Abstract
The nucleocapsid (NC) is an N-terminal protein derived from the HIV-1 Gag precursor polyprotein, pr55Gag. NC possesses key functions at several pivotal stages of viral replication. For example, an interaction between NC and the host double-stranded RNA-binding protein Staufen1 was shown to regulate several steps in the viral replication cycle, such as Gag multimerization and genomic RNA encapsidation. In this work, we observed that the overexpression of NC leads to the induction of stress granule (SG) assembly. NC-mediated SG assembly was unique as it was resistant to the SG blockade imposed by the HIV-1 capsid (CA), as shown in earlier work. NC also reduced host cell mRNA translation, as judged by a puromycylation assay of de novo synthesized proteins, and this was recapitulated in polysome profile analyses. Virus production was also found to be significantly reduced. Finally, Staufen1 expression completely rescued the blockade to NC-mediated SG assembly, global mRNA translation as well as virus production. NC expression also resulted in the phosphorylation of protein kinase R (PKR) and eIF2α, and this was inhibited with Staufen1 coexpression. This work sheds light on an unexpected function of NC in host cell translation. A comprehensive understanding of the molecular mechanisms by which a fine balance of the HIV-1 structural proteins NC and CA act in concert with host proteins such as Staufen1 to modulate the host stress response will aid in the development of new antiviral therapeutics.
Keywords: nucleocapsid, HIV-1, stress granules, mRNA translation, Staufen1
INTRODUCTION
The HIV-1 nucleocapsid (NC) is a highly versatile, 9 kDa protein that is intricately associated with the HIV-1 genomic viral RNA (vRNA), exerting an effect at both early and late steps of the HIV-1 replication cycle from reverse transcription (RT) and DNA integration to vRNA selection, packaging, and assembly (for review, see Darlix et al. 2014). It is a product of the proteolytic processing of the precursor Gag polyprotein (pr55Gag, referred to as Gag herein) and contains two CCHC zinc finger domains flanked by basic residues, all of which contribute to both sequence and non-sequence-specific nucleic acid binding activity. NC also possesses chaperone activity that facilitates the rearrangement of nucleic acids into thermodynamically more stable structures (South et al. 1990; Levin et al. 2005; Rein 2010; Bell and Lever 2013). NC recruits numerous host proteins to facilitate its functions and these include the double-stranded (ds) RNA-binding protein Staufen1, a host factor that is involved in mRNA trafficking and translation (Mouland et al. 2000; Kanai et al. 2004; Dugre-Brisson et al. 2005; Ricci et al. 2014). In our previous work, we have shown that Staufen1 regulates several events in the HIV-1 replication cycle by assembling large HIV-1-dependent ribonucleoprotein complexes (SHRNPs) and via its interactions with NC, affects various steps of virus assembly including Gag multimerization and vRNA encapsidation (Chatel-Chaix et al. 2004, 2007, 2008; Abrahamyan et al. 2010). Staufen1 also has been reported to play a role in modulating the cellular stress response (Thomas et al. 2009; Hanke et al. 2013; Dixit et al. 2016; Ravel-Chapuis et al. 2016).
To counteract conditions of stress, such as that of viral infection, the host mounts a cellular stress response that leads to the assembly of translationally silent ribonucleoprotein (RNP) complexes known as stress granules (SGs) (Anderson and Kedersha 2009; Thomas et al. 2011). Since viruses are obligate intracellular parasites that utilize the host cell machinery to facilitate their own gene expression, their replication can be markedly affected by an impediment to cellular mRNA translation. Therefore, viruses have developed the capability to circumvent this innate antiviral host cell response by numerous mechanisms (for review, see Valiente-Echeverria et al. 2012; Poblete-Duran et al. 2016). Two types of SGs have been described that differ in morphology, composition, and mechanism of assembly (Fujimura et al. 2012). In our previous work, we have shown that HIV-1 disrupts the canonical type I SG assembly in an eIF2α phosphorylation (eIF2α-P) independent manner via an interaction with the eukaryotic elongation factor eEF2 with the capsid (CA) domain on the Gag polyprotein (Abrahamyan et al. 2010; Valiente-Echeverria et al. 2014). We also demonstrated that Gag is able to block the assembly of type II, noncanonical SGs by reducing the amount of hypophosphorylated 4EBP1 associated with the 5′ cap potentially through an interaction with its target, eIF4E (Cinti et al. 2016). Interestingly, a recent study has reported that the expression of the HIV-1 NC alone leads to the assembly of SGs (Yu et al. 2016).
In this study, we have characterized NC-induced SGs and have elucidated the mechanism by which they assemble. Here, we demonstrate that NC induces the assembly of SGs, and although the composition resembles that of type I canonical SGs, they cannot be dissociated by HIV-1 Gag expression (Abrahamyan et al. 2010; Valiente-Echeverria et al. 2014). We also show that Staufen1, a host protein that has roles in stabilizing polysomes and SG dynamics (Thomas et al. 2009), is capable of inhibiting NC-induced SG assembly. We also demonstrate that Staufen1's F135 amino acid residue in its third dsRNA binding domain (dsRBD3) is critical for this activity. We also demonstrate that NC expression leads to the phosphorylation of protein kinase R (PKR) and eIF2α, resulting in hindered host cell mRNA translation, and that this impairs viral production; this can also be rescued by Staufen1 coexpression. This work sheds light on an unexpected function of NC on host cell mRNA translation and the mechanism by which it operates in concert with the host protein Staufen1 to modulate the host stress response.
RESULTS
NC induces the assembly of SGs containing TIAR1, G3BP1, eIF3, PABP, and poly(A) mRNAs
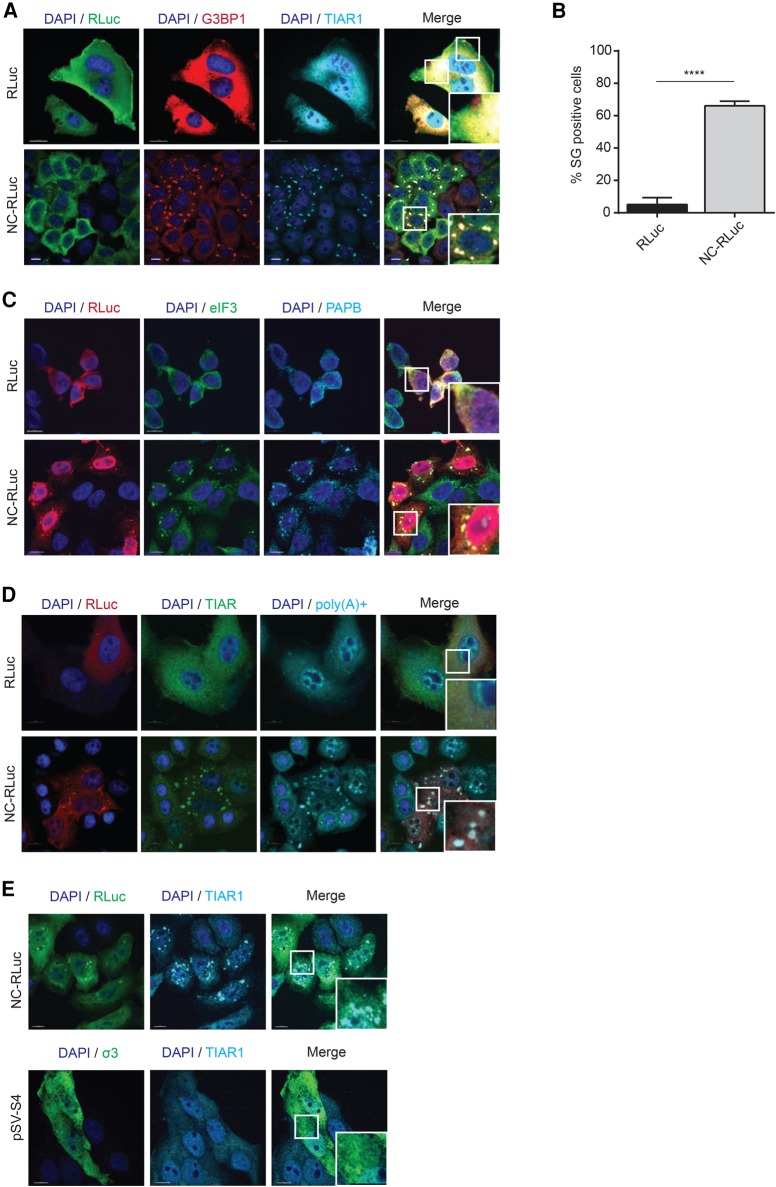
SGs are associated with silenced transcripts and many viruses are known to subvert the function of these RNA granules for their replicative advantage (Lloyd 2012). As NC expression has been recently demonstrated to lead to the assembly of SGs (Yu et al. 2016), we set out to quantify and describe this NC induced assembly of SGs. HeLa cells were either mock transfected with RLuc or transfected with NC-RLuc, fixed, and SG assembly was monitored by indirect immunofluorescence of Ras-GAP SH3 domain-binding protein 1 (G3BP1) and TIA-1-related RNA-binding protein (TIAR1). SGs were detected in 66.2 (SD ± 2.7)% of NC-expressing cells, in striking contrast compared to the 5.1 (SD ± 4.2)% in the RLuc-transfected cells (Fig. 1A,B).
FIGURE 1.
NC expression induces assembly of SG containing G3BP1, TIAR1, PABP, eIF3 and poly(A) mRNAs. (A) HeLa cells were transfected with RLuc or NC-RLuc, and 24 h later were stained for RLuc (green), G3BP1 (red), and TIAR1 (cyan). Scale bars are 10 µm. (B) Quantification of HeLa cells containing SGs transfected with RLuc or NC-RLuc from A. Error bars represent the standard deviation from three independent experiments with at least 150 cells counted per treatment. Asterisks represent statistically significant difference between RLuc and NC-RLuc-expressing cells (Student's t-test; P < 0.001). (C) HeLa cells transfected as in A were stained for RLuc (red), eIF3 (green), and PABP (cyan). Scale bars are 10 µm. (D) HeLa cells transfected as in A were stained for RLuc (green), TIAR (red), and poly(A) mRNAs (cyan). Scale bars are 10 µm. (E) Expression of CCHC-type zinc finger on a dsRNA binding protein does not lead to SG assembly. HeLa cells were transfected with pSV-S4 to express the Reovirus σ3 protein (which contains CCHC-zinc fingers). SG assembly was then monitored by staining the cells for TIAR1 (cyan).
Two distinct types of SGs have previously been characterized (Fujimura et al. 2012) that differ in their mechanism of assembly and localization, as well as in composition. The canonical type I SGs, such as those induced by Arsenite and Pateamine A, are larger and contain the eukaryotic initiation factors eIF4G, eIF4E, and eIF3, among several other components. In contrast, type II SGs, which are induced by Selenium, are smaller in size and do not contain eIF3. To determine which type of SGs are induced by NC, we performed indirect immunofluorescence analyses on NC-RLuc-transfected HeLa cells and probed them for eIF3 along with another SG marker Poly(A) binding protein (PABP). We observed that eIF3 is present in the NC induced SGs, indicating that they are likely to be the canonical, type I stress granules (Fig. 1C; Fujimura et al. 2012; Cinti et al. 2016). Thus, these newly characterized NC-SGs contain G3BP1, TIAR1, eIF3, and PABP.
Recent studies have demonstrated that some stresses such as ultraviolet irradiation and rocaglamide A (RocA) treatment assemble SG-like foci that do not contain poly(A) mRNAs (Aulas et al. 2017). In order to determine if the NC-induced SGs are bona fide SGs that contain polyadenylated mRNAs (Kedersha et al. 1999), we conducted FISH for poly(A) mRNA with an oligo(dT) probe in mock transfected and NC-expressing cells. It was observed that in the NC-expressing cells, the poly(A) mRNAs colocalized with the SG marker TIAR, indicating that NC expression leads to the assembly of bona fide SGs that contain mRNAs (Fig. 1D).
