Abstract
The transcription factor Sox2 controls the fate of pluripotent stem cells and neural stem cells. This gatekeeper function requires well-regulated Sox2 levels. We postulated that Sox2 regulation is partially controlled by the Sox2 overlapping long non-coding RNA (lncRNA) gene Sox2ot. Here we show that the RNA levels of Sox2ot and Sox2 are inversely correlated during neural differentiation of mouse embryonic stem cells (ESCs). Through allele-specific enhanced transcription of Sox2ot in mouse Sox2eGFP knockin ESCs we demonstrate that increased Sox2ot transcriptional activity reduces Sox2 RNA levels in an allele-specific manner. Enhanced Sox2ot transcription, yielding lower Sox2 RNA levels, correlates with a decreased chromatin interaction of the upstream regulatory sequence of Sox2 and the ESC-specific Sox2 super enhancer. Our study indicates that, in addition to previously reported in trans mechanisms, Sox2ot can regulate Sox2 by an allele-specific mechanism, in particular during development.
Introduction
Correct gene regulation, which relies on the temporally and spatially controlled expression of lineage specific transcription factors, determines the success of development. Sox2 is such a transcription factor key to development. Sox2 belongs to the family of high mobility group (HMG) DNA binding domain genes related to the sex determining gene Y (Sry) and together with Sox1 and Sox3, Sox2 forms the SoxB1 family. Sox2 exerts its cell type specific function by interaction with other homeodomain transcription factors, the POU domain protein Oct4, or the paired domain protein Pax61. An important function of Sox2 is maintaining the stem cell state of either naïve or primed pluripotent stem cells2. Reduction or overexpression of Sox2 in mouse and human embryonic stem cells (ESCs) induces the differentiation into primarily endoderm and trophoectoderm-like cells, respectively3–8. Endogenous Sox2 levels also influence the germ layer fate of pluripotent stem cells. High endogenous levels steer pluripotent cells into the (neural) ectodermal lineage, whereas low levels promote mesendodermal differentiation9. Sox2 fulfills a similar role in neural stem cells (NSCs) in vitro and in vivo. Overexpression of Sox2 in NSCs of the developing spinal cord represses differentiation by counteracting transcription factor driven proneural programs, whereas Sox2 protein inhibition enhances differentiation10,11. In the developing eye, retinal progenitor cells lose their proliferation and differentiation capacity after Sox2 ablation12. Reduced Sox2 levels (<40%) cause microphthalmia due to aberrant differentiation of the progenitor cells12. In addition, misexpression of Sox2 in astrocytes converts them into neuroblasts13, whereas it activates neural transcription programs in cells of mesodermal origin14,15. Thus, well-controlled and tightly-timed Sox2 activity appears to be important for correct neural development.
Sox2 activity is controlled by post-translational modifications, such as serine- and threonine phosphorylation, sumoylation, ubiquitination, and acytelation16. These modifications affect localization, DNA binding and stability. However, Sox2 activity is to a great extent controlled at the transcriptional level. The requirement for well-balanced, tightly controlled, and cell type specific expression explains the complex genomic architecture of the Sox2 locus. Multiple enhancer elements that drive tissue specific expression have been identified in the 200 kb region surrounding Sox2 17–20. Consequently, endogenous expression has only been fully recapitulated in transgenic mice through a knockin approach where one of the Sox2 alleles was replaced by a marker gene12,21,22 or through introduction of bacterial artificial chromosomes (BACs) containing >200 kb of Sox2 genomic sequences23.
Protein encoding genes like transcription factors and chromatin modifiers are key to transcription activation. However, RNA genes that do not encode proteins can fulfill transcriptional regulatory roles as well. Long non-coding RNAs (lncRNAs), which are >200 nucleotides in length, seem to have in particular evolved for controlling genes at a transcriptional level24. LncRNA-mediated transcription regulation is instructed in cis or in trans. Allele-specific in cis mechanisms include recruitment of chromatin modifying complexes repressing transcription25 or activating transcription26, transcriptional interference preventing transcription factor access27,28, or gene looping29. Recently, a lncRNA gene called Sox2 overlapping transcript (Sox2ot) that is transcribed in the same direction as Sox2 and is polyadenylated downstream of Sox2 was described30,31. To date several studies investigating the function of Sox2ot have been reported32–34. These studies utilized knockdown or overexpression of Sox2ot in cancer cell lines and the results have indicated a role of Sox2ot in regulating proliferation as well as regulating Sox2. Sox2ot levels were invariably positively correlated with Sox2, however, the underlying regulatory mechanism has remained unknown.
In this study we evaluated expression of Sox2ot during development and studied the effect of Sox2ot overexpression in modified mouse ESCs that allow discrimination between cis and trans regulatory effects. On basis of our data we propose that during development Sox2ot expression is mainly restricted to neural cell types and that, in contrast to previous reports, enhanced Sox2ot transcriptional activity negatively affects Sox2 RNA levels in an allele-specific manner.
Results
Characterization and conservation of Sox2ot transcripts
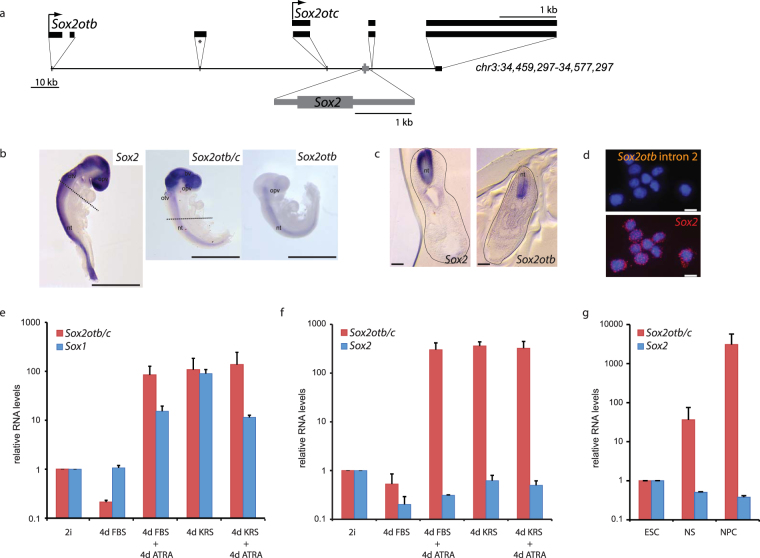
The Sox2 gene is a single exon gene that is located in a gene desert on mouse chromsosome 3 (Fig. 1a). Apart from Sox2 the only genes located within a 200 kb stretch of genomic DNA are presumably of non-coding nature. Two lncRNA genes (Sox2otb and Sox2otc) have been identified in this region31. The transcripts are initiated (~88 kb and ~11 kb) upstream of Sox2 and are terminated ~40 kb downstream of Sox2 (Fig. 1a). Transcriptome data, such as ESTs (expressed sequence tags) representing either Sox2ot transcript, have indicated that Sox2ot transcripts, like the flanking Sox2 gene, are predominantly present in brain as well as cell lines of neural origin. The expression pattern points to a function of Sox2ot in neural development and neural physiology, possibly through a Sox2-related mechanism. We first validated the transcription Sox2ot genes in neural progenitor cells (NPCs) derived from the lateral wall of the lateral ventricle in adult mouse. Primers recognizing an exon of Sox2otb that also is the first exon of Sox2otc could amplify Sox2ot transcripts in early passage neurospheres (data not shown and Fig. 1g), which is in agreement with two recent studies31,35. Using 5′ RLM-RACE we confirmed the 5′ ends of Sox2otb and Sox2otc (Supplementary Fig. S1a). Full-length cDNA sequence analysis showed extensive splicing, which is arguably random as almost any possible exon conjunction was retrieved. The splicing is largely conserved in other mammals as was recently shown34. We identified one previously undescribed exon located between Sox2otb exon 2 and Sox2otc exon 1 (Fig. 1a). We analyzed the cDNA sequences for the presence of open reading frames (ORFs) through Coding Potential Calculator36, NCBI’s ORFfinder, and a translation initiation prediction program (ATGpr) but the outcome underscored the non-coding nature of all Sox2otb and Sox2otc splice variants (Supplementary Fig. S1d,e, and f). To test whether the transcripts can be translated into a polypeptide we performed in vitro transcription/translation assays using the largest, multi-exonic, Sox2otb and Sox2otc cDNA sequences, but we could not detect any Sox2ot polypeptides (Supplementary Fig. S1g). This result indicates that Sox2otb and Sox2otc are likely of non-coding nature as was suggested before30,31. However, our analyses do not fully exclude the generation of very small peptides with a function, which can be produced from presumed non-coding RNA transcripts37.
Figure 1.
