Abstract
This meta-analysis of 23 eligible articles comprehensively and quantitatively evaluated the effects of tumor necrosis factor-α (TNF-α) rs1800629 and rs361525 polymorphisms on sepsis risk. We found that TNF-α rs1800629 was associated with increased sepsis risk in the overall population in four genetic models, including A vs. G (P<0.001, odds ratio (OR)=1.32), GA vs. GG (P<0.001, OR=1.46), GA+AA vs. GG (P<0.001, OR=1.46), and carrier A vs. carrier G (P<0.001, OR=1.32). Subgroup analyses showed a similar result for Asian patients (all P<0.05, OR>1). TNF-α rs361525 was also associated with increased sepsis risk in Asian patients in the four genetic models (all P<0.05, OR>1). Begg’s and Egger’s tests excluded large publication bias, and sensitivity analysis indicated stable results. Our results suggest that the G/A genotype of TNF-α rs1800629 and rs361525 increases sepsis risk in an Asian population.
Keywords: tumor necrosis factor-α, single nucleotide polymorphisms, sepsis, rs1800629, rs361525
INTRODUCTION
Sepsis consumes considerable health care resources and has a high mortality rate, especially in elderly patients and in infants born pre-term or with low birth weights [1, 2]. Lack of early detection is implicated in the high incidences of severe sepsis and septic shock [3, 4]. Thus, sepsis-related biomarkers and risk factors must be identified to improve early detection rates. Recent studies have addressed associations between various gene SNPs (single nucleotide polymorphisms) and sepsis risk. Sepsis risk was associated with TLR4 (toll like receptor 4) SNPs, rs4986790 and rs4986791 [5], but not the SERPINE1 [Serpin Peptidase Inhibitor, Clade E (Nexin, Plasminogen Activator Inhibitor Type 1), Member 1] rs1799768 polymorphism [6].
TNF-α (tumor necrosis factor-α) is important for normal body functions, but is also implicated in some disease mechanisms, including sepsis, diabetes mellitus, and cancer [7-11]. There are several known SNPs within the TNF-α gene, including rs1800629, rs361525, rs1800630, and others [12]. Associations between TNF-α SNPs and sepsis risk are still uncertain. TNF-α rs1800629 was reported as a sepsis risk factor in severely injured North Indian patients [13], critically ill Japanese patients [14], the Chinese Han population [15], and Turkish children [16]. However, TNF-α rs1800629 was also negatively correlated with sepsis susceptibility in preterm infants in Germany [17] and low-birth-weight infants in Hungary [18]. We did not obtain data from genome wide association studies (GWAS) of sepsis-associated SNPs. Thus, our meta-analysis is a relatively objective evaluation of TNF-α SNPs in sepsis risk. Our analysis focused on the genetic relationship between sepsis risk and the rs1800629 and rs361525 polymorphisms within the TNF-α promoter region, in that sufficient data was only obtained for the meta-analysis of rs1800629, rs361525 polymorphisms, after our data extraction.
RESULTS
Eligible studies
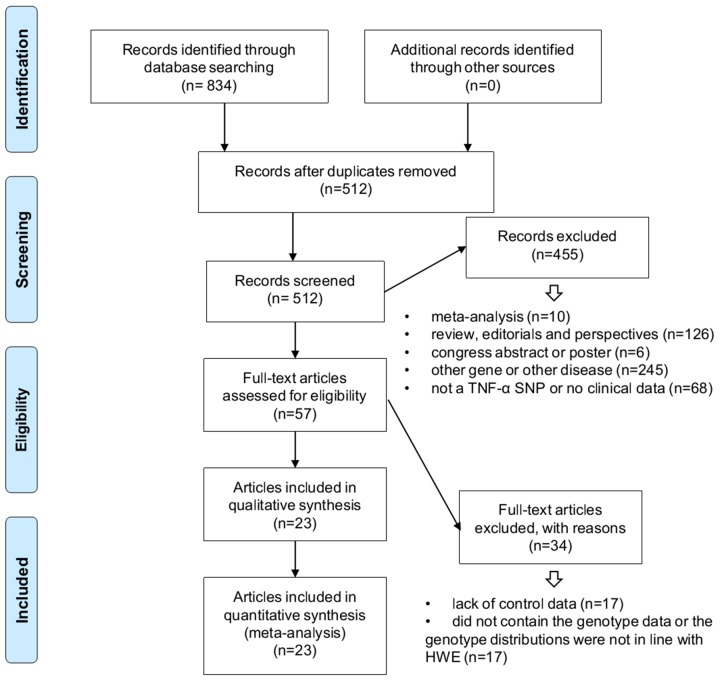
We identified a total of 834 records by searching six online databases, including PubMed, WOS (Web of Science), EMBASE (Excerpta Medica Database), CNKI (China National Knowledge Infrastructure), WANFANG, and Scopus, during April 2017 (Figure 1, Supplementary Table 1). We included 23 articles that fit our inclusion/exclusion criteria in our meta-analysis [13-35]. Case/control group characteristics and genotype frequencies are shown in Table 1 and Supplementary Table 2. The 23 articles included 15 high quality studies (NOS score >6 [14-19, 22, 24, 25, 27, 29, 30, 33-35]) and eight medium quality studies (NOS=5 [21, 23, 28]; NOS=6 [13, 20, 26, 31, 32]).
Figure 1. Records identification and study inclusion.
Table 1. Characteristics of case-control studies included in this meta-analysis.
| First author, year | Ethnicity | SNP | Case | Control | NOS | Genotyping assay |
|---|---|---|---|---|---|---|
| Azevedo, 2012 [19] | Caucasian | rs1800629 | 439 | 564 | 7 | TaqMan “Assay by Design” system |
| Balding, 2003 [20] | Caucasian | rs1800629 | 183 | 389 | 6 | PCR-RFLP |
| Davis, 2010 [21] | Caucasian | rs1800629 | 28 | 53 | 5 | Taqman SNP allele discrimination assay |
| Dou, 2007 [22] | Asian | rs1800629 | 45 | 60 | 9 | PCR-RFLP |
| Duan, 2011 [23] | Asian | rs1800629 | 131 | 174 | 5 | PCR-RFLP |
| Fu, 2016 [24] | Asian | rs1800629 | 115 | 108 | 7 | PCR-RFLP |
| rs361525 | 115 | 108 | 7 | PCR-RFLP | ||
| Gordon, 2004 [25] | Caucasian | rs1800629 | 212 | 354 | 8 | PCR-RFLP |
| rs361525 | 205 | 354 | 8 | End-labeled allele-specific probe hybridisation. | ||
| Gupta, 2015 [13] | Asian | rs1800629 | 25 | 89 | 6 | PCR-SSP |
| rs361525 | 25 | 89 | 6 | PCR-SSP | ||
| Majetschak, 2002 [26] | Caucasian | rs1800629 | 14 | 56 | 6 | Real-time PCR assay with specific fluorescence-labeled hybridization probes |
| Mira, 1999 [27] | Caucasian | rs1800629 | 81 | 78 | 7 | DGGE analysis |
| rs361525 | 59 | 72 | 7 | DGGE analysis | ||
| Nakada, 2005 [14] | Asian | rs1800629 | 86 | 214 | 7 | PCR-RFLP |
| O'Keefe, 2002 [28] | mixed | rs1800629 | 37 | 115 | 5 | Pyrosequencing/PCR-RFLP |
| rs361525 | 37 | 114 | 5 | Pyrosequencing | ||
| Peres, 2012 [29] | Caucasian | rs1800629 | 166 | 214 | 8 | PCR-RFLP |
| Phumeetham, 2012 [30] | Asian | rs1800629 | 66 | 101 | 8 | PCR-RFLP |
| Schaaf, 2003 [31] | Caucasian | rs1800629 | 28 | 50 | 6 | PCR-RFLP |
| Schueller, 2006 [17] | Caucasian | rs1800629 | 67 | 233 | 7 | PCR-RFLP |
| Sipahi, 2006 [16] | Caucasian | rs1800629 | 53 | 77 | 7 | PCR-RFLP |
| Sole, 2010 [32] | Caucasian | rs1800629 | 320 | 1152 | 6 | Rapid cycle real-time PCR |
| rs361525 | 320 | 1172 | 6 | Rapid cycle real-time PCR | ||
| Song, 2012 [15] | Asian | rs1800629 | 802 | 600 | 9 | gene sequencing |
| rs361525 | 803 | 598 | 9 | gene sequencing | ||
| Tian, 2015 [33] | Asian | rs1800629 | 32 | 50 | 9 | PCR-RFLP |
| rs361525 | 32 | 50 | 9 | PCR-RFLP | ||
| Treszl, 2003 [18] | Caucasian | rs1800629 | 33 | 35 | 7 | PCR-RFLP |
| Yu,B, 2003 [34] | Asian | rs1800629 | 40 | 100 | 9 | PCR-RFLP |
| Yu,D, 2007 [35] | Asian | rs1800629 | 56 | 60 | 9 | PCR-RFLP |
SNP: single nucleotide polymorphisms; NOS: Newcastle-Ottawa Scale; PCR-RFLP: polymerase chain reaction-restriction fragment length polymorphism; PCR-SSP: polymerase chain reaction-sequence specific primer; DGGE: denaturing gradient gel electrophoresis.
