ABSTRACT
Cadherin-based intercellular adhesions are essential players in epithelial homeostasis, but their dynamic regulation during tissue morphogenesis and remodeling remain largely undefined. Here, we characterize an unexpected role for the membrane-anchored metalloproteinase MT2-MMP in regulating epithelial cell quiescence. Following co-immunoprecipitation and mass spectrometry, the MT2-MMP cytosolic tail was found to interact with the zonula occludens protein-1 (ZO-1) at the apical junctions of polarized epithelial cells. Functionally, MT2-MMP localizes in the apical domain of epithelial cells where it cleaves E-cadherin and promotes epithelial cell accumulation, a phenotype observed in 2D polarized cells as well as 3D cysts. MT2-MMP-mediated cleavage subsequently disrupts apical E-cadherin-mediated cell quiescence resulting in relaxed apical cortical tension favoring cell extrusion and re-sorting of Src kinase activity to junctional complexes, thereby promoting proliferation. Physiologically, MT2-MMP loss of function alters E-cadherin distribution, leading to impaired 3D organoid formation by mouse colonic epithelial cells ex vivo and reduction of cell proliferation within intestinal crypts in vivo. Taken together, these studies identify an MT2-MMP–E-cadherin axis that functions as a novel regulator of epithelial cell homeostasis in vivo.
KEY WORDS: MT2-MMP, ZO-1, E-cadherin, Apical junction, Src, Epithelial cell proliferation
Summary: Epithelial tissues should permit cell proliferation without losing their integrity for homeostasis. MT2-MMP protease cleaves E-cadherin at apical junctions, promoting regulated exit from quiescence in epithelial cells in the intestine.
INTRODUCTION
Epithelial barriers maintain structural integrity by forming epithelial cell junctions that preserve tissue architecture and control barrier permeability (Macara et al., 2014). Epithelial junctions are established through E-cadherin-based interactions between adjacent cells and by their association with the cytoskeleton, particularly with the actomyosin machinery and junctional proteins such as zonula occludens-1 (ZO-1) (Wu and Yap, 2013). ZO-1 is specifically enriched at the apical tight junctions of epithelial cells where its three PDZ domains allow multiple molecular interactions (Fanning and Anderson, 2009). Dynamic changes in cell–cell adhesive interactions are required for the normal development and growth of epithelial tissues because proliferative responses must proceed while maintaining tissue integrity. By contrast, cell–cell adhesion changes are also associated with epithelial–mesenchymal transition (EMT) programs that decrease E-cadherin expression, thereby promoting epithelial cell migration and proliferation, particularly in tumors (Macara et al., 2014).
While E-cadherin is transcriptionally repressed during EMT (Nieto, 2011), cell surface levels of E-cadherin can also be reduced by proteolysis (Kowalczyk and Nanes, 2012). Matrix metalloproteinases (MMPs) are a family of secreted or membrane-tethered Zn-dependent endopeptidases that are able to proteolyze all extracellular matrix (ECM) components (Page-McCaw et al., 2007). In addition, MMP family members can modulate cell responses by cleaving transmembrane receptors as well as soluble and ECM-bound growth factors and chemokines (Page-McCaw et al., 2007). E-cadherin can be cleaved by secreted MMPs, including MMP-7, while Ras-induced E-cadherin cleavage by MMP-2 has been shown to drive epithelial cell extrusion (Grieve and Rabouille, 2014). Transmembrane type-MMPs (MT-MMPs), i.e. MT1-MMP, MT2-MMP, MT3-MMP and MT5-MMP, can also hydrolyze ECM components as well as a range of cell-membrane-associated targets. Interestingly, MT1-MMP and MT2-MMP are expressed in epithelial tissues such as the salivary and mammary glands, although their pattern of expression and their function appear to be tissue- and context-dependent (Feinberg et al., 2016; Rebustini et al., 2009). For example, in the submandibular gland, MT2-MMP, rather than MT1-MMP, is preferentially expressed in epithelial cells where it regulates branching morphogenesis by hydrolyzing type IV collagen in the underlying basement membrane (Rebustini et al., 2009). By contrast, both MT1-MMP and MT2-MMP are expressed in mammary gland epithelial cells, but neither MT1-MMP-null nor MT2-MMP-null mice play required roles in regulating branching activity (Feinberg et al., 2016). Independent of their respective tissue-specific roles, the catalytic domains of MT1-MMP and MT2-MMP share a number of structural and functional characteristics. However, we noted distinct features in their respective cytosolic tails and hypothesized that different molecular interactions within the cytoplasmic compartment might contribute to the functional selectivity of these homologous MT-MMPs, particularly in epithelial cells. In this regard, we now demonstrate that the MT2-MMP cytosolic domain specifically regulates its interactions with ZO-1 and controls epithelial cell homeostasis by cleaving apical-domain-localized E-cadherin both in vitro and in vivo.
RESULTS
MT2-MMP interacts with ZO-1 in polarized MDCK epithelial cells
While the cytosolic tail of MT1-MMP has been reported to interact with a number of intracellular binding partners that regulate proteinase function, binding partners for MT2-MMP have not been described previously. Therefore, using MT-MMP GST fusion proteins and pull-down assays followed by mass spectrometry (MS), we first sought to identify proteins interacting with the cytosolic domain of MT2-MMP, but not MT1-MMP. These analyses resulted in the identification of ZO-1 as a bona-fide binding partner of the MT2-MMP cytosolic tail (Fig. S1A). Next, using wild-type or mutant MT2-MMP peptides in tandem with ELISA, we found that the MT2-MMP cytosolic tail preferentially interacted with the ZO-1 PDZ1 and PDZ2 domains while the P-1 tryptophan (W) and the C-terminal valine (V), but not the P-2 glutamic acid (E), of the MT2-MMP tail are essential for MT2-MMP/ZO-1 interactions (Fig. S1B,C). Of note, P-1 tryptophan of M2-MMP was especially relevant as mutation of the P-1 lysine (K) in the MT1-MMP cytosolic tail to a tryptophan residue conferred the mutant with the ability to interact with ZO-1 (Fig. S1C). Indeed, the cytosolic tail of wild-type MT2-MMP, but not the MT2WK mutant, as well as the MT1KW mutant, but not wild-type MT1-MMP, interacted with ZO-1 in cell lysates, confirming that the P-1 tryptophan residue is essential for this association in a cellular context (Fig. S1D).
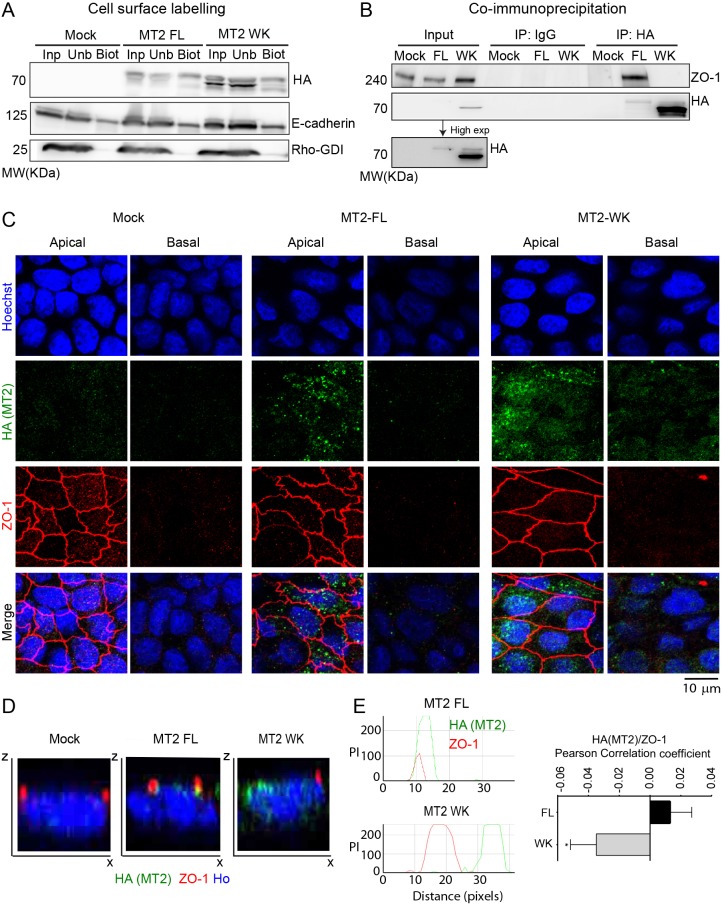
As ZO-1 is a key component of epithelial apical junctions, we next expressed full-length MT2-MMP (MT2FL) or mutant MT2WK in MDCK cells, an epithelial cell line commonly used for polarity studies (Simmons, 1982). Mature MT2FL and MT2WK were both expressed and functionally active at the cell surface as assessed by biotin labeling and fibrinogen zymography (Fig. 1A and data not shown). Cells expressing the WK mutant also displayed increased intracellular levels of a protein product likely corresponding to mistrafficked MT2WK arising as a consequence of the mutation occurring in juxtaposition to the C-terminal valine (Urena et al., 1999). The association of MT2-MMP, but not MT2WK, with ZO-1 in stable transfectants was confirmed by co-immunoprecipitation of both proteins from MDCK monolayers (Fig. 1B). Using polarized MDCK cells, MT2-MMP was also observed in intracellular vesicles enriched towards the apical plasma membrane where a pool of MT2-MMP co-localized with ZO-1 at epithelial apical junctions (Fig. 1C–E). By contrast, the intracellular distribution of MT2WK was more diffuse, and while present on the basal surface of MDCK cells, it failed to co-localize with ZO-1 (Fig. 1C–E). Similarly to MT2WK, MT1-MMP did not co-localize with ZO-1 in MDCK transfectants (Fig. S1E,F).
Fig. 1.
MT2-MMP associates with ZO-1 in polarized MDCK cells. (A) Western blot analysis of HA, E-cadherin and Rho-GDI in biotinylated cell lysates from Mock, MT2-MMP (MT2FL) and MT2-MMPWK (MT2WK) stable MDCK transfectants pulled down with streptavidin beads; input, unbound and bound fractions are shown (Inp, Unb, Biot). (B) Western blot analysis of ZO-1 and HA in cell lysates from Mock, MT2-MMP and MT2-MMPWK stable MDCK transfectants pulled down with anti-HA antibody; IgG immunoprecipitates and whole lysates (Input) are also shown as controls. A blot of the input lanes after a longer exposure is also shown. (C) Representative maximal projections from apical and basolateral stacks of confocal sections from polarized MDCK transfectants stained for HA (MT2-MMP, green), ZO-1 (red) and nuclei (Hoechst, blue). (D) Orthogonal x–z views of 3D confocal image stacks from C. (E) Representative peak intensity profiles from x–z views of 3D confocal image stacks from C. Graph to the right shows the quantification of MT2-MMP/ZO-1 Pearson correlation coefficient in polarized MT2-FL and MT2-WK MDCK transfectants. Values are mean±s.e.m. n=40 cells per condition in two independent experiments; *P<0.05.
