Abstract
The RecX protein, a very active natural RecA protein inhibitor, can completely disassemble RecA filaments at nanomolar concentrations that are two to three orders of magnitude lower than that of RecA protein. Based on the structure of RecX protein complex with the presynaptic RecA filament, we designed a short first in class α-helical peptide that both inhibits RecA protein activities in vitro and blocks the bacterial SOS-response in vivo. The peptide was designed using SEQOPT, a novel method for global sequence optimization of protein α-helices. SEQOPT produces artificial peptide sequences containing only 20 natural amino acids with the maximum possible conformational stability at a given pH, ionic strength, temperature, peptide solubility. It also accounts for restrictions due to known amino acid residues involved in stabilization of protein complexes under consideration. The results indicate that a few key intermolecular interactions inside the RecA protein presynaptic complex are enough to reproduce the main features of the RecX protein mechanism of action. Since the SOS-response provides a major mechanism of bacterial adaptation to antibiotics, these results open new ways for the development of antibiotic co-therapy that would not cause bacterial resistance.
INTRODUCTION
In addition to its role in recombinational DNA repair, bacterial RecA proteins are involved in the mechanism of bacterial resistance to antibiotics as the activator of a bacterial SOS-response. RecA is thus a good target for development of compounds blocking the bacterial SOS-response (1–4). In the past decade, several inhibitors of RecA activities in vitro have been discovered including metal cations (5), small organic (6) and inorganic compounds (7) as well as chemically modified peptides (8). Recently the first in vivo study of RecA inhibitors based on cyanine tetrasulfonate has been reported (9). However due to various reasons, still there is no reliable solution to the problem of combating bacterial antibiotic resistance by inhibiting RecA protein activities.
The RecX protein is known to be a very active natural RecA protein inhibitor. RecX will suppress various RecA activities including adenosine triphosphate (ATP) hydrolysis and DNA strand exchange reaction at concentrations hundreds of times smaller than that of the RecA itself. This is a direct result of RecX-mediated disassembly of RecA filaments in vitro (10–12). Since RecX is a small protein inhibiting RecA in the absence of any co-factors, it provides a good basis to develop a new class of peptide inhibitors of the protein. Over the past decade, new insights on the structure and function of RecX have been published and several mechanisms of RecA inhibition by RecX have been proposed (11,13–16). However, details of the mechanism of RecX action still remain unclear. For instance, RecX proteins are able to interact with DNA. However, it is not clear how these interactions relate to the RecX inhibition of RecA proteins. According to one model, RecX suppresses RecA filamentation on the DNA by finding a gap in the filament and binding to the nearest RecA monomer, thereby blocking monomer polymerization along the DNA strand (14). Another model suggests that RecX interacts with RecA monomers randomly along the whole filament and provokes RecA dissociation at the site of interaction (11). It was earlier obtained the spatial model of the nucleoprotein complex, where the RecX protein interacts both with DNA and RecA protein (16). Available data show that spatial structure of the RecX protein from Escherichia coli contains nine α-helices, one or more of which presumably bind within the RecA filament groove. Thus, α-helical peptides are good candidates to mimic RecX protein activities. Peptides are widely used to disrupt α-helix-mediated protein–protein interactions (17–21). However, typically α-helical peptides derived from α-helices of natural proteins are not stable enough and usually require additional stabilization using chemical modifications or special sequence motifs of high helical propensities.
Recently we have developed a method (SEQOPT) for the design of α-helices of maximum stability in short monomeric peptides (22–24) using global sequence optimization. Unlike other approaches used to increase conformational stability of protein α-helices by adding a few stabilizing interactions to the protein structure, this method deals with all possible sequences of 20 natural amino acids and selects the best one from them. The method can also account for pH, ionic strength, temperature and restrictions due to peptide solubility and known amino acid motifs required for desired activity.
In this work, using the RecX structure and SEQOPT, we designed and experimentally tested (both in vitro and in vivo) a novel peptide inhibitor with highly stable α-helical structure capable not only of inhibiting the RecA protein activities in vitro but also of suppressing the bacterial SOS-response in E. coli cells.
MATERIALS AND METHODS
Molecular modeling and bioinformatics
Molecular structure visualization, analysis and protein sequence alignment was carried out using the Molsoft ICM Pro software package (25). Peptides were designed using SEQOPT software for global sequence optimization to maximize α-helix stabilizing interactions of peptide (available on http://mml.spbstu.ru/seqopt/) (22–24). The SEQOPT method generates amino acid sequences with the maximum possible conformational stability at any given environmental conditions (temperature, pH and ionic strength) and arbitrary set of fixed amino acids if they are necessary for the functional activity of the peptide. The spatial structure of RecA:ssDNA:RecX complex was taken from the recently published work (16).
Circular dichroism (CD) measurements
Circular dichroism (CD) measurements of the peptides were done with JASCO J-815 spectropolarimeter. CD spectra were recorded at 22°C in a cell with a 1 cm path length. Each peptide was dissolved in Milli-Q water (stored in oxygen-free glovebox for 3 h before measurements) at a concentration of ∼10 μM. The estimations of α-helical content were obtained by deconvolution of the CD spectra using CDNN 2.1 software and its default set of parameters.
Strains and plasmids
AB1157 (thr-1 leuB6 ara14 proA2 hisG4 argE3 thi-1 supE44 rpsL31) and recombination deficient JC10289 (as AB1157 but Δ[recA-srlR306]::Tn10=ΔrecA306) and GY6871 (of AB1157 ancestry) with the relevant genotype sfiA::lacZ from R. Devoret’s collection. Genes recXEc or 4E1 were cloned in pET21b. Plasmid pT7 [original name is pT7POL26] codes for T7 RNA polymerase under the control of a lac promoter. This plasmid was used to overproduce the wild-type RecX or 4E1 proteins under conditions of lac promoter induction by 0.5 mM Isopropyl β-D-1-thiogalactopyranoside (IPTG). Plasmid carried 4E1 peptide was ordered from Eurogen company (Russia).
