Abstract
The proto-oncogene c-Maf is a transcription factor that plays a critical role in the differentiation of various T helper (TH) cell subsets. The amount of c-Maf increases after stimulation of the T cell receptor (TCR), which results in the production of multiple cytokines. We showed that two essential regulators of the transcription factor nuclear factor κB (NF-κB), the scaffold protein CARMA1 and the kinase IKKβ [inhibitor of NF-κB (IκB) kinase β], are also critical for the activation of c-Maf. Although CARMA1 deficiency did not affect the TCR-dependent increase in c-Maf abundance in T cells, CARMA1-dependent activation of the IKK complex was required for the nuclear translocation of c-Maf and its binding to the promoters of its target genes. Consistent with a role for c-Maf in the development of T follicular helper (TFH) cells, which provide help to B cells in the germinal centers of the spleen, CARMA1- or IKKβ-deficient mice immunized with peptide antigen had defects in the generation of TFH cells, formation of germinal centers, and production of antigen-specific antibodies. Together, these data suggest a mechanism by which c-Maf is regulated during T cell activation and differentiation.
INTRODUCTION
CD4+ T cells, also known as T helper (TH) cells, are key components of the adaptive immune response. Once activated through their cell surface T cell receptor (TCR), these cells differentiate into various TH subsets, including TH1, TH2, and TH17 cells, as well as regulatory T (Treg) cells, all of which have distinct transcription factors and secrete distinct cytokines (1). Initial stimulation of the TCR results in the rapid activation of multiple transcription factors, including nuclear factor κB (NF-κB), activator protein 1 (AP-1), and nuclear factor of activated T cells (NFAT), which leads to cell proliferation (2). TCR-induced NF-κB activity has been intensively studied, and multiple signaling molecules transduce signals from the TCR to this transcription factor (3, 4). Nuclear localization of NF-κB is prevented by its interaction with inhibitor of NF-κB (IκB) proteins. Upon TCR stimulation, the IκB kinase (IKK) complex, which consists of the kinases IKKα and IKKβ and the regulatory subunit IKKγ (also known as NEMO), phosphorylates IκB proteins, which target them for polyubiquitylation and proteasomal degradation (5).
TCR-mediated activation of the IKK complex is controlled by CARMA1 (also known as CARD11), a scaffold protein found exclusively in hematopoietic tissues (4). Loss of CARMA1 impairs TCR-dependent activation of IKK and the production of the cytokine interleukin-2 (IL-2) (6–8), which drives T cell proliferation. We previously showed that CARMA1 is indispensible for the TCR-dependent activation of the mitogen-activated protein kinase (MAPK) c-Jun N-terminal kinase 2 (JNK2), and that it is required for the accumulation of the transcription factors c-Jun and JunB (9, 10) in the nucleus, as well as for TH2 cell–associated inflammation (10).
Other studies indicated that CD4+ T cells localized in germinal centers (GCs), called T follicular helper (TFH) cells, are distinct from the previously defined TH1, TH2, and TH17 subsets (11), and that they are essential to provide cognate help to B cells for the generation of high-affinity plasma cells and memory cells that are crucial for long-term protection against infections (12). Although TFH cells produce IL-21, which is necessary for GC formation (11, 13–15), the development of TFH cells also requires IL-21 and the increased abundance of the transcription factor c-Maf (16). c-Maf, the cellular homolog of avian viral v-maf, is a member of the AP-1 family of proteins, and it contains basic region/leucine zipper domains (17). Initially, c-Maf was found to be required for the expression of Il4 in TH2 cells (17, 18), but later studies demonstrated that c-Maf protein abundance is also increased in other TH subsets, such as TH17 and TFH cells (12, 16, 19, 20). Loss of the gene encoding c-Maf results in impaired IL-21 production (16), whereas transduction of CD4+ T cells with retrovirus expressing c-Maf results in a marked increase in the numbers of IL-21–producing cells (20). A biochemical study further demonstrated that c-Maf directly activates Il21 expression (21); however, how TCR-dependent signaling regulates c-Maf and IL-21 in activated and differentiating T cells is unclear.
Here, we found that CARMA1-deficient CD4+ T cells produced less IL-4 and IL-21 than did wild-type cells because they had reduced activation of c-Maf. Although the TCR-dependent increase in c-Maf abundance was not affected by the loss of CARMA1, the extent of the nuclear localization of c-Maf was reduced, and its DNA binding ability was impaired. We found that CARMA1-dependent activation of the IKK complex was required for the nuclear translocation of c-Maf and for its binding to the promoters of target genes, such as Il4 and Il21. Consistent with previous findings that showed that c-Maf and IL-21 contribute to the development of TFH cells and the regulation of humoral immunity, we found that loss of CARMA1 or conditional deletion of IKKβ in CD4+ T cells resulted in the generation of decreased numbers of TFH cells and GC B cells, as well as impaired the production of antigen-specific antibodies upon immunization of mice with T cell–dependent antigen. Therefore, our study suggests a previously uncharacterized mechanism by which the CARMA1-IKKβ signalosome contributes to T cell differentiation and the immune response.
RESULTS
Loss of CARMA1 in CD4+ cells leads to reduced production of IL-21
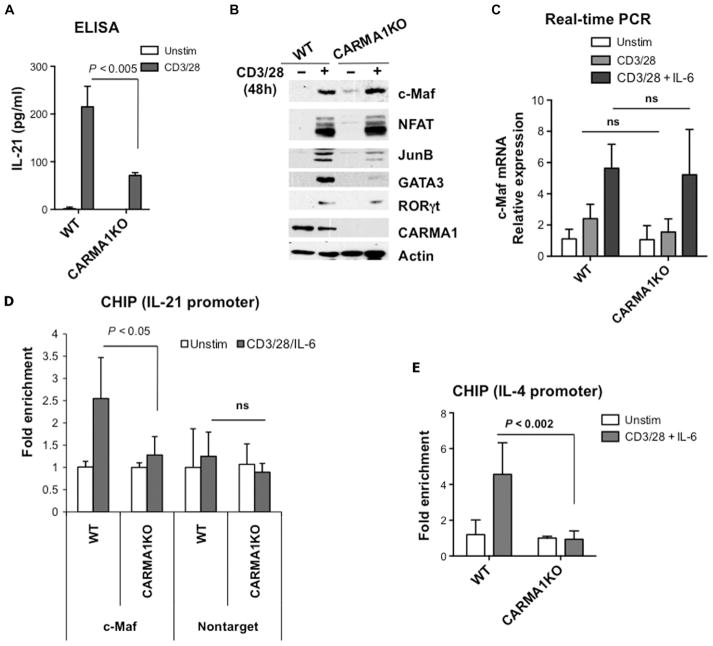
We previously showed that CARMA1 is required mainly for the initial production of IL-4, IL-5, IL-10, and IL-13 upon TCR stimulation, and that it directs the differentiation of TH2 cells (10). To further investigate the role of CARMA1 in TH cell differentiation, we analyzed the expression profiles of 26 cytokines and inflammatory proteins in TCR-stimulated CD4+ splenocytes isolated from CARMA1-deficient mice and wild-type control mice. We used the multiplex suspension array system formatted on magnetic beads, which enables the simultaneous detection of multiple biomarkers in the same sample and their quantification based on mean fluorescence intensity (MFI). We found that CARMA1 deficiency affected not only the initial (first 48 hours) release of IL-4 as reported previously (10) but also the release of IL-21 and CD40 ligand (CD40L) (fig. S1A). Although both of these proteins provide a costimulatory signal for B cells, IL-21 supports the development of TFH cells and the formation of GCs in response to T cell–dependent antigens (11, 13). Thus, we decided to determine the molecular mechanism by which CARMA1 regulated IL-21 production. First, we used enzyme-linked immunosorbent assay (ELISA) to investigate whether IL-21 production depended on CARMA1, and we found that CARMA1-deficient CD4+ splenocytes secreted reduced amounts of this cytokine compared to wild-type cells upon TCR stimulation (Fig. 1A). We also tested whether a deficiency in the direct CARMA1-binding partner, Bcl10, resulted in a similar defect. Indeed, TCR-stimulated CD4+ splenocytes isolated from Bcl10-deficient mice produced less IL-21 than did control cells from wild-type mice (fig. S1B).
Fig. 1. CARMA1-deficient CD4+ T cells show decreased recruitment of c-Maf to the Il21 promoter and reduced production of IL-21.
