SUMMARY
Detailed studies of the broadly neutralizing antibodies (bNAbs) that underlie the best available examples of the humoral immune response to HIV are providing important information for the development of therapies and prophylaxis for HIV-1 infection. Here, we report a CD4-binding site (CD4bs) antibody, named N6, that potently neutralized 98% of HIV-1 isolates, including 16 of 20 that were resistant to other members of its class. N6 evolved a mode of recognition such that its binding was not impacted by the loss of individual contacts across the immunoglobulin heavy chain. In addition, structural analysis revealed that the orientation of N6 permitted it to avoid steric clashes with glycans, which is a common mechanism of resistance. Thus, an HIV-1-specific bNAb can achieve potent, near-pan neutralization of HIV-1, making it an attractive candidate for use in therapy and prophylaxis.
In Brief
Detailed studies of broadly neutralizing antibodies are providing important information for the development of therapies, prophylaxis, and vaccines for HIV-1. Huang et al. report a CD4-binding-site antibody that evolved to circumvent common mechanisms of resistance, resulting in an antibody that achieves potent, near-pan neutralization of HIV-1.
INTRODUCTION
Generation of a humoral immune response capable of recognizing widely varied strains is considered a critical goal for vaccines, or passive or vectored prophylaxis, against HIV-1 (reviewed in Burton and Mascola, 2015). Passive or vectored transfer of neutralizing monoclonal antibodies can completely protect Rhesus macaques or humanized mice against lentiviral infection (reviewed in Stephenson and Barouch, 2016). The only target of neutralizing antibodies, the HIV envelope glycoprotein (Env), is extraordinarily variable both within a patient and, to an even greater degree, within a population. However, it is possible for the human immune response to bind and neutralize widely varied strains of HIV-1 (Doria-Rose et al., 2010; Rusert et al., 2016; Sather et al., 2009; Simek et al., 2009). Over the past six years, the establishment of cohorts of patients with broadly cross-neutralizing sera, advances in sorting and B cell culture, and isolation of immunoglobulin G (IgG) genes has permitted an extensive deconvolution of these responses through the isolation of monoclonal antibodies from such patients. This body of work has shown that within Env, which exists as a trimer of heterodimers of glycoprotein 120 (gp120) and gp41 on the viral and cell surfaces, there are areas of sufficient conservation where targeting by individual broadly neutralizing antibodies (bNAbs) results in neutralization of highly unrelated isolates (reviewed in Burton and Mascola, 2015). Several of these antibodies are now in development for use in passive prophylaxis or immunotherapies.
Highly detailed studies of the structure and function of these antibodies, and the means by which they develop in some individuals, are providing important information for the design of HIV vaccines and immunotherapies (Wu et al., 2015; Zhou et al., 2013; reviewed in Haynes, 2015). Among the broadest of these antibodies are members of a group that bind the CD4-binding site (CD4bs). Many of these antibodies share remarkably similar characteristics with regard to their immunoglobulin heavy chain variable gene (VH) usage and mode of binding and for this reason have been designated “VRC01-class” antibodies, after the first identified member (Zhou et al., 2015). Some recent work using next-generation sequencing (NGS) of immunoglobulin transcripts has also provided insight into how these potent or broad antibodies might develop in an individual. Such detailed information regarding the co-evolution of virus and antibody provides a potential pathway for generating such antibodies through vaccination.
Here, we report a new monoclonal CD4bs antibody, named N6, that achieved both potency and remarkable breadth. The antibody evolved by a pathway that diverged from an early precursor to other CD4bs antibodies in the patient. We generated structural, functional, and NGS data that showed that the activity of N6 was mediated through novel interactions between multiple domains of the antibody and HIV Env. Unlike other CD4bs antibodies, N6 had a unique mode of recognition that mediated extraordinary breadth by tolerating the absence of individual CD4bs antibody contacts across the length of the heavy chain. In addition, it was able to avoid steric clashes between the light chain and the highly glycosylated V5 region of Env, which are the major mechanism of resistance to VRC01-class antibodies. Understanding the development of this antibody and its mechanism of neutralization provides important information regarding how highly diverse viruses might be targeted in immunotherapies and vaccines.
RESULTS
N6 Mediated Broad and Potent Neutralization through a Unique Mode of CD4bs Recognition
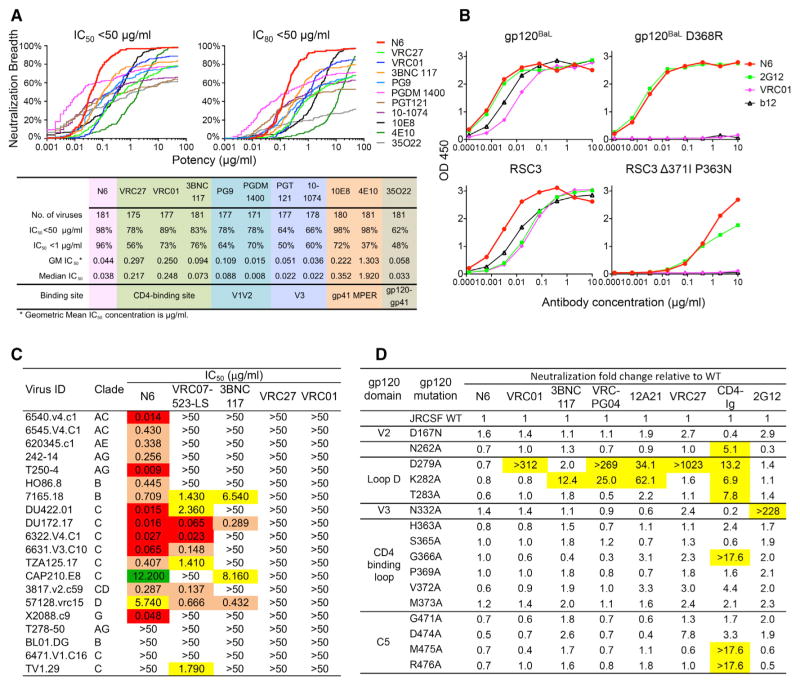
We sought to understand the specificities that might underlie the HIV-1-specific antibody response of a patient (Z258), whose serum was potent and broad (Figure S1A). The pattern of neutralization was similar to that of patient 45, from whom the well-known CD4bs antibody VRC01 was cloned. In addition, neutralization fingerprint analysis of this patient’s serum suggested that a CD4bs antibody was a dominant specificity (Figure S1B) (Georgiev et al., 2013). Consistent with this observation, a CD4bs antibody, named VRC27, was cloned from this patient via a probe sorting strategy. However, this antibody only neutralized 78% of isolates and was not potent (median 50% inhibitory concentration [IC50] = 0.217 μg/mL) (Figure 1A) and therefore did not appear to fully explain the breadth and potency of this patient’s serum. We therefore applied a technique to isolate monoclonal antibodies of interest from peripheral-blood B cells without prior knowledge of the target specificity (Huang et al., 2013). Peripheral-blood IgM−, IgA−, and IgD− memory B cells of patient Z258 were sorted and expanded. The supernatants of B cell microcultures were then screened for neutralizing activity, and IgG genes from wells with neutralizing activity were cloned and re-expressed. Three neutralizing antibodies were found and named N6, F8, and N17, among which the N6 antibody was the most potent and broad (Figures S1C and S1D).
Figure 1. N6 is a CD4bs Antibody with Extraordinary Neutralization Breadth and Potency.
(A) Neutralization potency and breadth of N6, in comparison to other bNAbs, against a 181-isolate Env-pseudovirus panel.
(B) ELISA binding of N6 to the indicated proteins.
(C) Neutralization profile of N6, in comparison to other VRC01-class antibodies, against 20 VRC01-resistant pseudoviruses. Values <0.1 μg/mL are highlighted in red, values between 0.1 and 1 μg/mL are highlighted in orange, values between 1 and 10 μg/mL are highlighted in yellow, and values between 10 and 50 μg/mL are highlighted in green.
(D) Neutralization of a panel of gp120JRCSF alanine mutants by N6, in comparison with other bNAbs. CD4-Ig and 2G12 were used as positive and negative controls respectively. The numbering of JRCSF mutants is based on the HXBC2 sequence. Neutralization fold change was calculated as follows: IC50 of the JRCSF mutant / IC50 of the JRCSF wild-type. Values >5 are highlighted in yellow. See also Figure S1 and Table S1.
Consistent with most other HIV-specific bNAbs, N6 was highly somatically mutated, in both heavy (31%) and light (25%) chains at the nucleotide level. The N6 antibody sequence also contained features that were consistent with a VRC01-class antibody (Zhou et al., 2013), such as a heavy chain derived from the VH1-2*02 germline gene and a light chain complementarity determining region 3 (CDR L3) composed of five amino acids (Figures S1D and S1E). The light chain was IGKV1-33 derived, similar to some other VRC01-class antibodies such as 12A21. Although they derived from the same patient, N6 was quite distinct from VRC27 and differed by 33% at the amino acid level of the heavy chain.
Using a 181-pseudovirus panel, we compared the neutralizing activity of N6 with that of VRC27, VRC01, and other bNAbs (Figure 1A and Table S1). N6 neutralized 98% of 181 pseudoviruses at an IC50 < 50 μg/mL. Although the breadth of many antibodies sharply declined at less than 1 μg/mL, at this level N6 still neutralized 96% of the tested isolates. The median IC50 was 0.038 μg/mL, among the most potent described thus far. In an extended panel of 173 clade C pseudoviruses, N6 also neutralized 98% of them at an IC50 < 50 μg/mL and a median IC50 of 0.066 μg/mL.
