SUMMARY
Trypanosoma cruzi is the kinetoplastid protozoan parasite that causes human Chagas disease, a chronic disease with complex outcomes including severe cardiomyopathy and sudden death. In mammalian hosts, T. cruzi colonizes a wide range of tissues and cell types where it replicates within the host cell cytoplasm. Like all intracellular pathogens, T. cruzi amastigotes must interact with its immediate host cell environment in a manner that facilitates access to nutrients and promotes a suitable niche for replication and survival. Although potentially exploitable to devise strategies for pathogen control, fundamental knowledge of the host pathways co-opted by T. cruzi during infection is currently lacking. Here we report, that intracellular T. cruzi amastigotes establish close contact with host mitochondria via their single flagellum. Given the key bioenergetic and homeostatic roles of mitochondria, this striking finding suggests a functional role for host mitochondria in the infection process and points to the T. cruzi amastigote flagellum as an active participant in pathogenesis. Our study establishes the basis for future investigation of the molecular and functional consequences of this intriguing host-parasite interaction.
INTRODUCTION
The successful colonization of mammalian cells by intracellular pathogens involves the remodeling of host metabolic and immune defense pathways to establish a permissive niche for pathogen replication and survival. To facilitate access to key nutrients that would otherwise be difficult to obtain, vacuole-dwelling pathogens intersect host membrane trafficking pathways (Pizarro-cerdá, Charbit, Enninga, & Lafont, 2016) and/or recruit host organelles to the outer vacuole membrane for nutrient scavenging (Plattner & Soldati-Favre, 2008). Striking examples of host organelle recruitment have been documented in cells infected with Chlamydia spp or Toxoplasma gondii (for a review see (Romano & Coppens, 2013)), both of which attract host endoplasmic reticulum, mitochondria (Matsumoto, Bessho, Uehira, & Suda, 1991), Golgi membranes, endo-lysosomes and lipid droplets (Kumar, Cocchiaro, & Valdivia, 2006; Nolan, Romano, & Coppens, 2017) to their vacuoles.
Unlike these examples, the kinetoplastid protozoan parasite Trypanosoma cruzi escapes its lysosome-derived parasitophorous vacuole (Ley, Robbins, Nussenzweig, & Andrews, 1990) to replicate as 'amastigotes' in the host cytosol. Fundamental knowledge of intracellular T. cruzi amastigote biology and how this cytosolic pathogen interacts with host organelles and subcellular structures to facilitate their intracellular growth and survival is lacking. In an effort to characterize the immediate intracellular environment inhabited by cytosolic T. cruzi amastigotes, we embarked on a systematic image-based study that examined the spatial positioning of the intracellular parasite relative to major host subcellular structures. We report the unexpected finding that cytosolic T. cruzi amastigotes are embedded within the host cell mitochondrial network with the distal part of the parasite flagellum establishing contact with the host outer mitochondrial membrane (OMM). The engagement of host mitochondria by this cytosolic pathogen suggests a key functional role of host mitochondrial metabolism and homeostatic functions in the intracellular T. cruzi infection process and highlights an unanticipated role for the T. cruzi amastigote flagellum in mediating host-parasite interactions.
RESULTS
T. cruzi amastigotes are non-randomly distributed within mammalian host cells
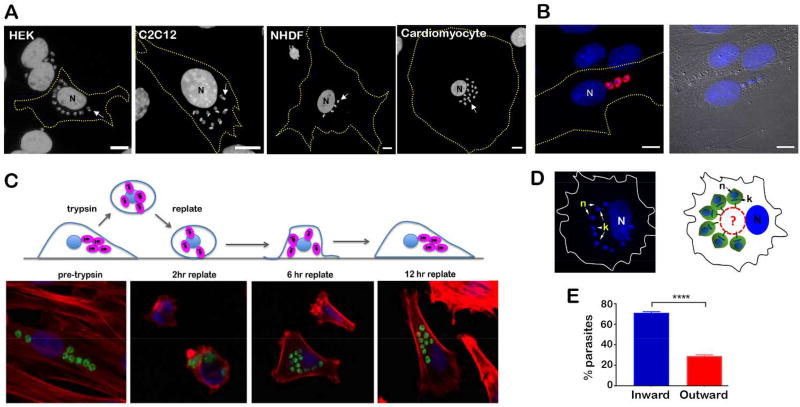
T. cruzi establishes intracellular infection in most nucleated mammalian cell types where, shortly after host cell entry by the trypomastigote form, the parasite develops into the 'amastigote' form that establishes cytosolic residence and replicates by binary fission. Microscopic examination of T. cruzi-infected mammalian cells reveals that the intracellular parasites are not randomly distributed throughout the host cell cytoplasm. In contrast, T. cruzi amastigotes, shown in several different mammalian cell types (Figure 1A), typically group together close to the host cell nucleus. For example, in ~95% of infected fibroblasts (NHDF) T. cruzi amastigotes are arranged in clusters and ~85% of these clusters are positioned next to the nucleus, as opposed to near the cell edges (data from 4 independent experiments; n=100 cells/experiment p<0.0001). The tendency of cytosolic amastigotes to group together near the host nucleus occurs prior to the first parasite cell division as exemplified in Figure 1B which shows a cell that was invaded by three separate parasites and imaged at 18 hours post-infection (18 hpi), a time point at which the parasites have escaped the vacuole but have not yet replicated (Caradonna, Engel, Jacobi, Lee, & Burleigh, 2013). Therefore, the clustering observed is not simply the result of parasite doublings, although the local production of non-motile daughter parasites likely contributes to the formation of amastigote clusters within the host cell cytoplasm.
Figure 1. T. cruzi intracellular amastigotes adopt a non-random spatial organization in mammalian cells.
