Abstract
Bacterial infections constitute an increasing problem to human health in response to build-up of resistance to present antibiotics and sluggish development of new pharmaceuticals. However, a means to address this problem is to pinpoint the drug delivery to - and into - the bacteria. This results in a high local concentration of the drug, circumventing the increasingly high doses otherwise necessary. Combined with other effectors, such as covalent attachment to carriers, rendering the drugs less degradable, and the combination with efflux inhibitors, old drugs can be revived. In this context, glyconanomaterials offer exceptional potential, since these materials can be tailored to accommodate different effectors. In this feature article, we describe the different advantages of glyconanomaterials, and point to their importance in antibiotic ‘revitalization’.
Graphical Abstract
Introduction
Bacterial infections
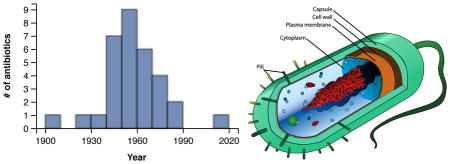
The success of combating bacterial infections relies to a large extent on the development of efficient antibiotics. However, in response to the increasing problem of antimicrobial resistance, many important drugs are no longer effective, resulting in a major public health risk. This problem was recently highligthed by the World Economic Forum, describing antibiotic resistance as “the greatest risk of hubris to human health”,[1] resulting not only in increasing human fatality rates but also accelerating health care costs.[2,3] Certain killer bacteria show emerging resistance to even the strongest antibiotics,[4,5] and current drug development in the field is slow. With the possible exception of the recently discovered teixobactin,[6] there have been no successful discoveries of new classes of antibiotics since 1987.[7] As a consequence, alternative methodologies are warranted to avoid a scenario in which all antibiotics would be rendered ineffective for treating even the most common infections.
Such methodologies should in principle address major causes for antibiotic resistance such as mutational alteration in the targeted proteins, periplasmic β-lactamase action, decreased drug uptake due to loss of membrane-bound porin, and drug extrusion via efflux pumps.[8–11] A complementary approach involves increasing the drug titer at or inside the bacterium, leading to higher therapeutic efficacy and lower systemic toxicity. This can in principle be addressed using different drug delivery protocols, furnishing the active pharmaceutical to the site of infection and creating a local concentration effect. This effect can in principle also lead to a revival of the relative antibiotic potency of the drug.
Nanomaterials are in this context highly attractive, designed to carry therapeutic agents to the specific disease sites while avoiding healthy tissue and cells.[12] The efficacies of such nanotherapeutics have been shown to be higher than for small molecule drugs, at the same time lowering the off-target toxicities.[13–17] Nanomaterials can furthermore be tailored to for example escape premature degradation and release of therapeutic agent, or to present targeting entities and/or imaging elements enabling controlled delivery and monitoring.[18,19] Functional nanocarriers presenting specific targeting entities have been shown to improve tissue specificity, leading to increased cellular uptake into target cells and higher intracellular drug concentrations. In addition, nanotherapeutics have been shown to overcome drug resistance due to the inability of efflux pumps to remove drug-nanoparticle complexes.[20] These materials can also be designed in many different forms with tailored properties, such as nanoemulsions, liposomes, virosomes, nanocrystals, polymer-based materials, nanoparticles, nanocomplexes, etc, specially adapted for targeted entities. These features have resulted in a wide range of applications, where more than 40 nanotherapeutic formulations have reached the market, and over 200 new candidates are undergoing clinical trials.[21–25]
Glyconanomaterials
Glyconanomaterials (GNMs) can be defined as nanomaterials incorporating carbohydrate entities, typically presenting interactive carbohydrate/glycan functionalities at their surfaces. As such, these materials mimic cell surfaces, which are generally decorated with different glycolipids and glycoproteins that enable recognition of carbohydrate-binding proteins such as lectins and antibodies, and thereby mediate a wide variety of cellular communication events. [26,27] Some cells, such as certain bacteria and cells involved in endothelial tissue, present a relatively thick “glycocalyx” layer at their surfaces involving carbohydrate-based cell-adhesion molecules.[28–30] These coating layers play multiple roles, for example to prevent adhesion and serving as sieves for specific molecules.
During the last decade, glyconanomaterials have become established as highly useful platforms for a range of applications.[31–38] A variety of materials have been developed and evaluated for various applications, where special focus has been put on inorganic nanoparticles. Early accounts based on gold nanoparticles describe GNMs functionalized with lactose and the LewisX antigen (Galβ1-4(Fucα1-3)GlcNAcβ1-R), monitoring carbohydrate-carbohydrate interactions (“glycosphingolipid cluster mimics”) by SPR, and controlling nanoparticle assembly.[39,40] Since then, a wide array of core materials, such as silver, iron oxide, silica, copper, bismuth, palladium, and platinum, have been used in addition to gold,[41–50] and miscellaneous cadmium/zinc chalcogen quantum dots have been evaluated.[51] A major reason for the interest in these materials is their many attractive physical features, such as plasmonic effects, luminescence, magnetism/magnetic susceptibility, etc, which render them especially useful for both analysis and therapy. However, organic materials have also witnessed increasing attention since early syntheses and applications of glycosylated C60, poly-L-lysine, and dendrons/dendrimers.[52–54] The range of carbon materials, such as fullerenes, carbon nanotubes, graphene, and nanodiamonds have for example been functionalized with carbohydrate structures and used in different situations,[55–59] and glyconanomaterials based on organic polymer-type materials have been continuously demonstrated, where both artificial and natural polymers have been used. In this context, the development of glycodendrimers has perhaps proven especially successful,[34] where the resulting entities are robust and structurally well defined. Finally, different supramolecular constructs, such as amphiphilic aggregates, liposomes and vesicles, virus-like particles, etc, have been reported.[60–63] Glycoliposomes and vesicles mimic cell membrane surfaces and show especially attractive features in terms of synthesis and biocompatibility. Their stability may however present an issue, and it can be difficult to obtain high densities of surface-exposed carbohydrates in certain cases.[64]
High carbohydrate surface density is otherwise a major advantage of glyconanomaterials, to some extent mimicking the carbohydrate landscape at cell surfaces. This feature leads to the establishment of multivalent ligand presentation, which upon realization of optimal geometries can result in very high binding affinities with carbohydrate-binding proteins compared to the free ligands.[37,45,65–67] This affinity enhancement effect, which can be several orders of magnitude, thus enables the use of very low concentrations of materials. In consequence to this and to other attractive features, glyconanomaterials have thus been tailored to act as integral binding entities in affinity-based methods.[31–38] For example, analyses of the interactions between the materials and specific proteins and other entities have been amply demonstrated for different systems, in part used to validate and optimize the various methods developed. Model systems are thus used to evaluate the engineered structures, and to further the understanding of intricate binding phenomena at the GNM surfaces. The results from such binding assays are also of particular interest for the parallel development of methodologies for sensing, imaging and diagnostics, where good results often are dependent on strong and selective binding in combination with efficient monitoring or signal transduction. These types of applications are obviously of very high importance, generating high-quality analytical data, but given the importance of carbohydrates in biology and medicine, the ultimate challenge for these materials lies in medical therapy. Development in this area is ongoing, where the carbohydrate structures of the materials often serve as targeting entities for delivery of therapeutic agents, as well as the materials themselves, to the site of disease. Through selective interactions with specific determinants at tissue and cell surfaces, improved therapy can be achieved.
