Abstract
Chlamydia is responsible for millions of new infections annually, and current efforts focus on understanding cellular immunity for targeted vaccine development. The Chlamydia-specific CD4 T cell response is characterized by the production of IFNγ, and polyfunctional Th1 responses are associated with enhanced protection. A major limitation in studying these responses is the paucity of tools available for detection, quantification, and characterization of polyfunctional, antigen-specific T cells. We addressed this problem by developing a TCR transgenic mouse with CD4 T cells that respond to a common antigen in Chlamydia muridarum and Chlamydia trachomatis. Using an adoptive transfer approach, we show that naïve transgenic CD4 T cells become activated, proliferate, migrate to the infected tissue, and acquire a polyfunctional Th1 phenotype in infected mice. Polyfunctional Tg Th1 effectors demonstrated enhanced IFNγ production compared to polyclonal cells, protected immune deficient mice against lethality, mediated bacterial clearance, and orchestrated an anamnestic response. Adoptive transfer of Chlamydia-specific CD4 TCR Tg T cells with polyfunctional capacity offers a powerful approach for analysis of protective effector and memory responses against chlamydial infection, and demonstrates that an effective monoclonal CD4 T cell response may successfully guide subunit vaccination strategies.
Introduction
CD4 T cells contribute to cell-mediated immunity through effector functions mediated by the production of cytokines. Polyfunctional T-helper 1 (Th1) cells can sequentially produce IFNγ, IL-2, and TNF in response to T-cell receptor (TCR) stimulation (1). This phenotype has been reported in a variety of infectious disease models, including Leishmania (2) tuberculosis (3), HIV (4), Plasmodium (5), and Chlamydia (6). Polyfunctional Th1 cells demonstrate enhanced protective efficacy in comparison to IFNγ monofunctional cells (3), potentially by producing higher levels of Th1 cytokines (7, 8). Th1 polyfunctionality represents a measure of immunogenicity in vaccine studies (9), and generation of durable polyfunctional Th1 memory will likely be critical for Chlamydia vaccine development (6).
Protective immunity against Chlamydia is primarily mediated through Th1 cells (10, 11), and the importance of Chlamydia-specific CD4 T cells has been demonstrated by adoptive transfer (12, 13) and depletion studies (14). Despite the importance of CD4 T cells in controlling chlamydial infection, little is known about the generation of polyfunctional Th1 cells and how they contribute to cell-mediated immunity. Previous studies showed that a Chlamydia-specific IFNγ monofunctional Th1 clone was not protective, whereas a clone producing IFNγ and TNF cleared C. muridarum infection in nude mice (15). Vaccine models have shown that antigens and adjuvants generating polyfunctional (IFNγ+ TNF+) Th1 cells were more protective than IFNγ monofunctional Th1 cells (16, 17), and this protection has been observed in immunogenicity studies investigating single (18, 19) or multiple chlamydial antigens (20–23).
We recently developed the first TCR transgenic (Tg) mouse specific for a conserved antigen in C. muridarum and C. trachomatis to investigate the polyfunctional Th1 response in vivo. Identification of a polyfunctional Th1 clone allowed us to isolate and clone the Chlamydia-specific TCR for Tg mouse generation. After adoptive transfer, naïve TCR Tg CD4 T cells proliferated in the iliac lymph nodes, migrated to the infected genital tract, and primarily differentiated into polyfunctional Th1 cells. Polyfunctional Tg Th1 cells exhibited enhanced effector function characterized by increased IFNγ production associated with improved bacterial clearance compared to polyclonal, predominately monofunctional, Th1 cells. These studies demonstrate the first transgenic TCR to protect against C. muridarum genital infection and exhibit C. trachomatis cross-reactivity, and further define antigen-specific, enhanced effector function afforded by Th1 polyfunctionality.
Materials and Methods
Strains, cell lines, and culture conditions
Chlamydia muridarum Nigg stock (AR Nigg) was obtained from Roger Rank at the University of Arkansas for Medical Sciences, and has been previously described (24). C. trachomatis D/UW-3/Cx (25) was obtained from the American Type Culture Collection (Manassas, VA) and plaque purified before use (24). Plaque-purified C. trachomatis D/UW-3/Cx, Nigg strain CM001 (26), and plasmid-deficient CM3.1 (27) were propagated in mycoplasma-free L929 cells (28), and titrated by plaque assay or as inclusion-forming units (29), using a fluorescently tagged anti-chlamydial lipopolysaccharide monoclonal antibody (Bio-Rad). UV-inactivated bacteria were prepared, as described (30).
Generation of Chlamydia-specific T Cell hybridoma and transgenic mice
Two eight-week-old female C57BL/6J mice were intravaginally infected with 3×105 inclusion forming units (IFU) of wild-type Chlamydia muridarum Nigg. Infected mice were allowed to resolve primary infection, and were re-challenged two months later. The spleen and lymph nodes were collected one-week post-secondary challenge, and single-cell suspensions were stimulated ex vivo with reticulate body (RB)-enriched Nigg (1μg/mL) (31) for 5 days prior to fusion with murine BW5147 T cell lymphoma cells (32) in 50% PEG solution. Fused cells were cultured in HAT medium for an additional 7 to 9 days. Hybridomas were screened and sorted based on CD3, CD4, CD8, and TCRβ expression. Sorted CD4 T cell hybridomas underwent limiting dilution and were co-cultured with irradiated syngeneic splenocytes in the presence of Nigg elementary bodies (1μg/mL) or RB (1μg/mL) for 24–48 hours at 37°C. Harvested supernatants were tested for IL-2 and IFNγ levels by enzyme-linked immunosorbent assay (ELISA) from R&D Systems. The CD4 T cell clone with the highest co-production of IL-2 and IFNγ in the presence of Nigg elementary bodies (EB) was harvested and cultured for cloning of TCRα and TCRβ cDNA.
RNA from the CD4 T cell clone was made using the Qiagen RNAeasy method, and TCRα and TCRβ cDNA was obtained using the SuperTCRExpress™ Mouse TCR Vα/Vβ Repertoire Clone Screening Assay Kit (BioMed Immunotech), which contains 5′ RACE primers for all TCR Vα/Vβ. The cDNAs were cloned into the TOPO vector (Promega), sequenced, and identified as Vα6 and Vβ10. Genomic sequences corresponding to the mRNA sequences were used to map the variable, joining, and constant regions in the sequence. Primers with flanking XmaI site and NotI site, GATCCCGGGCAGAGCTGCAGCCTTCCCAAGGCTC and CATGCGGCCGCAGTGCTAGGAAGGGCGGCCTGGAC were generated for amplifying the variable region of Vα6 from genomic DNA. Primers with flanking XhoI site and SacII site, TCCGCTCGAGCCTTGACCCAACTATGGGCTGT and ATTCCCGCGGCTGGTCTACTCCAAACTACTCCAGG were generated to amplify the variable region of Vβ10. Vα6 amplicon was cloned into the pTαcass and Vβ10 amplicon into pTβcass vectors (33), which contain the respective promoters for Vα and Vβ expression and provided the joining and constant region, as a genomic clone (Fig S1 and S2). DNA constructs were sequenced for confirmation, linearized at SalI (Vα6) and KpnI (Vβ10) sites, respectively, purified and injected into the pronuclei of (C57BL/6J × SJL/J) F2 fertilized eggs.
Animals
Female C57BL/6J (Stock No: 000664), B6.SJL-Ptprca Pepcb/BoyJ (CD45.1+; Stock No: 002014), B6.129S7-Rag1tm1Mom/J (Rag1−/−; Stock No: 002216), and B6.129S2-Tcratm1Mom/J (Tcra−/−; Stock No: 002116) mice were purchased from The Jackson Laboratory (Bar Harbor, ME). Mice were given food and water ad libitum in an environmentally controlled pathogen-free room with a cycle of 12 h of light and 12 h of darkness. TCR transgenic mice generated as described above at the University of Pittsburgh were subsequently backcrossed onto C57BL/6J for over 10 generations. Transgenic mice were screened for expression of Vα6 and Vβ10 on CD4+ T cells from peripheral blood by PCR and FACS. Experimental mice were age-matched and used between 8 and 12 weeks of age. All animal experiments were approved by the Institutional Animal Care and Use Committee at the University of Pittsburgh and University of North Carolina.
Generation of bone marrow-derived DCs
Dendritic cells were generated from the tibias/femurs of C57BL/6J mice as previously described (34). Briefly, erythrocytes were lysed with ACK lysis buffer, and bone marrow precursors were cultured for 7 days in complete media (RPMI containing 10% fetal bovine serum, 2 mM glutamine, 10 mM HEPES, pH 7.4, 100 μM nonessential amino acids, 1 mM sodium pyruvate, 50 μM β-mercaptoethanol, and 50 μg/ml gentamicin) supplemented with 1000 U/mL recombinant murine GM-CSF and 1000 U/mL recombinant murine IL-4 (both from Peprotech). CD11c+ DCs were isolated using specific beads (Miltenyi Biotech), according to the manufacturer’s protocol.
Antigen-specific T-cell proliferation, activation, and cytokine analysis
The spleens of littermate or TCR transgenic mice were processed into a single cell suspension, as described previously (35). Splenocytes (1×105 cells/well) were seeded in a 96-well flat-bottomed tissue culture plate in complete media with 5 μg/ml C. muridarum AR Nigg, plasmid-deficient CM 3.1, C. trachomatis D/UW-3/Cx, or recombinant ovalbumin (Sigma) for 6 days. Splenocytes were treated with 20 U/mL murine IL-2 (Peprotech) over the final 48 hours. Cells were treated with 20 μl of Alamar Blue (Biosource) 6 hours before the end of the 6-day culture, and proliferation was measured at 530-nm excitation/590-nm emission with a Biotek fluorescence microplate reader.