Protein purification
The wild-type E. coli RecA and RecX proteins were purified as previously described (14,26). Single-strand binding (SSB) protein was kindly provided by Prof. M. Cox (University of Wisconsin-Madison) Their concentrations were determined using native extinction coefficients: ϵ280 = 2.23 × 104 M−1cm−1 for RecA protein (27), and ϵ280 = 2.38 × 104 M−1cm−1 for SSB protein (28). The concentration of E. coli RecX was determined from the absorbance at 280 nm using the native extinction coefficient 2.57 × 104 M−1cm−1 (13). 4E1, and Pep2-4 peptides extinction coefficient calculated by ‘protein calculator v3.4’ was 0.384 × 104 M−1cm−1, and 0.128 × 104 M−1cm−1 respectively. Solid-phase synthesized peptides were ordered from Peptide 2.0 Inc. (USA).
ssDNA-dependent ATP hydrolysis (ATPase) assays
A coupled enzyme spectrophotometric assay was used to measure RecA-mediated ATP hydrolysis. The ADP generated by hydrolysis was converted back to ATP by a regeneration system of pyruvate kinase and phosphoenolpyruvate (PEP). The resultant pyruvate was converted to lactate by lactate dehydrogenase using NADH as a reducing agent. The conversion of NADH to NAD+ was monitored by decrease in absorbance at 380 nm. The amount of ATP hydrolyzed over time was calculated using the NADH extinction coefficient ϵ380 = 1.21 mM−1cm−1. The assays were carried out by Varian Cary 5000 spectrometer, with a temperature controller and path length 1 cm. All ssDNA-dependent ATPase assays contained a reaction solution of 25 mM Tris-OAc (pH 7.5, 80% cation), 10 mM MgOAc, 2 mM ATP, 3 mM potassium glutamate, 5% w/v glycerol, 1 mM dithiothreitol (DTT), 3 mM PEP, 30 U/ml pyruvate kinase, 30 U/ml lactate dehydrogenase, 4.5 mM NADH and 5 μM poly(dT). The wild-type RecA, (3 μM) was preincubated with ATP and 5 μm poly(dT) for 5 min before APTase measurements.
Single-molecule assay
Single-molecule DNA manipulation and measurement of RecA nucleoprotein filaments disassembly were performed using a custom-built dual-trap optical tweezers setup and a four-channel laminar flow cell that have been previously described (29). In brief, two optical traps were formed by splitting a laser beam from Nd:YVO4 1064 nm CW laser source (5 W, BL-106C, Spectra Physics) in two beams of orthogonal polarization and subsequent focusing with a high numerical aperture (NA) oil immersion objective (100×, 1.25 NA, LOMO). One of the traps can be positioned using a piezo-mirror in the range of tens of micrometres with nanometre accuracy. A four-channel flow cell was made of a 110 μm double-sided tape sandwiched between a microscope slide and a cover slip. A pattern representing four channels, each 1 mm wide, merged into a common channel, 4 mm wide, was cut manually out of a double-sided tape.
A 4-channel flow cell design allowed for efficient DNA catching and rapid changing of the reaction conditions. First channel contained 50 fM of streptavidin-coated polystyrene beads (2.1 μm, Spherotech), second channel—5 pM of Lambda DNA (New England Biolabs) end labeled with biotinylated nucleotides (Biotin-16-dCTP, Jena Bioscience) using Klenow fragment (Thermo Scientific), third channel—400 nM of RecA and 1 mM of ATP (Sigma), fourth channel—400 nM of RecA, 1 mM of ATP and 10 μM of peptide. Experiments were performed in a buffer containing 10 mM Tris–HCl pH 7.5, 1 mM MgCl2 at 37°C.
Two beads were optically trapped in the first channel of the flow cell and a single biotinylated DNA molecule was attached to the beads in the second channel. After a tether was formed, beads were moved to the third channel to form RecA–DNA nucleoprotein filaments. Since RecA nucleation on dsDNA is complicated by a large kinetic barrier (30), the DNA molecule was subjected to a 45 pN stretching force in order to facilitate RecA binding. RecA binding was registered as a DNA length extension. DNA was considered to be saturated with bound RecA when DNA length extension reached its maximum value (typically 1.43). Subsequently, stretching force was lowered down to 4 pN which leaded to DNA extension drop to the typical value of 1.35–1.37, DNA was moved to the channel containing peptide and the DNA length dynamics was registered.
Strand exchange reaction
Three-strand exchange reactions were carried out in 25 mM Tris-OAc buffer (pH 7.5, 80% cation), 1 mM dithiothreitol, 5% (w/v) glycerol, 3 mM potassium glutamate, 2 mM ATP and 10 mM of Mg(OAc)2. ATP regeneration system of 30 units/ml pyruvate kinase and 5 mM phosphoenolpyruvate was also included in reactions. All incubations were carried out at 37°C. The wild-type RecA (3 μM) were pre-incubated with ATP and 5 μm M13mp18 circular ssDNA for 5 min. SSB protein (0.5 μM) was then added, followed by another 5-min incubation. The reactions were initiated by the addition of M13mp18 linear dsDNA to 10 μM. Peptides (4 μM) were added 5 min before dsDNA. Aliquots of 20 μl of the reaction mixture were collected and stored on ice. Thereafter, the aliquots were incubated in 1% sodium dodecyl sulphate and proteinase K (2 mg/ml) for 30 min and analyzed in 0.9% agarose gel.
UV radiation sensitivity
Escherichia coli cells carrying plasmid 4E1 or RecX were grown in Lysogeny broth (LB) media with 100 mg/l ampicillin for OD600=0.4–0.6, followed by induction of protein synthesis with 0.5 mM IPTG and subsequent incubation for 1.5 h at 37°C. After that, 100 μl of appropriate dilutions were spread onto LB plates. Dilutions for samples/treatments were empirically determined. The plates were then exposed to ultraviolet (UV) in a calibrated Chromoscope UV to the dose indicated. After incubating at 37°C overnight, the colonies were counted and divided by the dilution factor to get cfu/ml. For percent survival, colony counts on the treated plates were divided by the counts on untreated plates.
SOS-response reaction activity
Induction of the SOS response was monitored using an assay measuring synthesis of β-galactosidase as previously described (31,32) using the strain GY6871 sfiA::lacZ carrying appropriate plasmids (33). In order to make these measurements, the β-galactosidase gene was integrated under the sfiA promoter so that SOS-response activity could be judged by the level of expression of the β-galactosidase caused by nalidixic acid. Cultures were grown in LB-media containing 100 μg/ml ampicillin up to OD600=0.4–0.6, followed by induction of protein synthesis with 0.07 mM of IPTG and subsequent incubation for 1 h at 37°C. After that fractions were divided and Nalidixic acid was added to the final concentration 40 μg/ml and following induction for 2 h. About 1 ml of the cells was added to 1 ml of Z-buffer and reaction was initiated by 0.4 ml (4 mg/ml) of ortho-Nitrophenyl-β-D-galactoside. The reaction was stopped by 1 ml 1 M Na2CO3 (pH 12) and the extract was clarified by centrifugation. The so-called Miller units are standard values to measure the protein activity and in that way bacterial SOS-response. Typically SOS-response increase Miller units in 7–100× relative to its baseline values (33,34).
