Abstract
In the context of seeking novel therapeutic targets for treating ischemic stroke, the preconditioning ischemia-induced brain ischemic tolerance has been used as a model of endogenously operative, broad-based neuroprotective mechanisms. Targeting such mechanisms is considered potentially less prone to adverse side effects, as those seen in many failed clinical trials that focus on single targets using exogenous compounds. Results from previous studies have revealed an overall decrease in potassium channel activity in tolerance development. The objective of this study is to identify ion channel genes that are differentially regulated under different brain ischemic conditions, as a mean to identify those ion channels that are associated with ischemic brain injury and ischemic tolerance. In mice in vivo, transient focal cerebral ischemia was induced by middle cerebral artery occlusion. In cultured neuronal cells in vitro, simulated ischemia was modeled by oxygen-glucose deprivation. For both in vivo and in vitro studies, three principal ischemic conditions were included: ischemic-preconditioned, injured and tolerant, respectively, plus appropriate controls. In these model systems, transcript levels of a panel of 84 neuronal ion channels genes were analyzed with a quantitative real-time PCR mini-array. The results showed that, both in vivo and in vitro, there was a predominant down regulation in neuronal ion channel genes under ischemic-tolerant conditions, and an up regulation in ischemic injury. Similar changes were observed among potassium, sodium and calcium channel genes. A number of regulated genes exhibited opposing changes under ischemic-injured and ischemic-tolerant conditions. This subset of ion channel genes exemplifies potentially novel leads for developing multi-factorial therapeutic targets for treating ischemic stroke.
Keywords: Ischemic stroke, neuroprotection, ion channels, neuronal excitability
Introduction
Stroke is the 5th leading cause of death in North America. Ischemic stroke accounts for approximately 87% of stroke cases [1]. However, to date, there has been only one FDA-approved therapy for ischemic stroke-the administration of thrombolytic recombinant tissue plasminogen activator (rtPA), which has severe limitations in that it is applicable to less than 5% of stroke patients, as it must be administered within the first few hours after the known onset of acute stroke symptoms, and may increase the risks of a secondary, hemorrhagic stroke [2-4]. The quest for effective neuroprotective agents for treating ischemic brains has had few successes in clinical trials, due to, at least partially, the fact that the therapeutic treatments often focus on a single target or single pathway (for example, using an antagonist against NMDA receptor) [4-7]. Such approaches are therapeutically limited and are prone to developing adverse side effects. A different approach is to consider identifying and targeting mechanisms that are broad-based, and are operative endogenously, which potentially can be more effective and also better tolerated. In this regard, induced ischemic tolerance provides an appealing model [8-10], whereby neuroprotection is achieved by molecular mechanisms that have already been encoded in the genome. Previously, using a mouse model of brain ischemic tolerance, we reported a predominant down regulation in genomic (transcriptional) changes [11] and an enriched presence of epigenetic gene repressor proteins, namely polycomb group (PcG) proteins, among proteomic changes [12]. PcG proteins are known to silence a broad range of genes in the genome. Their exact targets in ischemic brains, however, remain elusive. The genomic and proteomic characteristics of brain ischemic tolerance suggest the involvement of repressive responses of many potential PcG protein target genes. In the genomic study, using then available probes on Affymetrix genechips, a decrease in transcript levels of Kcna5, a potassium channel gene, was observed in ischemic tolerance. Interestingly, in cultured cortical neurons under simulated, ischemic-tolerant conditions, there is a decrease in the whole cell potassium currents [11]. Further, similar decreases in potassium currents can be achieved in neuronal cultures by over expressing a PcG protein without preconditioning [12]. In the literature, results from studies on other non-neuronal cell lines have suggested that PcG proteins may regulate the expression of tens of the known potassium channel genes [13,14]. These observations raise the question of whether there are wide-spread changes in potassium channel genes in brain ischemia, perhaps conferring endogenous neuroprotection.
Ion channels play pivotal roles in the pathology of ischemic stroke and post-stroke recovery. Extensive efforts have been made in targeting individual ion channels, especially those involved in excitatory neurotransmissions to alleviate excitotoxicity. For example, numerous pharmacological agents have been tested for their effects of reducing or blocking sodium channel activities in brain ischemia [7,15,16]. The roles of potassium channels in neuronal ischemia are more complex. While a decrease in potassium channel activity may potentially increase neuronal excitability, hence producing adverse consequences in neuronal ischemia, it can be beneficial by lowering energy demand [11,12,17]. Other ion channels, such as acid sensing ion channels (ASICs) [18,19] and transient receptor potentiation channels (TRPC) [20], may also regulate the outcome of ischemic stroke. In recent years, there has been an increased interest in the role of ionic homeostasis in ischemic brain injury [21,22] or preconditioning [23]. However, as mentioned earlier, the published studies in general address the roles of individual ion channel genes in brain ischemia, especially under injurious conditions. Little is known in regard to how many ion channel genes are categorically involved in ischemic injury or in its prevention (tolerance), and whether changes under different brain ischemic conditions may differ.
