ABSTRACT
Chemosensory systems (CSS) are among the most complex organizations of proteins functioning cooperatively to regulate bacterial motility and other cellular activities. These systems have been studied extensively in bacteria, and usually, they are present as a single system. Eight CSS, the highest number in bacteria, have been reported in Myxococcus xanthus DK1622 and are involved in coordinating diverse functions. Here, we have explored and compared the CSS in all available genomes of order Myxococcales. Myxococcales members contain 97 to 476 two-component system (TCS) proteins, which assist the bacteria in surviving and adapting to varying environmental conditions. The number of myxobacterial CSS ranges between 1 and 12, with the largest number in family Cystobacteraceae and the smallest in Nannocystaceae. CheA protein was used as a phylogenetic marker to infer evolutionary relatedness between different CSS, and six novel CSS (“extra CSS” [ECSS]) were thus identified in the myxobacteria besides the previously reported Che1 to Che8 systems from M. xanthus. Che1 to Che8 systems are monophyletic to deltaproteobacteria, whereas the newly identified ECSS form separate clades with different bacterial classes. The comparative modular organization was concordant with the phylogeny. Four clusters lacking CheA proteins were also identified via CheB-based phylogenetic analysis and were categorized as accessory CSS (ACSS). In Archangium, an orphan CSS was identified, in which both CheA and CheB were absent. The novel, accessory, and orphan multimodular CSS identified here suggest the emergence of myxobacterial CSS and could assist in further characterizing their roles.
IMPORTANCE This study is focused on chemosensory systems (CSS), which help the bacterium in directing its movement toward or away from chemical gradients. CSS are present as a single system in most of the bacteria except in some groups, including Myxococcus xanthus, which has 8 CSS, the highest number reported to date. This is the first comprehensive study carrying out a comparative analysis of the 22 available myxobacterial genomes, which suggests the evolutionary diversity of these systems. We are interested in understanding the distribution of CSS within all known myxobacteria and their probable evolution.
KEYWORDS: chemotaxis, modular organization, phylogeny, signal transduction, two-component system, evolution
INTRODUCTION
Myxobacteria, Gram-negative deltaproteobacteria, are aerobic gliding bacteria that form swarms (1, 2) and develop fruiting bodies under starvation conditions (3–5). These organisms exhibit a complex life cycle during which numerous cell-cell interactions define their peculiar physiological attributes, such as gliding motility (6, 7), biofilm production (8), and predatory characteristics (9). Furthermore, myxobacteria contain a large number of proteins involved in signal transduction pathways and transcriptional regulation (10–12). These proteins help in regulating cell-cell communication and coordinate social motility and fruiting body formation (3). The predation and fruiting body formation of myxobacteria depend on directed cell movement, controlled by chemosensory systems (13). On perceiving environmental stimuli, these organisms regulate their growth and enhance their fitness either by moving away or by adapting to the environmental change (1, 14).
In the bacterial kingdom, proteins of two-component systems (TCS) help in transducing the environmental stimuli through a phosphotransfer mechanism (11, 15). These TCS proteins harbor a sensor histidine kinase (HK), which responds to the stimulus by activating its autokinase domain, and a response regulator (RR), which on phosphorylation activates the output domain, leading to transcriptional control at either the DNA or RNA level (11, 16, 17). Bacteria harbor a number of TCS proteins to coordinate diverse functions in response to variable stimuli (18–20). Previous genome-wide distribution studies have reported TCS proteins in many bacterial groups (21). Similar studies reporting the distribution of TCS in myxobacteria were performed (10). The functional role of many of the TCS in Myxococcus xanthus has been predicted in sporulation and gliding motility of the bacteria (3). Further, the orphan TCS proteins present in M. xanthus were suggested to be involved in vegetative cell growth, developmental processes, and secondary metabolism (12).
A subset of the TCS proteins are involved in specialized multiprotein systems, the chemosensory systems (CSS), which help the bacterium in directing its movement toward or away from chemical gradients (14, 17, 22). In CSS, the extracellular signals are perceived by specialized chemoreceptors known as methyl-accepting chemotaxis proteins (MCPs) instead of HK as in the TCS (23). MCPs contain a sensory domain, a HAMP domain, and a CheA-interacting signaling domain (24). Scaffolding proteins CheW and CheV (a CheW-like protein) assist in the interaction of the MCP and CheA, which together form a signaling complex (17, 25). After receiving stimuli, CheA protein becomes autophosphorylated and further transfers the phosphoryl moiety to the response regulator CheY. Here CheA and CheY proteins behave like TCS proteins, functionally regulated by other chemotaxis proteins (23). The phosphorylated CheY protein interacts with the flagellum switch complex, which in turn drives the flagellar motor (22). All the components of the chemotaxis excitation pathway, viz., MCPs, CheW, CheA, and CheY, lead to signal interaction with the motility organelle (23, 24). In some chemotaxis systems, one or more phosphatases, like CheC and CheX, that dephosphorylate CheY are also present (24). Also, one methylesterase, CheB, and one methyltransferase, CheR, assist in memorizing the signals in the process of chemotaxis adaptation (24, 25). CheR and CheB work in an antagonistic manner, whereby CheR adds a methyl group to a specific glutamate residue at the C terminus of the MCP receptor whereas the phosphorylated CheA by phosphotransfer activates the CheB that removes the methyl group as methanol, to reset the transducer to the sensing mode (24). Also, CheD protein interacts with CheC to increase its dephosphorylation activity. This cascade of combinations of these eight proteins constitutes the most complex signal transduction system known in prokaryotes, i.e., CSS (13, 14, 23).
Various studies have reported the distribution of these systems across diverse bacteria that perform specific functions (26–30). Most of the bacterial species have one CSS, but certain species of Gammaproteobacteria, such as Pseudomonas aeruginosa, have four CSS, Sinorhizobium meliloti and Rhodobacter sphaeroides, which belong to Alphaproteobacteria, have two and three CSS, respectively, while Myxococcus xanthus, a deltaproteobacterium, has eight CSS (25). The presence of more than one CSS has been studied for their evolutionary pattern and functional relatedness (14, 28, 30). Typical CSS are found to be involved in chemotaxis and flagellum-based motility, but the presence of multiple CSS encoded in a genome has been related to alternative cellular functions such as biofilm formation, flagellum biosynthesis, developmental gene expression, etc. (14, 31, 32). As myxobacteria do not have flagella, CSS are involved in regulating motility via type IV pili. Various CSS in M. xanthus have been verified for their diverse roles in different cellular transduction processes, such as Che1, which is involved in regulation of reversal frequency (33–35), Che2, which is involved in regulation of EPS production (36, 37), Che3 in transcriptional regulation (26, 31), and Che7 in the regulation of M. xanthus development (32). In M. xanthus, many TCS proteins are essential for motility and fruiting body formation (3, 11, 38). Along with a large repertoire of TCS proteins and CSS, the M. xanthus DK1622 genome encodes 21 MCPs, which have been suggested to pass the environmental signal to multiple CSS modules (25). These modules work in a collaborative way to regulate complex behaviors such as swarm motility, biofilm formation, and various other alternative cellular functions (13, 25, 27, 32). Thus, to gain a comprehensive understanding of the number, types, and roles of the CSS in myxobacteria, we have performed a comparative analysis of CSS in all available complete and draft genomes of myxobacteria.
Definitions. (i) CSS.
CSS is a multiprotein two-component system involved in chemotaxis/motility and various alternative cellular functions, such as biofilm formation, flagellum biosynthesis, developmental gene expression, etc. In M. xanthus DK1622, eight CSS (Che1 to Che8) have been reported earlier.
(ii) ECSS.
Besides Che1 to Che8, this study also highlights the presence of additional CSS within myxobacteria, designated here “extra CSS” (ECSS). We identified ECSS1 to ECSS6 in this study. They should be functionally similar to CSS.
(iii) ACSS.
This study depicts some clusters within myxobacteria in which CheA protein, the major constituent of a CSS, is absent. Here these clusters are designated accessory CSS (ACSS). We identified ACSS1 to ACSS4 in this study. Owing to the absence of CheA protein, they may have lost their function.
RESULTS
Genomic characteristics of myxobacteria sequenced in this study.
High-quality draft genomes of Cystobacter fuscus DSM 2262T, Hyalangium minutum DSM 14724T, and Chondromyces apiculatus DSM 436 were assembled in 76 contigs (shortest sequence length at 50% of the genome [N50], 457,853 bp; size, 12.28 Mbp), 44 contigs (N50, 504,291 bp; size, 11.19 Mbp), and 182 contigs (N50, 120,949 bp; size, 11.58 Mbp), respectively (Table 1). High GC content, comparable to that of other myxobacteria (3, 39), was observed (GC contents in C. fuscus, H. minutum, and C. apiculatus, 68.2, 68.0, and 70.3%, respectively). In C. fuscus, 5,196 (49.42%) of 10,515 proteins are annotated as hypothetical proteins. In H. minutum and C. apiculatus genome annotations, 3,364 (36.93%) and 4,962 (54.90%) proteins of 9,109 and 9,038 were annotated as hypothetical proteins, respectively. These whole-genome shotgun projects have been deposited at DDBJ/EMBL/GenBank (see Materials and Methods). The genome overview of members of order Myxococcales used in this study is given in Table 2.
TABLE 1.
Assembly and annotation features of genomes sequenced with Illumina HiSeq 1000
| Feature (unit) | Cystobacter fuscus DSM 2262 | Hyalangium minutum DSM 14724 | Chondromyces apiculatus DSM 436 |
|---|---|---|---|
| Assembly method | CLC NGS Cell v. wb 6.0 | SPAdes v. 3.0 | CLC NGS Cell v. wb 6.0 |
| Total no. of reads | 91,397,672 | 20,783,476 | 43,878,720 |
| Total no. of bases | 9,231,164,872 | 2,099,131,076 | 4,431,750,720 |
| Avg reference coverage | 741× | 187× | 370× |
| BioProject no. | PRJNA177202 | PRJNA242458 | PRJNA192263 |
| NCBI accession no. | ANAH00000000 | JMCB00000000 | ASRX00000000 |
| No. of contigs | 76 | 44 | 182 |
| GC% | 68.2 | 68.0 | 70.3 |
| Genome size (Mbp) | 12.28 | 11.19 | 11.58 |
| N50 size (bp) | 457,853 | 504,291 | 120,949 |
| Longest contig size (bp) | 1,016,131 | 1,229,042 | 374,143 |
| No. of CDSa | 10,515 | 9,109 | 9,038 |
| % coding density | 89.09 | 89.57 | 87.90 |
| No. of CDS from (+) strand | 5,161 | 4,551 | 4,425 |
| No. of CDS from (−) strand | 5,354 | 4,558 | 4,613 |
| Maximum CDS length | 25,547 | 31,466 | 26,225 |
| Mean CDS length | 1,038 | 1,109 | 1,126 |
| No. of hypothetical proteins | 5,196 | 3,364 | 4,962 |
| % of hypothetical proteins | 49.42 | 36.93 | 54.90 |
| No. of tRNAs | 63 | 70 | 79 |
CDS, coding sequence.
TABLE 2.