TNF-α rs1800629 meta-analysis
We enrolled 27 case-control studies with 3,404 cases and 5,973 controls [13-35] in our TNF-α rs1800629 meta-analysis (Table 2). Sepsis risk was increased in the case group in four genetic models: A vs. G (P value from association test <0.001, odds ratio (OR)=1.32, 95% confidence interval (CI) =1.05–1.65); GA vs. GG (P<0.001, OR=1.46, 95% CI=1.19–1.79); GA+AA vs. GG (P<0.001, OR=1.46, 95% CI=1.20–1.78); carrier A vs. carrier G (P<0.001, OR=1.32, 95% CI=1.14–1.54), but not other models (all P>0.05), compared with controls. This suggested that the TNF-α rs1800629 G/A genotype was associated with sepsis risk in the overall population.
Table 2. Genetic relationship between TNF-α rs1800629 and sepsis risk.
| Comparison | Subgroup | Sample size | Association test | |||
|---|---|---|---|---|---|---|
| Studies | Case/control | z | P-value | OR (95% CI) | ||
| A vs. G | overall | 27 | 3,404/5,973 | 3.88 | <0.001 | 1.32 (1.05–1.65) |
| PB | 17 | 2,388/3,003 | 2.68 | 0.007 | 1.41 (1.10–1.80) | |
| HB | 9 | 796/1,818 | 2.56 | 0.010 | 1.44 (1.09–1.91) | |
| Caucasian | 15 | 1,883/4,191 | 1.87 | 0.062 | - | |
| Asian | 11 | 1,484/1,667 | 3.86 | <0.001 | 1.88 (1.36–2.59) | |
| Sepsis | 3 | 431/668 | 0.30 | 0.766 | - | |
| Severe sepsis | 9 | 903/2,934 | 2.20 | 0.027 | 1.55 (1.05–2.29) | |
| Septic shock | 6 | 450/2,203 | 0.60 | 0.549 | - | |
| AA vs. GG | overall | 20 | 3,047/5,320 | 1.89 | 0.058 | - |
| PB | 12 | 2131/2,517 | 1.51 | 0.132 | - | |
| HB | 7 | 696/1,651 | 1.21 | 0.228 | - | |
| Caucasian | 12 | 1,783/4,023 | 0.79 | 0.430 | - | |
| Asian | 7 | 1,227/1,182 | 2.20 | 0.028 | 2.25 (1.09–4.63) | |
| Sepsis | 2 | 403/650 | 1.26 | 0.208 | - | |
| Severe sepsis | 7 | 836/2,801 | 1.13 | 0.257 | - | |
| Septic shock | 6 | 450/2,203 | 0.44 | 0.657 | - | |
| GA vs. GG | overall | 27 | 3,404/5,973 | 3.67 | <0.001 | 1.46 (1.19–1.79) |
| PB | 17 | 2,388/3,003 | 2.34 | 0.019 | 1.42 (1.06–1.89) | |
| HB | 9 | 796/1,818 | 2.84 | 0.005 | 1.57 (1.15–2.15) | |
| Caucasian | 15 | 1,883/4,191 | 1.64 | 0.101 | - | |
| Asian | 11 | 1,484/1,667 | 3.74 | <0.001 | 1.96 (1.38–2.78) | |
| Sepsis | 3 | 431/668 | 1.04 | 0.298 | - | |
| Severe sepsis | 9 | 903/2,934 | 1.89 | 0.059 | - | |
| Septic shock | 6 | 450/2,203 | 0.57 | 0.566 | - | |
| GA+AA vs. GG | overall | 27 | 3,404/5,973 | 3.79 | <0.001 | 1.46 (1.20–1.78) |
| PB | 17 | 2,388/3,003 | 2.52 | 0.012 | 1.44 (1.08–1.91) | |
| HB | 9 | 796/1,818 | 2.70 | 0.007 | 1.55 (1.13–2.12) | |
| Caucasian | 15 | 1,883/4,191 | 1.74 | 0.082 | - | |
| Asian | 11 | 1,484/1,667 | 3.72 | <0.001 | 1.95 (1.37–2.78) | |
| Sepsis | 3 | 431/668 | 0.76 | 0.448 | - | |
| Severe sepsis | 9 | 903/2,934 | 2.06 | 0.039 | 1.55 (1.02–2.34) | |
| Septic shock | 6 | 450/2,203 | 0.57 | 0.572 | - | |
| AA vs. GG+GA | overall | 20 | 3,047/5,320 | 1.52 | 0.128 | - |
| PB | 12 | 2131/2,517 | 1.43 | 0.152 | - | |
| HB | 7 | 696/1,651 | 0.81 | 0.420 | - | |
| Caucasian | 12 | 1,783/4,023 | 0.53 | 0.594 | - | |
| Asian | 7 | 1,227/1,182 | 1.95 | 0.051 | - | |
| Sepsis | 2 | 403/650 | 1.35 | 0.176 | - | |
| Severe sepsis | 7 | 836/2,801 | 1.09 | 0.278 | - | |
| Septic shock | 6 | 450/2,203 | 0.49 | 0.621 | - | |
| carrier A vs. carrier G | overall | 27 | 3,404/5,973 | 3.60 | <0.001 | 1.32 (1.14–1.54) |
| PB | 17 | 2,388/3,003 | 2.41 | 0.016 | 1.33 (1.05–1.67) | |
| HB | 9 | 796/1,818 | 2.60 | 0.009 | 1.35 (1.08–1.70) | |
| Caucasian | 15 | 1,883/4,191 | 1.96 | 0.050 | - | |
| Asian | 11 | 1,484/1,667 | 3.90 | <0.001 | 1.75 (1.32–2.31) | |
| Sepsis | 3 | 431/668 | 0.33 | 0.742 | - | |
| Severe sepsis | 9 | 903/2,934 | 2.60 | 0.009 | 1.30 (1.07–1.58) | |
| Septic shock | 6 | 450/2,203 | 0.11 | 0.909 | - | |
PB: population-based control; HB: hospital-based control; OR: odd ratio; CI: confidence interval; -: ORs (95% CIs) not provided for Passociation >0.05.
