Abstract
Long non-coding RNAs (lncRNAs) are expressed in a highly tissue-specific manner and function in various aspects of cell biology, often as key regulators of gene expression. In this study, we established a role for lncRNAs in chondrocyte differentiation. Using RNA sequencing we identified a human articular chondrocyte repertoire of lncRNAs from normal hip cartilage donated by neck of femur fracture patients. Of particular interest are lncRNAs upstream of the master chondrocyte transcription factor SOX9 locus. SOX9 is an HMG-box transcription factor that plays an essential role in chondrocyte development by directing the expression of chondrocyte-specific genes. Two of these lncRNAs are upregulated during chondrogenic differentiation of mesenchymal stem cells (MSCs). Depletion of one of these lncRNAs, LOC102723505, which we termed ROCR (regulator of chondrogenesis RNA), by RNA interference disrupted MSC chondrogenesis, concomitant with reduced cartilage-specific gene expression and incomplete matrix component production, indicating an important role in chondrocyte biology. Specifically, SOX9 induction was significantly ablated in the absence of ROCR, and overexpression of SOX9 rescued the differentiation of MSCs into chondrocytes. Our work sheds further light on chondrocyte-specific SOX9 expression and highlights a novel method of chondrocyte gene regulation involving a lncRNA.
KEY WORDS: SOX9, LncRNA, Cartilage, Chondrogenesis, MSC, Epigenetics, Differentiation
Summary: This study identified a chondrocyte repertoire of lncRNAs and discovered that ROCR (regulator of chondrogenesis RNA) is important for MSC chondrogenesis and cartilage gene expression by promoting the expression of SOX9.
INTRODUCTION
Tens of thousands of long non-coding RNAs (lncRNAs) have been identified in the human genome through the use of RNA deep sequencing (RNA-Seq) (Iyer et al., 2015). lncRNAs are classified as >200-nucleotide RNAs that derive from both intergenic and overlapping protein-coding gene regions (Derrien et al., 2012). Detailed studies are beginning to ascribe functional roles for many of these lncRNAs, which appear to regulate numerous cell processes (Rinn and Chang, 2012). Indeed, lncRNAs have emerged as key regulators of gene expression transcriptionally and post-transcriptionally, acting through diverse mechanisms such as the regulation of epigenetic modifications and by acting as scaffolds for protein complex formation at gene loci (Rinn and Chang, 2012). lncRNAs display more tissue-specific expression patterns than protein-coding genes, and cell differentiation during development is particularly susceptible to experimental loss of lncRNAs (Derrien et al., 2012; Sauvageau et al., 2013; Fatica and Bozzoni, 2014). For example, lncRNAs play important roles in guiding limb development. In limb patterning, HOTTIP is required for specification of mesenchyme condensation sites through promotion of HOXA gene expression by a cis-regulatory mechanism, and HOTAIR is now also recognised for a similar trans-acting role in regulating HOXD gene expression during skeletal patterning (Wang et al., 2011; Li et al., 2013).
Little is known about the expression of lncRNAs in cartilage or in the development of the chondrocyte, the sole cartilage cell type. Chondrocytes develop from condensations of mesenchymal cells in a process known as chondrogenesis, which is essential for development of the endochondral skeleton (Onyekwelu et al., 2009). During chondrogenesis, cells of the mesenchyme commit to a chondrocyte differentiation programme then progress through multiple stages to specify the resting, proliferating and hypertrophic regions of the growth plate. They also constitute the articular cartilage at the ends of the long bones. This differentiation is a coordinated process determined by temporal and spatial expression of multiple growth factors and dependent on the specific activity of the HMG-box transcription factor SOX9 (Akiyama, 2008). SOX9 controls the expression of numerous chondrocyte genes, including its co-factors L-SOX5a and SOX6, and extracellular matrix genes such as type II collagen and the proteoglycan aggrecan. Experimental loss of SOX9 abrogates limb development in mice (Akiyama, 2008; Akiyama and Lefebvre, 2011) and mutations in the SOX9 coding sequence lead to the skeletal malformation syndrome campomelic dysplasia (CD) (Foster et al., 1994; Wagner et al., 1994). DNA alterations around the SOX9 locus can also lead to CD, highlighting the complex regulatory mechanisms governing SOX9 expression (Foster et al., 1994; Wagner et al., 1994).
SOX9 is found in a gene desert on chromosome 17, as is common for developmental transcription factors, surrounded by many potential regulatory regions. However, the cellular mechanisms for regulating SOX9 are not fully established. Analyses of CD patient chromosomal rearrangements and promoter yeast artificial chromosome transgenes suggest that certain chondrogenesis-specific enhancers lie in a region between 50 kb and 350 kb upstream of SOX9 (Foster et al., 1994; Wagner et al., 1994; Wunderle et al., 1998; Gordon et al., 2009). SOX9 also specifies the fate of other lineages, including Sertoli cells, neural stem cells, pancreas progenitor cells and neural crest, neuronal, glial, heart valve, gut and kidney cells (Pritchett et al., 2011). Again, tissue-specific enhancers have been demonstrated to regulate the expression in some of these tissues (Gordon et al., 2009).
cDNA cloning methods and in silico genome analysis have established that numerous expressed sequence tags and predicted transcripts are localised to these enhancer regions upstream of SOX9 but it is unclear which are expressed in particular tissues and whether any have a functional role in chondrocytes. We established a chondrocyte repertoire of lncRNAs and confirmed the presence of a number of transcripts around the SOX9 locus by whole transcriptome analysis of human articular cartilage RNA by RNA-Seq. We discovered a novel cartilage-specific 4-exon lncRNA corresponding to a 3-exon RefSeq transcript LOC102723505 (LINC02095, Ensembl transcript ENST00000430908) 94 kb upstream of SOX9, which we termed ROCR (regulator of chondrogenesis RNA). This lncRNA is required for successful differentiation of mesenchymal stem cells (MSCs) into chondrocytes where it appears to contribute to SOX9 expression. Thus, we have identified a previously unknown mechanism of SOX9 regulation involving a chondrocyte-specific lncRNA.
RESULTS
Human articular chondrocyte lncRNAs
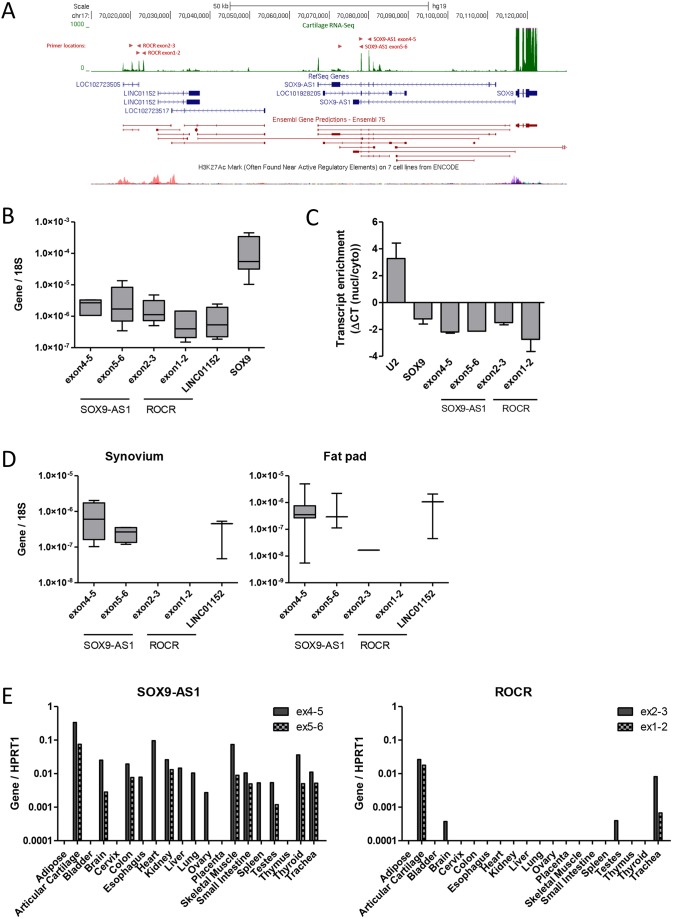
RNA-Seq was performed on normal human hip articular chondrocyte RNA obtained from female neck of femur (NOF) fracture patients to establish the adult chondrocyte transcriptome and its complement of lncRNAs (6 samples; median age=76 years). Of the 46,087 transcripts identified [fragments per kilobase of exon per million fragments mapped (FPKM)>1], 813 were annotated as lncRNAs (Table S2). Examination of cartilage RNA-Seq reads uploaded to the UCSC genome browser identified processed transcripts upstream of the SOX9 locus on chromosome 17, with robust expression of transcripts corresponding to SOX9-AS1 and LOC102723505 (Fig. 1A), exon/intron boundaries, and evidence of transcript start and end sites using cap analysis gene expression (CAGE) and PolyA-Seq data (ENCODE Project Consortium, 2012; Flicek et al., 2014) (Fig. S1). Proximal to the SOX9 locus transcript, variants of SOX9-AS1 were detected partially corresponding to Refseq and predicted Ensembl transcripts. 94 kb upstream of SOX9 we detected a novel 4-exon variant of an existing 3-exon RefSeq transcript LOC102723505. We designated the 3-exon LOC102723505 as ROCR (regulator of chondrogenesis RNA) transcript variant 1 and the novel 4-exon isoform ROCR transcript variant 2. We noted the presence of chromatin features of actively transcribed genes, such as histone H3 lysine 4 trimethylation (H3K4me3), at the presumed ROCR promoter and enhancer-like signatures based on histone lysine 27 acetylation (H3K27ac) states from ENCODE chromatin state data (Fig. 1A, Fig. S1) (Ernst et al., 2011). The ROCR locus is also notable for the expression of an additional lncRNA, LINC01152, albeit at very low levels in cartilage. In comparison with other coding transcripts, SOX9-AS1 and ROCR were moderately expressed in cartilage with FPKM in the range of 5-15, approximately 10% of the level of SOX9 itself (Table S2).
