Abstract
Formins are a conserved group of proteins that nucleate and processively elongate actin filaments. Among them, the formin homology domain–containing protein (FHOD) family of formins contributes to contractility of striated muscle and cell motility in several contexts. However, the mechanisms by which they carry out these functions remain poorly understood. Mammalian FHOD proteins were reported not to accelerate actin assembly in vitro; instead, they were proposed to act as barbed end cappers or filament bundlers. Here, we show that purified Drosophila Fhod and human FHOD1 both accelerate actin assembly by nucleation. The nucleation activity of FHOD1 is restricted to cytoplasmic actin, whereas Drosophila Fhod potently nucleates both cytoplasmic and sarcomeric actin isoforms. Drosophila Fhod binds tightly to barbed ends, where it slows elongation in the absence of profilin and allows, but does not accelerate, elongation in the presence of profilin. Fhod antagonizes capping protein but dissociates from barbed ends relatively quickly. Finally, we determined that Fhod binds the sides of and bundles actin filaments. This work establishes that Fhod shares the capacity of other formins to nucleate and bundle actin filaments but is notably less effective at processively elongating barbed ends than most well studied formins.
Keywords: actin, cytoskeleton, Drosophila, formin, processivity, Fhod, Fhos, TIRF
Introduction
Formins are a major, conserved group of proteins known for their ability to both nucleate new actin filaments and remain processively associated with fast-growing barbed ends via their formin homology 2 (FH2)3 domains. While bound to the barbed end, many formins accelerate elongation, using their proline-rich FH1 domains to recruit profilin-bound actin monomers to the barbed end. In addition to these classic activities, many formins have additional effects on actin filaments, such as severing, bundling, or cross-linking to microtubules (1). Animals have seven formin families, which share the same domain structure, including the highly conserved FH2 domains. Importantly, formins differ in their actin assembly activities and modes of regulation, allowing them to fulfill distinct cellular roles.
The formin homology domain–containing protein (FHOD) family of formins has two mammalian isoforms, FHOD1 and FHOD3, which assemble contractile actin structures in several contexts. FHOD1 is widely expressed and assembles stress fibers that contribute to the adhesion, spreading, and motility of numerous cell types (2–8). FHOD3 has also been implicated in the motility of some cancers (9, 10), but its expression is predominantly restricted to striated muscle (11, 12). In cardiomyocytes, FHOD3 localizes to the sarcomere and is required for its assembly and maintenance (12–15), whereas FHOD1 distinctly localizes to costameres and intercalated discs (16, 17). Changes in expression level and polymorphisms of both FHOD1 and FHOD3 are associated with cardiomyopathies (12, 17–20).
The mechanisms by which FHOD family members function remain poorly understood. Unlike other formins, purified FHOD1 and FHOD3 were shown to slow, rather than accelerate, actin assembly in vitro and have therefore been proposed to act as actin cappers or bundlers (13, 21). In contrast, the cell biology data suggest that FHOD1 and FHOD3 function as actin nucleators in vivo. Across a wide range of cell types, expression of constitutively active FHOD1 or FHOD3 is sufficient to induce the formation of stress fibers (2–5, 13). Furthermore, FHOD3 is required for sarcomere assembly following latrunculin washout (12). These data are most suggestive of actin nucleation, although FHOD1 and FHOD3 might instead function by stabilizing or reorganizing existing actin filaments.
Drosophila melanogaster has a single FHOD family member, referred to here as Fhod (it is also known as Fhos or knittrig). The Fhod gene is alternatively spliced to produce eight different isoforms, which maintain constant FH1 and FH2 domains but alter their regulatory N termini and C-terminal tails. The role of FHOD proteins in cell motility and contractility is well conserved in Drosophila, because Fhod contributes to motility in macrophages and tracheal tip cells (22), sarcomere organization in striated muscle (23, 24), and cardiac contractility (18).
Here, we show that purified Drosophila Fhod and human FHOD1 accelerate actin assembly by nucleation. The nucleation activity of FHOD1 is restricted to cytoplasmic actin, whereas Drosophila Fhod potently nucleates both cytoplasmic and sarcomeric actin isoforms. Drosophila Fhod remains processively associated with the barbed end, where it slows elongation in the absence of profilin and allows elongation, at rates similar to actin alone, in the presence of profilin. Although Fhod does not accelerate barbed-end elongation, we find that Fhod protects barbed ends from capping protein with a characteristic run length of ∼2 μm. Fhod additionally binds tightly to the sides of filaments and bundles filaments together.
Results
Fhod accelerates actin assembly
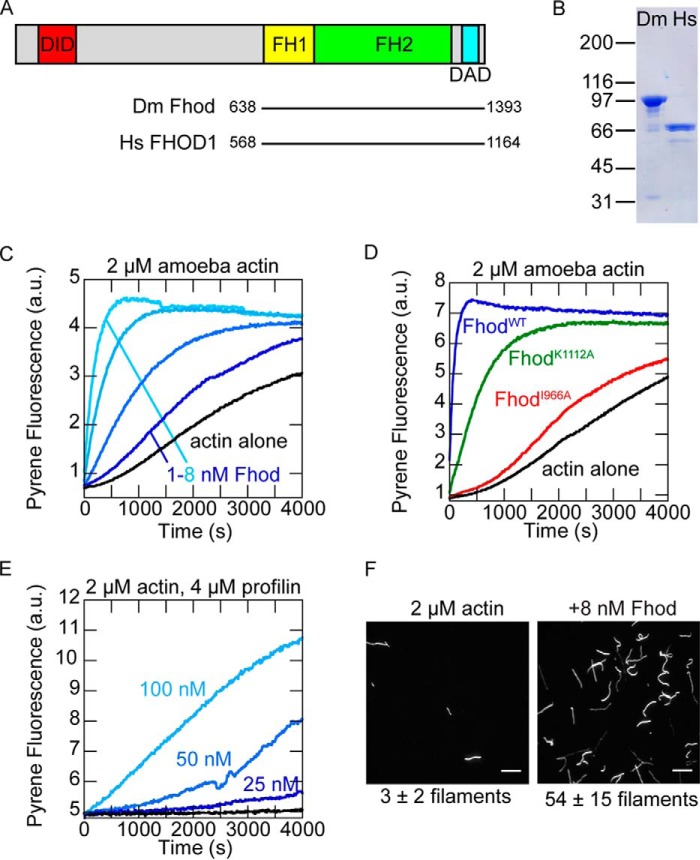
We purified the C-terminal half of Drosophila Fhod isoform A, encompassing the FH1 domain, FH2 domain, and C-terminal tail (Fig. 1, A and B). This isoform is sufficient to rescue viability in Fhod null flies (22), and its C terminus is identical to that of isoform H, which rescues sarcomere organization in indirect flight muscle (23). We first tested the effect of Fhod on the assembly of Acanthamoeba actin in bulk pyrene assays; Fhod potently accelerated actin assembly in a dose-dependent manner (Fig. 1C).
Figure 1.
The Drosophila formin Fhod accelerates actin assembly. A, domain structure of Drosophila Fhod isoform A. Fhod includes formin homology domains for actin assembly and the Diaphanous inhibitory domain (DID) and Diaphanous autoregulatory domain (DAD) for autoinhibition and regulation. The Fhod construct used in this work spans residues 638–1393 and includes the FH1 domain, FH2 domain, and tail. Human FHOD1 has a nearly identical domain structure. B, Coomassie-stained polyacrylamide gel showing purified Drosophila Fhod and human FHOD1 constructs. C, assembly of 2 μm A. castellanii actin (10% pyrene-labeled) with 1–8 nm Fhod. Fhod accelerates actin assembly in a dose-dependent manner. D, assembly of 2 μm amoeba actin (10% pyrene-labeled) in the presence of 16 nm wild-type (blue) or mutant (I966A, red; K1112A, green) Fhod. Actin assembly by Fhod is reduced moderately by the K1112A mutation and severely by the I966A mutation. E, assembly of 2 μm amoeba actin (5% pyrene-labeled) in the presence of 4 μm S. pombe profilin and the indicated concentrations of Fhod. Fhod accelerates actin assembly in the presence of profilin. F, 2 μm actin was polymerized in the presence or absence of 8 nm Fhod for 5 min, stabilized with Alexa Fluor 488-phalloidin, and imaged by TIRF microscopy. Scale bars, 10 μm. Numbers of filaments per field of view from three independent experiments are indicated below (means ± standard deviation).
