Abstract
Cullins, critical members of the cullin-RING ubiquitin ligases (CRLs), are often aberrantly expressed in different cancers. However, the underlying mechanisms regarding aberrant expression of these cullins and the specific substrates of CRLs in different cancers are mostly unknown. Here, we demonstrate that overexpressed CUL4B in human osteosarcoma cells forms an E3 complex with DNA damage binding protein 1 (DDB1) and DDB1- and CUL4-associated factor 13 (DCAF13). In vitro and in vivo analyses indicated that the CRL4BDCAF13 E3 ligase specifically recognized the tumor suppressor PTEN (phosphatase and tensin homolog deleted on chromosome 10) for degradation, and disruption of this E3 ligase resulted in PTEN accumulation. Further analyses indicated that miR-300 directly targeted the 3′ UTR of CUL4B, and DNA hypermethylation of a CpG island in the miR-300 promoter region contributed to the downregulation of miR-300. Interestingly, ectopic expression of miR-300 or treatment with 5-AZA-2′-deoxycytidine, a DNA methylation inhibitor, decreased the stability of CRL4BDCAF13 E3 ligase and reduced PTEN ubiquitination. By applying in vitro screening to identify small molecules that specifically inhibit CUL4B-DDB1 interaction, we found that TSC01131 could greatly inhibit osteosarcoma cell growth and could disrupt the stability of the CRL4BDCAF13 E3 ligase. Collectively, our findings shed new light on the molecular mechanism of CUL4B function and might also provide a new avenue for osteosarcoma therapy.
Keywords: miR-300, DNA methylation, CUL4B, CRL4B E3 ligase, PTEN, ubiquitination, small molecule, TSC01131, osteosarcoma
Introduction
Protein ubiquitination and degradation through the ubiquitin proteasome system (UPS) are conserved mechanisms in prokaryotic and eukaryotic organisms, and disruption of these processes is associated with the occurrence of many diseases, including cancer.1, 2, 3 Protein ubiquitination requires three critical enzymes: ubiquitin-activating enzymes (E1s), ubiquitin-conjugating enzymes (E2s), and ubiquitin ligases (E3s).4, 5 The human genome encodes at least 2 E1s, 38 E2s, and nearly 1,000 E3s, which further recognize thousands of substrates for modification in different biological processes.6, 7 Currently, four major classes of E3 ligases, including homologs to E6-AP carboxyl terminus (HECT), RING-finger, U-box, and plant homeodomain (PHD) finger, have been identified.4, 8 Of these, the RING-finger E3 ligases represent the largest family, which can be further classified into different subgroups, such as the anaphase-promoting complex (APC), cullin-RING ubiquitin ligases (CRLs), and the Skp1-cullin-F-box protein (SCF) complex, depending on the presence of different adapters.10, 11 CRLs, the largest class of RING-type E3 ligases, conservatively utilize seven cullins, including CUL1, CUL2, CUL3, CUL4A, CUL4B, CUL5, and CUL7, as scaffolds to facilitate the assembly of E3 ligase complexes and to transfer ubiquitin from E2 to the substrates.10, 12 Dysregulation of cullin members has been broadly reported to contribute to tumorigenesis through diverse mechanisms such as involvement in DNA damage and repair, cell-cycle progression, as well as participation in the ubiquitination of oncoproteins or tumor suppressors.13, 14 For example, CUL4A and CUL4B, two paralogous genes in the human genome that share 82% amino acid sequence identity, are highly expressed in different cancers, such as breast cancer,15 hepatocellular carcinoma, and osteosarcoma.16, 17 Both CUL4A and CUL4B have been shown to form CRL complexes with RING-box protein 1 (RBX1), DNA damage binding protein 1 (DDB1), and DDB1- and CUL4-associated factors (DCAFs).18, 19 CUL4-RBX is responsible for E3 ligase activities, whereas DCAFs are in charge of the specificity of substrates.18, 19 In the human genome, more than 100 DCAFs have been characterized according to the WD40 repeats that they contain.20 Through these DCAFs, the CRL4A-based E3 ligases are capable of recognizing a number of substrates, including DDB2,21 p21,22, 23 p27,24 CDT1 (chromatin licensing and DNA replication factor 1),24 Chk1 (checkpoint kinase 1),25 and STAT1 (signal transducer and activation of transcription 1),26 in different cancers. By contrast, although CUL4B is highly expressed in several cancer types, such as esophageal cancer and osteosarcoma,17, 27 the underlying mechanism of CUL4B overexpression and the specific substrates in these cancers are generally unknown.
Downregulation of tumor suppressors is a major factor that leads to tumorigenesis. Phosphatase and tensin homolog deleted on chromosome 10 (PTEN), a common tumor suppressor, the expression of which is often downregulated or even absent in the majority of human cancers, functions as a phosphatase to dephosphorylate phosphatidylinositol (3,4,5)-trisphosphate (PIP3), resulting in the inhibition of the phosphatidylinositol 3-kinase (PI3K)/Akt signaling pathway.28, 29, 30 The enzymatic activity of PTEN is crucial for the maintenance of its function because its inactivation increases cancer cell proliferation but attenuates cell death.28, 29, 30, 31 At present, PTEN is known to be regulated in many ways, including by microRNAs (miRNAs) at the transcriptional level and by phosphorylation and ubiquitination at the posttranslational level.32 In recent years, several miRNAs, including miR-23a,33 miR-26a,34 and miR-93,35 have been reported to directly target the 3′ UTR of PTEN and to negatively regulate its expression, eventually participating in different biological processes, including cell migration and invasion.33, 34, 35 In addition, multiple kinases, including CK2 (casein kinase 2),36 GSK3β (glycogen synthase kinase 3 beta),37 and PICT-1 (protein interacting with the C terminus-1),38 are capable of phosphorylating the C terminus of PTEN at the S380, T382, and T383 sites, and this phosphorylation facilitates the maintenance of PTEN stability and function.36, 37, 38, 39 Moreover, two PEST domains in PTEN associate with ubiquitin-dependent degradation.40 PTEN is able to be ubiquitinated by NEDD4-1 at several lysine residues, including K289.40 However, these different PTEN expression-modulating mechanisms in tumorigenesis and their relevance remain to be elucidated in more physiological environments.
Osteosarcoma remains the leading cause of cancer-related death in children and adolescents.41 Despite tremendous efforts to minimize osteosarcoma cancer deaths, the prognosis of osteosarcoma remains poor, with a 5-year survival rate of only 15%–30%.41 Common treatment approaches for osteosarcoma patients who are diagnosed at an early MSTS (Musculoskeletal Tumor Society) stage include surgery followed by chemotherapy.41 However, the majority of patients with advanced MSTS stages will eventually experience tumor progression and require further effective treatment.42 Thus, understanding the molecular basis for the progression of osteosarcoma is critical to improve the treatment and prognosis of osteosarcoma patients. Because our previous results revealed that CUL4B is overexpressed in osteosarcoma cells, and that this overexpression promotes cell proliferation and inhibits cell apoptosis,17 we further investigated the pathogenesis of CUL4B in this process. In this study, we first verified the formation of a CUL4B-based E3 ligase complex, followed by a demonstration of the mechanism of CUL4B overexpression in osteosarcoma cells. DNA methylation-mediated downregulation of miR-300 was found to be responsible for the entire regulatory process. Then, a small compound named TSC01131 was identified by screening small molecules that inhibited the CUL4B-DDB1 interaction in a sesterterpenoid pool, and this compound greatly inhibited osteosarcoma cell growth by disrupting the stability of CRL4BDCAF13 E3 ligase. Together, our results provide new insights into the understanding of the mechanisms underlying CUL4B overexpression and how CRL4BDCAF13 E3 ligase recognizes its substrate PTEN in osteosarcoma cells. More importantly, our findings provide an opportunity for the development of CRL4BDCAF13 E3 ligase-targeted therapeutics.
Results
CUL4B Formed a Complex with DDB1 and RBX1 in Human Osteosarcoma Cells
It is well known that CUL4B forms a complex with DDB1 and RBX1 in cells derived from different species, including humans.18 To determine whether CUL4B also interacts with DDB1 and RBX1 in human osteosarcoma cells, we constructed a pCDNA3-Flag-CUL4B vector and transfected it into U2OS osteosarcoma cells. After immunoprecipitation (IP) experiments with anti-Flag-Agarose, we detected whether CUL4B pulled down DDB1 and RBX1. As shown in Figure S1A, the anti-Flag immunocomplex from U2OS cells pulled down not only DDB1, but also RBX1. By contrast, we could not detect CUL4B, DDB1, or RBX1 in U2OS cells transfected with the negative vector pCDNA3-Flag. These in vivo data verified the formation of an RBX1-CUL4B-DDB1 complex in osteosarcoma cells. To further confirm this observation, we constructed yeast-two-hybrid (Y2H) vectors, including pGADT7-CUL4B, pGBKT7-DDB1, and pGBKT7-RBX1, and tested the direct interactions of CUL4B-DDB1 and CUL4B-RBX1 in yeast cells. As shown in Figure S1B, the in vitro results clearly demonstrated that CUL4B directly interacted with DDB1 and RBX1. Thus, our in vivo and in vitro results suggested that CUL4B formed a conserved complex with DDB1 and RBX1 in human osteosarcoma cells.
Components of CRL4BDCAF13 E3 Ligase Were Upregulated in Human Osteosarcoma Cells
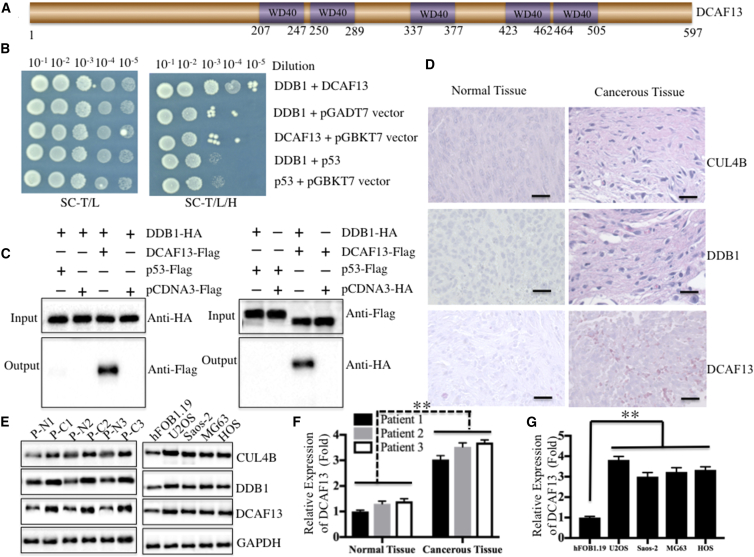
Because CUL4A and CUL4B form CRL4 E3 ligases with RING-box proteins, adaptor proteins, and substrate recognition receptors, we verified that CUL4B was able to interact with the RING-box protein RBX1 and the adaptor protein DDB1. However, the substrate recognition receptor DCAF is still lacking to form an E3 ligase. To identify the specific DCAFs for CRL4B-based E3 ligases in human osteosarcoma cells, we primarily constructed a human osteosarcoma cell-specific cDNA library in the pACT2 vector and then screened the DCAFs that interacted with DDB1 using pGBKT7-DDB1 as a bait vector. The transformed yeast cells were selected on synthetic dropout nutrient medium (SC/-Trp-Leu-His) containing X-α-Gal. The candidate genes that interacted with DDB1 were obtained from blue colonies after PCR amplification with pACT2-specific primers. We identified a DCAF protein, termed DCAF13, in our primary results. DCAF13 is a protein containing 597 amino acids with five WD40 repeats located in the middle portion of the molecule (Figure 1A); however, its function has not yet been demonstrated. To further verify the interaction between DDB1 and DCAF13, we performed in vitro Y2H and in vivo co-immunoprecipitation (Co-IP) experiments. For the Y2H assay, we constructed pGADT7-DCAF13, pGADT7-p53 (negative control), and pGBKT7-DDB1 vectors and tested the direct interactions of DCAF13-DDB1 and p53-DDB1 in yeast cells. The results indicated that DCAF13 was able to directly interact with DDB1, but not with p53 (Figure 1B). To determine whether DCAF13 formed a complex with DDB1 in human osteosarcoma cells, we performed reciprocal IP experiments in U2OS cells transfected with pCDNA3-HA-DDB1 + pCDNA3-Flag-p53, pCDNA3-HA-DDB1 + pCDNA3-Flag, pCDNA3-HA-DDB1 + pCDNA3-Flag-DCAF13, pCDNA3-HA + pCDNA3-Flag-p53, or pCDNA3-HA + pCDNA3-Flag-DCAF13. The anti-human influenza hemagglutinin (HA) immunocomplex from HADDB1 cells only pulled down DCAF13, but not p53 (Figure 1C, left panel). In this anti-Flag immunocomplex, only Flag-DCAF13, and not Flag-p53, was able to pull down DDB1 (Figure 1C, right panel). These results clearly demonstrated that CUL4B, RBX1, DDB1, and DCAF13 formed a CRL4BDCAF13 E3 ligase in human osteosarcoma cells.
