Abstract
Rationale: The severity of cystic fibrosis (CF) lung disease varies widely, even for Phe508del homozygotes. Heritability studies show that more than 50% of the variability reflects non-cystic fibrosis transmembrane conductance regulator (CFTR) genetic variation; however, the full extent of the pertinent genetic variation is not known.
Objectives: We sought to identify novel CF disease-modifying mechanisms using an integrated approach based on analyzing “in vivo” CF airway epithelial gene expression complemented with genome-wide association study (GWAS) data.
Methods: Nasal mucosal RNA from 134 patients with CF was used for RNA sequencing. We tested for associations of transcriptomic (gene expression) data with a quantitative phenotype of CF lung disease severity. Pathway analysis of CF GWAS data (n = 5,659 patients) was performed to identify novel pathways and assess the concordance of genomic and transcriptomic data. Association of gene expression with previously identified CF GWAS risk alleles was also tested.
Measurements and Main Results: Significant evidence of heritable gene expression was identified. Gene expression pathways relevant to airway mucosal host defense were significantly associated with CF lung disease severity, including viral infection, inflammation/inflammatory signaling, lipid metabolism, apoptosis, ion transport, Phe508del CFTR processing, and innate immune responses, including HLA (human leukocyte antigen) genes. Ion transport and CFTR processing pathways, as well as HLA genes, were identified across differential gene expression and GWAS signals.
Conclusions: Transcriptomic analyses of CF airway epithelia, coupled to genomic (GWAS) analyses, highlight the role of heritable host defense variation in determining the pathophysiology of CF lung disease. The identification of these pathways provides opportunities to pursue targeted interventions to improve CF lung health.
Keywords: cystic fibrosis, transcriptome, genome-wide association study, epithelia, genome
At a Glance Commentary
Scientific Knowledge on the Subject
Although candidate gene modifiers of cystic fibrosis lung disease severity have been identified through genome-wide association studies, the full extent of the pertinent genetic variation is not known.
What This Study Adds to the Field
We demonstrate that cystic fibrosis lung disease severity is associated with increased airway epithelial expression of genes under genomic (heritable) influence in pathways involving airway mucosal host defense.
Cystic fibrosis (CF) (Online Mendelian Inheritance in Man catalogue number 219700) is an autosomal recessive disorder caused by mutations in the cystic fibrosis transmembrane conductance regulator (CFTR) gene. More than 1,800 mutations have been described in CFTR (1), with the most common mutation, Phe508del, accounting for approximately 66% of CFTR mutations worldwide. Patients with CF experience multiorgan system dysfunction, but lung disease, characterized by chronic (bacterial) infection and inflammation, remains the most common cause of morbidity and mortality, and preserving lung function is a key therapeutic priority. The severity of CF lung disease varies widely, even among Phe508del homozygotes. Twin/sibling studies have demonstrated that more than 50% of the variation in CF lung disease severity reflects non-CFTR genetic variation, with environmental factors also having a role (2–4). The recognition of this heritable variability has led to the search for genetic modifiers, with the hope of identifying genes and gene networks, or pathways, that are harmful or protective, thus providing targets for novel therapeutics.
Such efforts have culminated in a recently reported metaanalysis of genome-wide association studies (GWAS) comprising 6,365 individuals with CF from the International CF Gene Modifier Consortium. CF GWAS (5, 6) employed a standardized Consortium lung phenotype, termed the “Kulich Normal Residual Mortality Adjusted (KNoRMA)” lung disease phenotype, which is a quantitative phenotype that uses 3 years of FEV1 measures per subject, normalized to a CF reference population (7), and also adjusts for disease survival (8). The development of the KNoRMA phenotype allowed for harmonization of lung disease severity across international cohorts and led to identification of five loci associated with severity of CF lung disease (5). Complementary studies of gene expression in lymphoblastoid cell lines from 754 patients with CF, using KNoRMA as an outcome phenotype, identified additional genetic signatures based on gene expression pathways associated with severity of CF lung disease (9). The success of these studies provides an opportunity for mechanistic exploration. However, GWAS associations account for only a small percentage of expected genetic influence, and gene expression studies in lymphoblastoid cell lines do not optimally reflect airway epithelial biology.
To build upon previous success, we sought to identify novel non-CFTR genetic modifiers of lung disease severity by directly assessing gene expression in respiratory epithelia. We used RNA sequencing (RNA-seq) of nasal epithelial tissue, a well-recognized surrogate for lower airway epithelial function (10–12), from 134 patients with CF with existing GWAS data and the quantitative KNoRMA lung phenotype. We hypothesized that differential gene expression associated with CF lung disease severity would reveal novel candidate gene networks. We also analyzed GWAS data to (1) identify associations of single-nucleotide polymorphism (SNP) variation with nasal epithelial gene expression (i.e., expression quantitative trait loci [eQTLs]), (2) determine overlap between nasal epithelial gene expression– and GWAS-associated gene networks (pathways), and (3) explore the link between significant GWAS loci and nasal epithelial gene expression pathways. Some of the results of this study were previously reported in the form of abstracts (13–15).
Methods
Study Population, Sampling, and RNA-Seq Pipeline
Extended methods for each aspect of the study and analysis plan are provided in the online supplement (see Figure E1). Briefly, we conducted a multicenter study of nasal mucosal curettage biopsies obtained from 134 GWAS subjects with CF (5, 6) with two pancreatic insufficient CFTR mutations (n = 122 Phe508del homozygotes) and a broad spectrum of age and lung disease severity (Table 1). To quantify mucosal inflammation at sampling, nasal lavage obtained just prior to biopsy was analyzed for cytokine levels (IL-8, IFN-γ-inducible protein 10, and IL-1Ra), and the first curettage sample was stained for differential cell counts. From the next nine curettages, we collected cells for RNA isolation. RNA was sequenced using the Illumina HiSeq 2000 sequencing system by Expression Analysis (currently Q2 Solutions) following standard library preparation and achieving at least 25 million reads per sample. Fragments per kilobase of transcript per million mapped reads (FPKM) values were determined as described in the online supplement, and gene expression values were included in the data analysis if they met a minimum mean expression threshold level of at least 1 FPKM, based on the 95th percentile of mean Y-chromosome–specific gene expression observed in female samples.
Table 1.
Characteristics of Study Subjects by Research Site
| Research Site* | No. of Subjects | Consortium Lung Phenotype† (Mean ± SD) | Age at Consortium Lung Phenotype† (yr) (Median; Range) | Male Sex (%) | BMI (Mean ± SD) | Pseudomonas aeruginosa Infection‡ (%) | CFRD§ (%) | European|| (%) | Phe508del Homozygous (%) |
|---|---|---|---|---|---|---|---|---|---|
| CWRU | 38 | 0.9 ± 0.9 | 24.3; 11.4–49.2 | 46 | 21.8 ± 3.3 | 90 | 48 | 98 | 79 |
| JHU | 17 | 1.2 ± 0.8 | 27.1; 18.3–47.3 | 49 | 22.2 ± 3.7 | 92 | 30 | 100 | 77 |
| TOR | 35 | 0.8 ± 0.6 | 23.1; 10.4–42.6 | 63 | 22.0 ± 3.8 | 78 | 23 | 97 | 100 |
| UNC | 44 | 0.7 ± 0.8 | 23.8; 11.6–49.5 | 53 | 21.9 ± 3.8 | 91 | 35 | 100 | 100 |
| Total | 134 | 0.8 ± 0.8 | 26.5; 10.4–49.5 | 52 | 21.8 ± 3.3 | 86 | 35 | 99 | 91 |
Definition of abbreviations: BMI = body mass index; CFRD = cystic fibrosis–related diabetes; CWRU = Case Western Reserve University; JHU = Johns Hopkins University; KNoRMA = Kulich Normal Residual Mortality Adjusted; TOR = University of Toronto; UNC = University of North Carolina.
See Methods section of online supplement for participating sites and enrollment information.
Subjects were defined by the quantitative Consortium lung phenotype (KNoRMA) value (8).
Positive lower respiratory culture within 2 years preceding study enrollment; percentage noted is based on data available for 94 subjects.
CFRD percentage noted is based on data available for 117 subjects.
Based on self-identified ancestry and principal component analysis via SNP genotypes.
Analyses
KNoRMA (Consortium lung phenotype), a standardized quantitative phenotype that uses 3 years of measures of FEV1, was used as the lung phenotype to quantitate lung disease severity, as previously described (5, 8, 9). Linear models of gene expression as response variables, with clinically relevant covariates (sex, two genotype principal components [PCs], nine expression PCs, transplant history, nasal steroid use, azithromycin use, CD45 expression, and D statistic [mean pair-wise FPKM r2 per sample]), were used to determine associations of differential gene expression with KNoRMA, as well as with risk alleles at the five previously identified significant GWAS loci (Table E1) (5). These studies were complemented with a surrogate variable approach (16) (Table E1). To identify eQTLs, we used SNPs with a minor allele frequency greater than 0.05 and gene expression data (FPKM ≥1) as inputs in the Matrix eQTL package (17), which establishes eQTL associations under false discovery rate (FDR) control. To identify pathways significantly associated with differential gene expression, we used Significance Analysis of Function and Expression (SAFE) (18) coupled to pathway annotation sources selected for coverage, accuracy, and relevance (see online supplement). SAFE uses a resampling-based method, testing gene expression association with phenotype through random permutation of phenotype, and performs multiple test correction over the number of pathways tested in each analysis. To test the heritability of genes in significant pathways, we tested the likelihood of genes enriched for significant pathways versus their estimated heritability score determined in an independent blood gene expression report (19). To identify pathways significantly associated with GWAS data, a gene- and pathway-testing approach (GeneSetScan version 0.021) was applied to GWAS data from previously genotyped individuals with CF (n = 5,659, including 134 individuals in the present study) (5). GeneSetScan provides resampling-based multiple comparison–corrected P values for the number of pathways tested. In all analyses, pathways were reported if the corrected P value was less than 0.15, an established threshold for hypothesis generation in the context of these studies (20).