(A) CD4+ splenocytes from wild-type (WT) and CARMA1-deficient (CARMA1KO) mice were stimulated with plate-bound anti-CD3ε (2 μg/ml) and anti-CD28 (1 μg/ml) antibodies for 48 hours. Secretion of IL-21 into the cell culture medium was determined by ELISA of triplicate cultures. Data are means ± SD. (B) TCR-induced expression of c-Maf does not require CARMA1. CD4+ T cells from the indicated mice were stimulated as described in (A). Whole-cell lysates were analyzed by Western blotting with antibodies specific for c-Maf, GATA3, JunB, NFATc1, RORγt, actin, and CARMA1. Western blots are representative of two independent experiments. (C) CD4+ T cells from the indicated mice were activated with plate-bound anti-CD3 and anti-CD28 antibodies in the presence or absence of IL-6 (20 ng/ml) for 18 hours. Total RNA was isolated and reverse-transcribed, and quantitative PCR was performed with the SYBR Green PCR Master Mix. The amounts of c-Maf transcript were normalized to those of Gapdh, and the amounts of c-Maf mRNA in stimulated cells relative to those in unstimulated cells were calculated. The experiment was performed in triplicate, and data are means ± SD. (D and E) Defective recruitment of c-Maf to the Il4 and Il21 promoters in CARMA1KO T cells. CD4+ splenocytes from the indicated mice were activated with plate-bound anti-CD3 and anti-CD28 antibodies in the presence of IL-6 for 18 hours, and then c-Maf–DNA complexes were precipitated with c-Maf antibody. Real-time PCR analysis for (D) the Il21 promoter and (E) the Il4 promoter was performed in triplicate, and results are means ± SD. Data are representative of two independent experiments. ns, not significant.
Previous studies indicated that Il21 expression is dependent on c-Maf (16, 21). Activation of the Il4 promoter also requires c-Maf (17, 18), and c-Maf–deficient mice have impaired IL-4 production compared to that of wild-type mice (18). Therefore, we hypothesized that CARMA1 might be involved in the regulation of c-Maf after TCR stimulation. Because naïve T cells do not have detectable amounts of c-Maf, and because c-Maf protein abundance is slowly increased after TCR stimulation, we activated CD4+ splenocytes with plate-bound anti-CD3 and anti-CD28 antibodies for 2 days. We found that c-Maf protein was similarly abundant in wild-type and CARMA1-deficient cells (Fig. 1B), which suggests that c-Maf abundance is regulated by a CARMA1-independent mechanism. These results were also confirmed by real-time polymerase chain reaction (PCR) analysis (Fig. 1C and fig. S2). Next, we performed a chromatin immunoprecipitation (ChIP) assay to examine the binding of c-Maf to the c-Maf response element (MARE) motif in the promoters of Il21 and Il4 in TCR-stimulated CARMA1-deficient cells. Because the expression of Maf gene is enhanced by IL-6 (Fig. 1C) (22), we activated CD4+ splenocytes with plate-bound anti-CD3 and anti-CD28 antibodies in the presence of IL-6 for 18 hours, precipitated c-Maf–DNA complexes with antibody against c-Maf, and then analyzed the samples by real-time PCR (Fig. 1, D and E). We found that recruitment of c-Maf to the promoters of both Il21 and Il4 was induced in stimulated wild-type cells, but not in CARMA1-deficient cells. These results suggest that the activity of c-Maf, but not its abundance, is dependent on CARMA1.
CARMA1 is required for the binding of c-Maf to DNA
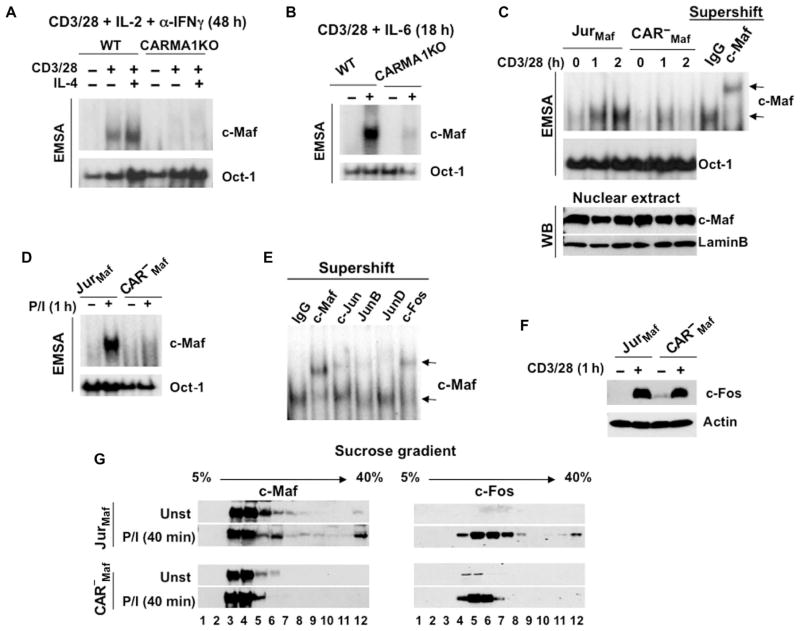
To further confirm the critical role of CARMA1 in c-Maf activation, we determined the extent of binding of this transcription factor to the MARE motif by electrophoretic mobility shift assay (EMSA). We activated CD4+ T cells under TH2-differentiating conditions for 48 hours (Fig. 2A) or stimulated the cells in the presence of IL-6 for 18 hours (Fig. 2B), and then prepared extracts of nuclear proteins. Consistent with the data obtained from the ChIP assays, we found that the DNA binding activity of c-Maf was markedly inhibited in CARMA1-deficient cells under all tested conditions. We performed similar experiments with cell lines stably transfected with plasmid encoding c-Maf (fig. S3). Under these conditions, we could reduce the stimulation time to 1 to 2 hours, and the cells were activated in the absence of exogenous and endogenous cytokines. Again, we showed that the DNA binding activity of c-Maf was defective in CARMA1-deficient cells stimulated with anti-CD3 and anti-CD28 antibodies or with phorbol 12-myristate 13-acetate (PMA) and ionomycin (P/I) (Fig. 2, C and D). The specificity of the binding of c-Maf to the probe was confirmed by supershift assay with a c-Maf–specific antibody (Fig. 2C). Further super-shift analysis revealed that c-Maf dimerized with other AP-1 transcription factors, mainly c-Fos (Fig. 2E), and that c-Fos abundance was intact in CARMA1-deficient cells (Fig. 2F). We also observed a similar distribution of c-Maf and c-Fos in sucrose gradients prepared from activated Jurkat cells (Fig. 2G). Furthermore, we found a signal-dependent enrichment of c-Maf in high–molecular mass fractions, and this effect was not observed in CARMA1-deficient cells. Together, our data suggest that c-Maf activity is dependent on TCR signaling mediated by CARMA1.
Fig. 2. CARMA1 is required for the activation of c-Maf.
(A and B) The DNA binding activity of c-Maf is defective in CARMA1-deficient CD4+ splenocytes. CD4+ T cells from the indicated mice were activated with plate-bound anti-CD3 and anti-CD28 antibodies in the presence of (A) IL-2 (30 U/ml), anti-IFNγ antibody (10 μg/ml), and recombinant murine IL-4 (10 ng/ml) for 48 hours or (B) IL-6 (20 ng/ml) for 18 hours. Nuclear extracts were prepared, and the binding activity of c-Maf was determined by EMSA. Data are representative of two experiments. (C and D) Jurkat cells (JurMaf) and CARMA1-deficient Jurkat cells (CAR−Maf) stably transfected with plasmid encoding c-Maf were stimulated with anti-CD3 and anti-CD28 antibodies or with P/I [PMA (20 ng/ml) and ionomycin (100 ng/ml)]. Nuclear extracts were prepared and analyzed by EMSA with 32P-labeled probes containing MARE motifs or Oct-1–binding sites. The specificity of c-Maf binding to the probe was confirmed by supershift with an anti–c-Maf antibody. Data are representative of three experiments. (E) Supershift of c-Maf dimers with different members of the AP-1 family. Jurkat cells stably expressing c-Maf were stimulated with soluble anti-CD3 and anti-CD28 antibodies. Nuclear extracts were incubated with the indicated antibodies for 30 min on ice and then were subjected to EMSA. (F) The abundance of c-Fos is intact in CARMA1-deficient cells. Jurkat cells and CARMA1-deficient Jurkat cells, both stably transfected with plasmid encoding c-Maf, were stimulated with anti-CD3 (3 μg/ml) and anti-CD28 (2 μg/ml) antibodies. Whole-cell lysates were subjected to Western blotting analysis with antibodies specific for c-Fos and actin. Data are representative of three experiments. (G) Fractionation of c-Maf–containing complexes. The indicated cells (20 × 106) were stimulated with P/I as described in (D), and whole-cell lysates were subjected to the sucrose density gradient centrifugation. Twelve fractions were collected and analyzed by Western blotting with antibodies specific for c-Maf or c-Fos. Blots are representative of two independent experiments.