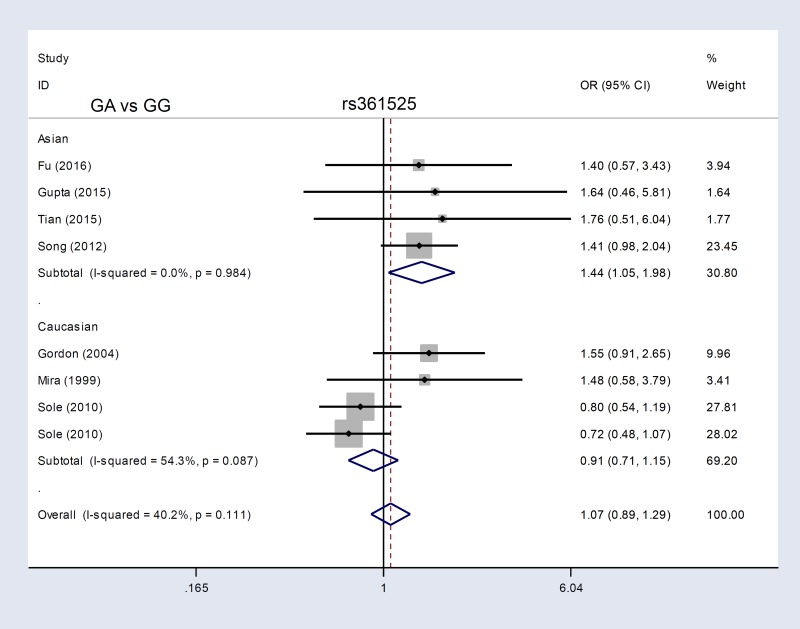
Next, we performed meta-analyses stratified as follows: PB (population-based)/HB (hospital-based), Caucasian/Asian, and sepsis/severe sepsis/septic shock. The PB, HB, and Asian patient groups differed from controls in all four models (A vs. G, GA vs. GG, GA+AA vs. GG, carrier A vs. carrier G) (Table 2, P<0.05, OR>1). These results showed a positive correlation between the TNF-α rs1800629 G/A genotype and sepsis risk in the Asian population. Additionally, in an analysis of eight articles [13, 15, 26-28, 31-33] stratified by sepsis severity, “severe sepsis” cases and controls differed in three models: A vs. G (P=0.027, OR=1.55), GA+AA vs. GG (P=0.039, OR=1.55), and carrier A vs. carrier G (P=0.009, OR=1.30) (Table 2). Figure 2 and Supplementary Figures 1–3 show forest plots of ethnicity subgroup analyses.
Figure 2. TNF-α rs1800629 subgroup analysis based on ethnicity using the GA vs. GG genetic model.
TNF-α rs361525 meta-analysis
We enrolled eight case-control studies containing 1,916 cases and 3,372 controls [13, 15, 24, 25, 27, 28, 32, 33] in our TNF-α rs361525 meta-analysis. Sepsis risk was increased in the AA vs. GG (P=0.001, OR=4.24) and AA vs. GG+GA (P=0.001, OR=4.24) genetic models, but not A vs. G, GA vs. GG, GA+AA vs. GG, or carrier A vs. carrier G (all P>0.05; Table 3 ). Similarly, for subgroup analyses of sepsis severity, increased risk of severe sepsis or septic shock was only observed in the AA vs. GG and AA vs. GG+GA models (Table 3, all P<0.05, OR>1). In contrast, PB subgroup analyses revealed differences in the A vs. G (P=0.001, OR=1.52, 95% CI=1.18–1.97), GA vs. GG (P=0.006, OR=1.46, 95% CI=1.12–1.91), GA+AA vs. GG (P=0.003, OR=1.51, 95% CI=1.15–1.97), and carrier A vs. carrier G (P=0.006, OR=1.45, 95% CI=1.11–1.89) models (Table 3). Asian patient subgroup analyses also showed differences in these four models (all P<0.05, OR>1). Figure 3 and Supplementary Figures 4–6 show forest plots for ethnicity subgroup analyses. These data suggested that the TNF-α rs361525 G/A genotype is associated with enhanced risk of sepsis in the Asian population.
Table 3. Genetic relationship between TNF-α rs361525 and sepsis risk.
| Comparison | Subgroup | Sample size | Association test | |||
|---|---|---|---|---|---|---|
| Studies | Case/control | z | Passociation | OR (95% CI) | ||
| A vs. G | overall | 9 | 1,916/3,372 | 1.82 | 0.069 | - |
| PB | 5 | 1,214/1,182 | 3.20 | 0.001 | 1.52 (1.18–1.97) | |
| HB | 3 | 382/1,018 | 0.56 | 0.574 | - | |
| Caucasian | 4 | 904/2,413 | 0.49 | 0.622 | - | |
| Asian | 4 | 975/845 | 2.68 | 0.007 | 1.52 (1.12–2.05) | |
| Severe sepsis | 5 | 817/2,749 | 0.17 | 0.863 | - | |
| Septic shock | 4 | 404/2,148 | 1.33 | 0.183 | - | |
| AA vs. GG | overall | 6 | 961/2,552 | 3.42 | 0.001 | 4.24 (1.85–9.69) |
| PB | 3 | 296/476 | 1.90 | 0.058 | - | |
| HB | 2 | 345/904 | 1.96 | 0.050 | - | |
| Caucasian | 4 | 904/2,413 | 2.86 | 0.004 | 3.81 (1.52–9.52) | |
| Asian | 2 | 57/139 | 1.89 | 0.059 | - | |
| Severe sepsis | 3 | 352/2,037 | 2.14 | 0.032 | 3.51 (1.11–11.05) | |
| Septic shock | 4 | 404/2,148 | 2.89 | 0.004 | 4.50 (1.62–12.51) | |
| GA vs. GG | overall | 9 | 1,916/3,372 | 0.53 | 0.595 | - |
| PB | 5 | 1,214/1,182 | 2.77 | 0.006 | 1.46 (1.12–1.91) | |
| HB | 3 | 382/1,018 | 1.66 | 0.098 | - | |
| Caucasian | 4 | 904/2,413 | 0.80 | 0.423 | - | |
| Asian | 4 | 975/845 | 2.25 | 0.024 | 1.44 (1.05–1.98) | |
| Severe sepsis | 5 | 817/2,749 | 0.95 | 0.342 | - | |
| Septic shock | 4 | 404/2,148 | 0.02 | 0.985 | - | |
| GA+AA vs. GG | overall | 9 | 1,916/3,372 | 1.17 | 0.243 | - |
| PB | 5 | 1,214/1,182 | 3.01 | 0.003 | 1.51 (1.15–1.97) | |
| HB | 3 | 382/1,018 | 1.14 | 0.254 | - | |
| Caucasian | 4 | 904/2,413 | 0.18 | 0.858 | - | |
| Asian | 4 | 975/845 | 2.49 | 0.013 | 1.49 (1.09–2.05) | |
| Severe sepsis | 5 | 817/2,749 | 0.59 | 0.558 | - | |
| Septic shock | 4 | 404/2,148 | 0.64 | 0.519 | - | |
| AA vs. GG+GA | overall | 6 | 961/2,552 | 3.41 | 0.001 | 4.24(1.85–9.72) |
| PB | 3 | 296/476 | 1.83 | 0.068 | - | |
| HB | 2 | 345/904 | 1.99 | 0.046 | 3.35 (1.02–10.99) | |
| Caucasian | 4 | 904/2,413 | 2.88 | 0.004 | 3.87 (1.54–9.69) | |
| Asian | 2 | 57/139 | 1.82 | 0.069 | - | |
| Severe sepsis | 3 | 352/2,037 | 2.19 | 0.029 | 3.63 (1.14–11.50) | |
| Septic shock | 4 | 404/2,148 | 2.85 | 0.004 | 4.45 (1.59–12.41) | |
| carrier A vs. carrier G | overall | 9 | 1,916/3,372 | 1.14 | 0.254 | - |
| PB | 5 | 1,214/1,182 | 2.73 | 0.006 | 1.45 (1.11–1.89) | |
| HB | 3 | 382/1,018 | 0.95 | 0.342 | - | |
| Caucasian | 4 | 904/2,413 | 0.07 | 0.948 | - | |
| Asian | 4 | 975/845 | 2.27 | 0.023 | 1.44 (1.05–1.97) | |
| Severe sepsis | 5 | 817/2,749 | 0.45 | 0.653 | - | |
| Septic shock | 4 | 404/2,148 | 0.65 | 0.517 | - | |
PB: population-based control; HB: hospital-based control; OR: odd ratio; CI: confidence interval; -: ORs (95% CIs) not provided for Passociation >0.05.