Genome overview of order Myxococcalesa
| Organism name | Sequencing center | NCBI accession no. | No. of contigs | N50 size (bp) | GC% | Size (Mb) | No. of genes |
|---|---|---|---|---|---|---|---|
| Vulgatibacter incomptus DSM 27710 | CSIR-IMTECH | CP012332.1 | 1 | 68.9 | 4.35 | 3,488 | |
| Anaeromyxobacter dehalogenans 2CP-1 | DOE-JGI | NC_011891.1 | 1 | 74.7 | 5.03 | 4,473 | |
| Archangium gephyra DSM 2261 | CSIR-IMTECH | CP011509.1 | 1 | 69.4 | 12.49 | 10,121 | |
| Cystobacter fuscus DSM 2262 | CSIR-IMTECH | ANAH00000000 | 76 | 457,853 | 68.2 | 12.28 | 10,515 |
| Cystobacter violaceus Cbvi76 | University of Notre Dame | JPMI00000000 | 431 | 75,432 | 68.9 | 12.54 | 8,373 |
| Hyalangium minutum DSM 14724 | CSIR-IMTECH | JMCB00000000 | 44 | 504,291 | 68.0 | 11.01 | 9,109 |
| Stigmatella aurantiaca DW4/3-1 | Max Planck, Marburg | NC_014623.1 | 1 | 67.5 | 10.26 | 8,444 | |
| Corallococcus coralloides DSM 2259 | Max Planck, Marburg | NC_017030.1 | 1 | 69.9 | 10.08 | 8,033 | |
| Myxococcus fulvus HW-1 | Shandong University | NC_015711.1 | 1 | 70.6 | 9.00 | 7,284 | |
| Myxococcus hansupus | CSIR-IMTECH | CP012109 | 1 | 69.2 | 9.44 | 7,666 | |
| Myxococcus stipitatus DSM 14675 | MPITM | NC_020126.1 | 1 | 69.2 | 10.35 | 8,043 | |
| Myxococcus xanthus DK 1622 | TIGR | NC_008095.1 | 1 | 68.9 | 9.14 | 7,316 | |
| Myxococcus xanthus DZF1 | University of Iowa | AOBT00000000 | 75 | 305,520 | 68.8 | 9.28 | 7,705 |
| Myxococcus xanthus DZ2 | University of Iowa | AKYI00000000 | 87 | 195,218 | 68.9 | 9.27 | 7,691 |
| Enhygromyxa salina DSM 15201 | CSIR-IMTECH | JMCC00000000 | 330 | 81,190 | 67.4 | 10.44 | 8,170 |
| Haliangium ochraceum SMP-2 DSM 14365 | DOE-JGI | NC_013440.1 | 1 | 69.5 | 9.45 | 6,719 | |
| Plesiocystis pacifica SIR-1 | JCVI | ABCS00000000 | 237 | 82,268 | 70.7 | 10.59 | 8,461 |
| Sandaracinus amylolyticus DSM 53668 | CSIR-IMTECH | CP011125.1 | 1 | 67.4 | 10.11 | 8,127 | |
| Chondromyces apiculatus DSM 436 | CSIR-IMTECH | ASRX00000000 | 182 | 120,949 | 70.3 | 11.58 | 9,038 |
| Sorangium cellulosum So0157-2 | Shandong University | NC_021658.1 | 1 | 72.1 | 14.78 | 10,400 | |
| Sorangium cellulosum Soce56 | Bielefeld University | NC_010162.1 | 1 | 71.4 | 13.03 | 9,375 | |
| Labilithrix luteola DSM 27648 | CSIR-IMTECH | CP012333.1 | 1 | 66.1 | 12.19 | 11,518 |
TIGR, The Institute for Genomic Research; DOE-JGI, Department of Energy, Joint Genome Institute; JCVI, J. Craig Venter Institute; MPITM, Max Planck Institute for Terrestrial Microbiology. Data in boldface pertain to organisms sequenced in this study.
Large-scale expansion of TCS proteins in the order Myxococcales.
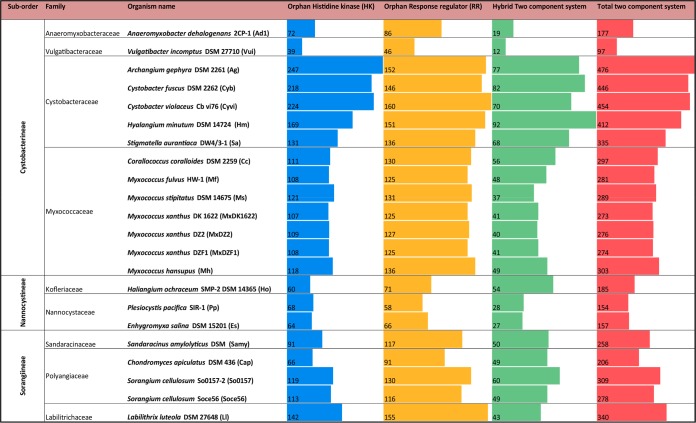
TCS proteins contain different combinations of constituent domains; HK, RR, input, and output domains. On the basis of these domain combinations, TCS proteins have been categorized as orphan HK, orphan RR, and hybrid (HK+RR) (40). In this study, TCS proteins were identified for myxobacterial genomes, and their distributions were compared (Fig. 1). Our study identified 97 to 476 TCS proteins distributed among members of order Myxococcales. For six genus Myxococcus genomes, the number of TCS proteins varies from 273 to 303 (Fig. 1). In the family Myxococcaceae, 303 TCS proteins were identified in Myxococcus hansupus, followed by 289 in Myxococcus stipitatus, 281 in Myxococcus fulvus HW-1, and 273 in M. xanthus DK1622. The members of family Cystobacteraceae have the largest numbers (from 335 in Stigmatella aurantiaca DW4/3-1 to 476 in Archangium gephyra DSM 2261T) of TCS proteins, compared to the numbers in Nannocystaceae (from 154 in Plesiocystis pacifica SIR-1 to 185 in Haliangium ochraceum SMP-2 DSM 14365T) and suborder Sorangiineae suborder (from 206 in C. apiculatus DSM 436 to 340 in Labilithrix luteola DSM 27648T). A recent study (10) identified 338 TCS proteins in S. aurantiaca DW4/3-1, 160 in P. pacifica SIR-1, 191 in H. ochraceum SMP-2 DSM 14365T, and 273 in Sorangium cellulosum Soce56, which is concordant with our findings. P. pacifica SIR-1, H. ochraceum SMP-2 DSM 14365T, and Enhygromyxa salina DSM 15201 are all marine myxobacteria with an ∼10-Mbp genome and have smaller sets of TCS proteins (154 to 185) than those (∼300) in soil myxobacteria with similar genome size.
FIG 1.
Categorization and distribution of TCS proteins involved in signal transduction among order Myxococcales genomes. Categorization was into three classes, orphan HK, orphan RR, and hybrid TCS proteins, based on their Pfam domain combinations. Their quantitative distribution among the given genomes is depicted by bar graphs in the columns.
Our study identified that the largest genome studied, that of S. cellulosum So0157-2 (14.78 Mbp), encodes 309 TCS, whereas the 12.49-Mbp A. gephyra DSM 2261T genome encodes the largest number of TCS (476 TCS). In Anaeromyxobacter dehalogenans 2CP-1 (5.03 Mbp) 177 TCS proteins were identified, whereas in Vulgatibacter incomptus DSM 27710T (4.35 Mbp), the smallest myxobacterium known, 97 TCS proteins were present. The genome size difference between A. dehalogenans 2CP-1 and V. incomptus DSM 27710T is 0.6 Mbp, but the number of TCS proteins in the former was approximately one-half of the number in the latter. The number of TCS in V. incomptus DSM 27710T is even greater than that of Escherichia coli (4.64 Mbp; 62 TCS proteins), an organism with an almost-equal genome size. In earlier studies, the number of TCS proteins has been shown to be correlated with genome size (25, 41–43), but in our analysis for myxobacterial genomes, no such direct correlation could be drawn (see Fig. S2 in the supplemental material).
Distribution of chemotaxis proteins.
The core modular organization of CSS comprises CheA, CheW, CheY, and MCP and may additionally contain CheR, CheB, CheX, CheC, and CheD. CSS identified in myxobacteria are composed of a maximum of one unit of CheA, CheB, and CheR proteins and multiple units of MCPs and CheW per system (Table 3). Variable distribution of CheB, CheD, and CheR modules was observed in myxobacterial CSS. In this study, a functional CSS (CSS and newly identified ECSS) corresponds to two or more adjacent Che proteins among which one protein is CheA, whereas a nonfunctional CSS (ACSS) may have any set of Che proteins.
TABLE 3.
Distribution of chemotaxis constituent proteins and CSS for order Myxococcales genomes
| Suborder and family | Organism name | No. of: |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CheA | CheB | CheBR | CheC | CheD | CheR | CheRD | CheW | CheX | MCPsignal | Che systems | ECSS | CheA alone | ACSS | ||
| Cystobacterineae | |||||||||||||||
| Anaeromyxobacteraceae | Anaeromyxobacter dehalogenans 2CP-1 | 7 | 6 | 1 | 2 | 8 | 12 | 1 | 17 | 4 | 3 | ||||
| Vulgatibacteraceae | Vulgatibacter incomptus DSM 27710 | 2 | 1 | 3 | 3 | 2 | 2 | ||||||||
| Cystobacteraceae | Archangium gephyra DSM 2261 | 10 | 8 | 1 | 1 | 12 | 18 | 32 | 7 | 2 | 1 | ||||
| Cystobacter fuscus DSM 2262 | 12 | 10 | 1 | 12 | 19 | 28 | 7 | 5 | 1 | ||||||
| Cystobacter violaceus Cb vi76 | 10 | 8 | 1 | 12 | 16 | 25 | 7 | 2 | 1 | ||||||
| Hyalangium minutum DSM 14724 | 10 | 15 | 2 | 1 | 1 | 12 | 18 | 26 | 8 | 2 | 3 | ||||
| Stigmatella aurantiaca DW4/3-1 | 10 | 12 | 1 | 1 | 1 | 12 | 18 | 22 | 8 | 2 | 3 | ||||
| Myxococcaceae | Corallococcus coralloides DSM 2259 | 10 | 9 | 1 | 11 | 15 | 21 | 8 | 2 | 2 | |||||
| Myxococcus fulvus HW-1 | 9 | 7 | 1 | 10 | 15 | 22 | 8 | 1 | 1 | ||||||
| Myxococcus stipitatus DSM 14675 | 7 | 5 | 1 | 7 | 13 | 19 | 7 | ||||||||
| Myxococcus xanthus DK 1622 | 8 | 7 | 1 | 9 | 14 | 21 | 8 | 1 | |||||||
| Myxococcus xanthus DZ2 | 8 | 7 | 1 | 9 | 14 | 21 | 8 | 1 | |||||||
| Myxococcus xanthus DZF1 | 8 | 7 | 1 | 9 | 14 | 21 | 8 | 1 | |||||||
| Myxococcus hansupus | 8 | 8 | 1 | 10 | 14 | 22 | 8 | 2 | |||||||
| Nannocystineae | |||||||||||||||
| Kofleriaceae | Haliangium ochraceum SMP-2 DSM 14365 | 2 | 5 | 6 | 4 | 3 | 4 | 1 | 1 | 2 | |||||
| Nannocystaceae | Plesiocystis pacifica SIR-1 | 1 | 1 | 1 | 2 | 2 | 1 | ||||||||
| Enhygromyxa salina DSM 15201 | 3 | 2 | 1 | 2 | 3 | 3 | 1 | 1 | 1 | ||||||
| Sorangiineae | |||||||||||||||
| Sandaracinaceae | Sandaracinus amylolyticus DSM 53668 | 5 | 5 | 4 | 7 | 5 | 3 | 2 | |||||||
| Polyangiaceae | Chondromyces apiculatus DSM 436 | 4 | 5 | 3 | 8 | 9 | 6 | 3 | 2 | 1 | |||||
| Sorangium cellulosum So0157-2 | 5 | 5 | 4 | 1 | 6 | 7 | 1 | 5 | 2 | 3 | 1 | ||||
| Sorangium cellulosum Soce56 | 5 | 2 | 2 | 1 | 4 | 6 | 2 | 5 | 2 | 3 | |||||
| Labilitrichaceae | Labilithrix luteola DSM 27648 | 2 | 3 | 1 | 3 | 1 | 5 | 2 | 6 | 1 | 1 | ||||
The CheA proteins identified in myxobacteria were marked by the presence of a histidine phosphotransfer domain, an ATPase domain, and a CheW scaffolding domain using the HMM profile from the Pfam database. H-kinase_dim in CheA proteins could not be traced using Pfam analysis owing to its poor domain model (24). Further, all the CheA protein sequences were aligned and we found that all four domains were well conserved (see Fig. S3 in the supplemental material).
