Abstract
This study was designed to examine the interaction of sulfamethoxazole (SMZ) with human serum albumin(HSA). Spectroscopic analysis of the emission quenching at different temperatures revealed that the quenching mechanism of human serum albumin by SMZ was static mechanism. The binding constant values for the SMZ–HSA system were obtained to be 22,500 L/mol at 288 K, 15,600 L/mol at 298 K, and 8500 L/mol at 308 K. The distance r between donor and acceptor was evaluated according to the theory of Föster energy transfer. The results of spectroscopic analysis and molecular modeling techniques showed that the conformation of human serum albumin had been changed in the presence of SMZ. The thermodynamic parameters, namely enthalpy change (∆H0) −36.0 kJ/mol, entropy change (∆S0) −41.3 J/mol K and free energy change (∆G0) −23.7 kJ/mol, were calculated by using van׳t Hoff equation. The effect of common ions on the binding of SMZ to HSA was tested.
Keywords: Human serum albumin, Sulfamethoxazole, Fluorescence quenching study, Static mechanism
1. Introduction
Human serum albumin (HSA), the most abundant human plasma protein, contains a single polypeptide chain of 585 amino acids. It is largely helical and is composed of three structurally homologous domains (I, II and III) [1]. Each domain contains 10 helices; helices 1–6 form the respective sub-domains A and helices 7–10 comprise sub-domains B. Aromatic and heterocyclic ligands are found to bind within two hydrophobic pockets in sub-domains IIA and IIIA, namely sites I and II. Seven binding sites are localized for fatty acids in sub-domains IB, IIIA, IIIB and on the sub-domain interfaces [2]. HSA has a high affinity metal binding site at the N-terminus [3]. The multiple binding sites underlie the exceptional ability of HSA to interact with many organic and inorganic molecules and make this protein an important regulator of intercellular fluxes, as well as the pharmacokinetic behavior of many drugs [1], [2]. Plasma protein binding of drugs assumes great importance since it influences their pharmacokinetic and pharmacodynamic properties, and may also cause interference with the binding of other endogenous and/or exogenous ligands as a result of overlap of binding sites and/or conformational changes. Therefore, detailed investigation of drug–protein interaction assumes significance for thorough understanding of the pharmacokinetic behavior of a drug and for the design of analogs with effective pharmacological properties. Hence, it becomes an important research field in chemistry, life sciences and clinical medicine.
Recently, many optical techniques have been carried out to investigate the interaction of proteins and drugs, because these methods are sensitive and relatively easy to use [4], [5]. Among these, fluorescence quenching is a useful method to study the reactivity of chemical and biological systems since it allows non-instrusive measurements of substances in low concentration under physiological conditions [6], [7], [8], [9], [10], [11]. It can reveal accessibility of quenchers to serum albumins fluorophores, which helps to understand serum albumin׳s binding mechanisms to compounds and provides clues to the nature of the binding phenomenon. Fourier transform infrared (FT-IR), a powerful technique for the study of hydrogen bonding, has recently become very popular for structural characterization of proteins. The most important advantage of FT-IR spectroscopy for biological studies is that spectra of almost any biological system can be obtained in a wide variety of environments.
Sulfamethoxazole (SMZ) [N-(5-methyl-3-iso-xazolyl) sulfanilamide] (Fig. 1) is a well known antibacterial drug. It is mainly used in treating urinary tract and lower respiratory tract infections and gets readily absorbed in the body [12]. The present study showed that there was a major change in the fluorescence and in structural properties of HSA upon binding to SMZ, which has been utilized for the characterization of different interaction parameters. In view of its biological importance, we found it worthwhile to study the binding of SMZ to HSA using various techniques viz., fluorescence, FT-IR, UV–vis and cyclic voltammetry, as it is not reported so far.
Fig. 1.
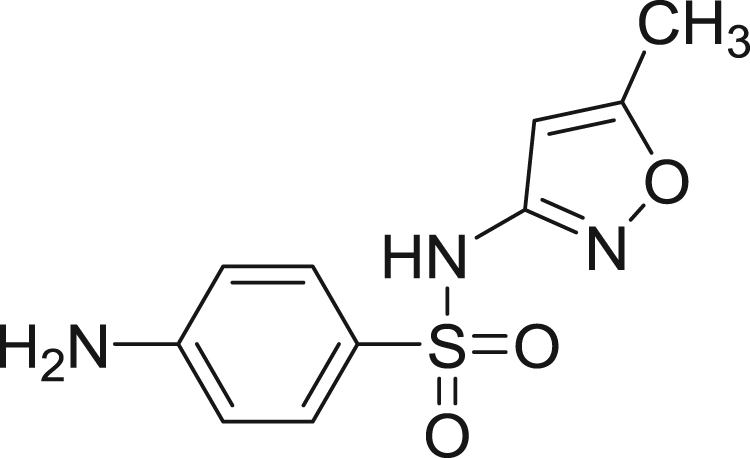
Structure of sulfamethoxazole.
2. Experimental
2.1. Equipments and spectral measurements
Fluorescence measurements were performed on a spectrofluorimeter Model F-2000 (Hitachi, Tokyo, Japan) equipped with a 150 W Xenon lamp and a slit width of 5 nm. The infrared spectra were acquired on a Thermo Nicolet- 5700 FT-IR spectrometer (Waltham, MA, USA) equipped with a germanium attenuated total reflection (ATR) accessory, a DTGS KBr detector and a KBr beam splitter. All spectra were recorded via the ATR method with a resolution of 4 cm−1 and 60 scans. The absorption spectra were recorded on a double beam UV–vis spectrophotometer (Varian CARY 50-BIO, Mulgrave Victoria, Australia) equipped with a 150 W Xenon lamp and a slit width of 5 nm.
2.2. Reagents and chemicals
Essentially fatty acid free HSA was obtained from Sigma Chemical Company (St. Louis, USA). SMZ was obtained from Sigma Chemical Company (St. Louis, USA). The solutions of SMZ and HSA were prepared in 0.1 M phosphate buffer of pH 7.4 containing 0.15 M NaCl. HSA solution was prepared based on its molecular weight of 66,500. The protein concentration was determined spectrophotometrically using the extention coefficient of 36,500 L/mol cm [13]. Salts of different metals, phosphate, acetate and urea were purchased from Merck (New Delhi, India). Site probes viz., warfarin, ibuprofen and digitoxin purchased from Sigma Chemical Company (St. Louis, USA) were initially dissolved in minimum amount of methanol and then diluted with distilled water. All other materials were of analytical reagent grade and double distilled water was used throughout.
2.3. Methods
2.3.1. SMZ–HSA interactions by fluorescence measurements
Appropriate volumes of the HSA solution (40 µL from 250 µM stock) and SMZ solution (40 µL from 250 µM stock) were transferred into a 5 mL conical flask. The mixtures were diluted with phosphate buffer solution (pH 7.4) to make the total volume (2 mL) and then were shaken. Fluorescence measurements were carried out by keeping the concentration of HSA fixed at 5 µM and that of SMZ varied from 5 to 60 µM. Fluorescence spectra were recorded at 288, 298 and 308 K in the range of 300–500 nm upon excitation at 280 nm in each case.
2.3.2. FT-IR measurements
The FT-IR spectra of HSA (5 µM) in the presence and absence of SMZ were recorded in the range of 1300–1800 cm−1. The molar ratio of HSA to drug was maintained at 1:4. The corresponding absorbance contributions of buffer and free SMZ solutions were recorded and digitally subtracted with the same instrumental parameters.
2.3.3. Energy transfer between protein and SMZ
The absorption spectrum of SMZ (5 µM) was recorded in the range of 300–500 nm. The emission spectrum of HSA (5 µM) was also recorded in the range of 300–500 nm. Then, the efficiency of transfer can be used to evaluate the distance between the ligand and the tryptophan residues in the protein.
