Summary
Precise control of gene expression during development is orchestrated by transcription factors and co-regulators including chromatin modifiers. How particular chromatin-modifying enzymes affect specific developmental processes is not well defined. Here, we report that GCN5, a histone acetyltransferase essential for embryonic development, is required for proper expression of multiple genes encoding components of the fibroblast growth factor (FGF) signaling pathway in early embryoid bodies (EBs). Gcn5−/− EBs display deficient activation of ERK and p38, mislocalization of cytoskeletal components, and compromised capacity to differentiate toward mesodermal lineage. Genomic analyses identified seven genes as putative direct targets of GCN5 during early differentiation, four of which are cMYC targets. These findings established a link between GCN5 and the FGF signaling pathway and highlighted specific GCN5-MYC partnerships in gene regulation during early differentiation.
Keywords: GCN5, MYC, FGF signaling, embryoid body, chromatin, histone H3, acetylation
Highlights
-
•
Gcn5 null embryoid bodies (EBs) display cytoskeletal abnormalities
-
•
Gcn5 null EBs are defective in mesoderm differentiation
-
•
Gcn5 loss affects fibroblast growth factor (FGF) signaling at multiple levels
-
•
Gcn5 loss impedes transcription of cMYC target genes associated with FGF signaling
Wang and colleagues show that the GCN5 lysine acetyltransferase regulates FGF signaling at multiple levels in early ESC differentiation and is essential for mesodermal lineage formation in vitro. Gcn5 loss leads to downregulation of specific genes involved in signaling and metabolism in an H3K9ac-dependent manner, including discrete MYC gene targets.
Introduction
Embryonic stem cells (ESCs) are derived from the inner cell mass (ICM) of blastocysts, which are early-stage preimplantation embryos. ESCs have the ability to remain pluripotent and self-renew or to differentiate into multiple cell lineages. ESC identity and subsequent differentiation are controlled by intricate networks of transcription factors and signaling pathways that drive precise gene expression programs. Diverse chromatin regulators play important roles in these networks (Chen and Dent, 2014, Lessard and Crabtree, 2010), but the roles of specific histone modifying enzymes in ESC self-renewal or lineage specification are poorly understood.
GCN5 was the first histone lysine acetyltransferase (HAT) to be linked to active gene transcription (Brownell et al., 1996). GCN5 functions within multimember protein complexes, including SAGA and ATAC in multicellular organisms, to coactivate transcription (Baker and Grant, 2007, Koutelou et al., 2010, Timmers and Tora, 2005). In yeast, Gcn5-containing complexes are recruited to target genes via interactions with specific DNA-binding factors, but only a few such partners, such as cMYC and nMYC, have been defined in mammalian cells (Hirsch et al., 2015, Martinez-Cerdeno et al., 2012, Zhang et al., 2008a). SAGA has also been suggested to act as a general transcription factor in yeast, widely enhancing expression of active genes (Baptista et al., 2017, Bonnet et al., 2014).
Genetic studies in mice revealed that both GCN5 and its catalytic activity are essential for normal development and embryo survival. Gcn5−/− embryos die soon after gastrulation and exhibit increased apoptosis in mesodermal lineages (Xu et al., 2000). Gcn5 catalytic mutant mice survive until mid-gestation but develop cranial neural tube closure defects (Bu et al., 2007) due to abnormal retinoic acid signaling involving a nonhistone substrate of GCN5 (Wilde et al., 2017). These findings indicate that GCN5-containing complexes have both HAT-dependent and -independent functions in early development. The phenotypes of Gcn5 mutant mice also support a selective role for this HAT in gene regulation, as loss of general transcription factors often leads to early lethality prior to embryo implantation (Tudor et al., 1999).
Our previous studies defined GCN5 as an important coactivator for MYC and E2F family transcription factors in the regulation of cell-cycle genes involved in ESC self-renewal (Hirsch et al., 2015) and pointed to the involvement of GCN5 in early ESC differentiation (Lin et al., 2007). However, early developmental processes and associated signaling pathways modulated by GCN5 have not yet been defined.
Fibroblast growth factor (FGF) signaling is required for multiple stages of early embryonic development, from segregation of trophectoderm and primitive endoderm from the ICM (Chazaud et al., 2006, Georgiades and Rossant, 2006, Kang et al., 2017, Yamanaka et al., 2010) to specification of primitive ectoderm and the primitive streak (Ciruna and Rossant, 2001), which in turn determine the fate of epiblast. ESC-based studies indicate that FGF signaling is involved in both pluripotency maintenance and lineage specification in vitro (Kunath et al., 2007, Lanner and Rossant, 2010, Ying et al., 2008). However, how epigenetic factors contribute to FGF-mediated gene regulation during early development is unclear.
Here, we use ESCs bearing a floxed allele of Gcn5 to define GCN5 functions in embryoid body (EB) differentiation. Morphological and molecular analyses of Gcn5fx/fx and Gcn5−/− EBs reveal an important role for GCN5 in the regulation of FGF signaling during early differentiation of EBs and confirm the importance of GCN5 for proper expression of select MYC target genes.
Results
Gcn5 Loss Leads to Epiblast Disorganization In Vitro
The early lethality of Gcn5−/− embryos poses challenges for detailed molecular studies, so we developed ESCs that carry a floxed allele of Gcn5 (Hirsch et al., 2015) to define pathways regulated by GCN5 during ESC differentiation in vitro. Before Cre-mediated recombination, the Gcn5 floxed allele behaves as wild-type, and mice homozygous for this allele (Gcn5fx/fx) show no overt phenotypes (Lin et al., 2008). After Cre exposure, exons 3–18 of Gcn5 are deleted, creating a null allele (Gcn5−/−).
ESCs readily aggregate when cultured in suspension without inhibitors of differentiation (leukemia inhibitory factor [LIF] and 2i) and undergo stepwise morphological changes to form distinct three-dimensional structures. Hallmark events of this process include visceral endoderm differentiation (VE) and basement membrane (BM) assembly (days 2–3), followed by polarized epiblast formation from the inner cells and the clearance of a central cavity (days 4–6) (Li et al., 2003). These sequential events recapitulate transitions from formation of the ICM through embryonic gastrulation, thereby providing opportunities to define molecular events in vitro that might contribute to the death of Gcn5−/− embryos shortly after gastrulation in vivo (Xu et al., 2000).
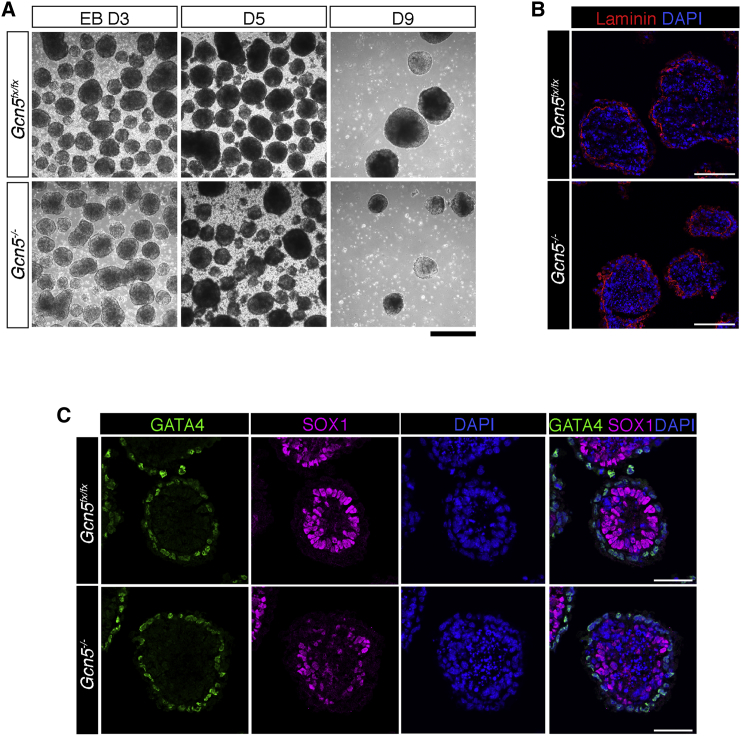
We initially observed that Gcn5−/− EBs were much smaller than Gcn5fx/fx EBs at day 9, when all three germ layers should be formed (Figure 1A). However, at day 3, Gcn5−/− EBs were not obviously different from controls. By day 5, Gcn5−/− EBs began to suffer more breakage than the control EBs (Figure 1A), suggesting that null EBs were structurally fragile or that they were in the process of dying. However, cleaved Lamin A levels were not increased in day 5 Gcn5−/− EBs (Figure S1A), indicating that Gcn5 loss did not induce apoptosis. Immunoblots of H3S10p (Figure S1A), a marker for mitotic cells, also indicated that deletion of Gcn5 did not inhibit cell proliferation. Global levels of H3K9ac were comparable in Gcn5fx/fx and Gcn5−/− EBs at day 5 (Figure S1B), consistent with the presence of other HATs (e.g., PCAF) in the EBs. Together these data indicate that GCN5 is required for normal morphology, but not proliferation or survival, in early stages of EB differentiation.
Figure 1.
