Abstract
The influenza M2 protein forms an acid-activated proton channel that is essential for virus replication. The transmembrane H37 selects for protons under low external pH (pHout) while W41 ensures proton conduction only from the N-terminus to the C-terminus and prevents reverse current under low internal pH (pHin). Here we address the molecular basis for this asymmetric conduction by investigating the structure and dynamics of a mutant channel, W41F, which permits reverse current under low pHin. Solid-state NMR experiments show that W41F M2 retains the pH-dependent α-helical conformations and tetrameric structure of the wild-type channel, but has significantly altered protonation and tautomeric equilibria at H37. At high pH, the H37 structure is shifted towards the π tautomer and less cationic tetrads, consistent with faster forward deprotonation to the C-terminus. At low pH, the mutant channel contains more cationic tetrads than the wild-type channel, consistent with faster reverse protonation from the C-terminus. 15N NMR spectra allow the extraction of four H37 pKa’s and show that the pKa’s are more clustered in the mutant channel compared to wild-type M2. Moreover, binding of the antiviral drug, amantadine, at the N-terminal pore at low pH did not convert all histidines to the neutral state, as seen in wild-type M2, but left half of all histidines cationic, unambiguously demonstrating C-terminal protonation of H37 in the mutant. These results indicate that asymmetric conduction in wild-type M2 is due to W41 inhibition of C-terminal acid activation by H37. When Trp is replaced by Phe, protons can be transferred to H37 bidirectionally with distinct rate constants.
Keywords: Magic-angle-spinning NMR, tautomeric equilibrium, proton dissociation equilibrium, ion channels, gating
Introduction
Membrane-bound ion channels require specific residues and structural features to achieve ion selectivity and to control channel opening and closing. Channels can be gated, i.e. opened and closed, by membrane potential, pH, ligand, mechanical activity, and temperature [1]. The influenza A virus M2 protein (AM2) forms a proton-selective, acid-activated, and asymmetrically conducting channel in the virus envelope that is essential for the virus lifecycle [2–4]. The acid activation is achieved by a single histidine, H37, in the transmembrane (TM) domain [5, 6] while the preferential conduction of protons from the N-terminus (outside) to the C-terminus (inside) is accomplished by a single tryptophan, W41 [7, 8]. This HxxxW motif is conserved among all influenza A and B viruses’ M2 sequences [2]. Asymmetric ion conduction is also known for some voltage-gated potassium channels, which use cytoplasmic factors such as polyamines to achieve inward rectification [9–11], and in voltage-gated proton channels of the Hv1 family, which use a conserved RxWRxxR motif to achieve inward rectification [12]. While pH-gated channels are often also voltage-gated in vivo [13], and whole-cell electrophysiological measurements of M2 proton conductance are carried out under both voltage and pH control, M2 is specifically activated by low pH of the external solution [14, 15].
Because M2 is an excellent model for larger ion channels and because it is also the target of the amantadine (Amt) family of antiviral drugs [16, 17], a large number of biochemical, biophysical and spectroscopic studies have been carried out to elucidate the structural mechanisms of proton conduction of AM2 [4]. These studies showed that the conduction-relevant structure of M2 depends markedly on the membrane environment [18], but the conduction properties are insensitive to the construct length beyond the TM domain if the same membrane environment is used [19]. Several M2 constructs have been used in biophysical studies because of the modular sequence and multifunctional nature of this protein: a highly conserved and disordered N-terminal ectodomain [20] mediates protein incorporation into the virion [21] and is the target of universal influenza vaccines [22]. The TM domain (residues 22-46) is necessary and sufficient to exhibit proton conductance that is similar to full-length M2 [19]. An amphipathic helix C-terminal to the TM helix mediates ESCRT-independent membrane scission during virus budding [23–25]. Finally a disordered cytoplasmic tail [26] is involved in M1 recognition and virus assembly [27]. The proton channel function is fully encapsulated in the TM domain based on proton-current measurements in vivo and in vitro. In oocytes, an epitope-tagged TM construct exhibits drug-sensitive single-channel activity that is indistinguishable from that of full-length (FL) M2 due to the low expression level of TM channels in the oocyte, and in lipid vesicles, the TM and FL protein exhibit the same drug-sensitive and pH-dependent proton conductance [19]. Molecular dynamics simulations [28] showed the same rate constants of proton conduction and gating between M2-TM and another commonly studied construct, M2-CD [29], which includes both the TM helix and the amphipathic helix and which exhibits similar chemical shifts as M2-FL [30]. Thus, the TM domain is sufficient and necessary for studying the mechanism of pH-dependent asymmetric conduction in M2.
While the conduction-relevant structure and dynamics of M2 is insensitive to the construct length, they depend markedly on the lipid composition of the membrane [18]. Higher membrane fluidity and negatively charged lipids favor H37 protonation [31–33]. Cholesterol promotes the α-helical conformation [34], immobilizes tetramer rotational diffusion [35], and stabilizes tetramer assembly [36, 37]. Membrane thickness affects the tilt angle of the TM helix [33, 38–40], and negative-curvature lipids can alter the tetrameric assembly of the protein [41]. Therefore, any structure-function studies of M2 by comparing wild-type and mutant sequences must be conducted in the same phospholipid membranes.
Solid-state NMR spectroscopy of AM2 bound to phospholipid bilayers has provided significant information about the proton conduction mechanism. It is now known that the H37 sidechain shuttles protons into the virion [42] by repeated protonation and deprotonation, as manifested by imidazole 15N chemical shift averaging at physiologically relevant acidic pH [43]. This proton shuttling is accompanied by microsecond-timescale imidazole ring reorientations, as seen by motionally averaged dipolar couplings [44], and by water-imidazole hydrogen bonding, as shown by 1H-15N correlation spectra [45, 46]. The protonation equilibria of the H37 tetrad have been measured in different membranes in the absence and presence of the cytoplasmic domain [31, 43, 47, 48]. While the exact proton-dissociation equilibrium constants depend on the membrane composition, all data indicate that the third protonation event, which gives rise to the +3 charged tetrad, correlates with channel activation. Amantadine binding to the channel lumen near Ser31 [17, 49, 50] prevents H37 protonation [44] and shifts the TM structural equilibrium towards the state that is populated at high pH [51, 52]. Further, W41 shows pH-dependent motion and proximity to H37: the indole ring acquires larger-amplitude motion at low pH and approaches the H37 imidazolium rings more closely [53]. The resulting cation-π interaction was proposed to explain why proton flux into the virion is much smaller than the His-water proton exchange rate [29]. Similar aromatic interactions have been proposed for the Hv1 channel between W207 and R211 [12], where W207 controls the gating kinetics, conduction activation energy, and the pH dependence of gating.
Although these studies have given rich insight into the acid activation and proton transfer mechanisms of AM2, the structural basis for asymmetric conduction, i.e. why outward proton conduction is prohibited even under the condition of low inside pH (pHin), is not yet well understood. Mutagenesis data suggested that this asymmetric conduction may be caused by pore-obstructing sidechain conformation of W41 [6, 7], which is stabilized by D44 [8]. Recent multiscale molecular dynamics (MD) simulations suggest that under low pHin and high pHout, reverse proton current is blocked by steric hindrance and desolvation penalty at W41 [28]. However, high-resolution crystal structures and SSNMR data show a t90 sidechain conformation of W41 [53, 54], which does not completely occlude the pore, and water cross peaks with C-terminal residues have been observed [55], indicating that the C-terminal region of the channel is hydrated. These results raise the question as to whether protons from the C-terminus may in fact protonate H37 at the atomic level in the presence of W41, but this protonation may have escaped detection. Due to the symmetric pH of phospholipid membranes employed in structural studies, it is so far not possible to uniquely attribute H37 protonation to the N-terminus when both sides of the membrane are acidic. This symmetric pH also makes it difficult to determine whether the TM conformation of AM2 differs between opposite proton concentration gradients as used in electrophysiological measurements, i.e. between low pHout/high pHin on the one hand and high pHout/low pHin on the other.
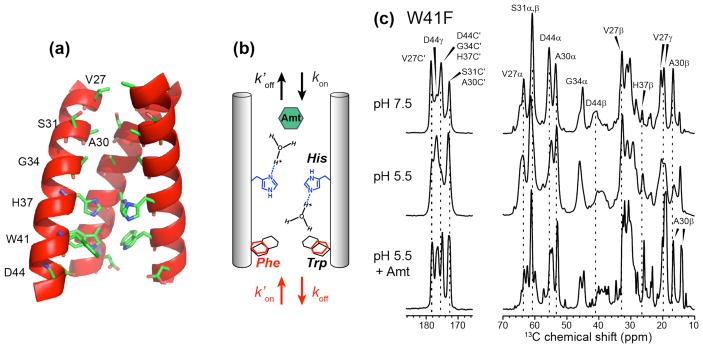
To better understand the structural basis for asymmetric conduction in this canonical proton channel, we have now investigated the conformation and proton-transfer dynamics of a W41F mutant of AM2 using solid-state NMR (Fig. 1a, b). Mutagenesis data show that substitution of the indole by the phenylene ring at residue 41 does not affect the tetrameric nature of the channel but changes proton conductance: the forward conductance increases by ~60% compared to the wild-type channel and reverse proton conductance becomes detectable. Under low pHin the reverse current is 25% of the forward current found under low pHout [6, 7]. The replacement of Trp by Phe is appealing for structural studies because by uncoupling the H37 tetrad from the W41 tetrad, we can separately probe the atomic processes of acid activation and asymmetric conduction.
Figure 1.
(a) Wild-type AM2-TM peptide structure (PDB: 3LBW) showing the locations of key residues examined in this study. One of the four chains is omitted for clarity. (b) Schematic of the channel topology, showing four possible rate constants for H37-mediated proton conduction: forward protonation (kon), forward deprotonation (koff), reverse protonation ( ), and reverse deprotonation ( ). Mutation of W41 changes the magnitude of these rate constants. (c) 13C MAS spectra of W41F AM2-TM bound to the VM+ membrane at pH 7.5, pH 5.5, and pH 5.5 with bound amantadine (Amt). The spectra were measured at 273 K.
We use the TM peptide of AM2 in this study to compare the W41F mutant structure with the large body of structural information available on the wild-type (WT) TM peptide. The TM peptide reproduces key channel properties of full-length M2 and at the same time is sufficiently small to allow the incorporation of site-specifically labeled residues using synthetic chemistry. We reconstitute the W41F mutant in a cholesterol-containing virus-mimetic membrane, which is the same membrane used in many previous studies of the proton conduction mechanism of WT M2-TM. This allows us to directly compare spectral differences between the two constructs. Using solid-state NMR, we show that the W41F mutant retains the α-helical conformation and tetrameric assembly of the channel, but has significantly different H37 protonation and tautomeric equilibria. Moreover, by binding Amt to the N-terminal pore at low pH, we have engineered a novel reverse proton concentration gradient, with low pHin and high pHout. In this drug-bound W41F channel, we found both cationic and neutral histidines, in contrast to the WT peptide, which exhibits only neutral histidines upon drug binding. These results show that asymmetric conduction is due to W41 inhibition of protonation of H37 from the C-terminus. Without the indole at residue 41, H37 can be protonated from both directions with distinct rate constants.
