, and receptor-mediated co-inhibition, respectively [14–16]. However, validation of these predictions for cDCs in vivo has not been undertaken because MHCII expression is required for cDC identification. To date, studies have been limited to in vitro analysis of myelomonocytic cell lines or cells derived from bone-marrow (BM) cultures treated with GM-CSF, so-called BMDCs [12, 14–17]. However, recent studies have challenged how well these cells reflect the in vivo properties of DCs [18, 19]. First, BMDCs are heterogeneous and include cells with both macrophage and DC properties [18]. Second, the transcriptional programs underlying some forms of antigen processing differ between BMDCs and in vivo cDCs [19]. In light of these findings, it appeared that the functional footprint of CIITA in DCs was in need of re-evaluation.
Expression of Zbtb46 is restricted to cDCs within the hematopoietic system [20, 21], and a Zbtb46gfp reporter allele [21] has been used to identify cDCs and their progenitors independently of conventional surface markers [22, 23]. Ciita was identified as being selectively induced along with Id2 and Batf3 in the specified progenitor of CD8α+ cDCs in BM [22], raising the question of whether it also acts in cDC development. In principle, conceivably it is possible to identify cDCs with a combination of surface markers without the use of MHCII. In practice, we are unaware of any published work that has used such a strategy to unequivocally identify cDCs in Ciita−/− mice. Here, we generated Zbtb46gfp Ciita−/− mice to test whether cDC lineages can develop without CIITA, and if so, to determine its transcriptional footprint in cDCs in vivo.
Results and Discussion
DC progenitors develop normally in Ciita−/− mice and can be identified using Zbtb46gfp expression
We previously identified committed BM progenitors for the two major lineages of cDCs, termed pre-CD4 DCs defined as Lin−CD135+CD11c+CD117−CD115+ BM cells, and pre-CD8 DCs defined as Lin−CD135+CD11c+CD117intMHCIIintZbtb46gfp+ BM cells [22]. Our previous definition of pre- CD8 DCs relied on MHCII expression, preventing its application in Ciita−/− mice. However, we asked whether we could use Zbtb46gfp expression to identify BM progenitors committed to this lineage in Ciita−/− mice (Fig. 1). BM cells that were Lin− SiglecH−CD135+CD115+CD11c+CD117− Zbtb46gfp+ were present in both Ciita+/+Zbtb46+/gfp and Ciita−/−Zbtb46+/gfp mice (Fig. 1A) in equal frequencies (Fig. 1B). Sorted populations generated CD172+ cDCs exclusively when cultured with Flt3L in vitro (Fig. 1C). Since we previously found that pre-CD8 DCs were largely CD115− [22], we analyzed BM cells that were Lin−SiglecH−CD135+CD115−CD11c+CD117intZbtb46gfp+, and found that they were present in both Ciita+/+Zbtb46+/gfp and Ciita−/−Zbtb46+/gfp mice in equal frequencies (Fig. 1A,B). These cells gave rise exclusively to CD24+CD172− cDCs (Fig. 1C). Differences in the efficiency of progenitor output between Ciita+/+Zbtb46+/gfp and Ciita−/− Zbtb46+/gfp mice were not observed, since each progenitor population gave rise to similar numbers of differentiated cells in Flt3L cultures. These results indicate that both pre-CD4 and pre-CD8 DC progenitors maintain their previously defined pattern of lineage commitment in Ciita−/− mice. However, since MHCII is the only marker currently known to separate differentiated cDCs from their progenitors in BM, populations analyzed here likely represent a mixture of committed progenitors and their differentiated counterparts.
Fig. 1. BM cDC progenitors can be identified without MHCII in Zbtb46+/GFP mice and their development is CIITA-independent.