Upon further analysis, MT2-MMP-positive intracellular vesicles were not only associated with ZO-1 at cell–cell junctions, but also in the cytosol (Fig. 1C), suggesting that interaction between the EWV motif and the PDZ domain may be initiated in the cytoplasm. Although MT2-MMP was scarcely present in intermediate/late endosomes (i.e. hepatocyte-growth-factor-regulated tyrosine kinase substrate, HGS+, and tumor susceptibility gene 101 protein, TSG101+), it was clearly observed in early (early endosome antigen 1, EEA1+) and recycling (transferrin receptor, TfR+) endosomes of MDCK stable transfectants where it co-localizes with ZO-1, particularly in peri-junctional EEA1+ vesicles (Fig. S2A and B). Indeed, inhibition of endosome recycling with endosidin-2 (Zhang et al., 2016) led to the accumulation of MT2-MMP/EEA1+ vesicles (Fig. S2C) with an extended apical distribution of MT2-MMP and ZO-1 beyond the non-treated cell junctions (Fig. S2D). Endosidin-2 treatment also resulted in lower MT2-MMP plasma membrane levels as assessed by biotin cell surface labeling (Fig. S2E). These findings suggest that a pool of MT2-MMP and ZO-1 traffic together during endosomal recycling towards the apical junctions.
MT2-MMP promotes apical accumulation of epithelial cells via E-cadherin cleavage
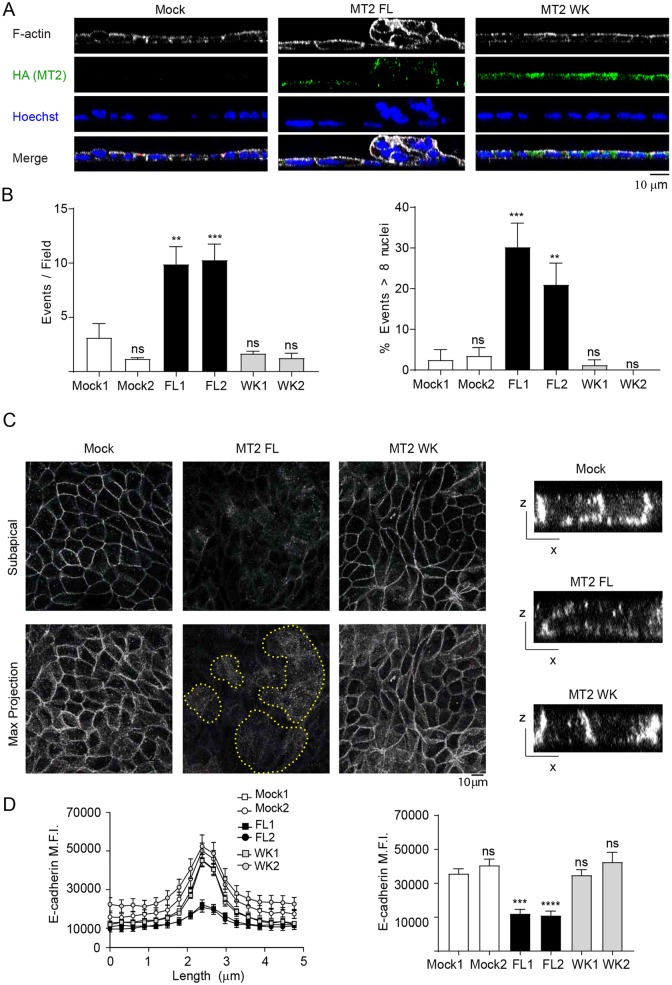
In our preliminary studies, we noted that MT2-MMP MDCK transfectants extruded cells onto the surface of the polarized monolayer over a 7 day period, with peak differences most prominent at day 3 (data not shown). Quantitative analysis of 3D stacks of MT2-MMP MDCK transfectants demonstrated a significant increase in the number of apical epithelial foci at day 3 as well as a significantly higher percentage of foci containing 8 or more cells relative to either mock or MT2-MMPWK MDCK transfectants (Fig. 2A,B). As intercellular junctions play an important role in epithelial cell quiescence and homeostasis, we next analyzed E-cadherin expression and localization in polarized monolayers formed by the MDCK stable transfectants. Despite no changes in total E-cadherin protein levels (Fig. S3A), quantitative image analysis demonstrated a significant reduction in E-cadherin intensity at epithelial junctions in MT2-MMP transfectants as compared to mock or MT2-MMPWK MDCK transfectants (Fig. 2C,D). E-cadherin reduction was apparent both when MDCK cells were plated individually and in mosaic experiments combining dual-labeled MT2-MMP and MT2WK MDCK transfectants (Fig. S3B).
Fig. 2.
MT2-MMP overexpression induces aberrant apical epithelial cell accumulation in polarized MDCK monolayers. (A) Orthogonal x–z projections of 3D confocal image stacks of MDCK transfectants stained for F-actin (Phalloidin, gray), HA (MT2-MMP, green) and Hoechst (nuclei, blue). Scale bar: 10 μm. (B) Quantification of apical epithelial foci per field (left) and the percentage of foci having more than 8 nuclei (right). 10 fields were counted per condition in n=4 independent experiments. (C) Representative maximal projections are shown from subapical and complete stacks of confocal sections from polarized MDCK transfectants stained for E-cadherin (gray). The dotted yellow line marks apical foci. Orthogonal x–z views are shown to the right. (D) Line and bar graphs show E-cadherin peak and average mean fluorescence intensity (MFI), respectively, around the junctions formed by MDCK transfectants. Data are represented as mean±s.e.m. and were tested by one-way ANOVA versus mock 1 followed by Dunnett's post-test in B and C. **P<0.01, ***P<0.001, ****P<0.001; ns, not significant.
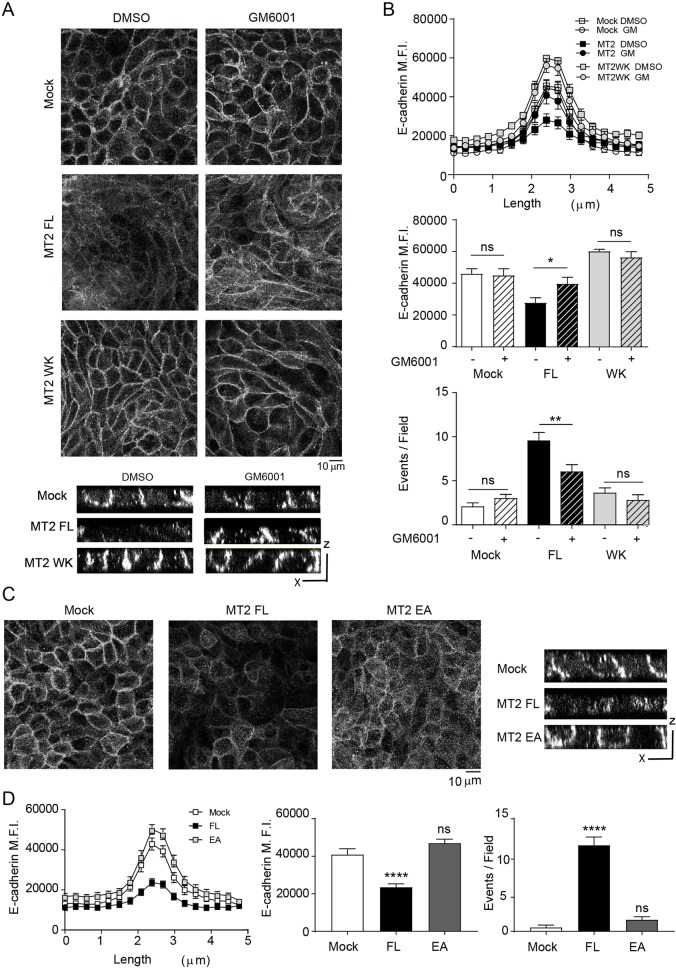
To next determine whether E-cadherin might undergo restricted proteolytic processing within apical junctional domains, MDCK transfectants were cultured in the presence of the broad range MMP inhibitor, GM6001. Interestingly, GM6001-treated MT2-MMP transfectants increased E-cadherin intensity at cell–cell junctions, thereby restoring E-cadherin levels to those observed in MDCK mock or MT2-MMPWK transfectants; this rescue was also observed in mosaic experiments with mixtures of differently colored MT2-MMP and MT2WK MDCK transfectants (Fig. 3A,B and Fig. S3C). GM6001 also decreased the number of epithelial cell foci accumulating on the apical face of MT2-MMP MDCK monolayers to levels comparable to those observed with MDCK mock or MT2WK-MDCK cells (Fig. 3B). To directly assess the role of MT2-MMP catalytic activity on junctional E-cadherin, we generated an MT2-MMP mutant construct where the glutamic acid at position 260 (required for MMP catalytic activity) was substituted with an alanine residue (MT2-MMPE260A, herein referred to as MT2-MMPEA). Following transfection, MT2-MMPEA was expressed at the cell membrane, displayed a similar subcellular distribution to that of wild-type MT2-MMP, co-localized with ZO-1 at epithelial junctions and interacted with ZO-1 in co-immunoprecipitation assays (Fig. S4A–C). Under these conditions, however, junctional E-cadherin intensity in MT2-MMPEA transfectants was restored to levels comparable to those found in mock cells and significantly increased relative to MT2-MMP MDCK cells despite no discernible change in total E-cadherin levels (Fig. 3C,D and Fig. S3A). Furthermore, the MT2-MMPEA transfectants did not display accumulation of apical epithelial foci relative to mock transfectants (Fig. 3D).
Fig. 3.
Apical epithelial cell accumulation depends on MT2-MMP catalytic activity. (A) Representative maximal projections from confocal sections of polarized MDCK transfectants stained for E-cadherin (gray) in the presence of GM6001 (50 μM) or vehicle (DMSO). Orthogonal x–z views are shown below. (B) Line and bar graphs show E-cadherin peak and average intensity, respectively, around the junctions formed by MDCK transfectants treated as in A. Bar graph at the bottom shows the number of apical events on the polarized MDCK monolayer in the presence or absence of DMSO. In the middle and bottom graphs, the difference between mock DMSO and MT2 FL were significant with P<0.01 and P<0.0001, respectively. (C) Representative maximal projections are shown from confocal sections of polarized MDCK transfectants (mock, MT2 and MT2EA) stained for E-cadherin (gray). Orthogonal x–z views are shown to the right. (D) Line and bar graphs show E-cadherin peak and average mean fluorescence intensity (MFI), respectively, around the junctions formed by MDCK transfectants shown in C. Bar graph on the right shows the number of apical events occurring in polarized MDCK monolayers. Data are represented as mean± s.e.m. and were tested by one-way ANOVA followed by Sidak post-test in B. Dunnett's post-test was used in D. *P<0.05, **P<0.01, ****P<0.001; ns, not significant.