Results
The W41F mutant preserves the conformational distribution and tetrameric structure of the wild-type TM domain
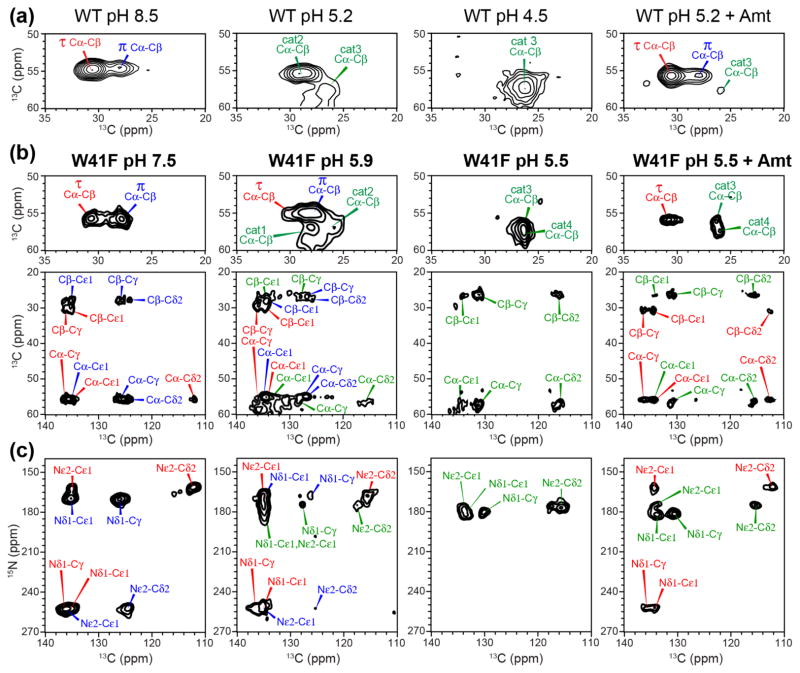
We first investigated the effects of W41F mutation on the global conformation of the TM domain by measuring chemical shifts. Fig. 1c shows 1D 13C MAS spectra of W41F-AM2 bound to a virus-mimetic membrane, VM+ [32], at high and low pH. Chemical shift assignment of the six 13C, 15N-labeled residues is obtained from 2D 13C-13C and 15N-13C correlation spectra (Fig. 2). At high pH, a single set of narrow peaks is observed for V27, A30, and D44, while H37 exhibits two sets of peaks, which can be assigned to τ and π tautomers based on the aromatic chemical shifts (vide infra). 15N-13C correlation spectra indicate the coexistence of a major and a minor conformer for S31 and G34 (Fig. 2a), with similar chemical shifts as in WT AM2 [33]. These two sets of chemical shifts can be assigned to a kinked Cclosed conformation for the major species and a straight Copen conformation for the minor species based on chemical shift comparisons with the WT peptide and the high-resolution crystal and SSNMR structures of AM2-TM solved under varying pH and membrane-mimetic environments [49, 54, 56]. The high-pH Cclosed conformation contains a helical kink at G34, causing the C-terminal half of the TM helices to be tightly packed [51], while the low-pH Copen conformation has relatively straight helices, facilitating proton release to the C-terminus [33, 57, 58]. Decreasing the pH to 5.5 broadened the resonances, indicating a larger conformational distribution (Fig. 2b). Two A30 Cα-Cβ cross peaks can be resolved at low pH, and the relative populations of the two S31 and G34 states have changed, with the Copen intensities dominating the Cclosed signals. The low-pH induced change of the backbone conformational equilibrium of W41F M2 is similar to the behavior of the WT peptide, indicating that the mutant retains the pH-dependent conformational distributions of the WT peptide, with acidic pH favoring the Copen conformation and high pH favoring the Cclosed state. This also indicates that Trp at residue 41 is not required for the peptide to adopt the Cclosed state, and that the W41F mutation does not cause a constitutively Copen conformation.
Figure 2.
Aliphatic regions of 2D 13C-13C (top) and 15N-13C (bottom) correlation spectra of W41F AM2-TM at (a) pH 7.5, (b) pH 5.5, and (c) pH 5.5 with bound Amt. Assignments for H37 cross peaks are shown in red, blue and green for the τ tautomer, π tautomer, and cationic states, respectively.
To determine whether the W41F mutation perturbs the tetrameric assembly of the channel, we carried out 19F CODEX experiments using 4-19F-Phe41-labeled W41F. Mono-substitution of a C-H bond by a C-F bond on the aromatic ring has little steric effect on proteins [59], but the electron-withdrawing fluorine makes the electrostatic potential of the phenylene π-face less negative. This has been calculated to reduce the cation-π binding energy by ~20% compared to the unsubstituted phenylene ring [60]. However, since the W41F mutation abolishes the asymmetric conductance of the channel, even unsubstituted Phe41 is not expected to have cation-π interactions with His37. Thus the para-fluorine substitution should have minimal perturbation on the mutant structure compared to unsubstituted Phe41-M2. This is also consistent with the large body of literature showing minimal effects of mono-fluorinated aromatic rings to protein structure [61].
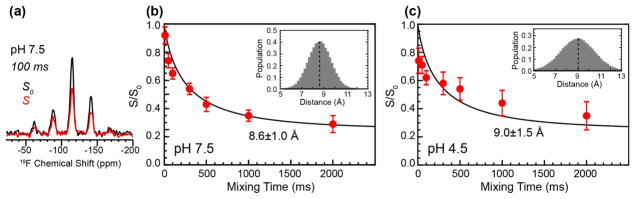
The 19F CODEX experiment measures the oligomerization state and intermolecular distances by detecting the equilibrium intensity of a stimulated echo [62, 63]. For an oligomeric membrane protein with n subunits, the equilibrium echo intensity, measured as the intensity ratio between an exchange experiment (S) and a T1-controlled experiment (S0), is 1/n. The mixing-time dependence of the intensity decay to equilibrium gives information on the inter-subunit distances [64, 65]. To obtain high sensitivity and resolution, we carried out these 19F CODEX experiments under 10 kHz magic-angle-spinning (MAS), which is faster than used in previous experiments. At this MAS frequency, the overlap integral, which is necessary for extracting distances from the spin diffusion data and which is MAS-frequency dependent, was found from model compound experiments (Fig. S1) to be 41 μs, similar to the previously measured value of 37μs [65].
Fig. 3 shows representative 19F control and exchange spectra of membrane-bound 4-19F-Phe41 labeled W41F M2 and the resulting CODEX decay curves for samples at pH 7.5 and pH 4.5. The exchange intensity equilibrates to ~0.25 for both samples, indicating that the W41F mutant retains the tetrameric structure. The best single-distance fit for the decay curves gives a nearest-neighbor distance of 8.6 Å at pH 7.5 and 9.0 Å at pH 4.5, assuming a symmetric tetramer. However, single-distance fits do not fully reproduce the more rapid initial decay compared to the long-time behavior. Thus, we used a Gaussian distribution model to fit the data. With this model, the high pH sample shows a distance distribution of σ=1.0 Å centered at 8.6 Å while the pH 4.5 sample shows a distance distribution of σ=1.5 Å centered at 9.0 Å. Therefore, the low-pH mutant is moderately expanded at the C-terminus compared to the high-pH mutant channel and also has a larger distribution of the interhelical packing. These results are in good agreement with the inter-helical distances measured for 5-19F-Trp41 in wild-type M2TM, which found nearest-neighbor distances of 11.4 Å at high pH and 12.4 Å at low pH [53]. These intermolecular 19F-19F distances depend on both the backbone helix orientation and the sidechain rotameric conformation; the latter depends upon the χ1 angle for 4-19F-Phe41 and (χ1, χ2) angles for 5-19F-Trp. For wild-type M2 harboring Trp41, helix orientation measurements by NMR [38, 57] and crystal structures [54, 66] have independently given high-resolution structures of the helix backbone at high and low pH, thus the 5-19F Trp41 inter-helical distances allowed the Trp41 rotamer to be determined, and the (χ1, χ2) angles were found to be modestly different (by ~20°) between high and low pH [53, 67]. For the W41F mutant, the backbone helix orientation is not known independently, but it is reasonable to assume that the 19F distance difference between high and low pH reflects a similar increase in the helix tilt angle to produce a more expanded channel [28, 68] at low pH.
Figure 3.
19F CODEX data of 4-19F-Phe41 in W41F AM2-TM at pH 7.5 and pH 4.5. (a) Representative 19F CODEX S0 and S spectra at a mixing time of 100 ms for the pH 7.5 sample. CODEX intensities as a function of mixing time are shown for the pH 7.5 sample (b) and for the pH 4.5 sample (c). Best fits used a Gaussian distribution of distances (inset). The mean and standard deviation of the distributions are 8.6 Å and 1.0 Å for (b) and 9.0 Å and 1.5 Å for (c).
The W41F mutation inverts the H37 tautomer equilibrium at high pH and increases the populations of highly cationic tetramers at low pH
Imidazole 13C and 15N chemical shifts provide detailed information about the histidine sidechain structure, and allow us to probe the effects of the W41F mutation on the protonation and tautomeric equilibria of H37. In WT AM2, 13C and 15N chemical shifts indicate the presence of both neutral tautomers at high pH, with the Nε2-protonated τ tautomer dominating over the Nδ1-protonated π tautomer by a 3 : 1 ratio [43, 44] (Fig. 4a). Interestingly, the W41F mutant shifted the tautomeric equilibrium towards the π form (Fig. 4b, c). Two Cα-Cβ peaks are resolved; the upfield and downfield Cβ peaks correlate with aromatic 13C chemical shifts that are indicative of the π and τ tautomers, respectively (Table S1). 15N-13C correlation spectra confirm the assignments by correlating the unprotonated 15N peak at 252 ppm with either Cδ2 of the π tautomer or Cγ of the τ tautomer. Overall, the mutant displays the same ensemble of chemical shifts as in WT AM2, but the π tautomer is the major species, with a τ : π population ratio of 2 : 3 based on the average intensities of Cα-Cβ, Nε2-Cδ2, and Nδ1-Cγ cross peaks.
Figure 4.