To understand the binding specificity of N6, we examined its ability to compete with other antibodies or bind gp120 mutants. Consistent with a CD4bs antibody, N6 competed with CD4Ig, and other CD4bs antibodies, for binding to gp120 (Figure S1F). We measured the binding affinity of N6 to three HIV gp120 proteins from strains 93TH057 (a strain sensitive to most VRC01-class antibodies), DU172 (a strain sensitive to some VRC01-class antibodies), and X2088 (which is only sensitive to N6). The N6 binding affinity to 93TH057 and DU172 was considerably higher than the binding affinity of VRC27 or VRC01 to these gp120 proteins (Figure S1G). Although VRC01 and VRC27 did not bind X2088, N6 bound it strongly. Unlike the other CD4bs antibodies tested by ELISA, N6 bound to CD4bs mutants: gp120 D368R and gp120 resurfaced stabilized core 3 (RSC3) Δ371I-P363N (Figure 1B). It is interesting to note that the RSC3 D368R mutant is commonly used to gate out non-CD4bs antibodies in probe-based sorting strategies. Therefore, N6 was most likely eliminated from analysis in prior efforts to recover CD4bs antibodies from this patient. In addition, N6 neutralized 16 of 20 VRC01-resistant isolates, and it did so potently (Figure 1C).
To more precisely map the epitope of N6 on HIV-1 gp120, we tested the binding of N6 to alanine scanning mutants in the context of monomeric gp120JRCSF. Members of the VRC01 antibody class are known to contact gp120 in three main areas: loop D, the CD4-binding loop (CD4 BLP), and the V5 region. Overall, we did not observe differences between N6 binding and other VRC01-class antibody binding to gp120JRCSF, as measured by ELISA (Figure S1H). However, binding to gp120 tested in an ELISA format might not accurately reflect the interaction between N6 and the intact functional trimer. To better assess this interaction, we used a panel of HIVJRCSF Env pseudo-virus alanine scanning mutants to examine the neutralization potency of N6 in comparison to those of other CD4bs antibodies (Figure 1D). In contrast to other CD4bs antibodies, single mutations in loop D, the CD4 BLP, or the V5 region of JRCSF showed no resistance to neutralization by N6 (Klein et al., 2014; Li et al., 2011; Lynch et al., 2015). These results, in addition to the binding to CD4bs mutants described above (Figure 1B), suggested that N6 had a unique mode of recognition of the CD4bs that permitted it to bind Envs not bound by other members of the VRC01 class.
Autoreactivity or polyreactivity is a property of several HIV-specific antibodies that could limit their use in therapies or prophylaxis. However, N6 did not bind Hep-2 epithelial cells (Figure S2A), nor did it bind cardiolipin (Figure S2B), a panel of autoantigens (Figure S2C), or 9,400 human proteins tested in a protein microarray (Figure S2D), suggesting that autoreactivity might not limit the potential use of N6 in HIV-1 prophylaxis, treatment, and prevention.
N6 Bound Envs Resistant to the VRC01 Class by Avoiding Steric Clashes
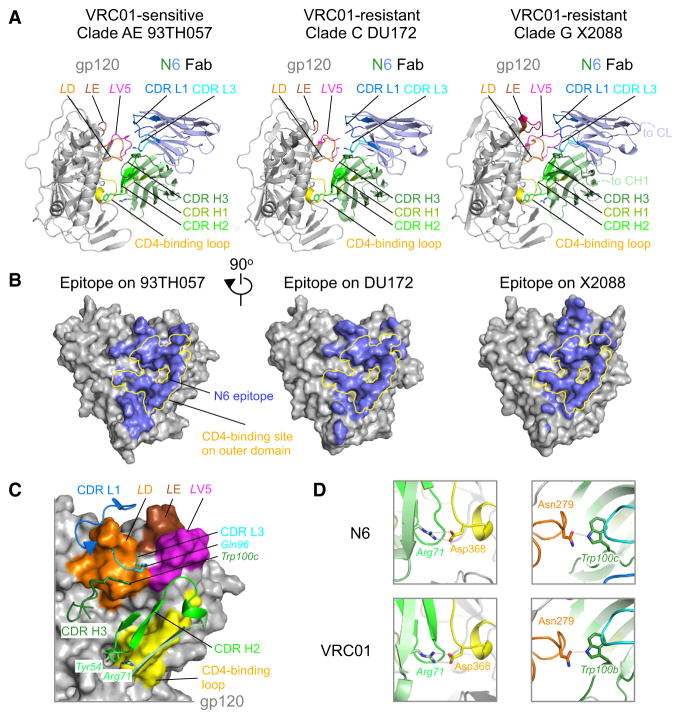
To define the mechanisms by which N6 might mediate such potency and breadth, we performed structural analyses of the antigen-binding fragment (Fab) of N6 in complex with HIV gp120 proteins from strains with varied sensitivities to VRC01-class antibodies (93TH057, DU172, and X2088, see above). The crystal structures of the complexes were determined to 2.40, 2.15, and 2.75 Å resolution, respectively (Table S2 and Figure 2A).
Figure 2. N6 Recognition of HIV-1 gp120 Has Features in Common with the VRC01 Class.
(A) Crystal structures of N6 in complex with gp120 proteins from three different HIV-1 clades that are sensitive or resistant to VRC01. The heavy chain is colored light green and the light chain is colored light blue.
(B) Epitopes of N6 on HIV-1 clade AE 93TH057, clade C DU172, and clade G X2088 are colored light blue on the gray gp120 surfaces. The initial binding site of CD4 on the outer domain of gp120 is highlighted in yellow.
(C) Interactions between HIV-1 gp120 and the complementarity determining regions of N6. HIV-1 gp120 is shown in surface representation in a 90°-view from (A). Key residues, such as heavy chain Tyr 54, Arg71 and Trp100c, light chain Gln96, are shown in sticks.
(D) Typical interactions between VRC01-class antibodies and HIV-1 gp120 are present with N6. Shown to the left are the conserved salt bridges between Arg71 and CD4-binding site Asp368, and to the right, the hydrogen bond between CDR H3 Trp and loop D Asn279. The color scheme in both (C) and (D) is the same as in (A). See also Table S4.
Analyses of the three structures revealed that N6 had several features in common with other members of the VRC01 class. The overall epitope of N6 was focused on the outer-domain CD4bs, with excursions to other areas (Figure 2B). All N6 CDRs except CDR H1 and CDR L2 were involved in gp120 recognition, with the CD4bs surface contacted by the N6 CDR H2 contributing ~50% of the gp120-binding surface (Figure 2C and Table S3). Several VRC01-class antibodies contain a large hydrophobic residue at heavy chain position 54 (54HC) (from this point forward, for clarity, residues are displayed with a subscript defining the molecule), which mimics the interaction of Phe43CD4 with gp120. This feature is found in antibodies such as VRC27, also isolated from patient Z258. Similarly, N6 had a pocket-filling Tyr54HC (Figure 2C). In addition, salt bridges between Arg71HC and Asp368gp120 were conserved in N6-gp120 complexes (Figure 2D and Table S4). Although the N6 CDR H3 had a different conformation from that of VRC01, the CDR H3 Trp100c (Figure S1D) was at a similar position to VRC01 Trp100b, which interacts with loop D Asp279gp120 (Figure 2D). Similarly to some other VRC01-class antibodies, N6 also contained the flexible Gly-x-Gly motif (residues 28–30) within the CDR L1 (Figure S1D), which permitted it to avoid steric clashes with the loop D glycan on Asn276.