To further compare Fhod to characterized formins, we introduced two classical mutations, I966A and K1112A, in conserved residues of the FH2 domain (25). The I966A mutation almost completely abolished activity, whereas the K1112A mutation markedly reduced, but did not eliminate, activity (Fig. 1D). We also tested the ability of Fhod to promote actin assembly in the presence of profilin, which binds most actin monomers in the cell and inhibits spontaneous nucleation. Profilin strongly impaired actin assembly by Fhod, but Fhod still accelerated actin assembly in a dose-dependent manner under these conditions (Fig. 1E).
Formins can promote both nucleation and elongation. To more directly assess nucleation activity, we imaged actin filaments polymerized in the absence or presence of Fhod. Fhod greatly increased the number of filaments per field of view, indicating that Fhod promotes actin assembly by nucleation (Fig. 1F). Fhod is thus similar to other studied formins in its ability to nucleate.
Human FHOD1 nucleates Acanthamoeba actin, but not rabbit skeletal actin
We compared Drosophila Fhod with human FHOD1, which was reported to inhibit actin assembly (21). Surprisingly, FHOD1 accelerated actin assembly in our hands, albeit weakly (Fig. 2A). Previous work with both FHOD1 and FHOD3 used actin from rabbit skeletal muscle (13, 21). Because formin activity can depend on the actin isoform,4 we asked whether our use of Acanthamoeba actin could account for these conflicting results. Indeed, human FHOD1 did not nucleate rabbit skeletal actin (Fig. 2B). As previously observed, actin assembly was inhibited over the first 1000 s (21). The pyrene trace suggests that FHOD1 does interact with rabbit skeletal actin, despite its inability to nucleate this isoform. In contrast, Drosophila Fhod nucleated both actin isoforms, with only slightly lower activity in the presence rabbit skeletal actin (Fig. 2, C and D). These results are consistent with the more restricted localization of human FHOD1 to cytoplasmic actin structures, versus the role of Drosophila Fhod in both sarcomeric and cytoplasmic structures (22, 23). Drosophila Fhod actin assembly activity is thus representative of FHOD family members, at least in non-sarcomere contexts. Although Drosophila Fhod appeared substantially more potent than human FHOD1, we note that human FHOD1 was highly prone to C-terminal truncation (Fig. 1B), which might reduce its nucleation activity (26–28). Unless otherwise indicated, all subsequent experiments were performed with Drosophila Fhod and Acanthamoeba actin.
Figure 2.
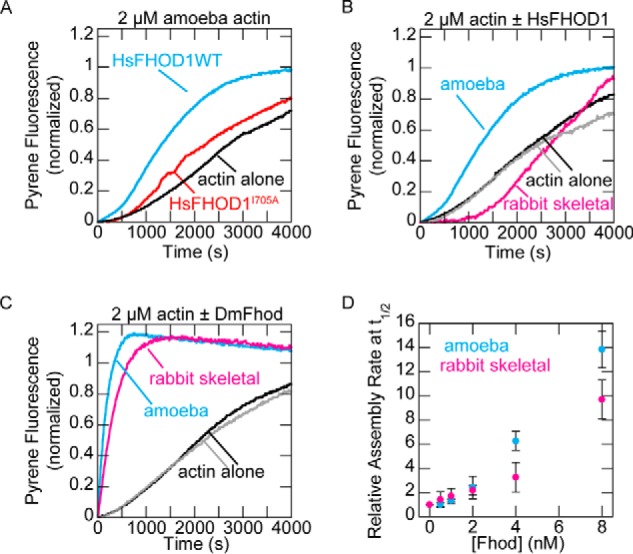
Human FHOD1 accelerates assembly of Acanthamoeba actin, but not rabbit skeletal actin. A, assembly of 2 μm A. castellanii actin (10% pyrene-labeled) alone or in the presence of 40 nm human FHOD1. B, assembly of 2 μm actin from Acanthamoeba or rabbit skeletal muscle (10% pyrene-labeled) alone or in the presence of 40 nm human FHOD1. C, assembly of 2 μm actin from Acanthamoeba or rabbit skeletal muscle (10% pyrene-labeled) alone or in the presence of 8 nm Drosophila Fhod. D, quantification of actin assembly rates from C. The data are means ± standard deviation from three independent experiments.
Fhod does not promote barbed-end elongation
Formins typically slow elongation in the absence of profilin and accelerate elongation in the presence of profilin. Using bulk seeded elongation assays, we found that Fhod slows barbed-end elongation in the absence of profilin with an affinity of 6 nm (Fig. 3, A and B). This effect was unchanged by the K1112A mutation but strongly reduced by the I966A mutation. Although high concentrations of I966A mutant appeared to slow elongation, we attribute this decrease to filament bundling (see “Fhod bundles actin filaments”) rather than a true effect on barbed-end elongation because the trend does not follow the expected dose-response curve, and substantial fluorescence artifacts were observed in other assays with high concentrations of Fhod.
Figure 3.
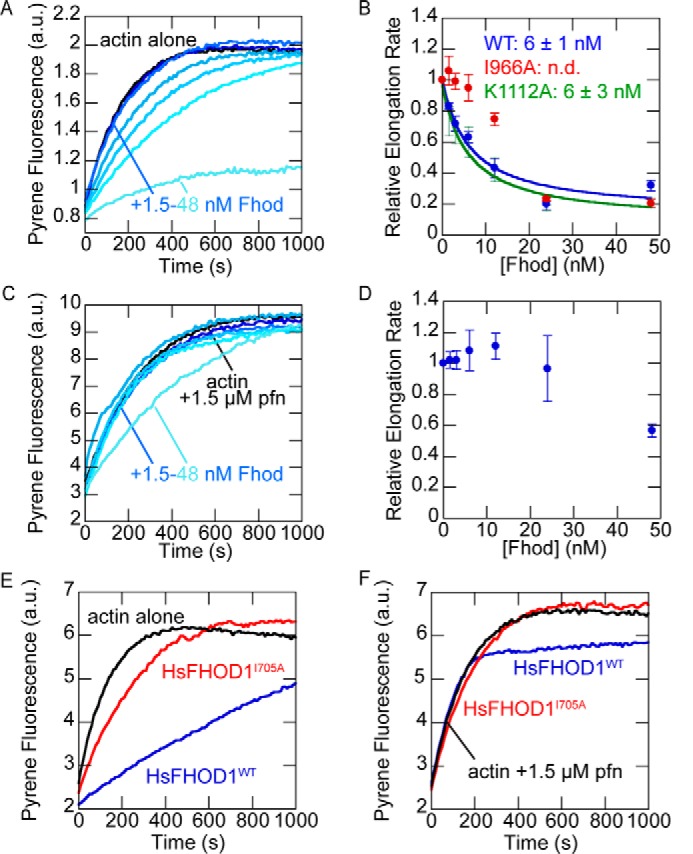
Fhod does not accelerate barbed-end elongation. A, actin elongation from preformed seeds. Final conditions were 0.25 μm F-actin seeds (∼0.4 nm barbed ends), 0.5 μm G-actin (10% pyrene-labeled), and 1.5–48 nm Fhod. Fhod slows barbed-end elongation in a dose-dependent manner. B, quantification of elongation rates from A. Elongation rates were measured as the initial slope over the first 90 s, relative to the slope of actin alone. The data are the means ± standard deviation from three independent experiments. The binding curves show the best fit to the average values. C, actin elongation from preformed seeds, as in A, with 1.5 μm S. pombe profilin. Fhod does not slow elongation in the presence of profilin. D, quantification of elongation rates from C. The data are the means ± standard deviation from three independent experiments. E, actin elongation from preformed seeds. Final conditions were 0.25 μm F-actin seeds (∼0.4 nm barbed ends), 0.5 μm G-actin (10% pyrene-labeled), and 40 nm human FHOD1. FHOD1 slows barbed-end elongation. F, actin elongation from preformed seeds, as in E, with 1.5 μm S. pombe profilin. FHOD1 allows barbed-end elongation at rates similar to actin alone.