Loss of Gcn5 Leads to Disorganization of Epiblast Cells during Early Differentiation
(A) Bright-field images of differentiating EBs at days 3, 5, and 9, comparing EB morphology between Gcn5fx/fx and Gcn5−/− EBs. Scale bar, 200 μM.
(B and C) Representative immunofluorescence/confocal images of EB architecture in day 5 Gcn5fx/fx and Gcn5−/− EBs, with (B) Laminin (red) staining for BM and (C) GATA4 (green) staining for VE, and SOX1 (magenta) and DAPI (blue) staining for nuclei of the epiblast. Scale bars, 100 μM in (B) and 50 μM in (C).
See also Figure S1.
To investigate whether the abnormal morphology of Gcn5−/− EBs reflects defective formation of VE or BM at day 5, we performed immunostaining of GATA4 (for VE) and Laminin (for BM). Since a majority of the inner cells express early neuroectodermal markers by day 5, we also used immunostaining of SOX1 to visualize the organization of epiblast cells. Laminin staining of the BM showed little difference between the null and control EBs (Figure 1B). GATA4 staining was also very similar in Gcn5fx/fx and Gcn5−/− EBs, indicating normal formation and organization of the VE (Figure 1C). In contrast, SOX1 staining revealed severe disorganization of the columnar epithelial morphology of the epiblast in Gcn5−/− EBs, indicating that GCN5 is required for normal epiblast formation and organization.
Compromised Differentiation Potential of Gcn5 Null EBs
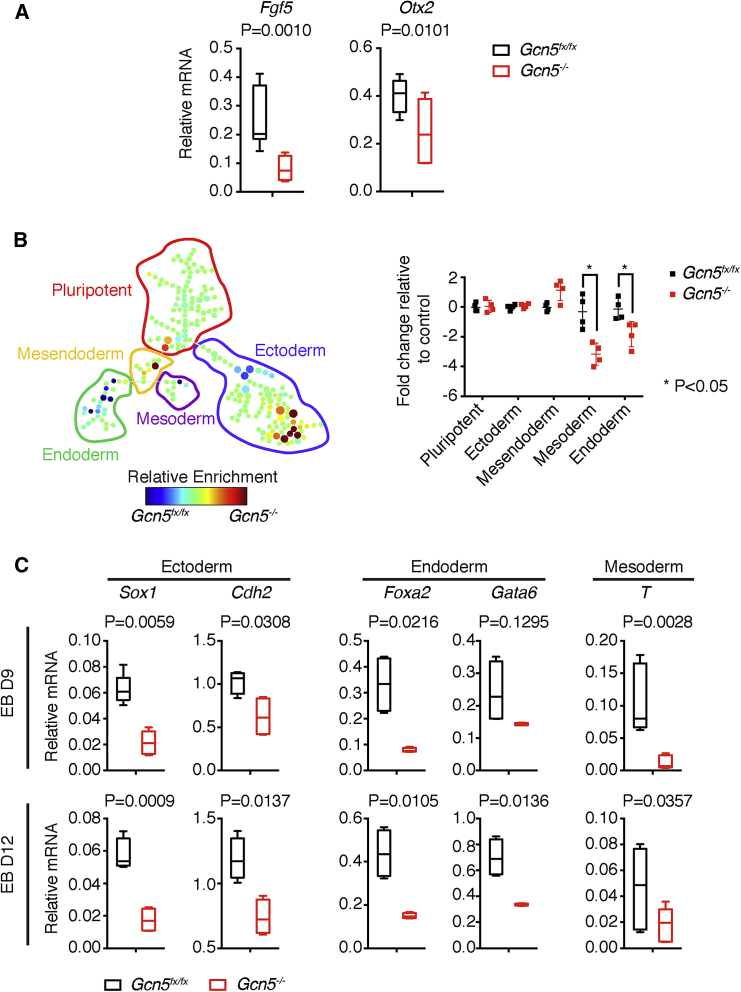
The epiblast gives rise to all three germ layers (Rivera-Perez and Hadjantonakis, 2014), so we asked whether Gcn5 loss affects lineage differentiation. We examined marker genes that normally exhibit peak expression in the embryonic epiblast, including Fgf5 and Otx2 (Kamiya et al., 2011, Kurokawa et al., 2004, Sumi et al., 2013, Yamanaka et al., 2010), and confirmed lower expression levels in the null epiblasts at day 5 (Figure 2A). We then used mass cytometry to more fully assess the composition of cell populations within the control and null EBs (Bendall and Nolan, 2012) (Figure S2 and Table S1). Day 5 EBs were dissociated and stained with antibodies specific for markers for either pluripotent cells or germ layer progenitors (Table S1). The antibodies were labeled with nonradioactive isotopes of rare earth metals with distinct atomic masses, which are distinguished by mass cytometry and serve as reporters for the labeled cells. This approach allowed us to delineate multiple lineages simultaneously and compare the compositions of heterogeneous populations in day 5 EBs. SPADE (spanning-tree progression analysis of density-normalized events) analysis was used to visualize and categorize populations based on combinatorial marker expression (Table S1) and to quantify the mass cytometry data. The results indicated that Gcn5−/− EBs contained significantly lower numbers of endodermal and mesodermal cells relative to Gcn5fx/fx EBs, whereas comparable numbers of cells were observed for pluripotent populations (Figures 2B, S3A, and S3B). Decreased mesoderm populations in the Gcn5−/− EBs were confirmed across four replicate experiments (Figures S3A and S3B). The ectoderm population in the null EBs was unchanged in cell number (Figure 2B, right), although the ectoderm region in the SPADE tree showed changes in some nodes (Figure 2B, left), likely due to specificity limitations of the antibodies used to define ectoderm (Figure S2). Decreased expression of all germ layer markers was observed in late-stage Gcn5−/− EBs (days 9 and 12) (Figure 2C), likely stemming from the earlier defects observed at day 5.
Figure 2.
Compromised Differentiation Potentials of Gcn5 Null EBs
(A) Gene expression analysis by quantitative real-time PCR for epiblast marker genes.
(B) Changes in population composition in Gcn5 null EBs at epiblast stage. Left: SPADE tree plot showing decreased endoderm and mesoderm populations in the Gcn5 null EBs at day 5; Right: quantitation of fold changes in cell numbers of a given population in Gcn5 null EBs relative to control.
(C) Quantitative real-time PCR plots showing decreased expression of marker genes for ectoderm (Sox1, Cdh2), endoderm (Foxa2, Gata6), and mesoderm (T) in late-stage EBs (days 9 and 12).
Data are presented as means ± SD from three (A and C) or four (B) independent experiments.
See also Figures S2 and S3 and Table S1.
To further confirm the effects of Gcn5 loss on mesoderm and endoderm formation, we utilized a monolayer differentiation protocol to direct ESCs toward these lineages (Villegas et al., 2013). Again we observed significantly decreased expression of mesoderm-specific genes (T, Flk1, and Pdgfrb), but in contrast to the results in EBs, expression of endoderm-specific genes was unaffected (Gata4) or upregulated (Sox17 and FoxA2) in Gcn5−/− cells differentiated in monolayer (Figure S3C). Altogether these results indicate that GCN5 is most important for mesoderm formation during ESC differentiation, reminiscent of our previous findings in Gcn5−/− embryos (Xu et al., 2000).
Expression Profiling Reveals a Regulatory Role for GCN5 in FGF Signaling
GCN5 acts as a transcriptional coactivator in the context of the SAGA and ATAC complexes (Koutelou et al., 2010, Spedale et al., 2012, Suganuma et al., 2008). To better understand the molecular basis underlying the defects caused by Gcn5 loss, we performed RNA sequencing (RNA-seq) to compare gene expression profiles in day 3 and day 5 control and null EBs. Total RNA from three technical replicates of Gcn5fx/fx and Gcn5−/− EBs were sequenced, and key gene expression changes were confirmed using a second biological replicate (EBs generated from a separate matched pair of Gcn5fx/fx and Gcn5−/− ESCs) by quantitative real-time polymerase chain reaction (PCR). These time points were chosen to define both early events (day 3) and events that coincide with the onset of the abnormal phenotype of Gcn5−/− EBs (day 5).