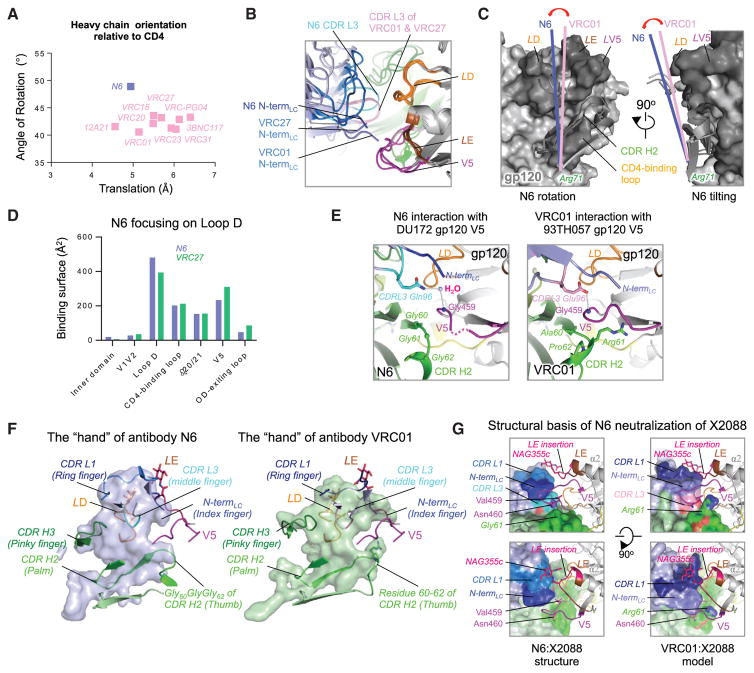
To further understand those features that distinguished N6 and enabled its breadth and potency, we aligned gp120 components from co-crystal structures to compare the binding modes of N6, CD4, and other VRC01-class antibodies. Relative to CD4, the binding angle of N6 was altered 5–8 degrees as compared to other VRC01-class antibodies (Figure 3A), and the translation distance was about 0.5 Å smaller than the average translation distance of other VRC01-class antibodies. Consequently, the N6 light chain was also rotated compared to VRC01 and VRC27 (Figure 3B), as indicated by the ~2.3 Å shift (Cα-Cα distance) of CDR L3 Gln96N6 from the position of Glu96VRC01. The axis of N6 was rotated about 10 degrees toward the conserved loop D and tilted ~6 degrees away from the gp120 surface compared to the axis of VRC01 (Figure 3C and Figure S3A). This increased the N6 binding surface on loop D by 20% and decreased that on variable loop V5 and the outer-domain-exiting loop by 25% (Figure 3D). The combination of rotation and tilting, pivoting around Arg71HC, caused the N6 light-chain N terminus and CDR L3 to move away from the highly variable loop V5 (Figure S3A). As a result, the conserved hydrogen bond between CDR L3 Glu/Gln and Gly459gp120 became a water-mediated hydrogen bond in the N6 structure (Figure 3E). The N terminus of the light chain and CDR H2 residues 60–62 of VRC01-class antibodies can be visualized as an index finger and thumb of a hand that grabs the gp120 variable loop V5 (Figure 3F). Changes in gp120 V5 that cause steric clashes with an antibody CDR H2, light-chain N terminus, or CDR L3 are well-described mechanisms of HIV-1 resistance to VRC01-class antibodies (Lynch et al., 2015). The N6 CDR H2 evolved to contain a Gly60GlyGly62 that is not present in any other previously isolated CD4bs antibodies (Figure 3F) to the best of our knowledge. This complete loss of side chains in CDR H2 residues 60–62, in addition to the rotation- and tilt-mediated retreat of the light-chain N terminus, shortened the thumb and index finger that permits N6 to accommodate variations at the gp120 V5 loop (Figure S3B). It is important to note that the unique orientation of the N6 light chain was not due to special features of the light chain, or heavy and light chain interface, given these overlapped with those of VRC27 when structurally aligned by antibody sequence (Figure S3B). Rather, it appears that the binding mode or orientation of the N6 heavy chain permitted this rotation of the light chain. Overall, N6 evolved a series of unique structural solutions to focus more binding surface on conserved loop D and tolerate changes in variable loop V5 that result in steric clashes with other VRC01-class antibodies.
Figure 3. N6 Avoids Steric Clashes Common to Other Members of the VRC01 Class.
(A) Antibody orientation relative to CD4 when structurally aligned on gp120. N6 is rotated more relative to gp120-bound CD4, compared to other VRC01-class antibodies.
(B) Unique position of N6 CDR L3 among the VRC01 class. Superposition of structures of gp120-VRC01-class antibodies indicated a unique position of N6 CDR L3. The color scheme is the same as in Figure 2A.
(C) When aligned on gp120, the binding axis of N6, defined as the line connecting Cα atoms of heavy chain Arg71 and light chain Glu/Gln96, is rotated toward loop D and tilted away from variable loop V5, as compared to other VRC01-class antibodies.
(D) Greater interaction of N6 with HIV-1 loop D, as compared to VRC27. When compared to VRC27 isolated from the same patient, N6 binding surface area on gp120 loop D increased ~20% and that on V5 and outer-domain-exiting loop decreased ~25% due to the rotation and tilting of the antibody.
(E) N6 interaction with gp120 V5. Conserved hydrogen bond between Glu/Gln96LC and Gly459gp120 becomes a water-mediated bond in N6. The CDR H2 Gly60GlyGly62 and the rotated light chain N terminus make more room on either to accommodate variations in loop V5.
(F) Comparison of the N6 paratope or that of VRC01 with morphology of a hand. The light chain N terminus and bulky side chains of CDR H2 residues 60–62 acts like an index finger and thumb grabbing HIV-1 variable loop V5. The rotation and tilting of light chain as well as the Gly60GlyGly62 mutation in the CDR H2 shortened the index finger and removed the thumb, leaving more space to accommodate variations in V5.
(G) Structural basis for N6 neutralization of clade G strain X2088. Rotation and tilting of N6 resolved potential clashes with loop E insertion, and the retreated light chain N terminus and CDR H2 Gly60GlyGly62 made space to accommodate bulky residues and glycosylation sites at base of V5. When modeled in VRC01, CDR L1 had a severe clash with the loop E insertion, and its light chain N terminus and CDR H2 Arg61 created a narrow path that could not accommodate the X2088 V5. See also Figure S3.
HIV-1 clade G strain X2088 is one of the viruses that is resistant to almost all CD4bs antibodies isolated so far. However, N6 was able to potently neutralize X2088 (IC50 = 0.048 μg/mL). Sequence analysis indicated that X2088 had a long V5 loop. In addition, although most easy-to-neutralize isolates have glycines at the base of V5 at positions 458 and 459, X2088 had a Val459 followed by a potential glycosylation site at Asn460X2088. Furthermore, there was a five-residue insertion in loop E, with a potential glycosylation site. The co-crystal structure of N6-X2088 gp120 revealed that N6 had developed multiple ways to accommodate these recognition constraints. The retreat of the N6 light-chain N terminus and water-mediated hydrogen bond between CDR L3 Gln96N6 and the main chain N of residue 459gp120 cleared space for these different features at the base of V5 (Figure 3G). Even though we did not observe a glycan attached to Asn460X2088, the N6 CDR H2 Gly60GlyGly62, toward which Asn460X2088 protrudes, would not clash with a glycan at this position should one occur (Figures 3E and 3G). The structural analysis showed that the loop E insertion added one full turn at the end of the α2 helix and an N-linked glycan in the middle that protruded directly toward the N6 CDR L1 (Figure 3G, left panels). However, the rotation and tilting of the N6 light chain moved its CDR L1 away to accommodate these features (Figure 3G, left panel). When VRC01 in its gp120-bound orientation was modeled into the N6-X2088 complex, the CDR L1 of VRC01 had a severe steric clash with this loop E insertion and glycan (Figure 3G, right panels, and Figure S3C). Moreover, the VRC01 light-chain N terminus and bulky CDR H2 Arg61 created a narrow path that could not accommodate X2088 (Figure 3F and 3G, right panels). Taken together, the evolved features of N6, such as the Gly60GlyGly62 motif in CDR H2 and the unique rotation and tilting of the light chain, combined to circumvent the mechanisms of resistance of X2088.
Epitope Mapping Reveals that N6 Tolerates Mutations in Known CD4bs Antibody Contacts
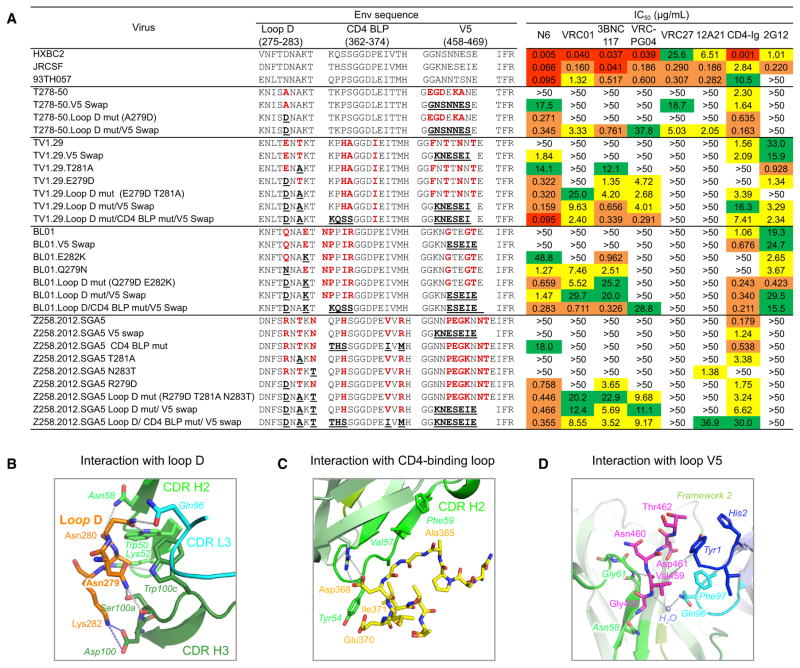
To better understand the relative role of contacts defined in the crystal structure, we analyzed the viral sequences with resistance to neutralization by N6. In prior work, resistance to VRC01-class antibodies has typically been mediated by mutations in known contact areas in loop D, the CD4 BLP, or V5 of gp120 (Lynch et al., 2015). Of 181 tested viruses, only four were highly resistant to N6, with an IC50 > 50 μg/mL (Figure S4A). Consistent with some prior observations (Doria-Rose et al., 2014; Lynch et al., 2015; Moore et al., 2012; Richman et al., 2003; Wu et al., 2012), pseudoviruses expressing autologous Envs were also resistant to N6-mediated neutralization (Table S5 and Figure S4A). Each of these viruses had mutations in loop D, the CD4 BLP, and the V5 region relative to the reference sequences (Figure 4A, Figure S4A, and Table S4). To determine the relative contributions of these mutations to resistance to N6, we reverse mutated resistant pseudo-viruses to a sensitive viral sequence, HIVJRCSF (Figure 4A). Only when sequences from HIVJRCSF were introduced into loop D did all the pseudoviruses become highly sensitive to neutralization by N6. In contrast, reverse mutations in loop D, the CD4 BLP, and the V5 region were required for full sensitivity to other CD4bs antibodies. Similarly, when tested by ELISA, reverse mutations at residues in loop D resulted in strong N6-mediated gp120 binding (Figure S4B), while reverse mutations in loop D, the CD4 BLP, and the V5 region were required for binding of the other CD4bs antibodies. Consistent with this result, substitution of the JRCSF loop D sequence by that of Z258.2012.SGA5 dramatically decreased the neutralization sensitivity to N6 by 11-fold (Figure S4C, right panel), whereas CD4 BLP and V5 swaps showed little or no significant change. Reverse mutations of individual positions revealed that only when the conserved residue Asp from HIVJRCSF was introduced into position 279 did most of the pseudoviruses become sensitive to neutralization by N6 (Figure 4A). The three N6-resistant pseudoviruses, T278-50, TV1.29, and BL01, and the autologous virus each have a mutation in position 279. The only exception was 6471.V1.C16, which has mutations at multiple positions within loop D, including the sequon predicted to add a glycan at Asn278 (Figure S4A). It is likely that resistance to N6 is mediated by the residues at position 279 that disrupt the contact with the CDR H3 Trp100c (Figure 4B). The mutagenesis suggested that resistance to N6 neutralization required mutations in loop D. However, unlike other CD4bs antibodies, N6 tolerated mutations in the CD4 BLP and V5 region (Figure 4B), including those that might cause a steric clash with the CDR H2 and CDR L3 of other VRC01-class antibodies.