In this work, using a limited array of quantitative reverse transcription PCR (RT-qPCR) (for simplicity, referred as mini-array hereafter), we analyzed expressional changes of a panel of eighty-four neuronal ion channel genes under ischemic-preconditioned, ischemic-injured and ischemic-tolerant conditions, in both mouse brains in vivo, and in cultured neuronal cells in vitro. The results showed a down regulation of most of the voltage-gated potassium channel genes on the panel in ischemic tolerance. Interestingly, a parallel down regulation in many of the sodium and calcium channel genes was also observed. Notably, a number of ion channel genes demonstrated opposing changes under ischemic-injured and ischemic-tolerant conditions. To the best of our knowledge, this is the first gene expression profiling focusing on neuronal ion channels under several different neuronal ischemic conditions.
Materials and methods
Focal cerebral ischemia in mice and simulated ischemia in neuronal cultures
Transient focal cerebral ischemia was induced in adult male C57BL/6J mice (20-25 g; Charles River Laboratories) by middle cerebral artery occlusion (MCAO) using the suture method, following previously described protocols [12]. All procedures were approved and monitored by the Institutional Animal Care and Use Committee at Morehouse School of Medicine. In vitro, simulated ischemia was modeled in cultured, differentiated neuroblastoma NS20Y cells by oxygen-glucose deprivation (OGD) [12], which was achieved by incubating cells in an anaerobic chamber (Forma Scientific, model 1025) equilibrated with 85% N2/5% CO2/10% H2, in a culture medium free of glucose, serum and glutamate, containing 5 mM Hepes (pH 7.4), 2 mM CaCl2, 135 mM NaCl, 5 mM KCl, 1X essential amino acids (without L-glutamine), 25 mM 2-deoxyglucose and penicillin/streptomycin. For both in vivo and in vitro studies, three principal ischemic conditions were included, as schematically illustrated in Figure 1.
Figure 1.
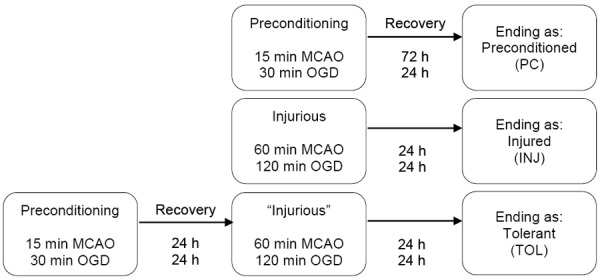
Experimental paradigm. The figure illustrates the experimental paradigm by which three principal ischemic conditions were modeled: Ischemic-preconditioned (PC), Ischemic-injured (INJ), and Ischemic-tolerant (TOL). In mice, the preconditioning or injurious ischemia was induced by 15 min or 60 min MCAO, respectively, followed by reperfusions for the numbers of hours noted in the figure (recovery), whereas ischemic tolerance was achieved by 15 min preconditioning MCAO followed by 72 h reperfusion, prior to the 60 min MCAO. In differentiated NS20Y cell cultures, the conditions of preconditioned, injured and tolerant were modeled by subjecting cells to OGD and subsequent recovery incubations for the noted periods of time, respectively. Controls (sham-operated animals or normoxic cell cultures) followed the time regimes of that of injurious groups, respectively.
Expression profiling of genes encoding neuronal ion channels
Total RNA was isolated from individual cortices (n=7 each animal group) or NS20Y cultures (three independent cultures per cell culture condition) using the SuperPrep RNA/DNA/Protein Purification kit (Fisher Scientific), following the manufacturer’s instruction. The RNA yield was determined on a NanoDrop spectrophotometer (Thermo Scientific). Complementary DNA (cDNA) was prepared from 0.7 µg (cortices) or 1 µg (NS20Y cells) of total RNA using the RT2 First Strand kit (Qiagen). Preparations of cDNAs from individual cortices or cell cultures were pooled according to their groups. The pooled cDNAs were then subjected to RT-qPCR mini-array analysis of a panel of eighty-four neuronal ion channel genes (complete list in Table 1) with a commercial real time RT2 Profiler PCR Array (Qiagen, Cat. No. PAMM-036Z), using the RT2 SYBR® Green qPCR Mastermix (Qiagen). The thermo steps were performed on a C1000 Thermal Cycler (Bio-Rad) as follows: hot start at 95°C for 10 min, followed by 40 cycles of 95°C for 15 sec and 60°C for 60 sec.