Fig. 1.
Expression of lncRNAs from the SOX9 locus. (A) UCSC genome browser schematic of cartilage RNA-Seq reads aligned to the human genome with RefSeq gene annotations, Ensembl gene predictions and active H3K27Ac chromatin marks. Reads are pooled from six neck of femur (NOF) fracture cartilage donors. Primer locations are indicated by red arrowheads. (B) Expression of SOX9 locus lncRNAs and SOX9 in RNA extracted from OA cartilage measured by real-time RT-PCR normalised to 18S. Values are mean±s.e.m. of data pooled from five separate donors. (C) Subcellular localisation of SOX9-AS1 and ROCR in comparison with small nuclear RNA U2 and SOX9 mRNA pooled from two OA HAC donors. Values are mean±s.e.m. of ΔCT between an equal fraction of nuclear and cytoplasmic RNA. (D) Expression of SOX9-AS1 and ROCR in RNA extracted from OA synovium and joint fat pad. Values are mean±s.e.m. of data pooled from eight separate synovium and fat pad donors. (E) Expression of SOX9-AS1 and ROCR in an RNA tissue panel. Values are the technical mean of data pooled from three donors per tissue.
We confirmed the expression of SOX9-AS1 and ROCR in human articular cartilage by qRT-PCR with two assays per transcript targeted to different exons (Fig. 1B). The ROCR exon1-2 assay detects only transcript variant 2. Rapid amplification of cDNA ends (RACE) confirmed the presence of this novel 4-exon 574-base ROCR transcript (variant 2) in cartilage (Fig. S2). We were also able to identify this ROCR variant by subsequent analysis of RNA-Seq data from knee cartilage RNA (Dunn et al., 2016). The majority of lncRNAs are considered to have nuclear functions and are often found enriched in the nucleus (Quinodoz and Guttman, 2014). In contrast to the nuclear enrichment of the small nuclear RNA U2, we found that both SOX9-AS1 and ROCR were enriched in the cytoplasm, comparable with the localisation of the SOX9 transcript itself (Fig. 1C). RNA fluorescence in situ hybridisation (RNA-FISH) analysis of ROCR in human articular chondrocytes (HACs) was unsuccessful owing to the relatively low expression and short transcript sequence, which limited the design of sufficient singly labelled Stellaris RNA-FISH probes (data not shown). In silico analysis indicates a lack of coding potential for both SOX9-AS1 and ROCR, with the existence of only very short open reading frames (ORF Finder) and codon substitution rates indicative of noncoding transcripts (CPAT, CPC and PhyloCSF) (Fig. S3). SOX9 is expressed in a variety of tissues but lncRNAs are reported to be more tissue specific (Derrien et al., 2012). Accordingly, we examined expression of SOX9-AS1 and ROCR in additional joint tissues extracted from osteoarthritis (OA) patients. SOX9-AS1 was also expressed in synovium and fat pad tissue but ROCR was largely undetected indicating that it might be specific to cartilage in the joint (Fig. 1D).
We further examined transcript expression bioinformatically using publicly available cell and tissue RNA-Seq databases. Reads corresponding to SOX9-AS1 were found in numerous cells types in both Human Protein Atlas (http://www.proteinatlas.org/) and Illumina BodyMap (ArrayExpress accession: E-MTAB-513; http://www.ebi.ac.uk/arrayexpress) sequence data (Table S3) (ENCODE Project Consortium, 2012; Krupp et al., 2012; Fagerberg et al., 2014; Flicek et al., 2014). Reads corresponding to the three exons of ROCR transcript variant 1 were found in pancreas and salivary gland tissue samples in the Human Protein Atlas RNA-Seq data and in breast tissue samples sequenced in the Illumina BodyMap data. Consistent with this analysis, further examination of expression by qRT-PCR across a 20-tissue RNA panel again identified the presence of SOX9-AS1 transcripts in a number of tissues (Fig. 1E), albeit in fewer tissues than SOX9 itself (Fig. 4). In contrast, detection of the novel ROCR transcript variant 2 was limited to chondrocytes alone (Fig. 1F), and the ROCR transcript variant 1 was additionally detected in brain and testis.
SOX9 locus lncRNA expression in MSC differentiation
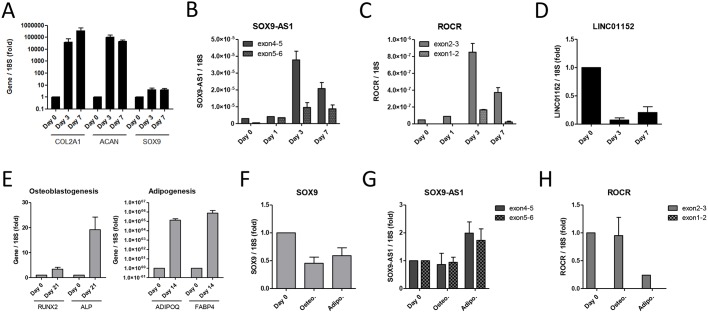
Considering the proximity of these transcripts to SOX9 and the potential chondrocyte specificity of ROCR, we sought to establish whether SOX9-AS1 and ROCR were regulated during chondrocyte development. Accordingly, we characterised expression of these lncRNAs using a robust transwell MSC chondrogenesis method, which produces a uniform cartilage disc with rapid and substantial induction of chondrocyte gene expression, albeit including the expression of chondrocyte hypertrophy genes, thus differing from articular cartilage (Fig. 2A) (Murdoch et al., 2007). SOX9 expression is upregulated during chondrogenesis. Similarly, the expression of both SOX9-AS1 and ROCR was induced during MSC chondrogenesis, paralleling the kinetics of SOX9 expression (Fig. 2B,C). In contrast, LINC01152, a potential testis-specific lncRNA (D43770 Genbank ID), was downregulated during MSC chondrogenesis (Fig. 2D) (Ninomiya et al., 1996). Interestingly, RACE for MSC RNA identified a further 624-base isoform with an alternative first exon, which we termed ROCR transcript variant 3 (Fig. S1), situated in a bidirectional promoter locus with LINC01152.
Fig. 2.
Expression of lncRNAs during MSC differentiation. (A) Expression of the indicated genes in RNA extracted from MSCs undergoing chondrogenic differentiation at the indicated time points between Day 0 and Day 7. (B-D) Expression of SOX9-AS1 (B), ROCR (C) and LINC01152 (D) during MSC chondrogenic differentiation. (E) Expression of the indicated genes in RNA extracted from MSCs undergoing osteoblastogenic and adipogenic differentiation. (F-H) Expression of SOX9 (F), SOX9-AS1 (G) and ROCR (H) during MSC osteoblastogenic (Osteo.) and adipogenic (Adipo.) differentiation. Values are mean± s.e.m. of data pooled from three or four MSC donors.
MSCs are capable of tri-lineage differentiation into chondrocytes, osteoblasts and adipocytes, dependent on specific differentiation factors (Pittenger et al., 1999). We differentiated MSCs into osteoblasts and adipocytes by established methods and confirmed the expression of the osteoblast-specific markers alkaline phosphatase (ALPL) and RUNX2, and the adipocyte-specific genes adiponectin and FABP4 (Fig. 2E). SOX9 was not upregulated during osteoblastogenesis or adipogenesis (Fig. 2F). Similarly, SOX9-AS1 and ROCR were not upregulated during MSC osteoblastogenesis (Fig. 2G,H). SOX9-AS1 was induced during MSC adipogenesis, in contrast to ROCR, but not to the level of chondrogenesis (Fig. 2G).
Role of ROCR in MSC chondrogenesis
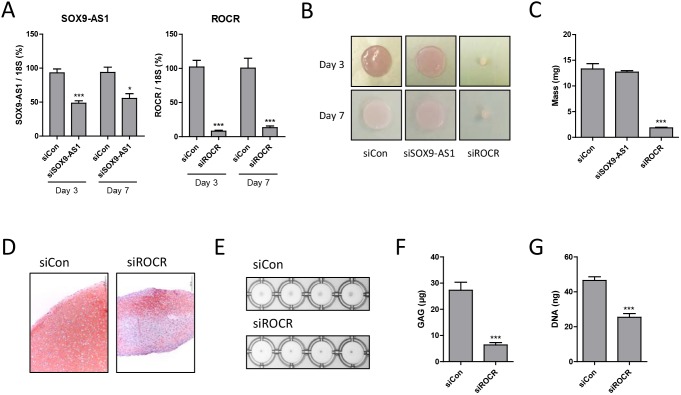
SOX9-AS1 and ROCR were both upregulated during chondrogenesis, with a profile similar to SOX9; therefore, we addressed their potential role during MSC chondrogenic differentiation by specific RNA interference (RNAi)-mediated depletion (Fig. 3A). Reduction of SOX9-AS1 expression had no effect on development of a cartilaginous disc (Fig. 3B,C). However, depletion of ROCR prevented disc formation (Fig. 3B) and caused a significant reduction in wet mass (Fig. 3C). Consistent with the disruption of disc formation following ROCR RNAi, matrix deposition in the form of glycosaminoglycan (GAG) polyanions was also reduced (Fig. 3D). In case the transwell chondrogenesis method was particularly susceptible to experimental manipulation, we also performed the traditional pellet chondrogenesis method and again found that ROCR was required for pellet development (Fig. 3E). Analysis of extracted sulphated GAG levels again indicated that ROCR is required for matrix GAG production (Fig. 3F). In addition, ROCR depletion reduced DNA levels suggesting it was required for MSC proliferation during the early stages of chondrocyte differentiation (Fig. 3G) (Murdoch et al., 2007).