The kinase activity of IKKβ is required for c-Maf activation
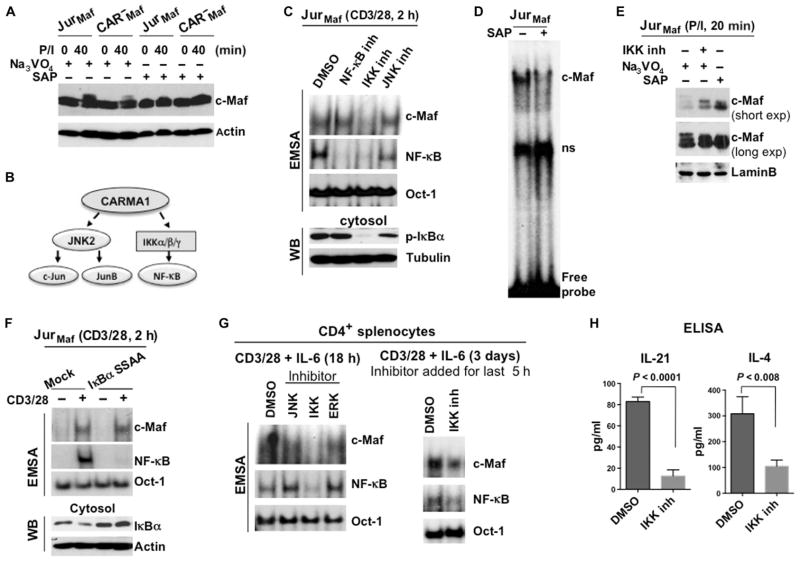
c-Maf, a member of the Maf protein family, is highly phosphorylated, and multiple phosphorylation events affect the stability, transactivation, and extent of DNA binding of Maf proteins (23–25). Although phosphorylation-dependent binding of c-Maf to DNA has not been shown previously, to our knowledge, one study reported that the dephosphorylation of MafA, another Maf family member, completely abolished its DNA binding activity (24). Therefore, we compared the c-Maf phosphorylation status in a c-Maf–expressing Jurkat cell line and in its CARMA1-deficient clone. To protect the phosphorylated c-Maf from endogenous phosphatases, we prepared whole-cell lysates in the presence of the phosphatase inhibitor sodium orthovanadate (Na3VO4) because a previous study indicated that Na3VO4 efficiently blocks endogenous phosphatases that target Maf (24). Upon treatment with P/I, we observed a substantial band shift in c-Maf in Jurkat cells, whereas there was a reduced band shift in CARMA1-deficient cells (Fig. 3A), suggesting that the modification of c-Maf was partially dependent on CARMA1. To confirm that the difference in the protein mobility was a result of phosphorylation, we treated the cell lysates with shrimp alkaline phosphatase (SAP) before performing Western blotting analysis. Indeed, this treatment reduced the c-Maf band shift in the stimulated samples (Fig. 3A).
Fig. 3. The kinase activity of IKK is required for the activation of c-Maf.
(A) Analysis of the c-Maf phosphorylation status in Jurkat and CARMA1-deficient cells stably expressing c-Maf. The indicated cells were stimulated with P/I, and cell lysates were treated with Na3VO4 or SAP. The band shift in c-Maf was determined by Western blotting analysis. Blots are representative of two independent experiments. (B) Scheme outlining the requirement of CARMA1 for TCR-dependent IKK and JNK2 activity. (C) Effects of JNK and IKK inhibitors on the DNA binding activity of c-Maf. Jurkat cells stably expressing c-Maf were stimulated with antibodies against CD3 and CD28 in the presence of inhibitors of JNK (SP600125), IKK (TPCA-1), or the translocation of NF-κB for 2 hours. Nuclear extracts were then subjected to EMSA (upper panels). Nuclear and cytoplasmic fractions from the same cells were also analyzed by Western blotting (WB) with antibodies against the indicated proteins (middle and lower panels). Data are representative of three independent experiments. (D) Jurkat cells stably expressing c-Maf were stimulated with P/I, and nuclear extracts were treated with SAP before being subjected to gel-shift analysis. (E) Jurkat cells stably expressing c-Maf were stimulated with P/I in the presence or absence of IKK inhibitor. The indicated lysates were also treated with Na3VO4 or SAP. c-Maf band shifts were determined by Western blotting analysis, and both short and long exposures of the Western blots for c-Maf are shown. Data are representative of two independent experiments. (F) Jurkat cells stably expressing c-Maf were transfected with plasmid encoding the super repressor IκBα mutant (IκBα-S32A/S36A). Forty-eight hours later, the cells were stimulated with antibodies against CD3 and CD28 for 2 hours, and nuclear extracts were prepared and subjected to EMSA. The DNA binding activities of c-Maf and NF-κB were analyzed by EMSA with 32P-labeled probes containing c-Maf–, NF-κB–, or Oct-1–binding sites. Data are representative of two independent experiments. (G) CD4+ splenocytes from WT mice were stimulated by antibodies against CD3 and CD28 together with IL-6 in the presence of inhibitors of the indicated kinases for 18 hours (left) or for 3 days with an IKK inhibitor (right), which was added for the last 5 hours. The DNA binding activities of c-Maf and NF-κB were then analyzed by EMSA. Data are representative of two independent experiments. (H) CD4+ splenocytes from WT mice were stimulated with plate-bound antibodies against CD3 and CD28 for 48 hours. An IKK inhibitor or vehicle [dimethyl sulfoxide (DMSO)] was added for the last 36 hours of the stimulation (triplicate cultures for each condition), and media were collected. The amounts of IL-4 and IL-21 were measured by ELISA and are presented as means ± SD.
CARMA1 mediates the activation of two kinases, IKK and JNK, which, in turn, activate NF-κB and the AP-1 family members c-Jun and JunB (Fig. 3B) (26). Therefore, we tested the effect of a JNK inhibitor (SP600125), a small-molecule inhibitor of IKKβ (TPCA-1), and an inhibitor of the nuclear translocation of NF-κB on the DNA binding activity of c-Maf. Gel-shift analysis revealed that the IKKβ inhibitor blocked the anti-CD3–and anti-CD28–dependent DNA binding activity of c-Maf in Jurkat cells, whereas the JNK and NF-κB inhibitors had no effect (Fig. 3C, upper panel). We also confirmed that only the IKK inhibitor blocked the kinase activity of IKK and suppressed the phosphorylation of IκBα (Fig. 3C, lower panel). These results suggest that the kinase activity of IKK is critical for the optimal phosphorylation of c-Maf and its DNA binding ability. Indeed, dephosphorylation of c-Maf with SAP completely diminished its binding to DNA (Fig. 3D). Western blotting analysis confirmed that both SAP and the IKK inhibitor reduced the extent of the band shift of c-Maf in acrylamide gels in samples from c-Maf–expressing Jurkat cells activated with P/I (Fig. 3E).
Our data also suggested that IKK regulated the activity of c-Maf in an NF-κB–independent manner. This conclusion was supported by experiments with cells expressing a mutant form of IκBα, known as a super-repressor (IκBα-S32A/S36A), which cannot be phosphorylated by the IKK complex, and thus cannot be degraded. This IκBα mutant completely blocked TCR-induced NF-κB activation but had no effect on the DNA binding activity of c-Maf (Fig. 3F). To further confirm our findings in primary T cells, we activated mouse CD4+ splenocytes with plate-bound anti-CD3 and anti-CD28 antibodies together with IL-6 in the presence of inhibitors of JNK, IKK, and extracellular signal–regulated kinase (ERK) for 18 hours (Fig. 3G). We included the ERK inhibitor PD98059 as a control because previous studies suggested that ERK phosphorylates Maf proteins (27) and that ERK activation does not depend on CARMA1 (7). Again, we found that the IKK inhibitor blocked the DNA binding activity of c-Maf in stimulated cells (Fig. 3G, left). Moreover, we observed a substantial reduction in the DNA binding ability of c-Maf in cells activated for 3 days and that had high amounts of c-Maf (Fig. 3G, right). As a consequence, prolonged suppression of c-Maf activity by the IKK inhibitor resulted in a substantial reduction in the amounts of IL-21 and IL-4 secreted by CD4+ T cells compared to those secreted by control cells (Fig. 3H).
CARMA1- and IKK-dependent modification of c-Maf contributes to its cellular localization
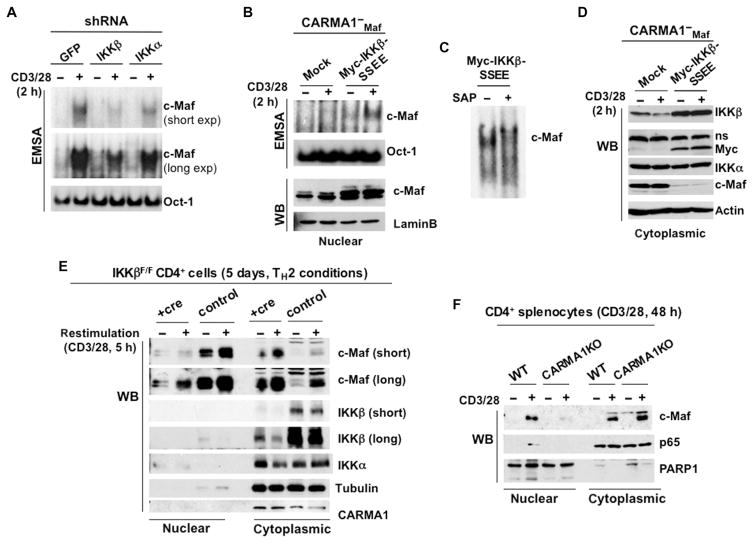
Our data suggest that IKK activity is involved in the activation of c-Maf. To support this finding, we decided to knock down the IKKα and IKKβ subunits of the IKK complex in c-Maf–expressing Jurkat cells and to determine the extent of the DNA binding activity of c-Maf in these cells upon TCR stimulation. We achieved knockdown of IKK subunits with several different specific short hairpin RNAs (shRNAs) in Jurkat cells (fig. S4), and we selected the cells with the greatest knockdown efficiency for the DNA binding experiments. We found that those cells in which IKKβ was knocked down also exhibited impaired DNA binding activity by c-Maf (Fig. 4A).