Results
Study Subjects and Evaluation of Inflammation in Nasal Mucosal Samples
Patients with CF tested in this study had a broad range of ages and lung disease severity (KNoRMA), and most had chronic lung infection with Pseudomonas aeruginosa (Table 1). Nasal curettage samples had a median of 87% epithelial cells (interquartile range, 77 to 94%) and a median of 12% neutrophils (interquartile range, 5 to 24%). To address the potential that subjects with more severe lung disease (low KNoRMA) might have more inflammation in the nasal mucosa and thus might confound the analyses, we tested for correlation of KNoRMA with degree of inflammation at the time of sample collection. We observed no significant correlation between KNoRMA and degree of inflammation in the nasal passages, as indexed by quantitative nasal mucosal examination scores, prebiopsy nasal lavage cytokine concentrations (IL-8, IFN-γ-inducible protein 10, and IL-1Ra), neutrophil counts derived from Diff-Quik stains, and CD45 expression (an indicator of inflammatory cells) in nasal mucosal RNA (Figures E2 and E3). Because there was strong correlation between CD45 expression and other measures of inflammation (cytokines, neutrophil counts) (Figure E3), CD45 expression was deemed a pertinent covariate in the analysis to adjust for overall inflammatory state.
Features of Gene Expression
Using the FPKM greater than or equal to 1 threshold for gene expression, 14,548 (52%) of 27,939 annotated genes were called as expressed and used in analyses. eQTLs with significant expression (FDR <0.15) were abundant (n = 14,098), with a preponderance of significant eQTLs within 1 Mb (cis) of the target gene (Table E2).
Relating Lung Disease Severity (KNoRMA) to Gene Expression
Linear models with covariates (see Methods section above and Table E1) were used to identify associations between gene expression level and KNoRMA. No individual gene met the level of statistical significance for association (Table E3). To detect coordinated networks of genes with pathophysiological relevance, we pursued rigorous pathway analysis to identify gene signatures. The analysis, using SAFE, identified pathways associated with lung disease severity with FDR less than 0.15, including viral infection, inflammatory signaling, lipid metabolism, macrophage function, and innate immunity (including HLA [human leukocyte antigen] genes) (Tables 2 and E4). Genes within pathways that contributed most robustly to the pathway significance (gene level P < 0.10) are provided in Table 2. (For a full listing of genes, see Table E5, tabs A and B.)
Table 2.
Gene Expression Pathways Significantly Associated with Consortium Lung Phenotype (KNoRMA)
| Pathways with FDR <0.15 |
Genes |
Statistics |
|||||
|---|---|---|---|---|---|---|---|
| Identifier | Name | No. of Genes | P Value* | Q Value† | Increased Expression‡ | Genes in the Pathway that Significantly Contribute to Pathway Signal (Gene-Level P < 0.10, Ordered by P Value)§ | |
| KEGG pathways, n = 329 tested |
|
||||||
| 05160 | Hepatitis C virus | 104 |
0.0004 | 0.0476 | Detrimental | CDKN1A, SCARB1, ARAF, STAT1, BRAF, NRAS, PIAS1, IRF9, TICAM1, NFKB1, CLDN3, PIK3R5, TLR3, TP53, MAPK9, OAS3, MAVS | |
| 05168 | Herpes simplex virus infection | 158 |
0.0005 | 0.0476 | Detrimental | TLR2, PML, JUN, HLA-DRB1, HLA-DMB, STAT1, CD74, HLA-A, TAF4B, HLA-G, IRF9, TICAM1, HLA-F, NFKB1, FOS, EP300, TLR3, HLA-DRA, TP53, HLA-DMA, CCL5, TAP1, MAPK9, OAS3, HLA-B, HLA-E, MAVS, HCFC2, C3 | |
| 04640 | Hematopoietic cell lineage | 56 |
0.0016 | 0.0960 | Detrimental | IL1R1, HLA-DRB1, ITGAM, CD7, CSF1, HLA-DRA, TFRC | |
| 04115 | p53 signaling pathway | 64 |
0.0017 | 0.0960 | Detrimental | CDKN1A, CCNG2, BID, GADD45A, TP73, SERPINB5, GADD45B, BBC3, TP53, TNFRSF10B, EI24 | |
| 00592 | α-Linolenic acid metabolism | 13 |
0.0007 | 0.0981 | Protective | PLA2G4F, PLA2G6 | |
| 00591‖ | |||||||
| 05322 | Systemic lupus erythematosus | 62 |
0.0022 | 0.1042 | Detrimental | HLA-DRB1, HLA-DMB, HIST1H2BG, C2, HLA-DRA, HLA-DMA, HIST1H2AE, HIST2H2BE, HIST4H4, HIST1H4H, TROVE2, C3 | |
| 04514 | Cell adhesion molecules (CAMs) | 86 |
0.0026 | 0.1063 | Detrimental | PTPRC, HLA-DRB1, HLA-DMB, ITGAM, HLA-A, CD276, HLA-G, HLA-F, CLDN3, ICAM1, HLA-DRA, HLA-DMA, ITGB8, HLA-B, HLA-E, ITGB2 | |
| 04930 | Type 2 diabetes mellitus | 28 |
0.0040 | 0.1356 | Detrimental | PIK3R5, PRKCD, MAPK9 | |
| 05219 | Bladder cancer | 36 |
0.0045 | 0.1356 | Detrimental | ARAF, BRAF, NRAS, DAPK1, TP53 | |
| 05161 | Hepatitis B virus | 120 |
0.0047 | 0.1356 | Detrimental | TLR2, CDKN1A, CREB3L2, JUN, STAT1, NRAS, TICAM1, NFKB1, FOS, EP300, PIK3R5, TLR3, TP53, MAPK9, CCNA2, SMAD4, MAVS | |
| 05323 | Rheumatoid arthritis | 69 |
0.0055 | 0.1445 | Detrimental | JUN, HLA-DRB1, HLA-DMB, CCL3L1, ATP6V0A4, FOS, CSF1, ICAM1, FLT1, CCL3, HLA-DRA, HLA-DMA, CCL5, TNFSF13B, ITGB2 | |
| 04620 | Toll-like receptor signaling pathway | 80 |
0.0068 | 0.1485 | Detrimental | TLR2, JUN, STAT1, CCL3L1, TICAM1, NFKB1, FOS, PIK3R5, TLR3, CCL3, CCL5, MAPK9, CCL4 | |
| 00061 | Fatty acid biosynthesis | 10 |
0.0072 | 0.1485 | Detrimental | ACSL1, ACACA, ACACB | |
| 05164 | Influenza A virus | 145 |
0.0074 | 0.1485 | Detrimental | PML, JUN, HLA-DRB1, HLA-DMB, STAT1, IRF9, TICAM1, NFKB1, EP300, PIK3R5, ICAM1, TLR3, HLA-DRA, HLA-DMA, SLC25A6, CCL5, MAPK9, TNFRSF10B, OAS3, MAVS | |
| M00034 | Methionine salvage pathway | 10 |
0.0083 | 0.1485 | Detrimental | AMD1, MTAP | |
| 05203 | Viral carcinogenesis | 167 |
0.0086 | 0.1485 | Detrimental | JUN, PRKACB, HLA-A, CDKN2B, NRAS, HDAC9, HLA-G, HIST1H2BG, IRF9, HLA-F, NFKB1, EP300, PIK3R5, TP53, HIST2H2BE, CCNA2, HIST4H4, HIST1H4H, HLA-B, HLA-E, C3 | |
| M00676 | PI3K-Akt signaling | 13 |
0.0086 | 0.1485 | Detrimental | PIK3R5, FOXO3 | |
| | |||||||
| GO biological process pathways, n = 4,228 tested |
|
||||||
| 0051591 | Response to cAMP | 57 |
0.0002 | 0.1261 | Detrimental | JUN, IGFBP5, STAT1, EGR1, SREBF1, APEX1, BRAF, JUNB, FOS, DUSP1, AKAP6, COL1A1, SPARC, FOSL2, AKAP7 | |
| 0014074‖ | |||||||
| 0046683‖ | |||||||
| 0070665 | Positive regulation of leukocyte proliferation | 79 |
0.0003 | 0.1261 | Detrimental | IGFBP2, PTPRC, HHLA2, CDKN1A, HLA-DMB, CD74, HLA-A, CD276, BST1, TICAM1, CSF1, CCL5, HLA-E, TNFSF13B | |
| 0032946‖ | |||||||
| 0050671‖ | |||||||
| 0051155 | Positive regulation of striated muscle cell differentiation | 24 |
0.0003 | 0.1261 | Detrimental | EDN1, FOXP1, CD53, AKAP6 | |
| 0033631 | Cell–cell adhesion mediated by integrin | 11 |
0.0004 | 0.1493 | Detrimental | FERMT3, CCL5 | |
| | |||||||
| GO molecular function pathways, n = 779 tested |
|
||||||
| 0050431 | Transforming growth factor-β binding | 15 |
0.0003 | 0.1048 | Bidirectional | LTBP1¶, VASN¶, CD109¶, HYAL2, ENG, CD36, LTBP4 | |
| | |||||||
| MetaMiner cystic fibrosis–specific pathways (GeneGo)**, n = 36 tested |
|
||||||
| Cholesterol and sphingolipid transport/distribution to the intracellular membrane compartments (normal and CF) | 11 |
0.0058 | 0.0909 | Bidirectional | STARD4¶, NPC1¶, NPC2¶, RAB7A¶ | ||
| | |||||||
| CF-relevant custom pathways††, n = 74 tested |
|
||||||
| EHF transcription factor–negative correlation; PMID 25414352 | 18 |