2.3.4. Displacement experiments
The displacement experiments were performed using the site probes (warfarin, ibuprofen and digitoxin) keeping the concentration of HSA and probe constant (each of 5 µM). SMZ was then gradually added to the HSA-site probe mixtures. The fluorescence quenching titration was used as before to determine the binding constants of SMZ–HSA systems in the presence of the above site probes, warfarin, ibuprofen and digitoxin for sites I, II and III [14].
2.3.5. Absorption measurements
The absorption spectra of HSA in the presence and absence of SMZ were noted in the range of 200–320 nm. HSA concentration was fixed at 5 µM while that of SMZ varied from 5 to 25 µM.
2.3.6. Voltametric interaction of SMZ with HSA
The concentration of SMZ was kept at 1×10−4 M and the HSA solution was added at different concentrations in the range of (1.25–12.5)×10−6 M. A given SMZ–HSA system was stirred for 10 s and then the cyclic voltammetry experiment was performed at 7.4 pH (scan rate=0.1 V/s).
2.3.7. Preparation of different conformers of HSA
HSA exists in different conformational states as N, F and I forms [15]. The N, F and I conformations were prepared by mixing 40 μL of HSA monomer stock solution (250 μM) with 1960 μL of pH 7 (60 mM phosphate), pH 3.5 (10 mM acetate) buffers, and 10 M urea was used as per the required concentration. The existence of different isomers in the experimental preparations was confirmed with various fluorescence properties of different forms. Different isomers in the experimental preparations were confirmed with various spectroscopic properties of different forms viz., max of N form=344 (ext. 295 nm), 339.3 (ext. 280 nm), max of F form=344 (ext. 295 nm), 334.6 (ext. 280 nm), max of I form=344 (ext. 295 nm), 341 (ext. 280 nm) and fluorescence intensities of N, F and I forms were 101, 115 and 71, respectively.
2.3.8. Effects of common ions
The effect of common ions viz. Mg2+, Ni2+, Co2+, V5+, NO3−, SO42− and CH3COO− on the binding constant of SMZ–HSA system was investigated at 298 K by recording the fluorescence intensity in the range of 300–500 nm upon excitation at 280 nm. The concentration of both HSA and common ion was fixed at 5 µM. The solutions of anions were prepared from sodium salts while chlorides of cations were used for preparing the solutions of cations except for V5+ solution where ammonium metavanadate was used. Under the experimental conditions, no cation gave precipitate in phosphate buffer.
3. Results and discussion
3.1. Analysis of fluorescence quenching of HSA by SMZ
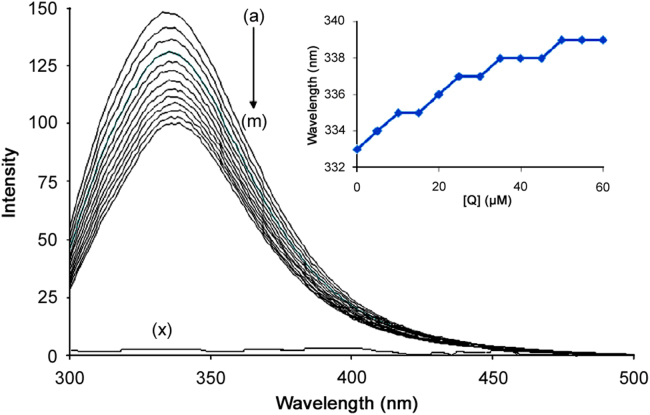
Fluorescence quenching is the decrease of the quantum yield of fluorescence from a fluorophore induced by a variety of molecular interactions with a quencher molecule. Generally, the fluorescence of HSA comes from tryptophan, tyrosine and phenylalanine residues. The change of intrinsic fluorescence intensity of HSA was due to tryptophan residue when small molecules bound to HSA. This viewpoint was well supported by the experimental observations of Sulkowska [16]. The fluorescence intensity of HSA gradually decreased with increasing the concentration of SMZ (Fig. 2), indicating that there was an interaction between SMZ and HSA. It was clear that HSA had a strong fluorescence emission band at 333 nm by fixing the excitation wavelength at 280 nm, while the drug SMZ had no intrinsic fluorescence (Fig. 2). A red shift in maximum emission wavelength of HSA (from 333 to 339 nm) was observed upon the addition of SMZ probably due to the loss of the compact structure of hydrophobic sub-domain where tryptophan was placed (inset Fig. 2) [11]. Also SMZ–HSA complex could quench the fluorescence of HSA and change the microenvironment of tryptophan residue. Hence, it was proposed that the binding took place near Trp-214 and led to a conformational change with a local perturbation of the IIA binding site in HSA. Under the experimental conditions, SMZ did not show any fluorescence intensity. SMZ caused a concentration-dependent quenching of the intrinsic fluorescence of HSA.
Fig. 2.
Fluorescence spectra of HSA in the presence of SMZ. HSA concentration was 5 µM (a), SMZ concentration was at 5 µM(b), 10 µM (c), 15 µM (d), 20 µM (e), 25 µM (f), 30 µM (g), 35 µM (h), 40 µM (i), 45 µM (j), 50 µM (k), 55 µM (l) and 60 µM (m).
3.2. Binding parameters and mechanism
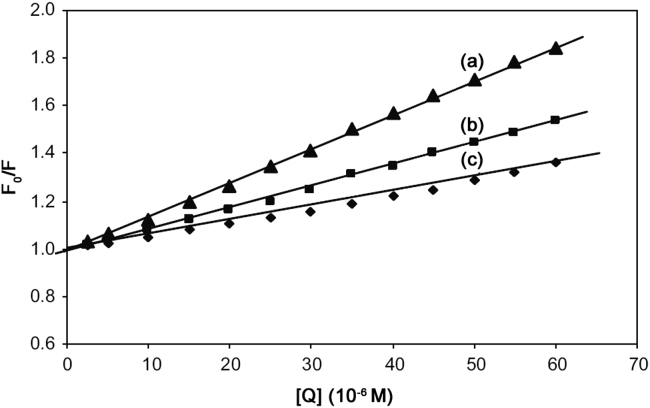
Quenching can be induced by dynamic and static processes. Dynamic and static quenching can be distinguished based on their differences in temperature dependence. Higher temperature results in faster diffusion and larger amounts of collisional quenching. It will typically lead to the dissociation of weakly bound complexes and smaller amounts of static quenching. Therefore, the quenching constant increases for dynamic quenching while it decreases for static quenching with an increase in temperature. The possible quenching mechanism can be deduced from the Stern–Volmer plot. The Stern–Volmer plot for a representative drug–HSA system is shown in Fig. 3. The Stern–Volmer plots were observed to be linear for SMZ–HSA with the slopes decreasing with an increase in temperatures. The values of KSV at different temperatures were evaluated as 1.440×104, 0.910×104 and 0.580×104 L/mol, respectively. The values of KSV at different temperatures indicated the presence of static quenching mechanism in the interaction between SMZ and HSA. In order to invoke this possibility, the mechanism was assumed to involve dynamic quenching. The equation for dynamic quenching is presented by Eq. (1) [17]
| (1) |
where F and F0 are the fluorescence intensities of HSA with and without quencher, respectively. Kq, KSV, τo and [Q] are the quenching rate constant, the dynamic quenching constant, the average lifetime of the biomolecule without quencher and the concentration of quencher, respectively.
| (2) |
Fig. 3.
The Stern–Volmer curves for the binding of SMZ to HSA at 288 K (a), 298 K (b), 308 K (c). λex=280 nm, λem=344 nm and CHSA=5 µM.
Since the average fluorescence lifetime of Trp in serum albumin without quencher is 10−8 s [18], the quenching constant (Kq, L/mol s) can be obtained by the slope using above equation, 0.91×1012 L/mol s (r=0.983) for HSA at 298 K. The values of Kq are given in Table 1. The maximum scatter collision quenching constant, Kq of various quenchers with the biopolymer [19], is reported to be 2×1010 L/mol s. The order of magnitude of Kq was calculated to be 1012 for SMZ–HSA system in the present study. So, the rate constant of the protein quenching procedure initiated by SMZ is greater than the value of Kq for the scatter mechanism. This implied that the quenching was not initiated by dynamic collision but originated from the formation of a complex.