Figure 3. TNF-α rs361525 subgroup analysis based on ethnicity using the GA vs. GG genetic model.
Heterogeneity, publication bias, and sensitivity analysis
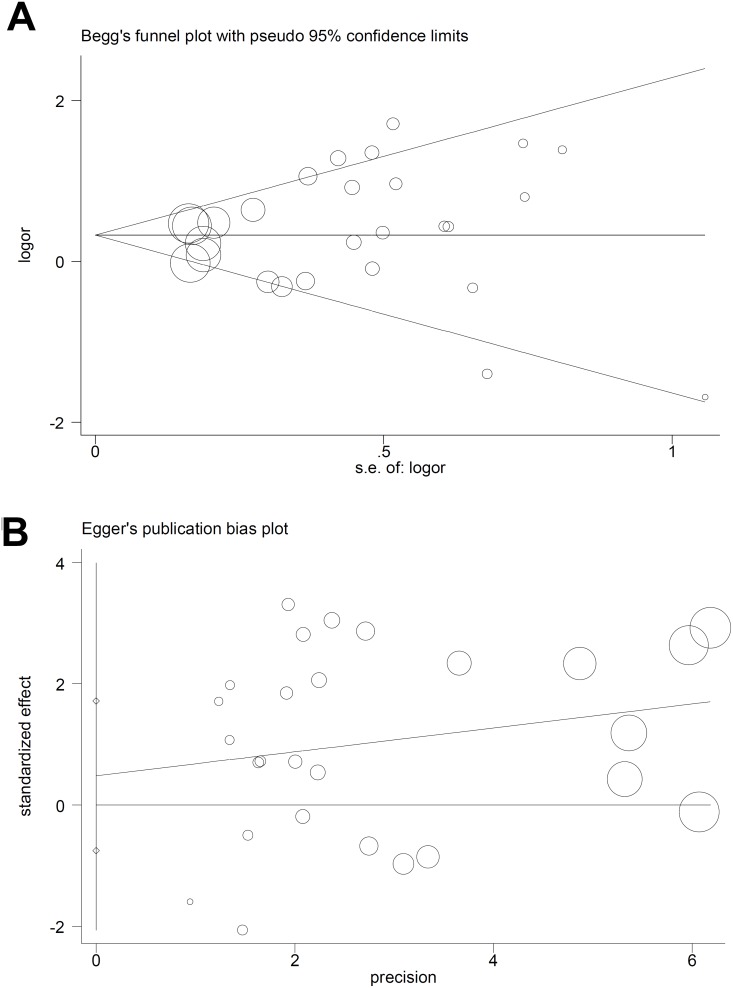
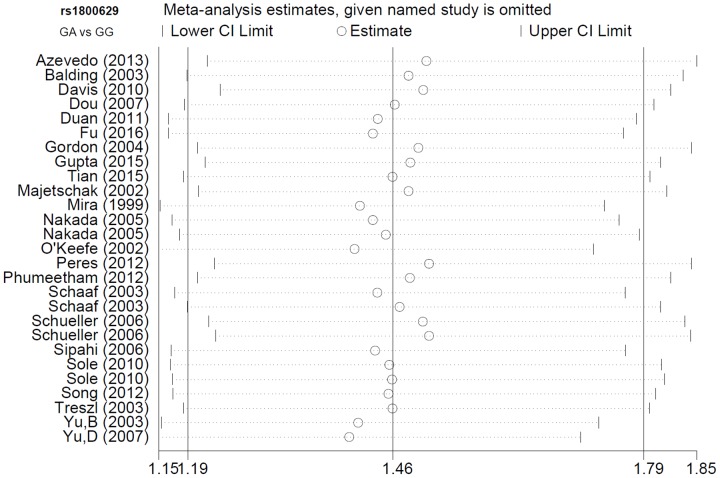
For TNF-α rs1800629, we applied the random-effect model in the allele, heterozygote, dominant, and carrier Mantel-Haenszel analyses, due to the following data (Table 4): A vs. G (I2=55.7%, heterogeneity P<0.001); GA vs. GG (I2=56.9%, P<0.001); GA+AA vs. GG (I2=58.2%, P<0.001); carrier A vs. carrier G (P<0.05). For TNF-α rs361525, the fixed-effected model was used for all comparisons (Table 4, all I2<50.0%, heterogeneity P>0.05). We performed Begg’s test and Egger’s test to evaluate publication bias. We did not observe any large publication bias (P>0.05 in both Begg’s test and Egger’s test), except in the rs1800629 Egger’s test in the AA vs. GG (P=0.042) and AA vs. GG+GA (P=0.041) models (Table 5). Figure 4 shows the Begg’s funnel plot and Egger’s publication bias plot for the GA vs. GG TNF-α rs1800629 meta-analysis. Similar pooled ORs in our sensitivity analysis suggested that our data were reliable (Figure 5 for the GA vs. GG model of TNF-α rs1800629; other data not shown).
Table 4. Heterogeneity evaluation.
| SNP | Comparison | I2 | P-value | Model |
|---|---|---|---|---|
| rs1800629 | A vs. G | 55.7% | <0.001 | Random |
| AA vs. GG | 0.0% | 0.828 | Fixed | |
| GA vs. GG | 56.9% | <0.001 | Random | |
| GA+AA vs. GG | 58.2% | <0.001 | Random | |
| AA vs. GG+GA | 0.0% | 0.892 | Fixed | |
| carrier A vs. carrier G | 37.1% | 0.029 | Random | |
| rs361525 | A vs. G | 43.6% | 0.077 | Fixed |
| AA vs. GG | 0.0% | 0.961 | Fixed | |
| GA vs. GG | 40.8% | 0.095 | Fixed | |
| GA+AA vs. GG | 42.3% | 0.085 | Fixed | |
| AA vs. GG+GA | 0.0% | 0.974 | Fixed | |
| carrier A vs. carrier G | 27.8% | 0.197 | Fixed |
SNP: single nucleotide polymorphisms.
Table 5. Publication bias evaluation.
| SNP | Comparison | Begg’s test | Egger’s test | ||
|---|---|---|---|---|---|
| z | P | t | P | ||
| rs1800629 | A vs. G | 1.17 | 0.243 | 1.68 | 0.106 |
| AA vs. GG | 1.46 | 0.144 | 2.19 | 0.042 | |
| GA vs. GG | 0.54 | 0.588 | 0.80 | 0.431 | |
| GA+AA vs. GG | 0.92 | 0.359 | 1.20 | 0.243 | |
| AA vs. GG+GA | 1.72 | 0.086 | 2.20 | 0.041 | |
| carrier A vs. carrier G | 1.13 | 0.260 | 1.49 | 0.148 | |
| rs361525 | A vs. G | 0.31 | 0.754 | 1.00 | 0.352 |
| AA vs. GG | 0.00 | 1.000 | 1.83 | 0.141 | |
| GA vs. GG | 0.31 | 0.754 | 0.44 | 0.673 | |
| GA+AA vs. GG | -0.10 | 1.000 | 0.76 | 0.469 | |
| AA vs. GG+GA | 0.00 | 1.000 | 1.59 | 0.186 | |
| carrier A vs. carrier G | -0.10 | 1.000 | 0.72 | 0.496 | |
SNP: single nucleotide polymorphisms.