Bacteria and glyconanomaterials
A wide range of nanomaterials have been applied to antibacterial activity, such as for example metal- and other inorganic nanoparticles, carbon-based structures, polysaccharides, nanoemulsions, liposomes, polymers and dendrimers.[68–70] Some metal salts and metal nanoparticles are well known to exert antibacterial properties,[71,72] where for example silver nanoparticles have proven active towards both Gram-positive and Gram-negative bacteria. [73–75] Also, different chitosan formulations have shown antimicrobial activity towards various bacterial strains.[76] Many of these structures display antibiotic activities per se through release of metal ions from the surface, generation of reactive oxygen species (ROS), and interaction with, and disruption of, the bacterial membranes. Other nanoparticle formulations such as liposomes and polymers act as more traditional carriers of encapsulated antibiotic drugs, thereby stabilizing the compounds from premature degradation and potentially resulting in controlled release. For example, different antitubercular drugs have been delivered into human monocyte-derived macrophages using polymeric nanoparticles.[77]
A variety of nanoparticle formulations have also been applied to delivery of antibiotic agents to bacteria via surface conjugation.[69] This is especially the case for the vancomycin-family of antibiotics, where different nanocarriers have been used. For example, vancomycin-presenting gold nanoparticles have been applied as rigid polyvalent inhibitors of vancomycin-resistant Enterococci (VRE) or E. coli,[78] and norvancomycin-conjugated silver nanoparticles have been used to induce aggregation in E. coli cell walls.[79] Vancomycin-presenting, magnetic FePt-nanoparticles have furthermore been demonstrated for detecting and capturing VRE and other Gram-positive and Gram-negative bacteria.[80,81] Other antibiotics and small molecules have also been evaluated for direct conjugation; for example, the in vitro antibacterial properties of emulsified polyacrylate nanoparticles conjugated with penicillin were evaluated against methicillin-susceptible and methicillin-resistant forms of S. aureus,[82] and isoniazid-conjugated poly(lactide-co-glycolide) particles were used for slow release to tuberculosis-affected bone tissue.[83] Combinatorial presentation of conjugated small ligands (not antibiotics) on gold nanoparticles have also been used to screen for antiobitic activity towards multiply drug resistant E. coli and K. pneumoniae.[84]
Table 1 lists an overview of reports where glyconanomaterials of different type have been applied to antibacterial action, where in some cases the more representative examples have been selected. As can be seen, different classes of bacteria have been targeted, including gram-positive- (Bacillus, Enterococcus, Listeria, Staphylococcus), gram-negative(Chlamydophila, Escherichia, Francisella, Klebsiella, Legionella) and myco-bacteria. Different strains of E. coli have been mostly studied with the materials, of which fimbric E. coli strains expressing carbohydrate-binding adhesins have been especially targeted. Already, a wide range of nanomaterial scaffolds have been used, with no apparent preference for specific type. Thus, both inorganic particles and organic materials have been applied to carbohydrate functionalization and further targeting of the microbes. In most cases, the nanomaterials have been conjugated with derivatized monosaccharide structures, linked to the GNM surfaces through suitable spacer units. However, disaccharides (trehalose, galabiose),[85,86] oligosaccharides (maltoheptaose),[87] and polysaccharides (kocuran),[88] have also been evaluated. Although monosaccharides show good effects in these systems, it can be expected that more complex carbohydrate structures will be increasingly probed in future developments. This will lead to both higher selectivities and enhanced affinities, of importance for improved therapeutic potential of the materials.
Table 1.
Selection of glyconanomaterials applied to antibacterial action.