In the presence of profilin, Fhod had a minimal effect on elongation at most concentrations we tested (Fig. 3, C and D). At high concentrations of Fhod, the elongation rate appeared to decrease, which we again attribute to bundling. To confirm that Fhod does not alter barbed-end elongation in the presence of profilin, we directly measured elongation rates using total internal reflection fluorescence (TIRF) microscopy. In agreement with our bulk assays, we detected no difference in elongation rates (Fig. 4, A and B). Human FHOD1 had a nearly identical effect on barbed-end elongation, slowing elongation in the absence of profilin and permitting elongation in the presence of profilin (Fig. 3, E and F).
Figure 4.
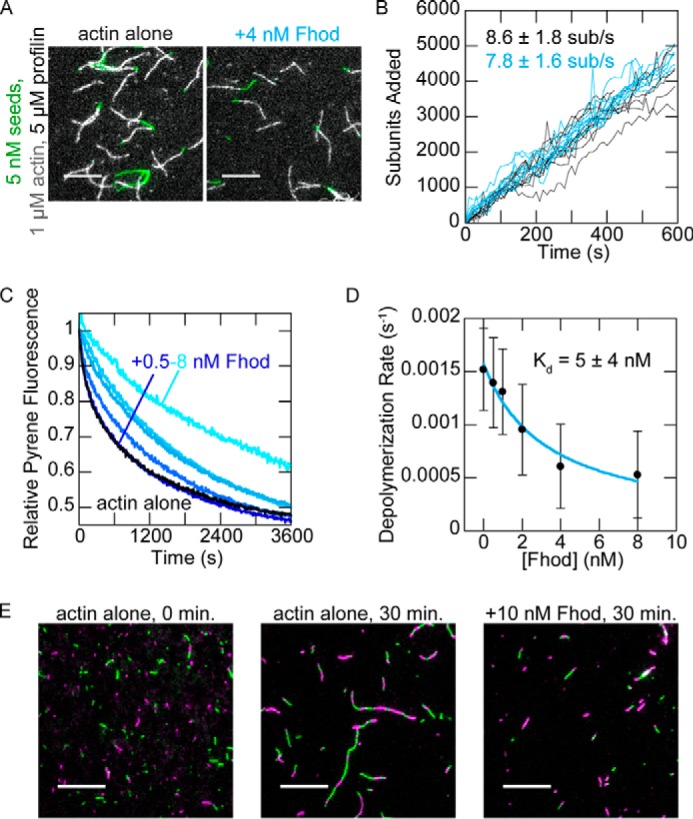
Drosophila Fhod binds barbed ends. A, direct observation of barbed-end elongation by TIRF microscopy with 5 nm F-actin seeds (1% biotinylated, labeled with Alexa Fluor 647-phalloidin), 1 μm G-actin (10% Alexa Fluor 594-labeled), and 5 μm S. pombe profilin, ± 4 nm Fhod. Images were taken 8 min after the start of polymerization. Scale bars, 10 μm. B, quantification of elongation from A. Ten representative traces from each condition are plotted. Elongation rates are average ± standard deviation from three flow chambers for each condition (n = 60, actin alone; n = 36, +4 nm Fhod). C, actin filaments depolymerized by dilution to 0.1 μm (70% pyrene-labeled) with varying concentrations of Fhod. The data were normalized against the initial pyrene fluorescence. D, quantification of depolymerization rates from C, represented by the initial slope over the first 90 s. Fhod binds barbed ends and slows depolymerization. The data and reported Kd are the means ± standard deviation from six independent experiments. The binding curve shows the best fit to the average values. E, reannealing of sheared actin filaments. Actin filaments were stabilized with Alexa Fluor 488- (green) or rhodamine- (magenta) phalloidin, sheared, and allowed to reanneal for 30 min at a concentration of 0.25 μm F-actin ± 10 nm Fhod. Fhod inhibits reannealing of the sheared filaments. Scale bars, 10 μm.
Because we did not observe clear evidence of processive elongation by Fhod, we used several additional assays to verify and characterize the interaction between Fhod and barbed ends. We first verified barbed end binding in bulk barbed end depolymerization assays. The dose dependence gives us an additional measure of the affinity between Fhod and barbed ends. Fhod inhibited actin depolymerization with a Kd of 5 nm (Fig. 4, C and D), similar to our measurement from the seeded elongation assays. We then used actin reannealing assays, in which two colors of sheared actin filaments were mixed and allowed to reanneal. Fhod inhibited actin reannealing, indicating that it binds barbed ends and can remain bound on the timescale of minutes (Fig. 4C). Thus Fhod binds barbed ends tightly, like other formins, but does not accelerate elongation, unlike most formins.
Fhod antagonizes capping protein
We measured the ability of Fhod to antagonize capping protein, which binds tightly to barbed ends and prevents elongation. In bulk seeded elongation assays, 6 nm capping protein was sufficient to completely abolish actin elongation. Fhod abrogated this effect when added to F-actin seeds prior to capping protein (Fig. 5A). We fit these data to a competition binding equation to determine that Fhod has an apparent Kd of 7 nm for growing barbed ends (Fig. 5B), consistent with our previous measurements. Recent evidence suggests that formins can antagonize capping protein not only by passive competition for the barbed end but also by binding capped barbed ends and actively displacing capping protein (29, 30). However, filaments did not grow when capping protein was added before Fhod (data not shown), indicating that the actin elongation we observed was due to Fhod processively protecting the barbed end and not due to Fhod actively displacing capping protein from barbed ends.
Figure 5.
