Abstract
Twin-arginine translocation (Tat) systems transport folded proteins across cellular membranes with the concerted action of mostly three membrane proteins: TatA, TatB, and TatC. Hetero-oligomers of TatB and TatC form circular substrate–receptor complexes with a central binding cavity for twin-arginine–containing signal peptides. After binding of the substrate, energy from an electro-chemical proton gradient is transduced into the recruitment of TatA oligomers and into the actual translocation event. We previously reported that Tat-dependent protein translocation into membrane vesicles of Escherichia coli is blocked by the compound N,N′-dicyclohexylcarbodiimide (DCCD, DCC). We have now identified a highly conserved glutamate residue in the transmembrane region of E. coli TatC, which when modified by DCCD interferes with the deep insertion of a Tat signal peptide into the TatBC receptor complex. Our findings are consistent with a hydrophobic binding cavity formed by TatB and TatC inside the lipid bilayer. Moreover, we found that DCCD mediates discrete intramolecular cross-links of E. coli TatC involving both its N- and C-tails. These results confirm the close proximity of two distant sequence sections of TatC proposed to concertedly function as the primary docking site for twin-arginine signal peptides.
Keywords: Escherichia coli (E. coli); mass spectrometry (MS); membrane protein; protein cross-linking; protein export; protein targeting; Dicyclohexylcarbodiimide (DCCD, DCC); TatC; twin-arginine translocation
Introduction
The twin-arginine translocation (Tat)3 system has the remarkable ability to transport folded proteins across cellular membranes It is found in the cytoplasmic membranes of bacteria and archaea and the thylakoid membrane of chloroplasts. Tat-substrates are characterized by the highly conserved consensus motif SRRXFLK in their signal peptides (reviewed in Refs. 1–6).
In Escherichia coli, the Tat-translocon consists of single spanning membrane proteins the TatA, TatB, and TatE and the hexahelical TatC. TatA and TatB share a similar core structure. A transmembrane helix (TM), too short to span the bilayer entirely, is linked through a short hinge region to an amphipathic helix that is followed by a C-terminal domain of different size (7–10). The six helices of TatC are tilted within the membrane and most of them are kinked forming the concave structure of a cupped hand (11, 12). It is not clear whether the cavity thus formed is filled with lipids or water.
A TatABCE complex was shown through fluorescence microscopy of living E. coli cells to assemble on demand (13–15). TatB and TatC interact in a 1:1 stoichiometry (16) and several of these TatBC protomers form a receptor complex for a Tat precursor (17, 18). Through TatB intercalating between two neighboring TatC monomers, circular TatBC receptor complexes are formed (19, 20), in which TatB was proposed to form the inner and TatC the outer shell of a dome-like structure. A current model of the TatBC complex is depicted in Fig. 1 (looking at its trans-sided surface; a side view of TatC and TatB molecules and their relative positions within the lipid bilayer is shown in Fig. 3a). Neighboring TatC monomers interact through the TM of TatB as well as via their periplasmic loops (19, 21–23). TatA is found at the periphery of the complex (19, 22, 24).
Figure 1.
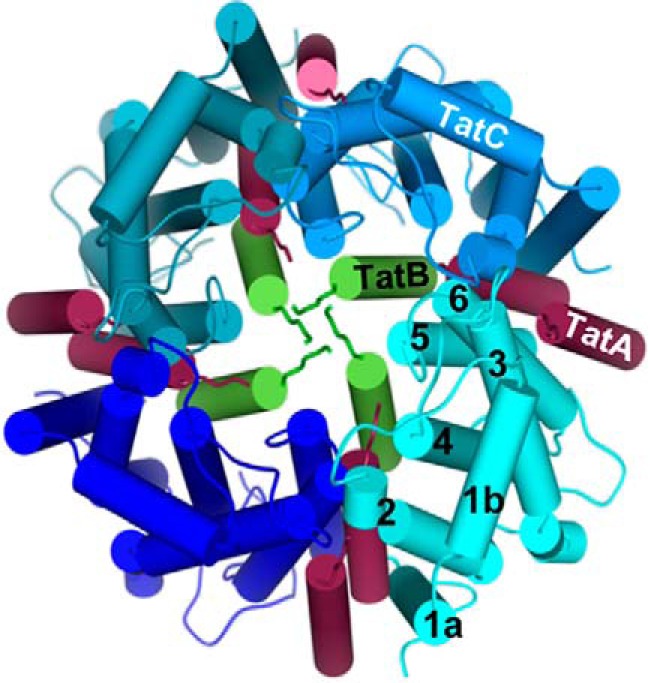
Model of the Tat translocase. The hexahelical X-ray crystallographic structures of four TatC monomers are shown in different shades of blue with the six transmembrane helices numbered for the monomer shown in cyan. A tetrameric structure was chosen for reasons of clarity, whereas experimental data for higher order assemblies have been provided. The isolated transmembrane helices of four TatB molecules (green) and eight TatA molecules (red) were modeled against the TatC structure according to experimentally verified contact sites. The side-by-side arrangement of two neighboring TatC monomers is also derived from cross-linking studies. View is from the trans-side down the membrane normal with substrates (not shown) approaching from below.
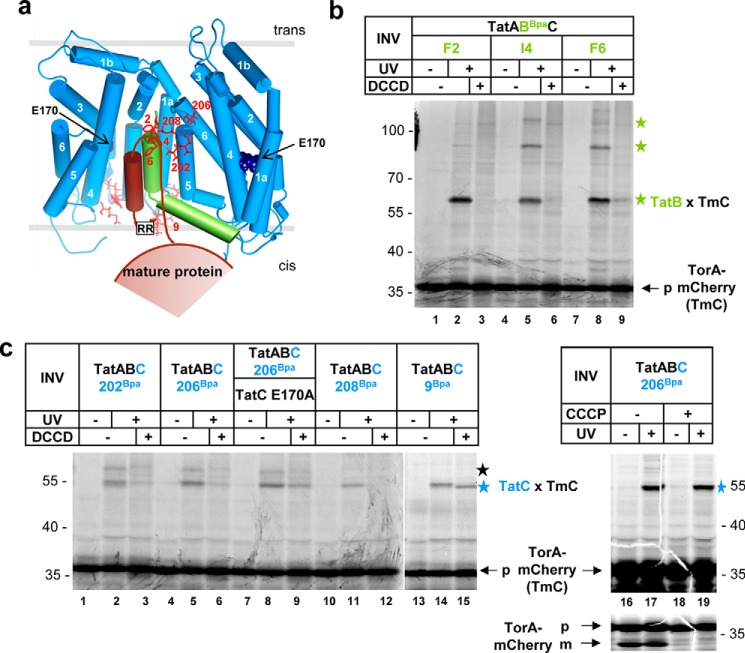
Figure 3.
Influence of DCCD on the binding of a Tat substrate to TatB and TatC. a, model of a signal peptide inserting into the TatBC complex according to Ref. 1. The Tat signal peptide attaches via its RR-consensus motif to the cis-sided signal peptide-binding site of the left TatC protomer represented by residue L9 and contacts the TM of TatB and TM5 of the adjacent TatC molecule. Labeled are residues of TatC and TatB that interact with the Tat substrate, as shown in b and c and previously (19, 22). The DCCD-sensitive TatC residue E170A is indicated. b, autoradiography. TorA–mCherry (TmC) was synthesized and radioactively labeled in vitro. Samples were either treated with DMSO or DCCD and vesicles (INV) harboring the indicated Bpa variants of TatB were added. For cross-linking, samples were irradiated with UV light. Green stars, TatB–TmC adducts. c as in b using INV with the indicated Bpa variants of TatC or in addition with the TatCE170A substitution. When indicated, samples were treated with 0.1 mm CCCP after synthesis. The lower panel of lanes 16–19 is a lighter representation of the upper autoradiograph with a better resolution of precursor (p) and mature forms (m) of TmC. Blue stars, TatC–TmC adducts; black star, unidentified adduct (19).
