Summary
CRISPR-Cas9 technology would be enhanced by the ability to inhibit Cas9 function spatially, temporally, or conditionally. Previously, we discovered small proteins encoded by bacteriophages that inhibit the CRISPR-Cas systems of their host bacteria. These “anti-CRISPRs” were specific to type I CRISPR-Cas systems that do not employ the Cas9 protein. We posited that nature would also yield Cas9 inhibitors in response to the evolutionary arms race between bacteriophages and their hosts. Here, we report the discovery of three distinct families of anti-CRISPRs that specifically inhibit the CRISPR-Cas9 system of Neisseria meningitidis. We show that these proteins bind directly to N. meningitidis Cas9 (NmeCas9), and can be used as potent inhibitors of genome editing by this system in human cells. These anti-CRISPR proteins now enable “off-switches” for CRISPR-Cas9 activity, and provide a genetically-encodable means to inhibit CRISPR-Cas9 genome editing in eukaryotes.
INTRODUCTION
CRISPR-Cas9 mediated genome editing has revolutionized biotechnology and holds immense promise for therapeutic applications. Cas9 is a nuclease that can be programmed with a guide RNA molecule to cut nearly any desired DNA sequence (Gasiunas et al., 2012; Jinek et al., 2012), enabling mutagenesis or editing at the site of cleavage (Cho et al., 2013; Cong et al., 2013; Hwang et al., 2013; Jiang et al., 2013; Jinek et al., 2013; Mali et al., 2013). This RNA-guided DNA editing technology is being developed for personalized gene therapy to correct inherited disease, for sequence-specific targeting of pathogens to treat infectious disease, and many other applications (Bikard et al., 2014; Ebina et al., 2013; Gomaa et al., 2014; Kaminski et al., 2016; Ousterout et al., 2015; Wu et al., 2013; Yin et al., 2014).
Although the utility of Cas9 DNA targeting is widely acknowledged, there are currently limited means to exert control over Cas9 activity once it has been activated or delivered, leading to practical difficulties and safety concerns. For example, off-target effects (cleavage and mutation at unintended, near-cognate genomic sites) are exacerbated by excessive or prolonged Cas9 activity (Fu et al., 2014; Hsu et al., 2013; Pattanayak et al., 2013). Many potential therapeutic applications of CRISPR-Cas9 only require editing in specific target tissues, and Cas9 activity in ancillary tissues is at best useless and at worst a safety risk. When zygotic injections of CRISPR-Cas9 components are used to generate mutant animals (Wang et al., 2013), Cas9 activity after the initial rounds of mitosis can give rise to mosaic genotypes (Yen et al., 2014). In applications that require homology-dependent repair (HDR) for precise editing, Cas9 activity during the G1 phase of the cell cycle (when HDR pathways are suppressed; (Orthwein et al., 2015)) increases the background of undesired imprecise edits. Recently, CRISPR-Cas9 gene drives (which cause desired genes to propagate throughout natural populations through non-Mendelian forced inheritance) have been developed, in part to advance the long-term goal of eradicating disease vectors such as mosquitos (Gantz et al., 2015; Hammond et al., 2016). A danger of this approach is that gene drives, once introduced into the environment, could be difficult to restrain, and could have unpredictable ecological consequences. Based on these and other considerations, the performance and safety of CRISPR-Cas9 applications could be greatly improved if Cas9 activity could be more effectively controlled. Several groups have devised methods to activate CRISPR-Cas9 genome editing in response to specific cues, including light-inducible and drug-inducible Cas9 activity (Nihongaki et al., 2015; Nunez et al., 2016; Wright et al., 2015). However, a robust, specific, and genetically-encodable “off-switch” for Cas9 activity has not yet been identified.
CRISPR-Cas9 technologies are derived from type II CRISPR-Cas adaptive immune systems of bacteria, which target and destroy foreign DNA entities such as bacteriophages (phages) and plasmids (Barrangou et al., 2007; Deltcheva et al., 2011). Although the Cas9 ortholog from Streptococcus pyogenes strain SF370 [SpyCas9, subtype II-A (Makarova et al., 2015)] is the most commonly used and the best understood, type II CRISPR-Cas systems from several other bacterial species have also been adapted for eukaryotic genome editing (Cong et al., 2013; Esvelt et al., 2013; Hirano et al., 2016; Hou et al., 2013; Lee et al., 2016; Muller et al., 2016; Ran et al., 2015). For example, Cas9 from Neisseria meningitidis (NmeCas9), which belongs to the CRISPR-Cas subtype II-C (Makarova et al., 2015), is an effective tool for human genome editing (Esvelt et al., 2013; Hou et al., 2013; Lee et al., 2016, Amrani et al., manuscript in preparation). NmeCas9 is hundreds of amino acids smaller than SpyCas9, facilitating viral delivery, and it is also less prone to off-target effects (Esvelt et al., 2013; Hou et al., 2013; Lee et al., 2016, Amrani et al., manuscript in preparation). “Dead” NmeCas9 (dNmeCas9), in which nuclease active-site residues have been mutated, has also proven to be an effective, specific RNA-guided genome binding platform (Esvelt et al., 2013; Hilton et al., 2015; Kearns et al., 2015; Ma et al., 2015b), similar to dSpyCas9 and other nuclease-inactivated orthologs (reviewed in (Dominguez et al., 2016; Wang et al., 2016)).
The goal of the work described here was to identify naturally occurring protein inhibitors of a CRISPR-Cas9 system. The rationale for this endeavour was our previous discovery of inhibitors of both the type I–E and type I–F CRISPR-Cas systems (Bondy-Denomy et al., 2013; Pawluk et al., 2014). These proteins, which we named anti-CRISPRs, are small proteins encoded by phages that allow a bacterial host’s CRISPR-Cas immune system to be evaded. Anti-CRISPRs function through a variety of mechanisms (Bondy-Denomy et al., 2015) and we recently discovered that multiple families of type I–F anti-CRISPRs occur widely in mobile genetic elements (MGEs, e.g. phages and conjugative elements) of diverse bacterial species (Pawluk et al., 2016). Although type I systems use Cas proteins that are completely unrelated to Cas9, we hypothesized that inhibitors of type II systems would also exist, since they would confer strong evolutionary advantages to MGEs encoding them. Thus, we employed the same bioinformatic approach that successfully identified type I anti-CRISPRs to search for inhibitors of Cas9. As described below, this effort led us to discover three distinct anti-CRISPR protein families that potently inhibit the N. meningitidis type II-C CRISPR-Cas system. These proteins directly interact with NmeCas9 and can function as off-switches for NmeCas9 genome editing activity in cultured human cells.
RESULTS
Three distinct anti-CRISPRs inhibit CRISPR-Cas activity in Neisseria meningitidis
A conserved feature of characterized anti-CRISPR (acr) genes is the presence of a downstream gene encoding a putative transcriptional regulator. In previous work, we identified two distinct families of these helix-turn-helix (HTH) containing anti-CRISPR associated (Aca) proteins, which we called Aca1 and Aca2. Identification of genes encoding Aca proteins in diverse bacterial species led us to discover five new families of type I–F acr genes encoded directly upstream of the aca genes, thereby providing precedent for the use of genomic localization to predict anti-CRISPR activity of novel, hypothetical protein families with high confidence (Pawluk et al., 2016). We reasoned that genes encoding inhibitors of type II CRISPR-Cas systems would be found upstream of aca genes in MGEs within species bearing type II systems. By conducting a series of BLAST searches with Aca1 and Aca2, we identified a candidate anti-CRISPR gene in a Brackiella oedipodis putative conjugative element that encoded a 91-residue hypothetical protein (accession WP_028357638.1) lying directly upstream of an aca2 gene (Figure 1A). This putative anti-CRISPR possessed several homologs encoded in MGEs of diverse Proteobacteria, and a distant, putative ortholog in a Firmicute, Fenollaria massiliensis (Figure S1). The most frequently observed CRISPR-Cas system among species encoding homologs of this protein was type II-C. Thus, we hypothesized that this putative anti-CRISPR family would inhibit the activity of one or more representative type II-C Cas9 orthologs. Because N. meningitidis strain 8013 harbors the best-established type II-C CRISPR-Cas system (Zhang et al., 2013; Zhang et al., 2015), and because we identified a strain of N. meningitidis among the genomes that contain an MGE encoding a member of this putative anti-CRISPR family (Figure 1A, Figure S1, Table S1, Table S2), we used NmeCas9 to test this hypothesis.
Figure 1. Identification and Validation of Type II-C Anti-CRISPRs.
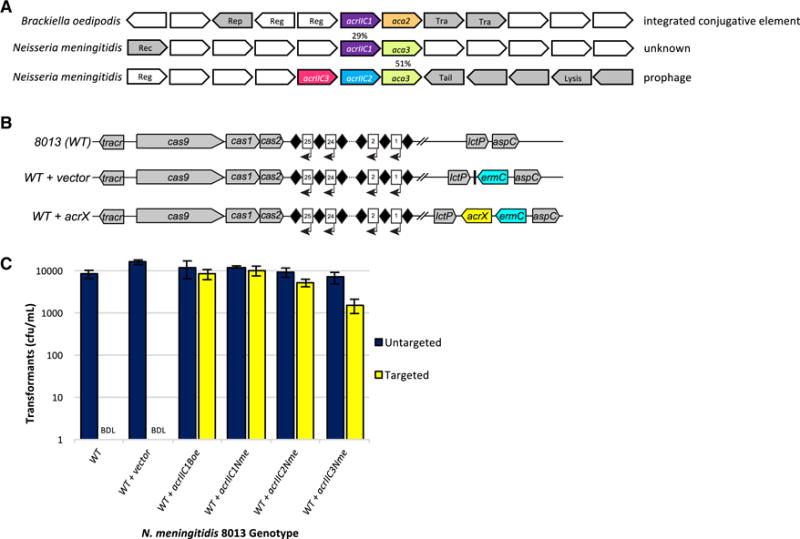
(A) Schematic representation of candidate type II-C acr and aca genes within putative MGEs in the genomes of strains of Brackiella oedipodis and Neisseria meningitidis. Homologous genes are color-matched, with percent amino acid identities indicated. Gene arrows are not drawn to scale. Any known, relevant gene product functions are annotated as follows: Rep, plasmid replication protein; Reg, transcriptional regulator; Tra, conjugal transfer protein; Rec, recombinase; Tail, phage tail structural protein; Lysis, phage lysis cassette. Genes colored in grey have MGE-related functions and/or show clear evidence of horizontal transfer.
(B) Schematic representation of genotypes in N. meningitidis strains used to test candidate anti-CRISPR function. Diamonds, CRISPR repeats; numbered rectangles, CRISPR spacers; arrows, CRISPR transcription. ermC, integrated erythromycin resistance cassette; acrX, integrated candidate anti-CRISPR cassette. Individual genetic elements are not to scale.
