Abstract
Approximately one-half of advanced (unresectable or metastatic) melanomas harbor a mutation in the BRAF gene, with V600E being the most common mutation. Targeted therapy with BRAF and MEK inhibitors is associated with significant long-term treatment benefit in patients with BRAF V600-mutated melanoma. Therefore, molecular testing for BRAF mutations is a priority in determining the course of therapy. A literature search was performed using MEDLINE/PubMed and scientific congress databases using the terms ‘BRAF,’ ‘mutation,’ and ‘cancer/tumor.’ These results were filtered to include manuscripts that focused on diagnostic tests for determining BRAF mutation status. Numerous BRAF testing methods were identified, including DNA-based companion diagnostic tests and DNA- and protein-based laboratory-developed tests. Herein we review the characteristics of each method and highlight the strengths and weaknesses that should be considered before use and when interpreting results for each patient. Molecular profiling has shown that mutation load increases with melanoma tumor progression and that unique patterns of genetic changes and evolutionary trajectories for different melanoma subtypes can occur. Discordance in the BRAF mutational status between primary and metastatic lesions, as well as intratumoral heterogeneity, is known to occur. Additionally, the development of acquired resistance to combination BRAF and MEK inhibitor therapy is still a formidable obstacle. Therefore, tumor heterogeneity and the development of acquired resistance have important implications for molecular testing and ultimately the treatment of patients with advanced-stage melanoma. Overall, this information may help community oncologists more accurately and effectively interpret results of diagnostic tests within the context of recent data characterizing melanoma tumor progression.
Melanoma represents a significant and increasing public health burden and an ongoing area of unmet need in oncology. Although melanoma accounts for only 1% of diagnosed skin cancers, it is the cause of most skin cancer-related deaths.1 In contrast to the incidence of other common cancer types, the incidence of melanoma is increasing.2 In the US, 87 110 new cases of melanoma and 9730 melanoma-related deaths are estimated in 2017.2 Until recently, patients with advanced melanoma had few effective treatment options; historically, response rates to conventional chemotherapy and immunomodulation therapy (interleukin 2 or interferon gamma) have been ~5–19%.3, 4, 5, 6 New therapeutic options include treatments targeted specifically to genetic mutations in patients’ tumors as well as immune checkpoint inhibitors.7, 8, 9, 10, 11, 12
Many different methods for BRAF testing are currently being used in the United States including US Food and Drug Administration (FDA)-approved companion diagnostic tests and various laboratory-developed tests.13, 14, 15 Information on FDA-approved tests for the detection of BRAF V600 mutations in melanoma is available at http://www.fda.gov/companiondiagnostics.16 Each BRAF testing method has its own unique strengths, weaknesses, and challenges that should be considered before using it to test a patient’s tumor sample.13 This review will discuss some of the currently available tests for determining BRAF mutation status in patients with advanced melanoma, focusing on considerations and approaches for accurate and effective testing in the community oncology setting.
BRAF-mutant metastatic melanoma: an overview
A recent whole-genome sequencing study analyzing mutation frequencies across several cancers identified melanoma as the most frequently mutated tumor type;17 significant variation in the mutation profiles for different types of melanoma (eg, cutaneous, acral) has also been demonstrated by whole-genome sequencing.18 The majority of melanomas have mutations associated with the mitogen-activated protein kinase (MAPK) pathway, an important signal transduction pathway involved in cell growth, proliferation, and survival.19 Oncogenic activation of the MAPK pathway can occur via multiple mechanisms, the most common of which in melanoma is constitutive activation of the BRAF kinase via mutation, which occurs in ~40–60% of cases. The second most common MAPK pathway aberration in melanoma is mutated NRAS, occurring in ~15–30% of cases.20, 21, 22, 23, 24, 25
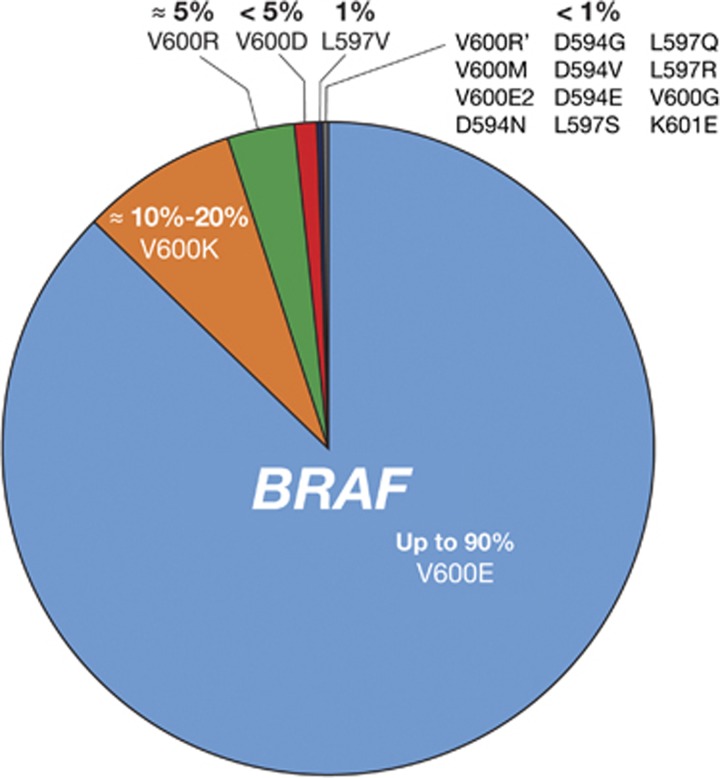
BRAF encodes a cytoplasmic serine–threonine kinase. More than 97% of BRAF mutations are located in codon 600 of the BRAF gene.13 The most common mutation (in up to 90% of cases) is the result of a transversion of T to A at nucleotide 1799 (T1799A), which results in a substitution of valine (V) for glutamic acid (E) at position 600.26 Less common are substitutions of V for lysine (V600K (~8–20%)), arginine (V600R (1%)), leucine (V600M (0.3%)), and aspartic acid (V600D (0.1%)).13, 24 Non-V600 mutations (eg, K601E, D594N) are also known to occur in a low percentage of patients.13, 27 Types and frequencies of BRAF mutations are summarized in Figure 1 and Table 1.26, 27
Figure 1.
BRAF mutation types in melanoma. Estimated incidence of BRAF mutation frequencies in patients with melanoma is shown.26
Table 1. Incidence of common BRAF mutations based on the COSMIC database 26, 27 .
| BRAF mutation | Mutation at codon 600 of BRAF gene | Incidence in BRAF-mutant melanoma, % |
|---|---|---|
| Common BRAF mutations | ||
| V600E | Valine → glutamic acid | 84.6 |
| V600K | Valine → lysine | 7.7 |
| Other BRAF mutations | ||
| V600R | Valine → arginine | 1 |
| V600M | Valine → leucine | 0.3 |
| V600D | Valine → aspartic acid | 0.1 |
| Non-V600 mutations (eg, K601E, D594N) | — | <1 |
BRAF-mutated melanoma tends to exhibit distinctive clinical features and is characterized by more aggressive biological behavior than BRAF wild-type (WT) melanoma. Compared with patients with BRAF WT melanoma, those with BRAF-mutated melanoma are more often younger and have tumors with superficial spreading or nodular histology and/or in anatomical regions without chronic sun damage.24 Furthermore, BRAF-mutant tumors are more likely to metastasize to the brain than BRAF WT tumors.25 BRAF-mutated melanoma has also been linked to shorter overall survival in patients with stage IV cancer than in those with BRAF WT disease.24, 25, 28 Given these features and the available effective therapies, it is crucial to quickly determine whether patients with melanoma have tumors harboring the BRAF mutation in order to select the optimal treatment.
The discovery that many melanomas harbor BRAF mutations29 led to the development of vemurafenib and dabrafenib, selective inhibitors of the BRAF V600-mutated kinase, and trametinib and cobimetinib, inhibitors of the downstream MEK kinase. Combination therapy with BRAF and MEK inhibitors (eg, dabrafenib plus trametinib, vemurafenib plus cobimetinib) has resulted in dramatic improvements in overall survival and progression-free survival rates in patients with BRAF V600-mutant advanced melanoma.30, 31, 32, 33, 34, 35 Three-year landmark analysis results have been presented from two randomized, phase 3 studies evaluating dabrafenib plus trametinib vs dabrafenib plus placebo (COMBI-d; NCT01584648) or single-agent vemurafenib (COMBI-v; NCT01597908) in patients with BRAF V600-mutant advanced melanoma.36, 37 The 3-year overall survival and progression-free survival rates were 44 and 22%, respectively, with dabrafenib plus trametinib vs 32% and 12%, respectively, with dabrafenib plus placebo in COMBI-d. In COMBI-v, overall survival and progression-free survival rates, respectively, at 3 years were 45 and 24% with dabrafenib plus trametinib vs 31 and 10% with vemurafenib. The safety profile of long-term combination therapy was similar to that in prior reports, with no additional safety signals observed. Three-year landmark data are also available from the phase 3 coBRIM study (NCT01689519), which evaluated cobimetinib plus vemurafenib vs placebo plus vemurafenib in patients with advanced BRAF V600-mutant melanoma; reported 3-year overall survival rates were 37.4 and 31.5%, respectively.38
Currently, BRAF mutation status is the only biomarker that predicts a therapeutic response in advanced melanoma.39, 40 Molecular testing for BRAF mutations in patients with advanced melanoma has become a standard for determining the course of therapy, and testing is recommended by the current National Comprehensive Cancer Network (NCCN) and European Society for Medical Oncology guidelines for melanoma.40, 41, 42 However, it is important to note that the NCCN Clinical Practice Guidelines in Oncology (2017) do not recommend testing the primary cutaneous melanoma for the BRAF mutation unless required to guide systemic therapy.40 BRAF inhibitors are approved only for use in patients with a BRAF V600 mutation detected by an FDA-approved test.43, 44 Patients with BRAF WT tumors may experience tumor promotion if treated with a BRAF inhibitor due to the paradoxical activity of the MAPK pathway in WT cells.43, 44 Per NCCN guidelines, first-line systemic treatment options for patients with metastatic or unresectable melanoma include immunotherapy (ie, anti-programmed death 1 (PD-1) receptor monotherapy (pembrolizumab or nivolumab) or nivolumab combined with the cytotoxic T-lymphocyte associated antigen 4 (CTLA4)-targeted antibody ipilimumab), targeted therapy (combination therapy with dabrafenib and trametinib or vemurafenib and cobimetinib) for patients with a BRAF V600 mutation, and enrollment in a clinical trial.40 In patients with a BRAF mutation, targeted therapy is preferred when an early response is clinically needed; however, the optimal sequence of BRAF inhibitors and immunotherapy is still under investigation. Because combination therapy with BRAF and MEK inhibitors has been shown to have the potential for significant long-term treatment benefit, identifying a patient’s BRAF mutation status should be a priority for the clinician.