CheW, CheB, and CheR proteins have only one domain each, PF01584 (CheW), PF01339 (CheB_methylest), and PF01739 (CheR). However, in some CheW proteins the respective domains conjugated with PF00072 (Response_reg) domains were identified, known as CheV proteins, which function in a fashion similar to that of CheW. Therefore, in this study, CheW and CheV proteins are grouped together. We also identified some CheB proteins in which PF00072 (Response_reg) was present. In some myxobacteria, CheB and CheR domains were fused together, representing a bifunctional protein with both methylesterase and methyltransferase functions. Fused CheBR were present in all members of order Myxococcales, with the exception of families Myxococcaceae and Vulgatibacteraceae. CheD and CheX homologs were reported previously to be present in beta- and gammaproteobacteria (17, 44) but were absent from M. xanthus DK1622 (25). Our study revealed that these modules were present in a few myxobacterial genomes; for example, CheD homologs were present in A. dehalogenans 2CP-1, H. minutum, S. aurantiaca DW4/3-1, and Sorangium spp., and CheX homologs were identified in A. dehalogenans 2CP-1, Sorangium spp., and L. luteola DSM 27648T. Fused CheR and CheD (as CheRD) were also present in the L. luteola DSM 27648T genome.
The myxobacterial MCP sequences were identified with different domain architectures in a combination of either a sensory domain (CHASE, PF03924; Cache_2, PF08269; and 4HB_MCP_1, PF12729), a HAMP linker domain (HAMP, PF00672), or a signaling domain (MCPsignal, PF00015) that interacts with CheA. In suborder Cystobacterineae, the number of MCPs was more than twice the number of CheA proteins, with the exception of V. incomptus DSM 27710T, in which only two MCPs were present, corresponding to the two CheA proteins. The approximately equal ratio of MCPs and CheA was observed in suborders Sorangiineae and Nannocystineae. As suggested in previous studies (24, 25), a high prevalence of MCPs in Cystobacterineae might be related to the broad spectra of stimuli received owing to their diverse habitats and complex life cycle.
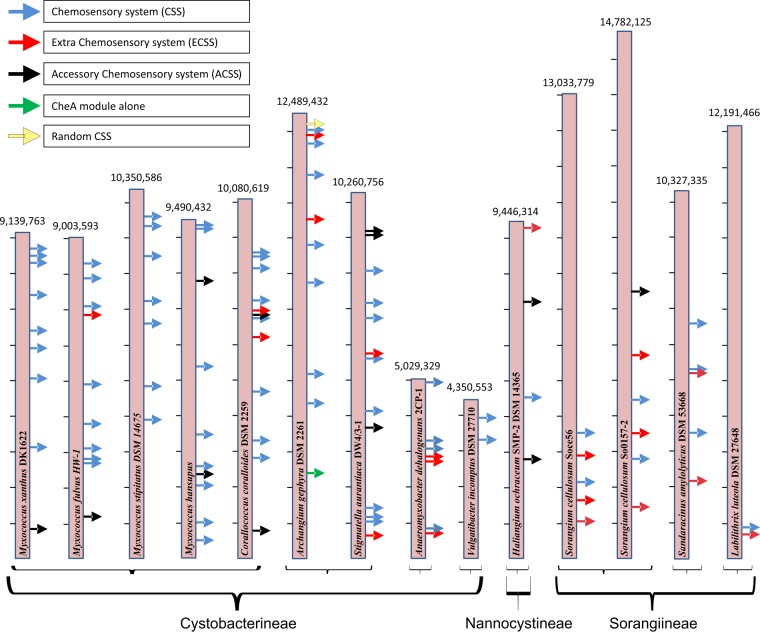
The number of CheA proteins is related to the number of CSS (denoted by the blue arrow in Fig. 2) present in an organism, but some CheA proteins are orphans (denoted by the green arrow in Fig. 2). No new CSS could be identified in M. xanthus DK1622, but we found differences within members of genus Myxococcus. We could trace seven CheA proteins and seven CSS in M. stipitatus and nine CheA proteins corresponding to nine CSS in M. fulvus. In M. hansupus, eight CheA proteins and corresponding eight CSS were identified, similar to the CSS distribution in M. xanthus DK1622 (25). For the genome of Corallococcus coralloides DSM 2259T, a close relative of the genus Myxococcus, 10 CheA proteins and their corresponding CSS were identified. In the Nannocystaceae family, only one CheA protein and one CSS in P. pacifica SIR-1 and two CheA proteins and corresponding two CSS in H. ochraceum SMP-2 DSM 14365T were identified. To our surprise, the suborder Sorangiineae, comprising members with the largest genomes, harbors just one-half the numbers of CSS found in suborder Cystobacterineae, which has the most abundant CSS; that is, S. cellulosum Soce56 and S. cellulosum So0157-2 genomes encode only five CheA proteins organized in five CSS compared to the average 10 CSS observed in suborder Cystobacterineae. In the members of subfamily Cystobacteraceae, more than 10 CheA proteins were encoded by all genomes whereas the maximum 12 CheA proteins, corresponding to 12 CSS, were identified for the C. fuscus draft genome reported here. Within this family, orphan CheA proteins that were not part of any CSS were also identified. In A. gephyra DSM 2261T, one CheA protein was present alone with no CSS constituent proteins present in the vicinity (denoted by the dark green arrow in Fig. 2). Similarly, for the draft genome of C. violaceus Cb vi76 (45) of subfamily Cystobacteraceae, a single CheA protein was observed apart from 10 complete CSS. E. salina DSM 15201, a member of suborder Nannocystineae, was found to have three CheA proteins that correspond to two CSS and an orphan CheA protein.
FIG 2.
Physical map showing the distribution of CSS in the myxobacterial complete genomes. The genome size and name are indicated for each bar. On the left side, each notch represents 1 Mbp of genome size. Arrows depict diverse categories of CSS: CSS, ECSS, ACSS, orphan CheA (CheA module alone), and unorganized CSS (Random CSS).
Apart from these complete CSS, a few other CSS that did not have CheA protein but had CheB and CheR modules were also observed. These systems are characterized as accessory CSS (ACSS) (denoted by a black arrow in Fig. 2) in this study. For the M. xanthus DK1622 genome, we identified one ACSS that stood apart from eight complete CSS in encoding no CheA, but it encoded CheB and CheR. Similar to what was observed in M. xanthus DK1622, ACSS were observed, ranging from zero to three, in other members of order Myxococcales. A single occurrence of ACSS was found in M. xanthus DK1622, M. fulvus, C. fuscus, C. apiculatus DSM 436, and S. cellulosum So0157-2, whereas two ACSS were found in C. coralloides DSM 2259T, M. hansupus, and H. ochraceum SMP-2 DSM 14365T and three ACSS in H. minutum and S. aurantiaca DW4/3-1. No ACSS was observed in M. stipitatus, A. dehalogenans, V. incomptus DSM 27710T, C. violaceus Cb vi76, S. cellulosum Soce56, Sandaracinus amylolyticus DSM 53668T, L. luteola DSM 27648T, P. pacifica SIR-1, and E. salina DSM 15201. These ACSS can be considered degraded CSS that likely play some other functional role. Similar to CSS, adjacent RR or HK or TCS proteins were also identified in ACSS. Apart from “extra CSS” (ECSS) and ACSS, one orphan CSS, A. gephyra CSS 10 (Ag-10), was identified with the modular organization CheR-W-MCP-MCP-W-x-x-Y, here called orphan, as both CheA and CheB were absent.
Evolutionary analysis of CSS systems in myxobacteria.
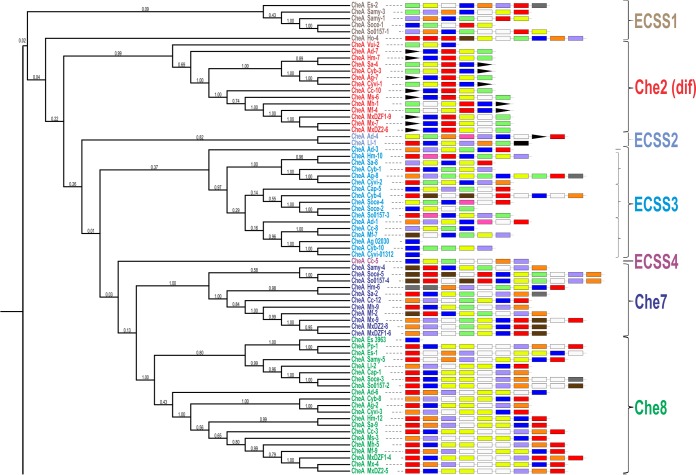
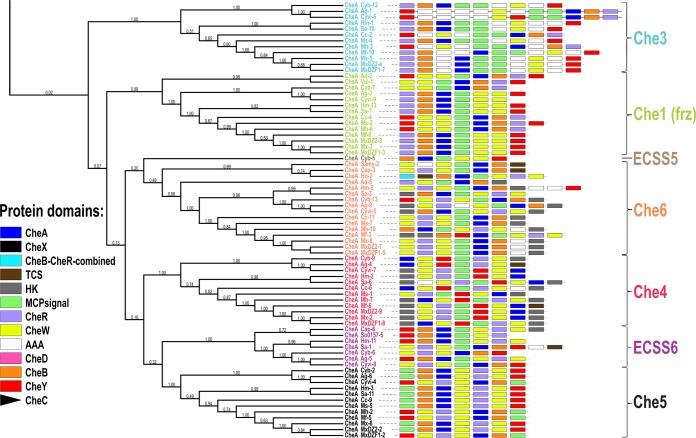
The genome-wide distribution of the CSS revealed the presence of a greater number of CSS in order Myxococcales members than the eight CSS in M. xanthus (25) and than those in other bacterial members (28, 30, 46). Myxobacterial CheA proteins were used for phylogenetic analysis, system modules architecture studies, and synteny analysis, in order to identify novel and diverse CSS. The phylogenetic tree of myxobacterial CheA proteins separates the tree into two large groups, group I and group II (Fig. 3). These groups comprise seven clades each, every demarcated clade having a unique organization of Che modules. Of 14 clades, 8 clades were classified corresponding to the eight previously characterized CSS in M. xanthus DK1622 (CheAMx) and the remaining 6 unmapped clades have been named ECSS (Table 4).
FIG 3.
CheA protein-based phylogeny for all CheA homologs in order Myxococcales genomes. The CheA protein-based phylogenetic tree is represented along with the respective CSS architecture (all CheA modules are portrayed here as having equal lengths, with color codes as shown in the bottom left corner) and name next to each terminal branch in each clade.
TABLE 4.