Alternatively, transgenic or polyclonal CD4+ T cells were isolated from the spleens of naïve TCR transgenic or wild-type C57BL/6J mice by negative magnetic selection (EasySep™ Mouse CD4 T cell). Isolated CD4+ T cells were co-cultured at a 1:5 ratio with bone marrow-derived dendritic cells (BMDCs) for 3 days in the presence or absence of C. muridarum AR Nigg (5μg/mL). Expression of CD69 and Ki67 was determined by FACS surface and nuclear staining respectively, as described previously (36). Supernatants from dendritic-cell stimulated CD4+ T cells were collected and IL-2 concentrations determined by ELISA.
Murine Chlamydia infection and monitoring
For genital tract infection, female mice at least 8 weeks old were s.c. injected with 2.5 mg medroxyprogesterone (Depo-Provera®; Upjohn) 5–7 days prior to infection to induce a state of anestrous (37). Mice were intravaginally inoculated with 3×105 inclusion-forming units (IFU) CM001 diluted in 30 μl sucrose-sodium phosphate-glutamic acid buffer. Mice were monitored for cervicovaginal shedding via endocervical swabs (38), and IFUs were calculated, as described previously (39). Prior to reinfection, mice were treated intraperitoneally with 0.3 mg doxycycline in 100 μl phosphate buffered saline for 5 days (40). Animal welfare was monitored daily. Genital tract gross pathology, including presence of hydrosalpinx, was examined and recorded at sacrifice.
Lymphocyte isolation and flow cytometry
Spleen, iliac lymph nodes, oviducts, uterine horns, and cervical tissues were isolated from sacrificed mice. Cervical tissue and uterine horns were minced separately and incubated with 1 mL of collagenase I (Sigma) for 20 minutes at 37°C before neutralization with EDTA (10 μM). Single-cell suspensions were prepared by dispersing tissues through a 70-micron tissue strainer (Falcon). Cell suspensions were treated with erythrocyte lysis buffer (VitaLyse®; BioE), incubated in Fc block (5 μg/ml) for 10 minutes, and stained with LIVE/DEAD Fixable Yellow (Life Technologies) plus various combinations of the following fluorochrome-labeled antibodies: anti-CD3 (clone 17A2) anti-CD3e (145-C211), anti-CD4 (GK1.5, RM4-5, H129.19), anti-CD8a (53-6.7), anti-TCRVβ10 (V21.5), anti-TCRβ (H57–597), anti-CD45 (30-F11) anti-CD45.1 (A20), anti-CD45.2 (104), anti-CD44 (IM7), anti-CD62L (MEL-14), and anti-CD69 (H1.2F3), from BD Biosciences. The samples were analyzed on a CyAN ADP (Beckman Coulter) or LSR II flow cytometer (BD Bioscience), and data were analyzed with FlowJo software.
CFSE-Labeling and adoptive transfer
Transgenic or polyclonal CD4+ T cells were negatively separated, and a sample of isolated cells was analyzed by flow cytometry to confirm >93% purity. The indicated numbers of transgenic or wild-type CD4+ T cells were injected i.v. into Depo-Provera®-treated CD45.1+, Rag1−/−, or Tcra−/− mice 5–6 days prior to intravaginal infection. In some experiments, Tg CD4+ T cells were labeled with 1 μM CFSE (Thermo Fisher) for 5 minutes at 37°C prior to intravenous transfer and analysis.
Intracellular cytokine detection
Lymphocytes isolated from infected mice as described above were incubated in a 96-well plate at a concentration of 1×106 cells per well in the presence of UV-irradiated C. muridarum AR Nigg (5 μg/ml) or media alone for 6 hours at 37°C; GolgiPlug (BD Biosciences) was added, and incubation was continued for an additional 12–16 hours. Control samples were stimulated for 4–6 hours in the presence of PMA/ionomycin (Cell Stimulation Cocktail; eBioscience) and GolgiPlug. Surface staining was performed as described above, and the cells were fixed in BD Bioscience Cytofix/Cytoperm for 20 minutes. For detection of intracellular cytokines, cells were incubated for 30 minutes in BD Bioscience Perm/Wash with various combinations of the following fluorochrome-labeled antibodies: anti-IL-2 (JES6-5H4), anti-TNFα (MP6-XT22), and anti-IFNγ (XMG1.2) from BD Bioscience.
Statistical analysis
Differences between the means of experimental groups after infection were calculated using two-way repeated measures (RM) ANOVA. Significant differences in flow cytometric data were determined by one-way and two-way ANOVA. Comparisons of animal survival were performed by an exact log rank test. Prism software (GraphPad Software) was utilized for statistical analyses, and values of P ≤ 0.05 were considered significant.
Results
Generation of a Chlamydia-specific TCR transgenic mouse
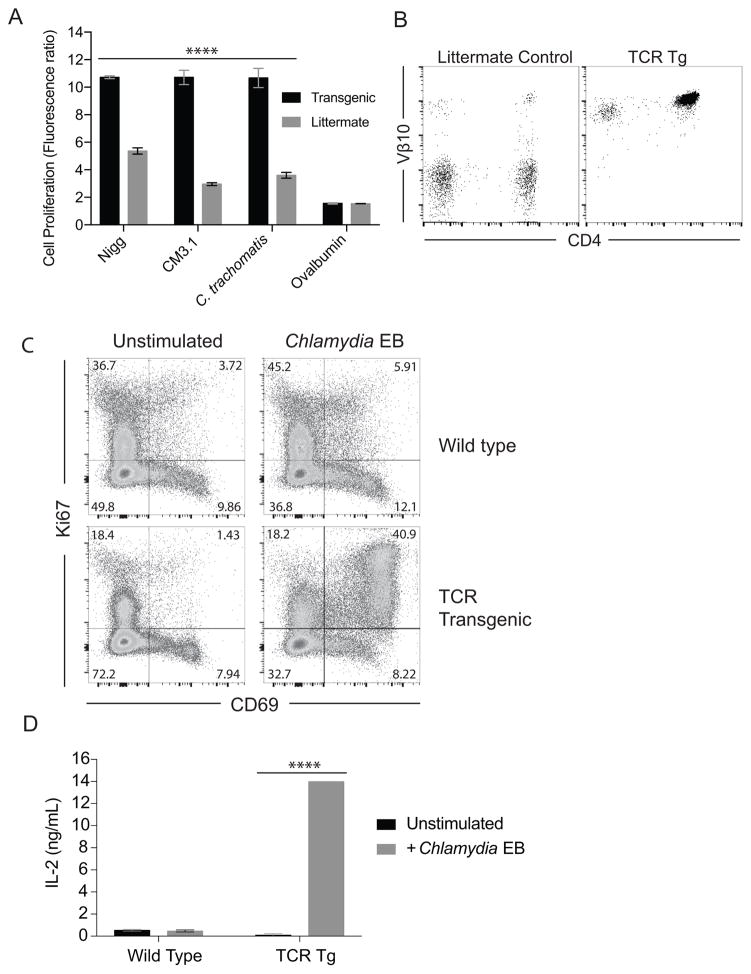
Previous studies demonstrate that the frequency of IFNγ-producing CD4 Th1 cells correlate with enhanced chlamydial clearance from the genital tract (10, 11, 13, 41). To directly monitor CD4 T cell responses during murine infection, we generated a Tg mouse strain with a TCR reactive with C. muridarum, which demonstrated cross-reactivity with C. trachomatis (Fig. 1A). The TCR genes were isolated from a hybridoma expressing Vα6 and Vβ10 chains specific for C. muridarum elementary and reticulate bodies. These genes (Fig. S1 and S2) were cloned into an expression vector used to generate germline Tg mice. Founder Tg mice almost uniformly express Vβ10 on the surface of autologous CD4 T cells (Fig. 1B), and were backcrossed to C57BL/6J mice for over ten generations. Tg mouse splenocytes demonstrated a 6–8-fold increase in Chlamydia-specific proliferation compared to littermates, with minimal proliferation induced by ovalbumin (Fig. 1A). C. muridarum Nigg plasmid-competent and -deficient (CM3.1) strains stimulated TCR Tg splenocytes equally. To further confirm the specificity of Tg CD4 T cells, we analyzed their ability to activate and proliferate in comparison to wild-type polyclonal CD4 T cells in vitro. Transgenic CD4 T cells demonstrated the ability to co-express high levels of CD69 and Ki67 when cultured with BMDC and C. muridarum elementary bodies, with over 40% being double-positive and ~50% expressing CD69 (Fig. 1C). In contrast, minimal activation was observed with polyclonal CD4 T cells. This enhanced proliferation was associated with significantly increased levels of IL-2 in the supernatants of stimulated TCR Tg CD4 T cells that were 29 times higher than polyclonal CD4 T cells (Fig. 1D). We next examined the ability of Tg CD4 T cells to become activated and proliferate in vivo.
Figure 1. Generation of a Chlamydia-specific TCR transgenic mouse.
(A) TCR Tg or littermate splenocytes were stimulated with 5 μg/ml C. trachomatis, C. muridarum AR Nigg, plasmid-deficient Nigg (CM 3.1), or recombinant ovalbumin, as indicated. Splenocytes were stimulated for 4 days, followed by 2 additional days in the presence of 20 U/mL IL-2. Change in proliferation was determined by the ratio of Alamar Blue fluorescent intensity compared to unstimulated controls (**** P < 0.0001 determined by two-way ANOVA). (B) Peripheral blood from C57BL/6J backcross progeny of Tg founder mice or littermate controls was stained with antibodies against CD3, CD4, and Vβ10. The right dot plot is representative of the Vβ10 expression on CD3+CD4+ T cells from Tg mice. (C) CD4+ T cells from Tg mice or wild type mice were incubated for 3 days with BMDC pulsed with and without 5 μg/ml C. muridarum. The right dot plots show CD69 and Ki67 expression from Tg and polyclonal CD4 T cells after stimulation. (D) Supernatants from dendritic cell-stimulated CD4 T cells were analyzed for IL-2 by ELISA (**** P < 0.0001 determined by two-way ANOVA).
