A modified virus vector permits better plant gene fragment retention and improves gene-silencing phenotypes in the host.
Abstract
Virus-induced gene silencing (VIGS) is used extensively for gene function studies in plants. VIGS is inexpensive and rapid compared with silencing conducted through stable transformation, but many virus-silencing vectors, especially in grasses, induce only transient silencing phenotypes. A major reason for transient phenotypes is the instability of the foreign gene fragment (insert) in the vector during VIGS. Here, we report the development of a Brome mosaic virus (BMV)-based vector that better maintains inserts through modification of the original BMV vector RNA sequence. Modification of the BMV RNA3 sequence yielded a vector, BMVCP5, that better maintained phytoene desaturase and heat shock protein70-1 (HSP70-1) inserts in Nicotiana benthamiana and maize (Zea mays). Longer maintenance of inserts was correlated with greater target gene silencing and more extensive visible silencing phenotypes displaying greater tissue penetration and involving more leaves. The modified vector accumulated similarly to the original vector in N. benthamiana after agroinfiltration, thus maintaining a high titer of virus in this intermediate host used to produce virus inoculum for grass hosts. For HSP70, silencing one family member led to a large increase in the expression of another family member, an increase likely related to the target gene knockdown and not a general effect of virus infection. The cause of the increased insert stability in the modified vector is discussed in relationship to its recombination and accumulation potential. The modified vector will improve functional genomic studies in grasses, and the conceptual methods used to improve the vector may be applied to other VIGS vectors.
RNA silencing (RNAi) is a powerful tool for reverse and forward genetic analyses in plants (Baulcombe, 2004; Brodersen and Voinnet, 2006; Eamens et al., 2008; Martínez de Alba et al., 2013). RNAi can be achieved in multiple ways, including stable transformation or transient expression of silence-inducing fragments with virus vectors (virus-induced gene silencing [VIGS]). VIGS is triggered by double-stranded RNAs that accumulate during a virus infection in the host and function as substrates for RNase III-like (dicer-like) enzymes to yield small interfering RNA (siRNA) duplexes (Baulcombe, 2004; Waterhouse and Fusaro, 2006; Ding and Voinnet, 2007). Single strands of these siRNA duplexes are then incorporated into RNAi-induced silencing complexes containing proteins encoded by the host ARGONAUTE gene family (Montgomery et al., 2008; Vaucheret, 2008; Poulsen et al., 2013; Fang and Qi, 2016). The single-stranded siRNAs serve as reverse complementary guides in RNAi-induced silencing complexes to bind mRNAs for targeted degradation. For many plant genes, knockdown of their expression leads to the onset of visible phenotypes in the plant (e.g. necrosis, photobleaching, and stunting). Because the preparation of a VIGS construct and plant inoculation are inexpensive and not labor intensive, and the silencing results can be achieved in weeks, VIGS technology for gene function studies in dicotyledonous and monocotyledonous plants has advantages over stable RNAi transformation, which requires more reagents and labor and months to complete (Robertson, 2004; Scofield and Nelson, 2009; Senthil-Kumar and Mysore, 2011; Ramanna et al., 2013; Lee et al., 2015).
Brome mosaic virus (BMV) has been used as a vector for VIGS in grasses over the last 11 years (Ding et al., 2006, 2007, 2010; Pacak et al., 2010; Martin et al., 2011; Shi et al., 2011; van der Linde et al., 2011, 2012; Benavente et al., 2012; Cao et al., 2012; Hemetsberger et al., 2012; Ramanna et al., 2013; Sun et al., 2013; Kong et al., 2014; Zhu et al., 2014; Zhan et al., 2016). BMV is a tripartite, single-stranded, positive-sense RNA virus (Noueiry and Ahlquist, 2003). BMV genomic RNA1 and RNA2 encode 1a and 2a protein, respectively, both required for virus replication (Kroner et al., 1990; Quadt and Jaspars, 1990; Kao and Ahlquist, 1992). BMV genomic RNA3 is dicistronic, encoding a 3a protein required for virus cell-to-cell movement in the plant and a capsid protein (CP) necessary for virion formation and virus movement in the plant (Dasgupta and Kaesberg, 1982; Ahlquist et al., 1984; Mise and Ahlquist, 1995; Schmitz and Rao, 1996). The CP is translated from a subgenomic RNA (RNA4). For both reported BMV vectors, foreign gene fragments are inserted into genomic RNA3 (Ding et al., 2006, 2010; Pacak et al., 2010; Sun et al., 2013).
Perhaps the most important factor during VIGS that ensures gene silencing in the most tissue for the longest period of time is the genetic stability of the foreign insert in a vector during infection (Scofield and Nelson, 2009; Ramanna et al., 2013; van der Linde and Doehlemann, 2013). Using Barley stripe mosaic virus, Bruun-Rasmussen et al. (2007) showed that inserts of 275, 400, and 584 nucleotides representing phytoene desaturase (PDS) were increasingly unstable in the vector in the inoculated plant. They concluded that this instability could explain the transient nature of silencing when using Barley stripe mosaic virus-based vector in barley (Hordeum vulgare). Using Potato virus X as a silencing vector in dicotyledonous species, inserts were lost with each subsequent passage to an uninfected Nicotiana benthamiana plant (Avesani et al., 2007). A Peanut stunt virus vector also displayed greater instability for maintaining larger PDS inserts (Yamagishi et al., 2015). Recently, Mei et al. (2016) demonstrated a good correlation between the loss of a PDS fragment insert in a Foxtail mosaic virus silencing vector and the loss of post-transcriptional silencing. For BMV, loss of the PDS insert from the BMV-based VIGS vector during virus infection in rice was correlated with a transient photobleaching phenotype affecting only two to three leaves (Ding et al., 2006).
Although the molecular mechanism underlying the deletion of foreign inserts is mostly unstudied during VIGS, it is known that replicase template switching is an accepted model for BMV recombination (Bujarski and Kaesberg, 1986; Nagy et al., 1998; Kim and Kao, 2001; Shapka and Nagy, 2004; Barr and Fearns, 2010; Simon-Loriere and Holmes, 2011; Bujarski, 2013; Rao and Kao, 2015). RNA recombination occurs within and between the BMV genomic RNAs (Sztuba-Solińska et al., 2011). Consequently, a foreign sequence introduced into the BMV RNA3 3′ untranslated region, the region we utilize as an insertion site in our BMV vector (Ding et al., 2006; Zhu et al., 2014), potentially could be removed through this activity, leading to the loss of silencing. Additionally, it has been suggested that loss of silencing during VIGS could be due to base pairing between the plant gene fragment present in BMV and the plant target mRNA, with the resulting double-stranded structure targeted for removal from the virus sequence by enzymes in the RNAi pathway (Pacak et al., 2010).
Here, we report our work directed toward producing a better BMV-based VIGS vector by improving the genetic stability of foreign inserts in BMV genomic RNA3. We demonstrated that our modified vector retained its foreign gene fragment insert longer in N. benthamiana and maize (Zea mays), and this observation is correlated with enhanced target gene silencing and more extensive visible silencing phenotypes, involving additional leaves and greater penetration of leaf tissue in maize.
RESULTS
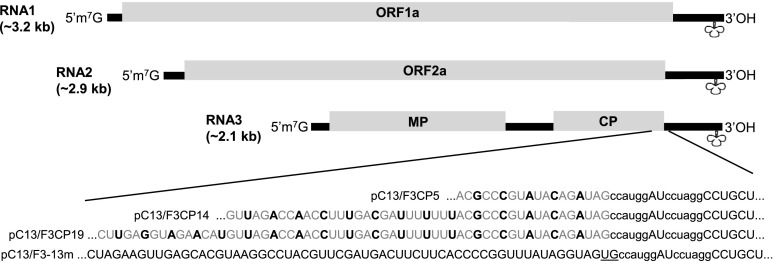
Modification of BMV RNA3 Sequences
In an effort to improve insert stability in the BMV VIGS vector during virus infection, we utilized the mFold Web Server to predict the folding of the full-length BMV RNA3 sequence from the original vector, BMVF13m, and identify secondary structure(s) in the region where we insert foreign gene fragments. Based on the prediction, a hairpin-like structure was found in the region (nucleotide positions 1,820–1,844) flanking the two insert cloning sites, NcoI and AvrII (Supplemental Fig. S2A, arrows). To alter this hairpin-like structure, we deleted nucleotides U and G present between the CP open reading frame (ORF) stop codon and the NcoI restriction site and made synonymous substitutions in the third nucleotide in each amino acid codon for the C-terminal five, 14, or 19 amino acids through mutagenic PCR of the plasmid containing the RNA3 sequence of the original vector (i.e. pC13/F3-13m; Fig. 1). The computer-predicted structures for RNA3 of one of the modified vectors, within plasmid pC13/F3CP5 of modified vector BMVCP5, showed that the region containing the original hairpin-like structure was modified to place the two insert cloning sites on the same side of a long stem loop-like structure (Supplemental Fig. S2, A and B, arrows). Considering that a gene fragment insert in BMV RNA3 is present during VIGS, we also modeled RNA3 (+) and (−) strand sequence structures for the BMVF13m and BMVCP5 vectors containing antisense gene fragments utilized in this work. Differences in the folded structure for both the full-length (+) and (−) strand sequences, primarily near the NcoI cloning site, were apparent for RNA3 of the modified vector BMVCP5 containing 250-bp inserts of ZmPDS, ZmHSP70-1, and GFPuv compared with the original vector with the same inserts (Supplemental Figs. S2, C–H, and S3, A–F). For the 250-bp insert representing ZmPDS, where the least difference in structure was apparent between the full-length (+) RNA3 sequence of the original and modified vectors with insert (Supplemental Fig. S2, C and D), the structure difference was enhanced when folding only the CP subgenomic RNA of each vector with insert (Supplemental Fig. S4, A and B). Thus, the folding predictions suggest that sequence modifications introduced into the original vector would alter the structures of (+) and (−) strands of RNA3 and the (+) strand of RNA4 in the modified BMV vector, BMVCP5, in the presence of inserts.
Figure 1.
Modification of the RNA3 sequence in the original BMV silencing vector. Portions of the transcribed plasmid sequences in pC13/F3-13m (plasmid containing BMV RNA3 from the original virus vector, BMVF13m) and pC13/F3CP5, pC13/F3CP14, or pC13/F3CP19 (plasmids containing BMV RNA3 from the modified virus vector BMVCP5, BMVCP14, or BMVCP19) proximal to the NcoI and AvrII cloning sites are shown. Nucleotide substitutions (black and boldface uppercase letters) in the sequence representing RNA3 from the modified plasmids pC13/F3CP5, pC13/F3CP14, and pC13/F3CP19 are shown. The sequence of the original RNA3 in pC13/F3-13m is shown for comparison (bottom). Lowercase letters represent the two cloning sites. Underlined uppercase letters are two nonviral nucleotides between the CP ORF and the NcoI restriction site in pC13/F3-13m. Nucleotide substitutions are shown in context with the full BMV genome (RNA1, RNA2, and RNA3). Within the genome schematic, lines represent untranslated regions and rectangular boxes represent ORFs. 1a and 2a, Proteins associated with virus accumulation; MP, movement protein; CP, coat protein. Cloverleaf shapes show the approximate location of the tRNA-like structure.
Local and Systemic Infection of the Original and Modified BMV Vectors in Their Host Plants
N. benthamiana plants Agrobacterium tumefaciens infiltrated with the original BMVF13m vector showed mild mosaic symptoms in their upper noninfiltrated young (i.e. systemic) leaves by 5 d post A. tumefaciens infiltration (dpai). N. benthamiana plants A. tumefaciens infiltrated with novel modified BMVCP5, BMVCP14, or BMVCP19 vectors developed mild mosaic symptoms in their systemic leaves by 5, 6, and 6 dpai, respectively. Through semiquantitative reverse transcription (RT)-PCR of extracts from A. tumefaciens-infiltrated leaves, it was determined that RNA3 of BMVF13m and BMVCP5 accumulated to similar levels by 4 dpai (Fig. 2A). This indicated that the sequence modification made within pC13/F3CP5 did not affect BMVCP5 RNA3 accumulation in the initial stages after infiltration of N. benthamiana.