Table 1.
Thermodynamic parameters of SMZ–HSA system.
| Temp. (K) | Binding constant (K)(×10−4 L/mol) | Kq (×10−12 L/mol s) | Number of binding sites (n) | ΔH0 (kJ/mol) | ΔS0 (J/mol K) | ΔG0(kJ/mol) |
|---|---|---|---|---|---|---|
| 288 | 2.250±0.001 | 1.440±0.006 | 1.04 | −36.0±3.0 | −41.3±5.0 | −23.7±3.0 |
| 298 | 1.560±0.003 | 0.910±0.005 | 1.04 | |||
| 308 | 0.850±0.002 | 0.580±0.008 | 1.02 |
3.3. Changes of the protein׳s secondary structure induced by drug binding
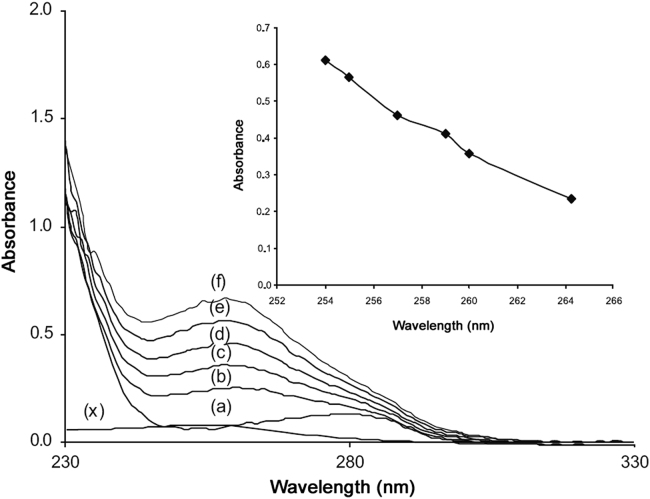
UV absorption spectrum is an important method and application to explore the structural change and to know the complex formation [20]. In order to obtain more information on SMZ–HSA interactions, UV absorption spectra of drug, HSA and drug–HSA systems were recorded. It was observed from Fig. 4 that the absorption of HSA increased regularly upon the successive addition of SMZ. The chromophore of SMZ–HSA gives very specific pattern of the UV–vis spectrum with slightly duel absorbance spectra at a higher concentration of SMZ in the system and the SMZ–HSA spectrum shifts towards shorter wave length. The evidence clearly indicated the interaction between SMZ and HSA. This was noticed due to formation of a complex between the drug and HSA [20]. This also indicated that the peptide strands of protein molecules extended further more upon the addition of SMZ to HAS [20], [21]. So, the binding of SMZ to protein molecule might lead to change in conformation of the protein [22].
Fig. 4.
Absorbance spectra of HSA, SMZ and HSA–SMZ system. HSA concentration was at 5 µM(a). SMZ concentration for SMZ–HSA system was at 5 (b), 10 (c), 15 (d), 20 (e), 25 (f), 30 (g), 35 (h) and 40 µM (i). A concentration of 5 µM SMZ (x) was used for SMZ only.
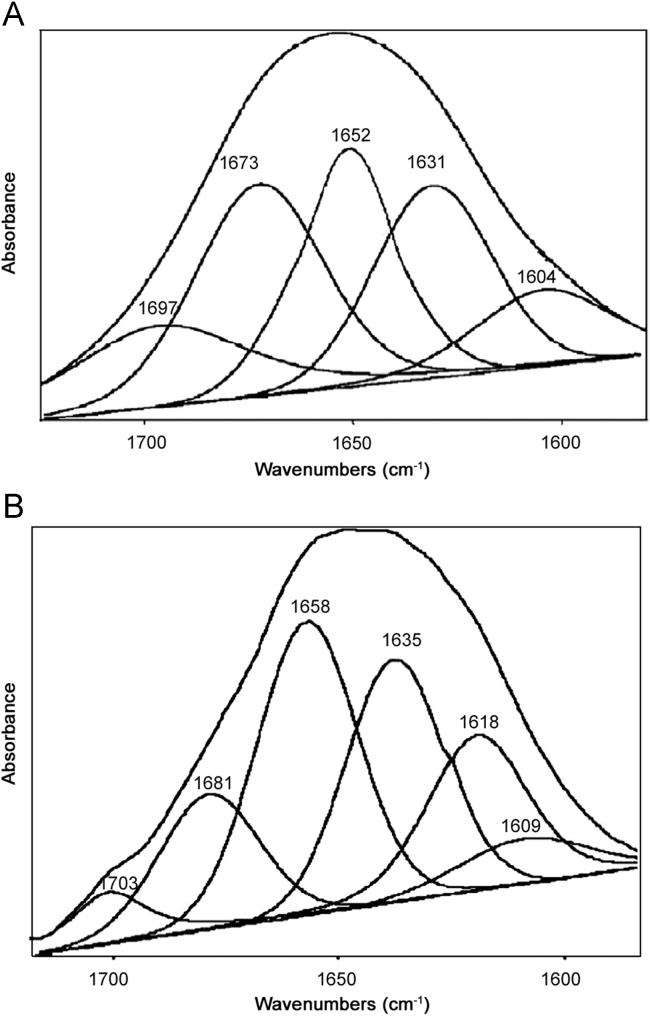
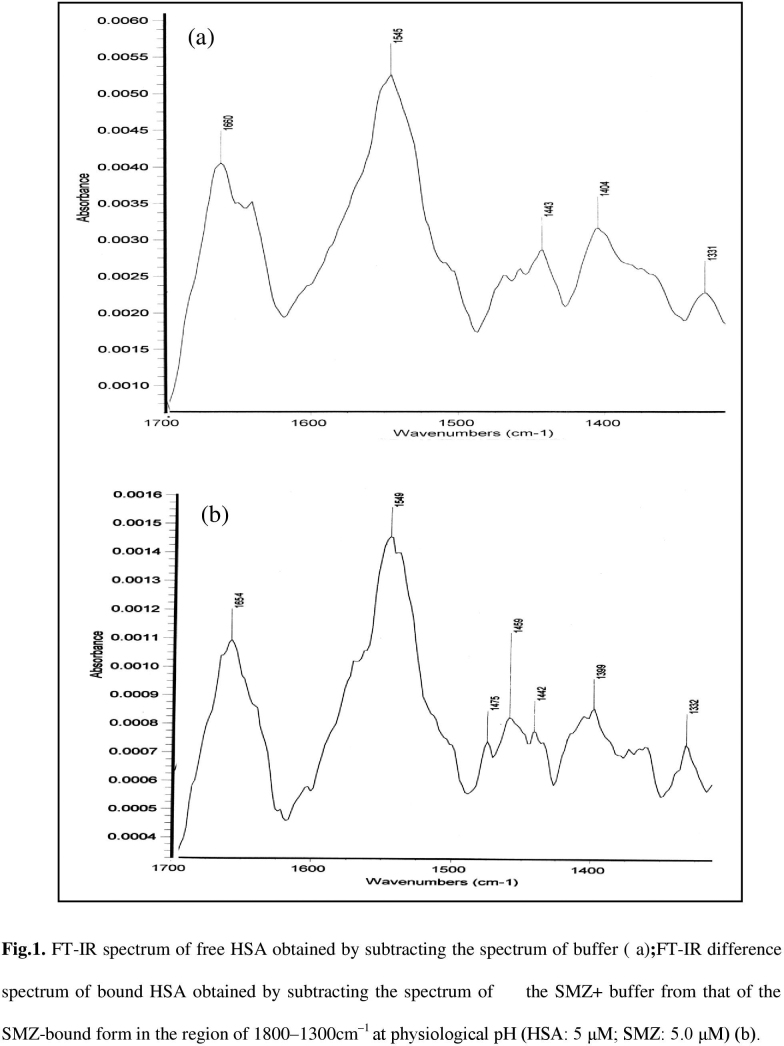
Additional evidence regarding SMZ–HSA interactions was obtained from FT-IR spectroscopic results. Infrared spectra of a protein exhibit a number of amide bands due to the different vibrations of the peptide moiety. Of all the amide modes of the peptide group, amide I band is more sensitive to the changes in protein secondary structure than amide II. Hence, the amide I band is more useful for studies of secondary structure [23], [24], [25], [26], [27]. The amides I and II peaks occur in the region of 1700–1600 cm−1 and 1600–1500 cm−1, respectively. Supplementary Fig. S1(a) and (b) shows the FT-IR spectra of the SMZ free and SMZ bound form of HSA with its difference absorption spectrum. Supplementary Fig. S1(a), the FT-IR spectra of free HSA (subtracting the absorption of the buffer solution from the spectrum of the protein solution) and Supplementary Fig. S1(b), the FT-IR difference spectra of HSA (subtracting the absorption of the SMZ–HSA free form that of SMZ–HSA bound form) in phosphate buffer, CHSA=5.0 and CSMZ=20 µM. The peak positions of amides I and II were found to be shifted upon the addition of SMZ to HSA. The amide I band was shifted from 1660 to 1654 cm−1 for SMZ while the amide II peak was shifted from 1545 to 1549 cm−1 for SMZ. These results indicated that both the SMZ interacted with the CO and CN groups in the protein polypeptides. The SMZ–protein complexes caused the rearrangement of the polypeptide carbonyl hydrogen-bonding network and finally the reduction of the protein helical structure. Supplementary Fig. S1 and Fig. 5 show a quantitative analysis of the protein secondary structure of HSA before and after the interaction with SMZ in phosphate buffer. According to the FT-IR spectra of amide I and the curve-fitted results (Fig. 5), the secondary