Abstract
Bronchiolitis obliterans syndrome (BOS), the main manifestation of chronic lung allograft dysfunction, leads to poor long-term survival after lung transplantation. Identifying predictors of BOS is essential to prevent the progression of dysfunction before irreversible damage occurs. By using a large set of 107 samples from lung recipients, we performed microarray gene expression profiling of whole blood to identify early biomarkers of BOS, including samples from 49 patients with stable function for at least 3 years, 32 samples collected at least 6 months before BOS diagnosis (prediction group), and 26 samples at or after BOS diagnosis (diagnosis group). An independent set from 25 lung recipients was used for validation by quantitative PCR (13 stables, 11 in the prediction group, and 8 in the diagnosis group). We identified 50 transcripts differentially expressed between stable and BOS recipients. Three genes, namely POU class 2 associating factor 1 (POU2AF1), T-cell leukemia/lymphoma protein 1A (TCL1A), and B cell lymphocyte kinase, were validated as predictive biomarkers of BOS more than 6 months before diagnosis, with areas under the curve of 0.83, 0.77, and 0.78 respectively. These genes allow stratification based on BOS risk (log-rank test p < 0.01) and are not associated with time posttransplantation. This is the first published large-scale gene expression analysis of blood after lung transplantation. The three-gene blood signature could provide clinicians with new tools to improve follow-up and adapt treatment of patients likely to develop BOS.
Keywords: lung transplantation, bronchiolitis obliterans syndrome, gene expression, biomarkers, blood
Introduction
Chronic lung allograft dysfunction is the main limitation of long-term survival after lung transplantation. It manifests mainly by abnormal remodeling of the small airways, resulting in progressive airflow obstruction called bronchiolitis obliterans syndrome (BOS) (1–3). The prevalence of BOS is around 35% at 5 years. Its late diagnosis, based on the irreversible decline of lung function, attests to irreversible and advanced degradation of the allograft. Prognosis is poor, with a 4-year median survival after disease diagnosis (4). Thus, there is an urgent need to identify the predictors of BOS, which would allow proactive and targeted strategies to slow disease progression before irreversible degradation of the allograft occurs.
Bronchiolitis obliterans syndrome is likely to arise from repeated injuries from both alloimmune and non-alloimmune mechanisms, generating fibrosis and airway obstruction (5). Tracking these inflammation and fibrotic processes has been used to identify early signs of the disease, and bronchoalveolar lavage neutrophilia, levels of regulatory T cells, chemokines/cytokines, or matrix metalloproteases have been proposed as early biomarkers of BOS (6–10). More recently, expression profiling of lung biopsies pinpointed fibrosis-associated genes for the diagnosis or prediction of BOS (11). Yet, these invasive lung-centered approaches remain hampered by the accessibility to such samples and are therefore limited for routine monitoring of lung transplant recipients (LTRs). In blood, circulating fibrocytes and cytokine concentration have been proposed as predictors of BOS (12–16). However, these studies used a limited number of patients and have yet to be confirmed by follow-up studies. Consequently, none of these attempts have demonstrated sufficient feasibility and robustness to achieve clinical acceptance and potential routine use in the future.
Large-scale gene expression profiling of peripheral blood represents a promising tool to identify transcriptomic markers associated with the natural history of an allograft (17). The technique has been successfully tested for monitoring kidney (18), heart (19), and liver (20) transplant recipients. For the first time, we used this non-invasive approach to identify blood biomarkers of BOS in a large set of samples from LTRs. By using an independent set of patients, we were able to validate a transcriptomic signature that predicts the occurrence of BOS more than 6 months before the clinical manifestations.
Materials and Methods
Patients
Lung transplant recipients were recruited from September 2009 to October 2013 within the multicentre Cohort of Lung Transplantation (COLT) cohort (NCT00980967). The local ethical committee (Comité de Protection des Personnes Ouest 1-Tours, 2009-A00036-51) approved the study, and all participants provided written informed consent. Inclusion criterion was at least 3 years of follow-up unless the diagnosis of BOS was made before (Figure S1 in Supplementary Material).
The eligible patients (n = 688) were phenotyped by a blind adjudication committee as described previously (21, 22), based on pulmonary function tests and chest imaging according to ISHLT/ERS/ATS guidelines (3, 23). Assessed pulmonary tests were performed before the transplantation, the day of the transplantation, 1 and 6 months after the transplantation and every 6 months thereafter up to 3 years posttransplantation. We excluded 265 patients because of other phenotypes or confounding factors, and 338 stable patients and 85 BOS patients were identified. Stable patients displayed no signs of chronic dysfunction for at least 3 years after lung transplantation. Stable and BOS patients were then further selected to constitute homogenous groups based on sample availability, absence of concurrent infection or acute rejection within 1 month before or after blood collection, and the quality of RNA (RNA integrity number ≥6.5). Eighty-nine patients (49 STA and 40 BOS) were included in the identification set and 25 in the validation set (13 STA and 12 BOS).
RNA Isolation
Peripheral blood samples were collected in PAXgene tubes (PreAnalytix, Qiagen, Hilden, Germany) and stored at −80°C. Total RNA was extracted using the PAXgene blood RNA system kit (Qiagen) with an on-column DNase digestion protocol according to the manufacturer’s instructions. Quantity and quality of total RNA were determined using a 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA, USA), and RNA samples with an RNA integrity number above 6.5 were selected for further analyses.
Gene Expression Microarray Analysis
Total RNA (100 ng) were labeled using the Two-Color Agilent Low Input Quick Amp Labeling Kit and hybridized on SurePrint G3 Human Gene Expression v3 8 × 60K Microarrays following the manufacturer’s instructions (Agilent Technologies). Data extraction of median feature intensity was performed with Feature Extraction software v10.7 (Agilent Technologies). To remove signal intensity bias between each array, median feature intensities were normalized with the lowess (locally weighted scatter plot smoothing) method, then spots for which half the samples exhibited a signal less than the mean of all median signals were removed. Correction between two microarray hybridization batches was performed on the 28,867 remaining spots with the Combat algorithm (24) available through the R package sva (25). These normalized microarray data were deposited in the Gene Expression Ominbus database (accession number GSE94557). Mean expression levels for the spots targeting the same genes were assessed, resulting in 16,128 unique genes. In the BOS PRED group, the closest time point from transplantation was selected in cases of several samples per patient, so that no patient duplicate was included. Genes with low variation (i.e., variance <0.2), and thus considered as invariants, were excluded, resulting in 6,581 analyzed genes. For the identification of differential genes, linear modeling with empirical Bayes statistical procedure was performed, comparing the STA group and each group of interest, using the limma package in R. Genes with p < 5% and fold change (FC) >1.5 were considered as differentially expressed. The biological significance of selected genes was assessed using GOminer software. Only gene ontology (GO) categories enriched with a false discovery rate (FDR) <10% and with at least five represented genes were selected. The cell type source of differential genes was evaluated using the web tool Enrichr (26). Relative estimation of cell abundances using gene expression data was performed through the CIBERSORT software (27), and gene set enrichment analysis (GSEA) was performed to highlight relevant biological processes using curated gene sets from the Molecular Signatures Database (1,000 permutations and FDR threshold of 25%) (28).
Quantitative PCR (qPCR) for Microarray Validation
After reverse transcription with Superscript III (Invitrogen, Carlsbad, CA, USA), real-time qPCR was performed on a Taqman StepOne plus real-time PCR system (Applied Biosystems, Foster City, CA, USA) using commercially available primers: HPRT1 (Hs99999909_m1), β2M (Hs00984230_m1), ACTB (Hs99999903_m1), CD19 (Hs99999192_m1), TCL1A (Hs00951350_m1), IGLL5 (Hs04330879_u1), POU2AF1 (Hs01573371_m1), and B cell lymphocyte kinase (BLK) (Hs01017452_m1). Samples were run in duplicate, and the geometric mean of quantification cycle values for HPRT1, β2M, and ACTB was used for normalization. Relative expression between a sample and a reference was calculated according to the 2−ΔΔCt method.
Gene Expression Data Sets
Gene expression values for the three genes of interest (BLK, POU2AF1, and TCL1A) from two public microarray data sets were collected from the Gene Expression Ominbus database: GSE38267 (29) and GSE28042 (30), corresponding to two studies analyzing blood gene expression in non-transplanted patients with terminal respiratory failure.
Statistical Analysis
Non-parametric Kruskal–Wallis tests with Dunn’s ad hoc pairwise comparisons, Mann–Whitney tests, ROC curves, log-ranked survival analyses, and Fisher’s exact test for categorical variables were performed using GraphPad Prism v. 6 (GraphPad Software, La Jolla, CA, USA). Time-to-event analysis was performed using Cox proportional analysis between gene expression and time to BOS with the R survival package (Coxph function, R software v. 3.3.2).