Figure 2.
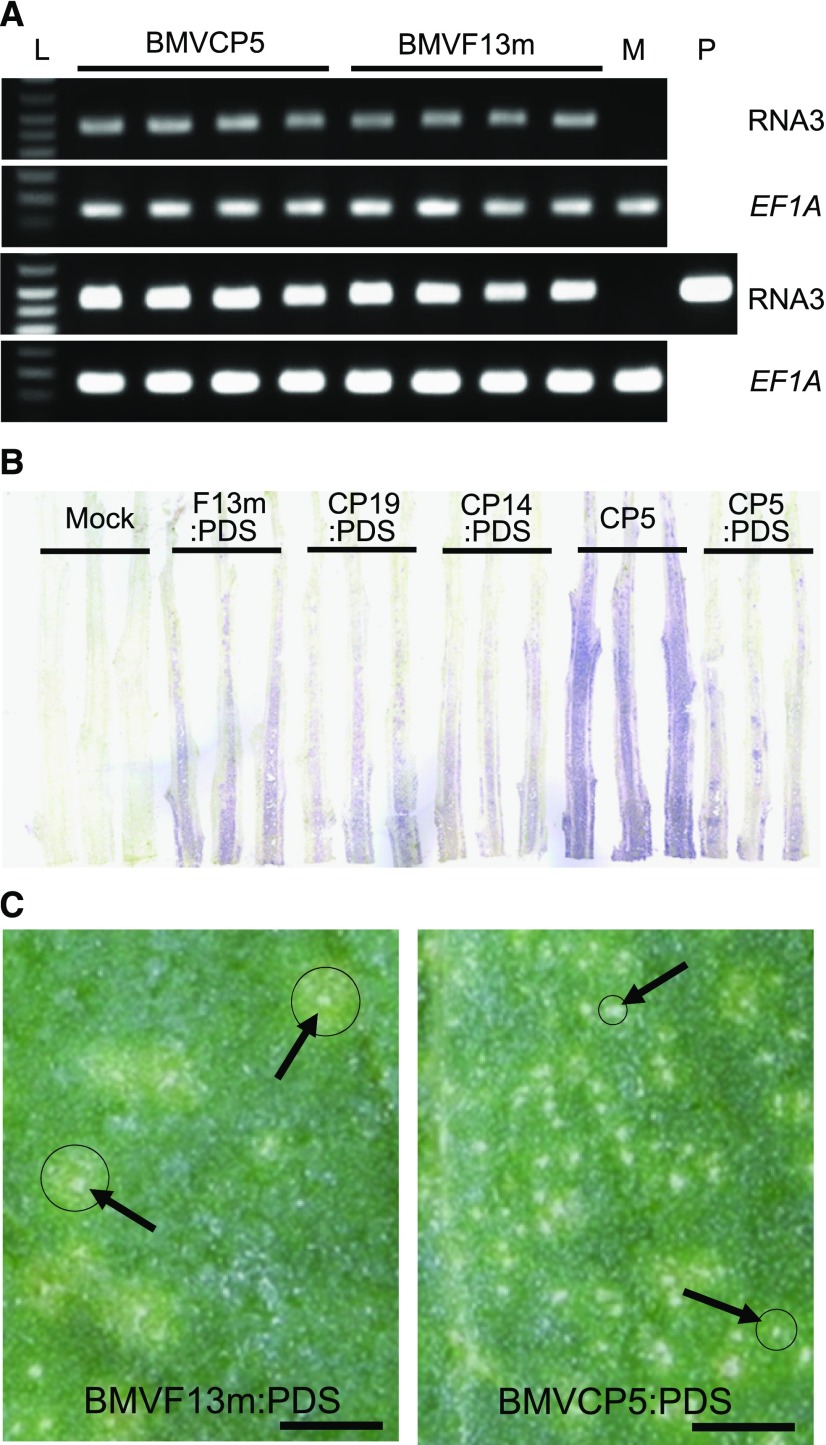
Modified BMV vectors can accumulate similarly to the original BMV vector in N. benthamiana after agroinfiltration. A, Accumulation of BMVCP5 and BMVF13m in N. benthamiana leaves at 4 dpai as measured through semiquantitative RT-PCR. PCR product from BMV genomic RNA3 after 25 cycles (top) and 30 cycles (bottom) is shown. Maize translation elongation factor 1A (ZmEF-1A) transcript levels were used as internal normalization controls. Each lane represents PCR product from an individual plant. Lane L is DNA ladder. Lane M represents amplification from extract of mock-inoculated tissue, as negative control. Lane P represents amplification from plasmid pC13/F3CP5 as positive control. B, Accumulation of BMV vectors in N. benthamiana stems at 10 dpai. BMV CP was detected through a tissue-print assay using a polyclonal antibody against the BMV CP. Purple color in stem longitudinal prints indicates positive detection of the virus CP. The experiment was repeated twice with similar results. C, Local lesions (noted by arrows) imaged at 6 dpi with BMVF13m:PDS or BMVCP5:PDS in leaves of C. amaranticolor. Strong chlorotic rings outside of necrotic lesions are marked with circles. Bars = 7 mm.
To determine whether the modified BMV vectors could be used as VIGS vectors in plants, a 250-bp ZmPDS sequence was cloned into the BMV RNA3 of the original or modified vectors. N. benthamiana plants infiltrated with A. tumefaciens containing BMVF13m:PDS displayed systemic mosaic symptoms by 5 dpai, while plants infiltrated with A. tumefaciens containing BMVCP5:PDS displayed systemic virus symptoms by 6 dpai. Plants infiltrated with A. tumefaciens containing BMVCP14:PDS or BMVCP19:PDS did not show systemic virus symptoms until 8 dpai. By 10 dpai, BMV CP was detected in stem sections infected with any of the vector constructs containing the PDS fragment, with the strongest and most extensive CP signal from the stems infected with BMVF13m:PDS (Fig. 2B). None of the vectors containing the PDS fragment accumulated as well as the modified virus vector without insert (BMVCP5), indicating that the foreign insert delayed systemic virus accumulation (Fig. 2B). When maize seedlings were inoculated with virion prepared from extracts of the N. benthamiana leaves infected with BMVF13m:PDS or BMVCP5:PDS, mosaic symptoms were observed in the base of the first systemically infected young leaves by 6 d post inoculation (dpi; BMVF13m:PDS) or 7 to 8 dpi (BMVCP5:PDS). Interestingly, maize seedlings inoculated with virion prepared from extracts of N. benthamiana leaves infected with BMVCP14:PDS or BMVCP19:PDS did not show systemic mosaic symptoms for approximately 2 weeks post virus inoculation, suggesting that the sequence modifications in these two vectors affected their ability to spread systemically or accumulate to levels capable of causing mosaic symptoms in maize. Consequently, only BMVF13m and BMVCP5 vectors were selected for further comparison in this study.
To investigate why systemic symptoms induced by BMVCP5:PDS were slightly delayed in these two host plants compared with BMVF13m:PDS, virion of these two viruses were inoculated to leaves of Chenopodium amaranticolor, a local lesion host of BMV. By 6 dpi, local lesions induced by BMVCP5:PDS were small and defined, while most local lesions induced by BMVF13m:PDS continued to expand and formed large chlorotic rings surrounding the initial necrotic lesions (Fig. 2C). The absence of large chlorotic rings around the BMVCP5:PDS-induced necrotic lesions suggested that intercellular spread was impeded, and this might be correlated with the maintenance of insert.
Stability of Foreign Inserts in the BMVF13m and BMVCP5 Vectors during Infection
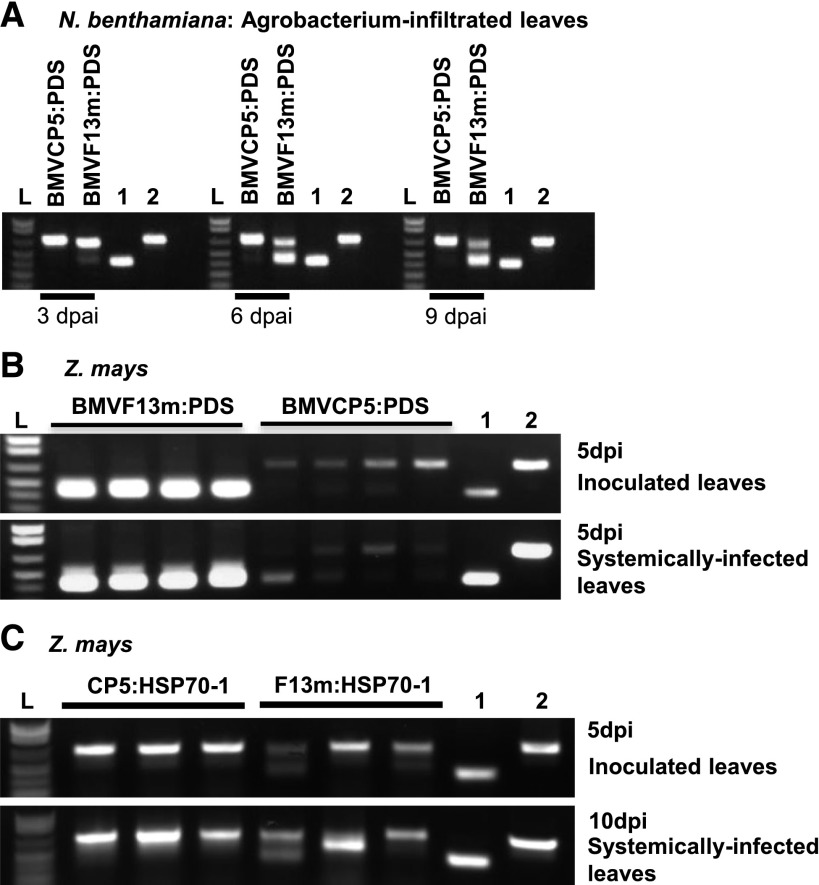
To compare insert stability within the BMVF13m and BMVCP5 vectors during infection in N. benthamiana, leaves infiltrated with A. tumefaciens containing BMVF13m:PDS or BMVCP5:PDS were harvested at 3, 6, and 9 dpai. Results from RT-PCR analysis of virion RNA from infiltrated leaves showed that BMVF13m:PDS had begun to lose the PDS fragment insert by 3 dpai, and by 6 dpai, most BMVF13m:PDS had lost its PDS insert, as indicated by the appearance of a strong and smaller sized band in the gel similar in size to the PCR product of pC13/F3CP5 without a PDS insert (Fig. 3A; Supplemental Fig. S5A). In contrast, the PCR products obtained from the BMVCP5:PDS-infiltrated leaves harvested at 3 to 9 dpai appeared mostly as a single band of similar size to the band amplified from pC13/F3CP5 with a PDS insert (pC13/F3CP5:PDS250). This result indicated that insert stability in the modified BMVCP5 vector was improved over the original vector in the N. benthamiana infiltrated leaves.
Figure 3.
BMVCP5 displays greater insert stability than BMVF13m in N. benthamiana and maize. A, N. benthamiana leaves were harvested at various dpai with BMVCP5:PDS or BMVF13m:PDS. Virion RNA was isolated and subjected to RT-PCR assays. RT-PCR products were obtained using pooled cDNA reactions from three plants representing leaves infiltrated with BMVCP5:PDS or BMVF13m:PDS, respectively. The experiment was repeated twice with similar results. B, RT-PCR products from extracts of individual inoculated (top) or systemically infected (bottom) maize leaves infected with BMVF13m:PDS or BMVCP5:PDS. Each lane represents extract from an individual plant. All samples were taken at 5 dpi. C, RT-PCR products from extracts of inoculated (top) or systemically infected (bottom) maize leaves infected with BMVF13m:HSP70-1 (F13m:HSP70-1) or BMVCP5:HSP70-1 (CP5:HSP70-1). Each lane represents extract from an individual plant. Samples were taken at 5 dpi (inoculated leaves) or 10 dpi (systemically infected leaves). All PCR products were visualized on 1% agarose gels after electrophoresis. Lanes labeled 1 and 2 are PCR products from pC13/F3CP5 or pC13/F3CP5:PDS plasmid DNA and show the positions of bands expected when the full-length insert is absent (410 bp; lane 1) or present (660 bp; lane 2). Lane L is 1 kb plus DNA ladder.
