Abstract
Defense responses of peanut (Arachis hypogaea) to biotic and abiotic stresses include the synthesis of prenylated stilbenoids. Members of this compound class show several protective activities in human disease studies, and the list of potential therapeutic targets continues to expand. Despite their medical and biological importance, the biosynthetic pathways of prenylated stilbenoids remain to be elucidated, and the genes encoding stilbenoid-specific prenyltransferases have yet to be identified in any plant species. In this study, we combined targeted transcriptomic and metabolomic analyses to discover prenyltransferase genes in elicitor-treated peanut hairy root cultures. Transcripts encoding five enzymes were identified, and two of these were functionally characterized in a transient expression system consisting of Agrobacterium-infiltrated leaves of Nicotiana benthamiana. We observed that one of these prenyltransferases, AhR4DT-1, catalyzes a key reaction in the biosynthesis of prenylated stilbenoids, in which resveratrol is prenylated at its C-4 position to form arachidin-2, whereas another, AhR3′DT-1, added the prenyl group to C-3′ of resveratrol. Each of these prenyltransferases was highly specific for stilbenoid substrates, and we confirmed their subcellular location in the plastid by fluorescence microscopy. Structural analysis of the prenylated stilbenoids suggested that these two prenyltransferase activities represent the first committed steps in the biosynthesis of a large number of prenylated stilbenoids and their derivatives in peanut. In summary, we have identified five candidate prenyltransferases in peanut and confirmed that two of them are stilbenoid-specific, advancing our understanding of this specialized enzyme family and shedding critical light onto the biosynthesis of bioactive stilbenoids.
Keywords: arachidin, hairy root, peanut, prenylation, stilbenoid, Arachidin, plant biochemistry, resveratrol, secondary metabolism, small molecule, stilbenoid, transcriptomics
Introduction
Stilbenoids are phenylpropanoid compounds that accumulate in response to biotic and abiotic stresses in a small number of plant families, including those of grape (Vitaceae), pine (Pinaceae), and peanut (Fabaceae). These compounds serve as phytoalexins and provide protection to the host plant against various microbial pathogens (1). Resveratrol (3,5,4′-trihydroxy-trans-stilbene) is the most studied compound in the stilbene family and has attracted great attention in the scientific community, not only because of its important role for pathogen defense in plants (1) but also because of its numerous bioactivities, including anticancer, cardioprotective, antioxidant, anti-inflammatory, and neuroprotective properties observed in human cell culture and in vivo (2–4). The extent of resveratrol bioavailability in medicinal contexts, however, remains unclear.
Resveratrol is synthesized in peanut, along with stilbenoids conjugated to a prenyl group, a notable modification not common in other stilbene-producing plants (5–7). Resveratrol and prenylated stilbenoids and analog, including arachidin-1, arachidin-2, arachidin-3, and isopentadienyl trihydroxystilbene, were the first peanut stilbenoids described. These were shown to accumulate in kernels, leaves, stems, pegs, and roots upon challenge with microorganisms (5, 6, 8, 9) and to inhibit spore germination of non-pathogenic and pathogenic fungi (5, 6, 8–10). Recently, deep sequencing analyses of the major peanut fungal pathogen Aspergillus flavus in response to resveratrol revealed that this stilbenoid can affect the expression of A. flavus genes leading to abnormal mycelial development (11).
To date, more than 45 prenylated stilbenoids and their derivatives have been detected in peanut tissues subjected to biotic or abiotic stress (7, 12–16). The prenylation of the stilbene backbone is the primary feature that contributes to the diversity of these peanut secondary metabolites. Differences occur in the position of prenylation as well as in subsequent modifications of the prenyl moiety, such as cyclization and hydroxylation. All prenylated stilbenoids and their derivatives in peanut appear to derive from a prenyl group at either position C-3′ or C-4 of the stilbene backbone (7, 12–16). Interestingly, the position of the prenyl group appears to affect the antimicrobial properties of the prenylated stilbenoid. Higher activity against Gram-positive bacteria was demonstrated when the prenyl group was present at stilbenoid position C-4 in contrast to C-3′ (17).
The prenylation of stilbenoids increases their lipophilicities and membrane permeabilities and may have additional impacts on bioactivities. Prenylated stilbenoids have shown equivalent or enhanced bioactivities relative to non-prenylated forms, such as resveratrol, in in vitro studies (18–20). The prenylated stilbenoids arachidin-1 and arachidin-3 showed favorable metabolic profiles when compared with their non-prenylated analogs piceatannol and resveratrol (21). Furthermore, arachidin-1 and arachidin-3 exhibited specific bioactivities not found in their non-prenylated forms, such as inhibiting the replication of rotavirus in HT29.f8 cells (22). Also, prenylated stilbenoids showed higher affinity to human cannabinoid receptors when compared with non-prenylated stilbenoids (21). A recent study comparing the bioactivities of different prenylated resveratrol analog identified arachidin-2 (i.e. 4-isopentenyl-3,5,4′-trihydroxystilbene) as a potential lead compound for Alzheimer's disease treatment due to its multitargeting properties. Arachidin-2 inhibited β-secretase and amyloid-β aggregation and showed antioxidant, neuroprotective, and neuritogenic properties, with no neurotoxicity (23).
Prenyltransferase(s) responsible for these stilbenoid modifications is/are crucial for the biosynthesis of peanut bioactive compounds of interest. To date, however, no gene encoding a stilbenoid-specific prenyltransferase has been identified in plants. Previously, we developed a peanut hairy root culture system to serve as a platform for sustainable production of prenylated stilbenoids (24, 25). Leveraging this system more recently, we characterized biochemically the first stilbenoid-specific prenyltransferase activity from the microsomal fraction of these elicited cultures (26). This prenyltransferase utilizes plastid-derived dimethylallyl pyrophosphate (DMAPP)2 to prenylate resveratrol or piceatannol into arachidin-2 or arachidin-5, respectively, and shares several features in common with flavonoid prenyltransferases reported in other legume species. For instance, all identified flavonoid and stilbenoid prenyltransferase enzymatic reactions require divalent cations as cofactors and show maximum activity at basic pH (26–32). These characteristics and others guided cloning of the peanut prenyltransferase genes described here. We took a dual approach, combining parallel targeted transcriptome and metabolome analyses to isolate prenyltransferase activities. We built a large database of transcript assemblies from RNAseq experiments designed to capture elicitor-induced mRNAs, and we used these sequences to clone potential prenyltransferase cDNAs. Here, we describe the first plant transcripts encoding stilbenoid-specific prenyltransferase enzymes. These catalyze two distinct dimethylallylation reactions in which resveratrol is prenylated at either the C-3′ or C-4 position. These prenylation steps appear to represent the first committed steps for the biosynthesis of the large diversity of prenylated stilbenoids and derivatives described in peanut. Functionalities of the enzymes are uncovered using a transient expression system of Agrobacterium-infiltrated Nicotiana benthamiana leaves, and their subcellular location is proposed from fluorescence microscopy imaging in particle-bombarded onion epidermal cells. We further use available genome assemblies of the two diploid progenitors of Arachis hypogaea to align the prenyltransferase transcripts and estimate their genomic structure.
Results
Identification of resveratrol prenyltransferase activities from the A. hypogaea hairy root transcriptome
The first flavonoid-specific prenyltransferase, SfN8DT-1, was cloned from a cDNA (EST) library of Sophora flavescens cell cultures, and its enzymatic activity was characterized using the microsomal fraction of recombinant yeast (27). Sequence homology to SfN8DT-1 was the basis for discovery of several other flavonoid prenyltransferases, such as SfiLDT and SfG6DT in S. flavescens (29) and LaPT1 in Lupinus albus (30). Our previous work had indicated that resveratrol prenyltransferase(s) in peanut are membrane-bound proteins that utilize DMAPP from the plastidic terpenoid pathway as the prenyl donor (26). These two key features are also observed in flavonoid-specific prenyltransferases identified from other legume species (27–32), suggesting that the sequence of stilbenoid prenyltransferase(s) may share similarity with flavonoid prenyltransferases genes.
To discover and clone peanut prenyltransferase genes, we first built a transcript sequence reference from RNA of our elicited hairy root culture system, and we annotated likely candidate stilbenoid prenyltransferase transcripts by alignment to well-characterized flavonoid prenyltransferases. The RNA-sequencing experiment was designed to capture mRNA temporally associated with stilbene accumulation in the hairy root cultures. We assembled and evaluated a variety of transcript sequences from RNA-Seq (33) reads sets (see “Experimental procedures”) and initially considered any whose translated product aligned to characterized flavonoid prenyltransferase sequences mentioned above. A total of 224 transcripts from our set of 2,591,753 transcript assemblies encoded full-length protein sequences of 101–432 amino acid residues that aligned to the set of flavonoid prenyltransferase sequences over a length of at least 100, with >80% sequence identity. As derived from a tetraploid, we anticipated that these sequences would represent transcripts of unique enzyme genes, as well as homolog, alleles, and potential assembly errors. The cultivated peanut genome is allotetraploid and thought to have been formed as the result of a single hybridization between two closely-related diploid species, Arachis duranensis and Arachis ipaensis. Reference sequence assemblies of the latter diploid genomes were reported recently (34), and these provided a draft proxy reference we used to evaluate and reduce our transcriptome to potentially unique genic loci.
Our previous work had shown that prenyltransferase activities of interest occurred in the plastid (26). We therefore reduced the candidate transcript sequences to 10 whose encoded proteins contain predicted chloroplast targeting peptide sequences according to analyses with both ChloroP (35) and iPSORT (36). All 10 transcripts showed expression in the 9-h elicitor-treated hairy root transcriptome; therefore, we used 9-h elicited hairy roots to clone their cDNAs. PCR using primers designed against the assembled transcript sequences resulted in amplification of three full-length cDNAs (AhR3′DT-1, AhRPT-10a4, and AhRPT-10d4) encoding stilbenoid prenyltransferase candidates (Table 1). We subcloned these candidates into yeast expression vector, pPICZ, for heterogeneous expression in Pichia pastoris. The microsomal fractions of the yeast cultures were employed for prenyltransferase assays. However, prenylation activity using resveratrol as substrate was not detected in any of the recombinant yeast cultures assayed (data not shown). In previous studies, researchers failed to detect flavonoid prenyltransferase activity when the full-length open reading frame (ORF) of GmG4DT was expressed in yeast, whereas the truncated form of GmG4DT without its N-terminal transit peptide showed genistein prenyltransferase activity (37). Similarly, the microsomal fraction of yeast expressing a truncated form of LaPT1, in which the first 44 amino acids were deleted, showed 6-fold higher activity than that of the full-length protein (30). Reasons for these observations may be low tolerance of plant transit peptides in yeast, correlated with incorrect folding and decreased stability of the prenyltransferases, resulting in low enzymatic activity. A coumarin-specific prenyltransferase, PcPT from Petroselinum crispum, failed to be expressed in yeast but was successfully expressed and functionally characterized using Agrobacterium-infiltrated Nicotiana benthamiana leaves (38). Here, we likewise shifted to a heterologous plant expression system to characterize the peanut enzymes.
Table 1.
Prenyltransferase transcripts described in this study
Reaction product of the enzyme encoded by each transcript and the loci of best alignment in diploid Arachis reference genomes are shown.
| cDNA clone | GenBankTM accession no. | Product | Alignment to proxy genome |
|---|---|---|---|
| AhR4DT-1 | KY565244 | Arachidin-2 | A. duranensis A08, 40,225,499–40,234,019, −8520 bp |
| AhRPT-9i2 | NDa | ||
| AhR3′DT-1 | KY565245 | 3-Methyl-2-butenyl-3′-resveratrol | A. ipaensis B08, 114,687,148–114,693,743, −6595 bp |
| AhRPT-10a4 | ND | ||
| AhRPT-10d4 | ND | ||
| AhR3′DT-2 | KY565246 | 3-Methyl-2-butenyl-3′-resveratrol | A. ipaensis B08, 119,033,572–119,042,841, +9269 bp |
| AhR3′DT-3 | KY565247 | 3-Methyl-2-butenyl-3′-resveratrol | |
| AhR3′DT-4 | KY565248 | 3-Methyl-2-butenyl-3′-resveratrol | A. duranensis A01, 2,413,393–2,427,986, −14.5 Kbp |
a ND means not detected.