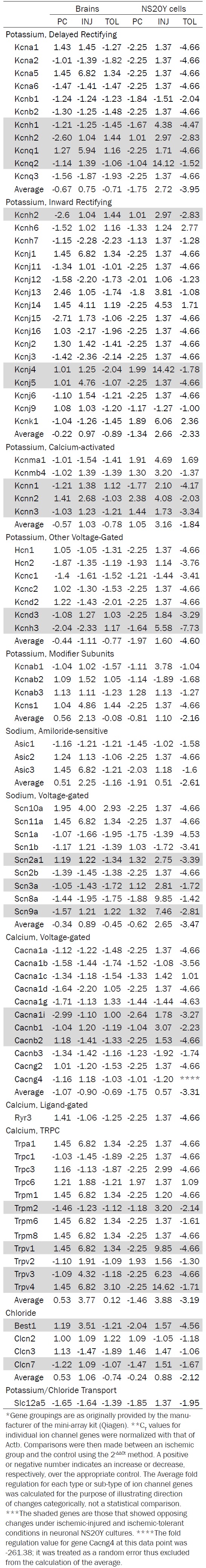
Table 1.
Ion channel genes included in the mini-array analysis and their fold regulations. *, **, ***
|
Data processing and statistics
Analysis of the cycle threshold (CT) values, threshold selection, quantitation with the 2-ΔΔ CT method, and establishment of fold regulations of ion channel genes in ischemic samples over controls were performed using the online tools provided by the manufacturer (Gene Globe Data Analysis Center, Qiagen). The quality of sample preparations (RT efficiency, reproducibility and DNA contamination) was assessed with manufacture’s online data analysis tools. The mini-array included five housekeeping genes (HKGs) as candidate internal standards, namely Actb, B2m, Gapdh, Gusb and Hsp90ab1. Prior to calculating the expressional changes in ion channel genes, relative standard deviation (RSD) of CT values were calculated for each of the five HKGs across all brain or cell culture conditions, respectively, to assist the selection of a HKG for data normalization. The commercial RT2 Profiler PCR Array does not include technical replications by design, and only pooled samples were analyzed. Accordingly, fold regulation values represent single entries. For ion channel genes, a fold regulation of 1.5 (approximately 3 times of the RSD of HKGs across different groups) or greater was accepted as a change. Hierarchical clustering of datasets that enlist fold regulations of the eighty-four ion channel genes under different ischemic conditions were performed using the ClustVis program [24] online (biit.cs.ut.ee/clustvis).
Results
First, we evaluated data for the HKGs on the mini-array panel as a quality control. Their RSD values, as calculated from the results of RT-qPCR analysis, were as follows (gene, brain/cell): Actb, 5.59/5.80; B2m, 4.28/6.73; Gapdh, 4.86/6.63; Gusb, 4.72/8.05; Hsp-90ab1, 4.79/5.82. Hence, the results demonstrated comparable expression levels for all of the five HKGs across experimental conditions, in samples from both mouse brains and neuronal NS20Y cultures. For the results described next, Atbc was used as the internal standard for data normalization and calculation of fold regulations of ion channel genes in ischemic groups over controls.
Wide-spread down regulation in ischemic tolerance
Values of fold regulations for all eighty-four ion channel genes under different ischemic conditions are provided in Table 1. Figure 2 presents heatmaps of expressional changes in neuronal ion channel genes under the three principal ischemic conditions that were included in this study. Noticeably, as summarized in Figure 3A, both in mouse brains in vivo and in neuronal NS20Y cultures in vitro, there were more down regulated ion channel genes than up regulated under ischemic-tolerant conditions, whereas under ischemic-injured conditions the changes were predominantly an up regulation. A greater number of ion channel genes showed changes in neuronal NS20Y cells than in mouse brains. For example, of the forty-three potassium channel genes on the panel, twelve and thirty three were down regulated in ischemic-tolerant mouse brains and neuronal NS20Y cells, respectively, while zero and four, respectively, showed an up regulation (Table 1).
Figure 2.