To compare insert stability within the BMVF13m:PDS and BMVCP5:PDS vectors during infection of maize, virion RNA was isolated from leaves inoculated with partially purified virion from N. benthamiana tissues or from the first systemically infected leaves of the same inoculated plants at 5 dpi. Through RT-PCR analysis, it was observed that, by 5 dpi, BMVF13m:PDS had lost its PDS insert in all inoculated leaf samples (Fig. 3B, top). In contrast, BMVCP5:PDS maintained a majority of full-length PDS insert (Fig. 3B, top). Also at 5 dpi, BMVCP5:PDS again maintained more full-length insert than BMVF13m:PDS in the first systemically infected leaves, although loss of full-length insert was more noticeable in these leaves than in the inoculated leaves for BMVCP5:PDS (Fig. 3B, bottom). In a separate experiment, improved insert maintenance was demonstrated from 3 to 5 dpi in inoculated leaves for BMVCP5:PDS compared with BMVF13m:PDS, although some loss of insert occurred at a higher frequency for BMVCP5:PDS than in the previously described experiment (compare Fig. 3B, top, with Supplemental Fig. S5B). Also, in an experiment conducted separately from all other experiments, BMVCP5:PDS maintained a high percentage of full-length insert in the second systemically infected leaves from three of four plants, while BMVF13m:PDS lost its PDS insert in all four analyzed plants at 7 dpi (Supplemental Fig. S5C). To determine if inserts from other host genes also were more stable in the BMVCP5 vector in infected maize plants, maize seedlings were inoculated with BMVF13m:HSP70-1 or BMVCP5:HSP70-1 virion (insert size, 250 nucleotides). The inoculated and the second systemically infected leaves were harvested at 5 and 10 dpi, respectively, for the analyses of insert stability. Through RT-PCR analyses, although BMVF13m:HSP70-1 displayed increased insert stability compared with BMVF13m:PDS, it was still less than that observed for BMVCP5:HSP70-1 in both inoculated and systemically infected leaves (Fig. 3C).
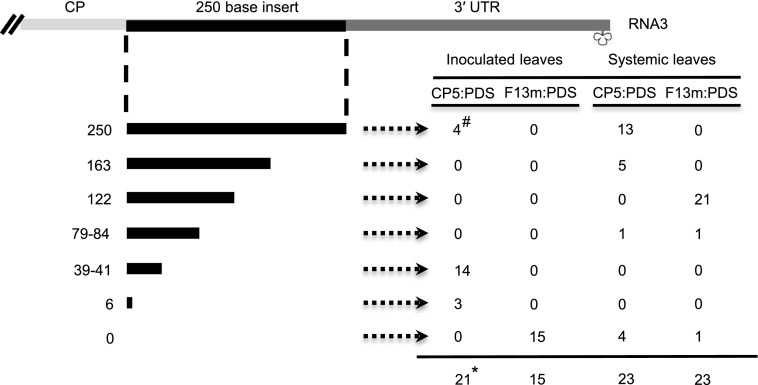
An additional experiment was conducted to further support these observations. Virus RT-PCR fragments from leaves inoculated or systemically infected with BMVF13m:PDS or BMVCP5:PDS were cloned and the plasmids were sequenced to determine the presence and the length of insert (Fig. 4). By 5 dpi, 100% of the clones from the leaves inoculated with BMVCP5:PDS retained some PDS insert sequence, with approximately 20% maintaining the full-length PDS insert. However, none of the clones from the leaves inoculated with BMVF13m:PDS maintained any PDS insert. In the second systemically infected leaves harvested at 7 dpi, 82% of the clones from the BMVCP5:PDS-infected tissues retained some PDS sequence, with 56% maintaining the full-length PDS insert. In contrast, none of the clones from the BMVF13m:PDS-infected tissues maintained a full-length insert, although a large percentage (96%) maintained a partial sequence. Results from all the experiments reported in this section indicated the increased stability of gene inserts in the BMVCP5 vector compared with the original BMVF13m vector.
Figure 4.
Stability of the PDS insert in the BMVF13m or BMVCP5 vector during infection in maize. Virion RNA was isolated from BMVF13m:PDS (F13m:PDS)- or BMVCP5:PDS (CP5:PDS)-inoculated leaves harvested at 5 dpi or from the second systemically infected leaves harvested at 7 dpi and used for RT-PCR. Three maize plants were used for each treatment, PCR products from the same treatment were gel purified, pooled, and cloned, and plasmids were sequenced for the presence of insert. The top diagram illustrates the RNA3 3′ sequence organization in the BMV vectors. The black lines and numbers in the far left column under the diagram show the lengths of retained PDS sequences in the progeny virus populations from the pooled samples for each treatment (250 nucleotides representing the full-length insert). #, Values in the table represent the number of clones from each treatment containing a particular length of insert; *, the total number of clones sequenced for the treatment. UTR, Untranslated region.
Visual Phenotypes and Target Gene Transcript Levels in Leaves Targeted for PDS or HSP70-1 Gene Silencing through VIGS
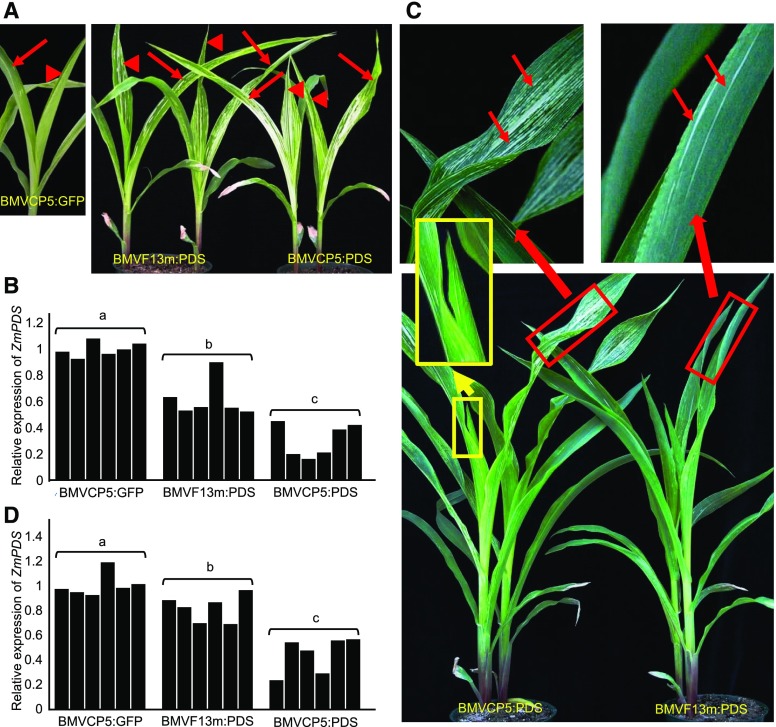
To demonstrate that the BMVCP5 vector could induce stronger and more uniform gene silencing in maize than the original BMVF13 vector, we analyzed maize leaves targeted for PDS silencing. Maize plants inoculated with BMVF13m:PDS virion isolated from A. tumefaciens-infiltrated N. benthamiana leaves at 4 dpai first developed multiple photobleaching streaks in the second systemically infected leaves by 10 dpi that were further apparent at 12 dpi (Fig. 5A, arrows, right). Photobleaching streaks continued to appear in the third systemically infected leaves, but the number decreased significantly (Fig. 5A, arrowheads). Maize plants inoculated with BMVCP5:PDS displayed numerous photobleaching streaks in the second and third systemically infected leaves by 12 dpi (Fig. 5A, right). Plants inoculated with BMVCP5:GFP showed only mosaic symptoms in their second and third systemically infected leaves (Fig. 5A, left). Knockdown of PDS expression in the second and third systemically infected leaves at 14 dpi with BMVCP5:PDS was approximately twice the relative levels observed with BMVF13m:PDS (Fig. 5B). Photobleaching streaks also were prominent in the fourth and fifth systemically infected leaves of plants at 30 dpi with BMVCP5:PDS (Fig. 5C). In contrast, very few photobleaching streaks were seen in the fourth systemically infected leaves and none in the fifth systemically infected leaves of plants inoculated with BMVF13m:PDS (Fig. 5C). The emerging sixth systemically infected leaf of plants inoculated with BMVCP5:PDS also displayed photobleaching streaks (Fig. 5C). Results from quantitative reverse transcription (qRT)-PCR analyses indicated that knockdown of PDS expression in BMVCP5:PDS-infected maize plants was about 50% to 70%, while knockdown of PDS expression in BMVF13m:PDS-infected maize plants was less than 20% at 30 dpi in the fourth and fifth systemically infected leaves (Fig. 5D).
Figure 5.
Silencing the PDS gene in maize using BMVF13m:PDS or BMVCP5:PDS. Similar amounts of BMVCP5:GFP, BMVF13m:PDS, or BMVCP5:PDS virion were inoculated to maize seedlings. A, Systemically infected leaves above the inoculated leaves from BMVCP5:GFP-inoculated plants (left image) or BMVF13m:PDS- or BMVCP5:PDS-inoculated plants (right image) were photographed at 12 dpi. Arrows indicate the second systemically infected leaves, and arrowheads indicate the third systemically infected leaves. Similar visual phenotypes were observed in two additional experiments. B, Relative expression levels of ZmPDS transcript in maize plants inoculated with BMVCP5:GFP, BMVF13m:PDS, or BMVCP5:PDS were determined through qRT-PCR of extracts from the second and third systemically infected leaves harvested 2 weeks post inoculation. The expression of the maize EF-1A gene in these plants was analyzed and used to normalize ZmPDS expression levels across treatments. Each bar represents expression from an individual plant. C, Maize plants inoculated with BMVF13m:PDS or BMVCP5:PDS virion were photographed at 30 dpi (bottom image). Images at top are magnifications of the blades from the fourth systemically infected leaves shown in the image below (red rectangular boxes). Photobleaching streaks are indicated with arrows. Images in yellow rectangular boxes indicate unmagnified and magnified images of the emerging sixth systemically infected leaf with photobleaching streaks. D, Relative expression levels of ZmPDS in maize plants inoculated with BMVCP5:GFP, BMVF13m:PDS, or BMVCP5:PDS were determined through qRT-PCR of extracts from the fourth and fifth systemically infected leaves harvested at 30 dpi. The qRT-PCR assay was done as described in B. For B and D, different letters above each treatment group indicate significant differences in values between those treatments at the 0.05 significance level as determined by ANOVA and lsd test.