Figure 4. N6 Tolerates Mutations in the CD4 BLP and V5.
(A) Neutralization of N6 against three N6-resistant and the Z258 autologous pseudoviruses. Neutralization of these pseudoviruses with reverse mutations in loop D, CD4 BLP, and V5 region are also shown. Sequence variations of gp120, as compared to reference sequences, are listed in bold red. Reverse mutations are highlighted in bold and underlined.
(B) Details of the structural interaction between N6 and HIV-1 gp120 loop D.
(C) Details of the structural interaction between N6 and HIV-1 gp120 CD4-binding loop C.
(D) Details of the structural interaction between N6 and HIV-1 gp120 V5.
N6 components are shown in ribbon with key residues highlighted in sticks. HIV-1 loop D, CD4-binding loop, and V5 are shown in sticks. Color scheme is the same as Figure 2A. See also Figure S4 and Table S5.
Breadth and Potency is Attributable to Multiple Elements within the N6 Heavy and Light Chains
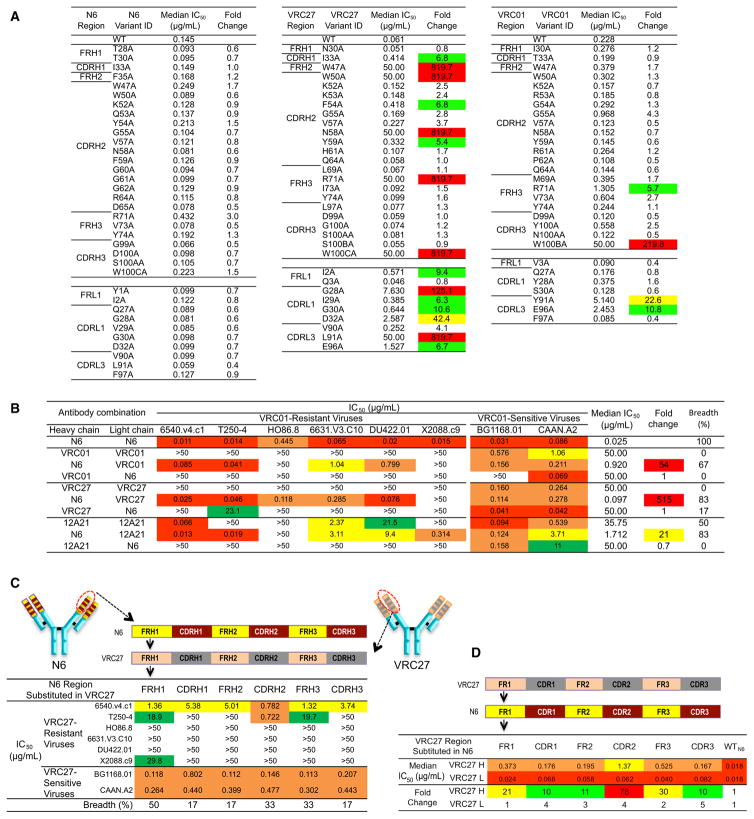
To understand the paratope of N6, we designed a panel of N6 alanine scanning mutants to examine the impact on the neutralization potency of N6 against six VRC01-sensitive pseudoviruses. Several changes made a greater than 5-fold decrease in average neutralization potency for VRC01 and VRC27 (Figure 5A and Table S6). The Trp100bVRC01 and Trp100cVRC27 alanine mutations within the CDR H3 had large effects presumably due to disruptions of interactions with Asp279gp120. In addition, scattered changes across the heavy and light chains also had large effects (Figure 5A and Table S6). However, no such effect was observed in alanine point mutations of N6. Thus, N6 was able to tolerate single mutations across the length of the heavy and light chains presumably because there was sufficient energy spread across the remaining contacts to mediate binding.
Figure 5. Features across the Length of the Heavy Chain Contribute to the Breadth and Potency of N6.
(A) Alanine scanning of N6, VRC01, and VRC27. Residues with a high buried surface area were selected for alanine scanning. Each alanine mutant was tested for neutralization with six VRC01-sensitive viruses. See Table S6. Neutralization fold change was calculated as follows: IC50 of antibody mutant / IC50 of antibody wild-type. Residues that resulted in fold change values >50 are highlighted in red, values between 20 and 50 are highlighted in yellow, and values between 5 and 20 are highlighted in green.
(B) Neutralization of cross-complemented antibodies, including the heavy and light chains of the N6, VRC01, VRC27, and 12A21 antibodies. IC50 values <0.1 μg/mL are highlighted in red, values between 0.1 and 1 μg/mL are highlighted in orange, values between 1 and 10 μg/mL are highlighted in yellow, and values between 10 and 50 μg/mL are highlighted in green. Neutralization fold change was calculated as follows: IC50 of original antibody / IC50 of the antibody combination. Median IC50 is calculated based on VRC01-resistant viruses only. A value of 50 was assigned to the resistant viruses with an IC50 >50. Breadth was calculated as the number of viruses sensitive to the combination antibody / total number of tested VRC01-resistant viruses.
(C) Neutralization of six VRC27-resistant and two VRC27-sensitive viruses by VRC27 mutants with substitutions of N6 heavy chain framework (FR) or CDRs.
(D) Neutralization IC50 of six VRC27-resistant viruses by N6 mutants with substitutions of VRC27 FR or CDRs. Neutralization fold change was calculated as follows: IC50 of N6 mutant / IC50 of N6. See also Figure S5 and Tables S6 and S7.
We also examined the neutralization potency of N6 alanine scanning mutants against six VRC01-resistant viruses, which contain mutations in loop D, the CD4 BLP, and the V5 loop. Mutation of Ile33, Trp47, Trp50, Arg71, and Trp 100c of the N6 heavy chain and Ile91 of the N6 light chain to alanine markedly diminished neutralization potency against these VRC01-resistant viruses (Table S7). However, contacts at these residues do not fully explain the activity of N6 against such viruses given that these residues at these positions are the same as those of VRC27.
Next, we sought to determine the regions of the N6 heavy or light chains that played an essential role in its neutralization breadth and potency. As shown in Figure 5B, the combination of N6 heavy chain and VRC01, VRC27, or 12A21 light chain increased the antibody neutralizing potency and breadth relative to VRC01, VRC27, or 12A21, respectively. However, the combination of N6 light chain and the VRC01, VRC27, or 12A21 heavy chain showed no change in breadth. This result is potentially consistent with the structural data that suggested that the ability of N6 to bind VRC01-resistant viruses, although permitted by the light chain orientation, is primarily mediated by the conformation and orientation adopted by the heavy chain.
To further understand the domains of N6 responsible for its breadth and potency, we also substituted portions of N6 into other antibodies of the VRC01-class. Substitution of the key areas of N6 interaction with gp120 into VRC01 did not increase its breadth or potency (Figure S5). Introduction of each of the N6 framework regions (FRs) and CDRs into VRC27 also did not have a dramatic effect (Figure 5C). Overall, we did not observe a large increase in activity among VRC01-class antibodies by substitution of sequence elements from N6.