(C) Candidate type II-C anti-CRISPRs inhibit CRISPR interference in N. meningitidis. Results of the transformation assay in N. meningitidis strain 8013, and isogenic derivatives with each indicated acr gene integrated at the nics locus (see B), are plotted. The CRISPR-targeted protospacer plasmid (yellow) cannot transform wild-type and empty vector-containing cells due to an active CRISPR-Cas system, resulting in zero transformants. BDL = below detection limit of this assay. Plasmid DNA that lacks a target protospacer sequence can transform all strains equally well (navy). Experiments were repeated three times and error bars represent the standard error of the mean (s.e.m) between three replicates. Cells were also plated on non-selective media and the total number of cfu/mL present was equivalent in each sample (data not shown).
We measured the ability of the candidate type II-C anti-CRISPR gene from B. oedipodis and its 29% identical homolog from N. meningitidis (Figure 1A) to inhibit type II-C CRISPR-Cas activity in its native context, using a previously described natural transformation assay in N. meningitidis 8013 (Zhang et al., 2013; Zhang et al., 2015). In this assay, the transformation frequency of a plasmid bearing a CRISPR-targeted protospacer sequence was compared to that of a control plasmid lacking the protospacer. We used the wild-type strain as well as isogenic derivatives with an integrated, empty nics (Neisseria intergenic complementation site) cassette, or the same cassette carrying genes encoding either of the two candidate anti-CRISPR proteins (Figure 1B), each driven by the N. meningitidis cas9 promoter. In wild-type cells and the empty-vector control, robust type II-C CRISPR-Cas activity resulted in a ≥104-fold decrease in the transformation frequency of CRISPR-targeted DNA (Figure 1C). Strikingly, expression of the putative anti-CRISPRs resulted in equal transformation frequencies when targeted or untargeted DNA was used (Figure 1C), reflecting a lack of CRISPR interference. These data implied that the type II-C CRISPR-Cas system of N. meningitidis was indeed inhibited by these putative anti-CRISPR genes, which we named acrIIC1Boe and acrIIC1Nme. Although acrIIC1Boe has presumably evolved to inhibit the Cas9 ortholog found in B. oedipodis (BoeCas9), NmeCas9 is 47% identical to BoeCas9, suggesting that this similarity is sufficient to account for the observed cross-species inhibition.
All of the identified acrIIC1 homologs except acrIIC1Boe were not found adjacent to aca1 or aca2 genes, but instead many were encoded upstream of a variety of genes encoding distinct HTH-containing proteins (Figure 1, Figure S1). We hypothesized that these could represent new families of aca genes, and could therefore lead to new anti-CRISPR candidates. Using BLAST searches, we determined that homologs of the HTH protein-coding gene downstream of acrIIC1Nme were the only putative aca genes found repeatedly in genomic regions displaying MGE-like properties. Of greatest interest, we identified members of this gene family in putative prophage elements in N. meningitidis strains, and in several cases the HTH-containing protein coding gene was immediately downstream of the same two small, uncharacterized open reading frames, neither of which exhibited detectable sequence similarity to acrIIC1Nme. We cloned these two distinct genes and tested each one for anti-CRISPR activity. Using the N. meningitidis transformation assay described above, we showed that both of these genes displayed robust anti-CRISPR activity (Figure 1B,C), and we named them acrIIC2Nme and acrIIC3Nme (Table S1, Table S2). Based on this result, we classified the HTH protein-coding gene as a bona fide aca gene, making it the third such family, hereafter referred to as aca3. Overall, these results demonstrate the existence of three distinct families of anti-CRISPR genes that are active against a type II CRISPR-Cas system.
Type II-C anti-CRISPR proteins interact directly with NmeCas9 to prevent DNA cleavage
To determine whether the type II-C anti-CRISPRs function by directly interacting with NmeCas9, we mixed purified, untagged anti-CRISPR proteins with purified, 6xHis-tagged NmeCas9 protein (preloaded with coexpressed sgRNA) and conducted nickel affinity chromatography to assess whether the anti-CRISPRs directly bound NmeCas9 in vitro. We found that AcrIIC1Nme, AcrIIC2Nme, and AcrIIC3Nme were all retained on the nickel column, reflecting association with NmeCas9. By contrast, a previously identified type I anti-CRISPR protein (AcrE2; (Pawluk et al., 2014)) did not associate with NmeCas9 (Figure 2A, Figure S2). In a parallel experiment, the anti-CRISPRs did not bind significantly to AnaCas9 (Figure 2B, Figure S2) (Jinek et al., 2014; Ma et al., 2015a), a distantly related type II-C Cas9 homolog with ~20% sequence identity to NmeCas9. These data demonstrate that the anti-CRISPRs identified in this study specifically bind to NmeCas9.
Figure 2. Anti-CRISPRs Bind Directly to NmeCas9:sgRNA.
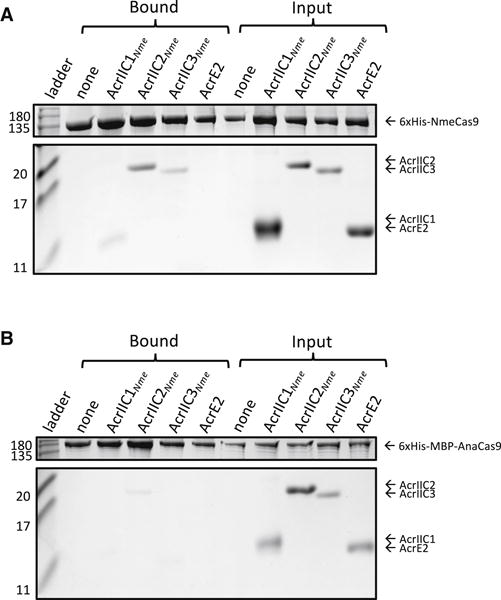
(A) Purified, untagged anti-CRISPR proteins were mixed with purified, 6xHis tagged NmeCas9:sgRNA in vitro. The input and elution fractions (before and after nickel affinity purification) are shown on the right and left sides of the Coomassie-stained SDS-PAGE gel, respectively. Mobilities of marker proteins (in kDa) are denoted on the left. AcrE2 is an inhibitor of the type I–E CRISPR-Cas system, and is included in this assay as a negative control. The gel image was cropped to conserve space and to remove irrelevant bands resulting from Cas9 degradation. The image is representative of at least three replicates. Uncropped gel images are presented in Figure S2.
(B) Binding assays were carried out between the same anti-CRISPRs tested in (A) and Cas9 from Actinomyces naeslundii (AnaCas9). AnaCas9 is a distantly related type II-C Cas9 protein (~20% amino acid sequence identity with NmeCas9). The image is representative of at least three replicates.
To assess the effect of the anti-CRISPRs on Cas9 enzymatic activity, in vitro DNA cleavage assays were performed. When purified NmeCas9 was loaded with in vitro transcribed sgRNA and then mixed with target DNA, robust and specific cleavage was observed (Figure 3), as described previously (Zhang et al., 2015). Cleavage was unaffected by prior incubation of NmeCas9 with increasing amounts of the control, type I-specific anti-CRISPR, AcrE2. In contrast, addition of the N. meningitidis anti-CRISPRs to these reactions resulted in inhibition of NmeCas9-catalyzed cleavage in a dose-dependent manner. Approximately 50% cleavage inhibition resulted when the anti-CRISPRs were added at a 1:1 molar ratio, and complete inhibition was seen at a 5:1 anti-CRISPR:NmeCas9 ratio (Figure 3). The DNA cleavage activity of S. pyogenes Cas9 (SpyCas9), which is the most commonly used Cas9 for genome editing, was not affected by addition of any of the anti-CRISPRs (Figure 3, lower panel). This result was expected because SpyCas9 belongs to the type II-A CRISPR-Cas subtype and is very distantly related to NmeCas9. Overall, these in vitro data clearly demonstrate that these anti-CRISPRs directly bind to and specifically inhibit the DNA cleavage activity of NmeCas9. The inhibitory effects of anti-CRISPRs on NmeCas9 in its sgRNA-loaded form imply that the natural protective functions of the anti-CRISPR proteins require inhibition of crRNA/tracrRNA-loaded NmeCas9 that is already present in the host cell at the time of phage infection.
Figure 3. Type II-C Anti-CRISPRs Specifically Block DNA Cleavage by NmeCas9 In Vitro.
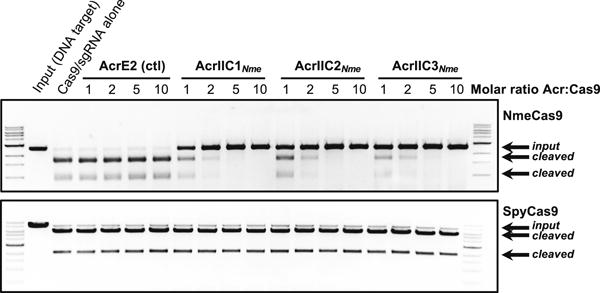
Linearized plasmid DNA bearing a protospacer adjacent to a PAM sequence was subjected to in vitro digestion by purified, recombinant, sgRNA-programmed NmeCas9 (upper panel) or SpyCas9 (lower panel). Where indicated at the top of each lane, Cas9 was pre-incubated with purified anti-CRISPR proteins as indicated with AcrE2 as a negative control. Molar equivalents of anti-CRISPR protein (relative to Cas9) are shown at the top of each lane, and mobilities of input and cleaved DNAs are denoted on the right. The NmeCas9 cleavage assays shown are representative of three independent replicates.
Anti-CRISPRs inhibit genome editing by NmeCas9 in cultured human cells
Our discovery of direct inhibitors of NmeCas9 activity raised the possibility that these anti-CRISPRs could be used as off-switches for CRISPR-Cas9 genome editing in mammalian cells. To address this, we co-transfected HEK293T cells with three plasmids: one expressing Cas9, one expressing a genome-targeting sgRNA, and one expressing an anti-CRISPR. Genome editing efficiency was determined using an established T7 endonuclease 1 (T7E1)-based protocol. Strikingly, we found that each of the anti-CRISPRs greatly decreased the ability of NmeCas9 to create genome lesions in cultured human cells (Figure 4, Figure S3, Table S3, Figure S4). Because the anti-CRISPRs are all below ~14 kDa (i.e., small enough to diffuse freely through nuclear pores), they inhibited NmeCas9 genome editing even without an appended, heterologous nuclear localization sequence (NLS). Plasmid titration experiments demonstrated that the three anti-CRISPR families could each completely inhibit editing, with AcrIIC3Nme appearing to be the most potent (Figure S4). The superior potency of AcrIIC3Nme anti-CRISPR activity in mammalian cells is noteworthy given that it was slightly less effective at inhibiting transformation interference in meningococcal cells (Figure 1C). The variations in activities of these anti-CRISPRs in mammalian cells are likely due to differences in expression or stability as they all displayed similar inhibitory activities in vitro (Figure 3). Consistent with our in vitro results, the anti-CRISPRs had no effect on editing mediated by SpyCas9 targeting the same genomic site (Figure 4, Figure S3). In addition, type I–E anti-CRISPR AcrE2 had no significant inhibitory effect in any of these experiments. In no instance did we observe any sign of cellular toxicity by any anti-CRISPR. In summary, these human cell experiments illustrate the potential application of these anti-CRISPRs for precise control of Cas9-mediated genome editing.
