SUMMARY
Despite tremendous progress made toward the identification of the molecular circuitry that governs cell fate in embryonic stem cells, genes controlling this process in the adult hematopoietic stem cell have proven to be more difficult to unmask. We now report the results of a novel gain-of-function screening approach, which identified a series of 18 nuclear factors that affect hematopoietic stem cell activity. Overexpression of ten of these factors resulted in an increased repopulating activity compared to unmanipulated cells. Interestingly, at least four of the 18 factors, Fos, Tcfec, Hmgb1, and Sfpi1, show non-cell-autonomous functions. The utilization of this screening method together with the creation of a database enriched for potential determinants of hematopoietic stem cell self-renewal will serve as a resource to uncover regulatory networks in these cells.
INTRODUCTION
The mature cell contingent of adult hematopoietic tissue is continuously replenished during the life span of an animal by the periodic supplies from hematopoietic stem cells (HSCs) that reside in a niche. To maintain blood homeostasis, these primitive cells rely on two critical properties, namely multipotency and self-renewal. The former enables differentiation into multiple lineages, while the latter ensures preservation of HSC fate upon cellular division. By definition, a self-renewal division implies that an HSC is permissive to cell cycle entry, while restrained from engaging in differentiation, apoptosis or senescence pathways. The transcriptional regulatory network of HSC self-renewal still remains largely undefined, an observation that contrasts with that of embryonic stem cells (ESCs), for which self-renewal is increasingly dissected molecularly (reviewed in Jaenisch and Young, 2008). Only a few nuclear factors have been documented as promoters of HSC expansion (reviewed in Hai-Jiang et al., 2008). Of these factors, Hoxb4 and its derivatives (Hoxa9, NA10HD) are among the most potent and best documented (Ohta et al., 2007; Thorsteinsdottir et al., 2002).
The differential pace of progress propelling the fields of ESC and HSC research reflects, at least in part, seminal discoveries that rendered ESCs more amenable to large-scale experiments. First, although only observable as a transient state in vivo, ESCs derived from the inner cell mass can be maintained in vitro as cell lines by the addition of serum (as a source of bone morphogenic protein [BMP]) and leukemia inhibitory factor (LIF). Attempts to maintain or expand HSCs ex vivo as homogenous populations have been modest, and successful development of cell lines have not been reported, hampering harvest of large numbers of HSCs. Second, a stringent surrogate marker to follow the HSC multipotent state, comparable to the pluripotency tags of Oct4, Nanog, AP, or SSEA1 for ESCs, is still lacking. Albeit yielding small numbers, current cell sorting strategies allow isolation of HSC populations to near purity (Kiel et al., 2005). However, shortly upon facing the selective pressures of in vitro culture conditions, changes in cell phenotype are observed (Uchida et al., 2004), impeding HSC tracking in this context. The gold standard to confirm HSC activity for cells kept in culture remains the in vivo competitive repopulation assay. Importantly, generation of retroviral vectors provides highly efficient tools to infect and thereby modulate gene expression in both ESCs and HSCs (Root et al., 2006).
The demonstration that in mouse fibroblasts a given nucleocytoplasmic configuration, or state, can be reverted to a stem cell phenotype by the enforced overexpression of four defined nuclear factors, i.e., Oct4, Sox2, c-Myc, and Klf4, stands as a conceptual breakthrough (Takahashi and Yamanaka, 2006). Indeed, the ability to create induced pluripotent stem cells, or iPS cells, suggests that a putative role for as yet unidentified nuclear factors in orchestrating HSC fate is probable. With this mindset, interrogation of stem cell expression profile databases was undertaken. From this data set, a listing and ranking of nuclear factors whose transcripts were abundant in stem cell-enriched subpopulations was generated. Over 100 of the highest-scoring candidates were then functionally tested in HSCs, using a high-throughput overexpression in vitro to in vivo assay tailored to circumvent current limitations imposed by the biology of HSCs. As detailed below, these studies serve as a further step forward into the exploration of the molecular circuitry that governs HSC self-renewal.
RESULTS
Selection and Ranking of Candidate Genes
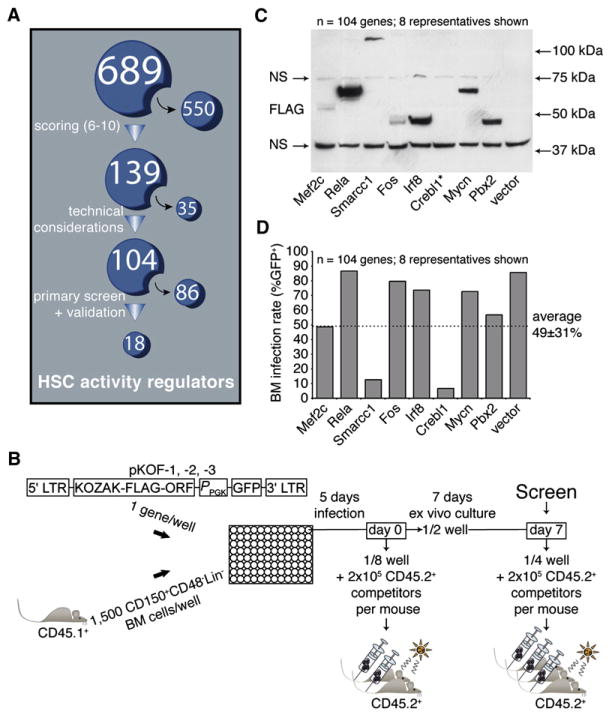
As a corollary of ESC studies, it can be proposed that HSC fate is also controlled by a series of master regulators analogous to Oct4 and several subordinate effectors, providing sound basis for the generation of a stem cell nuclear factors database. Toward this end, we created a database consisting of 689 nuclear factors (Figure 1A; Table S1 available online; see also http://www.bioinfo.iric.ca/self-renewal/) considered as candidate regulators of HSC activity. This list was predominantly derived from microarray gene expression profiling of normal and leukemia stem cells, including our recently generated FLA2 leukemia (1 in 1.5 cells is a leukemia stem cell, A.F. and G.S., unpublished data). Genes obtained after a review of the literature on stem cell self-renewal were also added to the list (see legend of Table S1 for references). Genes in this database were then ranked from 1 (lowest priority) to 10 (highest priority) on the basis of three factors: differential expression between primitive and more mature cell fractions (e.g., HSC-enriched), expression level (high levels were given highest priority), and the consistency of findings between data sets. Genes with a score of 6 and above (n = 139) were selected for functional studies, of which 104 were tested (Figure 1A; see also asterisks in Table S1). Interestingly, the list of selected candidates is highly enriched for bona fide regulators of hematopoiesis, namely Egr1, Gata2, Sfpi1 (PU.1), Foxo1, Meis1, Myb, Hoxa9, and Runx1 (Min et al., 2008; reviewed in Lessard et al., 2004). However, the majority of the other candidates have no reported function in primitive hematopoietic cells.
Figure 1. Experimental Design of Nuclear Factors Screening Strategy.
(A) A list of candidate genes (see Table S1 for the complete list) was generated as described in the Results. The 689 nuclear factors were subsequently ranked on the basis of an algorithm that stratifies them according to properties predictive of self-renewal regulation. The highest scoring candidates (n = 139) were further selected for functional assessment with a retroviral overexpression approach. Of these, 104 were tested (see “*” in Table S1), and the remaining 35 genes were excluded for technical reasons.