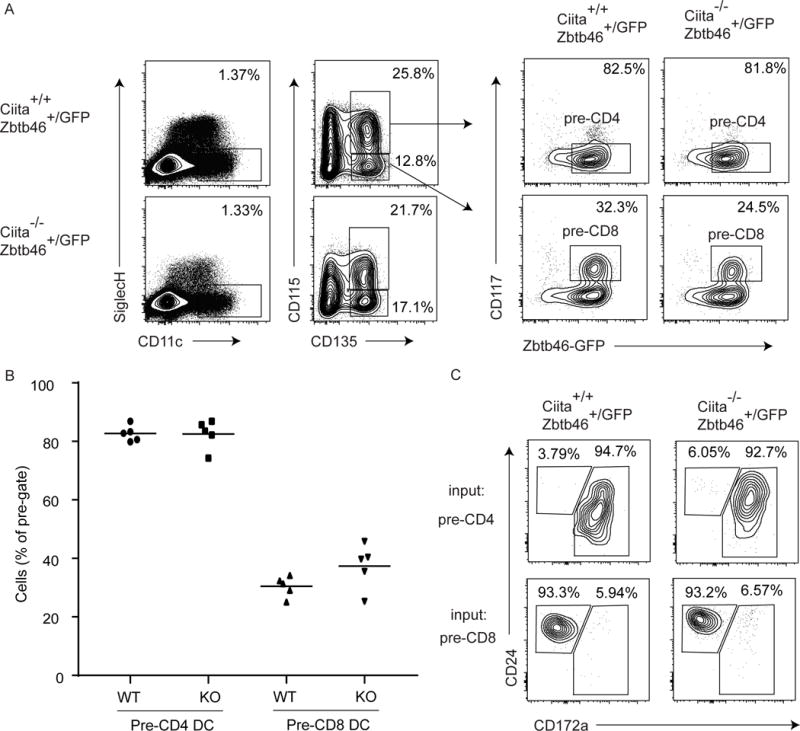
(A) Flow cytometry analysis of BM cDC progenitors from Ciita+/+Zbtb46+/GFP and Ciita−/−Zbtb46+/GFP mice defined as Lin−SiglechH− CD11c+CD135+CD115+CD117−Zbtb46gfp+ and Lin−SiglechH−CD11c+CD135+CD115−CD117int Zbtb46gfp+ for pre-CD4 and pre-CD8 DCs, respectively (n = 5). Contour plots are from a single experiment representative of 2 experiments with 2–3 mice per experiment. (B) Frequency of pre-CD4 and pre-CD8 DCs in vivo as defined in. Percent for pre-CD4 DC was based on a pre-gate defined as Lin−SiglechH−CD11c+CD135+CD115+cells. Percent for the pre-CD8 DC was based on a pre-gate defined as SiglechH−CD11c+CD135+CD115−cells. Data are shown as a dot plot with mean bar for two pooled experiments with 2–3 mice per experiment (n = 5). (C) Output of sorted pre-CD4 and pre-CD8 DC progenitors as defined in (A) after 5 days of Flt3L-treated cultures. Shown are contour plots from a single experiment pre-gated as B220−SiglecH−CD11c+Zbtb46gfp+ representative of 2 experiments with 2–3 mice per experiment (n = 5).
Peripheral cDCs develop normally in Ciita−/− mice and can be identified by Zbtb46gfp expression
We next analyzed mature cDC populations comparing Ciita+/+Zbtb46+/gfp and Ciita−/−Zbtb46+/gfp mice. First, we asked whether cDCs can be identified equivalently using either Zbtb46gfp or MHCII expression in the spleen (Fig. 2A). We compared two gating strategies that directly substitute MHCII with Zbtb46gfp. Using Ciita+/+ Zbtb46+/gfp mice, we compared SiglecH−B220−CD11c+ CD172a+ that were either MHCII+ or Zbtb46gfp+ to evaluate the DC2 lineage, and SiglecH−B220− CD11c+CD24+ cells that were either MHCII+ or Zbtb46gfp+ to evaluate the DC1 lineage (Fig. 2A). We found that both gating strategies identified the same percentage of total cDCs as a fraction of the total spleen, and also identified the same distribution of two major subsets of CD24+ DC1s and CD172a+ DC2s (Fig. 2A). Having established that Zbtb46gfp expression can be used in place of MHCII, we compared peripheral splenic DCs in Ciita+/+Zbtb46+/gfp and Ciita−/−Zbtb46+/gfp mice (Fig. 2B). In agreement with normal BM progenitor populations in Ciita−/−Zbtb46+/gfp mice, splenic cDC subsets also showed no difference between Ciita+/+Zbtb46+/gfp and Ciita−/−Zbtb46+/gfp mice (Fig. 2B). Although not defined by MHCII expression, we found no differences in B220+SiglecH+ plasmacytoid DCs (pDCs) between Ciita+/+Zbtb46+/gfp and Ciita−/−Zbtb46+/gfp mice (Fig. 2B). Therefore, DC lineage development in vivo is not dependent on CIITA.