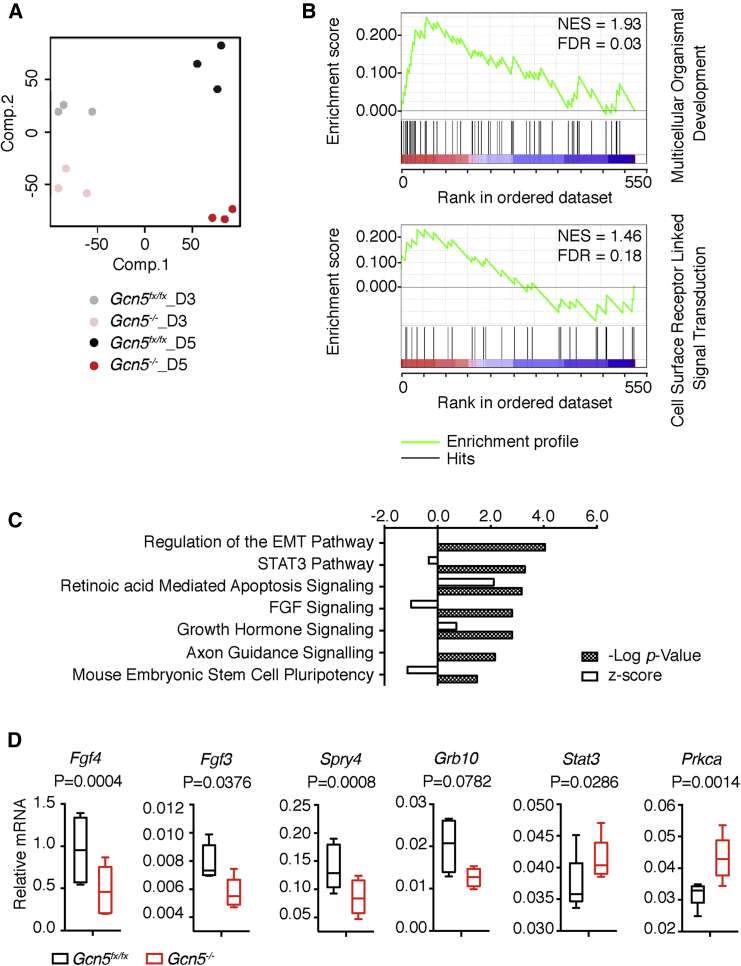
Principal component analysis revealed significant differences in gene expression profiles between day 3 and day 5 EBs, consistent with developmental progression over time. Of note, differences in the gene expression profiles between null and control EBs were less pronounced at day 3 than at day 5 (Figure 3A), with 754 genes exhibiting altered expression in day 5 null EBs, whereas 158 genes were affected in day 3 nulls (Figure S4A). These data indicate that Gcn5 loss more significantly affects gene expression programs at day 5, consistent with the timing of the onset of Gcn5 null morphological phenotype. Therefore, we focused further detailed analyses on differences in gene expression observed at day 5.
Figure 3.
Expression Profiling Points to a Regulatory Role of Gcn5 in FGF Signaling in Day 5 EBs
(A) Principal component analysis plots showing variance of expression profiles among replicative samples and increased differences in profiles between Gcn5fx/fx and Gcn5−/− EBs at day 5 compared with day 3 (n = 3).
(B) Example gene set enrichment analysis enrichment plots showing top processes enriched in the control EBs compared with Gcn5−/− EBs at day 5. Color code: red, positively correlated with Gcn5fx/fx; blue, negatively correlated with Gcn5−/−.
(C) Significantly altered pathways in the day 5 Gcn5 null EBs identified by IPA canonical pathway analysis.
(D) Quantitative real-time PCR validating the key genes in the FGF signaling identified by the RNA-seq using a second biological sample. Data are presented as means ± SD from three independent experiments.
FDR, false discovery rate; NES, normalized enrichment score. See also Figure S4, Tables S2 and S3.
Gene set enrichment analysis revealed significant downregulation of several biological processes in the null EBs at day 5, with multicellular organismal development (MOD) and cell surface receptor linked signal transduction (CSRLST) among the most affected (Figure 3B). In the MOD category, 15 of 50 genes were among the core enrichment group in the null EBs, including Pax5, Msx2, Gli1, Spry2, and Mest, which are all important for early development or ESC differentiation (Lee et al., 2016, Szabo et al., 2009, Tefft et al., 1999, Urbanek et al., 1997, Wu et al., 2015) (Figure S4B and Table S2). In the CSRLST category, 7 of 30 genes were in the core enrichment group, including Grb10, the most downregulated gene in the null EBs (Figure S4C and Table S3). Consistent with these results, Ingenuity Pathway Analysis (IPA) identified a number of signaling pathways to be significantly altered in the null EBs. Interestingly, four of the seven top-ranked affected pathways were intimately linked to FGF signaling, including regulation of epithelial-to-mesenchymal transition, STAT3, FGF, and growth hormone signaling pathways (Figure 3C).
FGF ligands and their receptor tyrosine kinases control multiple developmental processes, including cell proliferation, survival, differentiation, and migration (Brewer et al., 2016). We confirmed that expression of key genes in the FGF pathway, including Fgf3, Fgf4, and Spry4, was altered in the Gcn5−/− EBs using quantitative real-time PCR. We also confirmed that Grb10, an effector of insulin signaling (Desbuquois et al., 2013, Yu et al., 2011), was strongly downregulated. Conversely, effector genes such as Stat3 and Prkca were upregulated (Figure 3D).
Connections between Defective FGF Signaling and Morphological Abnormalities of Gcn5−/− EBs at Day 5
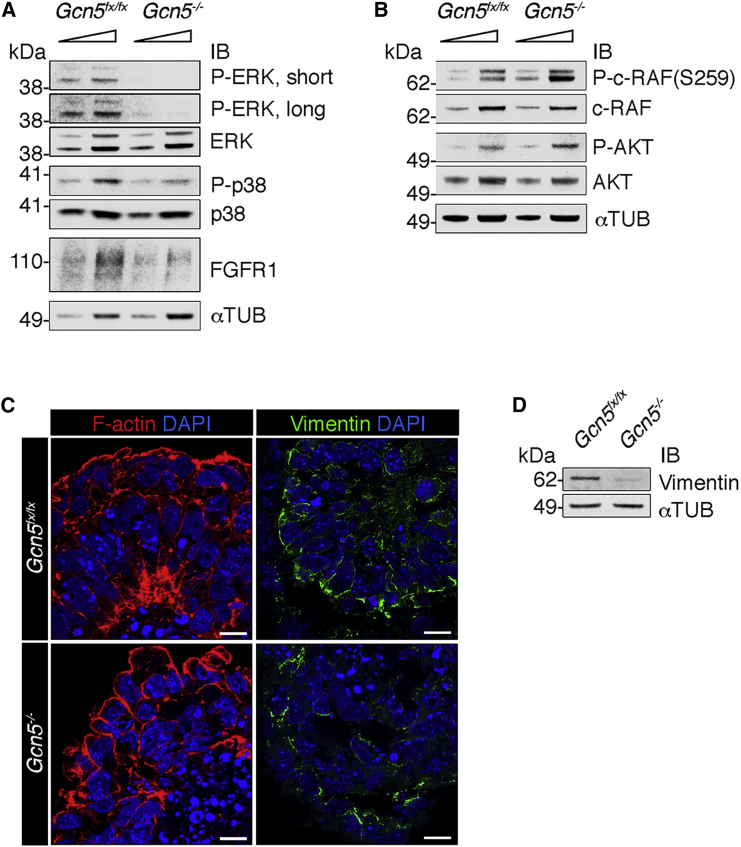
To link the above changes in gene expression to the status of FGF signaling intermediates in Gcn5−/− EBs, we examined levels of FGFR1 and phosphorylation of ERK, p38, c-RAF, and AKT by immunoblotting (Figures 4A and 4B). FGFR1 protein levels and activated, phosphorylated forms of both ERK and p38 (P-ERK and P-p38, respectively) were notably decreased in Gcn5−/− EBs, indicating deficient activation of the RAS/MAPK pathway. In contrast, levels of activated AKT (P-AKT) and P-c-RAF-Ser259 were unchanged or slightly increased. Levels of phospholipase C-γ did not change (data not shown), even though Prkca mRNA was upregulated in the null EBs.
Figure 4.
Abnormal Activities of the FGF Signaling Pathway in Day 5 Gcn5−/− EBs
(A) Representative immunoblots showing decreased phosphorylated forms of ERK and p38 in the Gcn5 null EBs at day 5.
(B) Representative immunoblots showing the AKT pathway is affected to a lesser degree in the null EBs at day 5.
(C) Zeiss LSM 880 confocal Airyscan images showing disorganization of F-actin (red) and vimentin (green) in day 5 null EBs. Scale bars, 10 μM.
(D) Representative immunoblots showing decreased vimentin in Gcn5−/− EBs at day 5.
To relate these molecular changes back to the morphological phenotypes of the Gcn5−/− EBs, we examined the organization of cytoskeletal components regulated by ERK and p38 signaling (Huang et al., 2004) using Airyscan (Zeiss) laser-scanning confocal microscopy. We first assessed filamentous actin (F-actin) in day 5 EBs using phalloidin staining. In epiblasts of EBs or early embryonic epithelial structures, F-actin localizes to the periphery of epithelial cells and is particularly enriched at apical sites of columnar epithelial cells (Loebel et al., 2011, Sakai et al., 2003). This staining pattern was observed as expected in Gcn5fx/fx EBs. In contrast, Gcn5−/− EBs displayed reduced apical distribution of F-actin, primarily in the inner cells (epiblast) (Figure 4C). Airyscan images of vimentin staining also indicated altered localization and decreased staining intensity of this intermediate filament protein in Gcn5−/− EBs (Figure 4C). Depletion of vimentin was further confirmed by immunoblotting (Figure 4D). These findings suggest that insufficient activation of ERK and p38 upon Gcn5 loss affects F-actin and vimentin organization during early differentiation, in concordance with disorganized epithelial architectures of the inner cell layer observed in the null EBs (Figure 1C). Defective ERK signaling is also consistent with decreased mesoderm differentiation (Binetruy et al., 2007) in Gcn5−/− EBs (Figure 2) and embryos (Xu et al., 2000).