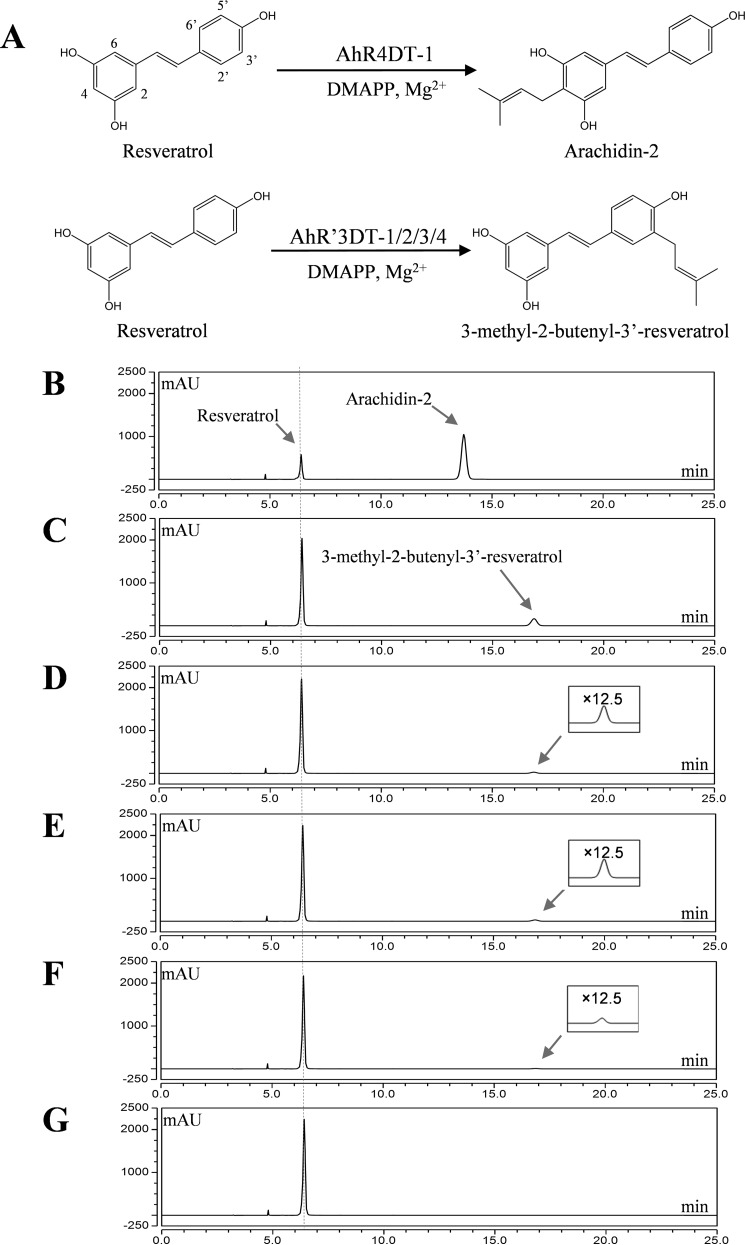
The full-length cDNAs of the three candidate stilbenoid prenyltransferases were subcloned into binary vectors under the control of the CaMV35S-TEV promoter and transiently expressed upon Agrobacterium-infiltration of N. benthamiana leaves. The crude cell extract of N. benthamiana leaves was incubated with DMAPP and resveratrol to test prenyltransferase activities. One of the cDNA products (amplified with primers PT-10-FW-NotI/PT-k-RV-KpnI, Table S1) showed resveratrol dimethylallyltransferase activity (Fig. 1). No activity was observed in the crude cell extract of N. benthamiana control leaves that were infiltrated with Agrobacterium harboring an empty binary vector (Fig. 1). Mass spectrometry analysis of the reaction product (m/z 297 [M + H]+) gave a primary fragment with m/z of 241 [M + H − 56]+ in MS2, which suggested the presence of a prenyl moiety (Fig. 2B; Table S2). After recovery of the reaction product from a large-scale enzymatic assay, it was purified by semi-preparative HPLC, and its structure was further elucidated by NMR analysis.
Figure 1.
Enzymatic characterization of recombinant stilbenoid prenyltransferases expressed in N. benthamiana. A, AhR4DT-1 and AhR′3DT-1/2/3/4 from peanut catalyzes the 4- and 3′-prenylation of resveratrol, respectively. B, enzymatic characterization of resveratrol prenyltransferase transiently expressed in N. benthamiana leaf. HPLCs (UV 320 nm) of ethyl acetate extract of 1 ml of reaction mixture of 100 μm resveratrol, 300 μm DMAPP, 10 mm MgCl2, and 10 mm DTT were incubated with 5 mg of crude protein of N. benthamiana leaf after vacuum infiltration with A. tumefaciens LBA4404 harboring pBIB-Kan-AhR4DT-1 (B), pBIB-Kan-AhR3′DT-1 (C), pBIB-Kan-AhR3′DT-2 (D), pBIB-Kan-AhR3′DT-3 (E), pBIB-Kan-AhR3′DT-4 (F), and pBIB-Kan binary vectors (G) in a pH 9.0 Tris-HCl buffer for 40 min. mAU, milli-absorbance units.
Figure 2.
Structural characterization of prenylated resveratrol synthesized by AhR3′DT-1. A, chemical structure of 3-methyl-2-butenyl-3′-resveratrol. B, mass spectrometry fragmentation analysis of prenylated product, 3-methyl-2-butenyl-3′-resveratrol. C–F, NMR spectra of 3-methyl-2-butenyl-3′-resveratrol isolated from a large-scale enzymatic assay. C, 1D 1H NMR; D, 1D 13C NMR spectrum; E, 2D 1H-13C HMBC; and F, 2D 1H-13C HSQC spectra obtained on 700 MHz Bruker Avance spectrometer dissolved in d6-acetone at 298K. The cross-peaks circled in red corresponds to the protons bonded to the carbon atoms at the positions (2/6, β, α, and 2′) on the resveratrol scaffold.
Because the purified compound was available in extremely low quantities, we continued performing each experiment (1H-13C HMBC and HSQC) over longer time intervals with an increased number of scans. Data obtained showed well-resolved peaks, forming the basis for our unambiguous assignment of both 1H and 13C chemical shifts. In both the 1H and 13C spectra, peaks assigned were in agreement with the original scaffold of resveratrol. To determine the position of the prenyl group on the resveratrol scaffold, the NMR predict tool was used to generate multiple 1H and 13C spectra for various combinations of prenyl positions, with reference to the 4′-hydroxyphenyl ring (39, 40). Two possible prenyl positions appear to be the highest probability and matched well with the experimental 1H and 13C NMR spectra (Fig. 2, C and D). We further narrowed down to the final conformation by examining the 1H signals at 6.3, 6.8, 7.1, and 7.3 ppm, and were they found to be consistent with the resveratrol scaffold. 1H peaks at 3.3 and 5.3 ppm matched well with the prenyl side chain (Fig. 2C). The experimental NMR data strongly support the predicted location of the prenyl at the ortho position of the 4′-hydroxyphenyl ring. The conformation of resveratrol is further corroborated by the 2D NMR that shows 13C peaks at 104.5, 122.8, and 129.0 ppm (Fig. 2D). From the 1H-13C HSQC and 1H-13C HMBC spectrum, the 1H signal at 7.3, 6.8, and 6.3 ppm correlates with protons at the ortho position on 1,3-benzenediol, ethenyl proton, and ortho-positioned proton on 4′-hydroxyphenyl ring, respectively. Their corresponding 13C signals were at 129.0, 128.8, and 128.6 ppm, respectively (Fig. 2, E and F). Analysis of the NMR data suggests that the prenyl moiety is attached to the C-3′ position of the resveratrol backbone to produce 3-methyl-2-butenyl-3′-resveratrol. Thus, the enzyme that catalyzed this reaction was named AhR3′DT-1 (A. hypogaea resveratrol 3′-dimethylallyltransferase).
Interestingly, this prenylated resveratrol was not the arachidin-2 we had expected, given our previous work identifying prenylated products in the reaction using microsomal fraction of elicited peanut hairy root (26). Moreover, the 3-methyl-2-butenyl-3′-resveratrol reaction product described here was not observed in the elicited hairy root culture of peanut. Our results indicate then that there are other prenyltransferase(s) responsible for prenylating resveratrol to arachidin-2 in peanut. To further search for additional resveratrol prenyltransferase(s) from the peanut hairy root transcriptome, other primer pairs were designed based on the alignment of the putative prenyltransferase transcripts (Table S1). Using this approach, additional PCR amplicons were amplified from the cDNA of 9-h-elicited peanut hairy roots, and five were subsequently subcloned into binary vectors and transiently expressed in N. benthamiana leaves for prenyltransferase activity assays using DMAPP and resveratrol as substrates. Four additional cDNAs were determined to encode resveratrol prenyltransferases. One cDNA clone (amplified with primers PT-9-FW-NotI/PT-b-RV-KpnI, Table S1) showed a clear dimethylallyltransferase activity for resveratrol and prenylated it at the C-4 position to form arachidin-2 (Fig. 1). This product was confirmed by comparison of its retention time, UV light spectrum, mass spectra, and fragmentation patterns obtained by tandem mass spectrometry (MS2 and MS3) with arachidin-2 purified from peanut hairy root culture (Fig. S1). Hence, this enzyme was designated AhR4DT-1 (A. hypogaea resveratrol 4-dimethylallyltransferase). The other three cDNA clones (two amplified with primers PT-4-FW-NotI/PT-e-RV-KpnI and one amplified with primers PT-5-FW-NotI/PT-m-RV-KpnI, Table S1) exhibited the same catalytic activities as AhR3′DT-1, converting resveratrol into 3-methyl-2-butenyl-3′-resveratrol using DMAPP as prenyl donor. Hence, we named these AhR3′DT-2, AhR3′DT-3, and AhR3′DT-4. Among these four isoenzymes, the highest amount of reaction product was found with AhR3′DT-1, and therefore this enzyme was used in further characterization studies.
Genomic and phylogenetic relationships of the prenyltransferases
We report here five active resveratrol prenyltransferases identified from biochemical analyses of eight peanut transcripts. Transcripts assembled from RNA-Seq reads and cloned as cDNAs each independently show that these encode polypeptides of 389–414 amino acids (Fig. 3). All possess nine transmembrane α-helices as predicted by TMHMM 2.0 (Figs. S2–S5) (41), as well as two aspartate-rich motifs, NQXXDXXXD in loop 2 and KD(I/L)XDX(E/D)GD in loop 6, that are also conserved in flavonoid prenyltransferases (Fig. 3).
Figure 3.
Primary structures of stilbenoid prenyltransferases. Two conserved NQXXDXXXD and KDXXDXEGD motifs are boxed in red.
The gene structure of peanut prenyltransferases was estimated by aligning the transcripts to available Arachis diploid progenitor genome sequence references (34). Four loci in each became apparent as candidate origins. Three of these are on pseudochromosome 8 in each genome, within a span containing notable gaps and described by Bertioli et al. (34) as effected by a genomic reduction during polyploidization. Only AhR3′DT-4 aligned completely to pseudochromosome 1 (Table 1). Eliminating poor alignments, we estimated that two loci in the A. duranensis subgenome and two in the A. ipaensis subgenome could explain the origin of the set of prenyltransferases we characterized in this study (Table 1). Transcript alignments showed that AhR3′DT-2 and -3 differ by a deletion in AhR3′DT-2 that encodes 16 amino acid residues in AhR3′DT-3 (Fig. 3). Although the genomic reference contained gaps in this region that prevented gene structure validation, we expect that these two transcripts may be expressed from the same gene in A. hypogaea as alternatively spliced forms (Table 1).
Among the eight peanut transcripts tested in N. benthamiana transient assays, three cDNAs failed to show resveratrol prenyltransferase activity. These appear to be alternative splice forms, or expressed from different alleles, of AhR4DT-1 (one transcript isoform) and of AhR3′DT-1 (two transcript isoforms). Observed variation among these may offer preliminary insights into structural requirements for the activities we characterize here. Interestingly, each of the three inactive cDNAs varies in length and sequence at the C-terminal end (Figs. S2 and S3). A nine-amino acid residue deletion at the C terminus of the inactive AhR4DT-1 transcript, AhRPT-9i2, likely reduces its encoded protein structure to eight transmembrane spans (Fig. S2) and suggests that the integrity of the nine transmembrane domains in AhR4DT-1 are essential for its activity. Two inactive transcripts of AhR3′DT-1 encode a C-terminal extension that does not appear to have transmembrane properties (Fig. S3). Each furthermore harbors several coding single-nucleotide polymorphisms and an eight-amino acid residue deletion (Δ41–48) that disrupts a region conserved in both active AhR4DT-1 and AhR3′DT-1 (Fig. 3). Interestingly, although not fully deleted in AhR3′DT-2, -3, and -4, this sequence is highly variable in these forms, suggesting this uncharacterized region may play a role in influencing the prenyltransferase active site.