Evolution from benign to malignant melanoma: primary and secondary mutations
Although the BRAF V600 mutation is an oncogenic driver mutation involved in cellular proliferation, it is considered insufficient to induce melanoma in the absence of other cytogenetic abnormalities. It has been known for some time that BRAF V600 mutations are frequently found in benign and dysplastic melanocytic nevi, indicating that additional cellular changes are needed for transformation.45 These changes—including loss of tumor suppressors, activation of the TERT promoter, inactivation of genes involved in DNA repair, or activation of other protein kinases and signaling cascades46—in essence release the ‘brakes,’ allowing cellular proliferation to occur.46, 47 Common genes in which loss-of-function mutations can occur are CDKN2A, PTEN, and BAP1. Mutation of CDKN2A, for example, removes a key element of the p53 tumor suppressor pathway.48 Dysfunction of this pathway eliminates a mechanism that would normally consign precancerous cells with preexisting germline mutations or local, somatic mutations to apoptosis or senescence. Thus, cells that have a driver mutation combined with the loss of a tumor suppressor are primed to undergo transformation and develop into melanoma.
Determining genetic heterogeneity of melanoma
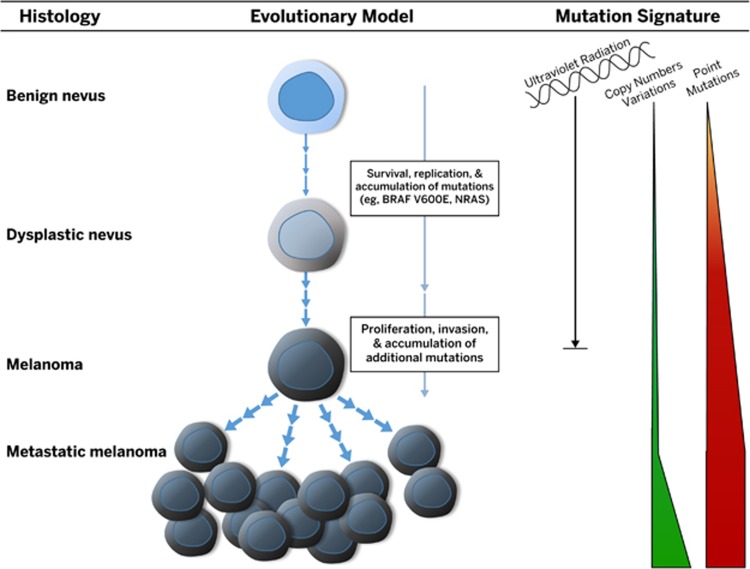
Data have shown that the molecular profile of a tumor changes over time due to various selection pressures.46, 47 Tumor heterogeneity in advanced-stage melanoma has important implications for molecular testing and thus treatment. Shain et al. examined 37 primary melanocytic neoplasms and their adjacent precursor lesions, ultimately microdissecting and sequencing 150 distinct areas46 (Figure 2). Interestingly, tumors that lacked malignant behavior harbored only BRAF V600E mutations. The mutation load increased with progression, and unique patterns of genetic changes, termed ‘evolutionary trajectories,’ for different melanoma subtypes were observed. Intermediate lesions were enriched for NRAS mutations and additional driver mutations. Progression was marked by the appearance of TERT promoter mutations and an increasing burden of NRAS mutations in intermediate lesions. This was followed by inactivation of CDKN2A or mutations in SWI/SNF chromatin remodeling genes in invasive melanomas and loss of PTEN and TP53 in advanced melanoma. In rare cases, lesions had both BRAF V600E and NRAS mutations identified. Although it is thought that BRAF and NRAS mutations are mutually exclusive, this study by Shain et al. and another recent study by Chiappetta et al. showed that these mutations can both be present, most likely due to clonal heterogeneity within the tumor.46, 49
Figure 2.
Genomic evolution of melanoma. Genetic alterations accumulate during melanoma progression, from benign lesions that carry the BRAF V600E mutation to intermediate lesions that have accumulated more genetic mutations to invasive melanoma that has acquired pathogenic mutations that confer the ability to metastasize. As melanoma progresses, distinct evolutionary trajectories for different melanoma subtypes develop.
Researchers have evaluated the discordance in BRAF mutation status between primary and matched metastatic lesions in relation to the potential implications for therapeutic success and treatment decision-making. Heinzerling et al. evaluated BRAF mutation heterogeneity between tumor samples isolated from the primary tumor and those from metastases from patients with melanoma.50 In this study, the tumors were not under the selection pressure of a BRAF inhibitor because tumor samples were collected prior to the availability of BRAF inhibitor therapy. A total of 53 of the 187 patients with melanoma had multiple tumor samples available for analysis. Ten of 53 patients (19%) had discordant tumor samples that harbored both BRAF WT and mutation-positive metastases. BRAF mutations were as likely to first appear in metastases derived from WT tumors as they were to be found in the primary tumor that subsequently had WT metastases. Additionally, similar to results reported earlier by Shain et al,46 considerable intratumoral heterogeneity was found using immunohistochemistry (IHC) staining for mutated BRAF, suggesting the appearance of de novo BRAF mutations in the metastases of some patients. The investigators speculated that BRAF mutations may arise spontaneously in the secondary tumor from a BRAF WT primary tumor. Alternatively, genetically heterogeneous metastatic tumors could derive from BRAF mutation-positive or BRAF WT primary tumor cells, leading to intrapatient discordance. It is also possible that these results may have been a result of sampling or the detection method used.50
Yancovitz et al. used mutation-specific real-time polymerase chain reaction (RT-PCR) to examine intra- and inter-tumoral heterogeneity of the BRAF mutation.51 To determine intratumoral heterogeneity, laser microdissection of three to five sites from nine primary melanomas was performed and showed variability in the percentages of BRAF-mutant cells across the tumor sites. Samples were classified as unlikely to be heterogeneous (three patients), likely to be heterogeneous (two patients), or having marked heterogeneity (four patients). To determine intertumoral heterogeneity, samples from 18 patients with matched primary and secondary tumors and 19 patients with tumors from more than one metastatic site were analyzed for concordance in BRAF mutations. Discordance was observed between primary and metastatic tumors in 8 of 18 patients (44%) and between metastatic tumors in 5 of 19 patients (26%). The investigators suggested that primary melanomas may contain mixed populations of BRAF-mutant and BRAF WT cells, both able to metastasize, and that resistance to targeted therapies may be mediated by these genetically distinct tumor subclones.51 Data in support of this idea were reported in a study by Colombino et al., who analyzed 291 samples from 132 patients with melanoma.20 Although BRAF mutations were detected in 43% of primary tumors, the rate of BRAF mutations in metastases was 5% greater (48%), consistent with accumulation of de novo genetic abnormalities as melanoma develops. Interestingly, BRAF mutation concordance varied by metastasis site: concordance with the primary tumor was highest in lymph node and visceral metastases (93 and 96%) and lowest in skin and brain metastases (75 and 80%).20
Bradish et al. evaluated the concordance in BRAF mutational status between primary and matched metastatic lesions collected from 25 patients with metastatic melanoma.52 Six of these patients also had multiple metastatic lesions available for testing. None had received treatment with a BRAF inhibitor. Using an RT-PCR kit able to detect five different somatic BRAF V600 mutations—including the most common V600E and less common V600K, V600D, and V600R—intertumoral discordance was detected in the tumors of four patients (16%). Of these four patients, two had BRAF WT primary lesions with BRAF V600-mutant metastatic lesions and the other two had BRAF V600-mutant primary lesions with BRAF WT metastatic lesions. Additionally, the discordance rate was higher in patients with multiple metastases (two of six (33%)) than in patients with a single metastasis (two of 19 (11%)), indicating that increasing the number of lesions tested increases the likelihood of a discordant finding.