Fig. 3.
Effect of lncRNA depletion on MSC chondrogenic differentiation. (A-D) MSCs were transfected for 2 days with SOX9-AS1 or ROCR-targeting or non-targeting control (siCon) siRNA prior to chondrogenic differentiation in hanging transwell inserts. (A) SOX9-AS1 and ROCR expression in RNA extracted from Day 3 and Day 7 chondrogenic discs. Expression is presented as a percentage of non-targeting control levels. (B) Representative Day 3 and 7 chondrogenic discs. (C) Wet mass of Day 7 chondrogenic discs. (D) Representative Safranin O staining of Day 7 chondrogenic discs. (E-G) MSCs were transfected for 1 day with ROCR-targeting or non-targeting control siRNA prior to chondrogenic differentiation in a V-bottom 96-well plate. (E) Representative Day 7 chondrogenic pellets. (F) GAG levels assayed by DMB assay in Day 7 chondrogenic pellets. (G) DNA quantification by PicoGreen assay in Day 7 chondrogenic pellets. Values are mean±s.e.m. of data pooled from three (A-D) or four (E-G) MSC donors. *P<0.05; ***P<0.001 for lncRNA siRNA versus non-targeting siRNA. Significant differences between sample groups were assessed by one-way analysis of variance followed by the Bonferroni post-hoc test for multiple comparisons or a two-tailed Student's t-test was performed for single comparisons.
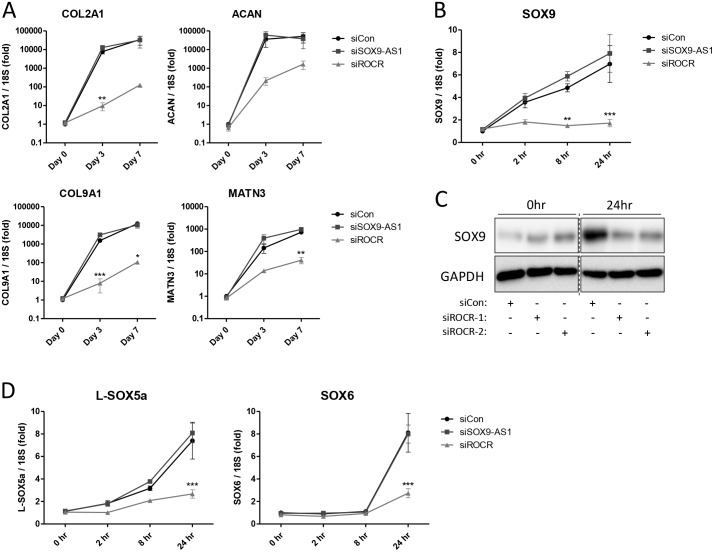
Examination of chondrocyte gene expression following SOX9-AS1 and ROCR RNAi indicated that depletion of ROCR also significantly abrogated the induction of cartilage extracellular matrix (ECM) genes including COL2A1 and ACAN (Fig. 4A). SOX9 is essential for cartilage matrix gene expression, so we assessed the impact of depletion of SOX9-AS1 and ROCR at earlier time points in the chondrogenesis time course. Following ROCR depletion, SOX9 mRNA (Fig. 4B) and protein (Fig. 4C) was significantly reduced after 1 day of MSC differentiation, and at even earlier time points the upregulation of SOX9 expression during MSC chondrogenesis was lost following ROCR depletion suggesting a crucial role for ROCR in SOX9 induction. During chondrogenesis SOX9 is required for expression of SOX5 and SOX6, which subsequently cooperate with SOX9 in directing chondrocyte gene expression (Akiyama et al., 2002). ROCR depletion also prevented the upregulation of the SOX9 target genes SOX5 and SOX6, which occurred after SOX9 induction (Fig. 4D).
Fig. 4.
Effect of lncRNA depletion on MSC chondrogenic gene expression. (A) MSCs were transfected for 2 days with SOX9-AS1 or ROCR-targeting or non-targeting control (siCon) siRNA prior to chondrogenic differentiation in hanging transwell inserts. RNA was extracted and expression of the indicated genes measured by real-time RT-PCR. (B-D) MSCs were transfected for 1 day with SOX9-AS1 or ROCR-targeting or non-targeting control siRNA prior to chondrogenic differentiation in a V-bottom 96-well plate for up to 24 h. RNA and protein was extracted and expression of SOX9 mRNA (B) or protein (C) measured by real-time RT-PCR or immunoblotting, respectively. (D) Expression of L-SOX5a and SOX6. Values are mean±s.e.m. of data pooled from three (A) or four (B-D) MSC donors. *P<0.05; **P<0.01; ***P<0.001 for lncRNA siRNA versus non-targeting siRNA. Significant differences between sample groups were assessed by one-way analysis of variance followed by the Bonferroni post-hoc test for multiple comparisons or a two-tailed Student's t-test was performed for single comparisons.
To complement the role identified by RNAi for ROCR in MSC chondrogenesis and SOX9 expression, we also used an LNA GapmeR approach to deplete cellular ROCR levels (Fig. S5). Again, the loss of ROCR resulted in a significant reduction in matrix GAG formation during MSC mini-pellet chondrogenesis with concomitant reduction in SOX9 and matrix gene expression (Fig. S5). ROCR transcript variants 2 (HAC) and 3 (MSCs) were cloned and overexpressed in MSCs and HAC by lentiviral transduction (Fig. S6). Overexpression of ROCR had no effect on SOX9 expression or induction of the cartilage ECM genes COL2A1 and ACAN during MSC chondrogenesis (Fig. S6A). Overexpression of ROCR had no effect on SOX9 expression in HAC (Fig. S6B).
Specificity of ROCR function to chondrogenesis
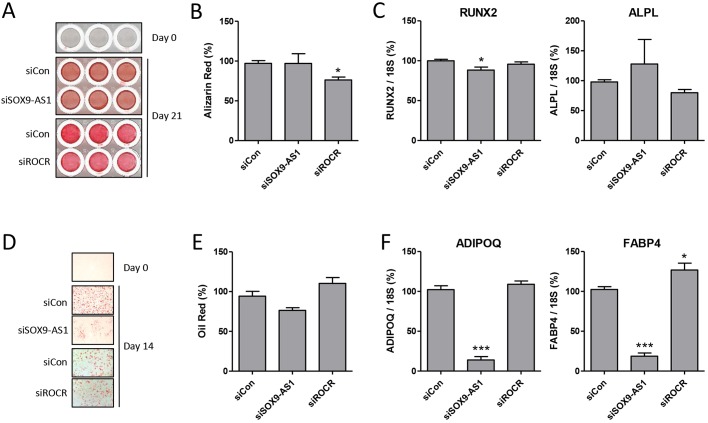
The above data suggested that ROCR is important for MSC chondrogenesis. We sought to establish whether the role of ROCR was specific to chondrocyte development consistent with its restricted expression profile. Accordingly, we also performed SOX9-AS1 and ROCR RNAi during MSC osteoblastogenesis and adipogenesis. Depletion of ROCR during osteoblast differentiation caused a partial decrease in matrix mineralisation (Fig. 5A,B), but no significant impact on RUNX2 or ALPL expression (Fig. 5C). During MSC adipogenesis ROCR depletion had little effect, whereas SOX9-AS1 depletion partially reduced fat droplet generation (Fig. 5D,E) and significantly decreased MSC adipogenic gene expression (Fig. 5F).
Fig. 5.
Effect of lncRNA depletion on MSC osteoblastogenic and adipogenic differentiation. (A-C) MSCs were transfected for 2 days with SOX9-AS1 or ROCR-targeting or non-targeting control (siCon) siRNA prior to osteoblastogenic differentiation. (A) Representative matrix mineralisation assayed by Alizarin Red staining after 21 days. (B) Quantification of Alizarin Red staining. (C) RNA was extracted and expression of the indicated genes at Day 7 measured by real-time RT-PCR. (D-F) MSCs were transfected for 2 days with SOX9-AS1 or ROCR-targeting or non-targeting control siRNA prior to adipogenic differentiation. (D) Representative fat droplet generation assayed by Oil Red O staining after 14 days. (E) Quantification of Oil Red O staining. (F) RNA was extracted and expression of the indicated genes at Day 7 measured by real-time RT-PCR. Values are mean±s.e.m. of data pooled from four MSC donors. *P<0.05; ***P<0.001 for lncRNA siRNA versus non-targeting siRNA. Significant differences between sample groups were assessed by one-way analysis of variance followed by the Bonferroni post-hoc test for multiple comparisons or a two-tailed Student's t-test was performed for single comparisons.