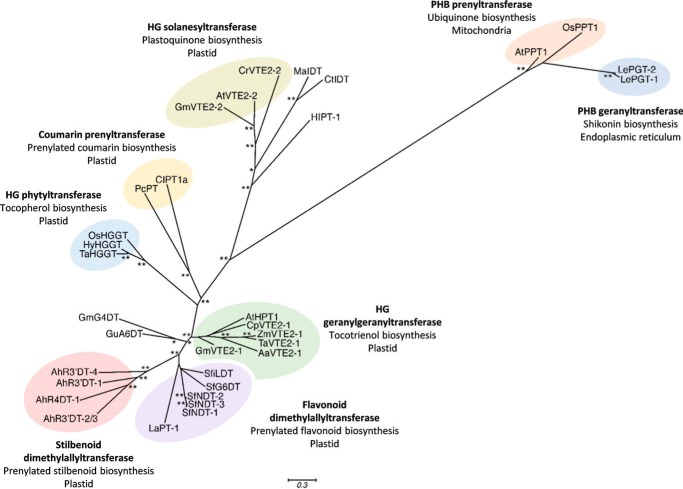
Phylogenetic analysis of these characterized prenyltransferase enzymes together showed that the peanut stilbenoid prenyltransferases form their own monophyletic group (Fig. 4). This clade is notably distinct from the flavonoid and coumarin prenyltransferases, as well as from homogentisate prenyltransferases involved in ubiquinone and shikonin biosynthesis, and p-hydroxybenzoate prenyltransferases involved in the tocotrienol, tocopherol, and plastoquinone biosynthetic pathways (Fig. 4).
Figure 4.
Phylogenetic relationship between peanut stilbenoid prenyltransferases and related prenyltransferases accepting aromatic substrates. Species abbreviations are: Aa, Allium ampeloprasum; Ah, A. hypogaea; At, Arabidopsis thaliana; Cl, Citrus limon; Cp, Cuphea avigera var. pulcherrima; Cr, Chlamydomonas reinhardtii; Ct, C. tricuspidata; Gm, Glycine max; Hl, Humulus lupulus; Hv, Hordeum vulgare; La, L. albus; Le, Lithospermum erythrorhizon; Ma, M. alba; Os, Oryza sativa; Pc, P. crispum; Sf, Sophora flavescens; Ta, Triticum aestivum; Zm, Zea mays. Homogentisate phytyltransferases (VTE2–1s) and homogentisate geranylgeranyltransferases, homogentisate solanesyltransferases (VTE2–2s), p-hydroxybenzoate geranyltransferase, and p-hydroxybenzoate polyprenyltransferases are involved in the biosynthesis of vitamin E, plastoquinone, shikonin, and ubiquinone, respectively. IDT is isoliquiritigenin dimethylallyltransferase. Accession numbers of these proteins are listed in Table S4.
Biochemical characterization of AhR4DT-1 and AhR3′DT-1
AhR3′DT-1 along with AhR4DT-1 were selected for further biochemical characterization. Similar to the previous resveratrol prenyltransferase activity characterized from peanut hairy roots (26), the specific activity in the microsomal fraction of N. benthamiana leaves expressing AhR4DT-1 or AhR3′DT-1 was about 10- and 15-fold higher than that in the crude cell-free extracts, respectively (Table 2). Therefore, we used the microsomal fraction enriched with AhR4DT-1 or AhR3′DT-1 for subsequent enzymatic assays. Reactions were incubated with resveratrol, DMAPP, and Mg2+ as cofactor. Although the optimum activities of AhR4DT-1 and AhR3′DT-1 were observed at 37 and 30 °C, respectively, in the microsomal fraction of N. benthamiana, all further prenylation assays were performed at 28 °C, to match the culture temperature of peanut hairy roots during the elicitor-induced production of prenylated stilbenoids (Fig. S6). The accumulation of the prenylated product produced by AhR4DT-1 or AhR3′DT-1 showed a linear relationship with the amount of the microsomal fraction (25∼75 μg) and with incubation time (30–120 min) (Fig. S7).
Table 2.
AhR4DT-1 and AhR3′DT-1 activity from N. benthamiana leaf fractions
The preparation of fractions and the resveratrol prenyltransferase assay are described under “Experimental procedures.” Values are the mean ± S.D. for three replicates.
| Enzyme solution | AhR4DT-1 | AhR3′DT-1 |
|---|---|---|
| pmol·s−1·mg−1 protein | pmol·s−1·mg−1 protein | |
| Crude cell-free extracts | 151 ± 7 | 17.0 ± 5.9 |
| 156,000 × g supernatant | 16.8 ± 4.2 | 2.02 ± 1.15 |
| Microsomal fraction | 1350 ± 70 | 236 ± 26 |
The effects of pH on AhR4DT-1 and AhR3′DT-1 activities were investigated using three buffers that spanned a pH range from 7.0 to 10.7. The optimum pH of AhR4DT-1 activity was 9.4 in glycine-NaOH buffer (relative activity of each prenyltransferase, 100%), 9.0 in Tris-HCl buffer (99.8%), and 9.7 (92.1%) in NaHCO3-Na2CO3 buffer (Fig. 5A). AhR3′DT-1 exhibited optimal activities at pH 9.0 in both Tris-HCl (relative activity, 100%) and glycine-NaOH (92.8%) buffers, but at pH 9.7 in NaHCO3-Na2CO3 buffer (50.6%) (Fig. 5B). In summary, for these two prenyltransferases, the optimum pH was ∼9.0, which is consistent with the basic pH of the chloroplast stroma (42). Therefore 100 mm Tris-HCl (pH 9.0) was used as a standard reaction buffer in subsequent prenyltransferase reactions.
Figure 5.
pH dependence of AhR4DT-1 and AhR3′DT-1 activity. AhR4DT-1 (A) and AhR3′DT-1 (B) activities at various pH values were measured in three different buffers: 100 mm Tris-HCl buffer at pH 7.0, 8.0, 8.6, and 9.0; 100 mm glycine-NaOH buffer at pH 8.6, 9.0, 9.4, 10.0, and 10.6; and 100 mm NaHCO3-Na2CO3 buffer at pH 9.2, 9.7, 10.2, and 10.7. All the reactions were performed at 28 °C for 40 min.
A variety of divalent cations other than Mg2+ were tested to determine their effects on AhR4DT-1 and AhR3′DT-1 activities. Mg2+ was the most effective (100%) for AhR4DT-1, followed by Mn2+ (83.9%) and Fe2+ (6.4%) (Fig. S8A). Interestingly, in AhR3′DT-1 reactions, Mn2+ (210%) provided about 2-fold higher efficiency than Mg2+ (100%). Mg2+ forms a bidentate complex with DMAPP, which becomes stabilized for an efficient transferase reaction. Fe2+ (34.8%) also appeared to contribute to AhR3′DT-1 activity (Fig. S8B). Trace amounts of AhR4DT-1 and AhR3′DT-1 activities (<0.5%) were detected with all other divalent cations (Ca2+, Co2+, Zn2+, Ni2+, and Cu2+) and in the control group in which no divalent cation was added, although no activity was detected in the EDTA-treated group (Fig. S8). The trace activities observed are likely due to the presence of low levels of Mg2+ in the leaf microsomal fractions, released from chlorophyll-containing plastids.
The apparent Km values of AhR4DT-1 for both resveratrol (99.5 ± 15.1 μm) and DMAPP (154 ± 27 μm) were somewhat similar and comparable with those of the resveratrol prenyltransferase we had purified from peanut hairy root cultures (Table 3). In contrast, AhR3′DT-1 exhibited a notably lower Km for resveratrol (17.7 ± 1.6 μm) but a much higher Km for DMAPP (>640 μm) (Table 3; Fig. S9). Piceatannol, a compound detected in peanut hairy roots and considered to be another putative substrate of the prenyltransferases, provided another contrast. The AhR4DT-1 Km for piceatannol was 311.4 ± 54.8 whereas that of AhR3′DT-1 was 50.3 ± 5.5 and 154 ± 27 μm (Table 3; Fig. S9). Importantly, AhR4DT-1 and AhR3′DT-1 each exhibited a higher Vmax/Km value for resveratrol than piceatannol, suggesting that both of these prenyltransferases prefer resveratrol over piceatannol as substrate.
Table 3.
Comparison of kinetic values of AhR4DT-1, AhR3′DT-1, and prenyltransferase activity from peanut hairy roots
The apparent Km and Vmax values of AhR4DT-1 and AhR3′DT-1 for resveratrol, piceatannol, and DMAPP were measured using the microsomal fraction of N. benthamiana leaves transiently expressing these two enzymes. Values are the mean ± S.D. of three replicates. The apparent Km and Vmax values of prenyltransferase activity identified from the microsomal fraction of peanut hairy roots for resveratrol and DMAPP were previously reported by Yang et al. (26).
| AhR4DT-1 |
AhR3′DT-1 |
Prenyltransferase from peanut hairy root |
||||
|---|---|---|---|---|---|---|
| Km | Vmax | Km | Vmax | Km | Vmax | |
| μm | pmol·s1·mg−1 | μm | pmol·s1·mg−1 | μm | pmol·s1·mg−1 | |
| Resveratrol | 99.5 ± 15.1 | 2950 ± 150 | 17.7 ± 1.6 | 296 ± 6 | 111 ± 40 | 710 ± 90 |
| Piceatannol | 311 ± 55 | 2580 ± 220 | 50.3 ± 5.5 | 372 ± 12 | ||
| DMAPP | 154 ± 27 | 3380 ± 230 | >640 | 573 ± 25 | 91.9 ± 7.0 | 687 ± 17 |
Substrate specificity of AhR4DT-1 and AhR3′DT-1
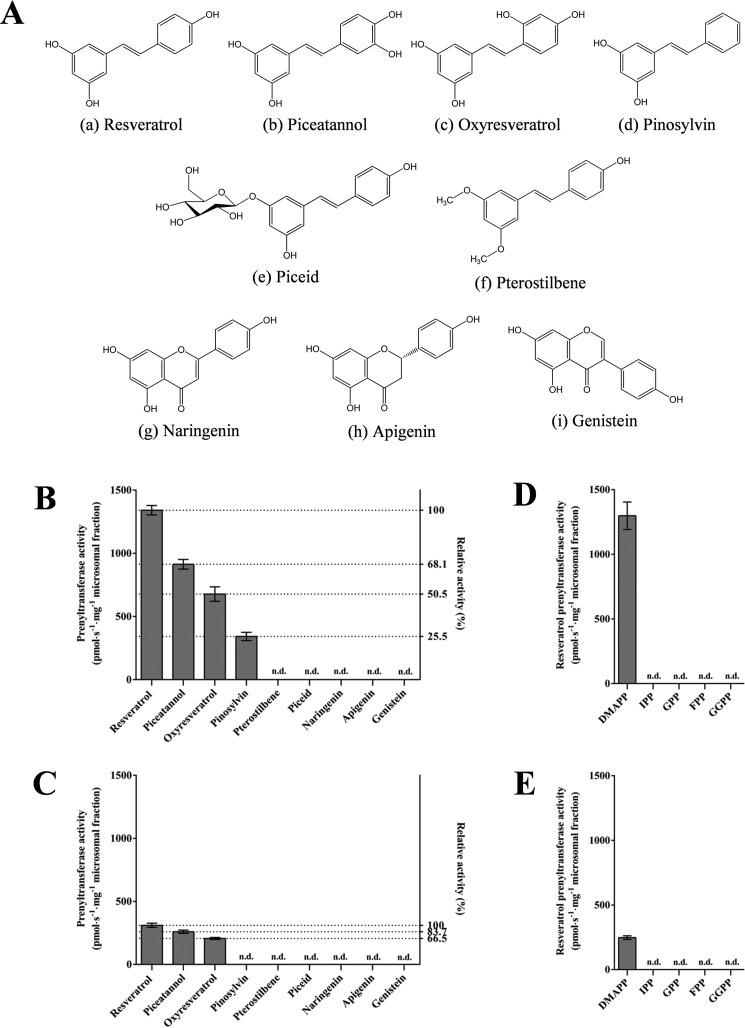
In addition to resveratrol, various other stilbenoids (piceatannol, oxyresveratrol, pinosylvin, pterostilbene, and piceid) and flavonoids (naringenin, apigenin, and genistein) were used as potential substrates to analyze the prenyl acceptor specificity of AhR4DT-1 and AhR3′DT-1. The results (Fig. 6, Figs. S10–S14, and Table S2) showed that AhR4DT-1 can selectively catalyze piceatannol, pinosylvin, and oxyresveratrol into arachidin-5, chiricanine A, and prenylated oxyresveratrol (the position of the prenyl moiety on the prenylated oxyresveratrol remains undetermined), respectively. In the reactions of AhR3′DT-1, the prenylated products of piceatannol and prenylated oxyresveratrol were identified by HPLC-PDA/ESI-MSnn analysis (Figs. S13 and S14 and Table S2), although the positions of their prenyl moiety have not been confirmed due to the insufficient amount of these products for further structural elucidation. Pterostilbene, which has two methoxy groups at the C-3 and C-5 positions, along with piceid, a resveratrol glucoside with a glycosidic group at the C-3 position, were not prenylated by either AhR4DT-1 or AhR3′DT-1 (Fig. 6), suggesting that either or both hydroxyl groups on C-3 and C-5 of the stilbene backbone might be crucial for substrate recognition by AhR4DT-1 and AhR3′DT-1. Interestingly, no prenylated pinosylvin was produced in the AhR3′DT-1 reaction (Fig. 6C), suggesting that other than the necessary 3- and 5-hydroxyl groups, a hydroxyl group at the C-4′ position might be an additional requirement for AhR3′DT-1 activity. Moreover, neither prenylated flavanone, prenylated flavone, nor prenylated isoflavone was detected in either AhR4DT-1 or AhR3′DT-1 reactions when flavonoid was used as substrate, indicating that both of these peanut prenyltransferases may be stilbenoid-specific prenyltransferases.