Together, these studies indicate that detection of a negative BRAF mutation in the primary tumor may not necessarily reflect the BRAF mutation status of metastases. Therefore, clinicians should consider repeat testing of the primary tumor and recent metastatic site(s) to determine the BRAF mutation status.52, 53, 54
Resistance to BRAF and MEK inhibitor therapy
MAPK pathway inhibitors are an option for patients with BRAF V600-mutant advanced melanoma.26, 40, 55 The introduction of BRAF inhibitor therapy was an important step forward in the treatment of BRAF-mutated melanoma. Yet up to one-half of patients with BRAF-mutated tumors did not respond to treatment in previous studies; in those who did respond, acquired resistance generally developed at a median of 6–7 months with single-agent BRAF inhibitors.56, 57 Combination treatment with BRAF and MEK inhibitors has been shown to significantly increase the proportion of patients with an objective response while also significantly delaying the development of acquired resistance.33, 36, 37, 58, 59 Therefore, combination therapy (dabrafenib plus trametinib or vemurafenib plus cobimetinib) is now considered a standard of care in patients with BRAF-mutated advanced melanoma.40
Mechanisms of de novo and acquired resistance to BRAF inhibitor monotherapy were recently described in a comprehensive review article by Manzano et al 60 In vitro and in vivo models have identified primary resistance mechanisms that drive proliferation despite inhibition of BRAF V600 mutations, including alterations in RAC1, loss of PTEN, changes in cyclin-dependent kinase 4 and cyclin D1, and stromal secretion of hepatocyte growth factor activating c-MET.60 Several mechanisms of acquired resistance have also been identified, including the selection of subclonal populations that do not express mutated BRAF (as previously discussed) or populations that have alterations or mutations in components of the MAPK (eg, NRAS mutations, upregulation of tyrosine kinase receptors) or PI3K/AKT pathways (eg, loss of PTEN) or changes in downstream targets (eg, overexpression of transcription factors). As with de novo resistance, these mechanisms can drive proliferation.61
Although significant improvement in survival has been achieved with combination BRAF and MEK inhibition, de novo and acquired resistance continue to be formidable obstacles.59 Patients with disease progression on BRAF inhibitor therapy who subsequently received combination BRAF and MEK inhibitor treatment had an overall response rate of 10–15%, indicating limited clinical benefit from combination therapy once resistance had developed.62, 63 Long et al. evaluated resistance mechanisms in 20 BRAF V600E-mutant tumor samples from 10 patients with early resistance (≤6 months of dabrafenib plus trametinib prior to progression). MAPK pathway reactivation, indicating an ‘addiction’ to the MAPK pathway, was observed in 9 of 10 tumors (90%). Resistance mechanisms included BRAF amplification as well as mutations in MEK1/2 and NRAS.64 In this and a second study by Rizos et al, the rate of MAPK pathway reactivation was higher than that previously observed in patients who progressed on a BRAF inhibitor alone (50%).64, 65
In vitro studies evaluating the pathophysiology of acquired resistance to combination therapy have shown that resistant melanoma cell lines exhibit a much greater degree of drug addiction than cell lines resistant to BRAF inhibitors alone.66 Similar to what has been observed in tumors of patients who progressed on a single BRAF inhibitor and were then treated with BRAF and MEK inhibitors, BRAF inhibitor-resistant cell lines demonstrated the ability to rapidly adapt to combination therapy. This is thought to be due to pre-existing resistance mechanisms. The researchers suggested that intermittent drug dosing may be a strategy to delay the development of acquired resistance, and further studies are currently testing this hypothesis.66
A reduced ability to mount an antitumor immune response has also been observed in patients who progressed on BRAF inhibitor monotherapy and then received the combination of BRAF and MEK inhibitor therapy.63 Chen et al evaluated 14 tumor samples from a cohort of 23 patients refractory to BRAF inhibitor therapy who were switched to combination therapy. Prior to being treated with combination therapy, the tumors showed a relatively low number of tumor-infiltrating CD8-positive T cells and limited expression of programmed death-ligand 1 (CD274; previously named PD-L1). When patients were switched to combination therapy, an increase in immune cell infiltrate into the tumor was observed in one of nine evaluable patients who achieved stable disease.63
There is also great interest in combining BRAF and MEK inhibitor therapy with immune checkpoint inhibitor therapy.67 Preliminary animal models evaluating the combination of MAPK pathway inhibitors with checkpoint inhibitors suggest that the combination may result in greater clinical benefit than what can be achieved using either agent alone. Working with tumor-bearing mice, Ebert et al showed that MEK inhibition stimulated the infiltration of antigen-specific CD8-positive T cells and prevented upregulation of programmed cell death protein 1 (PDCD1). Furthermore, combining MEK inhibition with anti-PDL1 therapy resulted in synergistic antitumor activity.68 Another animal study evaluated BRAF plus MEK inhibition combined with anti-PDCD1 therapy and demonstrated superior antitumor activity with the triple combination over combined BRAF plus MEK or PDCD1 inhibition alone.69 An ongoing clinical study (KEYNOTE-022; NCT02130466) is evaluating dabrafenib plus trametinib plus pembrolizumab in patients with BRAF-mutant advanced melanoma.67 Another ongoing study is evaluating sequencing of combination targeted therapy and checkpoint inhibitors (EA6134; NCT02224781).
BRAF V600 mutation diagnostic tests
Obtaining and Processing Materials for BRAF Testing
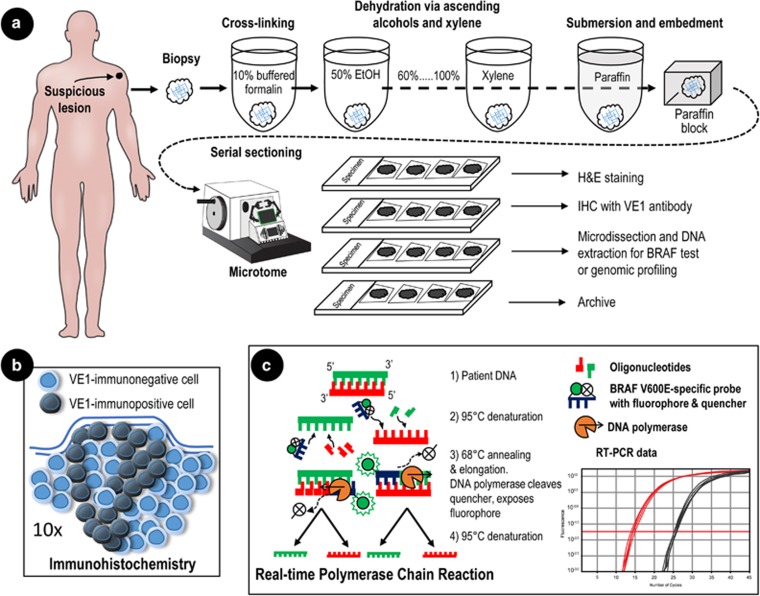
After a preliminary diagnosis of stage III or IV metastatic melanoma, the lesion(s) should be evaluated for BRAF mutational status.40 Cutaneous lesions suspected of being melanoma are typically resected with a wide margin, providing the entire lesion and its margins for analysis. Radiologically detected lesions within internal organs are sampled by core-needle and/or fine-needle aspiration biopsy. The postsurgical processing of surgical resections vs fine-needle aspirates is slightly different. Biopsy specimens are usually processed by formaldehyde fixation and paraffin embedding in blocks for subsequent IHC or DNA extraction for sequencing70, 71 (Figure 3).
Figure 3.
Processing of lesions for pathological analysis of BRAF V600 mutation. (a) The suspicious lesion is biopsied and prepared for formalin-fixed, paraffin-embedded tissue. The tissue sample is immersed in 10% buffered formalin for 4–6 h, then immersed in a series of increasingly concentrated ethanol to extract water. The final dehydration occurs in xylene, after which the tissue is immersed in a series of increasingly concentrated molten paraffin. Finally, the sample is embedded in a block of paraffin suitable for serial sectioning. Serial sectioning produces ‘ribbons’ of tissue sections that can be directly mounted onto slides, deparaffinized, and then stained. Every other section is stained, typically with hematoxylin and eosin, to identify tumor margins while unstained alternate sections are used for immunohistochemistry (IHC) or to obtain DNA for genetic analyses. In contrast to the process for surgical specimens, fine-needle aspirates are taken and transferred directly from the syringe to a slide, where the material is smeared across its surface, air dried, and fixed. (b) IHC reveals cellular expression of the BRAF kinase in BRAF mutation-positive (dark gray) cells in situ. (c) RT-PCR uses a fluorescent-labeled target-specific probe allowing amplification of the target sequence and real-time quantification of PCR products. EtOH, ethyl alcohol.
Alternatively, a blood sample (‘liquid biopsy’) may be assayed for circulating tumor DNA (ctDNA).72, 73 ctDNA is composed of small fragments of nucleic acids that are found in the circulation but are not associated with any cell or cell fragment. Although the mechanism by which ctDNA is released into the blood is unclear, its abundance seems to reflect the biological aggressiveness of the tumor.74, 75, 76, 77 Despite being an important innovation in cancer diagnostics, ctDNA detection has limitations compared with a tissue-based biopsy. Currently, ctDNA analysis is less sensitive than tissue biopsy. Santiago-Walker et al estimated the sensitivity of ctDNA testing for BRAF V600E to be 76% relative to standard PCR using a direct tumor biopsy. This was determined by calculating the concordance in BRAF V600E-positive mutation status between tissue biopsies and ctDNA (N=661).78 Furthermore, because blood lacks anatomical specificity, the presence of BRAF-mutant DNA in blood does not indicate the number of tumors or the tumor origin. Additional diagnostic tests are still required to identify the tumor site(s), which may add to the overall expense, unlike conducting a standard biopsy and mutational test.
