Abstract
Since the genomic region containing the Bcl-2-related ovarian killer (BOK) locus is frequently deleted in certain human cancers, BOK is hypothesized to have a tumor suppressor function. In the present study, we analyzed primary non-small cell lung carcinoma (NSCLC) tumors and matched lung tissues from 102 surgically treated patients. We show that BOK protein levels are significantly downregulated in NSCLC tumors as compared to lung tissues (P<0.001). In particular, we found BOK downregulation in NSCLC tumors of grades two (P=0.004, n=35) and three (P=0.031, n=39) as well as in tumors with metastases to hilar (pN1) (P=0.047, n=31) and mediastinal/subcarinal lymph nodes (pN2) (P=0.021, n=18) as opposed to grade one tumors (P=0.688, n=7) and tumors without lymph node metastases (P=0.112, n=51). Importantly, in lymph node positive patients, BOK expression greater than the median value was associated with longer survival (P=0.002, Mantel test). Using in vitro approaches, we provide evidence that BOK overexpression is inefficient in inducing apoptosis but that it inhibits TGFβ-induced migration and epithelial-to-mesenchymal transition (EMT) in lung adenocarcinoma-derived A549 cells. We have identified epigenetic mechanisms, in particular BOK promoter methylation, as an important means to silence BOK expression in NSCLC cells. Taken together, our data point toward a novel mechanism by which BOK acts as a tumor suppressor in NSCLC by inhibiting EMT. Consequently, the restoration of BOK levels in low-BOK-expressing tumors might favor the overall survival of NSCLC patients.
Keywords: Non-small cell lung carcinoma, BOK, apoptosis, epithelial-to-mesenchymal transition, Bcl-2 family
Introduction
Bcl-2-related ovarian killer (BOK) is a poorly understood member of the Bcl-2 protein family. It is similar in domain structure and homologous in amino acid sequence to the pro-apoptotic family members BAX and BAK. Accordingly, BOK was shown to have proapoptotic function upon overexpression (1–4). Even though it is known that overexpressed BOK leads to the release of cytochrome-c from mitochondria leading to procaspase-9 activation, exact mechanisms of this pro-death function are controversially discussed as is its pro-apoptotic function under physiological conditions (1, 2, 5–10). Intriguingly, most of the cytoplasmic BOK seems to be localized at the endoplasmic reticulum (2, 11, 12), by a mechanism requiring its C-terminal tail-anchor (2), where it strongly interacts with IP3 receptors and is subject to posttranslational regulation through ubiquitination and proteasomal degradation (1, 13, 14). Significant amounts of BOK are also found within the nuclear compartment (2, 11) and non-apoptotic roles in regulation of proliferation (12), and even protective roles in response to specific stressors or in specialized cell types have been proposed (2, 15).
Recently, it was shown that – unexpectedly - the BOK gene is relatively frequently deleted in human cancers (16). BOK might therefore have a previously unrecognized tumor suppressor function.
Lung cancers, 80% of which are NSCLC type tumors, remain the leading cause of cancer-related deaths worldwide (17). Importantly, NSCLCs are well-known to develop high resistance towards chemo- and radiotherapy and patients often relapse because of progression of the primary tumor to metastatic disease. In this study, we investigated the tumor suppressor potential of BOK and its possible molecular function(s) in NSCLC cells and tissues. The results of our study show that high levels of BOK in primary NSCLC tumors correlate with longer overall survival of NSCLC patients and that BOK plays a role in the epithelial-to-mesenchymal transition and migration of NSCLC cells whereas it might only play a minor role in apoptosis induction.
Materials and Methods
Reagents
High glucose Dulbecco’s Modified Eagle Medium (DMEM with GlutaMAX), trypsin-EDTA solution and penicillin/streptomycin stock solutions were purchased from Thermo Fisher Scientific (Waltham, MA, USA). Fetal calf serum (FCS, Sera Pro, ultra-low endotoxin) was purchased from Pan Biotech (Aidenbach, DE). RPMI 1640 AQmedia™ and 4-hydroxytamoxifen (4-OHT) were from Sigma Aldrich Chemie GmBH (Buchs, Switzerland).
NSCLC patients
Specimens of NSCLC tumors and matched non-transformed lung parenchyma were obtained from 102 patients who did not received radiotherapy nor chemotherapy before surgery for lung cancer. Patients are characterized in supplemental data S1. The study was approved by the local institutional ethical committee and was conducted in accordance with the Declaration of Helsinki. Signed written informed consent was obtained from each patient before entry to the study. Tissue samples (~1 g, wet mass) from non-necrotic parts of the tumor and from the lung at a site located as distantly as possible from the tumor, were excised from the resected lung lobe or lung immediately after surgery. All tissues were snap-frozen in liquid nitrogen and stored at −80 °C until protein and RNA extraction. The histopathological classification of the tumors was done according to the World Health Organization criteria (18) and tumor staging was performed according to the international pTNM system (19).
NSCLC cell lines
NSCLC cell lines used in the present study were derived from squamous cell lung carcinoma: CALU-1, NCI-H520, SKMES-1, lung adenocarcinoma: A549, SKLU-1, LXF-289, COLO-699, and large cell lung carcinoma: NCI-H1299, LCLC-103H, CORL23. CALU-1, SKMES-1, A549 and SKLU-1 cells were obtained from the European Collection of Cell Cultures (Salisbury, UK), LXF-289 and COLO-699 cells were obtained from The German Collection of Microorganisms and Cell Cultures (Braunschweig, Germany), and NCI-H520 and NCI-H1299 cells were obtained from the American Type Culture Collection (ATCC, Rockville, MD, USA). The cell lines were cultured in humidified atmosphere of 5% CO2 and air at 37 °C in DMEM or RPMI containing 10% heat-inactivated FCS, 105 IU/l penicillin-G and 100 mg/l streptomycin.
Quantitative Real time RT-PCR analysis
Total RNA isolation, reverse transcription and PCR amplification was performed as described earlier (20). The BOK (GeneBank accession no. NM_032515) mRNA-directed primers were: forward 5'-CAGTCTGAGCCTGTGGTGAC-3', reverse 5'-TGATGCCTGCAGAGAAGATG-3'. β-Actin was used as reference gene (GeneBank accession no. NM_001101) using the primers: forward: 5'-CTGGCACCCAGCACAATG-3', reverse: 5'-GGGCCGGACTCGTCATAC-3'. Cell cycle regulatory gene expression was assessed using the Human Cell Cycle Primer Library (Biomol GmbH, Hamburg, DE).
Protein extraction and immunoblotting
Protein lysates from NSCLC cell lines were prepared in RIPA buffer (50 mM Tris-HCl pH 8.0, 150 mM NaCl, 1% Triton-X100, 0.5% Na-deoxycholate, 0.1% SDS, 1 mM EDTA) supplemented with a protease inhibitor cocktail (Complete, Roche, Basel, Switzerland), 1 µg/ml pepstatin, 2 mM Na3VO4 and 50 mM NaF. Total protein was determined by BCA Assay (Thermo Fisher Scientific, Waltham, MA, USA). Protein extracts from frozen tissue samples were prepared as described previously (21). Protein extracts were boiled for 5 min in reducing (100 mM DTT) Laemmli buffer. 50 µg per lane were separated on denaturing 7.5–20% SDS-PAGE gradient gels and electro-transferred to PVDF membranes (0.45 µm, Merck Millipore, Zug, Switzerland). Membranes were probed with the following antibodies: mouse anti-Actin (clone C4/Actin) and mouse anti-PARP (clone C2–10) from BD Biosciences, rat anti-ATF4 (clone W16016A, BioLegend), rabbit monoclonal anti-BOK (clone 1–5 (2)), rabbit anti-E-cadherin (SC-7870) and mouse anti-vimentin (clone V9) from Santa Cruz Biotechnology (Dallas, TX, USA), mouse anti-Tubulin (clone B-5-1-2, Sigma-Aldrich) and mouse-anti-GAPDH (clone 6C5, Merck, Millipore, Zug, Switzerland). Secondary anti-mouse and anti-rabbit IgG antibodies were linked to near-infrared fluorescent dyes 680CW and 800CW, respectively (LI-COR, Bad Homburg, Germany). Quantification was performed using Odyssey software. Integrated intensity values with subtracted median background were imported into SigmaStat (Systat Software, Point Richmond, CA, USA) for statistical analysis.