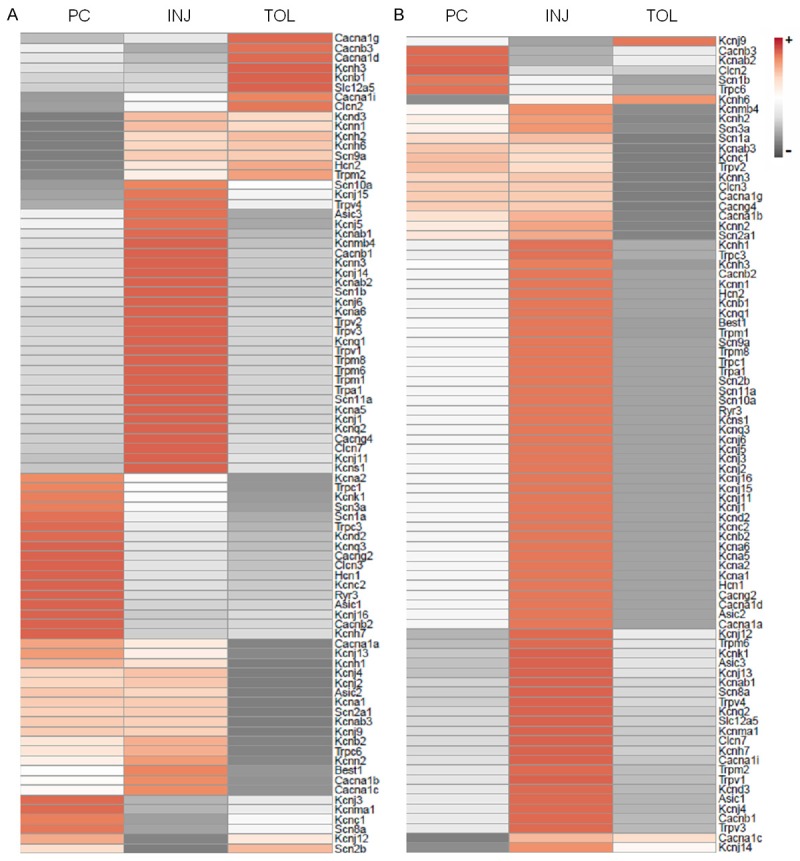
Heatmaps of expressional changes in neuronal ion channel genes under the three principal ischemic conditions. The heatmaps were generated using the fold regulations listed in Table 1 and arranged according to the tightest clustering. The scale bar indicates the direction of changes (+ and -, an increase or decrease, respectively, when ischemic groups were compared with the appropriate controls). A. Mouse brains; B. Cultured neuronal NS20Y cells. PC, INJ, and TOL: Same as in Figure 1.
Figure 3.
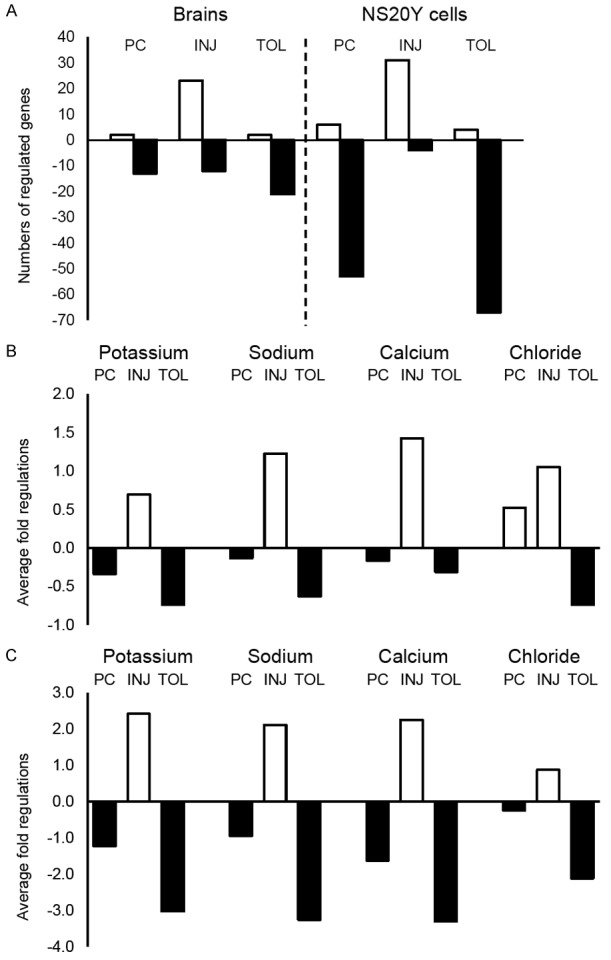
Numbers of regulated ion channel genes and directions of their changes. The bars represent numbers of regulated genes under different ischemic conditions (A), or average fold regulations for each of the noted ion channel types under different ischemic conditions (B. mouse brains; C. NS20Y cells). Positive (open bars) and negative (filled bars) values indicate up and down regulations, respectively. PC, INJ, and TOL: Same as in Figure 1.