We then examined whether the activity of N6 against resistant viruses was diminished by mutating the FR and CDRs of N6 to those of other members of the VRC01-class (Figure 5D and Figure S5). Substitution of Tyr54N6 with Gly54VRC01 and Gly60GlyGly62N6 with corresponding residues of VRC01 each caused large decreases in neutralization. However, the largest decrease in the activity against these viruses was caused by the substitutions of the N6 CDRs and FRs with those of VRC01 or VRC27 (Figure 5D and Figure S5). This was not due to a large disruption of function by the substitutions given that the neutralization of sensitive isolates was unchanged. Substitutions of N6 CDR L1 and FRL3 with those of VRC01 also dramatically decreased the antibody neutralizing activities. These results suggest that each of the N6 heavy chain CDRs and FRs contributed to the potency of N6 against VRC01-resistant viruses.
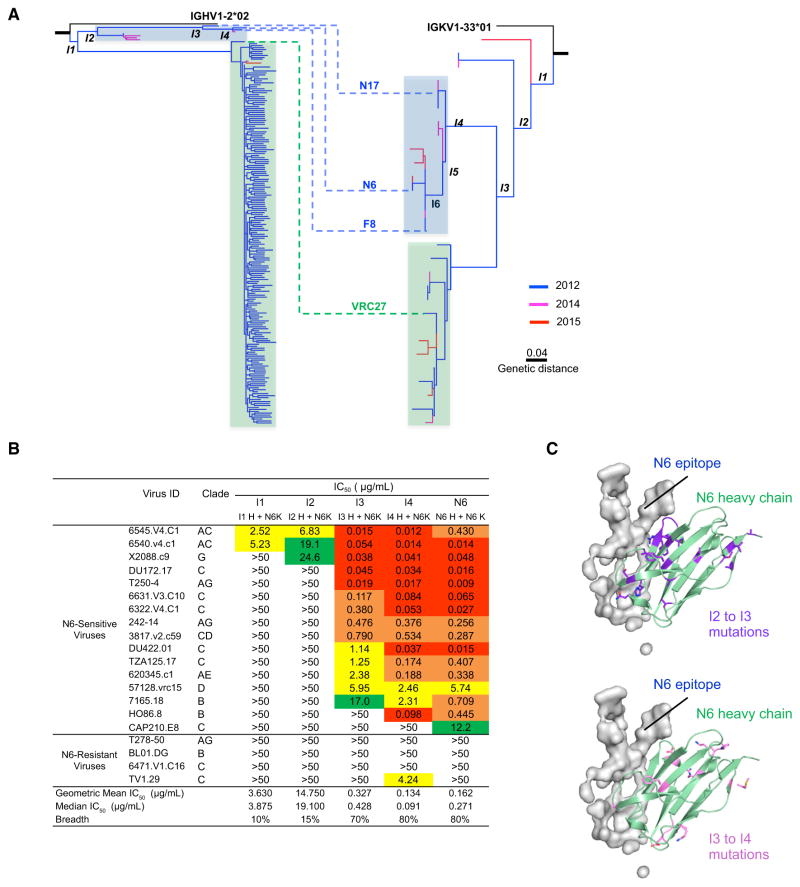
N6 Evolved from an Early Intermediate to Avoid Steric Clashes
To understand how N6 developed such extraordinary potency and breadth, we performed NGS of peripheral-blood memory B cell antigen receptors (BCRs) at three time points (2012, 2014, and 2015). The curated transcripts of both heavy and light chains showed high levels of somatic hypermutation (>20%) (Figure S6A). Nonetheless, heavy chain transcripts showing less somatic mutation (~23%) than either N6 or VRC27 were observed at the 2012 and 2014 time points. More lineage-related transcripts were observed from the 2012 time point than from 2014 or 2015, most likely because more B cells were used for the NGS experiment. N6-like light chain transcripts were observed at all three time points (Figure S6A), but no heavy chain transcripts with high similarity to N6 were observed in the 2012 or 2015 data. Overall, the vast majority of curated heavy chain sequences are closely related to VRC27 in the phylogenetic tree (Figure 6A). However, it should be noted that it is possible that some of the apparent diversity in the VRC27 branch might have been artificially generated by low sequence quality at the 3′ end of R2 and in the overlap between forward and reverse MiSeq reads. Despite this issue, the overall phylogenetic structure and inferred intermediates were robust. The genetic distance between the N6 and VRC27 clades and between early and matured members of the lineage was greater than 0.1. Becuase the inferred intermediates only contain mutations shared by many transcripts and the chance of two transcripts having identical sequencing errors is very low, the inferred intermediates should not be affected by sequencing errors.
Figure 6. N6 Evolved from an Early Intermediate to Circumvent Mechanisms of Resistance to the VRC01 Class.
(A) Paired phylogenetic tree of the N6 lineage. The curated heavy chain transcripts of the lineage-related members were identified using the sequence identity in CDR H3 to that of N6, VRC27, F8, or N17. The curated light chain transcripts of the lineage-related members were identified from next-generation sequencing transcripts originated from IGKV1-33*01 and contained the five-amino-acid CDR L3 signature of the VRC01-class antibodies.
(B) Neutralization by N6 intermediate antibodies against 20 VRC01-resistant pseudoviruses.
(C) Mapping of I2 to I3 and I3 to I4 mutations onto the N6 heavy chain. It is of note that most I2 to I3 mutations are close to the antibody:antigen interface, whereas the I3 to I4 mutations are distant from the interface. See also Figure S6 and Table S8.
To investigate the phylogenetic structure of the lineage, a maximum likelihood tree was constructed for the heavy chain and light chain (Figure 6A). The phylogenetic trees consistently showed that N6 and VRC27 formed two highly divergent groups. Both groups have similar heavy chain and light chain junctions and share 15 and 14 mutations in the heavy and light chains, respectively (Figure 6A and Figure S6B), confirming that they originated from a common precursor. The phylogenetic tree also showed that the two lineages diverged from a very early intermediate.
We then expressed the intermediates I1–I4 to determine the stage at which the N6 lineage gained its breadth and potency. We observed that a large increase in neutralizing activity against VRC01-resistant viruses occurred between I2 and I3 (Figure 6B). When mapped onto the N6 structure, most of the I2 to I3 mutations, which facilitated the gain of breadth, were at the N6 paratope that interacted directly with HIV gp120 (Figure 6C). In contrast, the I3 to I4 mutations, most likely accounting for the increases in potency, were located at non-paratope sites (Figure 6C).
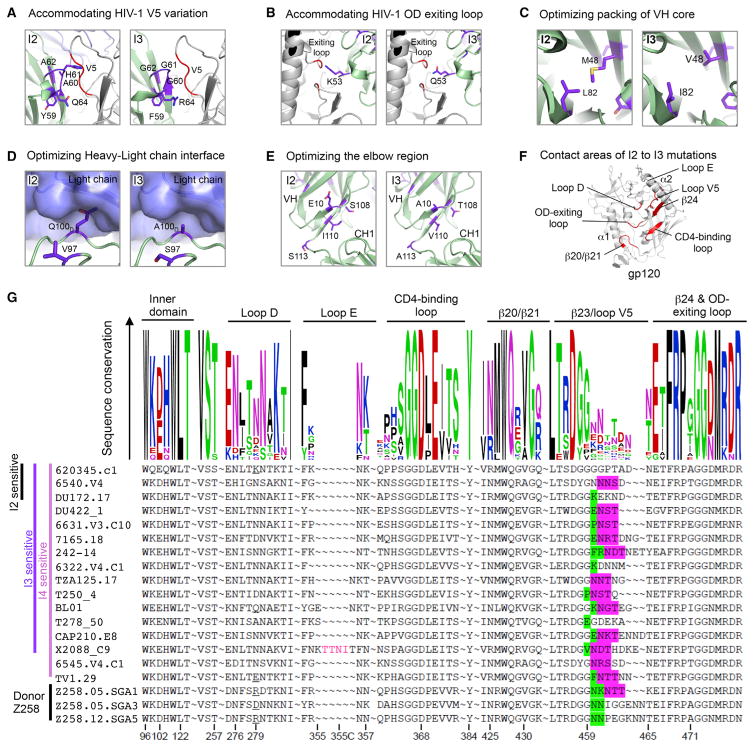
Additional modeling analysis indicated that most I2 to I3 mutations resolved potential steric clashes with HIV-1 gp120. This included the Gly60GlyGly62 motifs within the CDR H2 that enabled N6 to better accommodate changes in V5 (Figure 7A). Similarly, the replacement of Lys53, Met48, Leu82, and Gln100d either reduced the potential for steric clashes or improved the packing of the heavy chain or facilitated rotation of the light chain (Figures 7B–7D). A cluster of residues was also changed at the elbow region, which possibly increased elbow flexibility (Figure 7E). Notably, 7 out of 18 changes from I2 to I3 were mapped to the gp120-binding surface (Figure 7F). Most of these were concentrated in the CDR H2 interactions between N6 and V5.
Figure 7. Changes in V5 Induced Evolution of N6 Intermediates.
(A) Model showing key I2 to I3 mutations that increased I3 breadth by reducing CDR H2 clashes with the HIV-1 V5.
(B) Model showing key I2 to I3 mutations that increased I3 breadth by reducing CDR H2 clashes with the HIV-1 outer-domain-exiting loop.
(C) Model showing key I2 to I3 mutations that increased I3 breadth by improving the VH domain core packing.
(D) Model showing key I2 to I3 mutations that increased I3 breadth by removing heavy chain and light chain clashes.
(E) Model showing key I2 to I3 mutations that increased I3 breadth by optimizing the elbow region.