Co-expression of Sox2otb/c and Sox2 during mouse neural development. (a) Schematic representation of the Sox2 locus on mouse chromosome 3 (mm9 assembly). Depicted are the single exon gene Sox2, and the overlapping Sox2otb and Soxtotc genes. Sox2otb shares exons 4, 5 and 6 with Sox2otc. *Indicates a newly identified exon. (b) Whole mount RNA in situ hybridization of E9.25 mouse embryos using antisense Sox2, Sox2otbc and Sox2otb RNA probes. Scale bar represents 1 mm. otv, otic vesicle; opv, optic vesicle; nt, neural tube; bv, brain vesicle. (c) Transverse sections of the embryos depicted in b. Dashed line in b indicates the level of the transverse section. nt, neural tube. Scale bar represents 100 μm. (d) smFISH on mouse ESCs using Sox2otb intron 2 (upper panel) or Sox2 (lower panel) probe sets. Nuclei are stained with DAPI. (e) and (f) qRT-PCR analysis of Sox2otb/c and Sox1 (e), or Sox2otb/c and Sox2 (f) RNA levels during EB-mediated neural differentiation of mouse ESCs. Cells were cultured for 4 days in FBS or KSR containing medium followed by another 4 days in the same medium with 0.5 μM ATRA. (g) qRT-PCR analysis of Sox2otb/c and Sox2 RNA levels in mouse ESCs, ESC-derived radial glia-like NS cells and NPCs derived from the lateral wall of the lateral ventricle of the adult mouse. Expression was first normalized against β-Actin (e and f) or Myl6 (g), after which the relative expression to the expression in mouse ESCs was calculated. Values are mean + standard deviation (SD) of one representative out of 3 experiments and presented on a 10 log scale.
Sox2ot exonic and intronic sequences have been conserved between mammals and vertebrates (Supplementary Fig. S1c)31. The extent of conservation of genomic sequences between man and other vertebrates, like marsupials, is a measure of importance of these sequences for development. A larger evolutionary distance, i.e. between man and pufferfish (Fugu rubripes) diverging 450 million years ago, has been shown to be even more instrumental in uncovering coding as well as non-coding sequences crucial for proper development38. It was previously reported that the highest level of evolutionary conservation was observed in the promoter proximal regions of lncRNAs39–41. Likewise, the regions surrounding Sox2otb exon1 and Sox2otc exon 1, and not the exonic sequences, are highly conserved between man and Fugu. The high conservation of Sox2ot proximal promoter regions infers that Sox2ot sequences that govern transcription are more important during development than the transcript per se.
Expression of Sox2ot during neural development
Since previous studies have indicated that Sox2ot expression positively correlates with Sox2 RNA, we wished to test the correlative expression during neural development. We restricted the expression analysis to Sox2otb, Sox2otc and Sox2 only. First we analyzed expression of Sox2otb, Sox2otb and Sox2otc (from here on referred to as Sox2otb/c because the riboprobe contains Sox2otc exon 1 sequence, which is also present Sox2otb transcripts), and Sox2 in developing mouse embryos using RNA whole mount in situ hybridization (ISH). At 9.25 dpc Sox2 expression is mainly restricted to the neural tube, developing brain, nasal placodes, otic vesicles and optic vesicles (Fig. 1b, Supplementary Fig. S2a,b) (sense controls in Supplementary Fig. S2c). In contrast, a probe recognizing Sox2otb showed an expression pattern limited to the ventral part of the neural tube and optic vesicle, whereas a probe hybridizing to Sox2otb/c showed additional expression in the developing brain and otic vesicles (Fig. 1b). The spatial and temporal specific expression patterns of Sox2otb and Sox2otc during neural development indicate that the independent Sox2ot transcripts may have different roles. Although it is difficult to robustly interpret co-localization data at the single cell level on basis of RNA ISH using independent single probe hybridizations, the ISH data show that Sox2otb, Sox2otc and Sox2 are co-localized in tissues during neural development. To further investigate Sox2otb, Sox2otc and Sox2 coexpression we analyzed Sox2otb/c and Sox2 expression during the differentiation of mouse ESCs into neuroectoderm. In the tested feeder-independent and feeder-dependent wild type mouse ESC lines Sox2otb/c is very lowly expressed during maintenance. This is in sharp contrast with a previous study, which claimed abundant expression of Sox2ot in ESCs31. To further corroborate the low level of Sox2ot expression in ESCs we measured transcription of Sox2ot in mouse ESCs by single molecule FISH (smFISH) using a probe set lying in intron 2 of Sox2otb. smFISH has single molecule sensitivity42, yet, Sox2otb transcripts were very rare confirming the qRT-PCR results (Fig. 1d, positive control in Supplementary Fig. S2d). We observed a strong upregulation of Sox2otb/c upon neurectodermal differentiation using embryoid bodies (Fig. 1e, and Supplementary Fig. S2e). Upregulation coincides with the presence of neural progenitor/stem cells (NP/SCs) as measured through induction of Sox1, which is a very early and specific marker of the neuroectoderm lineage43. Sox2ot induction is all trans retinoic acid (ATRA) independent as neuroectodermal differentiation using knockout replacement serum (KRS) that is devoid of any form of retinol yielded a similar induction of Sox2otb/c (Fig. 1e). In more defined monolayer-based differentiation conditions Sox2otb/c was also induced upon neural differentiation (Supplementary Fig. S2d, and f), whereas BMP4-mediated differentiation towards mesendoderm failed to induce Sox2otb/c RNA levels (Supplementary Fig. S2g) indicating a primary role of Sox2ot in neural development. These results differ from the observations by Amaral et al., who have reported higher Sox2ot expression levels in mouse ESCs and enhanced Sox2ot transcription upon mesodermal commitment31. The discrepancies may be caused by differences in the used maintenance and differentiation protocols. Alternatively, a confounding factor may have been transcription initiation downstream of Sox2 in certain cell types, which yields transcripts that encompass Sox2ot exon 6 sequences.
ESC-based neural differentiation cultures are a mixture of distinct cell types, which include ESCs, NSCs/NPCs, and early neurons. During neural differentiation Sox2otb/c RNA levels were rather negatively correlated with Sox2 RNA levels (Fig. 1f) but the heterogeneic nature of the cultures thwarts to directly link Sox2otb/c levels to Sox2 levels. To investigate whether Sox2 levels are indeed negatively correlated with Sox2otb/c levels we measured the levels of Sox2 and Sox2otb/c in Sox2 heterozygous and homozygous ESC lines, in multiple monoclonal ESC-derived, radial glia-like neural stem (NS) cell lines generated from wild type mouse ESCs, and in neurosphere cultures of primary NPCs from the lateral ventricle of the adult mouse brain. NS cells express two to three-fold less Sox2 RNA44,45 (Fig. 1g) but contain higher levels of Sox2otb/c RNA in comparison with mouse ESCs. Primary NPCs contain higher Sox2otb/c RNA levels, whereas Sox2 levels are further reduced (Fig. 1g). In contrast to previous studies on Sox2ot expression in immortalized transformed cells32–34, we observed a negative correlation between Sox2otb/c and Sox2 RNA levels (Spearman r = −0,7857, P-value = 0.048)(Supplementary Fig. S2h).
Transcriptional activity of Sox2ot alters Sox2 RNA levels in cis
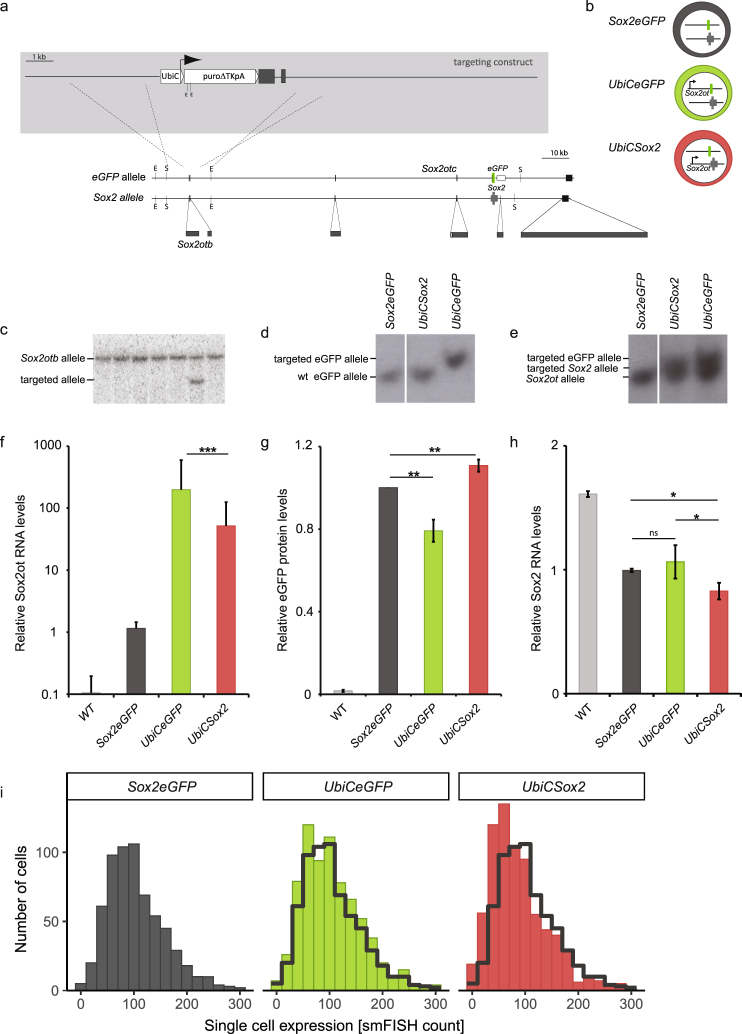
Next we wondered whether the negative correlation between Sox2ot and Sox2 is caused by a direct mechanism. Long non-coding RNAs are known to regulate neighboring genes in a variety of ways by either a cis (only the allele from which the lncRNA is transcribed is affected) or trans (the effect is independent of the allele from which the lncRNA is transcribed) mechanism. However, knocking out all three Sox2ot genes (Sox2otb, Sox2otc, and the 545 kb upstream of Sox2 located Sox2dot (Supplementary Fig. S1b)) simultaneously is extremely difficult. Moreover, such a strategy would likely perturb ordinary locus regulation as removal of critical Sox2ot promoter sequences may delete important regulatory sequences that are key for correct expression of neighboring genes. To circumvent these pitfalls, we opted to enhance the transcriptional activity of Sox2otb in Sox2 expressing cells that normally contain very low levels of Sox2ot. We introduced the human ubiquitin C (UbiC) promoter directly upstream of Sox2otb exon 1 by homologous recombination in mouse Sox2eGFP ESCs (Fig. 2a,c), which have one copy of Sox2 replaced by eGFP22. Three clones contained an insertion of the UbiC promoter into the eGFP allele (UbiCeGFP) and two into the Sox2 allele (UbiCSox2) (Fig. 2d, e). Sox2otb was highly transcribed in all targeted ESCs, albeit, levels were lower when the UbiC promoter was inserted into the Sox2 allele, hinting towards the existence of an allele-specific modulatory mechanism (Fig. 2f).