(B) The coding sequence of each tested candidate was subcloned into one out of three modified MSCV vectors, each containing a different reading frame (pKOF-1, -2 and -3). Respective retroviral producers were seeded in a single well of a 96-well plate and cocultured for 5 days with 1500 CD150+CD48−Lin− freshly sorted bone marrow CD45.1+ cells. Immediately upon infection (day 0), one-eighth of each well was transplanted into two congenic recipient mice along with 2 × 105 total BM cells (CD45.2+). A similar assay, this time with three recipient mice, was performed after an additional week of ex vivo culture (day 7), on which the screen was performed.
(C) Expression of candidate proteins in retroviral-producing cells was tested by western immunoblotting and revealed with an anti-FLAG antibody. A list of predicted and observed molecular weights for most proteins tested in this screen is available in Table S2. NS, nonspecific signal; *, example of a protein that could not be detected by western blot analysis (see also Table S2).
(D) Range of retroviral gene transfer efficiencies of sampled candidate genes on the basis of EGFP expression assessed at day 4 of HSC culture (only eight representatives shown; dashed line represents average on all 104 genes).
Design and Principle of the Screen
The screening protocol is outlined in Figure 1B. In brief, high-titer retroviruses were produced in 96-well plates seeded with viral producer cells using an optimized procedure. Protein extracts derived from producer cells in each of the 104 wells were analyzed by western blotting, which confirmed the presence of a FLAG protein in 89% of the cases (Figure 1C provides eight representative candidates; details for all 104 genes are listed in Table S2, sixth column), with 92% of these proteins showing the expected molecular size (Table S2, compare the fifth and sixth columns). CD150+CD48−Lin− mouse bone marrow (BM) cells were infected during 5 days and transplanted at two different time points (i.e., day 0 and day 7 in Figure 1B). Under these conditions, the average gene transfer to the cultured CD150+CD48−Lin− cells was at 49% ± 31% (Figure 1D provides eight representative candidates; details for all 104 genes are listed in Table S3, second column). Harvested cells from each well were transplanted into irradiated recipients together with 2 × 105 congenic BM cells. Donor-derived peripheral white blood cell reconstitution was assessed after short (4 and 8 weeks) and long (12 and 16 weeks) periods of time after transplantation.
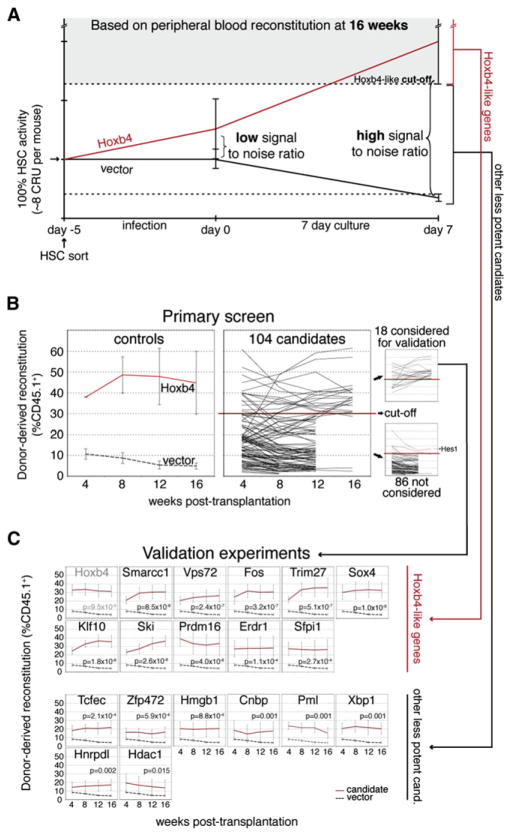
Previous results obtained from several in vivo transplantation experiments, using freshly transduced CD150+CD48−Lin− cells, revealed marked interrecipient heterogeneity in hematopoietic tissue reconstitution for a given candidate gene, thereby raising the critical issue of signal-to-noise discrimination. Optimization of this parameter was crucial for increasing the specificity of the screen while limiting to a minimum the number of mice that would be required. Toward this goal, we confirmed previous findings (Antonchuk et al., 2002) showing that the activity of Hoxb4-overexpressing HSCs is enhanced during short-term cultures (see red line in Figure 2A). For control (i.e., vector-transduced) HSCs, we also confirmed a noticeable decline in their activity during the 7 day culture (black line in Figure 2A). Interestingly, this HSC activity was preserved during the infection period (Figure 2A). Importantly, we found that recipients of these cultured control CD150+CD48−Lin− cells showed much less variation in blood cell reconstitution levels than those transplanted with “day 0” cells (compare error bars at day 0 versus day 7 on the black line in Figure 2A). In search for additional genes with Hoxb4-like activity, it appears that the signal-to-noise ratio is thus substantially enhanced by keeping cells in such cultures (Figure 2A). For this reason, the primary screen was performed with cells harvested at day 7 of the culture (see “Screen” in Figure 1B).
Figure 2. Identification of Positive Candidates with Hoxb4-like Activity.
(A) Estimation of HSC activity during viral transduction and culture. Lines show estimation of HSC activity in recipients of day −5, day 0, or day 7 bone marrow (BM) cells transduced with Hoxb4 (red) or control vector (black). Two independent experiments were performed with purified and whole bone marrow cells for Hoxb4, and three with purified BM cells for controls. Extrapolation of HSC numbers in recipients (y axis) was based on relative reconstitution levels observed in the CRU assay performed with our starting HSC subpopulation in Figures S2B–S2C. In all experiments, each mouse received eight equivalent day ~5 HSCs (representing 100% HSC activity and evaluated by CRU assay) and 20 equivalent fresh competitor HSCs (see legend of Figure S2 for details). Cutoff for identification of Hoxb4-like candidates used in the primary screen/ validation experiments (shaded area) is set on the basis of the standard deviation/error of the mean reconstitution level observed in multiple recipients of Hoxb4-transduced HSCs. Cutoff for other less potent candidates (area between dotted lines) is used only in validation experiments and is based on statistical difference between candidates and vector control.
(B) Graft-derived hematopoiesis was evaluated at 4 week intervals in recipients of cultured HSCs during the primary screen. As a set of reference values, the left panel indicates peripheral blood reconstitution levels from mice transplanted with cells from cultures initiated with a positive regulator of self-renewal (Hoxb4) in relation to values observed with control vectors (mean of pKOF-1, -2 and -3), all after 7 days of culture. Mean reconstitution level values for each of the 104 tested candidates (at day 7) are compiled and presented in the middle panel, with the established cutoff level for a gain-of-function readout. Candidates clustering above the cutoff level for identification of Hoxb4-like genes (shaded area in Figure 2A), corresponding to 30% CD45.1+ donor-derived cells in the primary screen, were selected for the validation experiments (upper-right panel), while those below were disregarded (lower-right panel). One candidate (Hes1) was eliminated on the basis of the marked reduction in repopulation noted between early and late time points (upper line in lower-right panel). Values are presented as mean ±SEM of independent experiments (n) for the left panel (n = 2 for Hoxb4 and n = 3 for control vector; mean of three mice per experiment) and as mean ±SD for the middle and right panels (n = 3 mice for each candidate cDNA). Note that several mice were eliminated at 12 or 16 weeks after transplantation because they did not meet our criteria for hit selection (see also Table S3, ninth and tenth columns).
(C) Validation experiments confirming ten Hoxb4-like genes and eight other less potent candidates. p values were established at the 16 weeks after transplantation time point. Values are shown as mean ±SEM. The number of independent experiments (n) per candidate gene equals four, except for control vectors (n = 8); Sox4, Zfp472, Xbp1, and Hnrpdl (n = 5); and Hoxb4, Cnbp, Ski, and Prdm16 (n = 3). For each experiment, a mean of three mice per gene was evaluated. cand., candidates.