| Bacterium | Carbohydrate | Nanomateriala | Application | Reference |
|---|---|---|---|---|
| B. anthracis | Manα1R | SWCNT | interaction | [89,90] |
| B. subtilis | GlcN | Cu | growth inhibition | [91] |
| B. subtilis | Manα1R | SWCNT | interaction | [89] |
| B. subtilis | Manα1R | SWCNT | binding, aggregation | [92] |
| C. penumoniae | Manα1R | Liposome | delivery | [93] |
| E. faecalis | GlcN | Cu | growth inhibition | [91] |
| E. coli | Manα1R | Cellulose | aggregation, imaging | [94] |
| E. coli | Manα1R | Au | binding, aggregation | [95] |
| E. coli | Manα1R | Chitin | aggregation, imaging | [96] |
| E. coli | (Glcα1-4)6Glc | SiO2 | binding, aggregation | [87] |
| E. coli | (Glcα1-4)6Glc | Fe3O4 | binding/internalizatn | [87] |
| E. coli | (Glcα1-4)6Glc | SiO2/Fe3O4 | binding, aggregation | [87] |
| E. coli | (Glcα1-4)6Glc | SiO2/CdSe/CdS/ZnS | aggregation, imaging | [87] |
| E. coli | Manα1CRb | Ag | growth inhibition | [97] |
| E. coli | Glcα1R | ND | aggregation, imaging | [56] |
| E. coli | Manα1R | ND | aggregation, imaging | [56] |
| E. coli | Man | SiO2 | aggregation, imaging | [42] |
| E. coli | Manα1R | Au | growth inhibition | [98] |
| E. coli | Manα1R | SiO2/Fe3O4 | aggregation, imaging | [99] |
| E. coli | Man | αFe2O3 | aggregation, removal | [46] |
| E. coli | Manα1R | Au | binding, aggregation | [100] |
| E. coli | Manα1R | CdS | aggregation, imaging | [101] |
| E. coli | Galβ1R | SWCNT | binding, aggregation | [92,102] |
| E. coli | Galα1-4Galβ1NRc | PDA | binding, detection | [86] |
| E. coli | Manα1R | Amphiphilic aggregates | aggregation, imaging | [61,103–105] |
| E. coli | Manα1R | Polystyrene | binding, aggregation | [106,107] |
| E. coli | Manα1R | Au | binding, aggregation | [108] |
| F. tularensis | Manα1R | Liposome | delivery | [93] |
| L. monocytogenes | Manα1R | Liposome | delivery | [93] |
| L. pneumophila | Manα1R | Liposome | delivery | [93] |
| M. avium | Manα1R | Liposome | delivery | [93] |
| M. intracellulare | Manα1R | Liposome | delivery | [93] |
| M. smegmatis | Trehalose | Fe3O4 | binding, aggregation | [85] |
| M. smegmatis | Trehalose | SiO2 | aggregation, imaging | [85] |
| M. smegmatis | Trehalose | SiO2 | binding, delivery | Submitted |
| M. tuberculosis | Manα1R | Liposome | delivery | [93] |
| S. aureus | Kocuran | Ag | growth inhibition | [88] |
SWCNT: single-wall carbon nanotubes; ND: nanodiamonds; PDA: polydiacetylene;
C-glycoside;
N-glycoside
Targeting
One of the most attractive properties of glyconanomaterials is their targeting potential, and this is one of the most studied effects to date. For example, a range of materials have targeted fimbric, generally uropathogenic E. coli strains, which express pili at their surfaces. These bacteria can adhere to carbohydrate determinants of tissue cells lining the urinary tract, which may lead to subsequent infection and colonization of the bladder.[109] Nanomaterials presenting mannose- or glucose-units at their surfaces have thus been evaluated for interaction with E. coli strains expressing the FimH lectin at type 1 pili (E coli strains ORN178, K12, DH5α, HB101 pPKL4, PKL1162).[42,46,56,61,87,94–101,103–108] This adhesin is selective for mannopyranosyl units, and the glyconanomaterials can serve as cell-surface mimics targeting the bacterial pili. This concept has proven highly efficient, and the materials developed have indeed shown efficient binding to the bacteria. The Shiga toxin-producing E. coli (STEC) strain O157:H7 has also been targeted.[86,92,102] This pathogen can bind galabiose (Galα1-4Gal) structures at host cells through interaction with the toxins’ pentameric B-unit. Nanomaterials functionalized with galabiose and galactose can thus bind to the pathogen, shown for photocrosslinked diacetylene-based nanoparticles and single-wall carbon nanotubes (SWCNTs). Carbon nanotubes functionalized with mannose- and galactose derivatives were also found to interact with Bacillus sp. spores.[89,90,92] Calcium-dependent aggregation was observed in the case of galactosylated nanotubes, as well as a reduction in colony-forming units. Since the spores express glycan structures, the authors suggested aggregation dependent on carbohydrate-carbohydrate interactions.
Targeting is the essential property for a wide range of applications involving analysis and detection of bacteria, and the current development has shown very strong potential in this area. Bacterial imaging has thus been demonstrated in several examples, either using the optical properties of the nanomaterial cores themselves, such as quantum dots and crosslinked polydiacetylene,[56,87,101] or by introducing fluorescent labels in the materials. [42,85,94,96] Furthermore, magnetic nanoparticles enable the use of magnetic resonance imaging.[99] These methodologies can thus in principle be used for detection and diagnostics of bacterial infections, where high affinities are crucial for assay sensitivities. Rapid and reliable diagnostic tools are of high importance not only to identify the infections, but also to avoid overuse of antibiotic agents. In addition to their imaging properties, magnetic nanoparticles have other potential advantages that can be employed. The particles are thus susceptible to interactions with external magnetic fields, which can lead to clustering and eventually removal of the bacteria from the solutions. Furthermore, alternating magnetic fields can induce local heat generation at the site of the nanomaterials (magnetic hyperthermia),[110] thereby causing damage to the surrounding microbial cells.
Carbohydrate-mediated targeting can also lead to growth inhibition effects, mainly exerted by the core materials. This has for example been shown with mannosylated silver and gold nanoparticles, interacting with fimbric E. coli strains.[97,98] Growth inhibition has otherwise been evaluated with glyconanomaterials where the carbohydrate entities were more likely to serve as stabilizing and solubilizing agents rather than targeting devices. For example, silver nanoparticles capped with the polysaccharide kocuran were probed against a range of bacterial strains, of which S. aurerus was affected the most.[88] Likewise, copper nanoparticles functionalized with glucosamine were primarily found to inhibit the growth of B. subtilis and E. faecalis.[91] In addition to the metal core, the glucosamine structure was in this case expected to exert reactive oxygen species in the solution.
For all these applications, selectivity is an important factor that needs further attention. If the materials are to be used in vivo, it is essential that the affinities to endogenous receptors are minimized. This will lower the clearance rate and ensure prolonged action time of the materials.
In addition, the selection between pathogenic and benign bacteria is of utmost importance to avoid unnecessary buildup of resistance. In light of the relatively simple systems developed to date, it is however very likely that selectivity can be substantially improved with the use of more complex carbohydrate structures in combination with other effectors.
Delivery
In combination with targeting, transport of potent antibiotic agents to the bacteria is a highly appealing application of glyconanomaterials. As has been shown with other nanocarriers, the drug substances can thus be protected from degradation and premature release leading to low efficacies. The selective release of the antimicrobial substances at the bacterial surfaces, or even potentially inside the bacteria, can thus lead to more efficient antimicrobial action compared to separate administration of the drugs. The localized release effect can furthermore result in relatively high local concentration of drugs at the bacterial dwelling area, thereby exceeding the levels of antibiotic resistance displayed by the microbial species. This effect can thus lead to ‘revitalization’ of otherwise less efficient antibiotic species, especilly if combined with other means of action, such as efflux inhibition, antimicrobial effects of the nanomaterials themselves, etc. Although this effect may be temporary, where the bacteria may continue to evolve in response to the higher efficacies witnessed, this can lead to prolongued effects of presently developed antibiotic agents.
