Abstract
Background:
High-risk non-muscle invasive bladder cancer (HR-NMIBC) is a clinically unpredictable disease. Despite clinical risk estimation tools, many patients are undertreated with intra-vesical therapies alone, whereas others may be over-treated with early radical surgery. Molecular biomarkers, particularly DNA methylation, have been reported as predictive of tumour/patient outcomes in numerous solid organ and haematologic malignancies; however, there are few reports in HR-NMIBC and none using genome-wide array assessment. We therefore sought to identify novel DNA methylation markers of HR-NMIBC clinical outcomes that might predict tumour behaviour at initial diagnosis and help guide patient management.
Patients and methods:
A total of 21 primary initial diagnosis HR-NMIBC tumours were analysed by Illumina HumanMethylation450 BeadChip arrays and subsequently bisulphite Pyrosequencing. In all, 7 had not recurred at 1 year after resection and 14 had recurred and/or progressed despite intra-vesical BCG. A further independent cohort of 32 HR-NMIBC tumours (17 no recurrence and 15 recurrence and/or progression despite BCG) were also assessed by bisulphite Pyrosequencing.
Results:
Array analyses identified 206 CpG loci that segregated non-recurrent HR-NMIBC tumours from clinically more aggressive recurrence/progression tumours. Hypermethylation of CpG cg11850659 and hypomethylation of CpG cg01149192 in combination predicted HR-NMIBC recurrence and/or progression within 1 year of diagnosis with 83% sensitivity, 79% specificity, and 83% positive and 79% negative predictive values.
Conclusions:
This is the first genome-wide DNA methylation analysis of a unique HR-NMIBC tumour cohort encompassing known 1-year clinical outcomes. Our analyses identified potential novel epigenetic markers that could help guide individual patient management in this clinically unpredictable disease.
Keywords: high-risk non-muscle invasive bladder cancer, epigenetics, methylation, HumanMethylation450 BeadChip array, prognostic biomarker
Introduction
Bladder cancer is a common and worldwide health problem.1 Most bladder cancers arise from the urothelium (urothelial cell carcinomas ‘UCC’), of which 70% to 80% are non-muscle invasive bladder cancers (NMIBC) at presentation.2 Grade 3 NMIBC is a clinically important sub-type of bladder UCC, accounting for approximately 10% to 15% of all NMIBCs at presentation and considered to be ‘high-risk’ NMIBC (HR-NMIBC).3,4 These tumours are more aggressive than their low- and intermediate-risk counterparts and manifest by higher rates of tumour recurrence, progression to muscle invasive bladder cancer (MIBC) and/or metastatic disease despite intra-vesical therapies.5,6 Although progression to MIBC is associated with poor outcomes, many HR-NMIBC tumours do not recur or progress. Therefore, immediate radical cystectomy based on estimated future risks may be considered ‘over-treatment’ with inherent morbidity and quality of life implications.3,5,7
As adverse patient outcomes may result from under-treatment with intra-vesical therapies alone, or from over-treatment with early radical surgery, additional methods of risk estimation are required. The European Association of Urology (EAU) recommends the use of the European Organisation for the Research and Treatment of Cancer (EORTC) risk estimation tool.3 This provides 1- to 5-year estimates of disease recurrence and progression each year. However, this tool generates estimates only and is based on 10-year-old data with recognised limitations.8 Therefore, additional methods of risk stratification to support clinical decision making (for the patient and surgeon) are required.9 In this regard, molecular markers are a key area of investigation.