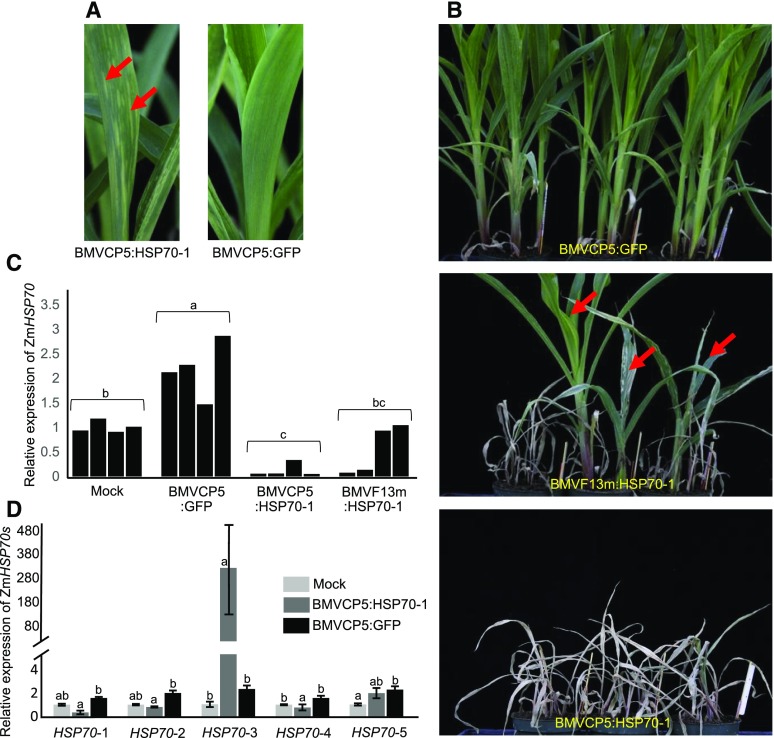
To knock down maize HSP70-1 gene expression through VIGS, we inoculated maize seedlings with BMVF13m:HSP70-1, BMVCP5:HSP70-1, or BMVCP5:GFP virion. The maize plants inoculated with BMVF13m:HSP70-1 started to show systemic mosaic symptoms by 6 dpi. By 2 weeks post inoculation, the second and third systemically infected leaves showed chlorotic streaks along the veins. These streaks continued to expand and later became necrotic, causing leaf collapse. By 1 month post inoculation, most of the plants inoculated with BMVF13m:HSP70-1 died while a few plants continued to grow, often with severe necrosis in their leaves and plant stunting (Fig. 6B). Plants inoculated with BMVCP5:HSP70-1 developed systemic mosaic by 7 to 8 dpi followed by the appearance of chlorotic streaks in their systemic leaves (Fig. 6A). By 1 month post inoculation, all the plants inoculated with BMVCP5:HSP70-1 died (Fig. 6B). Similar results were obtained in two additional experiments (Table I). In all three experiments, the BMVCP5:GFP-inoculated plants showed mosaic symptoms only in leaves and stunting of the plants compared with the mock-inoculated plants (Fig. 6B). qRT-PCR analyses demonstrated that BMVCP5:GFP infection in maize plants generally increased the expression of cytosolic HSP70 in systemically infected leaves compared with those from mock-inoculated plants (Fig. 6C). We observed a similar effect on the HSP70 protein in barley leaves infected with BMV, as determined through immunocytochemistry with leaf tissue sections and anti-BMV CP or anti-HSP70 antibodies (Supplemental Fig. S6). In spite of the potential of induced gene expression during BMV infection, inoculation of maize with BMVCP5:HSP70-1 resulted in less HSP70-1 expression in the upper leaves compared with the tissue infected with BMV expressing the GFP fragment or mock inoculated (Fig. 6C). An apparent bimodal expression of HSP70-1 was evident in tissues infected with BMVF13m:HSP70-1, with some extracts displaying transcript levels similar to those from mock-inoculated plants and others similar to those from plants infected with BMVCP5:HSP70-1. We considered that the inability to statistically separate results from BMVF13m:HSP70-1-infected tissue from those from BMVCP5:HSP70-1-infected and mock-inoculated tissues was due to this apparent bimodal distribution of HSP70-1 transcript expression between plants infected with BMVF13m:HSP70-1. Expression values for ZmHSP70 transcript from plants infected with BMVF13m:HSP70-1 were analyzed for unimodal expression using results from three experiments. A test for unimodal expression using Hartigans dip test (Maechler, 2015) showed at least bimodality (P = 0.086), with marginal significance for transcripts from this population. The bimodal expression distribution likely reflected enhanced selection against virus with an HSP70-1 insert compared with a PDS insert, since silencing HSP70-1 resulted in tissue death, while silencing PDS resulted in chlorotic, but living, cells that could continue to produce virus with insert.
Figure 6.
A more extensive necrosis phenotype is correlated with greater target transcript silencing in maize infected with BMVCP5:HSP70-1. Similar amounts of BMVCP5:GFP, BMVF13m:HSP70-1, or BMVCP5:HSP70-1 virion were inoculated to maize seedlings. A, Systemically infected leaves from maize plants inoculated with BMVCP5:HSP70-1 (left) or BMVCP5:GFP (right) were photographed 2 weeks post inoculation. Arrows indicate pale green streaks in the BMVCP5:HSP70-1-infected leaf. B, Chlorotic streaks continued to expand and eventually changed to necrosis, resulting in leaf death for many plants infected with BMVF13m:HSP70-1 and all plants infected with BMVCP5:HSP70-1 at 1 month post virus inoculation. Three plants inoculated with BMVF13m:HSP70-1 (arrows) showed necrosis and severe stunting but did not die. C, Relative expression levels of ZmHSP70-1 transcript in the mock-inoculated (Mock), BMVCP5:GFP-, BMVCP5:HSP70-1-, or BMVF13m:HSP70-1-inoculated maize plants were determined through qRT-PCR of extracts from second and third systemically infected leaves harvested at 18 dpi. Maize EF-1A transcript levels in these plants were determined and used to normalize ZmHSP70-1 expression levels across treatments. Each bar represents expression from a single plant. Different letters above each treatment group indicate significant differences in mean values between those treatments at the 0.05 significance level determined by ANOVA followed by lsd test. The experiment was repeated twice with identical findings. D, Analysis of transcript levels of five different ZmHSP70s in plants silenced for HSP70-1 at 18 dpi. Primers used for qRT-PCR are listed in Supplemental Table S1. The expression of maize EF-1A in these samples was used to normalize HSP70 expression levels across treatments for each HSP70 family member. Each bar and error bracket represents the mean value ± sd for results from three plants. Different letters above each treatment group indicate significant differences in values between those treatments at the 0.05 significance level determined by ANOVA followed by lsd test. The experiment was repeated with similar results.
Table I. Plant death caused by the knockdown of maize HSP70 using BMVF13m:HSP70-1 or BMVCP5:HSP70-1.
Maize seedlings were inoculated with same amount of virion of BMVCP5:GFP, BMVF13m:HSP70-1, or BMVCP5:HSP70-1. The number of dead plants was counted at 1 month post virus inoculation. The experiment was repeated twice. Values shown are numbers of dead plants/total plants inoculated.
| Virus | Experiment |
||
|---|---|---|---|
| I | II | III | |
| BMVF13m:HSP70-1 | 16/18 | 16/18 | 15/18 |
| BMVCP5:HSP70-1 | 18/18 | 18/18 | 18/18 |
| BMVCP5:GFP | 0/18 | 0/18 | 0/18 |
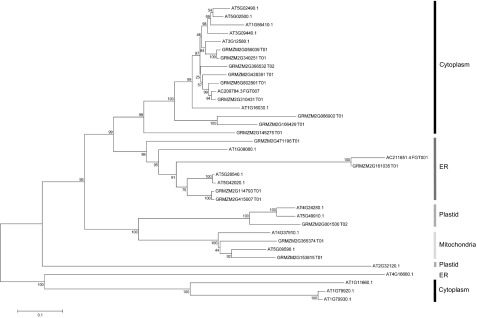
HSP70 is encoded by a gene family, and we were interested in determining whether other gene members also were altered in expression when we targeted HSP70-1 for silencing. A BLAST search of the maize genome sequence using the GRMZM2G428391 sequence (our HSP70-1 sequence) indicated that maize may have 18 HSP70 genes in its genome. Phylogenetic analysis against the known Arabidopsis (Arabidopsis thaliana) HSP70 protein sequences suggested that 10 of the maize HSP70s may be cytoplasmic (Fig. 7). Further analysis of the 10 possible maize cytoplasmic HSP70 nucleotide sequences showed that only HSP70-2, HSP70-3, HSP70-4, and HSP70-5 contained one or more stretches with more than 21 nucleotides identical to stretches in the HSP70-1 insert (Supplemental Fig. S1A). qRT-PCR analysis with primers designed to amplify individual HSP70-1 to HSP70-5 transcripts showed induction of each after infection with BMVCP5:GFP (Fig. 6D). qRT-PCR analysis of the tissues infected with BMVCP5-HSP70-1 using primers designed to amplify the sequence of maize HSP70-2 (GRMZM2G056039; Supplemental Fig. S1B) indicated that this gene was silenced when compared with the values from tissues infected with BMVCP5:GFP (Fig. 6D). A similar result was observed when the same tissues were analyzed using primers designed to amplify the sequence of HSP70-4, and a similar trend (not statistically significant within the sample size studied) was observed with primers specific for HSP70-5 (Fig. 6D). Interestingly, qRT-PCR analyses using the primers designed to amplify HSP70-3 (GRMZM2G366532; Supplemental Fig. S1B) indicated a large induction of expression for this gene in plants inoculated with BMVCP5:HSP70-1 compared with its expression in plants inoculated with BMVCP5:GFP or phosphate buffer only (Fig. 6D).
Figure 7.
Phylogenetic tree of HSP70 proteins from Arabidopsis and maize. The tree was constructed by aligning the maize HSP70 protein sequences with the protein sequences of Arabidopsis HSP70s using the MEGA 6.0 program. The percentage values of 1,000 bootstrap trials are indicated near the nodes. The subcellular localizations of maize HSP70s were determined based on clustering with the known Arabidopsis HSP70s (Lin et al., 2001). ER, Endoplasmic reticulum.
DISCUSSION
During VIGS, a poorly visible silencing phenotype, observed as less complete penetration within specific organs or decreased appearance in newly developing leaves, is often correlated with the loss of the foreign gene insert in the VIGS vector (Bruun-Rasmussen et al., 2007; Mei et al., 2016; Wang et al., 2016). Foreign gene insert loss can occur soon after virus inoculation or after serial passage through a host (Avesani et al., 2007; Igarashi et al., 2009; Yuan et al., 2011). The correlation of silencing phenotype with the presence of insert in the VIGS vector is consistent with the notion that the virus vector is the ultimate initial substrate for silencing pathway enzymes. Here, we modified our BMV VIGS vector to address this issue, creating a vector that maintains its insert for longer periods during infection in N. benthamiana and maize, therefore providing an increased tissue penetration and longer-lived silencing phenotype. We modified the BMV RNA3 sequence just 5′ of the restriction sites for foreign gene fragment insertion through a computer-assisted sequence modification strategy. The modified BMVCP5 vector has a deletion of two nucleotides immediately after the CP stop codon and synonymous substitutions in the third position nucleotides of the five C-terminal amino acid codons encoding the CP. This modification resulted in greater retention of gene fragments complementary to ZmPDS and ZmHSP70-1 during virus infection in N. benthamiana and maize (Fig. 3; Supplemental Fig. S5). We also observed greater retention of PDS insert within the BMV vectors having synonymous third position nucleotide substitutions in codons encoding the C-terminal 14 and 19 amino acids of the CP during their infection in N. benthamiana. The enhanced foreign insert retention in the BMVCP5 vector in maize was associated with a greater penetration of visible silencing phenotypes in leaf tissue and silencing in additional developing leaves compared with the original BMV vector (Figs. 5 and 6). Although the BMVCP5 vector retained inserts better than the original vector in N. benthamiana and maize, it had a 1- to 2-d delay in systemic infection in these two hosts compared with the original vector. We speculate that the slower systemic infection caused by BMVCP5:PDS or BMVCP5:HSP70-1 in plants was due mainly to the improved insert stability in the modified vectors, and from this, attenuated virus accumulation in systemically infected tissue often associated with nonviral inserted sequences (Bujarski and Kaesberg, 1986; Nagy and Bujarski, 1996; Bruun-Rasmussen et al., 2007)
Regarding the mechanism by which the new VIGS vector retains insert longer, decreased recombination by the modified vector is one possibility. It is known that many viruses undergo recombination during infection (Bujarski, 2013). BMV undergoes recombination during infection and is a model virus for RNA recombination studies (Bujarski and Kaesberg, 1986; Nagy and Bujarski, 1996; Bruyere et al., 2000; Alejska et al., 2005; Urbanowicz et al., 2005; Kwon and Rao, 2012; Kolondam et al., 2015). Our modeling of secondary structures for RNA3 from both vector species (original or modified) with inserts indicated that, regardless of the insert fragment modeled, there was a difference in structure between the original BMVF13m RNA3 vector sequence and the modified BMVCP5 RNA3 vector sequence (Supplemental Figs. S2, C–H, S3, A–F, and S4, A and B). It is generally considered that most RNA recombination events proceed through template switching and that RNA structures that cause the polymerase to pause may enhance its release from the donor template strand, leading to association with the acceptor template strand (Sztuba-Solińska et al., 2011). Olsthoorn et al. (2002) determined that insertion of structured stem-loop sequences into regions where recombination occurred could influence the accumulation of recombinants. It will be important to determine whether structural differences caused by the nucleotide deletions and substitutions in our modified vector altered stem loops in planta, leading to decreased recombination by this vector. Additionally, it would seem that altering RNAi-mediated deletion of the antisense foreign gene fragment within RNA3 of the virus vector when base paired with target host mRNA, a mechanism suggested by Pacak et al. (2010) to explain the loss of VIGS in some instances, is less likely to account for insert stability in our modified vector, since foreign gene fragment sequences in the original and modified vectors were identical. However, considering that the RNA structures involving foreign gene fragment sequences differ in the (+) strands between the original and modified vectors (Supplemental Fig. S2, C–G), there could be diminished base-pairing ability between host mRNA and the gene fragment in the modified vector in planta, supporting this explanation for insert stability. Further work is necessary to determine whether this or another mechanism explains why inserts are more stable within this modified VIGS vector.