Fig. 2. Splenic CD172a+ and CD24+ DCs can be identified without MHCII as a linage marker in Zbtb46+/GFP mice and develop in Ciita-deficient mice.
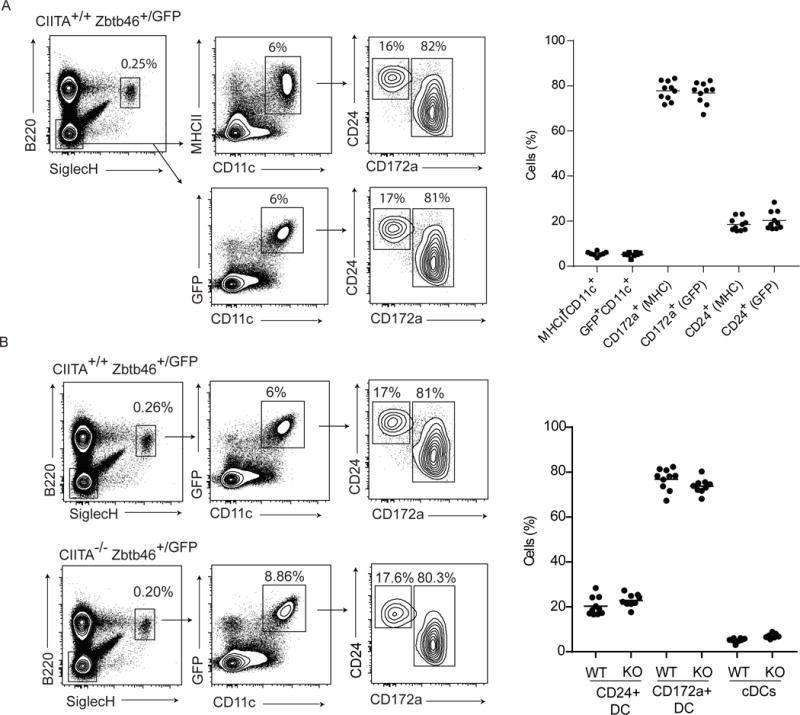
(A) Flow cytometry analysis of splenic CD172a+ and CD24+ DCs from Zbtb46+/gfp mice using either B220−SiglecH−CD11c+MHCII+ or B220−SiglecH−CD11c+Zbtb46gfp+ pre-gates. Contour plots on the left are from a single, representative experiment, and dot plots on the right pool 5 experiments with 1–3 mice per experiment with mean bars (n = 8–10). (B) Flow cytometry analysis of splenic cDC populations in Ciita+/+Zbtb46+/GFP and Ciita−/− Zbtb46+/gfp mice with populations using B220−SiglecH− CD11c+Zbtb46gfp+ pre-gates. Contour plots on the left are from a single, representative experiment and dot plots on the right pool 5 experiments with 1–3 mice per experiment (n = 7–10).
Transcriptional activity of Ciita is restricted to MHCII genes in cDCs
Next, we determined the functional impact of CIITA in cDCs both at homeostasis and after activation (Fig. 3). We sorted splenic DC1 and DC2 cells from Ciita+/+Zbtb46+/gfp and Ciita−/− Zbtb46+/gfp mice at homeostasis and compared their transcriptome using gene expression microarrays (Fig. 3A). Remarkably, there were very few genes expressed differently between Ciita+/+Zbtb46+/gfp and Ciita−/−Zbtb46+/gfp samples. Differentially expressed genes were limited to both chains of H2-A (H2-Aa1, H2-Ab1), the H2-E beta chain (H2-Eb1, H2-Eb2), both chains of H2-DO (H2-Oa, H2-Ob), and both chains of H2-DM (H2-DMa, H2-DMb2). These genes were all reduced in Ciita−/− Zbtb46+/gfp DCs compared with the corresponding Ciita+/+Zbtb46+/gfp DCs, and no genes were found that were substantially increased in Ciita−/−Zbtb46+/gfp DCs. In addition, we examined DC1 and DC2 cells that were generated from cultures of whole BM treated with Flt3L and activated with interferon-γ (IFN-γ) and lipopolysaccharide (LPS). Strikingly, even after activation, very few genes were changed between Ciita+/+Zbtb46+/gfp and Ciita−/−Zbtb46+/gfp samples. Differentially expressed genes included those mentioned above, and four additional genes, Pdia4, Creld2, Hyou1 and Sdf2l1, which were reduced in Ciita−/− Zbtb46+/gfp DCs DC1s but not DC2s (Fig. 3B). Among putative CIITA targets reported to date, only B2m, CD74, Rab4b, Psmd3, Trim26, Plxna1, Tpp1, Col1a2, and Rfx5 were expressed above baseline in splenic or in vitro activated Flt3L cDCs (linear signal > 100) (Fig. 3D). However, their expression is not controlled by CIITA in cDCs.