C. muridarum genital infection initiates TCR Tg CD4 T cell proliferation and activation in vivo
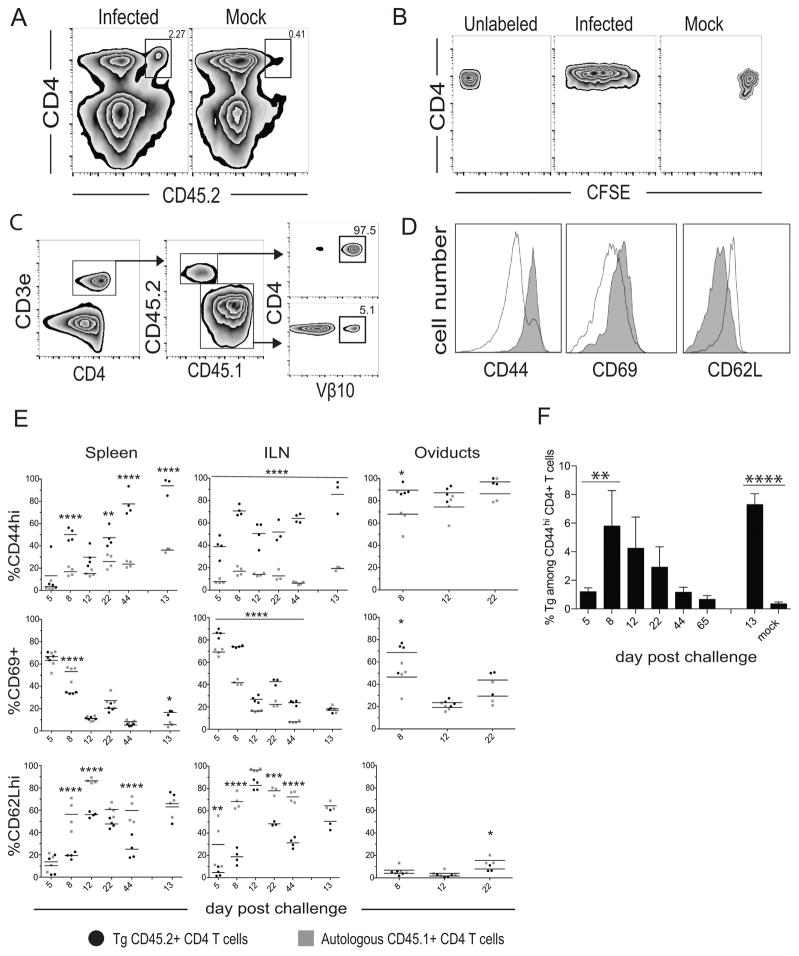
To determine the ability of TCR Tg CD4 T cells to proliferate and become activated in response to intravaginal C. muridarum infection, we utilized an adoptive transfer approach. To first test if these cells proliferate in vivo, we labeled naïve CD45.2+ TCR Tg CD4 T cells with CFSE and intravenously transferred 1×106 cells into congenic CD45.1+ mice. An increased percentage of CD45.2+ TCR Tg CD4 T cells were detectable on day 5 post-infection in the iliac lymph nodes compared to mock-infected controls (Fig. 2A), and infection resulted in their loss of CFSE expression consistent with proliferation (Fig. 2B). We then compared the activation state of endogenous and Tg CD4 T cells after infection by examining expression of the activation markers CD44, CD69, and CD62L on CD45.2+ TCR Tg and endogenous CD45.1+ CD4 T cells in the spleen, iliac lymph nodes, and oviducts. The gating strategy is shown in Fig. 2C. Comparison of the surface marker frequency between all CD45.1+ CD4 T cells or CD45.1+ CD4 T cells expressing the Vβ10 chain did not significantly alter the frequency of surface marker-positive endogenous cells (data not shown). TCR Tg CD4 T cells upregulated CD44 and CD69 concomitantly with down-regulation of CD62L by day 5 in the iliac lymph nodes and demonstrated greater percentages of activated cells in peripheral and secondary lymphoid organs (SLO) compared to the endogenous pool by day 8 post-infection (Fig. 2D, 2E). TCR Tg cells expressed significantly higher levels of CD69 in the ILN (iliac lymph node) on each day analyzed after primary infection, compared to endogenous cells. This was further associated with significantly decreased percentages of CD62Lhi TCR Tg cells on days 5, 8, 22, and 44 post-primary infection. Similar CD62L kinetics was observed in the spleen.
Figure 2. Proliferation and activation kinetics of TCR Tg CD4 T cells during C. muridarum genital tract infection.
One million CFSE-labeled Tg T cells were transferred into CD45.1+ female recipients, which were mock infected or infected with CM001. (A) Iliac lymph nodes from infected (top left) or mock-infected (top right) mice were examined for the presence of Tg T cells, and (B) Tg cells were examined for CFSE fluorescence. (C) Diagram of the flow cytometric gating strategy used to analyze CD62L, CD69, and CD44 expression by CD45.2+ Tg and CD45.1+ endogenous, polyclonal CD4 T cells. (D) Representative histograms comparing surface marker expression between Tg (grey) and endogenous (white) CD4 T cells during early infection. (E) Expression of CD62L, CD69, and CD44 on donor Tg and endogenous host CD4 T cells in the spleen, iliac lymph node, and oviducts on the indicated days post primary and secondary infection. Data points are representative of individual mice. Horizontal bars indicate the mean of 3–4 mice per group. Statistical significance was noted relative to autologous CD45.1+ CD4 T cells and indicated by asterisks: * P < 0.05, ** P < 0.01, *** P < 0.001, **** P < 0.0001 by two-way ANOVA. (F) Percentage of transgenic cells comprising the CD44hi CD4 T cell population from the spleen on the indicated days post-infection (** P < 0.01, **** P < 0.0001 by RM one-way ANOVA).
CD44 hi expression is used as a marker of T cell activation and Th1 memory (42, 43), and this memory phenotype was significantly increased among TCR Tg CD4 T cells in the iliac nodes throughout the course of primary infection, and on day 13 of secondary infection, compared to endogenous cells. Similar CD44 expression was observed for splenic TCR Tg cells. On day 13 post-secondary challenge, 85–98% of splenic Tg T cells were CD44hi, and these Tg cells comprised ~7% of the total splenic CD44hi CD4 T cell pool (Fig. 2F). The kinetics reflect the enhanced ability of Tg cells to adopt an activated effector and/or effector memory phenotype in the lymphoid tissues throughout infection compared to the endogenous T cell repertoire, and by day 8 in the infected peripheral tissues. These data collectively support other studies demonstrating Chlamydia-specific CD4 T cell priming and proliferation in the ILN (44, 45) and the presence of activated cells in the genital tract one-week post-infection (46).
Naïve TCR Tg CD4 T cells confer protection, acquire effector function, and demonstrate a recall response
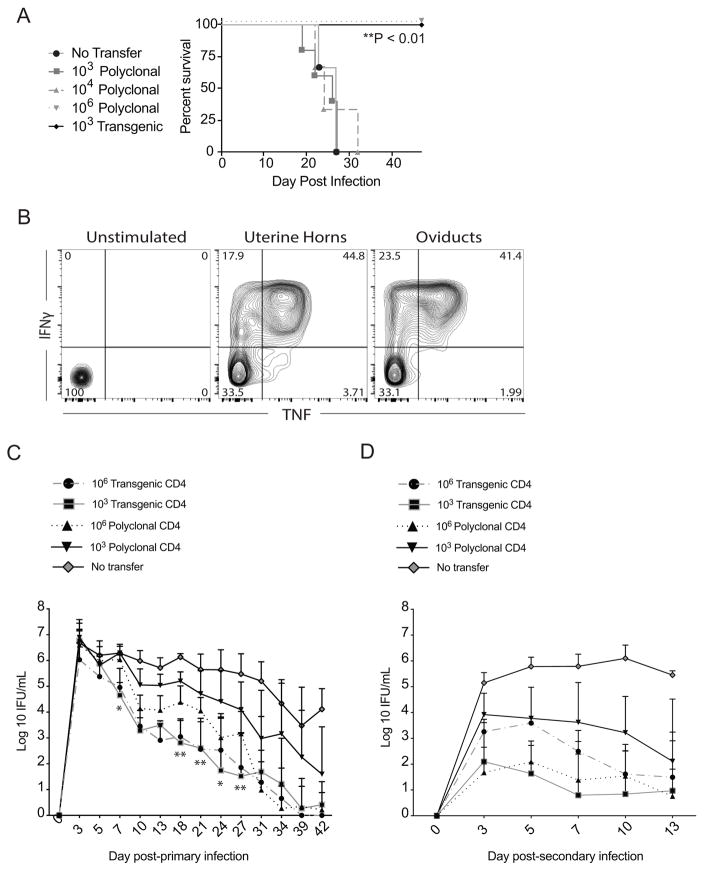
After demonstrating that TCR Tg CD4 T cells become activated, proliferate, and migrate to the infected genital tract, we investigated whether they would provide protection and produce IFNγ upon challenge. Prior data from our lab had revealed the plaque-purified strain of C. muridarum Nigg, CM001, resulted in disseminated lethal infection in Rag1−/− mice after intravaginal inoculation (26). Rag1−/− mice received adoptive transfers of 103 Tg or 103, 104, and 106 polyclonal CD4 T cells 5 days prior to infection (Fig. 3A). A precursor frequency of 106 polyclonal CD4 T cells was used as a positive control, based on previous observations that this dose conferred protection (data not shown). Mice receiving 103 Tg CD4 T cells survived infection, whereas 103 and 104 polyclonal CD4 T cells were not protective. TCR Tg CD4 T cells demonstrated a recall response characterized by the production of inflammatory cytokines. Transgenic CD4 T cells isolated from the uterine horns and oviducts on day 7 post-secondary challenge produced IFNγ and TNF in response to in vitro re-stimulation with UV-irradiated C. muridarum (Fig. 3B). These data indicate that TCR Tg CD4 T cells prevent death from disseminating infection, migrate to infected tissues, and acquire Th1 effector functions post-infection.