As HR-NMIBC tumours appear to be molecularly heterogeneous,10 previous investigations have failed to find common genetic changes as reliable biomarkers, either as stand-alone ‘tests’ or in combination with clinical parameters.9,11 However, epigenetic modifications, and particularly DNA methylation, have been identified as diagnostic and prognostic in numerous solid organ and haematologic malignancies, even in those considered particularly heterogeneous, for example, lung and malignant melanoma.12,13 Furthermore, DNA methylation changes are ideal for biomarker exploitation as they occur early in tumour development and are stable and readily measurable.14 However, although previous reports in HR-NMIBC have suggested correlations with clinical outcomes,14,15 few studies have sought to assess DNA methylation patterns as prognostic in this tumour type, with limitations in number of genes assessed, sample heterogeneity, and the presence of other bladder tumour types.16–19
To more comprehensively assess DNA methylation for biomarker potential in HR-NMIBC, we used Illumina HumanMethylation450 BeadChip array technology. Our ‘450K’ array interrogated a unique cohort of HR-NMIBC with 1-year clinical outcomes of ‘no recurrence’, ‘recurrence’, or ‘progression’. Through comparisons of DNA methylation patterns, we identified epigenetic differences between these outcome ‘sub-types’ of HR-NMIBC. We thus report potential prognostic methylation biomarkers that may guide patient management at initial diagnosis of this unpredictable disease.
Patients and Methods
Human tissue samples
Primary tumour and normal bladder tissues were provided by the Bladder Cancer Prognosis Programme (BCPP, National Research Ethics Service East Midlands – Derby 06/MRE04/65),20 the University of Birmingham Human Biomaterials Resource Centre (National Research Ethics Service [North West 5]: 09/H1010/75), and the University Hospitals of North Midlands NHS Trust (National Research Ethics Service [South Central – Oxford C]: 12/SC/0725). All samples were confirmed histologically as G3 T1 UCC (discovery cohort n = 21, validation cohort n = 32). All tumours were from initial presentation bladder tumours, in patients with no prior history of bladder cancer and intra-vesical therapies. As previously described,21 patients received repeat bladder tumour resection (TURBT [transurethral resection of bladder tumour]), intra-vesical therapy, and/or cystectomy as recommended by EAU guidelines.22 All samples (Supplemental Table S1) were stored at −80°C prior to nucleic acid extraction, as described below.
DNA extraction and bisulphite modification
Genomic DNA was isolated from tumour tissue using a standard phenol-chloroform extraction,23 then bisulphite converted using the EZ DNA Methylation Gold Kit (Zymo Research, Irvine, CA, USA) as previously described.21,24 Bisulphite conversion was confirmed by successful polymerase chain reaction with primers specific to bisulphite-converted DNA. To increase the amount and stability of bisulphite-converted DNA, whole-genome amplification was performed as previously described.21,24
Illumina 450K methylation bead array analyses
Bisulphite-converted DNA from 21 initial presentation bladder tumours and 3 normal bladder controls was hybridised to Infinium-based HumanMethylation450 BeadChip arrays (Illumina, San Diego, CA, USA). Arrays were processed according to the manufacturer’s instructions (performed by Barts and the London Genome Centre, UK).21,24 Raw array data were processed using GenomeStudio software and the bioinformatical platform ‘NIMBL’, as we and others have described previously.21,24,25 For each probe, methylation was reported as a ‘β-value’, where ‘β’ is defined as the ratio of the methylated signal intensity over the summed intensity of the methylated and unmethylated signals + 100.40 (β values range from 0 [unmethylated] to 1 [fully methylated]). NIMBL was used to perform ‘peak-based’ correction and to adjust for potential differences in array probe-type sensitivity previously reported26; all comparative analyses were performed on peak-based corrected β values, as described previously.21,24 Each array passed quality control assessment based on the performance of internal controls and the distribution of β values across all array CpGs.
As previously described,21,24 we excluded all CpGs for which any of the 24 samples displayed: (1) probe detection P values of >.05 (unreliable probe data) or (2) missing β values (preventing analyses of all samples). We also excluded all CpG loci on allosomes (reducing confounding sex-based methylation differences).
Technical validation of methylation bead chip array data
The correlation between ‘450K’ array and Pyrosequencing was confirmed across a total of 120 CpGs using Spearman rank correlation, as previously reported.21
Pyrosequencing of sodium bisulphite–converted DNA
Pyrosequencing of sodium bisulphite–converted DNA was used to validate the discovery cohort array data (21 tumours) and to assess methylation in the independent validation tumour cohort (32 tumours). A PyroMark Q24 Pyrosequencer, PyroMark Q24 Software 2.0, and PyroMark Gold Q24 Reagents were used, as previously described by us.21,24
Assessment of potential clinical performance
MedCalc Statistical Software (version 17.0.4; Ostend, Belgium; https://www.medcalc.org; 2017) was used to perform receiver operating characteristics analyses, area under the curve (AUC) calculations to determine sensitivity and specificity, and also to determine positive and negative predictive values.