Both TatB and TatC recognize a Tat-signal peptide in a concerted fashion (19, 25–29). The RR-motive is first recognized by the N-terminal domain and the TM2/TM3 loop of TatC (21, 25, 26, 30). Subsequently, a Tat-signal peptide inserts deeply into a TatB/TatC-walled cavity (19, 24, 27–29, 31, 32), the conformation of which in turn is influenced by the signal peptide itself (29).
Upon substrate binding, TatA is thought to promote the actual translocation step by either forming a translocation pore (reviewed in Ref. 3) or destabilizing the membrane (8, 33, 34). Both, recruitment of TatA oligomers as well as the thereby triggered translocation event require the proton-motive force (PMF) as sole energy source.
N,N′-Dicyclohexylcarbodiimide (DCCD, DCC) was previously shown to act as an inhibitor of the E. coli Tat-system by preventing the binding of a Tat-substrate to the Tat-translocase (35). In screening E. coli TatC for potential binding sites of DCCD, we now discovered that modification by DCCD of the highly conserved and deeply membrane-embedded glutamyl residue 170 interferes with the insertion of a Tat-signal peptide into the TatBC complex. In addition, DCCD-mediated intramolecular cross-linking of TatC revealed conformational details of the RR-recognition site of E. coli TatC.
Results
Glutamate 170 of E. coli TatC becomes quantitatively modified by DCCD
DCCD is known to modify carboxyl side chains that are located in hydrophobic regions of proteins giving rise to N-acyl urea adducts (36, 37) (Fig. S1a). To identify potential DCCD-reactive carboxyl side chains of TatC, E. coli membrane vesicles containing overexpressed TatA, TatB, and a His-tagged TatC variant were treated with DCCD in the absence of substrate, and TatC was subsequently purified by affinity chromatography and SDS-PAGE. Peptides derived from a combined digestion of monomeric TatC with trypsin and chymotrypsin were analyzed by liquid chromatography-tandem mass spectrometry (LC-MS/MS). Data analysis consisted of comparison of the data with known protein sequences and chromatographic peak integration using the MaxQuant program taking into account possible modifications by DCCD. The recovery of TatC peptides and their cumulative MS intensities are plotted in Fig. 2a along the E. coli TatC sequence, and the theoretical as well as the experimentally verified trypsin and chymotrypsin cleavage sites of TatC are depicted in Fig. S2. Sequence coverage of TatC was 90.3% with the three gaps indicated in Fig. 2a and Fig. S2. The first one flanked by Lys18 and Phe37 represents the hydrophobic stretch of TM1a and the third between Lys191 and Val196 is located at the beginning of TM5 (Fig. S2). Whereas these two sections of TatC were also missing in the MS/MS spectra obtained from an untreated TatC sample (not shown), the central gap (Lys101–Arg105) was due to the treatment with DCCD as demonstrated below. Except for Glu103, which is further discussed below, the non-recovered sequence sections of TatC were devoid of Asp and Glu residues as potential target sites for DCCD.
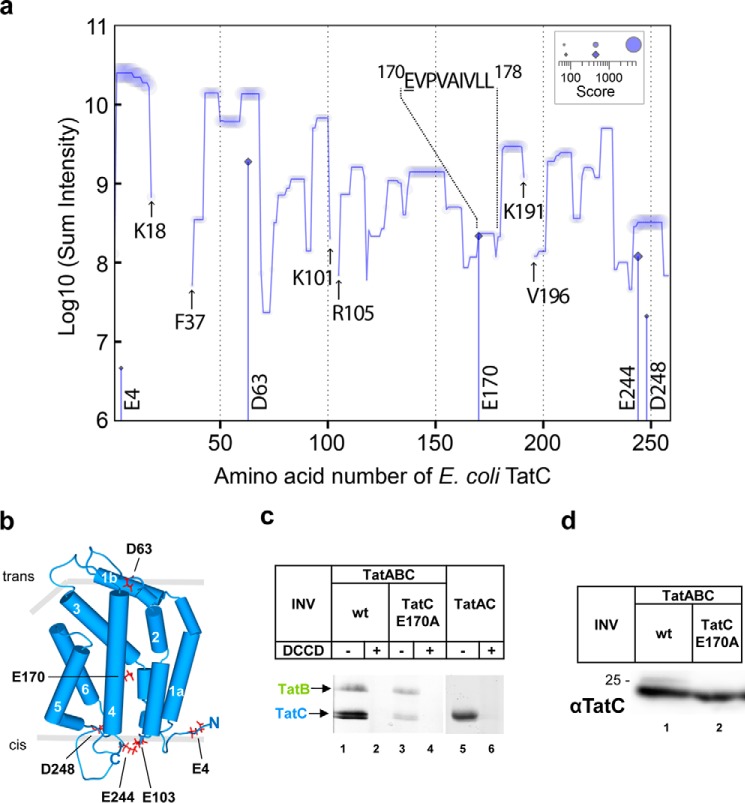
Figure 2.
Glutamate 170 of E. coli TatC becomes quantitatively modified by DCCD. a, sequence coverage and sites of modification by DCCD of E. coli TatC analyzed by quantitative mass spectrometry. For each amino acid along the sequence of TatC, MS intensities of peptides containing this residue were summed up and plotted against its sequence residue number. Relative quantification of peptides was performed based on detected ion intensities. Peptide scores are indicated and reflect the probability-based significance of peptide identifications based on matched fragment ions in MS/MS spectra. The sum of peptide scores per amino acid is decoded by the size of the symbols. Indicated are amino acids flanking the three sequence sections of DCCD-treated TatC that were not covered by the MS/MS analysis (arrows). Cumulative intensities of peptides identified with modification of aspartic or glutamic acid by DCU are plotted at positions of the modified sites (diamonds on vertical lines) according to the same logarithmic scale as the total sum of intensities including modified and unmodified peptides. Virtually 100% of the peptide 170EVPVAIVLL178 depicted was found modified by DCU. b, model of the E. coli TatC structure adapted from the crystal structure of A. aeolicus TatC (PDB codes 4B4A and 4HTT) (11, 12) with the six transmembrane helices and the N and C termini marked. Residue Glu170, which is virtually completely modified by DCCD, and the other DCCD sensitive residues are marked in red. The bilayer is outlined by gray bars, the cis-side corresponds to the cytoplasmic face of E. coli TatC. c, vesicles (INV) were prepared from E. coli strains overexpressing TatABC (wt), TatABCE170A, and TatAC. INV were either treated with DCCD or DMSO before incubating with the fluorescent DCCD analogue NCD-4. Proteins were separated by SDS-PAGE and analyzed under UV light. d, Western blot analysis using anti-TatC antibodies to demonstrate unimpaired expression of the E170A variant of TatC.
The vertical bars in Fig. 2a mark all amino acid residues of E. coli TatC that were found to carry the additional mass of dicyclohexylurea (DCU) (cf. Fig. S1a), namely Glu4, Asp63, Glu170, Glu244, and Asp248 (also highlighted in the structure representation of E. coli TatC shown in Fig. 2b) and the lengths of these bars indicate the cumulative MS intensities of peptides harboring the DCU modification at the respective positions. Relative to the intensities of the non-modified peptides, the lengths of the bars reflect the extent by which modification through DCCD occurred. Virtually 100% of the peptide 170EVPVAIVLL178 (Fig. 2a) contained the DCCD-derived modification, whereas modification of Glu244 was observed with less than 50% of total intensity of the respective peptide and those of the others with 10% or lower. The validity of the modification of Glu170 of TatC by DCCD is further documented in Fig. S3 depicting properties of the isolated peptide 170EVPVAIVLL and its MS/MS-generated fragments containing or lacking the DCU moiety. Collectively these data demonstrate that the TM4 residue Glu170 of TatC, which is positioned in the middle of the lipid bilayer (Fig. 2b), represents the predominant DCCD target of E. coli TatC.