NC is composed of an N-terminal basic region, two CCHC type zinc fingers (ZFs) and a basic linker region between the ZFs. In a previous study, it was observed that a loss of the ZFs of NC led to impaired SG assembly when compared to the wild-type NC (Yu et al. 2016), suggesting that the NC ZFs contribute to SG assembly. In order to test if the presence of a ZF from another virus can also elicit a stress response, we transfected cells with the plasmid pSV-S4 that encodes the Reovirus σ3 protein, which is a dsRNA binding protein that is a component of the reovirus outer capsid and contains CCHC type zinc fingers similar to NC (Mabrouk and Lemay 1994). The expression of this protein did not lead to SG induction, indicating that merely the presence of CCHC-type zinc fingers alone does not lead to SG assembly, and that this activity is specific to NC (Fig. 1E).
Neither Gag nor CA disassembles NC-induced SGs
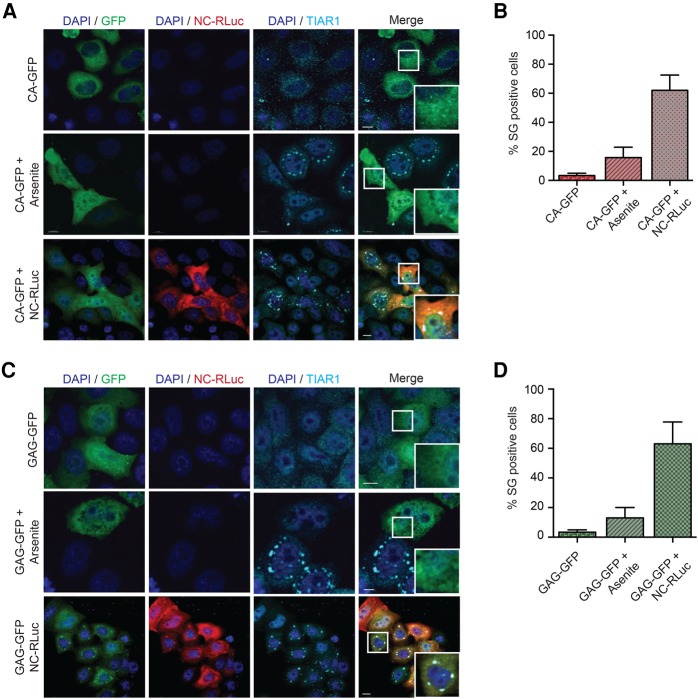
In our previous work, we have shown that Gag disassembles preformed type I SGs, irrespective of eIF2α phosphorylation, by interacting with the eukaryotic elongation factor eEF2 via the Gag capsid (CA) domain (Valiente-Echeverria et al. 2014). Therefore, we sought to determine if full-length Gag or CA dissociates NC-induced SGs. HeLa cells were transfected with either Gag-GFP or CA-GFP plasmids alone or with NC-RLuc. Twenty-four hours later, cells were left untreated or treated with arsenite and SGs were visualized by indirect immunofluorescence. Although Gag and CA were able to efficiently inhibit arsenite-induced SGs, neither Gag nor CA was capable of dissociating NC-induced SGs (Fig. 2A–D). These results suggest that the NC-induced SGs are of a different nature than the ones induced by arsenite, pateamine A, or selenite that Gag is able to dissociate (Valiente-Echeverria et al. 2014; Cinti et al. 2016).
FIGURE 2.
Gag and CA block Arsenite-induced SGs but cannot disrupt NC-induced SGs. (A) HeLa cells were transfected with CA-GFP and CA-GFP + NC-RLuc. Twenty-four hours later, cells were either untreated or treated with Arsenite and stained for RLuc (red) and TIAR1 (cyan). Scale bars are 10 µm. (B) Quantification of HeLa cells containing SGs from A. Only CA and NC expressing cells were considered for the quantification. Error bars represent the standard deviation from three independent experiments. (C) HeLa cells were transfected with GAG-GFP and GAG-GFP + NC-RLuc. Twenty-four hours later, cells were either untreated or treated with Arsenite and stained for RLuc (red) and TIAR1 (cyan). Scale bars are 10 µm. (D) Quantification of HeLa cells containing SGs from C. Only Gag and NC expressing cells were considered for the quantification. Error bars represent the standard deviation from three independent experiments.
NC-induced SG assembly and translation arrest are inhibited by Staufen1
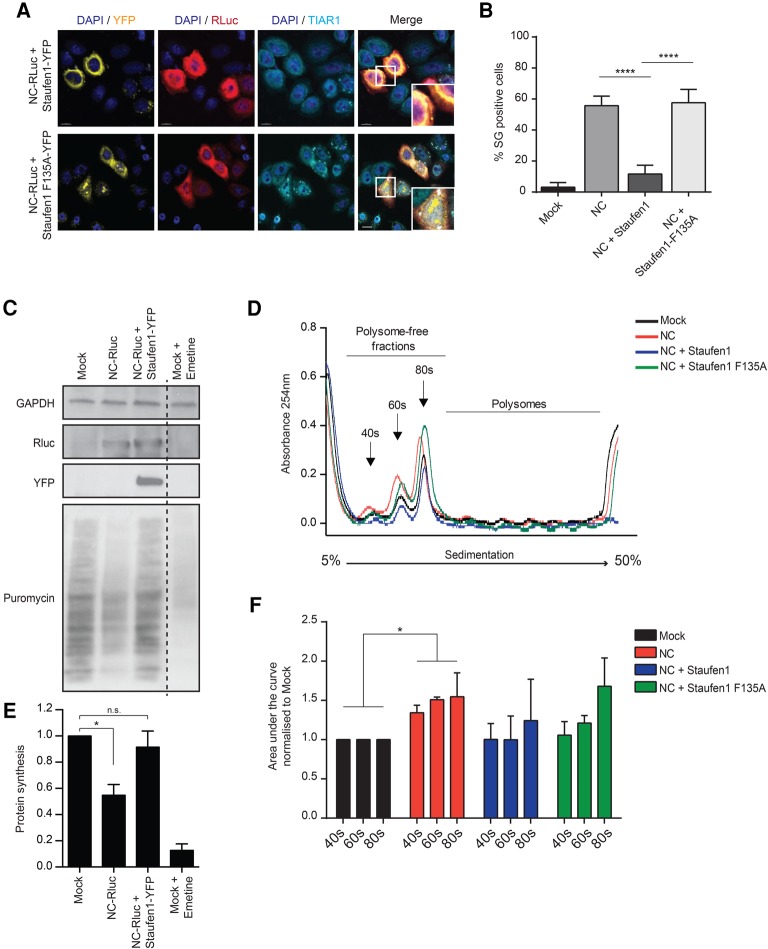
Staufen1 is a dsRNA binding protein that affects HIV-1 at multiple stages of its life cycle coinciding with many of the NC-associated functions in Gag multimerization and assembly, as well as in vRNA encapsidation (Mouland et al. 2000; Chatel-Chaix et al. 2007, 2008; Stopak et al. 2007; Milev et al. 2010). It exerts many of these functions by interacting with the zinc fingers of NC via its dsRBD3 domain, as shown in our previous work (Mouland et al. 2000; Chatel-Chaix et al. 2007, 2008). As a known interacting partner of NC with previously defined roles in the modulation of the stress response (Thomas et al. 2009), we therefore hypothesized that Staufen1 may be able to counteract NC-induced SG assembly. When HeLa cells were cotransfected with NC-RLuc and Staufen1-YFP, SGs were present only in 11.6 (SD ± 5.6)% of cotransfected cells, when compared to the 55.6 (SD ± 6.1)% of SG containing cells observed in the cells transfected with NC-RLuc only (Fig. 3A,B). To determine the mechanism of Staufen1-mediated disruption of NC-induced SGs, we co-transfected HeLa cells with NC-RLuc and Staufen1-F135A-YFP, that possesses a point mutation in the dsRBD3 domain which reduces Staufen1's capacity to bind both NC and RNA (Ramos et al. 2000; Chatel-Chaix et al. 2008). Under this condition, SGs were observed in 58.6 (SD ± 8.6)% of cotransfected cells, at levels comparable to the NC expressing cells alone (Fig. 3A,B).
FIGURE 3.
Staufen1 rescues NC-induced SG assembly and translation arrest. (A) HeLa cells were cotransfected with NC-RLuc and Staufen1-YFP or Staufen1-F135A-YFP and 24 h later were stained for RLuc (red) and TIAR1 (cyan). Scale bars are 10 µm. (B) Quantification of HeLa cells containing SGs from A. Error bars represent the standard deviation from three independent experiments with at least 150 cells counted per treatment. Asterisks represent statistically significant difference between groups (one-way ANOVA; P < 0.001). (C) Measurements of protein synthesis by puromycylation technique were performed by incubating mock, NC-RLuc, or NC-RLuc + Staufen1-YFP-transfected HeLa cells with medium containing puromycin as described in Materials and Methods. As positive control mock-transfected cells were incubated with 1 µM Emetine 1 h before the puromycin treatment. HeLa extracts were separated by denaturing electrophoresis and analyzed by western blot with antibody to puromycin (12D10). GAPDH immunoblot is shown as a loading control. (D) HeLa cells were mock-transfected or transfected with NC-RLuc, NC-RLuc + Staufen1-YFP, or NC-RLuc + Staufen1-F135A-YFP and 24 h later polysome fractionation and profiling was conducted. (E) Quantification of the puromycin-labeled peptides from C, values were normalized against mock cells extracts. Error bars represent the standard deviation from three independent experiments. Asterisks represent statistically significant difference between groups (one-way ANOVA; P < 0.05) (F) Area under the curve corresponding to 40s, 60s, and 80s peaks from D were quantified using GraphPad Prism 6. Error bars represent the standard deviation from three independent experiments. Asterisks represent statistically significant difference between groups (two-way ANOVA; P < 0.05).
To determine if de novo synthesis of proteins was reduced by NC expression, de novo synthesized proteins were labelled with puromycin in tissue culture. The puromycylation technique has been shown to be a valid alternative to the use of radioisotopes for measuring quantitative changes in protein synthesis in cell culture (Schmidt et al. 2009; Goodman et al. 2011). HeLa cells transfected with RLuc, NC-RLuc, or NC-RLuc and Staufen1-YFP were incubated with puromycin and then analyzed for the amount of de novo puromycin-labeled proteins by western blotting (Fig. 3D,F). As a positive control, RLuc-transfected cells were treated with emetine, a translation inhibitor (Fig. 3C,E). The results demonstrated that NC induced a twofold decrease in puromycin-labelled peptides, while coexpression of Staufen1 restored the protein synthesis to a level similar to mock transfected cells (Fig. 3C,E).
To confirm that NC-induced SG assembly has an effect on host cell translation and whether translation can be rescued by Staufen1 coexpression, we performed polysome profile analyses of cell lysates derived from cells that were either mock-transfected (RLuc-N1), transfected with NC-RLuc, NC-RLuc and Staufen1-YFP or Staufen1-F135A-YFP. An increase in the levels of RNA present in the polysome-free fractions implies an inhibition in host cell translation. When compared to mock-transfected cells, the expression of NC induced an increase in absorbance in polysome-free gradient fractions corresponding to the 40S, 60S ribosomal subunits and 80S ribosomes of the profile (Fig. 3D,F), thus indicating that in the presence of NC, there are increased free ribosomal subunits and monosomes. The presence of Staufen1 partially reversed the effects of NC expression on polysome profiles, but this ability, was lost when the Staufen1-F135A construct was coexpressed (Fig. 3D,F). These findings show that the proportion of free ribosomal subunits and monosomes was increased in the presence of NC, and this is relieved by Staufen1 coexpression, therefore indicating that NC reduces cellular mRNA translation.