Figure 2.
Allele-specific overexpression of endogenous Sox2otb. (a) Schematic view of the targeting strategy and targeting construct to generate allele-specific transcription of Sox2ot. R = EcoRV and S = SbfI restriction sites. (b) Illustration of the genetic possibilities after targeting the Sox2eGFP ESC line: Sox2eGFP (untargeted), UbiCeGFP (Sox2ot is expressed from the eGFP allele), or UbiCSox2 (Sox2ot is expressed from the Sox2 allele). (c) Southern blotting showing correctly recombined Sox2eGFP ESC clone using a 3′ probe (EcoRV restricted DNA). (d) and (e) Southern blot analysis showing correct targeting of the eGFP allele (UbiCeGFP) or Sox2 (UbiCSox2) allele using eGFP (d) or Sox2 (e) specific probes (SbfI restricted DNA). Full blots are shown in Supplementary Fig. S3a. (f) Sox2otb expression in Sox2eGFP, UbiCeGFP, and UbiCSox2 cells as measured by qRT-PCR. (g) eGFP expression measured by flow cytometry in Sox2eGFP, UbiCeGFP, and UbiCSox2 cells. (h) Sox2 RNA levels in Sox2eGFP, UbiCeGFP, and UbiCSox2 cells measured by qRT-PCR. (i) smFISH quantification of Sox2 RNA copies per single cell in Sox2eGFP, UbiCeGFP, and UbiCSox2 lines. The gray line depicts the distribution of Sox2 in Sox2eGFP cells. ***P value < 0.002, **P value < 0.01 *P value < 0.05. Results are from three independent experiments using (sub)clones of Sox2eGFP (n = 2), UbiCeGFP (n = 3), and UbiCSox2 (n = 2). Values are presented as mean +/− SD (g and h) or +SD (10 log scale (f)). qRT-PCR data were normalized against β-Actin, and relative levels to the levels in Sox2eGFP cells were determined. Statistical analysis was performed using the paired t-test, except for flow cytometry results (Wilcoxon signed-rank test) and smFISH results (Mann-Whitney U test).
If the negatively correlated expression of Sox2 and Sox2ot is an immediate consequence of Sox2ot expression, an effect on Sox2 as well as eGFP (trans regulation) or, on either Sox2 or eGFP (cis regulation) should be evident in the targeted cells. Indeed, Sox2ot transcription resulted in a 20–30% reduction in Sox2 or eGFP levels (Fig. 2g, h). However, reduced expression was solely observed for the gene (Sox2 or eGFP) that was located on the targeted allele. These data demonstrate that Sox2ot transcription regulates Sox2 transcription in cis. Although reductions were relatively moderate, a compensatory mechanism was activated in the ESCs that have decreased Sox2 levels as illustrated by enhanced eGFP levels. This is reminiscent of the results in hybrid ESCs, in which allele-specific reduction of Sox2 by deletion of the ESC prevalent transcriptional enhancer led to upregulation of Sox2 from the unmodified allele20.
To determine whether the Sox2 downregulation is specific for the whole population or whether only a proportion of the population contributed to the lower Sox2 levels we quantified Sox2 RNA at the single cell level by smFISH. smFISH allows us to count the expression of individual RNA molecules in individual cells, which reveals expression heterogeneity within the population. We measured Sox2 levels in 700 cells of each ESC line (Fig. 2i, and Supplementary Fig. S3b). Only when Sox2ot was expressed from the Sox2 allele we observed a ~20% reduction in the means (77 versus 96 (Sox2eGFP) or 98 (UbiCeGFP) transcripts). Moreover, the distribution of Sox2 gene expression in UbiCSox2 cells differed from UbiCeGFP and the parental Sox2eGFP cells (Mann-Whitney U; FDR = 3.19e-10 and FDR = 1.11e-10, respectively), whereas the distributions in UbiCeGFP and Sox2eGFP cells were comparable. This analysis confirmed that Sox2 RNA levels are decreased when Sox2ot is transcribed from the same allele and showed that this effect is likely not restricted to a subpopulation of cells (Fig. 2i).
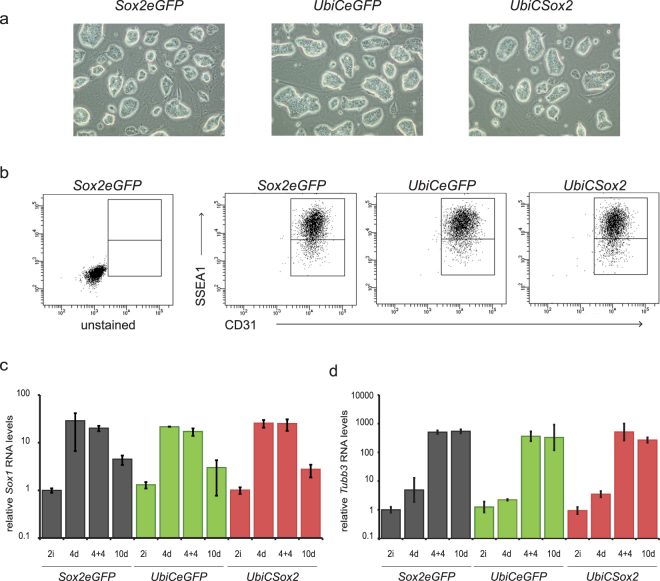
Mouse ESCs overexpressing Sox2ot are very similar to wild type ESCs
Next we investigated the effect of Sox2ot overexpression on the maintenance and differentiation of mouse ESCs. On basis of morphology we could not identify phenotypic differences between the parental Sox2eGFP ESCs and the Sox2ot overexpressing ESCs (Fig. 3a). The absence of a maintenance phenotype was underscored by the analysis of the expression of platelet endothelial cell activation marker CD31 (PECAM) and stage-specific embryonic antigen (SSEA1), which discriminates naïve and primed pluripotent cell states44,45. Sox2eGFP and Sox2ot overexpressing lines showed a similar and homogeneous CD31 expression profile, whereas SSEA1 was more heterogeneously expressed which is a normal feature of ESCs (Fig. 3b). Also the expression of other pluripotency genes like Nanog and Oct4 was not altered (Supplementary Fig. S4a, S4b). In addition, prolonged passaging at a constant splitting ratio did not reveal gross differences in the expansion rate between Sox2eGFP and Sox2otb overexpressing ESCs (data not shown). Possibly this is due to adaptation of the UbicSox2 ESCs to lower levels of Sox2 RNA by acquiring more normal SOX2 protein levels (Supplementary Fig. S4c). Since Sox2otb is induced during the differentiation of ESCs into neuroectoderm we also investigated the effect of Sox2otb overexpression on neuroectodermal differentiation. Using EB-based differentiation protocols we could not detect quantitative or temporal differences in the generation of either NSCs or more mature Tubb3 positive cells (Fig. 3c,d). In addition, the differentiation into mesendoderm as determined by Brachyury expression is largely unaltered (Supplementary Fig. S4d). Taken together these results indicate that enhanced Sox2ot levels do not majorly alter the phenotype of ESCs and do not exert gross effects on the EB-based differentiation of mouse ESCs.
Figure 3.
ESCs overexpressing endogenous Sox2otb are similar to Sox2eGFP ESCs. (a) Phase contrast pictures of Sox2eGFP, UbiCeGFP, and UbiCSox2 cells cultured in 2i medium (100x magnification). (b) SSEA1 and CD31 expression in Sox2eGFP, UbiCeGFP, and UbiCSox2 cells as measured by flow cytometry. (c and d) RNA levels of Sox1 (c) and Tubb3 (d) during EB-mediated neural differentiation of Sox2eGFP, UbiCeGFP, and UbiCSox2 cells as measured by qRT-PCR. RNA levels were normalized against β-actin. RNA levels relative to the levels in Sox2eGFP cells are depicted on a 10 log scale. The results of one representative experiment (out of three independent experiments) using (sub)clones of Sox2eGFP (n = 2), UbiceGFP (n = 3), and UbiCSox2 (n = 2) is depicted as mean +/− SD.