Labeling of TatCGlu-170 with DCCD could further be demonstrated using the fluorescent analogue of DCCD, NCD-4 (N-cyclohexyl-N′-(4-dimethylamino-α-naphthyl)carbodiimide) (Fig. S1b) (38). For this purpose, inside-out inner membrane vesicles (INV) of E. coli containing TatABC at overexpressed levels were treated with NCD-4, either directly or after preincubation with a 10-fold molar excess of DCCD. Membrane proteins were then separated by SDS-PAGE and inspected under UV-light. Two bands became fluorescently labeled with NCD-4, unless DCCD was also present, indicating DCCD-specific binding sites in both proteins (Fig. 2c, lanes 1 and 2). The lower of the two bands was of the size of TatC. Accordingly, its labeling with NCD-4 was drastically reduced when membrane vesicles were used that had Glu170, the major DCCD target of TatC, exchanged against alanine (lane 3). As demonstrated in Fig. 2d, this decrease in labeling with NCD-4 could not be accounted for by reduced TatC levels in the TatCE170A vesicles but must have been caused by the E170A mutation of TatC. These results therefore prove the identity of the lower band with TatC and confirm the accessibility of TatC residue Glu170 to DCCD and NCD-4. The residual labeling by NCD-4 of TatC carrying the E170A mutation (Fig. 2c, lane 3) is most likely due to binding of NCD-4 to one or more of the minor DCCD targets revealed by mass spectrometry. The upper band that became labeled by NCD-4 was of the size of TatB and in fact was not obtained for TatB-lacking membrane vesicles (lane 5). Thus obviously also TatB contains DCCD-sensitive residues but this is not subject of this study.
DCCD interferes with the proper accommodation of a Tat signal peptide within the TatBC-binding cavity
Upon binding, the RR-consensus motif of a Tat signal peptide is recognized by surface-exposed residues of the N-tail and the TM2/TM3 loop of TatC (12, 21, 22), whereas the downstream part of the signal peptide inserts as a hairpin into a TatBC-formed cavity (19, 24). In this cavity, it contacts the N terminus of TatB (19, 27, 28) as well as trans-sided residues of the TM5 of TatC (19, 24). In accordance with a current model (1), Fig. 3a illustrates how an RR precursor might be accommodated in the TatBC receptor complex. To understand how DCCD might affect this binding step, we synthesized and radioactively labeled the model Tat substrate TorA–mCherry (39) in vitro in the presence of TatABC-containing INV carrying the photo-activatable cross-linker p-benzoylphenylalanine (Bpa) either in the non-helical N-tail of TatB or at the internal binding site of TatC (Fig. 3a, residues marked in red). As shown in Fig. 3b, Bpa variants at positions Phe2, Ile4, and Phe6 of TatB when exposed to UV-light yielded a prominent radioactive 60-kDa product representing the adduct of one TatB molecule to the radioactively labeled 37-kDa precursor TorA–mCherry (green star, TatB × TmC). In addition, higher molecular mass adducts were obtained representing adducts between precursor and more than one TatB molecule (green stars). This follows from the fact that they carry the radioactive label of the precursor and must contain a Bpa-hosting protein, the only one of which is TatB in this setup (19). These adducts did not form, or were at least drastically diminished, in the presence of DCCD (Fig. 3b, lanes 3, 6, and 9). Similarly, when Bpa was replacing the trans-sided residues Val202, Leu206, and Thr208 in the TM5 of TatC, 1:1 cross-links between TorA–mCherry and TatC were obtained (Fig. 3c, blue star). Again DCCD considerably interfered with the formation of these adducts (lanes 3, 6, and 12). This was, however, not the case when Bpa had been incorporated into the superficial binding site for the consensus motif of Tat signal peptides at position L9 of TatC (lanes 14 and 15).
This latter result demonstrates that DCCD does not prevent binding of a Tat substrate to TatC but specifically seems to impair its hairpin-like insertion into the TatBC-binding cavity. Notably, when glutamate 170, the main target site of DCCD in TatC, had been mutated to alanine, DCCD hardly interfered with precursor binding to TatC206 (Fig. 3c, compare lanes 5 and 6 with 8 and 9). This result indicates that the major reason for DCCD blocking precursor insertion was modification of Glu170 of TatC. Different from DCCD, the protonophore CCCP (carbonyl cyanide m-chlorophenylhydrazone) did not prevent cross-linking between TorA–mCherry and residue 206 of TatC (Fig. 3c, compare lanes 17 and 19), although it efficiently blocked transport of TorA–mCherry into INV as indicated by the lack of signal sequence processing (m-form of TorA–mCherry, lanes 18 and 19). This rules out that DCCD inhibited the binding of TorA–mCherry to the internal binding site of TatC via dissipation of the PMF, which DCCD causes by default through blockage of the vesicle-bound F1FO-ATPase (40).
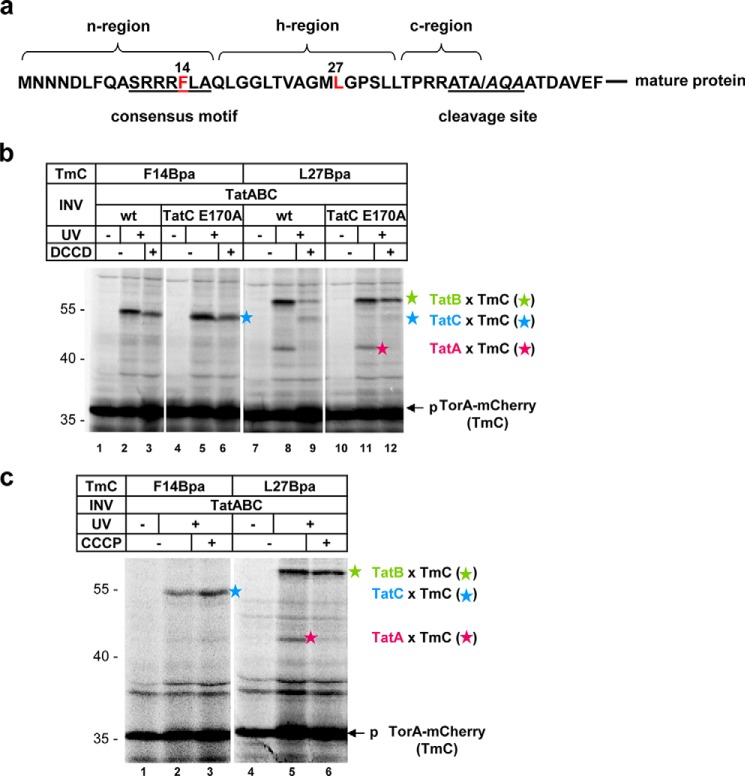
To further demonstrate that DCCD, by binding to Glu170 of TatC, interfered with the hairpin-like insertion of a Tat signal peptide into the TatBC-binding cavity, we analyzed the interaction between Tat precursor and TatBC also by incorporating the cross-linker Bpa into the TorA signal sequence at the two sites highlighted in Fig. 4a. As shown in numerous previous reports (19, 21, 25, 26, 35, 41), the consensus motif, represented in Fig. 4a by the Phe14-Bpa mutation of TorA–mCherry, cross-links to TatC (Fig. 4b, lane 2, blue star). On the contrary, the hydrophobic core of the signal peptide, represented in Fig. 4a by the Leu27-Bpa mutation, cross-links to TatB and to some degree also to TatA (Fig. 4b, lane 8, green and pink stars). DCCD did not interfere with cross-linking of the Phe14-Bpa variant of TorA–mCherry to TatC (Fig. 4b, compare lanes 2 and 3). This is totally consistent with the finding shown in Fig. 3c that binding of TorA–mCherry to Leu9 located within the RR-recognition site of TatC was unaffected by DCCD.