Figure 3. TCR Tg CD4 T cells mediate protection and demonstrate a recall response following challenge.
Rag1−/− mice (4–5 per group) were mock treated or intravenously injected with indicated numbers of CD4 T cells isolated from the spleens of naïve C57BL/6J wild type or Tg mice, and intravaginally inoculated with CM001 5 days later. Survival was monitored daily, and an exact log rank test was used to analyze survival differences between polyclonal and Tg groups (** P < 0.01). (B) Recall response of CD4+Vβ10+ Tg CD4 T cells to secondary infection. Following primary infection, Tg mice were treated with doxycycline, rested for 9 weeks, and re-challenged with CM001. On day 7 post-challenge, genital tract CD4 T cells were harvested, stimulated with 5 μg/ml C. muridarum, and analyzed for IFNγ and TNF production by intracellular cytokine staining. (C) Indicated numbers of naïve Tg or polyclonal CD4 T cells were adoptively transferred to Tcra−/− mice 5 days prior to intravaginal infection with CM001, and the course of primary infection was monitored by culture of lower genital tract swabs. Significance was determined by two-way RM ANOVA. Data represent the mean ± SEM of 10 mice per group. Comparison of individual days for 103 Tg versus 106 polyclonal: * P < 0.05, ** P < 0.01. Comparison of primary infection course between groups: P=NS for 106 Tg versus 103 Tg. *** P =0.0001 for 103 Tg versus 106 Polyclonal. **** P <0.0001 for all remaining group comparisons. (D) Immune mice were treated with doxycycline on days 52–56 post-infection, rested for 5 weeks, re-challenged with CM001, and infection monitored by culture of vaginal swabs. Significance was determined by two-way RM ANOVA. Data represent the mean + SD of 4–5 mice per group. Comparison of groups over primary infection course: P=NS for 103 Tg versus 106 polyclonal, 106 Tg versus 103 or 106 polyclonal. *P < 0.05 for 103 Tg versus 106 Tg. **** P <0.0001 for all remaining group comparisons.
TCR Tg CD4 T cells can mediate bacterial clearance during primary and secondary infection
We next investigated whether adoptively transferring TCR Tg CD4 T cells to αβ TCR-deficient mice would lead to clearance of primary genital tract infection and enable resistance to challenge infection. Mice that did not receive T cells failed to clear infection, whereas adoptive transfer of 103 or 106 TCR Tg CD4 T cells to Tcra−/− mice resulted in equivalent rates of infection clearance, with a 3.5-log reduction in shedding being detected by day 10 post-infection (Fig. 3C). In addition, infection clearance after adoptive transfer of 103 or 106 TCR Tg CD4 T cells was accelerated when compared to groups that received either 103 or 106 polyclonal CD4 T cells, indicating that the Tg CD4 T cells are more efficient effectors (Fig. 3C).
We also investigated if TCR Tg CD4 T cells would contribute to a recall response upon secondary challenge. Immune mice that had received 103 or 106 TCR Tg CD4 T cells prior to primary infection exhibited a 4.5- and 3-log reduction in shedding, respectively, on day 3 post-challenge compared to primary infection (Figs. 3C, 3D). Mice that received 103 Tg CD4 T cells prior to primary infection were more resistant to challenge when compared to mice that received 106 Tg CD4 T cells (Fig. 3D). This was consistent with findings in other TCR transgenic models, where lower numbers of adoptively transferred naïve Tg CD4 T cells induce better memory development. It is possible that decreased interclonal competition for antigen leads to enhanced differentiation of the fittest effectors into memory cells (47). In contrast, infectious burden during secondary infection was significantly lowered in mice receiving 106 but not 103 polyclonal CD4 T cells. In this instance, a broad array of antigen-specific cells avoids interclonal competition for peptide-MHC class II stimulation.
Although mice that were re-infected without prior receipt of adoptive T cells failed to exhibit any decline in infectious burden up to two weeks post inoculation, on day 3 post-challenge, their infectious burden was 2-log lower than that observed during primary infection. This transient protection may be a result of circulating T-cell independent antibody or memory γδ T cells that are not capable of clearing infection independent of conventional CD4 T cells.
TCR Tg CD4 T cells preferentially adopt a polyfunctional Th1 phenotype with increased IFNγ production
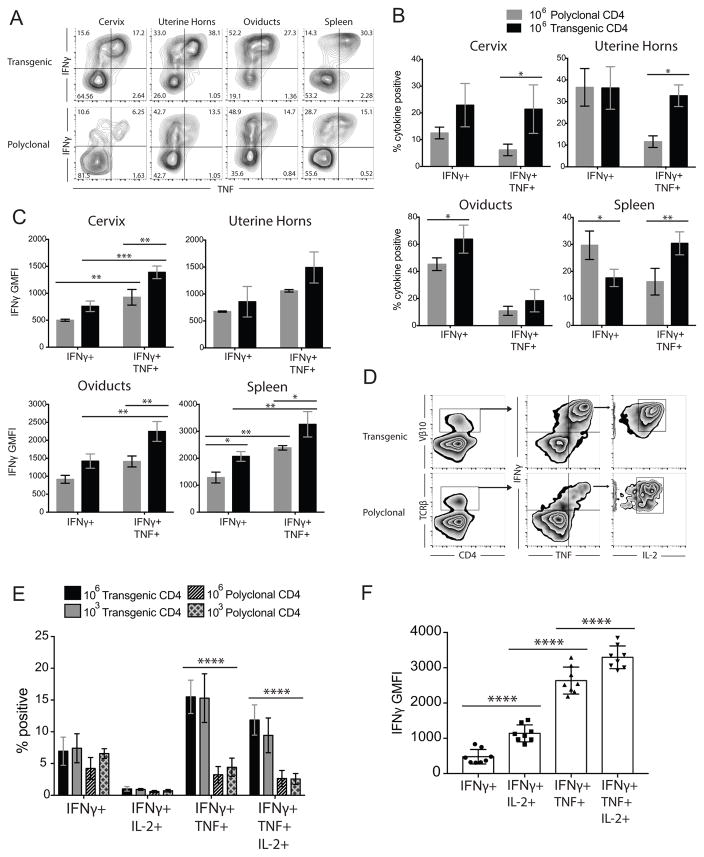
The TCR transgenic mouse was developed using a TCR that induced a Th1 response after chlamydial stimulation in vitro. We hypothesized that TCR Tg CD4 T cells would differentiate into polyfunctional Th1 cells in vivo since adoptive transfer of these cells led to enhanced chlamydial clearance during primary infection (Fig. 3C). We detected significantly increased percentages of IFNγ+TNF+ double-positive cells in the spleen and genital tissues of Tcra−/− mice receiving Tg CD4 T cells, on day 13 post-infection compared to mice receiving polyclonal CD4 T cells (Fig. 4A, 4B). Additionally, transgenic polyfunctional IFNγ+TNF+ CD4 T cells expressed significantly higher amounts of IFNγ compared to polyfunctional, polyclonal populations. TCR Tg polyfunctionality for TNF and IFNγ was associated with increased IFNγ production compared to cells singly positive for IFNγ (Fig. 4C). These data indicate that TCR Tg CD4 T cells preferentially adopt a polyfunctional phenotype characterized by high levels of IFNγ production. Furthermore, the percentage of triple-positive (IFNγ+TNF+IL-2+) CD4 T cells in the spleen was increased among the Tg CD4 T cell population when compared to polyclonal CD4 T cells on day 48 post-infection (Fig. 4D, 4E). Triple positive transgenic cells expressed significantly higher levels of IFNγ per cell compared with single- or double-positive cells (Fig. 4F), as previously described for pathogen-specific polyfunctional T cells in other models of infection (4, 8, 48). Triple positive cells also expressed significantly higher levels of TNF per cell compared to TNF monofunctional cells, but at a reduced magnitude (60% increase), and no differences were observed in IL-2 GMFI (data not shown). These data collectively show that Tg CD4 T cells have superior functional capacity with enhanced cytokine production, and this polyfunctional effector response is associated with enhanced bacterial clearance.
Figure 4. Naïve TCR Tg CD4 T cells differentiate into polyfunctional Th1 cells with increased IFNγ production.
Tcra−/− mice receiving polyclonal or Tg CD4 T cells were analyzed for polyfunctional Th1 responses on Day 13 (A–B) and Day 48 (D–E) post-infection. (A) CD4 T cells isolated from indicated tissues were stimulated with 5 μg/ml C. muridarum and evaluated for intracellular cytokine production. Contour plots show representative IFNγ and TNF co-production by Tg CD3e+CD4+Vβ10+ cells (top) and polyclonal CD3e+CD4+TCRβ+ cells (bottom). (B) Comparison of the percentage of cytokine positive cells between single positive (IFNγ+) and double-positive (IFNγ+ TNF+) polyclonal and Tg CD4 T cells (C) Associated IFNγ geometric mean fluorescent intensities (GMFI). Data represent the mean ± SD of 3 mice per group. * P < 0.05, ** P < 0.01, *** P < 0.001 determined by two-way ANOVA. (D) Diagram of the flow cytometric gating strategy used to analyze CD4 T cell polyfunctionality on day 48 post-infection. (E) The frequency of single positive (IFNγ+), double positive (IFNγ+ TNF+), and triple positive (IFNγ+ TNF+IL-2+) Tg or polyclonal CD4 T cells on day 48 post-infection. Data represent the mean ± SD of 4–5 mice per group (**** P < 0.0001). (F) Comparison of the IFNγ GMFI between spleen-isolated Tg CD4 T cell single, double, and triple-positive populations. Data represent the mean ± SD of 8 mice receiving 103 and 106 Tg cells. **** P < 0.0001 determined by two-way RM ANOVA.