Figure 6.
Substrate specificity of AhR4DT-1 and AhR3′DT-1. A, chemical structures of prenyl acceptors used for substrate specificity analysis and their prenylated products: stilbenoids (a, resveratrol; b, piceatannol; c, oxyresveratrol; d, pinosylvin; e, piceid; and f, pterostilbene), flavanone (g, naringenin), flavone (h, apigenin), and isoflavone (i, genistein). B and C, relative prenylation activity of AhR4DT-1 (B) and AhR3′DT-1 (C) with various prenyl acceptors were compared with that of resveratrol. D and E, the prenyl donor specificity of AhR4DT-1 (D) and AhR3′DT-1 (E) was tested using DMAPP, IPP, GPP, FPP, and GGPP with resveratrol as a prenyl acceptor. All these reactions were performed in 100 mm Tris-HCl buffer (pH 9.0) at 28 °C for 40 min. Values are the average of triplicate, and error bars represent standard deviation (n.d., not detected).
To address the prenyl donor specificity of AhR4DT-1 and AhR3′DT-1, in addition to DMAPP, other prenyl diphosphates, including isopentenyl pyrophosphate (IPP), geranyl pyrophosphate (GPP), farnesyl pyrophosphate (FPP), and geranylgeranyl pyrophosphate (GGPP), were examined with resveratrol as a prenyl acceptor. Neither AhR4DT-1 nor AhR3′DT-1 showed any detectable activity when these prenyl diphosphates were used as prenyl donor, suggesting that both of these prenyltransferases had strict specificity for DMAPP (Fig. 6, D and E).
Subcellular localization of AhR4DT-1 and AhR3′DT-1
In evaluating primary structures using available software, we found conflicting predictions for the subcellular localization of these enzymes. The iPSORT program predicted a chloroplast transit peptide (cTP) in AhR4DT-1 and a mitochondrial targeting peptide (mTP) in AhR3′DT-1. However, ChloroP and TargetP predictions suggested AhR4DT-1 contained neither cTP nor mTP, whereas AhR3′DT-1 contained an N-terminal cTP. To confirm their subcellular localizations experimentally, AhR4DT-1-GFP and AhR3′DT-1-GFP gene fusion constructs driven by the CaMV35S-TEV promoter were expressed transiently in onion epidermal cells via particle bombardment. As a positive control for plastid localization, we co-expressed a construct created by Nelson et al. (43) that features the transit peptide (1st 79 amino acids) of tobacco Rubisco small subunit fused to red fluorescent protein (RS-TP-mCherry). The green fluorescence signals of AhR4DT-1-GFP and AhR3′DT-1-GFP appeared in punctate patterns against organelles in the onion epidermal cells (Fig. 7), patterns highly similar to that of the red fluorescence derived from RS-TP-mCherry (Fig. 7). In contrast, control GFP driven by the Ca35S-TEV promoter was localized throughout the cytosol and nucleus (Fig. 7). These results strongly suggest that AhR4DT-1 and AhR3′DT-1 in peanut are localized to plastids, similar to flavonoid prenyltransferases such as SfN8DT-1, GmG4DT, and LaPT1 characterized in other plant species (27, 30, 37).
Figure 7.
Microscopic analysis of subcellular localization of AhR4DT-1 and AhR3′DT-1 fused with GFP in onion epidermal cells upon particle bombardment. Plasmids containing CaMV35S-TEV-GFP and RS-TP-mCherry (A and D), AhR4DT-1-GFP and RS-TP-mCherry (B and E), or AhR3′DT-1-GFP and RS-TP-mCherry (C and F) were mixed and introduced into onion epidermal cells by particle bombardment. Bright field images (A1–C1), expression pattern of mCherry from construct CaMV35S-RS-TP-mCherry (A2–C2), expression pattern of GFP from construct CaMV35S-TEV-GFP (A3), CaMV35S-TEV-AhR4DT-1-GFP (B3), and CaMV35S-TEV-AhR3′DT-1-GFP (C3), and merged images of green and red fluorescence (A4–C4) in the same single optical section were obtained with the confocal laser scanning microscope. Scale bars, A–C equal 100 μm. D1–F3 correspond to the close-up images of A2–C4. Scale bars, D–F equal 10 μm. CaMV35S, cauliflower mosaic virus 35S promoter; TEV, tobacco etch virus translational enhancer; GFP, green fluorescent protein; RS-TP, Rubisco small subunit transit peptide.
Expression of AhR4DT-1 and AhR3′DT-1 in peanut hairy roots during elicitation
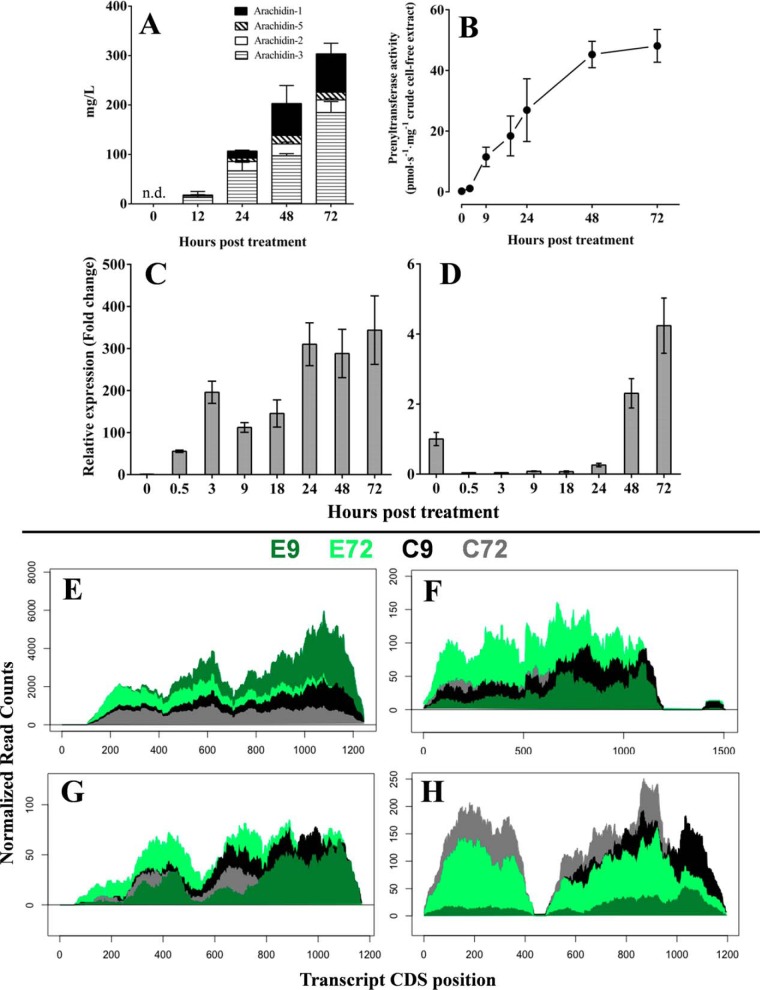
Our previous studies have shown that the accumulation of prenylated stilbenoids in peanut hairy root culture (Fig. 8A; Yang et al. (24, 26)) and the prenyltransferase activities from crude cell-free extracts of peanut hairy roots were up-regulated by elicitor treatments and increased with incubation time (26). We therefore hypothesized that the mRNA of enzymes involved in the prenylation of stilbenoids, i.e. AhR4DT-1 and AhR3′DT-1, may accumulate upon elicitation in peanut hairy roots. To test this hypothesis, transcript levels of AhR4DT-1 and AhR3′DT-1 during co-treatment of hairy root cultures with MeJA and CD were quantified using real-time PCR. A rapid up-regulation of AhR4DT-1 was observed after 0.5-h post-elicitation (Fig. 8C), and its expression pattern was consistent with the prenyltransferase activities assayed in the same system (Fig. 8B). Despite an apparent delay in accumulation as compared with AhR4DT-1, levels of AhR3′DT-1 mRNA increased as well after 48 h of elicitation in these experiments (Fig. 8D). Whereas qPCR detects a short sequence-unique portion of the target mRNA, comparative mapping of RNA-Seq reads can provide a more complete picture of differential expression. Assessment of differential expression of individual transcripts in this case is, however, confounded by the complexity of this tetraploid transcriptome, the large target enzyme family, and the lack of A. hypogaea genome sequence reference. Employing available sequence references of the peanut diploid progenitors (above), we mapped all assembled A. hypogaea transcripts, both to confirm singularity of genomic loci and to select transcript sequence of each prenyltransferase that is most sequence-inclusive to use as reference for quantification (Table 3). Fig. 8, E–H, represents sample-normalized counts of reads that mapped unambiguously to these transcript references. Not surprisingly, mock treatments (assayed at 9 and 72 h) that mechanically stimulate the hairy root cultures resulted in increases in all described transcripts. AhR4DT-1 mRNA stood out, however, as accumulating to 2–3-fold over control levels early in response to the addition of MeJA + CD (Fig. 8E). AhR3′DT-1, however, among the treatments assayed, reached its highest levels only 72 h post-elicitation (Fig. 8F). AhR3′DT-2/3 and -4 transcripts were clearly detectable by RNA-Seq across the time course but did not appear to change in response to elicitation (Fig. 8, G and H). These results indicate the activation of AhR4DT-1 and AhR3′DT-1 genes correlates with stress elicitation in peanut hairy root tissue, and the accumulation of mRNA encoding these two enzymes correlates temporally with their prenyltransferase activities observed and with their catalyzed product accumulation. In contrast, mRNAs of similar enzymes we show to exhibit activities that are not relevant, or less relevant, to arachidin-2 and 3-methyl-2-butenyl-3′-resveratrol production in peanut are not noticeably transcriptionally responsive to the elicitation. As products attributable to AhR4DT-1 and AhR3′DT-1 activities do not accumulate in peanut hairy root cultures in response to the control treatments used here (24), protein expression and/or localization are likely to be controlled by mechanisms beyond the transcript accumulation observed.
Figure 8.