Fig. 4. CARMA1-dependent IKKβ activity contributes to the nuclear localization of c-Maf.
(A) Jurkat cells stably expressing c-Maf were transfected with IKKα- or IKKβ-specific shRNAs and then were selected with puromycin. Nuclear extracts were prepared from the indicated stable cell lines, and the DNA binding activity of c-Maf was analyzed by EMSA with 32P-labeled probes containing c-Maf– or Oct-1–binding sites. Data are representative of three independent experiments. (B to D) CARMA1-deficient Jurkat cells expressing c-Maf were stably transfected with plasmid encoding constitutively active IKKβ (IKKβ-SSEE) and then were subjected to (B and C) EMSA (nuclear extracts) or (D) Western blotting analysis (cytoplasmic extracts) with antibodies against the indicated proteins. In (C), nuclear extracts from the stimulated cells were treated with SAP before the gel-shift analysis was performed. Blots are representative of two or three experiments. (E) CD4+ splenocytes from IKKβF/F mice were stimulated with plate-bound anti-CD3 and anti-CD28 antibodies in the presence of IL-4 for 5 days. On days 5 and 6, the cells were infected with RV-GFP-Cre virus (+cre) or RV-GFP control virus (control). GFP+ cells were sorted on day 7, rested overnight in complete medium, and restimulated with antibodies against CD3 and CD28 for 2 hours. Nuclear and cytoplasmic fractions were prepared and subjected to Western blotting analysis with antibodies against the indicated proteins. Blots are representative of two experiments. (F) CD4+ splenocytes from the indicated mice were stimulated with plate-bound antibodies against CD3 and CD28 in the presence of IL-2 for 48 hours. Nuclear and cytoplasmic extracts were prepared and analyzed by Western blotting with antibodies against the indicated proteins. The experiment was repeated twice with similar results.
To provide further evidence that IKK activity was required for the DNA binding ability of c-Maf, we stably expressed a constitutively active mutant IKKβ (IKKβ-SSEE) (28) in c-Maf–expressing, CARMA1-deficient Jurkat cells. Although expression of IKKβ-SSEE alone did not stimulate the binding of c-Maf to MARE motifs in CARMA1-deficient cells, it rescued the DNA binding activity of c-Maf in these cells upon stimulation with anti-CD3 and anti-CD28 antibodies (Fig. 4B). These results suggest that activated IKKβ was not sufficient to induce the binding of c-Maf to the DNA probe containing the MARE motif, and that other phosphorylation events may be required to fully induce binding of c-Maf to DNA upon costimulation through CD3 and CD28. Indeed, SAP treatment of nuclear extracts of stimulated cells expressing constitutively active IKKβ inhibited the binding of c-Maf to DNA (Fig. 4C).
We observed that CARMA1-deficient cells expressing IKKβ-SSEE had increased amounts of nuclear c-Maf (Fig. 4B) but lacked cytosolic c-Maf (Fig. 4D), which suggests that IKKβ might promote the translocation of c-Maf to the nucleus. To test this hypothesis, we examined the cellular localization of endogenous c-Maf upon depletion of IKKβ in Ikk2F/F CD4+ T cells. First, we stimulated the purified CD4+ cells with plate-bound anti-CD3 and anti-CD28 antibodies in the presence of IL-4 for 5 days. Conditional deletion of loxP-flanked Ikk2 alleles was performed in vitro by transduction of CD4+ splenocytes with a Cre-expressing retrovirus. We found that decreased IKKβ abundance correlated with the reduced amount of c-Maf in the nucleus (Fig. 4E). Next, we examined the amounts of endogenous c-Maf in the nuclear and cytoplasmic fractions of CARMA1-deficient CD4+ splenocytes (Fig. 4F). Consistent with our earlier data, we found that upon initial stimulation of CD4+ T cells with antibodies against CD3 and CD28, the translocation of c-Maf to the nucleus was more efficient in wild-type cells than in CARMA1-deficient cells. These results suggest that the CARMA1-mediated activation of IKKβ plays an important role in the proper cellular localization of c-Maf. However, the remaining question was whether c-Maf served as a direct substrate for IKKβ. To test this possibility, we performed an in vitro kinase assay (fig. S5). Because we did not observe c-Maf phosphorylation in the presence of activated IKKβ, we conclude that IKK regulates c-Maf indirectly and that other kinases may be involved in this process.
CARMA1 contributes to the development of TFH cells and formation of GCs after immunization with ovalbumin
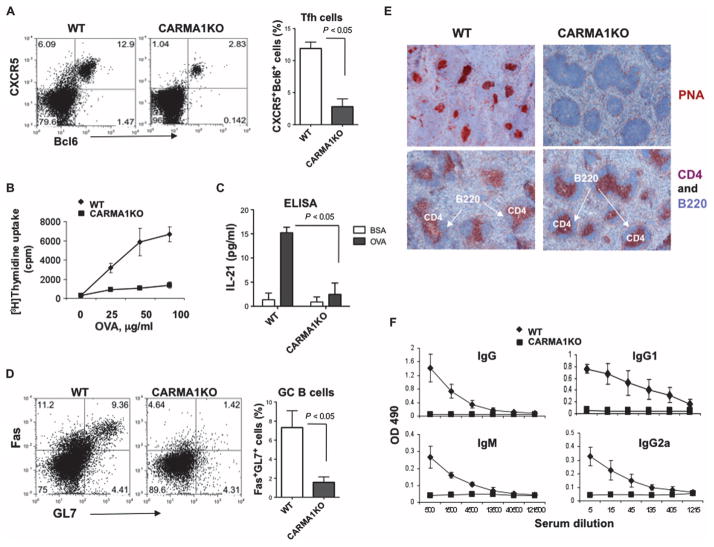
Our data collectively suggest that CARMA1 is likely involved in the generation of IL-4– and IL-21–producing T cells through c-Maf. Because c-Maf, as well as IL-21, plays an essential role in the generation of TFH cells (16), we hypothesized that CARMA1 might be required for the antigen-dependent development of these cells. To investigate this possibility, we compared the numbers of TFH cells, defined as CD4+CXCR5+Bcl6+ cells, in the spleens of wild-type and CARMA1-deficient mice immunized with ovalbumin (OVA) peptide (Fig. 5A). Indeed, we found that CARMA1-deficient mice exhibited substantially reduced percentages of TFH cells 7 days after immunization. Consistent with these data, CD4+ T cells from immunized CARMA1-deficient mice were unable to proliferate in response to OVA peptide in vitro (Fig. 5B) or to produce IL-21 upon restimulation with OVA peptide (Fig. 5C). These results suggest that TH cells from CARMA1-deficient mice may not be able to provide effective cognate help for B cells in GC formation or to support the development of humoral immunity. Indeed, we found that immunization of CARMA1-deficient mice resulted in the generation of fewer GC B cells, defined as B220+GL7+FasHigh cells, than were generated in immunized wild-type mice (Fig. 5D). Histological evaluation of spleen sections confirmed this finding by showing the lack of B220+PNA+ cells in CARMA1-deficient mice (Fig. 5E). In addition, we were unable to detect OVA-specific antibodies in sera of these mice 7 days after immunization (Fig. 5F).
Fig. 5. CARMA1 is required for the generation of TFH cells and the formation of GCs after immunization of mice with OVA peptide.