At present, only few examples of delivery using glyconanomaterials have been reported in the literature. Using large (~1000 nm) mannosylated liposomes, Chono and coworkers for example evaluated the delivery of ciprofloxacin to different bacterial strains involved in respiratory intracellular parasitic infections.[93] In this case, however, mannose-functionalization served as a potential targeting device for delivery of the liposomes to alveolar macrophages rather than the bacteria themselves. Nevertheless, it could be shown that the delivery increased the effect of ciprofloxacin compared to controls. In another example, we recently showed that trehalose-conjugated mesoporous silica nanoparticles can be used to deliver isoniazid to M. smegmatis (submitted), thereby leading to increased biocidal action compared to the free drug. In this case, the combination of the targeting feature of these materials to the mycobacterial surfaces with nanomaterial-based drug delivery, thus enhanced the isoniazid effect likely by resulting in higher local amount of drug near the bacteria. These examples show that drug delivery using glyconanomaterials can lead to improved antibiotic action of known drug substances, and it can therefore be expected that this line of research will be much developed in the years to come.
Internalization
Uptake of nanoparticles in eucaryotes generally occurs through endocytosis, a well-established pathway based on encapsulation by the plasma membrane followed by internalization into lysosomes or recycling to the cell surface. However, this mechanism is not common to bacteria or archae and has only recently been suggested in Planctomycetes,[111] a bacterial phylum that displays certain similarities, such as compartmentalization, to eucorytic cells. Internalization in bacteria has however been demonstrated for specific nanoparticles, thus following alternative mechanisms. Three different pathways have in this context been suggested: uptake due to nonspecific cell envelope disruption, nonspecific diffusion, and specific uptake. Of these, cell envelope disruption has been proposed for certain, reactive nanomaterials as mentioned above, such as silver nanoparticles and halogen-adsorbing structures.[75,112] Internalization through nonspecific uptake appears relatively unlikely, owing to the size of the nanoparticles. It has for example been shown that protein uptake in B. subtilis only occurs for structures <4 nm in diameter,[113] whereas most nanoparticle structures are generally larger. This size exclusion effect has been described to be due to so called crosslinked “tessera” structures in the peptidoglycan layer, forming small pores that makes the layer permeable to small molecules but not to larger entities.[114] For softer, elastic nanostructures, such as certain polymeric structures and dendrimers, this route may however offer a potential possibility. The third pathway, specific uptake, can in principle accommodate larger structures, and the pore sizes of certain holins, porins, permeases and secretins have been shown to be up to around 6 nm in diameter.[115–118] Uptake of functionalized nanoparticles of core sizes <6 nm have also been proposed.[87,119,120] For example, Nadeau and coworkers reported that adenine- and AMP-conjugated CdSe and CdSe/ZnS quantum dots were internalized in B. subtilis and E. coli, presumably through purine-dependent mechanisms.[120] Furthermore, Yan and coworkers showed that maltoheptaose-functionalized magnetic iron oxide nanoparticles appeared to be internalized in E. coli, also suggesting that translocation through the inner membrane is a major barrier.[87] Also, Chakrabarti et al. demonstrated uptake of polyethyleneimine-functionalized ZnO nanoparticles in H. pylori, however in this case presumably by partial disruption of the cell envelope by formation of reactive oxygen species.[119] In this context, it should be stressed that the analysis of internalization effect presents a significant challenge. The methods generally used, i.e. TEM and flow cytometry, can not easily distinguish between internalized and surface-bound nanoparticles, and the best method involves in this case cellular thin sectioning in combination with TEM. Recent advances in super-resolved fluorescence microscopy may however offer an alternative.
Conclusions
Recent developments have shown that glyconanomaterials offer a viable route to detect and combat various pathogenic bacteria. It has thus become well established that these materials can selectively interact with bacteria, thus serving as efficient tools for pathogen detection and diagnosis. Certain formulations furthermore show antibacterial activities, either through targeted delivery of antibiotic agents, or through effects from the materials themselves, such as formation of reactive oxygen species. In some cases, such targeted delivery can increase the efficacies of the antibiotics, thereby lowering the dosage needed, and revitalizing antimicrobial drugs to which the bacteria are otherwise resistant. In principle, the materials can also be infinitely varied, and be exclusively tailored in terms of size, shape, chemical composition, cargo, and targeting entities. They may thus offer a means to continuously address effects of antibiotic resistance, whereby the materials can be reoptimized towards certain pathogenic strains. A number of challenges remain however, especially related to delivery to and across the bacterial envelope. For example, efficient transport through protective layers, such as biofilms and granulocytes, is a prevailing problem, and internalization into the bacterial cytosol has proven difficult. Especially for antibiotic agents that target cytosolic proteins and DNA, this needs additional attention. Selective distinction between pathogenic and non-pathogenic bacteria is another challenge that needs to be further addressed. Good selectivity can both reduce excessive uses of antibiotic agents and thereby attenuate antibiotic resistance, and spare beneficial bacteria of for example the gut microbiota. The biocompatibility and ADMET properties of the materials furthermore need to be continuously evaluated. However, in step with the increasingly detailed knowledge of bacterial structures and behavior, and in light of the rapid progress seen with these materials, many of these challenges are likely to be met in the very near future.
Acknowledgments
This study was supported by the National Institutes of Health (R01GM080295), the National Science Foundation (CHE-1112436), and the European Union’s seventh framework for research, technological development and demonstration (289033).