Fig. 3. CIITA has a restricted transcriptional footprint in cDCs.
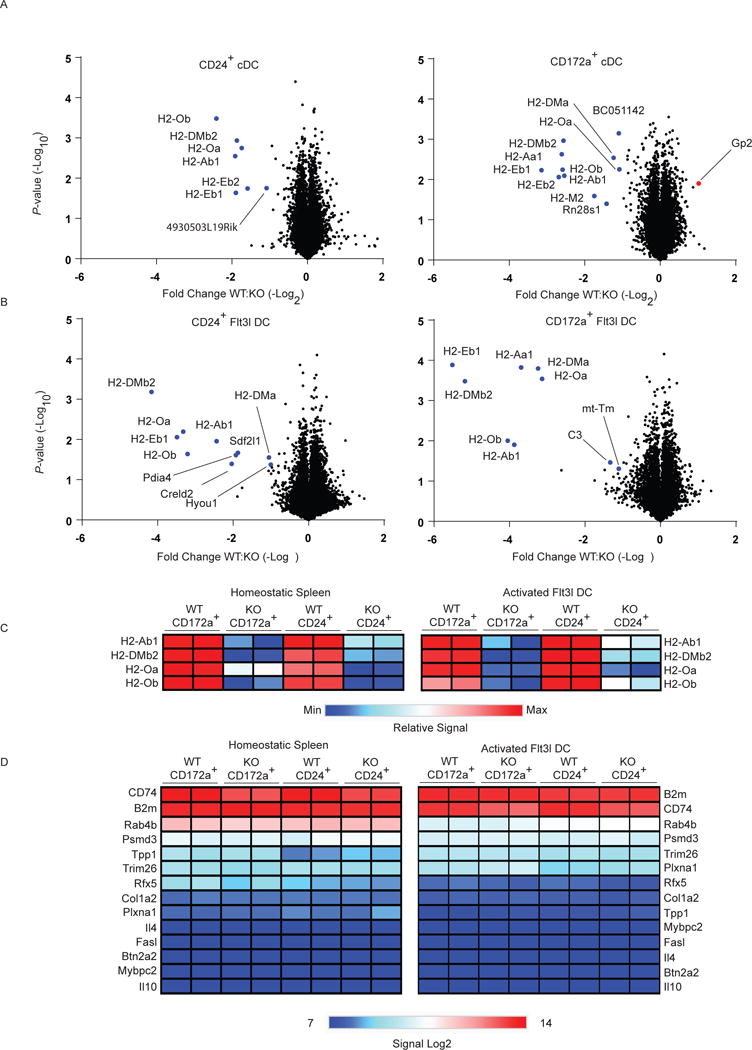
(A) Volcano plot showing differentially expressed genes in homeostatic splenic CD24+ (left) and CD172a+ (right) DCs sorted from Ciita+/+Zbtb46+/gfp and Ciita−/−Zbtb46+/gfp mice. (B) Volcano plot showing differentially expressed genes in CD24+ (left) and CD172a+ (right) DCs from whole BM Flt3L cultures derived from Ciita+/+Zbtb46+/gfp and Ciita−/−Zbtb46+/gfp mice and activated with IFN-γ and LPS. In (A,B) fold-change is derived from the mean difference in Log2 expression levels and P-values are derived from Welch’s t-test executed with Log2 expression levels of two biological replicates from one experiment (n = 2). (C) Heat map relative expression levels of select differentially expressed genes from (A) and (B). (D) Heat map of Log2 expression levels of selected previously reported CIITA target genes from (A) and (B).