Cell death analysis by flow cytometry
Viability was determined by flow cytometry using FITC-Annexin V/ PI exclusion as previously described (2). Data were acquired using FACS-Calibur flow cytometer (BD Biosciences) and analyzed using VenturiOne software (Applied Cytometry Systems).
Immunofluorescence
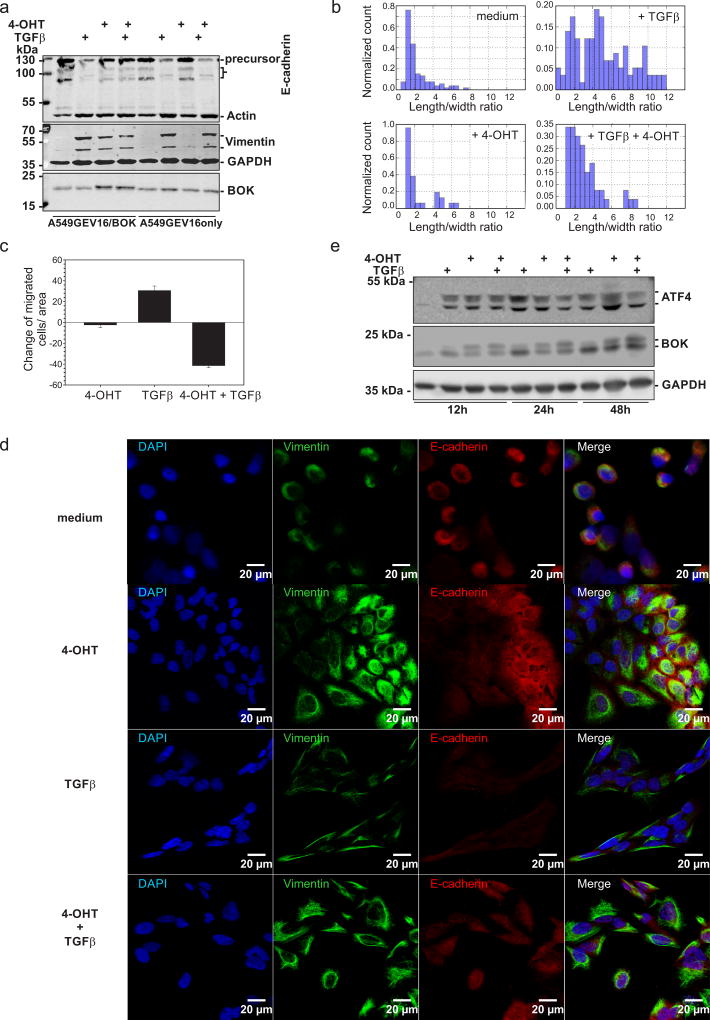
A549GEV16/BOK cells were seeded on sterile glass coverslips. After attachment, 100 nM of 4-OHT was added to induce BOK expression. The following day, cells were treated with or without 10 ng/ml of TGF-β2 for 48 hours. Immunofluorescence staining was performed as described in (2). Primary antibodies used were rabbit anti-E-cadherin (SC-7870) and mouse anti-vimentin (clone V9) from Santa Cruz Biotechnology (Dallas, TX. USA). Secondary antibodies were Alexa Fluor 488- or 555-conjugated goat anti-mouse or goat anti-rabbit IgG (f(ab’)2 specific). Cell microphotographs were taken with Zeiss LSM 5 confocal.
Boyden chamber assay
A549GEV16/BOK and A549GEV16only cells were treated with or without 100 nM of 4-OHT overnight to induce/mock BOK expression, following 10 ng/ml of TGF-β2/mock treatment for 48 h. Subsequently, 104 cells were added in FCS-free medium to the upper part of the Boyden chamber (Transwell, Costar), while the lower part contained medium with 10% FCS. After 16-hour incubation at 37°C and 5% CO2, inserts were washed twice in PBS, fixed in 4% paraformaldehyde and the non-transmigrated cells in the upper part of the 8-µm filter were scraped off with a cotton swab. The filter was mounted with ProLong Gold Antifade Reagent with DAPI (Life Technologies). Cell microphotographs were taken with Zeiss Axiovert 35 fluorescent microscope and at least 100 DAPI-stained nuclei in the filter were counted per condition.
Protein expression profile by flow cytometry
A549GEV16/BOK, cells were seeded in 10-cm dishes. After attachment, cells were treated with or without 100 nM of 4-OHT to induce/mock BOK expression. The next day, cells were treated with or without 10 ng/ml of TGF-β. After 48 hours, cells were trypsinized, washed with ice cold PBS and counted using ViCell (Beckman Coulter). Cells were fixed with 2% paraformaldehyde for 20 min and blocked with goat serum for 20 min. A minimum of 7×104 cells/well was added to a 96-well plate containing FITC, PE and APC-conjugated antibodies specific to 238 cell proteins (FACSCAP Lyoplate, BP80394, BD Biosciences, San Diego, CA, USA). Plates were incubated in dark and centrifuged at 240g for 7 min. Cells were permeabilized with 0.05% Saponin in immunofluorescence assay buffer (IFA, 4% calf serum, 5% human serum albumin, 2 mM EDTA in PBS without Ca2+ and Mg2+, pH 7.2) and washed twice with IFA. Finally, DAPI was added to the cells at a concentration of 8 µg/ml and plates were measured the next day on Fortessa flow cytometer (BD Biosciences). FITC, APC and PE Calibrite beads (BD Biosciences) were used for compensation settings along with cells stained with DAPI alone. Acquired data were analyzed using VenturiOne software (Applied Cytometry Systems) and clusters were generated in SysStat (San Jose, CA, USA).
CRSIPR/Cas9- and shRNA-mediated downregulation of BOK
The BOK genomic locus was edited using CRISPR/Cas9 technology producing H1299, A549 and LFX-289 cells devoid of detectable BOK protein expression. The lentiCRISPR v2 plasmid was a gift from Feng Zhang (Addgene plasmid #52961) (22). The following gRNA sequences targeting the exon 2 of human BOK was designed using the publicly available service at http://crispr.mit.edu: gRNA2: Fw 5’-caccgAAAGGCGTCCATGATCTCGG-3’ and Rev 5’-aaacCCGAGATCATGGACGCCTTTc -3’; gRNA3: Fw 5’-caccgGTCTGTGGGCGAGCGGTCAA-3’ and Rev 5’-aaacTTGACCGCTCGCCCACAGACc-3’. HEK 293T cells were transfected with the plasmids lentiCRISPR v2 (minus gRNA for control), pMD2GVSV-G, psPAX2 using X-tremeGENE HP DNA Transfection Reagent (Roche). Viruses were harvested after 24 h, filtered and freshly added to semi-confluent cultures of H1299 cells in the presence of 8 µg/mL polybrene, followed by 3 weeks of selection with 2 µg/mL puromycin. BOK downregulation was achieved using Mission™ lentiviral plasmids (Sigma-Aldrich) expressing (shRNA targeting human Bok (5’-CCGGCCGCTTCCTGAAGGCTGCCTTCTCGAGAAGGCAGCCTTCAGGAAGCGGTTTTTG-3’) and non-targeting shRNA control (shC002). Plasmids were transiently co-transfected with pMD2GVSV-G and pCMVδR8.2 into HEK293T cells using X-tremeGENE HP DNA Transfection Reagent (Roche Diagnostics). After 48 h, viral supernatant was harvested and freshly used to transduce H1299 cells in the presence of 8 µg/mL polybrene, followed by selection with 2 µg/mL puromycin for two weeks.