Results
Lung Transplant Recipients
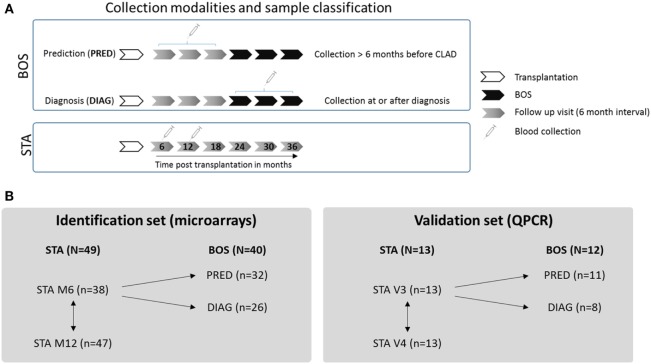
Lung transplant recipients included in this study were recruited from the multicentre COLT cohort, allowing for longitudinal follow-up and 6-month interval biocollection from transplantation. On the basis of this longitudinal follow-up, we retrospectively defined two classes of BOS samples depending on the time between blood collection and BOS diagnosis (defined as the time point with a decline of ≥20% in FEV1 from baseline; Figure 1A). Blood samples collected at least 6 months before BOS diagnosis were included in the prediction class (PRED), and blood samples collected at the time or after BOS diagnosis (up to 13 months after initial diagnosis) were included in the diagnosis class (DIAG). For the group of patients with stable graft function (STA), blood was collected 6 and 12 months after transplantation, and a comparison of these two time points was performed to exclude irrelevant genes possibly altered during this interval after transplantation (Figure 1B). No patient duplicate was included within any classes.
Figure 1.
Experimental design. (A) Collection modalities and sample classification. For bronchiolitis obliterans syndrome (BOS) patients, two classes of samples were defined depending on the time between blood collection and BOS diagnosis: a prediction class (PRED) combining blood samples collected at least 6 months before BOS diagnosis and a diagnosis class (DIAG) combining blood samples collected at or up to 13 months after BOS diagnosis. For STA patients, samples collected 6 and 12 months after LT were used. (B) Strategy for gene expression analysis. In both identification and validation sets, STA samples collected 6 months posttransplantation were compared with PRED and DIAG class samples. No patient duplicate was included in these groups. For the STA group, comparison between gene expression at 6 and 12 months posttransplantation was performed to exclude irrelevant genes modulated with time.
Clinical parameters of patients in the identification and validation sets are presented in Table 1. LTR groups were homogeneous regarding age, sex, BMI, type of transplantation, induction treatment, smoking status, and infection and rejection events. A significant difference in azithromycin exposure was observed in the identification set between the DIAG and STA and PRED (76.9 vs. 22.4 and 37.5%, p < 0.0001). Predicted FEV1 was significantly lower in the DIAG in both cohorts (p < 0.05). Time of blood collection differed between the DIAG and PRED (p < 0.001).
Table 1.
Patient characteristics.
| Identification set |
Validation set |
|||||||
|---|---|---|---|---|---|---|---|---|
| BOS (N = 40) |
BOS (N = 12) |
|||||||
| STA (N = 49) | DIAG (N = 26) | PRED (N = 32) | STA (N = 13) | DIAG (N = 8) | PRED (N = 11) | |||
| Mean age at Tx (range) | 40.2 (16–62) | 40.6 (15–65) | 43.3 (15–65) | ns | 38.8 (23–62) | 50.8 (29–63) | 49.5 (27–63) | ns |
| Sex | ns | ns | ||||||
| M (%) | 25 (51) | 14 (53.8) | 17 (53.1) | 7 (53.8) | 6 (75) | 8 (72.7) | ||
| F (%) | 24 (49) | 12 (46.2) | 15 (46.9) | 6 (46.2) | 2 (25) | 3 (27.3) | ||
| BMI | 20 | 20.5 | 21.2 | ns | 18.7 | 22.9 | 21.2 | ns |
| Pathology leading to Tx | ns | ns | ||||||
| Emphysema/COPD, n (%) | 11 (22.4) | 8 (30.8) | 9 (28.2) | 3 (23) | 3 (37.5) | 5 (45.4) | ||
| CF, n (%) | 29 (59.2) | 9 (30.6) | 10 (31.3) | 9 (69.2) | 2 (25) | 4 (36.4) | ||
| PAH, n (%) | 3 (6.1) | 2 (7.7) | 2 (6.3) | – | – | – | ||
| IPF, n (%) | 1 (2) | 2 (7.7) | 4 (12.5) | 1 (7.8) | 2 (25) | 1 (9.1) | ||
| Other, n (%) | 5 (10.2) | 5 (19.2) | 7 (21.9) | – | 1 (12.5) | 1 (9.1) | ||
| Type of Tx | ns | ns | ||||||
| Double lung, n (%) | 41 (82.5) | 19 (73.1) | 24 (75) | 11 (84.6) | 7 (87.5) | 10 (90.9) | ||
| Single lung, n (%) | 5 (12.5) | 6 (23.1) | 7 (21.9) | 2 (15.4) | – | – | ||
| Heart lung, n (%) | 2 (2.5) | 1 (3.8) | – | – | 1 (12.5) | 1 (9.1) | ||
| Lobar, n (%) | 1 (2.5) | – | 1 (3.1) | – | – | – | ||
| Induction | ns | ns | ||||||
| Basiliximab, n (%) | 17 (34.7) | 5 (19.2) | 9 (28.1) | 3 (23.1) | 1 (12.5) | 2 (18.2) | ||
| Thymoglobulin, n (%) | 17 (34.7) | 12 (46.2) | 16 (50) | 7 (53.8) | 1 (12.5) | 2 (18.2) | ||
| None, n (%) | 15 (30.6) | 9 (34.6) | 7 (21.9) | 3 (23.1) | 6 (75) | 7 (63.6) | ||
| Immunosuppressives | ||||||||
| Steroids, n (%) | 49 (100) | 26 (100) | 32 (100) | 13 (100) | 8 (100) | 11 (100) | ||
| Tacrolimus, n (%) | 38 (79.6) | 18 (69.2) | 23 (71.9) | 11 (84.6) | 3 (37.5) | 6 (54.5) | ||
| Cyclosporin, n (%) | 11 (22.4) | 8 (30.8) | 9 (28.1) | 2 (15.4) | 5 (62.5) | 5 (45.4) | ||
| MMF/MPA, n (%) | 47 (95.9) | 26 (100) | 29 (90.1) | 13 (100) | 7 (87.5) | 10 (90.1) | ||
| Azithromycin | 11 (22.4) | 20 (76.9)† | 12 (37.5) | p < 0.0001 | 6 (46.2) | 7 (87.5) | 6 (54.5) | |
| Acute cellular rejections | ns | ns | ||||||
| Ever A1 grade, n (%) | 12 (24.5) | 7 (26.9) | 2 (6.25) | 4 (30.7) | 5 (62.5) | 2 (18.2) | ||
| Ever ≥A2 grade, n (%) | 4 (8.2) | 4 (15.4) | 2 (6.25) | 1 (7.7) | 1 (12.5) | 1 (9.1) | ||
| Infectionsa | ns | |||||||
| Bacteria, n (%) | 39 (79.6) | 23 (88.5) | 28 (87.5) | 11 (84.6) | 6 (75) | 8 (72.7) | ||
| Fungi, n (%) | 28 (57.2) | 21 (80.8) | 21 (65.6) | 6 (46.2) | 4 (50) | 7 (63.6) | ||
| Virus, n (%) | 19 (38.8) | 16 (61.5) | 14 (43.8) | 3 (23) | 4 (50) | 3 (27.3) | ||
| Collection time post-Tx (days ± SD)b | 6 mo: 196 ± 31 | 642 ± 254# | 210 ± 70## | p < 0.0001 | 6 mo: 185 ± 12 | 738 ± 86# | 311 ± 39## | p < 0.0001 |
| 12 mo: 375 ± 28 | 12 mo: 364 ± 9 | |||||||
| FEV1 (% predicted) at collectionb | 6 mo: 74.5 ± 19.5 | 50.7 ± 15.5† | 72.5 ± 19.9 | p < 0.0001 | 6 mo: 74.7 ± 17.1 | 57.7 ± 18.9†† | 83.4 ± 14.6 | 0.032 |
| 12 mo: 80.6 ± 19.2 | 12 mo: 81.5 ± 18.4 | |||||||
| Smoking status | ns | ns | ||||||
| Non smoker | 30 | 12 | 17 | 9 | 3 | 5 | ||
| Weaned | 19 | 14 | 13 | 4 | 5 | 6 | ||
| Active smoker | 0 | 0 | 2 | 0 | 0 | 0 | ||
Clinical parameters of the patients included in the identification and validation sets. The p values were calculated with the Fisher’s exact test for categorical variables and Kruskal–Wallis test with Dunn’s ad hoc pairwise comparisons for continuous variables.
aNumber of patients with at least one event of infection during the follow-up.
bAt 6 months and 12 months posttransplantation.