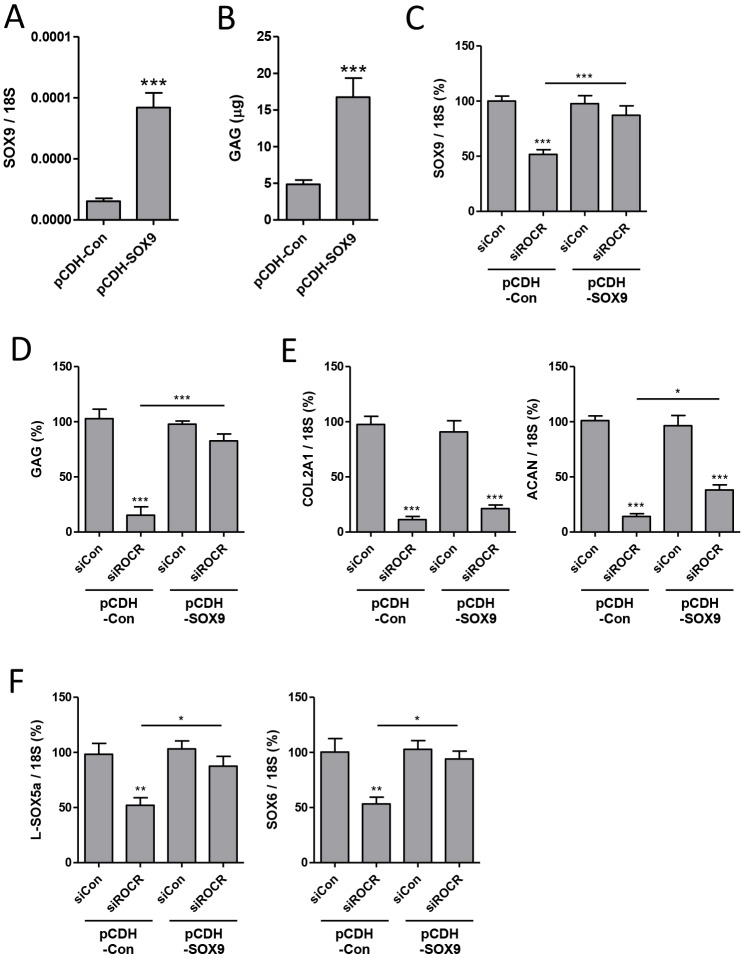
SOX9 is essential for chondrogenesis and as lncRNAs can contribute to the expression of neighbouring genes (Vance and Ponting, 2014) we reasoned that the primary role of ROCR is to promote SOX9 expression. Accordingly, overexpression of SOX9 would be expected to rescue the chondrogenesis impairment caused by ROCR depletion. Lentiviral overexpression of SOX9 successfully enhanced MSC chondrogenesis (Fig. 6A,B). By overexpressing SOX9 and thereby returning the levels of SOX9 to those of control (Fig. 6C) the significant reduction of cartilage matrix GAG levels following depletion of ROCR was almost fully reversed (Fig. 6D). Reduction of COL2A1 and ACAN by ROCR depletion was partially reversed by overexpression of SOX9 (Fig. 6E,F), and the levels of L-SOX5a and SOX6 were completely rescued (Fig. 6G,H).
Fig. 6.
Effect of SOX9 overexpression in combination with ROCR depletion on MSC chondrogenic differentiation and gene expression. MSCs were transduced with SOX9 or control lentivirus (pCDH) for 1 day then transfected for 1 day with ROCR-targeting or non-targeting control (siCon) siRNA prior to chondrogenic differentiation in a V-bottom 96-well plate. (A) Expression of SOX9 in only non-targeting control siRNA pellets at Day 3. (B) GAG levels assayed by DMB assay in only non-targeting control siRNA pellets at Day 7. (C) Expression of SOX9 at Day 3. (D) GAG levels assayed by DMB assay at Day 7. (E,F) Expression of the indicated genes at Day 3. (C-F) Expression is presented as a percentage of non-targeting control levels for cells transduced with each virus. Values are mean±s.e.m. of data pooled from four MSC donors. *P<0.05; **P<0.01; ***P<0.001 for lncRNA siRNA versus non-targeting siRNA, or for SOX9 versus control lentivirus where indicated above the chart. Significant differences between sample groups were assessed by one-way analysis of variance followed by the Bonferroni post-hoc test for multiple comparisons or a two-tailed Student's t-test was performed for single comparisons.
DISCUSSION
In this study, we established a panel of lncRNAs in normal human articular cartilage and identified two transcripts upstream of the SOX9 locus that were upregulated during MSC chondrogenesis. One of these, ROCR, is a functional cartilage-restricted lncRNA that appears to be important for chondrocyte differentiation where it might facilitate the induction of SOX9 itself. This study established that a lncRNA contributes to SOX9 expression during differentiation of MSCs into chondrocytes, thereby furthering our understanding of the key regulatory elements contained upstream of the SOX9 promoter.
SOX9 is the master transcription factor governing chondrocyte development, as confirmed by genetic studies (Akiyama et al., 2002). Regulation of SOX9 occurs at both the transcriptional and post-transcriptional levels. Phosphorylation of SOX9 regulates its DNA-binding activity and subcellular localisation, and numerous other interactions regulate SOX9 stability and facilitate its transcriptional activity (Kawakami et al., 2006; Akiyama, 2008). At the transcriptional level, induction of SOX9 occurs rapidly during mesenchyme condensation in cartilage development both in vivo and in vitro (Wright et al., 1995; Sekiya et al., 2002), a process regulated by an interplay between growth factor signals and cell-cell interactions (Chimal-Monroy et al., 2003; Yoon et al., 2005). Our data indicated that during in vitro chondrogenesis a lncRNA, ROCR, is also important for this process.
lncRNAs in chondrocytes and chondrogenesis
A number of lncRNAs have key roles in stem cell differentiation, including RMST in neuronal differentiation, Braveheart in cardiac differentiation and lnc-RAP1-10 in adipocyte differentiation (Klattenhoff et al., 2013; Ng et al., 2013; Perry and Ulitsky, 2016). Previously identified lncRNAs with a potential role in cartilage development include DA125942 and LncRNA-HIT (Maass et al., 2012; Carlson et al., 2015). DA125942, a lncRNA transcribed from the CISTR-ACT locus interacts in cis with PTHLH and in trans with SOX9 to organise chromatin structure and promote transcription in cartilage (Maass et al., 2012). No direct role for the lncRNA in chondrogenesis was explored although the lncRNA locus was active during mouse limb bud development. LncRNA-HIT, expressed in mouse limb mesenchyme from the Hoxa gene locus, is able to bind and regulate DNA regions surrounding a number of cartilage genes including the Hoxa genes themselves (Carlson et al., 2015). LncRNA-HIT may activate gene expression by binding to the p100/CBP complex and it contributes to micromass chondrogenic differentiation of murine MSCs. Interestingly, we detected no RNA expression from the CISTR-ACT locus in our human cartilage RNA-Seq data and the conserved regions of LncRNA-HIT in human corresponded to an isoform of HOXA13 with an extended 3′UTR rather than a lncRNA. It is possible that these lncRNAs might be developmental stage or MSC specific. The lncRNA DANCR may also promote chondrogenic differentiation of synovium-derived MSCs in concert with SOX4 (Zhang et al., 2015). Two recent reviews elaborate on the roles of these lncRNAs during chondrogenesis (Huynh et al., 2017; Lefebvre and Dvir-Ginzberg, 2017).
SOX9 is located in a ∼2 Mb gene desert on chromosome 17 in humans and lncRNA ROCR is expressed from a locus 94 kb upstream of SOX9. Chromosomal rearrangements within this region are associated with CD, a skeletal malformation syndrome, and Pierre Robin sequence (PRS), a craniofacial disorder. Such disruptions can occur in regions up to and greater than 1 Mb upstream of SOX9 (Gordon et al., 2009). Characterisation of these DNA alterations has indicated the presence of enhancer regions linked to the regulation of SOX9 expression. Breakpoints causing more severe forms of CD are found more proximal to SOX9 at locations 50-375 kb upstream (Leipoldt et al., 2007). Transgene and reporter experiments have also indicated that sequences in these locations are able to drive gene expression in vivo (Gordon et al., 2009). More recent analysis confirmed the presence of a murine enhancer element at −70 kb (−62 kb in human) capable of regulating SOX9 expression in a number of tissues (Mead et al., 2013), and three further enhancers with prominent activity in chondrocytes at −84 kb, −195 kb and −250 kb in mice (Yao et al., 2015).