Figure 1.
DCAF13 Was a Substrate Recognition Receptor for CUL4B-Based E3 Ligase, and Its Expression Was Elevated in Osteosarcoma Cells
(A) Schematic diagram of the WD40 domains of DCAF13 protein. Five WD40 repeats and their locations are indicated. (B) DCAF13 directly interacted with DDB1 in yeast cells. The pGADT7-DCAF13 or pGADT7-p53 plasmid was co-transformed with pGBKT7-DDB1 into AH109 cells. Growth of the transformed yeast cells was assayed on SC-T/L (left panel) or SC-H/T/L medium (right panel). (C) DCAF13 interacted with DDB1 in osteosarcoma cells. U2OS cells transfected with different combinations of plasmids (as indicated in the pictures) were incubated for 48 hr. The HA-tagged or Flag-tagged proteins were immunoprecipitated with anti-HA-Agarose or anti-Flag-Agarose, respectively. The pull-down products were then analyzed via immunoblotting with anti-Flag or anti-HA antibodies. (D) DCAF13 was upregulated in cancerous tissues from an osteosarcoma patient (MSTS stage III). H&E staining was performed to detect CUL4B, DDB1, or DCAF13 using their corresponding antibodies. Scale bars, 100 μm. (E) DCAF13 was upregulated in cancerous tissues from three osteosarcoma patients (MSTS stage II–IV: patient 1, P1, stage II; patient 2, P2, stage III; patient 3, P3, stage IV) and in osteosarcoma cell lines. Immunoblots were performed to detect the protein levels of CUL4B, DDB1, DCAF13, and GAPDH in the cancerous tissues (C) and normal tissues (N) from three osteosarcoma patients (P1, P2, and P3) and in four osteosarcoma cell lines, including U2OS, Saos-2, MG63, and HOS. (F) Protein level of DCAF13 in the left panel of (E) was normalized against GAPDH. (G) Protein level of DCAF13 in the right panel of (E) was normalized against GAPDH. **p < 0.001.
We previously showed that CUL4B was highly expressed in osteosarcoma cells;17 thus, it is highly possible that the other components of the CRL4BDCAF13 complex are also upregulated in osteosarcoma cells. To verify this hypothesis, we first analyzed histological changes of members of the CRL4BDCAF13 complex, including CUL4B, DDB1, and DCAF13, using H&E staining in tumor specimens derived from an osteosarcoma patient (MSTS stage III). The results confirmed that the expression of these three proteins was much higher in cancerous tissue compared with adjacent non-tumor tissue (Figure 1D). In addition, we assessed the protein levels of these proteins in three osteosarcoma patients under different MSTS stages (II–IV), as well as in four osteosarcoma cell lines, including U2OS, Saos-2, MG63, and HOS. As expected, in cancerous tissues and osteosarcoma cells, CUL4B, DDB1, and DCAF13 were upregulated (Figure 1E). The relative protein level of DCAF13 was also normalized against GAPDH, and a nearly 3-fold induction was identified in the cancerous tissues from patients and in the four osteosarcoma cell lines (Figures 1F and 1G).
CRL4BDCAF13 E3 Ligase Recognized PTEN as a Substrate in Human Osteosarcoma Cells
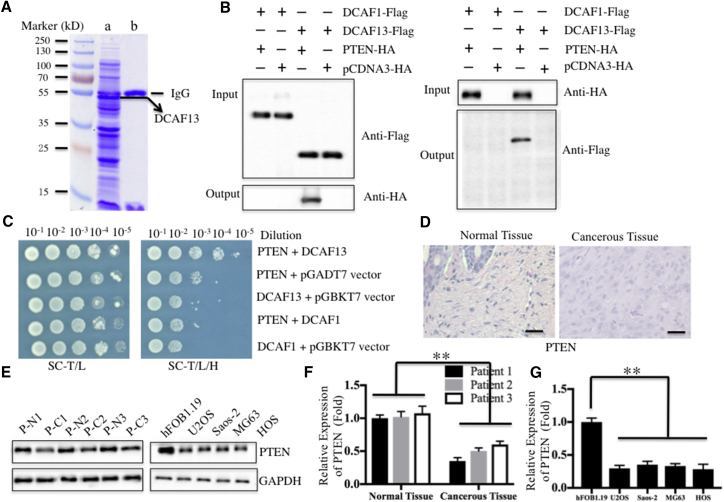
Previous investigations have identified a number of substrates of CRL4A E3 ligases, such as DDB2, p12, CDT1, p21, and p27.21, 22, 23, 24, 25 However, limited information is available for the substrates of CRL4B E3 ligases in different cancers. Given that the substrates of CRL4 E3 ligases are recognized by DCAFs, we first sought to identify the specific substrates of CRL4BDCAF13 E3 ligase in human osteosarcoma cells by screening proteins that interacted with DCAF13. Accordingly, we constructed a two-epitope-tagged vector containing DCAF13, called pCDNA3-Flag-HA-DCAF13, and then transfected it into U2OS cells. After a two-step purification method with anti-Flag-Agarose and anti-HA-Agarose, DCAF13-containing complexes were enriched (Figure 2A) and subjected to liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis. By blasting in the NCBI database, we obtained a total of 56 proteins that might interact with DCAF13 in our primary list (Table S1). Interestingly, those proteins included the tumor suppressor protein PTEN. To confirm that DCAF13 interacts with PTEN, we performed an in vivo Co-IP analysis and an in vitro Y2H assay. For Co-IP analysis, U2OS cells transfected with pCDNA3-HA-PTEN + pCDNA3-Flag-DCAF1, pCDNA3-HA + pCDNA3-Flag-DCAF1, pCDNA3-HA-PTEN + pCDNA3-Flag-DCAF13, or pCDNA3-HA + pCDNA3-Flag-DCAF13 were lysed and immunoprecipitated with either anti-HA-Agarose or anti-Flag-Agarose. In the anti-Flag immunocomplex, Flag-DCAF13, but not Flag-DCAF1, was able to pull down PTEN (Figure 2B, left panel). The anti-HA immunocomplex from HAPTEN cells brought down DCAF13, but not DCAF1 (Figure 2B, right panel). For the Y2H assay, we determined the direct interaction of DCAF13 and PTEN using constructs with pGADT7-DCAF13 + pGBKT7-PTEN, pGADT7-DCAF1 + pGBKT7-PTEN, and other negative control combinations, including pGADT7 + pGBKT7-PTEN, pGADT7-DCAF13 + pGBKT7, and pGADT7-DCAF1 + pGBKT7. These results also indicated that DCAF13, but not DCAF1, was able to directly interact with PTEN in yeast cells (Figure 2C). Thus, it is highly possible that PTEN is a substrate of CRL4BDCAF13 E3 ligase in human osteosarcoma cells. Components of CRL4BDCAF13 E3 were upregulated, suggesting that this complex should have increased E3 activity in osteosarcoma cells. Thus, we proposed that PTEN would be downregulated in cancerous tissues of osteosarcoma patients and in osteosarcoma cells if it was a substrate of CRL4BDCAF13 E3 ligase. To confirm this hypothesis, we detected PTEN expression by H&E staining and immunoblotting. As expected, PTEN was downregulated in cancerous tissues and cells (Figures 2D and 2E). After normalization against GAPDH, we found a nearly 2-fold decrease of PTEN protein levels in both cancerous tissues and cells (Figures 2F and 2G).
Figure 2.
DCAF13 Interacted with PTEN In Vivo and In Vitro
(A) In vivo pull-down of the Flag-HA-DCAF13-associated complexes. U2OS cells transfected with pCDNA3-Flag-HA-DCAF13 or pCDNA3-Flag-HA were subjected to IP analysis. The resulting complexes were loaded onto SDS-PAGE gel for separation and then stained with Coomassie Brilliant Blue R 250. The IgG and DCAF13 bands were indicated. (B) DCAF13 interacted with PTEN in vivo. U2OS cells co-transfected with different combinations of plasmids (as indicated in the pictures) were subjected to Co-IP analysis with anti-Flag-Agarose (left panel) or anti-HA-Agarose (right panel). The pull-down products were then analyzed via immunoblots with anti-HA and anti-Flag antibodies. (C) DCAF13 directly bound to PTEN in yeast. The pGBKT7-PTEN plasmid was co-transformed with the plasmid pGADT7-DCAF13, pGADT7-DCAF1, or pGADT7 empty vector into AH109 cells. The resulting yeast colonies were dotted on SC-T/L (left panel) or SC-H/T/L medium to determine their growth. (D) PTEN was downregulated in cancerous tissues from a patient with osteosarcoma (MSTS stage III). H&E staining was performed to detect PTEN levels using a specific antibody in slices from the same tissues as in Figure 1D. Scale bars, 100 μm. (E) PTEN was downregulated in cancerous tissues from three patients with osteosarcoma and in osteosarcoma cell lines. The same samples used in Figure 1E were used to detect the protein level of PTEN. (F and G) The protein level of PTEN in (E) was normalized against GAPDH. **p < 0.001.