Despite these limitations, ctDNA can provide useful information for treatment monitoring. The correlation between ctDNA abundance and disease stage has been examined in patients with melanoma. The quantity of BRAF V600E mutation-specific DNA has been shown to be positively correlated to tumor burden and disease stage.78, 79, 80 Accordingly, patients with BRAF-positive ctDNA had significantly shorter progression-free and overall survival than patients without detectable BRAF-positive ctDNA.78 The quantity of BRAF V600E-positive ctDNA was also shown to be greater in patients with more metastatic sites.81
BRAF Testing: Companion Diagnostic and Laboratory-Developed Tests
BRAF genotype tests are classified as either companion diagnostic or laboratory developed, and US and European regulatory agencies have issued guidelines on their use. The European Medicines Agency summary of product characteristics for vemurafenib states that before starting treatment, a validated test is required to identify BRAF mutation status but does not specify which test should be used. It does state that the cobas 4800 BRAF V600 Mutation Test (Roche Molecular Systems, Inc) was used to identify BRAF mutation-positive patients in the clinical trial on which marketing approval was based.82 In the US, companion diagnostic tests are often codeveloped with a drug or drug class and are recommended and approved by the FDA for informing the safe and effective use of its corresponding therapeutic product.83 Companion diagnostic tests undergo rigorous testing and are reviewed by the FDA and the US Centers for Medicare and Medicaid Services to ensure analytical and clinical validity. In contrast, laboratory-developed tests are typically designed, developed, and used by a single institution, such as a hospital or research laboratory.84, 85 Unlike companion diagnostic tests, laboratory-developed tests are not linked to a particular drug or drug class. Two companion diagnostic tests are commercially available for detection of BRAF mutations: the cobas 4800 BRAF V600 Mutation Test for vemurafenib plus cobimetinib and the THxID-BRAF kit (bioMérieux, Inc) for dabrafenib plus trametinib.16 In addition, laboratory-developed tests may be used to determine a patient’s BRAF mutation status, provided that the tests are reviewed and tested by the US Centers for Medicare and Medicaid Services and used in a Clinical Laboratory Improvement Amendments (CLIA)-certified laboratory.84, 86 In a recently published survey of laboratories participating in the College of American Pathologists proficiency testing program for molecular analysis of BRAF mutations, most evaluable reports (100 of 107 (94%)) included only laboratory-developed procedures (the remaining 7 included a companion diagnostic assay).87 Although the extent of regulatory oversight of laboratory-developed tests has been minimal, the FDA has developed more robust guidance as the complexity of tests has increased. The FDA issued draft guidance on the oversight of laboratory-developed tests in 2014 and issued a discussion paper in early 2017.84, 88, 89
Current and Developing Tests
Several BRAF mutation tests are currently in use; they are classified as DNA based or antibody based.90 DNA-based tests include those that use PCR to selectively amplify the mutant BRAF gene and others that directly determine the nucleotide sequence of the BRAF gene.26, 90 Of note, PCR and other DNA-based tests reveal the presence (or absence) of the gene but do not indicate whether the gene is being transcribed and translated into a protein.70, 71
Currently, the only available antibody-based test for mutant BRAF protein uses the monoclonal antibody VE1 to detect expression in tumor samples using IHC.26, 90, 91 This approach is effective in identifying a qualitative change (ie, presence or absence of the protein) but is less accurate in quantitating changes in expression than other antibody-based assays, such as the enzyme-linked immunosorbent assay.92
Regardless of the technical approach used, every test can be characterized by its sensitivity, specificity, and limit of detection. Sensitivity is the ability of the test to correctly identify BRAF mutations with a low rate of false negatives. Specificity is the ability of the test to correctly identify BRAF mutations with a low rate of false positives. Limit of detection is the threshold at which a signal (eg, DNA harboring the BRAF mutation) can be distinguished from the background (eg, BRAF WT DNA).93, 94 Tests also vary in terms of their selectivity.94, 95 For example, some tests can detect only the V600E mutation, while others can detect V600E and other mutations, such as V600K.14, 15, 91 This is important because some agents (vemurafenib plus cobimetinib, trametinib, and dabrafenib plus trametinib) have been shown to be effective in patients with V600E/K mutations.26, 31, 32, 33
PCR DNA-Based Tests
The cobas 4800 BRAF V600 mutation test and the THxID-BRAF kit both use RT-PCR to amplify and detect the mutant DNA sequence in a tumor sample. The major advantages of RT-PCR are faster performance, better reproducibility, and lower cost compared with traditional genomic sequencing methods.
The RT-PCR process is illustrated in Figure 3 and includes an example of a data readout from an RT-PCR assay. Briefly, the RT-PCR-based BRAF test uses one primer set to detect the BRAF mutation and another set to detect the WT sequence.14, 45 Each primer includes a distinct fluorophore and quencher. The quencher prevents the excitation/emission of light by the fluorophore as long as the primer is unbound to DNA. As the DNA polymerase incorporates the primer into a new DNA strand, it cleaves the quencher from the primer, freeing the fluorophore to emit narrow-bandwidth light that can be detected. As DNA is synthesized with each cycle, the quantity of light increases relative to the abundance of the DNA product, which is determined by the number of cycles necessary for the intensity of emitted light to cross an arbitrary threshold. The relative abundance of BRAF V600E is determined by comparing this threshold with the number of cycles necessary to detect WT BRAF.
It is recommended that patients undergo an FDA-approved test, such as the cobas 4800 BRAF V600 mutation test or THxID-BRAF kit, before starting treatment with vemurafenib plus cobimetinib or dabrafenib plus trametinib.14, 15 The cobas 4800 test has an analytical sensitivity of 95% for detecting the V600E mutation.82 However, the test shows limited cross-reactivity in detecting the V600D and K mutations, and the package insert states that the test does not detect non-V600E mutations reliably. The recommended DNA input is 125 ng total, since the test can detect the BRAF V600E mutation at ≥5% using the standard input of 125 ng/25 μl.15 The THxID-BRAF kit detects both V600E and K mutations. The kit has been validated for DNA input of 10 ng/μl to 350 ng/μl.14 In contrast to the cobas 4800 test, the THxID test has a high degree of sensitivity for both V600E and V600K (Table 2).
Table 2. Comparison of methods currently used for BRAF mutational analysis.
| THxID-BRAF kit | cobas 4800 BRAF V600 mutation test | Sanger | HRM | Pyrosequencing | IHC | |
|---|---|---|---|---|---|---|
| CDx or LDT | CDx | CDx | LDT | LDT | LDT | LDT |
| Sensitivity, % | >96 for V600E; >92 for V600K | ~97 for V600E; 66–70 for V600K | 92–98 | 98–100 | >98 | 85–100 |
| Specificity, % | 100 | >98 | 100 | 98–100 | 90–100 | 98–100 |
| Limit of detection, % | 5 for V600E, V600K | 5–7 for V600E; >35 for V600K | 6.6 | 6.6 | 5 | 5 |
| Mutations detected | Approved for V600E, V600K | Approved for V600E only | 99% of all detectable mutations | 99% of all detectable mutations | Assay optimized for V600 mutations is available | VE1 antibody specific for V600E |
| Sample | Tumor-derived DNA | Tumor-derived DNA | Tumor-derived DNA | Tumor-derived DNA | Tumor-derived DNA | Tissue |
| Cost | Medium | Medium | Medium | Low | High | Low |
Abbreviations: CDx, companion diagnostic; HRM, high-resolution melt; IHC, immunohistochemistry; LDT, laboratory-developed test.
Although the RT-PCR-based cobas 4800 and THxID assays are the companion diagnostic tests for BRAF plus MEK inhibitor use, many treatment centers use laboratory-developed tests based on other DNA sequencing methods.87, 90 The goal of genomic sequencing in the context of BRAF testing is to identify any one of the nucleic acid substitutions associated with the mutant BRAF gene (Table 2). The advantage of genomic sequencing is that it is not specific for a certain mutation; therefore, any change in the sequence may be identified. Three main methods are currently being used: Sanger sequencing, pyrosequencing, and high-resolution melt analysis/real-time PCR.87, 90, 96 Next generation sequencing platform is less frequently used in clinical labs and is beyond the scope of this review.90, 97
Sanger sequencing
The Sanger sequencing method, generally considered to be the gold standard, was developed in the 1970s and was the basis for the method used by Venter et al in the Human Genome Project to generate the first draft human genome sequence.98 The Sanger method relies on incorporation of deoxynucleotide triphosphates (dNTPs; dATP, dCTP, dGTP, and dTTP) into a complementary strand of DNA that exactly matches a denatured target strand. The nucleotide sequence is detected by chain termination, which results in DNA strands of variable length. Chain termination occurs due to the incorporation of 2′,3′-dideoxynucleotide triphosphates (ddNTPs), which lack a 3′-OH and thus cannot form a bond with the next dNTP.99 Reactions with each of four ddNTPs (ddATP, ddCTP, ddGTP, and ddTTP) that are tagged with a radioactive phosphate moiety or fluorophore are run in parallel and resolved by electrophoresis. The length of each DNA product indicates where the chain terminated and which nucleotide was the last to be incorporated. Advantages of Sanger sequencing include reliability and availability and affordability of the reagents; the primary disadvantage is its relatively low sensitivity.