0.0007 | 0.0237 | Detrimental | ACSL1, C10orf10, DMKN, ID2, H1F0 | ||
| Asthma-COPD (down); PMID 25611785 | 26 |
0.0023 | 0.0504 | Detrimental | CCDC81, PTGFR, FOLR1, STEAP2, DAPK1, LTF, CYP4X1 | ||
| Macrophage specific: M1 (classic) activation markers; PMID 25204199; and Macrophage activation: combined M1 and M2 markers‖; PMID 19635926 | 52 |
0.0038 | 0.0632 | Detrimental | TLR2, GBP3, IL1R1, GBP2, IL8, ICAM1, CCL3, CCL5, IL32, C3AR1, GBP5, CCL4, APOL3 | ||
| Hypoxia responses: HIF1 target hypoxia (up); PMID 19491311 | 188 |
0.0099 | 0.1219 | Detrimental | STARD4, KLHL24, IGFBP2, EDN1, NDRG1, CXCR4, BRAF, BCL2L11, GAPDH, PTGS2, FNDC3B, PSD3, ARL5B, GADD45B, FOXO3, ATF3, C1orf51, PLOD2 | ||
| Hypoxia responses: DC hypoxia (up); PMID 21148811 | 85 |
0.0117 | 0.1219 | Detrimental | TLR2, CHST15, LOC374443, PPIF, CD53, SYNJ2, GBP2, LGALS8, LCP2, CD109, CDCP1, SLC29A1, INSIG1, FCAR, ERRFI1 | ||
| Hypoxia responses: MCF7 hypoxia (up); PMID 16565084 | 163 |
0.0174 | 0.1355 | Detrimental | PLIN2, KLHL24, DSC2, SCARB1, JUN, HLA-DRB1, NDRG1, SOX9, CXCR4, IGFBP5, CCNG2, EGR1, ADM, DDR1, PLAUR, FLNB, FOS, CAV1, GADD45B, GJA1, ATF3, DUSP1, KLF7, ATXN1, EMR2 | ||
| Nasal scrape CF (down); PMID 16614352 | 29 |
0.0183 | 0.1355 | Detrimental | EPSTI1, CD74, PRKACB, HLA-G, HLA-F, RPS2 | ||
| |
|||||||
| CFTR interactome pathways (none), n = 11 tested |
|
||||||
| | |||||||
| HLA-specific pathways, n = 2 tested |
|||||||
| Class I and class II | 30 | 0.0853 | 0.0747 | Bidirectional | HLA-DRB1¶, HLA-DMB¶, HLA-H¶, HLA-A¶, HLA-G¶, HLA-F¶, HLA-DRA¶, HLA-DMA¶, TAP1¶, HLA-B¶, HLA-E¶, PSMB8¶ | ||
| Class I and class II | 30 | 0.0093 | 0.0080 | Detrimental | HLA-DRB1, HLA-DMB, HLA-H, HLA-A, HLA-G, HLA-F, HLA-DRA, HLA-DMA, TAP1, HLA-B, HLA-E, PSMB8 | ||
| Class II | 16 | 0.0968 | 0.0577 | Detrimental | HLA-DRB1, HLA-DMB, HLA-DRA, HLA-DMA | ||
Definition of abbreviations: CF = cystic fibrosis; CFTR = cystic fibrosis transmembrane conductance regulator; COPD = chronic obstructive pulmonary disease; DC = dendritic cell; EHF = ETS homologous factor; FDR = false discovery rate; GO = Gene Ontology Consortium; HIF1 = hypoxia-inducible factor 1; HLA = human leukocyte antigen; KEGG = Kyoto Encyclopedia of Genes and Genomes database; KNoRMA = Kulich Normal Residual Mortality Adjusted; PI3K = phosphoinositide 3-kinase; PMID = PubMed reference number.
Pathways limited to those with at least 10 but less than or equal to 200 genes.
SAFE (Significance Analysis of Function and Expression) analysis used 10,000 permutations to establish significance thresholds (18).
Benjamini-Hochberg FDR for pathway testing within each pathway set; Q values less than 0.15 were included.
Increased expression of genes in pathway are detrimental (associated with worse lung disease) or protective (associated with milder lung disease) or bidirectional (associated with either worse or milder lung disease).
See Table E5, tab A, for an inclusive list of genes for these pathways; see Table E3 for gene Online Mendelian Inheritance in Man catalogue numbers.
These pathways are statistically significant and carry robust overlap of genes with first-listed pathway; see Table E5, tab A, for an inclusive list of genes in pathways.
For bidirectional pathways, genes with increased expression associated with worse disease are noted.
MetaMiner CF-specific pathways represent a version of the Thomson Reuters (formerly GeneGo) MetaDiscovery suite that is enriched with content specific for cystic fibrosis.
CF-relevant custom pathways were developed (46) using human gene counterparts (Table E8).
Because multiple methods have been proposed to correct for uncontrolled technical and population stratification, we also performed a secondary analysis using two surrogate variables (16) in lieu of nine expression PCs (Table E1) to obtain gene-level data. Analyses of these gene-level data with SAFE methodology yielded pathways associated with KNoRMA (Table E6; Table E5, tab C), including pathways related to viral infection, inflammatory signaling, lipid metabolism, and innate immunity (including HLA genes), concordant with our primary findings. Restricting the study cohort to 122 Phe508del homozygous patients also supported the primary findings (Table E5, tabs D and E). Increased gene expression was associated with worse lung disease for a majority of the pathways (labeled “detrimental” in Table 2; Table E5, tab A), and two examples of this relationship are provided in genes (HLA-DRB1 and TLR2) that significantly contributed to pathway results (Figure E4).
Heritable Features of Nasal Epithelial Gene Expression
Many of the top-ranked pathways were related to infectious/environmental exposures, but these pathways also had genes with significant eQTLs, which suggested a heritable component. To test if the significant pathways showed evidence of underlying heritability, we performed logistic regression of gene membership in enriched pathways for lung disease phenotype versus estimated heritability (see Methods section in the online supplement). Using heritability estimates (or proportion of gene expression controlled by genetic variances) of blood gene expression from a previous twin-based study of individuals without CF (19), we demonstrated that genes in the enriched pathways with FDR less than 0.15 (Table 2) showed significantly greater evidence of being heritable than the complementary set of genes not represented in the pathways (P = 2.6 × 10−6). We conclude that lung disease severity is associated with gene expression pathways that reflect, in part, underlying heritable traits.
Repeatability of Sample Measures
We acknowledge that nasal gene expression is prone to dynamic changes related to environmental influences. To provide additional insights related to this issue, we obtained nasal mucosal biopsies in a random subset of the study cohort (n = 39) at a second study visit and obtained RNA-seq data. We tested sample–sample correlations across all genes in the 39 paired samples (mean r = 0.958), relative to a background distribution derived from all 8,911 unique pairwise combinations from the 134 unique samples (mean r = 0.924). We demonstrated (using t statistic and permutation testing to account for dependence) that the paired samples had significantly higher correlation than the unpaired samples (P < 0.0001) (Figure E5), confirming robust intrasubject correlation of nasal epithelial gene expression.
Relating Lung Disease Severity (KNoRMA) to GWAS Pathways
Gene analysis and pathway analysis (GeneSetScan version 0.021) (21) of GWAS data from the previously genotyped cohort (5) had not been performed, and we used this method to identify pathways arising from the GWAS associations with KNoRMA (Table 3; Table E5, tab F). Pathways identified in this analysis were related to airway mucosal host defense, including viral response, inflammation, mucin/goblet cell biology, and cilia function. Interestingly, several pathways with diverse functional annotations (goblet-cell–relevant pathways, cytokine production by Th17 cells, vasodilation, and CFTR interactome [22] pathways) contained CFTR itself.
Table 3.