Figure 4.
Influence of DCCD on signal peptide binding to the TatABC-receptor complex. a, amino acid sequence of the TorA-signal peptide. Amino acids that were replaced by Bpa are marked in red. b and c, autoradiographies. TorA–mCherry (TmC) variants harboring Bpa at the indicated positions were synthesized and radioactively labeled in vitro. Samples were treated with DMSO, DCCD, and CCCP as indicated prior to incubation with TatABC (wt) vesicles (INV) or TatABC INV harboring the TatCE170A mutation. For cross-linking, samples were irradiated with UV light. p, precursor of TmC; blue, green, and pink stars, adducts of TmC to TatC, TatB, and TatA, respectively.
In contrast, cross-linking of the Leu27-Bpa variant of TorA–mCherry to TatB, as well as to TatA, was strongly reduced when DCCD was added (Fig. 4b, lanes 8 and 9). Instead, an adduct of the size of TorA–mCherry cross-linked to TatC appeared (lane 9, blue star), similar to what we previously observed for INV that totally lacked TatB (19). This DCCD caused reversal of contacts between the signal peptide and TatB and TatC was largely abolished when membrane vesicles were used that contained the E170A variant of TatC (Fig. 4b, green and blue stars, compare lanes 9 and 12). These findings confirm that DCCD interferes with the proper insertion of a Tat substrate into the TatBC-binding cavity through modification of residue Glu170 of TatC. Again, DCCD did not cause these disturbances by dissipating the PMF. This follows from the data shown in Fig. 3c that in contrast to DCCD, the protonophore CCCP did not diminish cross-linking of the RR precursor to TatC (blue star) and TatB (green star), although it abolished the PMF-sensitive interaction of the Leu27-Bpa variant of TorA–mCherry with TatA (pink star).
Interference of DCCD with the proper insertion of a Tat signal peptide into the TatBC-binding cavity could further be demonstrated using a different strategy. In Fig. 5, TorA–mCherry was synthesized and radioactively labeled in vitro. In the presence of TatABC vesicles (Fig. 5a, lane 1), about half of the precursor of TorA–mCherry (p) was found processed to the mature form (m). Because of its resistance toward proteinase K (lane 2) the m-form must have been translocated into the lumen of the vesicles. By the same criterion, also some non-processed precursor was found translocated, although this fraction of precursor was partially digested by proteinase K removing a few amino acids from the N terminus of the membrane-embedded TorA signal peptide (22). Dissipation of the PMF by CCCP (lane 4) and impairment of signal peptide insertion by DCCD (lane 6) totally prevented the accumulation of any proteinase K-resistant p- and m-forms of TorA–mCherry. Similarly, when instead of TatABC vesicles, TatAC vesicles were used, translocation of TorA–mCherry was also completely abolished, now due to the missing TatB (lane 8).
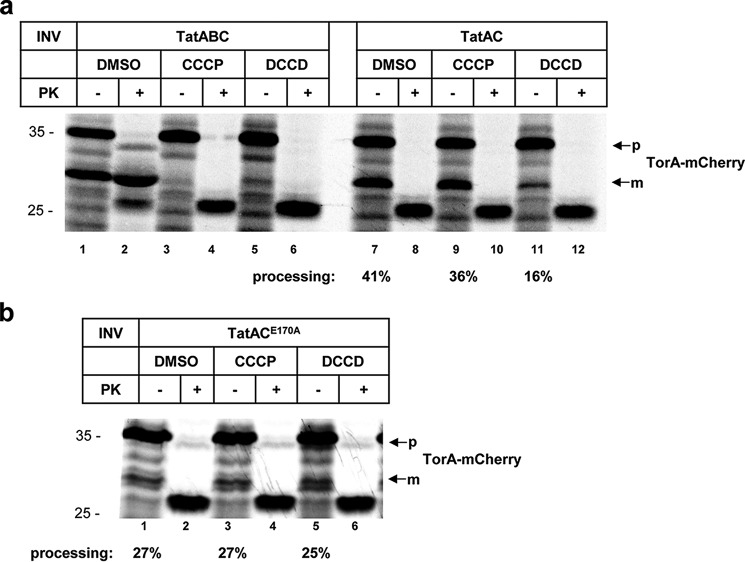
Figure 5.
Influence of DCCD on the premature cleavage of a Tat-signal peptide in the absence of TatB. Autoradiographies are shown. TorA–mCherry was synthesized and radioactively labeled in vitro. a, samples were treated with DMSO, CCCP, or DCCD as indicated, before adding vesicles (INV) containing either TatABC or only TatA and TatC. Upon addition of TatABC vesicles, the precursor of TorA–mCherry (p) is processed to the mature form (m). Transport into INV is indicated by proteinase K (PK) resistance. The PK-resistant precursor is of slightly reduced size because of the removal a few N-terminal amino acids by PK. When TatAC vesicles were used, digestion of precursor (p) and mature forms (m) by PK indicates a lack of transport due to the missing TatB. The translocation-independent processing of the precursor to the mature form was quantified from three independent experiments. b, samples were treated as in a before adding vesicles containing only TatA and the TatCE170A variant.
Nevertheless, TatB-deficient vesicles allowed for the appearance of the m-form of TorA–mCherry (lane 7). As previously reported (31), in the absence of TatB, TatC obviously inserts the TorA signal peptide across the membrane so that it becomes prematurely cleaved off by signal peptidase without prior translocation of the Tat substrate into the vesicles. Upon treatment of TatAC vesicles with DCCD, the premature cleavage of the TorA signal peptide dropped from 41 to 16% (compare lanes 7 and 11). Again, this inhibitory effect of DCCD was not based on DCCD dissipating the PMF of the vesicles, because the uncoupler CCCP did not affect processing of TorA–mCherry by TatAC vesicles to any significant degree (lane 9). However, in TatAC vesicles carrying the TatCE170A variant, DCCD did not any longer interfere with the premature processing of TorA–mCherry (Fig. 5b, compare lanes 1 and 6) indicating that also in the absence of TatB, DCCD impairs the insertion of a Tat signal peptide into the membrane via modifying the Glu170 residue of TatC.
Intramolecular cross-linking by DCCD reveals conformational details of the RR-recognition site of E. coli TatC
As mentioned above for our LC-MS/MS analysis of TatC, the linear peptide sequence His102-Glu103-Arg104 from the largely hydrophilic TM2/TM3 loop of TatC was not recovered when TatC had been treated with DCCD (Fig. 2a and Fig. S2). In theory, this could be explained if DCCD caused an intramolecular cross-link between the missing peptide and another part of the same TatC molecule thereby generating a branched peptide. As illustrated in Fig. S1a, adducts of DCCD to free carboxyl side chains form via a reactive intermediate. If this intermediate is attacked by a nearby primary amine, DCCD is released as dicyclohexylurea and an amide (isopeptide) bond between the original carboxyl group and the attacking amino group is generated. In fact, the LC-MS/MS analysis of DCCD-treated TatC revealed two branched peptides involving the 101KHER104 peptide sequence of the TM2/TM3 loop of TatC (Fig. 6a). One originated from a cross-link of Glu103 to the α-amino group of the N-terminal octapeptide 2SVEDTQPL9 of TatC, whereas the other encompassed the C-terminal tetradecapeptide 242NREEENDAEAESEK255 of TatC cross-linked via Glu244 to Lys101 of the KHER peptide. Fig. S4, a and b demonstrates the identification of both branched peptides via their MS/MS-generated fragments. Both products were not detected in the MS data obtained from non-treated TatC (Fig. 6a, green curve) demonstrating that they resulted from the cross-linking activity of DCCD.
Figure 6.