#p < 0.05 in DIAG vs. STA 6 mo and PRED.
##p < 0.05 in PRED vs. STA 12 mo.
†p < 0.0001 in DIAG vs. PRED and vs. STA.
††p < 005 in DIAG vs. STA 12 mo.
Identification of Gene Signatures Associated With BOS
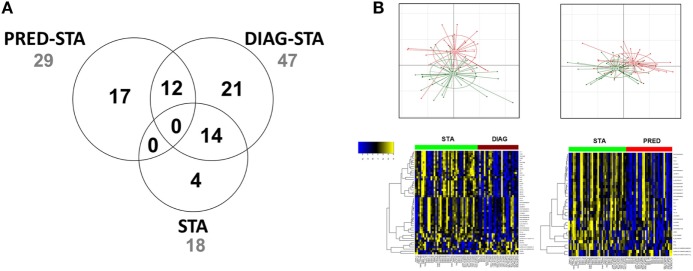
Gene expression profiling evidenced a total of 50 transcripts differentially expressed between STA and the 2 DIAG and PRED BOS groups, excluding differential transcripts between STA at the 2 time points (Figure 2; Table S1 in Supplementary Material). We identified 33 transcripts differentially expressed between STA and DIAG. While no GO term was significantly enriched, GSEA analysis highlighted 14 enriched gene sets (FDR < 25% and nominal enrichment <−2; Table S2 in Supplementary Material) among which 3 were related to B cell biology (31, 32) and one to upregulated genes in lung tissue of smokers with chronic obstructive pulmonary disease (COPD) (33).
Figure 2.
Differentially expressed genes. (A) Venn diagram of differentially expressed genes. Numbers of genes differentially expressed comparing STA vs. PRED, STA vs. DIAG, and STA at 6 and 12 months posttransplantation are indicated. (B) Principal component analysis and expression heat map (yellow and blue indicate high and low expression, respectively, and red and green indicate bronchiolitis obliterans syndrome and STA, respectively).
Twenty-nine transcripts were differentially expressed between STA and PRED. According to the GO analysis, five genes participated to the enrichment of the immune system process (GO:0002376, FDR = 0.064): CD19 (log2FCPRED/STA = −0.60), the major histocompatibility complex class II DQα1 (HLA-DQA1, log2FCPRED/STA = −0.67) and DQα2 (HLA-DQA2, log2FCPRED/STA = −0.66), POU class 2 associating factor 1 (POU2AF1, log2FCPRED/STA = −0.74), and Spi-B transcription factor (SPIB, log2FCBOS/STA = −0.6). Analysis using the Enrichr tool stressed the enrichment of genes related to CD19+ B cells including CD19, HLA-DQA1, POU2AF1, B lymphoid tyrosine kinase (BLK, log2FCPRED/STA = −0.62), and T-cell leukemia/lymphoma 1A (TCL1A, log2FCPRED/STA = −0.80). Based on unsupervised hierarchical clustering of expressed genes, these genes clustered with known B cell-related genes such as MS4A1 (membrane-spanning 4-domains, subfamily A, member 14 also called CD20 molecule), BANK1 (B cell scaffold protein with ankyrin repeats 1), and CD40, reinforcing the potential association of B cell-related genes with prediction of BOS. Estimation of memory and plasma B cells relative abundances with gene expression using CIBERSORT did not highlighted significant alteration, while naive CD4+ T cells abundance was decreased in PRED compared to STA (Figure S2 in Supplementary Material). It is noteworthy that 14 transcripts, including TCL1A, immunoglobulin lambda-like polypeptide 5 gene (IGLL5), and various immunoglobulin lambda and kappa light chain variable regions, were associated with both the DIAG and the PRED.
While these results highlight genes differentially expressed between STA and each of the two BOS groups, BOS is a time-dependent phenomenon after lung transplantation. Thus, we reanalyzed differential genes from the STA and PRED comparison using a time-dependent analysis, the Cox proportional hazards test. According to the univariate analysis, all 29 differentially expressed genes displayed a significant association with BOS occurrence with time (p value of Wald test <0.05; Table S1 in Supplementary Material).
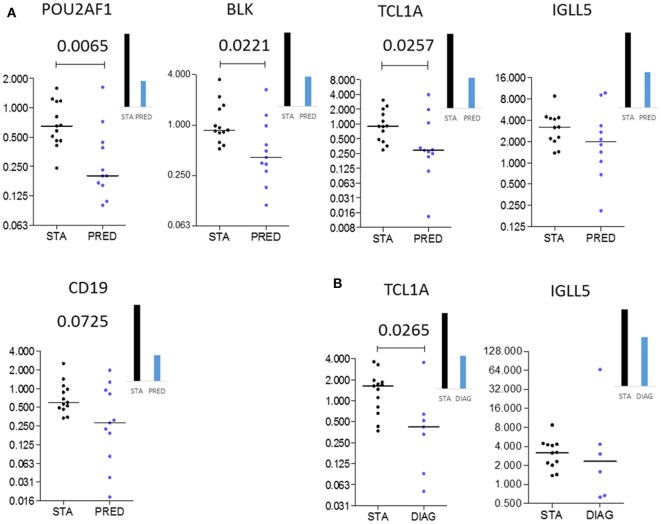
Validation of POU2AF1, TCL1A, and BLK As Predictive Biomarkers of BOS
Among the differentially expressed genes between STA and PRED, five were selected on the basis of their p values, FC magnitude, and expression level in microarrays. These five genes were measured by qPCR in an independent set of 25 patients (13 STA and 12 BOS), with 11 and 8 samples in the PRED and DIAG, respectively (Table 2). Downregulation of 3 genes, POU2AF1 (p = 0.0065), TCL1A (p = 0.0257), and BLK (p = 0.0221) in the PRED was validated by qPCR (Figure 3A). By contrast, the downregulation of CD19 and IGLL5 were not confirmed. Expression of POU2AF1, TCL1A, and BLK was constant in the STA between 6 and 12 months posttransplantation (Figure S3 in Supplementary Material) but was not significantly downregulated at the time of transplantation between STA and PRED (n = 6 and 9, respectively; Figure S4 in Supplementary Material). For diagnostic purposes, we confirmed the downregulation of TCL1A (p = 0.0265; Figure 3B).
Table 2.
Microarray-identified genes selected for quantitative PCR (qPCR) validation.
| Comparison | Symbol | Gene name | log2FC (BOS/STA) | Fold change (FC) [bronchiolitis obliterans syndrome (BOS)/STA] | p Value | Wald test |
|---|---|---|---|---|---|---|
| STA vs. PRED | CD19 | CD19 molecule | −0.60 | 0.66 | 0.003 | 0.006 |
| BLK | B lymphoid tyrosine kinase | −0.62 | 0.65 | 0.000 | 0.001 | |
| POU2AF1 | POU class 2 associating factor 1 | −0.74 | 0.60 | 0.001 | 0.003 | |
| TCL1A | T-cell leukemia/lymphoma 1A | −0.80 | 0.57 | 0.003 | 0.003 | |
| IGLL5 | Immunoglobulin lambda-like polypeptide 5 | −1.18 | 0.44 | 0.000 | 0.001 | |
| STA vs. DIAG | IGLL5 | Immunoglobulin lambda-like polypeptide 5 | −0.82 | 0.57 | 0.016 | |
| TCL1A | T-cell leukemia/lymphoma 1A | −0.79 | 0.58 | 0.006 | ||
Five genes were selected for qPCR validation based on their p values and FC magnitude. Gene name, function, p values, and FC values are presented.
Figure 3.
Independent validation. Microarray gene expression data (bar histograms) were validated by quantitative PCR in an independent set of patients (dot histograms) comparing STA and PRED (A) and STA and DIAG (B). Mann–Whitney p values are indicated.
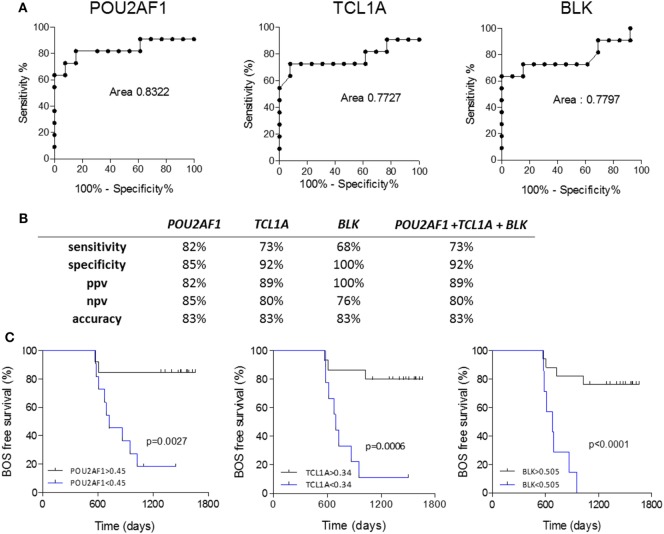
Because POU2AF1, TCL1A, and BLK were differentially expressed in the PRED, that is, more than 6 months before the clinical diagnosis of BOS, we evaluated the predictive performance of these three markers using ROC curve analysis. POU2AF1 (AUC = 0.832, 95% CI = 0.638–1.026), TCL1A (AUC = 0.773, 95% CI = 0.553–0.993), and BLK (AUC = 0.780, 95% CI = 0.569–0.991) discriminated well between STA and BOS patients (Figure 4A). Global performances of the prediction presented in Figure 4 show an accuracy higher than 80% for the three markers. Due to the high level of correlation between the three genes, performance of the prediction was not improved by their combination (Figure S5 in Supplementary Material).