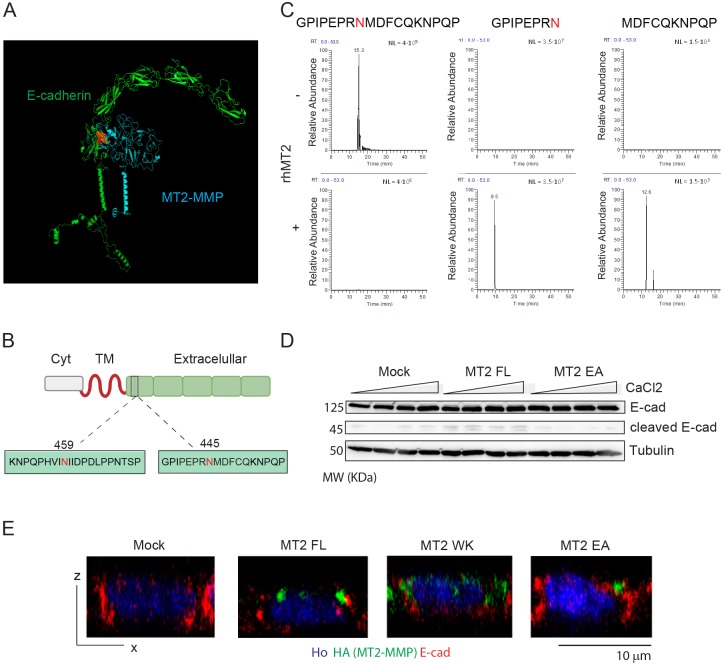
Given these results, we next sought to determine the possibility that E-cadherin might be targeted as an MT2-MMP substrate in epithelial cells. By in silico modeling and cleavage site prediction (http://cleavpredict.sanfordburnham.org/; Table S1), a potential docking site between the MT2-MMP catalytic domain and the EC5 loop of E-cadherin in a cis orientation was identified (Fig. 4A). The EC5 loop includes the sequence GPIPEPRN445 MDFCQKNPQP and KNPQPHVIN459IIDPDLPPNTSP with potential MT2-MMP cleavage sites identified after positions N445 and N459 (Fig. 4B). While both regions are accessible to the MT2-MMP catalytic domain, the GPIPEPRNMDFCQKNPQP yielded a more stable complex in the in silico model (Fig. 4A). To directly assess the ability of MT2-MMP to hydrolyze E-cadherin within this domain, canine E-cadherin peptides spanning the predicted cleavage sites were incubated with the human recombinant MT2-MMP catalytic domain and the obtained peptide fragments analyzed by MS. As predicted, MS identified specific cleavage after residue N445, yielding the fragments GPIPEPRN and MDFCQKNPQP (Fig. 4C), with no specific cleavage observed when MT2-MMP was incubated with KNPQPHVIN459IIDPDLPPNTSP (data not shown). Importantly, we confirmed that this cleavage occurred within intact cells as we detected a twofold increase in the abundance of a 45 kDa E-cadherin C-terminal fragment (compatible with the cleavage after N445) in lysates from MT2-MMP MDCK cells compared with mock, MT2EA or MT2WK transfectants (Fig. 4D and data not shown). Further, we verified the accessibility of E-cadherin to MT2-MMP cleavage at the apical junctions as assessed by co-immunostaining in MT2-MMP MDCK transfectants (Fig. 4E).
Fig. 4.
E-cadherin is cleaved by MT2-MMP after N445 in the EC5 loop. (A) In silico model of canine E-cadherin (green)/human MT2-MMP (blue) interactions in cis association at the plasma membrane; the catalytic MT2-MMP center and the E-cadherin peptide, GPIPEPRNMDFCQKNPQP, are shown in orange and red, respectively. (B) Scheme of E-cadherin structure with the peptide containing the predicted cleavage sites after N445 and N459 in the EC5 loop. (C) Representative extracted ion chromatograms of 3 independent experiments corresponding to the peptides detected following in in vitro digestion of the GPIPEPRNMDFCQKNPQP peptide in the absence or presence of the human MT2-MMP recombinant catalytic domain (rhMT2). (D) Western blot analysis of lysates recovered from MDCK transfectants cultured with different calcium concentrations. Results are representative of two independent experiments. (E) Representative orthogonal x–z views of confocal images for polarized MDCK transfectants co-immunostained for HA (MT2-MMP, green), E-cadherin (red) and nuclei (Hoechst, blue).
MT2-MMP disrupts apical E-cadherin-dependent signaling in epithelial cells
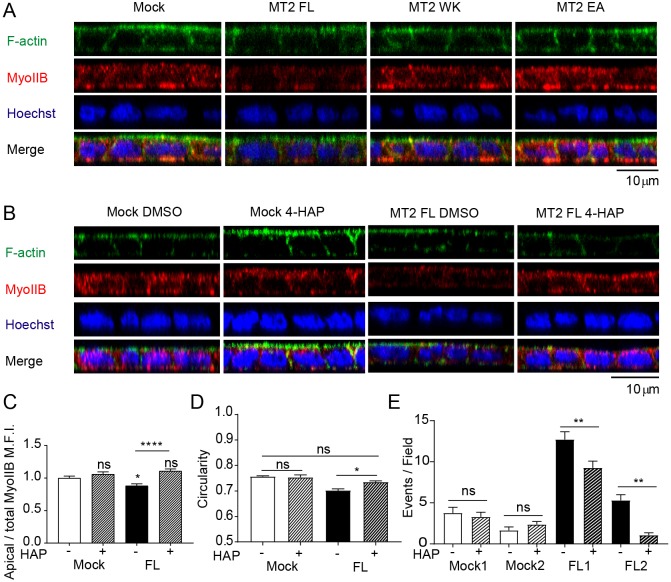
Apical junctions are essential for epithelial homeostasis maintenance (Baum and Georgiou, 2011). Given that MT2-MMP-mediated E-cadherin cleavage preferentially occurs at the apical junctions via ZO-1 interaction, we posited that apical junction integrity might be perturbed under these conditions. Indeed, MT2-MMP transfectants exhibited a decrease in other apical junctional markers such as β-catenin relative to mock, MT2WK or MT2EA transfectants (Fig. S5A). Moreover, MT2-MMP-MDCK transfectants showed significantly less apical myosin IIB staining and also decreased cell circularity (Fig. 5A–D), effects that are compatible with reduced cortical tension and a propensity for cell extrusion (Gu and Rosenblatt, 2012). Concomitantly, the apical actin organizer ezrin (Hughes and Fehon, 2007) was barely detected in MT2-MMP MDCK when compared with mock, MT2WK or MT2EA transfectants (Fig. S5B). To determine whether relaxed cortical contractility plays a role in the apical epithelial cell accumulation phenotype, MT2-MMP transfectants were incubated with 4-HAP, the active metabolite of carbamate-7, which increases myosin-dependent cortical tension (Surcel et al., 2015). Indeed, 4-HAP rescued the circularity defect and significantly reduced the number of apical epithelial cell foci in MT2-MMP MDCK transfectants as compared with mock-transfected MDCK cells (Fig. 5C–E).
Fig. 5.
MT2-MMP disrupts apical E-cadherin-mediated signals. (A) Orthogonal x–z projections of 3D confocal image stacks are shown of polarized MDCK transfectants stained for F-actin (Phalloidin, green), myosin IIB (red), and nuclei (Hoechst, blue). (B) Orthogonal x–z projections of 3D confocal image stacks are shown of polarized MDCK transfectants (mock and MT2-MMP) in the presence of 4-HAP (500 nM) or vehicle (DMSO) for 72 h and stained for F-actin (phalloidin, green), myosin IIB (red), and nuclei (Hoechst, blue). (C) Quantification of apical/total MFI of myosin IIB in polarized MDCK transfectants treated with 4-HAP (500 nM) or vehicle (DMSO); n=5 independent experiments. (D) Quantification of cell circularity in MDCK cells presented in C. 25 cells per field were counted in 2 images per condition in 6 independent experiments. (E) Quantification of the number of apical events on polarized MDCK cells presented in panel B. 10 fields were counted per condition in n=4 independent experiments. Difference between mock DMSO1 and MT2 FL1, and mock DMSO2 and MT2 FL2 were significant with P<0.0001 and P<0.05, respectively. Data are represented as mean±s.e.m. and were tested by one-way ANOVA followed by Sidak post-test. *P<0.05, **P<0.01, ****P<0.0001; ns, not significant.
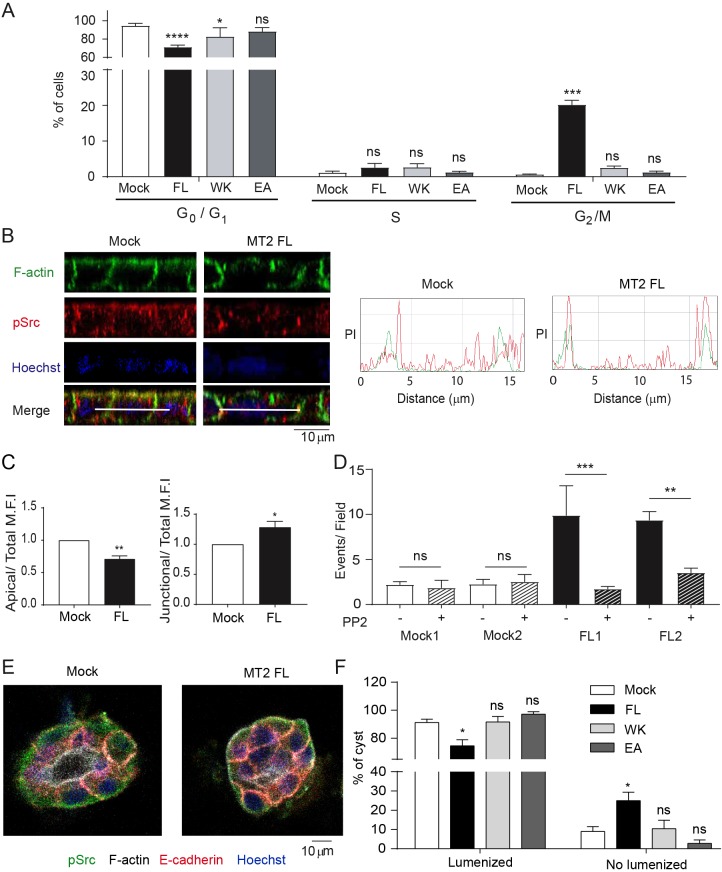
Consistent with the fact that the disrupted apical E-cadherin signals might break epithelial cell quiescence, accumulation of extruded foci in polarized MT2-MMP-MDCK cells (Fig. 2B) correlated with abnormal proliferative responses as growth-factor-deprived MT2-MMP-transfected MDCK cells displayed persistent entry into cell cycle with a significantly higher percentage of cells in the G2–M phase relative to mock, MT2WK or MT2EA mutants (Fig. 6A). As the disruption of apical E-cadherin-mediated signals can impact the localization and activity of Src kinase (Kourtidis et al., 2015), we next sought to determine whether MT2-MMP affected this signaling hub. Although no increase in total pSrc was detected in MT2-MDCK transfectants, its subcellular location was altered (Fig. 6B and Fig. S6A,B). Whereas high levels of pSrc are localized to the apical membrane of control transfectants, pSrc was alternatively enriched at cell–cell junctions in MT2-MMP-MDCK cells and reduced in the apical domain (Fig. 6B,C). To determine whether mislocalized pSrc played a direct role in driving cell extrusion and proliferation, MT2-MMP transfectants were next cultured with the Src inhibitor, PP2. Indeed, PP2 significantly decreased apical epithelial foci and reduced the number of foci harboring 8 or more cells in MT2-MMP MDCK cells (Fig. 6D and Fig. S6A–D). pSrc appears to act, at least partially, by regulating junctional E-cadherin as PP2 also restored E-cadherin localization to normal levels in MT2-MMP transfectants (Fig. S6C).