A. Fluorescence microscopy images of T. cruzi-infected human embryonic kidney cells (HEK), mouse myoblasts (C2C12), neonatal human dermal fibroblasts (NHDF) and human cardiomyocytes fixed at 48 hpi and stained with DAPI to visualize host nuclei (N) and parasite DNA (eg. white arrow). Host cell boundaries are depicted by yellow dashed lines. Scale bar = 10 µm. B. Immunofluorescence microscopy performed on fixed T. cruzi-infected NHDF (18 hpi). Intracellular amastigotes were visualized using rabbit anti-T. cruzi serum (red) and DNA is visualized with DAPI (blue). N = host cell nucleus. Scale bar = 10 µm. C. Upper panel: Schematic representation of cell shape and appearance of T. cruzi-infected cells at different times pre- and post-trypsinization and replating onto a tissue culture dish (grey); intracellular T. cruzi amastigotes (pink), host nucleus and host cell membrane (blue). Lower panel: Representative fluorescence images of infected NHDF fixed before (pre-trypsin) or at different times post trypsinization and replating. CFSE-stained parasites (green), host actin filaments (red) and DNA (blue). D. Left panel: Fluorescence image of DAPI-stained T. cruzi-infected NHDF showing parasite organization observed in 15% of the cells at 4 hr post-replating. Right panel: Schematic representation of amastigote orientation toward a non-stained area next to the host cell nucleus (N). k = kinetoplast, n = parasite nucleus. Host cell boundaries are depicted in both panels. E. Bar graph representing the percentage of individual parasites with kinetoplast DNA oriented toward an unstained juxtanuclear space in cells 4 hr post-replating. Only cells harboring parasites arranged in semi-circular pattern around juxtanuclear structures were counted. n = 300 parasites in each of the 3 independent experiments (unpaired Student's t-test, p < 0.0001).
To facilitate study of the spatio-temporal dynamics of intracellular T. cruzi amastigotes, a brief trypsinization of T. cruzi-infected NHDF monolayers was performed followed by immediate re-plating of cells onto new glass coverslips to promote dramatic reorganization of host cytoskeletal elements and retraction of organelles toward the host cell nucleus. Upon replating, the infected cells reattached and slowly re-spread on the culture dish (Figure 1C). Imaging of infected cells at time points pre- and post-trypsin/replating revealed that the parasites tend to remain grouped throughout this process, with no evidence of amastigote dispersal in the host cytoplasm in any instance. In the example shown (Figure 1C, lower panel), the approximate linear arrangement of intracellular T. cruzi amastigotes observed prior to trypsinization of the infected monolayer (Figure 1C; pre-trypsin) is re-established in cells that have re-spread to adopt an elongated morphology typical of fibroblasts in culture (Figure 1C; 12 hr replate). Even at early stages of host cell spreading post-replating (~2–4 hours), where there is significant heterogeneity with respect to host cell shape, we observe instances where the intracellular amastigotes are arranged in a semi-circle around a juxtanuclear space (not stained with DAPI, Figure 1D) with their anterior end, identified by DAPI-staining of kinetoplast DNA, oriented inward toward that space (Figure 1D). Quantification of this event in multiple infected cells revealed that the majority of amastigotes that are arranged in a semi-circular pattern are anteriorly oriented with their kinetoplast facing the center of the cell (Figure 1E). Combined, these observations suggest that intracellular T. cruzi amastigotes are subject to spatial constraints in mammalian host cells that may be due to physical association with host subcellular structures.
T. cruzi amastigotes maintain close proximity to the host mitochondrial network
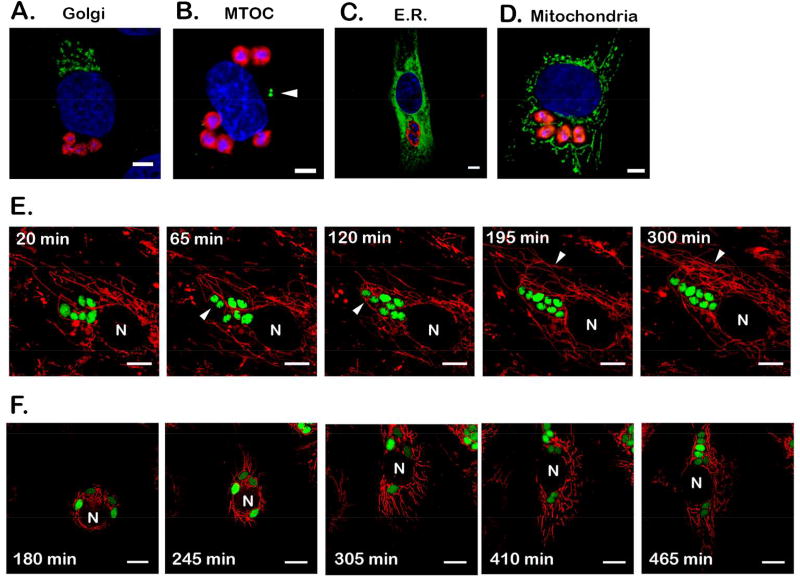
To explore the possibility that cytosolically-localized T. cruzi amastigotes might physically interact with host subcellular structures, imaging studies were undertaken to examine the intracellular positioning of amastigotes relative to several major host cell organelles and cytoskeletal structures that are enriched in the vicinity of the host cell nucleus (Figure 2A–D). Amastigotes did not consistently co-localize with the host Golgi apparatus (Figure 2A, Supplemental Fig. 1A) or the microtubule organizing center (MTOC, arrowhead; Figure 2B, Supplemental Fig. 1B). Cytosolic amastigotes were always found enveloped by the host endoplasmic reticulum (ER) (Figure 2C, Supplemental Fig. 1C) but without obvious enrichment of host ER around the intracellular T. cruzi amastigotes (Figure 2C) as seen with vacuolar pathogens that actively recruit host ER (Romano & Coppens, 2013). The ubiquitous nature of the ER makes it difficult to draw any conclusions regarding specificity of any interactions occurring between the parasites and this host organelle. Contrasting this pattern is the more limited, defined branching network of the host mitochondria, within which intracellular amastigotes were consistently found to be embedded (Figure 2D, Supplemental Figure 1D, E, F). The parasite-host mitochondria association is particularly obvious in instances where the mitochondria are unevenly distributed in a host cell (eg. Supplemental Figure 1G). In cells that are heavily infected (the example shown in Supplemental Fig. 1D), it is possible to see individual parasites within a larger cluster that are more distant from the host mitochondria, however, these represent a small fraction of the parasites in such groups (Supplemental Fig. 1D).