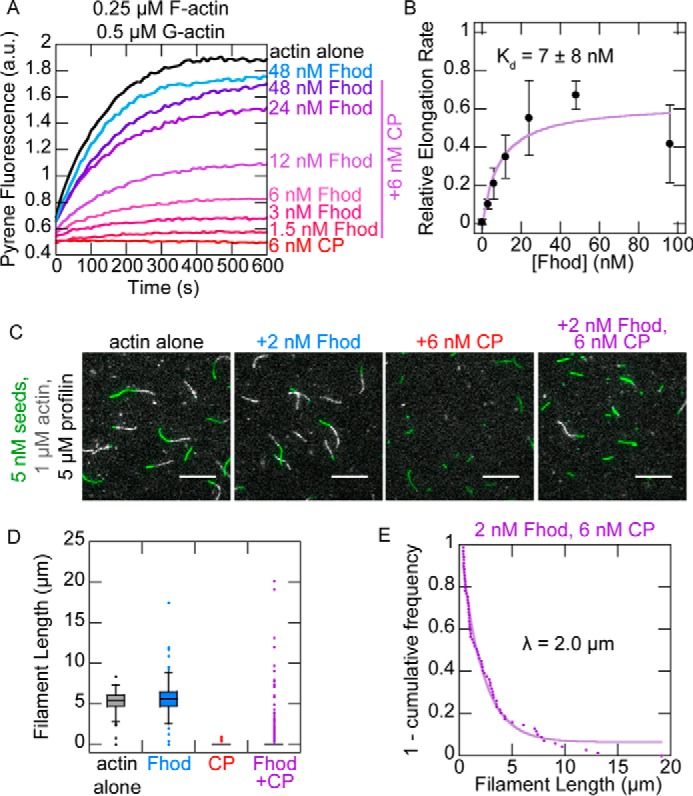
Fhod antagonizes capping protein. A, actin elongation from preformed seeds with a range of Fhod concentrations added before capping protein. Final conditions were 0.25 μm F-actin seeds (∼0.4 nm barbed ends), 0.5 μm G-actin (10% pyrene-labeled), 1.5 μm S. pombe profilin, ± 6 nm mouse capping protein and 1.5–48 nm Fhod. B, quantification of elongation rates from A, measured as the initial slope over the first 90 s relative to the slope of actin alone. Fhod antagonizes capping protein, allowing elongation. Data with Fhod and capping protein were fit to a competition binding model to determine the affinity of Fhod to barbed ends. The data and reported Kd are means ± standard deviation from four independent experiments. The binding curve shows the best fit to the average values. C, observation of actin elongation (white) from preformed seeds (green) with Fhod added before capping protein. Final conditions were 5 nm F-actin seeds (1% biotinylated, labeled with Alexa Fluor 647-phalloidin), 1 μm G-actin (10% Alexa Fluor 594-labeled), 5 μm S. pombe profilin, ± 2 nm Fhod, and 6 nm capping protein. Images were taken 5 min after initiation of polymerization. Scale bars, 10 μm. D, quantification of filament lengths from C. The data represent the amount of elongation from preformed seeds (n > 150 for each condition). At least five fields of view from one (actin alone) or two (all other conditions) flow chambers were analyzed for each condition. In conditions with capping protein, no box is visible because over 75% of seeds did not elongate. E, exponential fit of filament lengths in the presence of both Fhod and capping protein from D, excluding seeds that did not elongate (n = 69 filaments from two flow chambers). Fhod has a characteristic run length of 2.0 μm.
We used TIRF microscopy to observe competition between Fhod and capping protein on individual filaments. We incubated seeds with Fhod prior to adding capping protein and actin monomers and then measured how long Fhod could protect the growing barbed ends. Consistent with our bulk assays, we found that barbed ends were completely capped by capping protein in the absence of Fhod but were able to grow in the presence of Fhod. Because the vast majority of filaments were capped by the time we could start imaging (2–3 min after the start of polymerization), we measured filament lengths in static images taken 5 min after addition of actin monomers (Fig. 5, C and D). By fitting the filament lengths to a single exponential curve, we determined that Fhod has a characteristic run length of 2 μm (Fig. 5E). This provides us with an approximate measure of Fhod processivity, with two assumptions: 1) capping protein binds to barbed ends as soon as Fhod dissociates, and 2) capping protein does not cap barbed ends that are already bound by Fhod. The first assumption is consistent with the strong affinity (∼0.4 nm) of capping protein for barbed ends. However, the ability of capping protein to bind mDia1-bound barbed ends and displace mDia1 (29, 30) suggests that capping protein might also bind Fhod-bound barbed ends, which would make our measurement of Fhod processivity an underestimate.
Fhod bundles actin filaments
Finally, we asked whether Fhod shares the capacity of other formins to bind the sides of actin filaments and bundle them. In high-speed cosedimentation experiments, Fhod pelleted with F-actin, demonstrating that Fhod binds the sides of actin filaments, with a Kd of 180 nm (Fig. 6, A and B). In low-speed cosedimentation, Fhod increased the amount of F-actin in the pellet, indicating that it forms actin bundles (Fig. 6, C and D). The I966A and K1112A mutations had no detectable effect on bundling activity (Fig. 6D). We used TIRF microscopy to determine the polarity of actin bundles formed by Fhod. Actin bundles typically elongated from both ends, indicating that the filaments in these bundles were not arranged exclusively in parallel (Fig. 6E).
Figure 6.
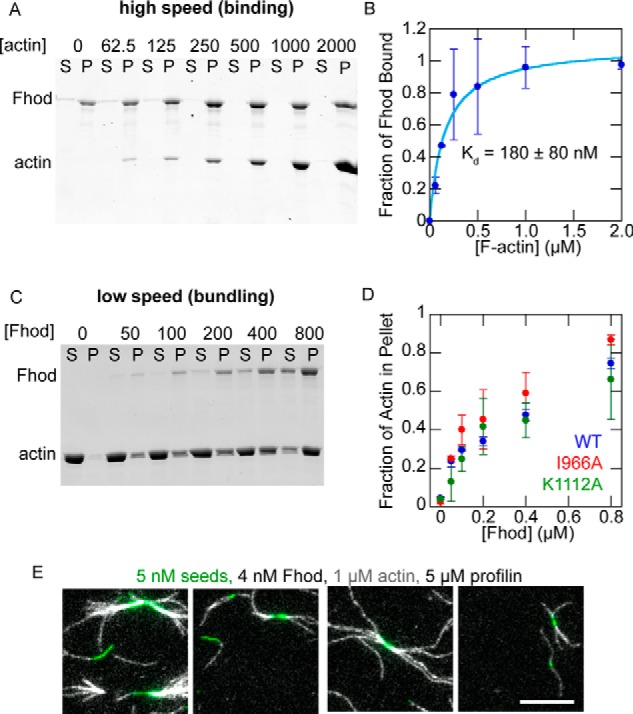
Fhod binds the sides of actin filaments and forms actin bundles. A, high-speed cosedimentation assay with 250 nm Fhod and the indicated concentrations of phalloidin-actin (in nm). B, quantification of A. The fraction of Fhod bound to F-actin was quantified by measuring the amount of Fhod in the pellet and correcting for the amount of Fhod that pellets in the absence of F-actin. The data and reported Kd are means ± standard deviation from three independent experiments. The binding curve shows the best fit to the average values. C, low-speed cosedimentation assay with 5 μm phalloidin-actin and the indicated concentrations of Fhod (in nm). D, quantification of C. The data are means ± standard deviation from two independent experiments. E, representative images of Fhod-induced actin bundles visualized by TIRF microscopy. Final conditions were 5 nm F-actin seeds (1% biotinylated, labeled with Alexa Fluor 647-phalloidin), 1 μm G-actin (10% Alexa Fluor 594-labeled), 5 μm S. pombe profilin, and 4 nm Fhod. Bundles elongate at both ends, indicating mixed or antiparallel polarity. Scale bar, 10 μm.
Discussion
Here, we show that Drosophila Fhod shares the classic activities of formins, actin nucleation and processive elongation, with the additional capacity to bundle actin filaments. Our observation of actin assembly with both Drosophila Fhod and human FHOD1 contrasts substantially with the previous reports that mammalian FHOD proteins slow actin assembly in vitro (13, 21). We resolved these conflicting results for FHOD1 by showing that FHOD1 nucleates actin from Acanthamoeba effectively but does not nucleate actin from rabbit skeletal muscle. Our group previously reported a similar preference for the formin Delphilin, which nucleates cytoplasmic actin isoforms much more effectively than actin from rabbit skeletal muscle.4 Although Delphilin is expressed exclusively in neurons and therefore unlikely to encounter sarcomeric actin isoforms, FHOD1 is expressed in both muscle and non-muscle cells. In cardiomyocytes, FHOD1 is largely excluded from the sarcomere, instead localizing primarily to the costamere and intercalated disc (16, 17). Therefore, the inability of FHOD1 to nucleate sarcomeric actin might be important to its function in this context.