(A) Immunized CARMA1-deficient mice generate reduced amounts of TFH cells. CARMA1KO mice and age-matched WT control mice (three mice per group) were immunized subcutaneously at the base of tail with OVA peptide emulsified in complete Freund’s adjuvant (CFA). Seven days after immunization, the mice were sacrificed and analyzed individually. TFH cells (CD4+CXCR5+Bcl6+ cells) were identified by flow cytometric analysis. Dot plots are from a representative experiment, and the histogram shows means ± SD from three mice of each genotype. (B) Splenocytes collected from each immunized mouse [from (A)] were stimulated with the indicated concentrations of OVA peptide for 48 hours. Cellular proliferation was then assayed by adding [3H]thymidine to the cultures for the last 8 hours. Data are means ± SD from three mice of each genotype. (C) Splenocytes from each of the immunized mice were restimulated with OVA peptide (100 μg/ml) for 72 hours, cell culture medium was collected, and the amounts of IL-21 produced were determined by ELISA. Data are means ± SD of triplicate cultures. (D) CARMA1 is required for the generation of GC B cells. CARMA1KO and WT control mice were immunized as described in (A), and splenocytes were isolated. GC B cells (B220+GL7+FasHigh) were determined by flow cytometry. Dot plots are from a single representative experiment, and the histogram shows means ± SD from three mice of each genotype. (E) Formation of GCs is reduced in immunized CARMA1KO mice. GCs in the spleens of the indicated mice immunized with OVA peptide were determined by immunohistochemical staining with anti-mouse PNA antibody (brown) and counterstained with hematoxylin. T cells and B cells were identified by staining with anti-CD4 (red) and anti-B220 (blue) antibodies, respectively. Images are from one experiment and are representative of three independent experiments with consistent results. (F) Defective production of OVA-specific antibodies in CARMA1KO mice. Sera from the indicated immunized mice were subjected to serial dilutions, and the concentrations of OVA-specific immunoglobulin M (IgM) and IgG antibodies were analyzed by ELISA and averaged for each group. Graphs show the means ± SD from three mice of each genotype.
Deficiency in IKKβ in CD4+ T cells reduces the activation of c-Maf and impairs the development of TFH cells
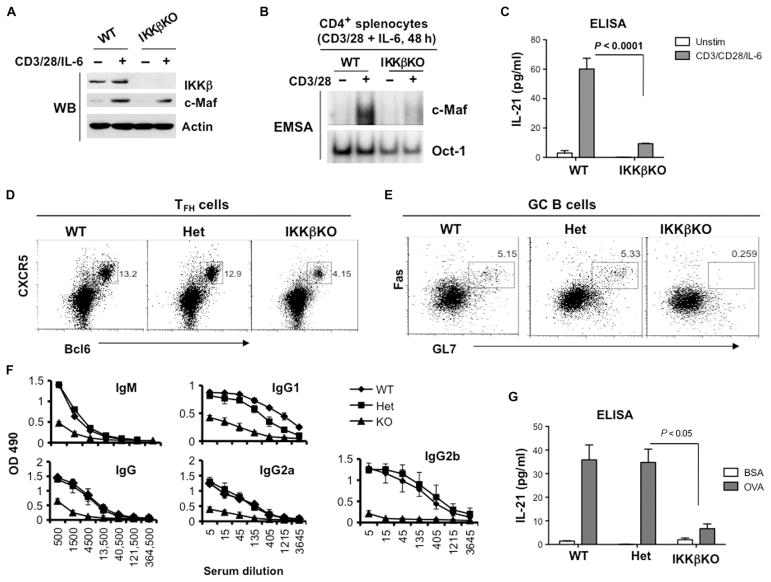
Thus far, our data suggest that CARMA1 is required for c-Maf activation in T cells in vitro and that it contributes to the development of TFH cells in vivo, most likely through IKKβ. To provide the genetic evidence that IKKβ is critical for these processes, we used mice with a T cell–specific loss of ikk2 alleles. Conditional deletion of loxP-flanked ikk2 alleles was achieved through the use of a CD4-Cre transgene. As expected, the loss of IKKβ did not affect the TCR-dependent increase in c-Maf abundance (Fig. 6A), but it reduced both the DNA binding activity of c-Maf as measured by EMSA (Fig. 6B) and the production of IL-21 (Fig. 6C). In contrast, conditional deletion of ikk1 alleles in CD4+ T cells (which leads to depletion of IKKα) did not have any effect on either the activation of c-Maf or the secretion of IL-21 (fig. S6).
Fig. 6. Loss of IKKβ in CD4+ T cells impairs the activation of c-Maf and inhibits the development of TFH cells in vivo.
(A) The TCR-dependent increase in c-Maf abundance does not require IKKβ. CD4+ splenocytes from the indicated mice were stimulated with plate-bound antibodies against CD3 and CD28 (2 and 1 μg/ml, respectively) in the presence of IL-6 (20 ng/ml) for 48 hours. Whole-cell lysates were then analyzed by Western blotting with antibodies specific for the indicated proteins. Blots are representative of two experiments. (B) CD4+ splenocytes from the indicated mice were activated as described in (A), and nuclear extracts were analyzed by EMSA with 32P-labeled probes containing c-Maf– or Oct-1–binding sites. Data are representative of three experiments. (C) The amounts of IL-21 secreted by splenocytes from the indicated mice were determined by the analysis of culture media by ELISA. Data are means ± SD from triplicate cultures and are representative of three independent experiments. (D to G) WT, IKKβ heterozygous (Het), and IKKβ-deficient (IKKβKO) mice (two mice per group) were immunized subcutaneously at the base of tail with OVA peptide emulsified in CFA. Seven days after immunization, the mice were sacrificed and analyzed individually. (D and E) IKKβ is required for the generation of TFH cells and GC B cells. Splenocytes were isolated from the indicated immunized mice, and TFH cells (CD4+CXCR5+Bcl6+ cells) and GC B cells (B220+GL7+FasHigh) were determined by flow cytometry. Dot plots are from a single representative experiment. (F) Defective production of OVA-specific antibodies in IKKβKO mice. Sera from the indicated immunized mice were subjected to serial dilutions, and the concentrations of OVA-specific IgM and IgG antibodies were analyzed by ELISA. Data are means ± SD from each experimental group calculated from triplicate measurements. (G) Splenocytes from the indicated immunized mice were restimulated with OVA peptide (100 μg/ml) for 48 hours, and IL-21 production was determined by ELISA. Data are means ± SD of triplicate cultures.
To test whether IKKβ played a role in TFH cell development or GC formation, we immunized wild-type, IKKβ heterozygous, and IKKβ-deficient mice with OVA peptide. Flow cytometric analysis of splenocytes isolated from mice 7 days after immunization revealed a marked reduction in the percentage of CXCR5+Bcl6+ cells (Fig. 6D) and GL7+FasHigh GC B cells (Fig. 6E) in IKKβ-deficient mice compared to those in wild-type mice. Consistent with these results, the IKKβ-deficient mice had reduced concentrations of OVA-specific antibodies in their sera (Fig. 6F), confirming that their humoral response was impaired. Moreover, the recall response of IKKβ-deficient T cells to OVA peptide did not result in the production of substantial amounts of IL-21 (Fig. 6G). Together, our results demonstrate that ablation of IKKβ in CD4+ T cells leads to defects similar to those observed in CD4+ T cells from CARMA1-deficient mice.
DISCUSSION
The IKK complex is a fundamental component of all signaling pathways that lead to activation of the transcription factor NF-κB. Although the role of IKK in NF-κB activation, as well as its molecular mechanism, is well established, our knowledge about its effect on other transcription factors is less clear. Here, we demonstrated that TCR-dependent IKK activation was critical to stimulate the activity and nuclear localization of c-Maf. Furthermore, this process required the scaffold protein CARMA1, but was independent on NF-κB activation.
c-Maf is a member of the Maf family of transcription factors, which consist of an N-terminal transactivation domain, a DNA binding domain, and a C-terminal leucine zipper dimerization domain (29). Deletion of either the transactivation or dimerization domain of Maf proteins abolishes their transcriptional activity (24, 29, 30). In addition, phosphorylation of the N terminus of Maf is required for its dimerization and DNA binding, whereas dephosphorylation of this region suppresses the DNA binding activity of Maf (24). Although all of the previous observations were made with overexpression system or recombinant proteins, our data are consistent with these previous findings. Consistent with these findings, we found that endogenous c-Maf was phosphorylated in activated cells and that loss of CARMA1 resulted in only a partial decrease in the extent of phosphorylation of c-Maf; however, loss of CARMA1 reduced the nuclear accumulation of c-Maf and suppressed its binding to DNA. These data suggest that c-Maf might be phosphorylated by multiple kinases and that at least one of them is not activated in the absence of CARMA1.
To date, several kinases have been suggested to phosphorylate Maf proteins, including ERK (27), p38 MAPK (25), and GSK-3β (glycogen synthase kinase 3β) (23). Because CARMA1 mediates the activation of two kinases, JNK and IKK, without affecting ERK and p38 (7), we focused on these two enzymes. Indeed, our data revealed that IKK activity was required for c-Maf activation because inhibition of IKK diminished the DNA binding ability of c-Maf in mouse CD4+ splenocytes and in Jurkat cells. Moreover, CARMA1-dependent activation of IKK promoted the nuclear translocation of c-Maf, and the TCR-dependent nuclear localization of c-Maf was reduced in the absence of CARMA1 or IKKβ. In contrast, expression of a constitutively active mutant IKKβ in CARMA1-deficient cells led to the enhanced nuclear translocation of c-Maf; however, we found some discrepancy between the results obtained from primary T cells and those from a Jurkat cell line overexpressing c-Maf. The increased abundance of c-Maf in the Jurkat cell line resulted in its nuclear localization even in the absence of CARMA1, suggesting that ectopic expression of Maf leads to its nuclear accumulation in a signal-independent manner.