Many silencing studies with BMV vector used N. benthamiana as an intermediate host to increase virus titer for reliable infection of the subsequent monocotyledonous hosts (Ding et al., 2007; van der Linde et al., 2011, 2012; Hemetsberger et al., 2012; Zhu et al., 2014; Zhan et al., 2016). Therefore, it was important to determine that, in addition to maintaining foreign gene fragment inserts, the modified vector accumulated well in this intermediate host. BMVCP5 accumulated to similar levels to the original vector, BMVF13m, in the infiltrated leaves of N. benthamiana (Fig. 2A). It should be noted, however, that the BMV vectors were infiltrated into large areas of the N. benthamiana leaves, negating much of the need for local intercellular spread within this tissue for infection. Additionally, tissue-print assays using BMVF13m:PDS, BMVCP5:PDS, BMVCP14:PDS, and BMVCP19:PDS showed that the three modified vectors accumulated less in the stems of A. tumefaciens-infiltrated N. benthamiana plants than the original BMVF13m:PDS vector, and the symptoms induced in systemically infected leaves of N. benthamiana and maize were delayed for all the modified vectors. Thus, it is possible that these vectors move less efficiently between cells, through the vasculature, or both, due to the maintenance of their inserts. It is known that a negative correlation exists between the length of a foreign insert and the accumulation of a BSMV-silencing vector in systemically infected leaves of barley (Bruun-Rasmussen et al., 2007). Our findings with C. amaranticolor may provide further explanation of this phenomenon in that lesions induced by BMVCP5:PDS on inoculated leaves stayed small while lesions induced by BMVF13m:PDS continued to expand and formed chlorotic rings surrounding the initial lesions, the latter correlated with the loss of insert. While the results from C. amaranticolor alone could not distinguish between poor accumulation per cell or poor intercellular movement of the improved vector, the results from the N. benthamiana infiltrated leaves suggest that accumulation per cell is similar between the two BMV vectors and that intercellular or vascular spread is delayed for the improved vector. This delay, which could lead to fewer rounds of virus replication and less opportunity for recombination, is modest for BMVCP5, however, and does not negate the benefit of the improved insert stability to induce target gene silencing by this vector.
Findings from our HSP70-silencing studies not only supported our PDS-silencing results, showing that the modified BMVCP5 vector induced better gene silencing in maize than our original vector, but also provided additional information for those interested in the regulation of genes in this family. HSP70 members are core components of the cellular chaperone complex, with roles in various cellular processes including protein folding, assembly, translocation, and degradation (Mayer and Bukau, 2005; Bukau et al., 2006; Park and Seo, 2015). They prevent the aggregation of denatured proteins and assist in protein refolding during stress conditions (Mayer and Bukau, 2005; Tyedmers et al., 2010). Plant HSP70s also are influenced by and influence virus infection in plants (Verchot, 2012; Park and Seo, 2015). Regarding the influence of virus infection on HSP70 expression, immature pea (Pisum sativum) cotyledons infected with four different plant RNA viruses displayed increased HSP70 transcript levels in the region where virus was rapidly accumulating (Aranda et al., 1996; Escaler et al., 2000). Aranda et al. (1996) also observed during infection with Pea seed-borne mosaic virus that one HSP70 family member, PsHCP71.2, was induced while another, PsHSC71.0, was marginally reduced in transcript level in the tissue where virus was replicating. In N. benthamiana, challenge with five RNA viruses showed significant increases in the expression of multiple HSP70 genes, many encoding proteins that resided in the cytoplasm (Chen et al., 2008; Alam and Rochon, 2015). HSP70 family members also were induced by multiple viruses in Arabidopsis (Whitham et al., 2003). Regarding HSP70 expression and its effect on virus accumulation or plant phenotype, when one of the HSP70 family members, NbHSP70c-1, was overexpressed, the accumulation of Tobacco mosaic virus was increased (Chen et al., 2008). In Chenopodium quinoa plants heat shocked to induce HSP70 expression and then challenged with CNV, viral genomic RNA accumulation increased (Alam and Rochon, 2015). Silencing the expression of cytosolic Nicotiana HSP70 family members in N. benthamiana using the TRV- or PVX-based VIGS vectors resulted in dwarfed and crinkled leaves or white spots near major veins in developing leaves (Chen et al., 2008; Wang et al., 2009). We previously observed a dwarfing phenotype after we silenced HSP70 in barley with our original BMV vector (Ramanna et al., 2013). Based on this finding, we speculated that silencing HSP70 with BMVF13m:HSP70-1 or BMVCP5:HSP70-1 would produce easily differentiated visible silencing phenotypes in maize by these vectors. Indeed, our new vector did produce a more dramatic visible stunting and death phenotype than the original vector (Fig. 6B). We also analyzed the expression patterns for HSP70-1 and four additional cytosolic HSP70 family members (HSP70-2 to HSP70-5) that were potential off targets during the silencing of HSP70-1. For all five HSP70 members, we observed increased transcript levels in plants infected with BMVCP5:GFP versus mock-inoculated plants (Fig. 6D). Additionally, we observed that infection with one of our BMV vectors induced a higher level of HSP70 protein in barley (Supplemental Fig. S6). Thus, BMV is similar to other viruses in its ability to induce the expression of this gene family during infection.
Interestingly, during the silencing of HSP70-1, we observed decreased levels of HSP70-2, HSP70-4, and HSP70-5 transcript, but an over 100-fold increase in HSP70-3 expression, compared with levels from tissue infected with BMVCP5:GFP (Fig. 6D). Sequence alignment of maize cytosolic HSP70s indicated that the four HSP70s (HSP70-2 to HSP70-5) harbored varying numbers and lengths of nucleotide stretches identical with the HSP70-1 insert in our BMV vector (one 32-nucleotide stretch in HSP70-2, 26- and 25-nucleotide stretches in HSP70-3, one 41-nucleotide stretch in HSP70-4, and 24-, 23-, and 41-nucleotide stretches in HSP70-5; Supplemental Fig. S1). HSP70-3, therefore, has the shortest stretch of identity with the HSP70-1 target sequence among all the HSP70s studied. We speculate from this that a portion of the over 100-fold increase of HSP70-3 expression in the BMVCP5:HSP70-1-inoculated maize plants could be due to inefficient off-target silencing during VIGS of HSP70-1. A further component of the increased HSP70-3 expression could be compensation by the host to balance the loss of expression of the other HSP70s due to silencing.
It is noteworthy that the two quantitative PCR (qPCR) primers used to amplify HSP70-1 had only one nucleotide difference from the corresponding regions in HSP70-5 (Supplemental Fig. S1B). From this, it could be considered that HSP70-5 transcript was amplified by the primers designed for HSP70-1, leading to an underestimate of the true silencing level of HSP70-1 expression. We consider this to be less likely, since qPCR primers designed for HSP70-4 transcript amplification had only two nucleotide differences (one in each primer and in similar locations within the primers to the position of the nucleotide difference between HSP70-1 and HSP70-4 transcript in the primer for HSP70-1) from the HSP70-3 sequence (Supplemental Fig. S1B), and we did not observe a large increase in HSP70-4 transcript from tissue infected with BMVCP5:HSP70-1, as would be expected considering the over 100× increase in HSP70-3 transcript levels compared with those of HSP70-4 (Fig. 6D).
Whether necrosis observed in HSP70-1-silenced maize plants was due to silencing the expression of multiple HSP70s or the induction of HSP70-3 expression requires further investigation. It was demonstrated that transient overexpression of a cytoplasmic pepper (Capsicum annuum) HSP70 (HSP70a) during heat stress or Xanthamonas campestris pv vesicatoria infection caused cell death in pepper leaves (Kim and Hwang, 2015). Chen et al. (2008) did not observe increased expression for any of the six HSP70 gene family members after infecting N. benthamiana plants with a PVX VIGS vector harboring a fragment of NbHSP70c-1. They did, however, observe a cell death phenotype in that study.
It is difficult to compare our findings showing the duration of the silencing phenotype with our modified BMV vector with the durations reported for other virus vectors infecting grasses. This is due to differences in growth conditions, different sizes or forms (i.e. antisense versus inverted repeat) of inserts, or different target genes and hosts, all of which affect virus accumulation and VIGS efficiencies (Senthil-Kumar and Mysore, 2011; Ramanna et al., 2013; Lee et al., 2015). With this caveat in mind and considering only vectors studied in maize, our modified vector, containing a 250-bp insert of PDS and inoculated by mechanical abrasion with partially purified vector virus from N. benthamiana onto the first two leaves of 8-d-old seedlings, induced visible silencing through the sixth systemically infected leaf at 30 dpi (Fig. 5), the last time point measured for visible phenotype or target transcript silencing. The maize-infecting Cucumber mosaic virus vector, containing a 215-bp fragment of IspH (a gene important for early stages of chloroplast development) and inoculated by a modified vascular puncture inoculation technique for maize seeds, induced silencing through the fifth systemically infected leaf and 60 dpi, again the last time points and growth stages measured (Wang et al., 2016). The Foxtail mosaic virus vector, containing a 313-bp insert of PDS and inoculated by DNA bombardment onto leaves of 7-d-old seedlings, induced photobleaching in most plants through the seventh systemically infected leaf, although virus harvested from leaves above sometimes contained full or partial inserts (Mei et al., 2016). It should be noted that all of these vectors utilize plasmids from which infectious virus is made in planta (Sun et al., 2013; Mei et al., 2016; Wang et al., 2016; this work). Thus, costs to conduct VIGS studies are reduced compared with vectors requiring in vitro transcription to produce infectious virus. Most importantly, from the findings reported in this study, it may be possible to apply the concept of modifying nucleotide sequences in the region of the insertion sites to limit virus recombination for each of these vectors. This is particularly relevant for virus vectors having multipartite genomes with an insertion site in or near a conserved domain between the genomes available for recombination. However, considering that the mechanism of recombination in this BMV system is not understood, this concept may still be relevant for monopartite viruses or multipartite viruses with insertion sites in nonconserved regions.
In summary, our findings demonstrated that modification of nucleotide sequences in a BMV-based VIGS vector can improve the stability of foreign inserts in the vector. Computer-assisted sequence modification methods may be considered to improve insert stability in other VIGS vectors, leading to enhanced gene-silencing phenotypes. The combination of vector improvement with other modifications, such as inoculation of assay plants with normalized, partially purified virus (van der Linde et al., 2011; Zhu et al., 2014), can improve the usefulness of VIGS for plant gene function studies in grasses.