Genome-Wide Association Study Data Pathways Significantly Associated with Consortium Lung Phenotype (KNoRMA)
| Pathway |
Genes (n) | Corrected P Value* | Genes with Gene-Level P Value <0.10 (Ordered by P Value) | |
|---|---|---|---|---|
| Identifier | Name | |||
| Analyses included all available pathways† |
||||
| KEGG pathways, n = 338 tested |
||||
| 00510 | N-glycan biosynthesis | 48 | 0.019 | ALG12, MAN1C1, MGAT5B, MGAT4C, MGAT4A, TUSC3, ALG14, MAN1A1, MAN2A1, GANAB, DPM3, ALG6 |
| 05168 | Herpes simplex virus infection | 173 | 0.030 | HLA-DQA1, HLA-DQB1, HLA-DRB1, PVRL2, PVRL1, PER2, CCL2, TLR2, SRSF2, TYK2, CCL5, POLR2A, IFNA6, TP53, C3, IFNA13, IFNA1, EIF2AK2, LTA, TNF, IFNA2, IFNA5, MCRS1, TBPL1, IFNA14, TLR3, IFNA8, TAF5, HLA-B, IFNA17, PPP1CC, HLA-DOB, TAP1, TAP2, MAPK9, HCFC2, ALYREF, TBPL2 |
| 00601 | Glycosphingolipid biosynthesis lacto and neolacto series | 24 | 0.030 | ST3GAL6, B3GALT5, FUT3, FUT5, FUT6, FUT2, B3GALT2, GCNT2, ST3GAL3, FUT4 |
| 05310 | Asthma | 23 | 0.102 | HLA-DQA1, HLA-DQB1, HLA-DRB1, FCER1A, IL13, IL4, TNF, CCL11, HLA-DOB, PRG2 |
| 04650 | Natural killer cell–mediated cytotoxicity | 123 | 0.120 | FCGR3B, ICAM1, PRKCB, KRAS, VAV2, VAV3, IFNA6, VAV1, PIK3R2, TNFSF10, IFNA13, IFNA1, NCR3, TNF, RAC2, IFNA2, IFNA5, HCST, TYROBP, PRF1, IFNA14, LCP2, IFNA8, MAPK3, HLA-B, IFNA17, PIK3R3, ULBP3, FCGR3A, RAET1L, RAF1 |
| | ||||
| GO cellular component pathways, n = 516 tested |
||||
| 0044448 | Cell cortex part | 114 | 0.057 | EXOC3, CAPZA2, GYS2, TCHP, CAPZB, PCLO, EXOC4, CORO1A, MYH2, SPTAN1, EXOC7, TRPV4, SPTBN4, EXOC3L2, SPTBN2, SPTA1, CDH1, LLGL1, ANK1, GYPC, PRKCZ, CALD1 |
| 0009898 | Cytoplasmic side of plasma membrane | 152 | 0.114 | FRK, TNK2, GNA12, ACP1, KRAS, TYK2, LDLRAP1, PTK6, LYN, GNAO1, NPHS2, GNG5, GNG7, RASA1, GNA14, CABP1, HTRA2, TEC, SRMS, SPTA1, PTPN7, CDH1, ALOX15, GNAI3 |
| 0098562‡ | ||||
| GO biological pathways, n = 4,670 tested |
||||
| 0032770 | Positive regulation of monooxygenase activity | 25 | 0.024 | AGTR2, APOE, KRAS, TNF, CALM1, POR, TERF2 |
| 0051000‡ | ||||
| 2000027 | Regulation of organ morphogenesis | 165 | 0.029 | AGTR2, MET, POU5F1, FOXP2, HNF1B, CNTF, SFRP2, SIX4, SMAD4, SNAI2, SOX17, MSX1, IFT88, MMP20, HGF, DMRT3, CTHRC1, SFRP1, FGFR2, CAV3, XBP1, SIX1, EDNRA, GPC3, TNF, WNT9B, ZNRF3, CDH1, EDN1, FGF1, POR, TBX5 |
| 0042311 | Vasodilation | 67 | 0.058 | AGTR2, APOE, MRVI1, NPR1, SMTNL1, ADCYAP1, CFTR, NPPB, UTS2B, ADORA1, MKKS, P2RY2, HMOX1, BDKRB2, NOS1 |
| 0035150‡ | ||||
| 0050880‡ | ||||
| 0003018‡ | ||||
| 0001711 | Endodermal cell fate commitment | 16 | 0.098 | POU5F1, HNF1B, SOX17, CDC73, EOMES |
| 0042659‡ | ||||
| 0031960 | Response to corticosteroid | 140 | 0.139 | AGTR2, TRH, S100B, KRAS, AQP1, CCL2, ALPL, ADCYAP1, GHRHR, SCGB1A1, STAR, BMP6, CASP9, SPARC, TNF, CALM1, ALDH3A1, GBA, TPH2, EDN1, SSTR3, ACADS, SLC18A2 |
| | ||||
| GO molecular function pathways, n = 910 tested |
||||
| 0044548 | S100 protein binding | 10 | 0.123 | S100B, AHNAK, S100A1, ATP2A2, FGF1 |
| 0032794 | GTPase-activating protein binding | 11 | 0.144 | PLCD1, TSC1, GNAO1, FMNL3, CDH1, GNAI3 |
| | ||||
| CF-relevant custom pathways, n = 72 tested |
||||
| Goblet cell relevant | 37 | 0.001 | MUC4, TFF2, CFTR, FUT6, GALNT12, SCGB1A1, ERN2, B4GALNT2, ST6GALNAC1, XBP1, MUC1, GCNT3, FUT4 | |
| Ciliary trafficking | 157 | 0.006 | RAB8B, TBC1D7, PTCH1, EFHC1, ARFGEF2, IFT88, TTC26, IFT74, KIF19, RAB4A, RP1, VMA21, GLI3, IFT122, TRAF3IP1, TRPV1, COPG2, DNAH2, MKKS, OFD1, HSPB11, ODF2, IFT81, SSTR3, PACS1, ARHGEF1, KLC3, PCM1, GLI2, SCLT1 | |
| Mucin Calu3 | 12 | 0.024 | MUC4, MUC20, MUC1 | |
| MCF7 hypoxia (down) | 162 | 0.054 | OSTM1, ADAT1, CORO1A, SNRNP40, GAS2L1, SPAG1, POLR3K, RAB35, EEF1E1, GPATCH2, CALM1, ADORA1, KPNA1, PPIF, GDPD3, SLCO3A1, GYG1, PIK3R3, ARHGDIA | |
| HIF1si (up)/MCF7 hypoxia (down)‡ | ||||
| Asthma-COPD (up) | 36 | 0.059 | CEP72, FAM110C, CD44, TMEM200A, S100A16, CSTA, GCNT3, IGF2BP3, CEACAM5, CDC42EP5 | |
| EHF positive correlation | 154 | 0.092 | MUC20, SLC44A4, SH3YL1, RAB25, LCN2, LIMA1, FUT3, STAP2, CTNND1, CEACAM6, FUT6, PTK6, CHMP4C, SH2D3A, SPAG1, PIGR, ST6GALNAC1, S100A14, MYH14, RIPK4, FUT2, SPINT2, CDH1, C10orf99, YAP1, CEACAM5, CGN, CDC42EP5, SLC44A3 | |
| COPD (up) | 50 | 0.099 | MUC4, CFB, NR4A1, LCN2, ARNTL2, FUT3, IRAK3, MTNR1A, GCNT3, IGF2BP3, CEACAM5 | |
| Airway epithelium T-helper type 2 | 92 | 0.115 | CEP72, SCGB2A1, TFF3, FAM110C, CD44, TMEM200A, S100A16, CSTA, GCNT3, ITLN1, IGF2BP3, ALOX15, CEACAM5, CDC42EP5, SLC18A2, SLC22A16 | |
| CFTR interactome pathways§, n = 11 tested |
||||
| loT1hr dCF; Table E7, 484 genes | 466 | 0.055 | LMNA, PRDX1, AHNAK, LIMA1, CAPZB, PDCD6, MCM6, CFTR, ACLY, STAU1, CLPTM1, PPP6R1, SDHA, MYH2, RDX, XRCC5, STRBP, SPTAN1, TPM1, TUBB6, ACSL4, TP53, RBBP4, C3, POLR2E, CNN2, UBR4, MYH13, MOV10, PPP1R12A, RPLP0, MMS19, YTHDF3, SAE1, CSTA, MYH14, SNX27 | |
| loT6hr dCF; Table E8, 618 genes | 592 | 0.059 | LMNB2, ICAM1, LMNA, PRDX1, AHNAK, LIMA1, CAPZB, PDCD6, MCM6, THADA, STAU1, CLPTM1, EXOC4, PPP6R1, SDHA, MYH2, RDX, XRCC5, STRBP, SPTAN1, TPM1, TUBB6, ACSL4, TP53, RBBP4, C3, POLR2E, SLC35E1, UBR4, SLC27A3, MYH13, MOV10, PPP1R12A, RPLP0, DARS, MMS19, NUP155, SAE1, MSN, MYH14, SNX27 | |
| Core CFTR interactome; Table E1, 638 genes | 620 | 0.088 | CAPZA2, LMNB2, HSPA1B, BLMH, HSPA1A, ICAM1, LMNA, PRDX1, AHNAK, LIMA1, CAPZB, MCM6, THADA, RGPD2, COG6, CFTR, ACLY, STAU1, SORCS1, CLPTM1, EXOC4, SDHA, MYH2, RDX, XRCC5, STRBP, SPTAN1, TPM1, TUBB6, EXOC7, ACSL4, TP53, RBBP4, C3, POLR2E, CNN2, YTHDF2, UBR4, MYH13, MOV10, PPP1R12A, RPLP0, DARS, MMS19, YTHDF3, SAE1, RAC2, CSTA, MSN, MYH14, SNX27 | |
| SAHA dCF‡; Table E11, 681 genes | ||||
| MetaMiner cystic fibrosis–specific pathways (GeneGo)‖, n = 36 tested |
||||
| Cytokine production by Th17 cells in CF | 41 | 0.090 | ICAM1, CFTR, RELB, IL12RB1, CXCL1, IL8, RORC, CXCL6 | |
| | ||||
| HLA-specific pathways, n = 2 tested |
||||
| Class I and class II | 18 | 0.095 | HLA-DQA1, HLA-DQB1, HLA-DRB1, HLA-B, HLA-DOB, PSMB8, PSMB9, TAP1, TAP2 | |
| Class II | 8 | 0.146 | HLA-DQA1, HLA-DQB1, HLA-DRB1, HLA-DOB | |
| | ||||
| | ||||
| Analyses confined to pathways significant for differential expression in nasal scrape samples¶, n = 37 tested |
||||
| KEGG pathways |
||||
| 05168 | Herpes simplex virus infection | 173 | 0.008 | HLA-DQA1, HLA-DQB1, HLA-DRB1, PVRL2, PVRL1, PER2, CCL2, TLR2, SRSF2, TYK2, CCL5, POLR2A, IFNA6, TP53, C3, IFNA13, IFNA1, EIF2AK2, LTA, TNF, IFNA2, IFNA5, MCRS1, TBPL1, IFNA14, TLR3, IFNA8, TAF5, HLA-B, IFNA17, PPP1CC, HLA-DOB, TAP1, TAP2, MAPK9, HCFC2, ALYREF, TBPL2 |
| 05164 | Influenza A virus | 165 | 0.080 | HSPA1B, HLA-DQA1, HLA-DQB1, HLA-DRB1, HSPA1A, HSPA1L, PABPN1L, ICAM1, PRKCB, CCL2, TYK2, CCL5, IFNA6, PIK3R2, TNFSF10, IFNA13, IFNA1, CASP9, EIF2AK2, TNF, IFNA2, IFNA5, IFNA14, TLR3, KPNA1, IFNA8, MAPK3, IFNA17, DDX39B, PIK3R3, HLA-DOB, MAPK9, RAF1 |
Definition of abbreviations: CF = cystic fibrosis; CFTR = cystic fibrosis transmembrane conductance regulator; COPD = chronic obstructive pulmonary disease; dCF = Phe508del; EHF = ETS homologous factor; GO = Gene Ontology Consortium; GTPase = GTP (guanosine triphosphate) enzyme; HIF1si = HIF-1α siRNA; HLA = human leukocyte antigen; KEGG = Kyoto Encyclopedia of Genes and Genomes database; KNoRMA = Kulich Normal Residual Mortality Adjusted; SAHA = suberoylanilide hydroxamic acid.