Fig. 6.
Mislocalization of pSrc in polarized MT2-MMP-MDCK cells contributes to apical cell accumulation. (A) Percentage of cells in G0/G1, S, and G2/M phases of the cell cycle analyzed by flow cytometry in propidium-iodide-stained MDCK transfectants after 72 h of serum deprivation. Means±s.e.m. are shown for 3 independent experiments. (B) Orthogonal x–z projections of 3D confocal image stacks of polarized MDCK transfectants (mock and MT2-MMP) stained for F-actin (Phalloidin, green), pSrc (red), and nuclei (Hoechst, blue). Representative peak intensity profiles are shown on the right for pSrc (red) and F-actin (green). (C) Bar graphs show the apical (left) and junctional (right) pSrc intensity, relative to total mean fluorescence intensity (MFI) in 6 independent experiments. (D) Number of apical events occurring in polarized MDCK cells treated with PP2 or vehicle (DMSO). 10 fields were counted per condition in 3 independent experiments. Differences between mock DMSO1 and MT2 FL1, and mock DMSO2 and MT2 FL2 were significant with P<0.001 and P<0.01, respectively. (E) Representative confocal images of 3D cysts formed by MDCK transfectants in Matrigel and stained for pSrc (green), F-actin (white), E-cadherin (red), and nuclei (Hoechst, blue). (F) Quantification of the percentage of lumenized cysts. Data are represented as the mean±s.e.m. and were tested by two-way ANOVA followed by Dunnett's post-test in A, two-tailed Welch-test comparison was used in B, and one-way ANOVA followed by Sidak post-test was used in C. *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001; ns, not significant.
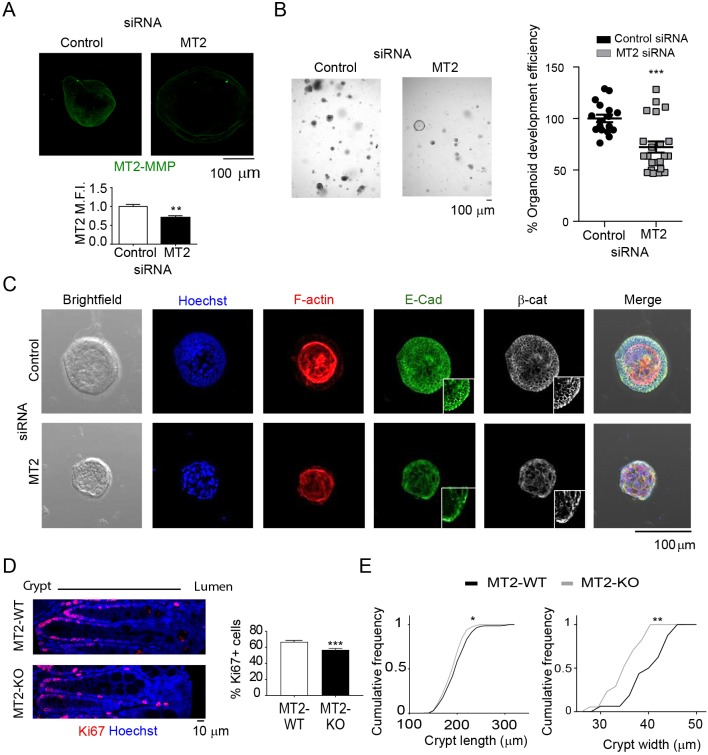
In 3D culture, MDCK cells form cystic structures that more closely recapitulate their tubulogenic properties (Montesano et al., 1991). To assess the impact of MT2-MMP expression under these conditions, MT2-MMP MDCK cells were next embedded in Matrigel (Martín-Belmonte et al., 2008). MT2-MMP was distributed in cysts similarly to polarized cells (Fig. S6E and Fig. 1A). Notably, cysts formed by MT2-MMP MDCK cells displayed impaired lumenization and contained more junctional pSrc relative to cysts formed by mock, MT2WK or MT2EA MDCK transfectants (Fig. 6E,F and Fig. S6F,G). While the basal extrusion of cells occurred more frequently in MT2-MMP MDCK cells relative to controls, such events occurred only rarely (data not shown). Nevertheless, these data confirm the ability of MT2-MMP to disrupt apical junction integrity in 3D microenvironments. The functional relevance of the identified MT2-MMP–E-cadherin axis was further validated by a loss-of-function approach wherein MT2-MMP expression was reduced by siRNA in polarized Caco2 cells (a human colon epithelial cancer cell line). Under these conditions, MT2-MMP silencing led to increased levels of E-cadherin at cell–cell junctions and more abundant myosin IIB and pSrc expression throughout the cell (Fig. S6H–K), especially at the apical domain, thereby resembling the phenotype observed in mock-transfected MDCK cells. By contrast, in the siRNA control cells, E-cadherin and myosin IIB were reduced and pSrc preferentially localized to cell–cell junctions in a manner similar to MT2-MMP MDCK transfectants (Figs S6H–K, Fig. 2A, Fig. 5A, Fig. 6B and Fig. S5A).
MT2-MMP loss of function impairs colon homeostasis
We finally evaluated the physiological impact of the MT2-MMP–E-cadherin axis in the normal epithelium. Fine-tune modulation of epithelial cell proliferation–differentiation balance is especially important near stem cell niches in tissues with high turnover rates, such as the intestine (Haegebarth and Clevers, 2009). Using mouse colonic epithelial cells cultured as 3D organoids formed ex vivo, endogenous MT2-MMP expression was confirmed (Fig. 7A). Importantly, silencing MT2-MMP expression significantly impaired the efficiency of organoid formation (∼27.8%; Fig. 7A,B). This phenotype was accompanied by altered F-actin, E-cadherin and β-catenin distribution as well as aberrant cell morphology in MT2-MMP-silenced crypts relative to siRNA control-treated crypts (Fig. 7C). Within mouse intestinal crypts, MT2-MMP was likewise detected in epithelial cells, particularly in apical domains as assessed by anti-MT2-MMP immunohistochemistry in wild-type mice (Fig. S7C). To determine the impact of MT2-MMP on colon epithelial cells in vivo, we next turned to a recently generated loss-of-function mouse model for the analysis of possible defects related to quiescence or proliferation (Feinberg et al., 2016). While colons recovered from MT2-MMP-null mice did not display major histological defects under steady-state conditions (data not shown), quantitative image analysis that focused on the crypt areas, uncovered an extended junctional overlapping of E-cadherin with ZO-1, a phenotype consistent with defective apical E-cadherin cleavage (Fig. S6B). Moreover, pSrc was largely confined to the intracellular compartment rather than junctional domains in the MT2-MMP-null epithelium (Fig. S6C), recapitulating the phenotype of control MDCK cells (Fig. 6B). Furthermore, altered E-cadherin and pSrc subcellular localization was associated with significantly lower numbers of proliferating epithelial cells (Ki67+) in colon crypts of MT2-MMP-null mice relative to wild-type controls (Fig. 7D). Impaired proliferation also correlated with an increased abundance of narrower and shorter crypts in colons from MT2-MMP-null mice compared with the wild type (Fig. 7E), a phenotype in line with impaired 3D organoid formation in MT2-MMP-silenced crypts. Taken together, these findings support a new model wherein MT2-MMP promotes the local proliferation of intestinal epithelial cells via E-cadherin cleavage at apical junctions – an event that is essential for proper intestinal homeostasis.
Fig. 7.
MT2-MMP deficiency alters junctional E-cadherin and leads to decreased 3D colon organoid formation ex vivo and smaller crypts in vivo. (A) Representative confocal images of MT2-MMP-silenced colon organoids stained for MT2-MMP (green; top). Graph shows the normalized MT2-MMP mean fluorescence intensity (MFI) in stained organoids 72 h after siRNA transfection (bottom), n=6 images per condition from 3 independent experiments; Mmp15 mRNA levels decreased ∼20% in silenced organoids. (B) Bright-field microscopy images of MT2-MMP-silenced colon organoids. Bar graph shows the percentage of organoid generation efficiency 48 h after siRNA transfection (right) in 3 independent experiments. (C) Representative confocal images of MT2-MMP-silenced colon organoids stained for nuclei (Hoechst/Ho, blue), F-actin (red), E-cadherin (green) and β-catenin (white); magnified views of E-cadherin and β-catenin staining are shown in insets. (D) Representative confocal images of colonic tissues recovered from wild-type or MT2-MMP-null mice, and stained for Ki67 (red) and nuclei (Hoechst, blue). On the right, graph shows the percentage of Ki67-positive cells per crypt. 9–15 crypts were quantified per condition in 3 images taken from 2 mice per genotype (top). (E) Quantification of the cumulative frequency of crypt length (left) and width (right). Data are represented as mean±s.e.m. and were tested by unpaired Student's t-test in A and B and by two-tailed Welch-test comparison in D and E. *P<0.05, **P<0.01, ***P<0.001; ns, not significant.
DISCUSSION
Herein, we have identified MT2-MMP as a novel regulator of epithelial cell homeostasis and proliferation in vitro as well as in vivo. In this model, the cytosolic domain of MT2-MMP binds ZO-1, thereby allowing the protease to cleave E-cadherin at epithelial junctions, while promoting epithelial cell proliferation. These findings highlight the relevance of selective molecular interactions to confer MT2-MMP with specific cellular functions. The cytosolic tails of MT2-MMP and MT1-MMP are highly conserved and in fact, interact with a common Golgi-associated protein partner, GRASP55 (Roghi et al., 2010). By contrast, we have identified the P-1 tryptophan (W) in the cytosolic tail of MT2-MMP, but not MT1-MMP, as a key determinant for its selective interaction with ZO-1, providing a possible mechanism for the previously reported distinct functions of these two proteases in epithelial cells (Hotary et al., 2000). Interestingly, the cytosolic tails of MT3-MMP and MT5-MMP also contain a P-1 W and bind ZO-1 (our unpublished data), supporting the key role of this interaction in distinct scenarios. Moreover, our data highlight the cooperative interaction of the different ZO-1 PDZ domains with MT2-MMP, as also shown for interactions between ABP and MT5-MMP (Monea et al., 2006). Of note, while both MT2-MMP and MT1-MMP contain basolateral sorting di-leucine motifs, the selective interaction of MT2-MMP with the apical protein ZO-1 might determine their co-trafficking from early or recycling endosomes towards apical junctions. The preferential localization of MT2-MMP to apical domains promotes its accessibility to junctional E-cadherin where proteolysis ensues. In contrast to MT1-MMP, which localizes to the basal compartments of MDCK cells where it promotes a collagenolytic phenotype (Weaver et al., 2014), the apical distribution and function of MT2-MMP persisted in 3D epithelial cysts, underlining the fact that this new action is conserved in 2D and 3D contexts. Cancer cells may also use MT2-MMP to cleave E-cadherin in other cell compartments. Indeed, unregulated MT2-MMP-dependent E-cadherin proteolysis has been postulated to drive an epithelial–mesenchymal transition program (Liu et al., 2016).