MATERIALS AND METHODS
Modification of BMV Vector RNA3
Construction of the DNA-based BMV-silencing two-part vector, pC13/F1+2 and pC13/F3-13m, was reported previously (Sun et al., 2013). To improve the genetic stability of foreign inserts in this VIGS vector during RNA silencing in plants, we first predicted the structure formed by the full-length BMV RNA3 (F3-13m) sequence in the pC13/F3-13m vector using the mFold Web Server (http://mfold.rna.albany.edu/). Based on our interpretation of the predicted BMV RNA3 folding structure (Fig. 1B), we substituted nucleotides in the third position of codons for the last five, 14, or 19 amino acids at the C terminus of the CP through PCR using three primer sets (i.e. B3-1490F and B3-CP19R, B3-1490F and B3-CP14R, and B3-1490F and B3-CP5R; Supplemental Table S1). None of the nucleotide changes modified the CP amino acid sequence. The resulting PCR fragments were digested with AflII and NcoI restriction enzymes (New England Biolabs) and individually ligated into the original pC13/F3-13m vector predigested with AflII and NcoI to generate pC13-F3CP5 (five codon modifications), pC13/F3CP14 (14 codon modifications), and pC13/F3CP19 (19 codon modifications), respectively. All the modified RNA3 constructs contained two unique cloning sites (NcoI and AvrII) immediately downstream of the stop codon for the CP ORF. The modified vectors were sequenced to verify the expected sequence modifications.
Insertion of Foreign Gene Fragments into RNA3 Vectors
To investigate insert stability in these modified BMV vectors during virus infection in Nicotiana benthamiana and maize (Zea mays), 250-bp fragments representing partial sequences of ZmPDS (GenBank accession no. NM_001111911.1) and ZmHSP70 (X73472.1; Bates et al., 1994) were amplified from a maize cv Va35 leaf cDNA (synthesized using oligo[dT] primer) with primer sets PDS-F and PDS-R or HSP70-1F and HSP70-1R (Supplemental Table S1). Through DNA sequencing and BLAST search of genomic sequences (Phytozome; http://www.phytozome.net), ZmHSP70 was determined to be a partial sequence of a cytoplasmic ZmHSP70 gene (GRMZM2G428391; here referred to as ZmHSP70-1). After digestion of the RT-PCR fragments with AvrII and NcoI restriction enzymes, each was ligated into the pC13/F3-13m or pC13/F3CP5 RNA3 vector predigested with AvrII and NcoI to generate pC13/F13m:PDS, pC13/F13m:HSP70-1, pC13/F3CP5:PDS, and pC13/F3CP5:HSP70-1. The PDS fragment also was inserted into the pC13/F3CP14 and pC13/F3CP19 RNA3 vectors to produce pC13/F3CP14:PDS and pC13/F3CP19:PDS. A control RNA3 vector plasmid containing a 250-bp insert from a variant of the GFP gene, GFPuv (Wang et al., 2007), was produced through PCR using primers GFP-F and GFP-R (Supplemental Table S1) followed by digestion and ligation of the fragment into the pC13/F3CP5 vector, as described above, to produce pC13/F3CP5:GFP. The GFP, HSP70-1, and PDS gene fragment inserts were all placed within the virus sequence so that their antisense orientation would be present in the infectious virus transcript.
Identification and Phylogenetic Analysis of Maize HSP70 Genes
Additional maize HSP70 genes were identified through a BLAST search of the maize genome using the GRMZM2G428391 sequence (Phytozome). To predict the subcellular localizations for these additional ZmHSP70 genes, we aligned their predicted protein sequences against the known Arabidopsis (Arabidopsis thaliana) HSP70 proteins (Lin et al., 2001) using the ClustalW algorithm (Larkin et al., 2007) and constructed a neighbor-joining phylogenetic tree based on 1,000 bootstrap trials using the MEGA 6.0 program (Tamura et al., 2013). Names and accession numbers of the analyzed Arabidopsis and maize HSP70s are shown in Table II.
Table II. Maize and Arabidopsis HSP70s.
Maize HSP70s were identified through BLAST search of the maize genome sequence at Phytozome (http://www.phytozome.net) using the HSP70-1 insert sequence. *The numbering of maize HSP70s is arbitrary.
| Maize |
Arabidopsis |
|||
|---|---|---|---|---|
| Gene Name | Accession No. | Gene Name | Accession No. | Protein Identifier No. |
| ZmHSP70-1* | GRMZM2G428391_T01 | AtHSP70-1 | AT5G02500.1 | CAB85987 |
| ZmHSP70-2 | GRMZM2G056039_T01 | AtHSP70-2 | AT5G02490.1 | CAB85986 |
| ZmHSP70-3 | GRMZM2G366532_T02 | AtHSP70-3 | AT3G09440.1 | AAF14038 |
| ZmHSP70-4 | GRMZM2G340251_T01 | AtHSP70-4 | AT3G12580.1 | BAB02269 |
| ZmHSP70-5 | GRMZM5G802801_T01 | AtHSP70-5 | AT1G16030.1 | AAF18501 |
| ZmHSP70-6 | AC209784.3_FGT007 | AtHSP70-6 | AT4G24280.1 | CAB45063 |
| ZmHSP70-7 | GRMZM2G310431_T01 | AtHSP70-7 | AT5G49910.1 | BAA97012 |
| ZmHSP70-8 | GRMZM2G106429_T01 | AtHSP70-8 | AT2G32120.1 | AAD15393 |
| ZmHSP70-9 | GRMZM2G066902_T01 | AtHSP70-9 | AT4G37910.1 | CAB37531 |
| ZmHSP70-10 | GRMZM2G145275_T01 | AtHSP70-10 | AT5G09590.1 | CAB89371 |
| ZmHSP70-11 | GRMZM2G415007_T01 | AtHSP70-11 | AT5G28540.1 | AAF88019 |
| ZmHSP70-12 | GRMZM2G365374_T01 | AtHSP70-12 | AT5G42020.1 | BAB08435 |
| ZmHSP70-13 | GRMZM2G153815_T01 | AtHSP70-13 | AT1G09080.1 | AAB70400 |
| ZmHSP70-14 | GRMZM2G471196_T01 | AtHSP70-14 | AT1G79930.1 | AAG52240 |
| ZmHSP70-15 | GRMZM2G415007_T01 | AtHSP70-15 | AT1G79920.1 | AAG52244 |
| ZmHSP70-16 | GRMZM2G114793_T01 | AtHSP70-16 | AT1G11660 | AAD30257 |
| ZmHSP70-17 | GRMZM2G161035_T01 | AtHSP70-17 | AT4G16660.1 | |
| ZmHSP70-18 | AC211651.4_FGT007 | AtHSP70-18 | AT1G56410.1 | AAG51503 |
Agrobacterium tumefaciens Transformation and Virus Inoculation and Propagation
To transform A. tumefaciens with plasmid containing the BMV RNA3 genome with inserts, approximately 3 µg of plasmid DNA containing a gene fragment insert (either GFP, PDS, or HSP70-1) within the sequence representing BMV RNA3 was mixed with 25 µL of A. tumefaciens strain C58C1 (OD600 = 20) in a 1.5-mL Eppendorf tube. The tube was incubated on ice for 10 min, plunged into liquid nitrogen for 10 s, and then incubated at 37°C for 5 min. One milliliter of Yeast Extract Peptone (YEP) liquid medium (20 g of peptone, 10 g of yeast extract, and 5 g of NaCl in 1 L of distilled water) was added to the tube followed by a 5-h incubation at 28°C with 250 rpm shaking. The cells were pelleted, resuspended in 100 µL of YEP, and spread onto a YEP/kanamycin plate (24 g of Bacto agar and 50 mg of kanamycin in 1 L of YEP solution). After 2 d of incubation at 28°C, a single C58/C1 colony was selected and grown overnight in 1 mL of YEP/kanamycin/rifampicin medium (10 mg of rifampicin in 1 L of YEP/kanamycin liquid medium), and the culture was mixed (1:1, v/v) with 50% glycerol and stored at −80°C as a stock. A. tumefaciens cultures containing sequences for BMV RNA1 and RNA2 and BMV RNA3 with gene inserts were initiated from glycerol stocks, grown to OD600 = 1, pelleted, resuspended in infiltration buffer to OD600 = 2, and then equal amounts of each A. tumefaciens culture were mixed together and infiltrated into leaves of 3-week-old N. benthamiana plants using a needleless syringe. The infiltrated plants were grown inside a greenhouse set at 22°C/20°C (day and night) with a light intensity of approximately 140 μmol photons m–2 s–1. The resulting viruses were referred to as BMVF13m:PDS, BMVF13m:HSP70-1, BMVCP5:PDS, BMVCP14:PDS, BMVCP19:PDS, BMVCP5:HSP70-1, and BMVCP5:GFP.
BMV virion was partially purified from A. tumefaciens-infiltrated N. benthamiana leaves harvested at various dpai and analyzed for maintenance of the gene insert, all as described (Zhu et al., 2014). Approximately 20 µg of BMV virion in 40 μL of 0.1 m phosphate buffer, pH 7, representing virus with a specific gene insert, was rub inoculated to both sides of two leaves on an 8-d-old maize seedling (cv Va35). The inoculated maize plants were covered with plastic domes to maintain high humidity and grown inside a growth chamber set at 20°C and 16 h of light and 8 h of dark for 1 week prior to transferring to a greenhouse for phenotype observations.
Virus Accumulation in N. benthamiana
To determine whether the modified BMV vectors can cause systemic infection in N. benthamiana plants, A. tumefaciens-infiltrated plants were harvested at 10 dpai and analyzed for virus accumulation in stems through an antibody-based tissue-printing assay described previously (Nelson et al., 1993). After removing all leaves, freehand longitudinal stem sections were made from each assayed plant starting from the shoot apex down to the stem node just below the top infiltrated leaf. Stem section prints were made on nitrocellulose membranes (Bio-Rad) by pressing the stem sections against the membranes for 10 s. The membranes were air dried and then probed with a BMV CP-specific antibody (Ding et al., 1999) followed by an alkaline phosphatase-conjugated goat anti-rabbit IgG antibody, as instructed by the manufacturer (Promega). The probed membranes were scanned using the Epson Perfection V700 Photo scanner, and the images were processed with Adobe Photoshop Elements 9.
The accumulation of BMVF13m and BMVCP5 in the A. tumefaciens-infiltrated N. benthamiana leaves was analyzed through semiquantitative RT-PCR. Initially, the two largest leaves of 3-week-old N. benthamiana plants were infiltrated with BMVF13m or BMVCP5. Four N. benthamiana plants were used for each virus, and the infiltrated leaves were harvested at 4 dpai. Total RNA was isolated from individual leaves using TRIzol Reagent (Life Technologies) and treated with DNase I (New England Biolabs). After phenol/chloroform extraction and ethanol precipitation, the quality and concentration of each total RNA sample were monitored with a TECAN Infinite M200 PRO instrument (Tecan Systems). cDNA synthesis was done using 0.5 µg of total RNA, 0.5 µL of 10 mm BMV R primer (Supplemental Table S1), 0.5 µL of Moloney murine leukemia virus reverse transcriptase, and 0.25 µL of RNase inhibitor (New England Biolabs) in a 10-µL reaction. The relative accumulation level of BMV RNA3 in each sample was determined using 2 µL of 20-fold diluted cDNA in a 20-µL PCR containing primers B3-633F and B3-1003R (Supplemental Table S1), specific for the BMV 3a gene, and 20, 25, and 30 reaction cycles. Virus RNA levels were normalized between samples by determining the expression level of N. benthamiana EF-1A in each sample using primers EF-F and EF-R (Supplemental Table S1). The PCR products were visualized on 1% agarose gels through electrophoresis.
Insert Stability in N. benthamiana and Maize
Foreign gene fragment insert stability in the BMV VIGS vectors was initially determined through RT-PCR of virion RNAs from extracts of A. tumefaciens-infiltrated N. benthamiana leaves harvested at 3, 6, and 9 dpai. BMV RNA isolation and cDNA synthesis were as described in the previous section. PCR was performed using primers B3-1564F and B3-1974R (Supplemental Table S1), and the products were visualized on 1% agarose gels through electrophoresis.