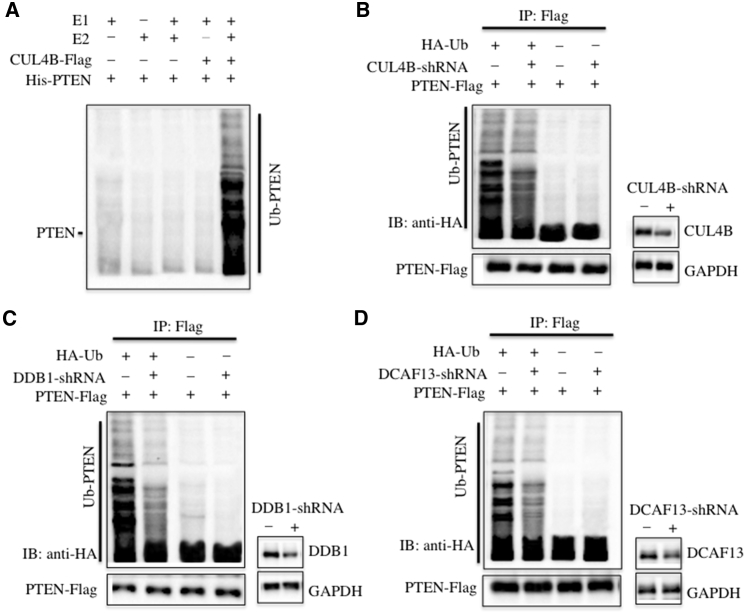
Next, we performed in vitro and in vivo experiments to detect whether CRL4BDCAF13 E3 ligase could ubiquitinate PTEN. For the in vitro analysis, we reconstituted the ubiquitination reaction using combinations of E1, E2, immunoprecipitated CUL4B-Flag complex, and purified recombinant His-PTEN protein, and we then detected the ubiquitination level by immunoblotting against PTEN. Our results showed that only the incubation of E1, E2, and the CUL4B-Flag complex could ubiquitinate PTEN, but that the reactions lacking E2 or the CUL4B-Flag complex could not (Figure 3A). For the in vivo analysis, we co-transfected pCDNA3-Flag-PTEN with or without pCDNA3-HA-Ubiquitin in U2OS cells with and without the suppression of CUL4B by its specific small hairpin RNA (shRNA). After IP with anti-Flag-Agarose, the ubiquitination of PTEN was examined by immunoblotting against HA. As shown in Figure 3B, inhibition of CUL4B expression significantly reduced PTEN ubiquitination, suggesting that CRL4BDCAF13 E3 ligase contributed to PTEN ubiquitination in vivo. Similarly, we also knocked down DDB1 and DCAF13 in U2OS cells with their corresponding shRNAs, and we then co-transfected pCDNA3-Flag-PTEN with or without pCDNA3-HA-Ubiquitin in these CUL4B downregulated cells or normal U2OS cells. Then, the same procedure was performed to detect PTEN ubiquitination. Our results indicated that either DDB1 or DCAF13 downregulation was able to decrease the PTEN ubiquitination level (Figures 3C and 3D). These results clearly demonstrated that CRL4BDCAF13 E3 ligase recognized PTEN as a substrate in osteosarcoma cells.
Figure 3.
PTEN Was a Substrate of CRL4BDACF13 E3 Ligase in Osteosarcoma Cells
(A) CRL4BDACF13 E3 ligase could ubiquitinate PTEN in vitro. The purified His-PTEN protein was incubated with E1 (+), with (+) or without (−) E2, and with (+) or without (−) CUL4B-Flag complex in reaction buffer, followed by immunoblotting for PTEN. (B–D) Knockdown of CUL4B, DDB1, or DCAF13 attenuated the ubiquitination level of PTEN in vivo. U2OS cells were first co-transfected with PTEN-Flag and HA-Ubiquitin, followed by retroviral infection of CUL4B-shRNA (B), DDB1-shRNA (C), or DCAF13-shRNA (D). After 48 hr of incubation, the cells were lysed, immunoprecipitated with anti-Flag antibody, and then probed with anti-HA antibody to detect the ubiquitinated PTEN. The protein levels of CUL4B, DDB1, and DCAF13 after their knockdown are indicated in the right panels of each figure.
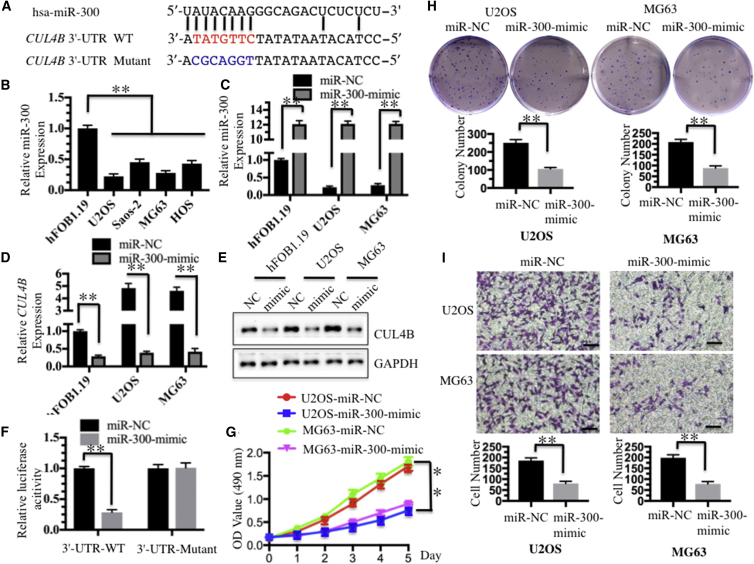
miR-300 Specifically Bound to the 3′ UTR of CUL4B and Negatively Regulated Its Expression in Human Osteosarcoma Cells
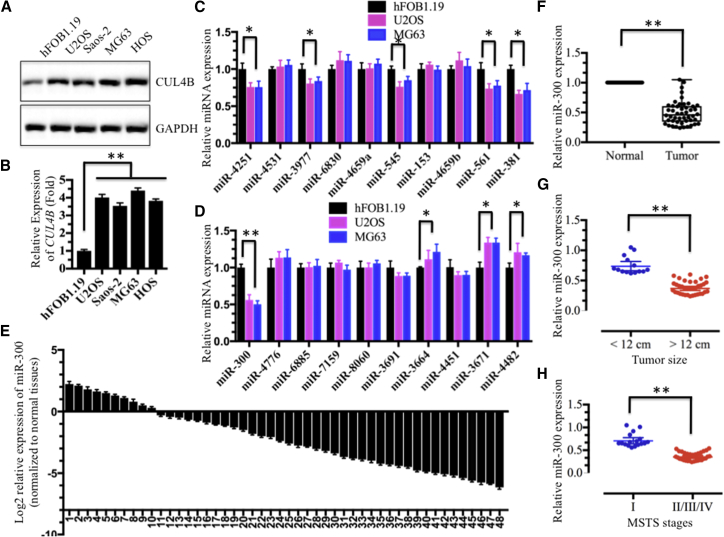
Given that a number of miRNAs play important regulatory roles in the pathogenesis of different diseases, including cancer, by targeting specific mRNAs for degradation or translation repression,43 we next sought to determine whether miRNAs participated in the regulation of CUL4B expression in human osteosarcoma cells. We first determined whether the overexpression of CUL4B was mediated transcriptionally or post-transcriptionally. Both the CUL4B mRNA and protein levels were detected in U2OS, Saos-2, MG-63, and HOS cells, and we found that both levels were clearly upregulated compared with those measured in hFOB1.19 cells (Figures 4A and 4B), suggesting that CUL4B was regulated at the transcriptional level. Then, we searched for miRNAs that might target the 3′ UTR of CUL4B in an online database (miRDB, http://www.mirdb.org) for miRNA target prediction and functional annotations. After inputting the CUL4B gene name, we found a total of 106 miRNAs predicted to target CUL4B, with target scores from 100 to 52 (Table S2). In this study, we selected only the top 20 predicted miRNAs with target scores from 100 to 90 to detect their expression in hFOB1.19, U2OS, and MG63 cells. Among these 20 miRNAs, 9 were downregulated in both U2OS and MG63 cells compared with hFOB1.19 cells (Figures 4C and 4D). These nine miRNAs included miR-4251 (target score 100), miR-3977 (target score 99), miR-545 (target score 97), miR-561 (target score 96), miR-381 (target score 96), miR-300 (target score 96), miR-3664 (target score 91), miR-3671 (target score 90), and miR-4482 (target score 90). Of these, the expression of miR-300 was much lower than the expression of the other eight miRNAs (Figures 4C and 4D). Thus, we focused our following studies on investigating the regulatory role of miR-300 for CUL4B.
Figure 4.
Expression of miR-300 Was Downregulated in Osteosarcoma Cell Lines and in the Majority of Cancerous Tissues from Patients with Osteosarcoma
(A and B) Both the CUL4B protein (A) and mRNA (B) levels were upregulated in osteosarcoma cell lines. The hFOB1.19, U2OS, Saos-2, MG63, and HOS cell lines were subjected to immunoblotting and qRT-PCR analyses to determine CUL4B protein and mRNA levels. (C and D) Expression of 20 miRNAs that were predicted to target the 3′ UTR of CUL4B in hFOB1.19, U2OS and MG63 cells. (C) miR-4251, miR-4531, miR-3977, miR-6830, miR-4659a, miR-545, miR-153, miR-4659b, miR-561 and miR-381. (D) miR-300, miR-4776, miR-6885, miR-7159, miR-8060, miR-3691, miR-3664, miR-4451, miR-3671 and miR-4482. (E and F) Expression of miR-300 in osteosarcoma cancerous tissues is shown. Relative expression of miR-300 in osteosarcoma tumors (n = 48) was normalized to corresponding adjacent normal tissues (n = 48). (E) Log2 fold change; (F) absolute fold change. p < 0.001. (G and H) The expression of miR-300 was negatively correlated with osteosarcoma tumor size and MSTS stage. The expression of miR-300 was significantly lower in larger tumors (tumor maximal diameter ≥ 12 cm) (G) and was significantly lower in patients with osteosarcoma with advanced MSTS stages (II/III/IV) than in those with an early MSTS stage (H). *p < 0.05; **p < 0.001.
To confirm that the expression of miR-300 was downregulated, we performed qRT-PCR analysis to detect miR-300 expression in 48 paired cancerous and non-tumor tissues from patients who were diagnosed with osteosarcoma at different MSTS stages (see detailed information in Table S3). Consistent with the results from the osteosarcoma cell analysis, the expression of miR-300 was also significantly downregulated in the majority of cancerous tissues (38/48) (Figures 4E and 4F). Moreover, the expression of miR-300 was negatively correlated with tumor size (Figure 4G) and was gradually decreased with the MSTS malignant degrees of patients with osteosarcoma (Figure 4H). Meanwhile, we also analyze CUL4B expression in these 48-paired cancerous and non-tumor tissues. In contrast with miR-300 expression, CUL4B was only downregulated in a small amount of cancerous tissues (10/48), but significantly overexpressed in the majority of cancerous tissues (38/48) (Figures S2A and S2B). The decreased expression of miR-300 in cancerous tissues was negatively correlated with CUL4B expression (p < 0.001) with a Pearson correlation coefficient (R) of −0.24 (Figure S2C). Overexpression of CUL4B in osteosarcoma patients was associated with larger tumor size (≥12 cm; p < 0.01) (Figure S2D) and higher MSTS stages (p < 0.01) (Figure S2E).