Figure 2. Intracellular T. cruzi amastigotes maintain close proximity with host mitochondria.
(A–D) Representative images of cytosolically-localized T. cruzi amastigotes in NHDF (48 hpi) and host subcellular structures visualized by immunofluorescence microscopy. Parasites are labeled with CFSE (A) or stained with anti-T. cruzi antibodies (red) (B–D). For consistency, parasite images were artificially colored in red and host structures in green. (A) Golgi apparatus (golgin-97 abs), (B) microtubule organizing center (MTOC, white arrowhead) (gamma-tubulin abs), (C) endoplasmic reticulum (sec61β-GFP) and (D) mitochondria (ATP5B abs). DNA is stained using DAPI (blue). Scale bar = 5 µm. The images showing Golgi apparatus (A) and ER (C) were acquired with a confocal microscope. E and F. Time-lapse imaging of unperturbed (E) or freshly replated (F) NHDF expressing mitochondrially-targeted mCherry (red) and infected with GFP-expressing T. cruzi (green) (36 hpi). N = host nucleus. Scale bar = 10 µm. In E, white arrowheads indicate dividing parasites (65 and 120min) and increase in mitochondrial density around parasites (195 and 300min). Time of recording is indicated at the top left corner of each frame with t = 0 arbitrarily set for the first frame of recording (~36 hpi). In F, recording was initiated 180 min post-replating, highlighting the dynamics of the host mitochondrial network in live cells (red) and the proximity between host mitochondria and intracellular amastigotes (green).
We then monitored the dynamics of the intracellular parasites relative to the host mitochondrial network by live fluorescence imaging of NHDF expressing mCherry in the mitochondrial matrix (mito-mCherry) infected with GFP-expressing T. cruzi parasites for 48 hpi. Representative time-lapse images of T. cruzi-infected cells (Figure 2E, Supplemental Movie 1 and 2) revealed a highly dynamic mitochondrial network in the fibroblasts as well as the close proximity that is maintained between the intracellular amastigotes and host mitochondria over the course of these imaging experiments. In infected host cells, amastigote clusters, and many of the individual parasites within these groups, were surrounded by the highly connected host mitochondrial network (eg. Supplemental Movie 1). These interactions appear to be highly dynamic. As the intracellular amastigotes undergo division (the average doubling time of this parasite strain is ~8 hr, where the first amastigote division does not occur until ~22 hpi), the parasite maintains close proximity to host mitochondria (eg. Figure 2E, white arrowheads 65 and 120min). Similar results were obtained when intracellular parasites were imaged in spreading cells following trypsin/re-plating (Figure 2F; Supplemental Movie 3 and 4). Although the host mitochondrial network undergoes remarkable reorganization in spreading cells, the close proximity between the amastigotes and host mitochondria is maintained throughout. Notably, mitochondria in the vicinity of dividing parasites appear to be highly branched (Figure 2F, 410 – 465min), a possible indication of increased mitochondrial fusion in this region of the host cell. Thus, our combined results suggest that the proximity observed between intracellular T. cruzi amastigotes and the host mitochondrial network may be of biologically relevant as opposed to incidental.
The interaction between the host mitochondria and T. cruzi amastigotes involves the parasite flagellum
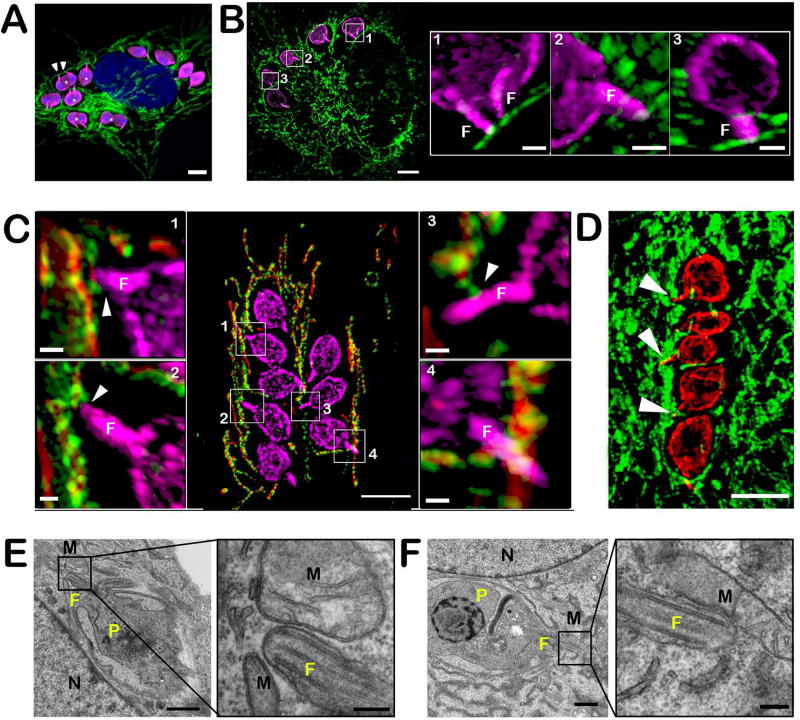
The semi-circular arrangement of T. cruzi amastigotes in host cells that are in the early stages of spreading following trypsinization/re-plating (~4 hr), where the parasite kinetoplast is oriented toward a juxtanuclear structure (Figure 1D,E), prompted us to ask whether the parasite flagellum, which protrudes from the basal body located at the anterior end of the parasite (Robinson & Gull, 1991), could mediate interactions with host subcellular structures such as the mitochondria. Using an antibody directed to the T. cruzi flagellar calcium binding protein (FCaBP) (Godsel, Tibbetts, Olson, Chaudoir, & Engman, 1995), which is concentrated in the parasite flagellum, but also found on the amastigote plasma membrane, we find that the amastigote flagellum is in very close