The use of rabbit skeletal actin in previous work is unlikely to explain why FHOD3 did not nucleate in vitro, because FHOD3 is expressed predominantly in striated muscle and required for sarcomere assembly. Given the conserved role of FHOD family members in striated muscle, Drosophila Fhod and mammalian FHOD3 likely share a common mechanism in assembling sarcomeric actin. Although we find the cellular data with FHOD3 most suggestive of nucleation (12, 13), it remains possible that FHOD proteins instead stabilize or bundle filaments that are polymerized by a different actin nucleator. Flies carrying the I966A mutation, which abolishes nucleation and barbed end binding while retaining bundling activity, have relatively mild defects in sarcomere organization (23), supporting the possibility that the side binding and bundling activities of Fhod are more critical than nucleation or barbed end binding.
We did not observe evidence of accelerated barbed-end elongation with either Fhod or human FHOD1. This is not unprecedented, because some formins such as Drosophila Daam (31) and mouse FMNL1 (32) either slow barbed-end elongation or leave the elongation rate unchanged in the presence of profilin. We expect that both the FH1 and FH2 domains contribute to the inability of Fhod to accelerate barbed-end elongation. The slow barbed-end elongation in the absence of profilin is suggestive of an FH2 domain that spends most of the time in a closed conformation, similar to Cdc12 (33). The addition of profilin restores the elongation rate to that of actin alone, indicating that the FH1 domain has some ability to recruit profilin-actin but perhaps not as effectively as the FH1 domains of other formins. The effectiveness of polyproline tracks in the FH1 domain depends on the number of prolines and their distance from the FH2 domain (34, 35). The polyproline tracks of the Fhod FH1 domain are located relatively far from the FH2 domain, with the closest track (PPPMMPP) located 31 residues from the FH2 domain. For comparison, the weak elongator Cdc12 has its closest polyproline track 26 residues away from the FH2 domain, whereas the closest polyproline tracks of the strong elongators Bni1 and mDia1 are only 22 and 16 residues away, respectively.
We approximate that Fhod has a characteristic run length of 2 μm, which is equivalent to a dissociation rate of ∼0.01 s−1 based on the elongation rate of 8 subunits/s. This dissociation rate is an order of magnitude faster than mDia1 and several orders of magnitude faster than mDia2, Bni1, Cdc12, and Capu (26, 36), which does not fit the general trend of faster elongation rates, resulting in faster dissociation rates (34). We observed evidence of Fhod protecting barbed ends only when experiments were performed in a tube, i.e. reannealing assays and when Fhod was incubated with seeds and actin monomers in a tube prior to introducing the mixture onto the surface. This suggests that the surface hinders Fhod processivity, making our measurement of Fhod processive elongation activity an underestimate. However, that Fhod is sensitive to conditions that do not perturb the processivity of other formins may indicate that processive elongation is not a critical activity of Fhod. FHOD family members generally localize to the relatively short actin filaments found in stress fibers and the sarcomere, which likely do not require accelerated barbed end growth. Therefore, Fhod nucleation and bundling activities might be more important in these contexts. We found that Fhod is indeed a potent actin bundler; its affinity of 0.18 μm for sides of actin filaments is comparable with the formins Fus1 (37) and AFH1 (38) and an order of magnitude stronger than mDia1 (39), Daam (31), and Capu (26). Alternatively, it is possible that Fhod accelerates actin elongation in vivo through collaborations with other proteins; for example, CLIP-170 was recently shown to augment the processive elongation of mDia1 (40).
Experimental procedures
Protein expression, purification, and labeling
cDNA for Drosophila Fhod isoform A (SD08909, obtained from the Drosophila Genomics Resource Center) and human FHOD1 (generous gift from T. Iskratsch) were used as templates to clone C-terminal constructs into a modified version of the pET-15b plasmid with an N-terminal His6 tag. Point mutations were generated by site-directed mutagenesis as described (41). Drosophila Fhod constructs were transformed in Rosetta (DE3) cells (Novagen), which were grown in 1 liter of Terrific Broth supplemented with 100 mg/liter ampicillin and 32 mg/liter chloramphenicol. Expression was induced at an OD of 0.6–0.8 by adding 0.25 mm isopropyl β-d-thiogalactoside and shaking overnight at 18 °C. The cells were harvested by centrifugation, washed in PBS, and flash frozen in liquid nitrogen. Human FHOD1 was expressed in Rosetta 2 (DE3) cells induced with 0.5 mm isopropyl β-d-thiogalactoside as above.
Cell pellets expressing Drosophila Fhod were thawed in extraction buffer (10 mm MOPS, pH 7, 0.5% Triton X-100, 1 mm DTT, 1 mm PMSF, 2 μg/ml DNaseI). All subsequent steps were performed on ice or at 4 °C. The cells were lysed by microfluidizing, cleared by centrifugation at 20,000 × g for 20 min, and then purified using a HitrapSP-FF cation exchange column (GE Life Sciences) with a gradient of 0.3–1 m NaCl over 16 column volumes. Pooled fractions were diluted at least 6-fold into 10 mm Tris, pH 8, 1 mm DTT and further purified on a MonoQ anion exchange column (GE Life Sciences) with a gradient of 40–500 mm NaCl over 50 column volumes. Peak fractions were exchanged into storage buffer (10 mm Tris, pH 8, 150 mm NaCl, 1 mm DTT, 0–20% glycerol), centrifuged at 20,000 × g for 20 min, flash frozen in liquid nitrogen, and stored at −80 °C.
Cell pellets expressing human FHOD1 were resuspended in extraction buffer (50 mm sodium phosphate, pH 8, 300 mm NaCl, 1 mm βME) supplemented with PMSF and DNaseI and lysed as above. Clarified lysates were nutated with 1 ml of Talon resin (Clontech) per liter of culture for 1 h. Resin was washed with 20 column volumes of extraction buffer, followed by 20 column volumes of wash buffer (50 mm sodium phosphate, pH 7, 300 mm NaCl, 1 mm βME). Resin was washed twice for 30 min each with 20 column volumes of wash buffer supplemented with 10 mm MgCl2 and 5 mm ATP. FHOD1 was eluted with 200 mm imidazole in wash buffer. Eluted protein was dialyzed into 10 mm MOPS, pH 7, 200 mm NaCl, 1 mm DTT, and then run on a Mono S cation exchange column (GE Life Sciences) with a gradient of 0.2–1 m NaCl over 32 column volumes. Peak fractions were dialyzed overnight into storage buffer (10 mm Tris, pH 8, 150 mm NaCl, 1 mm DTT), centrifuged at 20,000 × g for 20 min, flash frozen in liquid nitrogen, and stored at −80 °C. Actin assembly activity remained stable after freeze-thaw or up to 4 days on ice.
Drosophila Fhod protein concentrations were determined using the absorbance at 280 nm with an extinction coefficient of 122,840 m−1 cm−1 (ProtParam), which was verified by comparing the absorbances of native and denatured protein. The concentration of human FHOD1 was determined by quantitative Sypro Red staining. All Drosophila Fhod and human FHOD1 concentrations are reported in terms of dimer concentrations.
Acanthamoeba castellani actin was purified (42) and labeled with pyrene iodoacetamide (42), Alexa Fluor 594 succinimidyl ester (43), or EZ-link maleimide-PEG2-biotin (Thermo Scientific) (44) according to published protocols. Unlabeled and pyrene-labeled rabbit skeletal actin were kindly provided by the Reisler laboratory (University of California, Los Angeles). Schizosaccharomyces pombe profilin was purified as described (32) on a polyproline column provided by the Reisler laboratory. The concentration was determined using the absorbance at 280 nm with an extinction coefficient of 1.63 OD/mg/ml (45). Mouse capping protein was purified as described (46).