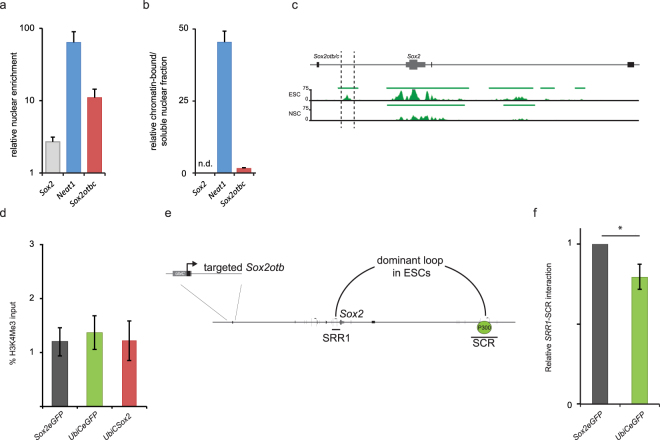
Sox2otb/c is enriched in the nucleus but not associated to chromatin
Many lncRNAs that regulate transcription are enriched in the nucleus. We therefore investigated the cellular localization of Sox2ot. As our Sox2ot exonic smFISH probe set was not specific enough, we analyzed the cellular localization of Sox2ot RNA by cell fractionation and qRT-PCR. Sox2ot RNA was 4 times more enriched in the nucleus than Sox2 RNA but 6 times less than Neat1, a lncRNA that is highly abundant in the nucleus46 (Fig. 4a). Next we examined whether Sox2ot is associated to the chromatin fraction. LncRNAs that function through a trans-acting mechanism are often found enriched in the chromatin fraction, like Neat1 46. In support of the observed in cis effect of Sox2otb/c we predominantly found Sox2ot RNA in the soluble nuclear fraction (Fig. 4b).
Figure 4.
Sox2 locus-specific H3K4 trimethylation and chromatin interactions in ESCs overexpressing Sox2otb. (a) Analysis of Sox2ot RNA localization in ESCs. Sox2ot is enriched in the nucleus when compared to β-Actin as determined by qRT-PCR after subcellular fractionation. The ratio (+SD) of nuclear/total RNA (200ng input) relative to that of β-actin is depicted on a 10 log scale. Neat1 is a lncRNA that is enriched in the nucleus, and which is predominantly associated to chromatin46. (b) Analysis of the nuclear localization of Sox2ot in ESCs by qRT-PCR. The depicted ratio of chromatin bound RNA (+SD) is relative to that of β-actin. (c) Genome browser view of H3K4me3 density signals in the regulatory Sox2 region of ESCs and ESC-derived NPCs67. For quantification of the difference see Supplementary Fig. 5Sa, b, and c. (d) H3K4me3 ChIP results for the region depicted between vertical lines in (c). Depicted H3K4me3 levels are relative to H3K4me3 levels of the housekeeping gene Myl6. (e) Schematic drawing of the dominant chromatin loop in ESCs formed by interaction of the Sox2 proximal region (Sox2 regulatory region 1) (SRR1) with a P300 bound super enhancer (SCR) located ~110 kb downstream of Sox2. HindIII fragments and primers used are shown. (f) 3C chromatin conformation capture of the SRR1-SCR interaction depicted in (e). Values are relative to interactions of the Sox2 intergenic region upstream of Sox2otc. Values are represented as mean +/− SD from three independent experiments (n = 10). *Paired t-test P value = 0.02.
H3K4 methylation is unaltered in Sox2otb overexpressing mouse ESCs
The allele-specific regulation of Sox2 prompted us to investigate the nature of this regulation. A large group of cis-acting lncRNA transcripts represses genes by recruiting chromatin-modifying proteins that install a repressive histone mark such as H3K27me3 or H3K9me3, or by controlling H3K4 methylation47. To gain evidence for the existence of a Sox2ot dependent chromatin-modifying mechanism we compared H3K4me1, H3K4me2, H3K4me3, H3K9me3, and H3K27me3 chromatin marks in the region between the first exon of Sox2otb and the last exon of Sox2otb/c in cells expressing Sox2 and Sox2ot at different ratios, i.e. ESCs and ESC-derived NPCs, using publicly available H3 methylation chromatin immunoprecipitation-sequencing (ChIP-seq) data sets (Fig. 4c, and Supplementary Fig. S5a and d). The only histone methylation profiles that are strongly altered between ESCs and ESC-derived NPCs are confined to a conserved region in the proximal enhancer/promoter region of Sox2 ~4 kb downstream of the first exon of Sox2otc (Supplementary Fig. S5a,b, and c). In this region H3K27me3 and H3K4me3 were high in ESCs indicating a bivalent chromatin signature, which is linked to key developmental genes48,49. The bivalent histone status is lost in this region in ESC-derived NPCs. We wondered whether overexpression of Sox2otb would change the ESC chromatin into a more NPC-like chromatin regarding H3K4me3. We performed H3K4me3 ChIP assays for this region but did not observe differences in H3K4me3 between the cell lines (Fig. 4d). Although we did not rule out the involvement of other epigenomic changes, we decided to investigate other candidate regulatory mechanisms.
Sox2otb transcription impairs the formation of the chromatin promoter-enhancer loop driving expression of Sox2
Development and homeostasis require coordinate regulation of neighboring genes through enhancers and locus control regions50. Chromatin looping enables transcription activation by juxtaposing locus control regions (LCRs), distal regulatory elements and promoter elements, and thus, function by bringing transcription factors, coactivators, and RNA polymerase II together. In ESCs multiple chromatin loops exist in the Sox2 locus51. The most prevalent chromatin interaction is formed by the Sox2 regulatory region 1 (SRR1) upstream of Sox2 and a 13 kb super enhancer termed Sox2 control region (SCR) located ~100 kb downstream of Sox2 20,52 (Fig. 4e). Deletion of this super enhancer decreases Sox2 levels in mouse ESCs 6 to 9 fold20,52. Thus, if a decrease in Sox2 levels were the consequence of Sox2otb mediated transcriptional interference the SRR1-SCR interaction would likely be diminished. Through chromosome conformation capture (3C) we analyzed whether the SRR1-SCR chromatin interaction was altered in Sox2otb overexpressing (UbiCeGFP) ESCs, which did not show altered Sox2 levels, compared to parental Sox2eGFP ESCs. We indeed observed a lower frequency of SRR1-SCR interactions in Sox2otb overexpressing cells versus Sox2eGFP cells (Fig. 4f). In summary, transcriptional activity of Sox2otb negatively correlates with Sox2 levels, and in addition, enhanced Sox2otb transcription correlates with reduced chromatin interactions between the upstream regulatory sequence of Sox2 and the super enhancer of Sox2 in mouse ESCs.
Discussion
Through transcription analysis in combination with genetic modification of the endogenous Sox2otb locus we identified that transcriptional activity of Sox2otb represses Sox2 expression in mouse ESCs. In contrast to our findings, previous studies in human cancer as well as cancer cell lines have demonstrated a positive correlation between Sox2ot and Sox2 in certain but not all cell types investigated32–34. A quantitative and qualitative comparison of the published expression data is rather difficult due to the genomic positions of the primers used as the applied primer pairs recognize either a variety of Sox2ot splice variants or amplify only Sox2ot sequences downstream of Sox2. Nevertheless, the positive co-regulation of Sox2 by Sox2ot has been strongly supported by ectopic overexpression or knockdown of Sox2ot pointing to a trans effect32–34. One may argue that transcription regulatory mechanisms of certain genes in human cells are different from those in murine cells, however, the strong conservation of the whole Sox2ot genomic region rather suggests a highly similar mode of operation. We believe that the disparities with the results obtained in this study are more likely caused by the differences in the cells analyzed, as gene regulation is very much cell type specific. In addition, cancer cells have undergone many epigenetic and genetic changes that interfere with the specificity and integrity of regular gene transcription programs53. Since we investigated early neural development using non-transformed mouse cells our data indicate that Sox2 regulation during stem cell maintenance and differentiation is completely different from Sox2 regulation in cancer cells.