NC and Staufen1 interact in situ and in vitro
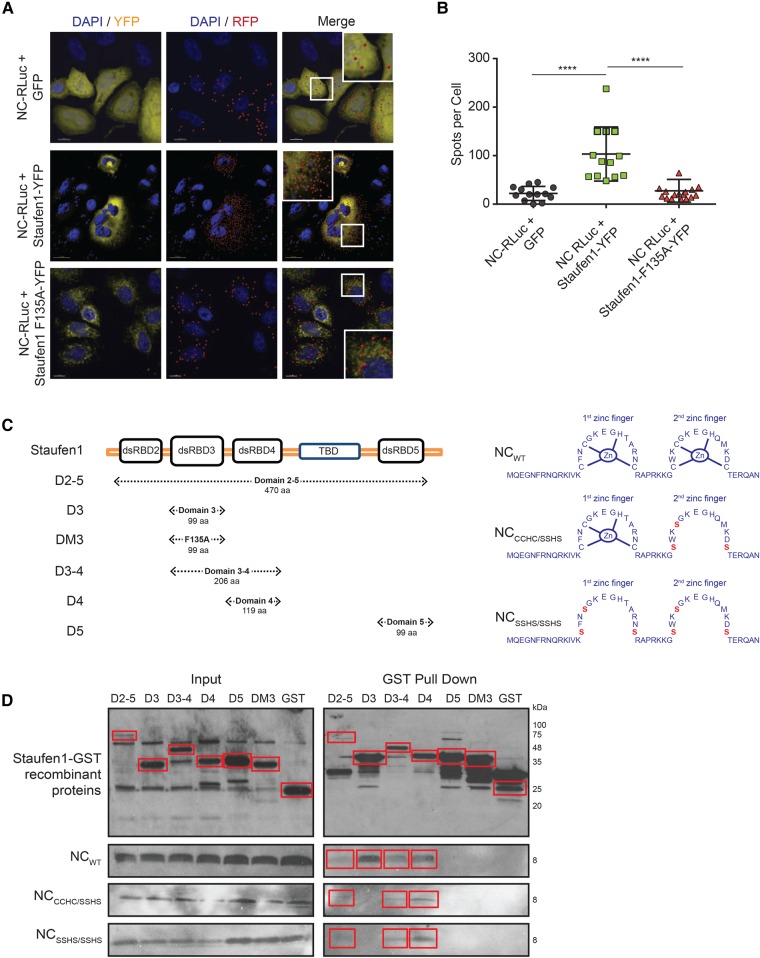
To further characterize the nature of the binding between Staufen1 and NC in host cells, we used a proximity ligation assay (PLA). This assay produces distinct countable spots that represent a single-molecule protein interaction ∼40 nm apart (Soderberg et al. 2006; Jarvius et al. 2007). In cells cotransfected with Staufen1-YFP and NC-RLuc, we confirmed a close localization between Staufen1 and NC (103.3, SD ± 16 spots per cell) (Fig. 4A,B), whereas there was little signal detected upon transfection of NC-RLuc together with Staufen1-F135A-YFP (19 ± SD 9.0 spots per cell), at levels that were comparable to the background PLA signal (22.1 ± SD 14.6 spots per cell) (Fig. 4A,B). These data indicate that Staufen1 is in close proximity to NC in situ, likely mediated via its dsRBD3.
FIGURE 4.
NC and Staufen1 interact in situ and in vitro. (A) HeLa cells were cotransfected with NC-RLuc and GFP or Staufen1-YFP or Staufen1-F135A-YFP and 24 h later were incubated with primary mouse and rabbit antibodies against RLuc and GFP. Coverslips were subsequently incubated with anti-mouse and anti-rabbit PLA probes. Each red signal corresponds to a single interaction event between NC and Staufen1. Nuclei were stained with DAPI (blue). Images shown are representative of >50 cells analyzed from two independent experiments. (B) The graph indicates the number of dots per cell. Asterisks represent statistically significant difference between groups (one-way ANOVA; P < 0.001). (C) Representation of Staufen1 and NC mutants used in GST pull down assays. (D) GST-Staufen1 mutants were incubated with GST SpinTrap columns in the presence of absence of NC mutants. After washing extensively, the proteins bound to the beads were detected by western blotting using anti-GST and anti-NC antibodies. Blot depicting GST tagged recombinant Staufen1 is a representative blot from three independent experiments using different NC constructs.
To determine if Staufen1 and NC interact by direct association and to precisely characterize the Staufen1 binding site on NC, we conducted in vitro GST-pull down assays. Full-length GST-tagged recombinant Staufen1 (D2-5), individual dsRBDs (D3, D3-4, D4, and D5; where D = dsRBD) as well as a dsRBD3 construct with point F135A mutation (DM3), used as a negative control, were incubated on GST-Spintrap columns (Fig. 4C). Recombinant, wild-type NC or recombinant mutated NC in one (CCHC-SSHS) or both (SSHS-SSHS) zinc fingers were added to the columns and eluted after washing (Fig. 4C). These assays are only qualitative, not quantitative as the expression levels of the recombinant Staufen1 proteins differed due to differences in solubility. As shown in Figure 4D, wild-type NC directly bound to the full-length Staufen1 (D2-5) as well as to the D3, D3-4, and D4 dSRBD truncations, but not to the D5, DM3, or GST only constructs. Furthermore, the binding of NC to D3 was lost when the two zinc finger mutants of NC were used (Fig. 4D). These data confirm the previously characterized binding of the Staufen1 dsRBD3 to the zinc fingers of NC (Chatel-Chaix et al. 2008), but also identify a novel zinc finger–independent binding site for Staufen1 via its dsRBD4. Taken together, these experiments indicate that Staufen1 is able to directly bind NC, both, in situ and in vitro in an RNA-independent manner and that this binding could lead to the sequestration of NC and a block to NC-induced SG assembly.
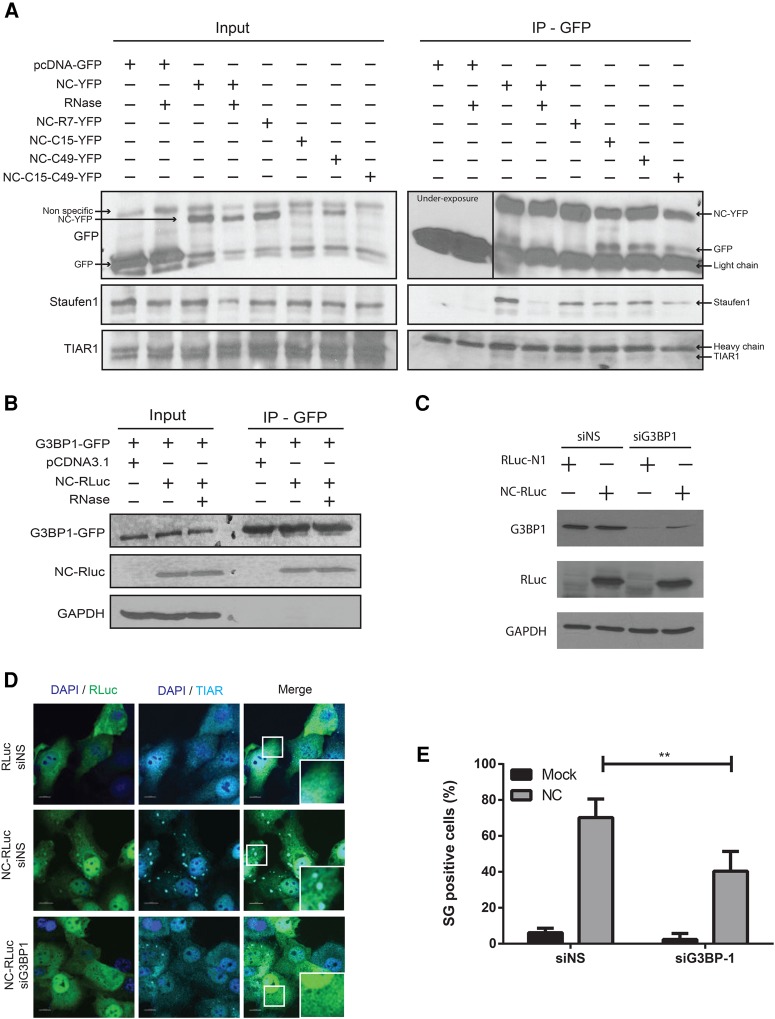
NC is found in a complex with SG components
To characterize a possible mechanism behind the NC-mediated SG assembly, we sought to determine the ability of NC to interact with components of SGs, by performing coimmunoprecipitation (co-IP) assays. HeLa cells were transfected with NC expressors that contained mutations in either the N-terminal region (NC-R7-YFP), the first zinc finger (ZF) (NC-C15-YFP), the second ZF (NC-C49-YFP), or both ZFs (NC-C14-C49-YFP) or mock transfected with GFP. As shown in Figure 5A, using anti-GFP beads, we demonstrated that TIAR1 and Staufen1 specifically interacted with NC-YFP, but not GFP alone, as well as with all the NC mutants tested (Fig. 5A). Additionally, the interactions were not dependent on RNA, as TIAR1 and Staufen1 still coimmunoprecipitated in the presence of RNAse, albeit to lower levels. This indicates that the binding of TIAR1 and Staufen1 to NC is enhanced in the presence of RNA, although RNA is not necessary for it (Fig. 5A). To determine if G3BP1 is a binding partner of NC, a U2OS cell line that constitutively expresses GFP-tagged G3BP1, was transfected with NC-RLuc. G3BP1-GFP was pulled down using anti-GFP beads and NC was found to specifically coimmunoprecipitate with it, even after RNase treatment (Fig. 5B). Taken together these results indicate that NC is capable of associating with a number of SG components even after RNase treatment, and suggest that the interaction with these factors could promote NC-induced SG assembly.
FIGURE 5.
NC coimmunoprecipitates with multiple SG markers. (A) HeLa cells were transfected with pEGFP-C1 or different NC-YFP mutants for 24 h. Cell lysates were collected, treated with RNase when indicated and subjected to anti-GFP immunoprecipitation. NC-associated proteins were processed for western blotting and probed for GFP, Staufen1, and TIAR1. Representative blots from three independent experiments are depicted. (B) U2OS cells stably expressing G3BP1-GFP were transfected with pcDNA3.1 or NC-RLuc for 24 h. Cell lysates were collected, treated with RNase when indicated and subjected to anti-GFP immunoprecipitation. G3BP1-associated proteins were processed for western blotting and probed for GFP, RLuc, and GAPDH. Representative blots from three independent experiments are depicted. (C) HeLa cells were transfected as indicated and cell lysates were processed for western blotting and probed for G3BP1, RLuc, and GAPDH. (D) Cells transfected as depicted were stained for RLuc (green) and TIAR1 (cyan). Scale bars are 10 µm. (E) Quantification of HeLa cells containing SGs from D. Error bars represent the standard deviation from three independent experiments with at least 100 cells counted per treatment. Asterisks represent statistically significant difference between groups (two-way ANOVA; P < 0.01).
A depletion of G3BP1 has been demonstrated to hinder the assembly of phospho-eIF2α dependent SGs (Kedersha et al. 2016). In order to determine if G3BP1 is required for the assembly of NC-induced SGs, cells were either treated with nonsilencing siRNA (siNS) or siRNA against G3BP1 (siG3BP1). The knockdown of G3BP1 was validated by western blot of cell lysates (Fig. 5C). They were either mock transfected or transfected with NC-RLuc and the assembly of SGs was determined by indirect immunofluorescence of the SG marker TIAR (Fig. 5D). It was observed that a knockdown of G3BP1 resulted in a significant decrease in the percentage of NC-induced SG assembly with only 40.42 (SD ± 10.96)% of cells displaying SG assembly when compared to 70.22 (SD ± 10.35)% of SG positive cells in the siNS treated cells (Fig. 5E). Thus, NC-induced SG assembly is impaired by the depletion of G3BP1.