Although some of the overexpressed c-Maf protein was localized in the nucleus in the absence of CARMA1, additional TCR stimulation was required to induce its DNA binding activity. This observation indicates that the nuclear localization of c-Maf alone is not sufficient to induce its binding to DNA, and suggests that another TCR-dependent modification of c-Maf is required. Therefore, we suggest that IKK is one of the signaling mediators that enhance the nuclear translocation of c-Maf and its DNA binding activity. Indeed, another study indicated that IKKβ physically interacts with and phosphorylates the forkhead transcription factor FOXO3a, which regulates its cellular localization (31). Also, SRC3 (steroid receptor coactivator 3) may be activated in an IKKβ-dependent manner (32). Thus, our future study will explore the detailed molecular mechanism that underlies the IKK–c-Maf interaction. Because we failed to demonstrate that c-Maf was a direct substrate for activated IKKβ in an in vitro kinase assay, we consider the possibility that IKK regulates c-Maf indirectly and that other kinases may be involved in this process.
A previous study indicated the important role of c-Maf in inducing the Il4 promoter (33). In CD4+ T cells, the c-Maf protein is synthesized upon TCR stimulation, and mice deficient in c-Maf have impaired IL-4 production (17, 18). Although c-Maf has been considered a TH2-associated transcription factor, other studies revealed its essential role in the generation of TFH cells (12, 16). Indeed, our work indicates that CARMA1 and IKKβ contribute to the generation of TFH cells, at least in part by regulating c-Maf activity. Genetic ablation of c-Maf impairs the generation of TFH cells in mice immunized with trinitrophenyl or OVA peptide (16). Here, we showed that CARMA1- and IKKβ-deficient mice had reduced numbers of TFH cells in response to immunization with OVA peptide. TFH cells are required for the formation and maintenance of GCs, as well as for the regulation of humoral immunity (12), and our results showed that CD4+ T cell–specific deletion of IKKβ resulted in reduced generation of GC B cells. Indeed, the concentrations of antigen-specific antibodies were decreased in the sera of immunized CD4-Cre/Ikk2F/F mice. These data are suggestive of a T cell–intrinsic role for the CARMA1-IKKβ signalosome in the initiation of the humoral response. Our observation is also consistent with a previous study that demonstrated that T cells from IKKβ-deficient mice fail to support GC reactions (34), suggesting that this defect is a result of the failure to activate c-Maf.
Although T cell–specific loss of IKKβ leads to a reduction in the numbers of natural killer T cells, Treg cells, and memory T cells (35), IKKβ-deficient cells can be activated in vitro, as well as proliferate and secrete cytokines in response to polyclonal stimulation (34, 36). Naïve IKKβ-deficient T cells produce slightly more interferon-γ (IFNγ) than do wild-type T cells during initial activation (the first 48 hours) (34), and the same observation has been reported for CARMA1-deficient cells (10), suggesting that these defects in T cells are not a result of a defect in T cell proliferation. Our data suggest that an IKKβ-dependent defect in c-Maf activation impairs the differentiation of TFH cells, which, in turn, affects the generation of GC B cells.
In summary, our study provides evidence for a molecular link between the TCR signaling cascade and the activation of c-Maf. We demonstrated that CARMA1 and IKKβ are critical signaling molecules in the regulation of the nuclear localization of c-Maf and its DNA binding activity. As a biological consequence, conditional deletion of IKKβ in CD4+ T cells resulted in the generation of reduced numbers of TFH cells, which further negatively affected the development of GC B cells and the quality of the humoral response to immunization with a T cell–dependent antigen.
MATERIALS AND METHODS
Reagents and plasmids
Antibodies specific for phosphorylated IκBα (pIκBα, Ser32/36) and CARMA1 were purchased from Cell Signaling. Antibodies specific for c-Maf, JunB, GATA3, NFAT, IKKβ, IKKα, IκBα, Myc, laminB, and actin were obtained from Santa Cruz Biotechnology. The JNK inhibitor SP600125 (#S5567), the ERK inhibitor PD98059 (#P215), and the IKKβ-specific small-molecule inhibitor TPCA-1 (#T1452) were obtained from Sigma. An inhibitor of the nuclear translocation of NF-κB [a synthetic inhibitor containing the nuclear localization sequence residues 360 to 369 of the NF-κB subunit p50] was purchased from Santa Cruz Biotechnology (#sc-3060). The retroviral construct GFP-RV-cMaf was described previously (37). The complementary DNA encoding constitutively active IKKβ (S177E S181E) (28) was amplified by PCR from pCMV IKKβ S177E S181E as a template (Addgene, http://www.addgene.org, plasmid 11105) and was subcloned into the Bam HI and Eco RI sites of the expression vector pRV3. The plasmid pBabe GFP-IκBα-S32A/S36A was obtained from Addgene (plasmid 15264). Lentiviral plasmids encoding shRNAs (pLKO.1-puro) were purchased from Sigma (NM_001278, NM_001556).
Cell lines and transfections
Jurkat cells deficient in CARMA1 were described previously (7). Jurkat cells and primary T cells were cultured in RPMI 1640 supplemented with 10% fetal bovine serum (FBS), penicillin, and streptomycin. Human embryonic kidney (HEK) 293T cells and Phoenix cells were maintained in Dulbecco’s modified Eagle’s medium supplemented with 10% FBS, penicillin, and streptomycin. Cells were transfected with the calcium phosphate coprecipitation method. Stable transfections were established by viral infection. First, HEK 293T cells were transfected with an expression vector (pRV3 IKK-SSEE) together with packaging plasmids (pHep and pEnv) with the calcium phosphate precipitation method. To generate retro-viruses, Phoenix cells were cotransfected with the plasmids GFP-RV-cMaf and pCL, and viral supernatants were collected 48 hours later. Jurkat cells were resuspended in viral supernatant in the presence of polybrene and were incubated for 6 hours. Cells were then washed and cultured in complete medium. The efficiency of viral infection was determined by flow cytometric or Western blotting analysis. Sorting of GFP+ cells was performed for cells infected with GFP-RV-cMaf.
Mice
CARMA1-deficient mice were described previously (6). Mice carrying the loxP-flanked Ikk2 and Ikk1 alleles were provided by M. Karin and M. Pasparakis (38). Animals were maintained under pathogen-free conditions in the institutional animal facility. All experiments were performed in compliance with institutional guidelines and according to the protocol approved by the Institutional Animal Use and Care Committee of The University of Texas MD Anderson Cancer Center.
Immunization of mice with OVA peptide
CARMA1-deficient mice, CD4-Cre/Ikk2-floxed and CD4-Cre/Ikk1-floxed mice, and their controls (7-week-old female mice) were immunized with 100 μg of OVA peptide (fraction V, Sigma) emulsified in CFA (0.5 mg/ml) at the base of tail (100 μl per mouse). Seven days after immunization, the mice were sacrificed, and lymphocytes from spleens and inguinal lymph nodes were obtained. TFH cells were identified by flow cytometric analysis with phycoerythrin (PE)–conjugated antibody specific for Bcl6 (K112-91, BD Bioscience) and biotinylated monoclonal antibody (mAb) against CXCR5 (clone 2G8, BD Biosciences) followed by allophycocyanin-labeled streptavidin (Jackson ImmunoResearch Laboratories) in the CD4+ cell population [peridinin chlorophyll protein (PerCP)–CD4 mAb, clone GK 1.5, BioLegend]. To analyze GC B cells, splenocytes were incubated with PerCP-conjugated antibody against B220 (clone RA3-6B2, BioLegend), fluorescein isothiocyanate–conjugated antibody against GL7 (also known as T and B cell–activated antigen) (clone GL7, BD Biosciences), and PE-conjugated antibody against Fas (CD95) (BD Bioscience). Serum samples were subjected to immunoglobulin analysis by ELISA. In brief, different dilutions of serum were added into wells of plates precoated with OVA peptide (10 μg/ml), and antigen-specific antibodies were detected with biotinylated anti-mouse IgG, horseradish peroxidase (HRP)–conjugated goat anti-mouse IgM, goat anti-mouse IgG1-HRP, and goat anti-mouse IgG2a-HRP antibodies (Southern Biotechnology Associates).
Immunohistochemical analysis
Spleen tissues were embedded in optimum cutting temperature freezing compound before sectioning was performed. Tissue blocks were sliced into 6-μm sections, air-dried, and fixed with cold acetone. Purified anti-mouse CD4 antibody (BD Pharmingen) or biotin-labeled PNA antibody (Vector Laboratories) was applied as primary antibody, followed by biotinylated anti-rat secondary antibody and avidin-peroxidase complex reagent. NovaRed was used as a substrate. For B cell staining, biotin-labeled anti-B220 antibody (BD Pharmingen) was used, followed by avidin-alkaline phosphatase complex reagent. Slides for PNA staining were counterstained with hematoxylin.