Primary Screen and Validation
The minimal cutoff level for selection of positive candidates in the primary screen was set on the basis of the standard deviation of the mean reconstitution level observed in multiple recipients of Hoxb4-transduced CD150+CD48−Lin− cells (Figure 2B, see also shaded area in Figure 2A). We therefore expected the newly identified candidates to be equivalent to, or more potent than, Hoxb4 in inducing enhanced HSC activity. With this criterion, a total of 18 hits were identified for a frequency of 17% (18/104; Figure 2B, upper-right panel; see also Table S3, tenth column). These 18 hits included Cnbp, Erdr1, Fos, Hdac1, Hmgb1, Hnrpdl, Klf10, Pml, Prdm16, Sfpi1 (PU.1), Ski, Smarcc1 (Baf155), Sox4, Tcfec, Trim27, Vps72, Xbp1, and Zfp472.
To validate the primary screen, we repeated the procedure described in Figure 1B in several independent experiments and confirmed ten primary hits (upper panels in Figure 2C; details in Table S3). From left to right and top to bottom, genes are presented on the basis of the level of statistical significance reached in these experiments, in comparison to control vector: Hoxb4 (positive control, gray in Figure 2C), Smarcc1, Vps72, Fos, Trim27, Sox4, Klf10, Ski, Prdm16, Erdr1, and Sfpi1, for a positive predictive value (PPV) of 56%. Although the other eight candidates failed to demonstrate Hoxb4-like activity (i.e., shaded area in Figure 2A), it is noteworthy that they all significantly enhanced HSC activity to level above that detected with vector-transduced cells (see “other less potent candidates” in Figures 2A and 2C).
We then explored whether the ten newly identified Hoxb4-like genes are endogenously expressed in populations highly enriched in HSCs. To test this, we sorted cells from hematopoietic tissues isolated from unmanipulated mice sacrificed at three different developmental stages including E14.5 fetal liver (HSC-enriched subset number 1) and postnatal bone marrow, where a switch between cycling (3 weeks, subset number 2) and quiescent (4 weeks, subset number 3) HSCs has been described (Bowie et al., 2006). In line with the selection of these factors in our database, all ten genes are highly expressed in HSC-enriched populations with endogenous levels exceeding those of TATA binding protein (TBP), used as endogenous control (see relative threshold cycle [Ct] values for all HSC-enriched subsets in Figure S1A). Interestingly, most of these genes are expressed at higher levels in HSC-enriched populations than in total tissues (total bone marrow or fetal liver, Figure S1B). This observation is most prominent in the fetal liver-derived HSC-enriched subset (upper panel Figure S1B). We next verified the level of overexpression achieved for each of these genes after retroviral infection in sorted CD150+CD48−Lin−cKit+Sca1+ BM cells and documented relative increases in mRNA levels that could be as low as 3-fold (e.g., Smarcc1) to as high as 1000-fold and above (e.g., Fos; Figure S1C).
Most Genes Identified Confer Enhancement in HSC Activity
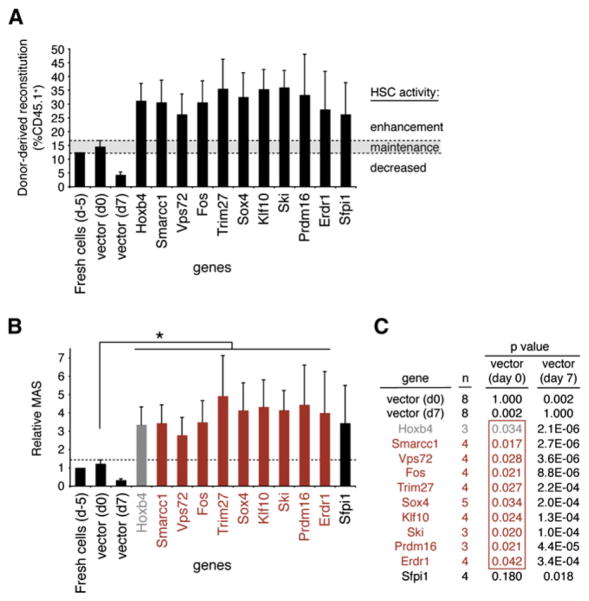
One important question is whether the newly identified genes enhance or simply maintain input stem cell activity. To address this point, we compared the reconstitution levels by donor cells isolated (at day 7) from test cultures to that observed in recipients which received an equivalent number of freshly purified CD150+CD48−Lin−cells. As shown in Figure 3A, baseline long-term HSC (LT-HSC) activity (Sauvageau et al., 2004), measured at ~12%–16% reconstitution level (see left black bars in Figure 3A), was essentially preserved during the 5 day infection period and set the baseline values for maintenance of input HSC activity (shaded area in Figure 3A). From this, we could determine that all validated hits conferred a net increase in HSC reconstitution activity above that determined for fresh cells (Figure 3A).
Figure 3. HSC Activity Is Enhanced by Overexpression of the Newly Identified Hoxb4-like Genes.
(A) Percentages of donor-derived blood cells at 16 weeks after transplantation in primary mice recipients of day 7 culture cells (experiments in Figure 2C) for the ten identified hits. Bars at the far left show values for freshly purified CD150+CD48−Lin− cells, day 0 control cells (empty vector), and day 7 control and Hoxb4-transduced cells, as references. The shaded area represents mean reconstitution levels observed in mice transplanted with day 0 control-transduced HSCs, ±1 SEM. Reconstitution values falling within this range are considered to reflect HSC activity derived from an injected dose of HSCs equivalent to culture input numbers, i.e., 8 competitive repopulation units (CRUs) (see Figure S2C). d-5, day −5; d0, day 0; d7, day 7. Note that two different cDNA were tested for Trim27 (see Figure S5).
(B) The mean activity of stem cells (MAS) was calculated from the data obtained in (A). As previously established, the MAS is equivalent to repopulation units (RUs) divided by CRUs (Ema and Nakauchi, 2000), where RU represents the donor-derived reconstitution level divided by the competitor-derived reconstitution level. MAS index was normalized to 1 for freshly sorted cells and excluded the gene transfer efficiency in its estimation for the candidate genes. In cases where gene transfer is low, this could lead to underestimated values. Grafts overexpressing genes shown in red and marked by an asterisk give rise to a MAS significantly higher than day 0 control grafts (p ≤0.05). Note that values are presented as the MAS relative to that of fresh cells (d −5), which was set at a value of 1. d-5, day −5; d0, day 0; d7, day 7.
(C) p values derived from MAS values presented in (B) comparing data obtained from control vectors (day 0 or 7) and validated genes. Day 0 p values framed in red (penultimate column) mirror the order and color code of candidate genes presented in (B) and are ≤0.05. In comparison, day 7 p values are listed in the adjacent column.; n, number of independent experiments; d0, day 0; d7, day 7.
In order to further quantitate the impact of our validated genes on HSC activity, we used the mean activity of stem cell (MAS) index as reported by Ema and Nakauchi (2000). This value, which provides a measure of the proliferative output per LT-HSC, is easily applicable to our culture condition since they were all initiated with a constant number of input LT-HSC measured at ~62 competitive repopulating units (CRU) per well (see Figures S2B–S2C for CRU assessment). The MAS was 3-fold higher for Hoxb4-transduced cells than for controls (Figure 3B). Similarly, the MAS varied between 2.8 (Vps72) and 4.9 (Trim27) for all ten hits identified in our screen (Figure 3B). Except for Sfpi1 (p = 0.18), all values reached statistical significance (see p values in red in Figure 3C).