NC expression leads to the phosphorylation of eIF2α by activating PKR
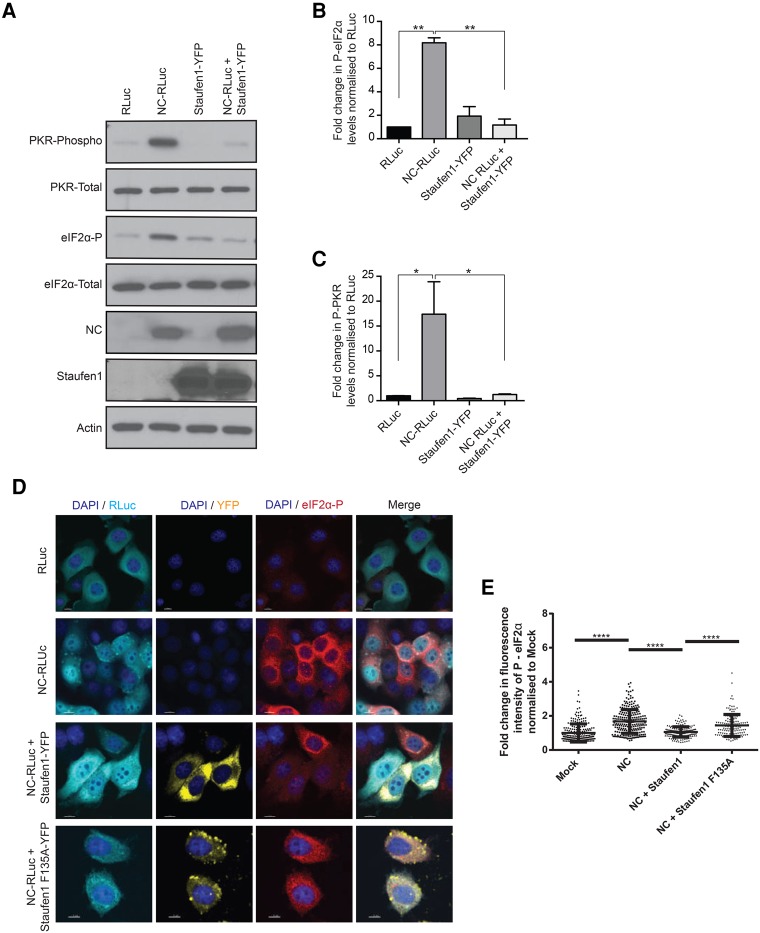
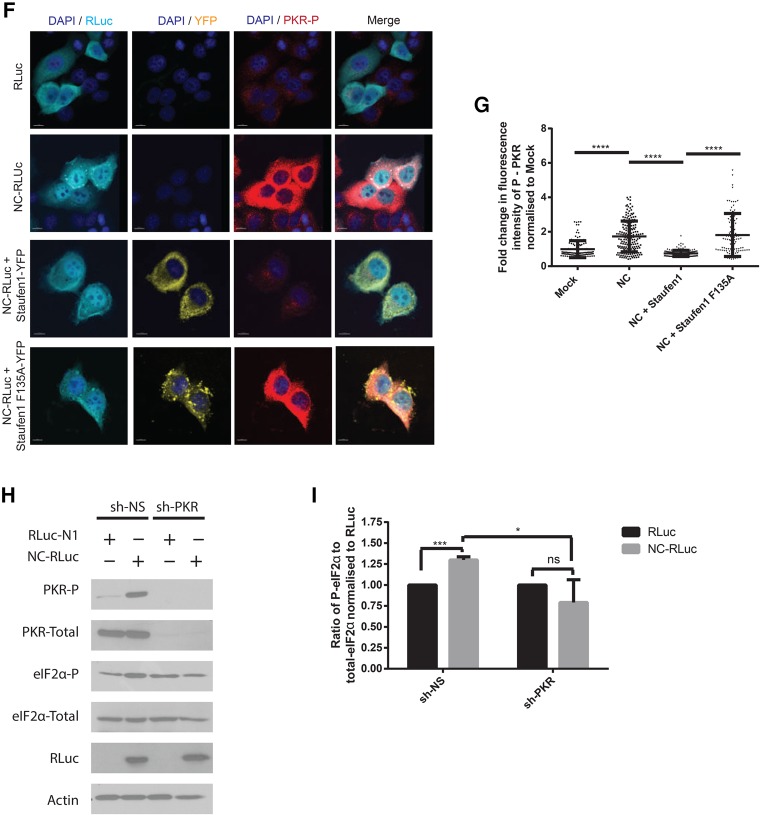
The phosphorylation of eIF2α is triggered by conditions of stress, thus blocking translation initiation and regulating SG assembly (Kedersha et al. 1999). However, the formation of type I SGs is either eIF2α phosphorylation-dependent or -independent (Dang et al. 2006). To determine whether NC-induced SG assembly is linked to the eIF2α phosphorylation status, cell lysates from mock transfected cells (pcDNA3.1) or from cells expressing NC (NC-RLuc) were analyzed by western blots using antibodies against total and phosphorylated forms of eIF2α. An eightfold increase in the amount of phosphorylated eIF2α was observed in the NC-expressing cells when compared to the mock-transfected cells (Fig. 6A,B). Furthermore, the coexpression of Staufen1 with NC significantly reduced the phosphorylation of eIF2α to levels comparable with mock-transfected cells (Fig. 6A,B). In order to determine the mechanism of eIF2α activation, we monitored PKR activation levels in the above conditions. PKR is an interferon-induced protein that senses dsRNA and its activation leads to the phosphorylation of eIF2α (Sadler and Williams 2007). PKR was activated in the NC expressing cells, but was inactive in mock and NC/Staufen1 coexpressing conditions (Fig. 6C). In order to determine whether Staufen1-F135A could also inhibit NC-mediated PKR and eIF2α phosphorylation, we either mock transfected cells or transfected them with NC-RLuc, NC-RLuc, and Staufen-YFP, or NC-RLuc and Staufen1-F135A-YFP. For each condition, indirect immunofluorescence was used to quantify the phosphorylation of eIF2α (Fig. 6D) and PKR (Fig. 6F). A significant increase in the fluorescence intensity of both the P-eIF2α (Fig. 6E) and P-PKR (Fig. 6G) was observed upon NC expression. This phosphorylation was reduced to levels comparable to wildtype upon NC/Staufen1 coexpression, but not in the NC/Staufen1-F135A coexpressing condition (Fig. 6E,G). Therefore, Staufen1, but not Staufen1-F135A, is capable of preventing NC-induced activation of PKR and eIF2α.
FIGURE 6.
NC induces PKR activation and eIF2α phosphorylation. (A) HeLa cells were transfected as indicated and 24 h later cell lysates were subjected to SDS-PAGE, immunoblotted, and probed to investigate eIF2α and PKR phosphorylation. (B) Densitometry quantification of P-eIF2α was determined by ImageJ analysis. Values presented in the graph are normalized against the total amount of eIF2α in the cell lysate and represent fold change with the RLuc-transfected cells being arbitrarily set to 1. Error bars represent the standard error of the mean from three independent experiments. Asterisks represent statistically significant difference between groups (one-way ANOVA; P < 0.01). (C) Densitometry quantification of P-PKR was determined by ImageJ analysis. Values presented in the graph are normalized against the total amount of PKR in the cell lysate and represent fold change with the RLuc-transfected cells being arbitrarily set to 1. Error bars represent the standard error of the mean from three independent experiments. Asterisks represent statistically significant difference between groups (one-way ANOVA; P < 0.05) (D) Cells were transfected as indicated and stained for RLuc (cyan) and P-eIF2α (red). Images shown are representative of >150 cells analyzed from three independent experiments. Scale bars represent 10 µm. (E) Quantification of the integrated density of p-eIF2α signal in cells from E from by ImageJ analysis. Each dot represents fluorescence intensity of a cell normalized to the mean fluorescence intensity of the mock transfected condition (arbitrarily set to 1). Error bars represent the standard error of the mean of cells from three independent experiments. Asterisks represent statistically significant difference between groups (one-way ANOVA; P < 0.0001). (F) Cells were transfected as indicated and stained for RLuc (cyan) and P-PKR (red). Images shown are representative of >150 cells analyzed from three independent experiments. Scale bars represent 10 µm. (G) Quantification of the integrated density of p-PKR signal in cells from F from by ImageJ analysis. Each dot represents fluorescence intensity of a cell normalized to the mean fluorescence intensity of the mock transfected condition (arbitrarily set to 1). Error bars represent the standard error of the mean of cells from three independent experiments. Asterisks represent statistically significant difference between groups (one-way ANOVA; P < 0.0001). (H) Cells were transfected as indicated and cell lysates were subjected to SDS-PAGE, immunoblotted, and probed to investigate eIF2α and PKR phosphorylation. (I) Densitometry quantification of P-eIF2α was determined by ImageJ analysis. Values presented in the graph are normalized against the total amount of eIF2α in the cell lysate and represent fold change with the RLuc-transfected cells being arbitrarily set to 1. Error bars represent the standard error of the mean from three independent experiments. Asterisks represent statistically significant difference between groups (one-way ANOVA; [*] P < 0.05, [***] P < 0.001).
The phosphorylation of eIF2α can be carried out by four kinases: PERK (PKR-like ER kinase), GCN2 (general control nonderepressible-2), HRI (heme-regulated inhibitor), and PKR (Donnelly et al. 2013). In order to ascertain that NC-mediated phosphorylation of eIF2α is via the activation of PKR and not another kinase, we knocked down PKR using an shRNA (shPKR) via lentiviral transduction and measured eIF2α phosphorylation. An shRNA with a scrambled sequence was used as a negative control (shNS). In the shNS condition, NC expression resulted in a significant increase in ratio of phosphor/total eIF2α (Fig. 6H,I). However, upon knockdown of PKR, no significant increase in eIF2α phosphorylation was observed in the NC-expressing cells (NC-RLuc) when compared to the mock treated cells in the same condition (RLuc-N1) (Fig. 6H,I). Thus, the NC-induced phosphorylation of eIF2α is dependent on the activation of PKR.
Staufen1 rescues the NC-mediated reduction of viral production
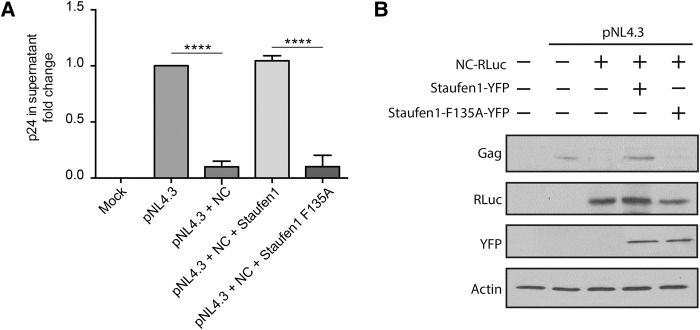
In order to determine whether the inhibition of global translation by NC can affect viral production, we transfected cells with either pNL4.3 alone, or cotransfected them with NC. The virus contained in the supernatants of these cells was then quantified by p24 ELISA. It was observed that an expression of NC led to a 10 (SD ± 0.3)-fold reduction of viral production when compared to the cells expressing pNL4.3 alone (Fig. 7A). To determine whether Staufen1 could rescue NC-induced inhibition of viral production, expression vectors encoding either Staufen1 or Staufen1-F135A were cotransfected with NC and pNL4.3. Staufen1 expression rescued viral production to levels comparable to pNL4.3 alone, whereas Staufen1-F135A was unable to do so (Fig. 7A). Cell lysates from the above conditions were analyzed by western blotting and decreased levels of Gag were observed in the NC-transfected cells when compared to pNL4.3 alone. Gag expression was rescued by Staufen1 coexpression, but not by F135A-Staufen1 coexpression (Fig. 7B). These results indicate that the decreased viral release is likely a result of the inhibition of mRNA translation thus resulting in reduced synthesis of Gag (Fig. 7B).
FIGURE 7.
NC-mediated reduction of viral production is rescued by Staufen1. (A) HIV-1 p24 in the supernatant of transfected HeLa cells was quantified via ELISA 48 h after transfection. Asterisks represent statistically significant difference between groups (one-way ANOVA; P < 0.001). (B) Cell lysates were subjected to SDS-PAGE, immunoblotted, and probed to investigate Gag production.