Jurkowska et al recently compared the reproducibility of Sanger sequencing with that of the cobas 4800 test. Among 236 formalin-fixed, paraffin-embedded tissue (FFPET) samples,100 Sanger sequencing identified the V600E mutation in 60.9% of samples and the cobas 4800 test identified it in 61%, with 95.2% agreement between the two tests. Sanger sequencing also detected 10 non-V600E mutations, while the cobas 4800 test detected 6. Six cases were unamplifiable with the standard PCR/sequencing method, five of which were identified as V600E positive by the cobas 4800 test.
Lopez-Rios et al also compared Sanger sequencing with the cobas 4800 test.101 The Sanger method produced invalid results in 8 of 116 samples vs 0 of 232 with the cobas 4800 test. The positive percent agreement with Sanger sequencing was 97.7% the negative percent agreement was 95.3%.
Qu et al performed a similar analysis and found lower sensitivity for the cobas 4800 test than for the Sanger method.102 In 275 FFPET samples, V600 mutations were identified in 35 and 43%, respectively. Compared with Sanger sequencing, the cobas 4800 test exhibited 80.5% sensitivity and 99.4% specificity. Of interest, 23 samples that were positive with Sanger sequencing were discordant with the cobas 4800 test. Based on these results, the authors suggested that Sanger sequencing should be used to test samples that are negative by the cobas 4800 test.
Pyrosequencing
In the early 2000s, multiple innovations in sequencing methods were introduced. One example is pyrosequencing. Like the Sanger method, pyrosequencing relies on DNA synthesis, but it detects the addition of new base pairs using an enzymatic reaction that ultimately produces light.103 The process starts when target DNA is mixed with one of four dNTPs. As the DNA polymerase incorporates the complementary dNTP (A to T and C to G) into the strand, a molecule of pyrophosphate is released. The presence of sulfurylase in the reaction converts pyrophosphate to adenosine triphosphate, which interacts with luciferase, producing light. Each flash of light can be detected with a charge coupled device camera, indicating the addition of specific dNTPs to the growing strand.104 As with the Sanger method, a separate reaction is required for each dNTP. However, four separate reactions can be conducted simultaneously, greatly speeding up the process.
Ihle et al used BRAF-specific pyrosequencing, high-resolution melt analysis, multigene next-generation sequencing, and IHC and compared these techniques with the Sanger method for sensitivity and specificity in detecting the BRAF mutation in FFPET samples.13 Pyrosequencing was found to have lower specificity than Sanger sequencing (90 vs 100%) but also a lower limit of detection. It detected mutant DNA in as few as 5% of copies compared with 6.6% with Sanger sequencing and 7% with RT-PCR (Cobas 4800).
As with the Sanger method, multiple biological steps are required for pyrosequencing. The process takes 2–4 days to complete, the run itself is 10 h, and additional time is required for bioinformatics analysis. A key advantage of pyrosequencing is that it has been adapted to a high-throughput platform, vastly increasing the amount of generated data.103 The disadvantages of pyrosequencing are high cost and increased complexity and time. A high-throughput pyrosequencing machine can cost >$600 000, and some reagents cost thousands of dollars.103 However, because of the longer reads made possible by pyrosequencing, the cost per base is lower than that for Sanger sequencing. In addition, sequencing the genome is unnecessary to determine the presence or absence of the BRAF V600E mutation, and the potential to detect multiple mutations with pyrosequencing has led to its increased use as a diagnostic tool.
High-Resolution Melt Analysis/Real Time PCR
High-resolution melt is a sequencing technique that incorporates aspects of PCR and sequencing by DNA synthesis. High-resolution melt is a relatively simple sequencing technique, as it is based on the principle that double-stranded DNA denatures (ie, ‘melts’) at a temperature that varies as a function of its G/C content, length, and overall nucleotide sequence.105 The process normally begins with amplification of the target DNA to nanogram quantities using PCR.105
Intercalating dyes (eg, SYBR Green, LC Green, SYTO 9, and EvaGreen) are included in the reaction, binding to double-stranded DNA but not single-stranded DNA.106 Thus, fluorescence will initially be high because the sample is composed solely of newly synthesized, double-stranded DNA. While fluorescence is continually monitored, the PCR-amplified sample is heated through a range of temperatures. As the temperature gradually increases and the strand begins to denature, the fluorescence slowly decreases. Then, at a temperature characteristic of the DNA strand’s G/C content, length, and overall nucleotide sequence, the fluorescence falls rapidly, indicating the complete separation of DNA into single strands. The substitution of a single base can lead to a 1 °C difference in the melting point, a difference easily detected by the melting curve shape. Data are usually presented as the derivative of the melting curve, showing the change in fluorescence over the change in temperature. The sensitivity of the technique is directly related to temperature control. The RapidCycler II (BioFire Diagnostics, Inc (formerly known as Idaho Technology, Inc)) and LightCycler 480 (Roche Diagnostics) melts samples at 0.3 °C/sec, whereas the Rotor-Gene 6000 (Corbett Life Science, a QIAGEN company) controls temperature at 0.017 °C/sec. The slower melting time results in higher resolution and better performance; however, the tradeoff is speed of data acquisition.105 The RapidCycler can acquire data in 1–2 min, whereas the Rotor-Gene device requires 20 min.105
The advantages of using high-resolution melt over other techniques include low cost and increased sensitivity and convenience. The PCR step and high-resolution melt steps can occur in the same tube or capillary, lowering the possibility of contamination and reducing the likelihood of operator error. The LightCycler 480 and RapidCycler II both provide integrated PCR and high-resolution melt analysis in one machine. Additionally, the technique is not destructive, allowing additional analyses on the same material. A disadvantage of high-resolution melt is that it does not permit the direct identification of the nucleotide sequence. However, from a community clinician’s perspective, this may not disqualify it as a technique since it screens for the presence of the BRAF mutation, which is sufficient for deciding whether to administer BRAF inhibitor therapy.
A meta-analysis of 14 studies that included 1324 samples from patients with melanoma and other cancers examined the sensitivity of high-resolution melting in detecting BRAF mutation.106 Most of the samples were obtained from FFPET, although some were from fresh-frozen tissue. The pooled value of the sensitivity of the high-resolution melting curve analysis was high: 0.99 (range, 0.93–1.00). The pooled specificity of the high-resolution melting curve analysis was also 0.99 (range, 0.88–1.00).
Richter et al compared the BRAF mutational testing accuracy of four techniques—high-resolution melt, single-strand conformation analysis, Sanger sequencing, and RT-PCR—in a blinded test across two laboratories of >90 FFPET samples from melanoma patients. They found 100% concordance between the Sanger and high-resolution melt techniques.96 Together, these results indicate a high level of overall accuracy for high-resolution melt.
Protein Based Analysis (immunohistochemistry)
Although molecular testing is currently considered to be the gold standard for the detection of BRAF mutations, the monoclonal antibody VE1, developed to specifically recognize a segment of the mutant BRAF V600E protein, is emerging as a reliable option.107, 108 Since BRAF has a role in intracellular signaling, BRAF staining is localized to the cytoplasm.109 There are both advantages and disadvantages to IHC detection of mutant BRAF.96 Advantages include visualization of BRAF V600E-mutant protein distribution at the single-cell level, semiquantitative readout of protein abundance, and high assay sensitivity and specificity. Because FFPET samples are usually available as part of a regular biopsy, testing by IHC should be relatively simple and inexpensive, and provide a fast turnaround on results. The main disadvantages of IHC include the possibility of false negatives due to high heterogeneity or low abundance of BRAF V600E and a lack of availability of antibodies for BRAF V600K or other variants. In addition, because IHC recognizes protein and not DNA, there may be discrepancies between IHC and DNA-based test results. Despite these drawbacks, reports have indicated that the sensitivity and selectivity of the VE1 antibody compare favorably with those of DNA-based tests.
Hugdahl et al used the VE1 antibody to assess the frequency of the mutant BRAF V600E protein in 248 nodular melanomas and investigate its prognostic value.28 They found BRAF V600E staining in 35% of the cases. Tumors with BRAF V600E staining were thicker and more likely to be ulcerated, and patients with these lesions had reduced survival. Despite these observations, BRAF V600E protein expression independently predicted reduced survival, separately, and distinctly from mitotic rate, presence of ulceration, and tumor thickness. The concordance between V600E staining and mutation status, as assessed by PCR, was 88%. In a comparison of Kaplan–Meier survival curves in patients grouped by BRAF protein expression (n=248) and those grouped by BRAF mutation status (n=191), the researchers found that expression of the mutant BRAF protein was a better prognostic indicator than presence of the mutant BRAF gene. However, caution should be taken when interpreting this finding, since the analysis based on BRAF protein expression included more patients than the analysis based on BRAF gene presence and thus had more statistical power.
Long et al compared the sensitivity and specificity of VE1 immunostaining with those of a DNA-based assay in 100 patients.110 VE1 immunostaining had a sensitivity of 97% (37 of 38 paired samples yielded identical results) and specificity of 98% (58 of 59) for detecting the presence of the BRAF V600E mutation. The authors noted that ‘…clinical use of the V600E BRAF antibody should be a valuable supplement to conventional mutation testing and allow V600E mutant metastatic melanoma patients to be triaged rapidly into appropriate treatment pathways.’ Further support for this conclusion was provided by a study by Pearlstein et al, who compared VE1 immunostaining results with pyrosequencing results in 76 patients with melanoma.111 A total of 27 patients (35%) had the V600E mutation, and another nine had an alternate mutation (V600K, V600R, or V600Q). Of these, there were five discordant cases in which pyrosequencing reported the presence of the V600E mutation but immunostaining was negative. Further analysis of these discordant findings showed that two of the five cases had a high percentage of V600E allele frequency while two others demonstrated a low allele frequency. The samples with a low allele frequency suggest the possibility of tumor heterogeneity. The final discordant case was reclassified as it was found to have a BRAF V600K rather than V600E mutation. Thus, the IHC test had a sensitivity of 85% and specificity of 100% compared with pyrosequencing.