Impact of Candidate Genes on Cell Proliferation, Death, and Differentiation In Vitro
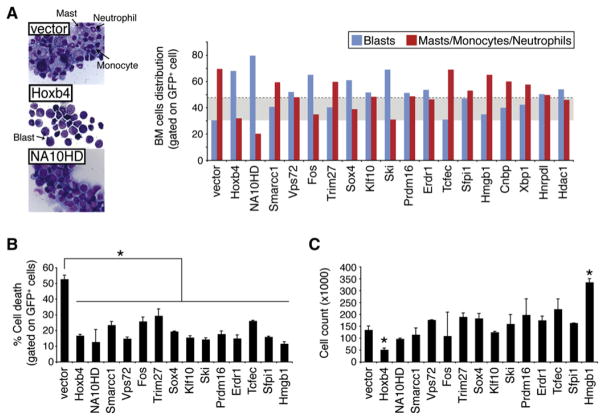
There is growing evidence to suggest that HSC self-renewal involves the active repression of a differentiation program, which is coupled to cell division (Cellot and Sauvageau, 2007). In support of this, we recently found that Hoxb4- or NA10HD-transduced cells, which actively undergo in vitro self-renewal divisions, show evidence of differentiation arrest (Figure 4A, left panels) (Cellot et al., 2007). We investigated whether our newly identified hits behave similarly. To achieve this, we first analyzed the cytological characteristics of transduced and sorted CD150+CD48−Lin− cells after a 7 day in vitro culture period (prior to their transplantation). In this context, cultures initiated with control vector-transduced cells contained 70% ± 8% of differentiated cells. These included neutrophils, monocytes, and mast cells (Figure 4A, arrows in upper-left panel with summary of results in right panel). Conversely, cellular differentiation was reduced in cultures initiated with HSCs transduced with most of the candidates (Figure 4A, right panel). The increase in the proportion of undifferentiated to differentiated cells was most important for Vps72, Fos, Sox4, Klf10, Ski, Prdm16, Erdr1, and Sfpi1 when compared to cultures initiated with control vector-transduced HSCs (see blue bars exceeding shaded area, representing mean of vector plus two standard deviations). Note that while some of these genes are as potent as Hoxb4 in this assay, none exceeds NA10HD in keeping cells undifferentiated.
Figure 4. In Vitro Differentiation, Survival, and Proliferation Profiles of Cultured Cells.
(A) Morphological analysis of cytological preparations of the starting HSC fraction overexpressing most of identified primary hits at day 7 of culture. Proportions of immature (blasts: black arrow in middle-left insert) versus terminally differentiated cells (neutrophils, monocytes and masts cells: black arrows in upper-left insert) for respective cultures are depicted in the right panel. A field comprising 100 cells was examined per independent experiment (n), and values are presented as mean; n = 3, except for vector (n = 6) and Hoxb4, Ski, Tcfec, Sfpi1, and Hmgb1 (n = 1). The shaded area represents the mean of vector plus two standard deviations. Note that a vast majority of the less significant hits described in Figure 2C are included in this study (i.e., Tcfec, Hmgb1, Cnbp, Xbp1, Hnrpdl, and Hdac1). BM, bone marrow.
(B) Annexin V and PI stains were used to determine the fraction of cell death in cultures initiated with our validated hits and the less potent gene candidates (Tcfec and Hmgb1) as well as empty vector, Hoxb4, or NA10HD, serving as controls. Assessment was performed at the day 4 time point of culture. Values are presented as mean ±SD of two independent experiments (n), where n represents the analysis of 30,000 cells. * p ≤ 0.05.
(C) Cell proliferation was assessed by flow cytometry-based cell counts (y axis) at day 4 of cultures. Analyses were performed for the same gene candidates as in (B). Values are presented as mean ±SD of two independent experiments. * p ≤ 0.05.
We also monitored transduced CD150+CD48−Lin− cells for cell death and proliferation. Figure 4B shows that all factors analyzed conferred a significant reduction in the proportion of dead cells harvested at day 4 of culture. Surprisingly, besides Hmgb1, there were no significant increases in total cell numbers in these cultures (Figure 4C). In fact, some genes such as Hoxb4, and possibly NA10HD, were associated with a reduction in total cell counts (Figure 4C).
Transduced HSCs Differentiate In Vivo
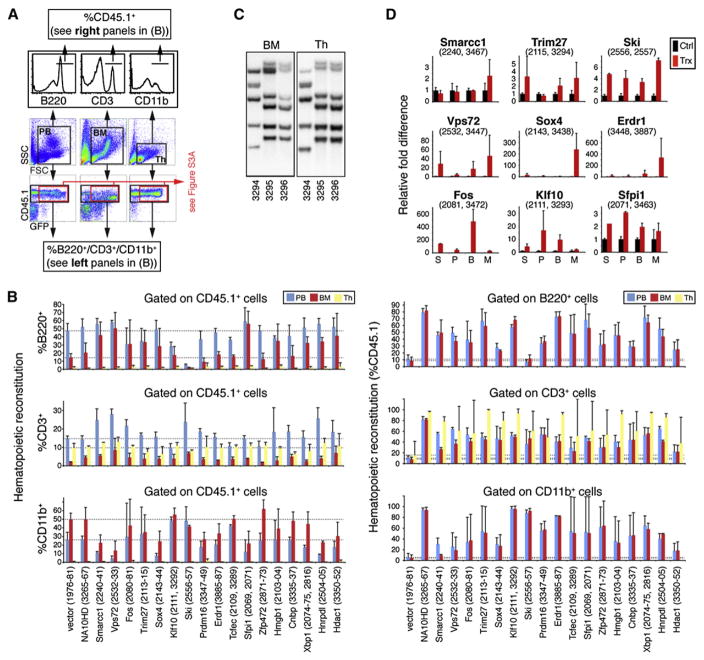
The in vitro differentiation arrest displayed by Hoxb4- or NA10HD-transduced HSCs is eventually reverted after their transplantation in vivo (Cellot et al., 2007; Ohta et al., 2007). Thus, depending on the environment, these two genes can either interfere with (e.g., in vitro in the presence of high levels of growth factors) or not affect (e.g., in vivo under steady state conditions) HSC differentiation. To determine whether the newly identified regulators of HSC activity are similarly permissive to HSC differentiation in vivo, we used four different approaches. First, we evaluated the general health, spleen size, and bone phenotype (white versus red) of each recipient. Except for the recipients of Prdm16-transduced cells, which eventually developed splenomegaly and myeloproliferation with white femurs at 20 weeks after transplantation (data not shown), none of the mice transplanted with cells expressing our nine other genes ever presented this or any other hematological phenotype. Second, we performed cytological evaluation of bone marrow and spleen derived from representative mice for each gene. Results from these analyses were normal for all groups, except for the Prdm16 cohort, which showed an excess of poorly differentiated myeloid cells in their bone marrow, as previously determined by others (Shing et al., 2007), and the Ski cohort, in which the number of lymphocytes in the bone marrow was reduced (data not shown). Besides recipients of Prdm16-transduced cells, spleens were never infiltrated with myeloid cells, nor did they include enhanced numbers of erythroblasts. To confirm this, we devised a third approach consisting of flow cytometry analysis on donor-derived (CD45.1+) cells. The results, presented in Figure 5A for the peripheral blood, bone marrow, and thymus of a representative mouse (Trim27) and summarized in Figure 5B for all groups, largely confirmed our cytological evaluation. Indeed, except for recipients of Ski-transduced cells, which showed a marked reduction in B lymphocytes in their peripheral blood and marrow with a compensatory increase in other cell types, most groups of mice either showed normal distributions of various cell types or presented some minor variations. We further extended this analysis by gating only on CD45.1+/GFP+ cells for genes in which this was possible and ended with the same conclusions, with the exception that B cell differentiation was not observed in Klf10-transduced cells (Figure S3A). Finally, clonal analyses of recipients that were reconstituted with retro-virally marked cells were performed on bone marrow (less than 5% T cells) and thymus (less than 5% non-T cells). A representative result is presented in Figure 5C for Trim27, showing that identical clones contributed to the reconstitution of these two tissues, thus reinforcing the finding that these transduced HSCs remain competent in T cell differentiation although they displayed enhanced reconstitution activity. This finding can be extended to all other genes except for Ski, Prdm16, and Erdr1, for which we cannot be certain that the same clone contributed to thymic and bone marrow reconstitution (Figure S3B).