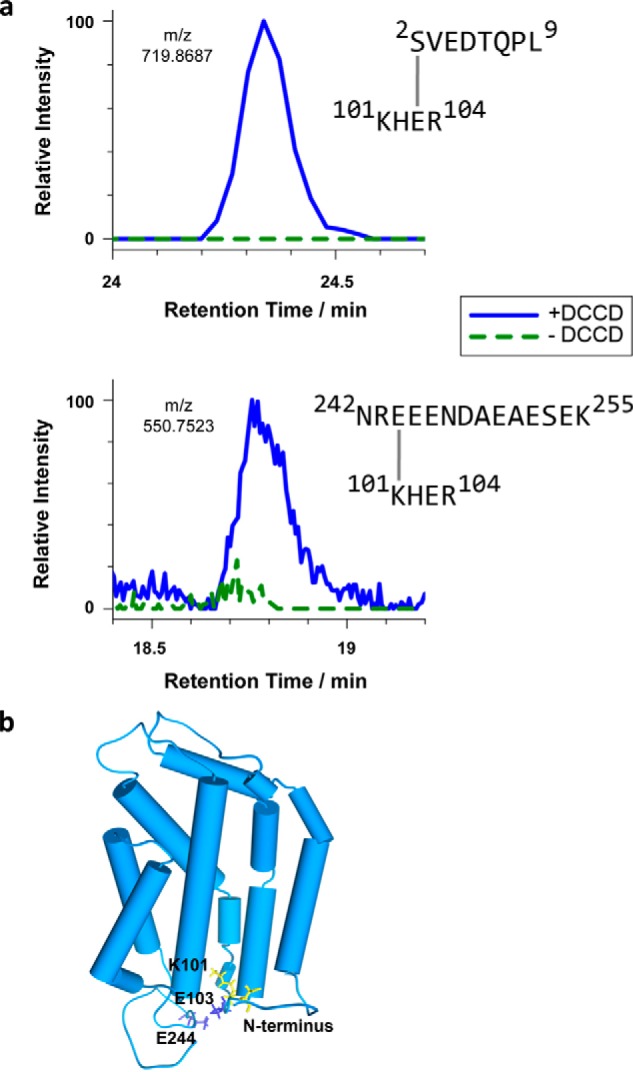
Intramolecular cross-linking by DCCD reveals conformational details of the RR-recognition site of TatC. a, extracted ion chromatograms of cross-linked TatC peptides analyzed by LC-MS. E. coli TatC treated with DCCD (blue curves) or mock-treated (dashed green curves) was isolated by affinity chromatography and SDS-PAGE prior to LC-MS. Identified cross-linked peptides represent intra-molecular TatC contacts between Ser2 and Glu103 (top), quantified on the 2+ charged precursor observed at m/z 719.8681 (molecular mass of 1437.7212 Da), and between Glu244 and Lys101 (bottom), quantified on the 4+ charged precursor at m/z 550.7523 (molecular mass of 2198.9788 Da). b, model of the E. coli TatC structure adapted from the crystal structure of A. aeolicus TatC (PDB codes 4B4A and 4HTT). Residues involved in the DCCD-mediated TatC cross-links specified in panel a are marked in blue (carboxylates) and yellow (amino groups). These intramolecular cross-links suggest an orientation of the N and C termini as modeled here, in which both cytosolic tails fold back toward the TM2/TM3 loop.
These findings indicate that besides the five carboxyl side chain residues Glu4, Asp63, Glu170, Glu244, and Asp248 of TatC, which became modified by DCCD (Fig. 2a), Glu103 is an additional target for DCCD. Moreover, the DCCD-mediated intramolecular cross-linking of TatC provides evidence for a close proximity of the cytosolic TM2/TM3 loop sequence 101KHE103 to both cytosolically oriented N- and C-tails of TatC. The conformations of the N- and C-tails of E. coli TatC, which are longer than those of the Aquifex aeolicus TatC, have not been ascertained thus far. The DCCD-mediated intramolecular TatC cross-links obtained now suggest that the N-terminal and C-terminal domains might fold back on the core structure of TatC (Fig. 6b) thereby contributing to the compact fold of the TatC molecule. Such an orientation is also supported by the identification of an isopeptide resulting from a DCCD-caused cross-link between S2 and Glu244 of TatC (Fig. S4c). Moreover, the juxtaposition of the N-tail and the TM2/TM3 loop is fully consistent with both cytosolic domains of TatC constituting the decoding area of TatC for the RR-pair of the Tat signal peptide (12, 21, 22).
Discussion
In trying to unravel how DCCD blocks binding of a Tat substrate to the Tat translocase, we screened TatC for carboxyl residues that became modified by DCCD and identified glutamate 170 as a major target of DCCD. DCCD treatment of TatC was performed in the absence of added substrate because of the insufficiently tight interaction of an RR-signal peptide with purified TatC (12) potentially causing heterogenous TatC populations. Modification of Glu170 by DCCD perturbed the signal peptide's interaction with trans-sided residues of TatB (N-tail) and TatC (distal part of TM5). Vice versa, in the presence of DCCD, the hydrophobic core of the TorA signal peptide (represented by Leu27) did not any more reach out to TatB but rather contacted TatC. Furthermore, modification of TatCGlu-170 by DCCD interfered with the insertase function of TatC, a property that can be experimentally demonstrated by use of membrane vesicles lacking TatB. In this artificial situation, TatC directly transfers RR precursors to the trans-sided signal peptidase resulting in a proteolytic removal of the signal peptide uncoupled from translocation (31). Importantly, all these DCCD-caused alterations were largely reversed by the TatCE170A mutation indicating that they directly resulted from DCCD modifying Glu170 of TatC.
The DCCD-sensitive residue Glu170 of TatC is highly conserved among bacterial TatCs (42). Mutational replacement of this glutamate residue impairs Tat-specific transport but does not eliminate it (22, 41, 43). A possible role of TatCGlu-170 in the binding of the SRRXFLK consensus motif has been discussed (2, 11). By contrast, cross-linking studies have not disclosed any vicinity of TatCGlu-170 neither to Tat substrates nor to TatB (22). In chloroplast TatC (cpTatC), the residue three positions upstream of the Glu170 equivalent yielded disulfide bonds with the TM of chloroplast TatA (Tha4) and this contact was dependent on the presence of a Tat substrate and the PMF (24). These results would be consistent with the idea that protonation events might allow TatCGlu-170 to form a hydrogen bond with the TM of TatA (12).
All studies performed thus far exclude Glu170 of TatC as a direct binding partner of a Tat signal peptide. Therefore impaired precursor binding to DCCD-modified TatC is unlikely to be caused by DCCD masking Glu170 as a possible interaction site of the cross-linker Bpa. We rather assume that the bulky DCU moiety when attached to TatCGlu-170 sterically blocks the insertion of an RR precursor into the TatBC-binding cavity. This is also suggested by the model depicted in Fig. 3a, where Glu170 of the left-hand TatC monomer would be close to the signal peptide shown, provided that our hand-crafted position of the signal peptide comes near to the actual molecular situation. In the crystal structure of TatC, Glu170 is predicted to form hydrogen bonds with TM2 and -3 in the back of the molecule (12). Obviously, the most prominent feature of TatCGlu-170 is its location in the interior of the bilayer, where according to MD simulations it is hydrated and thus perturbs the bilayer structure (11, 12). Conversely, modification of TatCGlu-170 by DCCD as established here would require that it is accommodated in a rather hydrophobic environment. Blocking insertion of a Tat signal peptide deeply into the membrane by modifying Glu170 by DCCD, suggests that the area around this glutamyl residue is part of, or at least very close to, the binding pocket for the Tat signal peptide, which TatC and TatB concertedly form. That at least discrete patches of this binding pocket are hydrophobic in nature is strongly suggested by recent findings indicating that the hydrophobic core of an RR-signal peptide significantly contributes to a productive interaction with the TatBC receptor complex (44).