Enzyme activities and transcript co-expression during elicitation time course in peanut hairy root cultures. A, accumulation of prenylated stilbenoids in the medium of peanut hairy root culture upon the MeJA and CD elicitation (n.d., not detected). B, prenyltransferase activities from crude cell-free extracts. C, relative transcript accumulation of AhR4DT-1; D, AhR3′DT-1 as determined by RT-qPCR. E, uniquely mapped RNA-Seq reads coverage over reference A. hypogaea transcripts AhR4DT-1. F, AhR3′DT-1; G, AhR3′DT-2/3; and H, AhR3′DT-4 as described. E9 and E72, 9 and 72 h of MeJA + CD treatment; C9 and C72, 9 and 72 h of control treatments.
Discussion
Stilbenoid-specific prenyltransferases from peanut
Prenylation of aromatic compounds plays an important role in the diversification of plant secondary metabolites and contributes to the enhancement of the biological activity of these polyphenolic compounds (37). To date, only a few flavonoid prenyltransferase genes have been identified, including SfN8DT-1, SfiLDT, SfG6DT, SfFPT, GmG4DT, GuA6DT, and LaPT1 cloned from legume species (27, 29–32, 37), along with MaIDT and CtIDT from non-legume species Morus alba and Cudrania tricuspidata, respectively (44). In the study reported here, we tested eight potential resveratrol prenyltransferase transcripts and five of them encoded enzymes with two distinct prenylation activities. The eight transcripts derived from four or more genes expressed in peanut hairy root cultures. Two of these, AhR4DT-1 and AhR3′DT-1, were characterized as stilbenoid-specific prenyltransferases.
The stilbenoid prenyltransferases described in this study share several common features with flavonoid prenyltransferases. First, each of these enzymes is a membrane-bound protein containing several putative transmembrane α-helices. The subcellular localization of the two stilbenoid prenyltransferases described here are primarily or exclusively in the plastid, as are the five flavonoid prenyltransferases previously characterized. Second, each enzyme contains two conserved aspartate-rich motifs. The observed divalent cation dependence of our prenylation reactions corroborates the proposed role of this structure in the active site, where the divalent cation and the prenyl diphosphate bind (45). Interestingly, with the exception of AhR3′DT-1 in which Mn2+ is most effective, all other flavonoid enzymes and the stilbenoid enzyme AhR4DT-1 show the highest activity in the present of Mg2+. Finally, most flavonoid prenyltransferases and the stilbenoid prenyltransferases identified in this study exhibit strict substrate specificity with respect to their prenyl acceptor and prenyl donor, a feature that contrasts sharply with the catalytically promiscuous aromatic prenyltransferases of fungi and bacteria. Despite sharing key features with flavonoid prenyltransferases, the stilbenoid prenyltransferases are monophyletic to other plant prenyltransferases accepting aromatic substrates (Fig. 4).
Involvement of AhR4DT-1 and AhR3′DT-1 in the biosynthesis of prenylated stilbenoids in peanut
AhR4DT-1 specifically transfers a 3,3-dimethylallyl group to the A-ring at the C-4 position of resveratrol, piceatannol, and pinosylvin. This enzyme exhibits biochemical properties that match well with the prenyltransferase activity identified from elicited peanut hairy roots (26), including Km values of resveratrol/DMAPP and identical preferences for prenyl acceptors and divalent cations. The consistency of all biochemical characteristics, along with our demonstration of transcript accumulation that is temporally correlated with C-4-prenylated stilbenoid (arachidin-1, arachidin-2, arachidin-3, and arachidin-5) accumulation, led us to propose AhR4DT-1 is responsible for the prenylation activity in the microsomal fraction of peanut hairy roots identified in our previous study (26).
The second stilbenoid-specific prenyltransferase characterized here was AhR3′DT-1 that recognizes 3,5,4′-trihydroxystilbene and adds a 3,3-dimethylallyl group to C-3′ of the B-ring. Notably, none of the prenylation products of resveratrol and piceatannol catalyzed by AhR3′DT-1 were detected in peanut hairy root culture or peanut hairy root tissue. When compared with AhR4DT-1, AhR3′DT-1 showed a lower Km value for resveratrol and piceatannol, indicating a higher affinity for these prenyl acceptor substrates. In contrast, its affinity for DMAPP was much lower than that of AhR4DT-1. Based on APCI-MSn analyses of stilbenes detected in a peanut root mucilage extract, Sobolev et al. (7) proposed several novel prenylated stilbenes with prenyl groups on both the A- and B-rings. It is possible that the prenylation product of AhR3′DT-1 becomes further prenylated to form a diprenylated stilbene in peanut, impeding our ability to detect the prenylation product of AhR3′DT-1 in the peanut hairy root culture even when the substrate for AhR3′DT-1 is present.
Under the co-treatment of 100 μm MeJA and 9 g/liter CD for 72 h, the peanut hairy root cultures secrete into the medium large amounts of resveratrol (44.6 ± 10.3 mg/liter), arachidin-1 (77.9 ± 21.5 mg/liter), and arachidin-3 (184 ± 22 mg/liter) and very low levels of piceatannol (4.02 ± 0.67 mg/liter) (24). Only moderate levels of arachidin-5 (15.5 ± 5.6 mg/liter) and arachidin-2 (25.6 ± 3.0 mg/liter) are found in these cultures (26). These observations suggest that arachidin-1 and arachidin-3 may be end products during the tested period of elicitation. Differing from arachidin-5, arachidin-2, and most other prenylated flavonoids that harbor a 3,3-dimethylallyl moiety, arachidin-1 and arachidin-3 have a unique 3-methyl-but-1-enyl moiety (Fig. 9). Until now, the biosynthesis pathway(s) of arachidin-1 and arachidin-3 have not been fully elucidated; however, several biosynthetic routes leading to their production could be proposed when considering results from our previous and current studies.
Figure 9.
Proposed pathway of prenylated stilbenoids in peanut. Stilbenoids identified from the medium of peanut hairy root culture upon elicitors treatment are in boldface type, and their proposed pathway is highlighted in yellow. Other prenylated stilbenoids identified in fungus-challenged peanut seeds are divided into three groups based on the prenyl unit and hydroxyl groups on their stilbene backbone. Prenylation reactions catalyzed by AhR4DT-1 and AhR3′DT-1 identified in this study are labeled with red solid arrows. Enzymatic reactions confirmed in peanut are marked with black solid arrow, whereas other proposed reactions are labeled in black arrows with dashed lines. *, pinosylvin, 3-methyl-2-butenyl-3′-resveratrol, and the prenylation product of piceatannol by AhR3′DT-1 have not been reported in peanut tissue.
In our previous study, it was demonstrated that exogenous resveratrol could be oxidized to piceatannol by an extract from the peanut hairy root tissue through a very efficient enzymatic reaction (26). With the abundance of resveratrol in the culture medium of peanut hairy roots, piceatannol generated from the oxidation of resveratrol could serve as a precursor, alternative to resveratrol, for prenylated stilbenoids in peanut. It appeared that this compound could be further metabolized into other derivatives, resulting in a relatively low yield of piceatannol in the peanut hairy root culture.
AhR4DT-1 identified here initiates the first step in the biosynthesis of prenylated stilbenoids in peanut by catalyzing the prenylation of resveratrol and piceatannol to form arachidin-2 and arachidin-5, respectively. Other than that derived from the prenylation of piceatannol, which is limited in the culture medium, it remains possible that arachidin-5 is also formed via hydroxylation of arachidin-2 by a stilbenoid monooxygenase (P450). Flavonoid 3′-monooxygenases, for example, are known to catalyze 3′-hydroxylation of the flavonoid backbone (46). Similarly, arachidin-3 might be hydroxylated by a monooxygenase to produce arachidin-1 (Fig. 9). Further enzyme discovery and testing are needed to explore these possibilities.
In the reactions with peanut hairy root microsomes, arachidin-2 and arachidin-5 prenylated from resveratrol and piceatannol, respectively, could be further converted into the derivatives of arachidin-2 and arachidin-5 we had detected in the medium upon the elicitor treatment (26). Notably, these derivatives were not detected in the AhR4DT-1 reactions (Fig. 9), indicating other enzyme(s) that modify arachidin-2 and arachidin-5 are present downstream of AhR4DT-1. To date, the structures of these arachidin derivatives have not been elucidated, and this incomplete information hindered the elucidation of additional enzyme(s) involved in peanut-prenylated stilbenoid biosynthesis in peanut.
In one proposed pathway for arachidin-1 and arachidin-3, the isopentenyl stilbenoids arachidin-2 and arachidin-5 might be directly converted to arachidin-3 and arachidin-1 by an isomerase that could shift the olefinic bond position on their prenylated moieties (Fig. 9). Alternatively, arachidin-2 or arachidin-5 might be converted into an intermediate product that is further modified into arachidin-3 or arachidin-1 through multiple enzymatic steps (Fig. 9). It is still unclear whether the arachidin-2 derivative and the arachidin-5 derivative found in peanut hairy root culture were intermediates involved in the biosynthesis of the predominant compounds arachidin-3 and arachidin-1 (Fig. 9). Alternatively, there remains a slight possibility that arachidin-3 and arachidin-1 were synthesized directly from resveratrol and piceatannol catalyzed by AhR4DT-1 or another specific prenyltransferase utilizing 3-methyl-but-1-enyl pyrophosphate as prenyl donor. As far as we know, however, this kind pyrophosphate has never been described in plants.
To date, over 45 prenylated stilbenoids and derivatives, including monomers and dimers, have been identified in peanut. Many of these chemical structures have been confirmed by NMR (Fig. 9) or predicted by mass spectrometry. Interestingly, all these stilbenoids can be divided into two groups, one showing a prenyl unit or a derivative at the C-4 position and a second group showing a prenyl unit or derivative at the C-3′ position (Fig. 9). These observations strongly suggest that the prenyltransferases encoded by AhR4DT-1 and AhR3′DT-1 act in the first committed steps that channel the diversification of non-prenylated stilbenoids into prenylated stilbenoids. As the first stilbenoid-specific prenyltransferases identified in a plant, these findings advance our understanding of this specialized gene family and the biosynthesis of important bioactive compounds in plant stress responses.
Experimental procedures
Plant materials and chemical reagents
Hairy roots of peanut cv. Hull line 3 were previously established by transforming peanut cotyledonary leaves with Agrobacterium rhizogenes strain ATCC 15834 and maintained in modified Murashige and Skoog (MSV) medium under continuous darkness at 28 °C as described before (25). The procedure of elicitation for stilbenoid production in peanut hairy root cultures was performed according to Yang et al. (24).
Authentic standards of resveratrol and piceatannol were obtained from Biophysica and Axxoram, respectively. Arachidin-1, arachidin-2, arachidin-3, and arachidin-5 standards were purified from elicited peanut hairy root cultures as described previously (26). Pinosylvin, oxyresveratrol, pterostilbene, naringenin, apigenin, genistein, IPP, GPP, FPP, and GGPP were purchased from Sigma. DMAPP used in this study was obtained from Isoprenoids.
RNA preparation
Nine-day-old peanut hairy root cultures were co-treated with 100 μm MeJA (Sigma) and 9 g/liter (6.87 mm) CD (Cavasal W7M) to induce the expression of genes involved in the biosynthesis of stilbenoids. Total RNA was extracted from the elicited root tissue at 0.5, 3, 9, 18, 24, and 72 h using TRIzol reagent (Life Technologies, Inc.), according to the manufacturer's instructions. For controls, RNA was likewise extracted from the roots prior to treatment (t = 0) and from non-elicited roots, collected 9 and 72 h after mock treatment (i.e. refreshed with fresh MSV medium).
Transcript sequencing and assembly
Strand-aware, poly(A)-enriched RNA libraries were prepared using Illumina TruSeq stranded mRNA sample preparation reagents with sequence-indexed adaptors, with inputs of 4 μg of total RNA per sample. Average insert size of indexed libraries was 300 bp according to Bioanalyzer (Agilent) evaluation. Libraries were sent to the Roy J. Carver Biotechnology Center at the University of Illinois at Urbana-Champaign, where they were quantified by qPCR and pooled together for sequencing of 2 × 101 paired cycles on an Illumina HiSeq2500 using TruSeq SBS sequencing kits version 3. Number of read pairs ranged from 34.8 to 58.9 m per sample. Data were processed and demultiplexed using Casava 1.8.2 (Illumina).