H37 chemical shifts in WT (a) and W41F (b, c) AM2-TM from 2D 13C-13C (a, b) and 15N-13C (c) correlation spectra as a function of pH. (a) H37 Cα-Cβ regions of the 2D 13C-13C spectra of the WT peptide. (b) H37 Cα-Cβ and aliphatic-aromatic regions of the 2D 13C-13C spectra of the W41F mutant. (c) Aromatic region of the 2D 15N-13C correlation spectra of the W41F mutant. The pH and drug binding state of the samples are indicated. The mutant channel shows higher π tautomer intensities at high pH and more cationic histidine peaks at low pH compared to the WT channel.
Decreasing the pH to 5.9 caused a mixture of neutral and cationic His peaks in W41F (Fig. 4b, c), with the π tautomer remaining as the dominant neutral species. Decreasing the pH further to 5.5 converted most H37 to the cationic state. Specifically, the Cα and Cβ chemical shifts match the previously assigned values for the +3 and +4 charge states of WT AM2, but the WT peptide exhibits these chemical shifts at a much lower bulk pH of 4.5. In other words, at low pH, the W41F mutant shifts the protonation equilibrium towards more cationic tetrads compared to the WT channel. Previous studies of AM2-TM indicated a stabilizing cation-π interaction between H37 and W41 [53], thus the W41F mutation might have been expected to destabilize cationic H37 at low pH. The fact that the opposite is observed, with higher populations of +3 and +4 states in the mutant than in WT M2, thus means that the cation-π interaction is less important than C-terminal water accessibility in controlling the protonation equilibria of H37. The increased cationicity of the H37 in the mutant is in excellent agreement with electrophysiology data, which show that the W41F mutant has larger proton conductance than the WT channel between pH 5 and 6 [6, 7] despite the loss of the cation-π interaction.
The W41F mutation clusters the proton-dissociation constants of H37
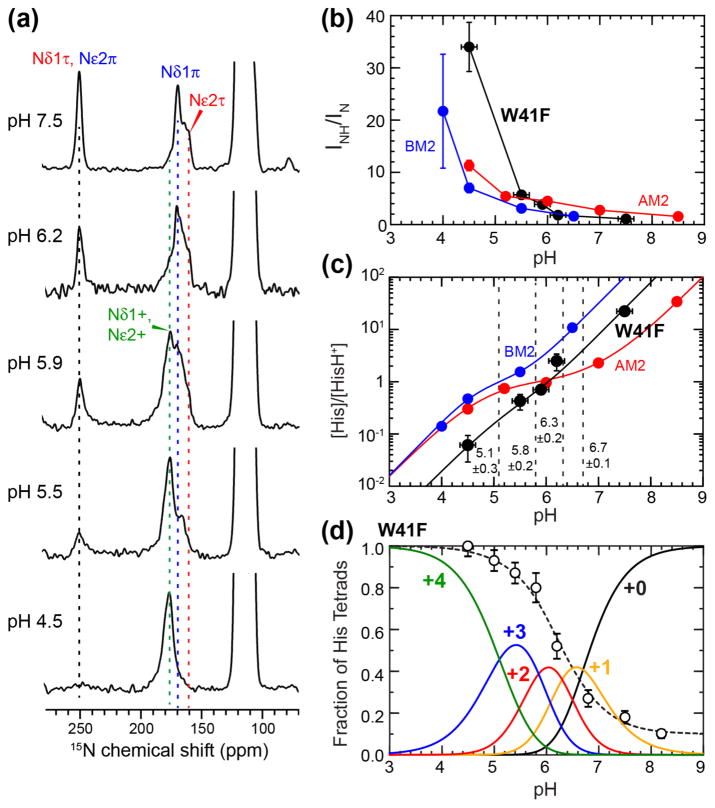
The 13C and 15N chemical shifts indicate altered H37 protonation equilibria in the W41F mutant compared to the WT channel. To quantify this equilibrium change, we measured 15N spectra of the H37 imidazole ring as a function of pH. The experiments were conducted at 243 K to suppress proton transfer dynamics, so that the 15N peak intensities reflect the relative populations of neutral and cationic histidines (Fig. S2). Although histidine and buffer ions have temperature dependent pKa’s, these W41F samples were measured under the same membrane composition, pH, and buffer conditions as wild-type AM2-TM and BM2-TM, thus the correction factors for the temperature dependence should be similar (see Materials and Methods), allowing us to compare the protonation equilibria of the different samples consistently.
The H37 tetrad has a maximum of four distinct pKa’s, corresponding to sequential transitions from the +0 to +4 charge states. Fig. 5a shows that, with decreasing pH, the 252-ppm unprotonated 15N peak intensity decreases while the 160–190 ppm protonated 15N peaks increase in intensity, as expected as the H37 tetrad becomes progressively protonated. The ratio of the protonated to unprotonated 15N intensity bands depends on the relative populations of neutral and cationic histidines. This ratio has a steeper rise at low pH for the W41F mutant than for WT AM2 and BM2 (Fig. 5b) [43, 69], indicating that the mutant has higher concentrations of cationic histidines at acidic pH. This increased charge state at low pH parallels the pH-dependent proton conductance of W41F measured in electrophysiological experiments [6, 7]. Using these 15N peak intensities, we calculated the concentration ratio of neutral to cationic histidine (Fig. 5c, Table S2), from which the pKa’s were extracted and found to be 6.7±0.1, 6.3±0.2, 5.8±0.2, and 5.1±0.3, where the standard deviations are obtained from a reduced χ2 analysis (Fig. S3 and Materials and Methods). Two adjacent pKa’s in this series have moderately negative correlations, with Pearson product-moment correlation coefficients of −0.36 to −0.41 (Table S3). In comparison, wild-type AM2-TM has little correlation among the three highest pKa’s and only negative correlation between the two lowest pKa’s.
Figure 5.
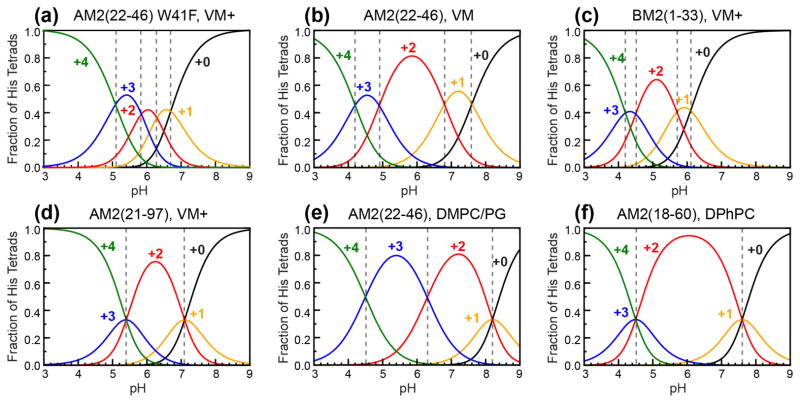
Quantification of H37 pKa’s in membrane-bound W41F AM2-TM by 15N NMR. (a) pH-dependent 15N spectra of H37 sidechains measured at 243–248 K. Assignment of protonated and unprotonated 15N peaks is based on 2D correlation spectra. (b) Integrated intensity ratios of protonated and unprotonated imidazole nitrogens as a function of pH. W41F AM2-TM (black) shows higher protonated nitrogen intensities at low pH compared to previously measured WT AM2-TM (red) [43] and BM2-TM results (blue) [69]. (c) Neutral-to-cationic histidine concentration ratios as a function of pH. Best fit of the data yield four pKa’s for W41F AM2-TM, which are indicated by dashed vertical lines. (d) Population distributions of charged tetrads of W41F AM2-TM. The +2 charged channel has the lowest population among the five charge states. Normalized proton conductance of W41F M2 from the literature [7] (open circles) is fit using the charge-state distribution to estimate the relative conductance of the different charge states.
Interestingly, this H37 pKa range of the W41F mutant is narrower than that of the WT channel, whose pKa’s span the range of 7.6 to 4.2 [43] (Table 1). In other words, the W41F channels are less cationic than the WT channels at high pH but more cationic at low pH, consistent with the 2D correlation spectra. Since the pKa is the pH value at which two adjacent charge states have equal populations, the elevation of the lowest pKa for the W41F mutant means that the +3 and +4 channels are equally populated at pH 5.1, while WT AM2-TM requires a lower pH of 4.2 to reach equimolar concentrations of the +3 and +4 charge states.
Table 1.
Comparison of H37 pKa’s in influenza A and B M2 proteins bound to various lipid membranes.
| M2 | Lipid membranes | pKa’s | Average pKa |
|---|---|---|---|
| W41F AM2(22-46) | POPC, POPE, SM, cholesterol | 6.7, 6.3, 5.8, 5.1 | 6.0 |
| AM2(22-46) | DPPC, DPPE, SM, cholesterol | 7.6, 6.8, 4.9, 4.2 | 5.9 |
| AM2(22-46) | DMPC, DMPG | 8.2, 8.2, 6.3, < 5.0 | < 6.9 |
| AM2(21-97) | POPC, POPE, SM, cholesterol | 7.1, 5.4 | 6.3 |
| AM2(18-60) | DPhPC | 7.6, 4.5 | 6.1 |
| BM2(1-33) | POPC, POPE, SM, cholesterol | 6.1, 5.7, 4.5, 4.2 | 5.1 |
From the pKa values, we calculated the charge state distributions of W41F channels as a function of pH (Fig. 5d). It can be seen that the +3 state dominates in the physiologically relevant acidic pH range of 5–6, while the +2 state, which has a high population in all other M2 peptides studied so far [31, 43, 47, 48, 69] (Fig. 6), is significantly suppressed. This is a manifestation of the clustering of the second and third protonation events, with pKa’s of 6.3 and 5.8. Since the +2 channel is known to be insufficient for channel activation [43, 47], the reduction of the +2 tetrad population and the increase of the +3 tetrad population in W41F explain the higher proton conductance of the mutant compared to the WT channel. The population distributions quantified from the pKa measurement at 243 K are in good qualitative agreement with the H37 chemical shifts seen in 2D correlation spectra measured at 273 K, indicating that the H37 protonation equilibria are similar in this experimental range.
Figure 6.
Comparison of pH-dependent populations of the five charge states of various M2 samples studied so far. (a) W41F AM2(22-46) in the VM+ membrane from this study. (b) WT AM2(22-46) in the VM membrane [43]. (c) BM2(1-33) in the VM+ membrane [69]. (d) AM2(21-97) in the VM+ membrane [31]. (e) AM2(22-46) in the DMPC/DMPG membrane [47]. (f) AM2(18-60) in the DPhPC membrane [48]. Dashed vertical lines indicate the intersection of two adjacent population curves, which correspond to the pKa’s.