structure compositions of HSA in the absence and in the presence of SMZ were estimated. The data obtained from Fig. 5 suggested that upon SMZ–HSA complexes, the α-helix structures were increased from 49.4% to 60.4%, β-sheet decreased slightly from 36.5% to 31.5%, and β-turn structures decreased from 14.1% to 8.1%. HSA and BSA are homolog proteins showing 78.1% of sequence identity. BSA contains two tryptophan residues (Trp-212 and Trp-134), while HSA has one tryptophan (Trp-214). The BSA three-dimensional structures are very similar to HSA. Based on our spectroscopic data, SMZ binding to BSA and HSA occurs via hydrophobic interactions. However, stronger SMZ–protein interaction was observed for HSA–SMZ with a binding constant (K) of (1.560±0.003)×104 L/mol at 298 K. The binding mode of SMZ–HSA was increased the percentage of protein α-helix from 49.4% (free HSA) to 60.4% in SMZ–HSA adduct. The increase in α-helix and decrease in β-sheet structure are evidence for some degree of stabilization of protein structure at a high SMZ concentration.
Fig. 5.
Curve-fitted amide I (1700–1600 cm−1) regions of free HSA (A) and SMZ–HSA (B) in buffer solution with CSMZ/CHSA=1/1.
3.4. Apparent binding constant and binding sites
When small molecules bind independently to a set of equivalent sites on a macromolecule, the equilibrium between free and bound molecules could be represented by the equation [20] as
| (3) |
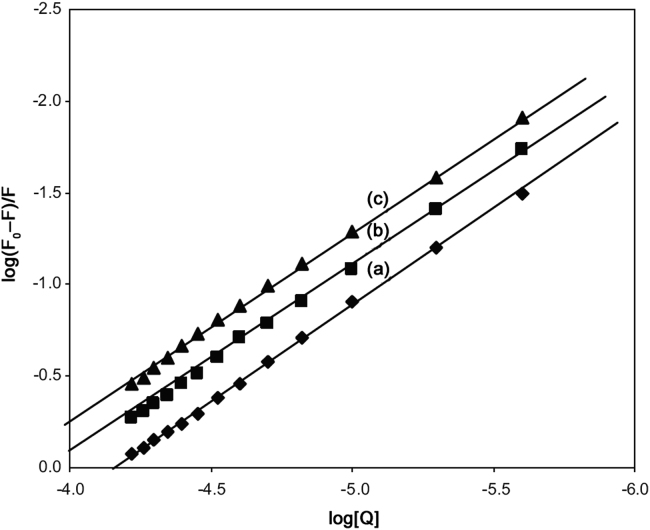
where K and n are the binding constant and number of binding sites, respectively. The values of K and n for SMZ–HSA system were calculated from the intercept and slope of the plot of log(F0−F)/F versus log[Q] and the same plot for a representative system, SMZ–HSA, is given in Fig. 6. The value of K is significant to understand the distribution of the drug in plasma since the weak binding can lead to a short lifetime or poor distribution, while strong binding can decrease the concentrations of free drug in plasma. The values of K and n are summarized in Table 1. It was noticed that the binding constant values decreased with an increase in temperature due to reduction of the stability of SMZ–HSA complexes. The values of n for SMZ–HSA were noticed to be almost unity, indicating that there was one independent class of binding sites on HSA for SMZ. Hence, SMZ is most likely to be bound to the hydrophobic pocket located in sub-domain IIA. That is to say, Trp-214 is near or within the binding site [28].
Fig. 6.
The plot of log(F0−F)/F vs. log[Q] for quenching of SMZ to HSA at 288 K (a), 298 K (b) and 308 K(c) (λex=280 nm, λem=344 nm, CSMZ=5–60 µM, CHSA=5 µM).
Sudlow et al. [14] suggested two main distinct binding sites (sites I and II) in HSA. Sites I and II of HSA show affinity to warfarin and ibuprofen, respectively. It is reported that digitoxin binding is independent of sites I and II and digitoxin binds to site III [29], [30]. In order to establish the binding site in HSA for SMZ, displacement experiments were performed using site probes, warfarin, ibuprofen and digitoxin. For this, emission spectra of ternary mixtures of SMZ, HSA and site probe were recorded. The binding constant values of SMZ–HSA after the addition of the site probes were evaluated to be (0.680±0.001)×10−4 L/mol, (1.540±0.005)×10−4 L/mol and (1.550±0.003)×10−4 L/mol respectively. The values of binding constant indicated that SMZ was not displaced by ibuprofen or by digitoxin. However, warfarin showed a significant displacement of SMZ, suggesting that SMZ binding site on HSA is site I. Hence, site I located in subdomain IIA near Trp-214 is the main binding site for SMZ in HSA.