Chromatin immunoprecipitation (ChIP)
1×107 cells treated with decitabine for 72 h, trichostatin A for 24 h, or a combination thereof, were fixed in 1% formaldehyde at room temperature. Crosslinking was stopped after 10 min with 0.125 M glycine. Cells were scraped off and washed twice with PBS. The pellet was either snap-frozen or directly lysed in SDS lysis buffer (50 mM Tris-HCl pH 8, 150 mM NaCl, 2 mM EDTA, 1% SDS containing protease and phosphatase inhibitors (Sigma, P8340, P2850, 9382)) and sonicated (Fisher Scientific Sonic Model 100 Dismembrator) to generate ~200 bp DNA fragments. Samples were centrifuged at 14’000g for 10 min and supernatant was divided for inputs, acetylation studies, polymerase studies and IgG controls. Chromatin was diluted 5-fold in ChIP Dilution Buffer (0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris-HCl, pH 8.1, 167 mM NaCl) and incubated with 2 µg of Histone H3K9ac antibody (61251, clone 1B10, Active Motif, Carlsbad, CA, USA) or 4 µg of Pol II antibody (sc-899X, Santa Cruz Biotechnology, Dallas, TX, USA). Samples were precipitated with Magna ChIP Protein A+G Magnetic beads (16–663, Millipore) at 4°C for 3 hours. Subsequently, the beads were washed once with low-salt buffer (0.1% SDS, 1% Triton X-100, 2mM EDTA, 20 mM Tris-HCl, pH 8.1, 150 mM NaCl), once with high-salt buffer (increasing NaCl to 550), once with LiCl wash buffer (0.25 M LiCl, 1% IGEPAL-CA630, 1% deoxycholic acid, 1mM EDTA, 10 mM Tris, pH 8.1) and twice with TE Buffer (10 mM Tris-HCl, 1 mM EDTA, pH 8.0). Immunocomplexes were eluted at 65°C for 2.5 hours in 200 µl ChIP Elution Buffer (1% SDS, 0.1 M NaHCO3−). Chromatin was reverse-crosslinked with 250 µM NaCl at 65°C for 4 hours. Beads were centrifuged and supernatant was incubated with 0.8 U of Proteinase K (New England Biolabs, Inc., Ipswich, MA) at 45°C for 1 hour. DNA was purified with GeneJET PCR Purification kit (0702, Thermo Fisher Scientific). 3 µl of DNA were used for qPCR in Fast Start Universal Sybr Green Master Mix (Roche Diagnostics) on a Stratagene Mx3005 analyzer (Agilent, Santa Clara, CA, USA). ROX was used as a reference dye. The primers used for the amplification were: BOK Promoter1 Fw: 5’-TCTAGGTCCCCACTGCTCTG-3’ and Rev: 5’-GAGACGCTTTCCGAGCTTC-3’; BOK Promoter2 Fw: 5’-TCGGGGAATGTCTGTAGCTG-3’ and Rev: 5’-GGGTCCGCTCTCGTTTTT-3’; BOK Promoter3 Fw: 5’-GCTCAGCCCAGATTTCAGTC-3’ and Rev: 5’-ACAAGGGTCACTGTGGAAGC-3’; BOK Promoter4 Fw: 5’-TGTTTATGTGATGCCCGAAG-3’ and Rev: 5’-CCACCGATTCAGTTTCACCT-3’; BOK Mid Fw: 5’-CACCCAAGCATGTGTTTTTG-3’ and Rev: 5’-AGCCTGGGCAATAGAGTGAA-3’ and BOK End Fw: 5’-GCATTCCCTTCATGCAATTT-3’ and Rev: 5’-GGGAAGAGGGAGACCGTTAG-3’. Fold enrichment was calculated using the 2(−ΔΔCT) method normalized to input and corrected for background using IgG controls.
Results
BOK protein is downregulated in NSCLC tumors as well as in poorly differentiated and lymph node positive NSCLC tumors compared to matched lung parenchyma
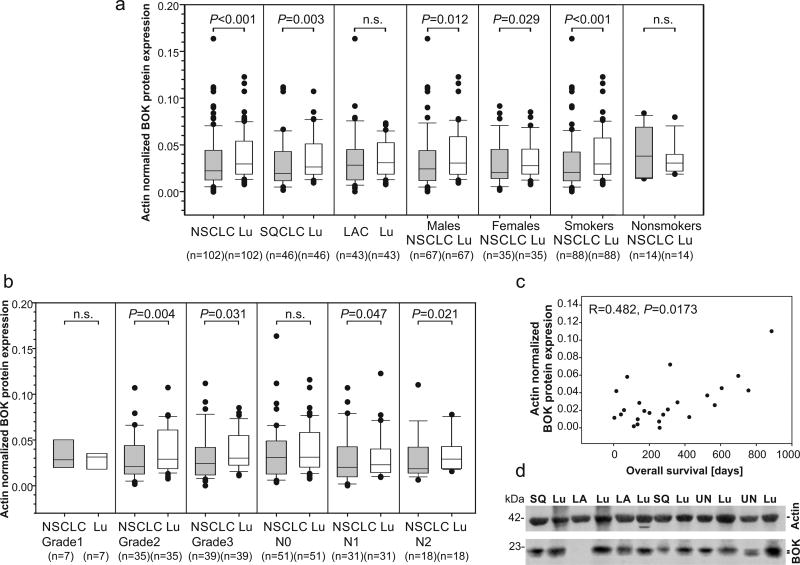
BOK protein expression was significantly downregulated in NSCLC tumors as compared to the paired grossly normal adjacent lung parenchyma (P<0.001, n=102, Wilcoxon signed rank test). Regarding the histopathological tumor type, BOK protein expression was particularly downregulated in squamous cell lung carcinomas (SQCLC) (P=0.003, n=46, Wilcoxon signed rank test), but not in lung adenocarcinomas (P=0.368, n=43, Wilcoxon signed rank test) (Figure 1a). A representative image of BOK protein expression using near-infrared fluorophores for quantitative western blot analysis is shown in Figure 1d. Additionally, BOK protein was equally downregulated in tumors in men (P=0.012; n=67, Wilcoxon signed rank test) as well as in women (P=0.029; n=35, Wilcoxon signed rank test) (Figure 1a). Of note, BOK protein was downregulated in smokers (P<0.001; n=88, Wilcoxon signed rank test) but not in non-smokers (P=0.326; n=14, Wilcoxon signed rank test), as compared to the paired lung tissue (Figure 1a). Moreover, BOK protein was decreased in less differentiated tumors i.e. grades 2 (P=0.004, n=35, Wilcoxon signed rank test) and 3 (P=0.031, n=39, Wilcoxon signed rank test) as compared to the well differentiated matched lungs.
Figure 1.
BOK protein expression in NSCLC tissues and adjacent lung tissue. (a and b) Expression data are shown as median with the upper ranges of 75% (box) and 90% (whisker) and the lower ranges of 25% (box) and 10% (whisker). Statistical differences were calculated by the Wilcoxon signed rank test. P values lower than 0.05 were considered statistically significant. (c) Spearman’s correlation between BOK protein expression in lymph node positive NSCLC and patients’ overall survival. (d) A representative image of quantitative western blot of total protein lysates from NSCLC and lung parenchyma. SQ - squamous cell lung carcinoma, LA - lung adenocarcinoma, UN- undifferentiated cell lung carcinoma, Lu - lung parenchyma.
We observed a decrease of BOK protein expression in stage III NSCLC as compared to the matched non-affected lungs (P=0.005; n=23; Wilcoxon signed rank test). Since staging is categorized according to the pTNM system (19), we analyzed whether BOK levels varied with tumor invasion to lymph nodes (pN). We found that in primary tumors with no spread to lymph nodes, there was no difference in BOK levels when compared to the control tissues (P=0.112, n=51, Wilcoxon signed rank test). However, lower BOK protein levels were detected in primary tumor cells spreading to nearby lymph nodes (pN1) (P=0.047, n=31, Wilcoxon signed rank test) and even less in tumors spreading to lymph nodes in mediastinum and/or around the carina (pN2) (P=0.021, n=18, Wilcoxon signed rank test) (Figure 1b).
Next, we analyzed the relationship between BOK expression and a prognostic relevance for patient survival. Interestingly, BOK protein was weakly correlated with overall survival of lymph node positive NSCLC patients (P=0.0173, n=24; Spearman’s correlation) (Figure 1c).