Concluding Remarks
In contrast to studies based on reporter or ChIP-seq analysis, our results imply a highly restricted transcriptional footprint for CIITA for cDCs in vivo. Studies using cell lines found that CIITA could activate promoters of genes that are highly expressed in cDCs, such the Ii (CD74) [24–26], consistent with reports that CIITA binds to the Ii locus [10, 12, 13]. By contrast, we find no regulation of Ii by CIITA in cDCs in vivo. While DOβ expression in a human B cell line was CIITA-independent [27], murine cDC expression of H2-Ob, H2-Oa, and H2-DMb2 was dependent on CIITA in vivo. Such discrepancies likely stem from previous use of in vitro-derived BMDCs or cell lines [18, 19] and our results do not exclude that putative targets may be regulated by CIITA in other immune cell lineages [14, 28–30]. Many previously suggested CIITA targets are not expressed in cDCs in vivo or in vitro. Btn2a2 was expressed in GM-CSF-derived BMDCs and regulated by CIITA at an SXY module [15]. But Btn2a2 is expressed very weakly in cDCs in vivo and is not altered in CIITA-deficient cDCs. Also, we find no regulation of cytokines by CIITA in cDCs, unlike suggestions that CIITA regulates IL-4 in T cells [28]. We also did not detect IL-10 expression in cDCs either under homeostatic conditions or upon activation, unlike the reported increase of IL-10 expression for Ciita−/− mice that was attributed to cDCs [17]. Finally, reports from human cell lines and monocytes suggested that CIITA controls B2m expression, but here we find its expression to be CIITA-independent in cDCs [2, 10]. Thus, at least in cDCs, CIITA appears to act in a highly restricted network of genes limited to MHC locus, consistent with its well-established role as a master regulator of MHCII expression.
Materials and Methods
Mice
Ciita−/− mice, B6.129S2-Ciitatm1Ccum/J, were purchased from Jackson Laboratories [25]. Zbtb46gfp mice were maintained in our lab on a C57BL/6J background [21]. Male and female Ciita+/+Zbtb46+/gfp and Ciita−/−Zbtb46+/gfp mice between 6 and 12 weeks of age were used for all experiments with littermate controls. Mice were maintained in pathogen-free facilities under institutional guidelines and protocols approved by the animal studies committee at Washington University in St. Louis.
Cell Isolation, Culture, and Treatment
BM cells were isolated by crushing pelvises, femurs and tibia in MACS buffer (0.5% BSA and 2mM EDTA in PBS) using a mortar and pestle, purified using Histopaque-119 gradient, and strained with 70 μm mesh. Splenic dendritic cells were isolated from spleens minced and digested with stirring at 37°C for 45 min in 5 mL complete IMDM with 30 U/mL of DNase I (Sigma-Aldrich) and 250 μg/mL of collagenase B (Roche). Red blood cells were lysed with buffer containing NH4Cl and KHCO3. For in vivo population analysis by flow cytometry, splenic DCs were identified as B220−SiglecH−CD11c+(GFP+ or MHCII+)CD172a+ for CD172a+ DC2s, B220− SiglechH−CD11c+(GFP+ or MHCII+)CD24+ for CD24+ DC1s, and B220+SiglechH+ for pDCs. BM progenitors were identified as Lin−SiglecH−CD135+CD115+CD11c+CD117−GFP+ for pre-CD4 DCs and Lin−SiglecH−CD135+CD115−CD11c+CD117intGFP+ for pre-CD8 DCs. BM cells were depleted by magnetic bead separation (Miltenyi) using lineage markers TER-119, CD19, B220, and Ly6G. In vitro activated Flt3L DCs used for microarray analysis were derived in complete IMDM supplemented with 100 ng/mL of Flt3L (PeproTech) at a cell density of 2×106 cells/mL. Cells were cultured for 9 days at 37°C and resuspended in fresh media with Flt3L as described above at a cell density of 2×106 cells/mL. To activate cells, cultures were resuspended in fresh, complete media supplemented with Flt3L (100 ng/mL) and IFNγ (0.1 ng/mL) (PeproTech) and incubated for 22h at 37°C, followed by the addition 1:10 volume of media supplemented with LPS (final concentration of 20 μg/mL, from E. coli 011:B4) (Sigma-Aldrich) for 4 h at 37°C. CD24+ DCs were identified and sorted as Lin−B220−SiglecH−CD11c+GFP+CD24+ and CD172a+ DCs were identified and sorted as Lin−B220−SiglecH−CD11c+GFP+Sirpα+ with 96.9 to 99.9 % purity. Splenocytes were depleted by magnetic bead separation prior to cell sorting using lineage markers TER-119, CD3, CD19, and Ly6G.