Figure 4.
Performance of POU2AF1, TCL1A, and B cell lymphocyte kinase (BLK) in prediction of bronchiolitis obliterans syndrome (BOS). (A) ROC curves for POU2AF1, TCL1A, and BLK for the prediction of BOS are displayed. (B) Discriminative characteristics of the three genes or the sum of expression of the three genes. (C) Kaplan–Meier analysis of BOS-free survival categorized by best expression thresholds of discrimination in ROC curves. Log-ranked p values are indicated.
In the survival analysis, these three genes were highly associated with BOS occurrence with time in the discovery set (BLK, p = 0.0013; POU2AF1, p = 0.0028; and TCL1A, p = 0.0031; Table S1 in Supplementary Material). The association between BOS-free survival and POU2AF1, TCL1A, and BLK was assessed through Kaplan–Meier analyses. As shown in Figure 4C, expression levels of POU2AF1, TCL1A, and BLK under 0.45, 0.34, and 0.505, respectively (corresponding to best expression thresholds according to ROC curves) were associated with significant reduction of BOS-free survival after lung transplantation (p < 0.01). In this validation cohort, 3 of 8 recipients developing BOS had donor-specific antibody (DSA) at blood collection vs. none of the 13 STA (Fisher p = 0.031). Using univariate logistic analysis, DSA was not significantly associated with BOS.
POU2AF1, TCL1A, and BLK Are Downregulated in Blood of Patients with End-Stage Chronic Respiratory Diseases
Given the relationship between POU2AF1, TCL1A, and BLK expression and the development of BOS, we investigated the expression of these three genes in public data sets from blood of patients with other causes of respiratory failure (29, 30). The three genes were downregulated in the blood of non-transplanted patients with end-stage chronic respiratory diseases (cystic fibrosis, idiopathic pulmonary fibrosis, or pulmonary hypertension) compared to healthy volunteers (Figure 5).
Figure 5.
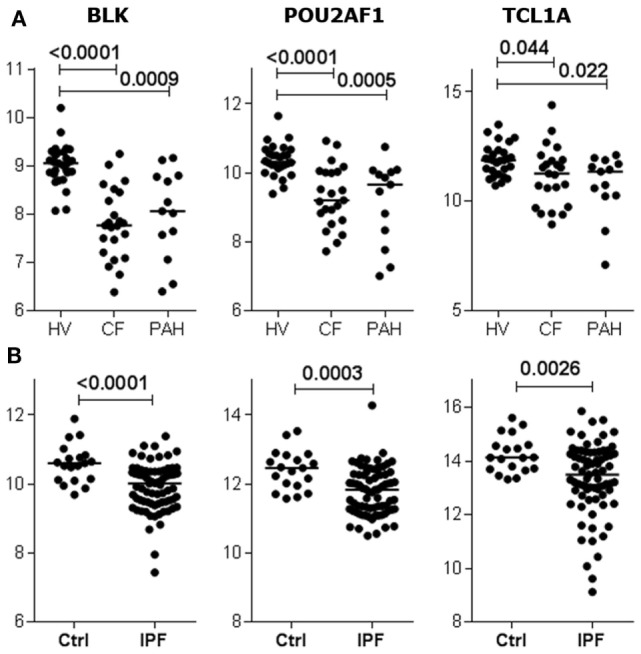
B cell lymphocyte kinase (BLK), POU2AF1, and TCL1A expression is downregulated by terminal respiratory failure. Expression of BLK, POU2AF1, and TCL1A in blood from non-transplanted patients with end-stage chronic respiratory diseases, displaying either cystic fibrosis (CF), pulmonary hypertension (PAH) (A), or idiopathic pulmonary fibrosis (IPF) (B) compared to healthy volunteers (HVs) (29, 30). The p values from Kruskal–Wallis tests with Dunn’s ad hoc pairwise comparisons or Mann–Whitney tests are indicated.
Discussion
Bronchiolitis obliterans syndrome is a major limitation of long-term survival after lung transplantation. Several previous attempts to identify early predictors of BOS remain limited by the absence of validation studies (6, 8–10, 12–14). Furthermore, they generally relied on invasive procedures incompatible with a routine clinical monitoring. We opted for a non-invasive large-scale molecular profiling approach to identify predictors of BOS in whole blood from 89 LTRs. A three-gene molecular signature differentiating BOS and STA was validated in an independent set of 25 patients. To the best of our knowledge, this is the first published study combining two independent cohorts for the identification and validation of predictors of BOS.
We showed that 29 and 33 transcripts were differentially expressed between STA and DIAG and STA and PRED. Twelve transcripts were common between both comparisons. This suggests a progression of the molecular profile during the development of the disease. Comparison of blood collected 6 and 12 months after transplantation for the STA group ruled out an early time-dependent alteration of gene expression. POU2AF1, TCL1A, and BLK were associated with the occurrence of BOS with time in a survival analysis and were validated as predictors of BOS more than 6 months before its diagnosis. The validation cohort used in our study revealed the strength of these biomarkers for BOS prediction. They displayed similar performances to predict BOS according to their correlation of expression, with AUC values reaching 0.83, 0.77, and 0.78, respectively, and exceeding the performance of biomarkers already published (14, 34, 35).
Recent literature highlights the correlation between de novo DSAs with the occurrence of chronic dysfunction (36–40). While in our validation cohort DSA was not significantly associated with BOS, an independent study is required to decipher whether these genes are associated with de novo DSA and BOS appearance and which parameter is the most predictive. Furthermore, our investigation focused on the BOS subtype, and further work will have to be conducted to identify predictors of the restrictive syndrome. Besides, stringent patient selection criteria were applied for a clear discrimination between groups, and all selected patients were not included in the analysis because of feasibility matters. This reduction in patient number may limit the generalizability of our results, whereas enough power was reached to highlight significantly statistical differences for the three genes. Although our predictions were confirmed on an independent set of patients, they will have to be validated on an external cohort as well.
Our primary objective was to identify a molecular biomarker for clinical monitoring in whole blood, which limits the interpretation of mechanistic investigations. Nevertheless, B cell signatures and the presence of B cells with suppressive properties are evidenced in other situations of transplantation and particularly in tolerance in kidney transplantation (41–43) and/or associated with long-term graft survival (44, 45). In a longitudinal study in lung transplantation, BOS appearance is associated with a lower level of CD24hiCD38hiIgDhiIgMhi transitional B cells with a regulatory phenotype 18 months after lung transplantation compared to STA (46). We also observed that 12 and 13 transcripts related to immunoglobulins are downexpressed in BOS (PRED and DIAG, respectively) compared to STA, while gene expression of BAFF (B cell activating factor coded by TNFSF13B) and BAFF-R (BAFF receptor coded by TNFRSF13C), required for B cell survival and maturation, were not different between the three situations (data not shown). These data suggest a dysregulation of B cells in the BOS situation.
In addition, several genes merit further exploration to better understand the underlying mechanism behind BOS. By using GO analysis, we evidenced differentially expressed genes between STA and PRED associated with immune system process, such as CD19, HLA-DQA1, and HLA-DQA2. Interestingly, the gene enrichment and unsupervised hierarchical clustering analyses reinforce this association of the B cell cluster with the development of BOS. POU2AF1 is a B cell transcriptional coactivator involved in B cell development and function (47). Yet, POU2AF1 is expressed in T cells as well, and recent evidence revealed its role in Th1–Th2 polarization (48) and in the mounting of T-cell-dependent B cell responses (49). BLK is a member of the Src family of tyrosine kinases and encodes a non-receptor protein tyrosine kinase involved in the regulation of B cell receptor signaling (50). We found a downregulation of TCL1A in the BOS before disease onset. TCL1A is notably expressed by B and T lymphocytes, where it promotes cell proliferation and survival (51). Interestingly, in renal transplantation, TCL1A is downregulated at the time of acute allograft rejection (52), whereas it is overexpressed in operationally tolerant patients, an ideal situation where recipients are off of immunosuppression with a functioning allograft (53). The exact contribution of these genes in the development of BOS remains to be investigated. Some polymorphisms have been described for BLK, POU2AF1, and TCL1A. BLK polymorphism is a risk factor for developing autoimmune diseases (54, 55). Genetic variations leading to reduced BLK expression are associated with several autoimmune diseases via lowering the threshold for B cell activation (54). A TCL1A single-nucleotide polymorphism is associated with downstream expression of cytokines and chemokines and the nuclear factor-κB transcriptional activity resulting in the modulation of inflammation and immune response (56). POU2AF1 polymorphism is associated with susceptibility to lymphoma (57), and mice deficient in POU2AF1 exhibit reduced numbers of mature B cells and defective immune responses to antigens (58, 59). The fact that none of our patients evidenced clinical symptoms or biological associated modifications (such as decrease of total B cells), in addition with the absence of differential expression of these three genes at the time of transplantation do not suggest existence of such polymorphisms in our study. Intriguingly, however, the analysis of gene expression data from two independent studies (29, 30) revealed that BLK, TCL1A, and POU2AF1 are downregulated in blood from patients with end-stage chronic respiratory diseases. GSEA analysis also highlighted the enrichment of a gene set related to COPD in lung from smokers in DIAG compared to STA, including the SPIB gene (33). Altogether, these results suggest common mechanisms between BOS and other chronic respiratory diseases, as previously noted (60), and support the need to further decipher the roles of POU2AF1, BLK, and TCL1A in the development of lung pathologies.