proximity to host mitochondria and often appears to be in contact (Figure 3A, white arrowheads). Approximately 76% of intracellular amastigotes exhibit flagellar contact with host mitochondria (n=452 flagella, 3 independent experiments) and in agreement with live fluorescence imaging data, the interaction between the parasite flagellum and the host mitochondria is maintained during amastigote cell division (Figure 3A, white asterisks). Using the same markers for parasite and mitochondria, SR-SIM analysis of freshly replated cells (6 hr post re-plating) with 3D reconstruction confirmed that this interaction is maintained following cell detachment/reattachment with no evident space between the two membranes (within the limits of spatial resolution for SR-SIM: ≤100nm) (Figure 3B). Moreover, the interaction between the parasite flagellum and the host mitochondria was demonstrated by SR-SIM for two additional mitochondrial markers in infected NHDF-mito-mCherry (i.e mCherry and TOM20) (Figure 3C). Although, most of the images presented here involved human fibroblasts, we confirmed that the T. cruzi amastigote flagellum-host mitochondria interaction occurs in other cell types as well including HeLa, C2C12 (not shown) and human cardiomyocytes, where amastigote flagella are buried within the dense mitochondrial network that is characteristic of high energy-demand cell types such as myocytes (Figure 3D, white arrowheads). Finally, transmission electron microscopy of infected NHDF demonstrates extremely close contact (<10nm) between the parasite flagellum and host mitochondria (Figure 3E, inset right panel) with direct membrane contacts observed between the two structures (Figure 3F, inset right panel). In these images, the interaction appears to occur mainly at the distal part of the flagellum, an observation that correlates with the results of fluorescence microscopy (Figures 3A, B and C). Experiments performed with Y and CL Brener strains of T. cruzi also show that, similar to the Tulahuen strain of T. cruzi studied here, amastigote parasite strains also establish contact with host mitochondria via their flagella (data not shown). In summary, our results provide clear evidence of a novel interaction between cytosolic T. cruzi amastigotes and the mitochondrial network of mammalian host cells that is mediated by the parasite flagellum.
Figure 3. Close interaction between T. cruzi amastigotes and host mitochondria is mediated by the parasite flagellum.
A. Immunofluorescence detection of T. cruzi amastigotes using anti-FCaBP (pink) and host mitochondrial network with anti-ATP5B antibody (green) in infected NHDF (48 hpi). DNA is stained with DAPI. Scale bar = 5 µm. Example of parasite flagella (white arrowheads). B. Super-resolution (SR-SIM) 3D projection of intracellular T. cruzi amastigotes in NHDF 6 hour post-replating following trypsin treatment (42 hpi). Parasites are stained with anti-FCaBP (pink) and the mitochondria is stained with ATP5B antibodies (green). Scale bar = 5 µm. Insets; 3D SR-SIM reconstruction of the framed areas using the 3D projection mode showing the zone of contact between the parasite flagellum (F) and the host mitochondria. Scale bar = 1 µm. C. Super-resolution (SR-SIM) 3D projection showing infected mito-mCherry NHDF. Parasites are stained with anti-FCaBP (pink) and the host mitochondria expressing mCherry fluorescent protein (red) are stained with TOM20 antibodies (green). Scale bar = 5 µm. Insets; 3D SR-SIM reconstruction of the framed area using the 3D projection mode showing the zone of contact between the parasite flagellum (F) and the host mitochondria indicated by white arrowheads. Scale bar = 0.5 µm. D. Super-resolution (SR-SIM) 3D projection of an infected human cardiomyocyte (iPSC-derived). Mitochondria are visualized using an anti-ATP5B antibody (green) and parasites are stained with an anti-FCaBP antibody (red). Flagella in contact with the host mitochondria are indicated by white arrowheads. Scale bar = 5 µm. E and F. TEM of T. cruzi intracellular amastigotes in NHDF cells (flat embedding). P, parasite; N, host nucleus; F, flagellum; M, mitochondria. Scale bar = 1 µm. Insets show the proximity between the amastigotes flagellum and the host mitochondria at high magnification. Scale bar = 200nm.