Classification and categorization of CSS, ECSS, and ACSS for order Myxococcales genomes by CheA-based phylogeny and modular organization
| CSS |
Cystobacterineae |
Sorangiineae |
Nannocystineae |
|||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
Myxococcaceae |
Anaeromyxobacteraceae: A. dehalogenans | Vulgatibacteraceae: V. incomptus |
Cystobacteraceae |
Polyangiaceae |
Sandaracinaceae: S. amylolyticus | Labilitrichaceae: L. luteola | Kofleriaceae: H. ochraceum |
Nannocystaceae |
||||||||||||
| M. xanthus DK1622 | M. fulvus | M. stipitatus | M. hansupus | C. coralloides | A. gephyra | C. fuscus | C. violaceus | H. minutum | S. aurantiaca | C. apiculatus | S. cellulosum 56 | S. cellulosum 0157 | P. pacifica | E. salina | ||||||
| Che1 (Frz) | Mx-3 | Mf-8 | Ms-2 | Mh-6 | Cc-4 | Ad-2 | Vui-1 | Ag-3 | Cyb-7 | Cyvi-9 | Hm-13 | Sa-7 | ||||||||
| Che2 (Dif) | Mx-7 | Mf-4 | Ms-6 | Mh-1 | Cc-10 | Ad-7 | Vui-2 | Ag-7 | Cyb-3 | Cyvi-1 | Hm-7 | Sa-4 | ||||||||
| Che3 | Mx-5 | Mf-10 | Ms-4 | Mh-3 | Cc-2 | Ag-1 | Cyb-12 | Cyvi-6 | Hm-1 | Sa-10 | ||||||||||
| Che4 | Mx-2 | Mf-6 | Ms-1 | Mh-7 | Cc-6 | Ag-4 | Cyb-9 | Cyvi-7 | Hm-2 | Sa-6 | ||||||||||
| Che5 | Mx-6 | Mf-5 | Ms-5 | Mh-2 | Cc-9 | Ag-6 | Cyb-2 | Cyvi-4 | Hm-3 | Sa-11 | ||||||||||
| Che6 | Mx-8 | Mf-3 | Ms-7 | Mh-10 | Cc-11 | Ad-5 | Ag-9 | Cyb-13 | Cyvi-5 | Hm-5 | Sa-3 | Cap-3 | Samy-2 | Ho-2 | ||||||
| Che7 | Mx-9 | Mf-2 | Mh-9 | Cc-12 | Hm-6 | Sa-2 | Cap-4a | Soce56-5 | So0157-4 | Samy-4 | ||||||||||
| Che8 | Mx-4 | Mf-9 | Ms-3 | Mh-5 | Cc-3 | Ad-6 | Ag-2 | Cyb-8 | Cyvi-3 | Hm-12 | Sa-9 | Cap-1 | Soce56-3 | So0157-2 | Samy-5 | Ll-2 | Pp-1 | Es-1, 3963b | ||
| ECSS1 | Soce56-1 | So0157-1 | Samy-3, Samy-1 | Ho-4 | Es-2 | |||||||||||||||
| ECSS2 | Ad-4 | Ll-1 | ||||||||||||||||||
| ECSS3 1 | Ad-3 | |||||||||||||||||||
| ECSS3 2 | Ag-8 | Cyb-1 | Cyvi-2 | Hm-10 | Sa-8 | |||||||||||||||
| ECSS3 3 | Cyb-4 | Cap-5 | Soce56-2, Soce56-4 | So0157-3 | ||||||||||||||||
| ECSS3 4 | Mf-7 | Cc-8 | Ad-1 | 2030b | Cyb-10 | 1312b | ||||||||||||||
| ECSS4 | Cc-5 | |||||||||||||||||||
| ECSS5 | Cyb-5 | |||||||||||||||||||
| ECSS6 | Ag-5 | Cyb-6 | Cyvi-8 | Hm-11 | Sa-1 | Cap-6 | So0157-5 | |||||||||||||
| ACSS1 | Hm-8 | |||||||||||||||||||
| ACSS2 | Cc-1 | Cyb-11 | Sa-12 | Cap-2 | So0157-6 | Ho-3 | ||||||||||||||
| ACSS3 | Mh-4 | Cc-7 | Hm-9 | Sa-5 | ||||||||||||||||
| ACSS4 | Mx-1 | Mf-1 | Mh-8 | Hm-4 | Sa-13 | Ho-1 | ||||||||||||||
Classified by CheB-based phylogeny.
Solitary CheA module present in genome (as depicted by green arrow in Fig. 2).
In addition, the CheA protein sequences from the different phyla were added to the myxobacterial CheA proteins to find the evolutionary relatedness of the CSS observed in myxobacteria to those of other bacteria. The phylogenetic analysis performed here was concordant to the myxobacterial CheA protein-based phylogeny. The CheA members corresponding to a unique taxonomic class were collapsed and are represented as a triangle (see Fig. S4 in the supplemental material) to depict the evolutionary relations.
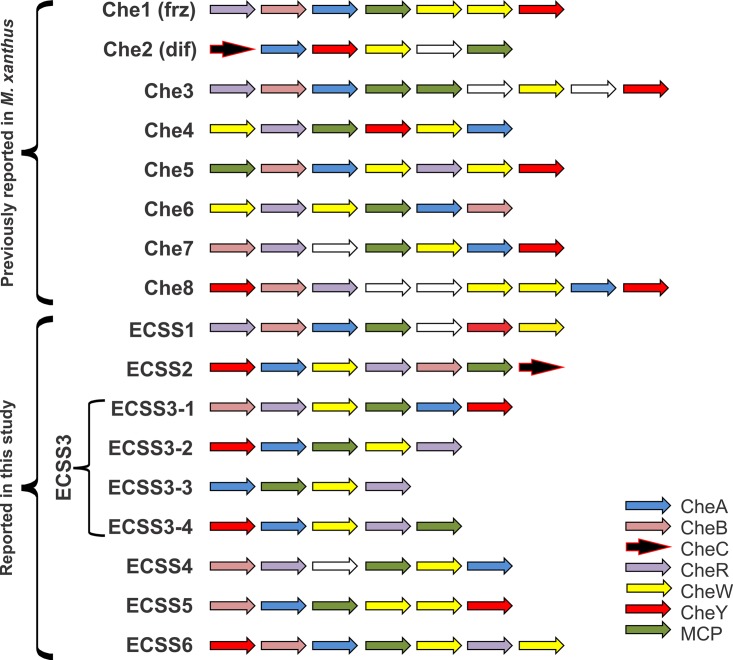
Concordant with the phylogenetic tree, the representative clades were named CSS and ECSS based on the clade demarcation and the modular architecture of the complete cluster. Group I is divided into seven clades, three of these clades having the characterized CheAMx belonging to Che2, Che7, and Che8 whereas the other four clades did not have the characterized CheA proteins and thus were designated ECSS1 to ECSS4. Similarly, group II of the tree topology is divided into seven clades among which five clades contained CheAMx (Che3, Che1, Che6, Che4, and Che5) and thus were classified accordingly, whereas two novel clades were identified (ECSS5 and ECSS6). Thus, overall, all of the eight characterized M. xanthus DK1622 CSS were identified as separate clades phylogenetically, and apart from these, six novel CSS were also identified as per the phylogenetic analysis and modular architecture of complete clusters. The modular distribution of each system having the respective CheA protein is depicted on the tree using iTol server (47) (Fig. 3), the overview is depicted in Fig. 4 and a system-wise depiction can be found in Fig. S5 in the supplemental material.
FIG 4.
Overview of the modular organization in identified CSS.
Group I.
The first cluster in group I tree topology was identified as ECSS1, where the modular organization is CheR(0,1)-B(0,1)-A-MCP-Y-W (Fig. S5; labeled as ECSS1). [The complete modular architectures of the various CSS are not identical for all genomes. Therefore, to depict the combination of different modules in a system, we have used this numbering for each module, e.g., CheR(0,1) means that either CheR is absent in a particular system or it is in one copy.] This cluster did not contain any CheAMx protein and thus was considered a novel class of CSS in myxobacteria. In this cluster, we found two Sandaracinus CheA homologs together, which also share the same modular architecture, suggesting a probable duplication event. The Soce56-1 system (the systems for each genome are named according to the locations of their coding sequences on the genome; Soce56-1 represents the first CSS encoded within the S. cellulosum Soce56 genome) does not have any CheB and CheR modules that are required for the CSS to memorize the signals (also known as the CheB-CheR adaptation pathway [17]), whereas these modules were present in S. cellulosum So0157-2-1. Transcriptional regulators adjacent to ECSS modules in H. ochraceum CSS 4 (Ho-4), Soce56-1, and S. cellulosum So0157-2-1 were identified, whereas the PilZ protein and the cell division FtsW protein were identified upstream to ECSS in S. aurantiaca DW4/3-1 CSS 1 (Samy-1) and Samy-3, respectively.
Similar to ECSS1, clusters 3, 4, and 5 did not contain CheAMx and thus were classified as ECSS2, ECSS3, and ECSS4. All the systems have a unique modular organization: ECSS2 has modules CheY-A-W-R-B-MCP-X-D(0,1) (Fig. S5; labeled as ECSS2), and ECSS4 has CheB-R-MCP-W-A (Fig. S5; labeled as ECSS4). ECSS3, the fourth cluster of group 1 topology, was divided into four subclades in which all four subclade members have a different modular organization.
ECSS3-1 has only A. dehalogenans CSS 3 (Ad-3) depicting a CheB-R-W-MCP-A-Y modular structure (Fig. S5; labeled as ECSS3-1). The second subclade, ECSS3-2, comprising CSS of family Cystobacteraceae (H. minutum, S. aurantiaca DW4/3-1, C. fuscus, A. gephyra DSM 2261T, and C. violaceus Cb vi76), has a modular organization of CheY(1,2)-A-MCP(0,1,3)-W(1,2)-R-B(0,1)-D(0,1) (Fig. S5; labeled as ECSS3-2). Adjacent to the Ag-8 and C. violaceus CCS 2 (Cyvi-2) systems, glutathione-regulated potassium efflux system protein KefB was found to be similar to Che6Mx (13). Third subclade ECSS3-3 was present in suborder Sorangiineae members along with C. fuscus (Cyb-4). This subclade had diverse organizations of chemotaxis modules, such as CheY(0,1,2)-A-MCP(0,1)-W(0,1)-R(0,1)-B(0,1)-D(0,1) (Fig. S5; labeled as ECSS3-3). The CheD module was present only in Sorangium species, whereas in Cyb-4 one CheB was present and none of the MCPs were identified. The fourth subclade of ECSS3, ECSS3-4, was present in some members of suborder Cystobacterineae with a CheR-MCP(0,1,2)-W-A-D(0,1)-Y(0,1) modular organization (Fig. S5; labeled as ECSS3-4). ECSS3-4 also contained orphan CheA proteins (Ag_2030 and Cyvi_1312; these numbers are RAST gene annotation for the respective genomes), and their functional roles have not been proposed yet. All these ECSS had no conserved synteny except Ad-4 (ECSS2) and Ad-3 (ECSS3-1), which exist adjacent to each other and are present inside the flagellar biosynthesis operon.
Clade 2 of group I was classified as Che2 (also known as the Dif system) based on the presence of CheA protein corresponding to the Che2Mx system. The Che2 system has the modules organized as CheC-A-Y-W-DifB-MCP (Fig. S5; labeled as Che2); the system lacks CheB and CheR homologs, hence impairing the chemotaxis adaptation pathway (24). Domain organization suggests that the CheC module is present exclusively in this system, with an exception in the V. incomptus CCS 2 (Vui-2) system (CheA-W-MCP). The DifB protein, a hypothetical protein within Che2 CSS, is present only in the genus Myxococcus. The Che2 system in proximity has gliding motility protein MglA and twitching motility protein PilT, except in Ad-7 and Vui-1, where MglA and the ferric siderophore transport system are present. The system is known to regulate M. xanthus motility and development processes, e.g., regulation of exopolysaccharide (EPS) production, a basic requirement for type IV pilus-based social motility in M. xanthus (36, 37, 48). This system controls EPS production, essential for fruiting body formation and sporulation in response to starvation (27).