The ROCR locus sits within these enhancer regions and it is attractive to suggest that the lncRNA might contribute to the regulation of SOX9 in vivo. Indeed, functional lncRNAs have been found to be enriched in genomic regions surrounding key developmental transcription factors (Ørom et al., 2010; Ulitsky et al., 2011). In addition to skeletal malformations, patients with CD often show XY sex reversal, with additional clinical features such as hearing loss, developmental delay, and occasional heart defects (Mansour et al., 2002). Consistent with this, genetic ablation of Sox9 in mice disrupts the differentiation of cells in the heart, central nervous system, testis, pancreas, gut and inner ear (Gordon et al., 2009). Tissue-specific enhancers regulate the expression of SOX9, for example the testis enhancer TES at −10 kb, and our analysis suggested that ROCR is restricted to certain cell types – cartilage, brain and testis – whereas ROCR variant 2 was only detected in cartilage. However, our work focussed on RNA extracted from aged NOF and OA tissue and further work is required to confirm the expression of ROCR in normal healthy tissues. In combination with tissue-specific enhancers ROCR might be required for the tightly coordinated spatiotemporal expression of SOX9 during development. The expression level of SOX9 in cartilage was one or two orders of magnitude higher than other tissues (Fig. S4) and we reasoned that ROCR might also contribute to the magnitude of SOX9 expression. But, in contrast to its role in chondrogenesis, we found no significant contribution by ROCR to SOX9 expression levels in adult articular chondrocytes (Fig. S7A). The role of ROCR in SOX9 expression might be in response to cues during chondrogenesis that are not present in cultured HAC, and ROCR might additionally regulate other genes/proteins. The induction of both SOX9-AS1 and ROCR paralleled the expression of SOX9. The activity of the aforementioned −70 kb, −84 kb and −195 kb SOX9 upstream enhancers is dependent on SOX9 in differentiated chondrocytes (Yao et al., 2015). Prior to the onset of chondrogenesis, SOX9 overexpression in MSCs did not significantly induce ROCR expression (Fig. S7B), but we cannot rule out the possibility that SOX9 promotes the expression of ROCR during chondrogenesis, or in adult chondrocytes. Despite knockdown of ROCR reducing SOX9 expression and cartilage gene expression in MSCs, reciprocal overexpression of ROCR had no effect. Overexpression from an artificial plasmid transcription start site is not entirely analogous to endogenous ROCR expression with potential alteration to secondary structure formation and cellular localisation of the RNA.
During skeletogenesis MSC condensation initiates the formation of multipotent osteochondroprogenitors the lineage fate of which is then determined by the combination of growth factor signals received. ROCR is only upregulated during chondrogenesis, not osteoblastogenesis, suggesting a key role in directing MSCs toward the chondrocyte lineage. Consistent with this, only a minor impact of ROCR depletion was observed during MSC osteoblastogenesis in contrast to its key requirement during chondrogenesis. During osteochondroprogenitor differentiation SOX9 has antagonistic effects on the osteoblast transcription factor RUNX2 in determining the specific differentiation into their respective chondrocyte and osteoblast lineages (Zhou et al., 2006). Owing to the lack of induction of ROCR during osteoblastogenesis, no effect would be expected. Interestingly, depletion of SOX9-AS1 significantly reduced the expression of adipogenic marker genes, confirming the efficacy of the SOX9-AS1 depletion and, given the role of SOX9 in adipogenic differentiation, suggests that SOX9-AS1 also contributes to the differentiation (Stockl et al., 2013).
Putative lncRNA function
We demonstrated that returning SOX9 levels to normal by overexpression could reverse the impaired chondrogenesis phenotype caused by depletion of ROCR. This indicated that SOX9 can largely replace ROCR during MSC chondrogenesis as SOX9 expression was sufficient to produce the cartilage matrix. Thus, suggesting ROCR is indirectly needed in chondrogenesis to establish the correct level of SOX9 expression in MSCs during differentiation. Both silencing and activating roles have been demonstrated for lncRNAs. XIST establishes X chromosome inactivation, whereas RMST facilitates SOX2 binding to promoter regions of neurogenic transcription factors (Vance and Ponting, 2014). In some cases, enhancer regions and the process of transcription at the lncRNA locus facilitate downstream gene expression rather than the lncRNA transcript itself (Engreitz et al., 2016). Our knockdown experiments indicate that ROCR transcript is functional, and the ROCR locus is considerably upstream from SOX9 (94 kb), but we cannot rule out the possibility that the ROCR locus might also function as an enhancer. Many of the identified functional lncRNA actions occur in the nucleus; however, ROCR appears to reside more in the cytoplasm than nucleus, indicating an indirect regulation of SOX9. Our coding analysis indicated that ROCR is unlikely to code for any significant peptide transcript, suggesting a role for the RNA in the cytoplasm. A number of cytoplasmic lncRNAs can regulate mRNA half-life and translation. TINCR is induced during epidermal differentiation and is required for stability of differentiation mediators (Kretz et al., 2013) and antisense Uchl1 lncRNA promotes translation of Uchl1 in mouse (Carrieri et al., 2012). Other factors also contribute to cartilage gene expression, such as SP1 and forkhead/winged-helix domain (FOX) proteins, and this could account for why, despite normal GAG levels, the expression of COL2A1 and ACAN was not completely restored during rescue by SOX9, again suggesting an indirect effect of ROCR (Liu et al., 2016). Or this might simply reflect the difference in sampling time for gene expression in relation to matrix GAG measurement. Almost all lncRNAs function through association with protein partners and, accordingly, RNA pulldown methods are commonly used to identify such interactions (Yang et al., 2015).
Conservation of lncRNAs across species is low, with less than 10% of all lncRNAs exhibiting regions of conservation compared with random control regions (Iyer et al., 2015), but there are key examples of conserved lncRNAs with crucial roles in mouse development having human counterparts (Sauvageau et al., 2013). By conducting a homology search for a mouse orthologue of ROCR, we identified a predicted noncoding RNA transcript (NR_024085/BC006965) with sequence similarity to exon 2 of ROCR transcript variant 1 (exon 3 of variants 2 and 3), but little mammalian sequence conservation in general (Fig. S8). Importantly, the transcripts are in syntenic regions (containing SOX9) of human chromosome 17q24 and mouse 11qE2. By real-time RT-PCR of mouse cartilage RNA we have now confirmed the expression of a murine multiple exon version of ROCR (Fig. S8). Further work will establish whether the murine transcript is regulated during chondrogenesis and contributes to chondrocyte development.
Conclusions
The cartilage transcriptome contains many lncRNA transcripts many of which may have important functions in cartilage biology. Our identification of cartilage lncRNAs complements the previous identification of inflammation-induced lncRNAs in chondrocytes (Pearson et al., 2016). This panel of chondocyte lncRNAs is specific to human aged hip cartilage and further work should establish the expression of lncRNAs specific to different zones of articular cartilage, as well as growth plate cartilage and to establish the impact of weight bearing, age and disease such as OA. Functional analysis indicated that ROCR was induced during chondrogenic differentiation and played an important role in the induction of SOX9 and, as a result, cartilage gene expression. Because SOX9-expressing cells are progenitors for numerous tissues, identifying chondrocyte-specific regulatory elements might aid our understanding of differentiation of chondrocytes from MSCs, which could be potentially useful in chondrocyte tissue-engineering applications.
MATERIALS AND METHODS
Human tissue isolation
Normal human articular cartilage was obtained from patients undergoing joint replacement surgery due to intracapsular neck of femur (NOF) fracture. OA human articular cartilage was obtained from knee joint replacement operations on patients diagnosed with osteoarthritis (OA). Synovium and infrapatellar fat pad were also collected from the knee of OA patients. All tissue was obtained with informed consent and ethics committee approval from the Newcastle and North Tyneside Health Authority. Scoring, extraction and patient information for the NOF samples are detailed in Xu et al. (2012). Briefly, joints were inspected macroscopically and scored by a blinded experienced orthopaedic surgeon to identify normal NOF cartilage. Cartilage, all zones, was collected within 2 h of surgery and stored at −80°C prior to RNA extraction.
Human bone marrow MSC culture
Human bone marrow MSCs (from seven donors, 18-25 years of age) were isolated from human bone marrow mononuclear cells (Lonza Biosciences) and cultured and phenotype-tested as described previously (Barter et al., 2015). Experiments were performed using cells between passage 2 and 7, and all experiments were repeated with cells from three or four donors.
Chondrogenic differentiation
MSCs were re-suspended in chondrogenic culture medium consisting of high-glucose DMEM containing 100 µg/ml sodium pyruvate, 10 ng/ml TGFβ3, 100 nM dexamethasone, 1× ITS-1 premix, 40 µg/ml proline and 25 µg/ml ascorbate-2-phosphate. MSCs (5×105 in 100 µl medium) were pipetted onto 6.5-mm diameter, 0.4-µm pore size polycarbonate Transwell filters (Merck Millipore), centrifuged at 200 g for 5 min, then 0.5 ml of chondrogenic medium was added to the lower well as described previously (Murdoch et al., 2007; Barter et al., 2015). For V-bottom 96-well-plate pellet chondrogenesis, 5×104 MSCs in 150 µl chondrogenic medium were pipetted into a UV-sterilised V-bottom 96-well plate and centrifuged at 500 g for 5 min. Media were replaced every 2 or 3 days for up to 7 days.
Osteoblast and adipocyte differentiation
MSCs were plated in 96-well plates at a density of 15,000/cm2 for 24 h then media were replaced with either osteoblastogenic culture medium consisting of DMEM supplemented with 10% (v/v) foetal bovine serum (FBS), 10 mM β-glycerol phosphate, 100 nM dexamethasone and 50 µg/ml ascorbic acid 2-phosphate, or adipogenic culture medium consisting of DMEM supplemented with 10% FBS, 1 μM dexamethasone, 10 μg/ml insulin, 0.5 mM IBMX, 60 μM indomethacin, 2 μM rosiglitazone and 20 nM IGF-1 (R&D Systems) (all Sigma unless specified). Media were replaced every 3 or 4 days. Seven days of differentiation was sufficient to assess gene expression changes in markers of differentiation. Cells were cultured for 21 days in osteoblastogenic medium to achieve fully mineralised cultures, and for 14 days in adipogenic medium for lipid production.