While blocking insertion of the signal peptide through interaction with Glu170, DCCD did not abolish every contact of the RR-signal sequence with the TatBC receptor complex. Cross-linking to position Leu9 in the N-terminal domain of TatC was not disturbed by DCCD, nor was cross-linking of the consensus motif of the TorA signal peptide via Phe14-Bpa to TatC. Thus, in contrast to the insertion of an RR precursor into the TatBC-binding cavity, DCCD did not negatively affect docking of an RR-signal peptide to the Tat translocase. Remarkably, this was the case, although DCCD formed an intramolecular cross-link between the N-tail and the TM2/TM3 loop of TatC. These two domains had previously been identified as interaction sites for RR-signal sequences through cross-linking studies as well as the mapping of E. coli tatC mutations, which suppress inactivating alterations in the RR motif. These studies had identified a number of residues in the N terminus (including Leu9 and going up to Gln22) and the TM2/TM3 loop of TatC as being directly or indirectly involved in interacting with RR-signal sequences (21, 22, 27–30). The exact residues within these two domains of TatC that directly interact with the twin-arginines of Tat signal peptides have not yet been established (11, 12). The composite nature of the superficial RR-recognition site involving the non-contiguous N terminus and TM2/TM3 loop of TatC, is, however, reinforced by our finding that the covalent fixation of both domains through DCCD does not negatively affect docking of an RR precursor.
About 50% of all TatC molecules that were digested with trypsin and chymotrypsin showed a modification of Glu244 by dicyclohexylurea (cf. Fig. 2a). Glu244 is located in the flexible C-tail of E. coli TatC. The finding that Glu244 becomes also cross-linked to Lys101 through DCCD suggests that this area of the C-terminal domain of E. coli TatC can move in close proximity to the TM2/TM3 loop. Moreover, the fact that DCCD attacks Glu244 to a significant extent indicates that this C-terminal stretch of E. coli TatC is located in a hydrophobic environment of the TatC molecule, which would be consistent with its vicinity to the membrane-enclosed (11, 12) TM2/TM3 loop. Such a conclusion is also supported by the DCCD modification of the nearby Asp248 residue, although this occurred to a considerably lower degree than that of Glu244 (cf. Fig. 2a). Collectively, these findings suggest that both, the N- and C-terminal ends of the E. coli TatC molecule are in close contact to the helical core of the molecule and that this conformation is compatible with its function as a substrate receptor.
Experimental procedures
Plasmids
Plasmids used in this study are listed in Table S1. Plasmids expressing Bpa variants of TatB and TatC have been described (19). Plasmid p8737 was used to introduce the Ala codon GCG into tatC and to add a His6 tag at the C terminus of TatC (p8737-TatABCHis) using the primers listed in Table S2. Plasmids were amplified using Pfu Ultra II Fusion HS DNA Polymerase (Agilent Technologies) according to the manufacturer's protocol. Amber stop codon mutations in the gene encoding the TorA–mCherry of plasmid pPJ3 have been described (19). T4 DNA ligase was purchased from Thermo Scientific. Gel extraction and DNA extraction kits (Qiagen) were used for DNA purification.
In vitro reactions
The RR precursor protein TorA–mCherry was synthesized and radioactively labeled by in vitro transcription/translation using plasmid pPJ3. Cell extracts used for the in vitro synthesis were prepared (45) from E. coli strain SL119 (46) or alternatively from Top10 (Invitrogen) transformed with plasmid pSup-BpaRS-6TRN(D286R) to express amber stop codon mutants of TorA–mCherry (32). Coupled transcription/translation reactions were performed in 50-μl aliquots as described (45). INV were added 10–15 min after starting the synthesis reaction and incubated for 20 min at 37 °C.
Assaying protein translocation into INV by proteinase K protection, addition of CCCP, and Bpa-dependent cross-linking by irradiating samples with UV-light for 20 min on ice have been described (39). DCCD was added to a final concentration of 0.5 mm before adding INV. SDS-PAGE using 10% gels was performed as described (45).
Membrane vesicles
INV were prepared as described (45) from E. coli strains BL21(DE3)* (Novagen) or BL21(DE3)ΔTat (kindly provided by B. Ize and T. Palmer) transformed with plasmid p8737 and derivatives thereof. TatABC-INV containing Bpa variants of TatA, TatB, and TatC were prepared as described (19).
Purification of DCCD-modified TatC
For mass spectrometry analysis of the DCCD-modified TatC, INV were prepared from E. coli strains BL21(DE3)* (Novagen) transformed with p8737-TatABCHis as described (45) except that vesicles were finally resuspended in buffer A (50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 5% glycerol) and diluted to a concentration of ∼10 mg of protein/ml. DCCD was added to a final concentration of 0.5 mm and incubation was performed overnight at 4 °C. Membrane proteins were solubilized for 1 h at 4 °C by NLS (N-lauroylsarcosine sodium salt, final concentration 0.33%) in the presence of 30 mm imidazole. Insoluble material was removed by centrifugation (30 min, 36,000 × g, 4 °C). Affinity purification of TatC was performed using an Äkta Prime System (Amersham Bioscience). The solubilized membrane proteins were loaded on a 5-ml HP His-trap column (GE Healthcare) equilibrated with buffer B (buffer A containing 0.17% NLS). Non-specifically bound material was removed by three washing steps each, using 30 mm and subsequently 50 mm imidazole in buffer B. Elution was performed by applying an imidazole gradient from 50 to 500 mm imidazole in buffer B. The eluate was concentrated using Amicon centrifugal tubes (30 kDa cutoff, Millipore) and separated by 10% SDS-PAGE.
In-gel digestion of membrane proteins
Protein-containing bands were excised from SDS-polyacrylamide gels, destained, and subjected to reduction of cysteine residues with 5 mm Tris(2-carboxyethyl)phosphine dissolved in 10 mm NH4HCO3 (incubation for 30 min at 37 °C) and subsequent alkylation of free thiol groups with 50 mm iodoacetamide in 10 mm NH4HCO3 (30 min at room temperature in the dark). Monomeric TatC was in-gel digested at 37 °C overnight using trypsin and chymotrypsin in 100 mm Tris-HCl, 10 mm CaCl2, pH 8, by adding 0.25 μg of each protease at the start and after 4 h of incubation.
LC-MS/MS
Peptide mixtures were analyzed in two biological replicates by UHPLC-MS/MS using an UltiMate 3000 RSLCnano coupled to a Q Exactive Plus (Thermo Fisher Scientific) mass spectrometer essentially as described (47), except for using a 45-min linear gradient for separation by C18 reversed-phase nano-LC. Peptides were identified by database searches using the MaxQuant program (version 1.5.5.1, (48)) and protein sequences for E. coli TatA, TatB, and TatC as well as for a set of common contaminants. A maximum of four missed sites for proteolytic cleavage by trypsin or chymotrypsin was allowed. Modification of aspartate or glutamate by DCU (+206.17830 Da, modification-specific neutral loss of −125.08406 Da) and oxidation of methionine were defined as variable and carbamidomethylation of cysteine as fixed modifications. Peptides were identified with a minimum length of six amino acids, a false discovery rate of <1%, and scores >17 (p value below 0.02) or >40 (p value below 0.0001) for unmodified and modified peptides, respectively. MS intensities and peptide scores were read out from the “evidence.txt” table and summed up per amino acid position. To estimate the proportion of proteins that are modified by DCU at a given site, intensities of DCU-modified peptides identified with a localization probability ≥0.9 were summed up per modified site. For identification of cross-linked peptides, the program pLink (49) was used essentially as described previously (50), however, taking into account zero-length cross-links (−18.0106 Da) between aspartate or glutamate and lysine or the N-terminal amino group as well as monolink modifications (+206.1783 Da) of aspartate or glutamate. Relative quantification comparing DCCD-treated versus untreated samples for visualization was done by extracted ion chromatograms integrating the first three isotope peaks (mass tolerance 5 ppm) of each precursor ion using Xcalibur Qual Browser software (version 2.2, Thermo Fisher Scientific).