Pyrene assays
Pyrene assays were performed essentially as described (47) on an Infinite 200 Pro plate reader (Tecan). In all assays, Fhod was diluted in storage buffer before addition to polymerization buffer to improve stability. The concentration of barbed ends was calculated from the slope (obtained by linear regression over 90 s) using the equation [be] = elongation rate/(k+[G-actin] − k−), where k+ = 11.6 μm−1 s−1, and k− = 1.4 s−1 (48). For seeded elongation assays, actin filaments were sheared by passing three times through a 24-gauge needle and then aliquoted into each well of a microplate. Proteins were added to the seeds and incubated for 2–4 min at room temperature; in experiments with both Fhod and capping protein, capping protein was added 2–4 min after addition of Fhod. Seeds and additional proteins in KMEH (10 mm HEPES, pH 7, 1 mm EGTA, 50 mm KCl, 1 mm MgCl2) were added to magnesium-actin to initiate actin elongation. Elongation rates were determined by linear regression over the first 90 s and normalized against the rate of actin alone in each experiment. For experiments without capping protein, the affinity of Fhod for barbed ends was determined by fitting the data to the simplified binding equation, r = [Fhod]/([Fhod] + Kd) * a + b, where r is the normalized elongation rate. The data for capping protein experiments were analyzed using a competition binding model (49). The following equation was used to determine the Kd of Fhod for growing barbed ends,
| (Eq. 1) |
where r is the normalized elongation rate, Kd1 is the affinity of capping protein for barbed ends (0.4 nm, measured in a seeded elongation assay in the absence of Fhod), Kd2 is the affinity of Fhod for barbed ends, and R0 is the concentration of free barbed ends when [Fhod] = 0. The total concentration of barbed ends was calculated from the initial slope of the polymerization trace for actin alone as described above.
For depolymerization assays, 1 μm F-actin (70% pyrene-labeled) was incubated for at least 15 min at room temperature and then depolymerized by diluting 10-fold in 1× KMEH with additional proteins. The depolymerization rate was determined by linear regression over the first 90 s. The affinity of Fhod for barbed ends was determined by fitting the data to the simplified binding equation as above.
TIRF microscopy
In nucleation assays, assembly of 2 μm actin was initiated by the addition of KMEH with or without Fhod. After 5 min, actin was removed from the reaction and stabilized by diluting 10-fold in 1× KMEH containing Alexa Fluor 488-phalloidin. Actin was incubated with phalloidin for 10 min, diluted 20-fold in 1× KMEH supplemented with 100 mm DTT, spotted on a poly-l-lysine-coated coverslip, and imaged. All steps were performed as delicately as possible with cut pipette tips to minimize shearing.
For elongation experiments, biotinylated coverslips were prepared as follows. Coverslips were rinsed three times in MilliQ water, placed in 2% Hellmanex (Hellma Analytics) at 60–65 °C for 2 h, and then rinsed another five times in MilliQ water. Once dry, the coverslips were silanized with (3-glycidyloxypropyl)trimethoxysilane for 1 h in a hybridization oven. Unreacted (3-glycidyloxypropyl)trimethoxysilane was removed by rinsing three times with acetone. Coverslips were then PEGylated with a mixture of methoxy-PEG-NHS and biotin-PEG-NHS as described (47).
Flow chambers of ∼15 μl were assembled on the slide using strips of double-sided tape. Flow chambers were prepared with the following steps: 1) block with 25 μl of 1% Pluronic F-127 (Sigma), 50 μg/ml casein, in PBS, for 2 min; 2) 25 μl of 1× KMEH; 3) 25 μl of 40 nm streptavidin in 1× KMEH; 4) 25 μl of 1× TIRF buffer (1× KMEH, 0.5% methylcellulose (400 cP, Sigma), 50 mm DTT, 0.2 mm ATP, 20 mm glucose); 5) 50 μl of magnesium-actin and additional proteins to be assayed, in 1× TIRF buffer supplemented with 5 nm F-actin seeds (1% biotinylated, stabilized with Alexa Fluor 647-phalloidin), 250 μg/ml glucose oxidase, 50 μg/ml catalase, and 50 μg/ml casein. Fhod was incubated with seeds for at least 30 s prior to addition of magnesium-actin; in experiments with both Fhod and capping protein, Fhod was incubated with seeds for 15 s prior to addition of capping protein, and magnesium-actin was added after an additional 30 s.
To determine the characteristic run length of Fhod on barbed ends in the presence of capping protein, 1 − cumulative frequency was treated as the fraction of filaments that were still elongating at a particular length. The data were fit to the exponential equation, 1 − cf = e−l/λ * a + b, where cf is the cumulative frequency, l is the filament length, and λ is the characteristic run length.
Reannealing assays were conducted essentially as described (50) using Alexa Fluor 488- or rhodamine-labeled phalloidin-actin, sheared by passing three times through a 27-gauge needle. The final concentrations were 250 nm F-actin and 10 nm Fhod. The samples were diluted 50-fold, spotted on poly-l-lysine-coated coverslips, and imaged.
In all experiments, actin filaments were visualized on a DMI6000 TIRF microscope (Leica) with an HCX PL APO objective (100× magnification, N.A. = 1.47), and an Andor DU-897 camera, using the Leica application suite advanced fluorescence software. The experiments were conducted at room temperature. The images were obtained at 10-s intervals for 10 min. The images were processed by applying rolling ball background subtraction and a Gaussian filter. Filament lengths were quantified using the JFilament plugin for Fiji (51).
Cosedimentation
For high-speed cosedimentation, 250 nm Fhod was incubated with varying concentrations of phalloidin-stabilized F-actin for 30 min at room temperature. Samples were centrifuged at 90,000 rpm for 25 min in a TLA-100 rotor. Pellets were concentrated by resuspending in one-fourth the original volume. The supernatants and pellets were analyzed with SDS-PAGE, and the gels were stained with SyproRed. The amount of Fhod in each fraction was quantified using QuantityOne software, dividing the intensities of pellet bands by four to correct for the 4-fold concentration of pellets with resuspension. The fraction of Fhod bound to F-actin was calculated by adjusting for the amount of Fhod that pellets in the absence of F-actin with the following equation: θ = (p − p0)/(1 − p0), where θ is the fraction of Fhod bound to F-actin, p is the fraction of Fhod in the pellet, and p0 is the fraction of Fhod in the pellet in the absence of F-actin. The affinity of Fhod for F-actin was determined by fitting the data to the binding equation, θ = [F-actin]/([F-actin] + Kd) * a + b.
For low-speed cosedimentation, Fhod was incubated with phalloidin-stabilized actin filaments (final concentration, 5 μm) for 1 h at room temperature and then centrifuged at 12,000 × g for 15 min. The amount of actin in the supernatants and pellets was quantified by Coomassie-staining SDS-PAGE gels.
Author contributions
A. A. P., Z. A. O. D., A. P. v. L., and M. E. Q. designed the experiments. A. A. P., Z. A. O. D., A. P. v. L., and K. V. B. performed the experiments. A. A. P. and M. E. Q. wrote the manuscript with input from all authors.
Acknowledgments
We thank members of the Quinlan lab for feedback, the Reisler labs for valuable discussions and reagents, and Thomas Iskratsch for human FHOD1 cDNA.
This work was supported by National Institutes of Health Grant NRSA F30 HL137263 (to A. A. P.), UCLA Medical Scientist Training Program T32 GM008042 (to A. A. P.), and R01 GM096133 (to M. E. Q.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
W.T. Silkworth, K.C. Kunes, G.C. Nickel, M.L. Phillips, M.E. Quinlan, C.L. Vizcarra, Mol. Biol. Cell, in press.
- FH2
- formin homology 2 domain
- FHOD
- formin homology domain–containing protein
- TIRF
- total internal reflection fluorescence.