STATA (version 8; Stata Corporation, College Station, TX, USA) was used to analyse associations between candidate methylation biomarkers and clinical or demographic variables. In these cases, P < .05 was considered statistically significant.
Results
Technical validation of array by pyrosequencing
As previously reported,21 and after array processing, normalisation, and peak-based correction, a technical validation confirmed a strong positive correlation between array- and Pyrosequencing-derived methylation values (Spearman rank correlation r = .912, P < .00001; data not shown).
Array filtering steps
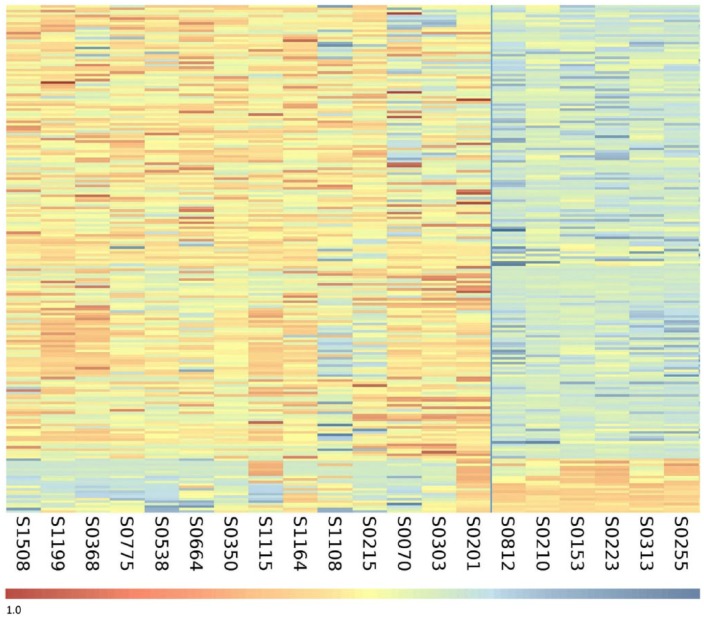
CpGs showing differential methylation between the HR-NMIBC no-recurrence tumours and the recurrence and/or progression tumours were included where 10 or more of 14 recurrence/progression tumours showed a ≥0.1 β value difference relative to all 7 of the no-recurrence tumours. Using these criteria, 206 differentially methylated CpGs were identified, as represented by heatmap in Figure 1 (cg identifier list of the 206 CpGs in Supplemental Table S2). In total, 186 were hypermethylated and 20 were hypomethylated in the recurrence and/or progression tumours relative to the no-recurrence tumours.
Figure 1.
Heatmap of the 206 differentially methylated CpG sites between the clinical outcomes of HR-NMIBC. Heatmap of the differentially methylated CpG sites identified by array analysis. The heatmap separates the 14 recurrence or progression tumours on the left (n = 14) from the no-recurrence tumours on the right (n = 7). Each row represents an individual CpG locus, and each column represents a tumour sample (listed beneath the heatmap). The colour scale beneath the heatmap represents methylation status: unmethylated is blue (β value = 0.0) and fully methylated is red (β value = 1.0).
Identification of potential prognostic biomarker candidates
To focus our assessment on targets with the greatest potential for clinical use, the top 20 CpG biomarker candidates (10 hypermethylated and 10 hypomethylated) were identified on the basis of the most frequent differential methylation (in 12 or more recurrence/progression tumours of 14) (listed in Table 1 left-hand panel). These putative biomarker candidates were subject to initial screening using MedCalc software (v17.0.4). This estimated the potential sensitivity, specificity, and positive and negative predictive values for tumour recurrence/progression of each CpG site in the 21 array tumours (Table 1).
Table 1.