ZO-1 is essential in preserving epithelial cell barrier integrity (Fanning and Anderson, 2009), and its redistribution is critical to cell ‘shedding’ in the intestine to assure epithelial barrier function (Guan et al., 2011). ZO-1 can also contribute to epithelial–mesenchymal transition as well as epithelial cell motility and cytokinesis via its association with α5β1 integrin (Hämälistö et al., 2013; Polette et al., 2007; Tuomi et al., 2009). The ZO-1-driven apical localization of MT2-MMP, and the consequent cleavage of E-cadherin, may confer ZO-1 with additional cellular functions. E-cadherin has previously been reported to undergo processing by MMP-7 and MMP-2, and also MT1-MMP (but not at apical-like lipid membrane microdomains in tumor cells) (Grieve and Rabouille, 2014; Lynch et al., 2010; Rozanov et al., 2004). Here, we demonstrate that E-cadherin is a novel cis substrate for MT2-MMP, in contrast to ADAM10, which cleaves E-cadherin in trans to mark boundaries for cell sorting (Solanas et al., 2011). Selective proteolytic events can therefore drive distinct epithelial cell responses in a tissue-context-dependent manner. In a physiological setting such as the intestinal niche, our data indicate a key role for MT2-MMP in driving the directional apical proliferation required for tissue homeostasis. These data support a model wherein MT-MMP cleavage of cadherins may serve as a general mechanism to promote regulated stem cell exit from quiescence in different contexts, expanding the previous report of N-cadherin cleavage by MT5-MMP in the neural niche (Porlan et al., 2014). Given our in vitro data, the association of MT5-MMP with ZO-1 may likewise regulate N-cadherin proteolysis.
MT2-MMP is expressed in epithelial tissues during embryogenesis (http://www.emouseatlas.org/) and our data demonstrate that in the adult organism, MT2-MMP is expressed in restricted locations in epithelial tissues, such as the intestine, that display rapid and regulated cell turnover. Despite the absence of gross intestinal defects in MT2-MMP-null mice, the absence of the protease results in crypt-specific alterations. First, in the absence of MT2-MMP, an extended overlap of E-cadherin and ZO-1 at cell–cell junctions occurs that is suggestive of an over accumulation of E-cadherin as a consequence of its impaired cleavage. Second, as a result of changes in E-cadherin localization and/or abundance, Src kinase was mislocalized to intracellular domain as opposed to the basolateral junctional compartment, an event correlating with reduced cell proliferation and morphological changes in intestinal crypts. The phenotype in the colon from null mice was also recapitulated ex vivo by impaired 3D organoid formation by colon epithelial cells devoid of MT2-MMP. A role for MT2-MMP in epithelial cell proliferation has previously been suggested by the drastic reduction in cell number and impaired branching observed in MT2-MMP-targeted salivary glands ex vivo. While decreased release of collagen IV-NC1 fragments was proposed as the mechanism of action (Rebustini et al., 2009), it seems likely that impaired E-cadherin cleavage also contributes to the proliferative phenotype. By contrast, MT2-MMP deletion in vivo did not affect epithelial cell proliferation in the mammary gland during the early postnatal period (Feinberg et al., 2016). However, whether a phenotype may be apparent in mammary glands during active lactation when proliferation is dramatically enhanced deserves further investigation. Interestingly, MT2-MMP overexpression has been reported to trigger the formation of a multilayered epithelium when MDCK cells are cultured on collagen gels, and although MT2-MMP-dependent collagenolysis was assumed to be the preferred mechanism of action, effects on proliferation or asymmetric division were not assessed (Hotary et al., 2000). In the intestinal stem niche, asymmetric cell division may be particularly important to maintain tissue fitness by balancing stemness and differentiation (De Mey and Freund, 2013). The apical location of MT2-MMP and the hypo-proliferative phenotype found in null intestinal crypts suggest a potential role for MT2-MMP-mediated E-cadherin cleavage in driving asymmetric cell division, perhaps also favoring epithelial cell polarized migration, as described recently in endothelial cells (Costa et al., 2016).
How does MT2-MMP drive apical epithelial cell accumulation? In Ras-overexpressing MDCK cells, MMP-dependent E-cadherin cleavage has been shown to promote epithelial cell extrusion (Grieve and Rabouille, 2014). Actomyosin-driven forces are essential for this process (Gu and Rosenblatt, 2012), and MT2-MMP-mediated cleavage of E-cadherin at ZO-1-positive apical junctional sites resulted in relaxation of the actomyosin cytoskeleton, supporting the contention that cell extrusion may contribute to the apical phenotype observed in MT2-MMP-transfected MDCK cells. Accordingly, activating cortical myosin tension with 4-HAP (Surcel et al., 2015) restored epithelial cell homeostasis. Apical accumulation of MT2-MMP-MDCK cells seems to also involve increased proliferation as supported by the observed persistent mitosis in the absence of growth factors. Disruption of apical E-cadherin could potentially decrease miR-30b, thereby increasing Snail 1 expression (Kourtidis et al., 2015), which could trigger epithelial cell proliferation (Tseng et al., 2016). However, we could not detect robust differences in miR-30b or Snail levels in MT2-MMP-MDCK cells (data not shown), suggesting that alternative mechanisms contribute to this phenotype. Upon apical disruption, the preserved basolateral E-cadherin pool could also promote anchorage-independent growth via basolateral junctional activation of Src. Indeed, inhibition of pSrc abolished epithelial cell accumulation and proliferation in MT2-MMP-expressing epithelial cells, pointing to Src activation as a key player. Moreover, Src inhibition also rescued E-cadherin levels at cell–cell junctions, potentially reflecting the Src-dependent regulation of E-cadherin endocytosis (Chen et al., 2016).
In sum, we have identified MT2-MMP as a specific molecular regulator of epithelial cell homeostasis and remodeling. The novel MT2-MMP–ZO-1–E-cadherin axis characterized herein controls epithelial cell proliferative responses that are essential for physiological cellular turnover.
MATERIALS AND METHODS
Antibodies
Antibodies used were against β-actin (Sigma-Aldrich, A5441), GST (Thermo Fisher Scientific, A5800), HA (Covance, MMS-101P), MT1-MMP (LEM2/63; Gálvez et al., 2001), MT2-MMP (R&D Systems, MAB9161; and a rabbit polyclonal antibody generated by our group at CNB, Madrid, Spain, directed against 16 aa of hMT2 DEPWTFSSTDLHGNNL), Tubulin (Sigma, T6074), pSrc (Cell Signaling, 2101), ZO-1 (Thermo Fisher Scientific, 40-2300), E-cadherin (Cell Signaling, 3195 and BD Biosciences, 610181), Hoechst 3342 (Thermo Fisher Scientific), Ki67 (Abcam, ab16667), Phalloidin 647 (Thermo Fisher Scientific, A22287), Myosin IIB (Santa Cruz Biotechnology, sc-15370), β-catenin (BD Biosciences, 610153), Rho-GDI (Santa Cruz Biotechnology, sc-360), Ezrin (Upstate, 07-130), EEA1 (Santa Cruz Biotechnology, sc-6415), TfR (Invitrogen, H68.4), HGS (Abcam, ab72053), TSG101 (Abcam, ab30871). All antibodies were used at 1:100 dilution.
Cell culture
Human umbilical vein endothelial cells (HUVECs) were obtained and cultured as described previously and used for proteomics assay (Gálvez et al., 2001). MDCK cells were provided by Fernando Martín-Belmonte from CBM (Madrid, Spain), and were cultured in MEM (Gibco, Thermo Fisher Scientific) supplemented with 5% FBS, 2 mM L-glutamine, 50 IU/ml penicillin, and 50 μg/ml streptomycin. In the case of the stable transfected MDCK 400 µg/ml neomycin (G418) (Sigma-Aldrich) was also added to the culture medium. Caco2 cells were provided by Francisco Real from CNIO (Madrid, Spain). All cell lines were routinely checked for mycoplasma infection.
MDCK stable transfection and clone selection
MDCK were transfected with pCR3.1 empty vector (mock) or pCR3.1 containing the cDNA fragments: MT1-MMP-HA, MT1-MMP-K581W-HA, MT2-MMP-HAFlag or MT2-MMP-W668K-HAFlag; the HA epitope YPYDVPDYA is located between the hemopexin 4 and the transmembrane domain, precisely after E584, in MT2-MMP. For transfection, FuGENE 6 (Roche) was used. Transfected MDCK cells were cultured in the presence of 400 µg/ml of G418 and selection of single cell colonies was performed by limited cell dilution in 96-well plates. Each cell clone was tested for the corresponding MT-MMP expression by immunofluorescence and western blot analysis. For MDCK polarization studies, cells were cultured on Transwell filters (Costar) for 3–5 days (Oztan et al., 2008). MDCK formation of 3D cysts in Matrigel was performed as previously described (Martín-Belmonte et al., 2008).
Expression vectors and plasmid construction
GST fusion proteins of MT1 and MT2 cytosolic tails were obtained by direct cloning. PGEX-4T2 vector was digested with BamHI and XhoI, and the DNA sequences corresponding to the cytosolic tails flanked with BamHI and XhoI sites were ordered. The oligonucleotides were heated for 5 min at 94°C and cooled slowly for hybridization. Hybridized oligonucleotides were added to digested pGEX-4T2 and ligation was performed by T4 ligase. The cytosolic tail mutants of MT1-MMP and MT2-MMP, K581W and W668K, respectively, were performed with a QuikChange Site-Directed Mutagenesis kit (Stratagene). The primers used for these mutations were: fw, 5′-CAGCGTTCCCTGCTGGACTGGGTCTGATAGAAGCCG-3′ and rv: 5′-CGGCTTCTATCAGACCCAGTCCAGCAGGGAACGCTG-3′ for MT2-MMP-K581W and fw: 5′-GCGC TCGCTGCAGGAGAAGGTCTGAAATTCTGCAG-3′ and rv: 5′-CTGCAGAATTTCAGACCTTCTCCTGCAGCGAGCGC-3′ for MT2-MMP-W668K. All constructs were finally sequenced at the Genomic Service of the Spanish National Cancer Research Centre (CNIO). To generate a catalytically inactive MT2-MMP, the E260 amino acid at its active site was changed to an alanine residue. This mutation was performed with a QuikChange Site-Directed Mutagenesis kit (Stratagene). The primers used for this mutation were; fw: 5′-CTGGTGGCAGTGCATGCGCTGGGCCACGCGCTG-3′ and rv: 5′-CAGCGCGTGGCCCAGCGCATGCACTGCCACCAG-3′.