(F) Contact regions of I2 to I3 mutations on HIV-1 gp120. Regions within 8.5 Å of the I2 to I3 mutation are colored red on the cartoon representation of gp120.
(G) Sequence comparison of I2, I3, and I4 sensitive HIV-1 strains that are resistant to VRC01. Residues within 8.5 Å of bound N6 antibody were shown in Weblogo representation (upper panel), with the degree of conservation represented by the height of the residue. Sequence alignment (lower panel) with the loop E insertion, bulky residues, and glycosylation sites at base of V5 highlighted.
To understand what triggered the I2 to I3 change that led to the development of breadth, we then analyzed the viral sequences of VRC01-resistant viruses to determine the common features of those that became sensitive to the lineage in the transition from I2 to I3. When mapped onto the N6 co-crystal structure with gp120, several I2 to I3 mutations interacted with HIV-1 V5 and the outer-domain-exiting loop. These viruses shared features of bulky residues, glycosylation, or insertions in V5 (Figure 7G). Similarly, the patient’s virus also had a V5 that was longer than reference sequences and a glycosylation site (Figure 7G). These changes in V5 most likely drove the substitution of the Gly60GlyGly62 into the CDR H2 of the N6 lineage. These changes in turn permit the N6 heavy chain to adopt its unique orientation among the VRC01-class such that it avoids steric clashes in V5, the major mechanism of resistance to VRC01-class antibodies.
DISCUSSION
The results of this study provide an understanding of the evolution and mechanism of action of the N6 antibody that developed from a lineage of CD4bs antibodies in vivo to mediate extraordinary breadth and potency. Our results show that N6 gained its breadth and potency relative to others in its lineage by two main mechanisms. First, N6 evolved such that its binding was relatively insensitive to the absence or loss of individual contacts typically found in the VRC01 class. Second, it was able to avoid the major mechanism of resistance to other VRC01-like antibodies, namely changes in the vicinity of V5, that result in steric clashes. The result of these changes is an antibody with a remarkable ability to avoid steric clashes with the glycosylated V5 and tolerate mutations that might disrupt contacts in V5 or the CD4 BLP. It is this combination of avoiding glycan clashes and ability to tolerate mutations that permitted the breadth of N6.
An understanding of the evolution and mechanisms of action of the best naturally occurring antibodies, similar to those described here, is currently driving advances in the design of the next generation of immunogens for HIV. Prior work has suggested that bNAbs arise through a complex co-evolution of the immune response and virus, involving somatic hypermutation, B cell selection and expansion, viral escape mutations, and viral compensatory mutations that restore viral fitness. The ontogeny analysis showed that the N6 and VRC27 clades diverged early and then evolved in parallel. A similar mode of evolution has been observed for other anti-HIV bNAb lineages, including VRC01, CAP256-VRC26 (targeting the V1V2 region), and PCDN (targeting glycan[s] in the V3 region) (Doria-Rose et al., 2014; MacLeod et al., 2016; Wu et al., 2015). This parallel evolution of antibody clades or sub-lineages might aid in the development of potency and breadth. A previous study on CD4bs antibodies showed that the breadth of an antibody lineage can be increased by development in the presence of a “helper” lineage targeting the same region (Gao et al., 2014). Similarly, the maturation of the VRC27 clade continued to select mutations in the V5 region, which in turn could have aided in development of the increased breadth of the N6 clade. Thus, cooperative evolution might play important roles in the development of breadth for an antibody lineage such as N6.
It is particularly interesting to note that through natural selection in vivo, N6 has features that have been engineered into other antibodies to increase potency or breadth or reduce the possibility of viral escape. Although the naturally occurring VH1-2*02-derived VRC01-class antibodies thus far described mimic the interaction of CD4 with gp120, the vast majority do not fill a hydrophobic pocket on gp120 typically filled by Phe43CD4. Substitutions of hydrophobic residues for Gly54 in the antibodies NIH45-46 (Diskin et al., 2011) and VRC07 (Rudicell et al., 2014) have resulted in large increases in potency. The N6 antibody, through natural selection in vivo, has a Tyr54N6 that binds in this pocket and contributes to the potency of N6. In addition, the light chain of a VRC01-class antibody, VRC07, has been engineered to avoid steric clashes with the V5 loop to produce VRC07-523 (Diskin et al., 2013; Li et al., 2011; Rudicell et al., 2014). However, many of these engineered changes in VRC01-class antibodies have come at a cost of increased autoreactivity (Rudicell et al., 2014; reviewed in Sievers et al., 2015). VRC07-523 has only slightly less overall potency and breadth than N6, although it is less active against resistant isolates. However, this engineered antibody is autoreactive in some assays. Although natural autoreactive antibodies occur, especially among HIV-1 bNAbs (Liu et al., 2015), thus far we have not detected autoreactivity for the N6 antibody in the assays used in this study. Through natural selection in vivo, it is likely that B cells expressing antigen receptors with particularly strong autoreactivity or immunogenicity are selected against, while breadth and potency are maintained or favored.
The N6 antibody also has several characteristics that make it a desirable candidate for clinical use in prophylaxis and therapy. Of those antibodies being considered for clinical development, there are examples of antibodies that are extremely broad but moderate in potency (e.g., 10E8 or VRC01) or extremely potent and less broad (e.g., PGT121 or PGDM1400). However, the discovery of the N6 antibody demonstrates that this new VRC01-class antibody can mediate both extraordinary breadth and potency even against isolates traditionally resistant to antibodies in this class. The potency of N6 might further increase the durability of a prophylactic or therapeutic benefit in the case of passive administration because less antibody is required to persist to mediate an effect. VRC01 is currently in phase IIb efficacy trials as an intravenous infusion for HIV prophylaxis. Antibodies such as N6, which is 5- to 10-fold more potent, offer the possibility of subcutaneous administration, a more feasible approach to immunoprophylaxis. This effect might be further extended by introducing mutations into N6 that are able to extend its half-life in vivo. In addition to its potency, use of an antibody with this breadth might dramatically limit the likelihood of transmission when used in prophylaxis or selection of escape mutations in the setting of therapy. The rare occurrence of N6 resistance mutations suggests that such mutations come at a relatively high fitness cost, which might represent a partial barrier to the selection of resistant mutants. In addition to potency and breadth, the ease with which an antibody selects resistant viruses and the replication capacity of those viruses could turn out to be important considerations for use of bNAbs in the therapeutic setting where highly diverse Envs exist.
Last, these results add to accumulating evidence suggesting that resistance to antibodies mediated by the addition of glycan, and the co-evolution of antibodies that avoid them, most likely extends to other antibodies to HIV and other viruses. Glycosylation of surface proteins is thought to play a dominant role in evasion of the humoral immune response in diverse human viral pathogens (Das et al., 2010; Helle et al., 2010; Sommerstein et al., 2015; Szakonyi et al., 2006; Wei et al., 2003; Zlateva et al., 2004). In the case of HIV-1, although some bNAbs target epitopes partially or completely composed of glycan, they can play an important role in immune evasion (Stewart-Jones et al., 2016; Wei et al., 2003). There is evidence that the humoral response preferentially targets glycan holes in a de novo response such as acute infection or vaccination (Crooks et al., 2015; Moore et al., 2012; Wibmer et al., 2013), suggesting that glycosylated areas are less immunogenic than those lacking glycans. Thus, mechanisms for steric accommodation of glycans are eagerly sought, as they might provide ways for antibodies to overcome glycan shielding. The developmental pathway and structural recognition by N6 exemplify one solution: a shift in antibody recognition to accommodate strain-specific glycans in the V5. The shift in orientation by N6 to avoid steric clashes is reminiscent of other shifts in orientation reported in the development pathways of other bNAbs, including the PGT121-family of antibodies (Garces et al., 2015). Parallels to N6 recognition are observed with the influenza hemagglutinin stem-region antibody CR9114, which also shifts its recognition relative to other members of the VH1-69 class to accommodate a group-2-specific glycan (Pappas et al., 2014). Insights from the structure and developmental pathway of N6 thus might be general to overcoming common mechanisms of neutralization resistance of viral surface glycoproteins in the case of HIV-1 and other diverse viruses.
EXPERIMENTAL PROCEDURES
Study Patients
HIV-1-infected patients were enrolled at the National Institute of Allergy and Infectious Diseases (NIAID) under a clinical protocol, and informed consent was approved by the NIAID institutional review board. At the time of leukapheresis, patient Z258 had been infected with HIV-1 for 21 years, had a CD4+ T cell count of 733 cells/μL and plasma HIV-1 RNA of 996 copies/mL, and was not on antiretroviral treatment.
Memory B Cell Staining, Sorting, and Antibody Cloning
Staining and single-cell sorting of memory B cells were performed according to a published protocol (Huang et al., 2013) (Supplemental Experimental Procedures). In brief, peripheral-blood switch-memory B cells were sorted, seeded into 384-well plates at 4 cells per well, and expanded with IL-2, IL-21, and CD40L for 13 days. From wells with supernatants with neutralizing activity, Ig genes were amplified by RT-PCR, cloned into expression vectors, re-expressed in 293T cells, and purified with a protein-A column (GE Healthcare).