Clades 6 and 7 of group I correspond to Che7 and Che8, as they have the CheAMx protein from Che7 and Che8, respectively. The modular organizations of Che7 and Che8 were CheB-R-x-MCP-W-A-Y (Fig. S5; labeled as Che7) and CheY-B-R-x-x-W-W-A-Y (Fig. S5; labeled as Che8), respectively, where x represents any protein inserted into the CSS. The Che7 system was completely absent in A. dehalogenans 2CP-1, V. incomptus DSM 27710T, and suborder Nannocystineae members. In subfamily Cystobacteraceae, this system was present as H. minutum CSS 6 (Hm-6) and S. aurantiaca CSS 2 (Sa-2), whereas it was absent in A. gephyra DSM 2261T and Cystobacter species. The Che7 system regulates starvation-induced development via interaction with a HEAT-repeat protein, Cpc7, which is similar to cyanobacterial phycocyanobilin lyases (32). This protein, Cpc7, is represented by “x” in the Che7 modular organization (Pfam, HEAT_EZ; PF13513, HEAT_PBS; PF03130). On the basis of the mutant analysis, it has been reported that the Che7 system might be important for resistance to temperature stress and viable spore production (25, 32). Che8 was the ubiquitous system with maximum representation from all the myxobacterial families with the exception of Vulgatibacteraceae and Kofleriaceae. Che8 CSS lacks the MCP module and contains two copies of CheW. The functional aspect of Che8 has not been explored yet, but the proteins involved in the biosynthesis of riboflavin, an indispensable constituent of metabolism, were found adjacent to the Che8 operon.
Phylogeny of group I CSS (Fig. S4) revealed the divergence of the Che systems from Deinococcus-Thermus, deltaproteobacteria, and gammaproteobacteria into the different Che systems studied in this group. The Che2 and Che8 clades are monophyletic with members of Deltaproteobacteria, whereas Che7 is monophyletic with both deltaproteobacteria and cyanobacteria. ECSS groups share their respective sister clades with members of different bacterial classes, e.g., Acidobacteria, Betaproteobacteria, Gammaproteobacteria, Cyanobacteria, and Firmicutes. ECSS4 represents a clade having Cyanobacteria with the parental taxa being Firmicutes. Similarly, within ECSS2, L. luteola DSM 27648T-1 represents a clade with gammaproteobacteria and Ad-4 forms its sister clade with Acidobacteria and Spirochaetes with an ancestral clade representing Deltaproteobacteria members. Ad-3, an ECSS-3-1 system, is monophyletic with Deltaproteobacteria. ECSS3-2, ECSS3-3, and ECSS3-4 clades have Acidobacteria homologs, and they are paraphyletic to various members of Gamma- and Betaproteobacteria.
Group II.
Group II was divided into seven clades. Clades 3 and 6 comprised novel CSS classified as ECSS5 and ECSS6. ECSS5, with a solitary representation from Cyb-5, had a modular organization of CheB-A-MCP-W-W-Y (Fig. S5; labeled as ECSS5). In myxobacterial CheA phylogeny, Cyb-5 is clustered with its sister clade of Che6, whereas after adding non-Myxococcales CheA homologs, Cyb-5 got clustered with ECSS1. We have considered Cyb-5 to be a separate subclade owing to its unique modular organization whereby CheR is absent, in contrast to ECSS1 and Che6. ECSS6 in clade 6 had the modular structure of CheY-B-A-MCP-W-R-W (Fig. S5; labeled as ECSS6), which is present only in the members of Cystobacteraceae and Polyangiaceae (with the exception of S. cellulosum Soce56). Compared to other ECSS6 members, the Cyb-6 system lacks MCP in its modular organization.
Clade1 in group II represents a Che3 system with the modular organization of CheR-B-A-MCP-MCP-CrdC-W-CrdB-CrdA (Fig. S5; labeled as Che3). The Che3 system is present in suborder Cystobacterineae, whereas it is absent in suborders Nannocystineae and Sorangiineae. The system was not identified in A. dehalogenans 2CP-1 and V. incomptus DSM 27710T, members of suborder Cystobacterineae. We found that the full cascade, which has been reported in M. xanthus to transduce the incoming signal to the transcription factor CrdA (26, 31), is well-conserved in all identified Che3 CSS.
Clade 2 of group II includes the CheAMx Frz (Che1) system and was therefore characterized as the Che1 system with a CheR-B-A-MCP-W-W-Y modular organization (Fig. S5; labeled as Che1). The Che1 system was well conserved in suborder Cystobacterineae members, whereas it is absent in suborders Sorangiineae and Nannocystineae. The response regulator CheY, which is an important part of the CSS, could not be retrieved in Cyb-7 and Cyvi-9. These systems are present at the contig boundaries of the draft assemblies, and complete genomes might help resolve this. The Frz system is involved in the regulation of cellular reversal frequency (35) mainly by the regulation of cell polarity (49, 50). This system has been recently shown to have accessory modules allowing signal amplification (51).
Clade 4 of group II is determined to be a Che6 operon based on its similarity to the CheAMx Che6 system and has a modular organization of CheW-R-W-MCP-A-B (Fig. S5; labeled as Che6), which helps in regulating cellular reversals and proper timing of development (52). CheY protein was absent in all but M. fulvus CSS 3 (Mf-3) and Cyb-13. Mutations in Che6 pathway proteins impair S motility, resulting in defects in reversal periods and changes in velocities, which can further display defects in M. xanthus DK1622 development (52). Though two CheW proteins were present in all Che6 systems in A. dehalogenans 2CP-1, only one CheW module was present. The Che6 system was present in all members of order Myxococcales except Sorangium spp., L. luteola DSM 27648T, V. incomptus DSM 27710T, and family Nannocystaceae members. Two type IV pilus assembly proteins (PilZ) were found adjacent to the Che6 system in Ag-9, Cyb-13, and Cyvi-5, while only one unit was present in the vicinity of Mf-3, M. xanthus CSS 8 (Mx-8), M. hansupus CSS 10 (Mh-10), and M. stipitatus CSS 3 (Ms-3). It has also been reported that a histidine kinase (SocD) and a potassium efflux pump homolog protein (KefC) (Pfam domain, PF00999/Na_H_Exchanger and PF02254/TrkA_N) are encoded by the Che6 system in M. xanthus DK1622 (13). The KefC homologs were found to be conserved and syntenic for C. fuscus, C. coralloides DSM 2259T, M. stipitatus, M. xanthus DK1622, and M. hansupus genomes, while these homologs were also a part of ECCS3-2 systems such as Ag-8 and Cyvi-2. Synteny studies (Fig. 2) suggested that in most of the members of suborder Cystobacterineae, Che6 and Che7 systems are present and adjacent to each other. Among the closely related Myxococcus genomes, the Che7 system was not identified in M. stipitatus. The Cpc7 protein (present in the Che7 system), which is similar to phycocyanobilin lyase protein in Cyanobacteria (32), was present in proximity to the Che6 system of M. stipitatus (see Fig. S6 in the supplemental material), suggesting the loss of the Che7 system in M. stipitatus.
Clade 5 of group II, containing a CheW-R-MCP-Y-W-A modular organization, has been characterized as the Che4 system (based on closeness to the CheAMx Che4 system), where CheB is absent (Fig. S5; labeled as Che4). The Che4 system was well conserved in members of suborder Cystobacterineae, with the exception of A. dehalogenans 2CP-1 and V. incomptus DSM 27710T. Also, the system was absent in suborder Nannocystineae and Sorangiineae members. A gliding motility protein, MglA, was found adjacent to all Che4 systems, probably regulating and helping the motility functions. The effect of deletion of the Che4 operon is visible only in S-motility strains (A−S+) and not in wild-type (A+S+) or A-motility strains (A+S−) (53). In S-motility strains, Che4 operon or CheY deletions cause increased vegetative swarming and inhibit aggregation and, therefore, sporulation (53).
Clade 7 of group II, having modular organization MCP-CheB-A-W-R-W-Y, is designated a Che5 system (Fig. S5; labeled as Che5). The Che5 system has CheB protein present, in contrast to a Che4 system, enabling members of this clade to adapt effectively to different stimuli and memorize those signals (17). Similar to the Che4 system, Che5 CSS was completely absent in suborder Nannocystineae and Sorangiineae members and present in the suborder Cystobacterineae members, with the exception of A. dehalogenans 2CP-1 and V. incomptus DSM 27710T. This operon was flanked by ATP-dependent Clp protease subunits ClpA and ClpS in the upstream and short-chain dehydrogenase and 2-oxoglutarate dehydrogenase E1 and E2 components in the downstream region. These flanking proteins along with the full operon were well conserved in families Myxococcaceae and Cystobacteraceae. The functional aspect of the Che5 system has not been explored yet, but the system might be hypothesized to be regulating ATP-dependent protein degradation.
Adding nonmyxobacterial CheA proteins to phylogeny revealed an organized pattern whereby the CheA proteins from deltaproteobacteria were found to be ancestral to all the clades. CheA homologs of Che1 (Frz), Che3, Che4, Che5, ECSS1, and ECSS5 were defined in separate subclades, and they did not show any closely related CheA homolog in the phylogeny. The Che6 clade had alphaproteobacterial homologs intermingled, whereas ECSS6 had an ancestral clade with betaproteobacteria and actinobacteria.
CheB phylogeny for CSS lacking the CheA module.
We have identified multiple clusters of Che proteins in which CheA proteins (the main marker of CSS) were absent; therefore, for their comparative analysis, we have used CheB as a phylogenetic marker due to its universal presence among all of these identified clusters. We have labeled these clusters here ACSS. Such systems had CheB and CheR modules along with other chemotaxis proteins, HK, TCS proteins, or RR, present in the vicinity. As CheA was absent from these systems, CheB protein sequences were fetched from all the myxobacterial proteomes to infer the phylogeny. A CheB protein-based phylogenetic tree was able to group Che1-8 and ECSS1-6 clusters in the respective groups concordant to a CheA-based phylogeny (see Fig. S7 in the supplemental material). CheB phylogeny included C. apiculatus CSS 4 (Cap-4) within the Che7 system, which was not clustered by CheA phylogeny owing to the absence of CheA protein in the system. The modular organization of Cap-4 was exactly similar to that of Che7 with the absence of CheA, which was not identified due to the C. apiculatus draft assembly, but CheB-based phylogenetic analysis and modular organization suggested that the Cap-4 system belongs to Che7 CSS. Perhaps the complete genome of C. apiculatus may reveal a Che7 system.
Besides the complete CSS, these ACSS have been grouped in four separate clusters, ACSS1 to ACSS4, on the basis of CheB phylogeny. In ACSS1, there is only one system, i.e., that present in Hm-8. The modular organization of the system comprised three consecutive CheB proteins with an adjacent HK protein. In ACSS2, CheB and CheR modules were present either as separate modules (as present in suborder Cystobacterineae) or as a combined module (CheBR) (as present in suborders Sorangiineae and Nannocystineae). CheB and CheR were accompanied by CheY in a few members, whereas a few were accompanied by HK or TCS proteins. Interestingly, in Ho-3, the fused CheBR module is accompanied by the CheB module, in which none of the HK or TCS proteins were identified. In the ACSS3 clade, some of the suborder Cystobacterineae members have CheB and CheR modules adjacent to TCS proteins, where no accessory domains are present in proximity. The ACSS4 clade is also limited to suborder Cystobacterineae (M. xanthus, M. fulvus, M. hansupus, H. minutum, and S. aurantiaca DW4/3-1) along with H. ochraceum SMP-2 DSM 14365T from suborder Nannocystineae. All these ACSS contain CheB, CheR, or CheY along with one or multiple TCS proteins (Fig. S5; labeled as ACSS). The functions of the ACSS have not been proposed, but these systems might be involved in some accessory functions or supporting functions to the intact Che systems.
DISCUSSION
The members of order Myxococcales have some of the largest genomes among the eubacteria. The increase in genome size is directly related to the increase in complexity influenced by environmental factors and occurrence of genetic events, viz., duplication and integration of foreign genes via horizontal gene transfer (54). The duplicated content in myxobacterial genomes has been suggested to help the organism adapt to diverse habitats and a complex life cycle (39). The interesting physiology and complex life cycle of myxobacteria have been of keen interest to researchers, but the public repositories have limited genome sequences in the order Myxococcales. Here we report the genomes of three myxobacteria from diverse families; Cystobacter fuscus DSM 2262T (ANAH00000000), Hyalangium minutum DSM 14724T (JMCB00000000), and Chondromyces apiculatus DSM 436 (ASRX00000000). The first two organisms belong to family Cystobacteraceae, and the last belongs to family Polyangiaceae.