To determine the insert stability during virus infection in maize, partially purified BMV virion was rub inoculated to the leaves of 8-d-old maize seedlings as described in the section, “Agrobacterium tumefaciens Transformation and Virus Inoculation and Propagation.” The virion-inoculated maize leaves were harvested individually at 5 dpi and rinsed five times in distilled water to remove the inoculated virion remaining on the leaf surface. The first or second systemically infected leaf of each assayed plant was harvested at various times (5–10 dpi). Virion RNA was isolated from each sample and analyzed for insert stability through RT-PCR as described for N. benthamiana in the previous section. For DNA sequencing, PCR products from three leaves inoculated or systemically infected with BMVF13m:PDS or BMVCP5:PDS were gel purified, pooled, and cloned into the pGEM-T Easy vector (Promega). After transformation into the JM109 competent cells (Promega), approximately 25 colonies were randomly selected from each treatment and plasmid DNA from these colonies was sequenced using primer B3-1564F (Supplemental Table S1) and the BigDye Terminator version 3.1 sequencing kit on an Applied Biosystem 3730 DNA Analyzer as per the manufacturer’s instructions (Life Technologies).
Gene-Silencing Assays in Maize
To silence ZmPDS or ZmHSP70-1 expression in maize, partially purified BMVF13m:PDS, BMVF13m:HSP70-1, BMVCP5:PDS, BMVCP5:HSP70-1, or BMVCP5:GFP virion was inoculated to leaves of maize seedlings as described in the section, “Agrobacterium tumefaciens Transformation and Virus Inoculation and Propagation.” The inoculated maize plants were photographed at various dpi. Gene knockdown efficiency by the original or the modified BMV vectors was determined through qRT-PCR as described previously (Bhat et al., 2013) with specific modifications. Total RNA from pooled second and third systemically infected maize leaves was extracted and treated with DNase I enzyme as described in the section, “Virus Accumulation in N. benthamiana.” First-strand cDNA was synthesized using an oligo(dT) primer, and the resulting cDNA diluted 20-fold in water was used for qPCR. qPCR was conducted using primers specific for ZmPDS (PDS-Fq and PDS-Rq) or ZmHSP70-1 (HSP70qF1 and HSP70qR1, designed based on the X73472.1 sequence). Because maize has multiple cytoplasmic HSP70 members (Fig. 7) and the insert we used to silence the ZmHSP70-1 gene in this host through VIGS might also knock down other ZmHSP70 family members based on sequence comparison (Supplemental Fig. S1A), we designed additional qPCR primers, HSP70qF2 and HSP70qR2 for HSP70-2, HSP70qF3 and HSP70qR3 for HSP70-3, HSP70qF4 and HSP70qR4 for HSP70-4, and HSP70qF5 and HSP70qR5 for HSP70-5 (Supplemental Table S1; Supplemental Fig. S1B), using Primer Express Software version 3.0.1 (Life Technologies). The relative expression level of each analyzed gene in individual samples was normalized across treatments by determining ZmEF-1A transcript levels in the same samples using primers EF1A-Fq and EF1A-Rq (Supplemental Table S1) and calculated using the 2−ΔΔCT method.
Statistics
ANOVA and lsd tests were conducted using either MSTAT-C (https://msu.edu/∼freed/mstatc.htm) or, respectively, lm function and the R/base package (R Core Team, 2016) and lsd.test function in the R/agricolae package (de Mendiburu, 2016).
To test unimodal expression, the expression values for ZmHSP70 transcript from plants infected with BMVF13m:HSP70-1 were standardized with median and median absolute deviation. The standardized expression values were analyzed for unimodality using dip.test function within the R/diptest package (Maechler, 2015).
Accession Numbers
Sequence data from this article can be found in Genbank/EMBL data libraries under accession numbers NM_001111911.1 (ZmPDS); U62636.1 (pDSK-GFPuv); X73472.1 (ZmHSP70); GRMZM2G428391_T01 (ZmHSP70-1); GRMZM2G056039_T01 (ZmHSP70-2); GRMZM2G366532_T02 (ZmHSP70-3); GRMZM2G340251_T01 (ZmHSP70-4); GRMZM5G802801_T01 (ZmHSP70-5).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Nucleotide sequence alignment for maize cytoplasmic HSP70 family members.
Supplemental Figure S2. Computer-predicted BMVF13m and BMVCP5 (+) strand RNA3 folding with and without gene fragment inserts.
Supplemental Figure S3. Computer-predicted BMVF13m and BMVCP5 (−) strand RNA3 folding with and without gene fragment inserts.
Supplemental Figure S4. Computer-predicted BMVF13m and BMVCP5 RNA4 folding with ZmPDS gene fragment insert.
Supplemental Figure S5. BMVCP5 displays greater insert stability than BMVF13m in N. benthamiana and maize.
Supplemental Figure S6. Detection of BMV CP and HSP70 protein in barley leaf cells.
Supplemental Table S1. Primer sequences used in this study.
Acknowledgments
We thank Jose Fonseca, Kiran Mysore, Phillip A. Harries, and Amr Ibrahim for critical review of the article, Frank Coker and Lynne Jacobs for maintaining plants in the greenhouse and growth chambers, and Kim Cooper and Darian Gonzales for technical assistance and text preparation, respectively.
Footnotes
This work was supported by the Samuel Roberts Noble Foundation and by the BioEnergy Science Center (DOE Office of Science BER DE-AC05-00OR22725) through a subcontract to the Noble Research Institute (subcontract no. 4000115929). The BioEnergy Science Center is a U.S. Department of Energy Bioenergy Research Center supported by the Office of Biological and Environmental Research in the DOE Office of Science.
Articles can be viewed without a subscription.
References
- Ahlquist P, Dasgupta R, Kaesberg P (1984) Nucleotide sequence of the brome mosaic virus genome and its implications for viral replication. J Mol Biol 172: 369–383 [DOI] [PubMed] [Google Scholar]
- Alam SB, Rochon D (2015) Cucumber necrosis virus recruits cellular heat shock protein 70 homologs at several stages of infection. J Virol 90: 3302–3317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alejska M, Figlerowicz M, Malinowska N, Urbanowicz A, Figlerowicz M (2005) A universal BMV-based RNA recombination system: how to search for general rules in RNA recombination. Nucleic Acids Res 33: e105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aranda MA, Escaler M, Wang D, Maule AJ (1996) Induction of HSP70 and polyubiquitin expression associated with plant virus replication. Proc Natl Acad Sci USA 93: 15289–15293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avesani L, Marconi G, Morandini F, Albertini E, Bruschetta M, Bortesi L, Pezzotti M, Porceddu A (2007) Stability of Potato virus X expression vectors is related to insert size: implications for replication models and risk assessment. Transgenic Res 16: 587–597 [DOI] [PubMed] [Google Scholar]
- Barr JN, Fearns R (2010) How RNA viruses maintain their genome integrity. J Gen Virol 91: 1373–1387 [DOI] [PubMed] [Google Scholar]
- Bates EEM, Vergne P, Dumas C (1994) Analysis of the cytosolic hsp70 gene family in Zea mays. Plant Mol Biol 25: 909–916 [DOI] [PubMed] [Google Scholar]
- Baulcombe D. (2004) RNA silencing in plants. Nature 431: 356–363 [DOI] [PubMed] [Google Scholar]
- Benavente LM, Ding XS, Redinbaugh MG, Nelson R, Balint-Kurti P (2012) Virus-induced gene silencing in diverse maize lines using the Brome mosaic virus-based silencing vector. Maydica 57: 206–214 [Google Scholar]
- Bhat S, Folimonova SY, Cole AB, Ballard KD, Lei Z, Watson BS, Sumner LW, Nelson RS (2013) Influence of host chloroplast proteins on Tobacco mosaic virus accumulation and intercellular movement. Plant Physiol 161: 134–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodersen P, Voinnet O (2006) The diversity of RNA silencing pathways in plants. Trends Genet 22: 268–280 [DOI] [PubMed] [Google Scholar]
- Bruun-Rasmussen M, Madsen CT, Jessing S, Albrechtsen M (2007) Stability of Barley stripe mosaic virus-induced gene silencing in barley. Mol Plant Microbe Interact 20: 1323–1331 [DOI] [PubMed] [Google Scholar]
- Bruyere A, Wantroba M, Flasinski S, Dzianott A, Bujarski JJ (2000) Frequent homologous recombination events between molecules of one RNA component in a multipartite RNA virus. J Virol 74: 4214–4219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bujarski JJ. (2013) Genetic recombination in plant-infecting messenger-sense RNA viruses: overview and research perspectives. Front Plant Sci 4: 68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bujarski JJ, Kaesberg P (1986) Genetic recombination between RNA components of a multipartite plant virus. Nature 321: 528–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukau B, Weissman J, Horwich A (2006) Molecular chaperones and protein quality control. Cell 125: 443–451 [DOI] [PubMed] [Google Scholar]
- Cao Y, Shi Y, Li Y, Cheng Y, Zhou T, Fan Z (2012) Possible involvement of maize Rop1 in the defence responses of plants to viral infection. Mol Plant Pathol 13: 732–743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Zhou T, Wu X, Hong Y, Fan Z, Li H (2008) Influence of cytoplasmic heat shock protein 70 on viral infection of Nicotiana benthamiana. Mol Plant Pathol 9: 809–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasgupta R, Kaesberg P (1982) Complete nucleotide sequences of the coat protein messenger RNAs of brome mosaic virus and cowpea chlorotic mottle virus. Nucleic Acids Res 10: 703–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Mendiburu F. (2016) Agricolae: statistical procedures for agricultural research. R package version 1.2-4. https://CRAN.R-project.org/package=agricolae (June 27, 2017)
- Ding SW, Voinnet O (2007) Antiviral immunity directed by small RNAs. Cell 130: 413–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding X, Ballard K, Nelson R (2010) Improving virus induced gene silencing (VIGS) in rice through Agrobacterium infiltration. In Antoun H, Avis T, Brisson L, Prevost D, Trepanier M, eds, Biology of Plant-Microbe Interactions, Vol 7 International Society for Molecular Plant-Microbe Interactions, St. Paul, MN. Paper no. 59 [Google Scholar]
- Ding XS, Flasinski S, Nelson RS (1999) Infection of barley by Brome mosaic virus is restricted predominantly to cells in and associated with veins through a temperature-dependent mechanism. Mol Plant Microbe Interact 12: 615–623 [Google Scholar]
- Ding XS, Rao CS, Nelson RS (2007) Analysis of gene function in rice through virus-induced gene silencing. Methods Mol Biol 354: 145–160 [DOI] [PubMed] [Google Scholar]
- Ding XS, Schneider WL, Chaluvadi SR, Mian MA, Nelson RS (2006) Characterization of a Brome mosaic virus strain and its use as a vector for gene silencing in monocotyledonous hosts. Mol Plant Microbe Interact 19: 1229–1239 [DOI] [PubMed] [Google Scholar]
- Eamens A, Wang MB, Smith NA, Waterhouse PM (2008) RNA silencing in plants: yesterday, today, and tomorrow. Plant Physiol 147: 456–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escaler M, Aranda MA, Thomas CL, Maule AJ (2000) Pea embryonic tissues show common responses to the replication of a wide range of viruses. Virology 267: 318–325 [DOI] [PubMed] [Google Scholar]
- Fang X, Qi Y (2016) RNAi in plants: an argonaute-centered view. Plant Cell 28: 272–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemetsberger C, Herrberger C, Zechmann B, Hillmer M, Doehlemann G (2012) The Ustilago maydis effector Pep1 suppresses plant immunity by inhibition of host peroxidase activity. PLoS Pathog 8: e1002684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igarashi A, Yamagata K, Sugai T, Takahashi Y, Sugawara E, Tamura A, Yaegashi H, Yamagishi N, Takahashi T, Isogai M, et al. (2009) Apple latent spherical virus vectors for reliable and effective virus-induced gene silencing among a broad range of plants including tobacco, tomato, Arabidopsis thaliana, cucurbits, and legumes. Virology 386: 407–416 [DOI] [PubMed] [Google Scholar]