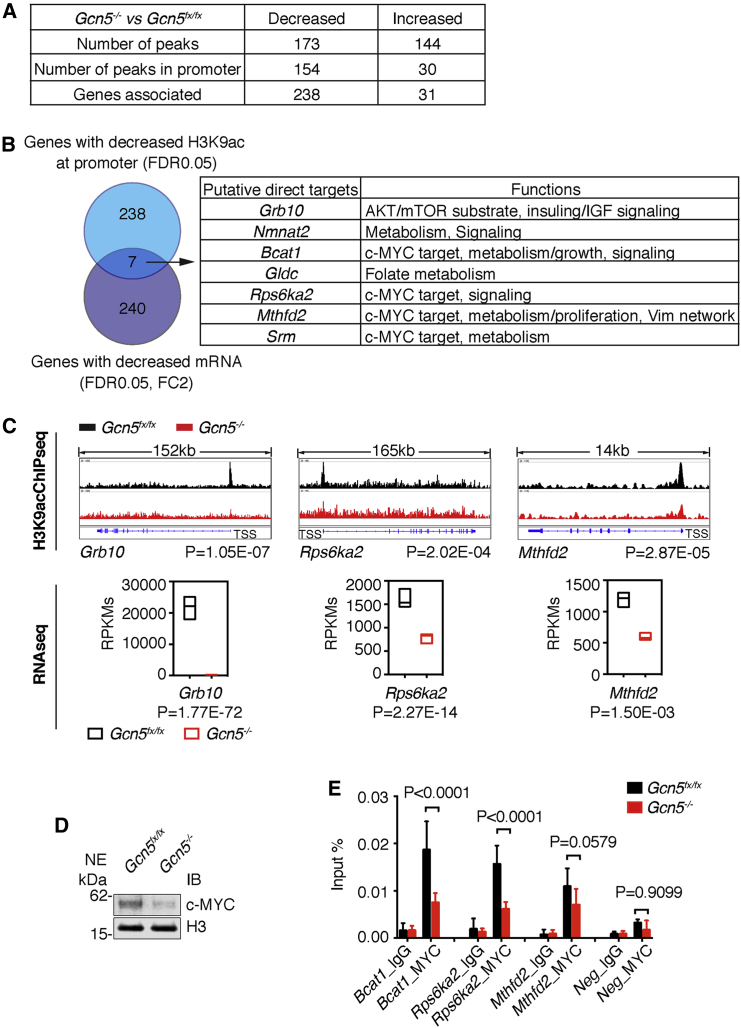
Decreased Gene Expression and Reduced H3K9ac at Gene Promoters Identified Likely Direct Targets of GCN5
To identify genes directly regulated by GCN5 in EBs at the epiblast stage, we attempted chromatin immunoprecipitation sequencing (ChIP-seq), but we were not successful using either commercially available antibodies for GCN5 or a biotin-tagging system (Hirsch et al., 2015) in differentiating ESCs. Since histone H3 lysine 9 (H3K9) is a well-characterized substrate for GCN5 (Jin et al., 2011, Kuo et al., 1996), we performed H3K9ac ChIP, using the same control and Gcn5−/− EBs used for RNA-seq. We reasoned that localized decreases in H3K9ac in the Gcn5−/− EBs might identify genes that uniquely require the presence of this HAT for proper regulation. As global levels of H3K9ac were not affected by Gcn5 loss (Figure S1B), regions identified by this approach will likely provide an underestimate of GCN5 targets due to redundancies with PCAF and possibly other HATs.
We identified 173 H3K9ac peaks that were decreased in the Gcn5−/− EBs, the majority (154) of which were located near gene promoters. Gene annotation identified 238 genes likely driven by these promoters, with occasions where H3K9ac peaks fell in putative promoter regions associated with more than one gene. We also uncovered 144 H3K9ac peaks that increased upon Gcn5 loss, yet only 30 of those peaks were near the promoters, associated with 31 genes. These data are consistent with the gene-specific coactivator role of GCN5 in gene transcription (Figure 5A). Comparison of genes identified in our analyses as having decreased H3K9ac promoter peaks with ENCODE ChIP-seq data (Auerbach et al., 2013) identified candidate transcription factors that might recruit GCN5 to these regions (Table S4). Top TFs identified by this approach include HCFC1 (Q value 1.25 × 10−77), a nuclear protein known to associate with GCN5-containing complexes (Wang et al., 2008), as well as TBP (Q value 1.69 × 10−59) and CTCF (Q value 3.77 × 10−59). Strikingly, a number of MYC family members, including MAX, MXI1, and cMYC, were also identified by this approach, consistent with our previous work connecting GCN5 to MYC functions in both ESCs and during somatic cell reprogramming.
Figure 5.
Identification of Direct GCN5 Target Genes at the Early Stage of Differentiation
(A) Comparing profiles of H3K9ac peaks and associated genes in Gcn5fx/fx and Gcn5−/− EBs at day 5. Loss of Gcn5 resulted in decreased H3K9ac peaks mostly in the promoter region. Most of the genes (238 of 269) with altered H3K9ac peaks exhibited a decreased level of this mark.
(B) Putative target genes directly regulated by GCN5. Venn diagram showing overlap between H3K9ac decreased genes and GCN5 induced genes. Overlapping genes are listed in the table with a general description of the reported functions.
(C) Examples of H3K9ac peak profiles (n = 4) and RNA transcripts (n = 3) for top targets of GCN5 in day 5 EBs. RNA transcript levels are presented as means ± SD.
(D) Representative immunoblots showing reduced levels of cMYC isolated from nuclei of day 5 Gcn5−/− EBs. NE, nuclear extracts.
(E) ChIPs for MYC reveal decreased MYC binding at the promoters of GCN5 target genes in day 5 Gcn5−/−EBs. Enrichment of MYC relative to input at each locus is presented as the mean ± SD from three independent experiments.
See also Table S4.
To better determine which of these regions might reflect genes directly activated by GCN5, we compared genes with decreased H3K9ac at their promoters with genes identified as downregulated more than 2-fold by RNA-seq. Only seven genes were both downregulated and decreased in H3K9ac. Three of these genes, Grb10, Gldc, and Nmnat, further reinforce the link among GCN5, metabolism, and signaling (Figures 5B and 5C). The other four genes (Rps6ka2, Mthfd2, Bcat1, and Srm) are directly regulated by cMYC (Ben-Yosef et al., 1998, Pikman et al., 2016, Snezhkina et al., 2016) (Figure 5B). Myc is induced by most mitogenic factors, including FGFs (Grandori et al., 2000). We observed decreased cMYC protein levels upon loss of Gcn5 (Figure 5D), consistent with a previous report in a different cellular system (Patel et al., 2004). ChIP-qPCR confirmed MYC recruitment to the promoter regions of Rps6ka2, Mthfd2, and Bcat1 in Gcn5fx/fx EBs and loss of MYC from these regions in Gcn5−/− EBs at day 5 (Figures 5C and 5E). These findings further highlight the GCN5-MYC partnership in gene regulation during early differentiation of EBs.
Discussion
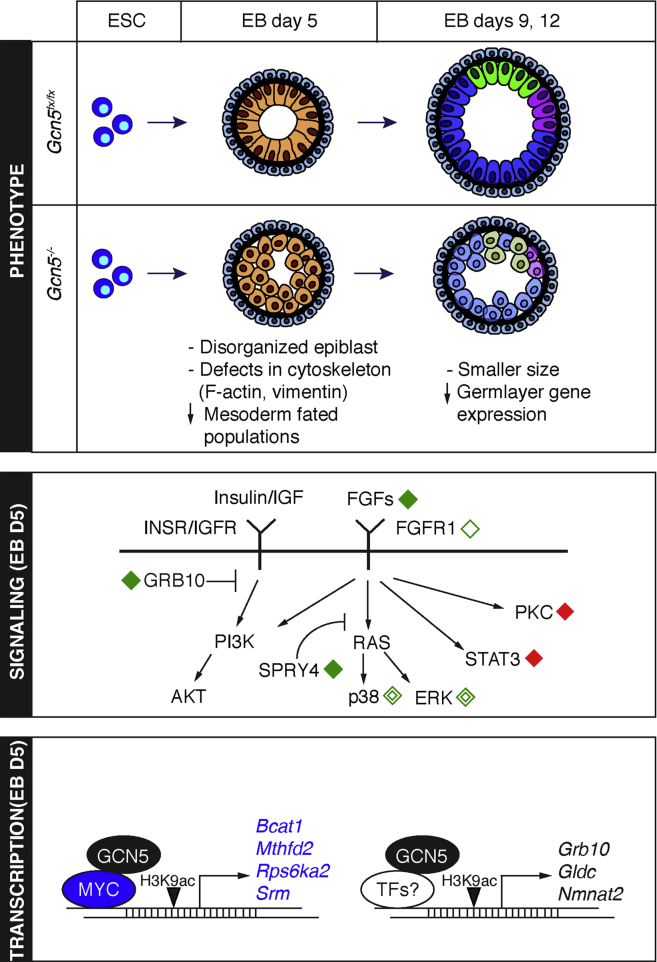
Our findings reveal that loss of Gcn5 strongly affects FGF signaling at multiple levels during early differentiation of EBs, with decreased expression of Fgfs and FGFR1 and deficient activation of ERK and p38. Moreover, Gcn5 loss leads to direct downregulation of specific genes involved in signaling and metabolism as well as discrete MYC gene targets (Figure 6).