Pull-down assays
For pull down with GST fusion proteins, HUVECs were lysed [lysis buffer: 50 mM Tris-HCl, pH 7.5, 1% Triton X-100, 0.1% SDS, 0.5% sodium deoxycholate, 500 mM NaCl, 10 mM MgCl2, proteinase inhibitor cocktail (Roche), 0.2 mg/ml PMSF, 25 mM NaF and 1 mM Na3VO4] and the lysates were precipitated with GST fusion proteins or GST alone. After washing (10–50 mM Tris-HCl, pH 7.5, 0.5% Triton X-100, 150 mM NaCl and 1 mM DTT), bound proteins were eluted by boiling in Laemmli buffer. Samples were resolved by SDS-PAGE and western blotting or protein identification by MS was performed. For pull down with biotinylated peptides, cells were lysed with 1% NP40-TBS [50 mM Tris-HCl pH 7.5, 150 mM NaCl with proteinase inhibitor cocktail (Roche), 0.2 mg/ml PMSF, 25 mM NaF and 1 mM Na3VO4] and N-terminal biotinylated peptides corresponding to the cytosolic tails of the different WT or MT-MMP mutants coupled with neutravidin beads were added. After overnight incubation, the precipitated was washed 6 times with 0.1% NP40-TBS and resuspended in Laemmli buffer for SDS-PAGE and western blot analysis.
Protein identification by mass spectrometry
Following SDS-PAGE, samples was stained with Coomassie Blue and the proteins corresponding to the observed bands, and the equivalent regions of the other lanes, were identified by liquid chromatography/mass spectrometry (LC-MS) in the CNIC Proteomics Unit. For protein identification, fragmentation spectra were searched against the MSDB database using the MASCOT program. Detailed analysis of mass spectra was carried out using Data Analysis (Bruker).
ELISA
Streptavidin binding plates (96 wells, Pierce) were incubated at 4°C for 3 h in 0.1 M Na2CO3 buffer (pH 9.6) containing 40 µM of biotinylated peptides (GenScript) corresponding to the MT-MMP cytosolic tails: MT1-MMPcyt (563RRHGTPRRLLYCQRSLLDKV582), MT1-MMPcyt-K581W, MT2-MMPcyt (650QRKGAPRVLLYCKRSLQEWV669), MT2-MMPcyt-E667A, MT2-MMPcyt-W668K, MT2-MMPcyt-669ΔV. To avoid non-specific binding to biotinylated peptides, samples were blocked with 2% BSA in TBS-Tween containing 10% FBS overnight at 4°C. GST-tagged recombinant ZO-1PDZ1-3, ZO-1PDZ1, ZO-1PDZ2, ZO-1PDZ3 or GST alone was added to the wells in equal concentrations and incubated at room temperature (RT) for 1 h. After extensive washing, GST protein binding to the cytosolic tail sequences was detected using anti-GST antibody and HRP-based detection. For analysis, the optical density of an empty well was subtracted from the absorbance data obtained.
Biotin labeling, co-immunoprecipitation and western blot assays
For biotin labeling, MDCK cells were incubated with biotin (EZ-Link Sulfo-NHS-Biotin, 21217 Thermo Fisher) for 30 min, and then washed with quenching solution (100 mM glycine). Cells were lysed in 1% NP40-TBS with proteinase and phosphatase inhibitor cocktail (Roche) followed by sonication. Lysates were incubated with streptavidin magnetic beads (Solulink M-1002-010) for 4 h and recovered with a magnet. For co-immunoprecipitation assays in MDCK transfectants, cell lysis was performed in 1% NP40-TBS with proteinase and phosphatase inhibitor cocktail (Roche) and with or without previous crosslinking (1% formaldehyde for 10 min) with similar results. After sonication for 5 min at low intensity and pre-clearing with Dynabeads (Protein G 10003D, Thermo Fisher), lysates were incubated for 3 h at 4°C with 2 µg of anti-HA antibody and then for 1.5 h with Dynabeads. Immunoprecipitates were washed 6 times with 0.1% NP40-TBS and resolved by 8% SDS-PAGE under reducing conditions. After transferring to a nitrocellulose membrane (Bio-Rad), western blot analysis was performed. Membranes were blocked with 5% BSA-PBS, and primary antibodies diluted in 2% BSA-PBS and were incubated for 1 h at RT. After membranes were washed with 0.2% Tween20-PBS, they were incubated for 1 h at RT with a horseradish peroxidase-conjugated secondary antibody (Jackson, 111-035-003 and 115-035-003). Protein bands were visualized by enhanced chemiluminescence (ECL; Amersham Pharmacia Biotech, Buckinghamshire, United Kingdom). Densitometric analysis of band intensity was done using ImageJ software (https://imagej.nih.gov/ij/).
Immunofluorescence microscopy
Cells were fixed with 4% paraformaldehyde in PBS for 10 min at RT, permeabilized for 15 min with 0.1% Triton X-100 in blocking buffer (2% BSA, 10% FBS, 1 mM MgCl2 and 1 mM EDTA in PBS) at 4°C and then blocked for 1 h in the same buffer. Primary antibodies and Alexa-conjugated secondary antibodies were incubated for 1 h at RT. For nuclear staining, Hoechst 33342 (Sigma) was used. Coverslips were mounted with Prolong mounting medium (Invitrogen). For tissue staining, paraffin-embedded colon slices obtained from 12-week-old wild-type and Mmp15(MT2-MMP)-null mice were deparaffinized, rehydrated, unmasked with citrate buffer, pH 6, and permeabilized for 1 h with 0.3% Triton X-100 in blocking buffer (5% BSA, 5% goat serum, in PBS). Primary and Alexa-Fluor-conjugated secondary antibodies were used as described above for cell staining. Images were acquired with objectives 40×/1.25 or 63×/1.4 in a LSM700 confocal microscope (Carl Zeiss). Images were converted to Tiff files with ZEN (Carl Zeiss) software and analyzed with ImageJ. For quantification of E-cadherin intensity in MDCK cells, a linear ROI was drawn, with the mid-point of the line in the junction between the cells, in order to also analyze the E-cadherin peak profile. For quantification of circularity, a polygonal ROI around the cell was drawn, and the ‘Shape description’ plugin was used. For analysis of pSrc distribution, orthogonal x–z views of the cells were obtained with ImageJ. Several polygonal ROIs in the F-actin channel were selected: the apical portion, the basolateral portion, the individual junctional portions, and one ROI of the total orthogonal view. Junctional intensity was shown as the average of individual junctional intensities. For RGB profile analysis, a ROI line was drawn from junction to junction inside one cell, and analyzed with the ‘RGB profile’ plugin. pSrc peaks were quantified in this RGB profile by hand. For quantification of E-cadherin overlapping with ZO-1, linear ROIs were drawn in 63× images from transversal colon slices, from the lumen to the basal portion of the sample along the junction, in the E-cadherin channel.
Prediction of cleavage sites and in silico protein modeling
For in silico protein modeling of canine E-cadherin, the Fasta sequence of extracellular domain (EC) of mature canine E-Cad protein (UniProt id: F1PAA9, residues 157–712) and the transmembrane and intra-cellular domain (TMIC; UniProt id: F1PAA9, residues 713–885) were submitted to a local implementation of I-Tasser software suite v4.4 (Yang et al., 2015) for threading modeling with homology. The best model with minimal energy and correct folding [best structural alignment to templates (cadherin domains)] with correct topology was selected for the EC domain. For the transmembrane and intracellular domain (TMIC) the model with minimal energy was selected and the dihedral angles were fixed according to the secondary structure predicted by the psi-pred program included in the I-Tasser suite to obtain a correct orientation of the TM domain (approximately normal to the membrane plane). Both models were aligned with pymol v1.8 program (www.pymol.org) and joined using the FG_MD tool (Zhang et al., 2011) to obtain a final refined model. A similar approach was used for human E-cadherin. For in silico modeling of human MT2-MMP/MMP15, Fasta sequence of extracellular domain (EC) of mature human MT2-MMP protein (Uniprot Id: P51511, residues 132-625) was submitted to a local implementation of I-Tasser software suite v4.4 (Yang et al., 2015) for threading modeling with homology. The best model with minimal energy and correct folding (best structural alignment to templates with a correct topology of the catalytic domain as previously described) was selected. For the TMIC of mature human MT2-MMP protein (UniProt id: P51511, residues 621–669), the process described above was used and the model with minimal energy and best topology compatible with a TM domain was selected. Both models were structurally aligned using pymol, and the new model generated, composed of the MT2-MMP EC and the MT2-MMP TMIC models, was used as template. The gap between them was closed using the loopmodel tool (Mandell et al., 2009; Stein and Kortemme, 2013) of Rosetta suite v3.5 release 2015.38.58158 (www.rosettacommons.org). The more stable model with topology compatible with a TM protein was selected as final model for MT2-MMP. For docking of both dog and human complexes, the MT2-MMP model and E-Cad model from dog or human were closely positioned with the only restriction marked by the membrane and the topology previously reported for normal TM proteins using the pymol v1.8 program (www.pymol.org) with the new dimeric model in each case used as the initial template. A new PDB file for the model where the membrane protein structure is transformed into PDB coordinates (z-axis is membrane normal) using the PPM server (http://opm.phar.umich.edu/server.php) was generated. To generate full (symmetric) and asymmetric spanfile from the PDB structure, the spanfile_from_pdb application from the membrane framework of Rosetta was used. 1000 complex models were generated using the mp_dock application (Alford et al., 2015) from the membrane framework of Rosetta and clustered. The model with minimal E and topology compatible with cleavage of E-Cad by MT2-MMP (active site of MT2-MMP close to E-Cad chain) was selected as the final model. In parallel, each E-Cad model (dog and human) was submitted to the CleavPredict server (http://cleavpredict.sanfordburnham.org/) to predict the putative cleavage site by MT2-MMP and the results were validated with MEROPS, the peptidase database (http://merops.sanger.ac.uk/). Comparing the predicted sites with the models of complexes obtained previously, two positions in E-Cad were marked as high probability (445 and 459) given that they were positioned more closely and accessible to the catalytic site of MT2-MMP. Both sites were ‘curated’ experimentally showing greater activity at position 445 than 459 (data not shown). In examining this difference, newly refined models for each position (445 and 459) and for both complexes (MT2-MMP plus E-Cad_dog or E-Cad_human) were modeled. In each case, using the complexes models obtained above, the active site of MT2-MMP was repositioned close to the 445 residue or the 459 residue of E-Cad protein (both dog and human) and a new cycle of docking with positional restrictions using the mp_dock application (Alford et al., 2015) from the membrane framework of Rosetta and cluster was made. In each case, the best model with minimal E and bigger probability was selected. A final cycle of relax for minimizing E and clashes using the model described above as template, and the full spanfile was made using the ‘relax’ application from Rosetta.