Figure 4. TNF-α rs1800629 publication bias analysis using the GA vs. GG genetic model.
Begg’s funnel plot (A) Egger’s publication bias plot (B).
Figure 5. TNF-α rs1800629 sensitivity analysis using the GA vs. GG genetic model.
DISCUSSION
This updated literature search and meta-analysis comprehensively reassessed the association between TNF-α polymorphisms and sepsis risk in Asian/Caucasian populations. There are several advantages in terms of database searching, screening strategy study inclusion, and sample size. To date, three related meta-analyses have been published [36-38]. Teuffel, et al. performed the first of these in 2010, and reported that the GA or AA TNF-α rs1800629 genotypes were associated with increased sepsis risk [37]. Twenty-five articles [14, 16, 18, 25-28, 31, 39-55] were included in this meta-analysis, however several articles [39-49, 51-55] did not contain sufficient genotype frequency data in the case and/or control groups, or were not in line with Hardy-Weinberg Equilibrium (HWE). Additionally, no or mild sepsis was set as the control group in one included article [40], which may not have been appropriate for our meta-analysis. Another meta-analysis by Srinivasan, et al. only investigated the association between TNF-α rs1800629 and neonatal sepsis risk [36]. After rigorous screening, we enrolled 23 articles with 3,404 cases and 5,973 controls [13-35] in our TNF-α rs1800629 meta-analysis.
Our study included articles from a variety of databases and we performed statistical analyses using six genetic models. Previous meta-analyses were not performed using different genetic models, and the roles of the GA and AA genotypes were thus not evaluated [37]. Another recent meta-analysis by Zhang, et al. assessed 26 articles [13-16, 18, 23-26, 28-32, 34, 41, 44, 47-49, 51, 53, 56-59] and associated both the TNF-α rs1800629 and rs361525 polymorphisms with increased sepsis risk [38]. Here, we included only moderate and high quality articles (NOS score >5) from 834 relevant articles from 2007 that contained complete case/control genotype data (GG, GA, AA). Genotype frequency distributions in controls must have been in line with HWE to be included in our analysis. We excluded three articles [41, 44, 57] inconsistent with HWE, and six [47-49, 51, 53, 59] that did not provide sufficient case/control genotype data. We excluded another article [56] that may have been the source of the high heterogeneity in the TNF-α rs1800629 Asian patient subgroup analysis. Additionally, our study included eight new articles [17, 19-22, 27, 33, 35] that were not assessed by Zhang, et al. [38].
Our stratified analysis of severe sepsis and septic shock included only articles that specified sepsis type. Our subgroup meta-analyses based on controls (HB/PB), showed that the TNF-α rs1800629 G/A genotype was linked to increased sepsis risk in both groups. However, TNF-α rs361525 and sepsis risk were positively correlated in the PB, but not HB group. Thus, the presence of other diseases may influence the genetic role of TNF-α rs361525, but not TNF- rs1800629 in sepsis risk.
Our study was subject to certain limitations. First, more studies with larger sample sizes and high qualities are needed for enhanced statistical power. We only observed the potential association between TNF-α rs1800629 and severe sepsis risk for the allele, dominant, and carrier models. TNF-α rs361525 was also found be slightly linked to the risk of severe sepsis or septic shock for the homozygote and recessive models. Thus, our lack of strong evidence for associations between the two SNPs and sepsis risk merits more case-control studies. Second, we found high heterogeneity between studies in some genetic comparisons. This may be caused at least in part by the complexity of the sepsis etiology and the non-uniformity of diagnostic criteria. Finally, more data are needed to clarify the genetic roles and prognostic significance of distinct cytokine gene combinations in sepsis risk. Different SNP linkages of the TNF-α gene should be considered as well.
In conclusion, our findings suggest a positive association between the G/A genotype of the TNF-α rs1800629 and rs361525 polymorphisms and sepsis risk in the Asian population, which is partly in line with the findings of Teuffel, et al. [37] and Zhang, et al. [38]. Abnormal TNF-α is implicated in the pathogenesis of sepsis. For example, TNF-α expression was closely related to neonatal sepsis in very low birth weight infants in Spain [60]. It may be that mutation in the TNF-α promoter region from common G (guanidine) to rare A (adenosine) at position -308 (rs1800629) or -238 (rs361525) affects normal TNF-α production, secretion, or function in sepsis patients [8].
MATERIALS AND METHODS
Records identification
We identified potential records from six databases (PubMed, WOS, EMBASE, CNKI, WANFANG, and Scopus). We performed a PRISMA (preferred reporting items for systematic reviews and meta-analyses [61])-compliant database search and study selection, and the relevant meta-analysis papers were referred [62-64]. Two authors (Yixin Zhang, Xiaoteng Cui) independently removed duplicate studies and assessed record eligibilities according to our exclusion/inclusion criteria. Exclusion criteria included: a) meta-analysis; b) review, editorials, and perspectives; c) congress abstract or poster; d) other gene or other disease; e) not a TNF-α SNP or no clinical data; f) lack of control data; g) did not contain genotype data or the genotype distributions were not in line with HWE.
According to Newcastle-Ottawa Scale (NOS) requirements (http://www.ohri.ca/programs/clinical_epidemiology/oxford.asp), we evaluated methodological quality independently. Included studies had a NOS score >5, and provide the following data: first author name, publication year, patient ethnicity, SNP, sample size, and genotype frequency within case-control studies and genotyping assays.
Quantitative synthesis and heterogeneity
We used the Mantel-Haenszel test to determine P-values, pooled ORs, and 95% CIs for the following six genetic models: A vs. G (allele); AA vs. GG (homozygote); GA vs. GG (heterozygote); GA+AA vs. GG (dominant); AA vs. GG+GA (recessive); and carrier A vs. carrier G (carrier). P<0.05 represented a statistically significant difference between case and control studies. We also assessed the heterogeneity between studies using the Q statistic and I2 test. High heterogeneity was likely when Q statistic P<0.05 or I2>50%. In this situation, we employed the random-effect model, not the fixed-effect model. We also performed a series of subgroup meta-analyses according to three factors: source of control [PB (population-based) or HB (hospital-based)], ethnicity (Caucasian or Asian), and sepsis severity (sepsis, severe sepsis, or septic shock).
Publication bias and sensitivity analysis
We evaluated publication bias using both Begg’s test and Egger’s test. P>0.05 in both tests would exclude a large publication bias. We also performed a sensitivity analysis to evaluate the robustness of our data. All analyses were performed using Stata/SE 12.0 software (StataCorp, USA).
SUPPLEMENTARY MATERIALS FIGURES AND TABLES
Footnotes
CONFLICTS OF INTEREST
The authors declare no conflicts of interest.