RESULTS
The peptide design
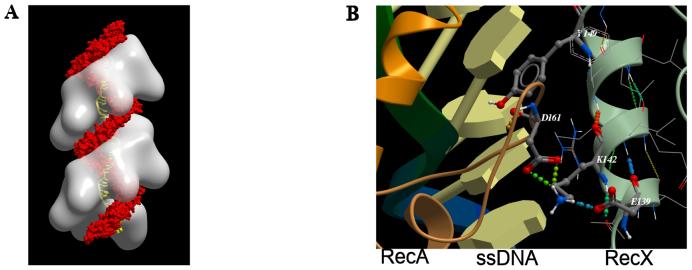
Analysis of the spatial structure of the RecA:ssDNA:RecX complex (16) (Figure 1A) showed that the most promising α-helix of RecX protein for the purposes of design of potent inhibitor of RecA protein was the one between amino acid positions #139 and #150 in the RecX primary structure. First of all, it is the most deeply situated α-helix inside the RecA filament groove structure as compared with other α-helices of the RecX protein. Additionally, this α-helix is involved in intermolecular hydrogen bonds with D161 of the RecA protein, known to be involved in RecA binding selectivity and intercalation between ssDNA bases (35).
Figure 1.
Molecular models of RecA:ssDNA:RecX filament (16). (A) Structure of RecA:ssDNA:RecX filament (RecA: white; RecX: red; DNA: yellow). (B) The fragment of full atomic model of RecA:ssDNA:RecX filament complex.
In order to elucidate a possible consensus motif of the peptide sequence, we analyzed RecX sequences available in the UniProt database (36). There are now 279 full sequences of RecX proteins identified in the database from different bacterial sources. Almost all RecX proteins exhibit a high degree of homology in the α-helix under consideration. However multiple sequence alignment of the dataset of RecX proteins showed that some proteins have an insertion in this part of their primary structure. Such proteins were removed from the dataset as well those having identical sequences in this α-helix. The remaining RecX proteins (a total of 109, not containing gaps in selected region) were analyzed for the presence frequency of 20 natural amino acid at different positions of the chosen α-helix. The consensus motif of the chosen RecX α-helix was found to be ‘*+*+∼**#L**+’, where *—any amino acid, +—positively charged amino acids (K or R), ∼—aliphatic amino acids (V, I, L, M or P) and #—aromatic amino acids (F, Y, W or H). These results show that many amino acids positions of the α-helix exhibit no preference for any specific amino acid.
The SEQOPT method is a new approach under development in our laboratory for global sequence optimization of protein α-helices which allows in silico generation of amino acid sequences of maximum possible conformational stability at any given set of conditions (temperature, pH and ionic strength). An arbitrary set of fixed amino acids can be included if they are necessary for the functional activity of the peptide (22–24). In this work SEQOPT was used to improve the stability of the α-helix sequence derived from the RecX protein (residues #136–153), containing 18 amino acid residues, where E139, K140, V141, K142, I143, R145, L147, L148, Y149 and R150 were fixed to maintain consensus motif and mentioned atoms interactions, and the remaining eight residues were allowed to vary by optimization algorithm in order to increase the α-helical conformational stability of the peptide structure while also providing for high solubility in aqueous solution (see Table 1). This was also expected to increase the fraction of active peptide and to protect it from degradation by proteases in vivo. In the course of optimization, the observed stabilizing interactions of side-chains Y149 (stacking interaction with DNA) and K142 of (H-bonding with RecA’s D161) were preserved (Figure 1B). Sequence optimization was done assuming a temperature of 310K, a pH of 7.5 and an ionic strength of 0.1 M. The sequence optimization resulted in a series of peptide sequences with high α-helical content and good solubility. From those we selected a sequence that we named 4E1 which also matches observed consensus motif of derived from RecX protein (see discussion above). For testing as potential negative controls, we selected three additional peptides (named Pep2, Pep3 and Pep4), also having high predicted stability, solubility and high similarity to 4E1 but not fully matching the consensus motif.
Table 1. The amino acid sequences of the peptides designed to inhibit RecA protein and theoretical and experimental estimates of their stability.
| Amino acid sequence (position # as in RecX from E. coli) | HC, % (theor.) | HC, % (exp.) | |
|---|---|---|---|
| #AA | 136-137-138-139-140-141-142-143-144-145-146-147-148-149-150-151-152-153 | ||
| RecX | Val-Phe-Ser-Glu-Lys-Val-Lys-Ile-Gln-Arg-Phe-Leu-Leu-Tyr-Arg-Gly-Tyr-Leu | 3.9 | N/S |
| 4E1 | Glu-Glu-Glu-Glu-Lys-Val-Lys-Ile-Leu-Arg-Tyr-Leu-Leu-Tyr-Arg-Leu-Ile-Tyr | 84.7 | 76 |
| Pep2 | Glu-Arg-Glu-Glu-Lys-Glu-Lys-Arg-Arg-Arg-Glu-Glu-Glu-Tyr-Arg-Arg-Arg-Met | 91.1 | 81 |
| Pep3 | Glu-Leu-Glu-Glu-Lys-Val-Lys-Arg-Leu-Arg-Glu-Glu-Leu-Tyr-Arg-Arg-Ile-Met | 84.9 | 81 |
| Pep4 | Glu-Glu-Glu-Glu-Lys-Arg-Lys-Arg-Leu-Arg-Glu-Glu-Leu-Tyr-Arg-Arg-Ile-Met | 93.1 | 86 |
RecX/#aa—the names and the position numbers of the RecX protein amino acid residues from E. coli. The amino acids that were fixed in the optimization are shown in bold.
CD measurements of peptide conformational stability
The sequences of all the peptides under consideration and their stability data are shown in Table 1. Experimental verification of their α-helix stability was done by CD spectroscopy (Figure 2). The CD spectra of all the peptides show typical α-helical shape with minima at 208 and 222 nm with high α-helical content. Since the CD spectra of a short peptide may contain contributions from several different conformations, the recorded CD spectra were analyzed using CDNN v.2.1 software. The results of this analysis are also shown in Table 1. As one can see from Figure 2 and Table 1, all peptides are highly helical with α-helical content approaching 80%. Theoretical predictions show a bit higher helical content to the peptides with optimized sequences, which is in agreement with our previously reported results (22). However, the correlation between theoretical and experimental data is very good (R = ∼0.8), indicating that the theoretical model accurately reproduces the stability of the peptides with optimized sequences under physiological conditions.
Figure 2.