Regulation of potassium, sodium and calcium channel genes
Ion channel genes on the mini-array included members of potassium, sodium, calcium and chloride channels (Table 1). Interestingly, as illustrated in Figure 2, the channel genes that were down regulated under ischemic-tolerant conditions comprise large numbers of both potassium and sodium channel genes, as well as calcium channel genes, especially in neuronal NS20Y cell cultures, which are homogeneous in cell populations. On average, under each of the three principal ischemic conditions, members of the aforementioned categories of ion channels demonstrated changes in the same directions: a decrease in ischemic tolerance and an increase in ischemic injury (Figure 3B and 3C). The changes in the preconditioned brains or neuronal cultures were moderate in general (i.e. the fold regulations for most genes did not meet the cut off values to be considered as changed), but nevertheless were in the same direction as that seen in ischemic tolerance. The most robust changes (greater fold regulations) were seen with voltage-gated ion channels (Table 1).
Differential changes under different ischemic conditions
We were particularly interested in identifying genes that are differentially regulated under different ischemic conditions. As shown in Table 1, in the homogenous neuronal NS20Y cultures, a number of genes (shaded) demonstrated opposing changes under ischemic-injured and ischemic-tolerant conditions. Such genes include potassium channel genes Kcnh1, Kcnh2, Kcnq1, Kcnq2, Kcnh2, Kcnj4, Kcnj5, Kcnn1, Kcnn2, Kcnn3, Kcnd3, Kcnh3; sodium channel genes Scn2a1, Scn3a, Scn9a; calcium channel genes Cacna1i, Cacnb1, Cacnb2; Trpm2, Trpv1, Trpv3, Trpv4; chloride channel genes Best1 and Clcn1. Worth-noting is that these genes represent members of all ion channel categories included in the mini-array.
Discussion
Besides considering thrombolytic or endovascular approaches for treatment of ischemic stroke, most therapeutic research efforts have focused on neuroprotective agents that may alleviate excitotoxicity, oxidative stress, inflammatory responses, or mitochondrial dysregulation after an ischemic insult to the brain [2,4-6,25-28]. As mentioned earlier, the therapeutic limitations of regulating a single target with exogenous drugs have promoted interests in defining the biology of ischemic tolerance. While the underlying mechanisms of tolerance are complex, the selective increases in multi-target, repressive PcG proteins, which in turn may lead to the characteristic transcriptional suppression, and their potential roles in regulating multiple ion channel genes, satisfy the criteria for an endogenous, broad-based neuroprotective mechanism in ischemic tolerance. Here for the first time we offer data focusing on regulation of a panel of neuronal ion channel genes under contrasting ischemic conditions, suggesting a role in tolerance induction.
In the present study, in both mouse brains in vivo and cultured neuronal cells in vitro, the ischemic-tolerant condition was marked with a down regulation of many potassium channel genes. This provides a possible explanation for our previously published finding on channel arrest in ischemic tolerance [11]. In the literature, reports of roles of potassium channels in ischemic stroke have generated controversy. Given their roles in establishing resting membrane potentials and regulating neuronal excitability, an increase in potassium channel activity has long been considered beneficial in ischemic stroke, as demonstrated by the neuroprotective effects of potassium channel openers [29-31]. However, in recent years, it has been recognized that a decrease in potassium channel activity may produce neuroprotection as well [12,32,33], simulating the conditions seen in hibernating animals [12,34,35]. The underlying protective mechanisms may include a reduced demand on energy metabolism to sustain ion gradients across the plasma membrane, prevention of adverse, potassium channel-mediated activation of microglia in ischemic brains [17,33], as well as limiting sodium influx [36].