Figure 5. In Vivo Differentiation Potential of HSCs Overexpressing Validated Hits.
(A) In vivo differentiation potential along the lymphomyeloid lineages was assessed in long-term recipients (20 weeks after transplantation) of HSCs transduced with Trim27, used as an example. Immunophenotypic analysis by flow cytometry was performed with specific antibodies against B, T, and myeloid cell surface markers (B220, CD3, and CD11b, respectively) on the CD45.1+ population of cells derived from the peripheral blood, bone marrow (BM), and thymus of these mice (and on CD45.1+/GFP+ cells in Figure S3A).
(B) Compilation of B-/T-lymphoid and myeloid cell percentages within the CD45.1+ population (left panels) and of CD45.1+ cell proportions in the B-lymphoid, T-lymphoid, or myeloid populations (right panels) of blood, thymus, and BM tissues, as gathered in (A), for most of the primary hits. Values are presented as mean ±SD. Mouse identification numbers are presented between brackets. Dashed lines indicate values obtained with control mice (vector) for the different hematopoietic tissues. Note that multilineage reconstitution was observed in all of mice analyzed except for those transplanted with cells transduced with Ski, which showed a block in B-lymphoid differentiation. The proportion of CD45.1+ cells within the B-lymphoid or myeloid populations in the thymus was not calculated in the right panels because these populations represent less than 5% of total cells in this tissue. Since weak donor-derived reconstitution levels were observed in mice transplanted with control vector cells at day 7 of culture (i.e., 4% in Figure 3A), we used mice transplanted with control vector cells at day 0 of culture in this case. Values pertaining to less statistically significant hits are also presented (Tcfec, Zfp472, Hmgb1, Cnbp, Xbp1, Hnrpdl, and Hdac1).
(C) Proviral integration pattern studies by Southern blot analysis performed on genomic DNA extracted from bone marrow (left panel) and thymus (right panel) of three long-term recipients of Trim27-overexpressing HSCs. Each lane represents a specific mouse (ID number below). For a given animal, identical integrations are found in both tissues, indicating a common precursor cell origin. Identical clones are also observed in two distinct recipients, retrospective molecular evidence for a self-renewal division in vitro. Multipotentiality (BM and thymus) analyses in reconstituted tissues harvested from primary recipients of culture cells overexpressing the various validated hits are shown in Figure S3B.
(D) Quantitative RT-PCR analysis of mRNA expression levels of nine of the ten newly identified Hoxb4-like gene transcripts. For technical reasons, Prdm16 was excluded from the analysis. RNA was extracted from distinct bone marrow cell fractions isolated from long-term, highly reconstituted recipients of day 7 culture cells (S, CD150+CD48−Lin−; P, CD150+CD48+Lin−; B, B220+; M, Mac+). Average ΔCt values (representative of expression levels) were determined with β-actin serving as an endogenous control to normalize levels of target gene expression. Relative fold differences were determined using control cells as a reference calibrator for each candidate gene. Reactions were done in triplicate; black bars represent mean endogenous expression levels ±SEM of three independent wild-type mice (Ctrl); red bars represent mean overexpression levels ±SEM of two transplanted mice (Trx) from two independent experiments. The identification numbers of mice used in this assay are presented between brackets. Note that control black bars on each panel are set at a relative fold difference value of 1.
We also used quantitative RT-PCR assays to monitor overexpression levels of the different transgenes in CD150+CD48−Lin−, CD150+CD48+Lin−, B, and myeloid cells isolated from bone marrow to verify whether the in vivo reversal of the differentiation arrest noted in vitro (as shown in Figure 4A) was associated with loss of transgene expression. Results presented in Figure 5D suggest that overexpression of Vps72, Fos, Sox4, Klf10, Ski, Erdr1, and Sfpi1 does not interfere with in vivo differentiation as several types of immature/differentiated cells still express the transgene. However, in vivo extinction of the transgene may have occurred for the three following genes: Smarcc1, Trim27, and Prdm16.
Together, these results confirm that the majority of the Hoxb4-like genes identified in our screen conferred enhanced HSC activity without causing hematological diseases or profoundly altering cell differentiation, while still overexpressing the transgenes at least until 20 weeks after transplantation. Prdm16 was a notable exception.
Evidence of Self-Renewal Divisions by Transduced HSCs
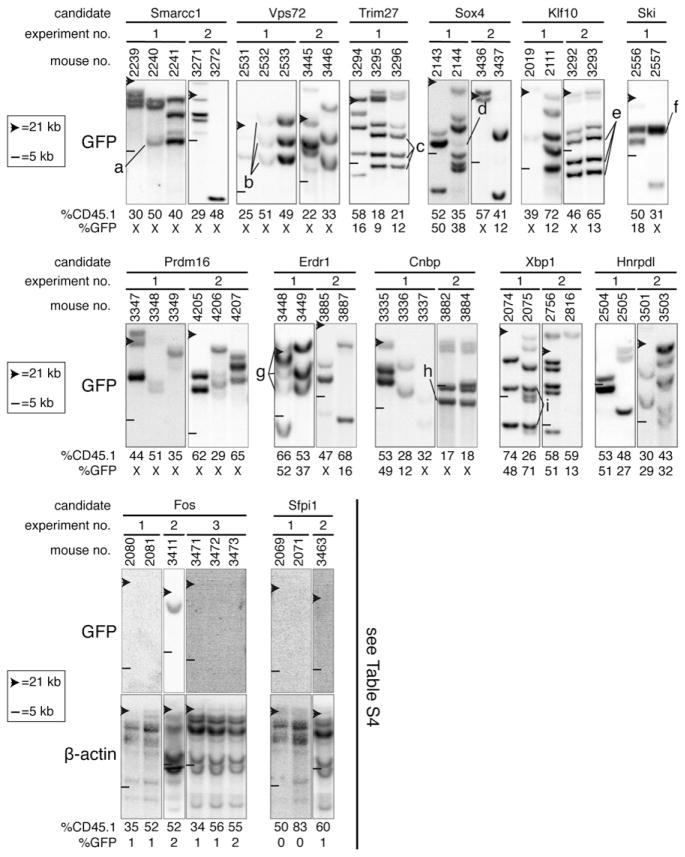
We next verified whether HSCs transduced with each of the confirmed hits remained capable of symmetrical self-renewal divisions in vitro. To address this, we performed clonal analysis (i.e., proviral integration pattern) of hematopoietic tissues derived from selected recipients that were highly reconstituted (17%–83% CD45.1+ cells) at 20 weeks after transplantation, a time point deemed sufficient for inferring that reconstitution is solely derived from the LT-HSC. For eight of the ten newly identified Hoxb4-like genes, namely Smarcc1, Vps72, Trim27, Sox4, Klf10, Ski, Prdm16, and Erdr1, we observed proviral DNA in the vast majority of mice that were analyzed (i.e., 43/49; Figure 6, two upper panels). Several different clones with long-term reconstitution ability contributed to hematopoiesis among different recipients from a given experiment, and from subsequent validation experiments. In several instances, we could identify the same proviral integrations in the DNA from two different mice reconstituted by cells derived from the same culture, demonstrating that LT-HSC self-renewal has indeed occurred in these cultures (see “a”–”g” in Figure 6). We also performed this analysis with the less potent candidates and found evidence for HSC self-renewal for Cnbp and Xbp1 (see clones “h” and “i” in Figure 6). Together, these data indicate that a significant proportion of HSCs engineered to overexpress our validated hits remained capable of symmetrical self-renewal divisions in vitro.