Discussion
Pathogen-specific TCR Tg mice have been utilized in a variety of infectious disease models (49–56), including NR1 mice that recognize C. trachomatis (46). Adoptive transfer of naïve TCR Tg cells is a superior approach to transfer of in vitro maintained T cell lines, since naïve TCR Tg cells allow analysis of the initial antigen encounter and phenotypic differences between in vivo derived effector and memory populations. We developed a TCR Tg mouse that recognizes a conserved antigen between C. trachomatis and C. muridarum. The TCR Tg cells of this mouse react to C. trachomatis serovars D, F, H, and L2 (data not shown), and preliminary biochemical analyses reveal that they recognize a soluble, secreted protein enriched in reticulate bodies (data not shown). We have excluded commonly studied immunogenic antigens such as MOMP, OmcB, HSP60, and PmpG based on their inability to stimulate TCR Tg cellular proliferation in vitro. This report demonstrates the first TCR to protect against C. muridarum genital infection, and allowed us to analyze enhanced effector function afforded by Th1 polyfunctionality at a level that had not been previously attainable. Generation of this mouse has allowed for the unique ability to adoptively transfer TCR Tg cells for investigation of antigen-specific T cell responses to both mouse and human chlamydial strains in the murine model of genital tract infection.
Development of the Chlamydia-specific TCR Tg mouse was based on selection of a Th1 clone specific for both C. muridarum elementary bodies and reticulate bodies. We used a non-biased approach, by analyzing T cell clones demonstrating the strongest IFNγ and IL-2 production, which has been shown to be effective in TCR Tg mouse development. Selecting clones reactive against whole organism or crude antigen preparations has resulted in Tg CD4 T cells with the capacity to mount robust effector and memory responses following infection and vaccination (55), compared to model antigens (57). Our studies reveal that the Tg CD4 T cells possess a TCR, which confers protection against intravaginal C. muridarum infection.
These Tg CD4 T cells become activated, proliferate extensively, and produce high levels of IL-2 when stimulated with C. muridarum. Naïve and memory CD4 T cells require TCR stimulation in combination with IL-2 signaling to proliferate (58), and TCR engagement upregulates IL-2R subunits (59). The strength of IL-2 signaling also correlates with the magnitude of proliferation in Th1 cells (60), and IFNγ expression increases with successive cell divisions (61). Furthermore, IL-2 signaling during priming enhances differentiation of the effector pool into memory (62).
Based on the ability of these cells to recognize chlamydia in vitro, we used an adoptive transfer approach to analyze the proliferation and activation kinetics in vivo. Similar to the C. trachomatis model, our approach revealed that C. muridarum infection induced significant TCR Tg CD4 T cell activation and expansion in the iliac lymph nodes by day 5 and Tg cells expressed an activated phenotype (CD44hiCD69+CD62Llo). The CD44hi CD62Llo phenotype was also observed in the infected oviducts. Expression of CD69 on Tg cells in the spleen and oviducts on day 8, in our model, is likely due to the ability of CM001 to quickly disseminate to the distal organs and rapidly ascend the genital tract. Increased CD69 expression on CD4 T cells early in CM001 infection may be a result of local priming events, prior to tissue infiltration of activated T cells primed in the ILN. At later time points, CD44 expression in SLOs steadily increased, particularly during re-infection, consistent with the formation of memory T cells. These kinetics are similar to other infectious disease models of CD4 T cell activation and memory (46, 51, 56, 63). After priming in the ILN, Tg cells made up ~7% of all CD44hi CD4 T cells in the spleen, which consistently decreased through infection, until mice received a secondary challenge. This is consistent with other systems demonstrating that peak CD4 T cell expansion typically occurs after one week (64), and is followed by CD4 T cell contraction over 1–2 weeks, where 90–95% of the expanded population undergoes cell death (65, 66). Late in the course of C. muridarum infection and during reinfection, a majority of Tg cells expressed high levels of the memory marker CD44. Additional phenotyping experiments are required to determine the proportions of Tg T cells in the terminal effector, effector memory, and central memory pools.
TCR Tg cells prevented death in immunocompromised mice infected with CM001, and these cells were recalled to the infected tissues and produced IFNγ and TNF upon secondary challenge. These results parallel other infectious disease models demonstrating that adoptive transfer of antigen-specific naïve CD4 T cells can protect against lethality (56). Adoptive transfer to T- and B-cell deficient Rag1−/− hosts illustrates the CD4 T helper-independent protective function of TCR Tg cells, likely mediated through their production of IFNγ (67). Furthermore, transfer of these cells into αβ TCR-deficient mice led to enhanced protection against primary infection and equivalent protection against a secondary challenge compared to the polyclonal response. Comparable levels of oviduct gross pathology were observed between the TCR Tg and polyclonal groups (100% and 95% hydrosalpinx, respectively), which was not surprising given the ability of CM001 to induce severe pathology in wild-type mice (68). Future studies utilizing vaccination or adoptive transfer of in vitro primed TCR Tg cells should help reveal their capacity to protect against oviduct pathology.
Reduced bacterial burden mediated by TCR Tg cells was associated with increased frequencies of double- and triple-positive Th1 populations producing higher levels of IFNγ compared to polyclonal CD4 T cells. IFNγ is a critical effector molecule for controlling chlamydial replication (13, 69–72), and enhanced frequencies of polyfunctional Tg cells producing IL-2 could allow for enhanced Th1 effector proliferation. Our TCR Tg cells clearly recognize an antigen that drives a favorable response that leads to enhanced bacterial clearance and resistance to challenge infection. Persistent antigen and antigen depots reduce the memory pool leading to non-protective responses from terminally differentiated, exhausted T cells. Removal of antigen drives T cell transition to memory (73), and these cells remain plastic and heterogeneous (74, 75). Thus, triple-positive Th1 Tg cells could be a consequence of improved effector function leading to lower bacterial load (76, 77). In addition, Tg cells may also demonstrate greater functional avidity, which has been linked with improved disease outcomes (78) and expression of decreased levels of inhibitory receptors (79). High avidity T cells are less susceptible to activation-induced cell death (80) and demonstrate increased polyfunctionality (81, 82).
Our analyses were limited to the study of CD4 T cells and focused on profiling three major Th1 cytokines. A comprehensive analysis of TCR Tg cell production of cytokines, chemokines, and cytotoxic effectors, as well as their helper function for antibody production by B cells is needed to fully delineate their protective, or pathological, mechanisms. Alternative effectors (83) and antibody (84, 85) have been shown to play a significant role in mediating chlamydial clearance. Additional analysis of the recall response is needed to determine the mechanisms whereby equivalent protection from reinfection occurred in Tcra−/− mice that had received 103 polyclonal T cells or 103 monoclonal TCR Tg T cells. Potentially, polyclonal, polyfunctional T cells were maintained and IFNγ monofunctional cells culled, or monofunctional cells responding to a variety of antigens elicit similar protection to polyfunctional TCR Tg cells recognizing a single antigen.
The TCR Tg C57BL/6 background presents a caveat. Some TCR Tg mice are backcrossed to Rag backgrounds to eliminate endogenous TCR subunit expression, however this can result in deletion of the TCR Tg cells due to the requirement of a second endogenous TCR alpha chain for thymic emigration (56). Endogenously activated CD4 T cells expressing a second TCR can be present in TCR Tg mice. However, the spleen population exists at a small frequency (86) and at similar numbers in wild-type mice (87). This phenomenon led us to utilize equal and greater numbers of wild-type CD4 T cells as controls for adoptive transfer experiments.
The finding that a single immunogenic antigen that elicits polyfunctional T cells can successfully induce a protective response is encouraging from a subunit vaccinology perspective. Viral models demonstrate that primary and secondary effectors share organ-specific expression patterns, but secondary effectors are more polyfunctional (triple positive); polyfunctional cells also express higher levels of genes associated with survival and migration (88). Whether CD4 T cell polyfunctionality can predict memory generation and subsequent, enhanced secondary effector functions is an important area to be addressed. Once we have identified the antigen recognized by the TCR Tg T cells, we can determine if vaccination with this antigen drives induction of a protective polyfunctional response, and whether adoptive transfer of in vitro antigen-primed Tg T cells can protect from infection and disease.
In conclusion, we have demonstrated that adoptive transfer of Tg CD4 T cells specific for a single Chlamydia antigen induces polyfunctional CD4 T cells that provide enhanced immunity against Chlamydia. Transgenic CD4 T cells specific for Chlamydia can be further used to directly monitor differentiation of antigen-specific effector and memory responses during infection and to better delineate protective responses upon challenge. The development of a successful vaccine will be facilitated by better understanding of how CD4 T cell polyfunctionality is generated, sustained, and provides immunity at the mucosal surface.
Supplementary Material
Acknowledgments
We thank the UNC Flow Cytometry Core Facility for technical assistance, and Dr. Chunming Bi and the staff of the University of Pittsburgh Transgenic and Gene Targeting Core Facility for assistance with transgenic mouse development.