Figure 6.
GCN5 Affects Multiple Components of the FGF Signaling Pathway and Activates Selective Targets during Early Differentiation
Top: the abnormal phenotype of Gcn5−/− EBs becomes evident early during EB differentiation (day 5). Loss of Gcn5 leads to disorganization of the epiblast architecture that is associated with defective cytoskeleton networks (F-actin and vimentin) and decreases in progenitors fated for mesoderm. At later stages of differentiation (days 9 and 12), Gcn5−/− EBs are smaller in size and express lower levels of marker genes for ectoderm (blue), endoderm (green), and mesoderm (magenta), compared with the controls. Lighter shades indicate decreased expression levels of marker genes for each population. Middle: at day 5, GCN5 affects expression of multiple genes encoding critical components in FGF signaling and for proper activation of ERK and p38 pathways. Solid diamond, up (red) or down (green) regulated transcripts; open diamond (green), decreased protein expression; double open diamond (green), decreased protein phosphorylation. Bottom: at day 5, GCN5 is required for activation of genes important for signaling through promoter-associated H3K9ac, including four cMYC targets (blue).
FGF signaling regulates both migration and patterning of mesoderm during gastrulation (Ciruna and Rossant, 2001). Failures in the execution of this pathway are consistent with the abnormal cytoskeletal organization and defective mesoderm formation observed in Gcn5−/− EBs. In addition, identification of Bcat1, Rps6ka2, Mthfd2, and Srm as likely direct targets of GCN5, which exhibit decreased expression upon Gcn5 loss along with reduced MYC binding and diminished H3K9ac at their promoters, provides further evidence that GCN5 is an important coactivator for MYC during early differentiation. These genes are known to be immediate-response, MYC target genes induced by FGF signaling.
Another notable common function of the direct targets of GCN5 identified here is that they regulate metabolism and growth downstream of AKT and/or ERK signaling, either positively (Nmnat2, Bcat1, Gldc, Mthfd2, and Srm) or negatively (Grb10, Bcat1, and Rps6ka2) (Figure 5B) (Gerdts et al., 2016, Pai et al., 2015, Pikman et al., 2016, Serra et al., 2013, She et al., 2007, Shi et al., 2012, Yu et al., 2011). Altered carbon metabolism induced by these gene expression changes could further derail differentiation (Garcia-Prat et al., 2017, Hu et al., 2016).
In addition to histones, GCN5 acetylates nonhistone proteins that may indirectly regulate gene transcription or function outside of gene regulation (Conacci-Sorrell et al., 2010, Jin et al., 2014, Wilde et al., 2017). For example, GCN5 pairs with MYC-nick, a CALPAIN-cleaved cytoplasmic derivative of MYC, to acetylate alpha-tubulin (ac-αTUB) and to regulate cytoskeleton organization and differentiation of myoblasts (Conacci-Sorrell et al., 2010). We found that total ac-αTUB levels in Gcn5 null EBs at day 5 were unchanged (data not shown), but we cannot exclude contributions of changes in acetylation of other targets to the developmental phenotypes we observe.
Our previous genetic studies showed that Gcn5−/−;Pcaf−/− embryos die earlier than Gcn5−/− embryos, even though Pcaf deletion on its own causes no abnormal phenotype (Xu et al., 2000, Yamauchi et al., 2000). These findings indicate that Gcn5 and Pcaf have shared functions during early development. The lack of global changes in H3K9ac and limited changes in H3K9ac at gene promoters observed in day 5 Gcn5−/− EBs is consistent with functional redundancy with PCAF and possibly other HATs. Nonetheless, compensatory H3K9ac by PCAF did not prevent defective morphology and signaling in the early Gcn5−/− EBs, suggesting that GCN5 and H3K9ac are uniquely required for regulation of specific genes during early differentiation.
GCN5 is most active in vivo when incorporated into SAGA and ATAC in mammals (Koutelou et al., 2010, Wang et al., 2008). Our studies do not differentiate the functions of the two complexes, and the phenotypes we observe may reflect loss of activity of both. However, knockout of Atac2, a component of ATAC but not SAGA, did not cause defects in lineage differentiation (Suganuma et al., 2008), as was observed in Gcn5−/− embryos, suggesting SAGA may be most important for these early developmental events.
Interestingly, more genes were upregulated upon loss of Gcn5 (at both days 3 and 5) than were downregulated, in contrast to the role of GCN5 as a coactivator of transcription. Many of these events are likely indirect effects of Gcn5 loss, although we cannot exclude the possibility that GCN5 may be involved in gene repression during early differentiation. Indeed, a recent study revealed that another HAT, TIP60, acts as a transcriptional repressor in ESCs (Fazzio et al., 2008). Notably, many of the genes upregulated upon Gcn5 loss are involved in acute phase response and interferon signaling (data not shown), consistent with our previous work in fibroblasts that indicated GCN5 and PCAF repress interferon-β expression by targeting a nonhistone substrate, TBK1 (Jin et al., 2014).
Abnormal regulation of growth-factor-driven pathways drive oncogenesis (Giancotti, 2014). Our findings here suggest that GCN5 may also be important in cancers associated with deregulation of FGFs. Future work will explore this possibility, as well as the therapeutic potential of targeting GCN5 to inhibit growth or progression of these cancers.
Experimental Procedures
ES Cell Culture and Differentiation
Gcn5fx/fx and Gcn5−/− ESC lines were generated and characterized previously (Hirsch et al., 2015). ESCs were routinely grown on gelatin-coated plates in Dulbecco’s modified Eagle’s medium (DMEM)/high glucose (HyClone, SH3002201) medium supplemented with 15% (v/v) ESC-screened fetal bovine serum (HyClone, SH3007003E), 2 mM L-glutamine (HyClone, SH3003401), 0.1 mM nonessential amino acids (Corning, MT25025CI), 1% (v/v) penicillin/streptomycin (HyClone, SV30010), 0.1 mM β-mercaptoethanol (BME) (Fisher, 03446I-100), 1000 U/mL LIF/ESGRO (Millipore, ESG1107), 1 μM PD0325901 (Sigma, PZ0162) and 3 μM 1-azakenpaullone (Sigma, A3734), and passaged every 2–3 days.
For EB differentiation, 3 × 105 cells/well were plated in ultra-low attachment six-well plates in differentiation medium without LIF or 2i-s. Media were replaced every other day by settling the EBs at low speed centrifugation (100 × g for 1 min). The differentiation medium was DMEM/high glucose:F12 (Cellgro, MT10080CV):neurobasal medium (Gibco, 21103049) (1:1:2) supplemented with 10% KnockOut Serum Replacement medium (Gibco, A3181502), 2 mM L-glutamine, 1% (v/v) penicillin/streptomycin, and 0.1 mM BME.
Immunofluorescence and Confocal Imaging
EBs were washed once in PBS/1% BSA and fixed in 4% paraformaldehyde (PFA) for 30 min at room temperature. The fixed EBs were incubated in 7.5% sucrose/PBS for at least 1 hr at room temperature, then in 15% sucrose/PBS at 4°C overnight. The EBs were then embedded in Tissue-Tek OCT compound (Electron Microscopy Sciences, 62550-12) and incubated for 10 min at room temperature with agitation before they were frozen in liquid nitrogen (LN2). Frozen sections (8 μM) were fixed in 2% PFA for 2 min then blocked with PBS containing 0.1% Tween 20 (Fisher, BP337-500) (PBT) and 5% normal donkey serum (Millipore, S30-100ML) for 30 min at room temperature. The blocked sections were incubated with primary antibodies diluted in blocking buffer overnight at 4°C. The slides were washed with PBT three times for a total 15 min, followed by incubations with fluorescence-conjugated secondary antibodies for 40 min at room temperature. DAPI staining was performed after washing off the secondary antibodies. The slides then were washed with PBT and mounted with coverslips using ProLong Gold Antifade mounting medium (Invitrogen, P36930). The slides were imaged on a Zeiss LSM 880 laser-scanning microscope. Airyscan detector array was used to image the cytoskeletons. Standard pinhole was used to image the markers for lineages, proliferation, and apoptosis. The antibodies used are listed in the Supplemental Experimental Procedures.
Expression Analysis
EBs were harvested and total RNA was isolated using an RNeasy mini plus kit (QIAGEN, 74134) following the manufacturer's instructions. Total RNA (10–20 ng) was used per reaction, and quantitative real-time PCRs were performed on a 7500 Fast Real-Time PCR System (Applied Biosystems, 4351107) using the Power SYBR Green RNA-to-CT1-Step Kit (Life Technologies, 4389986). Three technical replicates were performed for each gene target tested and Pbgd was used as a reference gene for quantification. A two-tailed Student’s t test was used for pairwise comparisons. Primers used are listed in Supplemental Experimental Procedures.