REFERENCES
- 1.Martin S, Perez A, Aldecoa C. Sepsis and immunosenescence in the elderly patient: a review. Front Med (Lausanne) 2017;4:20. doi: 10.3389/fmed.2017.00020. https://doi.org/10.3389/fmed.2017.00020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Perner A, Rhodes A, Venkatesh B, Angus DC, Martin-Loeches I, Preiser JC, Vincent JL, Marshall J, Reinhart K, Joannidis M, Opal SM. Sepsis: frontiers in supportive care, organisation and research. Intensive Care Med. 2017;43:496–508. doi: 10.1007/s00134-017-4677-4. https://doi.org/10.1007/s00134-017-4677-4. [DOI] [PubMed] [Google Scholar]
- 3.Ludwig KR, Hummon AB. Mass spectrometry for the discovery of biomarkers of sepsis. Mol Biosyst. 2017;13:648–64. doi: 10.1039/c6mb00656f. https://doi.org/10.1039/c6mb00656f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hotchkiss RS, Moldawer LL, Opal SM, Reinhart K, Turnbull IR, Vincent JL. Sepsis and septic shock. Nat Rev Dis Primers. 2016;2:16045. doi: 10.1038/nrdp.2016.45. https://doi.org/10.1038/nrdp.2016.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu R, Mo YY, Wang HL, Tan Y, Wen XJ, Deng MJ, Yan H, Li L. The relationship between toll like receptor 4 gene rs4986790 and rs4986791 polymorphisms and sepsis susceptibility: a meta-analysis. Sci Rep. 2016;6:38947. doi: 10.1038/srep38947. https://doi.org/10.1038/srep38947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shi Q, Mu X, Hong L, Zheng S. SERPINE1 rs1799768 polymorphism contributes to sepsis risk and mortality. J Renin Angiotensin Aldosterone Syst. 2015;16:1218–24. doi: 10.1177/1470320315614714. https://doi.org/10.1177/1470320315614714. [DOI] [PubMed] [Google Scholar]
- 7.Qiao YC, Chen YL, Pan YH, Tian F, Xu Y, Zhang XX, Zhao HL. The change of serum tumor necrosis factor alpha in patients with type 1 diabetes mellitus: a systematic review and meta-analysis. PLoS One. 2017;12:e0176157. doi: 10.1371/journal.pone.0176157. https://doi.org/10.1371/journal.pone.0176157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.El-Tahan RR, Ghoneim AM, El-Mashad N. TNF-alpha gene polymorphisms and expression. Springerplus. 2016;5:1508. doi: 10.1186/s40064-016-3197-y. https://doi.org/10.1186/s40064-016-3197-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lv B, Huang J, Yuan H, Yan W, Hu G. Tumor necrosis factor-alpha as a diagnostic marker for neonatal sepsis: a meta-analysis. 2014;2014:471463. doi: 10.1155/2014/471463. https://doi.org/10.1155/2014/471463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Patel HJ, Patel BM. TNF-alpha and cancer cachexia: molecular insights and clinical implications. Life Sci. 2017;170:56–63. doi: 10.1016/j.lfs.2016.11.033. https://doi.org/10.1016/j.lfs.2016.11.033. [DOI] [PubMed] [Google Scholar]
- 11.Kali A. TNFerade, an innovative cancer immunotherapeutic. Indian J Pharmacol. 2015;47:479–83. doi: 10.4103/0253-7613.165190. https://doi.org/10.4103/0253-7613.165190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Flori L, Delahaye NF, Iraqi FA, Hernandez-Valladares M, Fumoux F, Rihet P. TNF as a malaria candidate gene: polymorphism-screening and family-based association analysis of mild malaria attack and parasitemia in Burkina Faso. Genes Immun. 2005;6:472–80. doi: 10.1038/sj.gene.6364231. https://doi.org/10.1038/sj.gene.6364231. [DOI] [PubMed] [Google Scholar]
- 13.Gupta DL, Nagar PK, Kamal VK, Bhoi S, Rao DN. Clinical relevance of single nucleotide polymorphisms within the 13 cytokine genes in North Indian trauma hemorrhagic shock patients. Scand J Trauma Resusc Emerg Med. 2015;23:96. doi: 10.1186/s13049-015-0174-3. https://doi.org/10.1186/s13049-015-0174-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nakada TA, Hirasawa H, Oda S, Shiga H, Matsuda K, Nakamura M, Watanabe E, Abe R, Hatano M, Tokuhisa T. Influence of toll-like receptor 4, CD14, tumor necrosis factor, and interleukine-10 gene polymorphisms on clinical outcome in Japanese critically ill patients. J Surg Res. 2005;129:322–8. doi: 10.1016/j.jss.2005.05.020. https://doi.org/10.1016/j.jss.2005.05.020. [DOI] [PubMed] [Google Scholar]
- 15.Song Z, Song Y, Yin J, Shen Y, Yao C, Sun Z, Jiang J, Zhu D, Zhang Y, Shen Q, Gao L, Tong C, Bai C. Genetic variation in the TNF gene is associated with susceptibility to severe sepsis, but not with mortality. PLoS One. 2012;7:e46113. doi: 10.1371/journal.pone.0046113. https://doi.org/10.1371/journal.pone.0046113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sipahi T, Pocan H, Akar N. Effect of various genetic polymorphisms on the incidence and outcome of severe sepsis. Clin Appl Thromb Hemost. 2006;12:47–54. doi: 10.1177/107602960601200108. [DOI] [PubMed] [Google Scholar]
- 17.Schueller AC, Heep A, Kattner E, Kroll M, Wisbauer M, Sander J, Bartmann P, Stuber F. Prevalence of two tumor necrosis factor gene polymorphisms in premature infants with early onset sepsis. Biol Neonate. 2006;90:229–32. doi: 10.1159/000093605. https://doi.org/10.1159/000093605. [DOI] [PubMed] [Google Scholar]
- 18.Treszl A, Kocsis I, Szathmari M, Schuler A, Heninger E, Tulassay T, Vasarhelyi B. Genetic variants of TNF-[FC12]a, IL-1beta, IL-4 receptor [FC12]a-chain, IL-6 and IL-10 genes are not risk factors for sepsis in low-birth-weight infants. Biol Neonate. 2003;83:241–5. doi: 10.1159/000069484. [DOI] [PubMed] [Google Scholar]
- 19.Azevedo ZM, Moore DB, Lima FC, Cardoso CC, Bougleux R, Matos GI, Luz RA, Xavier-Elsas P, Sampaio EP, Gaspar-Elsas MI, Moraes MO. Tumor necrosis factor (TNF) and lymphotoxin-alpha (LTA) single nucleotide polymorphisms: importance in ARDS in septic pediatric critically ill patients. Hum Immunol. 2012;73:661–7. doi: 10.1016/j.humimm.2012.03.007. https://doi.org/10.1016/j.humimm.2012.03.007. [DOI] [PubMed] [Google Scholar]
- 20.Balding J, Healy CM, Livingstone WJ, White B, Mynett-Johnson L, Cafferkey M, Smith OP. Genomic polymorphic profiles in an Irish population with meningococcaemia: is it possible to predict severity and outcome of disease? Genes Immun. 2003;4:533–40. doi: 10.1038/sj.gene.6364020. https://doi.org/10.1038/sj.gene.6364020. [DOI] [PubMed] [Google Scholar]
- 21.Davis SM, Clark EA, Nelson LT, Silver RM. The association of innate immune response gene polymorphisms and puerperal group A streptococcal sepsis. Am J Obstet Gynecol. 2010;202:308.e1–8. doi: 10.1016/j.ajog.2010.01.006. https://doi.org/10.1016/j.ajog.2010.01.006. [DOI] [PubMed] [Google Scholar]
- 22.Dou L, Cao S, Wang Y, Wang J. Study on the relationships between tumor necrosis factor polymorphisms and sepsis susceptibility and factor productivity. Tianjin Med J. 2007;35:820–2. [Google Scholar]