Abbreviations
- IFNγ
interferon gamma
- TCR
T cell receptor
- Th1
T-helper 1
- Tg
transgenic
- IL-2
interleukin 2
- HIV
human immunodeficiency virus
- TNF
tumor necrosis factor
- EB
elementary body
- RB
reticulate body
- IFU
inclusion forming units
- HAT
hypoxanthine-aminopterin-thymidine
- PEG
polyethylene glycol
- ELISA
enzyme-linked immunosorbent assay
- CM
Chlamydia muridarum
- BMDC
bone marrow-derived dendritic cell
- ACK
ammonim-chloride-potassium
- UV
ultraviolet
- PMA
Phorbol 12-myristate 13-acetate
- SLO
secondary lymphoid organ
- ILN
iliac lymph node
- MHC
major histocompatibility complex
- GMFI
geometric mean fluorescent intensity
- MOMP
major outer membrane porin
- OmcB
outer membrane complex protein B
- HSP60
heat shock protein 60
- PmpG
polymorphic membrane protein G
Footnotes
This work was supported by National Institutes of Health-National Institute of Allergy and Infectious Diseases Grants R01 A105624 and U19 A1084024 (to T.D.).
Disclosures
The authors have no financial conflicts of interest.
References
- 1.Han Q, Bagheri N, Bradshaw EM, Hafler DA, Lauffenburger DA, Love JC. Polyfunctional responses by human T cells result from sequential release of cytokines. Proc Natl Acad Sci U S A. 2012;109:1607–1612. doi: 10.1073/pnas.1117194109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Darrah PA, Patel DT, De Luca PM, Lindsay RW, Davey DF, Flynn BJ, Hoff ST, Andersen P, Reed SG, Morris SL, Roederer M, Seder RA. Multifunctional TH1 cells define a correlate of vaccine-mediated protection against Leishmania major. Nat Med. 2007;13:843–850. doi: 10.1038/nm1592. [DOI] [PubMed] [Google Scholar]
- 3.Lindenstrom T, Agger EM, Korsholm KS, Darrah PA, Aagaard C, Seder RA, Rosenkrands I, Andersen P. Tuberculosis subunit vaccination provides long-term protective immunity characterized by multifunctional CD4 memory T cells. J Immunol. 2009;182:8047–8055. doi: 10.4049/jimmunol.0801592. [DOI] [PubMed] [Google Scholar]
- 4.Duvall MG, Precopio ML, Ambrozak DA, Jaye A, McMichael AJ, Whittle HC, Roederer M, Rowland-Jones SL, Koup RA. Polyfunctional T cell responses are a hallmark of HIV-2 infection. Eur J Immunol. 2008;38:350–363. doi: 10.1002/eji.200737768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Seder RA, Chang LJ, Enama ME, Zephir KL, Sarwar UN, Gordon IJ, Holman LA, James ER, Billingsley PF, Gunasekera A, Richman A, Chakravarty S, Manoj A, Velmurugan S, Li M, Ruben AJ, Li T, Eappen AG, Stafford RE, Plummer SH, Hendel CS, Novik L, Costner PJ, Mendoza FH, Saunders JG, Nason MC, Richardson JH, Murphy J, Davidson SA, Richie TL, Sedegah M, Sutamihardja A, Fahle GA, Lyke KE, Laurens MB, Roederer M, Tewari K, Epstein JE, Sim BK, Ledgerwood JE, Graham BS, Hoffman SL, Team VRCS. Protection against malaria by intravenous immunization with a nonreplicating sporozoite vaccine. Science. 2013;341:1359–1365. doi: 10.1126/science.1241800. [DOI] [PubMed] [Google Scholar]
- 6.Stary G, Olive A, Radovic-Moreno AF, Gondek D, Alvarez D, Basto PA, Perro M, Vrbanac VD, Tager AM, Shi J, Yethon JA, Farokhzad OC, Langer R, Starnbach MN, von Andrian UH. VACCINES. A mucosal vaccine against Chlamydia trachomatis generates two waves of protective memory T cells. Science. 2015;348:aaa8205. doi: 10.1126/science.aaa8205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boyd A, Almeida JR, Darrah PA, Sauce D, Seder RA, Appay V, Gorochov G, Larsen M. Pathogen-Specific T Cell Polyfunctionality Is a Correlate of T Cell Efficacy and Immune Protection. PLoS One. 2015;10:e0128714. doi: 10.1371/journal.pone.0128714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burel JG, Apte SH, Groves PL, McCarthy JS, Doolan DL. Polyfunctional and IFN-gamma monofunctional human CD4+ T cell populations are molecularly distinct. JCI Insight. 2017;2:e87499. doi: 10.1172/jci.insight.87499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Seder RA, Darrah PA, Roederer M. T-cell quality in memory and protection: implications for vaccine design. Nat Rev Immunol. 2008;8:247–258. doi: 10.1038/nri2274. [DOI] [PubMed] [Google Scholar]
- 10.Perry LL, Feilzer K, Caldwell HD. Immunity to Chlamydia trachomatis is mediated by T helper 1 cells through IFN-gamma-dependent and -independent pathways. J Immunol. 1997;158:3344–3352. [PubMed] [Google Scholar]
- 11.Gondek DC, Olive AJ, Stary G, Starnbach MN. CD4+ T cells are necessary and sufficient to confer protection against Chlamydia trachomatis infection in the murine upper genital tract. J Immunol. 2012;189:2441–2449. doi: 10.4049/jimmunol.1103032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Su H, Caldwell HD. CD4+ T cells play a significant role in adoptive immunity to Chlamydia trachomatis infection of the mouse genital tract. Infect Immun. 1995;63:3302–3308. doi: 10.1128/iai.63.9.3302-3308.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li W, Murthy AK, Guentzel MN, Seshu J, Forsthuber TG, Zhong G, Arulanandam BP. Antigen-specific CD4+ T cells produce sufficient IFN-gamma to mediate robust protective immunity against genital Chlamydia muridarum infection. J Immunol. 2008;180:3375–3382. doi: 10.4049/jimmunol.180.5.3375. [DOI] [PubMed] [Google Scholar]
- 14.Morrison SG, Su H, Caldwell HD, Morrison RP. Immunity to murine Chlamydia trachomatis genital tract reinfection involves B cells and CD4(+) T cells but not CD8(+) T cells. Infect Immun. 2000;68:6979–6987. doi: 10.1128/iai.68.12.6979-6987.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Igietseme JU, Ramsey KH, Magee DM, Williams DM, Kincy TJ, Rank RG. Resolution of murine chlamydial genital infection by the adoptive transfer of a biovar-specific, Th1 lymphocyte clone. Reg Immunol. 1993;5:317–324. [PubMed] [Google Scholar]
- 16.Yu H, Karunakaran KP, Kelly I, Shen C, Jiang X, Foster LJ, Brunham RC. Immunization with live and dead Chlamydia muridarum induces different levels of protective immunity in a murine genital tract model: correlation with MHC class II peptide presentation and multifunctional Th1 cells. J Immunol. 2011;186:3615–3621. doi: 10.4049/jimmunol.1002952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yu H, Karunakaran KP, Jiang X, Shen C, Andersen P, Brunham RC. Chlamydia muridarum T cell antigens and adjuvants that induce protective immunity in mice. Infect Immun. 2012;80:1510–1518. doi: 10.1128/IAI.06338-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jiang J, Liu G, Kickhoefer VA, Rome LH, Li LX, McSorley SJ, Kelly KA. A Protective Vaccine against Chlamydia Genital Infection Using Vault Nanoparticles without an Added Adjuvant. Vaccines (Basel) 2017:5. doi: 10.3390/vaccines5010003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hansen J, Jensen KT, Follmann F, Agger EM, Theisen M, Andersen P. Liposome delivery of Chlamydia muridarum major outer membrane protein primes a Th1 response that protects against genital chlamydial infection in a mouse model. J Infect Dis. 2008;198:758–767. doi: 10.1086/590670. [DOI] [PubMed] [Google Scholar]
- 20.Yu H, Jiang X, Shen C, Karunakaran KP, Jiang J, Rosin NL, Brunham RC. Chlamydia muridarum T-cell antigens formulated with the adjuvant DDA/TDB induce immunity against infection that correlates with a high frequency of gamma interferon (IFN-gamma)/tumor necrosis factor alpha and IFN-gamma/interleukin-17 double-positive CD4+ T cells. Infect Immun. 2010;78:2272–2282. doi: 10.1128/IAI.01374-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu H, Karunakaran KP, Jiang X, Brunham RC. Evaluation of a multisubunit recombinant polymorphic membrane protein and major outer membrane protein T cell vaccine against Chlamydia muridarum genital infection in three strains of mice. Vaccine. 2014;32:4672–4680. doi: 10.1016/j.vaccine.2014.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Olsen AW, Theisen M, Christensen D, Follmann F, Andersen P. Protection against Chlamydia promoted by a subunit vaccine (CTH1) compared with a primary intranasal infection in a mouse genital challenge model. PLoS One. 2010;5:e10768. doi: 10.1371/journal.pone.0010768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Picard MD, Cohane KP, Gierahn TM, Higgins DE, Flechtner JB. High-throughput proteomic screening identifies Chlamydia trachomatis antigens that are capable of eliciting T cell and antibody responses that provide protection against vaginal challenge. Vaccine. 2012;30:4387–4393. doi: 10.1016/j.vaccine.2012.01.017. [DOI] [PubMed] [Google Scholar]
- 24.O’Connell CM, Nicks KM. A plasmid-cured Chlamydia muridarum strain displays altered plaque morphology and reduced infectivity in cell culture. Microbiology. 2006;152:1601–1607. doi: 10.1099/mic.0.28658-0. [DOI] [PubMed] [Google Scholar]
- 25.Stephens RS, Kalman S, Lammel C, Fan J, Marathe R, Aravind L, Mitchell W, Olinger L, Tatusov RL, Zhao Q, Koonin EV, Davis RW. Genome sequence of an obligate intracellular pathogen of humans: Chlamydia trachomatis. Science. 1998;282:754–759. doi: 10.1126/science.282.5389.754. [DOI] [PubMed] [Google Scholar]