DISCUSSION
This investigation of the spatial positioning and intracellular dynamics of the protozoan parasite Trypanosoma cruzi in mammalian host cells led to the discovery of a close interaction between the cytosolically-localized amastigote forms of the parasite and host mitochondria. Whether there are functional consequences of this particular interaction for T. cruzi or its mammalian host cell is yet to be determined, host mitochondria are frequently targeted by intracellular pathogens to modulate key cellular functions including mitochondrial energetics, apoptosis, and innate immune activation to facilitate successful intracellular infection (Fielden, Kang, Newton, & Stojanovski, 2017). Some bacterial pathogens (Horwitz, 1983; Matsumoto et al., 1991; Mehlitz et al., 2014) and the protozoan parasites, Toxoplasma gondii and Neospora caninum (Sinai et al. 1997; Nolan et al. 2015), can recruit host mitochondria to the membrane-bound vacuoles inhabited by these microbes. While the vacuole membrane is well-recognized as the functional interface between intracellular pathogens and their host cells (Nolan et al., 2015), our observation that the cytosolic parasite T. cruzi establishes close contact with host mitochondria using the distal portion of its single flagellum is a unique variation on this theme. In addition to their role in propelling the motile life stages of kinetoplastid protozoan parasites, studies conducted largely in the extracellular parasite, Trypansoma brucei, reveal that the flagellum plays a key sensory role in the parasite life cycle, integrating signals from the extracellular environment to modulate parasite behavior (Lopez, Saada, & Hill, 2015; Oberholzer et al., 2011). Consistent with this role, the flagellar membrane of motile trypanosomatids is enriched in transporters and signaling proteins (Oberholzer et al., 2011) such the adenylate cyclases in T. brucei (Lopez et al., 2015) and glucose transporters in Leishmania promastigotes (Rodriguez-Contreras et al., 2015) that are developmentally-regulated.
In contrast, the truncated flagellum of the non-motile intracellular amastigote stages of Leishmania and T. cruzi has not been extensively studied. However, striking images of the attachment of Leishmania mexicana amastigotes to the luminal face of the phagolysosomal vacuole (Gluenz et al. 2010) provided a first indication that this 'remnant' structure in the mammalian infective stages of kinetoplastid protozoan parasites is not inert. In this context, our finding that T. cruzi amastigotes exploit their flagellum to establish close contact with host mitochondria re-inforce this idea. While the T. cruzi amastigote flagellar proteome has not been defined, the expression of FCaBP, a putative calcium-sensor (Buchanan et al., 2005) and the potassium channel TcCat (Jimenez & Docampo, 2012) in the T. cruzi amastigote flagellar membrane is consistent with the potential for environmental sensing by intracellular amastigotes with a key role for the parasite flagellum in this process.
While the functional consequences of this interaction for T. cruzi or its host cell are currently unknown, the role of host mitochondria as signaling platform gives ample opportunity to T. cruzi to exploit the many functions of this organelle. We can speculate that by contacting host mitochondria, T. cruzi amastigotes may be able to sense K+ and Ca2+-flux status in the infected host cell as well as a number of host metabolites, including ATP, ROS, cytochrome c, as a strategy to integrate information regarding the health of the host cell to regulate the parasite growth and differentiation cycle in mammalian cells. Proximity to host mitochondria may facilitate scavenging of nutrients or metabolites by the parasite. For example, heme, which is an essential co-factor that T. cruzi is not capable of making (Kořený, Lukeš, & Oborník, 2010; Salzman et al., 1982) is synthesized in the mitochondria of the mammalian host cell. Future work will focus on the molecular and functional characterization of this unique parasite-host interaction with the potential to identify novel mitochondrial functions that are exploited by intracellular pathogens and to better understand sensory and signaling mechanisms in ciliate and flagellates.
EXPERIMENTAL PROCEDURES
Plasmids
pROCK-GFP-NEO (DaRocha et al., 2004) was obtained from W. DaRocha (Universidade Federal do Paraná, Brazil). pcDNA3.1-Sec61β-GFP (Voeltz, Prinz, Shibata, Rist, & Rapoport, 2006) and mCherry-α-tubulin (Addgene #49149) (Friedman, Webster, Mastronarde, Verhey, & Voeltz, 2010) were obtained from Gia Voeltz. pLNCX-COX8A-mCherry (Jacobi et al., 2016) was a kind gift from Chih-Hao Lee (HSPH).
Cell culture and transfection
Mouse skeletal myoblast (C2C12; ATCC CRL-1772), African green monkey kidney epithelial cells (LLcMK2; ATCC CCL-7), HEK293T (ATCC CRL-3216) and neonatal human dermal fibroblast (NHDF; FGM-2, Lonza) were maintained as subconfluent monolayers in Dulbecco’s modified Eagle’s medium (DMEM, HyClone) supplemented with 10% fetal bovine serum (FBS, Invitrogen), 1% glutamine, 1% penicillin-streptomycin (Gibco) (DMEM-10). Human iCell cardiomyocytes, generously provided by Cellular Dynamics Inc., were cultured according to manufacturer’s protocols. NHDF were transfected with pcDNA3.1-Sec61β-GFP (1µg) or mCherry-α-tubulin (500ng) plasmids using an Amaxa Nucleofector TMII (Lonza) as directed by the manufacturer, with the exception that transfected cells were immediately resuspended in pre-warmed RPMI-10% FBS prior seeding onto glass coverslips in 24-well plates at a density of 1.5×105 cells/well. RPMI was replaced with DMEM-10 the following day and the cells were used for infection 2 days post-transfection. NHDF lines stably expressing mitochondrially-targeted mCherry were generated by retroviral-mediated gene delivery with the exception that the transfection into Phoenix™ packaging cell line was done with 10µg of pLNCX-COX8A-mCherry plasmid using TransIT-LT1 reagent (Mirus Bio) according to manufacturer’s protocol. Selection started two days post-transduction with 400µg/mL G418 until resistant cells were observed. Stable expression of mCherry in the mitochondria was verified by immunofluorescence using ATP5B antibody.
Parasite infection, transfection and CFSE staining
Maintenance of mammalian infective stages of Trypanosoma cruzi Tulahuén strain (from M. Perrin, Tufts University) and parasite infection were performed as previously described (Li et al., 2016). Unless otherwise stated, infections were performed at an MOI to achieve an average of ~1 parasite/infected cell prior to replication, empirically determined for each cell type. CFSE (CellTrace CFSE Cell Proliferation Kit (Molecular Probes)) labeling of T. cruzi trypomastigotes was performed as described (Caradonna, Engel, Jacobi, Lee, & Burleigh, 2013). T. cruzi parasites expressing GFP were generated using the plasmid pROCK-GFP-NEO for stable integration into the tubulin locus of the epimastigote stage as described (DaRocha et al., 2004). Expression of GFP was confirmed by microscopy in both the trypomastigote and amastigote stages.