Cis regulation of neighboring genes has been proposed to be an important function of many lncRNA genes, but up to now this has only been proven for a very small subset of lncRNAs predominantly involved in imprinting and X inactivation because of the more easily detectable allele-specific modifications47. In general, a major hurdle has been selection of an allele-specific genomic modification strategy to identify allele-specific differences that represent a bona fide phenotype. In addition, modification of lncRNA genes to study cis-acting mechanisms is rather challenging. Introducing single or small mutations that alter the function or expression of uncharacterized lncRNAs is very complicated due to the non-coding nature of these genes. Nevertheless, insertion of a strong polyadenylation signal that prematurely truncates the lncRNA transcript has been successfully exploited to gain insight into the requirement of the full-length lncRNA27. However, premature polyadenylation strategies do not allow analysis of the role of lncRNA transcription initiation or that of promoter/enhancer sequences. Instead deletion of presumed important regulatory regions may be considered to address their role. Recent genome editing advances using CRISPR/Cas9 have facilitated the deletion of genomic sequences54 but deletion of important promoter or exon sequences imposes the risk of removing important transcriptional regulatory regions of the neighboring genes, in particular, because lncRNAs are often transcribed from enhancer and promoter proximal sequences of adjacent genes. This may result in false attribution of the role of the modified lncRNA. As to Sox2ot the existence of at least three independent transcriptional initiation sites of Sox2ot, and possibly more as indicated by human CAGE datasets, would make the generation of a full knockout rather unrealistic. Moreover, one of the Sox2ot transcription initiation sites (that of Sox2otc) is located in regulatory sequences proximal of Sox2. Deletion of this genomic sequence may directly alter Sox2 transcription independent of Sox2otc. As feasible alternative we created a promoter insertion that drives transcription of only one of the Sox2ot genes to study the role of Sox2otb overexpression in development and the regulation of Sox2. Using this overexpression system we demonstrate that the reduction in Sox2 RNA levels is caused by allele-specific transcriptional activity of Sox2otb. The reduced levels of Sox2 did not exert a loss of pluripotent stem cell self-renewal phenotype, as may have been expected, likely due to adaptation of the ESCs to decreased Sox2 RNA levels. It is known that a decrease in Sox2 levels in ESCs activates a feedback mechanism enhancing expression of Sox2 20. Also in the UbicSox2 cells we observed upregulation of the other Sox2 allele (here eGFP allele) indicating the activation of such feedback loop and the importance of having higher levels of Sox2. However, since the other allele is non-functional, enhanced expression of the other allele was ineffective. Instead the UbicSox2 cells adapted to lower Sox2 levels by regaining SOX2 to a level similar to that of the parental Sox2eGFP cells.
Sox2 is also crucial for neuroectodermal differentiation of ESCs, and lower Sox2 levels favor mesendoderm commitment9. If the SOX2 protein levels would not have been enhanced upon adaptation a differentiation phenotype would have been expected in the cells that overexpress Sox2otb from the Sox2 allele. Although Sox2 adaptation may have obscured an early neuroectodermal, Sox2-dependent differentiation defect, a Sox2-independent trans effect was not observed. Thus our results indicate that the main function of Sox2otb is cis regulation of Sox2 rather than affecting cell physiology in trans via other routes. The importance of Sox2ot transcriptional activity is underscored by the genomic conservation of Sox2ot between mammals and fugu, which is much higher in Sox2ot promoter (proximal) sequences than exon sequences (Supplementary Fig. S1c).
The introduced Sox2otb transcriptional activity led to decreased Sox2 transcription and reduced interaction of the Sox2 proximal promoter region (SRR1) with the ESC-specific enhancer in this genomic region. However, we cannot rule out that other chromatin interactions are affected as well. In the presented heterozygous ESC model maximally 50% of a specific chromatin loop can be altered when considering an in cis effect. Therefore, only differences in very dominant chromatin loops, either the ones that are newly formed or the regular ones, are detectable. A hypothetical mechanism that would fit our observations is transcriptional repression by virtue of blocking recruitment of RNA polymerase II to the SSRI region (Fig. 5a). A very similar mechanism is exploited by Airn, which repress Igf2r by preventing RNA polymerase II recruitment to the Igf2r promoter27.
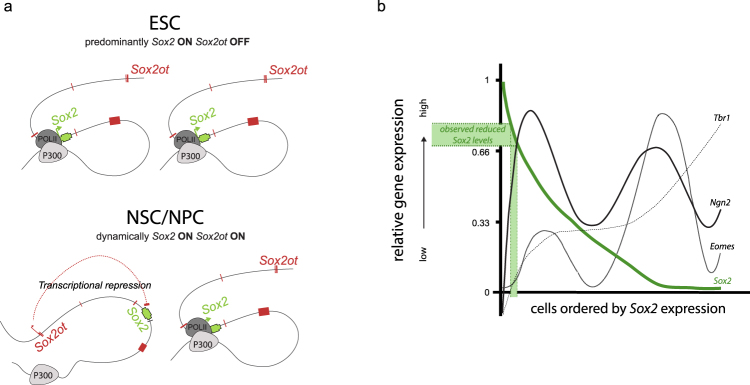
Figure 5.
Proposed model of transcriptional interference to modulate Sox2 levels during neural development. (a) Hypothetical model illustrating Sox2 and Sox2ot transcription in ESCs and NSCs/NPCs. In ESCs Sox2 (green) is predominantly transcribed, whereas Sox2ot (red) transcription is predominantly off. In NSCs Sox2 and Sox2ot are transcribed in the same cell. On basis of our data we propose a dynamic on/off situation. If Sox2ot is transcribed Sox2 transcription is repressed, and vice versa. (b) Adapted graph from a single cell RNA profiling study by Hagey and Muhr55 showing the influence of subtle reductions in Sox2 on the expression of neuronal genes. The observed 20–30% reduction in Sox2 transcription by Sox2ot transcription (indicated by green bars) lies at the threshold of the expression of neural genes Tbr1 and Eomes and corresponds to a steep rise in the expression of the proneural gene Ngn2 in cortical NSCs/NPCs.
As well-balanced Sox2 protein levels are crucial for correct development of the distinct subsets of neurons in the neural tube10,11, it is tempting to speculate that during development the main function of Sox2ot is controlling Sox2 levels. In this respect the 20–30% reduction in expression of Sox2 RNA that we have observed may seem irrelevant. However, recently it became clear from single cell RNA sequence analysis in primary mouse cortical NSCs/NPCs that Sox2 dosage regulates their division rate and controls their ability to maintain an undifferentiated state55. This study demonstrated that very modest decreases in Sox2 levels in NSCs/NPCs are accompanied by rapid increments of the neuronal specification factor Neurogenin2 (Fig. 5b). Moreover, an approximately 20% reduced expression of Sox2 (the same reduction as we observed in Sox2ot overexpressing ESCs) appeared to be a threshold for expression of the neural differentiation markers Eomes and Tbr1 (Fig. 5b). These data indicate that a subtle decrease of Sox2 may have a profound impact on the status of NSC/NPCs regarding their differentiation potential, and that Sox2ot transcription through the Sox2 gene may render NSCs/NPCs more susceptible to neural differentiation.
We believe that the here proposed role of Sox2ot is likely conserved in numerous loci containing key differentiation genes. Transcriptome data have revealed that analogous overlapping transcripts are present in the Sox1 and Sox4 loci. It will be interesting to learn the underlying nature of these Sox regulatory mechanisms, to what extent this regulation exists in the mammalian genome, and how disruptions disturb development.
Methods
Cell culture
Mouse ES cell lines (E14-cl2244, E14 subclone IB10, R1, CCE, and Sox2eGFP 22 (parental mouse ESCs as well as the targeted clones) were cultured feeder-free or on irradiated mouse embryonic fibroblasts (MEFs) on 0.1% gelatinized tissue culture surface in DMEM containing 1 mM L-glutamine, 1x non-essential amino acids (NEAA), PenStrep 1%, 1000 U/ml human LIF (Peprotech), 50 μM β-mercaptoethanol and 15% mouse ESC tested fetal bovine serum (FBS) (Life Technologies). Sox2eGFP ESCs were kindly provided by the late Dr. L. Pevny, University of North Carolina, Chapel Hill. For ChIP, 3C, differentiation and qRT-PCR cells were first cultured for 4 passages in 2i medium56 (DMEM/F12/NeuralBasal, Glutamax, PenStrep 1%, human LIF 1000 U/ml (Peprotech), 50 μM β-mercaptoethanol, 0.5x B27 plus vitamin A (ThermoFisher), 0.5x N2 (ThermoFischer), 1 μM PD0325901 (Axon Medchem) and 3 μM CHIR99021 (Axon Medchem) and a FBS percentage that was gradually decreased from 15% to 1%. Cells were passaged using Trypsin/EDTA (0.05%/0.02%). Cells were maintained at 37 °C and 5% CO2. Prior to the experiments the quality of the cells was analyzed by flow cytometric analysis using anti-mouse SSEA1-BV421 (BD) and anti-mouse CD31-PerCPefluor710 (eBioscience) antibodies. SOX2 was measured by flow cytometry using a goat anti-Sox2 polyclonal antibody (SantaCruz, Biotechnology, sc-17319), in combination with an anti-goat-Alexa568 secondary antibody (Thermo Fisher Scientific). Staining was performed using the fix & perm kit (Thermo Fisher Scientific) according the manufacturer’s instructions.