The neuronal ion channel panel analyzed in this study includes more than half of the known potassium channel genes, nine of the ten mammalian sodium channel genes, and members of voltage-gated calcium channel genes and TRPC channel genes. A down regulation in voltage-gated sodium or calcium channel genes or TRPC genes in ischemic tolerance was not surprising, as they have been considered therapeutic targets using inhibitory or antagonistic approaches. Of interest are the similar changes of these ion channel genes with those of potassium channel genes. This phenomenon, if validated with additional molecular biological (for gene transcript levels), biochemical (for protein levels) and physiological (for electrophysiological properties) analyses, would implicate a regulatory mechanism that operates genome-wide. In this regard, PcG protein-mediated epigenetic regulation, previously shown to be neuroprotective, is promising. Our recent results (Zhou et al, unpublished data) suggest a role of PcG proteins in repressing multiple ion channel genes and regulating sodium currents, following our previously published observations that an increase in cellular levels of PcG proteins, as seen in ischemic tolerance, can result in a decrease in whole cell potassium currents [12].
The present results from analyses of the whole cortices do not provide information about which cell populations in the brain may have had contributed to the observed changes under different brain ischemic conditions. It is likely that changes are not limited to neurons, but also occur in microglia and astrocytes. Changes may differ in direction in different cell populations. These assumptions are supported by the observations that more robust changes were seen in neuronal NS20Y cultures than in mouse brains (Figures 2 and 3, Table 1). Considering the regulatory roles of ion channels in the brain’s response to ischemia, it is reasonable to assume that ion channels that are instrumental in neuroprotection in ischemic tolerance may change in opposite direction in ischemic injury. Interestingly, the present results indeed showed such opposing changes in a number of ion channel genes, most of which are voltage-gated or TRPC channel genes (Table 1). Results of literature searches on those genes revealed that some of them have not been studied in the setting of brain ischemia. Hence, the results from the present study may direct future studies to establish panels of ion channels that are characteristics of brain ischemic injury or neuroprotection.
Conclusion
By means of mini-array RT-qPCR analysis, we show contrasting changes under different ischemic conditions, and note a predominant down regulation of multiple ion channel genes in ischemic tolerance and their upregulation in ischemic injury. The results support a potentially novel hypothesis that endogenous neuroprotection against ischemic injury is mediated by a broad-based repression of multiple neuronal ion channel genes.
Acknowledgements
Authors acknowledge grant supports from American Heart Association (GIA 17GRNT33700277, AZ) and National Institute of Health (R01NS073832 (AZ), U54NS083932 (Neuroscience Institute, Morehouse School of Medicine), S21MD000101 and G12-RP03034 (Morehouse School of Medicine)). We thank Tao Yang for preparation of ischemic mouse brains and Dr. Jason DeBruyne for advices on mini-array data processing.
References
- 1.Writing Group Members; Mozaffarian D, Benjamin EJ, Go AS, Arnett DK, Blaha MJ, Cushman M, Das SR, de Ferranti S, Després JP, Fullerton HJ, Howard VJ, Huffman MD, Isasi CR, Jiménez MC, Judd SE, Kissela BM, Lichtman JH, Lisabeth LD, Liu S, Mackey RH, Magid DJ, McGuire DK, Mohler ER 3rd, Moy CS, Muntner P, Mussolino ME, Nasir K, Neumar RW, Nichol G, Palaniappan L, Pandey DK, Reeves MJ, Rodriguez CJ, Rosamond W, Sorlie PD, Stein J, Towfighi A, Turan TN, Virani SS, Woo D, Yeh RW, Turner MB American Heart Association Statistics Committee; Stroke Statistics Subcommittee. Heart disease and stroke statistics-2016 update: a report from the american heart association. Circulation. 2016;133:e38–360. doi: 10.1161/CIR.0000000000000350. [DOI] [PubMed] [Google Scholar]