References
- 1. Goode B. L., and Eck M. J. (2007) Mechanism and function of formins in the control of actin assembly. Annu. Rev. Biochem. 76, 593–627 10.1146/annurev.biochem.75.103004.142647 [DOI] [PubMed] [Google Scholar]
- 2. Gasteier J. E., Madrid R., Krautkrämer E., Schröder S., Muranyi W., Benichou S., and Fackler O. T. (2003) Activation of the Rac-binding partner FHOD1 induces actin stress fibers via a ROCK-dependent mechanism. J. Biol. Chem. 278, 38902–38912 10.1074/jbc.M306229200 [DOI] [PubMed] [Google Scholar]
- 3. Schulze N., Graessl M., Blancke Soares A., Geyer M., Dehmelt L., and Nalbant P. (2014) FHOD1 regulates stress fiber organization by controlling the dynamics of transverse arcs and dorsal fibers. J. Cell Sci. 127, 1379–1393 10.1242/jcs.134627 [DOI] [PubMed] [Google Scholar]
- 4. Takeya R., Taniguchi K., Narumiya S., and Sumimoto H. (2008) The mammalian formin FHOD1 is activated through phosphorylation by ROCK and mediates thrombin-induced stress fibre formation in endothelial cells. EMBO J. 27, 618–628 10.1038/emboj.2008.7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Jurmeister S., Baumann M., Balwierz A., Keklikoglou I., Ward A., Uhlmann S., Zhang J. D., Wiemann S., and Sahin Ö. (2012) MicroRNA-200c represses migration and invasion of breast cancer cells by targeting actin-regulatory proteins FHOD1 and PPM1F. Mol. Cell. Biol. 32, 633–651 10.1128/MCB.06212-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Iskratsch T., Yu C. H., Mathur A., Liu S., Stévenin V., Dwyer J., Hone J., Ehler E., and Sheetz M. (2013) FHOD1 is needed for directed forces and adhesion maturation during cell spreading and migration. Dev. Cell 27, 545–559 10.1016/j.devcel.2013.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Koka S., Neudauer C. L., Li X., Lewis R. E., McCarthy J. B., and Westendorf J. J. (2003) The formin-homology-domain-containing protein FHOD1 enhances cell migration. J. Cell Sci. 116, 1745–1755 10.1242/jcs.00386 [DOI] [PubMed] [Google Scholar]
- 8. Gardberg M., Kaipio K., Lehtinen L., Mikkonen P., Heuser V. D., Talvinen K., Iljin K., Kampf C., Uhlen M., Grénman R., Koivisto M., and Carpén O. (2013) FHOD1, a formin upregulated in epithelial-mesenchymal transition, participates in cancer cell migration and invasion. PLoS One 8, e74923 10.1371/journal.pone.0074923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Paul N. R., Allen J. L., Chapman A., Morlan-Mairal M., Zindy E., Jacquemet G., Fernandez del Ama L., Ferizovic N., Green D. M., Howe J. D., Ehler E., Hurlstone A., and Caswell P. T. (2015) α5β1 integrin recycling promotes Arp2/3-independent cancer cell invasion via the formin FHOD3. J. Cell Biol. 210, 1013–1031 10.1083/jcb.201502040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Monzo P., Chong Y. K., Guetta-Terrier C., Krishnasamy A., Sathe S. R., Yim E. K., Ng W. H., Ang B. T., Tang C., Ladoux B., Gauthier N. C., and Sheetz M. P. (2016) Mechanical confinement triggers glioma linear migration dependent on formin FHOD3. Mol. Biol. Cell. 27, 1246–1261 10.1091/mbc.E15-08-0565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Krainer E. C., Ouderkirk J. L., Miller E. W., Miller M. R., Mersich A. T., and Blystone S. D. (2013) The multiplicity of human formins: Expression patterns in cells and tissues. Cytoskeleton 70, 424–438 10.1002/cm.21113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Iskratsch T., Lange S., Dwyer J., Kho A. L., dos Remedios C., and Ehler E. (2010) Formin follows function: a muscle-specific isoform of FHOD3 is regulated by CK2 phosphorylation and promotes myofibril maintenance. J. Cell Biol. 191, 1159–1172 10.1083/jcb.201005060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Taniguchi K., Takeya R., Suetsugu S., Kan-O M., Narusawa M., Shiose A., Tominaga R., and Sumimoto H. (2009) Mammalian formin Fhod3 regulates actin assembly and sarcomere organization in striated muscles. J. Biol. Chem. 284, 29873–29881 10.1074/jbc.M109.059303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fujimoto N., Kan-O M., Ushijima T., Kage Y., Tominaga R., Sumimoto H., and Takeya R. (2016) Transgenic expression of the formin protein Fhod3 selectively in the embryonic heart: role of actin-binding activity of Fhod3 and its sarcomeric localization during myofibrillogenesis. PLoS One 11, e0148472 10.1371/journal.pone.0148472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kan-o M., Takeya R., Taniguchi K., Tanoue Y., Tominaga R., and Sumimoto H. (2012) Expression and subcellular localization of mammalian formin Fhod3 in the embryonic and adult heart. PLoS One 10.1371/journal.pone.0034765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Al Haj A., Mazur A. J., Radaszkiewicz K., Radaszkiewicz T., Makowiecka A., Stopschinski B. E., Schönichen A., Geyer M., and Mannherz H. G. (2015) Distribution of formins in cardiac muscle: FHOD1 is a component of intercalated discs and costameres. Eur. J. Cell Biol. 94, 101–113 10.1016/j.ejcb.2014.11.003 [DOI] [PubMed] [Google Scholar]
- 17. Dwyer J., Pluess M., Iskratsch T., dos Remedios C. G., and Ehler E. (2014) The formin FHOD1 in cardiomyocytes. Anat. Rec. (Hoboken) 297, 1560–1570 10.1002/ar.22984 [DOI] [PubMed] [Google Scholar]
- 18. Wooten E. C., Hebl V. B., Wolf M. J., Greytak S. R., Orr N. M., Draper I., Calvino J. E., Kapur N. K., Maron M. S., Kullo I. J., Ommen S. R., Bos J. M., Ackerman M. J., and Huggins G. S. (2013) Formin homology 2 domain containing 3 variants associated with hypertrophic cardiomyopathy. Circ. Cardiovasc. Genet. 6, 10–18 10.1161/CIRCGENETICS.112.965277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Arimura T., Takeya R., Ishikawa T., Yamano T., Matsuo A., Tatsumi T., Nomura T., Sumimoto H., and Kimura A. (2013) Dilated cardiomyopathy-associated FHOD3 variant impairs the ability to induce activation of transcription factor serum response factor. Circ. J. 77, 2990–2996 10.1253/circj.CJ-13-0255 [DOI] [PubMed] [Google Scholar]
- 20. Esslinger U., Garnier S., Korniat A., Proust C., Kararigas G., Müller-Nurasyid M., Empana J.-P., Morley M. P., Perret C., Stark K., Bick A. G., Prasad S. K., Kriebel J., Li J., Tiret L., et al. (2017) Exome-wide association study reveals novel susceptibility genes to sporadic dilated cardiomyopathy. PLoS One 12, e0172995 10.1371/journal.pone.0172995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Schönichen A., Mannherz H. G., Behrmann E., Mazur A. J., Kühn S., Silván U., Schoenenberger C.-A., Fackler O. T., Raunser S., Dehmelt L., and Geyer M. (2013) FHOD1 is a combined actin filament capping and bundling factor that selectively associates with actin arcs and stress fibers. J. Cell Sci. 126, 1891–1901 10.1242/jcs.126706 [DOI] [PubMed] [Google Scholar]
- 22. Lammel U., Bechtold M., Risse B., Berh D., Fleige A., Bunse I., Jiang X., Klämbt C., and Bogdan S. (2014) The Drosophila FHOD1-like formin Knittrig acts through Rok to promote stress fiber formation and directed macrophage migration during the cellular immune response. Development 141, 1366–1380 10.1242/dev.101352 [DOI] [PubMed] [Google Scholar]
- 23. Shwartz A., Dhanyasi N., Schejter E. D., and Shilo B.-Z. (2016) The Drosophila formin Fhos is a primary mediator of sarcomeric thin-filament array assembly. Elife 5, e16540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kucherenko M. M., Marrone A. K., Rishko V. M., Magliarelli H. F., and Shcherbata H. R. (2011) Stress and muscular dystrophy: A genetic screen for Dystroglycan and Dystrophin interactors in Drosophila identifies cellular stress response components. Dev. Biol. 352, 228–242 10.1016/j.ydbio.2011.01.013 [DOI] [PubMed] [Google Scholar]