In addition, the sequence alignment also indicated that miR-300 was able to directly bind to the 3′ UTR of CUL4B mRNA (Figure 5A). Our previous study only detected expression of 20 miRNAs in U2OS and MG63 cells. To further determine whether miR-300 expression was commonly downregulated in human osteosarcoma cell lines, we measured its level by qRT-PCR in U2OS, Saos-2, MG63, and HOS cells. As expected, our results showed that miR-300 expression was significantly downregulated in all four cell lines compared with hFOB1.19 control cells (Figure 5B). Of these four malignant cell lines, U2OS and MG63 cells were selected to further analyze the role of miR-300. With the observation that miR-300 was dramatically suppressed in cancerous osteosarcoma tissues and cell lines, we hypothesized that miR-300 functioned as a tumor suppressor. To investigate whether higher levels of miR-300 had effects on CUL4B expression and osteosarcoma cell growth, we re-introduced miR-300-mimic into U2OS and MG63 cells, and the qRT-PCR results indicated that miR-300 was successfully overexpressed in these two cell lines (Figure 5C). Conversely, with the overexpression of miR-300, both the CUL4B mRNA and protein levels were significantly downregulated (Figures 5D and 5E). These results further suggested that CUL4B might be a direct target of miR-300. To validate this conclusion, we performed a luciferase reporter assay by which the wild-type (WT) and mutant 3′ UTRs of CUL4B were co-transfected with miR-300-mimic or negative control vector, miR-NC. Our results showed that the overexpression of miR-300 significantly decreased luciferase activity in the 3′ UTR WT but failed to repress luciferase activity in the 3′ UTR mutant (Figure 5F). These results suggested that mutation of the miR-300 binding site in the 3′ UTR of CUL4B abolished the transcriptional inhibition of miR-300. Moreover, cells transfected with miR-300-mimic were also used for cell proliferation assays, and the results indicated that overexpression of miR-300 significantly inhibited cell growth compared with cells transfected with miR-NC (Figure 5G). In addition, a significant decrease in colony formation rates was also observed in U2OS and MG63 cells overexpressing miR-300 (Figure 5H). Similarly, the invasiveness of U2OS and MG63 cells that were transfected with miR-300-mimic was also dramatically reduced (Figure 5I). We then performed rescue experiments to further validate that miR-300 exerted its role through targeting CUL4B in hFOB1.19 cells. Two CUL4B expression vectors, pCDNA3-CUL4B-3′ UTR (WT)-Flag and pCDNA3-CUL4B-3′ UTR (mutant)-Flag (the mutated sites were the same as Figure 5A), were used to examine cell proliferation, colony formation, and cell invasion abilities. As shown in Figure S3, the rescue experiments clearly indicated that hFOB1.19 cells transfected with miR-300-mimic + pCDNA3-Flag and miR-300-mimic + pCDNA3-CUL4B-3′ UTR (mutant)-Flag had similar phenotypes, whereas cells transfected with miR-300-mimic + pCDNA3-CUL4B-3′ UTR (WT)-Flag had similar phenotypes to hFOB1.19 cells. These results indicated that miR-300-mimic only bonds to the WT 3′ UTR of CUL4B, but not to its mutant. Because miR-300 was significantly downregulated in osteosarcoma cells, we next sought to investigate whether repression of miR-300 in non-cancerous cells could cause similar effects to osteosarcoma cells. Accordingly, we transfected miR-NC or anti-miR-300 into hFOB1.19 cells and then evaluated CUL4B expression, cell proliferation, colony formation, and cell invasion abilities. Interestingly, our results indicated that inhibition of miR-300 in hFOB1.19 cells resulted in similar phenotypes to U2OS cells (Figure S4). In addition, we also evaluated the in vivo tumor formation of miR-300 overexpression by injecting nude mice with U2OS-control (with pmR-ZsGreen1 empty vector), U2OS-miR-300 (with pmR-ZsGreen1-miR-300 vector), MG63-control, or MG63-miR-300 stable cell lines, then monitored the in vivo tumorigenesis for 30 days by measuring tumor volumes at 5-day intervals. Compared with mice injected with U2OS-control or MG63-control, mice injected with pmR-ZsGreen1-miR-300 vector showed significantly slower tumor growths and decreased tumor volumes (Figure S5). These results further demonstrated that miR-300 expression was important for osteosarcoma cell growth, and that increasing miR-300 expression may represent a promising strategy for osteosarcoma cancer therapy.
Figure 5.
miR-300 Directly Targeted CUL4B
(A) Schematic representation of the 3′ UTR of CUL4B shows a putative miR-300 binding site. The seed location for WT 3′ UTR of CUL4B is indicated in red, whereas the mutant 3′ UTR is indicated in blue. (B) The expression of miR-300 was decreased in osteosarcoma cell lines. The hFOB1.19, U2OS, Saos-2, MG63, and HOS cells were subjected to qRT-PCR to determine the miR-300 level. RNU6B was selected as an internal control for normalization, and the resulting ratio in hFOB1.19 cells was arbitrarily defined as 1-fold. (C) Detection of the overexpression of miR-300 in hFOB1.19, U2OS, and MG63 cells. The miR-300-mimic or its negative control miR-NC was transfected into hFOB1.19, U2OS, and MG63 cells for 24 hr. Expression of miR-300 was determined by qRT-PCR. (D and E) Overexpression of miR-300 caused the downregulation of CUL4B at both the mRNA and protein levels. Cells used in (C) were used to determine the expression of CUL4B by qRT-PCR (D) and western blot (E). (F) The CUL4B 3′ UTR mutant failed to bind miR-300. The miR-300-mimic or miR-NC was co-transfected with pMIR-CUL4B-3′-UTR-WT or pMIR-CUL4B-3′-UTR mutant into U2OS cells for 24 hr. Luciferase activity was quantified using the dual-luciferase assay reporter system by normalizing to Renilla activity. (G) Overexpression of miR-300 inhibited osteosarcoma cell proliferation. Cells used in (C) were subject to the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-2H-tetrazolium bromide (MTT) assay to evaluate the degree of cell proliferation, and cell viability was determined at 490 nm. (H) Overexpression of miR-300 inhibited osteosarcoma cell colony formation ability. Cells used in (C) were seeded onto six-well plates with a density of 1 × 103 cells/well and cultured with 0.1 mL of DMEM medium for 2 weeks. Then, the cells were stained with 0.5% crystal violet, and the number of colonies was counted. (I) Overexpression of miR-300 inhibited osteosarcoma cell invasion. Cells used in (C) were used to determine invasion ability with Boyden chamber assays. *p < 0.05; **p < 0.001.
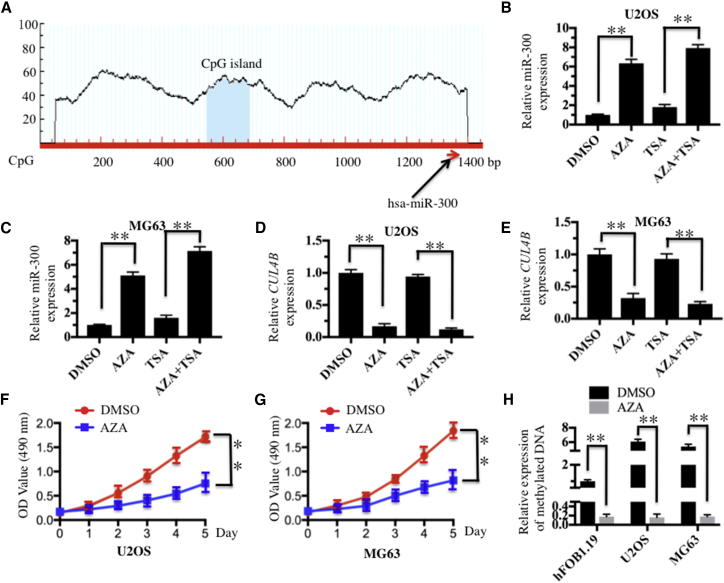
DNA Hypermethylation Was Responsible for the Downregulation of miR-300 in Osteosarcoma Cells
Given that miR-300 is a tumor suppressor miRNA in osteosarcoma cells, we next sought to reveal the mechanism of miR-300 downregulation. Several studies have revealed that DNA hypermethylation is a general mechanism for downregulating miRNA expression. Thus, we first tried to identify whether CpG islands existed upstream (−1,500) of the miR-300 genome locus. Fortunately, we found a CpG island that was located upstream (−800) of the miR-300 genomic locus (Figure 6A), which implied that DNA methylation might affect miR-300 expression. To verify this hypothesis, we treated U2OS and MG63 cells with the DNA methylation inhibitor 5-AZA-2′-deoxycytidine (AZA), the acetylation inhibitor trichostatin A (TSA), or with a combination of AZA and TSA; we then evaluated miR-300 expression by qRT-PCR analysis. We observed 6-fold or 8-fold induction of miR-300 expression in both U2OS and MG63 cells following AZA treatment or AZA + TSA treatment, respectively, but no obvious increase was observed after TSA treatment alone (Figures 6B and 6C). In contrast with miR-300 expression, the expression of CUL4B was significantly inhibited after treatment with AZA or AZA + TSA, although TSA alone did not affect the expression of CUL4B (Figures 6D and 6E). Importantly, AZA treatment could markedly inhibit U2OS and MG63 cell proliferation (Figures 6F and 6G). These results suggested that miR-300 expression in osteosarcoma cells might be regulated by DNA methylation. To confirm this hypothesis, we detected the DNA methylation status of this CpG island in hFOB1.19, U2OS, and MG63 cells with or without AZA treatment. Our results revealed that this CpG island was significantly methylated in U2OS and MG63 cells (a 6-fold induction) compared with hFOB1.19 cells without AZA treatment (Figure 6H). However, the DNA methylation level was dramatically decreased in all three cell lines with AZA treatment, but there were no obvious differences among them (Figure 6H). These results indicated that enhanced methylation of the CpG island located upstream of the miR-300 genomic locus correlated with the decrease of miR-300 expression in osteosarcoma cells, suggesting that the downregulation of miR-300 may be regulated, at least in part, by DNA methylation in osteosarcoma cells.
Figure 6.
DNA Hypermethylation Caused the Downregulation of miR-300 in Osteosarcoma Cells
(A) The promoter region of miR-300 contained one CpG island. CpG island prediction was performed using a database (http://www.urogene.org). The predicted CpG island and the miR-300 locus are indicated. (B and C) Effects of AZA and TSA on miR-300 expression in the U2OS and MG63 cell lines. U2OS (B) and MG63 (C) cells were primarily treated with DMSO, AZA (1 μM), or TSA (300 nM), followed by determination of miR-300 expression via qRT-PCR. (D and E) Effects of AZA and TSA on CUL4B expression in (D) U2OS and (E) MG63 cells. The cells used in (A) and (B) were used to determine the expression of CUL4B by qRT-PCR. (F and G) AZA treatment caused the inhibition of osteosarcoma cell proliferation. U2OS (F) and MG63 (G) cells were treated with DMSO or AZA (1 μM), followed by the determination of cell viability at 490 nm. (H) Expression levels of the methylated CpG island with or without AZA treatment. The hFOB1.19, U2OS, and MG63 cell lines were treated with or without 1 μM AZA; then the expression levels of the methylated CpG island were determined by qMSP analysis. *p < 0.05; **p < 0.001.