BOK expression is predictive of survival in lymph node positive patients
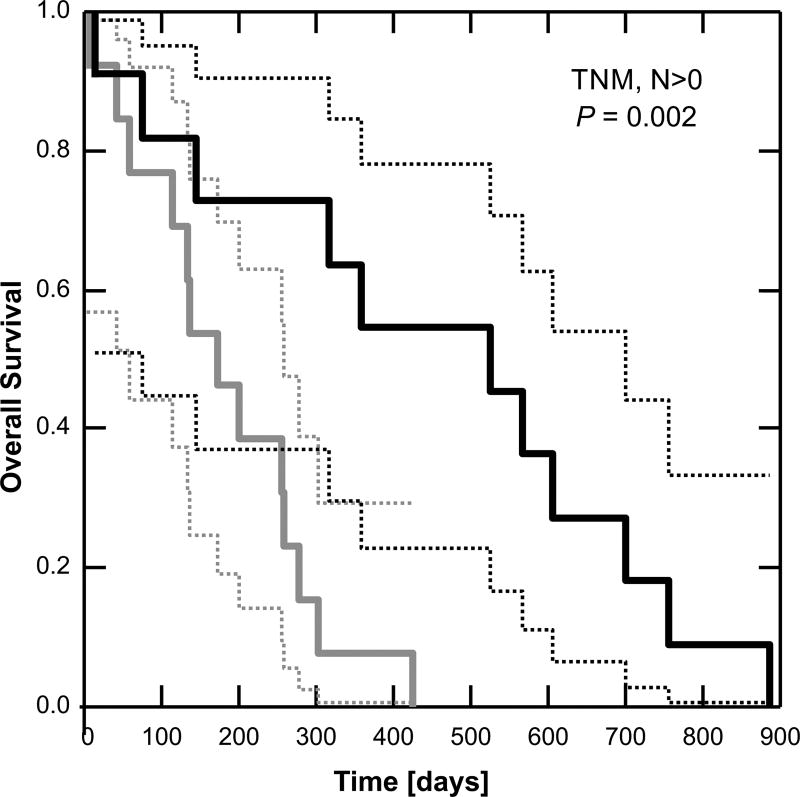
Since BOK expression was weakly correlated with survival time in our patient group (Figure 1c), we asked whether BOK level was predictive of survival. Patients (n = 39) were stratified by BOK expression: less than the median or equal to or greater than the median. When all patients were analyzed, the effect of BOK was not statistically significant (P = 0.62, Mantel test). However, when analysis was limited to patients with lymph node involvement (n = 24, Figure 2), those with BOK expression equal to or above the median value had significantly longer survival (450 [279, 621] days, mean [95% confidence intervals]) compared to those with BOK expression below the median (182 [124, 242] days, P = 0.002). The data suggest that BOK expression level is relevant to survival only in patients in whom the carcinoma has spread from the primary tumor.
Figure 2.
BOK expression level is predictive of survival in lymph node positive patients. Kaplan-Meier survival analysis: dashed lines = 95% confidence intervals. P-value was calculated according to the Mantel test. Grey < median BOK, Black ≥ median BOK.
BOK expression is regulated by epigenetic mechanisms in NSCLC cells
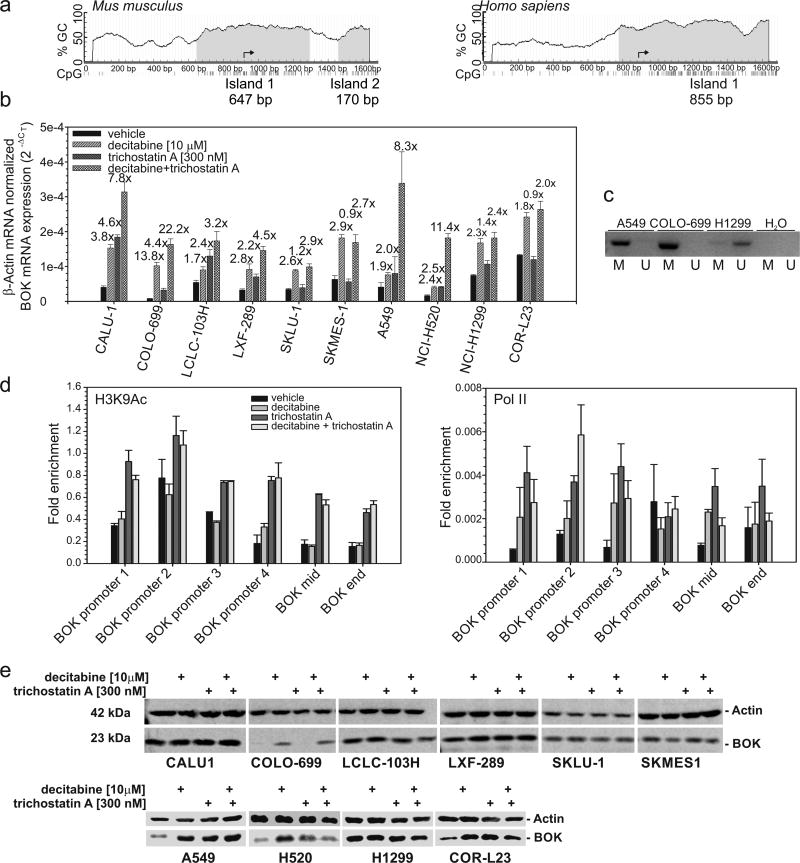
We further investigated whether the variable levels of BOK mRNA detected in NSCLC cell lines (Figure 3) might be due to transcriptional silencing via epigenetic mechanisms. Using the software MethPrimer (23), we found an evolutionary conserved CpG island in the BOK promoter region extending into the second exon of the BOK gene (Figure 3a). As expected from in silico analysis, treatment with the DNA methyltransferase inhibitor 5-aza-2’-deoxycytidine (decitabine) significantly increased BOK mRNA level in all 10 NSCLC cell lines studied (Figure 3b). Co-treatment with trichostatin A further upregulated BOK mRNA in 6 out of 10 cell lines (Figure 3b). By methylation specific PCR, we confirmed that methylation occurs directly in the BOK gene of NSCLC cells displaying low BOK levels (A549 and COLO-699), whereas no promoter methylation was detectable in the high BOK expressing H1299 cells (Figure 3c). ChIP analysis confirmed that the histone cores of the nucleosomes surrounding the BOK gene are deacetylated in A549 cells, which can be reversed by the histone class I and II deacetylase inhibitor trichostatin A (Figure 3d). The combined treatment with decitabine did not further increase the H3K9 acetylation (Figure 3d and Supplemental data S5) implying that BOK promoter methylation and histone deacetylation are not dependent on one another. This was confirmed by ChIP analysis of DNA-dependent RNA polymerase II, which revealed that single treatments with either decitabine or trichostatin A are able to re-express the BOK gene. However, the combined treatment had synergistic effects for the gene re-expression (Figure 3d), which correlated with mRNA levels (Figure 3b). BOK protein was markedly elevated in 5 of 10 studied cell lines upon decitabine treatment. In fact, BOK protein was upregulated more than 2 times in COLO-699, COR-L23, A549 and NCI-H520 cells (Figure 3e). Trichostatin A increased BOK protein in the same cells except for COLO-699 (Figure 3e).
Figure 3.
The BOK gene is epigenetically silenced in NSCLC cell lines. (a) Bioinformatic analysis of human BOK gene methylation. CpG island prediction was based on DNA regions > 100bp with a CG content > 0.5 and observed/expected CpG ratio above 0.6. An input sequence of 1600-base long DNA around the BOK transcriptional start (arrow) was searched. (b) Cells were treated as indicated with 10 µM of decitabine for 72h or 300 nM of trichostatin A for 24 h or a combination of both; mRNA expression was measured by RT-qPCR. Data represent mean ± S.E.M from 3 experiments. Fold change between control and treatment is indicated above the bar. (c) Methylation status of BOK promoter by methylation specific PCR in A549, COLO-699 and H1299 cells using primers for methylated (M) or unmethylated (U) bisulfite-modified DNA. Representative example from 3 independent experiments is shown. (d) Acetylated H3K9 active chromatin mark and RNA polymerase II binding throughout the BOK gene in A549 cells. A549 cells treated with decitabine (72h) and/or trichostatin A (24h) were analyzed by ChIP with anti-H3K9Ac, anti-Pol II and IgG, followed by qPCR analysis of amplicons in the promoter and in the body of the BOK gene (internal control). Error bars indicate the standard error of the mean (SEM) of two biological replicates. (e) NSCLC cells were treated as indicated in (b) and total protein lysates analyzed by western blotting.