The charge-state distribution further allows us to estimate the relative conductance of the differently charged channels by fitting the pH-dependent proton conductance of W41F [7]. We found that the +0, +1, +2, +3 and +4 charge states of the W41F tetramers have relative unitary conductance of 0.1 : 0.4 : 0.6 : 0.9 : 1.0. The similar conductance of the +3 and +4 channels is qualitatively different from the trend of WT AM2-TM channels, whose +3 state has 3- and 10-fold larger conductance than the +2 and +4 states, respectively [43].
Drug-bound W41F channel retains cationic histidines
WT AM2 is inhibited by the antiviral drug Amt, which binds to the N-terminal pore [32, 49, 50, 66, 70] to dehydrate the channel and prevent H37 protonation from the N-terminus. If the W41F mutant allows C-terminal proton transfer, then H37 would be expected to be accessible to protons even when the drug is bound at the N-terminal pore. To test this hypothesis, we measured 13C and 15N chemical shifts at low pH in the presence of drug. Four-fold excess drug was added to the membrane-bound peptides to saturate the channels. 1D and 2D spectra (Fig. 1c, 2c) show that Amt caused significant line narrowing and chemical shift perturbations. The peaks corresponding to Cclosed and Copen conformations, seen respectively at high and low pH in the apo samples, now appear with similar intensities in the drug bound sample. This is especially prominent for S31 and G34, which show two equal-intensity peaks in the 15N-13C correlation spectra, indicating a more bimodal distribution of the TM helix conformation (Fig. 2c). Three V27 N-Cα cross peaks can be resolved, with intensity ratios of 1 : 1 : 2. Close inspection of the drug-bound spectra reveals that the chemical shifts are similar but not identical to those of the apo spectra [49, 50, 71, 72]. For example, the two A30 Cβ chemical shifts are perturbed from the chemical shifts of the high and low pH apo samples (Fig. S4), and the two S31 N-Cα cross peaks have slightly different 15N chemical shifts from the apo values at high and low pH. These observations confirm that both the Cclosed and Copen conformations contain bound drugs.
Interestingly, drug binding to W41F at pH 5.5 created equal populations of the neutral τ tautomer and cationic histidines (Fig. 4b, c), as seen by the cross peak intensities in the 2D 13C-13C and 15N-13C correlation spectra. This differs from the apo W41F channel at the same pH, which is dominated (> 90%) by cationic histidine, and with drug-bound WT channel at low pH, which is dominated (> 90%) by neutral histidine. The fact that the drug-bound W41F channel retains some cationic H37 is unique to the mutant, since low-pH drug binding to the WT channel largely converts H37 to the neutral state. The latter can be understood because protons can neither access H37 from the N-terminus (due to drug blockage) nor the C-terminus (due to W41 blockage) in the drug-bound WT channel. Thus, the retention of cationic His in the drug-bound mutant channel definitively demonstrates the presence of C-terminal protonation of H37, as enabled by F41. Amt binding to both the mutant and WT channels at low pH is also consistent with electrophysiological data [17] and with the fact that the virus is inhibited by the drug at low pH.
Water accessibility and H37-water proton exchange of the W41F mutant channel
While H37 shuttles protons in M2 channels, water relays protons to His at the N-terminus and receives protons from His at the C-terminus. Thus, water-His proton exchange is integral to the proton conduction mechanism. To investigate channel hydration in W41F AM2-TM, we measured 2D water-peptide 1H-13C 2D correlation spectra at two mixing times, 4 ms and 100 ms (Fig. S5). The ratio of the two spectra (S/S0) gives information about the water accessibility of the various residues. Fig. 7 compares the S/S0 values of Cα and Cβ atoms of residues in W41F and WT AM2-TM to show the relative hydration of the two channels. Both peptides show larger S/S0 values at low pH than at high pH, indicating more hydrated channels at low pH. This is consistent with the increased diameter of the low-pH channel measured by the 19F CODEX experiment (Fig. 3). The S/S0 values also show residue-specific differences: pore-facing residues exhibit higher water-transferred intensities than lipid-facing residues [55]. Interestingly, the W41F mutant shows lower S/S0 values than the WT peptide at high pH and approximately the same S/S0 values at low pH, indicating lower average hydration at high pH for the mutant while similar hydration as the WT at low pH. More importantly, the mutant channel at low pH show similar water accessibilities for the C-terminal residues as the N-terminal residues, while the WT channel has decreasing water accessibilities from the N-terminus to the C-terminus. Thus, the W41F mutation preferentially increases C-terminal hydration at low pH. These results are consistent with the 2D correlation spectra (Fig. 4) that cationic H37 is favored in the W41F mutant than the wild-type peptide. The fact that the mutant channel is less hydrated at high pH compared to the WT is also consistent with the observation that at high pH, the H37 equilibrium is shifted towards the neutral form. We note that the lower average hydration of the mutant is not necessarily in conflict with the increased proton transfer kinetics (vide infra) from C-terminal water to H37, since water may be more dynamic in the mutant channel to facilitate H37-water proton transfer [56].
Figure 7.
Hydration of the W41F AM2-TM channel probed by 2D 1H-13C correlation spectra. (a) Cα S/S0 values of W41F M2. (b) Cα S/S0 values of WT M2. (c) Cβ S/S0 values of W41F M2. (d) Cβ S/S0 values of WT M2. Green squares, red diamonds, and black circles represent low pH, drug-bound low pH, and high pH data, respectively. Open symbols indicate values with greater uncertainty. Dashed green and black lines indicate the mean S/S0 value for low and high pH, respectively. The W41F mutant has lower average hydration than the WT channel at high pH and similar average hydration as the WT at low pH. However, the C-terminus of the WT peptide is less hydrated than the N-terminus at low pH, while the W41F mutant shows similar hydration of the two termini at low pH.
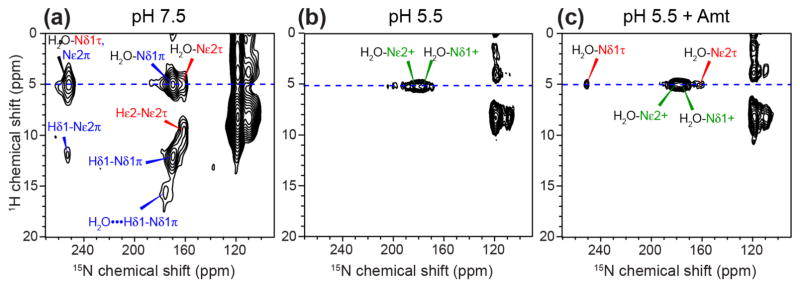
Water–H37 proton exchange in WT AM2-TM has been observed in 2D 1H-15N correlation spectra [45, 46]. To characterize this exchange process in the mutant and compare it with the WT channel, we measured 2D 1H-15N correlation spectra of W41F AM2-TM at 263 K (Fig. 8). All three samples show water 1H cross peaks at ~5 ppm with imidazole nitrogens, indicating His-water proton exchange, but the exchange rates differ. At high pH, the water cross peaks coexist with one-bond aromatic cross peaks, including Hε2-Nε2 of the τ tautomer, Hδ1-Nδ1 of the π tautomer, and a three-bond Hδ1-Nε2 cross peak of the π tautomer (Fig. 8a, S6). This coexistence indicates that His-water proton exchange at 263 K is significantly slower than the 1H chemical shift difference of ~4000 s−1 at the 1H Larmor frequency of 800 MHz used for these experiments. In addition, a (16 ppm, 175 ppm) cross peak is observed and can be assigned to a hydrogen-bonded Hδ1 correlated with the Nδ1 of the π tautomer. The lack of a second 15N cross peak at the same 1H chemical shift rules out another imidazole nitrogen as the hydrogen-bonding partner, but instead attributes the partner to water.
Figure 8.
2D 1H-15N correlation spectra of H37 in W41F AM2-TM at (a) pH 7.5, (b) pH 5.5, and (c) pH 5.5 with bound Amt. Water 1H cross peaks at 5.0 ppm with H37 imidazole nitrogens are detected in all three samples. A 16-ppm 1H chemical shift in the pH 7.5 sample can be assigned to the π tautomer with a strong hydrogen bond to water. The absence of HN cross peaks at pH 5.5 indicates rapid exchange of H37 with water at 263 K. Drug binding retains only water-H37 cross peaks, indicating fast exchange with C-terminal protons even when the N-terminus is blocked by drug.
Decreasing the pH to 5.5 suppressed the imidazolium N-H cross peaks in the spectrum while retaining the water-imidazolium cross peaks (Fig. 8b), indicating that the His-water exchange rate is much faster than 4000 s−1 at this pH, leading to a population-averaged 1H chemical shift of water. Interestingly, upon drug binding, the neutral histidines that exist under this condition still lack the one-bond N-H cross peaks (Fig. 8c), indicating that water-His proton exchange is fast for both neutral and cationic histidines even in the presence of drug. Since all channels are saturated by drug under the condition of our experiment, the presence of water-His cross peaks in this drug-bound sample provides further evidence for C-terminal protonation of H37, consistent with the increased C-terminal hydration of the channel at low pH.
Discussion
These data on the W41F mutant provide insights into the mechanism of asymmetric proton conductance of M2 as well as the dependence of drug binding on pH and helix conformation. Our study employs the TM peptide reconstituted into the same eukaryotic lipid mixture that has been used for studying WT AM2 and BM2 [32, 35], which enables us to compare the structure and dynamics of the mutant with the wild-type rigorously for this conformationally plastic protein and in addition allows us to translate the structural findings to full-length M2 for which the TM peptide is a fully functional analog.
Kinetic interpretation of the proton conduction equilibria of the W41F mutant and its implication on the asymmetric conductance of wild-type Trp41-M2
The observed pH-dependent H37 structural equilibria in W41F M2 can be understood by considering the proton-dissociation equilibrium constants in terms of rate constants of deprotonation and protonation. The channel can in principle conduct protons in the forward (N to C terminus) and reverse (C to N terminus) directions, with two rate constants in each direction. We define the protonation and deprotonation rate constants for the forward conduction as kon and koff and for the reverse conduction as k′on and k′off. Using the neutral to +1 charge state transition as an example, we can write the equilibrium reaction as
| (1) |
Assuming that protein conformational changes are not rate limiting, which is a reasonable assumption based on MD simulations [68], and the system is at equilibrium, then d[HisH+]/dt = 0, which leads to
| (2) |
Rearranging the above equation, we obtain
| (3) |
For structural studies, the proton concentrations are identical on the two sides of the membrane, . Thus, equation (3) simplifies to:
| (4) |
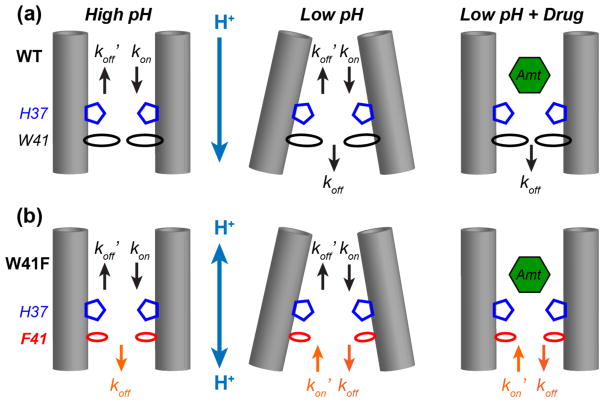
For the WT channel at high pH, forward protonation of H37 with a rate constant kon and reverse deprotonation with a rate constant are the two main pathways (Fig. 9a), since the forward protonation and reverse deprotonation free energy barriers in the N-terminal half of the TM helix are much lower than the forward deprotonation and reverse protonation barriers [28]. Thus the equilibrium constant simplifies to
Figure 9.