In vitro digestion and MS analysis
The catalytic domain of human recombinant MT2-MMP [expressed as a recombinant protein in E. coli (Merk Millipore, 475938)] and the peptide H-GPIPEPRNMDFCQKNPQP-OH corresponding to the sequence 438–455 of canine E-cadherin (synthesized by JPT Peptide Technologies) were incubated in assay buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 5 mM CaCl2) for 16 h at 37°C. The reaction mixture was analyzed on an Orbitrap Fusion Tribrid mass spectrometer (Thermo Scientific). Detailed analysis of MS/MS fragmentation spectra was carried out using Xcalibur software (Thermo Scientific).
siRNA transfection in Caco2 cells
Caco2 cells grown on Transwells were transfected in several rounds with human MT2-MMP siRNA oligonucleotides (AM16708, Thermo Fisher) according to the manufacturer's procedures with lipofectamine RNAimax (Invitrogen).
Mosaic cell cultures
MT2-FL and MT2-WK MDCK cells were infected with SFFVp lentivirus expressing GFP or mKate2 fluorescent proteins, respectively, at a MOI of 10. Twenty-four hours later, cells were trypsinized, mixed in a 1:1 proportion and seeded on Transwells for 3 days to allow for cell polarization.
Colon crypt isolation
Colon organoids were cultured as described (Sato et al., 2011). Briefly, distal portions of the colon (around 6 cm) were excised, flushed with cold PBS and opened longitudinally. Small pieces (2–4 mm in length) were cut with scissors and further washed with ice-cold PBS until the supernatant was clear. Next, tissue fragments were incubated in cold 2 mM EDTA chelation buffer for 30 min at 4°C. After removal of the EDTA buffer, tissue fragments were vigorously resuspended in cold chelation buffer using a 10 ml pipette to isolate intestinal crypts. The tissue fragments were allowed to settle under normal gravity for 1 min, and the supernatant was removed for inspection by inverted microscopy. The resuspension/sedimentation procedure was repeated typically 8 times and the supernatants containing crypts (from wash 1 to wash 8) were collected in 50 ml Falcon tubes coated with bovine serum albumin. Isolated crypts were pelleted, washed with cold chelation buffer, and centrifuged at 200 g for 3 min at 4°C to separate crypts from single cells. After centrifugation and supernatant removal, crypts were resuspended in Matrigel (Corning, 734-1101) and plated in 24-well plates (250 crypts/well).
Intestinal organoid culture
Organoids were cultured in basal crypt culture medium [advanced Dulbecco's modified Eagle medium supplemented with penicillin-streptomycin, 10 mM HEPES, 2 mM Glutamax, 1× N2 (ThermoFisher Scientific 100×, 17502048), 1× B-27 (ThermoFisher Scientific 50×, 17504044) and 1× N-acetyl-L-cysteine (Sigma, A7250)] and overlaid with medium containing 50 ng/ml murine EGF (Thermo Fisher Scientific, PMG8041), 20% R-spondin conditioned medium (CM) (kind gift from Calvin Kuo, Stanford University School of Medicine, Stanford, CA, USA), 10% Noggin-CM and 65% Wnt-CM (kind gifts from Hans Clevers, Hubrecht Institute, Utrecht, The Netherlands). Medium was renewed every other day. For passaging, organoid cultures were washed, then Matrigel and organoids were disrupted mechanically by strong pipetting, centrifuged at 200 g, 5 min at 4°C and resuspended in Matrigel prior to plating in 24-well or 8-well ibidi chambers.
siRNA transfection of cultured organoids
SiRNA transfection was performed in organoids as previously described (Zhang et al., 2015). Briefly, cultured organoids were manually disrupted by vigorous pipetting. Cells were pelleted by centrifugation at 200 g for 5 min, and medium was removed. Pelleted cells were suspended in 100 μl transfection mixture at 37°C for 20 min, after which cells were pelleted, suspended, and plated in Matrigel with culture medium as described above. Cells were imaged at 24, 48 and 72 h after transfection, and number of organoids/well quantified. After 72 h, cells were fixed to perform imaging analysis. The transfection mixture was prepared by mixing 50 μl Opti-MEM (Gibco, 11058021) containing 200 nM siRNA and 50 μl Opti-MEM mixed with 3 μl Lipofectamine siRNA Max transfection reagent (Invitrogen, 13778), Mock and Mmp15/MT2-MMP siRNA oligos were obtained from Dharmacon. Mock mouse oligos (ON-TARGETplus, non-targeting pool, D-001810-10-05) had the following target sequences: (1) 5′-UGGUUUACAUGUCGACUAA-3′, (2) 5′-UGGUUUACAUGUUGUGUGA-3′, (3) 5′-UGGUUUACAUGUUUUCUGA-3′ and (4) 5′-UGGUUUACAUGUUUUCCUA-3′. Mmp15/MT2-MMP mouse oligos [ON-TARGETplus Mouse Mmp15 (17388) siRNA, SMART pool] had the following target sequences: (1) 5′-GGCUAGAACACUCAAGUAA-3′, (2) 5′-GCACAGACAUCCCCUAUGA-3′, (3) 5′-GAGCGGAGGCUGACAUCAU-3′ and (4) 5′-CAACGAACGACUACGGAUG-3′.
Immunofluorescence staining of colon organoids
For immunofluorescence labeling and imaging, organoids were grown in Matrigel on eight-chamber μslides (Ibidi). Organoids were fixed in PBS containing 4% paraformaldehyde (pH 7.4) and 2% sucrose for 20 min, permeabilized (PBS, 0.15% Triton X-100), washed in PBS and blocked (PBS-Triton X-100 0.2%, 2% normal goat serum, 1% BSA). Then organoids were incubated with primary antibodies against the following antigens diluted in the same blocking buffer: MT2-MMP (1:100, rabbit polyclonal generated by our group; see Antibodies section), E-cadherin (1:100, rabbit monoclonal, Cell signaling, 3195) and β-catenin (1:100, mouse monoclonal, BD Biosciences, 610154) overnight at 4°C with slow agitation. Organoids were washed in PBS containing 0.2% Triton X-100 and incubated overnight in PBS with 0.2% Triton X-100, 1% normal goat serum and 0.5% BSA at 4°C with the appropriate Alexa Fluor secondary antibody (1:500) along with Hoechst 33342 (1:10,000) and Rhodamine-Phalloidin (1:300, Invitrogen R415) to counterstain nuclei and F-actin, respectively. Organoids were washed with PBS buffer with 0.2% Triton X-100 and mounted using Fluoromount G (ThermoFisher Scientific, 00-4958-02). Organoids were imaged with a Zeiss 510 Meta Live confocal microscope, using a 20× objective lens.
Statistical analysis
Test and control samples were compared for statistical significance by using the unpaired Student t-test or the Welch's t-test as indicated. For more than two populations, one-way ANOVA analysis with Dunnett's post-test was used when comparing samples against control; or with Sidak post-test when comparing different kinds of pairs. For cell cycle experiments, two-way ANOVA with Dunnett's post-test was used. Differences were considered statistically significant at P<0.05.
Supplementary Material
Acknowledgements
We thank Laura Balonga for technical support and Raquel Sánchez and Pilar Martín for performing qPCR assays.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: J.G.-E., V.M., M.M.-A., M.V.H.-R., M.J.O., A.G.A.; Methodology: J.G.-E., V.M., M.M.-A., M.V.H.-R., E. Camafeita, F.M., M.J.O.; Validation: J.G.-E., V.M., M.M.; Formal analysis: J.G.-E., V.M., M.M., E. Calvo, E. Camafeita, F.M.; Investigation: J.G.-E., V.M., M.M.-A., M.V.H.-R., A.C., E. Calvo, E. Camafeita, F.M.; Resources: T.F., S.J.W.; Writing - original draft: A.G.A.; Writing - review & editing: J.G.-E., V.M., M.M.-A., M.J.O., S.J.W.; Visualization: J.G.-E., V.M., M.M.-A., F.M.; Supervision: A.G.A.; Project administration: A.G.A.; Funding acquisition: M.J.O., S.J.W., A.G.A.
Funding
This work was supported by grants from the Ministerio de Economía y Competitividad [MEIC; RD12/0042/0023 (FEDER cofunded) and SAF2014-52050R to A.G.A.] and from Kreftforeningen (Norwegian Cancer Society; 182767-2016). J.G.-E. is funded by a fellowship from MEIC. S.J.W. is supported by the National Institutes of Health (CA088308) and the Breast Cancer Research Foundation. The CNIC is supported by MEIC and the Pro-CNIC Foundation, and is a Severo Ochoa Center of Excellence (MEIC award SEV-2015-0505). Deposited in PMC for release after 12 months.