Figure 4. Type II-C Anti-CRISPRs Specifically Block Genome Editing by NmeCas9 in Human Cells.
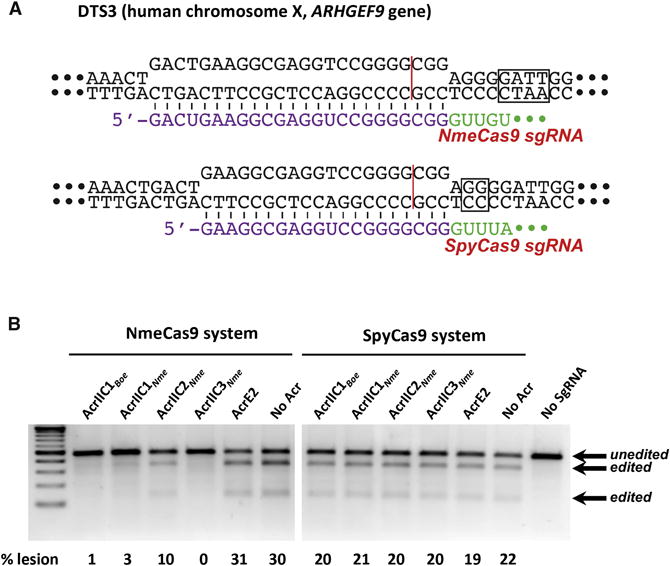
(A) Schematic representation of R-loop structures at a dual target site (DTS3) in the human genome that can be cleaved and edited by either SpyCas9 (top) or NmeCas9 (bottom). Guide sequences (purple), PAMs (boxed), and Cas9 cleavage sites (red line) are indicated.
(B) T7E1 assays of NmeCas9 or SpyCas9 editing efficiencies at DTS3 upon transient transfection of human HEK293T cells. Constructs encoding anti-CRISPR proteins were co-transfected as indicated at the top of each lane. Mobilities of T7E1-digested (edited) and -undigested (unedited) bands are indicated to the right, and editing efficiencies (“% lesion”) are given at the bottom of each lane. These images are representative of at least seven replicates.
AcrIIC3Nme prevents dNmeCas9 genome binding in cultured human cells
“Dead” Cas9 (dCas9) orthologs, including dNmeCas9 (Esvelt et al., 2013; Hilton et al., 2015; Kearns et al., 2015; Ma et al., 2015b), have proven to be exceptionally useful for RNA-guided DNA binding (without Cas9-catalyzed DNA cleavage), since a wide range of domains and functionalities can be fused or tethered to the DNA-bound dCas9/sgRNA complex (Dominguez et al., 2016; Wang et al., 2016). In principle, anti-CRISPR inhibition of sgRNA-guided NmeCas9 DNA cleavage (Figure 3) and genome editing (Figure 4) could reflect either inhibition upstream of stable R-loop formation, or inhibition of NmeCas9 catalytic activation after stable R-loop formation. In the former case, the anti-CRISPR could be used as an off-switch not only for genome editing, but also for dNmeCas9 DNA binding applications such as CRISPRi and CRISPRa (Dominguez et al., 2016; Wang et al., 2016). To determine whether our most potent genome editing inhibitor (AcrIIC3Nme) can prevent stable DNA binding by dNmeCas9 in mammalian cells, we used a previously developed system in which superfolder (sf) GFP-labeled dNmeCas9 and mCherry-labeled dSpyCas9 are simultaneously colocalized to telomeric loci by cognate sgRNAs upon co-transfection of their expression plasmids in U2OS cells (Ma et al., 2015b) (Figure 5A). We readily observed colocalizing telomeric dNmeCas9-(sfGFP)3 and dSpyCas9-(mCherry)3 foci as long as both of the telomere-directed sgRNAs were included for the two dCas9 orthologs (Figure 5, B–D), as reported previously (Ma et al., 2015b). We then repeated the experiment with the co-transfected, mTagBFP2-marked plasmid (Figure 5A, bottom) also carrying an anti-CRISPR expression cassette. AcrE2 had no effect on telomeric co-localization of dNmeCas9-(sfGFP)3 and dSpyCas9-(mCherry)3, as expected (Figure 5E). In contrast, co-expression of AcrIIC3Nme prevented the co-localization of dNmeCas9-(sfGFP)3 with the dSpyCas9-(mCherry)3 telomeric foci (Figure 5F). We then repeated this experiment in a blinded fashion, with unidentified samples that had been coded by a separate experimenter. Only cells that exhibited mTagBFP2 and sfGFP fluorescence as well as dSpyCas9-(mCherry)3 telomeric foci were assessed for the presence or absence of co-localizing dNmeCas9-(sfGFP)3 telomeric foci, and all such imaged cells were included in our quantitations. The results were tabulated, decoded, and plotted as a bar graph (Figure 5G). Telomeric dNmeCas9-(sfGFP)3 foci were observed in 94% (31 out of 33) of cells in the absence of any Acr protein, and 88% (31 out of 37) of cells in the presence of the negative control AcrE2 protein. By contrast, 0% of cells (0 out of 46) exhibited dNmeCas9-(sfGFP)3 telomeric foci when AcrIIC3Nme was coexpressed. These results confirm the robust inhibitory effect of AcrIIC3Nme on stable, sgRNA-programmed DNA binding by dNmeCas9, and indicate that it can be used as a potent off-switch not only for NmeCas9 genome editing, but also for dNmeCas9-based applications in mammalian cells.
Figure 5. AcrIIC3Nme Prevents DNA Binding by NmeCas9 in Human Cells.
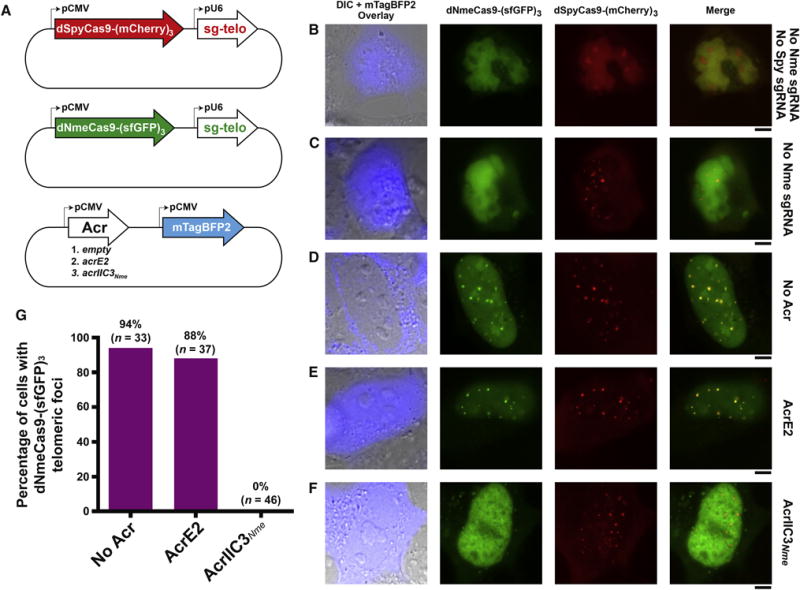
(A) Schematic representation of plasmids used for expression of dNmeCas9-(sfGFP)3, dSpyCas9-(mCherry)3, their respective telomeric sgRNAs, and anti-CRISPR protein. The plasmid encoding the anti-CRISPR protein is also marked with the blue fluorescent protein mTagBFP2.
(B-F) Fluorescence images of U2OS cells transiently transfected with plasmids depicted in (A). The specific version of each plasmid set (with or without sgRNAs, with or without anti-CRISPRs) is given to the right of each row. First column: differential interference contrast (DIC) and mTagBFP2 imaging, merged. Second column: dNmeCas9-(sfGFP)3. Third column: dSpyCas9-(mCherry)3. Fourth column: dNmeCas9-(sfGFP)3 and dSpyCas9-(mCherry)3, merged. Scale bars, 5 μm.
(G) Quantitation of dNmeCas9-(sfGFP)3 telomeric foci, as judged by co-localization with dSpyCas9-(mCherry)3 telomeric foci, in cells that express no anti-CRISPR, negative control anti-CRISPR (AcrE2), or AcrIIC3Nme. Foci were scored blind, i.e. without the experimenter knowing the sample identities (see STAR Methods). n represents the number of cells that were scored in each condition.
A wide range of type II-C CRISPR-Cas systems are likely susceptible to inhibition by anti-CRISPRs
CRISPR-Cas systems are divided into two broad classes, each encompassing several types and many subtypes. Class 1 systems employ multi-subunit surveillance complexes, whereas Class 2 systems have single, large effector proteins like Cas9 (Makarova et al., 2015). Our previous studies on anti-CRISPRs acting on type I systems (belonging to Class 1) suggest that each anti-CRISPR protein acts on a particular range of systems within one subtype due to the specificity of protein-protein interactions between the anti-CRISPR and Cas proteins (Bondy-Denomy et al., 2015; Bondy-Denomy et al., 2013; Pawluk et al., 2014; Pawluk et al., 2016). An anti-CRISPR gene will likely be selected for if it inhibits the CRISPR-Cas system of the bacterium in which it is found, as these genes are almost always located on MGEs that have successfully invaded a host. This principle was used to accurately predict that the anti-CRISPRs described here would block the type II-C CRISPR-Cas system of N. meningitidis.
To visualize the potential general impact of anti-CRISPR activity on type II CRISPR-Cas systems, we created a phylogenetic tree of Cas9, which is the sole effector protein of these systems and the direct binding target of the type II-C anti-CRISPRs (Figure 6). Bacterial genera in which known type II-C anti-CRISPR homologs are encoded are indicated in red. From this analysis, based on the phylogenetic breadth spanned by the anti-CRISPR putative orthologs, we propose that the majority of type II-C CRISPR-Cas diversity may be susceptible to at least one member of an anti-CRISPR family discovered in this study. These data, combined with our previous analysis of type I–F CRISPR-Cas systems and their cognate anti-CRISPRs, suggests that even the relatively small number of anti-CRISPR gene families discovered to date have a broad impact on CRISPR-Cas systems in bacteria. Now that anti-CRISPRs inhibiting both Class 1 and Class 2 CRISPR-Cas systems have been described, we expect that anti-CRISPRs able to inhibit all types and subtypes of CRISPR-Cas systems exist and await discovery.
Figure 6. Anti-CRISPRs Likely Have a Broad Impact on Diverse CRISPR-Cas Systems.