3.5. Thermodynamic parameters
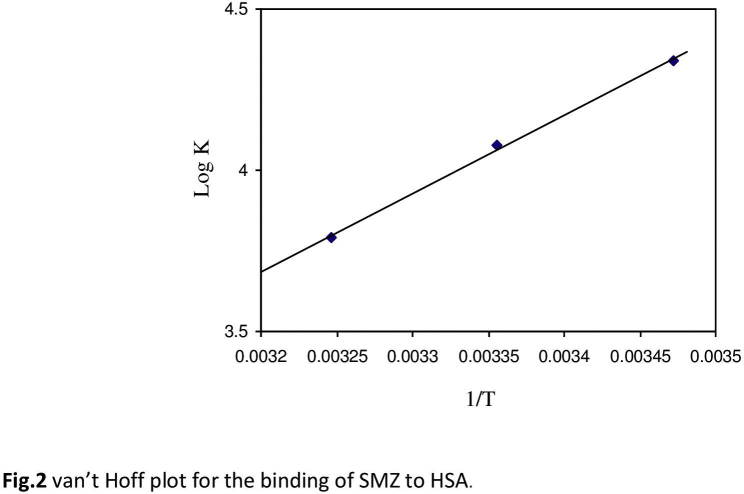
The thermodynamic parameters, enthalpy change (ΔH0) and entropy change (ΔS0), of SMZ–HSA interaction are important for confirming binding mode. For this purpose, the temperature dependence of binding constant was studied. Binding studies were carried out at 288, 298 and 308 K at which HSA did not undergo any structural degradation. The molecular forces contributing to protein interactions with small molecular substrates may include van der Waals interactions, hydrogen bonds, electrostatic and hydrophobic interactions and so on [20]. The thermodynamic parameters were evaluated using the following equations:
| (4) |
| (5) |
The plot of log K versus 1/T enabled the determination of the values of ΔH0 and ΔS0 (Supplementary Fig. S2). Ross et al. [31] have characterized the sign and magnitude of the thermodynamic parameters associated with various individual kinds of interactions. For typical hydrophobic interactions, both ΔH0 and ΔS0 are positive, while these are negative for van der Waals forces and hydrogen-bond formation in low dielectric media [31], [32]. Moreover, the specific electrostatic interaction between ionic species in an aqueous solution is characterized by positive ΔS0 value and negative ΔH0 value (small). From Table 1, it can be seen that both ΔH0 and ΔG0 have negative values, −36.0 kJ/mol and −23.7 kJ/mol, respectively. Negative values for ΔG0 and ΔH0 indicate that both binding processes occurred spontaneously and the formation of both SMZ–HSA complexes was exothermic. The negative values of ΔH0 (−36.0 kJ/mol) and ΔS0 (−41.3 J/mol K) for SMZ–HSA reveal that intermolecular forces such as hydrophobic interaction, hydrogen bonds, and van der Waals interaction are present in the system. In addition, the major contribution to ΔG0 arising from the ΔH0 term rather than from ΔS0 implies that the binding processes are enthalpy driven. Accordingly, it was not possible to account for the thermodynamic parameters of HSA–SMZ compound on the basis of a single interaction molecular force model. It was more likely that hydrophobic and hydrogen bond interactions were involved in the binding process [32]. But SMZ might be considered to be largely unionized under the experimental conditions, as can be expected from its structure. Hence, electrostatic interaction can be precluded from the binding process. Therefore, both hydrogen bonds and hydrophobic interactions are assumed to play a major role in the binding of SMZ to HSA.
3.6. Energy transfer from HSA to SMZ
Fluorescence resonance energy transfer (FRET) is a distance-dependent interaction between the different electronic excited states of molecules in which excitation energy is transferred from one molecule (donor) to another molecule (acceptor) without emission of a photon from the former molecular system. The efficiency of FRET depends mainly on the following factors: (i) the extent of overlap between the donor emission and the acceptor absorption, (ii) the orientation of the transition dipole of donor and acceptor and (iii) the distance between the donor and acceptor. The energy transfer effect is related not only to the distance between the acceptor and the donor, but also on the critical energy transfer distance R0. The spectral overlap between the fluorescence emission spectrum of free HSA and absorption spectrum of a drug, SMZ, is shown in Fig. 7. The efficiency of energy transfer, E, was determined according to Förster׳s energy transfer theory [33]. The value of E was calculated using the equation
| (6) |
where F and F0 are the fluorescence intensities of HSA in the presence and absence of SMZ, r is the distance between the acceptor and the donor, and R0 is the critical distance when the transfer efficiency is 50%. The value of R0 was evaluated using the equation
| (7) |
where k2 is the spatial orientation factor between the emission dipole of the donor and the absorption dipole of the acceptor. The dipole orientation factor, k2, is the least certain parameter in calculation of the critical transfer distance, R0. Although theoretically k2 can range from 0 to 4, the extreme values require very rigid orientations. If both the donor and acceptor are tumbling rapidly and free to assume any orientation, then k2 equals 2/3 [33]. If only the donor is free to rotate, then k2 can vary from 1/3 to 4/3 [34], [35], n is the refractive index of the medium, Φ is the fluorescence quantum yield of the donor and J is the overlap integral of the fluorescence emission spectrum of the donor and the absorption spectrum of the acceptor, given by
| (8) |
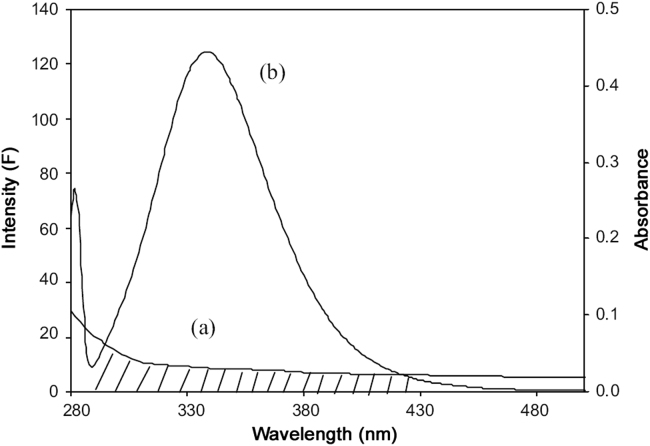
where F(λ) is the fluorescence intensity of the fluorescent donor in wavelength λ, and is dimensionless, ε(λ) is the molar absorption coefficient of the acceptor in wavelength λ and its unit is L/cm×mol. J can be evaluated by integrating the spectra in Fig. 7. It has been reported for BSA and HSA that k2=2/3, Φ=0.15 and n=1.36 [29]. From Eqs. (8), (9), (10), we could calculate that J=0.17×10−15 cm3·L/mol, E=0.1211, R0=2.79 nm, and r=3.885 nm. The average distance of 2–8 nm between a donor and acceptor indicated that the energy transfer from HSA to SMZ occurred with high probability [36]. The larger value of r compared to that of R0 in the present study also revealed the presence of static type quenching mechanism [16]. These values are rather approximate. However, the distances obtained this way agree well with literature values of substrate binding to HSA at site IIA [37], [38].
Fig. 7.
The overlap of the fluorescence spectrum of BSA and the absorbance spectrum of SMZ (λex=280 nm, λem=344 nm, CHSA/CSMZ=1:1). The absorption spectrum of SMZ (5 µM) (a) and the fluorescence spectrum of HSA (5 µM) (b).
3.7. Voltametric interaction of SMZ with HSA
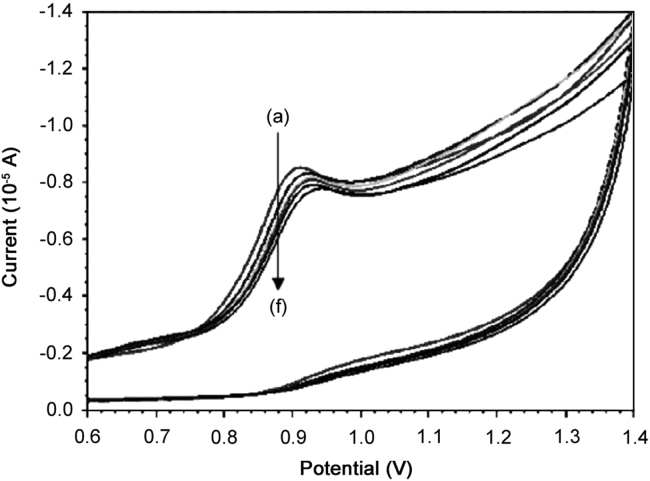
Cyclic voltammograms of SMZ in the presence of HSA showed that SMZ had an oxidation peak at 908 mV when HSA was added to the SMZ solution, the oxidation peak shifted towards high potential and decrease of the oxidation current was observed. Cyclic voltammograms of SMZ in the presence of HSA showed irreversible process (Fig. 8). At the HSA concentration of 1.25 µM, the oxidation peak shifted to 920 mV. Furthermore, the concentration of free SMZ at the electrode surface decreased during the experiment and the peak current was also reduced consequently, the observed potential can be attributed to the changes of the molecular environment of SMZ as a result of its interaction with HSA. Assuming that the interaction of SMZ and HSA produces only a single HSA/mSMZ molecular complex, the binding constant β can be determined as follows [39]:
Fig. 8.