CpG sites showing the greatest differential methylation between the no-recurrence and the recurrence/progression tumours.
| cg ID | Direction of methylation | Recurrence and/or progression | Sensitivity, % | Specificity, % | Positive predictive value, % | Negative predictive value, % |
|---|---|---|---|---|---|---|
| cg04415176 | Hyper | 12/14 | 85.7 | 100.0 | 100.0 | 77.8 |
| cg06391663 | Hyper | 12/14 | 85.7 | 100.0 | 100.0 | 77.8 |
| cg19457237 | Hyper | 12/14 | 85.7 | 100.0 | 100.0 | 77.8 |
| cg06607594 | Hyper | 12/14 | 85.7 | 100.0 | 100.0 | 77.8 |
| cg01392017 | Hyper | 12/14 | 85.7 | 100.0 | 100.0 | 77.8 |
| cg13322920 | Hyper | 12/14 | 85.7 | 100.0 | 100.0 | 77.8 |
| cg17180705 | Hyper | 12/14 | 85.7 | 100.0 | 100.0 | 77.8 |
| cg11850659 | Hyper | 13/14 | 92.9 | 100.0 | 100.0 | 87.5 |
| cg12228319 | Hyper | 13/14 | 92.9 | 100.0 | 100.0 | 87.5 |
| cg18916488 | Hyper | 12/14 | 85.7 | 100.0 | 100.0 | 77.8 |
| cg12539415 | Hypo | 13/14 | 92.9 | 100.0 | 100.0 | 87.5 |
| cg12050358 | Hypo | 12/14 | 85.7 | 100.0 | 100.0 | 77.8 |
| cg19182537a | Hypo | 11/14 | 78.6 | 100.0 | 100.0 | 70.0 |
| cg14729962 | Hypo | 12/14 | 85.7 | 100.0 | 100.0 | 77.8 |
| cg04382470 | Hypo | 12/14 | 85.7 | 100.0 | 100.0 | 77.8 |
| cg01149192 | Hypo | 12/14 | 85.7 | 100.0 | 100.0 | 77.8 |
| cg00397479 | Hypo | 12/14 | 85.7 | 100.0 | 100.0 | 77.8 |
| cg03540028 | Hypo | 12/14 | 85.7 | 100.0 | 100.0 | 77.8 |
| cg22328426 | Hypo | 13/14 | 92.9 | 100.0 | 100.0 | 87.5 |
| cg27084746 | Hypo | 12/14 | 85.7 | 100.0 | 100.0 | 77.8 |
Top 20 sites (with CG identifier) of differential methylation between the clinical outcomes of high-risk non-muscle invasive bladder cancer. The direction of methylation change in the recurrence/progression tumours is stated relative to the no-recurrence tumours, with the number of tumours showing differential methylation at each site shown. The values for sensitivity, specificity, and positive and negative predictive values of tumour recurrence/progression are given on the right side of the table.
cg19182537 was included with the candidates showing differential methylated 12 or more recurrence/progression tumours of 14, as methylation in one of the recurrence/progression tumours was very close to the differential methylation threshold used.
Validation of biomarker potential by pyrosequencing
The 6 best CpG biomarker candidates (cg12228319, cg19457237, cg11850659, cg22328426, cg12539415, and cg01149192) were identified based on biomarker potential (predictive values) suggested in Table 1, and by visual inspection of plotted array data, where discrimination of tumour/clinical outcome was most evident based on magnitude of differential methylation.
Methylation of these 6 candidates was confirmed by Pyrosequencing in the 21 array tumours, which showed good concordance with the corresponding array β values (data not shown). The 6 candidates were then assessed by Pyrosequencing in our independent tumour cohort of 32 HR-NMIBC tumours. In total, therefore, 53 tumours were investigated (24 no-recurrence and 29 recurrence/progression tumours). These analyses confirmed the array-identified patterns of differential methylation between the clinical outcome sub-types in most of the tumours. Based on the greatest discrimination between clinical outcome groups shown by these Pyrosequencing data, the top 2 performing prognostic biomarker candidates were identified (cg11850659 and cg01149192) (primer sequences: Supplemental Table S3).