Isolation and activation of CD4+ T cells
CD4+ T cells were isolated from mouse spleens and peripheral lymph nodes with magnetic beads (StemCell Technologies). The cells were purified by negative selection with a panel of biotinylated antibodies against CD8, CD11b, CD19, CD45R/B220, CD49b, TER119, and CD25. After 3 hours of rest, the cells were activated with plate-bound anti-CD3ε antibody (2 μg/ml) and anti-CD28 antibody (1 μg/ml) (both from BD Biosciences). To generate TH2 cells, anti-IFNγ antibody (10 μg/ml, XMG1.2) and recombinant murine IL-4 (10 ng/ml, PeproTech) were added to the cultures. Because CARMA1-deficient cells are defective in their ability to produce IL-2, some cell cultures were supplemented with recombinant IL-2 (30 U/ml).
Detection of secreted cytokines
CD4+ T cells were activated under different conditions, and secreted cytokines were detected in culture medium with the Bio-Plex Pro mouse cytokine panel B (Bio-Rad, #171-FA001M) and the Milliplex Map kit (Millipore, #MCYTOMAG-70K). All assays were performed according to the manufacturer’s protocols, and the MFIs were detected by the Luminex 200 system and were analyzed with Bio-Plex software (Bio-Rad). The concentrations of IL-21 and IL-4 in culture medium were also determined by sandwich ELISA (Ready-SET-GO kit, eBioscience). Results are presented as arithmetic means ± SD of triplicate cultures.
Western blotting analysis
Cells were lysed in buffer containing 50 mM Hepes (pH 7.4), 150 mM NaCl, 1% NP-40, 1 mM EDTA, 1 mM Na3VO4, 1 mM NaF, 1 mM phenyl-methylsulfonyl fluoride, 1 mM dithiothreitol, and a protease inhibitor cocktail (Roche Diagnostics). The samples were resolved by 10% SDS–polyacrylamide gel electrophoresis and were transferred onto nitrocellulose membranes. Western blots were incubated with specific primary antibodies, followed by HRP-conjugated secondary antibodies, and were developed with the enhanced chemiluminescence method according to the manufacturer’s protocol (Pierce).
Electrophoretic mobility shift assays
Nuclear proteins were extracted from 3 × 106 to 4 × 106 cells, as described previously (9). Nuclear extracts (4 μg) were incubated with a 32P-labeled, double-stranded, NF-κB–, c-Maf–, or Oct-1–specific probe for 15 min at room temperature, fractionated on a 5% polyacrylamide gel, and visualized by autoradiography. The NF-κB and Oct-1 probes were purchased from Promega. The sequence of the c-Maf–specific probe (the MARE consensus sequence is underlined) was 5′-GGAATTGCTGATTCAGCAACTTT-3′ (29).
ChIP assays
ChIPs were performed with a ChIP assay kit (Upstate Biotechnology). In brief, after being cross-linked with 1% formaldehyde for 10 min at room temperature, cells were lysed in SDS buffer [50 mM tris-HCl (pH 8.1), 10 mM EDTA, 1% SDS, and protease inhibitors]. Lysates were sonicated with a Bioruptor and were diluted with ChIP dilution buffer [20 mM tris-HCl (pH 8.1), 1 mM EDTA, 150 mM NaCl, and 0.3% Triton X-100]. Lysates were then precleared with protein G beads and incubated with anti–c-Maf antibody (M-153, Biothyl) or with control rabbit IgG at 4°C overnight. Protein G agarose was added to the lysates and incubated for 2 hours at 4°C. Antibody-protein-DNA complexes were eluted and incubated at 65°C overnight to reverse the formaldehyde cross-linking, and DNA was recovered with a PCR purification kit (Qiagen). DNA fragments were amplified by PCR with the following specific primers: mouse IL-21P, 5′-TGGTGAATGCT-GAAAACTGGAA-3′ (forward) and 5′-CCCATCTGCATCTTAGACAGGAA-3′ (reverse); mouse IL-4P, 5′-AGGGGTGTTTCATTTTCCAA-3′ (forward) and 5′-GTTGCTGAAACCAAGGGAAA-3′ (reverse).
Sucrose gradient fractionations
Cells (20 × 106) were stimulated with P/I [PMA (20 ng/ml) and ionomycin (100 ng/ml)] for 40 min, and whole-cell lysates were subjected to sucrose gradient centrifugation. In brief, each sample (300 μl) was loaded onto a 40 to 5% sucrose gradient (40% 0.2 ml; 30% 0.2 ml; 25% 0.5 ml; 20% 0.8 ml; 15% 0.8 ml; 10% 1.0 ml; 5% 1.0 ml) and centrifuged at 45,000 rpm for 20 hours at 4°C. Twelve fractions (400 μl per fraction) were collected from the top of the tube, and proteins were precipitated with trichloroacetic acid. Western blotting analysis was performed to detect c-Maf and c-Fos in the obtained fractions.
Statistical analysis
GraphPad Prism software was used for all statistical analyses. All in vivo and in vitro experiments were performed in triplicate. The histograms show means ± SD. The Student’s t test was used to evaluate the difference of two groups of data. P < 0.05 (using a two-tailed paired t test) was considered to be statistically significantly different.
Supplementary Material
Acknowledgments
We thank J. Penninger for the CARMA1-deficient mice, M. Karin and M. Pasparakis for the Ikk2- and Ikk1-floxed mice, and W. Hahn for the pBabe-GFP-IκBα-mutant plasmid. Funding: This work was partially supported by grants from the NIH (GM065899 and AI050848) and a pilot grant from the Center for Inflammation and Cancer of MD Anderson Cancer Center to X.L.
Footnotes
Author contributions: M.B. and X.L. designed the experiments, analyzed the data, and wrote the manuscript; M.B. performed most of the experiments; D.J. performed cloning and some biochemical experiments; R.I.N. performed animal experiments and flow cytometry; X.Z. assisted with animal experiments; P.C., S.-C.S., and C.D. provided critical reagents and critical comments.
Competing interests: The authors declare that they have no competing interests.
REFERENCES AND NOTES
- 1.Zhu J, Yamane H, Paul WE. Differentiation of effector CD4 T cell populations. Annu Rev Immunol. 2010;28:445–489. doi: 10.1146/annurev-immunol-030409-101212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Glimcher LH, Murphy KM. Lineage commitment in the immune system: The T helper lymphocyte grows up. Genes Dev. 2000;14:1693–1711. [PubMed] [Google Scholar]
- 3.Schulze-Luehrmann J, Ghosh S. Antigen-receptor signaling to nuclear factor κB. Immunity. 2006;25:701–715. doi: 10.1016/j.immuni.2006.10.010. [DOI] [PubMed] [Google Scholar]
- 4.Blonska M, Lin X. NF-κB signaling pathways regulated by CARMA family of scaffold proteins. Cell Res. 2011;21:55–70. doi: 10.1038/cr.2010.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hayden MS, Ghosh S. Shared principles in NF-κB signaling. Cell. 2008;132:344–362. doi: 10.1016/j.cell.2008.01.020. [DOI] [PubMed] [Google Scholar]
- 6.Hara H, Wada T, Bakal C, Kozieradzki I, Suzuki S, Suzuki N, Nghiem M, Griffiths EK, Krawczyk C, Bauer B, D’Acquisto F, Ghosh S, Yeh WC, Baier G, Rottapel R, Penninger JM. The MAGUK family protein CARD11 is essential for lymphocyte activation. Immunity. 2003;18:763–775. doi: 10.1016/s1074-7613(03)00148-1. [DOI] [PubMed] [Google Scholar]
- 7.Wang D, You Y, Case SM, McAllister-Lucas LM, Wang L, DiStefano PS, Nuñez G, Bertin J, Lin X. A requirement for CARMA1 in TCR-induced NF-κB activation. Nat Immunol. 2002;3:830–835. doi: 10.1038/ni824. [DOI] [PubMed] [Google Scholar]
- 8.Egawa T, Albrecht B, Favier B, Sunshine MJ, Mirchandani K, O’Brien W, Thome M, Littman DR. Requirement for CARMA1 in antigen receptor-induced NF-κB activation and lymphocyte proliferation. Curr Biol. 2003;13:1252–1258. doi: 10.1016/s0960-9822(03)00491-3. [DOI] [PubMed] [Google Scholar]