DISCUSSION
In this study, we have used NC as a tool to understand how HIV-1 modulates gene expression and have demonstrated a detrimental effect of NC expression on mRNA translation. Based on our results, we hypothesize that NC induces SG assembly by one of two ways. The first is linked to NC's molecular chaperone activity whereby it catalyzes the rearrangement of nucleic acids to more thermodynamically stable structures (Darlix et al. 1995; Cristofari and Darlix 2002; Levin et al. 2005). The interferon (IFN)-inducible PKR is a dsRNA sensor and is a key player in the innate antiviral immune response (Meurs et al. 1990; Garcia et al. 2006). Its activation leads to the phosphorylation of the eIF2α, thereby preventing translational initiation and inducing SG assembly (Sadler and Williams 2007). When NC is overexpressed it could aggregate cellular mRNAs (Stoylov et al. 1997; Le Cam et al. 1998; Mirambeau et al. 2006), thereby activating PKR (Fig. 6A,C). HIV-1 proteins like Tat have evolved countermeasures to block PKR activation by recruiting PKR Activator (PACT), adenosine deaminase acting on RNA (ADAR) 1 and TAR RNA binding protein (TRBP) (McMillan et al. 1995; Cai et al. 2000; Clerzius et al. 2013). However, in our experimental conditions NC was present in isolation, and therefore PKR activation could not be subverted. NC could then associate to SG components G3BP1 and TIAR1 (Fig. 5A,B), leading to SG assembly and the suppression of global host cell mRNA translation. The second mechanism of NC-induced SG assembly might also be a result of its nucleic acid binding property (Cruceanu et al. 2006). We observed an increase in the abundance of polysome-free mRNAs in NC-expressing cells (Fig. 3D,F). This suggests that NC is either preventing the attachment of the ribosomal subunits to the mRNA, probably due to steric hindrance as a result of its own binding to the mRNA; or NC is stalling the ribosomes due to NC's binding and aggregation of mRNA (Stoylov et al. 1997; Le Cam et al. 1998). Furthermore, the presence of the low complexity (LC) and intrinsically disordered (ID) regions in a protein can also promote SG assembly (Kedersha et al. 2013; Molliex et al. 2015). NC has been revealed to be a highly disordered protein (Xue et al. 2012) and this could contribute to its ability to induce SG assembly.
HIV-1 has developed strategies to subvert the host cellular stress response. In our previous work, we have shown that the capsid (CA) domain of Gag blocks SG assembly in an eIF2α phosphorylation-independent manner via an interaction with the eukaryotic elongation factor eEF2. This interaction is stabilized by a Gag–Cyclophilin A association and inhibits a later stage of SG assembly (Valiente-Echeverria et al. 2014). However, the NC-induced SGs are formed in part due to a dissociation or disruption of the attachment of the ribosomal subunits themselves, or an impediment to their translational initiation, steps upstream of eEF2 function. It is likely that for this reason, a coexpression of either CA or Gag with NC was unable to inhibit SG assembly (Fig. 2A,B).
Staufen1 is a host protein that has been reported to suppress SG assembly by binding the ribosomal subunits and stabilizing polysomes (Marion et al. 1999; Luo et al. 2002; Thomas et al. 2005, 2009). It also has been implicated in preventing the activation of PKR and the subsequent phosphorylation of eIF2α during hepatitis C virus infection (Dixit et al. 2016). As a known interacting partner of NC with previously characterized roles in the modulation of the stress response, we hypothesized that Staufen1 could block NC-induced PKR activation. Indeed, the coexpression of Staufen1 can prevent NC-induced activation of PKR and downstream phosphorylation of eIF2α (Fig. 6A–C). Staufen1 alone, unlike TRBP for instance, is not able to subvert PKR activity during HIV-1 infection (Ong et al. 2005; Abrahamyan et al. 2010; Clerzius et al. 2013). However, when in isolation or when bound to NC, the resulting suppression of PKR activation by Staufen1 is remarkable. The Staufen1-F135A can neither bind NC nor RNA. In this condition, NC is free to interact with cellular mRNAs and allows assembly of NC-induced SGs. Interestingly, the coexpression of the dsRBD3 binding mutant, F135A-Staufen1, with NC had little suppressive activity on PKR. This indicates that the efficient binding of NC to Staufen1 may be required to prevent PKR activation, or that the dsRBD3 and the ability to bind RNA is responsible for PKR down-regulation by Staufen1. Staufen1's ability to interact with RNA and stabilize polysomes by binding to ribosomes via its N-terminal domain (Thomas et al. 2009) may augment its ability to block NC-induced SG assembly. Staufen1 can prevent the dissociation of attached ribosomal subunits and facilitate mRNA translation, acting at a stage downstream from eIF2α phosphorylation. However, if this were the only method of Staufen1-mediated disruption of NC-SGs, then an increase in P-eIF2α would be observed in the NC/Staufen1 coexpressing cells. But our results show that Staufen1 can prevent eIF2α phosphorylation (Fig. 6A,B,D,E) and is therefore also acting upstream of polysome stabilization, probably by binding and sequestering NC. The F135A mutation in Staufen1 impairs RNA binding capability and this may hinder Staufen1's ability to stabilize polysomes, thereby exacerbating its inability to inhibit NC-induced SG-assembly. Staufen1 coexpression relieves the NC-induced global translation block as shown by polysome profile and puromycylation assays, resulting in enhanced virus production (Figs. 3E and 7A,B). Overall, NC induces the assembly of SGs by activating PKR and destabilizing polysomes. Staufen1 disrupts NC induced SG assembly by binding and sequestering NC and by binding to RNA and stabilizing polysomes. The model for Staufen1's blockade of NC-induced SG assembly is depicted in Figure 8.
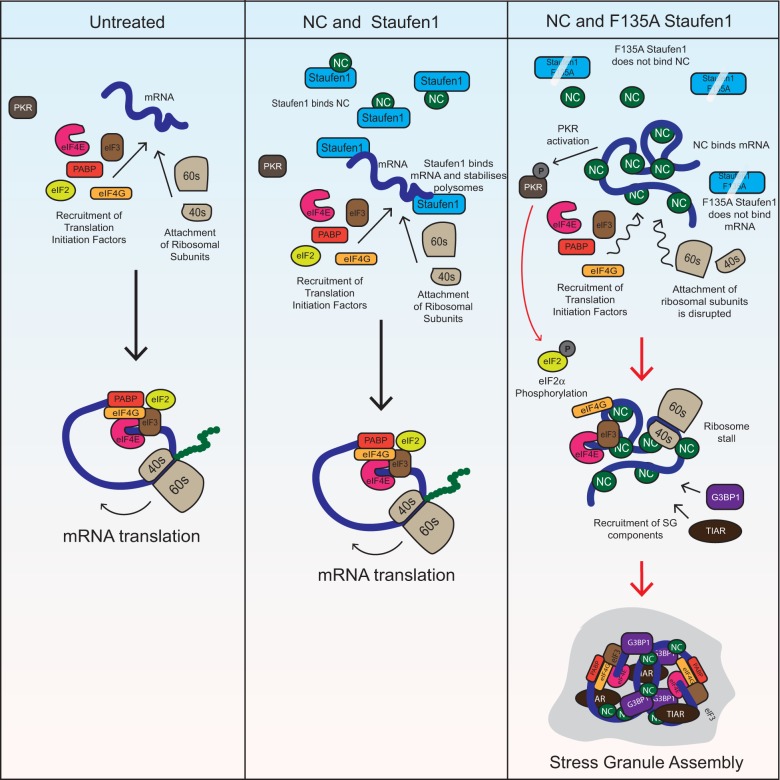
FIGURE 8.
Model of NC-induced SG assembly. Under untreated conditions, host cell translation progresses as normal. When NC is overexpressed, it binds cellular mRNAs, aggregates nucleic acids, and leads to PKR activation. NC also prevents ribosomal translocation, thereby leading to SG assembly. Staufen1 can bind and sequester NC as well as stabilize polysomes and disrupt NC-induced SG assembly; but not if it contains an F135A mutation by virtue of which it loses its ability to bind NC and RNA.
In our earlier work, we demonstrated that Staufen1 interacted with HIV-1 Gag precursor via the NC domain using a variety of in vitro and biophysical analyses. In this paper, we now show a direct association between NC and Staufen1 (Fig. 4D), a type of study that has largely been hampered by the solubility of recombinant Staufen1 proteins. Indeed, the full-length Staufen1 remains poorly soluble, but the data presented herein (Fig. 4D) indicate a rather selective association to the third dsRNA binding domain, as we have shown earlier (Chatel-Chaix et al. 2008). The results (Fig. 4D) also pronounce on an additional binding interaction between the dsRNA binding domain 4 and NC. The association of Gag to Staufen1 via the NC domain was shown to impact virus assembly (Chatel-Chaix et al. 2007, 2008), Gag and vRNA trafficking (Milev et al. 2010), vRNA encapsidation (Abrahamyan et al. 2010), and an antiviral stress response (Abrahamyan et al. 2010). These roles are likely to be coupled, such that the dsRNA binding protein Staufen1 likely functions by contacting the vRNA directly or as a component of a larger ribonucleoprotein as we and others have shown (Ajamian et al. 2008; Abrahamyan et al. 2010; Kula et al. 2011; Milev et al. 2012). A direct role has yet to be substantiated for Staufen1 in encapsidation (Mouland et al. 2000), but recent work in other viruses supports a role in this late step of virus assembly (Dixit et al. 2016).
During the late stages of the viral replication cycle, the detrimental effects of NC on host cell translation highlight the importance of timely Gag polyprotein processing. There is significant evidence that Gag polyprotein processing, and hence the generation of mature NC protein, takes place concomitantly or only shortly after budding (for review, see Sundquist and Krausslich 2012; Konvalinka et al. 2015). That is, under normal conditions of the viral life cycle during viral assembly, free NC is largely absent from the producer host cell. The premature precursor processing and the appearance of NC in the cytoplasm correlates with defects in virus assembly and production (Park and Morrow 1991), but also contributing to these parameters would be the marked decrease in mRNA translation, marked by the assembly of SGs. These observations are consistent with our previous work, where the presence of SGs in HIV-1 expressing cells decreased virus production and infectivity (Fig. 7A,B; Valiente-Echeverria et al. 2014), while more recently it was shown that G3BP1 can bind the HIV-1 vRNA in the cytoplasm of macrophages to inhibit viral replication (Cobos Jimenez et al. 2015). Furthermore, it has been demonstrated that the binding of the NC protein to the vRNA causes a rearrangement in vRNA secondary structure from the long distance interaction (LDI) to the branched structure with multiple hairpins (BMH) conformation, thus promoting dimer formation during virion assembly and reducing vRNA translation (Huthoff and Berkhout 2001).
Staufen1 was shown to suppress SG assembly during oxidative stress (Thomas et al. 2009), but it also assembles with Gag and vRNA to favor the assembly of another type of RNP, the Staufen1 HIV-1-dependent RNP (SHRNP) (Abrahamyan et al. 2010). SHRNPs are high molecular weight, detergent insoluble complexes containing Staufen1, among many other viral and cellular components (Mallardo et al. 2003; Chatel-Chaix et al. 2004; Milev et al. 2012; Tosar et al. 2012). Staufen1, likely in the context of SHRNPs, enhances Gag assembly and vRNA packaging (Abrahamyan et al. 2010; Milev et al. 2010), roles ascribed to the interaction of Staufen1 with the NC domain of Gag. Therefore, we speculate that the effects of Staufen1 on the rescue of NC-mediated translational arrest, viral assembly, and vRNA packaging are linked. This is supported by the recovery of viral production and Gag mRNA translation upon coexpression of Staufen1 following NC's inhibitory effects on host cell gene expression (Fig. 7A,B). Indeed, the link between translation and packaging has been explored in earlier work (Cimarelli and Luban 1999; Chamanian et al. 2013) and by studying whether translatable pools of vRNA were packageable or not (Butsch and Boris-Lawrie 2002; Poon et al. 2002).
This work sheds light on a novel function of NC on cellular mRNA translation and highlights how a tightly regulated balance of the HIV-1 proteins, NC and CA, act in concert with host proteins such as Staufen1 to modulate the host stress response to ensure viral gene expression. An elucidation of the molecular mechanisms of viral pathogenesis can identify novel targets for antiviral therapeutic interventions.
MATERIALS AND METHODS
Plasmids
The construction of pCMV-NC-RLuc, pCMV-NC-YFP, pCMV-NC-R7-YFP, pCMV-NC-C15S-YFP, pCMV-NC-C49S-YFP, pCMV- NC-C15S-C49S-YFP, CA-GFP pCMV-Staufen1-YFP, pCMV-Staufen1-F135A-YFP was described previously (Chatel-Chaix et al. 2004; 2007, 2008; Valiente-Echeverria et al. 2014). pcDNA3.1 was purchased from Invitrogen and pEGFP-C1 from Clontech. pGag-GFP was obtained from NIH AIDS Reference and Reagent Program. pSV-S4 was provided by Dr. Guy Lemay (Université de Montréal, Montréal, Québec) (Mabrouk and Lemay 1994).