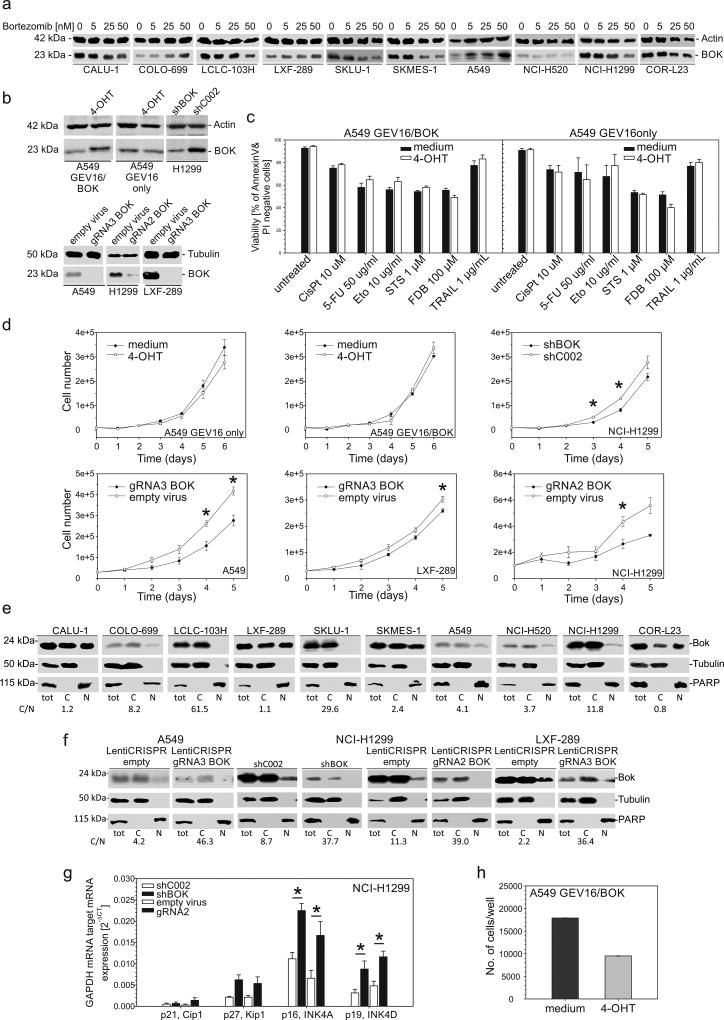
The lack of correlation between BOK mRNA and protein levels may be partially explained by a previously reported high BOK protein turnover (1). However, as shown in Figure 4a, treatment of NSCLC cell lines with bortezomib, a specific 26S proteasome inhibitor, led to stabilization of BOK protein only in 2 out of 10 NSCLC cell lines (A549 and COLO-699).
Figure 4.
Stabilization of BOK protein by bortezomib, cell death and proliferation analyses. (a) Cells were treated with bortezomib at the indicated concentration for 24 h; total protein lysates were analyzed by western blotting. (b) Western blot analysis of total protein lysates from A549GEV16/BOK and A549GEV16only cells treated with or without 100 nM 4-OHT for 48h and of H1299, A549 and LXF-289 cells with downregulated BOK using shRNA or CRISPR/Cas9 technology. (c) A549GEV16/BOK or A549GEV16only cells were pretreated with or without 100nM 4-OHT overnight and treated with cisplatin, 5-FU, etoposide, staurosporine, fludarabine or TRAIL for 48 h. Viability was assessed by FITC-AnnexinV/PI staining using flow cytometry. Data represent mean ± S.E.M. from 3–6 independent experiments. (d) Trypan-blue exclusion assay of cell proliferation. Data represent mean ± S.E.M. from 3 independent experiments. Statistical analysis was performed using Student’s t-test. Asterisks indicate a P value lower than 0.05. (e and f) Detection of nuclear BOK by western blot analysis. Nuclear and cytoplasmic fractions were prepared from NSCLC cells. The purity of fractions was confirmed by detection of PARP (nuclear) or Tubulin (cytoplasmic). The cytoplasmic/nuclear ratio of BOK protein expression (C/N) is shown below the blots. (g) mRNA expression of GAPDH-normalized cell cycle regulators by RT-qPCR. Data represent mean ± S.E.M. from 3 independent experiments. Asterisks indicate a P value lower than 0.05 analyzed by Student’s t-test. (h) Anchorage-independent growth of A549GEV16/BOK cells in soft agar for 7 days. Cells were treated as indicated with or without 100 nM 4-OHT.
BOK overexpression does not induce significant apoptosis and does not sensitize towards drug-induced apoptosis in NSCLC cells
To investigate the role of BOK in NSCLC cell death, we used a 4-hydroxytamoxifen (4-OHT) inducible system to overexpress BOK in A549 cells (2). Despite readily detectable protein levels (Figure 4b) enforced BOK expression in A549 cells did not induce cell death, nor did it sensitize to cell death induced by cisplatin, etoposide, staurosporine (STS), fludarabine (FDB), TRAIL or 5-fluorouracil (5-FU) as assessed by flow cytometry using FITC-AnnexinV/PI staining (Figure 4c) and enzymatic caspase-3/-7 activity assay (data not shown).
Loss of BOK negatively impacts on cellular proliferation rate in NSCLC cells
Interestingly, we noticed that BOK downregulation by RNA interference, as well as disruptive BOK gene editing using CRISPR/Cas9 (Figure 4b), decreased the proliferation rate of NSCLC cells, but not the number of dead cells as assessed by trypan blue exclusion (Figure 4d). Cells lacking BOK proliferated significantly slower, as confirmed by MTT assay (Supplemental data S2). Of note, besides prominent expression in the cytoplasm, we also detected significant amounts of BOK protein in purified nuclear fractions in 8 out of 10 cell lines (Figure 4e). Interestingly, upon downregulation of BOK, hardly any signal for the protein was detectable in the nuclear compartments (Figure 4f), which correlated with decreased proliferation. This was accompanied by a significant increase of p16INK4A and p19INK4D cell cycle inhibitors (Figure 4g). On the other hand, BOK overexpression did not influence the proliferation of A549 cells (Figure 4d and S2). Interestingly, however, BOK overexpression in A549 cells inhibited 7-day anchorage independent growth in soft agar (Figure 4h).
Role of BOK in epithelial-to-mesenchymal transition
To investigate a possible role of BOK in epithelial-to-mesenchymal transition (EMT), we treated cells with TGF-β2, an inducer of a mesenchymal cell type through the activation of SMAD2. We performed western blot and immunofluorescence analyses to verify the EMT marker expression. In A549 cells exposed to 10 ng/ml of TGF-β2 for 48 hours, the mesenchymal marker vimentin was upregulated (Figure 5a and 5d). On the other hand, an epithelial marker, E-cadherin, was downregulated (Figure 5a and 5d). However, A549 cells overexpressing BOK did not lose E-cadherin after TGF-β2 treatment (Figure 5a and 5d). Enforced expression of BOK in A549 cells treated with TGF-β2 did not only diminish expression of the full length 120 kDa E cadherin, but also of its 132 kDa precursor, suggesting not a proteolytic cleavage but rather regulation of its expression. These data indicate that BOK prevents the downregulation of E-cadherin. Moreover, morphological analysis revealed that BOK overexpression significantly abrogated TGF-β2-induced EMT (Figure 5b). Consistent with this interpretation, BOK inhibited TGF-β2-induced cell migration, as assessed by Boyden chamber migration assay (Figure 5c). Furthermore, we detected TGFβ2-induced increase of ATF4 as well as a shorter form of BOK that lacks the N-terminus due to alternative translation initiation at Met15 (13); ATF4, being required for cell invasion (24), was slightly diminished in TGFβ2-treated cell upon additional BOK overexpression and was accompanied by reduction of the shorter form of BOK (Figure 5e).
Figure 5.
BOK inhibits EMT. Cells were pretreated with or without 100 nM 4-OHT and treated with or without 10 ng/ml of TGFβ2 for 48 hours. (a) Whole cell lysates were analyzed by western blotting using anti-E-cadherin, anti-actin (loading control), anti-BOK (same membrane), anti-vimentin and anti-GAPDH (loading control) antibodies. (b) Histogram of morphological changes of A549GEV16/BOK cells: the length and width of at least 100 cells were measured per condition. (c) Boyden chamber migration assay (d) Confocal scanning micrographs of A549GEV16/BOK cells. Anti-vimentin (green) and anti-E-cadherin (red) antibodies were used. All cells were counterstained with DAPI (blue). The same exposure times and LUT settings were used. (e) Western blot analysis of A549GEV16/BOK whole cell lysates treated as indicated above for 12, 24 and 48 hours. Membrane was probed with anti-ATF4, anti-BOK and anti-GAPDH (loading control) antibodies. One representative immunoblot is shown from three independent experiments.