References
- 1.Howell L, editor. Global Risks 2013. World Economic Forum; Geneva: 2013. pp. 28–35. [Google Scholar]
- 2.Bush K, Courvalin P, Dantas G, Davies J, Eisenstein B, Huovinen P, Jacoby GA, Kishony R, Kreiswirth BN, Kutter E, et al. Nat Rev Microbiol. 2011;9:894–6. doi: 10.1038/nrmicro2693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Spellberg B, Blaser M, Guidos RJ, Boucher HW, Bradley JS, Eisenstein BI, Gerding D, Lynfield R, Reller LB, Rex J, et al. Clin Infect Dis. 2011;52(Suppl 5):S397–428. doi: 10.1093/cid/cir153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Phillips L. Nature. 2013;493:14–6. doi: 10.1038/493014a. [DOI] [PubMed] [Google Scholar]
- 5.Snitkin ES, Zelazny AM, Thomas PJ, Stock F, Henderson DK, Palmore TN, Segre JA. Sci Transl Med. 2012;4:148ra116–148ra116. doi: 10.1126/scitranslmed.3004129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ling LL, Schneider T, Peoples AJ, Spoering AL, Engels I, Conlon BP, Mueller A, Schäberle TF, Hughes DE, Epstein S, et al. Nature. 2015;517:455–459. doi: 10.1038/nature14098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Silver LL. Clin Microbiol Rev. 2011;24:71–109. doi: 10.1128/CMR.00030-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fernández L, Hancock REW. Clin Microbiol Rev. 2012;25:661–81. doi: 10.1128/CMR.00043-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hopkins KL, Davies RH, Threlfall EJ. Int J Antimicrob Agents. 2005;25:358–73. doi: 10.1016/j.ijantimicag.2005.02.006. [DOI] [PubMed] [Google Scholar]
- 10.Yang S, Clayton SR, Zechiedrich EL. J Antimicrob Chemother. 2003;51:545–556. doi: 10.1093/jac/dkg126. [DOI] [PubMed] [Google Scholar]
- 11.Mazzariol A, Tokue Y, Kanegawa TM, Cornaglia G, Nikaido H. Antimicrob Agents Chemother. 2000;44:3441–3. doi: 10.1128/aac.44.12.3441-3443.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang L, Gu FX, Chan JM, Wang AZ, Langer RS, Farokhzad OC. Clin Pharmacol Ther. 2007;83:761–769. doi: 10.1038/sj.clpt.6100400. [DOI] [PubMed] [Google Scholar]
- 13.Schroeder A, Heller DA, Winslow MM, Dahlman JE, Pratt GW, Langer R, Jacks T, Anderson DG. Nat Rev Cancer. 2012;12:39–50. doi: 10.1038/nrc3180. [DOI] [PubMed] [Google Scholar]
- 14.Lee J, Sohn JW, Zhang Y, Leong KW, Pisetsky D, Sullenger BA. Proc Natl Acad Sci U S A. 2011;108:14055–60. doi: 10.1073/pnas.1105777108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Park J-H, von Maltzahn G, Ong LL, Centrone A, Hatton TA, Ruoslahti E, Bhatia SN, Sailor MJ. Adv Mater. 2010;22:880–5. doi: 10.1002/adma.200902895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.von Maltzahn G, Centrone A, Park J-H, Ramanathan R, Sailor MJ, Hatton TA, Bhatia SN. Adv Mater. 2009;21:3175–3180. doi: 10.1002/adma.200803464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nie S, Xing Y, Kim GJ, Simons JW. Annu Rev Biomed Eng. 2007;9:257–88. doi: 10.1146/annurev.bioeng.9.060906.152025. [DOI] [PubMed] [Google Scholar]
- 18.Cheng Z, Al Zaki A, Hui JZ, Muzykantov VR, Tsourkas A. Science. 2012;338:903–10. doi: 10.1126/science.1226338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gabizon AA. Clin Cancer Res. 2001;7:223–5. [PubMed] [Google Scholar]
- 20.Yu YJ, Zhang Y, Kenrick M, Hoyte K, Luk W, Lu Y, Atwal J, Elliott JM, Prabhu S, Watts RJ, et al. Sci Transl Med. 2011;3:84ra44. doi: 10.1126/scitranslmed.3002230. [DOI] [PubMed] [Google Scholar]
- 21.Duncan R, Gaspar R. Mol Pharm. 2011;8:2101–41. doi: 10.1021/mp200394t. [DOI] [PubMed] [Google Scholar]
- 22.Benezra M, Penate-Medina O, Zanzonico PB, Schaer D, Ow H, Burns A, DeStanchina E, Longo V, Herz E, Iyer S, et al. J Clin Invest. 2011;121:2768–80. doi: 10.1172/JCI45600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miele E, Spinelli GP, Miele E, Tomao F, Tomao S. Int J Nanomedicine. 2009;4:99–105. doi: 10.2147/ijn.s3061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harisinghani MG, Saksena M, Ross RW, Tabatabaei S, Dahl D, McDougal S, Weissleder R. Urology. 2005;66:1066–71. doi: 10.1016/j.urology.2005.05.049. [DOI] [PubMed] [Google Scholar]
- 25.Safra T, Muggia F, Jeffers S, Tsao-Wei DD, Groshen S, Lyass O, Henderson R, Berry G, Gabizon A. Ann Oncol. 2000;11:1029–33. doi: 10.1023/a:1008365716693. [DOI] [PubMed] [Google Scholar]
- 26.Wang B, Boons G-J, editors. Carbohydrate Recognition. John Wiley & Sons; Weinheim, Germany: 2011. [Google Scholar]
- 27.Collins BE, Paulson JC. Curr Opin Chem Biol. 2004;8:617–25. doi: 10.1016/j.cbpa.2004.10.004. [DOI] [PubMed] [Google Scholar]
- 28.Alphonsus CS, Rodseth RN. Anaesthesia. 2014;69:777–84. doi: 10.1111/anae.12661. [DOI] [PubMed] [Google Scholar]