Reads were trimmed using Trimmomatic version 0.32 (47) with headcrop 12, sliding window 5, minimum quality 25. Parallel assemblies of each sample-specific reads set as well as a combination of all reads were generated using Trinity version 2013-11-10 (48), TransABySS 1.5.5 (49), Velvet-Oases 1.2.10 (50), and SOAPtrans 1.03 (51), yielding a total of 2.6 m putative complete and partial transcript sequences. Coding sequences were predicted and translated using CD-Hit (52) through scripts of EvidentialGenes (Don Gilbert, Indiana University, http://arthropods.eugenes.org/)3 to build the transcriptome BlastP and BLASTN databases.
Using GMAP 2016-04-04 (53), final transcript assemblies were aligned to both isolated and concatenated A. duranensis and A. ipaensis genomes available in PeanutBase (www.peanutbase.org and Bertioli et al. (34)), which confirmed clustering of highly similar forms. This allowed us to approximate a reduced A. hypogaea sequence reference against which we quantified reads coverage attributable to the enzyme transcripts under study. Reads mapped to the resulting four genomic loci were then isolated using Samtools 1.3.1 (54) and re-mapped to A. hypogaea transcript sequence references determined here. Mapping of RNA-Seq reads was performed using Tophat2, version 2.0.7 (55) using the genome-guided option. Uniquely mapped read counts from each sample were assessed using HTSeq version 0.6.1p1 (56).
Cloning of AhR4DT-1 and AhR3′DT-1 cDNA
The amino acid sequences of flavonoid prenyltransferases SfN8DT-1 (accession number BAG12671.1), SfG6DT (BAK52291.1), SfFPT (AHA36633.1), GmG4DT (BAH22520.1), GuA6DT (AIT11912.1), and LaPT1 (AER35706.1) were used as input to run BlastP (57) against our translated peanut hairy root transcriptome sequence database. Predictions of chloroplast transit peptides (cTP) were made using both ChloroP (http://www.cbs.dtu.dk/services/ChloroP/)3 and iPSORT (http://ipsort.hgc.jp/).3 To clone the full-length cDNA of these candidates, RNA of 9-h-elicited peanut hairy roots was prepared by TRIzol reagent, and the cDNA was synthesized using iScriptTM select cDNA synthesis kit (Bio-Rad) using oligo(dT) primer. One N-terminal primer and three C-terminal primers were synthesized with flanking NotI and KpnI restriction sites, respectively (Table S1), and PCR using these primers was performed with ExTaq DNA polymerase (Takara) following the program below: initial denaturation (3 min, 94 °C); 30 cycles (30 s, 94 °C; 30 s, 52 °C; 1 min 30 s, 72 °C); and a final extension step (10 min, 72 °C). Three amplicons (including AhR3′DT-1) were subcloned into a pGEM-T vector (Promega), and multiple clones from each amplicon were sequenced at University of Chicago Comprehensive Cancer Center. In the second screening, according to all candidate sequences, four N-terminal primers and five C-terminal primers with NotI/KpnI-flanking restriction sites (Table S1) were designed for prenyltransferase cloning, and another four amplicons (including AhR4DT-1) obtained from the same cDNA template were cloned into pGEM-T vector for sequencing validation.
Phylogenetic analysis
Protein sequences were aligned using MUSCLE (58), and a neighbor-joining phylogenetic tree was computed with PhyML (59) using the Dayhoff substitution model and 100 bootstrapped replicates.
Construction of binary vectors
The sequence of the double-enhanced cauliflower mosaic virus 35S promoter (CaMV35S) fused to the translational enhancer from tobacco etch virus (TEV) was amplified from plasmid pR8-2 (constructed by Medina-Bolívar and Cramer (60)) and subcloned into pGEM-T vector using Ca35S-FW-SalI-1/TEV-RW-NotI primers (Table S1) with SalI/NotI-flanking restriction sites. After validation of the sequences, the pGEM-CaMV35S-TEV and pGEM-T vectors containing putative prenyltransferase gene (PT) were digested with SalI/NotI and NotI/KpnI, respectively. A high copy number vector, pBC KS(−) digested with KpnI/SalI was used as a transition vector. Two fragments of full-length cDNA and CaMV35S-TEV promoter were ligated into the transition vector in a 16 °C overnight reaction with T4 ligase (New England Biolabs). Then the fragment of CaMV35S-TEV-PT from the transition vector digested by KpnI/SalI was subcloned into a binary vector pBIB-Kan, which was created by Becker (61). Eventually, the constructed binary vector with the putative prenyltransferase gene under the control of CaMV35S-TEV chimeric promoter and 3′-untranslated region (3′-UTR) of nopaline synthase from the original pBIB-Kan vector was transformed into Agrobacterium tumefaciens LBA4404 for stilbenoid prenylation activity screening.
Screening for stilbenoid prenylation activity
The engineered A. tumefaciens was grown in 5 ml of YEP medium, containing 50 mg/liter kanamycin (Sigma) and 30 mg/liter of streptomycin (Sigma) for antibiotic selection, at 28 °C on an orbital shaker at 200 rpm. After cultivation for 2 days, 5 ml of bacterial suspension was inoculated into 50 ml of fresh YEP medium containing the antibiotics and allowed to grow for 1 additional day under the same conditions. Bacteria were pelleted by centrifugation, resuspended in 500 ml of induction medium containing 10 mm MgCl2 with 1 mm acetosyringone, and incubated for 4 h at 28 °C under 200 rpm orbital shaking until their A600 reached to a range from 0.5 to 0.6. Before the infiltration, 0.005% of Tween 20, 0.005% of Triton X-100, and 0.005% of Silwet L-77 were added to bacterial cultures to enhance the efficiency of transformation in N. benthamiana leaves. Agrobacterium-mediated vacuum infiltration was performed on 4-week-old N. benthamiana following the methodology described previously (62).
After 48 h of post-infiltration, the “middle tier” of N. benthamiana leaves were harvested for prenylation activity screening. Five grams of leaf tissue (fresh weight) were ground and homogenized using a mortar with pestle in 10 ml of extraction buffer containing 100 mm Tris-HCl (pH 7.6) and 10 mm dithiothreitol (DTT). After removing the cell debris by centrifugation at 12,000 × g for 20 min at 4 °C, the crude cell-free extract was obtained by passing the 12,000 × g supernatant through a PD-10 desalting column (GE Healthcare) equilibrated with 100 mm Tris-HCl (pH 9.0) containing 10 mm DTT. The total protein concentration was determined by Coomassie protein assay (Thermo Fisher Scientific) using bovine serum albumin as standard.
The prenylation reactions contained resveratrol (100 μm), DMAPP (300 μm), MgCl2 (10 mm), and DTT (5 mm) in a Tris-HCl buffer (100 mm (pH 9.0)). After incubation (28 °C, 40 min) with the crude cell-free extract of N. benthamiana leaves (5 mg of total protein) in a total volume of 1 ml, the enzyme reaction was terminated by addition of HCl (6 m, 20 μl) and then the reaction mixture was extracted with ethyl acetate (1 ml). The ethyl acetate extract was dried under nitrogen gas and dissolved in 300 μl of methanol. The reaction product was identified and quantified using HPLC/ESI-MSn analysis as described previously (26). The reaction of crude cell-free extract of N. benthamiana leaves infiltrated with A. tumefaciens harboring the empty pBIB-Kan vector was used as control.
Enzymatic characterization of AhR4DT-1 and AhR3′DT-1
In preliminary experiments focused on studying the effect of the post-infiltration period on the prenylation activity in N. benthamiana, leaf tissues of plants, which transiently expressed AhR4D-1, were harvested 24, 48, 72, and 96 h post-infiltration. Among these reactions, the AhR4D-1 activity in the crude cell-free extract of N. benthamiana leaves increased with the post-infiltration period from 24 to 72 h, while the activity in the leaves harvested from 96-h post-infiltration was similar to 72 h (data not shown). In addition, because both AhR4DT-1 and AhR3′DT-1 were predicted as membrane-bound proteins by TMHMM 2.0, the microsomal fraction of N. benthamiana leaves at 72 h post-infiltration was used to study the biochemical properties of AhR4DT-1 or AhR3′DT-1. Ten grams of fresh leaves were homogenized using a mortar with pestle in 20 ml of extraction buffer (100 mm Tris-HCl (pH 7.6) containing 10 mm DTT). The homogenate was centrifuged at 12,000 × g for 20 min at 4 °C, and about 13.2 ml of the supernatant was centrifuged at 156,000 × g for 45 min at 4 °C to pellet the microsomal fraction, whereas the 156,000 × g supernatant was prepared by using a PD-10 desalting column (GE Healthcare) equilibrated with Tris-HCl buffer (100 mm (pH 9.0)) containing DTT (10 mm). The microsomal fraction was washed twice with Tris-HCl buffer (100 mm (pH 9.0)) containing DTT (10 mm) and resuspended in 1 ml of the same buffer.
The basic reaction and measurement for AhR4DT-1 and AhR3′DT-1 activities were the same as that for prenylation activity screening with exception of using 30 μg of microsomal fraction of N. benthamiana leaves as enzyme instead of crude cell-free extract of N. benthamiana leaves in 500 μl of reaction. To investigate the optimal pH, the enzymatic reactions were performed in Tris-HCl buffer (100 mm (pH 7.0 to 9.0)), glycine-NaOH buffer (100 mm (pH 8.6–10.6), and NaHCO3-Na2CO3 buffer (100 mm, pH 9.2–10.7). The optimal reaction temperatures for AhR4DT-1 and AhR3′DT-1 were tested at 20, 25, 28, 30, 37, 40, and 50 °C in Tris-HCl buffer (100 mm (pH 9.0)). For the divalent cation dependence study, 10 mm MnCl2, FeCl2, CaCl2, CoCl2, ZnCl2, NiCl2, or CuCl2 was added to the reaction mixture instead of MgCl2, and the enzyme activity was compared with the reaction containing MgCl2. The reactions without divalent cation and 10 mm EDTA instead of MgCl2 were used as controls.
For the kinetic study, increasing concentrations (10, 20, 40, 80, 160, 320, and 640 μm) of resveratrol or piceatannol with a fixed concentration of DMAPP (640 μm) and increasing concentrations (10, 20, 40, 80, 160, 320, and 640 μm) of DMAPP with a fixed concentration of resveratrol (640 μm) were incubated with 30 μg of microsomal fractions of N. benthamiana leaves expressing AhR4DT-1 or AhR3′DT-1 to calculate the apparent Km and Vmax values by non-linear regression analysis of the Michaelis-Menten equation using GraphPad Prism 6 software. The prenyl acceptor specificity of AhR4DT-1 and AhR3′DT-1 was tested using 100 μm of each stilbenoid (resveratrol, piceatannol, oxyresveratrol, pinosylvin, pterostilbene, and piceid), flavanone (naringenin), flavone (apigenin), and isoflavone (genistein) with 300 μm DMAPP as a prenyl donor, whereas the prenyl donor specificities of these two enzymes were tested using 300 μm prenyl diphosphates (DMAPP, IPP, GPP, FPP, and GGPP) with 100 μm resveratrol as a prenyl acceptor. All these reactions were performed in a total volume of 500 μl with 100 mm Tris-HCl buffer (pH 9.0) at 28 °C for 40 min.
NMR spectra
All NMR measurements were performed on a Bruker Avance 700 MHz and spectrometers at 298 K. The 1H-13C HMBC and 1H-13C HSQC spectra were collected in d6-acetone. The concentration of the sample was ∼1 mm. For 1H NMR analysis, 16 transients were acquired with a 1-s relaxation delay using 32,000 data points. The 90° pulse was 9.7 μs with a spectral width of 16 ppm. 1D 13C NMR spectra were obtained with a spectral width of 30 ppm collected with 64,000 data points. Two-dimensional spectra were acquired with 2048 data points for t2 and 256 for t1 increments. All NMR data were analyzed using Topspin version 2.0 and SPARKY version 3.0 software. Peaks were integrated and overlaid with the simulated spectra for different versions of the prenyl chain attached on the resveratrol compound.