Cell Staining and Flow Cytometry
Cells were stained at 4°C in CD16/32 block (2.4G2; BD) in MACS buffer. The following antibodies were used from Biolegend: B220 (RA3–6B2), MHCII (M5/114.15.2), SiglecH (551), CD115 (AFS98), CD3e (145–2C11), Ly6-G (1A8). The following were used from Tonbo: CD11c (N418), MHCII (M5/114.15.2). The following were used from BD: CD117 (2B8), CD135 (A2F10.1), TER-199 (TER-119), CD19 (1D3). The following were used from eBiosciences: CD172a (P84), CD24 (M1/69). Steptavidin-FITC and streptavidin-PE were used from BD. Cells were analyzed on a FACSCanto II or FACSAria Fusion flow cytometers (BD), and data analyzed with FlowJo software (TreeStar).
Expression Microarray Analysis
RNA was extracted from sorted cells using an EZNA MicroElute Total RNA Kit (Omega), followed by DNase treatment with TURBO DNA-free Kit (Invitrogen). RNA was amplified with the Ovation Pico WTA System (NuGEN) and hybridized to GeneChip Mouse Gene 1.0 ST microarray (Affymetrix). Data were processed using the robust multiarray average summarization method with quantile normalization using ArrayStar (DNASTAR). Expression levels were averaged for duplicate samples for all experiments. The NCBI GEO accession number for data reported in this paper is GSE96584.
Statistical Analysis
All statistical analysis was performed on Prism (GraphPad) as indicated in figure legends.
Acknowledgments
This work was supported by the Howard Hughes Medical Institute to K.M.M. and National Science Foundation Graduate Research Fellowship under Grant No. DGE-1143954 to D.A.A.
Abbreviations
- BM
bone marrow
- BMDCs
bone marrow-derived dendritic cell
- BSA
bovine serum albumin
- cDC
classical dendritic cell
- DC
dendritic cell
- DC1
Lineage to which CD24+ splenic DC is assigned
- DC2
Lineage to which CD172a+ splenic cDC is assigned
- EDTA
Ethylenediaminetetraacetic acid
- FACS
fluorescence-activated cell sorting
- IFN-γ
Interferon-γ
- IMDM
Iscove’s Modified Dulbecco’s Medium
- LPS
ipopolysaccharide
- MACS
magnetic activated cell sorting
- MHCII
major histocompatibility complex class II
- PBS
phosphate buffered saline
- pDC
Plasmacytoid DC
Footnotes
Conflicts of Interest: The authors declare no financial or commercial conflict of interest.