Targeting Sox2eGFP mouse ES cells
Two independent homologous recombination experiments were performed using Sox2OTb targeting vectors containing UbiCloxPHyTKpAloxP or UbiClox2272PurDTKpAlox2272 selection cassettes. The selection modules were inserted 9 nucleotides upstream of the identified Sox2OTb transcription start site (chr 3: 34,459,297 NCBI37/mm9) into the genomic sequence (chr 3: 34,453,460–34,463,055 NCBI37/mm9) that was amplified from 129Ola genomic DNA using Phusion polymerase (NEB). The knockin constructs were introduced into Sox2eGFP ESCs by electroporation, and drug resistant clones were selected using hygromycin (110 μg/ml) or puromycin (1.5 μg/ml). Homologous recombinants were identified by Southern blot analysis of EcoRV restricted genomic DNA using 32P labelled Sox2otb flanking probes. In total, 465 colonies were screened for correct homologous recombination. Five correctly recombined clones were further investigated to identify whether the Sox2 or eGFP allele was targeted. To this end SbfI restricted genomic DNA was separated by pulse field gel electroforesis (PFGE) and analyzed by Southern blotting using 32P labelled eGFP and Sox2 probes. Three clones contained an insertion of the UbiC promoter into the eGFP allele (UbiCeGFP) and two into the Sox2 allele (UbiCSox2).
RNA in situ hybridization
Whole mount in situ hybridization was performed according standard protocols. In short, dissected E9.25 embryos (C57Bl/6) were fixed in 4% PFA O/N. Fixed embryos were twice washed in PBS 0.1% Tween-20 (Sigma) (PBST), and dehydrated by subsequent methanol washing steps (25-50-75 and 100% methanol). Dehydrated embryos were slowly rehydrated (10′ per step) at RT while rotating. After rehydration the brain vesicle was punctured and the surround membrane ruptured to prevent trapping of the riboprobes. Embryos were treated with proteinase K (10 mg/ml) for 10′, and gently rinsed in PBST. Next embryos were again fixed in 4% PFA and 0.2% glutaraldehyde for 20′ while rotating, washed in PBST, and incubated in 50% PBT/50% hybridization solution (HS) (HS: 50% formamide (Sigma), 1.3x SSC, pH 5.0 (Ambion), 5 mM EDTA, pH 8.0 (Ambion), 50 mg/ml yeast tRNA (Sigma), 0.2% Tween-20 (Sigma), 0.5% CHAPS (Thermo Fisher Scientific), and 100 mg/mL Heparin (Sigma)), and subsequently 100% HS. Riboprobes, generated by T7 polymerase in vitro transcription (antisense and sense Sox2, Sox2OTb and Sox2OTb/c digoxigenin labeled RNA probes (sequences in Table S1)), were added to HS and incubated for 20 hours at 70 °C. Embryos were washed 3 times with 2x SSC, 0.1% CHAPS, three times with 0.2x SSC, 0.1% CHAPS, and twice with 1x KTBT (50 mM TrisHCl, pH 7.5, 150 mM NaCl, 10 mM KCl and 1% Triton X-100). Embryos were incubated with 10 ug/ml of RNase A in KTBT for 30 min. at 37 °C, blocked with 2% blocking solution (Roche), and 20% heat inactivated sheep serum, and subsequently O/N incubated with AP conjugated a-DIG, Fab fragment (Sigma) in the same blocking buffer at 4 °C. Embryos were 5 times washed in 0.1% Tween-20 and 1 mM levisamole (Roche) in ddH2O, and subsequently stained in 1x BM purple (Roche) plus 0.1% Tween, 1 mM levamisole. Reaction was stopped by washing in ddH2O.
Whole mount stained embryos, were embedded in 2% agarose and cross-sectioned on a vibratome (Leica). Mice were maintained under specific-pathogen-free conditions. All animal experiments were approved by the Animal Experiments Committee of the LUMC performed to the recommendations and guidelines set by the LUMC and by the Dutch Experiments on Animals Act that serves the implementation of guidelines on the protection of experimental animals by the Council of Europe.
RNA-linker mediated (RLM)-RACE and in vitro transcription translation
The used RLM-RACE procedure has been extensively described elsewhere57. Sox2ot reverse primers were located in exon 1 of Sox2otc. In vitro transcription/translation of human TP53 and the full-length Sox2otb and Sox2otc cDNA sequences was performed using TNT® Quick Coupled Transcription/Translation System (Promega) according the manufacturer’s protocol. 35M labeled proteins were separated on 5–15% and 20% polyacrylamide gels.
ESC differentiation
For embryoid body (EB) differentiation the original protocol was slightly adapted58. For neural differentiation: ESCs were seeded as a single cell suspension at a concentration of 100,000-200,000 cells/ml in ESC media containing FBS (as in the original protocol) or knockout serum replacement (KSR)59 lacking hLIF and 2i on ultra-low attachment plates (Corning). After 4 days of culture all trans retinoic acid (ATRA) (Sigma) or the synthetic substitute EC23 (Abcam) was added to the media at a concentration of 0.5 μM. Media was changed once every two days. For mesendodermal differentiation, aggregated ESCs were cultured in 2i media containing 3 μM CHIR99021 but without PD0325901, hLIF, and FBS as has been described for monolayer differentiation9. 3 ½ days after addition of CHIR99021 EBs were manually dissociated using the embryoid body dissociation kit (Miltenyi Biotech) according the manufacturer’s instructions. Cells were stained for Oct4 and Brachyury using mouse anti-Oct4-BV421 (BD) and goat anti-Brachyury (SC-17745, SantaCruz) and a secondary donkey anti goat Alexa568 antibody (Thermo Fisher Scientific) using the fix & perm kit (Thermo Fisher Scientific) according the manufacturer's protocol. Oct4 and Brachyury expression was measured on a LSRII flow cytometer (BD).
For monolayer differentiation we adapted the protocol used by Engberg et al.60. In brief, mouse ESCs were seeded at a density of 15,000 cells/cm2 onto 0.1% gelatin (Sigma) coated dishes in 2i media, lacking hLIF and 2i, but containing 1% FBS. Media was replaced with DMEM/F12/Neuralbasal containing L-glutamine (ThermoFisher), PenStrep 1%, 1x N2 (ThermoFisher), and 1x B27 without vitamin A (ThermoFisher), and ATRA (Sigma) or EC23 (Abcam), or hBMP4 (Peprotech) at the concentrations indicated, 12 hours after seeding the cells. Cells were cultured for the indicated periods and media was replaced every two days. NS cell lines were generated from different ESC lines using N2B27 media as described elsewhere61. One of the clones has been extensively characterized44.
RNA isolation and quantitative PCR analysis
Total RNA was isolated directly from the cells using Trizol (Life technologies) or NucleoSpin® columns (Macherey-Nagel). Following DNaseI treatment (Roche), cDNA was generated from 100–500 ng RNA using Transcriptor reverse transcriptase (Roche) and random hexamers or an oligod(T) primer according the manufacturer’s protocol. After the samples had been checked for genomic DNA contaminations, cDNA was measured quantitatively on a Bio-Rad CFX96 using SensiFASTTM Sybr green PCR mix (Bioline) and the primers listed in Supplementary Table S1. All primers were tested for a comparable and linear amplification efficiency using a dilution series of cDNA or gDNA. RNA levels were normalized against β-actin and 18 S, which yielded similar outcomes. For direct quantitative comparison of expression levels between ESCs and NS cells levels were normalized against housekeeping gene Myl6 because Myl6 expression is unaltered between ESCs and NSCs44. All measurements were performed in triplicate. Relative expression was calculated using the comparative Ct method, known as the 2–[delta][delta]Ct method, where [delta][delta]Ct = [delta]Ct(sample) - [delta]Ct(reference). Dependent on the experiment, the reference samples were the 2i samples (also described as day 0 of differentiation), or the parental ESC line Sox2eGFP.
Single molecule fluorescence in situ hybridization (smFISH)
Mouse ESCs were cultured in 2i medium or differentiated in N2B27 media without additives for 4 days as described above. Cells were detached with Accutase (Gibco), resuspended in serum containing medium, and fixed by adding paraformaldehyde to an end-concentration of 4% and subsequent incubation for 12 minutes at RT. Fixed cells were pelleted by a 3′ centrifugation and subsequently resuspended in 70% ethanol. Samples were stored at 4 °C until use. smFISH of Sox2 (Stellaris VSMF-3075-5-BS probe set) was performed exactly as before62 and signals were quantified using custom MATLAB scripts. Sox2ot transcription was determined using a custom probe set covering Sox2otb intron 2, which was designed by homemade MATLAB scripts.
Chromatin immunoprecipitation (ChIP) and 3C conformation capture
The chromatin of a single cell suspension of mouse ESCs was crosslinked in ESC medium containing 1% formaldehyde. Protocols used were previously described by Lee et al.63 (ChIP) and Stadhouders et al.64 (3C). For ChIP: the nuclear fraction was sonicated for 9 minutes (30″ on, 30″ off) using a Biorupter UCD-200 (Diagnode). After sonication, H3K4me3 chromatin was precipitated overnight at 4 °C in 0.1% fraction V BSA, protease inhibitors (Roche), 16.7 mM trisHCl, 167 mM NaCl, 1.25 mM EDTA, 0.01% SDS, 1% Triton X-100, Dynabeads Protein G (ThermoFisher) and 1 ug H3K4me3 rabbit polyclonal antibody (Diagenode). Chromatin was eluted in 1% SDS, and 0.1 M NaHCO3, de-crosslinked at 65 °C for 8–12 hours, treated with RNAse A and ProtK, and purified using phenol/chloroform extraction. Mouse insulin promoter primers and Myl6 primers were used as negative control and positive/normalization control, respectively. For 3C: chromatin was restricted with HindIII (Fermentas) for 24 hours and O/N ligated at 16 °C. Chromatin was de-crosslinked at 65 °C for 8–12 hours, treated with RNAse A and ProtK, and purified using phenol/chloroform extraction. Quality and quantity of DNA was checked by a linear amplification of Sox2UTR genomic sequences. Ligation efficiencies were checked through amplification of ESC-specific Dppa2 chromatin loop65.