Default parameters with 1,000 simulations were used, and pathways were limited to those that contained at least 10 but less than or equal to 200 genes. GeneSetScan uses mapping of genotyped single-nucleotide polymorphisms to 50 kb upstream and downstream of protein-coding genes based on ENSEMBL version 82 annotation and maps genes to annotated pathways and gene sets. CF relevant custom pathways were developed (46) using human gene counterparts (Table E8). See Table E3 for gene Online Mendelian Inheritance in Man catalogue numbers.
Multiple comparison corrected P values.
Gene level P values were calculated using family-wise error rate (all single-nucleotide polymorphisms, genes, and pathways tested) as provided by GeneSetScan. Pathways are listed if corrected P value is less than 0.15.
These pathways are statistically significant and carry robust overlap of genes with first-listed pathway; see Table E5, tab F, for complete listing of pathway genes.
For gene sets containing more than 200 genes, genes with P < 0.05 are listed; see Table E5, tab F, for complete list of pathway genes.
MetaMiner CF-specific pathways represent a version of the Thomson Reuters (formerly GeneGo) MetaDiscovery suite that is enriched with content specific for CF.
Pathways listed in Table 2 were evaluated for association with genotype.
Identification of Functional Overlap and Differences between Expression and GWAS Data
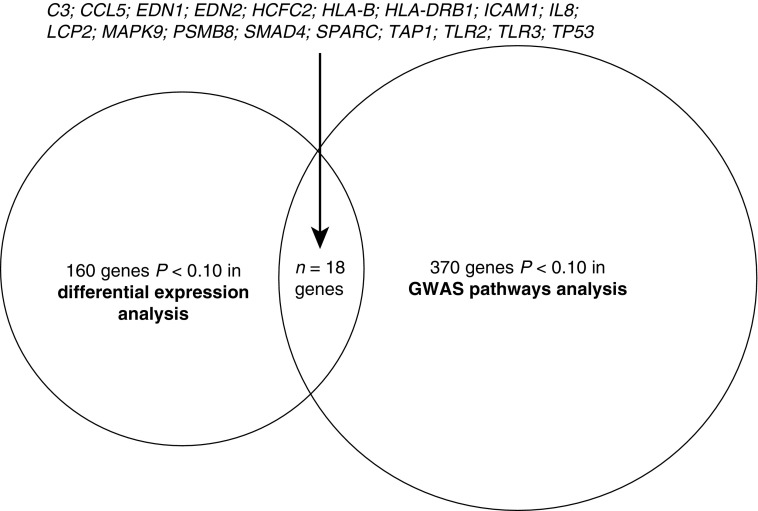
Pathways (and genes) identified in differential expression analysis (Table 2) were similar in many biological respects to those identified using GWAS data (Table 3). To determine the overlap of differential expression and GWAS results, we assessed those genes with P values less than 0.10 contributing to both expression (Table 2) and GWAS (Table 3) pathways. This yielded 18 genes (Figure 1), which is significantly greater than expected by random chance (P = 3.6 × 10−06). Strikingly, the biological functions of all 18 genes are highly reflective of the broader concept that airway mucosal host defense related to environmental stimuli contributes to lung phenotype (Table E7).
Figure 1.
Top-ranked genes (P < 0.10) common to significant pathways in both differential expression and genome-wide association study (GWAS) analyses. Eighteen genes with significance levels of P < 0.10 were observed in overlap of differential expression and GWAS analyses.
Integration of GWAS Signals with Nasal Epithelial Gene Expression
To further integrate GWAS signal with nasal epithelial gene expression, we tested risk alleles of SNPs at the top five loci in our GWAS (5) for association with gene expression pathways in our nasal epithelial RNA-seq data. We used SAFE and approximately 1,000 randomly selected SNPs to rigorously control for statistical error (Table 4; see also Methods section of online supplement). This analysis demonstrated a significant association between differential expression pathways and the risk allele at four of the five significant GWAS loci (chromosomes [chr] 11, 5, 6, and X). Notably, the chr11 top-ranked GWAS SNP (rs10742326) was significantly associated with multiple pathways relevant to CF pathogenesis (Table 4; Table E5, tab G), including two CFTR-related pathways (i.e., CFTR-dependent regulation of ion channels in airway epithelium and a CFTR interactome pathway specific to Phe508del) (22). HLA genes, lipid transport, and inflammatory signaling were also identified (Table 4).
Table 4.
Gene Expression Pathways Significantly Associated with Risk Alleles for Significant Cystic Fibrosis Genome-Wide Association Study Loci
| Chr | SNP rs Number | Pathway Identifier |
Genes (n) | Statistics |
Minor Allele | Risk Allele | Association with Risk Allele‡ | Genes with Gene-Level P Value <0.10 (Ordered by P Value)§ | ||
|---|---|---|---|---|---|---|---|---|---|---|
| Set | Name | P Value* | Q Value† | |||||||
| 11 | rs10742326 | CF-relevant custom pathways | COPD signature; PMID 23471465 | 66 | 0.0001 | 0.0035 | A | G | Decreased expression | MUC4, ATP10B, SAA2, TMPRSS11D, SLC26A2, SLC26A4, SLC5A8, SAA1, SAA4, IRAK3, C15orf48, SLCO1B3, SERPINB7, EPB41L2, TPRXL, TRIM31, CCDC81, MTNR1A |
| 11 | rs10742326 | CF-relevant custom pathways | Asthma nitric oxide gene cluster 3; PMID 25338189 | 48 | 0.0002 | 0.0046 | A | G | Decreased expression | DUOXA1, FER1L5, WDR90, C16orf93, STK36, ARHGAP33, CEP164, HGS, PDXDC2P, KIAA0895L, TMEM234, MAP4K4, FAM193B, FBXO31, LINC00479, SPPL2B, RAD9A, MYO15B |
| 11 | rs10742326 | CF-relevant custom pathways | CF MI Lasso; PMID 22466613 | 21 | 0.0005 | 0.0087 | A | G | Decreased expression | PROM1, SLC9A3, CD44, CTSB |
| 11 | rs10742326 | CF-relevant custom pathways | COPD up; PMID 23471465 | 49 | 0.0011 | 0.0153 | A | G | Decreased expression | MUC4, ATP10B, SAA2, TMPRSS11D, SLC26A2, SLC26A4, SLC5A8, SAA1, SAA4, IRAK3, C15orf48, SLCO1B3, TPRXL, TRIM31, MTNR1A |
| 11 | rs10742326 | MetaMiner cystic fibrosis–specific pathways (GeneGo) | CFTR-dependent regulation of ion channels in airway epithelium (normal and CF) | 33 | 0.0040 | 0.0605 | A | G | Decreased expression | ITPR3, ABCC9, WBP1, PRSS8, SLC9A3R1, GNA11, NEDD4, KCNN4, SCNN1A |
| 11 | rs10742326 | CFTR interactome pathways | Core increased dCF over WT; Table E6, 52 genes | 50 | 0.0337 | 0.0993 | A | G | Increased expression | PSMD3, PSMD4, UBXN1, PSMD8, PSMD11, PSMA2, LMAN2, UBAC2 |
| 6 | rs116003090 | HLA specific | HLA class II | 16 | 0.0626 | 0.0527 | C | C | Bidirectional | HLA-DQB1, HLA-DRB1‖, HLA-DRB4‖, HLA-DQA2‖, HLA-DQA1‖, HLA-DRB5, HLA-DOB‖, HLA-DQB2 |
| 6 | rs116003090 | CFTR interactome pathways | HDAC7 dCF; Table E13, 450 genes | 410 | 0.0202 | 0.0343 | C | C | Increased expression | RDX, APOL2, HSPH1, PPP2R2A, PPP2CA, DNAJA1, SLC25A22, SAMHD1, EZR, YWHAH, SPTLC2, HSPA8, ICAM1, LMNA, PHGDH, KRT7, YWHAE, DCTN2, GART, SFXN3, PPL, LGALS3BP, CDH1, TUBB6, PSMA4, ACTN4, TMEM40, RUVBL1, CAST, UBXN1, TPM4, TIMM50, HSPD1, KLHL22, PSMA6, LAMB3, ITGA3, TAPBP, VDAC2, ERAP1, TF, RAB18, PDXK, ILVBL, SFN, PSMA1, MARS, NCAPG2, AHSA1, YME1L1, CALR |
| loT6hr dCF; Table E8, 618 genes¶ | ||||||||||
| SAHA dCF; Table E11, 681 genes¶ | ||||||||||
| 6 | rs116003090 | CFTR interactome pathways | loT24hr dCF; Table E9, 199 genes | 175 | 0.0226 | 0.0343 | C | C | Increased expression | APOL2, HSPH1, PPP2CA, DNAJA1, YWHAH, HSPA8, KRT7, YWHAE, LMO7, LGALS3BP, TUBB6, ZW10, PSMA4, ACTN4, RUVBL1, TPM4, TIMM50, HSPD1, LAMB3, ERAP1, SFN, MARS |
| loT24hr rev dCF; Table E10, 199 genes¶ | ||||||||||
| 6 | rs116003090 | CFTR interactome pathways | Core dCF specific; Table E5, 208 genes | 193 | 0.0344 | 0.0404 | C | C | Increased expression | TMEM165, SAMHD1, CBR1, SEC24C, C9orf167, ICAM1, DCTN2, SFXN3, CDH1, MX1, ISG15, ZW10, PSMA4, TMEM40, CAST, UBXN1, MX2, MOV10, LAMB3, ITGA3, RFC2, PPA1, VDAC2, PDXK, AHSA1, YME1L1 |
| 5 | rs57221529 | MetaMiner cystic fibrosis–specific pathways (GeneGo) | Cholesterol and sphingolipids transport/distribution to the intracellular membrane compartments (normal and CF) | 11 | 0.0002 | 0.0032 | G | A** | Increased expression | RAB9A, SCP2 |
| 5 | rs57221529 | HLA specific | HLA class II | 16 | 0.0320 | 0.0268 | G | A** | Increased expression | HLA-DMA, HLA-DRA, HLA-DMB, HLA-DOA, HLA-DRB1, HLA-DPB1, HLA-DPA1 |
| 5 | rs57221529 | HLA specific | HLA class I and class II | 30 | 0.0957 | 0.0554 | G | A** | Increased expression | HLA-DMA, HLA-DRA, HLA-DMB, HLA-DOA, HLA-DRB1, HLA-DPB1, HLA-DPA1 |
| X | rs5952223 | KEGG: M00154 | Cytochrome c oxidase | 17 | 0.0007 | 0.0957 | T | C | Decreased expression | COX7A2L, COX6C, COX5B |
Definition of abbreviations: CF = cystic fibrosis; CFTR = cystic fibrosis transmembrane conductance regulator; COPD = chronic obstructive pulmonary disease; dCF = Phe508del; HDAC7 = histone deacetylase 7; KEGG = Kyoto Encyclopedia of Genes and Genomes database; MI = meconium ileus; PMID = PubMed reference number; SAHA = suberoylanilide hydroxamic acid; WT = wild type.