Schematic models of H37 protonation and deprotonation rate constants in WT (a) and W41F (b) M2 channels. Left: high pH; Middle: low pH; Right: low-pH with bound drug. For each scenario, the protonation equilibrium of the W41F mutant channel has an additional rate constant (orange) at the C-terminus compared to the WT channel. This suppresses the high pKa’s and increases the low pKa’s in the mutant compared to the WT channel, causing more clustered protonation events in the mutant.
| (5) |
Structurally, it is known that the neutral imidazole at high pH is oriented with Nδ1 pointing towards the N-terminus and Nε2 towards the C-terminus [44, 54, 56], and ring reorientation is absent [44]. Therefore, the N-terminus-facing Nδ1 is the site of both protonation and deprotonation in the WT channel at high pH. Forward protonation of Nδ1 gives rise to cationic His while reverse deprotonation gives rise to the τ tautomer. As a result, the small percentage of π tautomer in the high-pH WT channel indicates that the number of forward deprotonation events is low. This is consistent with functional data, which shows more rapid protonation rates from the viral exterior relative to the proton transmission rates to the viral interior [6], as well as with the high barrier of forward deprotonation from H37 [28].
The W41F mutation causes the high-pH channel to exhibit increased π tautomer population, which is consistent with increased forward deprotonation rate constant koff, suggesting that the mutant channel has increased solvent accessibility at the C-terminus. The equilibrium constant increases to
| (6) |
stabilizing neutral histidines (Fig. 9b, Table S4). The higher proton conductance towards the C-terminus at high pH is in excellent agreement with electrophysiology data, which show that the W41F mutant has five-fold higher normalized proton conductance than the WT channel between pH 8 and 6 [6, 7].
As the pH decreases, the WT channel can dissociate protons in both the forward and reverse directions, with the resulting equilibrium constant . In comparison, the W41F mutant can additionally allow reverse protonation due to increased solvent accessibility at the C-terminus, thus adding a term in the denominator,
| (7) |
The mutant equilibrium constant is thus lower than the WT channel at low pH, favoring more cationic tetrads. The forward deprotonation rate constant koff for the WT channel at low pH is likely different from the forward deprotonation rate constant of the mutant, , due to potential differences in free energy barriers [28, 73], but the direction of change between the two channels should be dominated by the larger total proton association rate constant of the mutant. Evidence for kon’ being of similar magnitude to kon at low pH in the W41F mutant is seen in the similar water-H37 exchange rates in the drug-bound and apo samples at pH 5.5.
The retention of cationic His in the drug-bound mutant channel definitively demonstrates the presence of C-terminal protonation of H37 that is enabled by F41 (Fig. 9b). The equilibrium constant of drug-bound channels can be expressed as for the WT channel, where ε is a small number indicating the residual forward protonation rate constant in the presence of drug. For the mutant, decreases to because of the reverse protonation pathway. Since kon’ is much larger than ε, the drug-bound equilibrium constant is smaller than that of the WT, shifting H37 to the cationic state in the mutant. In principle, amantadine can bind to each protonation state of the channel with different affinities and can perturb each of the four pKa’s differently. Here we do not attempt to extricate the complex interplay between the drug binding equilibria and the H37 protonation equilibria. Under the four-fold excess drug used in this study, the drug-binding equilibrium is shifted predominantly to the bound state for both the Copen and Cclosed conformations (Fig. 2c, Fig. S4), thus we can consider the direction of change in the H37 protonation equilibria. Together, the above analysis shows that this kinetic framework can account for all observed H37 pKa changes and tautomer equilibrium changes between the mutant and WT channels under both apo and drug-bound conditions.
While proton accessibility and proton transfer kinetics explain the H37 structural equilibria in the mutant, the Copen and Cclosed conformational distribution of the TM helix, as reported by the chemical shifts of key residues such as G34, S31 and H37, appear to be partly independent features. The mutant exhibits a similar Copen/Cclosed distribution as WT M2 (Fig. 2), even though the protonation equilibria differ. In the drug-bound W41F channel, both the neutral H37 peaks associated with the Cclosed conformation and the cationic H37 signal associated with the Copen conformation exhibit only cross peaks with water and no other cross peaks (Fig. 8c), indicating that the Copen and Cclosed conformations both undergo fast proton exchange with water from the C-terminus. Thus, the Copen conformation is not necessary for proton exchange.
The absence of strong imidazole-imidazolium hydrogen bonds in M2
Cross and co-workers proposed the existence of a pair of strong imidazole-imidazolium hydrogen bonds in the histidine tetrad at acidic pH to explain the conduction mechanism. The model was initially proposed based on TM peptide 15N spectra [47] to account for a +2 charged H37 tetrad at neutral pH, which was concluded from the measured pKa’s for the first two protonation events (both at 8.2). It was proposed that the strong imidazole-imidazolium hydrogen bonds stabilize the +2 charge in the middle of the membrane. Recently the model was extended to full-length M2 after the observation of a large 1H chemical shift of ~19 ppm correlated with an imidazole 15N peak [46]. However, the original pKa’s were extracted from partially overlapping 15N signals in a narrow range of 20 ppm, under conditions where the TM peptide has residual motion at the experimental temperature of 4°C in the DMPC/DMPG membrane used for reconstitution [47]. Thus, significant uncertainties exist in the pKa values. Indeed, subsequent measurements of 15N spectra of the TM peptide bound to a cholesterol-containing membrane at the low temperature of 243 K and analysis of the intensities of well-resolved unprotonated and protonated 15N peaks avoided these problems and showed that the two highest pKa’s are lower (7.6 and 6.8) and are resolved in the more biologically relevant membrane. Thus the average charge state of the tetrad at neutral pH is lower than originally thought [43]. Second, high-resolution crystal structures of the TM peptide showed a cluster of water molecules on both sides of the histidine tetrad, which delocalize the charge [54]. Thus, there is no energetic rationale for a strong imidazole-imidazolium hydrogen bond.
Moreover, all NMR data show that H37 in any construct of M2 is hydrogen-bonded to water and not to another histidine. 1) In the TM peptide the largest imidazole 15N and 1H chemical shifts observed for histidine at acidic pH are 178 ppm and 15 ppm [45], which are not sufficiently large to represent a strong hydrogen bond. Moreover, at high temperature under dynamic averaging, the 1H chemical shift changes to 5 ppm, definitively proving that the hydrogen-bonding partner of histidine is water rather than another histidine. 2) A strong N-H…N hydrogen bond should exhibit either a single 15N peak at the averaged chemical shift between N and NH (for equal-well potentials) or two 15N peaks centered around the averaged frequency (for unequal-well potentials) [74]. However, the His37 15N spectra of all M2 constructs only show a single peak that is away from the averaged chemical shift, thus ruling out an N-H…N hydrogen bond of any strength. 3) The 2D 1H-15N correlation spectra of His37 in DOPC/DOPE bound full-length M2 at pH 6.2 showed a 1H chemical shift of 19 ppm correlated with an imidazole 15N chemical shift of 190 ppm [46]. This cross peak, which represents less than 5% of the total spectral intensity, was interpreted as due to a strong imidazole-imidazolium hydrogen bond that is unique to full-length M2. However, the correlated 13C chemical shifts at 137 ppm indicate that these signals belong to neutral histidine rather than cationic histidine. In a strong N-H…N hydrogen bond, the neutral histidine must contribute the unprotonated nitrogen while the cationic histidine contributes the protonated nitrogen. The unprotonated nitrogen of a neutral histidine should have a 15N chemical shift larger than ~210 ppm, the average between the unprotonated and protonated chemical shifts. Therefore, the observed 1H-15N cross peak cannot be assigned to an N-H…N hydrogen bond. In addition, the 19-ppm 1H chemical shift lacks a second 15N cross peak, indicating that this proton is not shared by two similar-pKa nitrogens. 4) Finally, both high-resolution crystal structures [54] and directly measured His37 (χ1, χ2) angles in membrane-bound TM peptides [44] show that the H37 sidechain (χ1, χ2) angles are near 180°, which cannot direct the imidazole N-H groups in a geometry to form inter-histidine N-H…N hydrogen bonds. The current W41F mutant 2D 1H-15N correlation spectra (Fig. 8) are in excellent agreement with these WT observations. The largest 1H chemical shift in the W41F mutant is 16 ppm and occurs at pH 7.5. This 1H chemical shift is correlated with a single 15N chemical shift at 175 ppm, which can be assigned to Nδ1-Hδ1 of the π tautomer. The downfield 1H chemical shift indicates a relatively strong hydrogen bond to water, not to another histidine.
Conclusion
The W41F mutant data shown here provide molecular-level structural and dynamical signatures of the asymmetric conduction of influenza M2 channels. Replacing Trp by Phe at residue 41 caused pronounced changes in the structural equilibria of the proton-selective histidine. At high pH, the H37 structure is shifted towards the π tautomer and less cationic tetrads, indicating increased forward deprotonation compared to the WT channel. At low pH, highly charged tetrads are stabilized compared to the WT channel, indicating increased reverse protonation. The latter is consistent with the preferential hydration of the C-terminus of the mutant channel at low pH. The counter-directional changes of H37 chemical equilibria at high and low pH are quantitatively confirmed by the measured H37 pKa’s, which cluster to a narrower pH range than the WT AM2-TM channel. This pKa clustering reduces the population of the +2 charged channels in favor of the more cationic +3 and +4 channels, explaining the increased proton conductance of the mutant. The mutation does not weaken the tetrameric integrity of the channel, implying that the H37-W41 interaction is not necessary for tetramer assembly even though it is essential for asymmetric proton conduction. Chemical shift data indicate that the mutant TM helix has the same conformational dualism as the WT channel, and the relative populations of the Cclosed and Copen conformations at high and low pH are regulated by the H37 protonation state, regardless of whether protonation occurs from the N-terminus or the C-terminus. The fact that H37 can be protonated from the C-terminus is most conclusively manifested by the drug-bound W41F spectra, which show that half of the histidines remain cationic in the drug-bound state, in contrast to the WT channel, which contains only neutral histidines upon drug binding. Therefore, when the N-terminal pore is blocked by drug while pHin is low, H37 remains significantly protonatable in the mutant channel, indicating unambiguously that protons from the C-terminus can access H37.