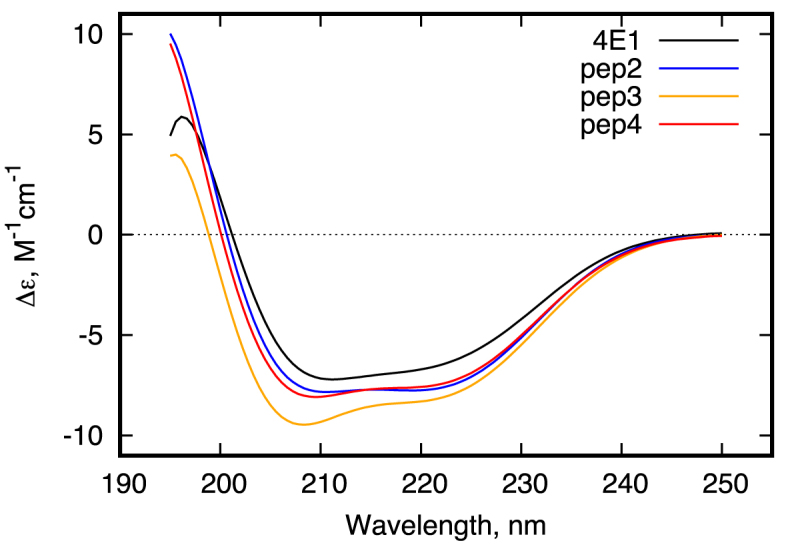
Far-UV CD spectra of 4E1 (black) and Pep2-4 (colored) peptides.
4E1 peptide inhibits RecA ATPase activity
Common strategies to characterize RecA protein activity in vitro include measurements of ATPase activity and DNA strand exchange. We also employed single-molecule observations using optical tweezers.
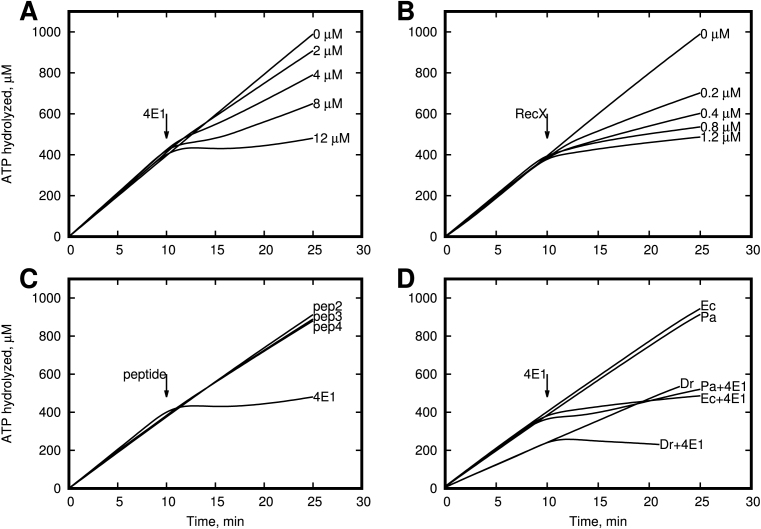
It is known that the RecA protein is a DNA dependent ATPase. The RecA protein hydrolyzes ATP when bound to DNA, and the rate of ATP hydrolysis generally correlates well to the amount of RecA bound to the DNA. Therefore, we checked the ability of the 4E1 and Pep2 peptides to inhibit RecA ATPase activity in the presence of ssDNA. We specially used poly-dT to avoid the additional complexity with natural DNA, which has a secondary structure. In addition, poly-dT eliminates the need for SSB protein, as has been shown previously is an antagonist of RecX protein (37) and can therefore make it more difficult to obtain a quantitative comparison of our peptides and RecX. This experiment was repeated three times with similar results. The time point of the 4E1 and RecX addition is indicated on Figure 3A and B with an arrow (10 min). As shown in Figure 3A, in the absence of any inhibitors the amount of hydrolyzed ATP increases proportionally to reaction time. Adding RecX protein (Figure 3B) or 4E1 peptide to the reaction mixture of RecA protein and poly (dT) leads to a dramatic reduction of the rate of ATP hydrolysis. Also, Figure 3A and B shows that the 4E1 peptide is a less effective inhibitor than RecX protein by a factor of ∼10- to 20-fold, assuming equal molecular concentrations. However, we have to note that unlike RecX protein, which binds to three consecutive RecA protomers, 4E1 peptide interacts with one RecA protomer only. Thus, the the first order estimation of effective difference in observed inhibitor efficiency of 4E1 peptide is about three times less relative to full-length RecX protein. As expected, we observed no effect with Pep2-4 peptides when they were employed in negative control experiments (Figure 3C). Additionally, we observe the ability of 4E1 peptide inhibit ATPase activity of RecA protein from Pseudomonas aeruginosa and Deinococcus radiodurans (Figure 3D). To elucidate the specific binding of 4E1 peptide with RecA filament, we performed experiments with concurrent RecA inhibition by the 4E1 peptide in the presence of 5 μM DinI protein which is known as strong RecX antagonist (10). As shown in Figure 4 we observed no 4E1 activity in the presence of DinI indicating that similar to RecX the 4E1 peptide and DinI protein compete for the same binding site in the RecA filament.
Figure 3.
Effect of α-helical peptide 4E1 on ATP activity of the RecA protein in vitro. (A) 4E1 peptide inhibits RecA ATPase activity on poly-(dT). In the absence of any inhibitors the amount of hydrolyzed ATP increases proportionally to reaction time. (B) RecX protein inhibits RecA ATPase activity on poly-(dT). In the absence of any inhibitors the amount of hydrolyzed ATP increases proportionally to reaction time. (C) Peptides Escherichia coli RecA ATPase activity on poly-(dT). In the absence of peptide 4E1 (12 μM) the amount of hydrolyzed ATP increases proportionally to reaction time. (D) RecA protein from E. coli, Deinococcus radiodurans and Pseudomonas aeruginosa ATPase activity on poly-(dT) inhibited by peptide 4E1 (12 μM).
Figure 4.
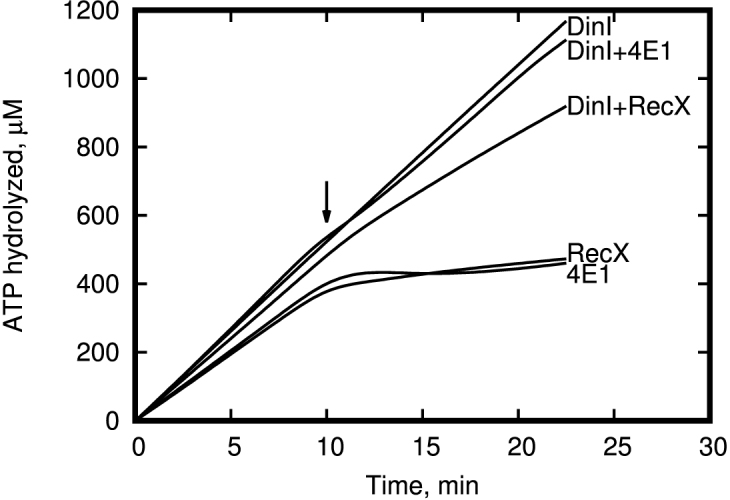
Effect of peptide 4E1 and RecX on ATP activity of the RecA+DinI complex in vitro. 4E1, RecX and DinI concentration was 12, 1.2 and 5 μM respectively.