A maximum likelihood phylogenetic tree of representative Cas9 protein sequences. Each protein is classified based on the CRISPR locus in which it resides as type II-A (blue), type II-B (yellow), or type II-C (purple). Cas9 proteins belonging to any genus that has a type II-C anti-CRISPR putative ortholog are coloured in red. With the assumption that a given anti-CRISPR ortholog inhibits the CRISPR-Cas system in the species where it is found, this visualization provides an estimate of the breadth of activity encompassed by the anti-CRISPR families discovered here. The position of notable Cas9 orthologs on the tree are indicated by asterisks.
DISCUSSION
In this work, we report that protein inhibitors of CRISPR interference, previously reported only for type I CRISPR-Cas systems, now extend into the type II systems that employ Cas9. Importantly, we show that the three different families of type II-C anti-CRISPRs that we have identified can be used to block genome editing by NmeCas9 in cultured human cells. Genetically encoded Cas9 inhibitors provide a means to spatially, temporally, or conditionally control Cas9 activity, thereby potentially allowing tissue-, cell cycle stage-, developmental stage-, or stimulus-specific inactivation of genome editing. Target site precision and tissue specificity are important safety concerns when considering CRISPR-Cas9 applications in gene therapy, and prolonged or misexpressed nuclease activity may exacerbate undesirable off-target effects. Effective Cas9 off-switches could ameliorate this difficulty through expression or delivery strategies that would enable anti-CRISPR proteins to accumulate whenever or wherever editing activity is unwanted. Finally, for gene drive applications employing CRISPR-Cas9 to force inheritance of desired alleles (e.g. in insect populations), possession of a functioning off-switch may provide a useful security or containment measure to avert unintended adverse consequences.
We have shown that members of all three anti-CRISPR families studied here bind directly to the NmeCas9/sgRNA complex and inhibit in vitro DNA cleavage. Given the completely unrelated sequences of these anti-CRISPRs, we expect that they may abrogate activity through different mechanisms, as was the case for three type I–F anti-CRISPRs previously characterized in our laboratory (Bondy-Denomy et al., 2015). We have already determined that AcrIIC3Nme prevents stable genomic localization of sgRNA-loaded dNmeCas9 in mammalian cells, indicating that it could be used as an off-switch for dNmeCas9-based applications. It remains possible that other anti-CRISPRs allow NmeCas9 DNA-binding activity but prevent catalytic activation. If so, this would effectively create an NmeCas9 complex with utility for modulation of transcription (Dominguez et al., 2016; Wang et al., 2016). We have identified a type I–F anti-CRISPR possessing this property (Bondy-Denomy et al., 2015).
Apart from their potential for biotechnological applications, the evolutionary implications of anti-CRISPRs are profound. CRISPR-Cas systems are present in approximately half of sequenced prokaryotic genomes and are widespread across diverse bacterial and archaeal lineages. The extreme diversity in and purifying selective pressure on CRISPR-Cas systems, combined with the co-occurrence of several different CRISPR-Cas system types in many genomes, is indicative of a dynamic co-evolutionary battle for survival between prokaryotes and parasitic MGEs (Makarova et al., 2015; Takeuchi et al., 2012). CRISPR-Cas systems are expected to pose a significant challenge to the process of horizontal gene transfer, especially given their ability to acquire heritable immunity against newly encountered threats and to upgrade their arsenal through new spacer acquisition (Barrangou et al., 2007; Fineran et al., 2014; Richter et al., 2014). However, recent studies have shown that the presence of a CRISPR-Cas system does not correlate with lower levels of HGT over evolutionary timescales, or with a lower number of acquired prophage elements (Gophna et al., 2015; Touchon et al., 2016). We propose that widespread MGE-encoded anti-CRISPRs could reconcile this paradox. Also from an evolutionary perspective, we note that Cas9 from type II-A systems is essential not only for the interference function of existing spacers, but also for the adaptive acquisition of new spacers (Heler et al., 2015; Wei et al., 2015). If this adaptation role of type II-A Cas9 extends to type II-C systems, as seems likely, then Cas9-associating anti-CRISPRs may prevent the acquisition of new spacers in response to ongoing invasions.
A recent in vitro evolution study showed that the only way for phages to escape CRISPR-mediated extinction is by the expression of an anti-CRISPR gene (van Houte et al., 2016). In strong accordance with the Red Queen theory, we have discovered a total of seventeen distinct anti-CRISPR protein families that are widespread among Proteobacteria, each inhibiting either type I-E, I-F, or II-C systems (Bondy-Denomy et al., 2013; Pawluk et al., 2014; Pawluk et al., 2016). The fact that anti-CRISPRs have evolved to inhibit both Class 1 and Class 2 CRISPR-Cas systems strongly suggests that they exist for other CRISPR-Cas types as well. We anticipate that anti-CRISPR activity has a large impact on CRISPR-Cas systems across prokaryotes and a profound effect on horizontal gene transfer.
STAR Methods
Contact for Reagent and Resource Sharing
Further information and requests for resources and reagents should be directed to the Lead Contact: Alan R. Davidson (alan.davidson@utoronto.ca).
The Lead Contact holds responsibility for responding to requests and providing reagents and information.
Experimental Model and Subject Details
Neisseria meningitidis strain 8013
Strains were grown on GC Medium Base (GCB) plates with Kellogg’s supplements (22.2 mM glucose, 0.68 mM glutamine, 0.45 mM cocarboxylase, 1.23 mM Fe(NO3)3, all from Sigma), with or without appropriate antibiotics (chloramphenicol, 2.5 μg/mL, erythromycin 2.5 μg/mL, both from Sigma). All solid cultures were incubated at 37 ˚C in a 5% CO2 humidified atmosphere.
Escherichia coli Rosetta (DE3)
E. coli Rosetta (DE3) cells were used for protein expression for in vitro studies. Cells were grown at 37 °C (unless otherwise indicated) in Terrific Broth (TB) medium supplemented with 34 μg/mL chloramphenicol, and, when appropriate, 100 μg/mL ampicillin for plasmid maintenance.
Escherichia coli BL21 (DE3)
This strain was used for recombinant anti-CRISPR protein expression for downstream use in in vitro assays. Cells were grown at 37 °C (unless otherwise indicated) in LB medium supplemented with 100 μg/mL ampicillin for plasmid maintenance.
HEK293T
Cells were cultured in 10 cm culture dish at 37 °C, 5% CO2 in complete DMEM in the presence of 10% FBS and 1% Penicillin/Streptomycin.
U2OS
Cells were cultured in 10 cm culture dish at 37 °C, 5% CO2 in complete DMEM in the presence of 10% FBS and 1% Penicillin/Streptomycin.
Method Details
Bioinformatics analysis
BLASTp searches for Aca2 were conducted with WP_019933869.1 from Oceanimonas smirnovii as the query (Pawluk et al., 2016). BLASTp searches for Aca3 were conducted with WP_049360086.1 from Neisseria meningitidis as the query. For the phylogenetic analysis of Cas9 protein sequences, a list of 257 representative Cas9 protein sequences was extracted from a previous analysis (Fonfara et al., 2014) and updated with newly deposited sequences in the NCBI Protein database. The list was manually trimmed so that only one representative from each species remained. After alignment of the sequences with MUSCLE (Edgar, 2004), FastTree was used to create an unrooted maximum likelihood tree (Price et al., 2009, 2010). Bootstrap values are shown at each node. Based on the data from Fonfara et al. (2014), each clade was classified into subtype II-A (blue), II-B (yellow), or II-C (purple). Clades on the tree are coloured in red if they belong to any genus where a validated type II-C anti-CRISPR gene or its homolog was found. Some noteworthy Cas9 proteins are highlighted on the tree by asterisks.
Plasmid Construction
Acr expression vectors for protein purification
DNA sequences encoding candidate anti-CRISPR proteins were synthesized by GenScript (Piscataway, NJ, USA) and subcloned into pHAT4 (Peranen et al., 1996) using NcoI-HindIII restriction sites. The gene encoding AcrE2 was amplified by PCR from Pseudomonas phage JBD88a and ligated into pHAT4 using NcoI-HindIII restriction sites. Table S1 contains the DNA and protein sequences of the anti-CRISPRs tested in this study. AcrIIC3Nme was found to be significantly more soluble upon addition of an N-terminal FLAG tag, so that construct was used for in vitro analyses.
Cas9:sgRNA vector for protein purification
DNA encoding a minimal T7 promoter upstream of an sgRNA (with a random sequence, i.e. no genomic target in E. coli: 5′-TGAGACCAGTCTCGGAAGCTCAAAGGTCTCGTTGTAGCTCCCTTTCTCATTTCGGAAACGAAATGAGAACCGTTGCTACAATAAGGCCGTCTGAAAAGATGTGCCGCAACGCTCTGCCCCTTAAAGCTTCTGCTTTAAGGGGCATCGTTTATTTCGGTTAAAAAATGCCGT-3′) was synthesized by GenScript (Piscataway, NJ, USA). This insert was cloned into the previously described pMCSG7-NmeCas9 expression vector (Zhang et al., 2015), downstream of the NmeCas9 protein-coding region, into the SalI-XhoI restriction sites.
Cas9/sgRNA mammalian expression vectors
For editing of DTS3 and DTS7 by both SpyCas9 and NmeCas9 (Figure 4 and Supplemental Figures S3 and S4), we used Cas9 expression vectors that were identical in all respects [plasmid backbone, CMV IE94 promoter (Villefranc et al., 2007), UTRs, terminal fusions of NLSs and epitope tags, etc.] except for the respective Cas9 ORFs. The SpyCas9 expression plasmid (pEJS24) has been described previously (Bolukbasi et al., 2015) and the NmeCas9 expression plasmid (pEJS424) was generated from (pEJS24) by Cas9 ORF replacement via Gibson assembly (New England Biolabs). Similarly, plasmids for the expression of sgRNAs for each Cas9 ortholog were also identical in all respects except for the sgRNA sequences themselves. The SpyCas9 sgRNA plasmid pLKO.1-puro has been described previously (Kearns et al., 2014), and the NmeCas9 sgRNA expression plasmid (pEJS333) was generated from it by Gibson assembly. The plasmids expressing NmeCas9 and its sgRNA are described in detail elsewhere (Amrani et al., manuscript in preparation).
For editing of the N-TS1C, N-TS4B, N-TS4C, N-TS7, N-TS8, N-TS11 and N-TS25 sites (Supplemental Figure S3), we used an all-in-one vector (pEJS15) expressing both NmeCas9 (under the control of the EF-1α promoter) and its sgRNA (under the control of the U6 promoter). This plasmid, which was derived from pSimpleII (Hou et al., 2013), is also described elsewhere (Amrani et al., manuscript in preparation). The 24-nt guide sequences for each distinct target site (see Supplemental Figure S3B and Supplemental Table S4 for target site sequences) were inserted into the sgRNA cassette of pEJS15 by the ligation of synthetic oligonucleotide duplexes into its BsmBI sites.