Ectopic Expression of miR-300 or AZA Treatment Suppressed PTEN Ubiquitination
Given that both ectopic expression of miR-300 and AZA treatment could inhibit CUL4B mRNA and protein levels, we next sought to determine whether these two treatments could affect the stability of CRL4BDCAF13 E3 ligase and PTEN ubiquitination. We primarily detected protein levels of CRL4BDCAF13 members in cells transfected with miR-300-mimic and in cells treated with AZA. CUL4B, DDB1, and DACF13 protein levels were significantly downregulated after these two treatments (Figures S6A and S6B), while the PTEN level was upregulated in these two conditions (Figures S6A and S6B). These results implied that ectopic expression of miR-300 and AZA treatment could decrease the stability of CRL4BDCAF13 E3 ligase, thereby attenuating the ubiquitination of PTEN. To validate this hypothesis, we transfected the following combinations of plasmids, including pCDNA3-Flag-PTEN + pCDNA3-HA-Ubiquitin + miR-NC, pCDNA3-Flag-PTEN + pCDNA3-HA-Ubiquitin + miR-300-mimic, pCDNA3-Flag-PTEN + miR-300-mimic, or pCDNA3-Flag-PTEN + miR-NC, into U2OS cells, followed by an in vivo ubiquitination analysis. Our results indicated that the PTEN ubiquitination level significantly decreased after transfection with miR-300-mimic or AZA treatment (Figures S6C and S6D).
TSC01131 Specifically Inhibited the CUL4B-DDB1 Interaction and Osteosarcoma Cell Growth
The identification of small molecules that disrupt protein-protein interaction by high-throughput screening (HTS) is a starting point for the development of novel therapeutic agents. Thus, we were interested in the identification of small molecules that specifically target the CUL4B-DDB1 interaction. To this end, we set up an in vitro HTS method in yeast cells to obtain small molecules that inhibited the growth of cells expressing DDB1 and CUL4B. Accordingly, equal volumes of AH109 yeast cells expressing pGADT7-CUL4B and pGBKT7-DDB1 were grown in 96-well plates supplemented with the individual compound in each well. The candidate compounds were selected based on cell density after 18 hr of incubation (Figure S7A). We identified a compound known as TSC01131 that showed strong cytotoxic activity against both U2OS and MG63 cells with an inhibitory concentration (IC50) growth of nearly 20 μM (Figure S7B), although much weaker cytotoxic activity was observed against hFOB1.19 cells (Figure S7B). Interestingly, this compound, also known as hyppospongide B, was a scalarane sesterterpenoid isolated from the sponge species Hippospongia, and it has been reported to show weak anticancer activity against human colon, breast, and leukemia cancer cells.43
To validate whether TSC01131 is able to specifically inhibit the CUL4B-DDB1 interaction, we treated hFOB1.19, U2OS, and MG63 cells with or without this compound. Then, the protein levels of CRL4BDCAF13 E3 ligase members were determined, and our results indicated that CUL4B, DDB1, and DCAF13 were significantly downregulated after TSC01131 treatment (Figure S7C). However, the PTEN protein levels were significantly upregulated (Figure S7C). These results were similar to those obtained from ectopic expression of miR-300 and AZA treatment (Figures S6A and S6B), implying that TSC01131 could also inhibit PTEN ubiquitination. To confirm this hypothesis, we transfected the following combinations of plasmids, including pCDNA3-Flag-PTEN + pCDNA3-HA-Ubiquitin, or pCDNA3-Flag-PTEN only. After 24 hr of incubation at 37°C, we treated cells with or without TSC01131, followed by in vivo ubiquitination analysis. Our results indicated that the PTEN ubiquitination level significantly decreased in a dose-dependent manner after TSC01131 treatment (Figure S7D). In addition, we also performed the in vitro ubiquitination assay to evaluate the inhibitory role of TSC01131. Our results indicated that TSC01131 significantly inhibited the ubiquitination of PTEN in vitro (Figure S8). These results present a new possibility for the development of a therapeutic agent targeting osteosarcoma cells by disrupting the CRL4B complex.
Discussion
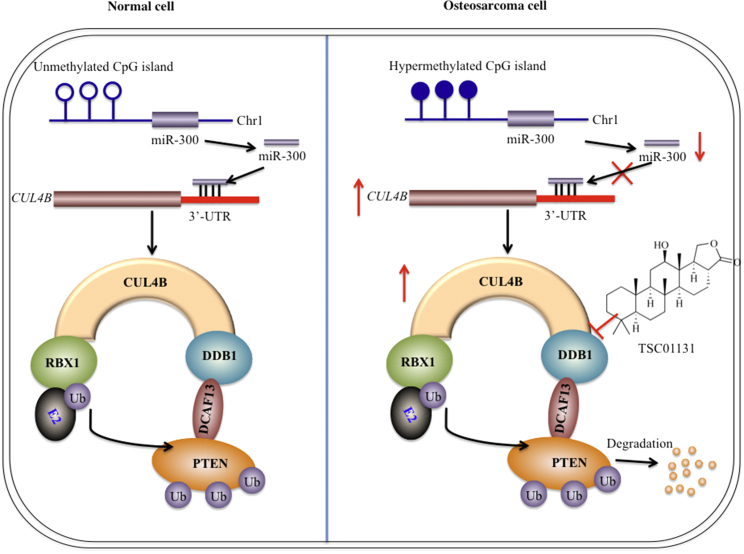
Cullin-based E3 ligases have been shown to play critical roles in tumorigenesis and to contribute to diverse biological processes in human cancers.18 CUL4A and CUL4B, two paralogs in the human genome, share high sequence similarity. The CUL4A-based E3 ligases have been reported to ubiquitinate a number of different substrates, including DDB2, p12, CDT1, p21, and p27.21, 22, 23, 24, 25 However, to date, the role of CUL4B in cancer, the substrates of CRL4B E3 ligases, and the underlying mechanism of CUL4B overexpression are far from being fully elucidated. In this study, we discovered that CUL4B formed an E3 ligase complex with DDB1, RBX1 and DCAF13, and the CRL4BDCAF13 E3 ligase recognized PTEN, a tumor suppressor, as a substrate for ubiquitination. Moreover, we also found that miR-300, an miRNA that directly targets the 3′ UTR of CUL4B, was significantly downregulated via a DNA hypermethylation mechanism in its promoter region in human osteosarcoma cells. The downregulation of miR-300 attenuated its transcriptional inhibition of CUL4B, thereby resulting in the overexpression of CUL4B. Either ectopic expression of miR-300 or treatment with the DNA methylation inhibitor AZA was capable of reducing PTEN ubiquitination, eventually resulting in the accumulation of PTEN. Importantly, we also identified a small molecule, TSC01131, which could directly abolish the CUL4B-DDB1 interaction, leading to reduced stability of CRL4BDCAF13 E3 ligase. Taken together, our results identified a new E3 ligase that recognized PTEN as a specific substrate in human osteosarcoma cells, and we also revealed the underlying mechanism for CUL4B overexpression and identified a small molecule that specifically targets the CRL4BDCAF13 E3 ligase (Figure 7). These results explain the mechanism of CUL4B in osteosarcoma and provide potential new therapeutic targets for future osteosarcoma treatment.
Figure 7.
Schematic Diagram of CRL4BDCAF13 E3 Ligase in Human Osteosarcoma Cells
In normal cells (left), the CpG island located in the promoter region of miR-300 is not subjected to methylation or is in a low methylation state; therefore, it does not inhibit the expression of miR-300. The normal expression of miR-300 further regulates CUL4B expression at the transcriptional level. In osteosarcoma cells (right), the CpG island is hypermethylated, resulting in downregulation of miR-300 expression, which weakens its inhibitory effect on CUL4B expression at the transcriptional level. This results in a significant increase in the expression of CUL4B, thereby increasing CRL4BDCAF13 activity and causing the degradation of PTEN, eventually resulting in a decrease in PTEN protein. The small molecule compound TSC01131 specifically inhibits the CUL4B-DDB1 interaction, which affects the stability or enzymatic activity of CRL4BDCAF13 ubiquitin ligase, thereby inhibiting the growth of osteosarcoma cells.
Although the amino acid sequences of CUL4A and CUL4B share 82% identity, the existing studies have shown that these two proteins do not have significant functional redundancy.18 Elevated expression of CUL4A has been observed in a variety of cancer cells, such as breast cancer,15 ovarian cancer,44 liver cancer,45 and medulloblastoma.46 CRL4A-based E3 ubiquitin ligase can recognize a series of substrate proteins, such as DDB2, XPC, UNG2, and SMUG1, as well as histones H2A, H3, and H4, which are widely involved in DNA damage and repair process.18 In addition, CRL4ACTD2 E3 ligase can recognize a number of cell-cycle regulatory proteins, such as CTD1, p21, and Chk1, as substrates to regulate cancer cell-cycle progression.18 Interestingly, CRL4A-based E3 ligases are also involved in the p53 and Wnt signaling pathways.18 However, the function of CUL4B in cancer remains unclear. Previous studies mainly focused on its function in cell development; for instance, CUL4B mutations lead to defects in nervous system and cardiac development.47, 48 In recent years, studies have also shown that CUL4B plays an important role in the process of tumorigenesis, and high expression of CUL4B has been reported in many cancers. For example, in esophageal cancer cells, CRL4B-based E3 ubiquitin ligase can ubiquitinate histone H2A and can coordinate with multi-comb inhibitory complex PRC2 to promote tumor cell proliferation and invasion.28 Silencing CUL4B in HeLa cells can lead to cyclin E aggregation and can cause cell-cycle arrest in S phase, thereby inhibiting cell proliferation.49 In prognosis studies of colorectal cancer patients, high expression of CUL4B was associated with a poor prognosis,50 but the molecular mechanism was unclear. Moreover, our previous results also showed that CUL4B is upregulated in human osteosarcoma cells,17 and its silencing could effectively inhibit osteosarcoma cell proliferation and induce apoptosis. In the current investigation, we identified the individual components of CRL4BDCAF13 E3 ligase through Y2H screening and LC-MS/MS methods, and we also identified the specific target of this E3 ligase: PTEN. By carefully analyzing the ubiquitination pattern of PTEN, we found that PTEN is modified by polyubiquitination through our in vitro and in vivo studies, indicating that more than one lysine (K) site is modified. Through amino acid sequence analysis, we found that PTEN contains a total of 33 K sites; however, these K sites are distributed everywhere, making it difficult to find the modified sites in our case. Some studies have noted that the C terminus of PTEN plays a crucial role in antagonizing NEDD4-1-mediated ubiquitination, and some K sites, such as K13 and K289, were reportedly modified in 293 cells.40 However, we cannot conclude which sites were modified in our system. We are currently mapping the modified sites through point mutagenesis.
In recent years, many studies have found that a series of miRNAs, such as miR-17-5p,51 miR-26a,52 miR-34a,53 miR-133b,54 miR-150,55 and miR-370,56 exhibit abnormal expression in osteosarcoma cells. In the investigation into the CUL4B overexpression mechanism, we found that 9 out of 20 miRNAs that were predicted to target the 3′ UTR of CUL4B showed downregulation in osteosarcoma cells. However, we focused our studies on the function of miR-300, the most downregulated miRNA. In addition, there were 86 miRNAs, the expression of which we did not detect in osteosarcoma cells. Therefore, we cannot conclude that miR-300 alone regulates the expression of CUL4B. However, there is no doubt that miR-300 plays a very important role in the regulation of miR-300 expression. For the mechanism of miRNA downregulation, DNA methylation and histone modification are the two major causes that inhibit miRNA expression. In this study, we primarily detected the effects of the DNA methylation inhibitor AZA and the acetylation inhibitor TSA on miR-300 expression. AZA treatment, but not TSA treatment, significantly inhibited miR-300 expression, suggesting the major role of DNA methylation is regulating miR-300 expression in osteosarcoma cells. Interestingly, both ectopic expression of miR-300 and AZA treatment showed similar effects on CRL4BDCAF13 E3 ligase stability and PTEN ubiquitination patterns, as well as on cell proliferation. These consistent results suggest that DNA hypermethylation of the CpG island upstream of the miR-300 genomic locus is the primary reason for tumorigenesis in human osteosarcoma cells.