4E1 peptide promotes disassembly of RecA–DNA nucleoprotein filaments
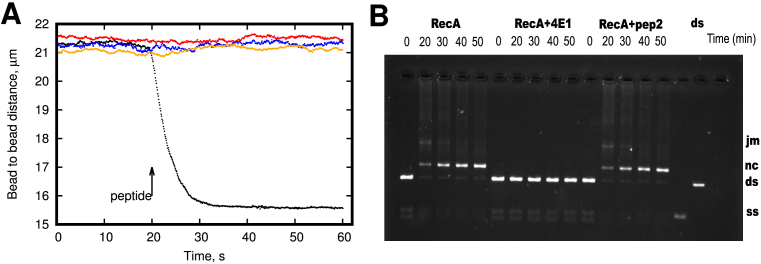
It is known that the RecA protein binds ssDNA and dsDNA and forms long helical nucleoprotein filaments, which is a critical step in homologous recombination and initiation of the bacterial SOS-response(38). In order to investigate the effect of the 4E1 peptide on RecA–DNA interaction, a single-molecule approach combining optical trapping and DNA manipulation in a laminar flow cell was utilized (29). A biotinylated dsDNA molecule was attached to two optically trapped streptavidin-coated beads and manipulated using dual-trap optical tweezers. First, DNA was introduced into the RecA containing channel supplied with Mg2 + and ATP. RecA nucleoprotein filaments assembly was initiated by applying a 45 pN stretching force to DNA, resulting in DNA length extension since DNA binding by RecA increases the DNA length by about 50% compared to the B-form helix. After saturation of DNA with bound RecA, a stretching force was reduced to about 4 pN and a peptide inhibitor was added to the reaction by moving the DNA–RecA complex into the channel containing RecA, Mg2 +, ATP and 4E1 peptide. In the presence of 4E1, the length of DNA was rapidly decreased to the value of bare DNA corresponding to the total dissociation of RecA from DNA. Negative controls using the Pep2-4 peptides produced no RecA dissociation under identical conditions (Figure 5A). Each experiment was repeated three times with identical results. Based on this observation we suggest that 4E1 peptide inhibits RecA DNA binding activity and promotes disassembly of RecA–DNA nucleoprotein filaments in vitro. This result indicates that 4E1 is not a RecA ATPase inhibitor, but instead acts as a stimulator of filament disassembly.
Figure 5.
Effect of α-helical peptides 4E1 and pep2-4 on RecA ATPase activity on poly-(dT) in vitro. (A) DNA length dynamics upon addition of 4E1 and Pep2-4 peptides. A single DNA molecule with prebound RecA is stretched by a low force (4 pN) and moved to the channel containing 10 μM 4E1 peptide (black dots). In the presence of 4E1 peptide DNA length rapidly decreases down to the value corresponding to the length of bare DNA showing inhibitory effect of 4E1 peptide on RecA–DNA interactions. Negative control showed no inhibition of DNA binding by RecA in the presence of the 20 μM Pep2-4 peptides (colored dots). (B) 4E1 peptide inhibits the RecA-mediated DNA strand-exchange reaction. In the absence of peptide 4E1 reaction completes in about 45 min—almost all DNA convert to nicked circular (nc). With peptide 4E1 (on the center) reaction is not initiated and no nicked circular DNA observed. Ds: double strand DNA, ss: single strand DNA.
4E1 peptide inhibits RecA-mediated DNA strand exchange
To investigate the functional significance of 4E1 peptide interaction with RecA, we measured the effect of 4E1 peptide on RecA recombinase activity in vitro. The agarose gel assay for DNA strand exchange between circular ssDNA and linear dsDNA allows for the separation of intermediate joint molecule DNA species from nicked circular heteroduplex DNA (final product)(26). This experiment was repeated five times with identical results. Figure 5B depicts the conversion of dsDNA to two intermediate DNA species and then into final products in the process catalyzed by RecA. RecA-promoted DNA strand exchange was measured in the presence or absence of 4E1 peptide. Peptide almost completely abolished formation of nicked circular DNA over a 45-min time course between homologous circular ssDNA and linear dsDNA molecules. This observation suggests that 4E1 peptide prevents the RecA–DNA complex formation and acts as an effective inhibitor of the strand exchange reaction. In the absence of 4E1 peptide, the RecA protein reaction is complete in about 45 min—converting almost all of the substrate DNA to a nicked circular (nc) product. When adding 4E1 peptide (on the center) before the double-stranded DNA, the reaction not initiated and joint-molecule (jm) and nc intermediates not formed.
4E1 peptide alters UV resistance
The above findings suggest that 4E1 peptide inhibits RecA activity in vitro. To explore the ability of the 4E1 peptide to work within bacterial cells we performed in vivo experiments.
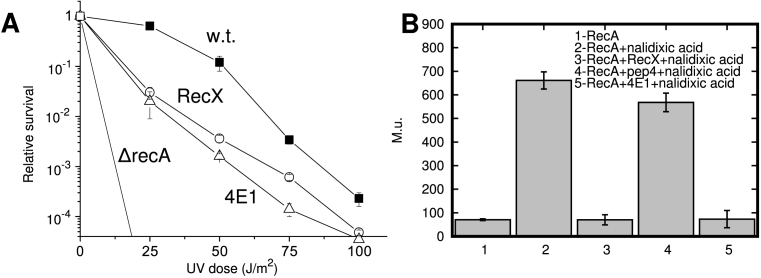
It is known that blocking the activity of RecA protein by high level expression of the RecX in E. coli cells dramatically reduces their ability to survive under the influence of high doses of ultraviolet radiation (34). When expressed from the chromosome, survival may increase slightly, but overexpression from a plasmid leads to a drop in survival of 2–2.5 orders of magnitude when cells are exposed to UV irradiation (34). Certainly, the maximal effect from UV is observed with a strain without recA gene (ΔrecA). However, Figure 6A shows that survival curve for the cells carrying the 4E1 peptide gene overexpressed from plasmid pET21b-4E1 is similar to that seen in cells in which the RecX gene is overexpressed from plasmid pET21b-recXEc. Calculations of uncertainty of relative survival values were determined as deviations from the average values by making use of the standard deviation and by inputting the values from independent repeats of three experiments. In this comparison, the inhibition activity of peptide 4E1 in vivo is very similar to that of RecX protein. It has been shown previously that depending on the amount of the expressed RecX, it can either increase cell survival or inhibit. Thus, this experiment demonstrated that the peptide is stable inside cells and interacted with a RecA–DNA filament with high efficacy. The experiment does not address the sensitivity of the peptide to proteolysis, but its quantity is sufficient for a significant effect on RecA function.