In conclusion, by using non-invasive whole blood profiling, we identified and validated POU2AF1, TCL1A, and BLK as three predictive biomarkers of BOS, more than 6 months before diagnosis. These genes allow stratification based on the BOS risk and could be easily monitored to provide clinicians with new tools to improve follow-up and adapt treatment of patient likely to develop BOS before clinical manifestations and allograft damage arise.
Cohort of Lung Transplantation
Cohort of Lung Transplantation, COLT (associating surgeons, anesthetists-intensivists, physicians, research staff). Bordeaux: J. Jougon, J.-F. Velly, H. Rozé, E. Blanchard, C. Dromer. Bruxelles: M. Antoine, M. Cappello, R. Souilamas, M. Ruiz, Y. Sokolow, F. Vanden Eynden, G. Van Nooten, L. Barvais, J. Berré, S. Brimioulle, D. De Backer, J. Créteur, E. Engelman, I. Huybrechts, B. Ickx, T. J. C. Preiser, T. Tuna, L. Van Obberghe, N. Vancutsem, J.-L. Vincent, P. De Vuyst, I. Etienne, F. Féry, F. Jacobs, C. Knoop, J. L. Vachiéry, P. Van den Borne, I. Wellemans, G. Amand, L. Collignon, M. Giroux. Grenoble: E. Arnaud-Crozat, V. Bach, P.-Y. Brichon, P. Chaffanjon, O. Chavanon, A. de Lambert, J. P. Fleury, S. Guigard, K. Hireche, A. Pirvu, P. Porcu, R. Hacini, P. Albaladejo, C. Allègre, A. Bataillard, D. Bedague, E. Briot, M. Casez-Brasseur, D. Colas, G. Dessertaine, M. Durand, G. Francony, A. Hebrard, M. R. Marino, B. Oummahan, D. Protar, D. Rehm, S. Robin, M. Rossi-Blancher, P. Bedouch, A. Boignard, H. Bouvaist, A. Briault, B. Camara, S. Chanoine, M. Dubuc, S. Lantuéjoul, S. Quétant, J. Maurizi, P. Pavèse, C. Pison, C. Saint-Raymond, N. Wion, C. Chérion. Lyon: R. Grima, O. Jegaden, J.-M. Maury, F. Tronc, C. Flamens, S. Paulus, J. F. Mornex, F. Philit, A. Senechal, J.-C. Glérant, S. Turquier, D. Gamondes, L. Chalabresse, F. Thivolet-Bejui, C. Barnel, C. Dubois, A. Tiberghien. Paris, Hôpital Européen Georges Pompidou: F. Le Pimpec-Barthes, A. Bel, P. Mordant, P. Achouh, V. Boussaud, R. Guillemain, D. Méléard, M. O. Bricourt, B. Cholley, V. Pezella. Marseille: M. Adda, M. Badier, F. Bregeon, B. Coltey, X. B. D’Journo, S. Dizier, C. Doddoli, N. Dufeu, H. Dutau, J. M. Forel, J. Y. Gaubert, C. Gomez, M. Leone, A. Nieves, B. Orsini, L. Papazian L. C. Picard, M. Reynaud-Gaubert, A. Roch, J. M. Rolain, E. Sampol, V. Secq, P. Thomas, D. Trousse, M. Yahyaoui. Nantes: O. Baron, P. Lacoste, C. Perigaud, J. C. Roussel, I. Danner, A Haloun, A. Magnan, A. Tissot, T. Lepoivre, M. Treilhaud, K. Botturi-Cavaillès, S. Brouard, R. Danger, J. Loy, M. Morisset, M. Pain, S. Pares, D. Reboulleau, P. J. Royer, E. Durand, A. Foureau. Le Plessis Robinson, Hôpital Marie Lannelongue: Ph. Dartevelle, D. Fabre, E. Fadel, O. Mercier, S. Mussot, F. Stephan, P. Viard, J. Cerrina, P. Dorfmuller, S. Feuillet, M. Ghigna, Ph. Hervén, F. Le Roy Ladurie, J. Le Pavec, V. Thomas de Montpreville, L. Lamrani. Paris, Hôpital Bichat: Y. Castier, P. Cerceau, F. Francis, G. Lesèche, N. Allou, P. Augustin, S. Boudinet, M. Desmard, G. Dufour, P. Montravers, O. Brugière, G. Dauriat, G. Jébrak, H. Mal, A. Marceau, A.-C. Métivier, G. Thabut, B. Ait Ilalne. Strasbourg: P. Falcoz, G. Massard, N. Santelmo, G. Ajob, O. Collange, O. Helms, J. Hentz, A. Roche, B. Bakouboula, T. Degot, A. Dory, S. Hirschi, S. Ohlmann-Caillard, L. Kessler, R. Kessler, A. Schuller, K. Bennedif, S. Vargas. Suresnes, Hôpital Foch: P. Bonnette, A. Chapelier, P. Puyo, E. Sage, J. Bresson, V. Caille, C. Cerf, J. Devaquet, V. Dumans-Nizard, M. L. Felten, M. Fischler, A. G. Si Larbi, M. Leguen, L. Ley, N. Liu, G. Trebbia, S. De Miranda, B. Douvry, F. Gonin, D. Grenet, A. M. Hamid, H. Neveu, F. Parquin, C. Picard, A. Roux, M. Stern, F. Bouillioud, P. Cahen, M. Colombat, C. Dautricourt, M. Delahousse, B. D’Urso, J. Gravisse, A. Guth, S. Hillaire, P. Honderlick, M. Lequintrec, E. Longchampt, F. Mellot, A. Scherrer, L. Temagoult, L. Tricot, M. Vasse, C. Veyrie, L. Zemoura. Toulouse: J. Berjaud, L. Brouchet, M. Dahan, F. Le Balle, O. Mathe, H. Benahoua, A. Didier, A. L. Goin, M. Murris, L. Crognier, O. Fourcade.
Swiss Transplant Cohort Study, STCS Genève-Lausanne. T. Krueger, H. B. Ris, M. Gonzalez, J.-D. Aubert, L. P. Nicod, B. J. Marsland, T. C. Berutto, T. Rochat, P. Soccal, Ph. Jolliet, A. Koutsokera, C. Marcucci, O. Manuel, E. Bernasconi, M. Chollet, F. Gronchi, C. Courbon, Zurich S. Hillinger, I. Inci, P. Kestenholz, W. Weder, R. Schuepbach, M. Zalunardo, C. Benden, U. Buergi, L. C. Huber, B. Isenring, M. M. Schuurmans, A. Gaspert, D. Holzmann, N. Müller, C. Schmid, B. Vrugt, T. Rechsteiner.
SME and Platforms. Biomax, Germany: A. Fritz, D. Maier. Finovatis, Lyon, France: K. Desplanche, D. Koubi. GATC, Germany: F. Ernst, T. Paprotka, M. Schmitt, B. Wahl. Novasdicovery, Lyon, France: J.-P. Boissel, G. Olivera-Botello. Prométhée Proteomics Platform, Grenoble, France: C. Trocmé, B. Toussaint, S. Bourgoin-Voillard, M. Séve. INSERM U823, Université Joseph Fourier, Grenoble, France, M. Benmerad, V. Siroux, R. Slama. European Institute for Systems Biology & Medicine, Lyon, France: C. Auffray, D. Charron, J. Pellet, C. Pison.
Ethics Statement
Lung transplant recipients were recruited from September 2009 to October 2013 within the multicentre COLT cohort (Cohort of Lung Transplantation; NCT00980967). The local ethical committee (Comité de Protection des Personnes Ouest 1-Tours, 2009-A00036-51) approved the study, and all participants provided written informed consent.