Manfredi et al compared RT-PCR and high-resolution melt analysis with IHC.112 Of 189 samples, 88 (47%) tested positive for BRAF V600E mutation by IHC. The V600E-positive samples were analyzed by genotyping of 74 samples, which yielded interpretable results. Of these, only two samples were not confirmed by both genotyping methods. Of the 87 samples that were determined to be BRAF V600E negative by IHC, 76 (87%) were determined to be BRAF WT by molecular testing. Ten of the remaining samples were determined to be other BRAF V600 variants. Only one was determined to be a true false-negative, as it was determined to be BRAF V600E following molecular testing. Overall, this analysis demonstrated that IHC testing with VE1 had high sensitivity (98.6%) and specificity (97.7%) for IHC.112
Another challenge introduced by IHC staining is the potential for subjective differences in interpretation between pathologists. A study by Eriksson et al evaluated VE1-stained FFPE melanoma samples and compared the reported interpretations with the results of DNA sequencing.109 They asked three observers to judge the staining intensity of samples on a three-point semiquantitative scale. This method resulted in a sensitivity of 96.7% and specificity of 94.5% for IHC with VE1. The level of agreement between the observers was not reported. A 2014 study by Marin et al illustrated the potential challenges associated with subjectivity introduced with interpretation of IHC.113 In that study, seven pathologists, two of whom had been previously trained to interpret V600E staining, reviewed VE1 staining in samples from 67 patients. Concordance between the seven pathologists was achieved only 79.1% of the time. This study emphasizes that VE1 staining has some diagnostic limitations and highlights the importance of training. Overall, the authors of this study and others conclude that IHC can serve as an important first step in determination of BRAF V600E mutation status.96
Conclusions
The introduction of combination BRAF and MEK inhibitor therapy has transformed treatment outcomes in patients with advanced-stage BRAF-mutated melanoma. Dramatic improvement in the treatment of patients with advanced-stage melanoma has occurred with the addition of checkpoint inhibitors. The use of targeted therapy with immunotherapy, either in sequence or in combination, will likely lead to further improvements in overall survival.
It is of paramount importance that a patient’s BRAF mutational status is promptly and accurately determined at the time of initial diagnosis, because it is currently the only reliable predictive biomarker that can influence the treatment of advanced melanoma. VE1 immunostaining for the detection of BRAF-mutated protein is a quick and inexpensive test that can be combined with other melanoma markers. However, since the only mutation that can be detected with the VE1 antibody by IHC is the most common V600E mutant, the presence or absence of other mutations is not determined by this diagnostic test; therefore, evaluation using DNA-based methods, such as the FDA-approved companion diagnostic tests, is still needed. Newer technologies are increasingly being used and refined (eg, evaluation of ctDNA from ‘liquid biopsy’ samples) and will help to address the shortcomings of tests that have been used to date.72, 73, 76, 77, 81 Furthermore, the molecular profile of a tumor has been shown to change over time, and inter- and intratumoral heterogeneity can make interpretation of test results challenging. Therefore, it is recommended that repeat testing of the putative primary tumor and recent metastatic sites be performed. Much progress has been made in tests for ctDNA, and this technology holds promise as a tool for early on-treatment monitoring as well as monitoring over time in the context of acquired resistance. It is expected that in this age of precision medicine, the future of BRAF biomarker testing will see more techniques move out of the research space and into the clinical practice setting.
Acknowledgments
We wish to acknowledge Amanda Kauffman, Heather Edens, and William Fazzone of ArticulateScience LLC, Hamilton, NJ, for providing editorial support and medical writing assistance, funded by Novartis Pharmaceuticals Corporation.
Footnotes
The authors declare no conflict of interest.
References
- American Cancer Society. Melanoma skin cancer: ACS. Available from http://www.cancer.org/acs/groups/cid/documents/webcontent/003120-pdf.pdf; Accessed 2017.
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin 2017;67:7–30. [DOI] [PubMed] [Google Scholar]
- Luke JJ, Flaherty KT, Ribas A et al. Targeted agents and immunotherapies: optimizing outcomes in melanoma. Nat Rev Clin Oncol 2017;14:463–482. [DOI] [PubMed] [Google Scholar]
- Atkins MB, Lotze MT, Dutcher JP et al. High-dose recombinant interleukin 2 therapy for patients with metastatic melanoma: analylsis of 270 patients treated between 1985 and 1993. J Clin Oncol 1999;17:2105–2116. [DOI] [PubMed] [Google Scholar]
- Schiller JH, Pugh M, Kirkwood JM et al. Eastern Cooperative Group trial of interferon gamma in metastatic melanoma: an innovative study design. Clin Cancer Res 1996;2:29–36. [PubMed] [Google Scholar]
- Parker BS, Rautela J, Hertzog PJ. Antitumor actions of interferons: implications for cancer therapy. Nat Rev Cancer 2016;16:131–144. [DOI] [PubMed] [Google Scholar]
- Shain AH, Bastian BC. From melanocytes to melanomas. Nat Rev Cancer 2016;16:345–358. [DOI] [PubMed] [Google Scholar]
- Bradish JR, Montironi R, Lopez-Beltran A et al. Towards personalized therapy for patients with malignant melanoma: molecular insights into the biology of BRAF mutations. Future Oncol 2013;9:245–253. [DOI] [PubMed] [Google Scholar]
- Ossio R, Roldan-Marin R, Martinez-Said H et al. Melanoma: a global perspective. Nat Rev Cancer 2017;17:393–394. [DOI] [PubMed] [Google Scholar]
- Schadendorf D, Fisher DE, Garbe C et al. Melanoma. Nat Rev Dis Primers 2015;1:15003. [DOI] [PubMed] [Google Scholar]
- Su MY, Fisher DE. Immunotherapy in the precision medicine era: melanoma and beyond. PLoS Med 2016;13:e1002196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merlino G, Herlyn M, Fisher DE et al. The state of melanoma: challenges and opportunities. Pigment Cell Melanoma Res 2016;29:404–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihle MA, Fassunke J, Konig K et al. Comparison of high resolution melting analysis, pyrosequencing, next generation sequencing and immunohistochemistry to conventional Sanger sequencing for the detection of p.V600E and non-p.V600E BRAF mutations. BMC Cancer 2014;14:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- THxID-BRAF [package insert]. Marcy l'Etoile, France: bioMérieux 2013.
- Roche Diagnostics Inc. cobas 4800 BRAF V600 Mutation Test. Available from https://www.accessdata.fda.gov/cdrh_docs/pdf11/P110020c.pdf (accessed 2017).
- US Food and Drug Administration. List of cleared or approved companion diagnostic devices (in vitro and imaging tools). Available from https://www.fda.gov/MedicalDevices/ProductsandMedicalProcedures/InVitroDiagnostics/ucm301431.htm (accessed 2017).
- Lawrence MS, Stojanov P, Polak P et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature 2013;499:214–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayward NK, Wilmott JS, Waddell N et al. Whole-genome landscapes of major melanoma subtypes. Nature 2017;545:175–180. [DOI] [PubMed] [Google Scholar]
- Burotto M, Chiou VL, Lee JM et al. The MAPK pathway across different malignancies: a new perspective. Cancer 2014;120:3446–3456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombino M, Capone M, Lissia A et al. BRAF/NRAS mutation frequencies among primary tumors and metastases in patients with melanoma. J Clin Oncol 2012;30:2522–2529. [DOI] [PubMed] [Google Scholar]
- Curtin JA, Fridlyand J, Kageshita T et al. Distinct sets of genetic alterations in melanoma. N Engl J Med 2005;353:2135–2147. [DOI] [PubMed] [Google Scholar]
- Hayward NK, Wilmott JS, Waddell N et al. Whole-genome landscapes of major melanoma subtypes. Nature 2017;11:175–180. [DOI] [PubMed] [Google Scholar]
- Fedorenko IV, Gibney GT, Smalley KS. NRAS mutant melanoma: biological behavior and future strategies for therapeutic management. Oncogene 2013;32:3009–3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long GV, Menzies AM, Nagrial AM et al. Prognostic and clinicopathologic associations of oncogenic BRAF in metastatic melanoma. J Clin Oncol 2011;29:1239–1246. [DOI] [PubMed] [Google Scholar]
- Ribas A, Flaherty KT. BRAF targeted therapy changes the treatment paradigm in melanoma. Nat Rev Clin Oncol 2011;24:426–433. [DOI] [PubMed] [Google Scholar]
- Bradish JR, Cheng L. Molecular pathology of malignant melanoma: changing the clinical practice paradigm toward a personalized approach. Hum Pathol 2014;45:1315–1326. [DOI] [PubMed] [Google Scholar]
- COSMIC. Catalogue of Somatic Mutations in Cancer. Available from http://cancer.sanger.ac.uk/cosmic (accessed 2017).