- 29.Weinbaum S, Tarbell JM, Damiano ER. 2007 doi: 10.1146/annurev.bioeng.9.060906.151959. [DOI] [PubMed] [Google Scholar]
- 30.Costerton JW, Irvin RT, Cheng KJ. Annu Rev Microbiol. 1981;35:299–324. doi: 10.1146/annurev.mi.35.100181.001503. [DOI] [PubMed] [Google Scholar]
- 31.Chen X, Ramström O, Yan M. Nano Res. 2014;7:1381–1403. doi: 10.1007/s12274-014-0507-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Barboiu M, Mouline Z, Silion M, Licsandru E, Simionescu BC, Mahon E, Pinteala M. Chem Eur J. 2014;20:6678–83. doi: 10.1002/chem.201402187. [DOI] [PubMed] [Google Scholar]
- 33.Bernardi A, Jiménez-Barbero J, Casnati A, De Castro C, Darbre T, Fieschi F, Finne J, Funken H, Jaeger K-E, Lahmann M, et al. Chem Soc Rev. 2013;42:4709–27. doi: 10.1039/c2cs35408j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chabre YM, Roy RR. Chem Soc Rev. 2013;42:4657–708. doi: 10.1039/c3cs35483k. [DOI] [PubMed] [Google Scholar]
- 35.Kennedy DC, Grünstein D, Lai C-H, Seeberger PH. Chem Eur J. 2013;19:3794–800. doi: 10.1002/chem.201204155. [DOI] [PubMed] [Google Scholar]
- 36.Marradi M, Chiodo F, García I, Penadés S. Chem Soc Rev. 2013;42:4728–45. doi: 10.1039/c2cs35420a. [DOI] [PubMed] [Google Scholar]
- 37.Wang X, Ramström O, Yan M. Adv Mater. 2010;22:1946–1953. doi: 10.1002/adma.200903908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang X, Liu L-H, Ramström O, Yan M. Exp Biol Med. 2009;234:1128–1139. doi: 10.3181/0904-MR-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.de la Fuente JM, Barrientos AG, Rojas TC, Rojo J, Cañada J, Fernández A, Penadés S. Angew Chem Int Ed. 2001;40:2257–2261. [PubMed] [Google Scholar]
- 40.Otsuka H, Akiyama Y, Nagasaki Y, Kataoka K. J Am Chem Soc. 2001;123:8226–8230. doi: 10.1021/ja010437m. [DOI] [PubMed] [Google Scholar]
- 41.Brown AL, Goforth AM. Chem Mater. 2012;24:1599–1605. [Google Scholar]
- 42.Wang X, Ramström O, Yan M. Chem Commun. 2011;47:4261–3. doi: 10.1039/c0cc05299j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang X, Ramström O, Yan M. Analyst. 2011;136:4174–4178. doi: 10.1039/c1an15469a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mahon E, Aastrup T, Barboiu M. Chem Commun. 2010;46:5491–5493. doi: 10.1039/c002652b. [DOI] [PubMed] [Google Scholar]
- 45.Wang X, Ramström O, Yan M. J Mater Chem. 2009;19:8944–8949. doi: 10.1039/B917900C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu L-H, Dietsch H, Schurtenberger P, Yan M. Bioconjug Chem. 2009;20:1349–55. doi: 10.1021/bc900110x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Park IY, Kim IY, Yoo MK, Choi YJ, Cho M-H, Cho CS. Int J Pharm. 2008;359:280–287. doi: 10.1016/j.ijpharm.2008.04.010. [DOI] [PubMed] [Google Scholar]
- 48.Horák D, Babič M, Jendelová P, Herynek V, Trchová M, Pientka Z, Pollert E, Hájek M, Syková E. Bioconjug Chem. 2007;18:635–644. doi: 10.1021/bc060186c. [DOI] [PubMed] [Google Scholar]
- 49.de la Fuente JM, Berry CC, Riehle MO, Curtis ASG. Langmuir. 2006;22:3286–3293. doi: 10.1021/la053029v. [DOI] [PubMed] [Google Scholar]
- 50.Huang H, Yuan Q, Yang X. Colloids Surfaces, B Biointerfaces. 2004;39:31–37. doi: 10.1016/j.colsurfb.2004.08.014. [DOI] [PubMed] [Google Scholar]
- 51.Chen Y, Ji T, Rosenzweig Z. Nano Lett. 2003;3:581–584. [Google Scholar]
- 52.Nishikawa M, Takemura S, Takakura Y, Hashida M. J Pharmacol Exp Ther. 1998;287:408–415. [PubMed] [Google Scholar]
- 53.Roy R, Zanini D, Meunier SJ, Romanowska A. J Chem Soc Chem Commun. 1993:1869. [Google Scholar]
- 54.Vasella A, Uhlmann P, Waldraff CAA, Diederich F, Thilgen C. Angew Chem Int Ed. 1992;31:1388–1390. [Google Scholar]
- 55.Kong N, Shimpi MR, Ramström O, Yan M. Carbohydr Res. 2014;405:33–38. doi: 10.1016/j.carres.2014.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hartmann M, Betz P, Sun Y, Gorb SN, Lindhorst TK, Krueger A. Chem Eur J. 2012;18:6485–6492. doi: 10.1002/chem.201104069. [DOI] [PubMed] [Google Scholar]
- 57.Chen Q, Wei W, Lin J-M. Biosens Bioelectron. 2011;26:4497–4502. doi: 10.1016/j.bios.2011.05.009. [DOI] [PubMed] [Google Scholar]
- 58.Nierengarten J-F, Iehl J, Oerthel V, Holler M, Illescas BM, Muñoz A, Martín N, Rojo J, Sánchez-Navarro M, Cecioni S, et al. Chem Commun. 2010;46:3860–3862. doi: 10.1039/c0cc00034e. [DOI] [PubMed] [Google Scholar]
- 59.Chen X, Lee GS, Zettl A, Bertozzi CR. Angew Chem Int Ed. 2004;43:6111–6116. doi: 10.1002/anie.200460620. [DOI] [PubMed] [Google Scholar]
- 60.Rhee JK, Baksh M, Nycholat C, Paulson JC, Kitagishi H, Finn MG. Biomacromolecules. 2012;13:2333–2338. doi: 10.1021/bm300578p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kim B-S, Hong D-J, Bae J, Lee M. J Am Chem Soc. 2005;127:16333–16337. doi: 10.1021/ja055999a. [DOI] [PubMed] [Google Scholar]