Figure 6.
Effect of α-helical peptide 4E1 on UV resistance of Escherichia coli and the bacterial SOS-response in vivo. (A) Escherichia coli cell survival, depending on the dose of UV irradiation in the absence and the presence of expression of RecX protein and peptide 4E1. (B) Effect of RecX and peptide 4E1 expression on the magnitude of SOS-response in E. coli GY6871 cells in vivo when exposed to nalidixic acid (bars #2–#5). In the absence of IPTG-dependent RecX induction (bar #2) or IPTG-induced pep4 expression (bar #4) SOS-response increased in eight to nine times relative to its baseline values (bar#1), as measured in Muller units(M.u.).
4E1 peptide blocks SOS response
There are 40 SOS response genes under the control of LexA repressor. Formation of RecA filaments triggers proteolysis of LexA and triggers the induction of these genes. The RecA pathway is the only known way in E. coli for SOS-response activation. We tested the ability of the 4E1 peptide to block bacterial SOS-response in vivo using the standard Miller method (31). Miller units are proportional to the increase of O-nitrophenol production by a bacterium in the presence nalidixic acid. Calculations of Muller units were determined as deviations from the average values by making use of the Student’s t-distribution and by inputting the values from independent repeats of three experiments.
Typically, expression of RecA results in an SOS-response of about 600 Miller units at a given concentration of nalidixic acid (40 μg/ml) and incubation at a predetermined time (2 h) and 0.07 mM IPTG. However, expression of both the RecX protein or the 4E1 peptide inhibited the activity of RecA and led to inhibition of the SOS response (Figure 6B, measurements number #3,5) almost to baseline value (bar #1). Baseline level (bar #1) was obtained using strain carrying pET21b-recXEc plasmid with no-IPTG and no nalidixic acid induction.
Based on this observation we suggest that 4E1 peptide effectively functions in vivo.
DISCUSSION
We report on a short α-helical peptide that reprises most of the regulatory functionality of the entire RecX protein in 20 amino acids. The choice of the α-helix for designing the peptide and its further modification was based on the previously published model of the RecX:RecA:ssDNA structure, in which RecX interacts with ssDNA inside the groove of the filament(37). The binding occurs mainly via Coulombic interactions between RecX and ssDNA. We selected the helix positioned most deeply within the RecA filament and in direct contact with DNA. Thus, the results provide experimental proof of the structure mentioned earlier. We also report on a consensus motif of the RecX α-helix between amino acids #139 and #150. Nevertheless, as have been shown, there are three positions in 4E1 peptide predominantly occupied by positively charged residues, two other positions for aromatic residues and other two are almost exclusively occupied by Leu and Gly. The presence of Gly would strongly decrease the stability of the α-helical conformation of a short peptide. However, our results indicate that the Gly is not essential at that position and can be effectively replaced with Leu (and maybe with some other amino acid residues) without affecting the peptide activity. Clearly, only future investigations can determine the set of essential amino acids for the activity of the peptide. It is clear from the ineffectiveness of peptides 2–4 that the activity of peptide 4E1 is sequence dependent.
According to our data, it is clear that interaction of 4E1 peptide with only one RecA monomer is sufficient for the inhibition process, whereas previously published structural data indicates that one RecX monomer interacts directly with three monomers of RecA (11,16). The role of the other α-helices in binding the two neighboring monomers remains beyond the frames of this project. We speculate that it is possible that RecX amino acid sequence redundancy is required for interaction with other regulatory proteins. For example, it has been previously shown that the RecX protein interacts with RecF (39). The possible role of the remaining part of the molecule is to stabilize the protein in the filament. Assuming that the peptide interacts with the filament in the same way that the amino acid segment 139–150 in RecX interacts with the filament, the 4E1 peptide leaves the inter-monomer interface zone intact. This fact implies a higher level of complexity of RecX function if compared to just inhibiting the filament polymerization in 5′-3′ direction (14). The result implies that the binding of this α-helical peptide deep within the filament groove is sufficient to substantially destabilize the RecA filament. While choosing an α-helix, we focused on the site where RecX is located most closely to the DNA. However, it certainly does not guarantee that other α-helices in the RecX structure, located far from the DNA, cannot also play the role in the inhibition process. Previously, it was shown that RecX itself is able to interact with DNA. Furthermore, SSB protein antagonizes RecX activity, although there is no evidence for any protein–protein interactions between them (37). Taken together, these data indicate that the importance of RecX–DNA interactions cannot be excluded from the mechanism model.
In summary, the results presented here show that α-helical peptide 4E1 with the amino acid sequence: ‘Glu-Glu-Glu-Glu-Lys-Val-Lys-Ile-Leu-Arg-Tyr-Leu-Leu-Tyr-Arg-Leu-Ile-Tyr’ is not only an effective inhibitor of RecA protein in vitro, but is also capable of blocking the bacterial SOS-response in vivo.
The results may suggest an approach to counter the development of antibiotic resistance in bacterial pathogens. Effective peptide delivery to the bacteria can be obtained using known penetrating peptide sequences for bacterial selectivity (40), combined with Exosomes carriers (41) or Cholestosome technology for oral delivery (42). The main drawback of using short peptides as drugs is in their lack of conformational stability. This results in a significant reduction in the specific activity of the peptides and their rapid degradation by proteases. SEQOPT optimization produces the amino acid sequences of the α-helix conformation with a very high stability similar to that in globular proteins even when a large part of the sequence is fixed to preserve its biological activity (22). Due to its clear effectiveness in vivo, peptide 4E1 clearly has sufficient stability and resistance to proteolysis in vivo to ensure function (Figure 6A).
A bacterial genome modification is a key factor involved in drug-resistant bacterial strains formation. Many antibiotics are in their third or fourth modification cycle and it is not clear how many more modifications will be permitted before these antibiotics are rendered ineffective (3). One of the possible ways to prolong antibiotic is co-therapy with SOS-response blocking agents. To our knowledge, 4E1 is the first peptide having such a range of activities both in vitro and in vivo. The 4E1 peptide contains only unmodified natural amino acids and therefore should not have a wide range of side effects typical for other known RecA inhibitors. This suggests a potential for minimal toxicity.
As have been shown, the 4E1 peptide inhibits the SOS-response and consequently the expression of all SOS-response genes. This includes expression of Polymerase V, the primary cause of elevated mutagenesis (43–45), which in turn is a material basis for bacterial adaptation to antibiotics (1,46). Despite the existence of some ‘small organic’ RecA inhibitors (9), there are some clear advantages of the peptide approach (17–21,47). One previous study presented peptides with the capacity to inhibit RecA protein activity in vitro (8). However, no data were reported on their activity in vivo. In addition, 4E1 peptide has a different sequence, structure and mechanism of action. We note that there is a wide range of proteins that interact with the RecA filament groove, including LexA, UmuD, RecX, the λ repressor and others (48). The availability of a peptide that binds deeply in the RecA filament groove may provide a valuable tool in the ongoing exploration of those interactions.