RNA Sequencing
Total RNA libraries were prepared from three independent experiments using the Illumina TruSeq stranded total RNA kit according to the manufacturer's protocol, except that the PCR amplification was reduced to eight cycles. Each library (10 pM) was sequenced on an Illumina HiSeq 2500. The reads were mapped to the mouse genome (mm10) by TopHat (version 2.0.10) (Kim et al., 2013) with an overall mapping rate of 84%–94%. DESeq (Anders and Huber, 2010) was used for differential gene expression analysis. Details of RNA-seq analysis are described in Supplemental Experimental Procedures.
ChIP, qPCR, and Deep Sequencing
Day 5 EBs (Gcn5fx/fx and Gcn5−/−) were washed once with PBS and dissociated using Accutase cell dissociation reagent (Gibco, A1110501). ChIPs were performed as previously described (Wen et al., 2014) with modifications in chromatin sonication. Details are included in Supplemental Experimental Procedures.
qPCRs were performed on a 7500 Fast Real-Time PCR System using ChIP DNA from three replicative experiments and Power SYBR Green PCR Master Mix (Life Technologies, 4367659). Student’s t test was used for pairwise comparisons. Primers used are listed in Supplemental Experimental Procedures.
ChIP DNA from four independent experiments, including two replicates for each isolate pair (Gcn5fx/fx and Gcn5−/−), were used for deep sequencing. Detailed ChIP library preparations are described in Supplemental Experimental Procedures. Each library (10 pM) was sequenced on an Illumina HiSeq 3000. Raw reads were aligned to mouse genome mm10 using bowtie (version 1.1.2) (Langmead et al., 2009). H3K9ac peaks were called by MACS (version 1.4.2) (Zhang et al., 2008b) using H3 as control. Detailed ChIP-seq data analysis is included in Supplemental Experimental Procedures.
Mass Cytometry and Data Analysis
Sample preparation for mass cytometry was performed as previously described (McCarthy et al., 2017a). Briefly, EBs were dissociated using Accutase, stained with cisplatin for viability at 25 μM for 1 min at room temperature, then quenched using PBS containing 1% BSA. The cells were fixed in 1.5% PFA for 10 min at room temperature, then permeabilized with methanol (1 mL per million cells) and incubated overnight at 4°C. Samples were barcoded (McCarthy et al., 2017b), pooled, and immunostained with the panel of antibodies shown in Supplemental Experimental Procedures. Cells were stained with 1:2000191/193 iridium (Ir) DNA intercalator (Fluidigm), 62.5 nM final, for 10 min at room temperature. The samples were combined with EQ Four Element Calibration Beads (Fluidigm) then diluted with water to a concentration of 5 × 105 cells/mL and run at 45 μL/min on a CyTOF 2 mass cytometer (Fluidigm). Data were normalized on bead passport using CyTOF software (v6.0.626; Fluidigm).
Initial data processing and gating was done with FlowJo vX10.0. EQ Four Element Calibration Beads were removed, and data were gated on singlets by Ir193 and Event Length parameters. Removal of dead cells was done in the Pt198 channel. SPADE analysis of the data was performed using SPADE V3.0 (Qiu et al., 2011) in MATLAB r2015b (Mathworks). SPADE tree construction was performed using agglomerative clustering on all markers listed in Table S1. Annotation of SPADE tree regions was done according to marker distribution as shown in Figure S2, and cell percentages in each region were calculated for all samples. Percentages were normalized relative to mean Gcn5fx/fx, and statistical significance was determined by the Wilcoxon rank-sum test performed in MATLAB using the ranksum function.
Immunoblotting
Whole-cell lysates (WCL) were prepared from D5 EBs using RIPA buffer. Nuclear extracts (NEs) were prepared following the Dignam-Roeder protocol (Dignam et al., 1983). All buffers contained protease inhibitor cocktail (Sigma, P8340) and phosphatase inhibitor cocktail (Roche, 04906845001). WCL or NE (10–20 μg) was resolved on 4%–12% Bis-Tris protein gels (WG1402BOX). Proteins were transferred to nitrocellulose membranes (Invitrogen, IB23001) using an iBlot2 (Life Technologies, IB21001), then blocked and incubated with primary antibodies following standard procedures. Primary antibodies were detected with peroxidase-conjugated secondary antibodies (1:8,000) and Amersham ECL prime western blotting detection reagent (GE Healthcare, RPN2232) following the manufacturer's instructions. The antibodies used are listed in the Supplemental Experimental Procedures.
Author Contributions
L.W., E.K., and S.Y.R.D. designed the study, analyzed the data, and wrote the paper. L.W. and E.K. planned all the molecular, cellular, and genomic studies, which were carried out by L.W.; C.H. provided the ESCs and performed some ChIP experiments; R.M. assisted with the mass cytometry experiments and performed data analysis; A.S. assisted with immunofluorescence/confocal experiments; K.L. and Y.L. performed bioinformatics analysis; C.J. and J.S. provided technical assistance for imaging and sequencing; M.C.B. supervised the cytometry experiments and provided expert advice; S.Y.R.D. supervised the overall research.
Acknowledgments
This work was largely supported by NIH R01 grant GM067718 to S.Y.R.D. The Science Park NGS Core performed the deep sequencing supported by CPRIT Core Facility Support Grants RP120348 and RP170002. Bioinformatics was made possible in part by support from the MDACC Center for Cancer Epigenetics (CCE). Immunofluorescence/confocal analysis was performed at the Science Park IF/imaging core, partially supported by CCE. We thank Andrew Salinger (A.S.) and Amanda Martin for critical material support in the lab, Hsueh-Ping Chao for help in bioinformatics, and Yoko Takata and Luis Coletta for help in NGS. We appreciate A.S. and Dr. Lisa Mustachio for helpful comments on the manuscript.
Published: December 14, 2017
Footnotes
Supplemental Information includes Supplemental Experimental Procedures, four figures, and four tables and can be found with this article online at https://doi.org/10.1016/j.stemcr.2017.11.009.
Accession Numbers
The accession number for the RNA-seq and H3K9ac ChIP-seq data reported in this paper is GEO: GSE104454.
Supplemental Information
References
- Anders S., Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auerbach R.K., Chen B., Butte A.J. Relating genes to function: identifying enriched transcription factors using the ENCODE ChIP-Seq significance tool. Bioinformatics. 2013;29:1922–1924. doi: 10.1093/bioinformatics/btt316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker S.P., Grant P.A. The SAGA continues: expanding the cellular role of a transcriptional co-activator complex. Oncogene. 2007;26:5329–5340. doi: 10.1038/sj.onc.1210603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baptista T., Grunberg S., Minoungou N., Koster M.J.E., Timmers H.T.M., Hahn S., Devys D., Tora L. SAGA is a general cofactor for RNA polymerase II transcription. Mol. Cell. 2017;68:130–143.e5. doi: 10.1016/j.molcel.2017.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Yosef T., Eden A., Benvenisty N. Characterization of murine BCAT genes: Bcat1, a c-Myc target, and its homolog, Bcat2. Mamm. Genome. 1998;9:595–597. doi: 10.1007/s003359900825. [DOI] [PubMed] [Google Scholar]
- Bendall S.C., Nolan G.P. From single cells to deep phenotypes in cancer. Nat. Biotechnol. 2012;30:639–647. doi: 10.1038/nbt.2283. [DOI] [PubMed] [Google Scholar]
- Binetruy B., Heasley L., Bost F., Caron L., Aouadi M. Concise review: regulation of embryonic stem cell lineage commitment by mitogen-activated protein kinases. Stem Cells. 2007;25:1090–1095. doi: 10.1634/stemcells.2006-0612. [DOI] [PubMed] [Google Scholar]
- Bonnet J., Wang C.Y., Baptista T., Vincent S.D., Hsiao W.C., Stierle M., Kao C.F., Tora L., Devys D. The SAGA coactivator complex acts on the whole transcribed genome and is required for RNA polymerase II transcription. Genes Dev. 2014;28:1999–2012. doi: 10.1101/gad.250225.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer J.R., Mazot P., Soriano P. Genetic insights into the mechanisms of Fgf signaling. Genes Dev. 2016;30:751–771. doi: 10.1101/gad.277137.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brownell J.E., Zhou J., Ranalli T., Kobayashi R., Edmondson D.G., Roth S.Y., Allis C.D. Tetrahymena histone acetyltransferase A: a homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell. 1996;84:843–851. doi: 10.1016/s0092-8674(00)81063-6. [DOI] [PubMed] [Google Scholar]