These results imply that, in the WT channel, while the four W41 indoles do not fully obstruct the pore, the sidechain conformation is sufficient to prevent proton access to H37 from the C-terminus at low pHin. Therefore, even though the membranes used in structural studies have symmetric pH on the two sides, acid activation of H37 in wild-type M2 occurs only from the N-terminus. In conclusion, asymmetric conduction in influenza M2 channels is due to W41 inhibition of C-terminal acid activation of H37, and is associated with the Cclosed conformation of the TM helix. These results may have implications to the mechanism of pH-induced asymmetric conduction in other channels such as voltage-gated proton channels.
Materials and Methods
Synthesis of isotopically labeled M2(22-46)
A peptide corresponding to the W41F M2(22-46) of the influenza A/Udorn/72 strain of M2 (SSDPLVVAASIIGILHLILFILDRL) was synthesized using Fmoc solid-phase peptide synthesis chemistry. Uniformly 13C, 15N-labeled V27, A30, S31, G34, H37, D44 and 4-19F-labeled F41 were introduced into the sequence (VASGHD). Rink Amide H-Rink amide ChemMatrix resin (Sigma) was swelled for 1 hour in 2 mL of N,N-dimethylformamide (DMF). Double coupling was carried out using 4 equivalents of amino acid, 4 equivalents of 1-[Bis(dimethylamino)methylene]-1H-1,2,3-triazolo[4,5-b]pyridinium 3-oxid hexafluorophosphate (HATU), and 8 equivalents of N,N-diisopropylethylamine (DIPEA). The reaction was carried out in 2 ml of DMF for 0.5 – 1 hour per coupling. Fmoc deprotection was carried out in 2 ml DMF containing 20% piperidine for 5 and 15 minutes. Cleavage of the peptide from the resin and sidechain deprotection were carried out in 88% trifluoroacetic acid (TFA), 5% water, 2% triisopropylsilane and 5% phenol for 3 hours at room temperature. The peptide was triturated three times with cold diethyl ether. The crude peptide was dissolved in a 50% acentonitrile solution containing 0.1% TFA and purified by preparative reversed-phase HPLC on a Varian ProStar 210 System using a Vydac C18 column (10 μm particle size, 2.2 × 25 cm2) with a linear gradient of 80–99% acetonitrile over 80 minutes at a flow rate of 10 mL/min. The mass (2737.26 Da) and purity (>95%) of the peptide was confirmed using MALDI-TOF mass spectrometry. A second peptide was synthesized and validated following the same procedure, but with uniformly 13C, 15N-labeled V28, A29, G34, L38, and I39 introduced into the sequence. Data collected using this second peptide is exclusively included in Figure 8.
Membrane protein sample preparation
Purified W41F AM2-TM peptide was reconstituted into a virus-mimetic (VM+) lipid membrane, which contains 1-palmitoyl-2-oleoyl-sn-glycero-3-phophocholine (POPC), 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine (POPE), sphingomyelin (SM) and cholesterol (Chol) at molar ratios of 25.6% : 25.6% : 25.6% : 23.2%. POPC, POPE and cholesterol were dissolved in chloroform, while SM was dissolved in a chloroform/methanol mixture. The peptide was dissolved in 300 μL of 2,2,2-trifluoroethanol (TFE), then mixed with the lipids at a peptide : lipid molar ratio of 1 : 12. The organic solvents were removed under a stream of nitrogen gas. The dried film was resuspended in appropriate buffer, vortexed and sonicated 3–4 times at 2 minutes each until a homogenous suspension was obtained. The mixture was centrifuged at 35,000 rpm at 4°C for 8 h to obtain a homogeneous membrane pellet. The pellet was equilibrated to a final hydration level of ~40% by mass, then transferred to a 3.2 mm or 4 mm MAS rotor for solid-state NMR experiments.
Five W41F AM2-TM membrane samples were prepared using the VASGHD labeled peptide, at pH 7.5 (20 mM Bis-Tris, 1 mM EDTA, 0.1 mM NaN3), pH 6.2 (20 mM Bis-Tris, 1mM EDTA, 0.1 mM NaN3), pH 5.9 (20 mM Bis-Tris, 1mM EDTA, 0.1 mM NaN3), pH 5.5 (20 mM citric acid/citrate, 1 mM EDTA, 0.1 mM NaN3), and pH 4.5 (20 mM citric acid/citrate, 1 mM EDTA, 0.1 mM NaN3). The pH was measured at three stages: 1) for the buffer before mixing with the peptide-lipid film, 2) for the uniform proteoliposome solution, and 3) for the supernatant after ultracentrifugation. Deuterated amantadine (d15-Amt) was titrated to a pH 5.5 sample at an drug : tetramer ratio of 8 : 1. Three of the above conditions were also used to make membrane samples of the VAGLI labeled peptide: pH 7.5, pH 5.5, and pH 5.5 with 8 : 1 d15-Amt.
Solid-state NMR experiments
Most solid-state NMR spectra were measured on Bruker 800 MHz (18.8 Tesla) and 900 MHz (21.1 Tesla) NMR spectrometers to obtain high resolution, except for low-temperature 1D 15N spectra for the pKa determination and 19F CODEX spectra, which were measured at 400 MHz. 3.2 mm or 4 mm MAS probes were used and MAS frequencies ranged from 7 kHz to 16 kHz. 13C chemical shifts were referenced externally to the adamantane CH2 chemical shift at 38.48 ppm on the tetramethylsilane (TMS) scale, while 15N chemical shifts were referenced to the 15N peak of N-acetylvaline at 122.0 ppm on the liquid ammonia scale. Sample temperatures are thermocouple-reported values.
1D 13C and 15N cross-polarization (CP) spectra were measured from 243 K to 298 K. 2D 13C-13C DARR and 15N -13C heteronuclear correlation (HETCOR) spectra were measured at 800 or 900 MHz using a DARR mixing time of 150 ms and a REDOR mixing time of 0.95 ms, respectively.
2D 1H-15N HETCOR spectra were measured at 800 MHz. A 1H-15N Lee-Goldburg (LG) CP period of 2.5 ms was applied to detect short-distance 1H-15N spin pairs, without 1H spin diffusion. 1H frequency-switched LG (FSLG) decoupling was applied during the 1H t1 evolution period to remove 1H-1H homonuclear coupling. The FSLG transverse field strength was 80 kHz, which corresponds to an effective field of 98 kHz tilted at the magic angle. The effective t1 dwell time was 23.58 μs, and 100 hypercomplex t1 slices were measured, giving a maximum evolution time of 1.12 ms.
2D 1H-13C HETCOR experiments with 1H spin diffusion were used to measure the hydration of W41F AM2-TM at 293 K on the 800 MHz spectrometer under 12 kHz MAS. A 1H T2 filter of 3–4 ms was used to suppress peptide magnetization before spin diffusion. Two 1H spin diffusion mixing times of 4 ms (S) and 100 ms (S0) were used. The intensity ratios (S/S0) of the resolved signals were analyzed from the water cross-section of each 2D spectrum to deduce the hydration levels of the different residues.
19F CODEX spectra were measured at 231 K on the 400 MHz spectrometer under 10 kHz MAS. Control (S0) and dephased (S) experiments were conducted within 1 hour of each other, and the intensity ratios (S/S0) were used to obtain distances. Mixing times of 10, 50, 100, 300, 500, 1000 and 2000 ms were used. Typical radiofrequency field strengths were 60 kHz for 19F and 80 kHz for 1H.
Extraction of H37 pKa’s and statistical analysis of the pKa uncertainties
H37 proton dissociation constants were extracted using a previously published procedure [43, 47]. Briefly, the protonated and unprotonated 15N regions at 160–190 ppm and 250 ppm were integrated and their intensity ratios, INH/IN, were read off and then converted to the concentration ratios [His]/[HisH+] between neutral and cationic histidines. This procedure does not require complete resolution of the different protonated nitrogen species between 160 and 190 ppm. The histidine concentration ratios were then fit to the following equation to obtain the four pKa’s:
| (8) |
In this equation, Ka4 describes the equilibrium constant of the first protonation event at the highest pH while Ka1 is the equilibrium constant of the last protonation event at the lowest pH. In the fit we assume that the proton affinity of the H37 tetrad is either the same or higher upon successive protonation events, i.e., pKa4 ≥ pKa3 ≥ pKa2 ≥ pKa1 from high to low pH. To obtain the best-fit pKa’s, we considered all combinations of pKa’s in the range between 9 and 2, sampled in 0.1 increments. For each combination of pKa’s, we calculated the reduced chi-square ( ) of the fit according to:
| (9) |
where yexp and ycalc are the experimental and calculated [His]/[HisH+] ratios, σexp is the experimental uncertainty in the [His]/[HisH+] ratio (Table S2), and v is the degree of freedom, which is the difference between the number of data points and the number of unknowns in the fit. Since five different pH samples were used for extracting four pKa’s, v = 1. The lowest values for the W41F mutant, WT AM2 and BM2 are 2.20, 0.82 and 0.28, respectively. We retained all fits with χv2 ≤ 3.84 (p < 0.05 for 1 degree of freedom) for the WT AM2 and BM2 datasets but a more conservative cutoff of χv2 ≤ 5.99 (p < 0.05 for 2 degrees of freedom) for the W41F sample because of its higher values. Using all fits within the cutoff, we computed the mean and standard deviation of the pKa’s.
To quantify the correlation among the four pKa’s, we computed the pairwise Pearson product-moment correlation coefficients rxy according to
| (10) |
where xi and yi are the i-th values of the two pKa’s, and x̄ and ȳ are the means of each pKa set. The distribution of the four pKa’s for the three constructs are shown in Fig. S3, while the correlation coefficient matrices are given in Table S4.
Temperature effects on the extracted pKa’s
Since pKa’s of histidine and buffer ions are generally temperature dependent, the pKa’s derived from low-temperature 15N NMR data will have a systematic difference from the physiological-temperature pKa’s. For example, a recent comparison of the BM2 His19 spectra at 263 K and the cryogenic temperature of 117 K [69] found that the low-temperature spectra showed a higher population of cationic histidine than the high-temperature spectra. This can be attributed to the fact that imidazole has a more negative temperature coefficient of pKa than the citrate buffer used to prepare the sample, which preferentially increased the histidine pKa’s relative to the citrate pKa at low temperature. Since the 15N spectra of the W41F mutant in the present study are measured under the same temperature, membrane and buffer conditions as wild-type AM2-TM and BM2-TM, the systematic correction factors for the temperature-dependent pKa’s should be similar, thus we can compare the protonation equilibria among these samples consistently. The W41F M2TM samples used here were prepared using Bis-Tris and citrate buffers. Based on their temperature coefficients relative to the temperature coefficient of the histidine pKa, the physiological temperature H37 tetrads are expected to be more cationic at high pH and more neutral at low pH compared to the 243 K situation, thus the pKa’s at high temperature may be more separated than at low temperature. However, this systematic difference should be relatively small due to the limited temperature range of 243 K and 310 K.