References
- 1.Steimle V, Otten LA, Zufferey M, Mach B. Complementation cloning of an MHC class II transactivator mutated in hereditary MHC class II deficiency (or bare lymphocyte syndrome) Cell. 1993;75:135–146. [PubMed] [Google Scholar]
- 2.Gobin SJ, Peijnenburg A, van Eggermond M, van Zutphen M, van den BR, van den Elsen PJ. The RFX complex is crucial for the constitutive and CIITA-mediated transactivation of MHC class I and beta2-microglobulin genes. Immunity. 1998;9:531–541. doi: 10.1016/s1074-7613(00)80636-6. [DOI] [PubMed] [Google Scholar]
- 3.Fontes JD, Jiang B, Peterlin BM. The class II trans-activator CIITA interacts with the TBP-associated factor TAFII32. Nucleic Acids Res. 1997;25:2522–2528. doi: 10.1093/nar/25.12.2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fontes JD, Kanazawa S, Jean D, Peterlin BM. Interactions between the class II transactivator and CREB binding protein increase transcription of major histocompatibility complex class II genes. Mol Cell Biol. 1999;19:941–947. doi: 10.1128/mcb.19.1.941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moreno CS, Beresford GW, Louis-Plence P, Morris AC, Boss JM. CREB regulates MHC class II expression in a CIITA-dependent manner. Immunity. 1999;10:143–151. doi: 10.1016/s1074-7613(00)80015-1. [DOI] [PubMed] [Google Scholar]
- 6.Gomez JA, Majumder P, Nagarajan UM, Boss JM. X box-like sequences in the MHC class II region maintain regulatory function. The Journal of Immunology. 2005;175:1030–1040. doi: 10.4049/jimmunol.175.2.1030. [DOI] [PubMed] [Google Scholar]
- 7.Muhlethaler-Mottet A, Krawczyk M, Masternak K, Spilianakis C, Kretsovali A, Papamatheakis J, Reith W. The S box of major histocompatibility complex class II promoters is a key determinant for recruitment of the transcriptional co-activator CIITA. J Biol Chem. 2004;279:40529–40535. doi: 10.1074/jbc.M406585200. [DOI] [PubMed] [Google Scholar]
- 8.Scholl T, Mahanta SK, Strominger JL. Specific complex formation between the type II bare lymphocyte syndrome-associated transactivators CIITA and RFX5. Proc Natl Acad Sci U S A. 1997;94:6330–6334. doi: 10.1073/pnas.94.12.6330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Westerheide SD, Louis-Plence P, Ping D, He XF, Boss JM. HLA-DMA and HLA-DMB gene expression functions through the conserved S-X-Y region. The Journal of Immunology. 1997;158:4812–4821. [PubMed] [Google Scholar]
- 10.Wong D, Lee W, Humburg P, Makino S, Lau E, Naranbhai V, Fairfax BP, et al. Genomic mapping of the MHC transactivator CIITA using an integrated ChIP-seq and genetical genomics approach. Genome Biol. 2014;15:494. doi: 10.1186/s13059-014-0494-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reith W, LeibundGut-Landmann S, Waldburger JM. Regulation of MHC class II gene expression by the class II transactivator. Nat Rev Immunol. 2005;5:793–806. doi: 10.1038/nri1708. [DOI] [PubMed] [Google Scholar]
- 12.Krawczyk M, Seguin-Estevez Q, Leimgruber E, Sperisen P, Schmid C, Bucher P, Reith W. Identification of CIITA regulated genetic module dedicated for antigen presentation. PLoS Genet. 2008;4:e1000058. doi: 10.1371/journal.pgen.1000058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Scharer CD, Choi NM, Barwick BG, Majumder P, Lohsen S, Boss JM. Genome-wide CIITA-binding profile identifies sequence preferences that dictate function versus recruitment. Nucleic Acids Res. 2015;43:3128–3142. doi: 10.1093/nar/gkv182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Krawczyk M, Leimgruber E, Seguin-Estevez Q, Dunand-Sauthier I, Barras E, Reith W. Expression of RAB4B, a protein governing endocytic recycling, is co-regulated with MHC class II genes. Nucleic Acids Res. 2007;35:595–605. doi: 10.1093/nar/gkl980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sarter K, Leimgruber E, Gobet F, Agrawal V, Dunand-Sauthier I, Barras E, Mastelic-Gavillet B, et al. Btn2a2, a T cell immunomodulatory molecule coregulated with MHC class II genes. J Exp Med. 2016;213:177–187. doi: 10.1084/jem.20150435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wong AW, Brickey WJ, Taxman DJ, van Deventer HW, Reed W, Gao JX, Zheng P, et al. CIITA-regulated plexin-A1 affects T-cell-dendritic cell interactions. Nat Immunol. 2003;4:891–898. doi: 10.1038/ni960. [DOI] [PubMed] [Google Scholar]