Construction of GFP fusion proteins
The nucleotide sequence of modified green fluorescence protein (mGFP5) was amplified from pR8-2 (60) using primers mgfp5-FW-BamHI/mgfp5-RW-KpnI (Table S1) and cloned into pGEM-T vector to give pGEM-mGFP5–1. PT-9b13-RV-BamHI or PT-10k1-RV-BamHI reverse primer with Ca35S-FW-SalI-2 forward primer were used to amplify the full-length of AhR4DT-1 or AhR3′DT-1 with the CaMV35S-TEV promoter region from pBC-CaMV35S-TEV-9b13 and pBC-CaMV35S-TEV-10k1 vector, which were created during the construction of the binary vector (Table S3). The PCR products were then cloned into pGEM-T vector for sequencing validation. After SalI/BamHI digestion, CaMV35S-TEV-AhR4DT-1 and CaMV35S-TEV-AhR3′DT-1 fragments were isolated and inserted into pGEM-mGFP5–1 to yield pGEM-CaMV35S-TEV-AhR4DT-1-GFP and pGEM-CaMV35S-TEV-AhR3′DT-1-GFP, respectively. Finally, the fragments of CaMV35S-TEV-AhR4DT-1-GFP and CaMV35S-TEV-AhR3′DT-1-GFP were excised with SalI/KpnI and ligated into binary vector pBIB-Kan to yield pBIBKan-AhR4DT-1-GFP and pBIBKan-AhR3′DT-1-GFP, respectively. For the GFP control construct, mGFP5 gene was amplified from pR8-2 using primers mgfp5-FW-NotI/mgfp5-RW-KpnI and cloned into pGEM-T vector to give pGEM-mGFP5-2. Two fragments CaMV35S-TEV digested from pGEM-CaMV35S-TEV by SalI/NotI and mGFP5 digested from pGEM-mGFP5–2 by NotI/KpnI were inserted into pBC KS(−) vector to form pBC-CaMV35S-TEV-GFP. The fragment of CaMV35S-TEV-GFP was eventually subcloned into binary vector pBIB-Kan to form pBIB-Kan-GFP.
Particle bombardment and microscopy
To investigate the subcellular localization of AhR4DT-1 and AhR3′DT-1, pBIB-Kan-AhR4DT-1-GFP, pBIB-Kan-AhR3′DT-1-GFP, and pBIB-Kan-GFP were co-bombarded with binary vector pt-rk (ABRC stock number CD3-999, Nelson et al. (43)) containing a plastid marker fused with red fluorescent protein into the onion epidermal peel cells by PDS-1000/HeTM systems (Bio-Rad) following the manufacturer's recommendations. In brief, 5 μg of target plasmid and 5 μg of pt-rk plasmid were together coated on 50 μl of 60 mg/ml tungsten particles (M17, 1 μm; Bio-Rad) in the presence of 1 m CaCl2 and 15 mm spermidine. After several ethanol washes, plasmid-coated particles were dried on plastic discs and accelerated with a helium burst at 1100 p.s.i. in a bombardment chamber. Bombarded onion epidermal peels were kept on plates containing MS medium for 60 h in the dark. The localization of the expressed proteins in the transformed cell was visualized with a Nikon Eclipse E800 microscope with a ×20/0.5W Fluor water immersion objective. Confocal fluorescence images were obtained by using Nikon digital eclipse C1 microscope system with 488-nm laser illumination and 525/50-nm filter for GFP fluorescence and 543-nm laser with 595/50-nm filter for red fluorescence protein.
Quantitative real-time PCR of AhR4DT-1 and AhR3′DT-1
Total RNA was isolated from 100 μm MeJA and 9 g/liter CD co-treated peanut hairy roots at 0.5, 3, 9, 18, 24, and 72 h using TRIzol reagent, and cDNA was synthesized using iScriptTM Select cDNA synthesis kit (Bio-Rad) with oligo(dT) primers following the manufacturer's instructions. Primers for AhR4DT-1 and AhR3′DT-1 were designed using Allele ID (PREMIER Biosoft). Two reference genes, ACT7 (encoding actin 7) and EFα1 (encoding elongation factor α1), were selected previously (63) and used to normalize the expression of AhR4DT-1 and AhR3′DT-1 in peanut hairy roots. qPCR were carried out using iQ SYBR Green Supermix (Bio-Rad), as described previously (24), and the expression of AhR4DT-1 and AhR3′DT-1 was analyzed by qbase+ (Biogazelle).
Accession numbers
The nucleotide sequences of AhR4DT-1, AhR3′DT-1, AhR3′DT-2, AhR3′DT-3, and AhR3′DT-4 have been deposited in the GenBankTM database under the accession numbers KY565244, KY565245, KY565246, KY565247, and KY565248, respectively.
Author contributions
T. Y., S. K. T., K. M., and F. M.-B. conceptualization; T. Y., S. S., S. K. T., K. M., and F. M.-B. data curation; T. Y., S. J., G. R., R. P., K. M., and F. M.-B. formal analysis; T. Y., L. F., S. S., S. J., G. R., R. P., S. K. T., K. M., and F. M.-B. investigation; T. Y., L. F., S. S., S. J., G. R., R. P., S. K. T., K. M., and F. M.-B. methodology; T. Y. and F. M.-B. writing-original draft; T. Y., S. K. T., K.M., and F. M.-B. writing-review and editing; S. K. T., K. M., and F.M.-B. resources; S. K. T., K. M., and F. M.-B. project administration; K. M. and F. M.-B. supervision; K. M. and F. M.-B. funding acquisition; F. M.-B. validation; F. M.-B. visualization.
Supplementary Material
Acknowledgments
The Center for Plant-Powered Production-P3 was funded by National Science Foundation-EPSCoR Grant EPS 0701890. The Indiana University, National Center for Genome Analysis Support was funded by the National Science Foundation Grants DBI-1458641 and ABI-1062432 (to Indiana University, National Center for Genome Analysis Support). Facility support was also received from the Arkansas Science and Technology Authority and the Arkansas Biosciences Institute.
This work was supported by United States Department of Agriculture–National Institute of Food and Agriculture Grant 2014-67014-21701. The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1–S14 and Tables S1–S4.
The nucleotide sequence(s) reported in this paper has been submitted to the DDBJ/GenBankTM/EBI Data Bank with accession number(s) KY565244, KY565245, KY565246, KY565247, and KY565248.
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party hosted site.
- DMAPP
- dimethylallyl pyrophosphate
- IPP
- isopentenyl pyrophosphate
- GPP
- geranyl pyrophosphate
- FPP
- farnesyl pyrophosphate
- GGPP
- geranylgeranyl pyrophosphate
- cTP
- chloroplast transit peptide
- mTP
- mitochondrial targeting peptide
- TEV
- tobacco etch virus
- RNA-Seq
- RNA-sequencing
- Rubisco
- ribulose-bisphosphate carboxylase/oxygenase
- qPCR
- quantitative real-time PCR
- MeJA
- methyl jasmonate
- CD
- methyl-β-cyclodextrin
- HMBC
- heteronuclear multiple bond correlation
- HSQC
- heteronuclear single-quantum correlation.
References
- 1. Ahuja I., Kissen R., and Bones A. M. (2012) Phytoalexins in defense against pathogens. Trends Plant Sci. 17, 73–90 10.1016/j.tplants.2011.11.002 [DOI] [PubMed] [Google Scholar]
- 2. Gambini J., Inglés M., Olaso G., Lopez-Grueso R., Bonet-Costa V., Gimeno-Mallench L., Mas-Bargues C., Abdelaziz K. M., Gomez-Cabrera M. C., Vina J., and Borras C. (2015) Properties of resveratrol: in vitro and in vivo studies about metabolism, bioavailability, and biological effects in animal models and humans. Oxid. Med. Cell Longev. 2015, 837042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Tomé-Carneiro J., Larrosa M., González-Sarrías A., Tomás-Barberán F. A., García-Conesa M. T., and Espín J. C. (2013) Resveratrol and clinical trials: the crossroad from in vitro studies to human evidence. Curr. Pharm. Des. 19, 6064–6093 10.2174/13816128113199990407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Baur J. A., and Sinclair D. A. (2006) Therapeutic potential of resveratrol: the in vivo evidence. Nat. Rev. Drug Discov. 5, 493–506 10.1038/nrd2060 [DOI] [PubMed] [Google Scholar]
- 5. Aguamah G. E., Langcake P., Leworthy D. P., Page J. A., Pryce R. J., and Strange R. N. (1981) Two novel stilbene phytoalexins from Arachis hypogaea. Phytochemistry 20, 1381–1383 10.1016/0031-9422(81)80044-1 [DOI] [Google Scholar]
- 6. Cooksey C., Garratt P., and Richards S. (1988) A dienyl stilbene phytoalexin from Arachis hypogaea. Phytochemistry 27, 1015–1016 10.1016/0031-9422(88)80263-2 [DOI] [Google Scholar]
- 7. Sobolev V. S., Potter T. L., and Horn B. W. (2006) Prenylated stilbenes from peanut root mucilage. Phytochem. Anal. 17, 312–322 10.1002/pca.920 [DOI] [PubMed] [Google Scholar]
- 8. Keen N. T., and Ingham J. L. (1976) New stilbene phytoalexins from American cultivars of Arachis hypogaea. Phytochemistry 15, 1794–1795 10.1016/S0031-9422(00)97495-8 [DOI] [Google Scholar]
- 9. Ingham J. L. (1976) 3,5,4′-Trihydroxystilbene as a phytoalexin from groundnuts (Arachis hypogaea). Phytochemistry 15, 1791–1793 10.1016/S0031-9422(00)97494-6 [DOI] [Google Scholar]
- 10. Wotton H. R., and Strange R. N. (1985) Circumstantial evidence for phytoalexin involvement in the resistance of peanuts to Aspergillus flavus. J. Gen. Microbiol. 131, 487–494 [DOI] [PubMed] [Google Scholar]
- 11. Wang H., Lei Y., Yan L., Cheng K., Dai X., Wan L., Guo W., Cheng L., and Liao B. (2015) Deep sequencing analysis of transcriptomes in Aspergillus flavus in response to resveratrol. BMC Microbiol. 15, 182 10.1186/s12866-015-0513-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wu Z., Song L., and Huang D. (2011) Food grade fungal stress on germinating peanut seeds induced phytoalexins and enhanced polyphenolic antioxidants. J. Agric. Food Chem. 59, 5993–6003 10.1021/jf200776w [DOI] [PubMed] [Google Scholar]
- 13. Sobolev V. S. (2013) Production of phytoalexins in peanut (Arachis hypogaea) seed elicited by selected microorganisms. J. Agric. Food Chem. 61, 1850–1858 10.1021/jf3054752 [DOI] [PubMed] [Google Scholar]
- 14. Sobolev V. S., Neff S. A., and Gloer J. B. (2010) New dimeric stilbenoids from fungal-challenged peanut (Arachis hypogaea) seeds. J. Agric. Food Chem. 58, 875–881 10.1021/jf903410e [DOI] [PubMed] [Google Scholar]
- 15. Sobolev V. S., Neff S. A., and Gloer J. B. (2009) New stilbenoids from peanut (Arachis hypogaea) seeds challenged by an Aspergillus caelatus strain. J. Agric. Food Chem. 57, 62–68 10.1021/jf802891v [DOI] [PubMed] [Google Scholar]
- 16. Sobolev V. S., Krausert N. M., and Gloer J. B. (2016) New monomeric stilbenoids from peanut (Arachis hypogaea) seeds challenged by an Aspergillus flavus strain. J. Agric. Food Chem. 64, 579–584 10.1021/acs.jafc.5b04753 [DOI] [PubMed] [Google Scholar]
- 17. Araya-Cloutier C., den Besten H. M., Aisyah S., Gruppen H., and Vincken J.-P. (2017) The position of prenylation of isoflavonoids and stilbenoids from legumes (Fabaceae) modulates the antimicrobial activity against Gram positive pathogens. Food Chem. 226, 193–201 10.1016/j.foodchem.2017.01.026 [DOI] [PubMed] [Google Scholar]
- 18. Huang C.-P., Au L.-C., Chiou R. Y.-Y., Chung P.-C., Chen S.-Y., Tang W.-C., Chang C.-L., Fang W.-H., and Lin S.-B. (2010) Arachidin-1, a peanut stilbenoid, induces programmed cell death in human leukemia HL-60 cells. J. Agric. Food Chem. 58, 12123–12129 10.1021/jf102993j [DOI] [PubMed] [Google Scholar]
- 19. Chang J.-C., Lai Y.-H., Djoko B., Wu P.-L., Liu C.-D., Liu Y.-W., and Chiou R. Y.-Y. (2006) Biosynthesis enhancement and antioxidant and anti-inflammatory activities of peanut (Arachis hypogaea L.) arachidin-1, arachidin-3, and isopentadienylresveratrol. J. Agric. Food Chem. 54, 10281–10287 10.1021/jf0620766 [DOI] [PubMed] [Google Scholar]