The pKa values allow the calculation of the charge state populations, Ni, according to previously described analytical equations [43]. The charge state populations were then fit to pH-dependent proton conductance g reported by Tang et al. (9) according to to determine the relative time-averaged unitary conductance, , of the five charge states.
Simulation of 19F CODEX exchange curves
The 19F CODEX exchange data were fit using a Python program that employs the exchange-matrix formalism to treat spin diffusion within a four-spin system. The 4 x 4 exchange matrix contains terms that are proportional to an overlap integral and the square of the pairwise 19F dipolar couplings. The overlap integral is calibrated to be 41 μs under 10 kHz MAS, based on experimental data of the model compound, 5-19F-Trp (Fig. S1). The Trp decay was fit using the same exchange-matrix formalism but for a two-spin system, using the known nearest-neighbor distance of 4.62 Å from the crystal structure. Best-fit distances were obtained by minimizing the uncertainty-weighted least squares RMSD between the calculated and experimental CODEX intensities. The Gaussian distributions were digitized into 0.2 Å bins between 5 and 13 Å, and the center of each bin was calculated to give the CODEX exchange curve.
Supplementary Material
Acknowledgments
This work is supported by NIH grant GM088204 to M.H. The 900 MHz spectra were measured at the MIT/Harvard Center for Magnetic Resonance, which is supported by NIH grant P41-EB-002026.
References
- 1.Hille B. Ionic channels of excitable membranes. 2. Sundeland: Sinauer Associates Inc; 1992. [Google Scholar]
- 2.Pinto LH, Lamb RA. The M2 proton channels of influenza A and B viruses. J Biol Chem. 2006;281:8997–9000. doi: 10.1074/jbc.R500020200. [DOI] [PubMed] [Google Scholar]
- 3.Cady SD, Luo WB, Hu FH, Hong M. Structure and Function of the Influenza A M2 Proton Channel. Biochemistry. 2009;48:7356–7364. doi: 10.1021/bi9008837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hong M, DeGrado WF. Structural basis for proton conduction and inhibition by the influenza M2 protein. Protein Sci. 2012;21:1620–1633. doi: 10.1002/pro.2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang C, Lamb RA, Pinto LH. Activation of the M(2) Ion-Channel of Influenza-Virus - a Role for the Transmembrane Domain Histidine Residue. Biophys J. 1995;69:1363–1371. doi: 10.1016/S0006-3495(95)80003-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Balannik V, Carnevale V, Fiorin G, Levine BG, Lamb RA, Klein ML, et al. Functional studies and modeling of pore-lining residue mutants of the influenza a virus M2 ion channel. Biochemistry. 2010;49:696–708. doi: 10.1021/bi901799k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tang Y, Zaitseva F, Lamb RA, Pinto LH. The gate of the influenza virus M2 proton channel is formed by a single tryptophan residue. J Biol Chem. 2002;277:39880–39886. doi: 10.1074/jbc.M206582200. [DOI] [PubMed] [Google Scholar]
- 8.Ma CL, Fiorin G, Carnevale V, Wang J, Lamb RA, Klein ML, et al. Asp44 Stabilizes the Trp41 Gate of the M2 Proton Channel of Influenza A Virus. Structure. 2013;21:2033–2041. doi: 10.1016/j.str.2013.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lopatin AN, Makhina EN, Nichols CG. The Mechanism of Inward Rectification of Potassium Channels - Long-Pore Plugging by Cytoplasmic Polyamines. J Gen Physiol. 1995;106:923–955. doi: 10.1085/jgp.106.5.923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smith PL, Baukrowitz T, Yellen G. The inward rectification mechanism of the HERG cardiac potassium channel. Nature. 1996;379:833–836. doi: 10.1038/379833a0. [DOI] [PubMed] [Google Scholar]
- 11.Fakler B, Brandle U, Glowatzki E, Weidemann S, Zenner HP, Ruppersberg JP. Strong Voltage-Dependent Inward Rectification of Inward Rectifier K+ Channels Is Caused by Intracellular Spermine. Cell. 1995;80:149–154. doi: 10.1016/0092-8674(95)90459-x. [DOI] [PubMed] [Google Scholar]
- 12.Cherny VV, Morgan D, Musset B, Chaves G, Smith SME, DeCoursey TE. Tryptophan 207 is crucial to the unique properties of the human voltage-gated proton channel, hH(V)1. J Gen Physiol. 2015;146:343–356. doi: 10.1085/jgp.201511456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Decoursey TE. Voltage-gated proton channels and other proton transfer pathways. Physiol Reviews. 2003;83:475–579. doi: 10.1152/physrev.00028.2002. [DOI] [PubMed] [Google Scholar]
- 14.Pinto LH, Holsinger LJ, Lamb RA. Influenza-Virus M2 Protein Has Ion Channel Activity. Cell. 1992;69:517–528. doi: 10.1016/0092-8674(92)90452-i. [DOI] [PubMed] [Google Scholar]
- 15.Chizhmakov IV, Geraghty FM, Ogden DC, Hayhurst A, Antoniou M, Hay AJ. Selective proton permeability and pH regulation of the influenza virus M2 channel expressed in mouse erythroleukaemia cells. J Physiol. 1996;494:329–336. doi: 10.1113/jphysiol.1996.sp021495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hay AJ, Wolstenholme AJ, Skehel JJ, Smith MH. The molecular basis of the specific anti-influenza action of amantadine. EMBO J. 1985;4:3021–3024. doi: 10.1002/j.1460-2075.1985.tb04038.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang C, Takeuchi K, Pinto LH, Lamb RA. Ion channel activity of influenza A virus M2 protein: characterization of the amantadine block. J Virol. 1993;67:5585–5594. doi: 10.1128/jvi.67.9.5585-5594.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou HX, Cross TA. Influences of membrane mimetic environments on membrane protein structures. Annu Rev Biophys. 2013;42:361–392. doi: 10.1146/annurev-biophys-083012-130326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ma CL, Polishchuk AL, Ohigashi Y, Stouffer AL, Schon A, Magavern E, et al. Identification of the functional core of the influenza A virus A/M2 proton-selective ion channel. Proc Natl Acad Sci USA. 2009;106:12283–12288. doi: 10.1073/pnas.0905726106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kwon B, Hong M. The Influenza M2 Ectodomain Regulates the Conformational Equilibria of the Transmembrane Proton Channel: Insights from Solid-State Nuclear Magnetic Resonance. Biochemistry. 2016;55:5387–5397. doi: 10.1021/acs.biochem.6b00727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Park EK, Castrucci MR, Portner A, Kawaoka Y. The M2 ectodomain is important for its incorporation into influenza A virions. J Virol. 1998;72:2449–2455. doi: 10.1128/jvi.72.3.2449-2455.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grandea AG, Olsen OA, Cox TC, Renshaw M, Hammond PW, Chan-Hui PY, et al. Human antibodies reveal a protective epitope that is highly conserved among human and nonhuman influenza A viruses. Proc Natl Acad Sci USA. 2010;107:12658–12663. doi: 10.1073/pnas.0911806107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rossman JS, Jing X, Leser GP, Lamb RA. Influenza virus M2 protein mediates ESCRT-independent membrane scission. Cell. 2010;142:902–913. doi: 10.1016/j.cell.2010.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cady SD, Wang T, Hong M. Membrane-dependent effects of a cytoplasmic helix on the structure and drug binding of the influenza virus M2 protein. J Am Chem Soc. 2011;133:11572–11579. doi: 10.1021/ja202051n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang T, Cady SD, Hong M. NMR Determination of Protein Partitioning into Membrane Domains with Different Curvatures and Application to the Influenza M2 Peptide. Biophys J. 2012;102:787–794. doi: 10.1016/j.bpj.2012.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kwon B, Tietze D, White PB, Liao SY, Hong M. Chemical Ligation of the Influenza M2 Protein for Solid-State NMR Characterization of the Cytoplasmic Domain Structure. Protein Sci. 2015;24:1087–1099. doi: 10.1002/pro.2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McCown MF, Pekosz A. Distinct domains of the influenza a virus M2 protein cytoplasmic tail mediate binding to the M1 protein and facilitate infectious virus production. J Virol. 2006;80:8178–8189. doi: 10.1128/JVI.00627-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liang R, Swanson JM, Madsen JJ, Hong M, DeGrado WF, Voth GA. Acid activation mechanism of the influenza A M2 proton channel. Proc Natl Acad Sci USA. 2016;113:E6955–E6964. doi: 10.1073/pnas.1615471113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sharma M, Yi M, Dong H, Qin H, Peterson E, Busath DD, et al. Insight into the mechanism of the influenza A proton channel from a structure in a lipid bilayer. Science. 2010;330:509–512. doi: 10.1126/science.1191750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miao YM, Qin HJ, Fu RQ, Sharma M, Can TV, Hung I, et al. M2 Proton Channel Structural Validation from Full-Length Protein Samples in Synthetic Bilayers and E. coli Membranes. Angew Chem Int Ed Engl. 2012;51:8383–8386. doi: 10.1002/anie.201204666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liao SY, Yang Y, Tietze D, Hong M. The influenza M2 cytoplasmic tail changes the proton-exchange equilibria and the backbone conformation of the transmembrane histidine residue to facilitate proton conduction. J Am Chem Soc. 2015;137:6067–6077. doi: 10.1021/jacs.5b02510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cady S, Wang T, Hong M. Membrane-dependent effects of a cytoplasmic helix on the structure and drug binding of the influenza virus M2 protein. J Am Chem Soc. 2011;133:11572–11579. doi: 10.1021/ja202051n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hu F, Luo W, Cady SD, Hong M. Conformational plasticity of the influenza A M2 transmembrane helix in lipid bilayers under varying pH, drug binding, and membrane thickness. Biochim Biophys Acta. 2011;1808:415–423. doi: 10.1016/j.bbamem.2010.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liao SY, Fritzsching KJ, Hong M. Conformational analysis of the full-length M2 protein of the influenza A virus using solid-state NMR. Protein Sci. 2013;22:1623–1638. doi: 10.1002/pro.2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Luo W, Cady SD, Hong M. Immobilization of the Influenza A M2 Transmembrane Peptide in Virus-Envelope Mimetic Lipid Membranes: A Solid-State NMR Investigation. Biochemistry. 2009;48:6361–6368. doi: 10.1021/bi900716s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cristian L, Lear JD, DeGrado WF. Use of thiol-disulfide equilibria to measure the energetics of assembly of transmembrane helices in phospholipid bilayers. Proc Natl Acad Sci USA. 2003;100:14772–14777. doi: 10.1073/pnas.2536751100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim SS, Upshur MA, Saotome K, Sahu ID, McCarrick RM, Feix JB, et al. Cholesterol-Dependent Conformational Exchange of the C-Terminal Domain of the Influenza A M2 Protein. Biochemistry. 2015;54:7157–7167. doi: 10.1021/acs.biochem.5b01065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cady SD, Goodman C, Tatko C, DeGrado WF, Hong M. Determining the orientation of uniaxially rotating membrane proteins using unoriented samples: a 2H, 13C, and 15N solid-state NMR investigation of the dynamics and orientation of a transmembrane helical bundle. J Am Chem Soc. 2007;129:5719–5729. doi: 10.1021/ja070305e. [DOI] [PubMed] [Google Scholar]