- Bu P., Evrard Y.A., Lozano G., Dent S.Y. Loss of Gcn5 acetyltransferase activity leads to neural tube closure defects and exencephaly in mouse embryos. Mol. Cell. Biol. 2007;27:3405–3416. doi: 10.1128/MCB.00066-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chazaud C., Yamanaka Y., Pawson T., Rossant J. Early lineage segregation between epiblast and primitive endoderm in mouse blastocysts through the Grb2-MAPK pathway. Dev. Cell. 2006;10:615–624. doi: 10.1016/j.devcel.2006.02.020. [DOI] [PubMed] [Google Scholar]
- Chen T., Dent S.Y. Chromatin modifiers and remodellers: regulators of cellular differentiation. Nat. Rev. Genet. 2014;15:93–106. doi: 10.1038/nrg3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciruna B., Rossant J. FGF signaling regulates mesoderm cell fate specification and morphogenetic movement at the primitive streak. Dev. Cell. 2001;1:37–49. doi: 10.1016/s1534-5807(01)00017-x. [DOI] [PubMed] [Google Scholar]
- Conacci-Sorrell M., Ngouenet C., Eisenman R.N. Myc-nick: a cytoplasmic cleavage product of Myc that promotes alpha-tubulin acetylation and cell differentiation. Cell. 2010;142:480–493. doi: 10.1016/j.cell.2010.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desbuquois B., Carre N., Burnol A.F. Regulation of insulin and type 1 insulin-like growth factor signaling and action by the Grb10/14 and SH2B1/B2 adaptor proteins. FEBS J. 2013;280:794–816. doi: 10.1111/febs.12080. [DOI] [PubMed] [Google Scholar]
- Dignam J.D., Lebovitz R.M., Roeder R.G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fazzio T.G., Huff J.T., Panning B. An RNAi screen of chromatin proteins identifies Tip60-p400 as a regulator of embryonic stem cell identity. Cell. 2008;134:162–174. doi: 10.1016/j.cell.2008.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Prat L., Sousa-Victor P., Munoz-Canoves P. Proteostatic and metabolic control of stemness. Cell Stem Cell. 2017;20:593–608. doi: 10.1016/j.stem.2017.04.011. [DOI] [PubMed] [Google Scholar]
- Georgiades P., Rossant J. Ets2 is necessary in trophoblast for normal embryonic anteroposterior axis development. Development. 2006;133:1059–1068. doi: 10.1242/dev.02277. [DOI] [PubMed] [Google Scholar]
- Gerdts J., Summers D.W., Milbrandt J., DiAntonio A. Axon self-destruction: new links among SARM1, MAPKs, and NAD+ metabolism. Neuron. 2016;89:449–460. doi: 10.1016/j.neuron.2015.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giancotti F.G. Deregulation of cell signaling in cancer. FEBS Lett. 2014;588:2558–2570. doi: 10.1016/j.febslet.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandori C., Cowley S.M., James L.P., Eisenman R.N. The Myc/Max/Mad network and the transcriptional control of cell behavior. Annu. Rev. Cell Dev. Biol. 2000;16:653–699. doi: 10.1146/annurev.cellbio.16.1.653. [DOI] [PubMed] [Google Scholar]
- Hirsch C.L., Coban Akdemir Z., Wang L., Jayakumaran G., Trcka D., Weiss A., Hernandez J.J., Pan Q., Han H., Xu X. Myc and SAGA rewire an alternative splicing network during early somatic cell reprogramming. Genes Dev. 2015;29:803–816. doi: 10.1101/gad.255109.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu C., Fan L., Cen P., Chen E., Jiang Z., Li L. Energy metabolism plays a critical role in stem cell maintenance and differentiation. Int. J. Mol. Sci. 2016;17:253. doi: 10.3390/ijms17020253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Jacobson K., Schaller M.D. MAP kinases and cell migration. J. Cell Sci. 2004;117:4619–4628. doi: 10.1242/jcs.01481. [DOI] [PubMed] [Google Scholar]
- Jin Q., Yu L.R., Wang L., Zhang Z., Kasper L.H., Lee J.E., Wang C., Brindle P.K., Dent S.Y., Ge K. Distinct roles of GCN5/PCAF-mediated H3K9ac and CBP/p300-mediated H3K18/27ac in nuclear receptor transactivation. EMBO J. 2011;30:249–262. doi: 10.1038/emboj.2010.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Q., Zhuang L., Lai B., Wang C., Li W., Dolan B., Lu Y., Wang Z., Zhao K., Peng W. Gcn5 and PCAF negatively regulate interferon-beta production through HAT-independent inhibition of TBK1. EMBO Rep. 2014;15:1192–1201. doi: 10.15252/embr.201438990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamiya D., Banno S., Sasai N., Ohgushi M., Inomata H., Watanabe K., Kawada M., Yakura R., Kiyonari H., Nakao K. Intrinsic transition of embryonic stem-cell differentiation into neural progenitors. Nature. 2011;470:503–509. doi: 10.1038/nature09726. [DOI] [PubMed] [Google Scholar]
- Kang M., Garg V., Hadjantonakis A.K. Lineage establishment and progression within the inner cell mass of the mouse blastocyst requires FGFR1 and FGFR2. Dev. Cell. 2017;41:496–510.e5. doi: 10.1016/j.devcel.2017.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., Salzberg S.L. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koutelou E., Hirsch C.L., Dent S.Y. Multiple faces of the SAGA complex. Curr. Opin. Cell Biol. 2010;22:374–382. doi: 10.1016/j.ceb.2010.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunath T., Saba-El-Leil M.K., Almousailleakh M., Wray J., Meloche S., Smith A. FGF stimulation of the Erk1/2 signalling cascade triggers transition of pluripotent embryonic stem cells from self-renewal to lineage commitment. Development. 2007;134:2895–2902. doi: 10.1242/dev.02880. [DOI] [PubMed] [Google Scholar]
- Kuo M.H., Brownell J.E., Sobel R.E., Ranalli T.A., Cook R.G., Edmondson D.G., Roth S.Y., Allis C.D. Transcription-linked acetylation by Gcn5p of histones H3 and H4 at specific lysines. Nature. 1996;383:269–272. doi: 10.1038/383269a0. [DOI] [PubMed] [Google Scholar]
- Kurokawa D., Takasaki N., Kiyonari H., Nakayama R., Kimura-Yoshida C., Matsuo I., Aizawa S. Regulation of Otx2 expression and its functions in mouse epiblast and anterior neuroectoderm. Development. 2004;131:3307–3317. doi: 10.1242/dev.01219. [DOI] [PubMed] [Google Scholar]
- Langmead B., Trapnell C., Pop M., Salzberg S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanner F., Rossant J. The role of FGF/Erk signaling in pluripotent cells. Development. 2010;137:3351–3360. doi: 10.1242/dev.050146. [DOI] [PubMed] [Google Scholar]
- Lee H.J., Choi N.Y., Lee S.W., Ko K., Hwang T.S., Han D.W., Lim J., Scholer H.R., Ko K. Epigenetic alteration of imprinted genes during neural differentiation of germline-derived pluripotent stem cells. Epigenetics. 2016;11:177–183. doi: 10.1080/15592294.2016.1146852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lessard J.A., Crabtree G.R. Chromatin regulatory mechanisms in pluripotency. Annu. Rev. Cell Dev. Biol. 2010;26:503–532. doi: 10.1146/annurev-cellbio-051809-102012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Edgar D., Fassler R., Wadsworth W., Yurchenco P.D. The role of laminin in embryonic cell polarization and tissue organization. Dev. Cell. 2003;4:613–624. doi: 10.1016/s1534-5807(03)00128-x. [DOI] [PubMed] [Google Scholar]
- Lin W., Srajer G., Evrard Y.A., Phan H.M., Furuta Y., Dent S.Y. Developmental potential of Gcn5(-/-) embryonic stem cells in vivo and in vitro. Dev. Dyn. 2007;236:1547–1557. doi: 10.1002/dvdy.21160. [DOI] [PubMed] [Google Scholar]
- Lin W., Zhang Z., Srajer G., Chen Y.C., Huang M., Phan H.M., Dent S.Y. Proper expression of the Gcn5 histone acetyltransferase is required for neural tube closure in mouse embryos. Dev. Dyn. 2008;237:928–940. doi: 10.1002/dvdy.21479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loebel D.A., Studdert J.B., Power M., Radziewic T., Jones V., Coultas L., Jackson Y., Rao R.S., Steiner K., Fossat N. Rhou maintains the epithelial architecture and facilitates differentiation of the foregut endoderm. Development. 2011;138:4511–4522. doi: 10.1242/dev.063867. [DOI] [PubMed] [Google Scholar]