Author Contributions
Conceived and designed the experiments/analyses: RD, PJR, SB, AM. Performed the experiments: RD, PJR, ED, JL. Analyzed the data: RD, PJR. Contributed samples/materials: DR, AT, PL, AR, MRG, CG, RK, SM, CD, OB, JFM, RG, MD, CK, KB, AF, CP, AK, LPN. Wrote the paper: RD, PJR, SB, AM. All authors approved the final version of the article.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are most grateful to the GenoBiRD Core Facility for its technical support and to the biological resource center for biobanking CHU Nantes, Hôtel Dieu, Centre de ressources biologiques (CRB), Nantes, F-44093, France (BRIF: BB-0033-00040).
Footnotes
Funding. This research program was supported by Vaincre la Mucoviscidose, Association Gregory Lemarchal, Agence de Biomédecine, INSERM, Région Pays de La Loire, Institut de Recherche en Santé Respiratoire des Pays de la Loire, Fonds de Recherche en Santé Respiratoire, Fondation du Souffle, and Fondation pour la Recherche Médicale. This work was realized in the context of the IHU-Cesti project, the DHU Oncogreffe and the LabEX IGO thanks to French government financial support managed by the National Research Agency via the “Investment into the Future” program (ANR-10-IBHU-005 and ANR-11- LABX-0016-01). The IHU-Cesti project is also supported by Nantes Métropole and Région Pays de la Loire. The research leading to these results has also received funding from the European Commission’s Seventh Framework Program FP7 VISICORT under grant agreement n 602470.
Contributor Information
The COLT and SysCLAD Consortia:
J. Jougon, J.-F. Velly, H. Rozé, E. Blanchard, C. Dromer, M. Antoine, M. Cappello, R. Souilamas, M. Ruiz, Y. Sokolow, F. Vanden Eynden, G. Van Nooten, L. Barvais, J. Berré, S. Brimioulle, D. De Backer, J. Créteur, E. Engelman, I. Huybrechts, B. Ickx, T. J. C. Preiser, T. Tuna, L. Van Obberghe, N. Vancutsem, J.-L. Vincent, P. De Vuyst, I. Etienne, F. Féry, F. Jacobs, C. Knoop, J. L. Vachiéry, P. Van den Borne, I. Wellemans, G. Amand, L. Collignon, M. Giroux, E. Arnaud-Crozat, V. Bach, P.-Y. Brichon, P. Chaffanjon, O. Chavanon, A. de Lambert, J. P. Fleury, S. Guigard, K. Hireche, A. Pirvu, P. Porcu, R. Hacini, P. Albaladejo, C. Allègre, A. Bataillard, D. Bedague, E. Briot, M. Casez-Brasseur, D. Colas, G. Dessertaine, M. Durand, G. Francony, A. Hebrard, M. R. Marino, B. Oummahan, D. Protar, D. Rehm, S. Robin, M. Rossi-Blancher, P. Bedouch, A. Boignard, H. Bouvaist, A. Briault, B. Camara, S. Chanoine, M. Dubuc, S. Lantuéjoul, S. Quétant, J. Maurizi, P. Pavèse, C. Pison, C. Saint-Raymond, N. Wion, C. Chérion, R. Grima, O. Jegaden, J.-M. Maury, F. Tronc, C. Flamens, S. Paulus, J. F. Mornex, F. Philit, A. Senechal, J.-C. Glérant, S. Turquier, D. Gamondes, L. Chalabresse, F. Thivolet-Bejui, C. Barnel, C. Dubois, A. Tiberghien, F. Le Pimpec-Barthes, A. Bel, P. Mordant, P. Achouh, V. Boussaud, R. Guillemain, D. Méléard, M. O. Bricourt, B. Cholley, V. Pezella, M. Adda, M. Badier, F. Bregeon, B. Coltey, X. B. D’Journo, S. Dizier, C. Doddoli, N. Dufeu, H. Dutau, J. M. Forel, J. Y. Gaubert, C. Gomez, M. Leone, A. Nieves, B. Orsini, L. Papazian, L. C. Picard, M. Reynaud-Gaubert, A. Roch, J. M. Rolain, E. Sampol, V. Secq, P. Thomas, D. Trousse, M. Yahyaoui, O. Baron, P. Lacoste, C. Perigaud, J. C. Roussel, I. Danner, A. Haloun, A. Magnan, A. Tissot, T. Lepoivre, M. Treilhaud, K. Botturi-Cavaillès, S. Brouard, R. Danger, J. Loy, M. Morisset, M. Pain, S. Pares, D. Reboulleau, P. J. Royer, E. Durand, A. Foureau, Ph. Dartevelle, D. Fabre, E. Fadel, O. Mercier, S. Mussot, F. Stephan, P. Viard, J. Cerrina, P. Dorfmuller, S. Feuillet, M. Ghigna, Ph. Hervén, F. Le Roy Ladurie, J. Le Pavec, V. Thomas de Montpreville, L. Lamrani, Y. Castier, P. Cerceau, F. Francis, G. Lesèche, N. Allou, P. Augustin, S. Boudinet, M. Desmard, G. Dufour, P. Montravers, O. Brugière, G. Dauriat, G. Jébrak, H. Mal, A. Marceau, A.-C. Métivier, G. Thabut, B. Ait Ilalne, P. Falcoz, G. Massard, N. Santelmo, G. Ajob, O. Collange, O. Helms, J. Hentz, A. Roche, B. Bakouboula, T. Degot, A. Dory, S. Hirschi, S. Ohlmann-Caillard, L. Kessler, R. Kessler, A. Schuller, K. Bennedif, S. Vargas, P. Bonnette, A. Chapelier, P. Puyo, E. Sage, J. Bresson, V. Caille, C. Cerf, J. Devaquet, V. Dumans-Nizard, M. L. Felten, M. Fischler, A. G. Si Larbi, M. Leguen, L. Ley, N. Liu, G. Trebbia, S. De Miranda, B. Douvry, F. Gonin, D. Grenet, A. M. Hamid, H. Neveu, F. Parquin, C. Picard, A. Roux, M. Stern, F. Bouillioud, P. Cahen, M. Colombat, C. Dautricourt, M. Delahousse, B. D’Urso, J. Gravisse, A. Guth, S. Hillaire, P. Honderlick, M. Lequintrec, E. Longchampt, F. Mellot, A. Scherrer, L. Temagoult, L. Tricot, M. Vasse, C. Veyrie, L. Zemoura, J. Berjaud, L. Brouchet, M. Dahan, F. Le Balle, O. Mathe, H. Benahoua, A. Didier, A. L. Goin, M. Murris, L. Crognier, O. Fourcade, T. Krueger, H. B. Ris, M. Gonzalez, J.-D. Aubert, L. P. Nicod, B. J. Marsland, T. C. Berutto, T. Rochat, P. Soccal, Ph. Jolliet, A. Koutsokera, C. Marcucci, O. Manuel, E. Bernasconi, M. Chollet, F. Gronchi, C. Courbon, Zurich S. Hillinger, I. Inci, P. Kestenholz, W. Weder, R. Schuepbach, M. Zalunardo, C. Benden, U. Buergi, L. C. Huber, B. Isenring, M. M. Schuurmans, A. Gaspert, D. Holzmann, N. Müller, C. Schmid, B. Vrugt, T. Rechsteiner, A. Fritz, D. Maier, K. Desplanche, D. Koubi, F. Ernst, T. Paprotka, M. Schmitt, B. Wahl, J.-P. Boissel, G. Olivera-Botello, C. Trocmé, B. Toussaint, S. Bourgoin-Voillard, M. Séve, M. Benmerad, V. Siroux, R. Slama, C. Auffray, D. Charron, J. Pellet, and C. Pison
Supplementary Material
The Supplementary Material for this article can be found online at http://www.frontiersin.org/articles/10.3389/fimmu.2017.01841/full#supplementary-material.