- 9.Blonska M, Pappu BP, Matsumoto R, Li H, Su B, Wang D, Lin X. The CARMA1-Bcl10 signaling complex selectively regulates JNK2 kinase in the T cell receptor-signaling pathway. Immunity. 2007;26:55–66. doi: 10.1016/j.immuni.2006.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blonska M, Joo D, Zweidler-McKay PA, Zhao Q, Lin X. CARMA1 controls Th2 cell-specific cytokine expression through regulating JunB and GATA3 transcription factors. J Immunol. 2012;188:3160–3168. doi: 10.4049/jimmunol.1102943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nurieva RI, Chung Y, Hwang D, Yang XO, Kang HS, Ma L, Wang YH, Watowich SS, Jetten AM, Tian Q, Dong C. Generation of T follicular helper cells is mediated by interleukin-21 but independent of T helper 1, 2, or 17 cell lineages. Immunity. 2008;29:138–149. doi: 10.1016/j.immuni.2008.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Crotty S. Follicular helper CD4 T cells (TFH) Annu Rev Immunol. 2011;29:621–663. doi: 10.1146/annurev-immunol-031210-101400. [DOI] [PubMed] [Google Scholar]
- 13.Vogelzang A, McGuire HM, Yu D, Sprent J, Mackay CR, King C. A fundamental role for interleukin-21 in the generation of T follicular helper cells. Immunity. 2008;29:127–137. doi: 10.1016/j.immuni.2008.06.001. [DOI] [PubMed] [Google Scholar]
- 14.Zotos D, Coquet JM, Zhang Y, Light A, D’Costa K, Kallies A, Corcoran LM, Godfrey DI, Toellner KM, Smyth MJ, Nutt SL, Tarlinton DM. IL-21 regulates germinal center B cell differentiation and proliferation through a B cell–intrinsic mechanism. J Exp Med. 2010;207:365–378. doi: 10.1084/jem.20091777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Linterman MA, Beaton L, Yu D, Ramiscal RR, Srivastava M, Hogan JJ, Verma NK, Smyth MJ, Rigby RJ, Vinuesa CG. IL-21 acts directly on B cells to regulate Bcl-6 expression and germinal center responses. J Exp Med. 2010;207:353–363. doi: 10.1084/jem.20091738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bauquet AT, Jin H, Paterson AM, Mitsdoerffer M, Ho IC, Sharpe AH, Kuchroo VK. The costimulatory molecule ICOS regulates the expression of c-Maf and IL-21 in the development of follicular T helper cells and TH-17 cells. Nat Immunol. 2009;10:167–175. doi: 10.1038/ni.1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ho IC, Hodge MR, Rooney JW, Glimcher LH. The proto-oncogene c-maf is responsible for tissue-specific expression of interleukin-4. Cell. 1996;85:973–983. doi: 10.1016/s0092-8674(00)81299-4. [DOI] [PubMed] [Google Scholar]
- 18.Kim JI, Ho IC, Grusby MJ, Glimcher LH. The transcription factor c-Maf controls the production of interleukin-4 but not other Th2 cytokines. Immunity. 1999;10:745–751. doi: 10.1016/s1074-7613(00)80073-4. [DOI] [PubMed] [Google Scholar]
- 19.Xu J, Yang Y, Qiu G, Lal G, Wu Z, Levy DE, Ochando JC, Bromberg JS, Ding Y. c-Maf regulates IL-10 expression during Th17 polarization. J Immunol. 2009;182:6226–6236. doi: 10.4049/jimmunol.0900123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kroenke MA, Eto D, Locci M, Cho M, Davidson T, Haddad EK, Crotty S. Bcl6 and Maf cooperate to instruct human follicular helper CD4 T cell differentiation. J Immunol. 2012;188:3734–3744. doi: 10.4049/jimmunol.1103246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hiramatsu Y, Suto A, Kashiwakuma D, Kanari H, Kagami S, Ikeda K, Hirose K, Watanabe N, Grusby MJ, Iwamoto I, Nakajima H. c-Maf activates the promoter and enhancer of the IL-21 gene, and TGF-β inhibits c-Maf-induced IL-21 production in CD4+ T cells. J Leukoc Biol. 2010;87:703–712. doi: 10.1189/jlb.0909639. [DOI] [PubMed] [Google Scholar]
- 22.Yang Y, Ochando J, Yopp A, Bromberg JS, Ding Y. IL-6 plays a unique role in initiating c-Maf expression during early stage of CD4 T cell activation. J Immunol. 2005;174:2720–2729. doi: 10.4049/jimmunol.174.5.2720. [DOI] [PubMed] [Google Scholar]
- 23.Guo S, Burnette R, Zhao L, Vanderford NL, Poitout V, Hagman DK, Henderson E, Ozcan S, Wadzinski BE, Stein R. The stability and transactivation potential of the mammalian MafA transcription factor are regulated by serine 65 phosphorylation. J Biol Chem. 2009;284:759–765. doi: 10.1074/jbc.M806314200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guo S, Vanderford NL, Stein R. Phosphorylation within the MafA N terminus regulates C-terminal dimerization and DNA binding. J Biol Chem. 2010;285:12655–12661. doi: 10.1074/jbc.M110.105759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sii-Felice K, Pouponnot C, Gillet S, Lecoin L, Girault JA, Eychène A, Felder-Schmittbuhl MP. MafA transcription factor is phosphorylated by p38 MAP kinase. FEBS Lett. 2005;579:3547–3554. doi: 10.1016/j.febslet.2005.04.086. [DOI] [PubMed] [Google Scholar]
- 26.Blonska M, Lin X. CARMA1-mediated NF-κB and JNK activation in lymphocytes. Immunol Rev. 2009;228:199–211. doi: 10.1111/j.1600-065X.2008.00749.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Benkhelifa S, Provot S, Nabais E, Eychène A, Calothy G, Felder-Schmittbuhl MP. Phosphorylation of MafA is essential for its transcriptional and biological properties. Mol Cell Biol. 2001;21:4441–4452. doi: 10.1128/MCB.21.14.4441-4452.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mercurio F, Zhu H, Murray BW, Shevchenko A, Bennett BL, Li J, Young DB, Barbosa M, Mann M, Manning A, Rao A. IKK-1 and IKK-2: Cytokine-activated IκB kinases essential for NF-κB activation. Science. 1997;278:860–866. doi: 10.1126/science.278.5339.860. [DOI] [PubMed] [Google Scholar]
- 29.Kataoka K, Noda M, Nishizawa M. Maf nuclear oncoprotein recognizes sequences related to an AP-1 site and forms heterodimers with both Fos and Jun. Mol Cell Biol. 1994;14:700–712. doi: 10.1128/mcb.14.1.700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cao S, Liu J, Song L, Ma X. The protooncogene c-Maf is an essential transcription factor for IL-10 gene expression in macrophages. J Immunol. 2005;174:3484–3492. doi: 10.4049/jimmunol.174.6.3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hu MC, Lee DF, Xia W, Golfman LS, Ou-Yang F, Yang JY, Zou Y, Bao S, Hanada N, Saso H, Kobayashi R, Hung MC. IκB kinase promotes tumorigenesis through inhibition of forkhead FOXO3a. Cell. 2004;117:225–237. doi: 10.1016/s0092-8674(04)00302-2. [DOI] [PubMed] [Google Scholar]
- 32.Wu RC, Qin J, Hashimoto Y, Wong J, Xu J, Tsai SY, Tsai MJ, O’Malley BW. Regulation of SRC-3 (pCIP/ACTR/AIB-1/RAC-3/TRAM-1) coactivator activity by IκB kinase. Mol Cell Biol. 2002;22:3549–3561. doi: 10.1128/MCB.22.10.3549-3561.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li B, Tournier C, Davis RJ, Flavell RA. Regulation of IL-4 expression by the transcription factor JunB during T helper cell differentiation. EMBO J. 1999;18:420–432. doi: 10.1093/emboj/18.2.420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schmidt-Supprian M, Tian J, Ji H, Terhorst C, Bhan AK, Grant EP, Pasparakis M, Casola S, Coyle AJ, Rajewsky K. IκB kinase 2 deficiency in T cells leads to defects in priming, B cell help, germinal center reactions, and homeostatic expansion. J Immunol. 2004;173:1612–1619. doi: 10.4049/jimmunol.173.3.1612. [DOI] [PubMed] [Google Scholar]
- 35.Schmidt-Supprian M, Tian J, Grant EP, Pasparakis M, Maehr R, OVAa H, Ploegh HL, Coyle AJ, Rajewsky K. Differential dependence of CD4+CD25+ regulatory and natural killer-like T cells on signals leading to NF-κB activation. Proc Natl Acad Sci USA. 2004;101:4566–4571. doi: 10.1073/pnas.0400885101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schmidt-Supprian M, Courtois G, Tian J, Coyle AJ, Israël A, Rajewsky K, Pasparakis M. Mature T cells depend on signaling through the IKK complex. Immunity. 2003;19:377–389. doi: 10.1016/s1074-7613(03)00237-1. [DOI] [PubMed] [Google Scholar]
- 37.Nurieva RI, Duong J, Kishikawa H, Dianzani U, Rojo JM, Ho I, Flavell RA, Dong C. Transcriptional regulation of Th2 differentiation by inducible costimulator. Immunity. 2003;18:801–811. doi: 10.1016/s1074-7613(03)00144-4. [DOI] [PubMed] [Google Scholar]
- 38.Pasparakis M, Courtois G, Hafner M, Schmidt-Supprian M, Nenci A, Toksoy A, Krampert M, Goebeler M, Gillitzer R, Israel A, Krieg T, Rajewsky K, Haase I. TNF-mediated inflammatory skin disease in mice with epidermis-specific deletion of IKK2. Nature. 2002;417:861–866. doi: 10.1038/nature00820. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.