Subcellular fractionation
Cell fractionation: mouse ESCs were divided into two fractions and used for either total RNA isolation or nuclear RNA isolation. Nuclear RNA was isolated as previously described57. In brief, cells were lysed and nuclei were pelleted after centrifugation (1350 g at 4 °C for 5 min). Cells (total RNA) or nuclei (nuclear RNA) were lysed using RA1 RNA lysis buffer (Macherey-Nagel) and RNA was isolated on NucleoSpin® columns (Macherey-Nagel) according to the manufacturer’s instructions. 200 ng of RNA was used in the reverse transcription reaction that was performed as described above.
Nuclear fractionation: Fractionation of the nucleus was performed as described by Werner et al.66 In brief, crude nuclei were resuspended in 250 μl NRB (20 mM HEPES pH 7.5, 50% Glycerol, 75 mM NaCl, 1 mM DTT, 1x protease inhibitor cocktail) and centrifuged for 5′ at 500 g at 4 °C. The pellet was again resuspended in 250 μl NRB and 1 volume of NUN buffer ((20 mM HEPES, 300 mM NaCl, 1 M Urea, 1% NP-40 Substitute, 10 mM MgCl2, 1 mM DTT) was added, followed by a 5′incubation on ice after which the suspension was centrifuged (1,200 g, 5 min, 4 °C). The soluble fraction supernatant was transferred to a tube and the pellet was resuspended in 1 ml buffer A (20 mM HEPES pH 7.5, 10 mM KCl, 10% glycerol, 340 mM sucrose, 4 mM MgCl2, 1 mM DTT, and 1x Protease Inhibitor Cocktail (Roche) and centrifuged (1,200 g, 5 min, 4 °C). The chromatin pellet was resuspended in 50 μl buffer A, and 500 μl Trizol (Life technologies) was added. Trizol was added as well to the soluble nuclear fraction. Subsequently, RNA was extracted following the manufacturer’s guidelines. cDNA was generated as described above.
Electronic supplementary material
Acknowledgements
This study was supported by the Dutch Brain foundation and ZonMW. We thank the late L. Pevny (University of North Carolina, Chapel Hill) for the Sox2eGFP cells, the LUMC sequence analysis support core for ChiPseq data alignments, and S. Chuva de Sousa Lopes (LUMC) for assessing the ISHs. The authors declare to have no competing financial interests.
Author Contributions
T.M., R.P., S.S., and H.M. designed the experiments. T.M., R.H. and H.M. wrote the paper. T.M., S.L., P.B., A.J., and H.M. performed the experiments. T.M., S.L., P.B., S.S., and H.M. analyzed the data.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-017-18649-4.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Kamachi Y, Kondoh H. Sox proteins: regulators of cell fate specification and differentiation. Development. 2013;140:4129–4144. doi: 10.1242/dev.091793. [DOI] [PubMed] [Google Scholar]
- 2.Nichols J, Smith A. Pluripotency in the embryo and in culture. Cold Spring Harb Perspect Biol. 2012;4:a008128. doi: 10.1101/cshperspect.a008128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ivanova N, et al. Dissecting self-renewal in stem cells with RNA interference. Nature. 2006;442:533–538. doi: 10.1038/nature04915. [DOI] [PubMed] [Google Scholar]
- 4.Kopp JL, Ormsbee BD, Desler M, Rizzino A. Small increases in the level of Sox2 trigger the differentiation of mouse embryonic stem cells. Stem Cells. 2008;26:903–911. doi: 10.1634/stemcells.2007-0951. [DOI] [PubMed] [Google Scholar]
- 5.Masui S, et al. Pluripotency governed by Sox2 via regulation of Oct3/4 expression in mouse embryonic stem cells. Nat Cell Biol. 2007;9:625–635. doi: 10.1038/ncb1589. [DOI] [PubMed] [Google Scholar]
- 6.Li J, et al. A dominant-negative form of mouse SOX2 induces trophectoderm differentiation and progressive polyploidy in mouse embryonic stem cells. J Biol Chem. 2007;282:19481–19492. doi: 10.1074/jbc.M702056200. [DOI] [PubMed] [Google Scholar]
- 7.Fong H, Hohenstein KA, Donovan PJ. Regulation of self-renewal and pluripotency by Sox2 in human embryonic stem cells. Stem Cells. 2008;26:1931–1938. doi: 10.1634/stemcells.2007-1002. [DOI] [PubMed] [Google Scholar]
- 8.Adachi K, Suemori H, Yasuda SY, Nakatsuji N, Kawase E. Role of SOX2 in maintaining pluripotency of human embryonic stem cells. Genes Cells. 2010;15:455–470. doi: 10.1111/j.1365-2443.2010.01400.x. [DOI] [PubMed] [Google Scholar]
- 9.Thomson M, et al. Pluripotency factors in embryonic stem cells regulate differentiation into germ layers. Cell. 2011;145:875–889. doi: 10.1016/j.cell.2011.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bylund M, Andersson E, Novitch BG, Muhr J. Vertebrate neurogenesis is counteracted by Sox1-3 activity. Nat Neurosci. 2003;6:1162–1168. doi: 10.1038/nn1131. [DOI] [PubMed] [Google Scholar]
- 11.Graham V, Khudyakov J, Ellis P, Pevny L. SOX2 functions to maintain neural progenitor identity. Neuron. 2003;39:749–765. doi: 10.1016/S0896-6273(03)00497-5. [DOI] [PubMed] [Google Scholar]
- 12.Taranova OV, et al. SOX2 is a dose-dependent regulator of retinal neural progenitor competence. Genes Dev. 2006;20:1187–1202. doi: 10.1101/gad.1407906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Niu W, et al. In vivo reprogramming of astrocytes to neuroblasts in the adult brain. Nat Cell Biol. 2013;15:1164–1175. doi: 10.1038/ncb2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oosterveen T, et al. SoxB1-driven transcriptional network underlies neural-specific interpretation of morphogen signals. Proc Natl Acad Sci USA. 2013;110:7330–7335. doi: 10.1073/pnas.1220010110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Su Z, Niu W, Liu ML, Zou Y, Zhang CL. In vivo conversion of astrocytes to neurons in the injured adult spinal cord. Nat Commun. 2014;5:3338. doi: 10.1038/ncomms4338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cai N, Li M, Qu J, Liu GH, Izpisua Belmonte JC. Post-translational modulation of pluripotency. J Mol Cell Biol. 2012;4:262–265. doi: 10.1093/jmcb/mjs031. [DOI] [PubMed] [Google Scholar]
- 17.Uchikawa M, Ishida Y, Takemoto T, Kamachi Y, Kondoh H. Functional analysis of chicken Sox2 enhancers highlights an array of diverse regulatory elements that are conserved in mammals. Dev Cell. 2003;4:509–519. doi: 10.1016/S1534-5807(03)00088-1. [DOI] [PubMed] [Google Scholar]
- 18.Catena R, et al. Conserved POU binding DNA sites in the Sox2 upstream enhancer regulate gene expression in embryonic and neural stem cells. J Biol Chem. 2004;279:41846–41857. doi: 10.1074/jbc.M405514200. [DOI] [PubMed] [Google Scholar]
- 19.Okamoto R, Uchikawa M, Kondoh H. Sixteen additional enhancers associated with the chicken Sox2 locus outside the central 50-kb region. Dev Growth Differ. 2015;57:24–39. doi: 10.1111/dgd.12185. [DOI] [PubMed] [Google Scholar]
- 20.Zhou HY, et al. A Sox2 distal enhancer cluster regulates embryonic stem cell differentiation potential. Genes Dev. 2014;28:2699–2711. doi: 10.1101/gad.248526.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Avilion AA, et al. Multipotent cell lineages in early mouse development depend on SOX2 function. Genes Dev. 2003;17:126–140. doi: 10.1101/gad.224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ellis P, et al. SOX2, a persistent marker for multipotential neural stem cells derived from embryonic stem cells, the embryo or the adult. Dev Neurosci. 2004;26:148–165. doi: 10.1159/000082134. [DOI] [PubMed] [Google Scholar]
- 23.Kang W, Hebert JM. A Sox2 BAC transgenic approach for targeting adult neural stem cells. PLoS One. 2012;7:e49038. doi: 10.1371/journal.pone.0049038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van Leeuwen S, Mikkers H. Long non-coding RNAs: Guardians of development. Differentiation. 2010;80:175–183. doi: 10.1016/j.diff.2010.07.003. [DOI] [PubMed] [Google Scholar]
- 25.Brockdorff N. Noncoding RNA and Polycomb recruitment. RNA. 2013;19:429–442. doi: 10.1261/rna.037598.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang KC, et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011;472:120–124. doi: 10.1038/nature09819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Latos PA, et al. Airn transcriptional overlap, but not its lncRNA products, induces imprinted Igf2r silencing. Science. 2012;338:1469–1472. doi: 10.1126/science.1228110. [DOI] [PubMed] [Google Scholar]
- 28.Martianov I, Ramadass A, Serra Barros A, Chow N, Akoulitchev A. Repression of the human dihydrofolate reductase gene by a non-coding interfering transcript. Nature. 2007;445:666–670. doi: 10.1038/nature05519. [DOI] [PubMed] [Google Scholar]