Figure 6. Clonal Analysis to Track HSC Self-Renewal Divisions.
Southern blot analyses of genomic DNA extracted from the bone marrow (BM) of selected long-term recipients (20 weeks after transplantation) of day 7 culture cells. Each blot was hybridized with a GFP-specific probe and systematically exposed for the same period of time. Each well was equally loaded, taking into consideration donor-derived peripheral blood reconstitution level of recipients as measured 16 weeks after transplantation (i.e., 17%–83% CD45.1+ cells). Top and bottom panels show gene sets for which proviral DNA was either detected or absent in the majority of recipients analyzed, respectively. For most bottom panels, the brightness and contrast was enhanced using Photoshop to further verify the absence of integrated proviral DNA, and a β-actin probe was also used to confirm the presence of genomic DNA. Clones that self-renewed during the 7 day culture, prior to transplantation, are labeled “a” to “i” and identified in more than one recipient. “X” indicates that GFP expression was not detectable for these constructs/clones. Note that panel 5, representing the Trim27 gene, is also included in Figure 5C (left panel).
Evidence of a Non-Cell-Autonomous Activity for Selected Genes
Surprisingly, we could not reveal any integrated provirus in the majority of recipients transplanted with cells transduced with the Hoxb4-like genes Fos and Sfpi1 (Figure 6, bottom panels). This suggests that untransduced HSCs in these cultures have favorably responded to some extrinsic factors. A detailed evaluation of recipients from which these observations are derived is provided in Table S4. Consistent with the presence of a non-cell-autonomous effect, we observed that upon exposure to Fos or Sfpi1 viral producer cells, HSC contribution to long-term repopulation increased from 5%–6% after 5 days of infection (i.e., day 0) to 26%–31% with 7 additional days of culture (Table S4, compare %CD45.1 at day 0 [seventh column] to that at day 7 [eighth column]). The absence of Fos and Sfpi1 proviruses in the hematopoietic system of long-term recipients is surprising considering the high level of gene transfer to transplanted cells (Table S4, third column). This possibly indicates that these genes also intrinsically interfere with HSC repopulation when overexpressed, leading to depletion of transduced HSCs. Similar observations were found with Tcfec and Hmgb1, members of the less potent category (data not shown).
To provide more-direct evidence of non-cell-autonomous activities for Fos, Sfpi1, Tcfec, and Hmgb1, we transduced non-viral-producing NIH 3T3 cells with each of these constructs and used them as feeder cells replacing the viral producers described in Figure 1B. Overexpression of each of these four factors in NIH 3T3 cells was verified by quantitative RT-PCR, with a relative fold difference above baseline values ranging from 3-fold (Hmgb1) to 18,000-fold (Tcfec) (data not shown). As per our experimental protocol, 1500 CD150+CD48−Lin− cells were seeded on non-viral-producing cells and maintained for 7 days prior to their transplantation into irradiated hosts. Strikingly, we found that three of the four genes, namely Fos, Tcfec, and Hmgb1, conferred a similar impact on HSC activity whether viral-producing or non-viral-producing cells were present in the cultures (see Table 1). Interestingly, background-level reconstitution was found in recipients of cells kept on Sfpi1-transduced NIH 3T3 cells, possibly indicating a more complex cellular network involved with this gene.
Table 1.
HSC Activity Is Enhanced Extrinsically
| GP+E86 (virus)
|
NIH 3T3 (no virus)a
|
|||
|---|---|---|---|---|
| %GFP | %CD45.1 | %GFP | %CD45.1 | |
| vector | 62 ± 26 | 4 ± 1 | 0 ± 0 | 5 ± 2 |
|
| ||||
| Fos | 71 ± 9 | 31 ± 8 | 0 ± 0 | 35 ± 14 |
|
| ||||
| Sfpi1 | 37 ± 17 | 26 ± 12 | 0 ± 0 | 2 ± 1 |
|
| ||||
| Tcfec | 48 ± 21 | 22 ± 8 | 0 ± 0 | 13 ± 5 |
|
| ||||
| Hmgb1 | 22 ± 14 | 20 ± 10 | 0 ± 0 | 14 ± 5 |
Donor-derived reconstitution levels (%CD45.1+) observed 16 weeks after transplantation in mice transplanted with cells cocultured either with virus-producing cells (GP+E-86) or similar cells not producing virus (NIH 3T3), both overexpressing selected genes. Values are presented as mean ±SEM of independent experiments (n) per candidate gene, where n = 4, except for control vector, where n = 8. For each independent experiment, a mean of three mice per gene was evaluated.
Mice were analyzed at 16 (experiments 1 and 2) and 8 (experiments 3 and 4) weeks after transplantation; %GFP, gene transfer assessed by flow cytometry.
DISCUSSION
In this study, we combined the power of expression profiling and functional studies to uncover candidate factors that impact on HSC activity. Our experimental procedure included an efficient production of high-titer retroviruses, a sharp and discriminating HSC gain-of-function signal optimized by an ex vivo culture step together with a robust and reliable in vivo assay (see importance of culture step for signal-to-noise discrimination in Figure S4). Using this strategy, we individually tested 104 preselected candidates revealing 18 genes that conferred a clear repopulation advantage to HSCs. Of these, ten displayed a Hoxb4-like effect, thereby greatly extending the repertoire of potential regulators of HSC activity. These factors, which are highly and in some case preferentially expressed in HSCs, are implicated in a diversity of processes, such as chromatin modification (e.g., Smarcc1 and Vps72), stress response (e.g., Fos), and gene transcription (e.g., Trim27 and Klf10) (see Table S4 and legend for more details on these Hoxb4-like factors). With this significant cohort of genes impacting HSC activity, it was possible to discern a subset of four factors (i.e., Fos, Tcfec, Sfpi1 and Hmgb1) that exerted their influence on stem cell function through a non-cell-autonomous phenomenon. Importantly, the non-cell-autonomous activity of three of these four factors was confirmed in gene transfer-free conditions.
Identified Hits Display Hoxb4-like HSC Activity
Similar to our previous results with Hoxb4-overexpressing HSCs, nine of the ten hits identified in this screen also significantly conferred increased HSC activity to levels above those observed with input cells (genes in red in Figures 3B–3C). Considering the ~50% average gene transfer level, not taken into consideration in the evaluation of the mean activity of stem cells (MAS) reported in Figure 3B, the impact of some of our confirmed hits was likely underestimated. Interestingly, our newly identified candidates also induced a maturation block in vitro, which was reversed in vivo. Moreover, and similar to Hoxb4, the majority of these genes failed to enhance cellular proliferation in vitro.