These results open a new very promising way to develop a wide range of bacteria-specific peptides capable to effectively block the SOS response in bacterial cells and its adaptation to new antibiotics. Also, our findings open a new way to discover numerous new targets, based on analyzing protein–protein complex structures and design the stable α-helices capable of preventing complex formation.
ACKNOWLEDGEMENTS
The authors would like to thank Prof. Michael M. Cox (University of Wisconsin–Madison) for SSB, DinI proteins and for proofreading the manuscript.
FUNDING
Russian Foundation for Basic Research [16-34-00518 mol_a to A.Y.].
Conflict of interest statement. It is noted that the subject matter of this publication forms the basis of a patent number RU 2 621 862, filled by authors on 7 July 2016.
REFERENCES
- 1. Cirz R.T., Chin J.K., Andes D.R., de Crécy-Lagard V., Craig W.A., Romesberg F.E.. Inhibition of mutation and combating the evolution of antibiotic resistance. PLoS Biol. 2005; 3:e176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Smith P.A., Romesberg F.E.. Combating bacteria and drug resistance by inhibiting mechanisms of persistence and adaptation. Nat. Chem. Biol. 2007; 3:549–556. [DOI] [PubMed] [Google Scholar]
- 3. McKenzie G.J., Harris R.S., Lee P.L., Rosenberg S.M.. The SOS response regulates adaptive mutation. PNAS. 2000; 97:6646–6651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Nautiyal A., Patil K.N., Muniyappa K.. Suramin is a potent and selective inhibitor of Mycobacterium tuberculosis RecA protein and the SOS response: RecA as a potential target for antibacterial drug discovery. J. Antimicrob. Chemother. 2014; 69:1834–1843. [DOI] [PubMed] [Google Scholar]
- 5. Lee A.M., Singleton S.F.. Inhibition of the Escherichia coli RecA protein: zinc(II), copper(II) and mercury(II) trap RecA as inactive aggregates. J. Inorg. Biochem. 2004; 98:1981–1986. [DOI] [PubMed] [Google Scholar]
- 6. Sexton J.Z., Wigle T.J., He Q., Hughes M.A., Smith G.R., Singleton S.F., Williams A.L., Yeh L.-A.. Novel Inhibitors of E. coli RecA ATPase Activity. Curr. Chem. Genomics. 2010; 4:34–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wigle T.J., Singleton S.F.. Directed molecular screening for RecA ATPase inhibitors. Bioorg. Med. Chem. Lett. 2007; 17:3249–3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cline D.J., Holt S.L., Singleton S.F.. Inhibition of Escherichia coli RecA by rationally redesigned N-terminal helix. Org. Biomol. Chem. 2007; 5:1525–1528. [DOI] [PubMed] [Google Scholar]
- 9. Alam M.K., Alhhazmi A., DeCoteau J.F., Luo Y., Geyer C.R.. RecA inhibitors potentiate antibiotic activity and block evolution of antibiotic resistance. Cell Chem. Biol. 2016; 23:381–391. [DOI] [PubMed] [Google Scholar]
- 10. Lusetti S.L., Drees J.C., Stohl E.A., Seifert H.S., Cox M.M.. The DinI and RecX proteins are competing modulators of RecA function. J. Biol. Chem. 2004; 279:55073–55079. [DOI] [PubMed] [Google Scholar]
- 11. Ragone S., Maman J.D., Furnham N., Pellegrini L.. Structural basis for inhibition of homologous recombination by the RecX protein. EMBO J. 2008; 27:2259–2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bakhlanova I.V., Dudkina A.V., Wood E.A., Lanzov V.A., Cox M.M., Baitin D.M.. DNA Metabolism in Balance: Rapid Loss of a RecA-Based Hyperrec Phenotype. PLoS ONE. 2016; 11:e0154137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Drees J.C., Lusetti S.L., Cox M.M.. Inhibition of RecA protein by the Escherichia coli RecX protein: modulation by the RecA C terminus and filament functional state. J. Biol. Chem. 2004; 279:52991–52997. [DOI] [PubMed] [Google Scholar]
- 14. Drees J.C., Lusetti S.L., Chitteni-Pattu S., Inman R.B., Cox M.M.. A RecA filament capping mechanism for RecX protein. Mol. Cell. 2004; 15:789–798. [DOI] [PubMed] [Google Scholar]
- 15. Yang C.-Y., Chin K.-H., Yang M.-T., Wang A.H.-J., Chou S.-H.. Crystal structure of RecX: A potent regulatory protein of RecA from Xanthomonas campestris. Proteins. 2009; 74:530–537. [DOI] [PubMed] [Google Scholar]
- 16. Shvetsov A.V., Lebedev D.V., Chervyakova D.B., Bakhlanova I.V., Yung I.A., Radulescu A., Kuklin A.I., Baitin D.M., Isaev-Ivanov V.V.. Structure of RecX protein complex with the presynaptic RecA filament: molecular dynamics simulations and small angle neutron scattering. FEBS Lett. 2014; 588:948–955. [DOI] [PubMed] [Google Scholar]
- 17. Estieu-Gionnet K., Guichard G.. Stabilized helical peptides: overview of the technologies and therapeutic promises. Expert Opin. Drug Discov. 2011; 6:937–963. [DOI] [PubMed] [Google Scholar]
- 18. Azzarito V., Long K., Murphy N.S., Wilson A.J.. Inhibition of α-helix-mediated protein–protein interactions using designed molecules. Nat. Chem. 2013; 5:161–173. [DOI] [PubMed] [Google Scholar]
- 19. Moon H., Lim H.-S.. Synthesis and screening of small-molecule α-helix mimetic libraries targeting protein–protein interactions. Curr. Opin. Chem. Biol. 2015; 24:38–47. [DOI] [PubMed] [Google Scholar]
- 20. Wilson A.J. Helix mimetics: Recent developments. Prog. Biophys. Mol. Biol. 2015; 119:33–40. [DOI] [PubMed] [Google Scholar]
- 21. Barnard A., Miles J.A., Burslem G.M., Barker A.M., Wilson A.J.. Multivalent helix mimetics for PPI-inhibition. Org. Biomol. Chem. 2015; 13:258–264. [DOI] [PubMed] [Google Scholar]
- 22. Petukhov M., Tatsu Y., Tamaki K., Murase S., Uekawa H., Yoshikawa S., Serrano L., Yumoto N.. Design of stable α-helices using global sequence optimization. J. Pept. Sci. 2009; 15:359–365. [DOI] [PubMed] [Google Scholar]
- 23. Yakimov A., Rychkov G., Petukhov M.. De Novo Design of Stable α-Helices. Methods Mol. Biol. 2014; 1216:1–14. [DOI] [PubMed] [Google Scholar]