Neutralization Assays
HIV-1 Env pseudoviruses were generated by co-transfection of 293T cells with an Env-deficient backbone (pSG3ΔEnv) and a second plasmid that expressed HIV-1 Env. Neutralization activity of monoclonal antibodies or serum was measured with single-round HIV-1 Env-pseudovirus infection of TZM-bl cells, according to standard, previously described protocols (Li et al., 2005). Heat inactivated patient serum or monoclonal antibodies were serially diluted 5-fold and incubated with pseudovirus for 30 min. TZM-bl cells, containing a Tat-responsive reporter gene, were then added and plates were incubated for 48 hr. Assays were developed with a luciferase assay system (Supplemental Experimental Procedures).
Binding Assays
Binding of N6 to HIVBAL26 gp120 monomers, gp120 RSC3 variants, and cross-competition was measured by standard ELISA (Supplemental Experimental Procedures). The 50% of maximal binding (EC50) was calculated as the antibody concentrations necessary to reach half of the maximum signal.
Autologous Viruses Sequencing and Cloning
Autologous viruses from donor Z258 were obtained by plasma viral RNA extraction and cDNA synthesis. The resulting cDNA was then distributed in replicates that were then diluted such that single HIV-1 Env genomes were amplified as previously described (Wu et al., 2012) (Supplemental Experimental Procedures).
Formation of Protein Complexes, Crystallization, and Data Collection
The HIV-1 gp120-antibody complexes were formed by mixing EndoH treated gp120 with antibody Fab in a 1:1.2 molar ratio and purified by size exclusion chromatography. Fractions with antibody in complex with gp120 were concentrated to ~10 mg/mL for crystallization experiments. Crystallization conditions for all gp120-Fab complexes were obtained robotically and were manually optimized in hanging drops. All structure was solved by molecular replacement with Phaser. Iterative model building and refinement procedures were carried out with Coot and Phenix (Supplemental Experimental Procedures).
NGS
NGS of donor Z258 PBMC memory B cells was performed with 5′ primer-adapters to avoid bias. The Illumina paired reads were assembled to single end transcripts with USEARCH and then the low-quality reads were filtered. The V, D, and J genes were assigned with an in-house bioinformatics pipeline described previously (Wu et al., 2015). The heavy and light chains of N6 lineage-related members were identified via CDR3 signatures. Then, the phylogenetic analysis of N6 lineage heavy chain and light chains were performed and developmental intermediates were inferred via MEGA6 (Supplemental Experimental Procedures).
Supplementary Material
Highlights.
CD4bs antibody N6 potently neutralizes 98% of HIV isolates
N6 potently neutralizes isolates resistant to other CD4bs antibodies
N6 evolved from an early intermediate within a VRC01-class antibody lineage
Unique structural features circumvent mechanisms of resistance to the VRC01 class
Acknowledgments
We thank the members of the Vaccine Research Center for discussions of the manuscript. We thank Lynn Morris for arranging the testing of N6 against an extended clade C pseudovirus panel. We thank J. Baalwa, D. Ellenberger, F. Gao, B. Hahn, K. Hong, J. Kim, F. McCutchan, D. Montefiori, L. Morris, J. Overbaugh, E. Sanders-Buell, G. Shaw, R. Swanstrom, M. Thomson, S. Tovanabutra, C. Williamson, and L. Zhang for contributing the HIV-1 envelope plasmids used in our neutralization panel. Support for this work was provided by the Intramural Research Program and the Vaccine Research Center, National Institute of Allergy and Infectious Diseases (NIAID), NIH. Use of sector 22 (Southeast Region Collaborative Access team) at the Advanced Photon Source was supported by the US Department of Energy, Basic Energy Sciences, Office of Science under contract number W-31-109-Eng-38.
Footnotes
ACCESSION NUMBERS
The accession numbers for the coordinates and structure factors for N6 in complex with HIV-1 gp120 are PDB: 5TE4, 5TE6, and 5TE7. The accession numbers for thenucleotide and protein sequences forvariable regions of N6 areGenBank: KX595119–KX595128. The accession numbers for autologous virus sequences are GenBank: KX595108–KX595118. NGS data for Z258 BCR transcripts have been deposited with the NCBI Short Reads Archives under accession numbers SRA: SRR4417615–SRR4417632. N6 plasmids and protein are available through the NIH AIDS Reagent Program (https://www.aidsreagent.org/).
Supplemental Information includes Supplemental Experimental Procedures, six figures, and eight tables and can be found with this article online at http://dx.doi.org/10.1016/j.immuni.2016.10.027.
AUTHOR CONTRIBUTIONS
M.C., J.H., B.H.K., E.I., T.Z., F.W., J.R.M., and P.D.K. each contributed to the design of the study, analysis of the data, and preparation of this manuscript. J.H., B.H.K., T.G., and E.I. performed B cell sorting, antibody cloning, neutralization assays, ELISA, epitope and paratope mapping, NGS, and antibody swaps. T.Z., A.Z., and P.D.K. performed the structural studies with assistance from M.G.J. and protein was expressed by A.D. N.A.D.-R. provided the VRC27 antibody and neutralization data. F.W. and V.H. performed the plasma virus sequence analysis. B.Z. produced alanine scanning mutants. G.-Y.C. compiled the virus panel for accessing alanine scanning mutants. I.S.G. analyzed the patient sera neutralization fingerprint. M.A., S.M.A., R.P., G.K., T.V.H., and B.F.H. performed the autoreactivity assays. S.A.M. led the clinical care of the patients. K.M., M.K.L., S.O.D., R.T.B., and M.S.S. conducted neutralization assays. A.R., S.D., and D.C.D performed NGS. Z.S., C.A.S., and L.S. conducted bioinformatics and phylogenetic analyses of NGS data.
References
- Burton DR, Mascola JR. Antibody responses to envelope glycoproteins in HIV-1 infection. Nat Immunol. 2015;16:571–576. doi: 10.1038/ni.3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooks ET, Tong T, Chakrabarti B, Narayan K, Georgiev IS, Menis S, Huang X, Kulp D, Osawa K, Muranaka J, et al. Vaccine-Elicited Tier 2 HIV-1 Neutralizing Antibodies Bind to Quaternary Epitopes Involving Glycan-Deficient Patches Proximal to the CD4 Binding Site. PLoS Pathog. 2015;11:e1004932. doi: 10.1371/journal.ppat.1004932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das SR, Puigbò P, Hensley SE, Hurt DE, Bennink JR, Yewdell JW. Glycosylation focuses sequence variation in the influenza A virus H1 hemagglutinin globular domain. PLoS Pathog. 2010;6:e1001211. doi: 10.1371/journal.ppat.1001211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diskin R, Scheid JF, Marcovecchio PM, West AP, Jr, Klein F, Gao H, Gnanapragasam PN, Abadir A, Seaman MS, Nussenzweig MC, Bjorkman PJ. Increasing the potency and breadth of an HIV antibody by using structure-based rational design. Science. 2011;334:1289–1293. doi: 10.1126/science.1213782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diskin R, Klein F, Horwitz JA, Halper-Stromberg A, Sather DN, Marcovecchio PM, Lee T, West AP, Jr, Gao H, Seaman MS, et al. Restricting HIV-1 pathways for escape using rationally designed anti-HIV-1 antibodies. J Exp Med. 2013;210:1235–1249. doi: 10.1084/jem.20130221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doria-Rose NA, Klein RM, Daniels MG, O’Dell S, Nason M, Lapedes A, Bhattacharya T, Migueles SA, Wyatt RT, Korber BT, et al. Breadth of human immunodeficiency virus-specific neutralizing activity in sera: clustering analysis and association with clinical variables. J Virol. 2010;84:1631–1636. doi: 10.1128/JVI.01482-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doria-Rose NA, Schramm CA, Gorman J, Moore PL, Bhiman JN, DeKosky BJ, Ernandes MJ, Georgiev IS, Kim HJ, Pancera M, et al. NISC Comparative Sequencing Program. Developmental pathway for potent V1V2-directed HIV-neutralizing antibodies. Nature. 2014;509:55–62. doi: 10.1038/nature13036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao F, Bonsignori M, Liao HX, Kumar A, Xia SM, Lu X, Cai F, Hwang KK, Song H, Zhou T, et al. Cooperation of B cell lineages in induction of HIV-1-broadly neutralizing antibodies. Cell. 2014;158:481–491. doi: 10.1016/j.cell.2014.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garces F, Lee JH, de Val N, de la Pena AT, Kong L, Puchades C, Hua Y, Stanfield RL, Burton DR, Moore JP, et al. Affinity Maturation of a Potent Family of HIV Antibodies Is Primarily Focused on Accommodating or Avoiding Glycans. Immunity. 2015;43:1053–1063. doi: 10.1016/j.immuni.2015.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgiev IS, Doria-Rose NA, Zhou T, Kwon YD, Staupe RP, Moquin S, Chuang GY, Louder MK, Schmidt SD, Altae-Tran HR, et al. Delineating antibody recognition in polyclonal sera from patterns of HIV-1 isolate neutralization. Science. 2013;340:751–756. doi: 10.1126/science.1233989. [DOI] [PubMed] [Google Scholar]