Pathways were limited to those with at least 10 but less than or equal to 200 genes. CF-relevant custom pathways were developed (46) using human gene counterparts (Table E8).
Significance Analysis of Function and Expression analysis used 10,000 permutations to establish significance thresholds (18).
Benjamini-Hochberg false discovery rate for pathway testing within each pathway set; Q values less than 0.15 were included.
Risk alleles may be associated with increased expression, decreased expression, or bidirectional expression of genes in pathway.
See Table E5, tab G, for an inclusive list of genes for these pathways; see Table E3 for gene Online Mendelian Inheritance in Man catalogue numbers.
For bidirectional pathways, genes with increased expression associated with CF genome-wide association study loci risk alleles are noted.
These pathways are statistically significant and carry robust overlap of genes with first-listed pathway; see Table E5, tab G, for a complete list of pathway genes.
For this study cohort, risk allele differs from that reported in broader CF genome-wide association studies (5).
Discussion
Using unbiased transcriptomic and integrative genomic approaches, we performed a comprehensive analysis to identify modifier genes and mechanistic pathways modulating CF lung disease severity. Although no single gene was statistically significant in isolation, the primary transcriptomic analysis identified differentially expressed genes in pathways (Table 2) under genomic (heritable) influence and relevant to airway mucosal host defense. The pathways that emerged from the analysis, particularly as related to viral infection, inflammation, apoptosis, lipid metabolism, and innate immune responses, including HLA genes, reflect the known complexity of CF pathophysiology. Importantly, the direction and content of differentially expressed genes in these pathways bear striking relevance to what is known about the pathogenesis of CF lung disease. Almost all of the significant pathways in the differential expression analysis demonstrate that increased gene expression is associated with worse lung disease (“detrimental”) (Table 2), which is congruent with the concept that persistent “hyperinflammatory” responses to environmental stimuli (such as viral or bacterial infection) contribute to more severe CF lung disease (23, 24). Indeed, viral infections in CF are known to lead to pulmonary exacerbations and decreased lung function (25, 26), and dysregulated inflammation is believed to adversely affect CF lung disease (23, 26). Our findings are congruent with a previous microarray analysis of nasal brushings in a small study of patients with CF (n = 12) which demonstrated that subjects with severe lung disease had increased expression of genes linked to viral infection, including STAT1 (Table 2), which is critical in the host response to viral infection and transcriptional activation of IFN-induced genes (27).
Pathway (GeneSetScan) analysis of genomic data in 5,659 patients with CF yielded significant pathways containing genes related to viral infection and innate immune response (Table 3), complementing the transcriptomic findings. Of the 18 top-ranked (P < 0.10) genes that were common to both results, nearly all were associated with airway mucosal host defense (Figure 1 and Table E7). Overlapping genes in these analyses, including ICAM1, IL8, and HCFC2, point to heritable variation in the inflammatory response to bacterial and viral infection yielding downstream effects on CF lung disease. Furthermore, IL8 has previously been implicated as a modifier gene in CF lung disease (28). Similarly, overlap of genes integral to the innate immune response (e.g., C3, HLA-B, HLA-DRB1, TLR2, and TLR3) demonstrates that heritable variation in expression of genes related to host defense plays a significant role in determining CF lung disease severity.
GWAS pathway analysis also identified additional host defense mechanisms related to lung disease severity, including goblet cell, mucin production, cilia trafficking, and CFTR interactome pathways (Table 3). Taken together, these pathways point to heritable variation in mucociliary clearance, a critical first-line innate airway defense mechanism involving the interaction of well-hydrated mucus with functional cilia, and a key mucosal defense mechanism regulating CF lung disease severity. Finally, pathways revealed by this GWAS analysis included genes located at significant CF GWAS loci (i.e., AGTR2, EXOC3, MUC20, MUC4, CD44, and HLA genes) (5). Genomic variation in regions near these genes is known to correlate with lung disease severity on the basis of our previously reported findings (5, 6); the gene networks identified in the present analysis provide new insight into potential mechanisms for effect of these candidate modifier genes on CF lung phenotype.
To further explore the mechanism of association between genomic variation at significant GWAS loci and lung disease severity, we used a novel approach to test for association of gene expression pathways with SNP variation at significant CF GWAS loci (5) (Table 4). The chr11p12-p13 GWAS locus is between EHF (an epithelial transcription factor) and APIP (an inhibitor of apoptosis as well as an enzyme in the methionine salvage pathway) (5, 6), and the association at this locus with lung disease severity is determined by Phe508del homozygotes (29); however, the mechanism by which the region produces its phenotypic effects is unresolved. The EHF transcription factor is implicated in recent reports (30, 31), as well as our findings (Tables 2 and 3), whereas other findings support a role for APIP by means of MTAP (Table 2, “Methionine salvage pathway” row) (32, 33). Our analysis demonstrated a significant association of the chr11 risk allele with decreased expression of genes involved in CFTR-dependent regulation of ion channels, as well as other CF-relevant pathways including genes pertinent to chronic obstructive pulmonary disease and asthma (Table 4). Importantly, both the chr11p12-p13 (rs10742326) and chr6p21.3 (near HLA; rs116003090) (5, 6) risk alleles were associated with increased expression of genes in CFTR interactome pathways (22). For the first time, to our knowledge, we demonstrate that in vivo networks, or pathways, of differentially expressed genes in airways are related to established genomic (SNP) variation, where risk alleles are associated with CF lung disease severity (5, 6). Importantly, to our knowledge, these findings represent the first described association of non-CFTR genomic variation with CFTR production, processing, and/or function itself. Coupled with GWAS (5, 6), gene expression networks associated with significant CF GWAS variants provide novel insight into potential mechanisms of effect for candidate modifier genes, and future research will benefit from exploration of these hypotheses.
Our integrated analysis also highlights the need for deep exploration of the HLA region. HLA genes have consistently been implicated across multiple studies of modifier genes in CF, including GWAS (5, 6), differential expression studies in transformed lymphocytes (9), and the nasal mucosal transcriptomic plus genomic pathway analyses described here. The association of genomic variation at the HLA chr6p21.3 (rs116003090) region with expression of genes regulating CFTR processing (Table 4) provides the first glimpse into a novel potential mechanism of action for genetic variation at the chr6 locus to modify CF lung disease, in addition to established roles of HLA in numerous inflammatory and pathogen response pathways (Table 2). The complexity of the HLA region has thus far denied the scientific community of a clear pathogenic mechanism for association with CF lung disease. It is now clear that detailed, integrated analysis of HLA genetic, allelic, and gene expression variability is a critical next step, with findings likely to be highly relevant to other chronic lung diseases, such as asthma, where GWAS signals also reside in the HLA region (34, 35).