Because of the important role of the DDB1-CUL4B interaction for CUL4B-based E3 ligases, disruption of the DDB1-CUL4B interaction could be an effective approach to treat cancer. Therefore, we developed an in vitro HTS assay based on yeast cell growth inhibition to identify compounds that could disrupt binding between DDB1 and CUL4B. Using a combination of a cell proliferation assay and in vivo ubiquitination analysis in osteosarcoma cells, we validated TSC01131 as an inhibitor of the DDB1-CUL4B complex. Owing to the much weaker cytotoxic activity against hFOB1.19 cells than U2OS and MG63 cells, TSC01131 may be a promising candidate for targeting CUL4B-based E3 ligases in the therapy of different cancers, not only in human osteosarcoma. In addition, we also obtained 21 other small molecules that also greatly inhibited yeast cell growth (data not shown). Future efforts will be focused on revealing the underlying mechanisms of these compounds.
In summary, this study uncovered three major findings: (1) we identified the individual members of the CRL4BDCAF13 E3 ligase, which could ubiquitinate the tumor suppressor PTEN in vitro and in vivo; (2) we revealed the mechanism of CUL4B overexpression, in which DNA hypermethylation attenuated the transcriptional inhibition of CUL4B by miR-300, leading to its upregulation; and (3) we obtained a small molecule, TSC01131, which could specifically disrupt the DDB1-CUL4B interaction and inhibit osteosarcoma cell proliferation. These results not only provide insight into how CUL4B specifically functions in osteosarcoma cells, but also identify new molecular targets and specific compounds for osteosarcoma therapy.
Materials and Methods
Cell Culture and Transfection
The human osteoblast cell line hFOB1.19 and human osteosarcoma cell lines U2OS, Saos-2, MG63, and HOS were obtained from the American Type Culture Collection (ATCC, USA). Cells were cultured in DMEM medium (GIBCO, USA) and supplemented with 10% heat-inactivated fetal bovine serum (FBS) and 100 U mL−1 penicillin-streptomycin (Thermo Fisher Scientific, USA), at 37°C with 5% CO2.
Plasmids were transfected into cultured cells with Lipofectamine 2000 reagent (Invitrogen, USA) strictly according to the manufacturer’s instructions. Then, the cells were incubated at 37°C for 48 hr and subjected to the required experiments. Specific knockdown of CUL4B, DDB1, or DCAF13 by shRNAs was carried out using a lentiviral system. In brief, cells were transfected with negative control shRNA, shCUL4B (TRCN0000006532; Sigma, USA), shDDB1 (TRCN0000082856; Sigma, USA), or shDCAF13 (TRCN0000138792; Sigma, USA) following the manufacturer’s instructions. Cells were then incubated at 37°C for 24 hr, followed by selection with puromycin (1 μg/mL) at 37°C for 48 hr before use in experiments.
Osteosarcoma Clinical Samples
Surgically dissected tumor tissues and their adjacent non-tumor tissues were obtained from 48 patients who underwent surgery and therapy from June 2013 to March 2015 at the Department of Orthopedics, Renji Hospital, School of Medicine, Shanghai Jiao Tong University, Shanghai, China. The clinical samples were immediately stored at −80°C until subjection to experiments. All patients were diagnosed with osteosarcoma under different MSTS stages dependent on histopathological features, which are summarized in Table S3. The clinical samples were acquired with written informed consent from all of the participants following protocols approved by the ethical board of Shanghai Jiao Tong University. All experimental procedures used in this study were carried out in accordance with the approved guidelines of the Ethics Committee of Shanghai Jiao Tong University.
Y2H Assay
For Y2H screening, cDNAs from U2OS cells were cloned into the pACT2 vector to generate an osteosarcoma-specific cDNA library. Plasmids from this cDNA library and the bait protein vector (pGBKT7-DDB1) were co-transformed into AH109 yeast cells. The transformed cells were selected on synthetic complete medium without Trp, Leu, and His amino acids (SC-T/L/H). DNA from the positive clones was amplified by PCR using specific primers for pACT2 (forward: 5′-CTATCTATTCGATGATGAAG-3′; reverse: 5′-ACAGTTGAAGTGAACTTGCG-3′). The inserted fragments were purified and sequenced; then the obtained sequences were blasted in the NCBI database.
To determine the interactions between DDB1 and DCAF13 and DCAF13 and PTEN in yeast, the full coding sequence of DDB1 or PTEN was fused into the pGBKT7 vector, while the full-length sequence of DCAF13, DCAF1, or p53 was cloned into the pGADT7 vector. The following plasmid combinations, including pGBKT7-DDB1 + pGADT7-DCAF13, pGBKT7-DDB1 + pGADT7, pGADT7-DCAF13 + pGBKT7, pGBKT7-DDB1 + pGADT7-p53, pGADT7-p53 + pGBKT7, pGBKT7-PTEN + pGADT7-DCAF13, pGBKT7-PTEN + pGADT7, pGADT7-DCAF13 + pGBKT7, pGBKT7-PTEN + pGADT7-DCAF1, pGADT7-DCAF1 + pGBKT7, were co-transformed into AH109 yeast cells. Transformed yeast cells were sprayed onto synthetic dropout medium (lacking Trp and Leu). The positive colonies were then used to determine interactions on SC-T/L/H medium.
Western Blot Analysis
Cells were lysed in Pierce radioimmunoprecipitation assay (RIPA) buffer (Thermo Scientific, USA) supplemented with 50 mM NaF, 1 mM Na3VO4, 1× complete protease cocktail inhibitor (Roche, USA), and 0.2% SDS. The whole-cell lysates were sonicated for 1 min at high intensity and then denatured at 95°C for 5 min. Equal amounts of proteins were loaded onto an SDS-PAGE gel for separation. The specific primary antibodies used for immunoblot were anti-GAPDH (mouse; Sigma, USA), anti-CUL4B (mouse; Sigma, USA), anti-DDB1 (rabbit; Sigma, USA), anti-Flag (mouse; Sigma, USA), anti-HA (mouse; Sigma, USA), anti-DCAF13 (rabbit; Sigma, USA), and anti-PTEN (mouse; Abcam, USA). The western blot signals were recorded using a ChemiDoc MP instrument (Bio-Rad, USA). The images represent three independent replicates.
Co-IP Assay
Cells transfected with pCDNA3-DDB1-HA + pCDNA3-p53-Flag, pCDNA3-DDB1-HA + pCDNA3-Flag, pCDNA3-DDB1-HA + pCDNA3-DCAF13-Flag, pCDNA3-HA + pCDNA3-p53-Flag, pCDNA3-HA + pCDNA3-DCAF13-Flag, pCDNA3-DCAF1-Flag + pCDNA3-PTEN-HA, pCDNA3-DCAF1-Flag + pCDNA3-HA, pCDNA3-DCAF13-Flag + pCDNA3-PTEN-HA, or pCDNA3-DCAF13-Flag + pCDNA3-HA were incubated at 37°C for 48 hr. Cells were then collected, lysed in 1 mL of RIPA buffer (Thermo Scientific, USA) supplemented with 50 mM NaF, 1 mM Na3VO4, and 1× complete protease cocktail inhibitor (Roche, USA), and centrifuged at 12,000 × g for 10 min at 4°C. Of the 1 mL supernatant, 50 μL was reserved as input, and the remaining volume was split into two equal parts. One part was incubated with anti-Flag-Agarose (Sigma, USA), and the other was incubated with anti-HA-Agarose (Sigma, USA) for 12 hr at 4°C. The beads were then washed five times with 1× PBS (pH 7.5). The immunoprecipitated proteins were analyzed by immunoblotting analysis.
Mass Spectrometry Analysis
Cells transfected with pCDNA3-Flag-HA-DCAF13 or pCDNA3-Flag-HA plasmid were lysed with Pierce IP lysis buffer (Thermo Fisher Scientific, USA) supplemented with 1× protease inhibitors (Roche, USA). Cell lysates were subjected to the IP procedure primarily using anti-Flag-Agarose (Sigma, USA) at 4°C for 4 hr. Then, the beads were washed five times with lysis buffer, followed by elution with Flag peptide (Sigma, USA) at room temperature for 2 hr. The resulting proteins were further bound with anti-HA-Agarose (Sigma, USA) at 4°C for another 4 hr. HA-tagged DCAF13 protein was loaded onto an SDS-PAGE gel for separation and then stained with Coomassie Brilliant Blue R 250, as previously described.57 The stained proteins were cut into small pieces, digested with trypsin, and analyzed by LC-MS/MS. The MS/MS spectra were blasted in the NCBI database using a MASCOT search engine (V.2.3).
Gene Expression Analysis by qRT-PCR
For CUL4B expression detection, mRNAs were extracted with TRIzol reagent (Invitrogen, USA) following the manufacturer’s guidelines, followed by removal of contaminating DNA with DNase I. A total of 1 μg of mRNA was used as the template to generate the first-strand cDNA with the M-MLV reverse transcriptase (Promega, USA). The resulting cDNAs were diluted 50-fold, and qRT-PCR analysis was performed on the Bio-Rad CFX96 real-time PCR system (Bio-Rad, USA) using the specific primers listed in Table S4. The PCR procedures consisted of 95°C for 3 min, followed by 45 cycles of 95°C for 15 s and 60°C for 30 s. The expression of CUL4B was normalized against β–Actin (ACTB) based on the 2−ΔCt method.58
For the quantitative analysis of miRNAs, total RNAs from the frozen clinical samples and cultured cells were extracted with the miRNeasy Mini Kit (QIAGEN, USA) following the manufacturer’s instructions. A total of 1 μg of RNA was used as the template to generate cDNAs using the TaqMan MicroRNA Reverse Transcription kit (Thermo Fisher Scientific, USA). The resulting cDNAs were used to determine the expression of individual miRNAs with the corresponding TaqMan MicroRNA Assay kit (Thermo Fisher Scientific, USA). The qRT-PCR program was the same as that described above. RNU6B was chosen as an internal control to normalize miRNA expression using the 2−ΔCt method.59 The data represent three independent experiments.
Cell Proliferation and Colony Formation Assays
Cells transfected with miR-300-mimic, anti-miR-300 or miR-NC, cells treated with or without AZA, and cells treated with or without TSC01131 were plated onto 96-well plates for 0, 24, 48, 72, 96, or 120 hr. Then, cell proliferation assays were analyzed with an MTT kit (Roche, USA) following the manufacturer’s protocol, and cell viability was determined at 490 nm. All experiments were performed in quadruplicate.
For the colony formation assay, cells transfected with miR-300-mimic, anti-miR-300, or miR-NC were diluted and seeded onto six-well plates with a density of approximately 1,000 cells/well. Cells were then incubated at 37°C for 14 days with a change of fresh medium every 3 days. Cell colonies were then fixed with 3.7% paraformaldehyde (PFA) for 5 min at room temperature, stained with 0.05% crystal violet for 30 min, and washed five times with double-distilled water (ddH2O) to remove excess dye. The plates were photographed, and the colony numbers were counted by a gel documentation system (Bio-Rad, USA).