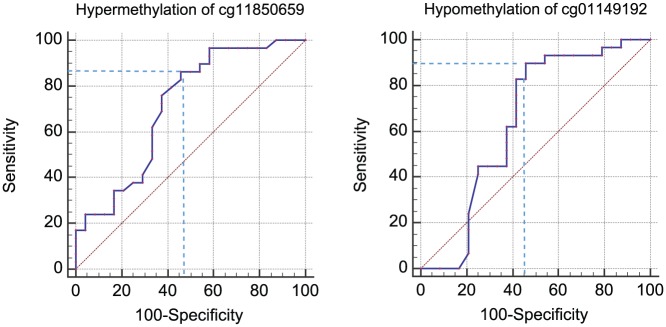
Again, based on methylation values across all 53 tumours, the biomarker potential of cg11850659 and cg01149192 was next determined by receiver operating characteristic (ROC) curve analyses. As shown in Figure 2, the AUC and the sensitivity and specificity values for tumour recurrence and/or progression were 0.71 (95% confidence interval [CI]: 0.57-0.83), 86.2% and 54.2% for cg11850659 (methylation values above 51%), and 0.64 (95% CI 0.50-0.77), 89.7% and 54.2% for cg01149192 (methylation values equal to or less than 41%) (ROC analyses: Supplemental Table S4). The combination of hypermethylation of cg11850659 and hypomethylation of cg01149192 was assessed using the threshold methylation values above in a 2 × 2 contingency table. This ‘combination’ biomarker (hypermethylation of cg11850659 and hypomethylation of cg01149192) showed a sensitivity of 82.8%, a specificity of 79.2%, a positive predictive value of 82.8%, and a negative predictive value of 79.2% for HR-NMIBC recurrence and/or progression at/within 1 year of initial diagnosis. The 10-fold cross-validation with 10% of the samples predicted outcome correctly 36 times of 50 (72%).
Figure 2.
Receiver operating characteristic (ROC) curves for cg11850659 and cg01149192. ROC curves for the 2 best performing biomarker candidates. Hypermethylation of CG11850659 (left) – AUC: 0.71 (95% CI: 0.57-0.83) and hypomethylation of CG01149192 (right) – 0.64 (95% CI: 0.50-0.77). AUC indicates area under the curve; CI, confidence interval.
Biomarker independence from demographic factors and treatment duration
To confirm that these 2 DNA methylation biomarker candidates (alone and in combination) were independent predictors of disease outcome, potential associations between methylation and other known factors were assessed. Multivariate regression did not identify any correlations between methylation with patient age, sex, or intra-vesical BCG treatment duration (Supplemental Table S1). However, data regarding smoking history, tumour size, and number, ethnicity, and occupational history were not available for these analyses.
Discussion
Similar to other solid organ and haematologic malignancies, patterns of DNA methylation correlate with clinical outcomes in bladder cancer.14,15 Despite the difficulty in predicting disease course, HR-NMIBC is rarely investigated as a discrete entity for subtype-specific DNA methylation.16,17 We therefore used HumanMethylation450 array technology in this tumour type to identify potential prognostic biomarkers. After the array data were confirmed reliable by technical validation, and similar to previous reports,27,28 we used a β value change of ≥0.1 to identify differential methylation. In this case, we grouped the recurrence and progression tumours together for assessment relative to the no-recurrence tumours. This grouping was considered appropriate as HR-NMIBC tumour recurrence or progression may prompt change(s) to the clinical management of patients, and both are associated with poorer prognosis than no (tumour) recurrence at 1 year.3,5,29 The number of differentially methylated CpG sites identified was broadly in keeping with similar studies in other tumour types,28,30 and comparable methylation patterns observed between array and independent tumour cohorts for our top 6 biomarkers suggested that our approach in identifying these candidates was robust.