Cyclic voltammograms of SMZ mixed with different concentrations of HSA. CSMZ=1×10−4 mol/L (a), CHSA=1.25×10−6 (b), 5×10−6 (c), 7.5×10−6 (d), 10×10−6 mol/L (e) and 12.5×10−6 mol/L (f).
Given that
| (9) |
by using the relationships (10), (11), (12), (13), Eq. (14) can be derived. These relate the changes in the measured current, ΔI, to the [SMZ]. From these equations, m and β may be extracted.
| (10) |
| (11) |
| (12) |
| (13) |
And the final equation is
| (14) |
where ΔI is the peak current change of the responses measured at the same concentration of SMZ in the absence or presence of HSA, ΔImax is the maximum peak current change. The concentration of HSA was kept constant at 1.0 µM and the concentration of added SMZ was in the range of 100–200 µM. Thus, the resulting linear plot (Fig. 9) gave an m value of 0.967±0.04 and β of (0.88±0.2)×104 L/mol.
Fig. 9.
The plot of log ΔI/(ΔImax−ΔI) vs. log[SMZ] of HSA–SMZ system at 288 K.
3.8. HSA unfolding pathways
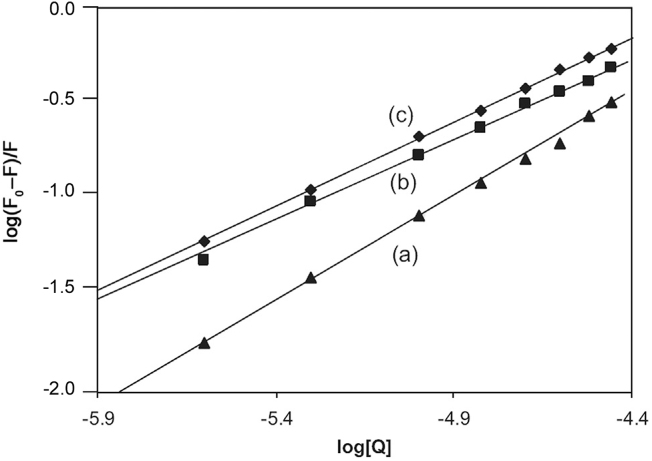
The acid- and urea-induced unfolding pathways were employed to locate the binding site for SMZ on HSA. HSA undergoes N-F and N-I transitions induced by the acidic pH between 7.0 and 3.5 and the urea concentration in the range of 4.8–5.2 M [40]. The F isomer which predominates at pH 3.5, is characterized by unfolding and separation of domain III and the I isomer is characterized by unfolding of domain III and partial but significant loss of native conformation of domain I. Domain II is known to persist in both N→F or N→I transitions. Fig. 10 shows conformation-dependent binding of SMZ to HSA. Hence, a small decrease in the number of binding sites of these conformational states compared to native (Table 2) is suggestive of the location of the binding sites for SMZ on domain II. The small decrease in the n and K values (Table 2) may be due to a loss of inter-domain interactions which stabilizes the protein domain structure. This is justified to predict that a domain in the presence of its other supportive unfolded domains cannot fully reflect its functional and its structural properties.
Fig. 10.
The plot of log(F0−F)/F vs. log[Q] for N (a), I (b) and F (c) conformation of HSA for binding constant and binding sites (λex=280 nm, λem=344 nm, CSMZ=2.5–40 µM and CHSA=5 µM).
Table 2.
Comparative assessment of quenching constants and SMZ binding parameters to HSA conformers.
| HSA | Binding constant (K) (×10−4 L/mol) | Number of binding sites (n) | ΔG0 (kJ/mol) |
|---|---|---|---|
| Native (N) | 1.732±0.005 | 1.06 | −24.2±5.0 |
| Fast moving (F) | 0.541±0.004 | 0.88 | −21.3±3.0 |
| Urea indused (I) | 0.330±0.001 | 0.86 | −20.1±1.0 |
3.9. Molecular modeling of SMZ binding to HSA
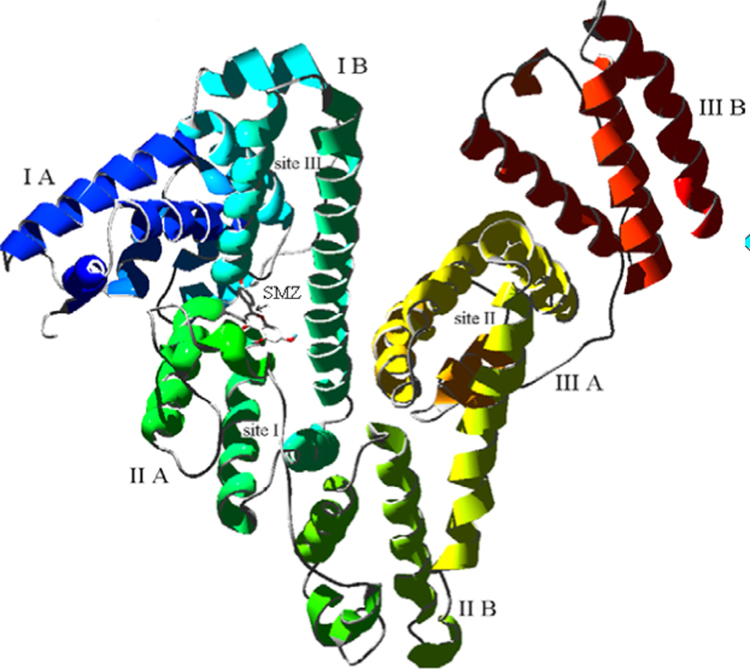
To further obtain the information of the right site and interaction forces of SMZ binding to HSA, Autodock 3.05 was applied to deduce the binding mode of SMZ in HSA. Crystal structure of HSA shows that HSA is a heart-shaped helical monomer of 66 kD composed of three similar homologous domains named I (residues 1–195), II (196–383) and III (384–585) and each domain includes two subdomains called A and B to form a cylinder and each subdomain involves 6 and 4 α-helixes, respectively, the only Trp residue (Trp214) is located in subdomain IIA [41]. Almost all hydrophobic amide acids are embedded in the cylinders and form hydrophobic cavities, which play an important role on absorption, metabolism, and transportation of biomolecules. The three domains have similar 3D structures, and their assembly is highly asymmetric. Domains I and II are perpendicular to each other, forming a T-shaped assembly in which the tail of subdomain IIA is attached to the interface region between subdomain IA and IB by hydrophobic and hydrogen bonds [42]. Despite the internal structural symmetry of the three domains, they have different capacities for binding drugs. In the structure of HSA, there are two primary sites called site I (classical example is warfarin) and site II (classical example is diazepam) for binding a wide variety of drugs. The inside wall of the pocket of subdomain IIA corresponding to the so-called site I is formed by hydrophobic side chains, whereas the entrance of the pocket is surrounded by positively charged residues such as Arg257, Arg222, Lys199, His242, Arg218, and Lys195. Site II corresponds to the pocket with hydrophobic side chains in subdomain IIIA, which is almost the same size as site I. The third site in D-shaped cavity of subdomain IB named site III is typical for binding hemin [43]. Unlike subdomain IIA and IIIA, subdomain IA has no pocket near its hydrophobic core. In the subdomain IA, the fourth helix is flabby, so it is cannot keep parallel to the third helix and bury the putative pocket, but thanks to the interaction of subdomain IIIA with them, the region is sealed to form site III. Although examples of drugs binding elsewhere on the protein have been reported, most investigations have focused on the three primary binding sites. HSA is a flexible protein that easily changes its molecular shape due to the relative motions of domain structures.