- Hugdahl E, Kalvenes MB, Puntervoll HE et al. BRAF-V600E expression in primary nodular melanoma is associated with aggressive tumour features and reduced survival. Br J Cancer 2016;114:801–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies H, Bignell GR, Cox C et al. Mutations of the BRAF gene in human cancer. Nature 2002;417:949–954. [DOI] [PubMed] [Google Scholar]
- Long GV, Weber JS, Infante JR et al. Overall survival and durable responses in patients with BRAF V600-mutant metastatic melanoma receiving dabrafenib combined with trametinib. J Clin Oncol 2016;34:871–878. [DOI] [PubMed] [Google Scholar]
- Long GV, Stroyakovskiy D, Gogas H et al. Dabrafenib and trametinib versus dabrafenib and placebo for Val600 BRAF-mutant melanoma: a multicentre, double-blind, phase 3 randomised controlled trial. Lancet 2015;386:444–451. [DOI] [PubMed] [Google Scholar]
- Robert C, Karaszewska B, Schachter J et al. Improved overall survival in melanoma with combined dabrafenib and trametinib. N Engl J Med 2015;372:30–39. [DOI] [PubMed] [Google Scholar]
- Ascierto PA, McArthur GA, Dréno B et al. Cobimetinib combined with vemurafenib in advanced BRAF(V600)-mutant melanoma (coBRIM): updated efficacy results from a randomised, double-blind, phase 3 trial. Lancet Oncol 2016;17:1248–1260. [DOI] [PubMed] [Google Scholar]
- McArthur GA, Chapman PB, Robert C et al. Safety and efficacy of vemurafenib in BRAF(V600E) and BRAF(V600K) mutation-positive melanoma (BRIM-3): extended follow-up of a phase 3, randomised, open-label study. Lancet Oncol 2014;15:323–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ugurel S, Rohmel J, Ascierto PA et al. Survival of patients with advanced metastatic melanoma: the impact of novel therapies. Eur J Cancer 2016;53:125–134. [DOI] [PubMed] [Google Scholar]
- Long GV, Flaherty KT, Stroyakovskiy D et al. Dabrafenib plus trametinib versus dabrafenib monotherapy in patients with metastatic BRAF V600E/K-mutant melanoma: long-term survival and safety analysis of a phase 3 study. Ann Oncol 2017;28:1631–1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert C, Karaszewska B, Schachter J et al. Three-year estimate of overall survival in COMBI-v, a randomized phase 3 study evaluating first-line dabrafenib (D)+trametinib (T) in patients (pts) with unresectable or metastatic BRAF V600E/K–mutant cutaneous melanoma. Ann Oncol 2016;27:LBA40. [Google Scholar]
- McArthur G, Dréno B, Atkinson VEfficacy of Long-term Cobimetinib Combined with Vemurafenib in Advanced BRAFV600-Mutated Melanoma: 3-year Follow-up of the Phase 3 coBRIM Study and 4-year Follow-up of the Phase 1b BRIM7 study Society for Melanoma Research Annual Meeting, Boston, MA, 2016. [Google Scholar]
- Dummer R, Hauschild A, Lindenblatt N et al. Cutaneous melanoma: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol 2017;26:v126–v132. [DOI] [PubMed] [Google Scholar]
- National Comprehensive Cancer Network (NCCN). Practice Guidelines in Oncology: Melanoma Version 1, Available from https://www.nccn.org/professionals/physician_gls/pdf/melanoma.pdf (accessed 2017).
- Dummer R, Hauschild A, Lindenblatt N et al. Cutaneous melanoma: ESMO clinical practice guidelines for diagnosis, treatment, and follow-up. Ann Oncol 2016;Suppl 5:126–132. [Google Scholar]
- Garbe C, Peris K, Hauschild A et al. Diagnosis and treatment of melanoma. European consensus-based interdisciplinary guideline-update 2016. Eur J Cancer 2016;63:201–217. [DOI] [PubMed] [Google Scholar]
- Zelboraf (vemurafenib) [package insert]. South San Francisco, CA: Genentech USA, Inc 2016.
- Tafinlar (dabrafenib) [package insert]. East Hanover, NJ: Novartis Pharmaceuticals Corporation 2016.
- Pollock PM, Harper UL, Hansen KS et al. High frequency of BRAF mutations in nevi. Nat Genet 2003;33:19–20. [DOI] [PubMed] [Google Scholar]
- Shain AH, Yeh I, Kovalyshyn I et al. The genetic evolution of melanoma from precursor lesions. N Engl J Med 2015;373:1926–1936. [DOI] [PubMed] [Google Scholar]
- Bastian BC. The molecular pathology of melanoma: an integrated taxonomy of melanocytic neoplasia. Annu Rev Pathol 2014;9:239–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang G, Rajadurai A, Tsao H. Recurrent patterns of dual RB and p53 pathway inactivation in melanoma. J Invest Dermatol 2005;125:1242–1251. [DOI] [PubMed] [Google Scholar]
- Chiappetta C, Proietti I, Soccodato V et al. BRAF and NRAS mutations are heterogeneous and not mutually exclusive in nodular melanoma. Appl Immunohistochem Mol Morphol 2015;23:172–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinzerling L, Baiter M, Kuhnapfel S et al. Mutation landscape in melanoma patients clinical implications of heterogeneity of BRAF mutations. Br J Cancer 2013;109:2833–2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yancovitz M, Litterman A, Yoon J et al. Intra- and inter-tumor heterogeneity of BRAF(V600E) mutations in primary and metastatic melanoma. PLoS One 2012;7:e29336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradish JR, Richey JD, Post KM et al. Discordancy in BRAF mutations among primary and metastatic melanoma lesions: clinical implications for targeted therapy. Mod Pathol 2015;28:480–486. [DOI] [PubMed] [Google Scholar]
- Fiskus W, Mitsiades N. B-raf inhibition in the clinic: present and future. Annu Rev Med 2016;67:29–43. [DOI] [PubMed] [Google Scholar]
- Xue Y, Marteloto L, Baslan T et al. An approach to suppress the evolution of resistance in BRAF V600E-mutant carrier. Nat Med 2017;23:929–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkel EA, Gerami P. Malignant melanoma of sun-protected sites: a review of clinical, histological, and molecular features. Lab Invest 2017;97:630–635. [DOI] [PubMed] [Google Scholar]
- Chapman PB, Hauschild A, Robert C et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med 2011;364:2507–2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauschild A, Grob JJ, Demidov LV et al. Dabrafenib in BRAF-mutated metastatic melanoma: a multicentre, open-label, phase 3 randomised controlled trial. Lancet 2012;380:358–365. [DOI] [PubMed] [Google Scholar]
- Long GV, Weber JS, Infante JR et al. Overall survival and durable responses in patients with BRAF V600-mutant metastatic melanoma receiving dabrafenib combined With trametinib. J Clin Oncol 2016;34:871–878. [DOI] [PubMed] [Google Scholar]
- Welsh SJ, Rizos H, Scolyer RA et al. Resistance to combination BRAF and MEK inhibition in metastatic melanoma: where to next? Eur J Cancer 2016;62:76–85. [DOI] [PubMed] [Google Scholar]
- Manzano JL, Layos L, Buges C et al. Resistant mechanisms to BRAF inhibitors in melanoma. Ann Transl Med 2016;4:237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiskus W, Mitsiades N. B-Raf inhibition in the clinic: present and future. Annu Rev Med 2016;67:29–43. [DOI] [PubMed] [Google Scholar]
- Johnson DB, Flaherty KT, Weber JS et al. Combined BRAF (dabrafenib) and MEK inhibition (trametinib) in patients with BRAFV600-mutant melanoma experiencing progression with single-agent BRAF inhibitor. J Clin Oncol 2014;32:3697–3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G, McQuade JL, Panka DJ et al. Clinical, molecular, and immune analysis of dabrafenib-trametinib combination treatment for BRAF inhibitor-refractory metastatic melanoma: a phase 2 clinical trial. JAMA Oncol 2016;2:1056–1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long GV, Fung C, Menzies AM et al. Increased MAPK reactivation in early resistance to dabrafenib/trametinib combination therapy of BRAF-mutant metastatic melanoma. Nat Commun 2014;5:5694. [DOI] [PubMed] [Google Scholar]
- Rizos H, Menzies AM, Pupo GM et al. BRAF inhibitor resistance mechanisms in metastatic melanoma: spectrum and clinical impact. Clin Cancer Res 2014;20:1965–1977. [DOI] [PubMed] [Google Scholar]
- Moriceau G, Hugo W, Hong A et al. Tunable-combinatorial mechanisms of acquired resistance limit the efficacy of BRAF/MEK cotargeting but result in melanoma drug addiction. Cancer Cell 2015;27:240–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long GV, Hamid O, Hodi FS et al. Phase 2 study of the safety and efficacy of pembrolizumab (pembro) in combination with dabrafenib (D) and trametinib (T) for advanced melanoma (KEYNOTE-022). J Clin Oncol 2016;34, (suppl TPS9596) [abstract]. [Google Scholar]
- Ebert PJ, Cheung J, Yang Y et al. MAP kinase inhibition promotes T cell and anti-tumor activity in combination with PD-L1 checkpoint blockade. Immunity 2016;44:609–621. [DOI] [PubMed] [Google Scholar]
- Hu-Lieskovan S, Mok S, Homet Moreno B et al. Improved antitumor activity of immunotherapy with BRAF and MEK inhibitors in BRAF(V600E) melanoma. Sci Transl Med 2015;7:279ra41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng L, Zhang DY, Eble JN. Molecular Genetic Pathology. New York, NY: Springer, 2013. [Google Scholar]
- Cheng L, Eble JN. Molecular Surgical Pathology. New York, NY: Springer, 2013. [Google Scholar]
- Siravegna G, Marsoni S, Siena S et al. Integrating liquid biopsies into the management of cancer. Nat Rev Clin Oncol 2017;14:531–548. [DOI] [PubMed] [Google Scholar]
- Wan JC, Massie C, Garcia-Corbacho J et al. Liquid biopsies come of age: towards implementation of circulating tumour DNA. Nat Rev Cancer 2017;17:223–238. [DOI] [PubMed] [Google Scholar]
- Thierry AR, Mouliere F, El Messaoudi S et al. Clinical validation of the detection of KRAS and BRAF mutations from circulating tumor DNA. Nat Med 2014;20:430–435. [DOI] [PubMed] [Google Scholar]
- Xi L, Pham TH, Payabyab EC et al. Circulating tumor DNA as an early indicator of response to T-cell transfer immunotherapy in metastatic melanoma. Clin Cancer Res 2016;22:5480–5486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynh K, Hoon DS. Liquid biopsies for assessing metastatic melanoma progression. Crit Rev Oncog 2016;21:141–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang SK, Hoon DS. Liquid biopsy utility for the surveillance of cutaneous malignant melanoma patients. Mol Oncol 2016;10:450–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago-Walker A, Gagnon R, Mazumdar J et al. Correlation of BRAF mutation status in circulating-free DNA and tumor and association with clinical outcome across four BRAFi and MEKi clinical trials. Clin Cancer Res 2016;22:567–574. [DOI] [PubMed] [Google Scholar]
- Ascierto PA, Minor D, Ribas A et al. Phase II trial (BREAK-2) of the BRAF inhibitor dabrafenib (GSK2118436) in patients with metastatic melanoma. J Clin Oncol 2013;31:3205–3211. [DOI] [PubMed] [Google Scholar]
- Calapre L, Warburton L, Millward M et al. Circulating DNA (ctDNA) as a liquid biopsy for melanoma. Cancer Lett 2017;404:62–69. [DOI] [PubMed] [Google Scholar]
- Girotti M, Gremel G, Lee R et al. Application of sequencing, liquid biopsies, and patient-derived xenografts for personalized medicine in melanoma. Cancer Discov 2016;6:286–299. [DOI] [PubMed] [Google Scholar]
- Zelboraf (vemurafenib)Summary of Product Characteristics. Welwyn Garden City, UK: Roche Registration Limited 2016.