Acr vectors for mammalian expression
To generate the Acr expression plasmids p427-AcrE2, p430-AcrIIC1Boe, p433-AcrIIC1Nme, p436-AcrIIC2Nme, and p443-AcrIIC3Nme, each ORF was synthesized as a gene block (Integrated DNA Technologies) flanked by XhoI and BstBI sites, with a Kozak consensus sequence upstream of the initiation codon. The synthetic Acr sequences (provided in Supplemental Table S1) were then inserted into the XhoI and BstBI sites of the pCS2-Dest vector (Addgene). The resulting plasmids placed the Acr-encoding genes under the control of the CMV IE94 promoter.
Vectors for fluorescence microscopy
pHAGE-TO-DEST dSpyCas9-(mCherry)3 and dNmeCas9-(sfGFP)3 plasmids (Ma et al., 2015b) were purchased from Addgene (#64108 and #64109, respectively) and used directly for no-sgRNA control experiments. We also modified each into an all-in-one version (pEJS466 and pEJS467, respectively) that also included an sgRNA-expressing cassette, with the sgRNAs targeted to telomeric repeats. For the latter, we first used the SpyCas9 sgRNA vector pLKO.1-puro (see above; Kearns et al., 2014) and the NmeCas9 sgRNA vector pEJS333 (see above) to generate the telomere-targeting sgRNAs, via insertion of synthetic oligonucleotide duplexes. We then inserted each U6 promoter/sg-telomere cassette into its cognate dCas9 plasmid via Gibson assembly to generate all-in-one plasmids, pEJS476 [for dNmeCas9-(sfGFP)3] and pEJS477 [dSpyCas9-(mCherry)3]. To make the Acr plasmids, we amplified an mTagBFP2 cassette and incorporated it into pEJS427 (expressing AcrE2) and pEJS443 (expressing AcrIIC3Nme) by Gibson assembly, yielding pEJS481 and pEJS482, respectively. To generate the control plasmid that lacks any Acr (pEJS507), we removed the AcrIIC3Nme cassette from pEJS482 by XhoI digestion followed by plasmid backbone purification and re-ligation.
Neisseria meningitidis natural transformation
Candidate anti-CRISPR genes with the native NmeCas9 promoter and Shine-Dalgarno sequence were cloned into pGCC2, a N. meningitidis vector containing homology arms for integration of the insert into the N. meningitidis chromosome at the nics locus, as described previously (Zhang et al., 2013). The pGCC2 constructs were transformed into N. meningitidis strain 8013, and erythromycin-resistant transformants were selected. Two or three representative transformants per reaction were verified by re-streaking on selective plates twice and then confirmed by PCR on purified genomic DNA. This procedure resulted in N. meningitidis strain 8013 derivatives with chromosomally integrated anti-CRISPR genes under the control of the native promoter of N. meningitidis Cas9. In all cases, we sequence-confirmed the CRISPR locus in the derived strains to ensure that the spacers to be tested for interference activity were intact. Transformation assays to assess CRISPR-Cas activity of these strains were completed as described previously (Duffin and Seifert, 2012; Zhang et al., 2013), with protospacer 25 (complementary to the crRNA derived from endogenous CRISPR spacer #25) as the target. Briefly, 150 ng of plasmids were used per transformation reaction and 10 μL of serial 10-fold dilutions were spotted on GCB plates in triplicate in the presence and absence of appropriate antibiotics. 200 μL from the undiluted final transformation mixture were also plated on GCB plates with appropriate antibiotics to enhance detection. Eight representative transformants per reaction were verified by re-streaking on selective plates twice and then verified by PCRs on cell extracts. Transformation frequencies were reported as antibiotic-resistant cfu/mL from at least three independent experiments (mean ± s.e.m.).
Cloning and purification of anti-CRISPR proteins
Anti-CRISPRs were purified from pHAT4 constructs expressed in E. coli BL21 (DE3) as described previously (Bondy-Denomy et al., 2015). After elution from Ni-NTA resin, anti-CRISPR proteins were dialyzed in 10 mM Tris pH 7.5, 250mM NaCl, and 5mM β-mercaptoethanol and incubated with His-tagged Tobacco Etch Virus (TEV) protease overnight at 4 °C. A second round of Ni-NTA purification was used to isolate successfully cleaved, untagged anti-CRISPRs by collecting the unbound fraction.
Purification of Cas9
6xHis-NmeCas9:sgRNA was expressed in E. coli Rosetta (DE3). Cells were grown in Terrific Broth (TB) medium at 37 °C to an optical density (OD600 nm) of 0.8 in the Lex Bubbling System (Structural Genomics Consortium, Toronto, Canada). Protein expression was induced by the addition of 1 mM IPTG for 16 h at 16 °C. Cells were lysed by sonication in 50 mM Tris pH 7.5, 500 mM NaCl, 20 mM imidazole, 0.5 mM DTT and 5% glycerol supplemented with 0.5 mM PMSF, lysozyme and protease inhibitor cocktail (Sigma). Clarified lysates were bound in batch to Ni-NTA agarose (Qiagen), and bound protein was eluted with 300 mM imidazole. Purified Cas9:sgRNA was dialyzed into 20 mM HEPES pH 7.5, 250 mM NaCl, 5% glycerol, 1 mM DTT and 1 mM PMSF) for protein interaction experiments. 6xHis-MBP-tagged AnaCas9 was purified from E. coli Rosetta (DE3) cells as described previously (Ma et al., 2015a).
Cas9-anti-CRISPR pulldown assays
Untagged anti-CRISPR proteins (after TEV cleavage) were incubated with and without NmeCas9 for 1 hour at 4 °C in binding buffer (20 mM HEPES pH 7.5, 250 mM NaCl, 5% glycerol, 5 mM imidazole), and input fractions were set aside for SDS–PAGE analysis. 50 μL 50% slurry Ni-NTA beads were added to each tube. After 30 minutes incubation at 4 °C with rotation, the beads were collected by centrifugation at 3000 rpm for 2 minutes. Beads were washed four times with 1 mL binding buffer supplemented with 20 mM imidazole and collected by centrifugation. Bound proteins were eluted with elution buffer (binding buffer containing 300 mM imidazole). The input and elution fractions were analyzed by SDS-PAGE followed by Coomassie staining.
In vitro DNA cleavage
NmeCas9 sgRNA derived from spacer 25 (Zhang et al., 2015) was generated by in vitro T7 transcription (Epicentre). NmeCas9 (500 nM) was incubated with purified, recombinant anti-CRISPR protein in cleavage buffer [20 mM HEPES-KOH (pH 7.5), 150 mM KCl, 10% glycerol, 1 mM DTT, and 10 mM MgCl2] for 10 minutes. Next, sgRNA (1:1, 500 nM) was added and the mixture was incubated for another 15 minutes. Plasmid containing the target protospacer 25 (pEJS560) was linearized by ScaI digestion. Linearized plasmid was added to the Cas9/sgRNA complex at ~5 nM final concentration. The reactions were incubated at 37 °C for 30 minutes and visualized after electrophoresis in a 1% agarose/1xTAE gel.
Mammalian genome editing
Plasmids for mammalian expression of NmeCas9, SpyCas9, their respective sgRNAs, and the anti-CRISPR proteins are listed in Supplemental Table S4. Approximately 1.5 × 105 mid-passage HEK293T cells [cultured at 37 °C, 5% CO2 in DMEM (Gibco) + 10% FBS(Sigma) + 1% Penicillin/Streptomycin (Sigma)] were transiently transfected with 150 ng Cas9-expressing plasmid and 150 ng sgRNA-expressing plasmid, using Polyfect transfection reagent (Qiagen) in 24-well plates according to the manufacturer’s instructions. Alternatively, 200 ng of an all-in-one plasmid expressing both NmeCas9 and the appropriate sgRNA (see Supplemental Experimental Procedures) was used for transfection. For experiments that included Acr protein expression, 100 ng of the Acr plasmid was included in the co-transfection mix.
72 hours after transfection, cells were harvested and genomic DNA was extracted with the DNeasy Blood and Tissue kit (Qiagen) according to the manufacturer’s instructions. 50 ng genomic DNA was used for PCR amplification [High Fidelity 2X PCR Master Mix (New England Biolabs)] with primers flanking the targeted site. 10 μl of each PCR product was heat-denatured, re-annealed, and digested with T7 Endonuclease I (New England Biolabs). The samples were fractionated in a 2.5% agarose/1xTAE gel and quantified with the ImageMaster-TotalLab program. Indel percentages (“% lesion” in the figures) were calculated as previously described (Guschin et al., 2010).
Fluorescence microscopy of dNmeCas9
U2OS cells were cultured at 37 °C (5% CO2) in DMEM (Gibco) supplemented with 10% FBS (Sigma) and 1% Pen/Strep (Sigma). For imaging, cells were grown on 170 μm, 35 × 10mm glass-bottom dishes (Eppendorf). Cells were cotransfected with 300 ng of all-in-one plasmids (150 ng of each dNmeCas9 and dSpyCas9 plasmid), an additional 600 ng of sgRNA-expressing plasmids, and 100ng of anti-CRISPR/mTagBFP2 plasmid using PolyFect (Qiagen) according to the manufacturer’s instructions. The additional sgRNA-only plasmid was included because we found the levels of sgRNAs expressed from the all-in-one plasmid alone to be subsaturating, relative to the amount of dCas9 that was expressed from the same plasmid. For the no-sgRNA control experiments, the additional sgRNA-only plasmids were excluded, and the sgRNA cassette was also excluded from the cognate dCas9-expressing plasmid. The total amount of DNA was equal in all transfections (e.g., for the no-sgRNA controls, the sgRNA-expressing plasmids were replaced with the same mass of an irrelevant plasmid). After 24 hours of incubation, live cells were imaged with a Leica DMi8 microscope equipped with a Hamamatsu camera (C11440-22CU), a 63× oil objective lens, and Microsystems software (LASX). Further imaging processing was done with Fiji-ImageJ. For the “blind” experiments (Figure 5G), cells from each condition were coded by one experimenter and then scored by another who did not know which set of cells were from which condition. Only cells that exhibited mTagBFP2 and sfGFP fluorescence as well as dSpyCas9-(mCherry)3 telomeric foci were assessed for the presence or absence of co-localizing dNmeCas9-(sfGFP)3 telomeric foci, and all such imaged cells were included in the quantifications.
Quantification and Statistical Analysis
Neisseria meningitidis transformation efficiency
Cfu/mL were counted manually and are reported as the mean ± s.e.m of at least three biological replicates.
Genome editing efficiency
Efficiency of genome editing in mammalian cells was calculated based on fraction of cleaved DNA as detected by ImageMaster TotalLab V2.0.
The gel images shown for these experiments are representative of at least seven replicates.