Through molecular modeling by Autodock 3.05, the optimum binding mode and site are displayed in Fig. 11. It can be seen that the entrance of site I, a small cavity with positive charged residues formed by interaction between subdomain IIA and IB, is the possible binding site. But SMZ might be considered to be unionized under the experimental conditions, as can be expected from its structure. Thus, electrostatic interaction cannot play a major role during the binding and SMZ bound to HSA is mainly based on the hydrogen bonding and hydrophobic interaction. In the region of SMZ (Fig. 11), there are 16 amino acids residues. Among them only two were hydrophobic residues (Pro147, Phe149), five residues had positive charge (Lys106, Arg197, Lys199, His242, Arg257) and the rest were polar residues (Gln29, Tyr148, Tyr150, Gln196, Cys200, Cys245, Cys246, Gly248, Cys253). On account of the unionized property of SMZ under the physiological condition, it can be concluded that hydrogen bonding is one of the major forces of interaction between SMZ and HSA.
Fig. 11.
Modeling of X-ray crystallographic structure of HSA complexed with SMZ. The SMZ was shown in the entrance of site I. The domains and subdomains were displayed with different colors, the every subdomain and classical binding site were marked in the corresponding location.
Furthermore, owing to the phenyl moiety of Phe149 exposed close to the A ring in SMZ with additional aliphatic contact from Arg197, His242 and Arg257, in addition, phenyl of Tyr148 approaches the ring B of SMZ at nearly vertical orientation and Tyr150 is close tightly to the ring B of SMZ at an angle of about 60° from another side of SMZ, it is concluded that hydrophobic force is another major force on the binding between SMZ and HSA. Attributed to SMZ binding to nearby Tyr148 and Tyr150, the phenyl of Trp214 faced slantingly to the hydrophobic part on the side of A-ring in SMZ, which suggests that when SMZ moves close enough to Trp214 during the dynamic equilibrium, the fluorescence energy of Trp can transfer to SMZ to bring the quenching, so the fluorescence quenching of HSA induced by binding of SMZ resulted from not only Tyr148 and Tyr150 but also Trp214, which agreed with the result of the synchronous fluorescence. According to references a wide range of biomolecules can be accommodated in site I, and the interaction between biomolecules and HSA appears to be dominated by hydrophobic contacts, but there are a number of specific electrostatic interactions [44]. The results suggested unanimously that hydrophobic and hydrogen bonding plays an important role in the interaction between SMZ and HSA.
3.10. Effects of other ions on the interactions of SMZ with HSA
In plasma, there are some metal ions which can affect the reactions of the drugs and serum albumins. Trace metal ions, especially the bivalent type, are essential in the human body and play an important structural role in many proteins. It was reported [45] that Mg2+, Ni2+, Co2+ and V5+ and other metal ions could form complexes with serum albumins. Hence, the effects of some metal ions: Mg2+, Ni2+, Co2+, V5+, NO3−, SO42− and CH3COO− on the binding of SMZ to HSA were investigated in the present study. The binding constants of SMZ–HSA in the presence of above ions were evaluated and these results are shown in Table 3. The binding constants of SMZ–HSA systems decreased in various degrees in the presence of Mg2+, Ni2+, Co2+, V5+, NO3−, SO42− and CH3COO−. This was likely to be caused by a conformational change in the vicinity of the binding site. The decrease in the binding constant in the presence of the above metal ions would shorten the storage time of the drug in blood plasma and hence more amount of free drug would be available in plasma [20]. This led to the need for more doses of SMZ to achieve the desired therapeutic effect in the presence of the above ions.
Table 3.
Effect of anions and cations on binding constant of HSA–SMZ system.
| Systems | Binding constant (L/mol) |
|---|---|
| HSA+SMZ | (1.560±0.003)×104 |
| HSA+SMZ+Mg2+ | (2.230±0.008)×103 |
| HSA+SMZ+Co2+ | (8.371±0.006)×102 |
| HSA+SMZ+Ni2+ | (1.890±0.003)×103 |
| HSA+SMZ+V5+ | (7.691±0.003)×102 |
| HSA+SMZ+K+ | (1.071±0.004)×103 |
| HSA+SMZ+NO3− | (1.030±0.001)×102 |
| HSA+SMZ+SO42− | (1.112±0.006)×102 |
| HSA+SMZ+CH3COO− | (3.463±0.005)×103 |
4. Conclusions
In the present work, the binding of SMZ to HSA was carried out employing spectroscopic techniques under physiological conditions. Various binding parameters have been evaluated. The decreasing values of K with an increase in temperature indicated the presence of static quenching mechanism. The values of n revealed the presence of a single class of binding site on HSA for SMZ. Thermodynamic parameters indicated that the hydrogen bonding and weak van der Waals interaction were the predominant intermolecular force stabilizing the complex. The distance r between the donor and the acceptor was obtained based on fluorescence resonance energy transfer. The spectroscopic results revealed the changes in the secondary structure of HSA in the presence of SMZ. The decreasing K values of SMZ–HSA complexes in the presence of common ions indicated the availability of higher concentrations of free drug. Since pharmaceutical firms need standardized screens for HSA binding in the first step of new drug design, this kind of study on interaction between HSA and SMZ would be useful in pharmaceutical industry, chemistry, life sciences and clinical medicine.
Acknowledgments
One of the authors (P.N. Naik) thanks UGC, New Delhi, for the award of Research Fellowship in Science for Meritorious Students.
Footnotes
Peer review under responsibility of Xi׳an Jiaotong University.
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.jpha.2015.01.003.
Appendix A. Supplementery materials
Fig. S1.
Fig. S2.
References
- 1.He H.M., Carter D.C. Atomic structure and chemistry of human serum albumin. Nature. 1992;358:209–215. doi: 10.1038/358209a0. [DOI] [PubMed] [Google Scholar]
- 2.Peters T. Academic Press; San Diego, California: 1996. All About Albumin. Biochemistry Genetics and Medical Applications. pp. 9–54. [Google Scholar]
- 3.Carter D.C., Ho J.X. Structure and ligand binding properties of human serum albumin. Adv. Protein Chem. 1994;45:153–203. doi: 10.1016/s0065-3233(08)60640-3. [DOI] [PubMed] [Google Scholar]
- 4.Barbero N., Barni E., Barolo C. A study of the interaction between fluorescein sodium salt and bovine serum albumin by steady-state fluorescence. Dyes Pigments. 2009;80:307–313. [Google Scholar]
- 5.Guo X.J., Zhang L., Sun X.D. Spectroscopic studies on the interaction between sodium ozagrel and bovine serum albumin. J. Mol. Struct. 2009;928:114–120. [Google Scholar]
- 6.Naik P.N., Chimatadar S.A., Nandibewoor S.T. Pharmacokinetic study on the mechanism of interaction of sulfacetamide sodium with bovine serum albumin: a spectroscopic method. Biopharm. Drug Dispos. 2010;31:120–128. doi: 10.1002/bdd.696. [DOI] [PubMed] [Google Scholar]
- 7.Gu A.Q., Zhu X.S., Hu Y.Y. A fluorescence spectroscopic study of the interaction between epristeride and bovin serum albumine and its analytical application. Talanta. 2007;73:668–673. doi: 10.1016/j.talanta.2007.04.041. [DOI] [PubMed] [Google Scholar]
- 8.Charbonneau D., Beauregard M., Tajmir-Riahi H.A. Structural analysis of human serum albumin complexes with cationic lipids. J. Phys. Chem. B. 2009;113:1777–1784. doi: 10.1021/jp8092012. [DOI] [PubMed] [Google Scholar]