Trypsin / Re-plating procedure for T. cruzi-infected cells
T. cruzi-infected NHDF (~40 hpi) were washed twice with PBS (Gibco) and detached from the flask by briefly incubating the monolayer with 0.25% trypsin-EDTA solution (Gibco) at 37°C. Cells were resuspended in prewarmed (37°C) DMEM-10 and reseeded on glass-bottom imaging dishes (Ibidi GmbH) or onto 12mm2 glass coverslips (EMS) for the indicated times prior to performing live imaging or immunostaining as indicated.
Indirect immunofluorescence
T. cruzi-infected cells were fixed with 3.7% paraformaldehyde (EMS) in PBS for 20 min (for actin or mitochondrial staining) or with 2% paraformaldehyde and 0.2% glutaraldehyde for 20 min followed by a quenching step with 1mg/mL of sodium borohydride for 5 min (for ER visualization) prior to permeabilization using 0.3% (v/v) Triton X-100/PBS for 10 min. For MTOC and Golgi staining, cells were fixed in 100% cold methanol at −20°C for 8 min. Following a blocking step in 3% BSA/PBS for 1hr, cells were incubated with primary Abs diluted in 1% BSA/PBS for 1hr, washed, and then incubated with secondary antibody diluted in 1% BSA/PBS for 1 hr. The following primary antibodies were used: actin - Alexa598-phalloidin (1/1000; Invitrogen), MTOC: anti-γ-tubulin (1/200; Sigma), Golgi: anti-golgin-97 abs (1/500; Abcam), mitochondrial matrix: anti-ATP5B (1/1000; 3D5 Abcam) and OMM: anti-TOM20 (1/500; F-10 Santa Cruz),T. cruzi surface: rabbit anti-T. cruzi (1/5000) (Woolsey, 2003) or flagellum: anti-FCaBP (1/5000) (Godsel et al., 1995). For secondary detection, Alexa Fluor conjugated antibodies (Alexa Fluor 488, 594 or 647; ThermoFisher Scientific) were used. DNA staining was performed by incubating the samples 5 min in 5µg/mL 4,6-Diamidino-2-phenylindole/PBS (DAPI, Sigma-Aldrich) and samples were mounted using ProLong AntiFade reagent (ThermoFisher Scientific). Images were acquired using an inverted Axioimager Observer Z1 microscope equipped with an AxioCam MRm (Zeiss) and processed using Zen (Zeiss) or ImageJ (NIH) software. Confocal images and Super-Resolution Structured Illumination Microscopy (SR-SIM) images where acquired using a LSM880 confocal microscope (Zeiss) and an ELYRA super-resolution microscope (Zeiss) respectively at the Harvard Center for Biological Imaging platform (HCBI). 3D reconstruction and projections were performed using Zen Black software (Zeiss) at the HCBI. Some images were contrast enhanced for figure presentation.
Time-lapse imaging
T. cruzi-infected NHDF cultivated in 35-mm glass bottom μ-dishes (Ibidi GmbH) were imaged in phenol red-free DMEM-10 (Life Technologies) buffered with 25mM HEPES, pH 7.0 and in the presence of Prolong Live antifade reagent (1/100; Life Technologies). Time-lapse images were acquired using a DeltaVision PersonalDV microscope equipped with an automated stage enclosed in an environmental chamber warmed to 37°C and under 5% CO2 (Applied Precision, Inc.) at 60× (Plan APO NA 1.42). Focus was maintained using the Ultimate Focus System (Applied Precision, Inc.). Cells were illuminated with the emission filters FITC for GFP and TRITC for mCherry (2% laser power) (Spectral Applied Research) and recorded with a CoolSnap HQ2 camera (Photometric). Images were acquired every five minutes for a period of 6 to 12 hours and deconvolved using Softworx software (Applied Precision Inc.). Some images were contrast enhanced for figure presentation.
Quantitative analysis
Evaluation of parasite proximity to host subcellular structures was performed with images of infected cells fixed at 48 hpi (8–10 parasites/cell) imported into the ImageJ software (imagej.nih.gov/ij). The proximity of any parasite cluster to a host subcellular structure was determined by measuring the distance between the closest parasite in the cluster to the host organelle of interest and scored with a binary outcome of: 'proximal' (p; minimal distance < 5µm) or 'distant' (d; minimal distance > 5 µm). For host ER and mitochondria a minimal distance of 2 µm was used as the threshold to score as proximal (<2 µm) or distant (>2 µm). Parasite flagella were considered to be 'in contact' with host mitochondria when the distance between the two staining was 0 µm - subject to the resolution of the method (epifluorescence microscopy or SR-SIM).
Transmission Electron Microscopy
NHDF, grown on Aclar (Ted Pella Inc.) filmed plastic coverslips and infected with T. cruzi for 48 hours, were fixed with 1.25% formaldehyde, 2.5 % glutaraldehyde and 0.03% picric acid in 0.1 M sodium cacodylate buffer, pH 7.4 for 1 hour and then washed 3 times in 0.1M sodium cacodylate buffer (pH 7.4) prior to the post-fixation processing step of 1% osmium tetroxide/1.5% potassium ferrocyanide in distilled water 30 minutes on ice. Following 3 washes in distilled water, coverslips were incubated overnight with 1% aqueous uranyl acetate at 4°C in the dark. Samples were rinsed in water and dehydrated in a graded ethanol series using the progressive lowering of temperature method. After a final dip in fresh 100% ethanol then 100% propylene oxide, they were infiltrated with solutions 2:1, 1:2 of propylene oxide:epon araldite 30 minutes each, then 100% Epon araldite for one hour, then mounted for polymerization at 65°C for 48 hours. Ultrathin sections (about 60nm) were cut on a Reichert Ultracut-S microtome (Leica), picked up on to copper grids stained with lead citrate and examined in a TecnaiG2 Spirit BioTWIN (FEI Company) and images were recorded with an AMT 2k CCD camera (Advanced Microscopy Techniques).