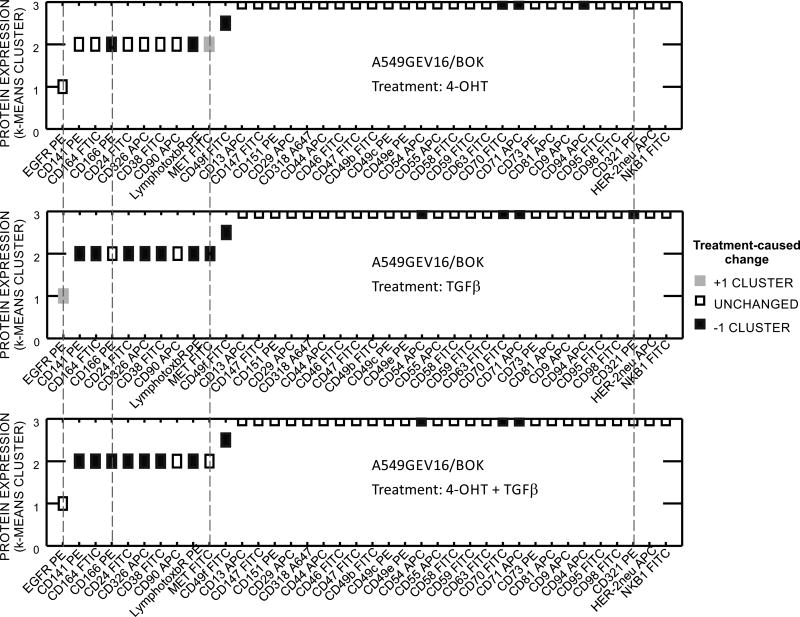
Given the possible involvement of BOK in antagonizing EMT, we performed protein expression profiling by flow cytometry. We screened A549GEV16/BOK cells treated with or without TGF-β and with or without 4-OHT for the expression of 238 surface proteins (supplemental data S3). Flow cytometry data were analyzed as previously described (25) with minor modifications. We used standardized percent of positive events and standardized mean fluorescence intensity of positive events for K-means clustering algorithm. Further, double weight was given to percentage of positive events. Flow cytometry data for the expression of proteins that changed with the treatments are shown in supplemental data S4. Specifically, TGF-β alone upregulated EGFR and downregulated CD141, CD164, CD24, CD326 (EpCAM), CD38, Lymphotoxinβ receptor, MET, CD49f, CD54, CD70, CD71, CD321 (F11 Receptor) (Figure 6).
Figure 6.
Cell surface protein expression by flow cytometry. Thirty-eight markers shown were selected from 238 measured specificities on the basis of strength of expression or extent of modulation. Cells were treated as indicated with or without 100 nM 4-OHT overnight, following addition of 10 ng/ml of TGFβ or vehicle and incubated for 48 h. K-means cluster 0: no expression, 1: low expression, 2: moderate expression, 3: high expression. Y axis: Cell surface marker expression in untreated A549GEV16/BOK. Color coded boxes indicate changes between treatments (4-OHT and/or TGFβ) and control cells (vehicle-treated). Dashed lines indicate markers that were modulated by BOK overexpression.
However, enforced BOK expression abrogated TGFβ-induced upregulation of EGFR, and downregulation of MET and CD321. BOK alone upregulated MET and had a downmodulating effect on the expression of CD166, CD49f, Lymphotoxinβ Receptor, CD70, CD71 (TfR1) and CD94 (Figure 6).
Discussion
We and others have recently shown that BOK is widely expressed and detectable at the protein level in the mouse, with highest levels found in reproductive organs, brain and gastrointestinal tract, but also in the lung (1, 2, 9). On the other hand, BOK mRNA expression seems to be downregulated in many human cancer cell lines (26, 27, 28). Interestingly, Beroukhim and colleagues identified somatic copy number alterations (deletions) of the BOK gene locus in a wide screen of human primary cancers and cancer cell lines (16). These findings point toward a possible tumor suppressor function of BOK. In the present study, we found that BOK is not deleted at the genomic level in any of the 10 NSCLC cell lines studied. However, in BOK low-expressing cell lines we identified an epigenetic silencing mechanism. Specifically, the DNA-methyltransferase inhibitor decitabine upregulated BOK mRNA in all 10 cell lines tested up to 13.8-fold (Figure 3b). Further, combination of decitabine with the histone deacetylase inhibitor trichostatin A had a synergistic effect on BOK mRNA induction in 6 out of 10 cells (Figure 3b). Our ChIP analysis revealed that the combined treatment of decitabine and trichostatin A has no synergistic effect on histone acetylation (Figure 3d and Supplemental data S5), implying that repression of the BOK gene occurs directly by methylation of CpG dinucleotides near the transcriptional start site of BOK (Figure 3b), thereby blocking the binding of transcription factors and not by recruitment of transcriptional repressors such as methyl-binding proteins that are associated with histone deacetylases.
Such epigenetic silencing might contribute to tumorigenesis as already shown for some BH3-only proteins (29), and also to tumor metastasis. While BOK had no effect on acceleration of Eμ-myc induced pre-B/B cell lymphoma, which is likely explained by the very low expression of BOK in lymphocytes (9), loss of BOK may slow NSCLC tumorigenesis as BOK downregulation by RNA interference or CRISPR/Cas9 mediated gene editing resulted in significantly slower proliferation of NSCLC cells, correlating with loss of detection of nuclear BOK and increase of cell cycle inhibitors p16INK4A and p19INK4D (Figure 4d–g and S2). Given the prominent BOK localization in the nucleus (2,11) it is therefore attractive to speculate that the nuclear form BOK may interact with the cell cycle machinery, as proposed by Ray et al. in the context of trophoblast proliferation (12). Interestingly, the observed role of BOK in regulating proliferation opposes the cell cycle specific roles described for the anti-apoptotic BCL-2 family members BCL-2 and BCL-XL, which were shown to inhibit cell cycle progression to G1 by upregulation of p27 (30, 31).
On the other hand, we show that BOK may have the potential to inhibit metastatic spread of developing NSCLC tumor cells by interfering with EMT (Figure 5). Unfortunately, the majority of NSCLC patients are diagnosed with advanced disease (17, 32). Patients with high-grade disease more often have a higher stage at diagnosis, a greater probability of recurrence or metastasis, and an overall shorter survival time after diagnosis (19). Although our patient population was heterogeneous, with patients ranging from Stage IA to IV, all were considered candidates for surgical resection. BOK expression level only appeared to impact survival in patients with lymph node involvement at the time of surgery. In this patient group, increased BOK protein expression was associated with significantly longer survival times (Figure 2). Importantly, we found that BOK levels were significantly decreased in tumors of lymph node positive patients and in less differentiated tumors compared to adjacent lung parenchyma (Figure 1b). This phenotype might be favoured by epigenetic mechanisms during tumor progression (Figure 3, 33) and/or by proteasome-dependent regulation of BOK levels (1, 13). Importantly, we found a poor correlation between BOK mRNA and protein levels. However, our data indicate that mechanisms other than proteasomal turnover may be crucial to control BOK levels and activity in NSCLC cells (Figure 4a).
The process by which cancer cells metastasize is thought to involve the transition of tumor cells with an epithelial phenotype to cells with mesenchymal traits. This process is accompanied with loss of tight junction proteins such as E-cadherin and upregulation of transcriptional repressors of tight junction proteins such as Twist, Snail or Slug. The resulting loss in polarity enables tumor cells to migrate and seed distant lesions. There is evidence concerning the involvement of the tumor microenvironment in EMT (34, 35). It was previously shown, that certain cytokines such as IL-6, IL-8 or TGFβ initiate and/or sustain the mesenchymal cancer cell state (36, 37, 38). We used TGFβ to induce EMT in A549 cells, which express low levels of endogenous BOK. When exogenous BOK expression was induced in those cells, TGFβ–induced phenotypic change was prevented (Figure 5). Along the same line, enforced BOK expression significantly diminished anchorage-independent growth in soft agar (Figure 4h), and reduced TGFβ-induced transmigration (Figure 5c). Importantly, these effects are independent of BOK-induced cell death, as we showed that overexpression of BOK does not induce apoptosis in A549 cells (Figure 4d and S2). It is also worthwhile mentioning that A549 cells have acquired a dysfunction in the apoptosome machinery in the procaspase-9 activation probably by inhibitory phosphorylation of procaspase-9 (39). We and others have shown that BOK mainly localizes to the endoplasmic reticular membrane and BOK has accordingly been implicated with the regulation of ER stress, ERAD pathway and unfolded protein response (UPR) (1, 2, 6). Whereas we did not find BOK to affect the ATF6 or IRE1α arms of the UPR (not shown), our data show that BOK is likely involved in the modulation of the PERK>ATF4 arm in the context of TGFβ induced EMT, supporting studies that link PERK to EMT (24). Furthermore, the quantitative surface proteomics profiling showed upregulation of EGFR by TGFβ (Figure 6) suggesting the involvement of JUN N-terminal kinases pathway leading to JUN transcription factor activation (40) that binds to EGFR responsive element. Although enforced expression of BOK abrogated EGFR upregulation (Figure 6), which is indispensable for AKT activation leading to cell survival, BOK also abrogated TGFβ-mediated MET downregulation. Thus, BOK overexpression contributes to cell survival by inhibition of EGFR and upregulation of MET. However, sustained expression of MET and CD321 in TGFβ-treated BOK overexpressing cells shows epithelial state of the cells, thus confirming the BOK role in EMT inhibition. Moreover, overexpression of BOK downregulated membrane expression of CD166/ALCAM (Figure 6), which is implicated in NSCLC cancer cells migration and invasion in vitro (41) and its reduced expression inhibited skeletal metastasis in prostate cancer in vivo (42).