Supplementary information
Supplementary information available online at http://jcs.biologists.org/lookup/doi/10.1242/jcs.203687.supplemental
References
- Alford R. F., Koehler Leman J., Weitzner B. D., Duran A. M., Tilley D. C., Elazar A. and Gray J. J. (2015). An integrated framework advancing membrane protein modeling and design. PLoS Comput. Biol. 11, e1004398 10.1371/journal.pcbi.1004398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum B. and Georgiou M. (2011). Dynamics of adherens junctions in epithelial establishment, maintenance, and remodeling. J. Cell Biol. 192, 907-917. 10.1083/jcb.201009141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H.-R., Yeh Y.-C., Liu C.-Y., Wu Y.-T., Lo F.-Y., Tang M.-J. and Wang Y.-K. (2016). DDR1 promotes E-cadherin stability via inhibition of integrin-beta1-Src activation-mediated E-cadherin endocytosis. Sci. Rep. 6, 36336 10.1038/srep36336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa G., Harrington K. I., Lovegrove H. E., Page D. J., Chakravartula S., Bentley K. and Herbert S. P. (2016). Asymmetric division coordinates collective cell migration in angiogenesis. Nat. Cell Biol. 18, 1292-1301. 10.1038/ncb3443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Mey J. R. and Freund J.-N. (2013). Understanding epithelial homeostasis in the intestine: an old battlefield of ideas, recent breakthroughs and remaining controversies. Tissue Barriers 1, e24965 10.4161/tisb.24965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanning A. S. and Anderson J. M. (2009). Zonula occludens-1 and -2 are cytosolic scaffolds that regulate the assembly of cellular junctions. Ann. NY Acad. Sci. 1165, 113-120. 10.1111/j.1749-6632.2009.04440.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg T. Y., Rowe R. G., Saunders T. L. and Weiss S. J. (2016). Functional roles of MMP14 and MMP15 in early postnatal mammary gland development. Development 143, 3956-3968. 10.1242/dev.136259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gálvez B. G., Matias-Román S., Albar J. P., Sánchez-Madrid F. and Arroyo A. G. (2001). Membrane type 1-matrix metalloproteinase is activated during migration of human endothelial cells and modulates endothelial motility and matrix remodeling. J. Biol. Chem. 276, 37491-37500. 10.1074/jbc.M104094200 [DOI] [PubMed] [Google Scholar]
- Grieve A. G. and Rabouille C. (2014). Extracellular cleavage of E-cadherin promotes epithelial cell extrusion. J. Cell Sci. 127, 3331-3346. 10.1242/jcs.147926 [DOI] [PubMed] [Google Scholar]
- Gu Y. and Rosenblatt J. (2012). New emerging roles for epithelial cell extrusion. Curr. Opin. Cell Biol. 24, 865-870. 10.1016/j.ceb.2012.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Y., Watson A. J. M., Marchiando A. M., Bradford E., Shen L., Turner J. R. and Montrose M. H. (2011). Redistribution of the tight junction protein ZO-1 during physiological shedding of mouse intestinal epithelial cells. Am. J. Physiol. Cell Physiol. 300, C1404-C1414. 10.1152/ajpcell.00270.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haegebarth A. and Clevers H. (2009). Wnt signaling, lgr5, and stem cells in the intestine and skin. Am. J. Pathol. 174, 715-721. 10.2353/ajpath.2009.080758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hämälistö S., Pouwels J., de Franceschi N., Saari M., Ivarsson Y., Zimmermann P., Brech A., Stenmark H. and Ivaska J. (2013). A ZO-1/alpha5beta1-integrin complex regulates cytokinesis downstream of PKCepsilon in NCI-H460 cells plated on fibronectin. PLoS ONE 8, e70696 10.1371/journal.pone.0070696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotary K., Allen E., Punturieri A., Yana I. and Weiss S. J. (2000). Regulation of cell invasion and morphogenesis in a three-dimensional type I collagen matrix by membrane-type matrix metalloproteinases 1, 2, and 3. J. Cell Biol. 149, 1309-1323. 10.1083/jcb.149.6.1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes S. C. and Fehon R. G. (2007). Understanding ERM proteins–the awesome power of genetics finally brought to bear. Curr. Opin. Cell Biol. 19, 51-56. 10.1016/j.ceb.2006.12.004 [DOI] [PubMed] [Google Scholar]
- Kourtidis A., Ngok S. P., Pulimeno P., Feathers R. W., Carpio L. R., Baker T. R., Carr J. M., Yan I. K., Borges S., Perez E. A. et al. (2015). Distinct E-cadherin-based complexes regulate cell behaviour through miRNA processing or Src and p120 catenin activity. Nat. Cell Biol. 17, 1145-1157. 10.1038/ncb3227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalczyk A. P. and Nanes B. A. (2012). Adherens junction turnover: regulating adhesion through cadherin endocytosis, degradation, and recycling. Subcell. Biochem. 60, 197-222. 10.1007/978-94-007-4186-7_9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Sun X., Feng J., Deng L.-L., Liu Y., Li B., Zhu M., Lu C. and Zhou L. (2016). MT2-MMP induces proteolysis and leads to EMT in carcinomas. Oncotarget 7, 48193-48205. 10.18632/oncotarget.10194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch C. C., Vargo-Gogola T., Matrisian L. M. and Fingleton B. (2010). Cleavage of E-cadherin by matrix metalloproteinase-7 promotes cellular proliferation in nontransformed cell lines via activation of RhoA. J. Oncol. 2010, 530745 10.1155/2010/530745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macara I. G., Guyer R., Richardson G., Huo Y. and Ahmed S. M. (2014). Epithelial homeostasis. Curr. Biol. 24, R815-R825. 10.1016/j.cub.2014.06.068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandell D. J., Coutsias E. A. and Kortemme T. (2009). Sub-angstrom accuracy in protein loop reconstruction by robotics-inspired conformational sampling. Nat. Methods 6, 551-552. 10.1038/nmeth0809-551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín-Belmonte F., Yu W., Rodríguez-Fraticelli A. E., Ewald A. J., Werb Z., Alonso M. A. and Mostov K. (2008). Cell-polarity dynamics controls the mechanism of lumen formation in epithelial morphogenesis. Curr. Biol. 18, 507-513. 10.1016/j.cub.2008.02.076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monea S., Jordan B. A., Srivastava S., DeSouza S. and Ziff E. B. (2006). Membrane localization of membrane type 5 matrix metalloproteinase by AMPA receptor binding protein and cleavage of cadherins. J. Neurosci. 26, 2300-2312. 10.1523/JNEUROSCI.3521-05.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montesano R., Schaller G. and Orci L. (1991). Induction of epithelial tubular morphogenesis in vitro by fibroblast-derived soluble factors. Cell 66, 697-711. 10.1016/0092-8674(91)90115-F [DOI] [PubMed] [Google Scholar]
- Nieto M. A. (2011). The ins and outs of the epithelial to mesenchymal transition in health and disease. Annu. Rev. Cell Dev. Biol. 27, 347-376. 10.1146/annurev-cellbio-092910-154036 [DOI] [PubMed] [Google Scholar]
- Oztan A., Rondanino C. and Apodaca G. (2008). Transcytosis of polymeric immunoglobulin a in polarized Madin-Darby canine kidney cells. Methods Mol. Biol. 440, 157-170. 10.1007/978-1-59745-178-9_12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page-McCaw A., Ewald A. J. and Werb Z. (2007). Matrix metalloproteinases and the regulation of tissue remodelling. Nat. Rev. Mol. Cell Biol. 8, 221-233. 10.1038/nrm2125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polette M., Mestdagt M., Bindels S., Nawrocki-Raby B., Hunziker W., Foidart J.-M., Birembaut P. and Gilles C. (2007). Beta-catenin and ZO-1: shuttle molecules involved in tumor invasion-associated epithelial-mesenchymal transition processes. Cells Tissues Organs 185, 61-65. 10.1159/000101304 [DOI] [PubMed] [Google Scholar]
- Porlan E., Martí-Prado B., Morante-Redolat J. M., Consiglio A., Delgado A. C., Kypta R., López-Otín C., Kirstein M. and Fariñas I. (2014). MT5-MMP regulates adult neural stem cell functional quiescence through the cleavage of N-cadherin. Nat. Cell Biol. 16, 629-638. 10.1038/ncb2993 [DOI] [PubMed] [Google Scholar]
- Rebustini I. T., Myers C., Lassiter K. S., Surmak A., Szabova L., Holmbeck K., Pedchenko V., Hudson B. G. and Hoffman M. P. (2009). MT2-MMP-dependent release of collagen IV NC1 domains regulates submandibular gland branching morphogenesis. Dev. Cell 17, 482-493. 10.1016/j.devcel.2009.07.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roghi C., Jones L., Gratian M., English W. R. and Murphy G. (2010). Golgi reassembly stacking protein 55 interacts with membrane-type (MT) 1-matrix metalloprotease (MMP) and furin and plays a role in the activation of the MT1-MMP zymogen. FEBS J. 277, 3158-3175. 10.1111/j.1742-4658.2010.07723.x [DOI] [PubMed] [Google Scholar]
- Rozanov D. V., Deryugina E. I., Monosov E. Z., Marchenko N. D. and Strongin A. Y. (2004). Aberrant, persistent inclusion into lipid rafts limits the tumorigenic function of membrane type-1 matrix metalloproteinase in malignant cells. Exp. Cell Res. 293, 81-95. 10.1016/j.yexcr.2003.10.006 [DOI] [PubMed] [Google Scholar]
- Sato T., Stange D. E., Ferrante M., Vries R. G., Van Es J. H., Van den Brink S., Van Houdt W. J., Pronk A., Van Gorp J., Siersema P. D. et al. (2011). Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett's epithelium. Gastroenterology 141, 1762-1772. 10.1053/j.gastro.2011.07.050 [DOI] [PubMed] [Google Scholar]
- Simmons N. L. (1982). Cultured monolayers of MDCK cells: a novel model system for the study of epithelial development and function. Gen. Pharmacol. 13, 287-291. 10.1016/0306-3623(82)90047-7 [DOI] [PubMed] [Google Scholar]
- Solanas G., Cortina C., Sevillano M. and Batlle E. (2011). Cleavage of E-cadherin by ADAM10 mediates epithelial cell sorting downstream of EphB signalling. Nat. Cell Biol. 13, 1100-1107. 10.1038/ncb2298 [DOI] [PubMed] [Google Scholar]
- Stein A. and Kortemme T. (2013). Improvements to robotics-inspired conformational sampling in rosetta. PLoS ONE 8, e63090 10.1371/journal.pone.0063090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surcel A., Ng W. P., West-Foyle H., Zhu Q., Ren Y., Avery L. B., Krenc A. K., Meyers D. J., Rock R. S., Anders R. A. et al. (2015). Pharmacological activation of myosin II paralogs to correct cell mechanics defects. Proc. Natl. Acad. Sci. USA 112, 1428-1433. 10.1073/pnas.1412592112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng C.-Y., Kao S.-H. and Hsu H.-J. (2016). Snail controls proliferation of Drosophila ovarian epithelial follicle stem cells, independently of E-cadherin. Dev. Biol. 414, 142-148. 10.1016/j.ydbio.2016.04.022 [DOI] [PubMed] [Google Scholar]
- Tuomi S., Mai A., Nevo J., Laine J. O., Vilkki V., Ohman T. J., Gahmberg C. G., Parker P. J. and Ivaska J. (2009). PKCepsilon regulation of an alpha5 integrin-ZO-1 complex controls lamellae formation in migrating cancer cells. Sci. Signal. 2, ra32 10.1126/scisignal.2000135 [DOI] [PubMed] [Google Scholar]
- Urena J. M., Merlos-Suarez A., Baselga J. and Arribas J. (1999). The cytoplasmic carboxy-terminal amino acid determines the subcellular localization of proTGF-(alpha) and membrane type matrix metalloprotease (MT1-MMP). J. Cell Sci. 112, 773-784. [DOI] [PubMed] [Google Scholar]
- Weaver S. A., Wolters B., Ito N., Woskowicz A. M., Kaneko K., Shitomi Y., Seiki M. and Itoh Y. (2014). Basal localization of MT1-MMP is essential for epithelial cell morphogenesis in 3D collagen matrix. J. Cell Sci. 127, 1203-1213. 10.1242/jcs.135236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S. K. and Yap A. S. (2013). Patterns in space: coordinating adhesion and actomyosin contractility at E-cadherin junctions. Cell Commun. Adhes. 20, 201-212. 10.3109/15419061.2013.856889 [DOI] [PubMed] [Google Scholar]
- Yang J., Yan R., Roy A., Xu D., Poisson J. and Zhang Y. (2015). The I-TASSER Suite: protein structure and function prediction. Nat. Methods 12, 7-8. 10.1038/nmeth.3213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Liang Y. and Zhang Y. (2011). Atomic-level protein structure refinement using fragment-guided molecular dynamics conformation sampling. Structure 19, 1784-1795. 10.1016/j.str.2011.09.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Pan Y., Yan R., Zeng B., Wang H., Zhang X., Li W., Wei H. and Liu Z. (2015). Commensal bacteria direct selective cargo sorting to promote symbiosis. Nat. Immunol. 16, 918-926. 10.1038/ni.3233 [DOI] [PubMed] [Google Scholar]
- Zhang C., Brown M. Q., van de Ven W., Zhang Z.-M., Wu B., Young M. C., Synek L., Borchardt D., Harrison R., Pan S. et al. (2016). Endosidin2 targets conserved exocyst complex subunit EXO70 to inhibit exocytosis. Proc. Natl. Acad. Sci. USA 113, E41-E50. 10.1073/pnas.1521248112 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.