Our study has some limitations. First, whereas we characterized the percentage of participants known to have chronic P. aeruginosa infection near the time of sampling, we cannot entirely eliminate infection status as a confounding factor, because microbial culture was not conducted at the sampling date, and this was coupled to our inability to access all possible infections known to have roles in CF lung disease (36). Furthermore, the study does not include a replication cohort or functional validation of any specific pathway. However, validation across analyses for certain genes/pathways (Figure 1) provided evidence of robust signatures that serve as a basis for future replication/functional validation. Future investigations should consider use of effect sizes demonstrated in this study (Table E3).
Despite recent advances in the development of CFTR correctors and potentiators for treatment of CF (37, 38), there remains a critical need for antiinflammatory therapies to ameliorate/optimize airway mucosal host defense that can be applied broadly to patients with CF (39). Currently, there are extensive ongoing efforts to develop such “antiinflammatory” therapies (40), and the genetic and genomic data presented here provide compelling support for these efforts. We highlight one example where the gene expression results have potential therapeutic relevance. Transcriptomic evidence of increased inflammatory signaling in the methionine salvage pathway (Table 2) includes increased gene expression of AMD1, MTAP, and APIP. The expected increases in enzymatic activity of these genes would reduce levels of the antiinflammatory metabolite methylthioadenosine while generating proinflammatory polyamines (32, 41, 42). Recent mass spectrometric metabolomic analysis of bronchoalveolar lavage fluid from children with CF has shown that increased polyamine levels correlate with neutrophilic inflammation and worse lung function (43), and the direction of this finding is congruent with our gene expression findings. Because pharmacologic inhibitors of this pathway are available (44), we have begun exploring this pathway in animal studies to provide proof-of-concept support for such inhibitors as a CF therapy (45).
In conclusion, this study represents a rigorous effort to use gene expression data from the highly CF-relevant airway (nasal) epithelial cell, complemented by extensive genetic data, to identify modifying pathways relevant to CF lung disease severity. The transcriptomic data we report provide unique evidence of increased airway epithelial gene expression in biologically informative pathways, congruent with underlying concepts that hyperinflammatory responses are deleterious to CF lung disease. The presence of genes in both expression- and genomics-based analyses (GWAS and SNP pathway analyses) provides support for the genomic basis of modifier genes, even when mediated through changes in expression. Although association studies of differential gene expression cannot establish cause and effect, genes in our significant pathways demonstrate robust heritability. Taken together with the results of our heritability analysis, these findings suggest that heritable traits linked to increased expression of non-CFTR genes, particularly those regulating inflammatory responses to environmental stimuli, play a key role in CF lung disease severity. Candidate pathways and genes identified by these studies offer novel targets for precision therapies directed toward genes with heritable effects on lung disease severity in CF.
Acknowledgments
Acknowledgment
The authors thank the research coordinators at participating sites for their efforts: Julie Avolio, Colette Bucur, Erin Felling, and Douglas Walker. The authors also thank Anthony T. Dang and Michael V. Patrone for their contributions in data analysis support; Alison Williams, Sarah N. Dalrymple, and Hemant Kelkar and Airong Xu (University of North Carolina Center for Bioinformatics) for data management support; Farnoosh Abbas Aghababazadeh for assistance in figure formatting; and Xueliang Guo for thoughtful discussions. Last, the authors thank the Cystic Fibrosis Foundation for the use of its CF Patient Registry data, as well as the participants with CF, their families, their care providers, and the clinic coordinators for their contributions to the registry. The authors are most grateful to every patient and family that participated in this study.
Footnotes
Supported by NHLBI grants HL095396 and HL068890, National Institute of Diabetes and Digestive and Kidney Diseases grant P30 DK065988, National Human Genome Research Institute grant R21HG007840, Canadian Institutes of Health Research Open Operating Grants Program (MOP) grant 258916, Cystic Fibrosis Canada (CFC) grant 2626, Genome Canada through the Ontario Genomics Institute (2004-OGI-3-05), Cystic Fibrosis Foundation grants POLINE09FO and BOUCHE15R0, and the Gilead Sciences Research Scholars Program in Cystic Fibrosis.
Author Contributions: D.P., L.C.J., R.G.P., J.R.S., L.A.C., H.C., G.R.C., M.L.D., L.J.S., P.R.D., J.F.C., W.K.O’N., and M.R.K.: conceived of and designed the experiments; D.P., L.C.J., R.G.P., J.R.S, L.A.C., J.E.K., M.P.B., P.R.D, J.F.C., and M.R.K.: performed the experiments; D.P., H.D., P.J.G., Y.-H.Z., F.Z., and F.A.W.: performed statistical analysis; D.P., H.D., P.J.G., L.C.J., R.G.P., J.R.S., J.E.K., Y.-H.Z., H.C., G.R.C., M.L.D., L.J.S., F.Z., F.A.W., W.K.O’N., and M.R.K.: analyzed the data; D.P., H.D., R.G.P., J.R.S., F.A.W., W.K.O’N., and M.R.K.: wrote the manuscript; M.P.B., P.R.D., J.F.C., W.K.O’N., and M.R.K.: jointly supervised the research. All authors read and approved the submitted manuscript.
This article has an online supplement, which is accessible from this issue’s table of contents at www.atsjournals.org.
Originally Published in Press as DOI: 10.1164/rccm.201701-0134OC on August 30, 2017
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Cystic Fibrosis Foundation Patient Registry. 2015 Annual data report. Bethesda, MD: Cystic Fibrosis Foundation; 2016 [accessed 2016 Dec 5]. Available from: https://www.cff.org/Our-Research/CF-Patient-Registry/2015-Patient-Registry-Annual-Data-Report.pdf.
- 2.Vanscoy LL, Blackman SM, Collaco JM, Bowers A, Lai T, Naughton K, et al. Heritability of lung disease severity in cystic fibrosis. Am J Respir Crit Care Med. 2007;175:1036–1043. doi: 10.1164/rccm.200608-1164OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mekus F, Ballmann M, Bronsveld I, Bijman J, Veeze H, Tümmler B. Categories of ΔF508 homozygous cystic fibrosis twin and sibling pairs with distinct phenotypic characteristics. Twin Res. 2000;3:277–293. doi: 10.1375/136905200320565256. [DOI] [PubMed] [Google Scholar]
- 4.Collaco JM, Blackman SM, McGready J, Naughton KM, Cutting GR. Quantification of the relative contribution of environmental and genetic factors to variation in cystic fibrosis lung function. J Pediatr. 2010;157:802–807.e3. doi: 10.1016/j.jpeds.2010.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Corvol H, Blackman SM, Boëlle PY, Gallins PJ, Pace RG, Stonebraker JR, et al. Genome-wide association meta-analysis identifies five modifier loci of lung disease severity in cystic fibrosis. Nat Commun. 2015;6:8382. doi: 10.1038/ncomms9382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wright FA, Strug LJ, Doshi VK, Commander CW, Blackman SM, Sun L, et al. Genome-wide association and linkage identify modifier loci of lung disease severity in cystic fibrosis at 11p13 and 20q13.2. Nat Genet. 2011;43:539–546. doi: 10.1038/ng.838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kulich M, Rosenfeld M, Campbell J, Kronmal R, Gibson RL, Goss CH, et al. Disease-specific reference equations for lung function in patients with cystic fibrosis. Am J Respir Crit Care Med. 2005;172:885–891. doi: 10.1164/rccm.200410-1335OC. [DOI] [PubMed] [Google Scholar]
- 8.Taylor C, Commander CW, Collaco JM, Strug LJ, Li W, Wright FA, et al. A novel lung disease phenotype adjusted for mortality attrition for cystic fibrosis genetic modifier studies. Pediatr Pulmonol. 2011;46:857–869. doi: 10.1002/ppul.21456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.O’Neal WK, Gallins P, Pace RG, Dang H, Wolf WE, Jones LC, et al. Gene expression in transformed lymphocytes reveals variation in endomembrane and HLA pathways modifying cystic fibrosis pulmonary phenotypes. Am J Hum Genet. 2015;96:318–328. doi: 10.1016/j.ajhg.2014.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rowe SM, Liu B, Hill A, Hathorne H, Cohen M, Beamer JR, et al. VX06-770-101 Study Group. Optimizing nasal potential difference analysis for CFTR modulator development: assessment of ivacaftor in CF subjects with the G551D-CFTR mutation. PLoS One. 2013;8:e66955. doi: 10.1371/journal.pone.0066955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McDougall CM, Blaylock MG, Douglas JG, Brooker RJ, Helms PJ, Walsh GM. Nasal epithelial cells as surrogates for bronchial epithelial cells in airway inflammation studies. Am J Respir Cell Mol Biol. 2008;39:560–568. doi: 10.1165/rcmb.2007-0325OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Knowles MR, Clarke LL, Boucher RC. Activation by extracellular nucleotides of chloride secretion in the airway epithelia of patients with cystic fibrosis. N Engl J Med. 1991;325:533–538. doi: 10.1056/NEJM199108223250802. [DOI] [PubMed] [Google Scholar]
- 13.Polineni D, Dang H, Pace RG, Stonebraker JR, Guo XL, Jones LC, et al. Correlation of inflammatory markers in human nasal epithelia in vivo informs transcriptomic studies in cystic fibrosis (CF) complementary to genomic variation studies. Am J Respir Crit Care Med. 2014;189:A5519. (abstract) [Google Scholar]
- 14.Polineni D, Dang H, Gallins PJ, Pace R, Stonebraker JR, Jones LC, et al. In vivo human nasal epithelia inform transcriptomic studies in cystic fibrosis (CF) complementary to genomic variation studies. Pediatr Pulmonol. 2014;49:272. (abstract) [Google Scholar]
- 15.Polineni D, Dang H, Pace RG, Stonebraker JR, Guo XG, Jones LC, et al. In vivo cystic fibrosis (CF) nasal epithelial transcriptomic studies reveal non-CFTR differential gene expression associated with CF lung disease severity. Am J Respir Crit Care Med. 2016;193:A6458. (abstract) [Google Scholar]
- 16.Leek JT, Johnson WE, Parker HS, Jaffe AE, Storey JD. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics. 2012;28:882–883. doi: 10.1093/bioinformatics/bts034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shabalin AA. Matrix eQTL: ultra fast eQTL analysis via large matrix operations. Bioinformatics. 2012;28:1353–1358. doi: 10.1093/bioinformatics/bts163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barry WT, Nobel AB, Wright FA. Significance analysis of functional categories in gene expression studies: a structured permutation approach. Bioinformatics. 2005;21:1943–1949. doi: 10.1093/bioinformatics/bti260. [DOI] [PubMed] [Google Scholar]
- 19.Wright FA, Sullivan PF, Brooks AI, Zou F, Sun W, Xia K, et al. Heritability and genomics of gene expression in peripheral blood. Nat Genet. 2014;46:430–437. doi: 10.1038/ng.2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gene Set Enrichment Analysis (GSEA)/Broad Institute. GSEA user guide, version 5.1 [accessed 2016 Apr 4]. Available from: http://software.broadinstitute.org/gsea/doc/GSEAUserGuideFrame.html.