Supplementary Material
Acknowledgments
We would like to thank D. Engman (Cedars-Sinai Medical Center, Los Angeles, CA) for the generous gift of FCaBP antibodies, T. Rapoport for providing the Sec61β-GFP plasmid, C-H Lee and N. Knudsen for providing the mCherry pLNCX2 plasmid and delivery system. We thank the HCBI for excellent training and support and acknowledge NIH-SIG award (1S10RR029237) used to acquire the ELYRA microscope. We thank the professionals at the HMS Electron Microscopy Facility, particularly to M. Ericsson and M. Coughlin for their expert help and advice as well as to S. Fortune and J. Kester (HSPH) for training and access to the DeltaVision PersonalDV microscope. Thank to the members of the Burleigh laboratory for their helpful discussions and suggestions. This work was funded by the National Institutes of Health grant R21 AI113121 awarded to B.A.B.
References
- Buchanan KT, Ames JB, Asfaw SH, Wingard JH, Olson CL, Campana PT, Engman DM. A Flagellum-specific calcium sensor. Journal of Biological Chemistry. 2005;280(48):40104–40111. doi: 10.1074/jbc.M505777200. http://doi.org/10.1074/jbc.M505777200. [DOI] [PubMed] [Google Scholar]
- Caradonna KL, Engel JC, Jacobi D, Lee CH, Burleigh BA. Host metabolism regulates intracellular growth of trypanosoma cruzi. Cell Host and Microbe. 2013 doi: 10.1016/j.chom.2012.11.011. http://doi.org/10.1016/j.chom.2012.11.011. [DOI] [PMC free article] [PubMed]
- DaRocha WD, Silva RA, Bartholomeu DC, Pires SF, Freitas JM, Macedo AM, Teixeira SMR. Expression of exogenous genes in Trypanosoma cruzi: Improving vectors and electroporation protocols. Parasitology Research. 2004;92(2):113–120. doi: 10.1007/s00436-003-1004-5. http://doi.org/10.1007/s00436-003-1004-5. [DOI] [PubMed] [Google Scholar]
- Fielden LF, Kang Y, Newton HJ, Stojanovski D. Targeting mitochondria: how intravacuolar bacterial pathogens manipulate mitochondria. Cell and Tissue Research. 2017;367(1):141–154. doi: 10.1007/s00441-016-2475-x. http://doi.org/10.1007/s00441-016-2475-x. [DOI] [PubMed] [Google Scholar]
- Friedman JR, Webster BM, Mastronarde DN, Verhey KJ, Voeltz GK. ER sliding dynamics and ER-mitochondrial contacts occur on acetylated microtubules. Journal of Cell Biology. 2010;190(3):363–375. doi: 10.1083/jcb.200911024. http://doi.org/10.1083/jcb.200911024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gluenz E, Höög JL, Smith AE, Dawe HR, Shaw MK, Gull K. Beyond 9+0: noncanonical axoneme structures characterize sensory cilia from protists to humans. The FASEB Journal. 2010;24(9):3117–3121. doi: 10.1096/fj.09-151381. http://doi.org/10.1096/fj.09-151381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godsel LM, Tibbetts RS, Olson CL, Chaudoir BM, Engman DM. Utility of recombinant flagellar calcium-binding protein for serodiagnosis of Trypanosoma cruzi infection. Utility of Recombinant Flagellar Calcium-Binding Protein for Serodiagnosis of Trypanosoma cruzi Infection. Journal of Clinical Microbiology. 1995;33(8):2082–2085. doi: 10.1128/jcm.33.8.2082-2085.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horwitz MA. Formation of a novel phagosome by the Legionnaires’ disease bacterium (Legionella pneumophila) in human monocytes. The Journal of Experimental Medicine. 1983 Oct;158 doi: 10.1084/jem.158.4.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobi D, Liu S, Burkewitz K, Kory N, Knudsen NH, Ryan K, Lee C. Hepatic Bmal1 regulates rhythmic mitochondrial dynamics and promotes metabolic fitness. 2016;22(4):709–720. doi: 10.1016/j.cmet.2015.08.006. http://doi.org/10.1016/j.cmet.2015.08.006.Hepatic. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez V, Docampo R. Molecular and Electrophysiological Characterization of a Novel Cation Channel of Trypanosoma cruzi. PLoS Pathog. 2012;8(6) doi: 10.1371/journal.ppat.1002750. http://doi.org/10.1371/journal.ppat.1002750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kořený L, Lukeš J, Oborník M. Evolution of the haem synthetic pathway in kinetoplastid flagellates: An essential pathway that is not essential after all? International Journal for Parasitology. 2010;40(2):149–156. doi: 10.1016/j.ijpara.2009.11.007. http://doi.org/10.1016/j.ijpara.2009.11.007. [DOI] [PubMed] [Google Scholar]
- Kumar Y, Cocchiaro J, Valdivia RH. The Obligate Intracellular Pathogen Chlamydia trachomatis Targets Host Lipid Droplets. Current Biology. 2006;16(16):1646–1651. doi: 10.1016/j.cub.2006.06.060. http://doi.org/10.1016/j.cub.2006.06.060. [DOI] [PubMed] [Google Scholar]