Antibodies
A rabbit anti-Staufen1 antiserum generated to the full-length recombinant protein was produced and purified at the McGill University Cell Imaging and Analysis Network (Montréal, Québec, Canada). Hybridoma cell lines producing mouse anti-σ3 (4F2) have been described before (Virgin et al. 1991) and were a kind gift from Dr. Guy Lemay (Université de Montréal, Montréal, Québec). Anti-Staufen1 was used for western blotting at a dilution of 1:1000; rabbit or mouse anti-G3BP1 (Santa Cruz Biotechnology) was used for indirect immunofluorescence microscopy at a dilution of 1:1000 and for western blotting at a dilution of 1:10,000; goat anti-eIF3 (Abcam) was used for indirect immunofluorescence microscopy at a dilution of 1:500; goat anti-TIAR1 (Santa Cruz Biotechnology) was used for indirect immunofluorescence microscopy at a dilution of 1:500 and for western blotting at a dilution of 1:2000; mouse anti-PABP (Sigma-Aldrich) was used for indirect immunofluorescence microscopy at a dilution of 1:200; rabbit anti-RLuc (MBL) was used for indirect immunofluorescence microscopy at a dilution of 1:500 and for western blotting at a dilution of 1:1000; mouse anti RLuc (Abcam) was used for indirect immunofluorescence microscopy at a dilution of 1:500; mouse anti-σ3 (4F2) was used for indirect immunofluorescence microscopy at a dilution of 1:2; rabbit anti-phospho-eIF2α (Ser51) (Cinti et al. 2016) (Cell Signaling Technology) was used for indirect immunofluorescence microscopy at a dilution of 1:200 and for western blotting at a dilution of 1:1000; mouse anti-eIF2α (Cell Signaling Technology) was used for western blotting at a dilution of 1:1000; rabbit anti-P-PKR (Abcam) was used for indirect immunofluorescence at a concentration of 1:300 and for western blotting at a dilution of 1:1000; mouse anti-PKR 71-10 (Laurent et al. 1985) was used for western blotting at a concentration of 1:1000 and was provided by Dr. Anne Gatignol (McGill University); rabbit anti-GST (Sigma-Aldrich) was used for western blotting at a concentration of 1:2000; goat anti-NC, a kind gift from Dr. Robert Gorelick (National Cancer Institute, Frederick, MD, USA; ACVP #77, lot R196099), was used for western blotting at a dilution of 1:1000 (Wu et al. 2013); mouse anti-GFP (Sigma) was used for western blotting at a dilution of 1:10,000; mouse anti-actin (Abcam) was used for western blotting at a dilution of 1:10,000; and mouse anti-GAPDH (Abcam) was used for western blotting at a dilution of 1:5000. Horseradish peroxidase-conjugated secondary antibodies were purchased from Rockland Immunochemicals, while AlexaFluor secondary antibodies were from Life Technologies.
Cell culture and transfection conditions
HeLa cells, HEK293T cells and U2OS cells were maintained in DMEM (Life Technologies) supplemented with 1% penicillin/streptomycin and 10% fetal bovine serum (Hyclone). Cells were transfected with 1 µg of total DNA per 4 × 105 cells, unless indicated otherwise, using JetPrime (PolyPlus transfections) according to the manufacturer's instructions. If more than one plasmid was used to transfect cells, the amount of each plasmid used per transfection reaction was constant. Twenty-four hours after transfection, cells were fixed or lysed. For siRNA transfection, 20 nM of siRNA was used to transfect 150,000 cells using Lipofectamine 2000 (Invitrogen) reagent according to manufacturer's instructions. Cells were treated with 500 mM arsenite (Sigma-Aldrich) for 1 h and with 1 µM Emetine (Sigma-Aldrich) for 50 min (Cinti et al. 2017).
siRNAs
siRNA duplexes were purchased from QIAGEN-Xeragon. siNS is commercially available nonsilencing control duplex (QIAGENXeragon) and siG3BP1 is an siRNA targeting G3BP1 (SI00300265).
Viral transduction
psPAX2, pMD2.G, pLKO-shPKR#2 (TRCN0000196400) expression vector containing shRNA to PKR (target sequence GCTGAACTTCTTCATGTATGT) and a lentiviral control vector containing scrambled non-target shRNA used as a negative control were kind gifts from Dr. Marc Fabian (McGill University). A total of 2,000,000 HEK293T cells were plated in six-well plates 1 d prior to transfection. HEK293 were cotransfected with either scrambled shRNA (shNS) or shPKR expressing lentivirus, psPAX2, and pMD2.G. Supernatants were collected 48 h post-transfection, passed through a 0.45-µM nitrocellulose filter, supplemented with 5 µg/mL polybrene, and applied to HeLa cells at ∼40% confluency. Cells were selected with puromycin (10 µg/mL, Sigma-Aldrich) for 2 d, following which they were transfected with plasmids of interest.
Western blotting
Cells were collected after transfection, washed with DPBS (Corning), and lysed in ice-cold lysis buffer (100 mM NaCl, 10 mM Tris, pH 7.5, 1 mM EDTA, 0.5% Nonidet P-40, protease and phosphatase inhibitor cocktail [Roche]). Cell lysates were quantified by the Bradford assay (Bio-Rad) and 20 µg of lysates were denatured in Laemmli sample buffer and incubated for 5 min at 95°C. The proteins were separated by SDS-PAGE and transferred onto nitrocellulose membranes (Bio-Rad). Membranes were blocked with 5% nonfat milk in Tris-buffered saline pH 7.4 and 0.5% Tween 20 (TBST) and then incubated with primary antibodies. After washes with TBST, the membranes were incubated with horseradish peroxidase-conjugated secondary antibodies (Rockland Immunochemicals) and detected using Western Lightning Plus-ECL reagent (Perkin-Elmer). Signal intensity and densitometry analyses were conducted using ImageJ (NIH).
Immunofluorescence and imaging analyses
After transfection, cells were washed once in Dulbecco's phosphate-buffered saline (DPBS) (Thermo Fisher Scientific) and fixed with 4% paraformaldehyde for 20 min. Cells were then washed with DPBS, incubated in 0.1 M glycine for 10 min, washed with DPBS, incubated in 0.2% Triton X-100 for 5 min, and washed in DPBS. Primary antibodies were applied for 1 h at 37°C, and then washed for 10 min in DPBS followed by secondary antibodies for 1 h. Cells were washed for 20 min in DPBS before being mounted on glass slides using ProLong Gold Antifade Reagent with DAPI (Life Technologies). Negative isotype-matched antibody was used to control staining specificity. Poly(A) mRNAs were detected by in situ hybridization assay. Briefly, cells were fixed with 4% PFA, treated with 0.1 M glycine and permeabilized with 0.5% Triton-X, washed 2× with DPBS and hybridized with Cy5-conjugated oligo(dT)(40) probe (0.2 µM) overnight at 37°C. Subsequently, cells were further processed for immunofluorescence for other proteins of interest. Confocal laser scanning microscopy was performed using a Leica DM16000B microscope equipped with a WaveFX spinning disk confocal head (Quorum Technologies), and images were acquired with a Hamamatsu ImageEM EM-charge coupled device camera. Scanning was performed and digitized at a resolution of 1024× 1024 pixels. Filter sets and laser wavelengths were described earlier (Monette et al. 2011; Valiente-Echeverria et al. 2014). Image processing and analyses were performed by Imaris software (version 8.4.1 Bitplane/Andor) or by MetaXpress software (Molecular Devices). All imaging experiments were performed at least three times. The observed phenotypes were representative of n > 100 cells per condition in each experiment. SGs were defined as large G3BP1 or TIAR1 foci measuring >0.5 µm and a cell was deemed as SG positive if it exhibited at least three or more SGs (Gilks et al. 2004). For fluorescence intensity quantitation, the fluorescence intensity of each cell was determined using the ImageJ program (NIH) and then normalized to the mock transfected control.
Immunoprecipitation (IP) assays
HeLa cells were transfected with pCMV NC-YFP, wild-type and NC zinc fingers mutants or pCMV-GFP, U2OS cells stably expressing G3BP1-GFP were transfected with pCMV NC-RLuc and 24 h later cells were solubilized with NP-40 lysis buffer (50 mM Tris HCl pH 8.0, 150 mM NaCL, 0.5 mM EDTA, and 0.5% NP-40). For immunoprecipitation, 500 µg of protein lysates were incubated with 25 µL of GFP-beads (Life Technologies) for 1 h at room temperature. Beads were washed with NP40 lysis buffer three times before being eluted with 1× Laemmli sample buffer. Samples were resolved by SDS-PAGE and probed using antibodies against GFP, Staufen1, and TIAR1 by western blot analysis.
In situ protein–protein interaction assay (DuoLink)
HeLa cells were transfected with NC-RLuc + pEGFP-E1, pCMV NC-RLuc + pCMV Staufen1, or pCMV NC-RLuc + pCMV Staufen1-F135A-YFP and, 24 h later, processed for in situ proximity ligation assay (PLA) using the DUOLINK II In Situ kit (Duolink) following the manufacturer's instructions as previously described (Valiente-Echeverria et al. 2014; Le Sage et al. 2017). Primary antibodies were mouse anti-RLuc and rabbit anti-GFP, which were detected using the DuoLink II Detection Reagent Red, Duolink II PLA Probe Anti-Mouse MINUS, and DuoLink II PLA Probe Anti-Rabbit PLUS. The NC-RLuc + pEGFP-C1 condition was used to measure background PLA signals for the above antibody combination. Imaging was performed as described above. The Spots Tool on Imaris software was used to quantify the number of spots per cell (Valiente-Echeverria et al. 2014; Le Sage et al. 2017).
In vitro binding assay
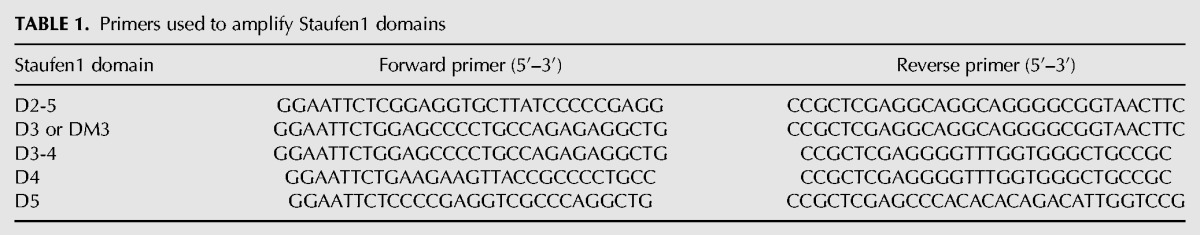
To generate Staufen1–glutathione S-transferases (GSTs) recombinant proteins, the hemagglutinin (HA)-tagged Staufen1 cDNA was PCR amplified from pcDNA3-RSV-Staufen1-HA (Wickham et al. 1999) with the primers described in Table 1. The resulting PCR products were digested with EcoRI and XhoI (New England Biolabs) and cloned in the pGEX-4t-2 vector and transformed into E. coli BL21 cells. The colonies that contained the plasmid + insert were grown in LB broth, 0.1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) was added to the bacterial culture to induce the expression of the GST fusion protein, and cells were solubilized with NP-40 lysis buffer (50 mM Tris–HCl pH 8.0, 150 mM NaCl, 0.5 mM EDTA, and 0.5% NP-40) 6 h after the addition of IPTG. These cell lysates were incubated in GST SpinTrap columns (GE Healthcare) for 30 min at room temperature. Columns were washed six times with TEN100 buffer (20 mM Tris pH 7.4, 0.1 mM EDTA, and 100 mM NaCl) to remove unbound proteins and subsequently incubated with 2 µg of recombinant NC protein for 2 h at 4°C. Captured complexes were washed three times with TEN100 buffer and elution was performed using elution buffer (50 mM Tris–HCl, pH8 and 10 mM glutathione). Samples were resolved by SDS-PAGE and probed using rabbit polyclonal antibodies against Staufen1 and NC by western blot analysis.
TABLE 1.