Generation of miR-300 Overexpression Stable Cell Lines and In Vivo Tumor Formation Assay
The miR-300 overexpressing vector (pmR-ZsGreen1-miR-300) was constructed into pmR-ZsGreen1 (Clontech, CA, USA) with the following primers: forward: 5′-CGGAATTCGGCTATGGAAGGAATAGATGTGTG-3′, reverse: 5′-CGGGATCCGGCCAAGGTAGGCCCTTTTTG-3′ (501 bp). The resulting plasmid was transfected into U2OS or MG63 cells, and the pmR-ZsGreen1 empty vector was also transfected into these two cell lines to use as a negative control. The transfected cells were then selected with G418 for 2 weeks; then the stable clones overexpressing miR-300 were collected.
For in vivo tumor formation assay, a total of 1 × 106 cells from each stable cell line were injected into the posterior flank of a 6-week-old male nude mouse (Shanghai SLAC Laboratory Animal, Shanghai, China). Twelve mice were used for each cell line, and tumor formation was monitored over 30 days. Tumor volume was measured with fine calipers at 5-day intervals and calculated with the formula: Volume = (length × width2)/2.
In Vitro and In Vivo Ubiquitination Analysis
For the in vitro PTEN ubiquitination assay, immunoprecipitated CUL4B-Flag was washed with Pierce IP lysis buffer (Thermo Fisher Scientific, USA). The His-tagged PTEN recombinant protein was purified with Ni-NTA magnetic agarose beads (Clontech, USA) according to the manufacturer’s protocol. Ubiquitination reactions were carried out with 0.1 μM E1 (Sigma, USA), 0.4 μM E2 (Sigma, USA), 10 μg CRL4B complex (E3), 20 μg His-PTEN (substrate), and 10 μg bovine ubiquitin (Sigma, USA) in 100 μL of ubiquitination buffer supplemented with 2 mM ATP and 0.6 mM DTT at 37°C for 90 min, followed by the termination of reactions by adding 2× SDS-PAGE loading buffer and boiling at 95°C for 10 min. The ubiquitination pattern was determined by immunoblot with the anti-PTEN antibody.
For the in vivo ubiquitination assay, cells co-expressing Flag-tagged PTEN and HA-ubiquitin were grown at 37°C for 48 hr. The cells were then lysed with buffer containing 2% SDS, 150 mM NaCl, and 10 mM Tris-HCl (pH 8.0). Cell lysates were incubated with anti-Flag-Agarose at 4°C for 2 hr. Then, the beads were washed five times with lysis buffer, followed by boiling in 2× SDS-PAGE loading buffer. The ubiquitination of PTEN was analyzed by immunoblotting with an anti-HA antibody.
Quantitative Methylation-Specific PCR
Potential CpG island in the promoter region of the miR-300 genomic locus was identified using an online database (http://www.urogene.org), and one CpG island was found. The specific primers used for quantitative methylation-specific PCR (qMSP) detection of DNA methylation of this CpG island were designed using Methyl Primer Express v1.0 (Thermo Fisher Scientific, USA) and are listed in Table S5. In brief, genomic DNAs from hFOB1.19, U2OS, and MG63 cells treated with or without AZA were modified with sodium bisulfite using the EpiTect Bisulfite Kit (QIAGEN, USA) following the manufacturer’s instructions. The resulting DNAs were subjected to qRT-PCR analysis on the Bio-Rad CFX96 real-time PCR system (Bio-Rad, USA) using KAPA SYBR FAST qPCR kits (Kapa Biosystems, USA). The methylated DNA was normalized against GAPDH using the 2−ΔCt method. All reactions were conducted in triplicate.
Small Molecule Screening
We constructed our own small molecule compound library containing 40,000 terpenoids sourced from plants and sponges. The compounds in this library were all obtained from natural sources with an average mass of 300 Da, with a range of 200–500 Da, and showed high chemical structural diversity. Individual compounds were arranged in 96-well plates as 5 mM solutions in DMSO, which were further diluted 1:5 with DMSO to 1 mM working stocks. AH109 yeast cells expressing pGADT7-CUL4B and pGBKT7-DDB1 were grown in liquid SC-T/H/L medium to mid-log phase (OD600 = 1.0). Then, the cells were diluted 1:10 (OD600 = 0.1) with fresh SC-T/H/L medium, and equal volumes of cells (49 μL) were transferred into individual wells of 96-well plates. Specifically, 1.0 μL of each compound was then independently added into each well at a final concentration at 20 μM. The 96-well plates were sealed with Parafilm to prevent evaporation and then incubated in a 30°C incubator for 18 hr. The cell density in each well was measured at 600 nm with the Synergy HTX Multi-Mode Reader (BioTek, USA). Compounds that significantly inhibited cell growth (OD600 < 0.2) were selected for secondary screening, using the same procedures as in the primary screening.
Statistical Analysis
All experiments were independently replicated at least three times. Statistical analyses of data were performed using a two-sided Student’s t test. Significance was set at p < 0.05.
Author Contributions
C.Z. and H.S. designed the research. Z.C., W.Z., and K.J. performed the major parts of the experiments. B.C., K.W., L.L., C.H., and F.W. performed parts of the research. Z.C., W.Z., and K.J. analyzed the data, tested statistics, and coordinated the figures. C.Z. and H.S. wrote the article. Z.C., W.Z., and K.J. revised the article.
Acknowledgments
This study was supported by two grants from the National Natural Science Foundation of China (81672203 and 81602354) and a grant from the Emerging Frontier Technology Joint Research Program of Shanghai Shen-Kang Hospital Development Center (SHDC12015103).
Conflicts of Interest
The authors declare no competing financial, professional, or personal interests that might have influenced the performance or presentation of the work described in this manuscript.
Footnotes
Supplemental Information includes eight figures and five tables and can be found with this article online at https://doi.org/10.1016/j.omtn.2017.12.010.
Contributor Information
Caiguo Zhang, Email: caiguo.zhang@ucdenver.edu.
Hongxing Shen, Email: shenhxgk@126.com.
Supplemental Information
References
- 1.Chen D., Dou Q.P. The ubiquitin-proteasome system as a prospective molecular target for cancer treatment and prevention. Curr. Protein Pept. Sci. 2010;11:459–470. doi: 10.2174/138920310791824057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Saez I., Vilchez D. The mechanistic links between proteasome activity, aging and age-related diseases. Curr. Genomics. 2014;15:38–51. doi: 10.2174/138920291501140306113344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mani A., Gelmann E.P. The ubiquitin-proteasome pathway and its role in cancer. J. Clin. Oncol. 2005;23:4776–4789. doi: 10.1200/JCO.2005.05.081. [DOI] [PubMed] [Google Scholar]
- 4.Metzger M.B., Pruneda J.N., Klevit R.E., Weissman A.M. RING-type E3 ligases: master manipulators of E2 ubiquitin-conjugating enzymes and ubiquitination. Biochim. Biophys Acta. 2014;1843:47–60. doi: 10.1016/j.bbamcr.2013.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pickart C.M., Eddins M.J. Ubiquitin: structures, functions, mechanisms. Biochim. Biophys. Acta. 2004;1695:55–72. doi: 10.1016/j.bbamcr.2004.09.019. [DOI] [PubMed] [Google Scholar]
- 6.Ye Y., Rape M. Building ubiquitin chains: E2 enzymes at work. Nat. Rev. Mol. Cell Biol. 2009;10:755–764. doi: 10.1038/nrm2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hou X., Zhang W., Xiao Z., Gan H., Lin X., Liao S., Han C. Mining and characterization of ubiquitin E3 ligases expressed in the mouse testis. BMC Genomics. 2012;13:495. doi: 10.1186/1471-2164-13-495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Metzger M.B., Hristova V.A., Weissman A.M. HECT and RING finger families of E3 ubiquitin ligases at a glance. J. Cell Sci. 2012;125:531–537. doi: 10.1242/jcs.091777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bulatov E., Ciulli A. Targeting Cullin-RING E3 ubiquitin ligases for drug discovery: structure, assembly and small-molecule modulation. Biochem. J. 2015;467:365–386. doi: 10.1042/BJ20141450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lipkowitz S., Weissman A.M. RINGs of good and evil: RING finger ubiquitin ligases at the crossroads of tumour suppression and oncogenesis. Nat. Rev. Cancer. 2011;11:629–643. doi: 10.1038/nrc3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhao Y., Sun Y. Cullin-RING Ligases as attractive anti-cancer targets. Curr. Pharm. Des. 2013;19:3215–3225. doi: 10.2174/13816128113199990300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sang Y., Yan F., Ren X. The role and mechanism of CRL4 E3 ubiquitin ligase in cancer and its potential therapy implications. Oncotarget. 2015;6:42590–42602. doi: 10.18632/oncotarget.6052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tu Y., Chen C., Pan J., Xu J., Zhou Z.G., Wang C.Y. The Ubiquitin Proteasome Pathway (UPP) in the regulation of cell cycle control and DNA damage repair and its implication in tumorigenesis. Int. J. Clin. Exp. Pathol. 2012;5:726–738. [PMC free article] [PubMed] [Google Scholar]
- 15.Chen L.C., Manjeshwar S., Lu Y., Moore D., Ljung B.M., Kuo W.L., Dairkee S.H., Wernick M., Collins C., Smith H.S. The human homologue for the Caenorhabditis elegans cul-4 gene is amplified and overexpressed in primary breast cancers. Cancer Res. 1998;58:3677–3683. [PubMed] [Google Scholar]
- 16.Yasui K., Arii S., Zhao C., Imoto I., Ueda M., Nagai H., Emi M., Inazawa J. TFDP1, CUL4A, and CDC16 identified as targets for amplification at 13q34 in hepatocellular carcinomas. Hepatology. 2002;35:1476–1484. doi: 10.1053/jhep.2002.33683. [DOI] [PubMed] [Google Scholar]
- 17.Chen Z., Shen B.L., Fu Q.G., Wang F., Tang Y.X., Hou C.L., Chen L. CUL4B promotes proliferation and inhibits apoptosis of human osteosarcoma cells. Oncol. Rep. 2014;32:2047–2053. doi: 10.3892/or.2014.3465. [DOI] [PubMed] [Google Scholar]
- 18.Chen Z., Sui J., Zhang F., Zhang C. Cullin family proteins and tumorigenesis: genetic association and molecular mechanisms. J. Cancer. 2015;6:233–242. doi: 10.7150/jca.11076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zimmerman E.S., Schulman B.A., Zheng N. Structural assembly of cullin-RING ubiquitin ligase complexes. Curr. Opin. Struct. Biol. 2010;20:714–721. doi: 10.1016/j.sbi.2010.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang C., Zhang F. The multifunctions of WD40 proteins in genome integrity and cell cycle progression. J Genomics. 2015;3:40–50. doi: 10.7150/jgen.11015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sugasawa K., Okuda Y., Saijo M., Nishi R., Matsuda N., Chu G., Mori T., Iwai S., Tanaka K., Tanaka K., Hanaoka F. UV-induced ubiquitylation of XPC protein mediated by UV-DDB-ubiquitin ligase complex. Cell. 2005;121:387–400. doi: 10.1016/j.cell.2005.02.035. [DOI] [PubMed] [Google Scholar]