- 26.Sullivan J, FL, Darville T, O’Connell C. C. muridarum Nigg Contains Multiple Variants Exhbiting Different Phenotypic Properties. Proceedings of the Thirteenth International Symposium on Human Chlamydial Infections. International Chlamydia Symposium; Pacific Grove, California. 2014. pp. 93–96. [Google Scholar]
- 27.O’Connell CM, Ingalls RR, Andrews CW, Jr, Scurlock AM, Darville T. Plasmid-deficient Chlamydia muridarum fail to induce immune pathology and protect against oviduct disease. J Immunol. 2007;179:4027–4034. doi: 10.4049/jimmunol.179.6.4027. [DOI] [PubMed] [Google Scholar]
- 28.O’Connell CM, AbdelRahman YM, Green E, Darville HK, Saira K, Smith B, Darville T, Scurlock AM, Meyer CR, Belland RJ. Toll-like receptor 2 activation by Chlamydia trachomatis is plasmid dependent, and plasmid-responsive chromosomal loci are coordinately regulated in response to glucose limitation by C. trachomatis but not by C. muridarum. Infect Immun. 2011;79:1044–1056. doi: 10.1128/IAI.01118-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kelly KA, Robinson EA, Rank RG. Initial route of antigen administration alters the T-cell cytokine profile produced in response to the mouse pneumonitis biovar of Chlamydia trachomatis following genital infection. Infect Immun. 1996;64:4976–4983. doi: 10.1128/iai.64.12.4976-4983.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Darville T, O’Neill JM, Andrews CW, Jr, Nagarajan UM, Stahl L, Ojcius DM. Toll-like receptor-2, but not Toll-like receptor-4, is essential for development of oviduct pathology in chlamydial genital tract infection. J Immunol. 2003;171:6187–6197. doi: 10.4049/jimmunol.171.11.6187. [DOI] [PubMed] [Google Scholar]
- 31.Skipp P, Robinson J, O’Connor CD, Clarke IN. Shotgun proteomic analysis of Chlamydia trachomatis. Proteomics. 2005;5:1558–1573. doi: 10.1002/pmic.200401044. [DOI] [PubMed] [Google Scholar]
- 32.Katz DH, Bechtold TE, Altman A. Construction of T cell hybridomas secreting allogeneic effect factor. J Exp Med. 1980;152:956–968. doi: 10.1084/jem.152.4.956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kouskoff V, Signorelli K, Benoist C, Mathis D. Cassette vectors directing expression of T cell receptor genes in transgenic mice. Journal of immunological methods. 1995;180:273–280. doi: 10.1016/0022-1759(95)00002-r. [DOI] [PubMed] [Google Scholar]
- 34.Tatsumi T, Huang J, Gooding WE, Gambotto A, Robbins PD, Vujanovic NL, Alber SM, Watkins SC, Okada H, Storkus WJ. Intratumoral delivery of dendritic cells engineered to secrete both interleukin (IL)-12 and IL-18 effectively treats local and distant disease in association with broadly reactive Tc1-type immunity. Cancer Res. 2003;63:6378–6386. [PubMed] [Google Scholar]
- 35.Darville T, Andrews CW, Jr, Laffoon KK, Shymasani W, Kishen LR, Rank RG. Mouse strain-dependent variation in the course and outcome of chlamydial genital tract infection is associated with differences in host response. Infect Immun. 1997;65:3065–3073. doi: 10.1128/iai.65.8.3065-3073.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim KH, Sederstrom JM. Assaying Cell Cycle Status Using Flow Cytometry. Curr Protoc Mol Biol. 2015;111:28 26 21–11. doi: 10.1002/0471142727.mb2806s111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tuffrey M, Taylor-Robinson D. Progesterone as a key factor in the development of a mouse model for genital-tract infection with Chlamydia trachomatis. FEMS Microbiol Let. 1981;12:111–115. [Google Scholar]
- 38.Kelly KA, Robinson EA, Rank RG. Initial route of antigen administration alters the T-cell cytokine profile produced in response to the mouse pneumonitis biovar of Chlamydia trachomatis following genital infection [published erratum appears in Infect Immun 1997 Jun;65(6):2508] Infection and Immunity. 1996;64:4976–4983. doi: 10.1128/iai.64.12.4976-4983.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Darville T, Andrews CW, Jr, Laffoon KK, Shymasani W, Kishen LR, Rank RG. Mouse strain-dependent variation in the course and outcome of chlamydial genital tract infection is associated with differences in host response. Infection and Immunity. 1997;65:3065–3073. doi: 10.1128/iai.65.8.3065-3073.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Riley MM, Zurenski MA, Frazer LC, O’Connell CM, Andrews CW, Jr, Mintus M, Darville T. The recall response induced by genital challenge with Chlamydia muridarum protects the oviduct from pathology but not from reinfection. Infect Immun. 2012;80:2194–2203. doi: 10.1128/IAI.00169-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shaw JH, Grund VR, Durling L, Caldwell HD. Expression of genes encoding Th1 cell-activating cytokines and lymphoid homing chemokines by chlamydia-pulsed dendritic cells correlates with protective immunizing efficacy. Infect Immun. 2001;69:4667–4672. doi: 10.1128/IAI.69.7.4667-4672.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pure E, Cuff CA. A crucial role for CD44 in inflammation. Trends Mol Med. 2001;7:213–221. doi: 10.1016/s1471-4914(01)01963-3. [DOI] [PubMed] [Google Scholar]
- 43.Baaten BJ, Li CR, Deiro MF, Lin MM, Linton PJ, Bradley LM. CD44 regulates survival and memory development in Th1 cells. Immunity. 2010;32:104–115. doi: 10.1016/j.immuni.2009.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cain TK, Rank RG. Local Th1-like responses are induced by intravaginal infection of mice with the mouse pneumonitis biovar of Chlamydia trachomatis. Infect Immun. 1995;63:1784–1789. doi: 10.1128/iai.63.5.1784-1789.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li LX, McSorley SJ. B cells enhance antigen-specific CD4 T cell priming and prevent bacteria dissemination following Chlamydia muridarum genital tract infection. PLoS Pathog. 2013;9:e1003707. doi: 10.1371/journal.ppat.1003707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Roan NR, Gierahn TM, Higgins DE, Starnbach MN. Monitoring the T cell response to genital tract infection. Proc Natl Acad Sci U S A. 2006;103:12069–12074. doi: 10.1073/pnas.0603866103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Blair DA, Lefrancois L. Increased competition for antigen during priming negatively impacts the generation of memory CD4 T cells. Proc Natl Acad Sci U S A. 2007;104:15045–15050. doi: 10.1073/pnas.0703767104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Maybeno M, Redeker A, Welten SP, Peters B, Loughhead SM, Schoenberger SP, Sette A, Arens R. Polyfunctional CD4+ T cell responses to immunodominant epitopes correlate with disease activity of virulent Salmonella. PLoS One. 2012;7:e43481. doi: 10.1371/journal.pone.0043481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Butz EA, Bevan MJ. Massive expansion of antigen-specific CD8+ T cells during an acute virus infection. Immunity. 1998;8:167–175. doi: 10.1016/s1074-7613(00)80469-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sano G, Hafalla JC, Morrot A, Abe R, Lafaille JJ, Zavala F. Swift development of protective effector functions in naive CD8(+) T cells against malaria liver stages. J Exp Med. 2001;194:173–180. doi: 10.1084/jem.194.2.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Roman E, Miller E, Harmsen A, Wiley J, Von Andrian UH, Huston G, Swain SL. CD4 effector T cell subsets in the response to influenza: heterogeneity, migration, and function. J Exp Med. 2002;196:957–968. doi: 10.1084/jem.20021052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Coles RM, Mueller SN, Heath WR, Carbone FR, Brooks AG. Progression of armed CTL from draining lymph node to spleen shortly after localized infection with herpes simplex virus 1. J Immunol. 2002;168:834–838. doi: 10.4049/jimmunol.168.2.834. [DOI] [PubMed] [Google Scholar]
- 53.McSorley SJ, Asch S, Costalonga M, Reinhardt RL, Jenkins MK. Tracking salmonella-specific CD4 T cells in vivo reveals a local mucosal response to a disseminated infection. Immunity. 2002;16:365–377. doi: 10.1016/s1074-7613(02)00289-3. [DOI] [PubMed] [Google Scholar]
- 54.Reiley WW, Calayag MD, Wittmer ST, Huntington JL, Pearl JE, Fountain JJ, Martino CA, Roberts AD, Cooper AM, Winslow GM, Woodland DL. ESAT-6-specific CD4 T cell responses to aerosol Mycobacterium tuberculosis infection are initiated in the mediastinal lymph nodes. Proc Natl Acad Sci U S A. 2008;105:10961–10966. doi: 10.1073/pnas.0801496105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wuthrich M, Hung CY, Gern BH, Pick-Jacobs JC, Galles KJ, Filutowicz HI, Cole GT, Klein BS. A TCR transgenic mouse reactive with multiple systemic dimorphic fungi. J Immunol. 2011;187:1421–1431. doi: 10.4049/jimmunol.1100921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stephens R, Albano FR, Quin S, Pascal BJ, Harrison V, Stockinger B, Kioussis D, Weltzien HU, Langhorne J. Malaria-specific transgenic CD4(+) T cells protect immunodeficient mice from lethal infection and demonstrate requirement for a protective threshold of antibody production for parasite clearance. Blood. 2005;106:1676–1684. doi: 10.1182/blood-2004-10-4047. [DOI] [PubMed] [Google Scholar]