- 39.Duong-Ly KC, Nanda V, DeGrado WF, Howard KP. The conformation of the pore region of the M2 proton channel depends on lipid bilayer environment. Protein Sci. 2005;14:856–861. doi: 10.1110/ps.041185805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Saotome K, Duong-Ly KC, Howard KP. Influenza A M2 protein conformation depends on choice of model membrane. Biopolymers. 2015;104:405–411. doi: 10.1002/bip.22617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Andreas LB, Reese M, Eddy MT, Gelev V, Ni QZ, Miller EA, et al. Structure and Mechanism of the Influenza A M218-60 Dimer of Dimers. J Am Chem Soc. 2015;137:14877–14886. doi: 10.1021/jacs.5b04802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pinto LH, Dieckmann GR, Gandhi CS, Papworth CG, Braman J, Shaughnessy MA, et al. A functionally defined model for the M-2 proton channel of influenza A virus suggests a mechanism for its ion selectivity. Proc Natl Acad Sci USA. 1997;94:11301–11306. doi: 10.1073/pnas.94.21.11301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hu F, Schmidt-Rohr K, Hong M. NMR detection of pH-dependent histidine-water proton exchange reveals the conduction mechanism of a transmembrane proton channel. J Am Chem Soc. 2012;134:3703–3713. doi: 10.1021/ja2081185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hu F, Luo W, Hong M. Mechanisms of proton conduction and gating in influenza M2 proton channels from solid-state NMR. Science. 2010;330:505–508. doi: 10.1126/science.1191714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hong M, Fritzsching KJ, Williams JK. Hydrogen-bonding partner of the proton-conducting histidine in the influenza M2 proton channel revealed from 1H chemical shifts. J Am Chem Soc. 2012;134:14753–14755. doi: 10.1021/ja307453v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Miao Y, Fu R, Zhou HX, Cross TA. Dynamic Short Hydrogen Bonds in Histidine Tetrad of Full-Length M2 Proton Channel Reveal Tetrameric Structural Heterogeneity and Functional Mechanism. Structure. 2015;23:2300–2308. doi: 10.1016/j.str.2015.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hu J, Fu R, Nishimura K, Zhang L, Zhou HX, Busath DD, et al. Histidines, heart of the hydrogen ion channel from influenza A virus: toward an understanding of conductance and proton selectivity. Proc Natl Acad Sci USA. 2006;103:6865–6870. doi: 10.1073/pnas.0601944103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Colvin MT, Andreas LB, Chou JJ, Griffin RG. Proton association constants of His 37 in the Influenza-A M218-60 dimer-of-dimers. Biochemistry. 2014;53:5987–5994. doi: 10.1021/bi5005393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cady SD, Schmidt-Rohr K, Wang J, Soto CS, Degrado WF, Hong M. Structure of the amantadine binding site of influenza M2 proton channels in lipid bilayers. Nature. 2010;463:689–692. doi: 10.1038/nature08722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cady SD, Wang J, Wu Y, DeGrado WF, Hong M. Specific binding of adamantane drugs and direction of their polar amines in the pore of the influenza M2 transmembrane domain in lipid bilayers and dodecylphosphocholine micelles determined by NMR spectroscopy. J Am Chem Soc. 2011;133:4274–4284. doi: 10.1021/ja102581n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hu J, Asbury T, Achuthan S, Li C, Bertram R, Quine JR, et al. Backbone structure of the amantadine-blocked trans-membrane domain M2 proton channel from Influenza A virus. Biophys J. 2007;92:4335–4343. doi: 10.1529/biophysj.106.090183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Luo W, Hong M. Conformational changes of an ion channel membrane protein detected through water-protein interactions using solid-state NMR spectroscopy. J Am Chem Soc. 2010;132:2378–2384. doi: 10.1021/ja9096219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Williams JK, Zhang Y, Schmidt-Rohr K, Hong M. pH-Dependent Conformation, Dynamics, and Aromatic Interaction of the Gating Tryptophan Residue of the Influenza M2 Proton Channel from Solid-State NMR. Biophys J. 2013;104:1698–1708. doi: 10.1016/j.bpj.2013.02.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Acharya A, Carnevale V, Fiorin G, Levine BG, Polishchuk A, Balannick V, et al. Structure and mechanism of proton transport through the transmembrane tetrameric M2 protein bundle of the influenza A virus. Proc Natl Acad Sci U S A. 2010;107:15075–15080. doi: 10.1073/pnas.1007071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Williams JK, Hong M. Probing membrane protein structure using water polarization transfer solid-state NMR. J Magn Reson. 2014;247:118–127. doi: 10.1016/j.jmr.2014.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Thomaston JL, Alfonso-Prieto M, Woldeyes RA, Fraser JS, Klein ML, Fiorin G, et al. High-resolution structures of the M2 channel from influenza A virus reveal dynamic pathways for proton stabilization and transduction. Proc Natl Acad Sci USA. 2015;112:14260–14265. doi: 10.1073/pnas.1518493112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang J, Kim S, Kovacs F, Cross TA. Structure of the transmembrane region of the M2 protein H+ channel. Protein Sci. 2001;10:2241–2250. doi: 10.1110/ps.17901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li C, Qin H, Gao FP, Cross TA. Solid-state NMR characterization of conformational plasticity within the transmembrane domain of the influenza A M2 proton channel. Biochim Biophys Acta. 2007;1768:3162–3170. doi: 10.1016/j.bbamem.2007.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.O’Hagan D. Understanding organofluorine chemistry. An introduction to the C-F bond. Chem Soc Rev. 2008;37:308–319. doi: 10.1039/b711844a. [DOI] [PubMed] [Google Scholar]
- 60.Mecozzi S, West AP, Dougherty DA. Cation-pi interactions in aromatics of biological and medicinal interest: Electrostatic potential surfaces as a useful qualitative guide. Proc Natl Acad Sci U S A. 1996;93:10566–10571. doi: 10.1073/pnas.93.20.10566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Salwiczek M, Nyakatura EK, Gerling UI, Ye S, Koksch B. Fluorinated amino acids: compatibility with native protein structures and effects on protein-protein interactions. Chem Soc Rev. 2012;41:2135–2171. doi: 10.1039/c1cs15241f. [DOI] [PubMed] [Google Scholar]
- 62.deAzevedo ER, Bonagamba TJ, Hu W, Schmidt-Rohr K. Centerband-only detection of exchange: efficient analysis of dynamics in solids by NMR. J Am Chem Soc. 1999;121:8411–8412. [Google Scholar]
- 63.deAzevedo ER, Bonagamba TJ, Hu W, Schmidt-Rohr K. Principle of centerband-only detection of the exchange and extension to a four-time CODEX. J Chem Phys. 2000;112:8988–9001. [Google Scholar]
- 64.Buffy JJ, Waring AJ, Hong M. Determination of Peptide Oligomerization in Lipid Membranes with Magic-Angle Spinning Spin Diffusion NMR. J Am Chem Soc. 2005;127:4477–4483. doi: 10.1021/ja043621r. [DOI] [PubMed] [Google Scholar]
- 65.Luo W, Hong M. Determination of the oligomeric number and intermolecular distances of membrane protein assemblies by anisotropic 1H-driven spin diffusion NMR spectroscopy. J Am Chem Soc. 2006;128:7242–7251. doi: 10.1021/ja0603406. [DOI] [PubMed] [Google Scholar]
- 66.Stouffer AL, Acharya R, Salom D, Levine AS, Di Costanzo L, Soto CS, et al. Structural basis for the function and inhibition of an influenza virus proton channel. Nature. 2008;451:596–599. doi: 10.1038/nature06528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Luo W, Mani R, Hong M. Sidechain conformation and gating of the M2 transmembrane peptide proton channel of influenza A virus from solid-state NMR. J Phys Chem. 2007;111:10825–10832. doi: 10.1021/jp073823k. [DOI] [PubMed] [Google Scholar]
- 68.Khurana E, Dal Peraro M, DeVane R, Vemparala S, DeGrado WF, Klein ML. Molecular dynamics calculations suggest a conduction mechanism for the M2 proton channel from influenza A virus. Proc Natl Acad Sci USA. 2009;106:1069–1074. doi: 10.1073/pnas.0811720106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Williams JK, Tietze D, Lee M, Wang J, Hong M. Solid-State NMR Investigation of the Conformation, Proton Conduction, and Hydration of the Influenza B Virus M2 Transmembrane Proton Channel. J Am Chem Soc. 2016;138:8143–8155. doi: 10.1021/jacs.6b03142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rosenberg MR, Casarotto MG. Coexistence of two adamantane binding sites in the influenza A M2 ion channel. Proc Natl Acad Sci USA. 2010;107:13866–13871. doi: 10.1073/pnas.1002051107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cady SD, Hong M. Amantadine-Induced Conformational and Dynamical Changes of the Influenza M2 Transmembrane Proton Channel. Proc Natl Acad Sci US A. 2008;105:1483–1488. doi: 10.1073/pnas.0711500105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cady SD, Mishanina TV, Hong M. Structure of amantadine-bound M2 transmembrane peptide of influenza A in lipid bilayers from magic-angle-spinning solid-state NMR: the role of Ser31 in amantadine binding. J Mol Biol. 2009;385:1127–1141. doi: 10.1016/j.jmb.2008.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Liang R, Li H, Swanson JM, Voth GA. Multiscale simulation reveals a multifaceted mechanism of proton permeation through the influenza A M2 proton channel. Proc Natl Acad Sci USA. 2014;111:9396–9401. doi: 10.1073/pnas.1401997111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.White PB, Hong M. (15)N and (1)H Solid-State NMR Investigation of a Canonical Low-Barrier Hydrogen-Bond Compound: 1,8-Bis(dimethylamino)naphthalene. J Phys Chem B. 2015;119:11581–11589. doi: 10.1021/acs.jpcb.5b06171. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.