The tumours investigated were initial presentation and intra-vesical BCG (treatment) naïve; the associated clinical outcomes were recorded prospectively.20 Overall, 45 of 53 patients received at least 6 intra-vesical instillations of BCG (induction) within the first year after tumour resection. As such, we reasoned that the methylation patterns identified might hold promise as ‘at diagnosis’ predictors of patient/tumour outcome despite standard treatment, similar to prognostic/treatment-response methylation biomarkers in other tumour types.31,32 However, although DNA methylation patterns have been described as sensitive and specific for HR-NMIBC diagnosis, methylation has not been previously described as reliably predictive of outcome in this tumour type when considered separately from low- and intermediate-risk NMIBC and/or bladder carcinoma in situ, an aggressive tumour type often associated with but histologically and molecularly distinct from HR-NMIBC.16–19,33
As described in similar reports,34 our biomarker candidate methylation data were used for ROC and AUC analyses to estimate sensitivity, specificity, and positive and negative predictive values for tumour recurrence or progression within 1 year of initial diagnosis. Hypermethylation of cg11850659 and hypomethylation of cg01149192 were the best predictors of tumour recurrence/progression; however, individually, their specificity (54.2%) was considered inadequate for a clinically usable test. Hypermethylation of cg11850659 and hypomethylation of cg01149192 in combination, however, demonstrated favourable sensitivity, specificity, and positive and negative predictive values. Furthermore, these values are in keeping with those reported in similar studies of, for example, breast and cervical cancers.35,36 Although these data therefore suggest the exciting clinical potential of our novel prognostic methylation biomarkers in HR-NMIBC, there are no comparable studies in this tumour type, and as such, our ability to interpret results in the context of previously published data is limited.
Although abnormal methylation at these 2 CpG sites has not been previously described as predictive of clinical outcome in any tumour type, cg11850659 (chr 6: 164 254 857; open sea) has been found hypomethylated in hepatocellular carcinoma,37 whereas cg01149192 (chr 5: 180 231 058; lying within an MGAT1 promoter-associated CpG island) has been found hypermethylated in head and neck squamous cell carcinoma.38 In this case, hypermethylation of cg01149192 was also associated with changes in transcript expression; however, methylation as a potential causal mechanism of altered gene expression and tumour development was not assessed. It is possible therefore that DNA methylation at one or more of our identified novel potential biomarker CpG sites may be contributory/causal or recurrent/progressive disease, rather than just predictive of these outcomes.
Conclusions and Limitations
In summary, we have presented the first 450K array DNA methylation assessment of initial presentation and treatment-naïve HR-NMIBC tumours associated with divergent clinical outcomes. Our analyses suggested multiple differentially methylated CpG sites between recurrence/progression tumours and their less aggressive no-recurrence counterparts. Assessment of these predictive methylation biomarkers suggests their exciting clinical potential to support clinical decision making in this unpredictable tumour type.
We recognise that our data represent ‘proof of principle’ and that further studies are required to validate our data. Specifically, we plan to assess these potential biomarkers in prospective studies including larger tumour cohorts, which will also aid more reliable assessments of associations with clinical and demographic variables than was possible in this study. We also recognise that the prevalence of our assessed outcomes (no-recurrence and recurrence/progression) differs slightly within our tumour cohort and therefore we advocate caution in the interpretation of the predictive values presented.
Supplementary Material
Footnotes
Funding:The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This study was funded in part by Cancer Research UK, the North Staffordshire Medical Institute, and the University Hospitals of North Midlands NHS Trust Charitable Foundation.