Histology and biochemical analysis
Transwell discs were stained as described (Barter et al., 2015). Chondrogenic pellets and transwell discs were digested with papain (10 U/ml) at 60°C (Murdoch et al., 2007). The sulphated glycosaminoglycan (GAG) content was measured by 1,9-dimethylmethylene blue (DMB) binding (Sigma) using chondroitin sulphate (Sigma) as standard (Farndale et al., 1982), and the DNA content was measured with PicoGreen (Invitrogen) intercalating dye following the manufacturer's instructions. Cells undergoing osteoblast differentiation were fixed in 70% cold ethanol (5 min, −20°C). After drying the wells to reveal calcium-rich mineralisation deposits, the cells were incubated at room temperature with a solution of Alizarin Red (Sigma) (40 mM, pH 4.2) for 20-30 min. For quantification the staining was extracted with 10% (w/v) cetylpyridinium (Sigma) solubilised in 10 mM sodium phosphate buffer (pH 7) and the absorbance measured at 620 nM. Cells undergoing adipogenesis were fixed with formalin for 1 h, washed with distilled water and 60% isopropanol then dried. To reveal the presence of lipid droplets, the cells were stained with a 21% (w/v) solution of Oil Red O for 10 min. For quantification the staining was extracted with 100% isopropanol and the absorbance measured at 500 nM. Stained cells were washed with distilled water prior to image acquisition.
RNA extraction and real-time reverse transcription PCR
Cartilage, synovium and fat pad samples were ground into powder and homogenised using Invitrogen TRIzol Reagent (Life Technologies) prior to RNA purification using the Qiagen RNeasy mini kit (Qiagen) essentially as previously described (Xu et al., 2012). MSC chondrogenic transwell discs were disrupted in TRIzol (for real-time RT-PCR) using a small disposable plastic pestle and an aliquot of Molecular Grinding Resin (G-Biosciences/Genotech). MSC chondrogenesis pellets were disrupted in Ambion Cells-to-cDNA II Cell Lysis buffer (Life Technologies). Total RNA was then extracted and converted to cDNA using MMLV reverse transcriptase (Invitrogen) and TaqMan real-time RT-PCR was performed and gene expression levels were calculated as described previously (Barter et al., 2010). Nuclear and cytoplasmic RNA fractions were separated using the CelLytic NuCLEAR Extraction Kit (Sigma) supplemented with RNaseOUT ribonuclease inhibitor (Life Technologies). All values are presented as the mean±s.e.m. of replicates in pooled experiments. lncRNA real-time RT-PCR amplification products were sequence verified by cloning into the pCR4-TOPO vector (Life Technologies). The Ambion FirstChoice Human Total RNA Survey Panel (AM6000) contains pools of total RNA from 20 different normal human tissues, each pool consisting of RNA from at least three tissue donors. Primer sequences are listed in Table S1.
RNA-Seq and analysis
RNA integrity was checked using an Agilent Bioanalyzer 2100 (Agilent Technologies); RNA samples with an RNA Integrity Number (RIN)≥7 were selected. For each sample, cDNA libraries were prepared for sequencing from 5 µg of total RNA using Illumina TrueSeq mRNA kits with the manufacturers' protocols. mRNA-enriched RNA was initially purified using polydT oligo-attached magnetic beads using two rounds of purification. During the second elution the RNA was fragmented and random primed for cDNA synthesis. After the addition of a single ‘A’, base adaptors were annealed, and the products purified and enriched with PCR to create a final cDNA library. No indexing (barcoding) was performed. Library DNA size was checked using the Agilent Bioanalyzer and quantified using the Kapa Library Quant kits (Kapa Biosciences). A 7.5 pM solution of each library was loaded onto each lane of an Illumina Genome Analyzer IIa and 78-base paired-end sequencing performed. On average, each sample gave 28 million read pairs. Sample quality control was performed using FastQC (Babraham Bioinformatics). Reads were aligned to the reference genome using TopHat, specifying mate inner distance (mean inner distance between mate pairs) and standard deviation for each sample (Trapnell et al., 2012). Mapped reads were then assembled into complete transcripts using the splice junction mapping tool Cufflinks, with option –G, which utilises the Ensembl reference gene track to improve mapping. Cuffmerge was used to merge the assembled transcripts into a consensus gene track from the all of the mapped samples. Ensembl transcript biotypes were applied to identify lncRNAs (biotype lincRNA). The coding potential of lncRNAs was assessed with ORFfinder (NCBI), Coding Potential Assessment Tool (CPAT), Coding Potential Calculator (CPC) and PhyloCSF (Kong et al., 2007; Lin et al., 2011; Wang et al., 2013). RNA sequencing data have been uploaded to Gene Expression Omnibus (GEO).
RNA-mediated interference, GapmeR transfection and lentiviral transduction
For siRNA transfection, 50 nM siRNA was transfected into 40-50% confluent MSCs using Dharmafect 1 lipid reagent (Thermo Fisher). 50 nM siRNA Dharmacon siGENOME and ON-TARGET+ siRNA (Thermo Fisher Scientific) were used to target SOX9-AS1 and ROCR. Depletion of gene-specific mRNA levels was calculated by comparison of expression levels with cells transfected with 50 nM siCONTROL (non-targeting siRNA 2; 001210-02, Dharmacon). For GapmeR transfection, 100 nM Antisense LNA GapmeR (Exiqon) targeting ROCR or non-targeting control (Negative Control A; 300610) were transfected as for siRNAs. siRNA and GapmeR sequences are listed in Table S1. pCDH-EF1-MCS-IRES-copGFP lentivirus expression vector (System Biosciences) containing SOX9 was generated by cloning SOX9 from pUT-FLAG-SOX9 (Lefebvre et al., 1997). pCDH-EF1-MCS lentivirus expression vectors containing ROCR transcript variant 2 and transcript variant 3 were generated by cloning gBlock gene fragments (IDT) containing the sequences specified in Fig. S1. Lentiviruses expressing SOX9, ROCR or control empty vector lentivirus were generated by transfecting HEK-293T cells with pCDH plasmids, together with packaging plasmids pCMV-VSV-G (Addgene plasmid #8454, deposited by Bob Weinberg) and psPAX2 (Addgene plasmid #12260, deposited by Didier Trono). The virus-containing culture media were collected every 24 h for 3 days and concentrated (10×) with Clontech Lenti-X Concentrator into PBS (Takara). MSCs and HAC were transduced with the lentivirus-containing PBS plus 8 μg/ml polybrene. A Promega CytoTox 96 cytotoxicity assay was used to assess cell viability following siRNA treatment (Fig. S9).
Rapid amplification of cDNA ends (RACE)
5′RACE was performed on RNA extracted from human articular cartilage or MSCs using the Invitrogen 5′ RACE System for Rapid Amplification of cDNA Ends (Life Technologies). Primer sequences are listed in Table S1. PCR amplification products were electrophoresed on agarose gels, cloned into the pCR4-TOPO vector and Sanger sequenced. The sequences have been uploaded to GenBank.
Immunoblotting
Lysates from MSCs were prepared as described previously (Barter et al., 2010). Lysates were immunoblotted with the following antibodies: SOX9 (AB5535, 1:2000) and GAPDH (AB2302, 1:40,000) (both Merck Millipore). Secondary anti-rabbit antibodies were from Dako and chemiluminescent images were captured using a G:BOX Chemi system (Syngene).
Statistical analysis
Data from each donor were individually analysed for gene expression and the values from each donor were then pooled to generate the mean±s.e.m. Significant differences between sample groups were assessed by one-way analysis of variance followed by the Bonferroni post-hoc test for multiple comparisons or by two-tailed Student's t-test for single comparisons.
Supplementary Material
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: M.J.B., D.A.Y.; Validation: D.A.Y.; Formal analysis: M.J.B., R.G.; Investigation: M.J.B., R.G., S.H.; Data curation: M.J.B., K.C., A.J.S., Y.X.; Writing - original draft: M.J.B., D.A.Y.; Writing - review & editing: I.M.C., D.A.Y.; Visualization: M.J.B., R.G.; Supervision: M.J.B., I.M.C., D.A.Y.; Project administration: D.A.Y.; Funding acquisition: I.M.C., D.A.Y.
Funding
This work was funded by Arthritis Research UK (grant 19424), the JGW Patterson Foundation, Newcastle upon Tyne Hospitals NHS Foundation Trust, the National Institute for Health Research Newcastle Biomedical Research Centre, Instituto de Salud Carlos III and the Medical Research Council. Deposited in PMC for immediate release.
Data availability
RNA-Seq data are available in Gene Expression Omnibus under accession number GSE107308. ROCR transcript variant sequence are available in GenBank under accession numbers MG018800 and MG018801.