- 29.Crevillen P, Sonmez C, Wu Z, Dean C. A gene loop containing the floral repressor FLC is disrupted in the early phase of vernalization. EMBO J. 2013;32:140–148. doi: 10.1038/emboj.2012.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fantes J, et al. Mutations in SOX2 cause anophthalmia. Nat Genet. 2003;33:461–463. doi: 10.1038/ng1120. [DOI] [PubMed] [Google Scholar]
- 31.Amaral PP, et al. Complex architecture and regulated expression of the Sox2ot locus during vertebrate development. RNA. 2009;15:2013–2027. doi: 10.1261/rna.1705309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Askarian-Amiri ME, et al. Emerging role of long non-coding RNA SOX2OT in SOX2 regulation in breast cancer. PLoS One. 2014;9:e102140. doi: 10.1371/journal.pone.0102140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hou Z, et al. A long noncoding RNA Sox2ot regulates lung cancer cell proliferation and is a prognostic indicator of poor survival. Int J Biochem Cell Biol. 2014;53:380–388. doi: 10.1016/j.biocel.2014.06.004. [DOI] [PubMed] [Google Scholar]
- 34.Shahryari A, et al. Two novel splice variants of SOX2OT, SOX2OT-S1, and SOX2OT-S2 are coupregulated with SOX2 and OCT4 in esophageal squamous cell carcinoma. Stem Cells. 2014;32:126–134. doi: 10.1002/stem.1542. [DOI] [PubMed] [Google Scholar]
- 35.Tosetti V, et al. Transcriptional role of androgen receptor in the expression of long non-coding RNA Sox2OT in neurogenesis. PLoS One. 2017;12:e0180579. doi: 10.1371/journal.pone.0180579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hanada K, et al. sORF finder: a program package to identify small open reading frames with high coding potential. Bioinformatics. 2010;26:399–400. doi: 10.1093/bioinformatics/btp688. [DOI] [PubMed] [Google Scholar]
- 37.Smith JE, et al. Translation of small open reading frames within unannotated RNA transcripts in Saccharomyces cerevisiae. Cell Rep. 2014;7:1858–1866. doi: 10.1016/j.celrep.2014.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Woolfe A, et al. Highly conserved non-coding sequences are associated with vertebrate development. PLoS Biol. 2005;3:e7. doi: 10.1371/journal.pbio.0030007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Carninci P, Hayashizaki Y. Noncoding RNA transcription beyond annotated genes. Curr Opin Genet Dev. 2007;17:139–144. doi: 10.1016/j.gde.2007.02.008. [DOI] [PubMed] [Google Scholar]
- 40.Marques AC, Ponting CP. Catalogues of mammalian long noncoding RNAs: modest conservation and incompleteness. Genome Biol. 2009;10:R124. doi: 10.1186/gb-2009-10-11-r124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chodroff RA, et al. Long noncoding RNA genes: conservation of sequence and brain expression among diverse amniotes. Genome Biol. 2010;11:R72. doi: 10.1186/gb-2010-11-7-r72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Raj A, van den Bogaard P, Rifkin SA, van Oudenaarden A, Tyagi S. Imaging individual mRNA molecules using multiple singly labeled probes. Nat Methods. 2008;5:877–879. doi: 10.1038/nmeth.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wood HB, Episkopou V. Comparative expression of the mouse Sox1, Sox2 and Sox3 genes from pre-gastrulation to early somite stages. Mech Dev. 1999;86:197–201. doi: 10.1016/S0925-4773(99)00116-1. [DOI] [PubMed] [Google Scholar]
- 44.Duinsbergen D, Eriksson M, t Hoen PA, Frisen J, Mikkers H. Induced pluripotency with endogenous and inducible genes. Exp Cell Res. 2008;314:3255–3263. doi: 10.1016/j.yexcr.2008.06.024. [DOI] [PubMed] [Google Scholar]
- 45.Silva J, Chambers I, Pollard S, Smith A. Nanog promotes transfer of pluripotency after cell fusion. Nature. 2006;441:997–1001. doi: 10.1038/nature04914. [DOI] [PubMed] [Google Scholar]
- 46.West JA, et al. The long noncoding RNAs NEAT1 and MALAT1 bind active chromatin sites. Mol Cell. 2014;55:791–802. doi: 10.1016/j.molcel.2014.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rinn JL, Chang HY. Genome Regulation by Long Noncoding RNAs. Annu Rev Biochem. 2012;81:145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bernstein BE, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 49.Voigt P, Tee WW, Reinberg D. A double take on bivalent promoters. Genes Dev. 2013;27:1318–1338. doi: 10.1101/gad.219626.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dean A. On a chromosome far, far away: LCRs and gene expression. Trends Genet. 2006;22:38–45. doi: 10.1016/j.tig.2005.11.001. [DOI] [PubMed] [Google Scholar]
- 51.Zhang Y, et al. Chromatin connectivity maps reveal dynamic promoter-enhancer long-range associations. Nature. 2013;504:306–310. doi: 10.1038/nature12716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li Y, et al. CRISPR reveals a distal super-enhancer required for Sox2 expression in mouse embryonic stem cells. PLoS One. 2014;9:e114485. doi: 10.1371/journal.pone.0114485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lee TI, Young RA. Transcriptional regulation and its misregulation in disease. Cell. 2013;152:1237–1251. doi: 10.1016/j.cell.2013.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Maggio I, Goncalves MA. Genome editing at the crossroads of delivery, specificity, and fidelity. Trends Biotechnol. 2015;33:280–291. doi: 10.1016/j.tibtech.2015.02.011. [DOI] [PubMed] [Google Scholar]
- 55.Hagey DW, Muhr J. Sox2 acts in a dose-dependent fashion to regulate proliferation of cortical progenitors. Cell Rep. 2014;9:1908–1920. doi: 10.1016/j.celrep.2014.11.013. [DOI] [PubMed] [Google Scholar]
- 56.Ying QL, et al. The ground state of embryonic stem cell self-renewal. Nature. 2008;453:519–523. doi: 10.1038/nature06968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Messemaker TC, et al. A novel long non-coding RNA in the rheumatoid arthritis risk locus TRAF1-C5 influences C5 mRNA levels. Genes Immun. 2016;17:85–92. doi: 10.1038/gene.2015.54. [DOI] [PubMed] [Google Scholar]
- 58.Bain G, Kitchens D, Yao M, Huettner JE, Gottlieb DI. Embryonic stem cells express neuronal properties in vitro. Dev Biol. 1995;168:342–357. doi: 10.1006/dbio.1995.1085. [DOI] [PubMed] [Google Scholar]
- 59.Verma I, Rashid Z, Sikdar SK, Seshagiri PB. Efficient neural differentiation of mouse pluripotent stem cells in a serum-free medium and development of a novel strategy for enrichment of neural cells. Int J Dev Neurosci. 2017;61:112–124. doi: 10.1016/j.ijdevneu.2017.06.009. [DOI] [PubMed] [Google Scholar]
- 60.Engberg N, Kahn M, Petersen DR, Hansson M, Serup P. Retinoic acid synthesis promotes development of neural progenitors from mouse embryonic stem cells by suppressing endogenous, Wnt-dependent nodal signaling. Stem Cells. 2010;28:1498–1509. doi: 10.1002/stem.479. [DOI] [PubMed] [Google Scholar]
- 61.Conti L, et al. Niche-independent symmetrical self-renewal of a mammalian tissue stem cell. PLoS Biol. 2005;3:e283. doi: 10.1371/journal.pbio.0030283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Semrau S, et al. FuseFISH: robust detection of transcribed gene fusions in single cells. Cell Rep. 2014;6:18–23. doi: 10.1016/j.celrep.2013.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lee TI, Johnstone SE, Young RA. Chromatin immunoprecipitation and microarray-based analysis of protein location. Nat Protoc. 2006;1:729–748. doi: 10.1038/nprot.2006.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Stadhouders R, et al. Multiplexed chromosome conformation capture sequencing for rapid genome-scale high-resolution detection of long-range chromatin interactions. Nat Protoc. 2013;8:509–524. doi: 10.1038/nprot.2013.018. [DOI] [PubMed] [Google Scholar]
- 65.Denholtz M, et al. Long-range chromatin contacts in embryonic stem cells reveal a role for pluripotency factors and polycomb proteins in genome organization. Cell Stem Cell. 2013;13:602–616. doi: 10.1016/j.stem.2013.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Werner MS, Ruthenburg AJ. Nuclear Fractionation Reveals Thousands of Chromatin-Tethered Noncoding RNAs Adjacent to Active Genes. Cell Rep. 2015;12:1089–1098. doi: 10.1016/j.celrep.2015.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mikkelsen TS, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.