- US Food and Drug Administration. Principles for codevelopment of an in vitro companion diagnostic device with a therapeutic product: Draft guidance for industry and Food and Drug. p1. Available from https://www.fda.gov/downloads/MedicalDevices/DeviceRegulationandGuidance/GuidanceDocuments/UCM510824.pdf (accessed 2017).
- Miller VA. FDA regulation of laboratory-developed tests. Clin Adv Hematol Oncol 2016;14:305–306. [PubMed] [Google Scholar]
- US Food and Drug Administration. Laboratory Developed Tests. p1. Available from https://www.fda.gov/MedicalDevices/ProductsandMedicalProcedures/InVitroDiagnostics/LaboratoryDevelopedTests/default.htm (accessed 2017).
- US Food and Drug Administration. CLIA and LDT FAQs. pp1–3. Available from https://www.cms.gov/Regulations-and-Guidance/Legislation/CLIA/Downloads/LDT-and-CLIA_FAQs.pdf (accessed 2017).
- Treece AL, Gulley ML, Vasalos P et al. Reporting results of molecular tests: a retrospective examination of BRAF mutation reporting. Arch Pathol Lab Med 2017;141:658–665. [DOI] [PubMed] [Google Scholar]
- US Food and Drug Administration. Discussion Paper on Laboratory Development Tests (LDTs). pp 1–12. Available from https://www.fda.gov/downloads/MedicalDevices/ProductsandMedicalProcedures/InVitroDiagnostics/LaboratoryDevelopedTests/UCM536965.pdf. (accessed 2017).
- Gatter K. FDA oversight of laboratory-developed tests: where are we now? Arch Pathol Lab Med 2017;141:746–748. [DOI] [PubMed] [Google Scholar]
- Sholl LM, Andea A, Bridge JA et al. Template for reporting results of biomarker testing of specimens from patients with melanoma. Arch Pathol Lab Med 2016;140:355–357. [DOI] [PubMed] [Google Scholar]
- Ventana Medical Systems Inc. White paper. Evaluation of BRAF (V600E) mutation by immunohistochemical staining with anti-BRAF V600E (VE1) antibody: a comparison with Sanger sequencing. Available from http://www.ventana.com/documents/N4850A_Evaluation_of_BRAF_WP.pdf (accessed 2017).
- Dunstan RW, Wharton KA Jr., Quigley C et al. The use of immunohistochemistry for biomarker assessment—can it compete with other technologies? Toxicol Pathol 2011;39:988–1002. [DOI] [PubMed] [Google Scholar]
- Armbruster DA, Pry T. Limit of blank, limit of detection and limit of quantitation. Clin Biochem Rev 2008;29(suppl 1):S49–S52. [PMC free article] [PubMed] [Google Scholar]
- Chau CH, Rixe O, McLeod H et al. Validation of analytic methods for biomarkers used in drug development. Clin Cancer Res 2008;14:5967–5976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Gramont A, Watson S, Ellis LM et al. Pragmatic issues in biomarker evaluation for targeted therapies in cancer. Nat Rev Clin Oncol 2015;12:197–212. [DOI] [PubMed] [Google Scholar]
- Colomba E, Helias-Rodzewicz Z, Von Deimling A et al. Detection of BRAF p.V600E mutations in melanomas: comparison of four methods argues for sequential use of immunohistochemistry and pyrosequencing. J Mol Diagn 2013;15:94–100. [DOI] [PubMed] [Google Scholar]
- Goodwin S, McPherson JD, McCombie WR. Coming of age: ten years of next-generation sequencing technologies. Nat Rev Genet 2016;17:333–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venter JC, Adams MD, Myers EW et al. The sequence of the human genome. Science 2001;291:1304–1351. [DOI] [PubMed] [Google Scholar]
- Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A 1977;74:5463–5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurkowska M, Gos A, Ptaszyński K et al. Comparison between two widely used laboratory methods in BRAF V600 mutation detection in a large cohort of clinical samples of cutaneous melanoma metastases to the lymph nodes. Int J Clin Exp Pathol 2015;8:8487–8493. [PMC free article] [PubMed] [Google Scholar]
- Lopez-Rios F, Angulo B, Gomez B et al. Comparison of testing methods for the detection of BRAF V600E mutations in malignant melanoma: pre-approval validation study of the companion diagnostic test for vemurafenib. PLoS ONE 2013;8:e53733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu K, Pan Q, Zhang X et al. Detection of BRAF V600 mutations in metastatic melanoma: comparison of the Cobas 4800 and Sanger sequencing assays. J Mol Diagn 2013;15:790–795. [DOI] [PubMed] [Google Scholar]
- Harrington CT, Lin EI, Olson MT et al. Fundamentals of pyrosequencing. Arch Pathol Lab Med 2013;137:1296–1303. [DOI] [PubMed] [Google Scholar]
- Agah A, Aghajan M, Mashayekhi F et al. A multi-enzyme model for pyrosequencing. Nucleic Acids Res 2004;32:e166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed GH, Kent JO, Wittwer CT. High-resolution DNA melting analysis for simple and efficient molecular diagnostics. Pharmacogenomics 2007;8:597–608. [DOI] [PubMed] [Google Scholar]
- Chen D, Wang YY, Chuai ZR et al. High-resolution melting analysis for accurate detection of BRAF mutations: a systematic review and meta-analysis. Sci Rep 2014;4:4168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirosi L, Strippoli S, Gaudio F et al. Is immunohistochemistry of BRAF V600E useful as a screening tool and during progression disease of melanoma patients? BMC Cancer 2016;16:905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anwar MA, Murad F, Dawson E et al. Immunohistochemistry as a reliable method for detection of BRAF-V600E mutation in melanoma: a systematic review and meta-analysis of current published literature. J Surg Res 2016;203:407–415. [DOI] [PubMed] [Google Scholar]
- Eriksson H, Zebary A, Vassilaki I et al. BRAFV600E protein expression in primary cutaneous malignant melanomas and paired metastases. JAMA Dermatol 2015;151:410–416. [DOI] [PubMed] [Google Scholar]
- Long GV, Wilmott JS, Capper D et al. Immunohistochemistry is highly sensitive and specific for the detection of V600E BRAF mutation in melanoma. Am J Surg Pathol 2013;37:61–65. [DOI] [PubMed] [Google Scholar]
- Pearlstein MV, Zedek DC, Ollila DW et al. Validation of the VE1 immunostain for the BRAF V600E mutation in melanoma. J Cutan Pathol 2014;41:724–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manfredi L, Meyer N, Tournier E et al. Highly concordant results between immunohistochemistry and molecular testing of mutated V600E BRAF in primary and metastatic melanoma. Acta Derm Venereol 2016;96:630–634. [DOI] [PubMed] [Google Scholar]
- Marin C, Beauchet A, Capper D et al. Detection of BRAF p.V600E mutations in melanoma by immunohistochemistry has a good interobserver reproducibility. Arch Pathol Lab Med 2014;138:71–75. [DOI] [PubMed] [Google Scholar]