Identification of DCCD-binding sites
The binding of DCCD was detected directly by MS analysis (see above) or indirectly using the fluorescent DCCD analogue NCD-4 (Synchem). To this end, 2.5 μl of each INV preparation (∼100 A280 units/ml) was diluted with 97.5 μl of INV buffer (45) and treated with either 5 mm DCCD (diluted from a 0.5 m stock solution in DMSO) or DMSO for 10 min at 37 °C before adding 0.5 mm NCD-4 (using a 50 mm solution in DMSO that had been diluted from a 1 m stock solution prepared in tetrahydrofurane) to each sample. Samples were incubated at 37 °C for 30 min. Proteins were precipitated with 5% TCA, redissolved in 25 μl of SDS-loading buffer, and separated by SDS-PAGE (12% gel). After electrophoresis, gels were irradiated with UV light. NCD-4 emits light at ∼450 nm if it is stimulated with UV light. The emission was detected using a Fusion FX-7 Spectra instrument (Vilber) with a F440 emission filter (Vilber).
Author contributions
Acquisition and analysis of data, A. S. B., F. D., B. K., J. F., E. E.; conception and design, A. S. B., F. D., B. W., J. F., M. M.; drafting and revision of article, A. S. B., F. D., B. W., J. F., M. M.; final approval, J. F., M. M.
Supplementary Material
Acknowledgment
We gratefully acknowledge MuDe Zou for excellent technical assistance.
This work was supported by Deutsche Forschungsgemeinschaft Grant SFB 746 and Grant FOR 1905 and the Excellence Initiative of the German Federal & State Governments (GSC-4, Spemann Graduate School, and EXC-294, BIOSS). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1–S4 and Tables S1 and S2.
- Tat
- twin-arginine translocation
- PMF
- proton-motive force
- DCCD
- N,N′-dicyclohexylcarbodiimide
- TM
- transmembrane helix
- LC-MS/MS
- liquid chromatography-tandem mass spectrometry
- DCU
- dicyclohexylurea
- NCD-4
- N-cyclohexyl-N′-(4-dimethylamino-α-naphthyl)carbodiimide
- INV
- inside-out inner membrane vesicles
- CCCP
- carbonyl cyanide m-chlorophenylhydrazone
- p
- precursor of TorA–mCherry
- m
- mature form of TorA–mCherry
- NLS
- N-lauroylsarcosine
- Bpa
- p-benzoylphenylalanine
- RR
- twin arginine.
References
- 1. Cline K. (2015) Mechanistic aspects of folded protein transport by the twin-arginine translocase (Tat). J. Biol. Chem. 290, 16530–16538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Berks B. C., Lea S. M., and Stansfeld P. J. (2014) Structural biology of Tat protein transport. Curr. Opin. Struct. Biol. 27, 32–37 [DOI] [PubMed] [Google Scholar]
- 3. Fröbel J., Rose P., and Müller M. (2012) Twin-arginine-dependent translocation of folded proteins. Philos. Trans. R. Soc. Lond. B Biol. Sci. 367, 1029–1046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Palmer T., and Berks B. C. (2012) The twin-arginine translocation (Tat) protein export pathway. Nat. Rev. Microbiol. 10, 483–496 [DOI] [PubMed] [Google Scholar]
- 5. Patel R., Smith S. M., and Robinson C. (2014) Protein transport by the bacterial Tat pathway. Biochim. Biophys. Acta 1843, 1620–1628 [DOI] [PubMed] [Google Scholar]
- 6. Goosens V. J., Monteferrante C. G., and van Dijl J. M. (2014) The Tat system of Gram-positive bacteria. Biochim. Biophys. Acta 1843, 1698–1706 [DOI] [PubMed] [Google Scholar]
- 7. Hu Y., Zhao E., Li H., Xia B., and Jin C. (2010) Solution NMR structure of the TatA component of the twin-arginine protein transport system from Gram-positive bacterium Bacillus subtilis. J. Am. Chem. Soc. 132, 15942–15944 [DOI] [PubMed] [Google Scholar]
- 8. Rodriguez F., Rouse S. L., Tait C. E., Harmer J., De Riso A., Timmel C. R., Sansom M. S., Berks B. C., and Schnell J. R. (2013) Structural model for the protein-translocating element of the twin-arginine transport system. Proc. Natl. Acad. Sci. U.S.A. 110, E1092–E1101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Walther T. H., Grage S. L., Roth N., and Ulrich A. S. (2010) Membrane alignment of the pore-forming component TatA (d) of the twin-arginine translocase from Bacillus subtilis resolved by solid-state NMR spectroscopy. J. Am. Chem. Soc. 132, 15945–15956 [DOI] [PubMed] [Google Scholar]
- 10. Zhang Y., Wang L., Hu Y., and Jin C. (2014) Solution structure of the TatB component of the twin-arginine translocation system. Biochim. Biophys. Acta 1838, 1881–1888 [DOI] [PubMed] [Google Scholar]
- 11. Ramasamy S., Abrol R., Suloway C. J., and Clemons W. M. Jr. (2013) The glove-like structure of the conserved membrane protein TatC provides insight into signal sequence recognition in twin-arginine translocation. Structure 21, 777–788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Rollauer S. E., Tarry M. J., Graham J. E., Jääskeläinen M., Jäger F., Johnson S., Krehenbrink M., Liu S. M., Lukey M. J., Marcoux J., McDowell M. A., Rodriguez F., Roversi P., Stansfeld P. J., Robinson C. V., et al. (2012) Structure of the TatC core of the twin-arginine protein transport system. Nature 492, 210–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Eimer E., Fröbel J., Blümmel A. S., and Müller M. (2015) TatE as a regular constituent of bacterial twin-arginine protein translocases. J. Biol. Chem. 290, 29281–29289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Alcock F., Baker M. A., Greene N. P., Palmer T., Wallace M. I., and Berks B. C. (2013) Live cell imaging shows reversible assembly of the TatA component of the twin-arginine protein transport system. Proc. Natl. Acad. Sci. U.S.A. 110, E3650–E3659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rose P., Fröbel J., Graumann P. L., and Müller M. (2013) Substrate-dependent assembly of the Tat translocase as observed in live Escherichia coli cells. PLoS ONE 8, e69488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bolhuis A., Mathers J. E., Thomas J. D., Barrett C. M., and Robinson C. (2001) TatB and TatC form a functional and structural unit of the twin-arginine translocase from Escherichia coli. J. Biol. Chem. 276, 20213–20219 [DOI] [PubMed] [Google Scholar]
- 17. Cline K., and Mori H. (2001) Thylakoid DeltapH-dependent precursor proteins bind to a cpTatC-Hcf106 complex before Tha4-dependent transport. J. Cell Biol. 154, 719–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Behrendt J., and Brüser T. (2014) The TatBC complex of the Tat protein translocase in Escherichia coli and its transition to the substrate-bound TatABC complex. Biochemistry 53, 2344–2354 [DOI] [PubMed] [Google Scholar]
- 19. Blümmel A. S., Haag L. A., Eimer E., Müller M., and Fröbel J. (2015) Initial assembly steps of a translocase for folded proteins. Nat. Commun. 6, 7234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Alcock F., Stansfeld P. J., Basit H., Habersetzer J., Baker M. A., Palmer T., Wallace M. I., and Berks B. C. (2016) Assembling the Tat protein translocase. Elife 5, e20718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ma X., and Cline K. (2013) Mapping the signal peptide binding and oligomer contact sites of the core subunit of the pea twin arginine protein translocase. Plant Cell 25, 999–1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zoufaly S., Fröbel J., Rose P., Flecken T., Maurer C., Moser M., and Müller M. (2012) Mapping precursor-binding site on TatC subunit of twin arginine-specific protein translocase by site-specific photo cross-linking. J. Biol. Chem. 287, 13430–13441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Cléon F., Habersetzer J., Alcock F., Kneuper H., Stansfeld P. J., Basit H., Wallace M. I., Berks B. C., and Palmer T. (2015) The TatC component of the twin-arginine protein translocase functions as an obligate oligomer. Mol. Microbiol 98, 111–129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Aldridge C., Ma X., Gerard F., and Cline K. (2014) Substrate-gated docking of pore subunit Tha4 in the TatC cavity initiates Tat translocase assembly. J. Cell Biol. 205, 51–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Alami M., Lüke I., Deitermann S., Eisner G., Koch H. G., Brunner J., and Müller M. (2003) Differential interactions between a twin-arginine signal peptide and its translocase in Escherichia coli. Mol. Cell 12, 937–946 [DOI] [PubMed] [Google Scholar]