- 2.Sahota P, Savitz SI. Investigational therapies for ischemic stroke: neuroprotection and neurorecovery. Neurotherapeutics. 2011;8:434–451. doi: 10.1007/s13311-011-0040-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bansal S, Sangha KS, Khatri P. Drug treatment of acute ischemic stroke. Am J Cardiovasc Drugs. 2013;13:57–69. doi: 10.1007/s40256-013-0007-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen X, Wang K. The fate of medications evaluated for ischemic stroke pharmacotherapy over the period 1995-2015. Acta Pharm Sin B. 2016;6:522–530. doi: 10.1016/j.apsb.2016.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheng YD, Al-Khoury L, Zivin JA. Neuroprotection for ischemic stroke: two decades of success and failure. NeuroRx. 2004;1:36–45. doi: 10.1602/neurorx.1.1.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reis C, Akyol O, Ho WM, Araujo C, Huang L, Applegate R II, Zhang JH. Phase i and phase ii therapies for acute ischemic stroke: an update on currently studied drugs in clinical research. Biomed Res Int. 2017;2017:4863079. doi: 10.1155/2017/4863079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Waszkielewicz AM, Gunia A, Szkaradek N, Sloczynska K, Krupinska S, Marona H. Ion channels as drug targets in central nervous system disorders. Curr Med Chem. 2013;20:1241–1285. doi: 10.2174/0929867311320100005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deryagin OG, Gavrilova SA, Gainutdinov KL, Golubeva AV, Andrianov VV, Yafarova GG, Buravkov SV, Koshelev VB. Molecular bases of brain preconditioning. Front Neurosci. 2017;11:427. doi: 10.3389/fnins.2017.00427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gidday JM. Cerebral preconditioning and ischaemic tolerance. Nat Rev Neurosci. 2006;7:437–448. doi: 10.1038/nrn1927. [DOI] [PubMed] [Google Scholar]
- 10.Thushara Vijayakumar N, Sangwan A, Sharma B, Majid A, Rajanikant GK. Cerebral ischemic preconditioning: the road so far. Mol Neurobiol. 2016;53:2579–2593. doi: 10.1007/s12035-015-9278-z. [DOI] [PubMed] [Google Scholar]
- 11.Stenzel-Poore MP, Stevens SL, Xiong Z, Lessov NS, Harrington CA, Mori M, Meller R, Rosenzweig HL, Tobar E, Shaw TE, Chu X, Simon RP. Effect of ischaemic preconditioning on genomic response to cerebral ischaemia: similarity to neuroprotective strategies in hibernation and hypoxia-tolerant states. Lancet. 2003;362:1028–1037. doi: 10.1016/S0140-6736(03)14412-1. [DOI] [PubMed] [Google Scholar]
- 12.Stapels M, Piper C, Yang T, Li M, Stowell C, Xiong ZG, Saugstad J, Simon RP, Geromanos S, Langridge J, Lan JQ, Zhou A. Polycomb group proteins as epigenetic mediators of neuroprotection in ischemic tolerance. Science Signaling. 2010;3:ra15. doi: 10.1126/scisignal.2000502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bracken AP, Dietrich N, Pasini D, Hansen KH, Helin K. Genome-wide mapping of polycomb target genes unravels their roles in cell fate transitions. Genes Dev. 2006;20:1123–1136. doi: 10.1101/gad.381706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schuettengruber B, Chourrout D, Vervoort M, Leblanc B, Cavalli G. Genome regulation by polycomb and trithorax proteins. Cell. 2007;128:735–745. doi: 10.1016/j.cell.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 15.Carter AJ, Grauert M, Pschorn U, Bechtel WD, Bartmann-Lindholm C, Qu Y, Scheuer T, Catterall WA, Weiser T. Potent blockade of sodium channels and protection of brain tissue from ischemia by biii 890 cl. Proc Natl Acad Sci U S A. 2000;97:4944–4949. doi: 10.1073/pnas.040577097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Urenjak J, Obrenovitch TP. Pharmacological modulation of voltage-gated na+ channels: a rational and effective strategy against ischemic brain damage. Pharmacol Rev. 1996;48:21–67. [PubMed] [Google Scholar]
- 17.Ortega FJ, Gimeno-Bayon J, Espinosa-Parrilla JF, Carrasco JL, Batlle M, Pugliese M, Mahy N, Rodríguez MJ. Atp-dependent potassium channel blockade strengthens microglial neuroprotection after hypoxia-ischemia in rats. Exp Neurol. 2012;235:282–296. doi: 10.1016/j.expneurol.2012.02.010. [DOI] [PubMed] [Google Scholar]
- 18.Xiong ZG, Zhu XM, Chu XP, Minami M, Hey J, Wei WL, MacDonald JF, Wemmie JA, Price MP, Welsh MJ, Simon RP. Neuroprotection in ischemia: blocking calcium-permeable acid-sensing ion channels. Cell. 2004;118:687–698. doi: 10.1016/j.cell.2004.08.026. [DOI] [PubMed] [Google Scholar]
- 19.Xiong ZG, Chu XP, Simon RP. Acid sensing ion channels--novel therapeutic targets for ischemic brain injury. Front Biosci. 2007;12:1376–1386. doi: 10.2741/2154. [DOI] [PubMed] [Google Scholar]