In conclusion, we found that alteration of BOK expression in NSCLC tumors could account for differences in tumour invasion and thus overall survival of NSCLC patients.
Supplementary Material
Novelty and Impact.
BOK is one of the most frequently deleted BCL-2 family members in human cancer. Here we show a possible cell death-independent tumor suppressor function of BOK in NSCLC. We found that, in vitro, BOK is epigenetically silenced and that its overexpression decreases anchorage-independent growth, TGF-beta induced migration and epithelial-to-mesenchymal transition. In vivo, BOK levels are predictive of survival in lymph node positive patients. BOK expression might therefore serve as prognostic marker in later stage NSCLC.
Acknowledgments
This work was supported by Sciex-NMSch (13.107; to EM), Ruth Crawford Mitchell Fellowship (2015; to EM), United States Department of Defense (BC132245_W81XWH-14-025; to VSD), National Cancer Institute (RO1-CA114246, R21-CA191647; to ADD), Swiss Cancer League (No. KFS-3014-08-2012; to TK), Swiss National Science Foundation (SNSF, grant No. 310030E-150805/1, part of the D-A-CH initiative from the SNSF and the Deutsche Forschungsgemeinschaft (DFG), FOR-2036; to TK). The UPCI Cytometry Facility is supported by CCSG P30CA047904.
The authors would like to thank Dr. Jan Cermak (Department of Pneumology and Thoracic Surgery, Hospital Bulovka, Prague), Dr. Ilona Rousalova (General University Hospital in Prague) for obtaining tissue samples, clinico-pathological evaluation and data from the patients entering the study. We also thank Dr. Christian T Carson and BD Biosciences (BD Biosciences, San Diego, CA, USA) for providing the Lyoplates and Dr. Juraj Adamik for technical feedback for ChIP analysis.
Abbreviations
- 4-OHT
4-hydroxytamoxifen
- BOK
BCL-2 ovarian killer
- EMT
epithelial-to-mesenchymal transition
- LAC
lung adenocarcinoma
- LCLC
large cell lung carcinoma
- NSCLC
non-small cell lung carcinoma
- SLC
sarcomatoid lung carcinoma
- SQCLC
squamous cell lung carcinoma
- UNDIF
undifferentiated lung carcinoma
Footnotes
Conflict of Interest
The authors declare no conflicts of interest
References
- 1.Llambi F, Wang YM, Victor B, Yang M, Schneider DM, Gingras S, Parsons MJ, Zheng JH, Brown SA, Pelletier S, Moldoveanu T, Chen T, et al. BOK Is a Non-canonical BCL-2 Family Effector of Apoptosis Regulated by ER-Associated Degradation. Cell. 2016;165(2):421–33. doi: 10.1016/j.cell.2016.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Echeverry N, Bachmann D, Ke F, Strasser A, Simon HU, Kaufmann T. Intracellular localization of the BCL-2 family member BOK and functional implications. Cell Death Differ. 2013;20(6):785–99. doi: 10.1038/cdd.2013.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hsu SY, Kaipia A, McGee E, Lomeli M, Hsueh AJ. Bok is a pro-apoptotic Bcl-2 protein with restricted expression in reproductive tissues and heterodimerizes with selective anti-apoptotic Bcl-2 family members. Proc Natl Acad Sci U S A. 1997;94(23):12401–6. doi: 10.1073/pnas.94.23.12401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Inohara N, Ekhterae D, Garcia I, Carrio R, Merino J, Merry A, Chen S, Nunez G. Mtd, a novel Bcl-2 family member activates apoptosis in the absence of heterodimerization with Bcl-2 and Bcl-XL. J Biol Chem. 1998;273(15):8705–10. doi: 10.1074/jbc.273.15.8705. [DOI] [PubMed] [Google Scholar]
- 5.Einsele-Scholz S, Malmsheimer S, Bertram K, Stehle D, Johanning J, Manz M, Daniel PT, Gillissen BF, Schulze-Osthoff K, Essmann F. Bok is a genuine multi-BH-domain protein that triggers apoptosis in the absence of Bax and Bak. J Cell Sci. 2016;129(11):2213–23. doi: 10.1242/jcs.181727. [DOI] [PubMed] [Google Scholar]
- 6.Carpio MA, Michaud M, Zhou W, Fisher JK, Walensky LD, Katz SG. BCL-2 family member BOK promotes apoptosis in response to endoplasmic reticulum stress. Proc Natl Acad Sci U S A. 2015;112(23):7201–6. doi: 10.1073/pnas.1421063112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ke F, Grabow S, Kelly GL, Lin A, O'Reilly LA, Strasser A. Impact of the combined loss of BOK, BAX and BAK on the hematopoietic system is slightly more severe than compound loss of BAX and BAK. Cell Death Dis. 2015;6:e1938. doi: 10.1038/cddis.2015.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ke F, Bouillet P, Kaufmann T, Strasser A, Kerr J, Voss AK. Consequences of the combined loss of BOK and BAK or BOK and BAX. Cell Death Dis. 2013;4:e650. doi: 10.1038/cddis.2013.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ke F, Voss A, Kerr JB, O'Reilly LA, Tai L, Echeverry N, Bouillet P, Strasser A, Kaufmann T. BCL-2 family member BOK is widely expressed but its loss has only minimal impact in mice. Cell Death Differ. 2012;19(6):915–25. doi: 10.1038/cdd.2011.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fernandez-Marrero Y, Bleicken S, Das KK, Bachmann D, Kaufmann T, Garcia-Saez AJ. The membrane activity of BOK involves formation of large, stable toroidal pores and is promoted by cBID. FEBS J. 2017;284(5):711–24. doi: 10.1111/febs.14008. [DOI] [PubMed] [Google Scholar]
- 11.Bartholomeusz G, Wu Y, Ali SM, Xia W, Kwong KY, Hortobagyi G, Hung MC. Nuclear translocation of the pro-apoptotic Bcl-2 family member Bok induces apoptosis. Mol Carcinog. 2006;45(2):73–83. doi: 10.1002/mc.20156. [DOI] [PubMed] [Google Scholar]
- 12.Ray JE, Garcia J, Jurisicova A, Caniggia I. Mtd/Bok takes a swing: proapoptotic Mtd/Bok regulates trophoblast cell proliferation during human placental development and in preeclampsia. Cell Death Differ. 2010;17(5):846–59. doi: 10.1038/cdd.2009.167. [DOI] [PubMed] [Google Scholar]
- 13.Schulman JJ, Wright FA, Han X, Zluhan EJ, Szczesniak LM, Wojcikiewicz RJ. The Stability and Expression Level of Bok Are Governed by Binding to Inositol 1,4,5-Trisphosphate Receptors. J Biol Chem. 2016;291(22):11820–8. doi: 10.1074/jbc.M115.711242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schulman JJ, Wright FA, Kaufmann T, Wojcikiewicz RJ. The Bcl-2 protein family member Bok binds to the coupling domain of inositol 1,4,5-trisphosphate receptors and protects them from proteolytic cleavage. J Biol Chem. 2013;288(35):25340–9. doi: 10.1074/jbc.M113.496570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.D'Orsi B, Engel T, Pfeiffer S, Nandi S, Kaufmann T, Henshall DC, Prehn JH. Bok Is Not Pro-Apoptotic But Suppresses Poly ADP-Ribose Polymerase-Dependent Cell Death Pathways and Protects against Excitotoxic and Seizure-Induced Neuronal Injury. J Neurosci. 2016;36(16):4564–78. doi: 10.1523/JNEUROSCI.3780-15.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Beroukhim R, Mermel CH, Porter D, Wei G, Raychaudhuri S, Donovan J, Barretina J, Boehm JS, Dobson J, Urashima M, Mc Henry KT, Pinchback RM, et al. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463(7283):899–905. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66(1):7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 18.Travis WD, Brambilla E, Nicholson AG, Yatabe Y, Austin JH, Beasley MB, Chirieac LR, Dacic S, Duhig E, Flieder DB, Geisinger K, Hirsch FR, et al. The 2015 World Health Organization Classification of Lung Tumors: Impact of Genetic, Clinical and Radiologic Advances Since the 2004 Classification. J Thorac Oncol. 2015;10(9):1243–60. doi: 10.1097/JTO.0000000000000630. [DOI] [PubMed] [Google Scholar]