- 23.Duan ZX, Gu W, Zhang LY, Jiang DP, Zhou J, Du DY, Zen L, Chen KH, Liu Q, Jiang JX. Tumor necrosis factor alpha gene polymorphism is associated with the outcome of trauma patients in Chinese Han population. J Trauma. 2011;70:954–8. doi: 10.1097/TA.0b013e3181e88adf. https://doi.org/10.1097/TA.0b013e3181e88adf. [DOI] [PubMed] [Google Scholar]
- 24.Fu Y, Chen Y, Bai N, Liu R, Li D. Correlation of TNF-alpha gene polymorphisms with sepsis susceptibility. Int J Clin Exp Pathol. 2016;9:2335–9. [Google Scholar]
- 25.Gordon AC, Lagan AL, Aganna E, Cheung L, Peters CJ, McDermott MF, Millo JL, Welsh KI, Holloway P, Hitman GA, Piper RD, Garrard CS, Hinds CJ. TNF and TNFR polymorphisms in severe sepsis and septic shock: a prospective multicentre study. Genes Immun. 2004;5:631–40. doi: 10.1038/sj.gene.6364136. https://doi.org/10.1038/sj.gene.6364136. [DOI] [PubMed] [Google Scholar]
- 26.Majetschak M, Obertacke U, Schade FU, Bardenheuer M, Voggenreiter G, Bloemeke B, Heesen M. Tumor necrosis factor gene polymorphisms, leukocyte function, and sepsis susceptibility in blunt trauma patients. Clin Diagn Lab Immunol. 2002;9:1205–11. doi: 10.1128/CDLI.9.6.1205-1211.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mira JP, Cariou A, Grall F, Delclaux C, Losser MR, Heshmati F, Cheval C, Monchi M, Teboul JL, Riche F, Leleu G, Arbibe L, Mignon A, et al. Association of TNF2, a TNF-alpha promoter polymorphism, with septic shock susceptibility and mortality: a multicenter study. JAMA. 1999;282:561–8. doi: 10.1001/jama.282.6.561. [DOI] [PubMed] [Google Scholar]
- 28.O'Keefe GE, Hybki DL, Munford RS. The G-->A single nucleotide polymorphism at the -308 position in the tumor necrosis factor-alpha promoter increases the risk for severe sepsis after trauma. J Trauma. 2002;52:817–25. doi: 10.1097/00005373-200205000-00001. discussion 25–6. [DOI] [PubMed] [Google Scholar]
- 29.Peres Wingeyer SD, Cunto ER, Nogueras CM, San Juan JA, Gomez N, de Larranaga GF. Biomarkers in sepsis at time zero: intensive care unit scores, plasma measurements and polymorphisms in Argentina. J Infect Dev Ctries. 2012;6:555–62. doi: 10.3855/jidc.2108. [DOI] [PubMed] [Google Scholar]
- 30.Phumeetham S, Chat-Uthai N, Manavathongchai M, Viprakasit V. Genetic association study of tumor necrosis factor-alpha with sepsis and septic shock in Thai pediatric patients. J Pediatr (Rio J) 2012;88:417–22. doi: 10.2223/JPED.2216. https://doi.org/doi:10.2223/JPED.2216. [DOI] [PubMed] [Google Scholar]
- 31.Schaaf BM, Boehmke F, Esnaashari H, Seitzer U, Kothe H, Maass M, Zabel P, Dalhoff K. Pneumococcal septic shock is associated with the interleukin-10-1082 gene promoter polymorphism. Am J Respir Crit Care Med. 2003;168:476–80. doi: 10.1164/rccm.200210-1164OC. https://doi.org/10.1164/rccm.200210-1164OC. [DOI] [PubMed] [Google Scholar]
- 32.Sole-Violan J, de Castro F, Garcia-Laorden MI, Blanquer J, Aspa J, Borderias L, Briones ML, Rajas O, Carrondo IM, Marcos-Ramos JA, Ferrer Aguero JM, Garcia-Saavedra A, Fiuza MD, et al. Genetic variability in the severity and outcome of community-acquired pneumonia. Respir Med. 2010;104:440–7. doi: 10.1016/j.rmed.2009.10.009. https://doi.org/10.1016/j.rmed.2009.10.009. [DOI] [PubMed] [Google Scholar]
- 33.Tian H, Wei D, He X. Relationships of TNF-α gene polymorphism with susceptibility to sepsis and its infections degrees. Shandong Med J. 2015;55:17–9. [Google Scholar]
- 34.Yu BJ, Li JS, Zhang DL, Tang XM. The associations of the single nucleotide polymorphisms on TNF and CD14 promoters with the mortality of infection, systematic inflammatory response syndromec and sepsis in surgical patients. Zhonghua Yi Xue Za Zhi. 2003;83:2132–6. [PubMed] [Google Scholar]
- 35.Yu DX, Wang KP, Yang J. Association between gene polymorphisms of tumor necrosis factor and the susceptibility and prognosis of severe sepsis. Chin J Crit Care Med. 2007;27:221–4. [Google Scholar]
- 36.Srinivasan L, Swarr DT, Sharma M, Cotten CM, Kirpalani H. Systematic review and meta-analysis: gene association studies in neonatal sepsis. Am J Perinatol. 2017;34:684–92. doi: 10.1055/s-0036-1597132. https://doi.org/10.1055/s-0036-1597132. [DOI] [PubMed] [Google Scholar]
- 37.Teuffel O, Ethier MC, Beyene J, Sung L. Association between tumor necrosis factor-alpha promoter -308 A/G polymorphism and susceptibility to sepsis and sepsis mortality: a systematic review and meta-analysis. Crit Care Med. 2010;38:276–82. doi: 10.1097/CCM.0b013e3181b42af0. https://doi.org/10.1097/CCM.0b013e3181b42af0. [DOI] [PubMed] [Google Scholar]
- 38.Zhang M, Zhao Y, Liu Q. Tumor necrosis factor-alpha -308G/A and -238G/A polymorphisms are associated with increased risks of sepsis: evidence from an updated meta-analysis. APMIS. 2017 doi: 10.1111/apm.12661. https://doi.org/10.1111/apm.12661. [DOI] [PubMed] [Google Scholar]
- 39.Appoloni O, Dupont E, Vandercruys M, Andriens M, Duchateau J, Vincent JL. Association of tumor necrosis factor-2 allele with plasma tumor necrosis factor-alpha levels and mortality from septic shock. Am J Med. 2001;110:486–8. doi: 10.1016/s0002-9343(01)00656-8. [DOI] [PubMed] [Google Scholar]
- 40.Barber RC, Aragaki CC, Rivera-Chavez FA, Purdue GF, Hunt JL, Horton JW. TLR4 and TNF-alpha polymorphisms are associated with an increased risk for severe sepsis following burn injury. J Med Genet. 2004;41:808–13. doi: 10.1136/jmg.2004.021600. https://doi.org/10.1136/jmg.2004.021600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Calvano JE, Um JY, Agnese DM, Hahm SJ, Kumar A, Coyle SM, Calvano SE, Lowry SF. Influence of the TNF-alpha and TNF-beta polymorphisms upon infectious risk and outcome in surgical intensive care patients. Surg Infect (Larchmt) 2003;4:163–9. doi: 10.1089/109629603766956951. https://doi.org/10.1089/109629603766956951. [DOI] [PubMed] [Google Scholar]
- 42.Dumon K, Rossbach C, Harms B, Gorelov V, Gross-Weege W, Schneider EM, Goretzki PE, Roher HD. [Tumor necrosis factor-alpha (TNF-alpha) gene polymorphism in surgical intensive care patients with SIRS]. [Article in German] Langenbecks Arch Chir Suppl Kongressbd. 1998;115:387–90. [PubMed] [Google Scholar]