References
- 1.Royer PJ, Olivera-Botello G, Koutsokera A, Aubert JD, Bernasconi E, Tissot A, et al. Chronic lung allograft dysfunction: a systematic review of mechanisms. Transplantation (2016) 100(9):1803–14. 10.1097/TP.0000000000001215 [DOI] [PubMed] [Google Scholar]
- 2.Sato M. Chronic lung allograft dysfunction after lung transplantation: the moving target. Gen Thorac Cardiovasc Surg (2013) 61(2):67–78. 10.1007/s11748-012-0167-3 [DOI] [PubMed] [Google Scholar]
- 3.Meyer KC, Raghu G, Verleden GM, Corris PA, Aurora P, Wilson KC, et al. An international ISHLT/ATS/ERS clinical practice guideline: diagnosis and management of bronchiolitis obliterans syndrome. Eur Respir J (2014) 44(6):1479–503. 10.1183/09031936.00107514 [DOI] [PubMed] [Google Scholar]
- 4.Sato M, Waddell TK, Wagnetz U, Roberts HC, Hwang DM, Haroon A, et al. Restrictive allograft syndrome (RAS): a novel form of chronic lung allograft dysfunction. J Heart Lung Transplant (2011) 30(7):735–42. 10.1016/j.healun.2011.01.712 [DOI] [PubMed] [Google Scholar]
- 5.Sato M, Keshavjee S. Bronchiolitis obliterans syndrome: alloimmune-dependent and -independent injury with aberrant tissue remodeling. Semin Thorac Cardiovasc Surg (2008) 20(2):173–82. 10.1053/j.semtcvs.2008.05.002 [DOI] [PubMed] [Google Scholar]
- 6.Devouassoux G, Drouet C, Pin I, Brambilla C, Brambilla E, Colle PE, et al. Alveolar neutrophilia is a predictor for the bronchiolitis obliterans syndrome, and increases with degree of severity. Transpl Immunol (2002) 10(4):303–10. 10.1016/S0966-3274(02)00074-6 [DOI] [PubMed] [Google Scholar]
- 7.Neurohr C, Huppmann P, Samweber B, Leuschner S, Zimmermann G, Leuchte H, et al. Prognostic value of bronchoalveolar lavage neutrophilia in stable lung transplant recipients. J Heart Lung Transplant (2009) 28(5):468–74. 10.1016/j.healun.2009.01.014 [DOI] [PubMed] [Google Scholar]
- 8.Reynaud-Gaubert M, Marin V, Thirion X, Farnarier C, Thomas P, Badier M, et al. Upregulation of chemokines in bronchoalveolar lavage fluid as a predictive marker of post-transplant airway obliteration. J Heart Lung Transplant (2002) 21(7):721–30. 10.1016/S1053-2498(02)00392-3 [DOI] [PubMed] [Google Scholar]
- 9.Hübner RH, Meffert S, Mundt U, Böttcher H, Freitag S, El Mokhtari NE, et al. Matrix metalloproteinase-9 in bronchiolitis obliterans syndrome after lung transplantation. Eur Respir J (2005) 25(3):494–501. 10.1183/09031936.05.00091804 [DOI] [PubMed] [Google Scholar]
- 10.Bhorade SM, Chen H, Molinero L, Liao C, Garrity ER, Vigneswaran WT, et al. Decreased percentage of CD4+FoxP3+ cells in bronchoalveolar lavage from lung transplant recipients correlates with development of bronchiolitis obliterans syndrome. Transplantation (2010) 90(5):540–6. 10.1097/TP.0b013e3181e8dabe [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jonigk D, Izykowski N, Rische J, Braubach P, Kühnel M, Warnecke G, et al. Molecular profiling in lung biopsies of human pulmonary allografts to predict chronic lung allograft dysfunction. Am J Pathol (2015) 185:3178–88. 10.1016/j.ajpath.2015.08.016 [DOI] [PubMed] [Google Scholar]
- 12.Shah RJ, Bellamy SL, Lee JC, Cantu E, Diamond JM, Mangalmurti N, et al. Early plasma soluble receptor for advanced glycation end-product levels are associated with bronchiolitis obliterans syndrome. Am J Transplant (2013) 13(3):754–9. 10.1111/ajt.12062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Salama M, Jaksch P, Andrukhova O, Taghavi S, Klepetko W, Aharinejad S. Endothelin-1 is a useful biomarker for early detection of bronchiolitis obliterans in lung transplant recipients. J Thorac Cardiovasc Surg (2010) 140(6):1422–7. 10.1016/j.jtcvs.2010.08.046 [DOI] [PubMed] [Google Scholar]
- 14.Paantjens AWM, Kwakkel-van Erp JM, Van Ginkel WGJ, Van Kessel DA, Van Den Bosch JMM, Van De Graaf EA, et al. Serum thymus and activation regulated chemokine levels post-lung transplantation as a predictor for the bronchiolitis obliterans syndrome. Clin Exp Immunol (2008) 154(2):202–8. 10.1111/j.1365-2249.2008.03764.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.LaPar DJ, Burdick MD, Emaminia A, Harris DA, Strieter BA, Liu L, et al. Circulating fibrocytes correlate with bronchiolitis obliterans syndrome development after lung transplantation: a novel clinical biomarker. Ann Thorac Surg (2011) 92(2):470–7. 10.1016/j.athoracsur.2011.04.065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weigt SS, Wang X, Palchevskiy V, Gregson AL, Patel N, DerHovanessian A, et al. Gene expression profiling of bronchoalveolar lavage cells preceding a clinical diagnosis of chronic lung allograft dysfunction. PLoS One (2017) 12(1):e0169894. 10.1371/journal.pone.0169894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Anglicheau D, Suthanthiran M. Noninvasive prediction of organ graft rejection and outcome using gene expression patterns. Transplantation (2008) 86(2):192–9. 10.1097/TP.0b013e31817eef7b [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brouard S, Mansfield E, Braud C, Li L, Giral M, Hsieh SC, et al. Identification of a peripheral blood transcriptional biomarker panel associated with operational renal allograft tolerance. Proc Natl Acad Sci U S A (2007) 104(39):15448–53. 10.1073/pnas.0705834104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen Y, Zhang H, Xiao X, Jia Y, Wu W, Liu L, et al. Peripheral blood transcriptome sequencing reveals rejection-relevant genes in long-term heart transplantation. Int J Cardiol (2013) 168(3):2726–33. 10.1016/j.ijcard.2013.03.095 [DOI] [PubMed] [Google Scholar]
- 20.Kurian SM, Fouraschen SMG, Langfelder P, Horvath S, Shaked A, Salomon DR, et al. Genomic profiles and predictors of early allograft dysfunction after human liver transplantation. Am J Transplant (2015) 15(6):1605–14. 10.1111/ajt.13145 [DOI] [PubMed] [Google Scholar]
- 21.Pain M, Royer PJ, Loy J, Girardeau A, Tissot A, Lacoste P, et al. T cells promote bronchial epithelial cell secretion of matrix metalloproteinase-9 via a C-C chemokine receptor type 2 pathway: implications for chronic lung allograft dysfunction. Am J Transplant (2016) 17(6):1502–14. 10.1111/ajt.14166 [DOI] [PubMed] [Google Scholar]
- 22.Koutsokera A, Royer PJ, Antonietti JP, Fritz A, Benden C, Aubert JD, et al. Development of a multivariate prediction model for early-onset bronchiolitis obliterans syndrome and restrictive allograft syndrome in lung transplantation. Front Med (2017) 4:109. 10.3389/fmed.2017.00109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Verleden GM, Raghu G, Meyer KC, Glanville AR, Corris P. A new classification system for chronic lung allograft dysfunction. J Heart Lung Transplant (2014) 33(2):127–33. 10.1016/j.healun.2014.01.316 [DOI] [PubMed] [Google Scholar]
- 24.Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics (2007) 8(1):118–27. 10.1093/biostatistics/kxj037 [DOI] [PubMed] [Google Scholar]
- 25.Leek JT, Johnson WE, Parker HS, Jaffe AE, Storey JD. The SVA package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics (2012) 28(6):882–3. 10.1093/bioinformatics/bts034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z, et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res (2016) 44(W1):W90–7. 10.1093/nar/gkw377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Newman AM, Liu CL, Green MR, Gentles AJ, Feng W, Xu Y, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods (2015) 12(5):453–7. 10.1038/nmeth.3337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A (2005) 102(43):15545–50. 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chesne J, Danger R, Botturi K, Reynaud-Gaubert M, Mussot S, Stern M, et al. Systematic analysis of blood cell transcriptome in end-stage chronic respiratory diseases. PLoS One (2014) 9(10):e109291. 10.1371/journal.pone.0109291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Herazo-Maya JD, Noth I, Duncan SR, Kim S, Ma SF, Tseng GC, et al. Peripheral blood mononuclear cell gene expression profiles predict poor outcome in idiopathic pulmonary fibrosis. Sci Transl Med (2013) 5(205):205ra136. 10.1126/scitranslmed.3005964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Basso K, Margolin AA, Stolovitzky G, Klein U, Dalla-Favera R, Califano A. Reverse engineering of regulatory networks in human B cells. Nat Genet (2005) 37(4):382–90. 10.1038/ng1532 [DOI] [PubMed] [Google Scholar]