- 19.Detterbeck FC, Boffa DJ, Tanoue LT. The new lung cancer staging system. Chest. 2009;136(1):260–71. doi: 10.1378/chest.08-0978. [DOI] [PubMed] [Google Scholar]
- 20.Gurzeler U, Rabachini T, Dahinden CA, Salmanidis M, Brumatti G, Ekert PG, Echeverry N, Bachmann D, Simon HU, Kaufmann T. In vitro differentiation of near-unlimited numbers of functional mouse basophils using conditional Hoxb8. Allergy. 2013;68(5):604–13. doi: 10.1111/all.12140. [DOI] [PubMed] [Google Scholar]
- 21.Krepela E, Dankova P, Moravcikova E, Krepelova A, Prochazka J, Cermak J, Schutzner J, Zatloukal P, Benkova K. Increased expression of inhibitor of apoptosis proteins, survivin and XIAP, in non-small cell lung carcinoma. Int J Oncol. 2009;35(6):1449–62. doi: 10.3892/ijo_00000464. [DOI] [PubMed] [Google Scholar]
- 22.Sanjana NE, Shalem O, Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods. 2014;11(8):783–4. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li LC, Dahiya R. MethPrimer: designing primers for methylation PCRs. Bioinformatics. 2002;18(11):1427–31. doi: 10.1093/bioinformatics/18.11.1427. [DOI] [PubMed] [Google Scholar]
- 24.Feng YX, Sokol ES, Del Vecchio CA, Sanduja S, Claessen JH, Proia TA, Jin DX, Reinhardt F, Ploegh HL, Wang Q, Gupta PB. Epithelial-to-mesenchymal transition activates PERK-eIF2alpha and sensitizes cells to endoplasmic reticulum stress. Cancer Discov. 2014;4(6):702–15. doi: 10.1158/2159-8290.CD-13-0945. [DOI] [PubMed] [Google Scholar]
- 25.Donnenberg AD, Meyer EM, Rubin JP, Donnenberg VS. The cell-surface proteome of cultured adipose stromal cells. Cytometry A. 2015;87(7):665–74. doi: 10.1002/cyto.a.22682. [DOI] [PubMed] [Google Scholar]
- 26.Reference Database for Gene Expression Analysis. [Accessed: September 30, 2016]; available at: http://sbmdb.genome.rcast.u-tokyo.ac.jp/refexa/main_search.jsp.
- 27.Gao S, Fu W, Durrenberger M, De Geyter C, Zhang H. Membrane translocation and oligomerization of hBok are triggered in response to apoptotic stimuli and Bnip3. Cell Mol Life Sci. 2005;62(9):1015–24. doi: 10.1007/s00018-005-4543-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Petryszak R, Keays M, Tang YA, Fonseca NA, Barrera E, Burdett T, Fullgrabe A, Fuentes AM, Jupp S, Koskinen S, Mannion O, Huerta L, et al. Expression Atlas update--an integrated database of gene and protein expression in humans, animals and plants. Nucleic Acids Res. 2016;44(D1):D746–D752. doi: 10.1093/nar/gkv1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Garrison SP, Jeffers JR, Yang C, Nilsson JA, Hall MA, Rehg JE, Yue W, Yu J, Zhang L, Onciu M, Sample JT, Cleveland JL, et al. Selection against PUMA gene expression in Myc-driven B-cell lymphomagenesis. Mol Cell Biol. 2008;28(17):5391–402. doi: 10.1128/MCB.00907-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Greider C, Chattopadhyay A, Parkhurst C, Yang E. BCL-x(L) and BCL2 delay Myc-induced cell cycle entry through elevation of p27 and inhibition of G1 cyclin-dependent kinases. Oncogene. 2002;21(51):7765–75. doi: 10.1038/sj.onc.1205928. [DOI] [PubMed] [Google Scholar]
- 31.Pierce RH, Vail ME, Ralph L, Campbell JS, Fausto N. Bcl-2 expression inhibits liver carcinogenesis and delays the development of proliferating foci. Am J Pathol. 2002;160(5):1555–60. doi: 10.1016/S0002-9440(10)61101-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Molina JR, Yang P, Cassivi SD, Schild SE, Adjei AA. Non-small cell lung cancer: epidemiology, risk factors, treatment, and survivorship. Mayo Clin Proc. 2008;83(5):584–94. doi: 10.4065/83.5.584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sarkar S, Horn G, Moulton K, Oza A, Byler S, Kokolus S, Longacre M. Cancer development, progression, and therapy: an epigenetic overview. Int J Mol Sci. 2013;14(10):21087–113. doi: 10.3390/ijms141021087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lu H, Clauser KR, Tam WL, Frose J, Ye X, Eaton EN, Reinhardt F, Donnenberg VS, Bhargava R, Carr SA, Weinberg RA. A breast cancer stem cell niche supported by juxtacrine signalling from monocytes and macrophages. Nat Cell Biol. 2014;16(11):1105–17. doi: 10.1038/ncb3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vizoso M, Puig M, Carmona FJ, Maqueda M, Velasquez A, Gomez A, Labernadie A, Lugo R, Gabasa M, Rigat-Brugarolas LG, Trepat X, Ramirez J, et al. Aberrant DNA methylation in non-small cell lung cancer-associated fibroblasts. Carcinogenesis. 2015;36(12):1453–63. doi: 10.1093/carcin/bgv146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shintani Y, Fujiwara A, Kimura T, Kawamura T, Funaki S, Minami M, Okumura M. IL-6 Secreted from Cancer-Associated Fibroblasts Mediates Chemoresistance in NSCLC by Increasing Epithelial-Mesenchymal Transition Signaling. J Thorac Oncol. 2016;11(9):1482–92. doi: 10.1016/j.jtho.2016.05.025. [DOI] [PubMed] [Google Scholar]
- 37.Kim YE, Kim JO, Park KS, Won M, Kim KE, Kim KK. Transforming Growth Factor-beta-Induced RBFOX3 Inhibition Promotes Epithelial-Mesenchymal Transition of Lung Cancer Cells. Mol Cells. 2016;39(8):625–30. doi: 10.14348/molcells.2016.0150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li L, Qi L, Liang Z, Song W, Liu Y, Wang Y, Sun B, Zhang B, Cao W. Transforming growth factor-beta1 induces EMT by the transactivation of epidermal growth factor signaling through HA/CD44 in lung and breast cancer cells. Int J Mol Med. 2015;36(1):113–22. doi: 10.3892/ijmm.2015.2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moravcikova E, Krepela E, Prochazka J, Benkova K, Pauk N. Differential sensitivity to apoptosome apparatus activation in non-small cell lung carcinoma and the lung. Int J Oncol. 2014;44(5):1443–54. doi: 10.3892/ijo.2014.2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sun C, Wang L, Huang S, Heynen GJ, Prahallad A, Robert C, Haanen J, Blank C, Wesseling J, Willems SM, Zecchin D, Hobor S, et al. Reversible and adaptive resistance to BRAF(V600E) inhibition in melanoma. Nature. 2014;508(7494):118–22. doi: 10.1038/nature13121. [DOI] [PubMed] [Google Scholar]
- 41.Ishiguro F, Murakami H, Mizuno T, Fujii M, Kondo Y, Usami N, Taniguchi T, Yokoi K, Osada H, Sekido Y. Membranous expression of activated leukocyte cell adhesion molecule contributes to poor prognosis and malignant phenotypes of non-small-cell lung cancer. J Surg Res. 2013;179(1):24–32. doi: 10.1016/j.jss.2012.08.044. [DOI] [PubMed] [Google Scholar]
- 42.Hansen AG, Arnold SA, Jiang M, Palmer TD, Ketova T, Merkel A, Pickup M, Samaras S, Shyr Y, Moses HL, Hayward SW, Sterling JA, et al. ALCAM/CD166 is a TGF-beta-responsive marker and functional regulator of prostate cancer metastasis to bone. Cancer Res. 2014;74(5):1404–15. doi: 10.1158/0008-5472.CAN-13-1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.