- 62.Hara T, Aramaki Y, Takada S, Koike K, Tsuchiya S. Gene. 1995;159:167–174. doi: 10.1016/0378-1119(95)00100-k. [DOI] [PubMed] [Google Scholar]
- 63.Reichert A, Nagy JO, Spevak W, Charych D. J Am Chem Soc. 1995;117:829–830. [Google Scholar]
- 64.Marradi M, Di Gianvincenzo P, Enríquez-Navas PM, Martínez-Ávila OM, Chiodo F, Yuste E, Angulo J, Penadés S. J Mol Biol. 2011;410:798–810. doi: 10.1016/j.jmb.2011.03.042. [DOI] [PubMed] [Google Scholar]
- 65.Wang X, Matei E, Gronenborn AM, Ramström O, Yan M. Anal Chem. 2012;84:4248–4252. doi: 10.1021/ac3006632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang X, Matei E, Deng L, Ramström O, Gronenborn AM, Yan M. Chem Commun. 2011;47:8620–8622. doi: 10.1039/c1cc12981c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang X, Ramström O, Yan M. Anal Chem. 2010;82:9082–9089. doi: 10.1021/ac102114z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Abed N, Couvreur P. Int J Antimicrob Agents. 2014;43:485–96. doi: 10.1016/j.ijantimicag.2014.02.009. [DOI] [PubMed] [Google Scholar]
- 69.Ray PC, Khan SA, Singh AK, Senapati D, Fan Z. Chem Soc Rev. 2012;41:3193. doi: 10.1039/c2cs15340h. [DOI] [PubMed] [Google Scholar]
- 70.Huh AJ, Kwon YJ. J Control Release. 2011;156:128–45. doi: 10.1016/j.jconrel.2011.07.002. [DOI] [PubMed] [Google Scholar]
- 71.Lemire JA, Harrison JJ, Turner RJ. Nat Rev Microbiol. 2013;11:371–84. doi: 10.1038/nrmicro3028. [DOI] [PubMed] [Google Scholar]
- 72.Hajipour MJ, Fromm KM, Ashkarran AA, Jimenez de Aberasturi D, de Larramendi IR, Rojo T, Serpooshan V, Parak WJ, Mahmoudi M. Trends Biotechnol. 2012;30:499–511. doi: 10.1016/j.tibtech.2012.06.004. [DOI] [PubMed] [Google Scholar]
- 73.Eckhardt S, Brunetto PS, Gagnon J, Priebe M, Giese B, Fromm KM. 2013 doi: 10.1021/cr300288v. [DOI] [PubMed] [Google Scholar]
- 74.Veerapandian M, Lim S, Nam H, Kuppannan G, Yun K. Anal Bioanal Chem. 2010;398:867–876. doi: 10.1007/s00216-010-3964-5. [DOI] [PubMed] [Google Scholar]
- 75.Morones JR, Elechiguerra JL, Camacho A, Holt K, Kouri JB, Ramírez JT, Yacaman MJ. Nanotechnology. 2005;16:2346–53. doi: 10.1088/0957-4484/16/10/059. [DOI] [PubMed] [Google Scholar]
- 76.Qi L, Xu Z, Jiang X, Hu C, Zou X. Carbohydr Res. 2004;339:2693–2700. doi: 10.1016/j.carres.2004.09.007. [DOI] [PubMed] [Google Scholar]
- 77.Anisimova Y, Gelperina S, Peloquin C, Heifets L. J Nanoparticle Res. 2000;2:165–171. [Google Scholar]
- 78.Gu H, Ho PL, Tong E, Wang L, Xu B. Nano Lett. 2003;3:1261–1263. [Google Scholar]
- 79.Wei Q, Ji J, Fu J, Shen J. Sci China Ser B Chem. 2007;50:418–424. [Google Scholar]
- 80.Kell AJ, Stewart G, Ryan S, Peytavi R, Boissinot M, Huletsky A, Bergeron MG, Simard B. 2008 doi: 10.1021/nn700183g. [DOI] [PubMed] [Google Scholar]
- 81.Gu H, Ho P-L, Tsang KWT, Wang L, Xu B. J Am Chem Soc. 2003;125:15702–3. doi: 10.1021/ja0359310. [DOI] [PubMed] [Google Scholar]
- 82.Turos E, Reddy GSK, Greenhalgh K, Ramaraju P, Abeylath SC, Jang S, Dickey S, Lim DV. Bioorg Med Chem Lett. 2007;17:3468–72. doi: 10.1016/j.bmcl.2007.03.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Huang D, Li D, Wang T, Shen H, Zhao P, Liu B, You Y, Ma Y, Yang F, Wu D, et al. Biomaterials. 2015;52:417–25. doi: 10.1016/j.biomaterials.2015.02.052. [DOI] [PubMed] [Google Scholar]
- 84.Bresee J, Bond CM, Worthington RJ, Smith CA, Gifford JC, Simpson CA, Carter CJ, Wang G, Hartman J, Osbaugh NA, et al. J Am Chem Soc. 2014;136:5295–300. doi: 10.1021/ja408505n. [DOI] [PubMed] [Google Scholar]
- 85.Jayawardana KW, Jayawardena HSN, Wijesundera Sa, De Zoysa T, Sundhoro M, Yan M. Chem Commun. 2015:1–4. doi: 10.1039/c5cc04251h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Nagy JO, Zhang Y, Yi W, Liu X, Motari E, Song JC, Lejeune JT, Wang PG. Bioorg Med Chem Lett. 2008;18:700–703. doi: 10.1016/j.bmcl.2007.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Jayawardena HSN, Jayawardana KW, Chen X, Yan M. Chem Commun. 2013;49:3034–6. doi: 10.1039/c3cc40491a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kumar CG, Sujitha P. Nanotechnology. 2014;25:325101. doi: 10.1088/0957-4484/25/32/325101. [DOI] [PubMed] [Google Scholar]
- 89.Luo PGJG, Wang HF, Gu LR, Lu FS, Lin Y, Christensen KA, Yang S-TT, Sun Y-PP. ACS Nano. 2009;3:3909–3916. doi: 10.1021/nn901106s. [DOI] [PubMed] [Google Scholar]
- 90.Wang HF, Gu LR, Lin Y, Lu FS, Meziani MJ, Luo PGJ, Wang W, Cao L, Sun YP. J Am Chem Soc. 2006;128:13364–13365. doi: 10.1021/ja065455o. [DOI] [PubMed] [Google Scholar]