Fluorescence imaging
Blind scoring was performed by having one researcher label plates of cells from each condition arbitrarily. A second researcher then collected and scored the images for presence of telomeric foci, and then the labels were decoded to yield the data presented in Figure 5G. These measures were taken to avoid bias. All imaged cells that exhibited mTagBFP2 and sfGFP fluorescence as well as dSpyCas9-(mCherry)3 telomeric foci were included in the quantification. In Figure 5G, n refers to the total number of cells that were scored in each indicated condition.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, Peptides, and Recombinant Proteins | ||
| T7 Endonuclease 1 | New England Biolabs | #M0302L |
| NcoI-HF | New England Biolabs | #R3193S |
| HindIII-HF | New England Biolabs | #R3104S |
| SalI | New England Biolabs | #R0138S |
| XhoI | New England Biolabs | #R0146S |
| BsmBI | New England Biolabs | #R0580S |
| BstBI | New England Biolabs | #R0519S |
| ScaI-HF | New England Biolabs | #R3122S |
| Tobacco Etch Virus (TEV) protease | A. Davidson Lab | |
| Ni-NTA agarose resin | Qiagen | #30210 |
| cOmplete, Mini, EDTA-free Protease Inhibitor Cocktail | Sigma Aldrich | #11836170001 |
| DNeasy Blood and Tissue Kit | Qiagen | #69504 |
| QIAamp DNA Mini Kit | Qiagen | #51304 |
| GC Medium Base | Difco | #DF0289-17-3 |
| DMEM (Medium for mammalian cell culture) | Gibco | #11965092 |
| Fetal Bovine Serum (For mammalian cell culture) | Sigma Aldrich | #F4135 |
| Penicillin-Streptomycin (For mammalian cell culture) | Sigma Aldrich | #P4333 |
| High Fidelity 2X PCR Master Mix | New England Biolabs | #M0541S |
| PolyFect transfection reagent | Qiagen | 3011 |
| AmpliScribe T7-Flash Transcription kit | Epicentre | ASF3507 |
| Gibson Assembly Master mix | New England Biolabs | E2611S |
| Experimental Models: Cell Lines | ||
| Human: HEK293T | ATCC | ATCC CRL-3216 |
| Human: U2OS | ATCC | ATCC HTB96 |
| Experimental Models: Organisms/Strains | ||
| Neisseria meningitidis strain 8013 | E. Sontheimer Lab | N/A |
| Escherichia coli Rosetta (DE3) | Thermo Fisher Scientific | # 709544 |
| Escherichia coli BL21 (DE3) | A. Davidson Lab | N/A |
| Recombinant DNA | ||
| N. meningitidis interference assays | ||
| pGCC2 | Zhang et al 2013 | N/A |
| pGCC2/ Pcas9+AcrIIC1Boe (For strain nics::Pcas9- acrIIC1Boe) | This study | N/A |
| pGCC2/ Pcas9+AcrIIC1Nme (For strain nics::Pcas9-acrIIC1Nme) | This study | N/A |
| pGCC2/ Pcas9+AcrIIC2Nme (For strain nics::Pcas9-acrIIC2Nme) | This study | N/A |
| pGCC2/ Pcas9+AcrIIC3Nme (For strain nics::Pcas9-acrIIC3Nme) | This study | N/A |
| In vitro protein-protein interactions | ||
| pHAT4 | Peranen et al 1996 | N/A |
| pHAT4-AcrE2 | This study | N/A |
| pHAT4-AcrIIC1Boe | This study | N/A |
| pHAT4-AcrIIC1Nme | This study | N/A |
| pHAT4-AcrIIC2Nme | This study | N/A |
| pHAT4-AcrIIC3Nme | This study | N/A |
| pEJS561:sgRNA (see pEJS561 below) | This study; derived from Zhang et al 2015 | N/A |
| In vitro DNA cleavage assays | ||
| pEJS560 (Protospacer 25 in pUC19) | E. Sontheimer Lab | N/A |
| pEJS561 (6XHis-TEV-WtNmeCas9 in pMCSG7) | Zhang et al 2015 | N/A |
| Genome Editing | ||
| pEJS24 (pCSDest2-SpyCas9-NLS-3XHA-NLS) | S. Wolfe Lab | N/A |
| pEJS424 (pCSDest2-NmeCas9-NLS-3XHA-NLS) | E. Sontheimer Lab | N/A |
| pEJS427 (pCSDest2-AcrE2) | This study | N/A |
| pEJS430 (pCSDest2-AcrIIC1Boe) | This study | N/A |
| pEJS433 (pCSDest2-AcrIIC1Nme) | This study | N/A |
| pEJS436 (pCSDest2-AcrIIC2Nme) | This study | N/A |
| pEJS443 (pCSDest2-AcrIIC3Nme) | This study | N/A |
| pEJS333 (pLKO.1-puro U6 Nme-sgRNA BfuAI stuffer) | S. Wolfe Lab | N/A |
| pEJS334 (pLKO.1-puro U6 Spy-sgRNA BfuAI stuffer) | S. Wolfe Lab | N/A |
| pEJS15 (pSimpleII-NmeCas9-sgRNA/Empty) | E. Sontheimer Lab | N/A |
| Fluorescence Imaging | ||
| pEJS333 (pLKO.1-puro U6 Nme-sgRNA BfuAI stuffer) | S. Wolfe Lab | N/A |
| pEJS334 (pLKO.1-puro U6 Spy-sgRNA BfuAI stuffer) | S. Wolfe Lab | N/A |
| pEJS466 (pHAGE-TO-Nme dCas9-3xGFP) | Addgene #64109 | N/A |
| pEJS467 (pHAGE-TO-Spy dCas9-3xmCherry) | Addgene #64108 | N/A |
| pEJS468 (pLK.O1-NmeSgRNA/DTS13-Telomere) | This study | N/A |
| pEJS469 (pLK.O1-SpySgRNA/DTS13-Telomere) | This study | N/A |
| pEJS476 (pHAGE-TO-Nme dCas9 3XGFP-SgRNA/Telomere-All-in-one) | This study | N/A |
| pEJS477 (pHAGE-TO-Spy dCas9 3XmCherry-SgRNA/Telomere-All-in-one) | This study | N/A |
| pEJS478 (pIRES-mTagBFP2) | D. Grünwald Lab | N/A |
| pEJS507 (pCDest2-noAcr-mTagBFP2-IRES) | This study | N/A |
| pEJS481 (pCDest2-AcrE2-mTagBFP2-IRES) | This study | N/A |
| pEJS482 (pCDest2-AcrIIC3Nme-mTagBFP2-IRES) | This study | N/A |
| Sequence-Based Reagents | ||
| sgRNA for coexpression with
NmeCas9 (used to create pEJS561:sgRNA)
5′-TGAGACCAGTCTCGGAAGCTCAAAGGTCTC GTTGTAGCTCCCTTTCTCATTTCGGAAACGAAA TGAGAACCGTTGCTACAATAAGGCCGTCTGAAA AGATGTGCCGCAACGCTCTGCCCCTTAAAGCTT CTGCTTTAAGGGGCATCGTTTATTTCGGTTAAA AAATGCCGT-3′ |
Genscript | N/A |
| See Supplemental Table S3 for sequences of oligonucleotides used in this study | ||
| Software and Algorithms | ||
| FastTree | http://www.genome.jp/tools/fasttree/ | N/A |
| ImageMaster TotalLab V2.0 | N/A | N/A |
Supplementary Material
Supplemental Figure 1: AcrIIC1 Putative Orthologs Are Widely Dispersed in MGEs of Different Species. Related to Figure 1.
(A) Schematic representation of AcrIIC1 putative orthologs identified by PSI-BLAST and their genomic contexts. The species in which each is found and its predicted genomic region classification (i.e. prophage, integrated conjugative element) are indicated. Gene arrows are not drawn to scale. Grey arrows represent genes that have a clear connection to mobile DNA; either by function (i.e. integrase) or by evidence of horizontal transfer as determined by BLAST search. Known, relevant gene functions are indicated by labels: Rep, plasmid replication protein; Reg, transcriptional regulator; Tra, conjugal transfer protein; Par, plasmid partitioning protein; H-NS, histone-like nucleoid-structuring protein; HTH, helix-turn-helix DNA-binding protein; Transp, transposase; Lysis, phage lysis cassette; Nuc, nuclease; Met, methyltransferase; RM, restriction-modification system.
(B) Protein alignment of AcrIIC1 homologs identified by PSI-BLAST searches, using the ClustalX color scheme.
Supplemental Figure 2: Anti-CRISPRs Interact Specifically with NmeCas9. Related to Figure 2.
Uncropped images of the Coomassie-stained SDS-PAGE analysis from Figure 2 are shown.
Supplemental Figure 3: AcrIIC1Boe Blocks NmeCas9-Mediated Genome Editing at Multiple Human Genome Sites. Related to Figure 4.
(A) T7E1 assays of NmeCas9 editing efficiencies at multiple sites, with canonical (N4GATT) or variant (N4GTTT, N4GTCT) PAMs, upon transient transfection of human HEK293T cells. Plasmid encoding AcrIIC1Boe proteins was co-transfected as indicated at the top of each lane. For the D-TS7 target site, SpyCas9 editing (with and without AcrIIC1Boe) was also tested. Editing efficiencies (“% lesion”) are given at the bottom of each lane.
(B) For the D-TS3 site tested in Figure 4, and for each site tested in (A), the NmeCas9 sgRNA spacer sequences (5’ to 3’) and DNA target sites (non-complementary strand, 5’ to 3’) are listed.
Supplemental Figure 4: Plasmid Titration of Anti-CRISPRs in Human Genome Editing. Related to Figure 4.
(A) T7E1 assays of NmeCas9 editing efficiencies at DTS3 upon transient transfection of human HEK293T cells. Constructs encoding anti-CRISPRs were co-transfected as indicated at the top of each lane. The total amount of anti-CRISPR proteins was held constant at 100 ng per well, but the relative amount of a negative control anti-CRISPR (AcrE2) and test anti-CRISPR (AcrIIC1Boe) was varied. Editing efficiencies (“% lesion”) are given at the bottom of each lane. (B-E) As in (A), except that AcrIIC1Nme (B), AcrIIC2Nme (C), and AcrIIC3Nme (D and E) were used. In (D), because inhibition was nearly complete even at the lowest dose (10 ng) of AcrIIC3Nme plasmid, we repeated the titration with lower levels of plasmid in (E), revealing the dose-dependence of inhibition.
Supplemental Table 1: Anti-CRISPR DNA and Protein Sequences Used in this Study. Related to Figure 1.
Supplemental Table 2: Information About Anti-CRISPR Homologs. Related to Figure 1, Figure S1.
For each anti-CRISPR family discovered in this study, a list of homologs identified by PSI-BLAST are listed with their accession numbers. The table indicates the species from which each homolog originated, the predicted genome region in which it is found, and its length in amino acids. The diversity of each family is described by the “% ID to *” column, which lists the pairwise percent identity between each homolog and the first listed member of the family.