- 26. Gérard F., and Cline K. (2006) Efficient twin arginine translocation (Tat) pathway transport of a precursor protein covalently anchored to its initial cpTatC binding site. J. Biol. Chem. 281, 6130–6135 [DOI] [PubMed] [Google Scholar]
- 27. Kreutzenbeck P., Kröger C., Lausberg F., Blaudeck N., Sprenger G. A., and Freudl R. (2007) Escherichia coli twin arginine (Tat) mutant translocases possessing relaxed signal peptide recognition specificities. J. Biol. Chem. 282, 7903–7911 [DOI] [PubMed] [Google Scholar]
- 28. Lausberg F., Fleckenstein S., Kreutzenbeck P., Fröbel J., Rose P., Müller M., and Freudl R. (2012) Genetic evidence for a tight cooperation of TatB and TatC during productive recognition of twin-arginine (Tat) signal peptides in Escherichia coli. PLoS ONE 7, e39867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Huang Q., Alcock F., Kneuper H., Deme J. C., Rollauer S. E., Lea S. M., Berks B. C., and Palmer T. (2017) A signal sequence suppressor mutant that stabilizes an assembled state of the twin arginine translocase. Proc. Natl. Acad. Sci. U.S.A. 114, E1958–E1967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Strauch E. M., and Georgiou G. (2007) Escherichia coli tatC mutations that suppress defective twin-arginine transporter signal peptides. J. Mol. Biol. 374, 283–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Fröbel J., Rose P., Lausberg F., Blümmel A. S., Freudl R., and Müller M. (2012) Transmembrane insertion of twin-arginine signal peptides is driven by TatC and regulated by TatB. Nat. Commun. 3, 1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Maurer C., Panahandeh S., Jungkamp A. C., Moser M., and Müller M. (2010) TatB functions as an oligomeric binding site for folded Tat precursor proteins. Mol. Biol. Cell 21, 4151–4161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Brüser T., and Sanders C. (2003) An alternative model of the twin arginine translocation system. Microbiol. Res. 158, 7–17 [DOI] [PubMed] [Google Scholar]
- 34. Aldridge C., Storm A., Cline K., and Dabney-Smith C. (2012) The chloroplast twin arginine transport (tat) component, tha4, undergoes conformational changes leading to tat protein transport. J. Biol. Chem. 287, 34752–34763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Panahandeh S., Maurer C., Moser M., DeLisa M. P., and Müller M. (2008) Following the Path of a twin-arginine precursor along the TatABC translocase of Escherichia coli. J. Biol. Chem. 283, 33267–33275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hassinen I. E., and Vuokila P. T. (1993) Reaction of dicyclohexylcarbodiimide with mitochondrial proteins. Biochim. Biophys. Acta 1144, 107–124 [DOI] [PubMed] [Google Scholar]
- 37. Valeur E., and Bradley M. (2009) Amide bond formation: beyond the myth of coupling reagents. Chem. Soc. Rev. 38, 606–631 [DOI] [PubMed] [Google Scholar]
- 38. Musser S. M., Larsen R. W., and Chan S. I. (1993) Fluorescence quenching of reconstituted NCD-4-labeled cytochrome c oxidase complex by DOXYL-stearic acids. Biophys. J. 65, 2348–2359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Fröbel J., Rose P., and Müller M. (2011) Early contacts between substrate proteins and TatA translocase component in twin-arginine translocation. J. Biol. Chem. 286, 43679–43689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Müller M., Fisher R. P., Rienhöfer-Schweer A., and Hoffschulte H. K. (1987) DCCD inhibits protein translocation into plasma membrane vesicles from Escherichia coli at two different steps. EMBO J. 6, 3855–3861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Holzapfel E., Eisner G., Alami M., Barrett C. M., Buchanan G., Lüke I., Betton J. M., Robinson C., Palmer T., Moser M., and Müller M. (2007) The entire N-terminal half of TatC is involved in twin-arginine precursor binding. Biochemistry 46, 2892–2898 [DOI] [PubMed] [Google Scholar]
- 42. Simone D., Bay D. C., Leach T., and Turner R. J. (2013) Diversity and evolution of bacterial twin arginine translocase protein, TatC, reveals a protein secretion system that is evolving to fit its environmental niche. PLoS ONE 8, e78742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Buchanan G., de Leeuw E., Stanley N. R., Wexler M., Berks B. C., Sargent F., and Palmer T. (2002) Functional complexity of the twin-arginine translocase TatC component revealed by site-directed mutagenesis. Mol. Microbiol. 43, 1457–1470 [DOI] [PubMed] [Google Scholar]
- 44. Ulfig A., Fröbel J., Lausberg F., Blümmel A. S., Heide A. K., Müller M., and Freudl R. (2017) The h-region of twin-arginine signal peptides supports productive binding of bacterial Tat precursor proteins to the TatBC receptor complex. J. Biol. Chem. 292, 10865–10882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Moser M., Panahandeh S., Holzapfel E., and Müller M. (2007) In vitro analysis of the bacterial twin-arginine-dependent protein export. Methods Mol. Biol. 390, 63–79 [DOI] [PubMed] [Google Scholar]
- 46. Lesley S. A., Brow M. A., and Burgess R. R. (1991) Use of in vitro protein synthesis from polymerase chain reaction-generated templates to study interaction of Escherichia coli transcription factors with core RNA polymerase and for epitope mapping of monoclonal antibodies. J. Biol. Chem. 266, 2632–2638 [PubMed] [Google Scholar]
- 47. Reimann L., Wiese H., Leber Y., Schwäble A. N., Fricke A. L., Rohland A., Knapp B., Peikert C. D., Drepper F., van der Ven P. F., Radziwill G., Fürst D. O., and Warscheid B. (2017) Myofibrillar Z-discs are a protein phosphorylation hot spot with protein kinase C (PKCα) modulating protein dynamics. Mol. Cell. Proteomics 16, 346–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Tyanova S., Temu T., and Cox J. (2016) The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat. Protoc. 11, 2301–2319 [DOI] [PubMed] [Google Scholar]
- 49. Yang B., Wu Y. J., Zhu M., Fan S. B., Lin J., Zhang K., Li S., Chi H., Li Y. X., Chen H. F., Luo S. K., Ding Y. H., Wang L. H., Hao Z., Xiu L. Y., et al. (2012) Identification of cross-linked peptides from complex samples. Nat. Methods 9, 904–906 [DOI] [PubMed] [Google Scholar]
- 50. Chan A., Schummer A., Fischer S., Schröter T., Cruz-Zaragoza L. D., Bender J., Drepper F., Oeljeklaus S., Kunau W. H., Girzalsky W., Warscheid B., and Erdmann R. (2016) Pex17p-dependent assembly of Pex14p/Dyn2p-subcomplexes of the peroxisomal protein import machinery. Eur. J. Cell Biol. 95, 585–597 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.