Supplementary information
Supplementary information available online at http://dev.biologists.org/lookup/doi/10.1242/dev.152504.supplemental
References
- Akiyama H. (2008). Control of chondrogenesis by the transcription factor Sox9. Mod. Rheumatol. 18, 213-219. 10.3109/s10165-008-0048-x [DOI] [PubMed] [Google Scholar]
- Akiyama H. and Lefebvre V. (2011). Unraveling the transcriptional regulatory machinery in chondrogenesis. J. Bone Miner. Metab. 29, 390-395. 10.1007/s00774-011-0273-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akiyama H., Chaboissier M. C., Martin J. F., Schedl A. and de Crombrugghe B. (2002). The transcription factor Sox9 has essential roles in successive steps of the chondrocyte differentiation pathway and is required for expression of Sox5 and Sox6. Genes Dev. 16, 2813-2828. 10.1101/gad.1017802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barter M. J., Hui W., Lakey R. L., Catterall J. B., Cawston T. E. and Young D. A. (2010). Lipophilic statins prevent matrix metalloproteinase-mediated cartilage collagen breakdown by inhibiting protein geranylgeranylation. Ann. Rheum. Dis. 69, 2189-2198. 10.1136/ard.2010.129197 [DOI] [PubMed] [Google Scholar]
- Barter M. J., Tselepi M., Gómez R., Woods S., Hui W., Smith G. R., Shanley D. P., Clark I. M. and Young D. A. (2015). Genome-wide microRNA and gene analysis of mesenchymal stem cell chondrogenesis identifies an essential role and multiple targets for miR-140-5p. Stem Cells 33, 3266-3280. 10.1002/stem.2093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson H. L., Quinn J. J., Yang Y. W., Thornburg C. K., Chang H. Y. and Stadler H. S. (2015). LncRNA-HIT functions as an epigenetic regulator of chondrogenesis through its recruitment of p100/CBP complexes. PLoS Genet. 11, e1005680 10.1371/journal.pgen.1005680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrieri C., Cimatti L., Biagioli M., Beugnet A., Zucchelli S., Fedele S., Pesce E., Ferrer I., Collavin L., Santoro C. et al. (2012). Long non-coding antisense RNA controls Uchl1 translation through an embedded SINEB2 repeat. Nature 491, 454-457. 10.1038/nature11508 [DOI] [PubMed] [Google Scholar]
- Chimal-Monroy J., Rodriguez-Leon J., Montero J. A., Ganan Y., Macias D., Merino R. and Hurle J. M. (2003). Analysis of the molecular cascade responsible for mesodermal limb chondrogenesis: sox genes and BMP signaling. Dev. Biol. 257, 292-301. 10.1016/S0012-1606(03)00066-6 [DOI] [PubMed] [Google Scholar]
- ENCODE Project Consortium E. P. (2012). An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57-74. 10.1038/nature11247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derrien T., Johnson R., Bussotti G., Tanzer A., Djebali S., Tilgner H., Guernec G., Martin D., Merkel A., Knowles D. G. et al. (2012). The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 22, 1775-1789. 10.1101/gr.132159.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn S. L., Soul J.-M., Anand S., Schwartz J.-M., Boot-Handford R. P. and Hardingham T. E. (2016). Gene expression changes in damaged osteoarthritic cartilage identify a signature of non-chondrogenic and mechanical responses. Osteoarthritis Cartilage 24, 1431-1440. 10.1016/j.joca.2016.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engreitz J. M., Haines J. E., Perez E. M., Munson G., Chen J., Kane M., McDonel P. E., Guttman M. and Lander E. S. (2016). Local regulation of gene expression by lncRNA promoters, transcription and splicing. Nature 539, 452-455. 10.1038/nature20149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst J., Kheradpour P., Mikkelsen T. S., Shoresh N., Ward L. D., Epstein C. B., Zhang X., Wang L., Issner R., Coyne M. et al. (2011). Mapping and analysis of chromatin state dynamics in nine human cell types. Nature 473, 43-49. 10.1038/nature09906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagerberg L., Hallström B. M., Oksvold P., Kampf C., Djureinovic D., Odeberg J., Habuka M., Tahmasebpoor S., Danielsson A., Edlund K. et al. (2014). Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol. Cell. Proteomics 13, 397-406. 10.1074/mcp.M113.035600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farndale R. W., Sayers C. A. and Barrett A. J. (1982). A direct spectrophotometric microassay for sulfated glycosaminoglycans in cartilage cultures. Connect. Tissue Res. 9, 247-248. 10.3109/03008208209160269 [DOI] [PubMed] [Google Scholar]
- Fatica A. and Bozzoni I. (2014). Long non-coding RNAs: new players in cell differentiation and development. Nat. Rev. Genet. 15, 7-21. 10.1038/nrg3606 [DOI] [PubMed] [Google Scholar]
- Flicek P., Amode M. R., Barrell D., Beal K., Billis K., Brent S., Carvalho-Silva D., Clapham P., Coates G., Fitzgerald S. et al. (2014). Ensembl 2014. Nucleic Acids Res. 42, D749-D755. 10.1093/nar/gkt1196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster J. W., Dominguez-Steglich M. A., Guioli S., Kwok C., Weller P. A., Stevanovic M., Weissenbach J., Mansour S., Young I. D., Goodfellow P. N. et al. (1994). Campomelic dysplasia and autosomal sex reversal caused by mutations in an SRY-related gene. Nature 372, 525-530. 10.1038/372525a0 [DOI] [PubMed] [Google Scholar]
- Gordon C. T., Tan T. Y., Benko S., Fitzpatrick D., Lyonnet S. and Farlie P. G. (2009). Long-range regulation at the SOX9 locus in development and disease. J. Med. Genet. 46, 649-656. 10.1136/jmg.2009.068361 [DOI] [PubMed] [Google Scholar]
- Huynh N. P. T., Anderson B. A., Guilak F. and McAlinden A. (2017). Emerging roles for long noncoding RNAs in skeletal biology and disease. Connect. Tissue Res. 58, 116-141. 10.1080/03008207.2016.1194406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer M. K., Niknafs Y. S., Malik R., Singhal U., Sahu A., Hosono Y., Barrette T. R., Prensner J. R., Evans J. R., Zhao S. et al. (2015). The landscape of long noncoding RNAs in the human transcriptome. Nat. Genet. 47, 199-208. 10.1038/ng.3192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami Y., Rodriguez-León J. and Izpisua Belmonte J. C. I. (2006). The role of TGFbetas and Sox9 during limb chondrogenesis. Curr. Opin. Cell Biol. 18, 723-729. 10.1016/j.ceb.2006.10.007 [DOI] [PubMed] [Google Scholar]
- Klattenhoff C. A., Scheuermann J. C., Surface L. E., Bradley R. K., Fields P. A., Steinhauser M. L., Ding H., Butty V. L., Torrey L., Haas S. et al. (2013). Braveheart, a long noncoding RNA required for cardiovascular lineage commitment. Cell 152, 570-583. 10.1016/j.cell.2013.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong L., Zhang Y., Ye Z.-Q., Liu X.-Q., Zhao S.-Q., Wei L. and Gao G. (2007). CPC: assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res. 35, W345-W349. 10.1093/nar/gkm391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kretz M., Siprashvili Z., Chu C., Webster D. E., Zehnder A., Qu K., Lee C. S., Flockhart R. J., Groff A. F., Chow J. et al. (2013). Control of somatic tissue differentiation by the long non-coding RNA TINCR. Nature 493, 231-235. 10.1038/nature11661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krupp M., Marquardt J. U., Sahin U., Galle P. R., Castle J. and Teufel A. (2012). RNA-Seq Atlas--a reference database for gene expression profiling in normal tissue by next-generation sequencing. Bioinformatics 28, 1184-1185. 10.1093/bioinformatics/bts084 [DOI] [PubMed] [Google Scholar]
- Lefebvre V. and Dvir-Ginzberg M. (2017). SOX9 and the many facets of its regulation in the chondrocyte lineage. Connect. Tissue Res. 58, 2-14. 10.1080/03008207.2016.1183667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebvre V., Huang W., Harley V. R., Goodfellow P. N. and de Crombrugghe B. (1997). SOX9 is a potent activator of the chondrocyte-specific enhancer of the pro alpha1(II) collagen gene. Mol. Cell. Biol. 17, 2336-2346. 10.1128/MCB.17.4.2336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leipoldt M., Erdel M., Bien-Willner G. A., Smyk M., Theurl M., Yatsenko S. A., Lupski J. R., Lane A. H., Shanske A. L., Stankiewicz P. et al. (2007). Two novel translocation breakpoints upstream of SOX9 define borders of the proximal and distal breakpoint cluster region in campomelic dysplasia. Clin. Genet. 71, 67-75. 10.1111/j.1399-0004.2007.00736.x [DOI] [PubMed] [Google Scholar]