- 24. Yakimov A.P., Afanaseva A.S., Khodorkovskiy M.A., Petukhov M.G.. Design of stable α-helical peptides and thermostable proteins in biotechnology and biomedicine. Acta Nat. 2016; 8:70–81. [PMC free article] [PubMed] [Google Scholar]
- 25. Abagyan R., Totrov M., Kuznetsov D.. ICM—a new method for protein modeling and design: applications to docking and structure prediction from the distorted native conformation. J. Comput. Chem. 1994; 15:488–506. [Google Scholar]
- 26. Cox M.M., McEntee K., Lehman I.R.. A simple and rapid procedure for the large scale purification of the recA protein of Escherichia coli. J. Biol. Chem. 1981; 256:4676–4678. [PubMed] [Google Scholar]
- 27. Craig N.L., Roberts J.W.. Function of nucleoside triphosphate and polynucleotide in Escherichia coli recA protein-directed cleavage of phage lambda repressor. J. Biol. Chem. 1981; 256:8039–8044. [PubMed] [Google Scholar]
- 28. Lohman T.M., Green J.M., Beyer R.S.. Large-scale overproduction and rapid purification of the Escherichia coli ssb gene product. Expression of the ssb gene under λPL control. Biochemistry. 1986; 25:21–25. [DOI] [PubMed] [Google Scholar]
- 29. Pobegalov G., Cherevatenko G., Alekseev A., Sabantsev A., Kovaleva O., Vedyaykin A., Morozova N., Baitin D., Khodorkovskii M.. Deinococcus radiodurans RecA nucleoprotein filaments characterized at the single-molecule level with optical tweezers. Biochem. Biophys. Res. Commun. 2015; 466:426–430. [DOI] [PubMed] [Google Scholar]
- 30. Fu H., Le S., Muniyappa K., Yan J.. Dynamics and Regulation of RecA Polymerization and De-Polymerization on Double-Stranded DNA. PLoS One. 2013; 8:e66712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Miller J. Experiments in Molecular Genetics, Bacterial Genetics—E. coli. 1972; NY: Cold Spring Harbor Laboratory. [Google Scholar]
- 32. Bakhlanova I.V., Ogawa T., Lanzov V.A.. Recombinogenic activity of chimeric recA genes (Pseudomonas aeruginosa/Escherichia coli): a search for RecA protein regions responsible for this activity. Genetics. 2001; 159:7–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Eldin S., Forget A.L., Lindenmuth D.M., Logan K.M., Knight K.L.. Mutations in the N-terminal region of RecA that disrupt the stability of free protein oligomers but not RecA-DNA complexes. J. Mol. Biol. 2000; 299:91–101. [DOI] [PubMed] [Google Scholar]
- 34. Stohl E.A., Brockman J.P., Burkle K.L., Morimatsu K., Kowalczykowski S.C., Seifert H.S.. Escherichia coli RecX inhibits RecA recombinase and coprotease activities in vitro and in vivo. J. Biol. Chem. 2003; 278:2278–2285. [DOI] [PubMed] [Google Scholar]
- 35. Shinohara T., Ikawa S., Iwasaki W., Hiraki T., Hikima T., Mikawa T., Arai N., Kamiya N., Shibata T.. Loop L1 governs the DNA-binding specificity and order for RecA-catalyzed reactions in homologous recombination and DNA repair. Nucleic Acids Res. 2015; 43:973–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. UniProt Consortium UniProt: a hub for protein information. Nucleic Acids Res. 2014; 43:D204–D212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Baitin D.M., Gruenig M.C., Cox M.M.. SSB antagonizes RecX-RecA interaction. J. Biol. Chem. 2008; 283:14198–14204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Stasiak A., Egelman E.H.. Structure and function of RecA-DNA complexes. Experientia. 1994; 50:192–203. [DOI] [PubMed] [Google Scholar]
- 39. Lusetti S.L., Hobbs M.D., Stohl E.A., Chitteni-Pattu S., Inman R.B., Seifert H.S., Cox M.M.. The RecF protein antagonizes RecX function via direct interaction. Mol. Cell. 2006; 21:41–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Rajarao G.K., Nekhotiaeva N., Good L.. Peptide-mediated delivery of green fluorescent protein into yeasts and bacteria. FEMS Microbiol. Lett. 2002; 215:267–272. [DOI] [PubMed] [Google Scholar]
- 41. Shtam T.A., Kovalev R.A., Varfolomeeva E., Makarov E.M., Kil Y.V., Filatov M.V.. Exosomes are natural carriers of exogenous siRNA to Human Cells In Vitro. Cell Commun. Signal. 2013; 11:88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Schentag J., McCourt M., Mielnicki L., Hughes J.. Cholestosome vesicles for incorporation of molecules into chylomicrons. 2016; US Patent App. 14/776,308.
- 43. Robinson A., McDonald J.P., Caldas V.E.A., Patel M., Wood E.A., Punter C.M., Ghodke H., Cox M.M., Woodgate R., Goodman M.F., van Oijen A.M.. Regulation of mutagenic DNA polymerase V activation in space and time. PLOS Genet. 2015; 11:e1005482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Patel M., Jiang Q., Woodgate R., Cox M.M., Goodman M.F.. A new model for SOS-induced mutagenesis: how RecA protein activates DNA polymerase V. Crit. Rev. Biochem. Mol. Biol. 2010; 45:171–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Goodman M.F. The discovery of error-prone DNA Polymerase V and its unique regulation by RecA and ATP. J. Biol. Chem. 2014; 289:26772–26782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Bellio P., Brisdelli F., Perilli M., Sabatini A., Bottoni C., Segatore B., Setacci D., Amicosante G., Celenza G.. Curcumin inhibits the SOS response induced by levofloxacin in Escherichia coli. Phytomedicine. 2014; 21:430–434. [DOI] [PubMed] [Google Scholar]
- 47. Müller M. “Non-chemical” drugs: biologicals, protein therapeutics, vaccines and antisense therapeutics. Clinical Pharmacology: Curr. Topics and Case Studies. 2010; Vienna: Springer; 309–321. [Google Scholar]
- 48. Adikesavan A.K., Katsonis P., Marciano D.C., Lua R., Herman C., Lichtarge O.. Separation of Recombination and SOS Response in Escherichia coli RecA Suggests LexA Interaction Sites. PLoS Genet. 2011; 7:e1002244. [DOI] [PMC free article] [PubMed] [Google Scholar]