- 21.Schaid DJ, Sinnwell JP, Jenkins GD, McDonnell SK, Ingle JN, Kubo M, et al. Using the gene ontology to scan multilevel gene sets for associations in genome wide association studies. Genet Epidemiol. 2012;36:3–16. doi: 10.1002/gepi.20632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pankow S, Bamberger C, Calzolari D, Martínez-Bartolomé S, Lavallée-Adam M, Balch WE, et al. ∆F508 CFTR interactome remodelling promotes rescue of cystic fibrosis. Nature. 2015;528:510–516. doi: 10.1038/nature15729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bruscia EM, Bonfield TL. Innate and adaptive immunity in cystic fibrosis. Clin Chest Med. 2016;37:17–29. doi: 10.1016/j.ccm.2015.11.010. [DOI] [PubMed] [Google Scholar]
- 24.Adam D, Roux-Delrieu J, Luczka E, Bonnomet A, Lesage J, Mérol JC, et al. Cystic fibrosis airway epithelium remodelling: involvement of inflammation. J Pathol. 2015;235:408–419. doi: 10.1002/path.4471. [DOI] [PubMed] [Google Scholar]
- 25.Flight WG, Bright-Thomas RJ, Tilston P, Mutton KJ, Guiver M, Morris J, et al. Incidence and clinical impact of respiratory viruses in adults with cystic fibrosis. Thorax. 2014;69:247–253. doi: 10.1136/thoraxjnl-2013-204000. [DOI] [PubMed] [Google Scholar]
- 26.Hendricks MR, Bomberger JM. Digging through the obstruction: insight into the epithelial cell response to respiratory virus infection in patients with cystic fibrosis. J Virol. 2016;90:4258–4261. doi: 10.1128/JVI.01864-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wright JM, Merlo CA, Reynolds JB, Zeitlin PL, Garcia JG, Guggino WB, et al. Respiratory epithelial gene expression in patients with mild and severe cystic fibrosis lung disease. Am J Respir Cell Mol Biol. 2006;35:327–336. doi: 10.1165/rcmb.2005-0359OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hillian AD, Londono D, Dunn JM, Goddard KAB, Pace RG, Knowles MR, et al. CF Gene Modifier Study Group. Modulation of cystic fibrosis lung disease by variants in interleukin-8. Genes Immun. 2008;9:501–508. doi: 10.1038/gene.2008.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dang H, Gallins PJ, Pace RG, Guo XL, Stonebraker JR, Corvol H, et al. Novel variation at chr11p13 associated with cystic fibrosis lung disease severity Hum Genome Var 2016316020[Published erratum appears in Hum Genome Var 2017;4:17016.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stolzenburg LR, Yang R, Kerschner JL, Fossum S, Xu M, Hoffmann A, et al. Regulatory dynamics of 11p13 suggest a role for EHF in modifying CF lung disease severity. Nucleic Acids Res. 2017;45:8773–8784. doi: 10.1093/nar/gkx482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fossum SL, Mutolo MJ, Tugores A, Ghosh S, Randell SH, Jones LC, et al. Ets homologous factor (EHF) has critical roles in epithelial dysfunction in airway disease. J Biol Chem. 2017;292:10938–10949. doi: 10.1074/jbc.M117.775304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang L, Ko ER, Gilchrist JJ, Pittman KJ, Rautanen A, Pirinen M, et al. Wellcome Trust Case Control Consortium 2; Kenyan Bacteraemia Study Group. Human genetic and metabolite variation reveals that methylthioadenosine is a prognostic biomarker and an inflammatory regulator in sepsis. Sci Adv. 2017;3:e1602096. doi: 10.1126/sciadv.1602096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ko DC, Gamazon ER, Shukla KP, Pfuetzner RA, Whittington D, Holden TD, et al. Functional genetic screen of human diversity reveals that a methionine salvage enzyme regulates inflammatory cell death. Proc Natl Acad Sci USA. 2012;109:E2343–E2352. doi: 10.1073/pnas.1206701109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kontakioti E, Domvri K, Papakosta D, Daniilidis M. HLA and asthma phenotypes/endotypes: a review. Hum Immunol. 2014;75:930–939. doi: 10.1016/j.humimm.2014.06.022. [DOI] [PubMed] [Google Scholar]
- 35.Lasky-Su J, Himes BE, Raby BA, Klanderman BJ, Sylvia JS, Lange C, et al. SHARP investigators. HLA-DQ strikes again: genome-wide association study further confirms HLA-DQ in the diagnosis of asthma among adults. Clin Exp Allergy. 2012;42:1724–1733. doi: 10.1111/cea.12000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Quinn RA, Whiteson K, Lim YW, Zhao J, Conrad D. LiPuma JJ, Rohwer F, Widder S. Ecological networking of cystic fibrosis lung infections. NPJ Biofilms Microbiomes. 2016;2:4. doi: 10.1038/s41522-016-0002-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wainwright CE, Elborn JS, Ramsey BW, Marigowda G, Huang X, Cipolli M, et al. TRAFFIC Study Group. TRANSPORT Study Group. Lumacaftor–ivacaftor in patients with cystic fibrosis homozygous for Phe508del CFTR. N Engl J Med. 2015;373:220–231. doi: 10.1056/NEJMoa1409547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ramsey BW, Davies J, McElvaney NG, Tullis E, Bell SC, Dřevínek P, et al. VX08-770-102 Study Group. A CFTR potentiator in patients with cystic fibrosis and the G551D mutation. N Engl J Med. 2011;365:1663–1672. doi: 10.1056/NEJMoa1105185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Torphy TJ, Allen J, Cantin AM, Konstan MW, Accurso FJ, Joseloff E, et al. Antiinflammatory Therapy Working Group. Considerations for the conduct of clinical trials with antiinflammatory agents in cystic fibrosis: a Cystic Fibrosis Foundation Workshop report. Ann Am Thorac Soc. 2015;12:1398–1406. doi: 10.1513/AnnalsATS.201506-361OT. [DOI] [PubMed] [Google Scholar]
- 40.Nichols DP, Chmiel JF. Inflammation and its genesis in cystic fibrosis. Pediatr Pulmonol. 2015;50:S39–S56. doi: 10.1002/ppul.23242. [DOI] [PubMed] [Google Scholar]
- 41.Mary C, Duek P, Salleron L, Tienz P, Bumann D, Bairoch A, et al. Functional identification of APIP as human mtnB, a key enzyme in the methionine salvage pathway. PLoS One. 2012;7:e52877. doi: 10.1371/journal.pone.0052877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kang W, Hong SH, Lee HM, Kim NY, Lim YC, Le TM, et al. Structural and biochemical basis for the inhibition of cell death by APIP, a methionine salvage enzyme. Proc Natl Acad Sci USA. 2014;111:E54–E61. doi: 10.1073/pnas.1308768111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Esther CR, Jr, Coakley RD, Henderson AG, Zhou YH, Wright FA, Boucher RC. Metabolomic evaluation of neutrophilic airway inflammation in cystic fibrosis. Chest. 2015;148:507–515. doi: 10.1378/chest.14-1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Basu I, Locker J, Cassera MB, Belbin TJ, Merino EF, Dong X, et al. Growth and metastases of human lung cancer are inhibited in mouse xenografts by a transition state analogue of 5′-methylthioadenosine phosphorylase. J Biol Chem. 2011;286:4902–4911. doi: 10.1074/jbc.M110.198374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Esther CR, O’Neal W, Polineni D, Mahon A, Isaacman S, Knowles M. A combined ’omics strategy identifies the methionine salvage pathway as a novel therapeutic target for cystic fibrosis airways disease. Am J Respir Crit Care Med. 2017;195:A4852. (abstract) [Google Scholar]
- 46.Saini Y, Dang H, Livraghi-Butrico A, Kelly EJ, Jones LC, O’Neal WK, et al. Gene expression in whole lung and pulmonary macrophages reflects the dynamic pathology associated with airway surface dehydration. BMC Genomics. 2014;15:726. doi: 10.1186/1471-2164-15-726. [DOI] [PMC free article] [PubMed] [Google Scholar]