- 20. Sobolev V. S., Khan S. I., Tabanca N., Wedge D. E., Manly S. P., Cutler S. J., Coy M. R., Becnel J. J., Neff S. A., and Gloer J. B. (2011) Biological activity of peanut (Arachis hypogaea) phytoalexins and selected natural and synthetic stilbenoids. J. Agric. Food Chem. 59, 1673–1682 10.1021/jf104742n [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Brents L. K., Medina-Bolivar F., Seely K. A., Nair V., Bratton S. M., Ñopo Olazabal L., Patel R. Y., Liu H., Doerksen R. J., Prather P. L., and Radominska-Pandya A. (2012) Natural prenylated resveratrol analogs arachidin-1 and -3 demonstrate improved glucuronidation profiles and have affinity for cannabinoid receptors. Xenobiotica 42, 139–156 10.3109/00498254.2011.609570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ball J. M., Medina-Bolivar F., Defrates K., Hambleton E., Hurlburt M. E., Fang L., Yang T., Nopo-Olazabal L., Atwill R. L., Ghai P., and Parr R. D. (2015) Investigation of stilbenoids as potential therapeutic agents for rotavirus gastroenteritis. Adv. Virol. 2015, 1–10 10.1155/2015/293524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Puksasook T., Kimura S., Tadtong S., Jiaranaikulwanitch J., Pratuangdejkul J., Kitphati W., Suwanborirux K., Saito N., and Nukoolkarn V. (2017) Semisynthesis and biological evaluation of prenylated resveratrol derivatives as multi-targeted agents for Alzheimer's disease. J. Nat. Med. 71, 665–682 10.1007/s11418-017-1097-2 [DOI] [PubMed] [Google Scholar]
- 24. Yang T., Fang L., Nopo-Olazabal C., Condori J., Nopo-Olazabal L., Balmaceda C., and Medina-Bolivar F. (2015) Enhanced production of resveratrol, piceatannol, arachidin-1, and arachidin-3 in hairy root cultures of peanut co-treated with methyl jasmonate and cyclodextrin. J. Agric. Food Chem. 63, 3942–3950 10.1021/jf5050266 [DOI] [PubMed] [Google Scholar]
- 25. Condori J., Sivakumar G., Hubstenberger J., Dolan M. C., Sobolev V. S., and Medina-Bolivar F. (2010) Induced biosynthesis of resveratrol and the prenylated stilbenoids arachidin-1 and arachidin-3 in hairy root cultures of peanut: Effects of culture medium and growth stage. Plant Physiol. Biochem. 48, 310–318 10.1016/j.plaphy.2010.01.008 [DOI] [PubMed] [Google Scholar]
- 26. Yang T., Fang L., Rimando A. M., Sobolev V., Mockaitis K., and Medina-Bolivar F. (2016) A stilbenoid-specific prenyltransferase utilizes dimethylallyl pyrophosphate from the plastidic terpenoid pathway. Plant Physiol. 171, 2483–2498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sasaki K., Mito K., Ohara K., Yamamoto H., and Yazaki K. (2008) Cloning and characterization of naringenin 8-prenyltransferase, a flavonoid-specific prenyltransferase of Sophora flavescens. Plant Physiol. 146, 1075–1084 10.1104/pp.107.110544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Akashi T., Sasaki K., Aoki T., Ayabe S., and Yazaki K. (2008) Molecular cloning and characterization of a cDNA for pterocarpan 4-dimethylallyltransferase catalyzing the key prenylation step in the biosynthesis of glyceollin, a soybean phytoalexin. Plant Physiol. 149, 683–693 10.1104/pp.108.123679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sasaki K., Tsurumaru Y., Yamamoto H., and Yazaki K. (2011) Molecular characterization of a membrane-bound prenyltransferase specific for isoflavone from Sophora flavescens. J. Biol. Chem. 286, 24125–24134 10.1074/jbc.M111.244426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Shen G., Huhman D., Lei Z., Snyder J., Sumner L. W., and Dixon R. A. (2012) Characterization of an isoflavonoid-specific prenyltransferase from Lupinus albus. Plant Physiol. 159, 70–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Li J., Chen R., Wang R., Liu X., Xie D., Zou J., and Dai J. (2014) GuA6DT, a regiospecific prenyltransferase from Glycyrrhiza uralensis, catalyzes the 6-prenylation of flavones. ChemBioChem 15, 1673–1681 10.1002/cbic.201402160 [DOI] [PubMed] [Google Scholar]
- 32. Chen R., Liu X., Zou J., Yin Y., Ou B., Li J., Wang R., Xie D., Zhang P., and Dai J. (2013) Regio- and stereospecific prenylation of flavonoids by Sophora flavescens prenyltransferase. Adv. Synth. Catal. 355, 1817–1828 10.1002/adsc.201300196 [DOI] [Google Scholar]
- 33. Mortazavi A., Williams B. A., McCue K., Schaeffer L., and Wold B. (2008) Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 5, 621–628 10.1038/nmeth.1226 [DOI] [PubMed] [Google Scholar]
- 34. Bertioli D. J., Cannon S. B., Froenicke L., Huang G., Farmer A. D., Cannon E. K., Liu X., Gao D., Clevenger J., Dash S., Ren L., Moretzsohn M. C., Shirasawa K., Huang W., Vidigal B., et al. (2016) The genome sequences of Arachis duranensis and Arachis ipaensis, the diploid ancestors of cultivated peanut. Nat. Genet. 48, 438–446 10.1038/ng.3517 [DOI] [PubMed] [Google Scholar]
- 35. Emanuelsson O., Nielsen H., and von Heijne G. (1999) ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci. 8, 978–984 10.1110/ps.8.5.978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bannai H., Tamada Y., Maruyama O., Nakai K., and Miyano S. (2002) Extensive feature detection of N-terminal protein sorting signals. Bioinformatics 18, 298–305 10.1093/bioinformatics/18.2.298 [DOI] [PubMed] [Google Scholar]
- 37. Yazaki K., Sasaki K., and Tsurumaru Y. (2009) Prenylation of aromatic compounds, a key diversification of plant secondary metabolites. Phytochemistry 70, 1739–1745 10.1016/j.phytochem.2009.08.023 [DOI] [PubMed] [Google Scholar]
- 38. Karamat F., Olry A., Munakata R., Koeduka T., Sugiyama A., Paris C., Hehn A., Bourgaud F., and Yazaki K. (2014) A coumarin-specific prenyltransferase catalyzes the crucial biosynthetic reaction for furanocoumarin formation in parsley. Plant J. 77, 627–638 10.1111/tpj.12409 [DOI] [PubMed] [Google Scholar]
- 39. Banfi D., and Patiny L. (2008) Resurrecting and processing NMR spectra on-line. Chim. Int. J. Chem. 62, 280–281 10.2533/chimia.2008.280 [DOI] [Google Scholar]
- 40. Castillo A. M., Patiny L., and Wist J. (2011) Fast and accurate algorithm for the simulation of NMR spectra of large spin systems. J. Magn. Reson. 209, 123–130 10.1016/j.jmr.2010.12.008 [DOI] [PubMed] [Google Scholar]
- 41. Krogh A., Larsson B., von Heijne G., and Sonnhammer E. L. (2001) Predicting transmembrane protein topology with a hidden markov model: application to complete genomes. J. Mol. Biol. 305, 567–580 10.1006/jmbi.2000.4315 [DOI] [PubMed] [Google Scholar]
- 42. Hauser M., Eichelmann H., Oja V., Heber U., and Laisk A. (1995) Stimulation by light of rapid pH regulation in the chloroplast stroma in vivo as indicated by CO2 solubilization in leaves. Plant Physiol. 108, 1059–1066 10.1104/pp.108.3.1059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Nelson B. K., Cai X., and Nebenführ A. (2007) A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J. 51, 1126–1136 10.1111/j.1365-313X.2007.03212.x [DOI] [PubMed] [Google Scholar]
- 44. Wang R., Chen R., Li J., Liu X., Xie K., Chen D., Yin Y., Tao X., Xie D., Zou J., Yang L., and Dai J. (2014) Molecular characterization and phylogenetic analysis of two novel regio-specific flavonoid prenyltransferases from Morus alba and Cudrania tricuspidata. J. Biol. Chem. 289, 35815–35825 10.1074/jbc.M114.608265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Huang H., Levin E. J., Liu S., Bai Y., Lockless S. W., and Zhou M. (2014) Structure of a membrane-embedded prenyltransferase homologous to UBIAD1. PLos Biol. 12, e1001911 10.1371/journal.pbio.1001911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Tanaka Y., and Brugliera F. (2013) Flower colour and cytochromes P450. Philos. Trans. R. Soc. B Biol. Sci. 368, 2012.0432 10.1098/rstb.2012.0432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Bolger A. M., Lohse M., and Usadel B. (2014) Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Haas B. J., Papanicolaou A., Yassour M., Grabherr M., Blood P. D., Bowden J., Couger M. B., Eccles D., Li B., Lieber M., Macmanes M. D., Ott M., Orvis J., Pochet N., Strozzi F., et al. (2013) De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 8, 1494–1512 10.1038/nprot.2013.084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Robertson G., Schein J., Chiu R., Corbett R., Field M., Jackman S. D., Mungall K., Lee S., Okada H. M., Qian J. Q., Griffith M., Raymond A., Thiessen N., Cezard T., Butterfield Y. S., et al. (2010) De novo assembly and analysis of RNA-seq data. Nat. Methods 7, 909–912 10.1038/nmeth.1517 [DOI] [PubMed] [Google Scholar]
- 50. Schulz M. H., Zerbino D. R., Vingron M., and Birney E. (2012) Oases: Robust de novo RNA-seq assembly across the dynamic range of expression levels. Bioinformatics 28, 1086–1092 10.1093/bioinformatics/bts094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Li R., Yu C., Li Y., Lam T. W., Yiu S. M., Kristiansen K., and Wang J. (2009) SOAP2: An improved ultrafast tool for short read alignment. Bioinformatics 25, 1966–1967 10.1093/bioinformatics/btp336 [DOI] [PubMed] [Google Scholar]
- 52. Li W., and Godzik A. (2006) Cd-hit: A fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22, 1658–1659 10.1093/bioinformatics/btl158 [DOI] [PubMed] [Google Scholar]
- 53. Wu T. D., and Watanabe C. K. (2005) GMAP: A genomic mapping and alignment program for mRNA and EST sequences. Bioinformatics 21, 1859–1875 10.1093/bioinformatics/bti310 [DOI] [PubMed] [Google Scholar]
- 54. Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., and 1000 Genome Project Data Processing Subgroup. (2009) The sequence alignment/map (SAM) format and SAMtools. Bioinformatics 25, 2078–2079 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., and Salzberg S. L. (2013) TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14, R36 10.1186/gb-2013-14-4-r36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Anders S., Pyl P. T., and Huber W. (2015) HTSeq-A Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 10.1093/bioinformatics/btu638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Altschul S. F., Gish W., Miller W., Myers E. W., and Lipman D. J. (1990) Basic local alignment search tool. J. Mol. Biol. 215, 403–410 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- 58. Edgar R. C. (2004) MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5, 113 10.1186/1471-2105-5-113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Guindon S., and Gascuel O. (2003) A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52, 696–704 10.1080/10635150390235520 [DOI] [PubMed] [Google Scholar]
- 60. Medina-Bolívar F., and Cramer C. (2004) Production of recombinant proteins by hairy roots cultured in plastic sleeve bioreactors. Methods Mol. Biol. 267, 351–363 [DOI] [PubMed] [Google Scholar]
- 61. Becker D. (1990) Binary vectors which allow the exchange of plant selectable markers and reporter genes. Nucleic Acids Res. 18, 203 10.1093/nar/18.1.203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Medrano G., Reidy M. J., Liu J., Ayala J., Dolan M. C., and Cramer C. L. (2009) Rapid system for evaluating bioproduction capacity of complex pharmaceutical proteins in plants. Methods Mol. Biol. 483, 51–67 10.1007/978-1-59745-407-0_4 [DOI] [PubMed] [Google Scholar]
- 63. Condori J., Nopo-Olazabal C., Medrano G., and Medina-Bolivar F. (2011) Selection of reference genes for qPCR in hairy root cultures of peanut. BMC Res. Notes 4, 392 10.1186/1756-0500-4-392 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.