Our study identified that the model myxobacterial organism M. xanthus has 273 TCS, whereas 97 to 476 TCS proteins were distributed across order Myxococcales genomes. The lowest number of TCS proteins, 97, was in V. incomptus DSM 27710T, the smallest myxobacterium known to date. Even though it has a genome size similar to that of Escherichia coli, V. incomptus DSM 27710T has a larger number of TCS proteins. We found that within myxobacterial genomes there is no correlation between the number of TCS and the genome size. Even though A. dehalogenans 2CP-1 is just ∼0.6 Mbp larger than V. incomptus DSM 27710T, it contains about double the number of TCS. Similarly, members of families Kofleriaceae and Nannocystaceae (genome size, ∼10 Mbp, and marine habitat) have one-half the number of TCS proteins found in other soil myxobacteria with similar genome sizes. This lack of correlation might be due to various environmental factors such as habitat (soil or marine environment), temperature, etc., for each organism.
At present, among myxobacteria, only M. xanthus DK1622 CSS have been studied extensively (13, 25). In this study, we have identified their widespread distribution and performed a comparative analysis across all the available myxobacterial genomes. Like TCS, we found that the number of CSS is not directly related to genome size. Sorangium species, which have the largest genomes (13 to 14.5 Mbp), encode five CSS, compared to seven CSS in A. dehalogenans 2CP-1 (5.03 Mbp). The smallest myxobacterium, V. incomptus DSM 27710T (4.35 Mbp), encodes only 2 CSS, whereas H. ochraceum SMP-2 DSM 14365T (9.45 Mbp), L. luteola DSM 27648T (12.19 Mbp), and E. salina DSM 15201 (10.98 Mbp) also encode 1, 2, and 2 CSS, respectively. CSS modules were present in larger number in suborder Cystobacterineae (except V. incomptus DSM 27710T) than in suborders Sorangiineae and Nannocystineae. Following similar patterns, we found a large number of MCPs in suborder Cystobacterineae members. Owing to their complex life cycle, it was not surprising to find a large number of CSS in myxobacteria.
The CSS modular organization and CheA protein-based phylogeny revealed the presence of six novel ECSS apart from the previously known eight Che systems. Comparative modular organization and synteny studies of identified myxobacterial CSS revealed conserved synteny in Che1 to Che8 systems, but no such conserved synteny was observed in the ECSS. The Che1 system regulates reversal frequency (33, 35); the Che2 system regulates EPS production (36, 48); the Che3 system regulates developmental gene expression (26); the Che4 system controls reversal frequency (53); the Che6 system regulates cellular reversals and proper timing of development (52); and the Che7 system regulates starvation-induced development (32). The functional aspects of Che5 and Che8 have been partially characterized (25), but no precise function is known to date. The presence of ATP-dependent Clp protease ATP-binding subunit ClpA, ATP-dependent Clp protease adaptor protein ClpS, and NifU-like proteins in the vicinity of the functionally uncharacterized Che5 system was observed. Similarly, adjacent to Che8 systems we found 6,7-dimethyl-8-ribityllumazine synthase (Pfam, DMRL synthase; PF00885) and riboflavin synthase (Pfam, Lum_binding; PF00677) in most members of the order Myxococcales and terpene synthase (Pfam, Terpene_synth_C; PF03936) in A. gephyra DSM 2261T and C. fuscus. We therefore speculate that the Che5 and Che8 systems might be involved in the regulation of ATP-dependent Clp protease and riboflavin synthesis, respectively. In addition, CheB phylogeny was used to identify four ACSS where CSS phylogenetic marker protein CheA was absent. Also, one orphan system was identified in A. gephyra DSM 2261T wherein neither CheA nor CheB protein was present. The other systems might be involved in regulating the alternative cellular functions or assisting the CSS in their functions.
We observed that the outgroup species are not grouped within any clade, suggesting the absence of closely related homologs and high divergence from their respective ancestors. The deltaproteobacteria were found to be monophyletic to most of the identified Che systems, i.e., Che7, Che2, and Che8, probably suggesting their emergence through lateral gene transfer. Che1, Che3, Che4, Che5, and Che6 were more conserved and do not have homologous systems in any known species, suggesting their novel origin within myxobacteria only. ECSS members were more diverged and had a closeness with representatives of Acidobacteria, Betaproteobacteria, Gammaproteobacteria, Cyanobacteria, and Firmicutes, suggesting their probable horizontal transfer from the last taxon. Conclusively, we can say that Che6, Che7, and Che8 are present in most of the myxobacteria, suggesting them to be core Myxococcales CSS; most of the CSS are accessory, whereas some newly identified systems, such as ECSS3-1, ECSS4, ECSS5, and ACSS1, are unique to one organism only (Table 4).
This is first comprehensive study of CSS from order Myxococcales, well known for their large genomes and peculiar physiological characteristics. The TCS protein repertoire involved in signal transduction for all known myxobacterial genomes is reported here. All putative myxobacterial CSS have been identified in this study along with their classification, comparative analysis, and putative evolution.
Conclusion.
The draft genomes of Cystobacter fuscus DSM 2262T, Hyalangium minutum DSM 14724T, and C. apiculatus (DSM 436) from diverse families within order Myxococcales were sequenced and assembled. TCS and CSS proteins were identified and analyzed among all available myxobacterial genomes. Although the distributions of TCS and CSS in the myxobacteria are independent of the genome size, the expansion of both the TCS and CSS in myxobacteria is likely to be governed by the habitat and environmental stimuli. CheA protein-based phylogeny and modular organization assisted us in identifying six novel ECSS apart from the previously known Che systems, i.e., Che1 to Che8. Putative functions of Che5 and Che8 systems were identified based on the conserved synteny across all myxobacteria. Phylogenetic studies revealed the monophyletic nature of Che1 to Che8 systems with deltaproteobacteria, whereas the ECSS were more diverged, as they were sharing clades with different bacterial classes such as Acidobacteria, Betaproteobacteria, Gammaproteobacteria, Cyanobacteria, and Firmicutes. Based on modular organization and CheB protein-based phylogeny, four types of chemosensory clusters, classified here as ACSS, in which CheA protein was absent were identified. Thus, the current study highlights the distribution and comparison between various CSS present in order Myxococcales and sets the foundation for experimental characterization of these systems to understand the complex signal transduction mechanisms in myxobacteria.
MATERIALS AND METHODS
Culturing and DNA isolation.
C. fuscus DSM 2262T, H. minutum DSM 14724T, and C. apiculatus DSM 436 strains were procured from the Deutsche Sammlung von Mikroorganismen und Zellkulturen (DSMZ) culture collection as strains DSM 2262, DSM 14724, and DSM 436. These cultures were grown on VY/2 agar and SP agar medium plates as a pure culture. C. fuscus (see Fig. S1A in the supplemental material) and H. minutum (Fig. S1B) grow as a stiff slime sheet with fine radially distributed veins all over the plate and are red-brown and tinted yellow-brown, respectively. C. apiculatus swarms grow as light orange colonies and form shallow depressions in the agar (Fig. S1C) (55). For each strain, whole-genomic DNA was isolated from the pure culture using ZR Fungal/Bacterial DNA MicroPrep and the phenol-chloroform-based manual DNA isolation method. 16S rRNA sequencing of the isolated DNA from each sample was performed using universal bacterial primers at our in-house Sanger sequencing facility. The obtained sequence was subjected to BLASTn against bacterial 16S rRNA database for strain identification.
Whole-genome sequencing, assembly, and annotation.
Whole-genome sequencing of the isolated genomic DNA from each sample was performed using the Illumina HiSeq 1000 technology. The library preparation was carried out as per the TruSeq DNA Sample Prep protocol (Illumina, Inc., San Diego, CA) at C-CAMP, Bangalore, India. Raw sequencing reads (average insert size, 350 bp; read length, 101 bp) were subjected to quality filtering (PHRED quality score, 20; minimum read length, 70) using NGSQC Toolkit v2.2.3 (56) and were then assembled de novo using SPAdes v3.0 (57), CLC Genomics Workbench 6.0 (http://www.clcbio.com/), SOAPdenovo-1.05 (58), and Velvet-1.2.07 (59). The best assembly of each assembler was selected based on the N50 value, the number of contigs, and the assembled genome length. Each assembly was subjected to SSPACE (60), which scaffolds contigs using filtered paired-end reads (minimum number of read pairs [k], 5; maximum link ratio [a], 0.7; and minimum read overlap [n], 15). Postscaffolding, physical gaps were filled using GAPfiller (61) (minimum overlap [m], 90 bp; sequence overlap [n], 10). The final gap-filled assemblies were then subjected to the CLC Microbial Genome Finishing Module to join contigs. Gene prediction and functional annotation were performed by rapid annotation using subsystem technology (RAST) (62). RNAmmer 1.2 (63, 64) and tRNAscan-SE-1.23 (63) were used to predict rRNA and tRNA genes, respectively.
Data sources for comparative genome analysis.
The genomes of M. hansupus (CP012109) (65), M. xanthus DK1622 (NC_008095.1) (66), M. fulvus HW-1 (NC_015711.1) (67), M. stipitatusT (NC_020126.1) (68), M. xanthus DZ2 (AKYI00000000) (69), M. xanthus DZF1 (AOBT00000000) (70), A. dehalogenans 2CP-1 (NC_011891.1), A. gephyra DSM 2261T (CP011509.1) (79), C. fuscus DSM 2262T (ANAH00000000), C. violaceus Cb vi76 (JPMI000000000) (45), H. minutum DSM 14724T (JMCB00000000), S. aurantiaca DW4/3-1 (NC_014623.1) (3), C. coralloides DSM 2259T (NC_017030.1) (71), H. ochraceum SMP-2 DSM 14365T (NC_013440.1) (72), E. salina DSM 15201 (JMCC00000000), P. pacifica SIR-1 (ABCS00000000), C. apiculatus DSM 436 (ASRX00000000), S. cellulosum So0157-2 (NC_021658.1) (73), S. cellulosum Soce56 (NC_010162.1) (74), L. luteola DSM 27648T (CP012333.1), V. incomptus DSM 27710T (CP012332.1), and S. amylolyticus DSM 53668T (CP011125.1) (75) were downloaded from NCBI for this study. For all these genomes, gene prediction and functional annotation were performed using RAST (62).
Identification of TCS and CSS.
The proteomes of all the members of order Myxococcales were scanned against the Pfam-A v29.0 database (76) with an E value threshold of 1e−5 using hmmscan program of HMMER suite (http://hmmer.janelia.org/) (77) and further parsed using hmmscan-parser.sh to identify functional domains. To compare the proteins involved in signal transduction among all the genomes, domains known to be present in TCS proteins and chemotaxis (as mentioned in MISTdb2.2 [40]) were identified from the Pfam database. Representative domains in TCS proteins are as follows: Response_reg, PF00072; HATPase_c, PF02518; HATPase_c_2, PF13581; HWE_HK, PF07536; HisKA, PF00512; HisKA_2, PF07568; HisKA_3, PF07730; His_kinase, PF06580; and Hpt, PF01627. Representative domains in CSS proteins are Pfam domains CheB_methylest, PF01339; CheC, PF04509; CheD, PF03975; CheR, PF01739; CheW, PF01584; CheX, PF13690; CheZ, PF04344; and MCPsignal, PF00015. Proteins exhibiting the presence of Hpt, H-kinase_dim, HATPase_c, and CheW domains all together were identified as CheA.
Phylogenetic analysis of CSS.