- 25. Xu Y., Moseley J. B., Sagot I., Poy F., Pellman D., Goode B. L., and Eck M. J. (2004) Crystal structures of a formin homology-2 domain reveal a tethered dimer architecture. Cell 116, 711–723 10.1016/S0092-8674(04)00210-7 [DOI] [PubMed] [Google Scholar]
- 26. Vizcarra C. L., Bor B., and Quinlan M. E. (2014) The role of formin tails in actin nucleation, processive elongation, and filament bundling. J. Biol. Chem. 289, 30602–30613 10.1074/jbc.M114.588368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gould C. J., Maiti S., Michelot A., Graziano B. R., Blanchoin L., and Goode B. L. (2011) The formin DAD domain plays dual roles in autoinhibition and actin nucleation. Curr. Biol. 21, 384–390 10.1016/j.cub.2011.01.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ramabhadran V., Gurel P. S., and Higgs H. N. (2012) Mutations to the formin homology 2 domain of INF2 protein have unexpected effects on actin polymerization and severing. J. Biol. Chem. 287, 34234–34245 10.1074/jbc.M112.365122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bombardier J. P., Eskin J. A., Jaiswal R., Corrêa I. R. Jr., Xu M.-Q., Goode B. L., and Gelles J. (2015) Single-molecule visualization of a formin-capping protein “decision complex” at the actin filament barbed end. Nat. Commun. 6, 8707 10.1038/ncomms9707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Shekhar S., Kerleau M., Kühn S., Pernier J., Romet-Lemonne G., Jégou A., and Carlier M.-F. (2015) Formin and capping protein together embrace the actin filament in a ménage à trois. Nat. Commun. 6, 8730 10.1038/ncomms9730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Barkó S., Bugyi B., Carlier M. F., Gombos R., Matusek T., Mihály J., and Nyitrai M. (2010) Characterization of the biochemical properties and biological function of the formin homology domains of Drosophila DAAM. J. Biol. Chem. 285, 13154–13169 10.1074/jbc.M109.093914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Harris E. S., Li F., and Higgs H. N. (2004) The mouse formin, FRLα, slows actin filament barbed end elongation, competes with capping protein, accelerates polymerization from monomers, and severs filaments. J. Biol. Chem. 279, 20076–20087 10.1074/jbc.M312718200 [DOI] [PubMed] [Google Scholar]
- 33. Kovar D. R., Kuhn J. R., Tichy A. L., and Pollard T. D. (2003) The fission yeast cytokinesis formin Cdc12p is a barbed end actin filament capping protein gated by profilin. J. Cell Biol. 161, 875–887 10.1083/jcb.200211078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Paul A. S., and Pollard T. D. (2008) The role of the FH1 domain and profilin in formin-mediated actin-filament elongation and nucleation. Curr. Biol. 18, 9–19 10.1016/j.cub.2007.11.062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Courtemanche N., and Pollard T. D. (2012) Determinants of formin homology 1 (FH1) domain function in actin filament elongation by formins. J. Biol. Chem. 287, 7812–7820 10.1074/jbc.M111.322958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kovar D. R., Harris E. S., Mahaffy R., Higgs H. N., and Pollard T. D. (2006) Control of the assembly of ATP- and ADP-actin by formins and profilin. Cell. 124, 423–435 10.1016/j.cell.2005.11.038 [DOI] [PubMed] [Google Scholar]
- 37. Scott B. J., Neidt E. M., and Kovar D. R. (2011) The functionally distinct fission yeast formins have specific actin-assembly properties. Mol. Biol. Cell. 22, 3826–3839 10.1091/mbc.E11-06-0492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Michelot A., Guérin C., Huang S., Ingouff M., Richard S., Rodiuc N., Staiger C. J., and Blanchoin L. (2005) The formin homology 1 domain modulates the actin nucleation and bundling activity of Arabidopsis FORMIN1. Plant Cell 17, 2296–2313 10.1105/tpc.105.030908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Li F., and Higgs H. N. (2003) The mouse formin mDia1 is a potent actin nucleation factor regulated by autoinhibition. Curr. Biol. 13, 1335–1340 10.1016/S0960-9822(03)00540-2 [DOI] [PubMed] [Google Scholar]
- 40. Henty-Ridilla J. L., Rankova A., Eskin J. A., Kenny K., and Goode B. L. (2016) Accelerated actin filament polymerization from microtubule plus ends. Science 352, 1004–1009 10.1126/science.aaf1709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Liu H., and Naismith J. H. (2008) An efficient one-step site-directed deletion, insertion, single and multiple-site plasmid mutagenesis protocol. BMC Biotechnol. 8, 91 10.1186/1472-6750-8-91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zuchero J. B. (2007) In vitro actin assembly assays and purification from Acanthamoeba. Methods Mol. Biol. 370, 213–226 10.1007/978-1-59745-353-0_15, [DOI] [PubMed] [Google Scholar]
- 43. Grintsevich E. E., Yesilyurt H. G., Rich S. K., Hung R. J., Terman J. R., and Reisler E. (2016) F-actin dismantling through a redox-driven synergy between Mical and cofilin. Nat. Cell Biol. 18, 876–885 10.1038/ncb3390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Roth-Johnson E. A., Vizcarra C. L., Bois J. S., and Quinlan M. E. (2014) Interaction between microtubules and the Drosophila formin cappuccino and its effect on actin assembly. J. Biol. Chem. 289, 4395–4404 10.1074/jbc.M113.499921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lu J., and Pollard T. D. (2001) Profilin binding to poly-l-proline and actin monomers along with ability to catalyze actin nucleotide exchange is required for viability of fission yeast. Mol. Biol. Cell. 12, 1161–1175 10.1091/mbc.12.4.1161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Palmgren S., Ojala P. J., Wear M. A., Cooper J. A., and Lappalainen P. (2001) Interactions with PIP2, ADP-actin monomers, and capping protein regulate the activity and localization of yeast twinfilin. J. Cell Biol. 155, 251–260 10.1083/jcb.200106157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Bor B., Vizcarra C. L., Phillips M. L., and Quinlan M. E. (2012) Autoinhibition of the formin Cappuccino in the absence of canonical autoinhibitory domains. Mol. Biol. Cell. 23, 3801–3813 10.1091/mbc.E12-04-0288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Pollard T. D. (1986) Rate constants for the reactions of ATP-and ADP-actin with the ends of actin filaments. J. Cell Biol. 103, 2747–2754 10.1083/jcb.103.6.2747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Vinson V. K., De La Cruz E. M., Higgs H. N., and Pollard T. D. (1998) Interactions of Acanthamoeba profilin with actin and nucleotides bound to actin. Biochemistry 37, 10871–10880 10.1021/bi980093l [DOI] [PubMed] [Google Scholar]
- 50. Vizcarra C. L., Kreutz B., Rodal A. A., Toms A. V., Lu J., Zheng W., Quinlan M. E., and Eck M. J. (2011) Structure and function of the interacting domains of Spire and Fmn-family formins. Proc. Natl. Acad. Sci. U.S.A. 108, 11884–11889 10.1073/pnas.1105703108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Smith M. B., Li H., Shen T., Huang X., Yusuf E., and Vavylonis D. (2010) Segmentation and tracking of cytoskeletal filaments using open active contours. Cytoskeleton 67, 693–705 10.1002/cm.20481 [DOI] [PMC free article] [PubMed] [Google Scholar]