- Li L., Liu B., Wapinski O. L., Tsai M.-C., Qu K., Zhang J., Carlson J. C., Lin M., Fang F., Gupta R. A. et al. (2013). Targeted disruption of Hotair leads to homeotic transformation and gene derepression. Cell Rep. 5, 3-12. 10.1016/j.celrep.2013.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin M. F., Jungreis I. and Kellis M. (2011). PhyloCSF: a comparative genomics method to distinguish protein coding and non-coding regions. Bioinformatics 27, i275-i282. 10.1093/bioinformatics/btr209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C. F., Samsa W. E., Zhou G. and Lefebvre V. (2016). Transcriptional control of chondrocyte specification and differentiation. Semin. Cell Dev. Biol. 63, 34-49. 10.1016/j.semcdb.2016.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maass P. G., Rump A., Schulz H., Stricker S., Schulze L., Platzer K., Aydin A., Tinschert S., Goldring M. B., Luft F. C. et al. (2012). A misplaced lncRNA causes brachydactyly in humans. J. Clin. Invest. 122, 3990-4002. 10.1172/JCI65508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansour S., Offiah A. C., McDowall S., Sim P., Tolmie J. and Hall C. (2002). The phenotype of survivors of campomelic dysplasia. J. Med. Genet. 39, 597-602. 10.1136/jmg.39.8.597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mead T. J., Wang Q., Bhattaram P., Dy P., Afelik S., Jensen J. and Lefebvre V. (2013). A far-upstream (-70 kb) enhancer mediates Sox9 auto-regulation in somatic tissues during development and adult regeneration. Nucleic Acids Res. 41, 4459-4469. 10.1093/nar/gkt140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murdoch A. D., Grady L. M., Ablett M. P., Katopodi T., Meadows R. S. and Hardingham T. E. (2007). Chondrogenic differentiation of human bone marrow stem cells in transwell cultures: generation of scaffold-free cartilage. Stem Cells 25, 2786-2796. 10.1634/stemcells.2007-0374 [DOI] [PubMed] [Google Scholar]
- Ng S.-Y., Bogu G. K., Soh B. S. and Stanton L. W. (2013). The long noncoding RNA RMST interacts with SOX2 to regulate neurogenesis. Mol. Cell 51, 349-359. 10.1016/j.molcel.2013.07.017 [DOI] [PubMed] [Google Scholar]
- Ninomiya S., Isomura M., Narahara K., Seino Y. and Nakamura Y. (1996). Isolation of a testis-specific cDNA on chromosome 17q from a region adjacent to the breakpoint of t(12;17) observed in a patient with acampomelic campomelic dysplasia and sex reversal. Hum. Mol. Genet. 5, 69-72. 10.1093/hmg/5.1.69 [DOI] [PubMed] [Google Scholar]
- Onyekwelu I., Goldring M. B. and Hidaka C. (2009). Chondrogenesis, joint formation, and articular cartilage regeneration. J. Cell. Biochem. 107, 383-392. 10.1002/jcb.22149 [DOI] [PubMed] [Google Scholar]
- Ørom U. A., Derrien T., Beringer M., Gumireddy K., Gardini A., Bussotti G., Lai F., Zytnicki M., Notredame C., Huang Q. et al. (2010). Long noncoding RNAs with enhancer-like function in human cells. Cell 143, 46-58. 10.1016/j.cell.2010.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson M. J., Philp A. M., Heward J. A., Roux B. T., Walsh D. A., Davis E. T., Lindsay M. A. and Jones S. W. (2016). Long intergenic noncoding RNAs mediate the human chondrocyte inflammatory response and are differentially expressed in osteoarthritis cartilage. Arthritis Rheumatol. 68, 845-856. 10.1002/art.39520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry R. B.-T. and Ulitsky I. (2016). The functions of long noncoding RNAs in development and stem cells. Development 143, 3882-3894. 10.1242/dev.140962 [DOI] [PubMed] [Google Scholar]
- Pittenger M. F., Mackay A. M., Beck S. C., Jaiswal R. K., Douglas R., Mosca J. D., Moorman M. A., Simonetti D. W., Craig S. and Marshak D. R. (1999). Multilineage potential of adult human mesenchymal stem cells. Science 284, 143-147. 10.1126/science.284.5411.143 [DOI] [PubMed] [Google Scholar]
- Pritchett J., Athwal V., Roberts N., Hanley N. A. and Hanley K. P. (2011). Understanding the role of SOX9 in acquired diseases: lessons from development. Trends Mol. Med. 17, 166-174. 10.1016/j.molmed.2010.12.001 [DOI] [PubMed] [Google Scholar]
- Quinodoz S. and Guttman M. (2014). Long noncoding RNAs: an emerging link between gene regulation and nuclear organization. Trends Cell Biol. 24, 651-663. 10.1016/j.tcb.2014.08.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn J. L. and Chang H. Y. (2012). Genome regulation by long noncoding RNAs. Annu. Rev. Biochem. 81, 145-166. 10.1146/annurev-biochem-051410-092902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauvageau M., Goff L. A., Lodato S., Bonev B., Groff A. F., Gerhardinger C., Sanchez-Gomez D. B., Hacisuleyman E., Li E., Spence M. et al. (2013). Multiple knockout mouse models reveal lincRNAs are required for life and brain development. Elife 2, e01749 10.7554/eLife.01749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekiya I., Vuoristo J. T., Larson B. L. and Prockop D. J. (2002). In vitro cartilage formation by human adult stem cells from bone marrow stroma defines the sequence of cellular and molecular events during chondrogenesis. Proc. Natl. Acad. Sci. USA 99, 4397-4402. 10.1073/pnas.052716199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockl S., Bauer R. J., Bosserhoff A. K., Gottl C., Grifka J. and Grassel S. (2013). Sox9 modulates cell survival and adipogenic differentiation of multipotent adult rat mesenchymal stem cells. J. Cell Sci. 126, 2890-2902. 10.1242/jcs.124305 [DOI] [PubMed] [Google Scholar]
- Trapnell C., Roberts A., Goff L., Pertea G., Kim D., Kelley D. R., Pimentel H., Salzberg S. L., Rinn J. L. and Pachter L. (2012). Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7, 562-578. 10.1038/nprot.2012.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulitsky I., Shkumatava A., Jan C. H., Sive H. and Bartel D. P. (2011). Conserved function of lincRNAs in vertebrate embryonic development despite rapid sequence evolution. Cell 147, 1537-1550. 10.1016/j.cell.2011.11.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vance K. W. and Ponting C. P. (2014). Transcriptional regulatory functions of nuclear long noncoding RNAs. Trends Genet. 30, 348-355. 10.1016/j.tig.2014.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner T., Wirth J., Meyer J., Zabel B., Held M., Zimmer J., Pasantes J., Bricarelli F. D., Keutel J., Hustert E. et al. (1994). Autosomal sex reversal and campomelic dysplasia are caused by mutations in and around the SRY-related gene SOX9. Cell 79, 1111-1120. 10.1016/0092-8674(94)90041-8 [DOI] [PubMed] [Google Scholar]
- Wang K. C., Yang Y. W., Liu B., Sanyal A., Corces-Zimmerman R., Chen Y., Lajoie B. R., Protacio A., Flynn R. A., Gupta R. A. et al. (2011). A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature 472, 120-124. 10.1038/nature09819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Park H. J., Dasari S., Wang S., Kocher J. P. and Li W. (2013). CPAT: coding-potential assessment tool using an alignment-free logistic regression model. Nucleic Acids Res. 41, e74 10.1093/nar/gkt006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright E., Hargrave M. R., Christiansen J., Cooper L., Kun J., Evans T., Gangadharan U., Greenfield A. and Koopman P. (1995). The Sry-related gene Sox9 is expressed during chondrogenesis in mouse embryos. Nat. Genet. 9, 15-20. 10.1038/ng0195-15 [DOI] [PubMed] [Google Scholar]
- Wunderle V. M., Critcher R., Hastie N., Goodfellow P. N. and Schedl A. (1998). Deletion of long-range regulatory elements upstream of SOX9 causes campomelic dysplasia. Proc. Natl. Acad. Sci. USA 95, 10649-10654. 10.1073/pnas.95.18.10649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y., Barter M. J., Swan D. C., Rankin K. S., Rowan A. D., Santibanez-Koref M., Loughlin J. and Young D. A. (2012). Identification of the pathogenic pathways in osteoarthritic hip cartilage: commonality and discord between hip and knee OA. Osteoarthritis Cartilage 20, 1029-1038. 10.1016/j.joca.2012.05.006 [DOI] [PubMed] [Google Scholar]
- Yang Y., Wen L. and Zhu H. (2015). Unveiling the hidden function of long non-coding RNA by identifying its major partner-protein. Cell Biosci. 5, 59 10.1186/s13578-015-0050-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao B., Wang Q., Liu C.-F., Bhattaram P., Li W., Mead T. J., Crish J. F. and Lefebvre V. (2015). The SOX9 upstream region prone to chromosomal aberrations causing campomelic dysplasia contains multiple cartilage enhancers. Nucleic Acids Res. 43, 5394-5408. 10.1093/nar/gkv426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon B. S., Ovchinnikov D. A., Yoshii I., Mishina Y., Behringer R. R. and Lyons K. M. (2005). Bmpr1a and Bmpr1b have overlapping functions and are essential for chondrogenesis in vivo. Proc. Natl. Acad. Sci. USA 102, 5062-5067. 10.1073/pnas.0500031102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Chen S., Bao N., Yang C., Ti Y., Zhou L. and Zhao J. (2015). Sox4 enhances chondrogenic differentiation and proliferation of human synovium-derived stem cell via activation of long noncoding RNA DANCR. J. Mol. Histol. 46, 467-473. 10.1007/s10735-015-9638-z [DOI] [PubMed] [Google Scholar]
- Zhou G., Zheng Q., Engin F., Munivez E., Chen Y., Sebald E., Krakow D. and Lee B. (2006). Dominance of SOX9 function over RUNX2 during skeletogenesis. Proc. Natl. Acad. Sci. USA 103, 19004-19009. 10.1073/pnas.0605170103 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.