- Martinez-Cerdeno V., Lemen J.M., Chan V., Wey A., Lin W., Dent S.R., Knoepfler P.S. N-Myc and GCN5 regulate significantly overlapping transcriptional programs in neural stem cells. PLoS One. 2012;7:e39456. doi: 10.1371/journal.pone.0039456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy R.L., Duncan A.D., Barton M.C. Sample preparation for mass cytometry analysis. J. Vis. Exp. 2017 doi: 10.3791/54394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy R.L., Mak D.H., Burks J.K., Barton M.C. Rapid monoisotopic cisplatin based barcoding for multiplexed mass cytometry. Sci. Rep. 2017;7:3779. doi: 10.1038/s41598-017-03610-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pai Y.J., Leung K.Y., Savery D., Hutchin T., Prunty H., Heales S., Brosnan M.E., Brosnan J.T., Copp A.J., Greene N.D. Glycine decarboxylase deficiency causes neural tube defects and features of non-ketotic hyperglycinemia in mice. Nat. Commun. 2015;6:6388. doi: 10.1038/ncomms7388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel J.H., Du Y., Ard P.G., Phillips C., Carella B., Chen C.J., Rakowski C., Chatterjee C., Lieberman P.M., Lane W.S. The c-MYC oncoprotein is a substrate of the acetyltransferases hGCN5/PCAF and TIP60. Mol. Cell. Biol. 2004;24:10826–10834. doi: 10.1128/MCB.24.24.10826-10834.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pikman Y., Puissant A., Alexe G., Furman A., Chen L.M., Frumm S.M., Ross L., Fenouille N., Bassil C.F., Lewis C.A. Targeting MTHFD2 in acute myeloid leukemia. J. Exp. Med. 2016;213:1285–1306. doi: 10.1084/jem.20151574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu P., Simonds E.F., Bendall S.C., Gibbs K.D., Jr., Bruggner R.V., Linderman M.D., Sachs K., Nolan G.P., Plevritis S.K. Extracting a cellular hierarchy from high-dimensional cytometry data with SPADE. Nat. Biotechnol. 2011;29:886–891. doi: 10.1038/nbt.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera-Perez J.A., Hadjantonakis A.K. The dynamics of morphogenesis in the early mouse embryo. Cold Spring Harb. Perspect. Biol. 2014;7 doi: 10.1101/cshperspect.a015867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai T., Li S., Docheva D., Grashoff C., Sakai K., Kostka G., Braun A., Pfeifer A., Yurchenco P.D., Fassler R. Integrin-linked kinase (ILK) is required for polarizing the epiblast, cell adhesion, and controlling actin accumulation. Genes Dev. 2003;17:926–940. doi: 10.1101/gad.255603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serra V., Eichhorn P.J., Garcia-Garcia C., Ibrahim Y.H., Prudkin L., Sanchez G., Rodriguez O., Anton P., Parra J.L., Marlow S. RSK3/4 mediate resistance to PI3K pathway inhibitors in breast cancer. J. Clin. Invest. 2013;123:2551–2563. doi: 10.1172/JCI66343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- She P., Reid T.M., Bronson S.K., Vary T.C., Hajnal A., Lynch C.J., Hutson S.M. Disruption of BCATm in mice leads to increased energy expenditure associated with the activation of a futile protein turnover cycle. Cell Metab. 2007;6:181–194. doi: 10.1016/j.cmet.2007.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi C., Welsh P.A., Sass-Kuhn S., Wang X., McCloskey D.E., Pegg A.E., Feith D.J. Characterization of transgenic mice with overexpression of spermidine synthase. Amino Acids. 2012;42:495–505. doi: 10.1007/s00726-011-1028-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snezhkina A.V., Krasnov G.S., Lipatova A.V., Sadritdinova A.F., Kardymon O.L., Fedorova M.S., Melnikova N.V., Stepanov O.A., Zaretsky A.R., Kaprin A.D. The dysregulation of polyamine metabolism in colorectal cancer is associated with overexpression of c-Myc and C/EBPbeta rather than enterotoxigenic Bacteroides fragilis infection. Oxid. Med. Cell. Longev. 2016;2016:2353560. doi: 10.1155/2016/2353560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spedale G., Timmers H.T., Pijnappel W.W. ATAC-king the complexity of SAGA during evolution. Genes Dev. 2012;26:527–541. doi: 10.1101/gad.184705.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suganuma T., Gutierrez J.L., Li B., Florens L., Swanson S.K., Washburn M.P., Abmayr S.M., Workman J.L. ATAC is a double histone acetyltransferase complex that stimulates nucleosome sliding. Nat. Struct. Mol. Biol. 2008;15:364–372. doi: 10.1038/nsmb.1397. [DOI] [PubMed] [Google Scholar]
- Sumi T., Oki S., Kitajima K., Meno C. Epiblast ground state is controlled by canonical Wnt/beta-catenin signaling in the postimplantation mouse embryo and epiblast stem cells. PLoS One. 2013;8:e63378. doi: 10.1371/journal.pone.0063378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo N.E., Zhao T., Cankaya M., Theil T., Zhou X., Alvarez-Bolado G. Role of neuroepithelial Sonic hedgehog in hypothalamic patterning. J. Neurosci. 2009;29:6989–7002. doi: 10.1523/JNEUROSCI.1089-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tefft J.D., Lee M., Smith S., Leinwand M., Zhao J., Bringas P., Jr., Crowe D.L., Warburton D. Conserved function of mSpry-2, a murine homolog of Drosophila sprouty, which negatively modulates respiratory organogenesis. Curr. Biol. 1999;9:219–222. doi: 10.1016/s0960-9822(99)80094-3. [DOI] [PubMed] [Google Scholar]
- Timmers H.T., Tora L. SAGA unveiled. Trends Biochem. Sci. 2005;30:7–10. doi: 10.1016/j.tibs.2004.11.007. [DOI] [PubMed] [Google Scholar]
- Tudor M., Murray P.J., Onufryk C., Jaenisch R., Young R.A. Ubiquitous expression and embryonic requirement for RNA polymerase II coactivator subunit Srb7 in mice. Genes Dev. 1999;13:2365–2368. doi: 10.1101/gad.13.18.2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbanek P., Fetka I., Meisler M.H., Busslinger M. Cooperation of Pax2 and Pax5 in midbrain and cerebellum development. Proc. Natl. Acad. Sci. USA. 1997;94:5703–5708. doi: 10.1073/pnas.94.11.5703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villegas S.N., Rothova M., Barrios-Llerena M.E., Pulina M., Hadjantonakis A.K., Le Bihan T., Astrof S., Brickman J.M. PI3K/Akt1 signalling specifies foregut precursors by generating regionalized extra-cellular matrix. Elife. 2013;2:e00806. doi: 10.7554/eLife.00806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y.L., Faiola F., Xu M., Pan S., Martinez E. Human ATAC Is a GCN5/PCAF-containing acetylase complex with a novel NC2-like histone fold module that interacts with the TATA-binding protein. J. Biol. Chem. 2008;283:33808–33815. doi: 10.1074/jbc.M806936200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen H., Li Y., Xi Y., Jiang S., Stratton S., Peng D., Tanaka K., Ren Y., Xia Z., Wu J. ZMYND11 links histone H3.3K36me3 to transcription elongation and tumour suppression. Nature. 2014;508:263–268. doi: 10.1038/nature13045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilde J.J., Siegenthaler J.A., Dent S.Y., Niswander L.A. Diencephalic size is restricted by a novel interplay between gcn5 acetyltransferase activity and retinoic acid signaling. J. Neurosci. 2017;37:2565–2579. doi: 10.1523/JNEUROSCI.2121-16.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Q., Zhang L., Su P., Lei X., Liu X., Wang H., Lu L., Bai Y., Xiong T., Li D. MSX2 mediates entry of human pluripotent stem cells into mesendoderm by simultaneously suppressing SOX2 and activating NODAL signaling. Cell Res. 2015;25:1314–1332. doi: 10.1038/cr.2015.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W., Edmondson D.G., Evrard Y.A., Wakamiya M., Behringer R.R., Roth S.Y. Loss of Gcn5l2 leads to increased apoptosis and mesodermal defects during mouse development. Nat. Genet. 2000;26:229–232. doi: 10.1038/79973. [DOI] [PubMed] [Google Scholar]
- Yamanaka Y., Lanner F., Rossant J. FGF signal-dependent segregation of primitive endoderm and epiblast in the mouse blastocyst. Development. 2010;137:715–724. doi: 10.1242/dev.043471. [DOI] [PubMed] [Google Scholar]
- Yamauchi T., Yamauchi J., Kuwata T., Tamura T., Yamashita T., Bae N., Westphal H., Ozato K., Nakatani Y. Distinct but overlapping roles of histone acetylase PCAF and of the closely related PCAF-B/GCN5 in mouse embryogenesis. Proc. Natl. Acad. Sci. USA. 2000;97:11303–11306. doi: 10.1073/pnas.97.21.11303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying Q.L., Wray J., Nichols J., Batlle-Morera L., Doble B., Woodgett J., Cohen P., Smith A. The ground state of embryonic stem cell self-renewal. Nature. 2008;453:519–523. doi: 10.1038/nature06968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y., Yoon S.O., Poulogiannis G., Yang Q., Ma X.M., Villen J., Kubica N., Hoffman G.R., Cantley L.C., Gygi S.P. Phosphoproteomic analysis identifies Grb10 as an mTORC1 substrate that negatively regulates insulin signaling. Science. 2011;332:1322–1326. doi: 10.1126/science.1199484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X.Y., Varthi M., Sykes S.M., Phillips C., Warzecha C., Zhu W., Wyce A., Thorne A.W., Berger S.L., McMahon S.B. The putative cancer stem cell marker USP22 is a subunit of the human SAGA complex required for activated transcription and cell-cycle progression. Mol. Cell. 2008;29:102–111. doi: 10.1016/j.molcel.2007.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W. Model-based analysis of ChIP-seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.