- 91.Veerapandian M, Sadhasivam S, Choi J, Yun K. Chem Eng J. 2012;209:558–567. [Google Scholar]
- 92.Gu L, Luo PG, Wang H, Meziani MJ, Lin Y, Veca LM, Cao L, Lu F, Wang X, Quinn RA, et al. Biomacromolecules. 2008;9:2408–2418. doi: 10.1021/bm800395e. [DOI] [PubMed] [Google Scholar]
- 93.Chono S, Tanino T, Seki T, Morimoto K. J Control Release. 2008;127:50–58. doi: 10.1016/j.jconrel.2007.12.011. [DOI] [PubMed] [Google Scholar]
- 94.Zhou J, Butchosa N, Jayawardena HSN, Park J, Zhou Q, Yan M, Ramström O. Biomacromolecules. 2015;16:1426–1432. doi: 10.1021/acs.biomac.5b00227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Richards S-J, Fullam E, Besra GS, Gibson MI. J Mater Chem B. 2014;2:1490. doi: 10.1039/c3tb21821j. [DOI] [PubMed] [Google Scholar]
- 96.Zhou J, Butchosa N, Jayawardena HSN, Zhou Q, Yan M, Ramström O. Bioconjug Chem. 2014;25:640–3. doi: 10.1021/bc500004c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ramtenki V, Raju D, Mehta UJ, Ramana CV, Prasad BLV. New J Chem. 2013;37:3716. [Google Scholar]
- 98.Tseng Y-TT, Chang H-TT, Chen C-HC-TTH, Chen C-HC-TTH, Huang C-CC. Biosens Bioelectron. 2011;27:95–100. doi: 10.1016/j.bios.2011.06.021. [DOI] [PubMed] [Google Scholar]
- 99.El-Boubbou K, Zhu DC, Vasileiou C, Borhan B, Prosperi D, Li W, Huang X. J Am Chem Soc. 2010;132:4490–4499. doi: 10.1021/ja100455c. [DOI] [PubMed] [Google Scholar]
- 100.Huang C-C, Chen C-T, Shiang Y-C, Lin Z-H, Chang H-T. Anal Chem. 2009;81:875–882. doi: 10.1021/ac8010654. [DOI] [PubMed] [Google Scholar]
- 101.Mukhopadhyay B, Martins MB, Karamanska R, Russell DA, Field RA. Tetrahedron Lett. 2009;50:886–889. [Google Scholar]
- 102.Gu L, Elkin T, Jiang X, Li H, Lin Y, Qu L, Tzeng T-RJ, Joseph R, Sun Y-P. Chem Commun. 2005:874–876. doi: 10.1039/b415015e. [DOI] [PubMed] [Google Scholar]
- 103.Ryu J-H, Lee E, Lim Y-b, Lee M. J Am Chem Soc. 2007;129:4808–4814. doi: 10.1021/ja070173p. [DOI] [PubMed] [Google Scholar]
- 104.Lim Y-b, Park S, Lee E, Ryu J-H, Yoon Y-R, Kim T-H, Lee M. Chem Asian J. 2007;2:1363–1369. doi: 10.1002/asia.200700163. [DOI] [PubMed] [Google Scholar]
- 105.Lim Y-b, Lee M. Org Biomol Chem. 2007;5:401–405. doi: 10.1039/b615744k. [DOI] [PubMed] [Google Scholar]
- 106.Qu L, Luo PG, Taylor S, Lin Y, Huang W, Anyadike N, Tzeng T-RJ, Latour RA, Sun Y-P. J Nanosci Nanotechnol. 2005;5:319–322. doi: 10.1166/jnn.2005.043. [DOI] [PubMed] [Google Scholar]
- 107.Qu L, Gu L, Li H, Taylor S, Elkin T, Luo PG, Tzeng T-RJ, Jiang X, Latour RA, Stutzenberger F, et al. J Biomed Nanotechnol. 2005;1:61–67. [Google Scholar]
- 108.Lin C-C, Yeh Y-C, Yang C-Y, Chen C-L, Chen G-F, Chen C-C, Wu Y-C. J Am Chem Soc. 2002;124:3508–3509. doi: 10.1021/ja0200903. [DOI] [PubMed] [Google Scholar]
- 109.Pera NP, Pieters RJ. Medchemcomm. 2014;5:1027. [Google Scholar]
- 110.Fortin JP, Wilhelm C, Servais J, Ménager C, Bacri JC, Gazeau F. J Am Chem Soc. 2007;129:2628–2635. doi: 10.1021/ja067457e. [DOI] [PubMed] [Google Scholar]
- 111.Lonhienne TGA, Sagulenko E, Webb RI, Lee K-C, Franke J, Devos DP, Nouwens A, Carroll BJ, Fuerst JA. Proc Natl Acad Sci U S A. 2010;107:12883–8. doi: 10.1073/pnas.1001085107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Stoimenov PK, Klinger RL, Marchin GL, Klabunde KJ. Langmuir. 2002;18:6679–6686. [Google Scholar]
- 113.Demchick P, Koch A. J Bacteriol. 1996;178:768–773. doi: 10.1128/jb.178.3.768-773.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Koch AL. Clin Microbiol Rev. 2003;16:673–687. doi: 10.1128/CMR.16.4.673-687.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Saier MH, Reddy BL. J Bacteriol. 2015;197:7–17. doi: 10.1128/JB.02046-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Disconzi E, Guilvout I, Chami M, Masi M, Huysmans GHM, Pugsley AP, Bayan N. J Bacteriol. 2014;196:121–128. doi: 10.1128/JB.00750-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Galdiero S, Falanga A, Cantisani M, Tarallo R, Della Pepa ME, D’Oriano V, Galdiero M. Curr Protein Pept Sci. 2012;13:843–54. doi: 10.2174/138920312804871120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Wang H-W, Chen Y, Yang H, Chen X, Duan M-X, Tai PC, Sui S-F. Proc Natl Acad Sci U S A. 2003;100:4221–4226. doi: 10.1073/pnas.0737415100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Chakraborti S, Bhattacharya S, Chowdhury R, Chakrabarti P. PLoS One. 2013;8:e70776. doi: 10.1371/journal.pone.0070776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Kloepfer JA, Mielke RE, Nadeau JL. Appl Environ Microbiol. 2005;71:2548–57. doi: 10.1128/AEM.71.5.2548-2557.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]