Primers used to amplify Staufen1 domains
Measurement of protein synthesis
Protein synthesis during NC expression was measured by the incorporation of puromycin into peptide chains (Schmidt et al. 2009; Goodman et al. 2011; Cinti et al. 2017). Briefly, pCMV NC-RLuc, pCMV NC-RLuc + pCMV Staufen1-YFP, and pCMV-RLuc transfected HeLa cells were incubated with 10 µg/mL puromycin (MilliporeSigma) for 10 min before cell lysis. Cell extracts were blotted with anti-Puromycin antibody (12D10, MilliporeSigma) and puromycin incorporation was assessed by summating the immunoblot intensity of all protein bands and subtracting background (Cinti et al. 2017).
Polysome profile analysis
Polysome profile analysis experiments were performed as described previously (Gandin et al. 2014; Valiente-Echeverria et al. 2014; Ajamian et al. 2015). Continuous sucrose density gradients (5%–50% w/v) were prepared in buffer containing 100 mM KCl, 5 mM MgCl2, 20 mM HEPES (pH 7.6), 100 µg/mL cycloheximide, 1× protease inhibitor, and 100 units/mL RNase Out (Invitrogen). Gradients were prepared in 5 mL polyallomer tubes by gently layering 2.2 mL of 5% sucrose in buffer over 2.2 mL of 50% sucrose in buffer. Tubes were then sealed and turned on their sides to generate a continuous gradient overnight at 4°C. HeLa cells were mock transfected or transfected with NC-RLuc, NC-RLuc + Staufen1 YFP, or NC-RLuc + Staufen1 F135A-YFP. Twenty-four hours post-transfection, cells were incubated with 100 µg/mL cycloheximide in growth media for 5 min and then washed twice with ice-cold PBS containing 100 µg/mL cycloheximide. Cells were scraped and collected by centrifugation al 200g for 5 min at 4°C. Supernatant was removed and the cells were resuspended and lysed in hypotonic Buffer (5 mM Tris–HCl [pH 7.5], 1.5 mM KCl, 2.5 mM MgCl2, 1× protease inhibitor, 200 units/mL RNase Out, 2 mM DTT, 150 µg/mL cycloheximide, 0.5% Triton X-100, and 0.5% SDS). Cell lysates were spun at 16,000g for 5 min at 4°C, and supernatants were transferred to new prechilled tubes. Five hundred microliters of sample lysate (containing equal quantities of material as normalized by spectrophotometry, λ = 260 nm) was layered gently on to the gradients and ultracentrifuged in a Beckman Ti55 swing rotor at 222,000g for 2 h at 4°C. Continuous OD254 readings for gradients were read from the bottom and fractions were collected using an ISCO fractionator (Teledyne, ISCO), as described in Gordon et al. (2013), Valiente-Echeverria et al. (2014), and Ajamian et al. (2015).
Quantification of virus in supernatants
Cells were transfected as described above and 48 h after transfection using 12 µg total DNA per 10 cm dish with each plasmid present in equal amounts. Culture supernatants were harvested and passed through a 0.2 µm filter (VWR) to remove cellular debris and centrifuged at 20,000 rpm for 1 h. The pellet containing the virus was resuspended in 200 µL RPMI and the levels of p24 were determined by enzyme-linked immunosorbent assay (ELISA) (PerkinElmer).
ACKNOWLEDGMENTS
We thank the late Mark Wainberg, Guy Lemay, Anne Gatignol, Marc Fabian, Tamiko Nishimura, Nancy Kedersha, and Robert Gorelick for generous provision of reagents; Maureen Oliviera and Ilinca Ibanescu for assay development; Hugues de Rocquigny for helpful discussions; and Fernando Valiente-Echeverria and Luc DesGroseillers for critical reading of the manuscript. This study was supported by Canadian Institutes of Health Research (CIHR) grants MOP-38111 and MOP-56974 (to A.J.M.) and by The Canadian HIV Cure Enterprise Team grant HIG-133050 (to A.J.M.) from the CIHR, in partnership with the Canadian Foundation for HIV-1/AIDS Research and the International AIDS Society.
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.064618.117.
REFERENCES
- Abrahamyan L, Chatel-Chaix L, Ajamian L, Milev M, Monette A, Clément JF, Song R, Lehmann M, DesGroseillers L, Laughrea M, et al. 2010. Novel Staufen1 ribonucleoproteins prevent formation of stress granules but favour encapsidation of HIV-1 genomic RNA. J Cell Sci 123: 369–383. [DOI] [PubMed] [Google Scholar]
- Ajamian L, Abrahamyan L, Milev M, Ivanov PV, Kulozik AE, Gehring NH, Mouland AJ. 2008. Unexpected roles for UPF1 in HIV-1 RNA metabolism and translation. RNA 14: 914–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ajamian L, Abel K, Rao S, Vyboh K, Garcia-de-Gracia F, Soto-Rifo R, Kulozik AE, Gehring NH, Mouland AJ. 2015. HIV-1 recruits UPF1 but excludes UPF2 to promote nucleocytoplasmic export of the genomic RNA. Biomolecules 5: 2808–2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson P, Kedersha N. 2009. RNA granules: post-transcriptional and epigenetic modulators of gene expression. Nat Rev Mol Cell Biol 10: 430–436. [DOI] [PubMed] [Google Scholar]
- Aulas A, Fay MM, Lyons SM, Achorn CA, Kedersha N, Anderson P, Ivanov P. 2017. Stress-specific differences in assembly and composition of stress granules and related foci. J Cell Sci 130: 927–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell NM, Lever AM. 2013. HIV Gag polyprotein: processing and early viral particle assembly. Trends Microbiol 21: 136–144. [DOI] [PubMed] [Google Scholar]
- Butsch M, Boris-Lawrie K. 2002. Destiny of unspliced retroviral RNA: ribosome and/or virion? J Virol 76: 3089–3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai R, Carpick B, Chun RF, Jeang KT, Williams BR. 2000. HIV-I TAT inhibits PKR activity by both RNA-dependent and RNA-independent mechanisms. Arch Biochem Biophys 373: 361–367. [DOI] [PubMed] [Google Scholar]
- Chamanian M, Purzycka KJ, Wille PT, Ha JS, McDonald D, Gao Y, Le Grice SF, Arts EJ. 2013. A cis-acting element in retroviral genomic RNA links Gag-Pol ribosomal frameshifting to selective viral RNA encapsidation. Cell Host Microbe 13: 181–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatel-Chaix L, Clement JF, Martel C, Beriault V, Gatignol A, DesGroseillers L, Mouland AJ. 2004. Identification of Staufen in the human immunodeficiency virus type 1 Gag ribonucleoprotein complex and a role in generating infectious viral particles. Mol Cell Biol 24: 2637–2648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatel-Chaix L, Abrahamyan L, Frechina C, Mouland AJ, DesGroseillers L. 2007. The host protein Staufen1 participates in human immunodeficiency virus type 1 assembly in live cells by influencing pr55Gag multimerization. J Virol 81: 6216–6230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatel-Chaix L, Boulay K, Mouland AJ, Desgroseillers L. 2008. The host protein Staufen1 interacts with the Pr55Gag zinc fingers and regulates HIV-1 assembly via its N-terminus. Retrovirology 5: 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimarelli A, Luban J. 1999. Translation elongation factor 1-α interacts specifically with the human immunodeficiency virus type 1 Gag polyprotein. J Virol 73: 5388–5401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cinti A, Le Sage V, Ghanem M, Mouland AJ. 2016. HIV-1 Gag blocks selenite-induced stress granule assembly by altering the mRNA cap-binding complex. MBio 7: e00329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cinti A, Le Sage V, Milev MP, Valiente-Echeverria F, Crossie C, Miron MJ, Pante N, Olivier M, Mouland AJ. 2017. HIV-1 enhances mTORC1 activity and repositions lysosomes to the periphery by co-opting Rag GTPases. Sci Rep 7: 5515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clerzius G, Shaw E, Daher A, Burugu S, Gelinas JF, Ear T, Sinck L, Routy JP, Mouland AJ, Patel RC, et al. 2013. The PKR activator, PACT, becomes a PKR inhibitor during HIV-1 replication. Retrovirology 10: 96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobos Jimenez V, Martinez FO, Booiman T, van Dort KA, van de Klundert MA, Gordon S, Geijtenbeek TB, Kootstra NA. 2015. G3BP1 restricts HIV-1 replication in macrophages and T-cells by sequestering viral RNA. Virology 486: 94–104. [DOI] [PubMed] [Google Scholar]
- Cristofari G, Darlix JL. 2002. The ubiquitous nature of RNA chaperone proteins. Prog Nucleic Acid Res Mol Biol 72: 223–268. [DOI] [PubMed] [Google Scholar]
- Cruceanu M, Urbaneja MA, Hixson CV, Johnson DG, Datta SA, Fivash MJ, Stephen AG, Fisher RJ, Gorelick RJ, Casas-Finet JR, et al. 2006. Nucleic acid binding and chaperone properties of HIV-1 Gag and nucleocapsid proteins. Nucleic Acids Res 34: 593–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang Y, Kedersha N, Low WK, Romo D, Gorospe M, Kaufman R, Anderson P, Liu JO. 2006. Eukaryotic initiation factor 2α-independent pathway of stress granule induction by the natural product pateamine A. J Biol Chem 281: 32870–32878. [DOI] [PubMed] [Google Scholar]
- Darlix JL, Lapadat-Tapolsky M, de Rocquigny H, Roques BP. 1995. First glimpses at structure-function relationships of the nucleocapsid protein of retroviruses. J Mol Biol 254: 523–537. [DOI] [PubMed] [Google Scholar]
- Darlix JL, de Rocquigny H, Mauffret O, Mely Y. 2014. Retrospective on the all-in-one retroviral nucleocapsid protein. Virus Res 193: 2–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixit U, Pandey AK, Mishra P, Sengupta A, Pandey VN. 2016. Staufen1 promotes HCV replication by inhibiting protein kinase R and transporting viral RNA to the site of translation and replication in the cells. Nucleic Acids Res 44: 5271–5287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnelly N, Gorman AM, Gupta S, Samali A. 2013. The eIF2α kinases: their structures and functions. Cell Mol Life Sci 70: 3493–3511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dugre-Brisson S, Elvira G, Boulay K, Chatel-Chaix L, Mouland AJ, DesGroseillers L. 2005. Interaction of Staufen1 with the 5′ end of mRNA facilitates translation of these RNAs. Nucleic Acids Res 33: 4797–4812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimura K, Sasaki AT, Anderson P. 2012. Selenite targets eIF4E-binding protein-1 to inhibit translation initiation and induce the assembly of non-canonical stress granules. Nucleic Acids Res 40: 8099–8110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandin V, Sikström K, Alain T, Morita M, McLaughlan S, Larsson O, Topisirovic I. 2014. Polysome fractionation and analysis of mammalian translatomes on a genome-wide scale. J Vis Exp 87: e51455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia MA, Gil J, Ventoso I, Guerra S, Domingo E, Rivas C, Esteban M. 2006. Impact of protein kinase PKR in cell biology: from antiviral to antiproliferative action. Microbiol Mol Biol Rev 70: 1032–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilks N, Kedersha N, Ayodele M, Shen L, Stoecklin G, Dember LM, Anderson P. 2004. Stress granule assembly is mediated by prion-like aggregation of TIA-1. Mol Biol Cell 15: 5383–5398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman CA, Mabrey DM, Frey JW, Miu MH, Schmidt EK, Pierre P, Hornberger TA. 2011. Novel insights into the regulation of skeletal muscle protein synthesis as revealed by a new nonradioactive in vivo technique. FASEB J 25: 1028–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon H, Ajamian L, Valiente-Echeverria F, Levesque K, Rigby WF, Mouland AJ. 2013. Depletion of hnRNP A2/B1 overrides the nuclear retention of the HIV-1 genomic RNA. RNA Biol 10: 1714–1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanke K, Hohn O, Liedgens L, Fiddeke K, Wamara J, Kurth R, Bannert N. 2013. Staufen-1 interacts with the human endogenous retrovirus family HERV-K(HML-2) rec and gag proteins and increases virion production. J Virol 87: 11019–11030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huthoff H, Berkhout B. 2001. Two alternating structures of the HIV-1 leader RNA. RNA 7: 143–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvius M, Paulsson J, Weibrecht I, Leuchowius KJ, Andersson AC, Wahlby C, Gullberg M, Botling J, Sjoblom T, Markova B, et al. 2007. In situ detection of phosphorylated platelet-derived growth factor receptor β using a generalized proximity ligation method. Mol Cell Proteomics 6: 1500–1509. [DOI] [PubMed] [Google Scholar]