Supplemental Table 3: Templates for Transcription of sgRNAs that Target Several Human Genome Sites. Related to Figure 4, Supplemental Figure 3, and STAR Methods.
Acknowledgments
The authors would like to thank Sabrina Stanley for technical assistance. The AnaCas9 expression construct was a generous gift from Lucas Harrington, Enbo Ma, and Jennifer Doudna. We are grateful to David Grünwald and members of his lab for help and advice with fluorescence imaging, and to Andrew Franck for help and advice with NmeCas9 purification. We thank Scot Wolfe and Wen Xue for helpful discussions and comments on the manuscript. This work was supported by a Canadian Institutes of Health Research Doctoral Award to A.P., NIH grant K99 GM117268 to Y.Z., NIH grant R01 GM115911 to E.J.S., and Canadian Institutes of Health Research grants to A.R.D (MOP-130482) and K.L.M (MOP-136845). E.J.S. is a co-founder and scientific advisor of Intellia Therapeutics.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
A.P. conducted bioinformatics analysis, designed experiments, purified proteins, performed Cas9-anti-CRISPR interaction experiments, and wrote the manuscript. N.A. designed and performed human genome editing experiments. Y.Z. designed and performed N. meningitidis transformation experiments. B.G. and Y.H-R. purified proteins and performed Cas9-anti-CRISPR interaction experiments. J.L. performed fluorescence live-cell imaging experiments. A.E. purified NmeCas9 and conducted in vitro DNA cleavage experiments. M.S. assisted in anti-CRISPR and NmeCas9 protein purification. E.J.S. supervised experimental work and wrote the manuscript. K.L.M. supervised experimental work and wrote the manuscript. A.R.D. supervised experimental work and wrote the manuscript.
References
- Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- Bikard D, Euler CW, Jiang W, Nussenzweig PM, Goldberg GW, Duportet X, Fischetti VA, Marraffini LA. Exploiting CRISPR-Cas nucleases to produce sequence-specific antimicrobials. Nat Biotechnol. 2014;32:1146–1150. doi: 10.1038/nbt.3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolukbasi MF, Gupta A, Wolfe SA. Creating and evaluating accurate CRISPR-Cas9 scalpels for genomic surgery. Nat Methods. 2015;13:41–50. doi: 10.1038/nmeth.3684. [DOI] [PubMed] [Google Scholar]
- Bondy-Denomy J, Garcia B, Strum S, Du M, Rollins MF, Hidalgo-Reyes Y, Wiedenheft B, Maxwell KL, Davidson AR. Multiple mechanisms for CRISPR-Cas inhibition by anti-CRISPR proteins. Nature. 2015 doi: 10.1038/nature15254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bondy-Denomy J, Pawluk A, Maxwell KL, Davidson AR. Bacteriophage genes that inactivate the CRISPR/Cas bacterial immune system. Nature. 2013;493:429–432. doi: 10.1038/nature11723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho SW, Kim S, Kim JM, Kim JS. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat Biotechnol. 2013;31:230–232. doi: 10.1038/nbt.2507. [DOI] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deltcheva E, Chylinski K, Sharma CM, Gonzales K, Chao Y, Pirzada ZA, Eckert MR, Vogel J, Charpentier E. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature. 2011;471:602–607. doi: 10.1038/nature09886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez AA, Lim WA, Qi LS. Beyond editing: repurposing CRISPR-Cas9 for precision genome regulation and interrogation. Nat Rev Mol Cell Biol. 2016;17:5–15. doi: 10.1038/nrm.2015.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffin PM, Seifert HS. Genetic transformation of Neisseria gonorrhoeae shows a strand preference. FEMS Microbiol Lett. 2012;334:44–48. doi: 10.1111/j.1574-6968.2012.02612.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebina H, Misawa N, Kanemura Y, Koyanagi Y. Harnessing the CRISPR/Cas9 system to disrupt latent HIV-1 provirus. Sci Rep. 2013;3:2510. doi: 10.1038/srep02510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esvelt KM, Mali P, Braff JL, Moosburner M, Yaung SJ, Church GM. Orthogonal Cas9 proteins for RNA-guided gene regulation and editing. Nat Methods. 2013;10:1116–1121. doi: 10.1038/nmeth.2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fineran PC, Gerritzen MJ, Suarez-Diez M, Kunne T, Boekhorst J, van Hijum SA, Staals RH, Brouns SJ. Degenerate target sites mediate rapid primed CRISPR adaptation. Proc Natl Acad Sci U S A. 2014;111:E1629–1638. doi: 10.1073/pnas.1400071111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonfara I, Le Rhun A, Chylinski K, Makarova KS, Lecrivain AL, Bzdrenga J, Koonin EV, Charpentier E. Phylogeny of Cas9 determines functional exchangeability of dual-RNA and Cas9 among orthologous type II CRISPR-Cas systems. Nucleic Acids Res. 2014;42:2577–2590. doi: 10.1093/nar/gkt1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Sander JD, Reyon D, Cascio VM, Joung JK. Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat Biotechnol. 2014;32:279–284. doi: 10.1038/nbt.2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gantz VM, Jasinskiene N, Tatarenkova O, Fazekas A, Macias VM, Bier E, James AA. Highly efficient Cas9-mediated gene drive for population modification of the malaria vector mosquito Anopheles stephensi. Proc Natl Acad Sci U S A. 2015;112:E6736–6743. doi: 10.1073/pnas.1521077112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasiunas G, Barrangou R, Horvath P, Siksnys V. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc Natl Acad Sci U S A. 2012;109:E2579–2586. doi: 10.1073/pnas.1208507109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomaa AA, Klumpe HE, Luo ML, Selle K, Barrangou R, Beisel CL. Programmable removal of bacterial strains by use of genome-targeting CRISPR-Cas systems. MBio. 2014;5:e00928–00913. doi: 10.1128/mBio.00928-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gophna U, Kristensen DM, Wolf YI, Popa O, Drevet C, Koonin EV. No evidence of inhibition of horizontal gene transfer by CRISPR-Cas on evolutionary timescales. ISME J. 2015;9:2021–2027. doi: 10.1038/ismej.2015.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guschin DY, Waite AJ, Katibah GE, Miller JC, Holmes MC, Rebar EJ. A rapid and general assay for monitoring endogenous gene modification. Methods Mol Biol. 2010;649:247–256. doi: 10.1007/978-1-60761-753-2_15. [DOI] [PubMed] [Google Scholar]
- Hammond A, Galizi R, Kyrou K, Simoni A, Siniscalchi C, Katsanos D, Gribble M, Baker D, Marois E, Russell S, et al. A CRISPR-Cas9 gene drive system targeting female reproduction in the malaria mosquito vector Anopheles gambiae. Nat Biotechnol. 2016;34:78–83. doi: 10.1038/nbt.3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heler R, Samai P, Modell JW, Weiner C, Goldberg GW, Bikard D, Marraffini LA. Cas9 specifies functional viral targets during CRISPR-Cas adaptation. Nature. 2015;519:199–202. doi: 10.1038/nature14245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilton IB, D’Ippolito AM, Vockley CM, Thakore PI, Crawford GE, Reddy TE, Gersbach CA. Epigenome editing by a CRISPR-Cas9-based acetyltransferase activates genes from promoters and enhancers. Nat Biotechnol. 2015;33:510–517. doi: 10.1038/nbt.3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano H, Gootenberg JS, Horii T, Abudayyeh OO, Kimura M, Hsu PD, Nakane T, Ishitani R, Hatada I, Zhang F, et al. Structure and Engineering of Francisella novicida Cas9. Cell. 2016;164:950–961. doi: 10.1016/j.cell.2016.01.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Z, Zhang Y, Propson NE, Howden SE, Chu LF, Sontheimer EJ, Thomson JA. Efficient genome engineering in human pluripotent stem cells using Cas9 from Neisseria meningitidis. Proc Natl Acad Sci U S A. 2013;110:15644–15649. doi: 10.1073/pnas.1313587110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu PD, Scott DA, Weinstein JA, Ran FA, Konermann S, Agarwala V, Li Y, Fine EJ, Wu X, Shalem O, et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol. 2013;31:827–832. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang WY, Fu Y, Reyon D, Maeder ML, Kaini P, Sander JD, Joung JK, Peterson RT, Yeh JR. Heritable and precise zebrafish genome editing using a CRISPR-Cas system. PLoS One. 2013;8:e68708. doi: 10.1371/journal.pone.0068708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W, Bikard D, Cox D, Zhang F, Marraffini LA. RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat Biotechnol. 2013;31:233–239. doi: 10.1038/nbt.2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, East A, Cheng A, Lin S, Ma E, Doudna J. RNA-programmed genome editing in human cells. Elife. 2013;2:e00471. doi: 10.7554/eLife.00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, Jiang F, Taylor DW, Sternberg SH, Kaya E, Ma E, Anders C, Hauer M, Zhou K, Lin S, et al. Structures of Cas9 endonucleases reveal RNA-mediated conformational activation. Science. 2014;343:1247997. doi: 10.1126/science.1247997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaminski R, Chen Y, Fischer T, Tedaldi E, Napoli A, Zhang Y, Karn J, Hu W, Khalili K. Elimination of HIV-1 Genomes from Human T-lymphoid Cells by CRISPR/Cas9 Gene Editing. Sci Rep. 2016;6:22555. doi: 10.1038/srep22555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearns NA, Genga RM, Enuameh MS, Garber M, Wolfe SA, Maehr R. Cas9 effector-mediated regulation of transcription and differentiation in human pluripotent stem cells. Development. 2014;141:219–223. doi: 10.1242/dev.103341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearns NA, Pham H, Tabak B, Genga RM, Silverstein NJ, Garber M, Maehr R. Functional annotation of native enhancers with a Cas9-histone demethylase fusion. Nat Methods. 2015;12:401–403. doi: 10.1038/nmeth.3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CM, Cradick TJ, Bao G. The Neisseria meningitidis CRISPR-Cas9 System Enables Specific Genome Editing in Mammalian Cells. Mol Ther. 2016;24:645–654. doi: 10.1038/mt.2016.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma E, Harrington LB, O’Connell MR, Zhou K, Doudna JA. Single-Stranded DNA Cleavage by Divergent CRISPR-Cas9 Enzymes. Mol Cell. 2015a;60:398–407. doi: 10.1016/j.molcel.2015.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H, Naseri A, Reyes-Gutierrez P, Wolfe SA, Zhang S, Pederson T. Multicolor CRISPR labeling of chromosomal loci in human cells. Proc Natl Acad Sci U S A. 2015b;112:3002–3007. doi: 10.1073/pnas.1420024112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Wolf YI, Alkhnbashi OS, Costa F, Shah SA, Saunders SJ, Barrangou R, Brouns SJ, Charpentier E, Haft DH, et al. An updated evolutionary classification of CRISPR-Cas systems. Nat Rev Microbiol. 2015;13:722–736. doi: 10.1038/nrmicro3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller M, Lee CM, Gasiunas G, Davis TH, Cradick TJ, Siksnys V, Bao G, Cathomen T, Mussolino C. Streptococcus thermophilus CRISPR-Cas9 Systems Enable Specific Editing of the Human Genome. Mol Ther. 2016;24:636–644. doi: 10.1038/mt.2015.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nihongaki Y, Kawano F, Nakajima T, Sato M. Photoactivatable CRISPR-Cas9 for optogenetic genome editing. Nat Biotechnol. 2015;33:755–760. doi: 10.1038/nbt.3245. [DOI] [PubMed] [Google Scholar]