- 32.Haddad R, Guardiola P, Izac B, Thibault C, Radich J, Delezoide AL, et al. Molecular characterization of early human T/NK and B-lymphoid progenitor cells in umbilical cord blood. Blood (2004) 104(13):3918–26. 10.1182/blood-2004-05-1845 [DOI] [PubMed] [Google Scholar]
- 33.Ning W, Li CJ, Kaminski N, Feghali-Bostwick CA, Alber SM, Di YP, et al. Comprehensive gene expression profiles reveal pathways related to the pathogenesis of chronic obstructive pulmonary disease. Proc Natl Acad Sci U S A (2004) 101(41):14895–900. 10.1073/pnas.0401168101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Budding K, van de Graaf EA, Kardol-Hoefnagel T, Kwakkel-van Erp JM, Luijk BD, Oudijk E-JD, et al. Soluble CD59 is a novel biomarker for the prediction of obstructive chronic lung allograft dysfunction after lung transplantation. Sci Rep (2016) 6:26274. 10.1038/srep26274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jaksch P, Taghavi S, Klepetko W, Salama M. Pretransplant serum human chitinase-like glycoprotein YKL-40 concentrations independently predict bronchiolitis obliterans development in lung transplant recipients. J Thorac Cardiovasc Surg (2014) 148(1):273–81. 10.1016/j.jtcvs.2014.02.059 [DOI] [PubMed] [Google Scholar]
- 36.Roux A, Bendib Le Lan I, Holifanjaniaina S, Thomas KA, Hamid AM, Picard C, et al. Antibody-mediated rejection in lung transplantation: clinical outcomes and donor-specific antibody characteristics. Am J Transplant (2016) 16(4):1216–28. 10.1111/ajt.13589 [DOI] [PubMed] [Google Scholar]
- 37.Le Pavec J, Suberbielle C, Lamrani L, Feuillet S, Savale L, Dorfmüller P, et al. De-novo donor-specific anti-HLA antibodies 30 days after lung transplantation are associated with a worse outcome. J Heart Lung Transplant (2016) 35(9):1067–77. 10.1016/j.healun.2016.05.020 [DOI] [PubMed] [Google Scholar]
- 38.Tikkanen JM, Singer LG, Kim SJ, Li Y, Binnie M, Chaparro C, et al. De novo DQ donor-specific antibodies are associated with chronic lung allograft dysfunction after lung transplantation. Am J Respir Crit Care Med (2016) 194(5):596–606. 10.1164/rccm.201509-1857OC [DOI] [PubMed] [Google Scholar]
- 39.Morrell MR, Pilewski JM, Gries CJ, Pipeling MR, Crespo MM, Ensor CR, et al. De novo donor-specific HLA antibodies are associated with early and high-grade bronchiolitis obliterans syndrome and death after lung transplantation. J Heart Lung Transplant (2014) 33(12):1288–94. 10.1016/j.healun.2014.07.018 [DOI] [PubMed] [Google Scholar]
- 40.Safavi S, Robinson DR, Soresi S, Carby M, Smith JD. De novo donor HLA-specific antibodies predict development of bronchiolitis obliterans syndrome after lung transplantation. J Heart Lung Transplant (2014) 33(12):1273–81. 10.1016/j.healun.2014.07.012 [DOI] [PubMed] [Google Scholar]
- 41.Newell KA, Asare A, Kirk AD, Gisler TD, Bourcier K, Suthanthiran M, et al. Identification of a B cell signature associated with renal transplant tolerance in humans. J Clin Invest (2010) 120(6):1836–47. 10.1172/JCI39933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sagoo P, Perucha E, Sawitzki B, Tomiuk S, Stephens DA, Miqueu P, et al. Development of a cross-platform biomarker signature to detect renal transplant tolerance in humans. J Clin Invest (2010) 120(6):1848–61. 10.1172/JCI39922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pallier A, Hillion S, Danger R, Giral M, Racape M, Degauque N, et al. Patients with drug-free long-term graft function display increased numbers of peripheral B cells with a memory and inhibitory phenotype. Kidney Int (2010) 78(5):503–13. 10.1038/ki.2010.162 [DOI] [PubMed] [Google Scholar]
- 44.Heidt S, Hester J, Shankar S, Friend PJ, Wood KJ. B cell repopulation after alemtuzumab induction-transient increase in transitional B cells and long-term dominance of naive B cells. Am J Transplant (2012) 12(7):1784–92. 10.1111/j.1600-6143.2012.04012.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shabir S, Girdlestone J, Briggs D, Kaul B, Smith H, Daga S, et al. Transitional B lymphocytes are associated with protection from kidney allograft rejection: a prospective study. Am J Transplant (2015) 15(5):1384–91. 10.1111/ajt.13122 [DOI] [PubMed] [Google Scholar]
- 46.Brosseau C, Durand M, Durand E, Loy J, Foureau A, Lacoste P, et al. Characterization of a B lymphocyte signature predictive of chronic lung allograft dysfunction. Am J Transplant (2017) 17(Suppl 3). 10.1111/ajt.14304 [DOI] [Google Scholar]
- 47.Luo Y, Fujii H, Gerster T, Roeder RG. A novel B cell-derived coactivator potentiates the activation of immunoglobulin promoters by octamer-binding transcription factors. Cell (1992) 71(2):231–41. 10.1016/0092-8674(92)90352-D [DOI] [PubMed] [Google Scholar]
- 48.Brunner C, Sindrilaru A, Girkontaite I, Fischer K-D, Sunderkötter C, Wirth T. BOB.1/OBF.1 controls the balance of TH1 and TH2 immune responses. EMBO J (2007) 26(13):3191–202. 10.1038/sj.emboj.7601742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stauss D, Brunner C, Berberich-Siebelt F, Höpken UE, Lipp M, Müller G. The transcriptional coactivator Bob1 promotes the development of follicular T helper cells via Bcl6. EMBO J (2016) 35(8):881–98. 10.15252/embj.201591459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dymecki SM, Niederhuber JE, Desiderio SV. Specific expression of a tyrosine kinase gene, blk, in B lymphoid cells. Science (1990) 247(4940):332–6. 10.1126/science.2404338 [DOI] [PubMed] [Google Scholar]
- 51.Laine J, Künstle G, Obata T, Sha M, Noguchi M. The protooncogene TCL1 is an Akt kinase coactivator. Mol Cell (2000) 6(2):395–407. 10.1016/S1097-2765(00)00039-3 [DOI] [PubMed] [Google Scholar]
- 52.Heidt S, Vergunst M, Anholts JD, Reinders ME, de Fijter JW, Eikmans M, et al. B cell markers of operational tolerance can discriminate acute kidney allograft rejection from stable graft function. Transplantation (2014) 99(5):1058–64. 10.1097/00007890-201407151-03002 [DOI] [PubMed] [Google Scholar]
- 53.Danger R, Chesneau M, Paul C, Guerif P, Durand M, Newell KA, et al. A composite score associated with spontaneous operational tolerance in kidney transplant recipients. Kidney Int (2017) 91(6):1473–81. 10.1016/j.kint.2016.12.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Simpfendorfer KR, Armstead BE, Shih A, Li W, Curran M, Manjarrez-Orduno N, et al. Autoimmune disease-associated haplotypes of BLK exhibit lowered thresholds for B cell activation and expansion of Ig class-switched B cells. Arthritis Rheumatol (2015) 67(11):2866–76. 10.1002/art.39301 [DOI] [PubMed] [Google Scholar]
- 55.Zeng C, Fang C, Weng H, Xu X, Wu T, Li W. B-cell lymphocyte kinase polymorphisms rs13277113, rs2736340, and rs4840568 and risk of autoimmune diseases: a meta-analysis. Medicine (2017) 96(36):e7855. 10.1097/MD.0000000000007855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ho MF, Ingle JN, Bongartz T, Kalari KR, Goss PE, Shepherd LE, et al. TCL1A single-nucleotide polymorphisms and estrogen-mediated toll-like receptor-MYD88-dependent nuclear factor-kappaB activation: single-nucleotide polymorphism- and selective estrogen receptor modulator-dependent modification of inflammation and immune response. Mol Pharmacol (2017) 92(2):175–84. 10.1124/mol.117.108340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhai K, Chang J, Hu J, Wu C, Lin D. Germline variation in the 3’-untranslated region of the POU2AF1 gene is associated with susceptibility to lymphoma. Mol Carcinog (2017) 56(8):1945–52. 10.1002/mc.22652 [DOI] [PubMed] [Google Scholar]
- 58.Nielsen PJ, Georgiev O, Lorenz B, Schaffner W. B lymphocytes are impaired in mice lacking the transcriptional co-activator Bob1/OCA-B/OBF1. Eur J Immunol (1996) 26(12):3214–8. 10.1002/eji.1830261255 [DOI] [PubMed] [Google Scholar]
- 59.Schubart DB, Rolink A, Kosco-Vilbois MH, Botteri F, Matthias P. B-cell-specific coactivator OBF-1/OCA-B/Bob1 required for immune response and germinal centre formation. Nature (1996) 383(6600):538–42. 10.1038/383538a0 [DOI] [PubMed] [Google Scholar]
- 60.Fernandez IE, Heinzelmann K, Verleden S, Eickelberg O. Characteristic patterns in the fibrotic lung. Comparing idiopathic pulmonary fibrosis with chronic lung allograft dysfunction. Ann Am Thorac Soc (2015) 12(Suppl 1):S34–41. 10.1513/AnnalsATS.201410-476MG [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.