- Kanai Y, Dohmae N, Hirokawa N. 2004. Kinesin transports RNA: isolation and characterization of an RNA-transporting granule. Neuron 43: 513–525. [DOI] [PubMed] [Google Scholar]
- Kedersha NL, Gupta M, Li W, Miller I, Anderson P. 1999. RNA-binding proteins TIA-1 and TIAR link the phosphorylation of eIF-2α to the assembly of mammalian stress granules. J Cell Biol 147: 1431–1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N, Ivanov P, Anderson P. 2013. Stress granules and cell signaling: more than just a passing phase? Trends Biochem Sci 38: 494–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N, Panas MD, Achorn CA, Lyons S, Tisdale S, Hickman T, Thomas M, Lieberman J, McInerney GM, Ivanov P, et al. 2016. G3BP-Caprin1-USP10 complexes mediate stress granule condensation and associate with 40S subunits. J Cell Biol 212: 845–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konvalinka J, Krausslich HG, Muller B. 2015. Retroviral proteases and their roles in virion maturation. Virology 479-480: 403–417. [DOI] [PubMed] [Google Scholar]
- Kula A, Guerra J, Knezevich A, Kleva D, Myers MP, Marcello A. 2011. Characterization of the HIV-1 RNA associated proteome identifies Matrin 3 as a nuclear cofactor of Rev function. Retrovirology 8: 60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurent AG, Krust B, Galabru J, Svab J, Hovanessian AG. 1985. Monoclonal antibodies to an interferon-induced Mr 68,000 protein and their use for the detection of double-stranded RNA-dependent protein kinase in human cells. Proc Natl Acad Sci 82: 4341–4345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Cam E, Coulaud D, Delain E, Petitjean P, Roques BP, Gerard D, Stoylova E, Vuilleumier C, Stoylov SP, Mely Y. 1998. Properties and growth mechanism of the ordered aggregation of a model RNA by the HIV-1 nucleocapsid protein: an electron microscopy investigation. Biopolymers 45: 217–229. [DOI] [PubMed] [Google Scholar]
- Le Sage V, Cinti A, McCarthy S, Amorim R, Rao S, Daino GL, Tramontano E, Branch DR, Mouland AJ. 2017. Ebola virus VP35 blocks stress granule assembly. Virology 502: 73–83. [DOI] [PubMed] [Google Scholar]
- Levin JG, Guo J, Rouzina I, Musier-Forsyth K. 2005. Nucleic acid chaperone activity of HIV-1 nucleocapsid protein: critical role in reverse transcription and molecular mechanism. Prog Nucleic Acid Res Mol Biol 80: 217–286. [DOI] [PubMed] [Google Scholar]
- Lloyd RE. 2012. How do viruses interact with stress-associated RNA granules? PLoS Pathog 8: e1002741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo M, Duchaine TF, DesGroseillers L. 2002. Molecular mapping of the determinants involved in human Staufen-ribosome association. Biochem J 365: 817–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mabrouk T, Lemay G. 1994. Mutations in a CCHC zinc-binding motif of the reovirus σ3 protein decrease its intracellular stability. J Virol 68: 5287–5290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallardo M, Deitinghoff A, Muller J, Goetze B, Macchi P, Peters C, Kiebler MA. 2003. Isolation and characterization of Staufen-containing ribonucleoprotein particles from rat brain. Proc Natl Acad Sci 100: 2100–2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marion RM, Fortes P, Beloso A, Dotti C, Ortin J. 1999. A human sequence homologue of Staufen is an RNA-binding protein that is associated with polysomes and localizes to the rough endoplasmic reticulum. Mol Cell Biol 19: 2212–2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMillan NA, Chun RF, Siderovski DP, Galabru J, Toone WM, Samuel CE, Mak TW, Hovanessian AG, Jeang KT, Williams BR. 1995. HIV-1 Tat directly interacts with the interferon-induced, double-stranded RNA-dependent kinase, PKR. Virology 213: 413–424. [DOI] [PubMed] [Google Scholar]
- Meurs E, Chong K, Galabru J, Thomas NS, Kerr IM, Williams BR, Hovanessian AG. 1990. Molecular cloning and characterization of the human double-stranded RNA-activated protein kinase induced by interferon. Cell 62: 379–390. [DOI] [PubMed] [Google Scholar]
- Milev MP, Brown CM, Mouland AJ. 2010. Live cell visualization of the interactions between HIV-1 Gag and the cellular RNA-binding protein Staufen1. Retrovirology 7: 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milev MP, Ravichandran M, Khan MF, Schriemer DC, Mouland AJ. 2012. Characterization of staufen1 ribonucleoproteins by mass spectrometry and biochemical analyses reveal the presence of diverse host proteins associated with human immunodeficiency virus type 1. Front Microbiol 3: 367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirambeau G, Lyonnais S, Coulaud D, Hameau L, Lafosse S, Jeusset J, Justome A, Delain E, Gorelick RJ, Le Cam E. 2006. Transmission electron microscopy reveals an optimal HIV-1 nucleocapsid aggregation with single-stranded nucleic acids and the mature HIV-1 nucleocapsid protein. J Mol Biol 364: 496–511. [DOI] [PubMed] [Google Scholar]
- Molliex A, Temirov J, Lee J, Coughlin M, Kanagaraj AP, Kim HJ, Mittag T, Taylor JP. 2015. Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell 163: 123–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monette A, Pante N, Mouland AJ. 2011. HIV-1 remodels the nuclear pore complex. J Cell Biol 193: 619–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouland AJ, Mercier J, Luo M, Bernier L, DesGroseillers L, Cohen EA. 2000. The double-stranded RNA-binding protein Staufen is incorporated in human immunodeficiency virus type 1: evidence for a role in genomic RNA encapsidation. J Virol 74: 5441–5451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong CL, Thorpe JC, Gorry PR, Bannwarth S, Jaworowski A, Howard JL, Chung S, Campbell S, Christensen HS, Clerzius G, et al. 2005. Low TRBP levels support an innate human immunodeficiency virus type 1 resistance in astrocytes by enhancing the PKR antiviral response. J Virol 79: 12763–12772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J, Morrow CD. 1991. Overexpression of the gag-pol precursor from human immunodeficiency virus type 1 proviral genomes results in efficient proteolytic processing in the absence of virion production. J Virol 65: 5111–5117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poblete-Duran N, Prades-Perez Y, Vera-Otarola J, Soto-Rifo R, Valiente-Echeverria F. 2016. Who regulates whom? An overview of RNA granules and viral infections. Viruses 8: E180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon DT, Chertova EN, Ott DE. 2002. Human immunodeficiency virus type 1 preferentially encapsidates genomic RNAs that encode Pr55Gag: functional linkage between translation and RNA packaging. Virology 293: 368–378. [DOI] [PubMed] [Google Scholar]
- Ramos A, Grunert S, Adams J, Micklem DR, Proctor MR, Freund S, Bycroft M, St Johnston D, Varani G. 2000. RNA recognition by a Staufen double-stranded RNA-binding domain. EMBO J 19: 997–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravel-Chapuis A, Klein Gunnewiek A, Belanger G, Crawford Parks TE, Cote J, Jasmin BJ. 2016. Staufen1 impairs stress granule formation in skeletal muscle cells from myotonic dystrophy type 1 patients. Mol Biol Cell 27: 1728–1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rein A. 2010. Nucleic acid chaperone activity of retroviral Gag proteins. RNA Biol 7: 700–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricci EP, Kucukural A, Cenik C, Mercier BC, Singh G, Heyer EE, Ashar-Patel A, Peng L, Moore MJ. 2014. Staufen1 senses overall transcript secondary structure to regulate translation. Nat Struct Mol Biol 21: 26–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadler AJ, Williams BR. 2007. Structure and function of the protein kinase R. Curr Top Microbiol Immunol 316: 253–292. [DOI] [PubMed] [Google Scholar]
- Schmidt EK, Clavarino G, Ceppi M, Pierre P. 2009. SUnSET, a nonradioactive method to monitor protein synthesis. Nat Methods 6: 275–277. [DOI] [PubMed] [Google Scholar]
- Soderberg O, Gullberg M, Jarvius M, Ridderstrale K, Leuchowius KJ, Jarvius J, Wester K, Hydbring P, Bahram F, Larsson LG, et al. 2006. Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nat Methods 3: 995–1000. [DOI] [PubMed] [Google Scholar]
- South TL, Blake PR, Sowder RC III, Arthur LO, Henderson LE, Summers MF. 1990. The nucleocapsid protein isolated from HIV-1 particles binds zinc and forms retroviral-type zinc fingers. Biochemistry 29: 7786–7789. [DOI] [PubMed] [Google Scholar]
- Stopak KS, Chiu YL, Kropp J, Grant RM, Greene WC. 2007. Distinct patterns of cytokine regulation of APOBEC3G expression and activity in primary lymphocytes, macrophages, and dendritic cells. J Biol Chem 282: 3539–3546. [DOI] [PubMed] [Google Scholar]
- Stoylov SP, Vuilleumier C, Stoylova E, De Rocquigny H, Roques BP, Gerard D, Mely Y. 1997. Ordered aggregation of ribonucleic acids by the human immunodeficiency virus type 1 nucleocapsid protein. Biopolymers 41: 301–312. [DOI] [PubMed] [Google Scholar]
- Sundquist WI, Krausslich HG. 2012. HIV-1 assembly, budding, and maturation. Cold Spring Harb Perspect Med 2: a006924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas MG, Martinez Tosar LJ, Loschi M, Pasquini JM, Correale J, Kindler S, Boccaccio GL. 2005. Staufen recruitment into stress granules does not affect early mRNA transport in oligodendrocytes. Mol Biol Cell 16: 405–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas MG, Martinez Tosar LJ, Desbats MA, Leishman CC, Boccaccio GL. 2009. Mammalian Staufen 1 is recruited to stress granules and impairs their assembly. J Cell Sci 122: 563–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas MG, Loschi M, Desbats MA, Boccaccio GL. 2011. RNA granules: the good, the bad and the ugly. Cell Signal 23: 324–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tosar LJ, Thomas MG, Baez MV, Ibanez I, Chernomoretz A, Boccaccio GL. 2012. Staufen: from embryo polarity to cellular stress and neurodegeneration. Front Biosci (Schol Ed) 4: 432–452. [DOI] [PubMed] [Google Scholar]
- Valiente-Echeverria F, Melnychuk L, Mouland AJ. 2012. Viral modulation of stress granules. Virus Res 169: 430–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valiente-Echeverria F, Melnychuk L, Vyboh K, Ajamian L, Gallouzi IE, Bernard N, Mouland AJ. 2014. eEF2 and Ras-GAP SH3 domain-binding protein (G3BP1) modulate stress granule assembly during HIV-1 infection. Nat Commun 5: 4819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virgin HW IV, Mann MA, Fields BN, Tyler KL. 1991. Monoclonal antibodies to reovirus reveal structure/function relationships between capsid proteins and genetics of susceptibility to antibody action. J Virol 65: 6772–6781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham L, Duchaine T, Luo M, Nabi IR, DesGroseillers L. 1999. Mammalian staufen is a double-stranded-RNA- and tubulin-binding protein which localizes to the rough endoplasmic reticulum. Mol Cell Biol 19: 2220–2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Mitra M, McCauley MJ, Thomas JA, Rouzina I, Musier-Forsyth K, Williams MC, Gorelick RJ. 2013. Aromatic residue mutations reveal direct correlation between HIV-1 nucleocapsid protein's nucleic acid chaperone activity and retroviral replication. Virus Res 171: 263–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue B, Mizianty MJ, Kurgan L, Uversky VN. 2012. Protein intrinsic disorder as a flexible armor and a weapon of HIV-1. Cell Mol Life Sci 69: 1211–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu KL, Lee SH, Lee ES, You JC. 2016. HIV-1 nucleocapsid protein localizes efficiently to the nucleus and nucleolus. Virology 492: 204–212. [DOI] [PubMed] [Google Scholar]