- 57.Wuthrich M, Ersland K, Pick-Jacobs JC, Gern BH, Frye CA, Sullivan TD, Brennan MB, Filutowicz HI, O’Brien K, Korthauer KD, Schultz-Cherry S, Klein BS. Limited model antigen expression by transgenic fungi induces disparate fates during differentiation of adoptively transferred T cell receptor transgenic CD4+ T cells: robust activation and proliferation with weak effector function during recall. Infect Immun. 2012;80:787–797. doi: 10.1128/IAI.05326-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gesbert F, Moreau JL, Theze J. IL-2 responsiveness of CD4 and CD8 lymphocytes: further investigations with human IL-2Rbeta transgenic mice. Int Immunol. 2005;17:1093–1102. doi: 10.1093/intimm/dxh289. [DOI] [PubMed] [Google Scholar]
- 59.Liao W, Lin JX, Leonard WJ. Interleukin-2 at the crossroads of effector responses, tolerance, and immunotherapy. Immunity. 2013;38:13–25. doi: 10.1016/j.immuni.2013.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pepper M, Pagan AJ, Igyarto BZ, Taylor JJ, Jenkins MK. Opposing signals from the Bcl6 transcription factor and the interleukin-2 receptor generate T helper 1 central and effector memory cells. Immunity. 2011;35:583–595. doi: 10.1016/j.immuni.2011.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bird JJ, Brown DR, Mullen AC, Moskowitz NH, Mahowald MA, Sider JR, Gajewski TF, Wang CR, Reiner SL. Helper T cell differentiation is controlled by the cell cycle. Immunity. 1998;9:229–237. doi: 10.1016/s1074-7613(00)80605-6. [DOI] [PubMed] [Google Scholar]
- 62.Dooms H, Wolslegel K, Lin P, Abbas AK. Interleukin-2 enhances CD4+ T cell memory by promoting the generation of IL-7R alpha-expressing cells. J Exp Med. 2007;204:547–557. doi: 10.1084/jem.20062381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Srinivasan A, Foley J, Ravindran R, McSorley SJ. Low-dose Salmonella infection evades activation of flagellin-specific CD4 T cells. J Immunol. 2004;173:4091–4099. doi: 10.4049/jimmunol.173.6.4091. [DOI] [PubMed] [Google Scholar]
- 64.Marsden VS, Strasser A. Control of apoptosis in the immune system: Bcl-2, BH3-only proteins and more. Annu Rev Immunol. 2003;21:71–105. doi: 10.1146/annurev.immunol.21.120601.141029. [DOI] [PubMed] [Google Scholar]
- 65.Ahmed R, Gray D. Immunological memory and protective immunity: understanding their relation. Science. 1996;272:54–60. doi: 10.1126/science.272.5258.54. [DOI] [PubMed] [Google Scholar]
- 66.Swain SL, Croft M, Dubey C, Haynes L, Rogers P, Zhang X, Bradley LM. From naive to memory T cells. Immunol Rev. 1996;150:143–167. doi: 10.1111/j.1600-065x.1996.tb00700.x. [DOI] [PubMed] [Google Scholar]
- 67.Teijaro JR, Verhoeven D, Page CA, Turner D, Farber DL. Memory CD4 T cells direct protective responses to influenza virus in the lungs through helper-independent mechanisms. J Virol. 2010;84:9217–9226. doi: 10.1128/JVI.01069-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.O’Connell CM, Ingalls RR, Andrews CW, Jr, Skurlock AM, Darville T. Plasmid-deficient Chlamydia muridarum fail to induce immune pathology and protect against oviduct disease. Journal of Immunology. 2007;179:4027–4034. doi: 10.4049/jimmunol.179.6.4027. [DOI] [PubMed] [Google Scholar]
- 69.Naglak EK, Morrison SG, Morrison RP. IFNgamma is Required for Optimal Antibody-Mediated Immunity against Genital Chlamydia Infection. Infect Immun. 2016 doi: 10.1128/IAI.00749-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nagarajan UM, Sikes J, Prantner D, Andrews CW, Jr, Frazer L, Goodwin A, Snowden JN, Darville T. MyD88 deficiency leads to decreased NK cell gamma interferon production and T cell recruitment during Chlamydia muridarum genital tract infection, but a predominant Th1 response and enhanced monocytic inflammation are associated with infection resolution. Infect Immun. 2011;79:486–498. doi: 10.1128/IAI.00843-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gondek DC, Roan NR, Starnbach MN. T cell responses in the absence of IFN-gamma exacerbate uterine infection with Chlamydia trachomatis. J Immunol. 2009;183:1313–1319. doi: 10.4049/jimmunol.0900295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang S, Fan Y, Brunham RC, Yang X. IFN-gamma knockout mice show Th2-associated delayed-type hypersensitivity and the inflammatory cells fail to localize and control chlamydial infection. Eur J Immunol. 1999;29:3782–3792. doi: 10.1002/(SICI)1521-4141(199911)29:11<3782::AID-IMMU3782>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 73.Swain SL, Hu H, Huston G. Class II-independent generation of CD4 memory T cells from effectors. Science. 1999;286:1381–1383. doi: 10.1126/science.286.5443.1381. [DOI] [PubMed] [Google Scholar]
- 74.Jameson SC, Masopust D. Diversity in T cell memory: an embarrassment of riches. Immunity. 2009;31:859–871. doi: 10.1016/j.immuni.2009.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Moulton VR, Bushar ND, Leeser DB, Patke DS, Farber DL. Divergent generation of heterogeneous memory CD4 T cells. J Immunol. 2006;177:869–876. doi: 10.4049/jimmunol.177.2.869. [DOI] [PubMed] [Google Scholar]
- 76.Sakai S, Kauffman KD, Schenkel JM, McBerry CC, Mayer-Barber KD, Masopust D, Barber DL. Cutting edge: control of Mycobacterium tuberculosis infection by a subset of lung parenchyma-homing CD4 T cells. J Immunol. 2014;192:2965–2969. doi: 10.4049/jimmunol.1400019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kaushal D, Foreman TW, Gautam US, Alvarez X, Adekambi T, Rangel-Moreno J, Golden NA, Johnson AM, Phillips BL, Ahsan MH, Russell-Lodrigue KE, Doyle LA, Roy CJ, Didier PJ, Blanchard JL, Rengarajan J, Lackner AA, Khader SA, Mehra S. Mucosal vaccination with attenuated Mycobacterium tuberculosis induces strong central memory responses and protects against tuberculosis. Nat Commun. 2015;6:8533. doi: 10.1038/ncomms9533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Vingert B, Perez-Patrigeon S, Jeannin P, Lambotte O, Boufassa F, Lemaitre F, Kwok WW, Theodorou I, Delfraissy JF, Theze J, Chakrabarti LA, AEHC, Group S. HIV controller CD4+ T cells respond to minimal amounts of Gag antigen due to high TCR avidity. PLoS Pathog. 2010;6:e1000780. doi: 10.1371/journal.ppat.1000780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Billeskov R, Wang Y, Solaymani-Mohammadi S, Frey B, Kulkarni S, Andersen P, Agger EM, Sui Y, Berzofsky JA. Low Antigen Dose in Adjuvant-Based Vaccination Selectively Induces CD4 T Cells with Enhanced Functional Avidity and Protective Efficacy. J Immunol. 2017;198:3494–3506. doi: 10.4049/jimmunol.1600965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Black CM, Armstrong TD, Jaffee EM. Apoptosis-regulated low-avidity cancer-specific CD8(+) T cells can be rescued to eliminate HER2/neu-expressing tumors by costimulatory agonists in tolerized mice. Cancer Immunol Res. 2014;2:307–319. doi: 10.1158/2326-6066.CIR-13-0145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Almeida JR, Price DA, Papagno L, Arkoub ZA, Sauce D, Bornstein E, Asher TE, Samri A, Schnuriger A, Theodorou I, Costagliola D, Rouzioux C, Agut H, Marcelin AG, Douek D, Autran B, Appay V. Superior control of HIV-1 replication by CD8+ T cells is reflected by their avidity, polyfunctionality, and clonal turnover. J Exp Med. 2007;204:2473–2485. doi: 10.1084/jem.20070784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Almeida JR, Sauce D, Price DA, Papagno L, Shin SY, Moris A, Larsen M, Pancino G, Douek DC, Autran B, Saez-Cirion A, Appay V. Antigen sensitivity is a major determinant of CD8+ T-cell polyfunctionality and HIV-suppressive activity. Blood. 2009;113:6351–6360. doi: 10.1182/blood-2009-02-206557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Johnson RM, Kerr MS, Slaven JE. Plac8-dependent and inducible NO synthase-dependent mechanisms clear Chlamydia muridarum infections from the genital tract. J Immunol. 2012;188:1896–1904. doi: 10.4049/jimmunol.1102764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Morrison SG, Morrison RP. A predominant role for antibody in acquired immunity to chlamydial genital tract reinfection. The Journal of Immunology. 2005;175:7536–7542. doi: 10.4049/jimmunol.175.11.7536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Su H, Feilzer K, Caldwell HD, Morrison RP. Chlamydia trachomatis genital tract infection of antibody-deficient gene knockout mice. Infect Immun. 1997;65:1993–1999. doi: 10.1128/iai.65.6.1993-1999.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Saparov A, Kraus LA, Cong Y, Marwill J, Xu XY, Elson CO, Weaver CT. Memory/effector T cells in TCR transgenic mice develop via recognition of enteric antigens by a second, endogenous TCR. Int Immunol. 1999;11:1253–1264. doi: 10.1093/intimm/11.8.1253. [DOI] [PubMed] [Google Scholar]
- 87.Lee WT, Cole-Calkins J, Street NE. Memory T cell development in the absence of specific antigen priming. J Immunol. 1996;157:5300–5307. [PubMed] [Google Scholar]
- 88.Strutt TM, McKinstry KK, Kuang Y, Bradley LM, Swain SL. Memory CD4+ T-cell-mediated protection depends on secondary effectors that are distinct from and superior to primary effectors. Proc Natl Acad Sci U S A. 2012;109:E2551–2560. doi: 10.1073/pnas.1205894109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.