- Nunez JK, Harrington LB, Doudna JA. Chemical and Biophysical Modulation of Cas9 for Tunable Genome Engineering. ACS Chem Biol. 2016;11:681–688. doi: 10.1021/acschembio.5b01019. [DOI] [PubMed] [Google Scholar]
- Orthwein A, Noordermeer SM, Wilson MD, Landry S, Enchev RI, Sherker A, Munro M, Pinder J, Salsman J, Dellaire G, et al. A mechanism for the suppression of homologous recombination in G1 cells. Nature. 2015;528:422–426. doi: 10.1038/nature16142. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Ousterout DG, Kabadi AM, Thakore PI, Majoros WH, Reddy TE, Gersbach CA. Multiplex CRISPR/Cas9-based genome editing for correction of dystrophin mutations that cause Duchenne muscular dystrophy. Nat Commun. 2015;6:6244. doi: 10.1038/ncomms7244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattanayak V, Lin S, Guilinger JP, Ma E, Doudna JA, Liu DR. High-throughput profiling of off-target DNA cleavage reveals RNA-programmed Cas9 nuclease specificity. Nat Biotechnol. 2013;31:839–843. doi: 10.1038/nbt.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawluk A, Bondy-Denomy J, Cheung VH, Maxwell KL, Davidson AR. A new group of phage anti-CRISPR genes inhibits the type I–E CRISPR-Cas system of Pseudomonas aeruginosa. MBio. 2014;5:e00896. doi: 10.1128/mBio.00896-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawluk A, Staals RHJ, Taylor C, Watson BNJ, Saha S, Fineran PC, Maxwell KL, Davidson AR. Inactivation of CRISPR-Cas systems by anti-CRISPR proteins in diverse bacterial species. Nature Microbiology. 2016 doi: 10.1038/nmicrobiol.2016.85. [DOI] [PubMed] [Google Scholar]
- Peranen J, Rikkonen M, Hyvonen M, Kaariainen L. T7 vectors with modified T7lac promoter for expression of proteins in Escherichia coli. Anal Biochem. 1996;236:371–373. doi: 10.1006/abio.1996.0187. [DOI] [PubMed] [Google Scholar]
- Price MN, Dehal PS, Arkin AP. FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol Biol Evol. 2009;26:1641–1650. doi: 10.1093/molbev/msp077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price MN, Dehal PS, Arkin AP. FastTree 2–approximately maximum-likelihood trees for large alignments. PLoS One. 2010;5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran FA, Cong L, Yan WX, Scott DA, Gootenberg JS, Kriz AJ, Zetsche B, Shalem O, Wu X, Makarova KS, et al. In vivo genome editing using Staphylococcus aureus Cas9. Nature. 2015;520:186–191. doi: 10.1038/nature14299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter C, Dy RL, McKenzie RE, Watson BN, Taylor C, Chang JT, McNeil MB, Staals RH, Fineran PC. Priming in the Type I–F CRISPR-Cas system triggers strand-independent spacer acquisition, bi-directionally from the primed protospacer. Nucleic Acids Res. 2014;42:8516–8526. doi: 10.1093/nar/gku527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi N, Wolf YI, Makarova KS, Koonin EV. Nature and intensity of selection pressure on CRISPR-associated genes. J Bacteriol. 2012;194:1216–1225. doi: 10.1128/JB.06521-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touchon M, Bernheim A, Rocha EP. Genetic and life-history traits associated with the distribution of prophages in bacteria. ISME J. 2016 doi: 10.1038/ismej.2016.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Houte S, Ekroth AK, Broniewski JM, Chabas H, Ben A, Bondy-Denomy J, Gandon S, Boots M, Paterson S, Buckling A, et al. The diversity-generating benefits of a prokaryotic adaptive immune system. Nature. 2016 doi: 10.1038/nature17436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villefranc JA, Amigo J, Lawson ND. Gateway compatible vectors for analysis of gene function in the zebrafish. Dev Dyn. 2007;236:3077–3087. doi: 10.1002/dvdy.21354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, La Russa M, Qi LS. CRISPR/Cas9 in Genome Editing and Beyond. Annu Rev Biochem. 2016;85:227–264. doi: 10.1146/annurev-biochem-060815-014607. [DOI] [PubMed] [Google Scholar]
- Wang H, Yang H, Shivalila CS, Dawlaty MM, Cheng AW, Zhang F, Jaenisch R. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell. 2013;153:910–918. doi: 10.1016/j.cell.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y, Terns RM, Terns MP. Cas9 function and host genome sampling in Type II-A CRISPR-Cas adaptation. Genes Dev. 2015;29:356–361. doi: 10.1101/gad.257550.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright AV, Sternberg SH, Taylor DW, Staahl BT, Bardales JA, Kornfeld JE, Doudna JA. Rational design of a split-Cas9 enzyme complex. Proc Natl Acad Sci U S A. 2015;112:2984–2989. doi: 10.1073/pnas.1501698112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Liang D, Wang Y, Bai M, Tang W, Bao S, Yan Z, Li D, Li J. Correction of a genetic disease in mouse via use of CRISPR-Cas9. Cell Stem Cell. 2013;13:659–662. doi: 10.1016/j.stem.2013.10.016. [DOI] [PubMed] [Google Scholar]
- Yen ST, Zhang M, Deng JM, Usman SJ, Smith CN, Parker-Thornburg J, Swinton PG, Martin JF, Behringer RR. Somatic mosaicism and allele complexity induced by CRISPR/Cas9 RNA injections in mouse zygotes. Dev Biol. 2014;393:3–9. doi: 10.1016/j.ydbio.2014.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin H, Xue W, Chen S, Bogorad RL, Benedetti E, Grompe M, Koteliansky V, Sharp PA, Jacks T, Anderson DG. Genome editing with Cas9 in adult mice corrects a disease mutation and phenotype. Nat Biotechnol. 2014;32:551–553. doi: 10.1038/nbt.2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Heidrich N, Ampattu BJ, Gunderson CW, Seifert HS, Schoen C, Vogel J, Sontheimer EJ. Processing-independent CRISPR RNAs limit natural transformation in Neisseria meningitidis. Mol Cell. 2013;50:488–503. doi: 10.1016/j.molcel.2013.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Rajan R, Seifert HS, Mondragon A, Sontheimer EJ. DNase H Activity of Neisseria meningitidis Cas9. Mol Cell. 2015;60:242–255. doi: 10.1016/j.molcel.2015.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1: AcrIIC1 Putative Orthologs Are Widely Dispersed in MGEs of Different Species. Related to Figure 1.
(A) Schematic representation of AcrIIC1 putative orthologs identified by PSI-BLAST and their genomic contexts. The species in which each is found and its predicted genomic region classification (i.e. prophage, integrated conjugative element) are indicated. Gene arrows are not drawn to scale. Grey arrows represent genes that have a clear connection to mobile DNA; either by function (i.e. integrase) or by evidence of horizontal transfer as determined by BLAST search. Known, relevant gene functions are indicated by labels: Rep, plasmid replication protein; Reg, transcriptional regulator; Tra, conjugal transfer protein; Par, plasmid partitioning protein; H-NS, histone-like nucleoid-structuring protein; HTH, helix-turn-helix DNA-binding protein; Transp, transposase; Lysis, phage lysis cassette; Nuc, nuclease; Met, methyltransferase; RM, restriction-modification system.
(B) Protein alignment of AcrIIC1 homologs identified by PSI-BLAST searches, using the ClustalX color scheme.
Supplemental Figure 2: Anti-CRISPRs Interact Specifically with NmeCas9. Related to Figure 2.
Uncropped images of the Coomassie-stained SDS-PAGE analysis from Figure 2 are shown.
Supplemental Figure 3: AcrIIC1Boe Blocks NmeCas9-Mediated Genome Editing at Multiple Human Genome Sites. Related to Figure 4.
(A) T7E1 assays of NmeCas9 editing efficiencies at multiple sites, with canonical (N4GATT) or variant (N4GTTT, N4GTCT) PAMs, upon transient transfection of human HEK293T cells. Plasmid encoding AcrIIC1Boe proteins was co-transfected as indicated at the top of each lane. For the D-TS7 target site, SpyCas9 editing (with and without AcrIIC1Boe) was also tested. Editing efficiencies (“% lesion”) are given at the bottom of each lane.
(B) For the D-TS3 site tested in Figure 4, and for each site tested in (A), the NmeCas9 sgRNA spacer sequences (5’ to 3’) and DNA target sites (non-complementary strand, 5’ to 3’) are listed.
Supplemental Figure 4: Plasmid Titration of Anti-CRISPRs in Human Genome Editing. Related to Figure 4.
(A) T7E1 assays of NmeCas9 editing efficiencies at DTS3 upon transient transfection of human HEK293T cells. Constructs encoding anti-CRISPRs were co-transfected as indicated at the top of each lane. The total amount of anti-CRISPR proteins was held constant at 100 ng per well, but the relative amount of a negative control anti-CRISPR (AcrE2) and test anti-CRISPR (AcrIIC1Boe) was varied. Editing efficiencies (“% lesion”) are given at the bottom of each lane. (B-E) As in (A), except that AcrIIC1Nme (B), AcrIIC2Nme (C), and AcrIIC3Nme (D and E) were used. In (D), because inhibition was nearly complete even at the lowest dose (10 ng) of AcrIIC3Nme plasmid, we repeated the titration with lower levels of plasmid in (E), revealing the dose-dependence of inhibition.
Supplemental Table 1: Anti-CRISPR DNA and Protein Sequences Used in this Study. Related to Figure 1.
Supplemental Table 2: Information About Anti-CRISPR Homologs. Related to Figure 1, Figure S1.
For each anti-CRISPR family discovered in this study, a list of homologs identified by PSI-BLAST are listed with their accession numbers. The table indicates the species from which each homolog originated, the predicted genome region in which it is found, and its length in amino acids. The diversity of each family is described by the “% ID to *” column, which lists the pairwise percent identity between each homolog and the first listed member of the family.
Supplemental Table 3: Templates for Transcription of sgRNAs that Target Several Human Genome Sites. Related to Figure 4, Supplemental Figure 3, and STAR Methods.