- 20.Huang J. Trpc channels and stroke. Adv Exp Med Biol. 2017;976:61–71. doi: 10.1007/978-94-024-1088-4_6. [DOI] [PubMed] [Google Scholar]
- 21.Song M, Yu SP. Ionic regulation of cell volume changes and cell death after ischemic stroke. Transl Stroke Res. 2014;5:17–27. doi: 10.1007/s12975-013-0314-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chao D, Wang Q, Balboni G, Ding G, Xia Y. Attenuating ischemic disruption of k+ homeostasis in the cortex of hypoxic-ischemic neonatal rats: dor activation vs. acupuncture treatment. Mol Neurobiol. 2016;53:7213–7227. doi: 10.1007/s12035-015-9621-4. [DOI] [PubMed] [Google Scholar]
- 23.Cuomo O, Vinciguerra A, Cerullo P, Anzilotti S, Brancaccio P, Bilo L, Scorziello A, Molinaro P, Di Renzo G, Pignataro G. Ionic homeostasis in brain conditioning. Front Neurosci. 2015;9:277. doi: 10.3389/fnins.2015.00277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Metsalu T, Vilo J. Clustvis: a web tool for visualizing clustering of multivariate data using principal component analysis and heatmap. Nucleic Acids Res. 2015;43:W566–570. doi: 10.1093/nar/gkv468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grossman AW, Broderick JP. Advances and challenges in treatment and prevention of ischemic stroke. Ann Neurol. 2013;74:363–372. doi: 10.1002/ana.23993. [DOI] [PubMed] [Google Scholar]
- 26.Majid A. Neuroprotection in stroke: past, present, and future. ISRN Neurol. 2014;2014:515716. doi: 10.1155/2014/515716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stocchetti N, Taccone FS, Citerio G, Pepe PE, Le Roux PD, Oddo M, Polderman KH, Stevens RD, Barsan W, Maas AI, Meyfroidt G, Bell MJ, Silbergleit R, Vespa PM, Faden AI, Helbok R, Tisherman S, Zanier ER, Valenzuela T, Wendon J, Menon DK, Vincent JL. Neuroprotection in acute brain injury: an up-to-date review. Crit Care. 2015;19:186. doi: 10.1186/s13054-015-0887-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ohya S, Kito H, Hatano N, Muraki K. Recent advances in therapeutic strategies that focus on the regulation of ion channel expression. Pharmacol Ther. 2016;160:11–43. doi: 10.1016/j.pharmthera.2016.02.001. [DOI] [PubMed] [Google Scholar]
- 29.Heurteaux C, Bertaina V, Widmann C, Lazdunski M. K+ channel openers prevent global ischemia-induced expression of c-fos, c-jun, heat shock protein, and amyloid beta-protein precursor genes and neuronal death in rat hippocampus. Proc Natl Acad Sci U S A. 1993;90:9431–9435. doi: 10.1073/pnas.90.20.9431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Judge SI, Smith PJ. Patents related to therapeutic activation of k(atp) and k(2p) potassium channels for neuroprotection: ischemic/hypoxic/anoxic injury and general anesthetics. Expert Opin Ther Pat. 2009;19:433–460. doi: 10.1517/13543770902765151. [DOI] [PubMed] [Google Scholar]
- 31.Kumar P, Kumar D, Jha SK, Jha NK, Ambasta RK. Ion channels in neurological disorders. Adv Protein Chem Struct Biol. 2016;103:97–136. doi: 10.1016/bs.apcsb.2015.10.006. [DOI] [PubMed] [Google Scholar]
- 32.Folyovich A, Biro E, Orban C, Bajnok A, Vasarhelyi B, Toldi G. Kv1.3 lymphocyte potassium channel inhibition as a potential novel therapeutic target in acute ischemic stroke. CNS Neurol Disord Drug Targets. 2014;13:801–806. doi: 10.2174/1871527313666140711090509. [DOI] [PubMed] [Google Scholar]
- 33.Schlichter LC, Kaushal V, Moxon-Emre I, Sivagnanam V, Vincent C. The ca2+ activated sk3 channel is expressed in microglia in the rat striatum and contributes to microglia-mediated neurotoxicity in vitro. J Neuroinflammation. 2010;7:4. doi: 10.1186/1742-2094-7-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dave KR, Christian SL, Perez-Pinzon MA, Drew KL. Neuroprotection: lessons from hibernators. Comp Biochem Physiol B Biochem Mol Biol. 2012;162:1–9. doi: 10.1016/j.cbpb.2012.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee YJ, Hallenbeck JM. Insights into cytoprotection from ground squirrel hibernation, a natural model of tolerance to profound brain oligaemia. Biochem Soc Trans. 2006;34:1295–1298. doi: 10.1042/BST0341295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen YJ, Hai MN, O’Donnell ME, Wulff H. Potassium channels in ischemic stroke. FASE. 2016;30 Suppl 1224.1219. [Google Scholar]