- Kao CC, Ahlquist P (1992) Identification of the domains required for direct interaction of the helicase-like and polymerase-like RNA replication proteins of brome mosaic virus. J Virol 66: 7293–7302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MJ, Kao C (2001) Factors regulating template switch in vitro by viral RNA-dependent RNA polymerases: implications for RNA-RNA recombination. Proc Natl Acad Sci USA 98: 4972–4977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim NH, Hwang BK (2015) Pepper heat shock protein 70a interacts with the type III effector AvrBsT and triggers plant cell death and immunity. Plant Physiol 167: 307–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolondam B, Rao P, Sztuba-Solinska J, Weber PH, Dzianott A, Johns MA, Bujarski JJ (2015) Co-infection with two strains of Brome mosaic bromovirus reveals common RNA recombination sites in different hosts. Virus Evol 1: vev021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong L, Wu J, Lu L, Xu Y, Zhou X (2014) Interaction between Rice stripe virus disease-specific protein and host PsbP enhances virus symptoms. Mol Plant 7: 691–708 [DOI] [PubMed] [Google Scholar]
- Kroner PA, Young BM, Ahlquist P (1990) Analysis of the role of brome mosaic virus 1a protein domains in RNA replication, using linker insertion mutagenesis. J Virol 64: 6110–6120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon SJ, Rao AL (2012) Emergence of distinct brome mosaic virus recombinants is determined by the polarity of the inoculum RNA. J Virol 86: 5204–5220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23: 2947–2948 [DOI] [PubMed] [Google Scholar]
- Lee WS, Hammond-Kosack KE, Kanyuka K (2015) In planta transient expression systems for monocots. In Azhakanandam K, Silverstone A, Daniell H, Davey MR, eds, Recent Advancements in Gene Expression and Enabling Technologies in Crop Plants. Springer, New York, pp 391–422 [Google Scholar]
- Lin BL, Wang JS, Liu HC, Chen RW, Meyer Y, Barakat A, Delseny M (2001) Genomic analysis of the Hsp70 superfamily in Arabidopsis thaliana. Cell Stress Chaperones 6: 201–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maechler M. (2015) Diptest: Hartigan’s Dip test statistic for unimodality-corrected. R package version 0.75-7. https://CRAN.R-project.org/package=diptest (June 27, 2017)
- Martin T, Biruma M, Fridborg I, Okori P, Dixelius C (2011) A highly conserved NB-LRR encoding gene cluster effective against Setosphaeria turcica in sorghum. BMC Plant Biol 11: 151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez de Alba AE, Elvira-Matelot E, Vaucheret H (2013) Gene silencing in plants: a diversity of pathways. Biochim Biophys Acta 1829: 1300–1308 [DOI] [PubMed] [Google Scholar]
- Mayer MP, Bukau B (2005) Hsp70 chaperones: cellular functions and molecular mechanism. Cell Mol Life Sci 62: 670–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei Y, Zhang C, Kernodle BM, Hill JH, Whitham SA (2016) A Foxtail mosaic virus vector for virus-induced gene silencing in maize. Plant Physiol 171: 760–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mise K, Ahlquist P (1995) Host-specificity restriction by bromovirus cell-to-cell movement protein occurs after initial cell-to-cell spread of infection in nonhost plants. Virology 206: 276–286 [DOI] [PubMed] [Google Scholar]
- Montgomery TA, Howell MD, Cuperus JT, Li D, Hansen JE, Alexander AL, Chapman EJ, Fahlgren N, Allen E, Carrington JC (2008) Specificity of ARGONAUTE7-miR390 interaction and dual functionality in TAS3 trans-acting siRNA formation. Cell 133: 128–141 [DOI] [PubMed] [Google Scholar]
- Nagy PD, Bujarski JJ (1996) Homologous RNA recombination in brome mosaic virus: AU-rich sequences decrease the accuracy of crossovers. J Virol 70: 415–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy PD, Zhang C, Simon AE (1998) Dissecting RNA recombination in vitro: role of RNA sequences and the viral replicase. EMBO J 17: 2392–2403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson RS, Li GX, Hodgson RAJ, Beachy RN, Shintaku MH (1993) Impeded phloem-dependent accumulation of the masked strain of tobacco mosaic virus. Mol Plant Microbe Interact 6: 45–54 [Google Scholar]
- Noueiry AO, Ahlquist P (2003) Brome mosaic virus RNA replication: revealing the role of the host in RNA virus replication. Annu Rev Phytopathol 41: 77–98 [DOI] [PubMed] [Google Scholar]
- Olsthoorn RCL, Bruyere A, Dzianott A, Bujarski JJ (2002) RNA recombination in brome mosaic virus: effects of strand-specific stem-loop inserts. J Virol 76: 12654–12662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacak A, Strozycki PM, Barciszewska-Pacak M, Alejska M, Lacomme C, Jarmołowski A, Szweykowska-Kulińska Z, Figlerowicz M (2010) The brome mosaic virus-based recombination vector triggers a limited gene silencing response depending on the orientation of the inserted sequence. Arch Virol 155: 169–179 [DOI] [PubMed] [Google Scholar]
- Park CJ, Seo YS (2015) Heat shock proteins: a review of the molecular chaperones for plant immunity. Plant Pathol J 31: 323–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulsen C, Vaucheret H, Brodersen P (2013) Lessons on RNA silencing mechanisms in plants from eukaryotic argonaute structures. Plant Cell 25: 22–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quadt R, Jaspars EMJ (1990) Purification and characterization of brome mosaic virus RNA-dependent RNA polymerase. Virology 178: 189–194 [DOI] [PubMed] [Google Scholar]
- R Core Team (2016) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna. https://www.R-project.org/ (June 27, 2017)
- Ramanna H, Ding XS, Nelson RS (2013) Rationale for developing new virus vectors to analyze gene function in grasses through virus-induced gene silencing. Methods Mol Biol 975: 15–32 [DOI] [PubMed] [Google Scholar]
- Rao AL, Kao CC (2015) The brome mosaic virus 3′ untranslated sequence regulates RNA replication, recombination, and virion assembly. Virus Res 206: 46–52 [DOI] [PubMed] [Google Scholar]
- Robertson D. (2004) VIGS vectors for gene silencing: many targets, many tools. Annu Rev Plant Biol 55: 495–519 [DOI] [PubMed] [Google Scholar]
- Schmitz I, Rao AL (1996) Molecular studies on bromovirus capsid protein. I. Characterization of cell-to-cell movement-defective RNA3 variants of brome mosaic virus. Virology 226: 281–293 [DOI] [PubMed] [Google Scholar]
- Scofield SR, Nelson RS (2009) Resources for virus-induced gene silencing in the grasses. Plant Physiol 149: 152–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senthil-Kumar M, Mysore KS (2011) Virus-induced gene silencing can persist for more than 2 years and also be transmitted to progeny seedlings in Nicotiana benthamiana and tomato. Plant Biotechnol J 9: 797–806 [DOI] [PubMed] [Google Scholar]
- Shapka N, Nagy PD (2004) The AU-rich RNA recombination hot spot sequence of Brome mosaic virus is functional in tombusviruses: implications for the mechanism of RNA recombination. J Virol 78: 2288–2300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Qin Y, Cao Y, Sun H, Zhou T, Hong Y, Fan Z (2011) Influence of an m-type thioredoxin in maize on potyviral infection. Eur J Plant Pathol 131: 317 [Google Scholar]
- Simon-Loriere E, Holmes EC (2011) Why do RNA viruses recombine? Nat Rev Microbiol 9: 617–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Zhang H, Li D, Huang L, Hong Y, Ding XS, Nelson RS, Zhou X, Song F (2013) Functions of rice NAC transcriptional factors, ONAC122 and ONAC131, in defense responses against Magnaporthe grisea. Plant Mol Biol 81: 41–56 [DOI] [PubMed] [Google Scholar]
- Sztuba-Solińska J, Urbanowicz A, Figlerowicz M, Bujarski JJ (2011) RNA-RNA recombination in plant virus replication and evolution. Annu Rev Phytopathol 49: 415–443 [DOI] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30: 2725–2729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyedmers J, Mogk A, Bukau B (2010) Cellular strategies for controlling protein aggregation. Nat Rev Mol Cell Biol 11: 777–788 [DOI] [PubMed] [Google Scholar]
- Urbanowicz A, Alejska M, Formanowicz P, Blazewicz J, Figlerowicz M, Bujarski JJ (2005) Homologous crossovers among molecules of brome mosaic bromovirus RNA1 or RNA2 segments in vivo. J Virol 79: 5732–5742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Linde K, Doehlemann G (2013) Utilizing virus-induced gene silencing for the functional characterization of maize genes during infection with the fungal pathogen Ustilago maydis. Methods Mol Biol 975: 47–60 [DOI] [PubMed] [Google Scholar]
- van der Linde K, Hemetsberger C, Kastner C, Kaschani F, van der Hoorn RA, Kumlehn J, Doehlemann G (2012) A maize cystatin suppresses host immunity by inhibiting apoplastic cysteine proteases. Plant Cell 24: 1285–1300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Linde K, Kastner C, Kumlehn J, Kahmann R, Doehlemann G (2011) Systemic virus-induced gene silencing allows functional characterization of maize genes during biotrophic interaction with Ustilago maydis. New Phytol 189: 471–483 [DOI] [PubMed] [Google Scholar]
- Vaucheret H. (2008) Plant ARGONAUTES. Trends Plant Sci 13: 350–358 [DOI] [PubMed] [Google Scholar]
- Verchot J. (2012) Cellular chaperones and folding enzymes are vital contributors to membrane bound replication and movement complexes during plant RNA virus infection. Front Plant Sci 3: 275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Kang L, Anand A, Lazarovits G, Mysore KS (2007) Monitoring in planta bacterial infection at both cellular and whole-plant levels using the green fluorescent protein variant GFPuv. New Phytol 174: 212–223 [DOI] [PubMed] [Google Scholar]
- Wang R, Yang X, Wang N, Liu X, Nelson RS, Li W, Fan Z, Zhou T (2016) An efficient virus-induced gene silencing vector for maize functional genomics research. Plant J 86: 102–115 [DOI] [PubMed] [Google Scholar]
- Wang RYL, Stork J, Nagy PD (2009) A key role for heat shock protein 70 in the localization and insertion of tombusvirus replication proteins to intracellular membranes. J Virol 83: 3276–3287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse PM, Fusaro AF (2006) Viruses face a double defense by plant small RNAs. Science 313: 54–55 [DOI] [PubMed] [Google Scholar]
- Whitham SA, Quan S, Chang HS, Cooper B, Estes B, Zhu T, Wang X, Hou YM (2003) Diverse RNA viruses elicit the expression of common sets of genes in susceptible Arabidopsis thaliana plants. Plant J 33: 271–283 [DOI] [PubMed] [Google Scholar]
- Yamagishi M, Masuta C, Suzuki M, Netsu O (2015) Peanut stunt virus-induced gene silencing in white lupin (Lupinus albus). Plant Biotechnol J 32: 181–191 [Google Scholar]
- Yuan C, Li C, Yan L, Jackson AO, Liu Z, Han C, Yu J, Li D (2011) A high throughput barley stripe mosaic virus vector for virus induced gene silencing in monocots and dicots. PLoS ONE 6: e26468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan B, Lang F, Zhou T, Fan Z (2016) Nuclear import of Maize chlorotic mottle virus capsid protein is mediated by importin-α. Eur J Plant Pathol 146: 881–892 [Google Scholar]
- Zhu M, Chen Y, Ding XS, Webb SL, Zhou T, Nelson RS, Fan Z (2014) Maize Elongin C interacts with the viral genome-linked protein, VPg, of Sugarcane mosaic virus and facilitates virus infection. New Phytol 203: 1291–1304 [DOI] [PMC free article] [PubMed] [Google Scholar]