- 22.Hall J.R., Bereman M.S., Nepomuceno A.I., Thompson E.A., Muddiman D.C., Smart R.C. C/EBPα regulates CRL4(Cdt2)-mediated degradation of p21 in response to UVB-induced DNA damage to control the G1/S checkpoint. Cell Cycle. 2014;13:3602–3610. doi: 10.4161/15384101.2014.962957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Abbas T., Sivaprasad U., Terai K., Amador V., Pagano M., Dutta A. PCNA-dependent regulation of p21 ubiquitylation and degradation via the CRL4Cdt2 ubiquitin ligase complex. Genes Dev. 2008;22:2496–2506. doi: 10.1101/gad.1676108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Higa L.A., Yang X., Zheng J., Banks D., Wu M., Ghosh P., Sun H., Zhang H. Involvement of CUL4 ubiquitin E3 ligases in regulating CDK inhibitors Dacapo/p27Kip1 and cyclin E degradation. Cell Cycle. 2006;5:71–77. doi: 10.4161/cc.5.1.2266. [DOI] [PubMed] [Google Scholar]
- 25.Higa L.A., Mihaylov I.S., Banks D.P., Zheng J., Zhang H. Radiation-mediated proteolysis of CDT1 by CUL4-ROC1 and CSN complexes constitutes a new checkpoint. Nat. Cell Biol. 2003;5:1008–1015. doi: 10.1038/ncb1061. [DOI] [PubMed] [Google Scholar]
- 26.Sertic S., Evolvi C., Tumini E., Plevani P., Muzi-Falconi M., Rotondo G. Non-canonical CRL4A/4B(CDT2) interacts with RAD18 to modulate post replication repair and cell survival. PLoS ONE. 2013;8:e60000. doi: 10.1371/journal.pone.0060000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Precious B., Childs K., Fitzpatrick-Swallow V., Goodbourn S., Randall R.E. Simian virus 5 V protein acts as an adaptor, linking DDB1 to STAT2, to facilitate the ubiquitination of STAT1. J. Virol. 2005;79:13434–13441. doi: 10.1128/JVI.79.21.13434-13441.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hu H., Yang Y., Ji Q., Zhao W., Jiang B., Liu R., Yuan J., Liu Q., Li X., Zou Y. CRL4B catalyzes H2AK119 monoubiquitination and coordinates with PRC2 to promote tumorigenesis. Cancer Cell. 2012;22:781–795. doi: 10.1016/j.ccr.2012.10.024. [DOI] [PubMed] [Google Scholar]
- 29.Abdulkareem I.H., Blair M. Phosphatase and tensin homologue deleted on chromosome 10. Niger. Med. J. 2013;54:79–86. doi: 10.4103/0300-1652.110033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Carnero A., Blanco-Aparicio C., Renner O., Link W., Leal J.F. The PTEN/PI3K/AKT signalling pathway in cancer, therapeutic implications. Curr. Cancer Drug Targets. 2008;8:187–198. doi: 10.2174/156800908784293659. [DOI] [PubMed] [Google Scholar]
- 31.Chalhoub N., Baker S.J. PTEN and the PI3-kinase pathway in cancer. Annu. Rev. Pathol. 2009;4:127–150. doi: 10.1146/annurev.pathol.4.110807.092311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Georgescu M.M. PTEN tumor suppressor network in PI3K-Akt pathway control. Genes Cancer. 2010;1:1170–1177. doi: 10.1177/1947601911407325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bermúdez Brito M., Goulielmaki E., Papakonstanti E.A. Focus on PTEN regulation. Front. Oncol. 2015;5:166. doi: 10.3389/fonc.2015.00166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tian K., Di R., Wang L. MicroRNA-23a enhances migration and invasion through PTEN in osteosarcoma. Cancer Gene Ther. 2015;22:351–359. doi: 10.1038/cgt.2015.27. [DOI] [PubMed] [Google Scholar]
- 35.Li B., Sun H. MiR-26a promotes neurite outgrowth by repressing PTEN expression. Mol. Med. Rep. 2013;8:676–680. doi: 10.3892/mmr.2013.1534. [DOI] [PubMed] [Google Scholar]
- 36.Fu X., Tian J., Zhang L., Chen Y., Hao Q. Involvement of microRNA-93, a new regulator of PTEN/Akt signaling pathway, in regulation of chemotherapeutic drug cisplatin chemosensitivity in ovarian cancer cells. FEBS Lett. 2012;586:1279–1286. doi: 10.1016/j.febslet.2012.03.006. [DOI] [PubMed] [Google Scholar]
- 37.Torres J., Pulido R. The tumor suppressor PTEN is phosphorylated by the protein kinase CK2 at its C terminus. Implications for PTEN stability to proteasome-mediated degradation. J. Biol. Chem. 2001;276:993–998. doi: 10.1074/jbc.M009134200. [DOI] [PubMed] [Google Scholar]
- 38.Jang H.D., Noh J.Y., Shin J.H., Lin J.J., Lee S.Y. PTEN regulation by the Akt/GSK-3β axis during RANKL signaling. Bone. 2013;55:126–131. doi: 10.1016/j.bone.2013.02.005. [DOI] [PubMed] [Google Scholar]
- 39.Okahara F., Ikawa H., Kanaho Y., Maehama T. Regulation of PTEN phosphorylation and stability by a tumor suppressor candidate protein. J. Biol. Chem. 2004;279:45300–45303. doi: 10.1074/jbc.C400377200. [DOI] [PubMed] [Google Scholar]
- 40.Wang X., Trotman L.C., Koppie T., Alimonti A., Chen Z., Gao Z., Wang J., Erdjument-Bromage H., Tempst P., Cordon-Cardo C. NEDD4-1 is a proto-oncogenic ubiquitin ligase for PTEN. Cell. 2007;128:129–139. doi: 10.1016/j.cell.2006.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ward E., DeSantis C., Robbins A., Kohler B., Jemal A. Childhood and adolescent cancer statistics, 2014. CA Cancer J. Clin. 2014;64:83–103. doi: 10.3322/caac.21219. [DOI] [PubMed] [Google Scholar]
- 42.Durfee R.A., Mohammed M., Luu H.H. Review of osteosarcoma and current management. Rheumatol. Ther. 2016;3:221–243. doi: 10.1007/s40744-016-0046-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang C., Liu Y. Targeting cancer with sesterterpenoids: the new potential antitumor drugs. J. Nat. Med. 2015;69:255–266. doi: 10.1007/s11418-015-0911-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ha T.Y. MicroRNAs in human diseases: from cancer to cardiovascular disease. Immune Netw. 2011;11:135–154. doi: 10.4110/in.2011.11.3.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Birner P., Schoppmann A., Schindl M., Dinhof C., Jesch B., Berghoff A.S., Schoppmann S.F. Human homologue for Caenorhabditis elegans CUL-4 protein overexpression is associated with malignant potential of epithelial ovarian tumours and poor outcome in carcinoma. J. Clin. Pathol. 2012;65:507–511. doi: 10.1136/jclinpath-2011-200463. [DOI] [PubMed] [Google Scholar]
- 46.Pan Y., Wang B., Yang X., Bai F., Xu Q., Li X., Gao L., Ma C., Liang X. CUL4A facilitates hepatocarcinogenesis by promoting cell cycle progression and epithelial-mesenchymal transition. Sci. Rep. 2015;5:17006. doi: 10.1038/srep17006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Michiels E.M., Weiss M.M., Hoovers J.M., Baak J.P., Voûte P.A., Baas F., Hermsen M.A. Genetic alterations in childhood medulloblastoma analyzed by comparative genomic hybridization. J. Pediatr. Hematol. Oncol. 2002;24:205–210. doi: 10.1097/00043426-200203000-00009. [DOI] [PubMed] [Google Scholar]
- 48.Nakagawa T., Xiong Y. X-linked mental retardation gene CUL4B targets ubiquitylation of H3K4 methyltransferase component WDR5 and regulates neuronal gene expression. Mol. Cell. 2011;43:381–391. doi: 10.1016/j.molcel.2011.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kerzendorfer C., Whibley A., Carpenter G., Outwin E., Chiang S.C., Turner G., Schwartz C., El-Khamisy S., Raymond F.L., O’Driscoll M. Mutations in Cullin 4B result in a human syndrome associated with increased camptothecin-induced topoisomerase I-dependent DNA breaks. Hum. Mol. Genet. 2010;19:1324–1334. doi: 10.1093/hmg/ddq008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xu B., Sun Z., Liu Z., Guo H., Liu Q., Jiang H., Zou Y., Gong Y., Tischfield J.A., Shao C. Replication stress induces micronuclei comprising of aggregated DNA double-strand breaks. PLoS ONE. 2011;6:e18618. doi: 10.1371/journal.pone.0018618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jiang T., Tang H.M., Wu Z.H., Chen J., Lu S., Zhou C.Z., Yan D.W., Peng Z.H. Cullin 4B is a novel prognostic marker that correlates with colon cancer progression and pathogenesis. Med. Oncol. 2013;30:534. doi: 10.1007/s12032-013-0534-7. [DOI] [PubMed] [Google Scholar]
- 52.Wang W., Zhang L., Zheng K., Zhang X. miR-17-5p promotes the growth of osteosarcoma in a BRCC2-dependent mechanism. Oncol. Rep. 2016;35:1473–1482. doi: 10.3892/or.2016.4542. [DOI] [PubMed] [Google Scholar]
- 53.Qu F., Li C.B., Yuan B.T., Qi W., Li H.L., Shen X.Z., Zhao G., Wang J.T., Liu Y.J. MicroRNA-26a induces osteosarcoma cell growth and metastasis via the Wnt/β-catenin pathway. Oncol. Lett. 2016;11:1592–1596. doi: 10.3892/ol.2015.4073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pei H., Jin Z., Chen S., Sun X., Yu J., Guo W. MiR-135b promotes proliferation and invasion of osteosarcoma cells via targeting FOXO1. Mol. Cell. Biochem. 2015;400:245–252. doi: 10.1007/s11010-014-2281-2. [DOI] [PubMed] [Google Scholar]
- 55.Qu Y., Pan S., Kang M., Dong R., Zhao J. MicroRNA-150 functions as a tumor suppressor in osteosarcoma by targeting IGF2BP1. Tumour Biol. 2016;37:5275–5284. doi: 10.1007/s13277-015-4389-8. [DOI] [PubMed] [Google Scholar]
- 56.Duan N., Hu X., Yang X., Cheng H., Zhang W. MicroRNA-370 directly targets FOXM1 to inhibit cell growth and metastasis in osteosarcoma cells. Int. J. Clin. Exp. Pathol. 2015;8:10250–10260. [PMC free article] [PubMed] [Google Scholar]
- 57.Zhou H., Lin H., Chen S., Becker K., Yang Y., Zhao J., Kudla J., Schumaker K.S., Guo Y. Inhibition of the Arabidopsis salt overly sensitive pathway by 14-3-3 proteins. Plant Cell. 2014;26:1166–1182. doi: 10.1105/tpc.113.117069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang Y., Li H., Zhang C., An X., Liu L., Stubbe J., Huang M. Conserved electron donor complex Dre2-Tah18 is required for ribonucleotide reductase metallocofactor assembly and DNA synthesis. Proc. Natl. Acad. Sci. USA. 2014;111:E1695–E1704. doi: 10.1073/pnas.1405204111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chen X., Chen X.G., Hu X., Song T., Ou X., Zhang C., Zhang W., Zhang C. MiR-34a and miR-203 inhibit survivin expression to control cell proliferation and survival in human osteosarcoma cells. J. Cancer. 2016;7:1057–1065. doi: 10.7150/jca.15061. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.