Declaration of conflicting interests:The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
References
- 1. Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359–E366. [DOI] [PubMed] [Google Scholar]
- 2. Kirkali Z, Chan T, Manoharan M, et al. Bladder cancer: epidemiology, staging and grading, and diagnosis. Urology. 2005;66:4–34. [DOI] [PubMed] [Google Scholar]
- 3. Sylvester RJ, van der Meijden AP, Oosterlinck W, et al. Predicting recurrence and progression in individual patients with stage Ta T1 bladder cancer using EORTC risk tables: a combined analysis of 2596 patients from seven EORTC trials. Eur Urol. 2006;49:466–475 (discussion 475-477). [DOI] [PubMed] [Google Scholar]
- 4. Boustead GB, Fowler S, Swamy R, et al. Stage, grade and pathological characteristics of bladder cancer in the UK: British Association of Urological Surgeons (BAUS) urological tumour registry. BJU Int. 2014;113:924–930. [DOI] [PubMed] [Google Scholar]
- 5. Cambier S, Sylvester RJ, Collette L, et al. EORTC Nomograms and risk groups for predicting recurrence, progression, and disease-specific and overall survival in non-muscle-invasive stage Ta-T1 urothelial bladder cancer patients treated with 1-3 years of maintenance Bacillus Calmette-Guérin. Eur Urol. 2016;69:60–69. [DOI] [PubMed] [Google Scholar]
- 6. Vedder MM, Márquez M, de Bekker-Grob EW, et al. Risk prediction scores for recurrence and progression of non-muscle invasive bladder cancer: an international validation in primary tumours. PLoS ONE. 2014;9:e96849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fritsche HM, Burger M, Svatek RS, et al. Characteristics and outcomes of patients with clinical T1 grade 3 urothelial carcinoma treated with radical cystectomy: results from an international cohort. Eur Urol. 2010;57:300–309. [DOI] [PubMed] [Google Scholar]
- 8. Fernandez-Gomez J, Madero R, Solsona E, et al. The EORTC tables overestimate the risk of recurrence and progression in patients with non-muscle-invasive bladder cancer treated with bacillus Calmette-Guérin: external validation of the EORTC risk tables. Eur Urol. 2011;60:423–430. [DOI] [PubMed] [Google Scholar]
- 9. Kluth LA, Black PC, Bochner BH, et al. Prognostic and prediction tools in bladder cancer: a comprehensive review of the literature. Eur Urol. 2015;68:238–253. [DOI] [PubMed] [Google Scholar]
- 10. Iyer G, Al-Ahmadie H, Schultz N, et al. Prevalence and co-occurrence of actionable genomic alterations in high-grade bladder cancer. J Clin Oncol. 2013;31:3133–3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kim PH, Cha EK, Sfakianos JP, et al. Genomic predictors of survival in patients with high-grade urothelial carcinoma of the bladder. Eur Urol, 2015;67:198–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. West L, Vidwans SJ, Campbell NP, et al. A novel classification of lung cancer into molecular subtypes. PLoS ONE. 2012;7:e31906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sigalotti L, Covre A, Fratta E, et al. Whole genome methylation profiles as independent markers of survival in stage IIIC melanoma patients. J Transl Med. 2012;10:185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wolff EM, Chihara Y, Pan F, et al. Unique DNA methylation patterns distinguish noninvasive and invasive urothelial cancers and establish an epigenetic field defect in premalignant tissue. Cancer Res. 2010;70:8169–8178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Catto JW, Azzouzi AR, Rehman I, et al. Promoter hypermethylation is associated with tumor location, stage, and subsequent progression in transitional cell carcinoma. J Clin Oncol. 2005;23:2903–2910. [DOI] [PubMed] [Google Scholar]
- 16. Kitchen MO, Bryan RT, Haworth KE, et al. Methylation of HOXA9 and ISL1 predicts patient outcome in high-grade non-invasive bladder cancer. PLoS ONE. 2015;10:e0137003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Alvarez-Mugica M, Cebrian V, Fernández-Gómez JM, et al. Myopodin methylation is associated with clinical outcome in patients with T1G3 bladder cancer. J Urol. 2010;184:1507–1513. [DOI] [PubMed] [Google Scholar]
- 18. Alvarez-Mugica M, Fernández-Gómez JM, Cebrian V, et al. Polyamine-modulated factor-1 methylation predicts Bacillus Calmette-Guérin response in patients with high-grade non-muscle-invasive bladder carcinoma. Eur Urol. 2013;63:364–370. [DOI] [PubMed] [Google Scholar]