- 43.Gallagher PM, Lowe G, Fitzgerald T, Bella A, Greene CM, McElvaney NG, O'Neill SJ. Association of IL-10 polymorphism with severity of illness in community acquired pneumonia. Thorax. 2003;58:154–6. doi: 10.1136/thorax.58.2.154. https://doi.org/10.1136/thorax.58.2.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Garnacho-Montero J, Aldabo-Pallas T, Garnacho-Montero C, Cayuela A, Jimenez R, Barroso S, Ortiz-Leyba C. Timing of adequate antibiotic therapy is a greater determinant of outcome than are TNF and IL-10 polymorphisms in patients with sepsis. Crit Care. 2006;10:R111. doi: 10.1186/cc4995. https://doi.org/10.1186/cc4995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jaber BL, Rao M, Guo D, Balakrishnan VS, Perianayagam MC, Freeman RB, Pereira BJ. Cytokine gene promoter polymorphisms and mortality in acute renal failure. Cytokine. 2004;25:212–9. doi: 10.1016/j.cyto.2003.11.004. https://doi.org/10.1016/j.cyto.2003.11.004. [DOI] [PubMed] [Google Scholar]
- 46.Jessen KM, Lindboe SB, Petersen AL, Eugen-Olsen J, Benfield T. Common TNF-alpha, IL-1 beta, PAI-1, uPA, CD14 and TLR4 polymorphisms are not associated with disease severity or outcome from Gram negative sepsis. BMC Infect Dis. 2007;7:108. doi: 10.1186/1471-2334-7-108. https://doi.org/10.1186/1471-2334-7-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McDaniel DO, Hamilton J, Brock M, May W, Calcote L, Tee LY, Vick L, Newman DB, Vick K, Harrison S, Timberlake G, Toevs C. Molecular analysis of inflammatory markers in trauma patients at risk of postinjury complications. J Trauma. 2007;63:147–57. doi: 10.1097/TA.0b013e31806bf0ab. discussion 57-8https://doi.org/10.1097/TA.0b013e31806bf0ab. [DOI] [PubMed] [Google Scholar]
- 48.Menges T, Konig IR, Hossain H, Little S, Tchatalbachev S, Thierer F, Hackstein H, Franjkovic I, Colaris T, Martens F, Weismuller K, Langefeld T, Stricker J, et al. Sepsis syndrome and death in trauma patients are associated with variation in the gene encoding tumor necrosis factor. Crit Care Med. 2008;36:1456–62, e1-6. doi: 10.1097/CCM.0B013E318170ABB6. https://doi.org/10.1097/ccm.0b013e318170abb6. [DOI] [PubMed] [Google Scholar]
- 49.Nuntayanuwat S, Dharakul T, Chaowagul W, Songsivilai S. Polymorphism in the promoter region of tumor necrosis factor-alpha gene is associated with severe meliodosis. Hum Immunol. 1999;60:979–83. doi: 10.1016/s0198-8859(99)00073-7. [DOI] [PubMed] [Google Scholar]
- 50.Stuber F, Udalova IA, Book M, Drutskaya LN, Kuprash DV, Turetskaya RL, Schade FU, Nedospasov SA. -308 tumor necrosis factor (TNF) polymorphism is not associated with survival in severe sepsis and is unrelated to lipopolysaccharide inducibility of the human TNF promoter. J Inflamm. 1995;46:42–50. [PubMed] [Google Scholar]
- 51.Tang GJ, Huang SL, Yien HW, Chen WS, Chi CW, Wu CW, Lui WY, Chiu JH, Lee TY. Tumor necrosis factor gene polymorphism and septic shock in surgical infection. Crit Care Med. 2000;28:2733–6. doi: 10.1097/00003246-200008000-00008. [DOI] [PubMed] [Google Scholar]
- 52.Watanabe E, Hirasawa H, Oda S, Shiga H, Matsuda K, Nakamura M, Abe R, Nakada T. Cytokine-related genotypic differences in peak interleukin-6 blood levels of patients with SIRS and septic complications. J Trauma. 2005;59:1181–9. doi: 10.1097/00005373-200511000-00025. discussion 9–90. [DOI] [PubMed] [Google Scholar]
- 53.Waterer GW, Quasney MW, Cantor RM, Wunderink RG. Septic shock and respiratory failure in community-acquired pneumonia have different TNF polymorphism associations. Am J Respir Crit Care Med. 2001;163:1599–604. doi: 10.1164/ajrccm.163.7.2011088. https://doi.org/10.1164/ajrccm.163.7.2011088. [DOI] [PubMed] [Google Scholar]
- 54.Zhang D, Li J, Jiang ZW, Yu B, Tang X. Association of two polymorphisms of tumor necrosis factor gene with acute severe pancreatitis. J Surg Res. 2003;112:138–43. doi: 10.1016/s0022-4804(03)00085-4. [DOI] [PubMed] [Google Scholar]
- 55.Zhang DL, Li JS, Jiang ZW, Yu BJ, Tang XM, Zheng HM. Association of two polymorphisms of tumor necrosis factor gene with acute biliary pancreatitis. World J Gastroenterol. 2003;9:824–8. doi: 10.3748/wjg.v9.i4.824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Allam G, Alsulaimani AA, Alzaharani AK, Nasr A. Neonatal infections in Saudi Arabia: association with cytokine gene polymorphisms. Cent Eur J Immunol. 2015;40:68–77. doi: 10.5114/ceji.2015.50836. https://doi.org/10.5114/ceji.2015.50836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Baghel K, Srivastava RN, Chandra A, Goel SK, Agrawal J, Kazmi HR, Raj S. TNF-alpha, IL-6, and IL-8 cytokines and their association with TNF-alpha-308 G/A polymorphism and postoperative sepsis. J Gastrointest Surg. 2014;18:1486–94. doi: 10.1007/s11605-014-2574-5. https://doi.org/10.1007/s11605-014-2574-5. [DOI] [PubMed] [Google Scholar]
- 58.Kothari N, Bogra J, Abbas H, Kohli M, Malik A, Kothari D, Srivastava S, Singh PK. Tumor necrosis factor gene polymorphism results in high TNF level in sepsis and septic shock. Cytokine. 2013;61:676–81. doi: 10.1016/j.cyto.2012.11.016. https://doi.org/10.1016/j.cyto.2012.11.016. [DOI] [PubMed] [Google Scholar]
- 59.Paskulin DD, Fallavena PR, Paludo FJ, Borges TJ, Picanço JB, Dias FS, Alho CS. TNF -308G > A promoter polymorphism (rs1800629) and outcome from critical illness. Braz J Infect Dis. 2011;15:231–8. doi: 10.1016/s1413-8670(11)70181-7. https://doi.org/10.1016/S1413-8670(11)70181-7. [DOI] [PubMed] [Google Scholar]
- 60.Cernada M, Serna E, Bauerl C, Collado MC, Perez-Martinez G, Vento M. Genome-wide expression profiles in very low birth weight infants with neonatal sepsis. Pediatrics. 2014;133:e1203–11. doi: 10.1542/peds.2013-2552. https://doi.org/10.1542/peds.2013-2552. [DOI] [PubMed] [Google Scholar]
- 61.Moher D, Liberati A, Tetzlaff J, Altman DG. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS Med. 2009;6:e1000097. doi: 10.1371/journal.pmed.1000097. https://doi.org/10.1371/journal.pmed.1000097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Su WH, Shi ZH, Liu SL, Wang XD, Liu S, Ji Y. Updated meta-analysis of the role of APOE epsilon2/epsilon3/epsilon4 alleles in frontotemporal lobar degeneration. Oncotarget. 2017;8:43721–32. doi: 10.18632/oncotarget.17341. https://doi.org/10.18632/oncotarget.17341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kim HS, Jeong CW, Kwak C, Kim HH, Ku JH. Association between demographic factors and prognosis in urothelial carcinoma of the upper urinary tract: a systematic review and meta-analysis. Oncotarget. 2017;8:7464–76. doi: 10.18632/oncotarget.10708. https://doi.org/10.18632/oncotarget.10708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhai Y, Dai Z, He H, Gao F, Yang L, Dong Y, Lu J. A PRISMA-compliant meta-analysis of MDM4 genetic variants and cancer susceptibility. Oncotarget. 2016;7:73935–44. doi: 10.18632/oncotarget.12558. https://doi.org/10.18632/oncotarget.12558. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.