- Ley V, Robbins ES, Nussenzweig V, Andrews NW. The exit of Trypanosoma cruzi from the phagosome is inhibited by raising the pH of acidic compartment. J. Exp. MED. 1990 Feb;171:401–413. doi: 10.1084/jem.171.2.401. http://doi.org/10.1084/jem.171.2.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Shah-Simpson S, Okrah K, Belew AT, Choi J, Caradonna KL, Burleigh BA. Transcriptome Remodeling in Trypanosoma cruzi and Human Cells during Intracellular Infection. PLoS Pathogens. 2016;12(4):e1005511. doi: 10.1371/journal.ppat.1005511. http://doi.org/10.1371/journal.ppat.1005511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez MA, Saada EA, Hill KL. Insect stage-specific adenylate cyclases regulate social motility in African trypanosomes. Eukaryotic Cell. 2015;14(1):104–112. doi: 10.1128/EC.00217-14. http://doi.org/10.1128/EC.00217-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto A, Bessho H, Uehira K, Suda T. Morphological studies of the association of mitochondria with chlamydial inclusions and the fusion of chlamydial inclusions. Journal of Electron Microscopy. 1991;40(5):356–63. [PubMed] [Google Scholar]
- Mehlitz A, Karunakaran K, Herweg JA, Krohne G, van de Linde S, Rieck E, Rudel T. The chlamydial organism Simkania negevensis forms ER vacuole contact sites and inhibits ER-stress. Cellular Microbiology. 2014;16(8):1224–1243. doi: 10.1111/cmi.12278. http://doi.org/10.1111/cmi.12278. [DOI] [PubMed] [Google Scholar]
- Nolan SJ, Romano JD, Coppens I. Host lipid droplets : An important source of lipids salvaged by the intracellular parasite Toxoplasma gondii. 2017 doi: 10.1371/journal.ppat.1006362. http://doi.org/10.1371/journal.ppat.1006362. [DOI] [PMC free article] [PubMed]
- Nolan SJ, Romano JD, Luechtefeld T, Coppens I. Neospora caninum recruits host cell structures to its parasitophorous vacuole and salvages lipids from organelles. Eukaryotic Cell. 2015;14(5):454–473. doi: 10.1128/EC.00262-14. http://doi.org/10.1128/EC.00262-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberholzer M, Langousis G, Nguyen HT, Saada Ea, Shimogawa MM, Jonsson ZO, Hill KL. Independent analysis of the flagellum surface and matrix proteomes provides insight into flagellum signaling in mammalian-infectious Trypanosoma brucei. Molecular & Cellular Proteomics : MCP. 2011;10(10) doi: 10.1074/mcp.M111.010538. M111.010538. http://doi.org/10.1074/mcp.M111.010538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pizarro-cerdá J, Charbit A, Enninga J, Lafont F. Manipulation of host membranes by the bacterial pathogens Listeria, Francisella, Shigella and Yersinia. Seminars in Cell and Developmental Biology. 2016;60:155–167. doi: 10.1016/j.semcdb.2016.07.019. http://doi.org/10.1016/j.semcdb.2016.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plattner F, Soldati-favre D. Hijacking of host cellular functions by the Apicomplexa. Annual Review of Microbiology. 2008;62:471–87. doi: 10.1146/annurev.micro.62.081307.162802. http://doi.org/10.1146/annurev.micro.62.081307.162802. [DOI] [PubMed] [Google Scholar]
- Robinson DR, Gull K. Basal body movements as a mechanism for mitochondrial genome segregation in the trypanosome cell cycle. Nature. 1991 doi: 10.1038/352731a0. http://doi.org/10.1038/352731a0. [DOI] [PubMed]
- Rodriguez-Contreras D, Aslan H, Feng X, Tran K, Yates PA, Kamhawi S, Landfear SM. Regulation and biological function of a flagellar glucose transporter in Leishmania mexicana: a potential glucose sensor. The FASEB Journal. 2015;29(1):11–24. doi: 10.1096/fj.14-251991. http://doi.org/10.1096/fj.14-251991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano JD, Coppens I. Host Organelle Hijackers: A similar modus operandi for Toxoplasma gondii and Chlamydia trachomatis -Co-infection model as a tool to investigate pathogenesis. Pathogens and Disease. 2013;69(2) doi: 10.1111/2049-632X.12057. http://doi.org/10.1111/2049-632X.12057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzman TA, Stella AM, Wider de Xifra EA, Batlle AMDC, Docampo R, Stoppani AOM. Porphyrin biosynthesis in parasitic hemoflagellates: Functional and defective enzymes in Trypanosoma cruzi. Comparative Biochemistry and Physiology -- Part B: Biochemistry and. 1982;72(4):663–667. doi: 10.1016/0305-0491(82)90523-5. http://doi.org/10.1016/0305-0491(82)90523-5. [DOI] [PubMed] [Google Scholar]
- Sinai AP, Webster P, Joiner KA. Association of host cell endoplasmic reticulum and mitochondria with the Toxoplasma gondii parasitophorous vacuole membrane: a high affinity interaction. Journal of Cell Science. 1997;110(Pt 1):2117–28. doi: 10.1242/jcs.110.17.2117. [DOI] [PubMed] [Google Scholar]
- Voeltz GK, Prinz WA, Shibata Y, Rist JM, Rapoport TA. A class of membrane proteins shaping the tubular endoplasmic reticulum. Cell. 2006;124(3):573–586. doi: 10.1016/j.cell.2005.11.047. http://doi.org/10.1016/j.cell.2005.11.047. [DOI] [PubMed] [Google Scholar]
- Woolsey AM. Novel PI 3-kinase-dependent mechanisms of trypanosome invasion and vacuole maturation. Journal of Cell Science. 2003;116(17):3611–3622. doi: 10.1242/jcs.00666. http://doi.org/10.1242/jcs.00666. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.