- 19. Agundez M, Grau L, Palou J, et al. Evaluation of the methylation status of tumour suppressor genes for predicting bacillus Calmette-Guérin response in patients with T1G3 high-risk bladder tumours. Eur Urol. 2011;60:131–140. [DOI] [PubMed] [Google Scholar]
- 20. Zeegers MP, Bryan RT, Langford C, et al. The West Midlands bladder cancer prognosis programme: rationale and design. BJU Int. 2010;105:784–788. [DOI] [PubMed] [Google Scholar]
- 21. Kitchen MO, Bryan RT, Emes RD, et al. Quantitative genome-wide methylation analysis of high-grade non-muscle invasive bladder cancer. Epigenetics. 2016;11:237–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Babjuk M, Burger M, Zigeuner R, et al. ; European Association of Urology. EAU guidelines on non-muscle-invasive urothelial carcinoma of the bladder: update 2013. Eur Urol. 2013;64:639–653. [DOI] [PubMed] [Google Scholar]
- 23. Blin N, Stafford DW. A general method for isolation of high molecular weight DNA from eukaryotes. Nucleic Acids Res. 1976;3:2303–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Glossop JR, Nixon NB, Emes RD, et al. Epigenome-wide profiling identifies significant differences in DNA methylation between matched-pairs of T- and B-lymphocytes from healthy individuals. Epigenetics. 2013;8:1188–1197. [DOI] [PubMed] [Google Scholar]
- 25. Wessely F, Emes RD. Identification of DNA methylation biomarkers from Infinium arrays. Front Genet. 2012;3:161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Dedeurwaerder S, Defrance M, Calonne E, Denis H, Sotiriou C, Fuks F. Evaluation of the Infinium Methylation 450K technology. Epigenomics. 2011;3:771–784. [DOI] [PubMed] [Google Scholar]
- 27. Meller S, Zipfel L, Gevensleben H, et al. CDO1 promoter methylation is associated with gene silencing and is a prognostic biomarker for biochemical recurrence-free survival in prostate cancer patients. Epigenetics. 2016;11:871–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Heyn H, Carmona FJ, Gomez A, et al. DNA methylation profiling in breast cancer discordant identical twins identifies DOK7 as novel epigenetic biomarker. Carcinogenesis. 2013;34:102–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Babjuk M, Böhle A, Burger M, et al. EAU guidelines on non-muscle-invasive urothelial carcinoma of the bladder: update 2016. Eur Urol. 2017;71:447–461. [DOI] [PubMed] [Google Scholar]
- 30. Li X, Zhou F, Jiang C, et al. Identification of a DNA methylome profile of esophageal squamous cell carcinoma and potential plasma epigenetic biomarkers for early diagnosis. PLoS ONE. 2014;9:e103162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gonzalo V, Lozano JJ, Alonso-Espinaco V, et al. ; Gastrointestinal Oncology Group of the Spanish Gastroenterological Association. Multiple sporadic colorectal cancers display a unique methylation phenotype. PLoS ONE. 2014;9:e91033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Martino D, Maksimovic J, Joo JH, Prescott SL, Saffery R. Genome-scale profiling reveals a subset of genes regulated by DNA methylation that program somatic T-cell phenotypes in humans. Genes Immun. 2012;13:388–398. [DOI] [PubMed] [Google Scholar]
- 33. Roperch JP, Grandchamp B, Desgrandchamps F, et al. Promoter hypermethylation of HS3ST2, SEPTIN9 and SLIT2 combined with FGFR3 mutations as a sensitive/specific urinary assay for diagnosis and surveillance in patients with low or high-risk non-muscle-invasive bladder cancer. BMC Cancer. 2016;16:704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lasseigne BN, Burwell TC, Patil MA, Absher DM, Brooks JD, Myers RM. DNA methylation profiling reveals novel diagnostic biomarkers in renal cell carcinoma. BMC Med. 2014;12:235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bediaga NG, Acha-Sagredo A, Guerra I, et al. DNA methylation epigenotypes in breast cancer molecular subtypes. Breast Cancer Res. 2010;12:R77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Liu P, Iden M, Fye S, et al. Targeted, deep sequencing reveals full methylation profiles of multiple HPV types and potential biomarkers for cervical cancer progression. Cancer Epidemiol Biomarkers Prev. 2017;26:642–650. [DOI] [PubMed] [Google Scholar]
- 37. Zhang C, Li J, Huang T, et al. Meta-analysis of DNA methylation biomarkers in hepatocellular carcinoma. Oncotarget. 2016;7:81255–81267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Teh MT, Gemenetzidis E, Patel D, et al. FOXM1 induces a global methylation signature that mimics the cancer epigenome in head and neck squamous cell carcinoma. PLoS ONE. 2012;7:e34329. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.