- Haynes BF. New approaches to HIV vaccine development. Curr Opin Immunol. 2015;35:39–47. doi: 10.1016/j.coi.2015.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helle F, Vieyres G, Elkrief L, Popescu CI, Wychowski C, Descamps V, Castelain S, Roingeard P, Duverlie G, Dubuisson J. Role of N-linked glycans in the functions of hepatitis C virus envelope proteins incorporated into infectious virions. J Virol. 2010;84:11905–11915. doi: 10.1128/JVI.01548-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Doria-Rose NA, Longo NS, Laub L, Lin CL, Turk E, Kang BH, Migueles SA, Bailer RT, Mascola JR, Connors M. Isolation of human monoclonal antibodies from peripheral blood B cells. Nat Protoc. 2013;8:1907–1915. doi: 10.1038/nprot.2013.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein F, Nogueira L, Nishimura Y, Phad G, West AP, Jr, Halper-Stromberg A, Horwitz JA, Gazumyan A, Liu C, Eisenreich TR, et al. Enhanced HIV-1 immunotherapy by commonly arising antibodies that target virus escape variants. J Exp Med. 2014;211:2361–2372. doi: 10.1084/jem.20141050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Gao F, Mascola JR, Stamatatos L, Polonis VR, Koutsoukos M, Voss G, Goepfert P, Gilbert P, Greene KM, et al. Human immunodeficiency virus type 1 env clones from acute and early subtype B infections for standardized assessments of vaccine-elicited neutralizing antibodies. J Virol. 2005;79:10108–10125. doi: 10.1128/JVI.79.16.10108-10125.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, O’Dell S, Walker LM, Wu X, Guenaga J, Feng Y, Schmidt SD, McKee K, Louder MK, Ledgerwood JE, et al. Mechanism of neutralization by the broadly neutralizing HIV-1 monoclonal antibody VRC01. J Virol. 2011;85:8954–8967. doi: 10.1128/JVI.00754-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Yang G, Wiehe K, Nicely NI, Vandergrift NA, Rountree W, Bonsignori M, Alam SM, Gao J, Haynes BF, Kelsoe G. Polyreactivity and autoreactivity among HIV-1 antibodies. J Virol. 2015;89:784–798. doi: 10.1128/JVI.02378-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch RM, Wong P, Tran L, O’Dell S, Nason MC, Li Y, Wu X, Mascola JR. HIV-1 fitness cost associated with escape from the VRC01 class of CD4 binding site neutralizing antibodies. J Virol. 2015;89:4201–4213. doi: 10.1128/JVI.03608-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLeod DT, Choi NM, Briney B, Garces F, Ver LS, Landais E, Murrell B, Wrin T, Kilembe W, Liang CH, et al. IAVI Protocol C Investigators & The IAVI African HIV Research Network. Early Antibody Lineage Diversification and Independent Limb Maturation Lead to Broad HIV-1 Neutralization Targeting the Env High-Mannose Patch. Immunity. 2016;44:1215–1226. doi: 10.1016/j.immuni.2016.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore PL, Gray ES, Wibmer CK, Bhiman JN, Nonyane M, Sheward DJ, Hermanus T, Bajimaya S, Tumba NL, Abrahams MR, et al. Evolution of an HIV glycan-dependent broadly neutralizing antibody epitope through immune escape. Nat Med. 2012;18:1688–1692. doi: 10.1038/nm.2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pappas L, Foglierini M, Piccoli L, Kallewaard NL, Turrini F, Silacci C, Fernandez-Rodriguez B, Agatic G, Giacchetto-Sasselli I, Pellicciotta G, et al. Rapid development of broadly influenza neutralizing antibodies through redundant mutations. Nature. 2014;516:418–422. doi: 10.1038/nature13764. [DOI] [PubMed] [Google Scholar]
- Richman DD, Wrin T, Little SJ, Petropoulos CJ. Rapid evolution of the neutralizing antibody response to HIV type 1 infection. Proc Natl Acad Sci USA. 2003;100:4144–4149. doi: 10.1073/pnas.0630530100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudicell RS, Kwon YD, Ko SY, Pegu A, Louder MK, Georgiev IS, Wu X, Zhu J, Boyington JC, Chen X, et al. NISC Comparative Sequencing Program. Enhanced potency of a broadly neutralizing HIV-1 antibody in vitro improves protection against lentiviral infection in vivo. J Virol. 2014;88:12669–12682. doi: 10.1128/JVI.02213-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusert P, Kouyos RD, Kadelka C, Ebner H, Schanz M, Huber M, Braun DL, Hozé N, Scherrer A, Magnus C, et al. Swiss HIV Cohort Study. Determinants of HIV-1 broadly neutralizing antibody induction. Nat Med. 2016 doi: 10.1038/nm.4187. advance online publication. [DOI] [PubMed] [Google Scholar]
- Sather DN, Armann J, Ching LK, Mavrantoni A, Sellhorn G, Caldwell Z, Yu X, Wood B, Self S, Kalams S, Stamatatos L. Factors associated with the development of cross-reactive neutralizing antibodies during human immunodeficiency virus type 1 infection. J Virol. 2009;83:757–769. doi: 10.1128/JVI.02036-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers SA, Scharf L, West AP, Jr, Bjorkman PJ. Antibody engineering for increased potency, breadth and half-life. Curr Opin HIV AIDS. 2015;10:151–159. doi: 10.1097/COH.0000000000000148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simek MD, Rida W, Priddy FH, Pung P, Carrow E, Laufer DS, Lehrman JK, Boaz M, Tarragona-Fiol T, Miiro G, et al. Human immunodeficiency virus type 1 elite neutralizers: individuals with broad and potent neutralizing activity identified by using a high-throughput neutralization assay together with an analytical selection algorithm. J Virol. 2009;83:7337–7348. doi: 10.1128/JVI.00110-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommerstein R, Flatz L, Remy MM, Malinge P, Magistrelli G, Fischer N, Sahin M, Bergthaler A, Igonet S, Ter Meulen J, et al. Arenavirus Glycan Shield Promotes Neutralizing Antibody Evasion and Protracted Infection. PLoS Pathog. 2015;11:e1005276. doi: 10.1371/journal.ppat.1005276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephenson KE, Barouch DH. Broadly Neutralizing Antibodies for HIV Eradication. Curr HIV/AIDS Rep. 2016;13:31–37. doi: 10.1007/s11904-016-0299-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart-Jones GB, Soto C, Lemmin T, Chuang GY, Druz A, Kong R, Thomas PV, Wagh K, Zhou T, Behrens AJ, et al. Trimeric HIV-1-Env Structures Define Glycan Shields from Clades A, B, and G. Cell. 2016;165:813–826. doi: 10.1016/j.cell.2016.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szakonyi G, Klein MG, Hannan JP, Young KA, Ma RZ, Asokan R, Holers VM, Chen XS. Structure of the Epstein-Barr virus major envelope glycoprotein. Nat Struct Mol Biol. 2006;13:996–1001. doi: 10.1038/nsmb1161. [DOI] [PubMed] [Google Scholar]
- Wei X, Decker JM, Wang S, Hui H, Kappes JC, Wu X, Salazar-Gonzalez JF, Salazar MG, Kilby JM, Saag MS, et al. Antibody neutralization and escape by HIV-1. Nature. 2003;422:307–312. doi: 10.1038/nature01470. [DOI] [PubMed] [Google Scholar]
- Wibmer CK, Bhiman JN, Gray ES, Tumba N, Abdool Karim SS, Williamson C, Morris L, Moore PL. Viral escape from HIV-1 neutralizing antibodies drives increased plasma neutralization breadth through sequential recognition of multiple epitopes and immunotypes. PLoS Pathog. 2013;9:e1003738. doi: 10.1371/journal.ppat.1003738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Wang C, O’Dell S, Li Y, Keele BF, Yang Z, Imamichi H, Doria-Rose N, Hoxie JA, Connors M, et al. Selection pressure on HIV-1 envelope by broadly neutralizing antibodies to the conserved CD4-binding site. J Virol. 2012;86:5844–5856. doi: 10.1128/JVI.07139-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Zhang Z, Schramm CA, Joyce MG, Kwon YD, Zhou T, Sheng Z, Zhang B, O’Dell S, McKee K, et al. NISC Comparative Sequencing Program. Maturation and Diversity of the VRC01-Antibody Lineage over 15 Years of Chronic HIV-1 Infection. Cell. 2015;161:470–485. doi: 10.1016/j.cell.2015.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T, Zhu J, Wu X, Moquin S, Zhang B, Acharya P, Georgiev IS, Altae-Tran HR, Chuang GY, Joyce MG, et al. NISC Comparative Sequencing Program. Multidonor analysis reveals structural elements, genetic determinants, and maturation pathway for HIV-1 neutralization by VRC01-class antibodies. Immunity. 2013;39:245–258. doi: 10.1016/j.immuni.2013.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T, Lynch RM, Chen L, Acharya P, Wu X, Doria-Rose NA, Joyce MG, Lingwood D, Soto C, Bailer RT, et al. NISC Comparative Sequencing Program. Structural Repertoire of HIV-1-Neutralizing Antibodies Targeting the CD4 Supersite in 14 Donors. Cell. 2015;161:1280–1292. doi: 10.1016/j.cell.2015.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zlateva KT, Lemey P, Vandamme AM, Van Ranst M. Molecular evolution and circulation patterns of human respiratory syncytial virus subgroup a: positively selected sites in the attachment g glycoprotein. J Virol. 2004;78:4675–4683. doi: 10.1128/JVI.78.9.4675-4683.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.