The increase in HSC repopulation potential (i.e., activity) observed with Hoxb4 overexpression involves a net expansion (self-renewal) of these cells (Antonchuk et al., 2002). Although our validated candidates show Hoxb4-like effects and we could document self-renewal divisions in vitro for several HSCs engineered to express these genes (Figure 6), it is important to stress that the formal proof for HSC expansion (enhancement in self-renewal divisions) as the driving force behind these effects will require intensive investigations that combine serial determinations of HSC numbers at consecutive time points (e.g., ΔCRU assay) with large-scale clonal analyses as previously reported for the Hoxb4 gene (Antonchuk et al., 2002; Cellot et al., 2007). Indeed, among several possibilities, enhancement in proliferative potential combined with in vitro HSC maintenance could explain the Hoxb4-like effects observed with our validated hits. Secondary transplantation assays performed with selected candidates (Vps72, Klf10, Erdr1, and Fos, Table S5) argue against, but do not refute, such a possibility.
The eventual identification of shared target genes between these factors would further strengthen this argumentation. Of interest, attempts to build transcriptional networks, or hubs, among the hit genes have currently proven inconclusive, when most of the respective mRNA expression levels were assessed in HSCs freshly transduced with each of the candidate genes (see http://www.bioinfo.iric.ca/self-renewal/Network/).
Non-Cell-Autonomous Enhancement in HSC Activity
The non-cell-autonomous influence exerted by Fos, Tcfec, and Hmgb1 was confirmed in cultures with NIH 3T3 support, confirming that gene transfer to sorted HSCs was not required for these three genes. In line with these findings, the proportion of GFP-positive cells was 0% in recipients of cells kept for 7 days in NIH 3T3-containing cultures (see column four in Table 1). It is also important to stress that these so-called “non-cell-autonomous” factors were identified in a context of overexpression, which in some cases (i.e., Fos) was outstanding (e.g., 38,000-fold above endogenous levels in some experiments, data not shown), potentially resulting in toxicities to transduced HSCs. It is therefore likely that several of these factors normally perform cell-autonomous activity in steady-state hematopoiesis. Sfpi1 is a notable example for this (Rosenbauer et al., 2006).
The results obtained with Sfpi1 during the screen and several additional experiments could not be confirmed in viral-free cultures. Reasons for this remain unclear. Several possibilities exist, including the presence of non-cell-autonomous contribution by some differentiated hematopoietic cells found in our culture or by essential cofactors unique to GP+E-86 viral producer cells.
These results thus raise critical questions about a possible, and even essential, contribution by the feeder cells (in this case NIH 3T3) to the observed effects on HSC activity. While this level of characterization is beyond the scope of our paper, this set of experiments exposes a complexity, often underestimated, when packaging (feeder) cell lines are used for cocultures in gene transfer studies. Awareness of this potentially confounding factor must therefore be taken into consideration in future experiments.
Although the demonstration of the non-cell-autonomous activity is relatively straightforward (e.g., three of the four factors described above), the proof for cell-autonomous function (e.g., the other newly identified factors) is more difficult to obtain within the scope of our experimental design. Thus, it is possible that some of the eight other Hoxb4-like genes (Smarcc1, Vps72, Trim27, Sox4, Klf10, Ski, Prdm16, and Erdr1) also or only display non-cell-autonomous activity.
Future of the Resource
Entries pertaining to candidate nuclear factors implicated in stem cell activity are stored in an open-access database (http://www.bioinfo.iric.ca/self-renewal/), as part of a building block in creating an interactive resource for the scientific community. Information thus far includes mRNA expression profiles, together with specific probe and primer sets, blood reconstitution patterns over time with possibilities of data reanalysis, and the complete nuclear factor list with ranking criteria. This database will eventually include similar data emerging from ongoing gain- and loss-of-function screens from different laboratories that will exploit this resource.
Thus far, nuclear factor candidates clustering in the uppermost levels (eight to ten) of our classification harbor a 23% primary hit rate, compared to 12% for those scoring lower (six to seven). This suggests that while positive HSC regulators tend to segregate in the top ranks, other downstream genes listed in our database (scores 1–5, n = 585 candidates) also include potential determinants of HSC self-renewal, with putative derivation of cross-regulation nodes as mentioned.
Our understanding of HSC self-renewal will mirror advancements in the rapidly evolving field of cellular therapy. As pioneered with Hoxb4 (Krosl et al., 2003a), recombinant TAT-fusion proteins could be envisioned and therapeutically tested on human cells. Moreover, findings from this work will potentially pave the way to identify the mediator(s) that link these non-cell-autonomous candidates to enhanced HSC activity, thereby bypassing the requirement for retroviral vectors in the goal of expanding HSCs for clinical purposes.
EXPERIMENTAL PROCEDURES
Retroviral Vectors
MSCV-Hoxb4-IRES-GFP and MSCV-NUP98-HOXA10HD-IRES-GFP (NA10HD) were previously described (Antonchuk et al., 2001; Ohta et al., 2007). For all candidate genes, single open reading frames (ORFs) were amplified by PCR (see Table S2 and the Supplemental Experimental Procedures for details).
Animals
(C57Bl/6J-CD45.2 x C3H/HeJ) F1 recipient mice and (C57Bl/6J-CD45.1-Pep3b x C3H/HeJ) F1 congenic donor mice were bred at a specific pathogen-free (SPF) animal facility at IRIC in Montreal.
CRU Assay on HSC-Enriched Populations
CRU assay and calculation were performed as described originally (Szilvassy et al., 1990) with modifications as in Sauvageau et al. (1995). Recipients were considered reconstituted (i.e., positive) when ≥1% of their peripheral blood leukocytes were of donor (CD45.1+) origin at 18–20 weeks after transplantation. Two CRU assays were performed with CD150+CD48−Lin− cells and one CRU assay for the CD150+CD48−Lin−cKit+Sca1+ subpopulation.
Bone Marrow Cell Culture, Retroviral Infection, and Transplantation
Generation of retrovirus-producing GP+E-86 cells, or gene-overexpressing NIH 3T3, were performed as previously described (Krosl et al., 2003b) and seeded in a 96-well plate format, enabling production of a single ecotropic pseudotyped retrovirus per well (for GP+E-86). See the Supplemental Experimental Procedures for details.
Flow Cytometry Assessment of Donor-Derived Hematopoiesis
The contribution of donor cells to peripheral blood reconstitution was determined at regular intervals after transplantation in individual recipients. See the Supplemental Experimental Procedures for details.
Analysis of Proliferation and Cell Death
Transduced cells harvested at day 4 of culture with trypsinization were counted with BD Trucount™ Tubes (BD Biosciences, San Jose, CA) according to manufacturer’s guidelines, or stained with Alexa350-annexin V (Invitrogen Molecular Probes, Eugene, OR) and propidium iodide (50 μg/ml) in accordance with the manufacturer’s instructions. Individual well contents were analyzed by flow cytometry. Gates were set to exclude GP+E86 retroviral producers by forward- and side-scatter criteria.
Resource Database and Web Application
The open-source postgreSQL relational database engine was used to store the accumulated data, and the web application was built with the open-source Webware application server framework. The application is designed to house results from other ongoing screens from our lab as well as screens performed elsewhere. As such, a sections-based authentication system has been implemented in order to restrict access to sensitive or nonpublished results. We expect this website to become an open-access resource for teams working on deciphering the molecular basis for stem cell self-renewal.
Statistical Analysis
The significance of differences was determined by a two-tailed Student’s t test.