- 9.Froehlich E., Jennings C.J., Sedaghat-Herati M.R. Dendrimers bind human serum albumin. J. Phys. Chem. B. 2009;113:6986–6993. doi: 10.1021/jp9011119. [DOI] [PubMed] [Google Scholar]
- 10.Agudelo D., Bourassa P., Bruneau J. Probing the binding sites of antibiotic drugs doxorubicin and N-(trifluoroacetyl) doxorubicin with human and bovine serum albumins. PLoS One. 2012;7(8):e43814. doi: 10.1371/journal.pone.0043814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Beauchemin R., N’soukpoé-Kossi C.N., Thomas T.J. Polyamine analogs bind human serum albumin. Biomacromolecules. 2007;8:3177–3183. doi: 10.1021/bm700697a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bamigboye, Oluwaseyi M., Obaleye Synthesis, characterization and antimicrobial activity of some mixed sulfamethoxazole-cloxacillin metal drug complexes. Int. J. Chem. 2012;2:105–108. [Google Scholar]
- 13.Khan S.N., Islam B., Khan A.U. Probing midazolam interaction with human serum albumin and its effect on structural state of protein. Int. J. Integ. Biol. 2007;1:102–112. [Google Scholar]
- 14.Sudlow G., Birkett D.J., Wade D.N. Further characterization of specific drug binding sites on human serum albumin. Mol. Pharmacol. 1976;12:1052–1061. [PubMed] [Google Scholar]
- 15.Ahmad B., Parveen S., Khan R.H. Effect of albumin conformation on the binding of ciprofloxacin to human serum albumin: a novel approach directly assigning binding site. Biomacromolecules. 2006;4:1350–1356. doi: 10.1021/bm050996b. [DOI] [PubMed] [Google Scholar]
- 16.Sulkowska A. Interaction of drugs with bovine and human serum albumin. J. Mol. Struct. 2002;614:227–232. [Google Scholar]
- 17.Lakowicz J.R. 3rd ed. Plenum Press; New York: 2006. Principles of Fluorescence Spectroscopy. pp. 277–286. [Google Scholar]
- 18.Chen G.Z., Huang X.Z., Xu J.G. 2nd ed. Science Press; Beijing: 1990. The Methods of Flourescence Analysis. pp. 2–39. [Google Scholar]
- 19.Lakowicz J.R., Weber G. Quenching of fluorescence by oxygen. Probe for structural fluctuations in macromolecules. Biochemistry. 1973;12:4161–4170. doi: 10.1021/bi00745a020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ulrich K.H. Molecular aspects of ligand binding to serum albumin. Pharmacol. Rev. 1981;33:17–53. [PubMed] [Google Scholar]
- 21.Tao W.S. The People׳s Education Press; Beijing: 1981. Protein Molecular Basic. p. 256. [Google Scholar]
- 22.Cui F.L., Fan J., Li J.P. Interactions between 1- benzoyl-4-p-chlorophenyl thiosemicarbazide and serum albumin: investigation by fluorescence spectroscopy. Bioorg. Med. Chem. 2004;12:151–157. doi: 10.1016/j.bmc.2003.10.018. [DOI] [PubMed] [Google Scholar]
- 23.Wi S., Pancoka P., Keiderling T.A. Predictions of protein secondary structures using factor analysis on Fourier transform infrared spectra: effect of Fourier self deconvolution of the amide I and amide II bands. Biospectroscopy. 1998;4:93–106. doi: 10.1002/(sici)1520-6343(1998)4:2<93::aid-bspy2>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- 24.Rahmelow R., Hubner W. Secondary structure determination of proteins in aqueous solution by infrared spectroscopy: a comparison of multivariate data analysis methods. Anal. Biochem. 1996;241:5–13. doi: 10.1006/abio.1996.0369. [DOI] [PubMed] [Google Scholar]
- 25.Lin S.Y., Wei Y.S., Li M.J. Effect of ethanol or/and captopril on the secondary structure of human serum albumin before and after protein binding. Eur. J. Pharm. Biopharm. 2004;57:457–464. doi: 10.1016/j.ejpb.2004.02.005. [DOI] [PubMed] [Google Scholar]
- 26.Brauner J.W., Flach C.R., Mendeisohn R. A quantitative reconstruction of the amide I contour in the IR spectra of globular proteins: from structure to spectrum. J. Am. Chem. Soc. 2005;127:100–109. doi: 10.1021/ja0400685. [DOI] [PubMed] [Google Scholar]
- 27.Bummer P.M. An FT-IR study of the structure of human serum albumin adsorbed to polysulfone. Int. J. Pharm. 1996;132:143–151. [Google Scholar]
- 28.Hong G., Liandi L., Jiaqin L. The study on the interaction between human serum albumin and a new reagent with antitumour activity by spectrophotometric methods. J. Photochem. Photobiol. Part A. 2004;167:213–221. [Google Scholar]
- 29.Sjoholm I., Ekman B., Kober A. Binding of drugs to human serum albumin: XI. The specificity of three binding sites as studied with albumin immobilized in microparticles. Mol. Pharmacol. 1979;16:767–777. [PubMed] [Google Scholar]
- 30.Zhang Y.Z., Dai J., Xiang X. Studies on the interaction between benzidine and bovine serum albumin by spectroscopic methods. Mol. Biol. Rep. 2010;37:1541–1549. doi: 10.1007/s11033-009-9555-x. [DOI] [PubMed] [Google Scholar]
- 31.Ross P.D., Subramanian S. Thermodynamics of protein association reactions: forces contributing to stability. Biochemistry. 1981;20:3096–3102. doi: 10.1021/bi00514a017. [DOI] [PubMed] [Google Scholar]
- 32.Jin J., Zhang X. Spectrophotometric studies on the interaction between pazufloxacin mesilate and human serum albumin or lysozyme. J. Lumin. 2008;128:81–86. [Google Scholar]
- 33.F¨orster T., Sinanoglu O., editors. vol. 3. Academic Press; New York: 1996. p. 93. (Modern Quantum Chemistry). [Google Scholar]
- 34.Wu C.W., Stryer L. Proximity relationships in rhodopsin. Proc. Natl. Acad. Sci. USA. 1972;69:1104–1108. doi: 10.1073/pnas.69.5.1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lakowicz J.R. Plenum Press; New York: 1983. Principles of Fluorescence Spectroscopy. [Google Scholar]
- 36.Hu Y.J., Liu Y., Zhang L.X. Studies of interaction between colchicine and bovine serum albumin by fluorescence quenching method. J. Mol. Struct. 2005;750:174–178. [Google Scholar]
- 37.Liu J., Tian J.N., Zhang J. Interaction of magnolol with bovine serum albumin: a fluorescence-quenching study. Anal. Bioanal. Chem. 2003;376:864–867. doi: 10.1007/s00216-003-2017-8. [DOI] [PubMed] [Google Scholar]
- 38.Analytical Methods Committee Recommendations for the definition, estimation and use of the detection limit. Analyst. 1987;112:199–204. [Google Scholar]
- 39.Sun W., Jiao K. Linear sweep voltammetric determination of protein based on its interaction with Alizarin Red S. Talanta. 2002;56:1073–1080. doi: 10.1016/s0039-9140(01)00628-2. [DOI] [PubMed] [Google Scholar]
- 40.Khan M.Y. Direct evidence for the involvement of domain III in the N–F transition of bovine serum albumin. Biochemistry. 1986;236:307–310. doi: 10.1042/bj2360307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tang J.H., Luan F., Chen X.G. Binding analysis of glycyrrhetinic acid to human serum albumin: fluorescence spectroscopy, FT-IR, and molecular modeling. Bioorg. Med. Chem. 2006;14:3210–3217. doi: 10.1016/j.bmc.2005.12.034. [DOI] [PubMed] [Google Scholar]
- 42.Sugio S., Kashima A., Mochizuki S. Crystal structure of human serum albumin at 2.5 Å resolution. Protein Eng. 1999;12:439–446. doi: 10.1093/protein/12.6.439. [DOI] [PubMed] [Google Scholar]
- 43.Zunszain P.A., Ghuman J., Kornatsu T. Crystal structural analysis of human serum albumin complexed with hemin and fatty acid. BMC Struct. Biol. 2003;3:6–9. doi: 10.1186/1472-6807-3-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Petitpas I., Bhattacharya A.A., Twine S. Crystal structure analysis of warfarin binding to human serum albumin: anatomy of drug site I. J. Boil. Chem. 2001;276:22804–22809. doi: 10.1074/jbc.M100575200. [DOI] [PubMed] [Google Scholar]
- 45.Zhang G., Wang Y., Zhang H. Human serum albumin interaction with paraquat studied using spectroscopic methods. Pest. Biochem. Physiol. 2007;87:23–29. [Google Scholar]