- 17.Yee CS, Yao Y, Xu Q, McCarthy B, Sun-Lin D, Tone M, Waldmann H, et al. Enhanced production of IL-10 by dendritic cells deficient in CIITA. The Journal of Immunology. 2005;174:1222–1229. doi: 10.4049/jimmunol.174.3.1222. [DOI] [PubMed] [Google Scholar]
- 18.Helft J, Bottcher J, Chakravarty P, Zelenay S, Huotari J, Schraml BU, Goubau D, et al. GM-CSF Mouse Bone Marrow Cultures Comprise a Heterogeneous Population of CD11c(+)MHCII(+) Macrophages and Dendritic Cells. Immunity. 2015;42:1197–1211. doi: 10.1016/j.immuni.2015.05.018. [DOI] [PubMed] [Google Scholar]
- 19.Briseno CG, Haldar M, Kretzer NM, Wu X, Theisen DJ, KC W, Durai V, et al. Distinct Transcriptional Programs Control Cross-Priming in Classical and Monocyte-Derived Dendritic Cells. Cell Rep. 2016;15:2462–2474. doi: 10.1016/j.celrep.2016.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meredith MM, Liu K, Darrasse-Jeze G, Kamphorst AO, Schreiber HA, Guermonprez P, Idoyaga J, et al. Expression of the zinc finger transcription factor zDC (Zbtb46, Btbd4) defines the classical dendritic cell lineage. Journal of Experimental Medicine. 2012;209:1153–1165. doi: 10.1084/jem.20112675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Satpathy AT, KC W, Albring JC, Edelson BT, Kretzer NM, Bhattacharya D, Murphy TL, et al. Zbtb46 expression distinguishes classical dendritic cells and their committed progenitors from other immune lineages. Journal of Experimental Medicine. 2012;209:1135–1152. doi: 10.1084/jem.20120030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grajales-Reyes GE, Iwata A, Albring J, Wu X, Tussiwand R, KC W, Kretzer NM, et al. Batf3 maintains autoactivation of Irf8 for commitment of a CD8alpha(+) conventional DC clonogenic progenitor. Nat Immunol. 2015;16:708–717. doi: 10.1038/ni.3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu X, Briseno CG, Durai V, Albring JC, Haldar M, Bagadia P, Kim KW, et al. Mafb lineage tracing to distinguish macrophages from other immune lineages reveals dual identity of Langerhans cells. J Exp Med. 2016;213:2553–2565. doi: 10.1084/jem.20160600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cao ZA, Moore BB, Quezada D, Chang CH, Jones PP. Identification of an IFN-gamma responsive region in an intron of the invariant chain gene. Eur J Immunol. 2000;30:2604–2611. doi: 10.1002/1521-4141(200009)30:9<2604::AID-IMMU2604>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 25.Chang CH, Guerder S, Hong SC, van Ewijk W, Flavell RA. Mice lacking the MHC class II transactivator (CIITA) show tissue-specific impairment of MHC class II expression. Immunity. 1996;4:167–178. doi: 10.1016/s1074-7613(00)80681-0. [DOI] [PubMed] [Google Scholar]
- 26.Chin KC, Li GG, Ting JP. Importance of acidic, proline/serine/threonine-rich, and GTP-binding regions in the major histocompatibility complex class II transactivator: generation of transdominant-negative mutants. Proc Natl Acad Sci U S A. 1997;94:2501–2506. doi: 10.1073/pnas.94.6.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Taxman DJ, Cressman DE, Ting JP. Identification of class II transcriptional activator-induced genes by representational difference analysis: discoordinate regulation of the DN alpha/DO beta heterodimer. The Journal of Immunology. 2000;165:1410–1416. doi: 10.4049/jimmunol.165.3.1410. [DOI] [PubMed] [Google Scholar]
- 28.Gourley T, Roys S, Lukacs NW, Kunkel SL, Flavell RA, Chang CH. A novel role for the major histocompatibility complex class II transactivator CIITA in the repression of IL-4 production. Immunity. 1999;10:377–386. doi: 10.1016/s1074-7613(00)80037-0. [DOI] [PubMed] [Google Scholar]
- 29.Gourley TS, Patel DR, Nickerson K, Hong SC, Chang CH. Aberrant expression of Fas ligand in mice deficient for the MHC class II transactivator. The Journal of Immunology. 2002;168:4414–4419. doi: 10.4049/jimmunol.168.9.4414. [DOI] [PubMed] [Google Scholar]
- 30.Sisk TJ, Gourley T, Roys S, Chang CH. MHC class II transactivator inhibits IL-4 gene transcription by competing with NF-AT to bind the coactivator CREB binding protein (CBP)/p300. The Journal of Immunology. 2000;165:2511–2517. doi: 10.4049/jimmunol.165.5.2511. [DOI] [PubMed] [Google Scholar]