Supplementary Material
Acknowledgments
The authors thank J. Krosl, R. Bisaillon and B.T. Wilhelm for technical help, C. Charbonneau from IRIC imagery platform, D. Gagné from IRIC flow cytometry platform, and P. Chagnon and R. Lambert from IRIC genomic platform for their expertise with quantitative RT-PCR. We also want to acknowledge C. Perreault, M. Therrien, K.J. Hope, and R.K. Humphries for discussions and critical comments about the manuscript. This work was supported by the Canadian Institute of Health Research (CIHR) Team Grant in Hematopoietic Stem Cell Self-Renewal: From Genes to Bedside (Grant number 154290, 2006–2011). G.S. holds a Canada Research Chair on molecular genetics of stem cells; E.D. and S.C. are recipients of a CIHR studentship and Clinician Scientist award, respectively; J.C. holds an American Society of Hematology fellowship; M.S. holds a National Canadian Institute of Cancer studentship; and S.B.T. is the recipient of National Health Medical Research Council and a Royal Australian College of Physicians fellowships. IRIC is supported in part by the Canadian Center of Excellence in Commercialization and Research (CECR), the Canada Foundation for Innovation (CFI), and the Fonds de Recherche en Santé du Québec (FRSQ).
Footnotes
Supplemental Data include Supplemental Experimental Procedures, five figures, and six tables and can be found with this article online at http://www.cell.com/supplemental/S0092-8674(09)00325-0.
References
- Antonchuk J, Sauvageau G, Humphries RK. HOXB4 overexpression mediates very rapid stem cell regeneration and competitive hematopoietic repopulation. Exp Hematol. 2001;29:1125–1134. doi: 10.1016/s0301-472x(01)00681-6. [DOI] [PubMed] [Google Scholar]
- Antonchuk J, Sauvageau G, Humphries RK. HOXB4-induced expansion of adult hematopoietic stem cells ex vivo. Cell. 2002;109:39–45. doi: 10.1016/s0092-8674(02)00697-9. [DOI] [PubMed] [Google Scholar]
- Bowie MB, McKnight KD, Kent DG, McCaffrey L, Hoodless PA, Eaves CJ. Hematopoietic stem cells proliferate until after birth and show a reversible phase-specific engraftment defect. J Clin Invest. 2006;116:2808–2816. doi: 10.1172/JCI28310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cellot S, Sauvageau G. Zfx: at the crossroads of survival and self-renewal. Cell. 2007;129:239–241. doi: 10.1016/j.cell.2007.04.002. [DOI] [PubMed] [Google Scholar]
- Cellot S, Krosl J, Chagraoui J, Meloche S, Humphries RK, Sauvageau G. Sustained in vitro trigger of self-renewal divisions in Hoxb4hiPbx1(10) hematopoietic stem cells. Exp Hematol. 2007;35:802–816. doi: 10.1016/j.exphem.2007.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ema H, Nakauchi H. Expansion of hematopoietic stem cells in the developing liver of a mouse embryo. Blood. 2000;95:2284–2288. [PubMed] [Google Scholar]
- Hai-Jiang W, Xin-Na D, Hui-Jun D. Expansion of hematopoietic stem/progenitor cells. Am J Hematol. 2008;83:922–926. doi: 10.1002/ajh.21262. [DOI] [PubMed] [Google Scholar]
- Jaenisch R, Young R. Stem cells, the molecular circuitry of pluripotency and nuclear reprogramming. Cell. 2008;132:567–582. doi: 10.1016/j.cell.2008.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiel MJ, Yilmaz OH, Iwashita T, Terhorst C, Morrison SJ. SLAM family receptors distinguish hematopoietic stem and progenitor cells and reveal endothelial niches for stem cells. Cell. 2005;121:1109–1121. doi: 10.1016/j.cell.2005.05.026. [DOI] [PubMed] [Google Scholar]
- Krosl J, Austin P, Beslu N, Kroon E, Humphries RK, Sauvageau G. In vitro expansion of hematopoietic stem cells by recombinant TAT-HOXB4 protein. Nat Med. 2003a;9:1428–1432. doi: 10.1038/nm951. [DOI] [PubMed] [Google Scholar]
- Krosl J, Beslu N, Mayotte N, Humphries RK, Sauvageau G. The competitive nature of HOXB4-transduced HSC is limited by PBX1: the generation of ultra-competitive stem cells retaining full differentiation potential. Immunity. 2003b;18:561–571. doi: 10.1016/s1074-7613(03)00090-6. [DOI] [PubMed] [Google Scholar]
- Lessard J, Faubert A, Sauvageau G. Genetic programs regulating HSC specification, maintenance and expansion. Oncogene. 2004;23:7199–7209. doi: 10.1038/sj.onc.1207940. [DOI] [PubMed] [Google Scholar]
- Min IM, Pietramaggiori G, Kim FS, Passegue E, Stevenson KE, Wagers AJ. The transcription factor EGR1 controls both the proliferation and localization of hematopoietic stem cells. Cell Stem Cell. 2008;2:380–391. doi: 10.1016/j.stem.2008.01.015. [DOI] [PubMed] [Google Scholar]
- Ohta H, Sekulovic S, Bakovic S, Eaves CJ, Pineault N, Gasparetto M, Smith C, Sauvageau G, Humphries RK. Near-maximal expansions of hematopoietic stem cells in culture using NUP98-HOX fusions. Exp Hematol. 2007;35:817–830. doi: 10.1016/j.exphem.2007.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Root DE, Hacohen N, Hahn WC, Lander ES, Sabatini DM. Genome-scale loss-of-function screening with a lentiviral RNAi library. Nat Methods. 2006;3:715–719. doi: 10.1038/nmeth924. [DOI] [PubMed] [Google Scholar]
- Rosenbauer F, Owens BM, Yu L, Tumang JR, Steidl U, Kutok JL, Clayton LK, Wagner K, Scheller M, Iwasaki H, et al. Lymphoid cell growth and transformation are suppressed by a key regulatory element of the gene encoding PU.1. Nat Genet. 2006;38:27–37. doi: 10.1038/ng1679. [DOI] [PubMed] [Google Scholar]
- Sauvageau G, Thorsteinsdottir U, Eaves CJ, Lawrence HJ, Largman C, Lansdorp PM, Humphries RK. Overexpression of HOXB4 in hematopoietic cells causes the selective expansion of more primitive populations in vitro and in vivo. Genes Dev. 1995;9:1753–1765. doi: 10.1101/gad.9.14.1753. [DOI] [PubMed] [Google Scholar]
- Sauvageau G, Iscove NN, Humphries RK. In vitro and in vivo expansion of hematopoietic stem cells. Oncogene. 2004;23:7223–7232. doi: 10.1038/sj.onc.1207942. [DOI] [PubMed] [Google Scholar]
- Shing DC, Trubia M, Marchesi F, Radaelli E, Belloni E, Tapinassi C, Scanziani E, Mecucci C, Crescenzi B, Lahortiga I, et al. Overexpression of sPRDM16 coupled with loss of p53 induces myeloid leukemias in mice. J Clin Invest. 2007;117:3696–3707. doi: 10.1172/JCI32390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szilvassy SJ, Humphries RK, Lansdorp PM, Eaves AC, Eaves CJ. Quantitative assay for totipotent reconstituting hematopoietic stem cells by a competitive repopulation strategy. Proc Natl Acad Sci USA. 1990;87:8736–8740. doi: 10.1073/pnas.87.22.8736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Thorsteinsdottir U, Mamo A, Kroon E, Jerome L, Bijl J, Lawrence HJ, Humphries K, Sauvageau G. Overexpression of the myeloid leukemia-associated Hoxa9 gene in bone marrow cells induces stem cell expansion. Blood. 2002;99:121–129. doi: 10.1182/blood.v99.1.121. [DOI] [PubMed] [Google Scholar]
- Uchida N, Dykstra B, Lyons K, Leung F, Kristiansen M, Eaves C. ABC transporter activities of murine hematopoietic stem cells vary according to their developmental and activation status. Blood. 2004;103:4487–4495. doi: 10.1182/blood-2003-11-3989. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.