CheA proteins identified from all myxobacterial genomes were aligned using the MUSCLE program of MEGA v6.0 (78). The alignment was further subjected to MEGA v6.0 to generate a maximum likelihood (ML) phylogeny using the JTT matrix model with 100 bootstrap values. The obtained ML tree was visualized in iTOL (47), and the modular organization was mapped onto the tree. Further homologs of myxobacterial CheA proteins were searched by subjecting these proteins to BLASTp (BLAST 2.2.29+) against a nonredundant database (NR; downloaded in October 2015) with an E value cutoff of 1e−5. The resulting homologs were screened, and the top 10 unique homologs except the ones belonging to order Myxococcales were fetched. These sequences were clustered (at 80% sequence identity and 80% length coverage) using BlastClust (BLAST 2.2.20). These outgroup sequences along with myxobacterial CheA proteins were subjected to phylogeny construction with parameters similar to the ones mentioned above. CheB protein-based phylogeny was also drawn for all CSS identified in myxobacteria using similar parameters.
Data availability.
Genome information for the draft assemblies of Cystobacter fuscus DSM 2262T, Hyalangium minutum DSM 14724T, and Chondromyces apiculatus DSM 436 were deposited in GenBank under the accession numbers ANAH00000000, JMCB00000000, and ASRX00000000 with Bio-Project numbers PRJNA177202, PRJNA242458, and PRJNA192263, respectively.
Supplementary Material
ACKNOWLEDGMENTS
This work is supported in part by the Department of Biotechnology (project BTISNET; GAP001) and project “Expansion and modernization of Microbial Type Culture Collection and Gene Bank (MTCC) jointly supported by Council of Scientific and Industrial Research (CSIR) grant no. BSC0402 and Department of Biotechnology (DBT) Govt. of India grant no. BT/PR7368/INF/22/177/2012.” G.S. and I.K. are supported by research fellowships from the CSIR and University Grants Commission (UGC), respectively.
We thank the C-CAMP (http://www.ccamp.res.in/) genomics facility for help in obtaining NGS data. We thank S. Mayilraj and T. N. C. Ramya for providing help and lab space during the culturing and purifying of myxobacterial genomic DNA.
G.S. and S.S. conceived and designed the work. G.S. performed the experiments. G.S., I.K., and S.S. analyzed the data. G.S. and I.K. contributed reagents, materials, and analysis tools. G.S., I.K., and S.S. wrote the manuscript.
We declare that we do not have any conflict of interest.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JB.00620-17.
REFERENCES
- 1.Velicer GJ, Vos M. 2009. Sociobiology of the myxobacteria. Annu Rev Microbiol 63:599–623. doi: 10.1146/annurev.micro.091208.073158. [DOI] [PubMed] [Google Scholar]
- 2.Shimkets LJ. 1990. Social and developmental biology of the myxobacteria. Microbiol Rev 54:473–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huntley S, Hamann N, Wegener-Feldbrugge S, Treuner-Lange A, Kube M, Reinhardt R, Klages S, Muller R, Ronning CM, Nierman WC, Sogaard-Andersen L. 2011. Comparative genomic analysis of fruiting body formation in Myxococcales. Mol Biol Evol 28:1083–1097. doi: 10.1093/molbev/msq292. [DOI] [PubMed] [Google Scholar]
- 4.Sliusarenko O, Zusman DR, Oster G. 2007. Aggregation during fruiting body formation in Myxococcus xanthus is driven by reducing cell movement. J Bacteriol 189:611–619. doi: 10.1128/JB.01206-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reichenbach H. 1984. Myxobacteria: a most peculiar group of social prokaryotes, p 1–50. In Rosenberg E. (ed), Myxobacteria. Springer Series in Molecular Biology. Springer, New York, NY. [Google Scholar]
- 6.Mauriello EM, Mignot T, Yang Z, Zusman DR. 2010. Gliding motility revisited: how do the myxobacteria move without flagella? Microbiol Mol Biol Rev 74:229–249. doi: 10.1128/MMBR.00043-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hodgkin J, Kaiser D. 1979. Genetics of gliding motility in Myxococcus xanthus (Myxobacterales): two gene systems control movement. Mol Gen Genet 171:177–191. doi: 10.1007/BF00270004. [DOI] [Google Scholar]
- 8.Wu Y, Jiang Y, Kaiser AD, Alber M. 2011. Self-organization in bacterial swarming: lessons from myxobacteria. Phys Biol 8:055003. doi: 10.1088/1478-3975/8/5/055003. [DOI] [PubMed] [Google Scholar]
- 9.Berleman JE, Kirby JR. 2009. Deciphering the hunting strategy of a bacterial wolfpack. FEMS Microbiol Rev 33:942–957. doi: 10.1111/j.1574-6976.2009.00185.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Whitworth DE. 2015. Genome-wide analysis of myxobacterial two-component systems: genome relatedness and evolutionary changes. BMC Genomics 16:780. doi: 10.1186/s12864-015-2018-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Whitworth DE, Cock PJ. 2008. Two-component systems of the myxobacteria: structure, diversity and evolutionary relationships. Microbiology 154:360–372. doi: 10.1099/mic.0.2007/013672-0. [DOI] [PubMed] [Google Scholar]
- 12.Shi X, Wegener-Feldbrugge S, Huntley S, Hamann N, Hedderich R, Sogaard-Andersen L. 2008. Bioinformatics and experimental analysis of proteins of two-component systems in Myxococcus xanthus. J Bacteriol 190:613–624. doi: 10.1128/JB.01502-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zusman DR, Scott AE, Yang Z, Kirby JR. 2007. Chemosensory pathways, motility and development in Myxococcus xanthus. Nat Rev Microbiol 5:862–872. doi: 10.1038/nrmicro1770. [DOI] [PubMed] [Google Scholar]
- 14.Kirby JR. 2009. Chemotaxis-like regulatory systems: unique roles in diverse bacteria. Annu Rev Microbiol 63:45–59. doi: 10.1146/annurev.micro.091208.073221. [DOI] [PubMed] [Google Scholar]
- 15.Cozzone AJ. 1988. Protein phosphorylation in prokaryotes. Annu Rev Microbiol 42:97–125. doi: 10.1146/annurev.mi.42.100188.000525. [DOI] [PubMed] [Google Scholar]
- 16.Whitworth DE, Cock PJ. 2009. Evolution of prokaryotic two-component systems: insights from comparative genomics. Amino Acids 37:459–466. doi: 10.1007/s00726-009-0259-2. [DOI] [PubMed] [Google Scholar]
- 17.Wuichet K, Zhulin IB. 2010. Origins and diversification of a complex signal transduction system in prokaryotes. Sci Signal 3:ra50. doi: 10.1126/scisignal.2000724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alm E, Huang K, Arkin A. 2006. The evolution of two-component systems in bacteria reveals different strategies for niche adaptation. PLoS Comput Biol 2(11):e143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koretke KK, Lupas AN, Warren PV, Rosenberg M, Brown JR. 2000. Evolution of two-component signal transduction. Mol Biol Evol 17:1956–1970. doi: 10.1093/oxfordjournals.molbev.a026297. [DOI] [PubMed] [Google Scholar]
- 20.Borland S, Oudart A, Prigent-Combaret C, Brochier-Armanet C, Wisniewski-Dyé F. 2015. Genome-wide survey of two-component signal transduction systems in the plant growth-promoting bacterium Azospirillum. BMC Genomics 16:1–17. doi: 10.1186/1471-2164-16-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Capra EJ, Laub MT. 2012. Evolution of two-component signal transduction systems. Annu Rev Microbiol 66:325–347. doi: 10.1146/annurev-micro-092611-150039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Falke JJ, Bass RB, Butler SL, Chervitz SA, Danielson MA. 1997. The two-component signaling pathway of bacterial chemotaxis: a molecular view of signal transduction by receptors, kinases, and adaptation enzymes. Annu Rev Cell Dev Biol 13:457–512. doi: 10.1146/annurev.cellbio.13.1.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wadhams GH, Armitage JP. 2004. Making sense of it all: bacterial chemotaxis. Nat Rev Mol Cell Biol 5:1024–1037. doi: 10.1038/nrm1524. [DOI] [PubMed] [Google Scholar]
- 24.Wuichet K, Alexander RP, Zhulin IB. 2007. Comparative genomic and protein sequence analyses of a complex system controlling bacterial chemotaxis. Methods Enzymol 422:1–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moine A, Agrebi R, Espinosa L, Kirby JR, Zusman DR, Mignot T, Mauriello EM. 2014. Functional organization of a multimodular bacterial chemosensory apparatus. PLoS Genet 10:e1004164. doi: 10.1371/journal.pgen.1004164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kirby JR, Zusman DR. 2003. Chemosensory regulation of developmental gene expression in Myxococcus xanthus. Proc Natl Acad Sci U S A 100:2008–2013. doi: 10.1073/pnas.0330944100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.He K, Bauer CE. 2014. Chemosensory signaling systems that control bacterial survival. Trends Microbiol 22:389–398. doi: 10.1016/j.tim.2014.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hamblin PA, Maguire BA, Grishanin RN, Armitage JP. 1997. Evidence for two chemosensory pathways in Rhodobacter sphaeroides. Mol Microbiol 26:1083–1096. doi: 10.1046/j.1365-2958.1997.6502022.x. [DOI] [PubMed] [Google Scholar]
- 29.Xu Q, Black WP, Cadieux CL, Yang Z. 2008. Independence and interdependence of Dif and Frz chemosensory pathways in Myxococcus xanthus chemotaxis. Mol Microbiol 69:714–723. doi: 10.1111/j.1365-2958.2008.06322.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guvener ZT, Tifrea DF, Harwood CS. 2006. Two different Pseudomonas aeruginosa chemosensory signal transduction complexes localize to cell poles and form and remould in stationary phase. Mol Microbiol 61:106–118. doi: 10.1111/j.1365-2958.2006.05218.x. [DOI] [PubMed] [Google Scholar]
- 31.Willett JW, Kirby JR. 2011. CrdS and CrdA comprise a two-component system that is cooperatively regulated by the Che3 chemosensory system in Myxococcus xanthus. mBio 2(4):e00110-11. doi: 10.1128/mBio.00110-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Darnell CL, Wilson JM, Tiwari N, Fuentes EJ, Kirby JR. 2014. Chemosensory regulation of a HEAT-repeat protein couples aggregation and sporulation in Myxococcus xanthus. J Bacteriol 196:3160–3168. doi: 10.1128/JB.01866-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bustamante VH, Martinez-Flores I, Vlamakis HC, Zusman DR. 2004. Analysis of the Frz signal transduction system of Myxococcus xanthus shows the importance of the conserved C-terminal region of the cytoplasmic chemoreceptor FrzCD in sensing signals. Mol Microbiol 53:1501–1513. doi: 10.1111/j.1365-2958.2004.04221.x. [DOI] [PubMed] [Google Scholar]
- 34.Mignot T, Merlie JP Jr, Zusman DR. 2005. Regulated pole-to-pole oscillations of a bacterial gliding motility protein. Science 310:855–857. doi: 10.1126/science.1119052. [DOI] [PubMed] [Google Scholar]
- 35.Blackhart BD, Zusman DR. 1985. “Frizzy” genes of Myxococcus xanthus are involved in control of frequency of reversal of gliding motility. Proc Natl Acad Sci U S A 82:8767–8770. doi: 10.1073/pnas.82.24.8767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Black WP, Yang Z. 2004. Myxococcus xanthus chemotaxis homologs DifD and DifG negatively regulate fibril polysaccharide production. J Bacteriol 186:1001–1008. doi: 10.1128/JB.186.4.1001-1008.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Black WP, Xu Q, Yang Z. 2006. Type IV pili function upstream of the Dif chemotaxis pathway in Myxococcus xanthus EPS regulation. Mol Microbiol 61:447–456. doi: 10.1111/j.1365-2958.2006.05230.x. [DOI] [PubMed] [Google Scholar]
- 38.Boysen A, Ellehauge E, Julien B, Sogaard-Andersen L. 2002. The DevT protein stimulates synthesis of FruA, a signal transduction protein required for fruiting body morphogenesis in Myxococcus xanthus. J Bacteriol 184:1540–1546. doi: 10.1128/JB.184.6.1540-1546.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Goldman B, Bhat S, Shimkets LJ. 2007. Genome evolution and the emergence of fruiting body development in Myxococcus xanthus. PLoS One 2:e1329. doi: 10.1371/journal.pone.0001329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ulrich LE, Zhulin IB. 2010. The MiST2 database: a comprehensive genomics resource on microbial signal transduction. Nucleic Acids Res 38:D401–D407. doi: 10.1093/nar/gkp940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Galperin MY. 2005. A census of membrane-bound and intracellular signal transduction proteins in bacteria: bacterial IQ, extroverts and introverts. BMC Microbiol 5:35. doi: 10.1186/1471-2180-5-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ulrich LE, Koonin EV, Zhulin IB. 2005. One-component systems dominate signal transduction in prokaryotes. Trends Microbiol 13:52–56. doi: 10.1016/j.tim.2004.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Galperin MY, Higdon R, Kolker E. 2010. Interplay of heritage and habitat in the distribution of bacterial signal transduction systems. Mol Biosyst 6:721–728. doi: 10.1039/b908047c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Muff TJ, Foster RM, Liu PJ, Ordal GW. 2007. CheX in the three-phosphatase system of bacterial chemotaxis. J Bacteriol 189:7007–7013. doi: 10.1128/JB.00896-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stevens DC, Young J, Carmichael R, Tan J, Taylor RE. 2014. Draft genome sequence of gephyronic acid producer Cystobacter violaceus strain Cb vi76. Genome Announc 2(6):e01299-14. doi: 10.1128/genomeA.01299-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tran HT, Krushkal J, Antommattei FM, Lovley DR, Weis RM. 2008. Comparative genomics of Geobacter chemotaxis genes reveals diverse signaling function. BMC Genomics 9:471. doi: 10.1186/1471-2164-9-471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Letunic I, Bork P. 2011. Interactive Tree Of Life v2: online annotation and display of phylogenetic trees made easy. Nucleic Acids Res 39:W475–W478. doi: 10.1093/nar/gkr201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bonner PJ, Xu Q, Black WP, Li Z, Yang Z, Shimkets LJ. 2005. The Dif chemosensory pathway is directly involved in phosphatidylethanolamine sensory transduction in Myxococcus xanthus. Mol Microbiol 57:1499–1508. doi: 10.1111/j.1365-2958.2005.04785.x. [DOI] [PubMed] [Google Scholar]
- 49.Zhang Y, Guzzo M, Ducret A, Li YZ, Mignot T. 2012. A dynamic response regulator protein modulates G-protein-dependent polarity in the bacterium Myxococcus xanthus. PLoS Genet 8:e1002872. doi: 10.1371/journal.pgen.1002872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Keilberg D, Wuichet K, Drescher F, Sogaard-Andersen L. 2012. A response regulator interfaces between the Frz chemosensory system and the MglA/MglB GTPase/GAP module to regulate polarity in Myxococcus xanthus. PLoS Genet 8:e1002951. doi: 10.1371/journal.pgen.1002951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Guzzo M, Agrebi R, Espinosa L, Baronian G, Molle V, Mauriello EM, Brochier-Armanet C, Mignot T. 2015. Evolution and design governing signal precision and amplification in a bacterial chemosensory pathway. PLoS Genet 11:e1005460. doi: 10.1371/journal.pgen.1005460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Catherine SJ. 2011. Che6 chemosensory regulation of multicellularity in Myxococcus xanthus. PhD dissertation The University of Iowa, Ames, IA. [Google Scholar]
- 53.Vlamakis HC, Kirby JR, Zusman DR. 2004. The Che4 pathway of Myxococcus xanthus regulates type IV pilus-mediated motility. Mol Microbiol 52:1799–1811. doi: 10.1111/j.1365-2958.2004.04098.x. [DOI] [PubMed] [Google Scholar]
- 54.Wang Y, Yang JK, Lee OO, Li TG, Al-Suwailem A, Danchin A, Qian P-Y. 2011. Bacterial niche-specific genome expansion is coupled with highly frequent gene disruptions in deep-sea sediments. PLoS One 6:e29149. doi: 10.1371/journal.pone.0029149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Whitworth DE. 2008. Myxobacteria: multicellularity and differentiation. ASM Press, Washington, DC. [Google Scholar]
- 56.Patel RK, Jain M. 2012. NGS QC Toolkit: a toolkit for quality control of next generation sequencing data. PLoS One 7:e30619. doi: 10.1371/journal.pone.0030619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Luo R, Liu B, Xie Y, Li Z, Huang W, Yuan J, He G, Chen Y, Pan Q, Liu Y, Tang J, Wu G, Zhang H, Shi Y, Liu Y, Yu C, Wang B, Lu Y, Han C, Cheung DW, Yiu SM, Peng S, Xiaoqian Z, Liu G, Liao X, Li Y, Yang H, Wang J, Lam TW, Wang J. 2012. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. Gigascience 1:18. doi: 10.1186/2047-217X-1-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res 18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Boetzer M, Henkel CV, Jansen HJ, Butler D, Pirovano W. 2011. Scaffolding pre-assembled contigs using SSPACE. Bioinformatics 27:578–579. doi: 10.1093/bioinformatics/btq683. [DOI] [PubMed] [Google Scholar]
- 61.Boetzer M, Pirovano W. 2012. Toward almost closed genomes with GapFiller. Genome Biol 13:R56. doi: 10.1186/gb-2012-13-6-r56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, Meyer F, Olsen GJ, Olson R, Osterman AL, Overbeek RA, McNeil LK, Paarmann D, Paczian T, Parrello B, Pusch GD, Reich C, Stevens R, Vassieva O, Vonstein V, Wilke A, Zagnitko O. 2008. The RAST server: rapid annotations using subsystems technology. BMC Genomics 9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lagesen K, Hallin P, Rodland EA, Staerfeldt HH, Rognes T, Ussery DW. 2007. RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res 35:3100–3108. doi: 10.1093/nar/gkm160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lowe TM, Eddy SR. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res 25:955–964. doi: 10.1093/nar/25.5.0955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sharma G, Narwani T, Subramanian S. 2016. Complete genome sequence and comparative genomics of a novel myxobacterium Myxococcus hansupus. PLoS One 11:e0148593. doi: 10.1371/journal.pone.0148593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Goldman BS, Nierman WC, Kaiser D, Slater SC, Durkin AS, Eisen JA, Ronning CM, Barbazuk WB, Blanchard M, Field C, Halling C, Hinkle G, Iartchuk O, Kim HS, Mackenzie C, Madupu R, Miller N, Shvartsbeyn A, Sullivan SA, Vaudin M, Wiegand R, Kaplan HB. 2006. Evolution of sensory complexity recorded in a myxobacterial genome. Proc Natl Acad Sci U S A 103:15200–15205. doi: 10.1073/pnas.0607335103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li ZF, Li X, Liu H, Liu X, Han K, Wu ZH, Hu W, Li FF, Li YZ. 2011. Genome sequence of the halotolerant marine bacterium Myxococcus fulvus HW-1. J Bacteriol 193:5015–5016. doi: 10.1128/JB.05516-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Huntley S, Kneip S, Treuner-Lange A, Sogaard-Andersen L. 2013. Complete genome sequence of Myxococcus stipitatus strain DSM 14675, a fruiting myxobacterium. Genome Announc 1:e0010013. doi: 10.1128/genomeA.00100-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Muller S, Willett JW, Bahr SM, Darnell CL, Hummels KR, Dong CK, Vlamakis HC, Kirby JR. 2013. Draft genome sequence of Myxococcus xanthus wild-type strain DZ2, a model organism for predation and development. Genome Announc 1(3):e00217-13. doi: 10.1128/genomeA.00217-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Muller S, Willett JW, Bahr SM, Scott JC, Wilson JM, Darnell CL, Vlamakis HC, Kirby JR. 2013. Draft genome of a type 4 pilus defective Myxococcus xanthus strain, DZF1. Genome Announc 1(3):e00392-13. doi: 10.1128/genomeA.00392-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Huntley S, Zhang Y, Treuner-Lange A, Kneip S, Sensen CW, Sogaard-Andersen L. 2012. Complete genome sequence of the fruiting myxobacterium Corallococcus coralloides DSM. 2259. J Bacteriol 194:3012–3013. doi: 10.1128/JB.00397-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ivanova N, Daum C, Lang E, Abt B, Kopitz M, Saunders E, Lapidus A, Lucas S, Glavina Del Rio T, Nolan M, Tice H, Copeland A, Cheng JF, Chen F, Bruce D, Goodwin L, Pitluck S, Mavromatis K, Pati A, Mikhailova N, Chen A, Palaniappan K, Land M, Hauser L, Chang YJ, Jeffries CD, Detter JC, Brettin T, Rohde M, Goker M, Bristow J, Markowitz V, Eisen JA, Hugenholtz P, Kyrpides NC, Klenk HP. 2010. Complete genome sequence of Haliangium ochraceum type strain (SMP-2). Stand Genomic Sci 2:96–106. doi: 10.4056/sigs.69.1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Han K, Li ZF, Peng R, Zhu LP, Zhou T, Wang LG, Li SG, Zhang XB, Hu W, Wu ZH, Qin N, Li YZ. 2013. Extraordinary expansion of a Sorangium cellulosum genome from an alkaline milieu. Sci Rep 3:2101. doi: 10.1038/srep02101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Schneiker S, Perlova O, Kaiser O, Gerth K, Alici A, Altmeyer MO, Bartels D, Bekel T, Beyer S, Bode E, Bode HB, Bolten CJ, Choudhuri JV, Doss S, Elnakady YA, Frank B, Gaigalat L, Goesmann A, Groeger C, Gross F, Jelsbak L, Jelsbak L, Kalinowski J, Kegler C, Knauber T, Konietzny S, Kopp M, Krause L, Krug D, Linke B, Mahmud T, Martinez-Arias R, McHardy AC, Merai M, Meyer F, Mormann S, Munoz-Dorado J, Perez J, Pradella S, Rachid S, Raddatz G, Rosenau F, Ruckert C, Sasse F, Scharfe M, Schuster SC, Suen G, Treuner-Lange A, Velicer GJ, Vorholter FJ, Weissman KJ, Welch RD, Wenzel SC, Whitworth DE, Wilhelm S, Wittmann C, Blöcker H, Pühler A, Müller R. 2007. Complete genome sequence of the myxobacterium Sorangium cellulosum. Nat Biotechnol 25:1281–1289. doi: 10.1038/nbt1354. [DOI] [PubMed] [Google Scholar]
- 75.Sharma G, Khatri I, Subramanian S. 2016. Complete genome of the starch-degrading myxobacteria Sandaracinus amylolyticus DSM 53668T. Genome Biol Evol 8:2520–2529. doi: 10.1093/gbe/evw151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Finn RD, Bateman A, Clements J, Coggill P, Eberhardt RY, Eddy SR, Heger A, Hetherington K, Holm L, Mistry J, Sonnhammer EL, Tate J, Punta M. 2014. Pfam: the protein families database. Nucleic Acids Res 42:D222–D230. doi: 10.1093/nar/gkt1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Eddy SR. 2011. Accelerated profile HMM searches. PLoS Comput Biol 7:e1002195. doi: 10.1371/journal.pcbi.1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sharma G, Subramanian S. 2017. Unravelling the complete genome of Archangium gephyra DSM 2261T and evolutionary insights into myxobacterial chitinases. Genome Biol Evol 9:1304–1311. doi: 10.1093/gbe/evx066. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Genome information for the draft assemblies of Cystobacter fuscus DSM 2262T, Hyalangium minutum DSM 14724T, and Chondromyces apiculatus DSM 436 were deposited in GenBank under the accession numbers ANAH00000000, JMCB00000000, and ASRX00000000 with Bio-Project numbers PRJNA177202, PRJNA242458, and PRJNA192263, respectively.